Abstract
In most flowering plants, the female germline is initiated in the subepidermal L2 layer of ovule primordia forming a single megaspore mother cell (MMC). How signaling from the L1 (epidermal) layer could contribute to the gene regulatory network (GRN) restricting MMC formation to a single cell is unclear. We show that EPIDERMAL PATTERNING FACTOR-like (EPFL) peptide ligands are expressed in the L1 layer, together with their ERECTA family (ERf) receptor kinases, to control female germline specification in Arabidopsis thaliana. EPFL-ERf dependent signaling restricts multiple subepidermal cells from acquiring MMC-like cell identity by activating the expression of the major brassinosteroid (BR) receptor kinase BRASSINOSTEROID INSENSITIVE 1 and the BR-responsive transcription factor BRASSINOZOLE RESISTANT 1 (BZR1). Additionally, BZR1 coordinates female germline specification by directly activating the expression of a nucleolar GTP-binding protein, NUCLEOSTEMIN-LIKE 1 (NSN1), which is expressed in early-stage ovules excluding the MMC. Mutants defective in this GRN form multiple MMCs resulting in a strong reduction of seed set. In conclusion, we uncovered a ligand/receptor-like kinase-mediated signaling pathway acting upstream and coordinating BR signaling via NSN1 to restrict MMC differentiation to a single subepidermal cell.
A ligand/receptor-like kinase-mediated signaling pathway coordinates brassinosteroid signaling via the GTP-binding protein NUCLEOSTEMIN-LIKE 1 to restrict megaspore mother cell differentiation to a single subepidermal cell.
IN A NUTSHELL.
Background: In most flowering plants, only a single female germline precursor cell-also called megaspore mother cell (MMC)-is formed in the subepidermal L2 layer of ovule primordia. This cell initiates a developmental program that ultimately leads to the production of two female gametes required for the double fertilization process that is characteristic for flowering plants. It is highly critical to suppress other subepidermal L2 layer cells from acquiring MMC cell fate as the formation of multiple MMCs usually leads to sterility. In a recent study, we showed that a brassinosteroid (BR) gradient established in the distal nucellus of ovule primordia with a maximum in epidermal L1 layer cells is required to restrict female germline cell fate to a single L2 layer nucellus cell.
Question: How signaling from the L1 (epidermal) layer could contribute to the gene regulatory network (GRN) restricting MMC formation to a single cell in the L2 (subepidermal) layer?
Findings: We showed that a group of EPFL peptide ligands, expressed in the L1 layer of ovules, and ERECTA family (ERf) receptor kinases acted together to control female germline specification. The redundant ligand-receptor complex restricts multiple subepidermal cells in the L2 layer from acquiring MMC-like cell identity by activating expressions of the BR receptor BRI1 and BZR1 family transcription factor genes. Moreover, BZR1 family transcription factors coordinate the regulation of female germline specification by directly activating expression of a nucleolar GTP-binding protein gene, named NSN1, which is expressed in early-stage ovules excluding the MMC. This activation depends on EPFL-ERf signaling. This report links peptide-mediated EPFL-ERf signaling with BR hormone signaling and demonstrates how these pathways are interconnected to restrict MMC differentiation to a single subepidermal cell.
Next steps: In a future study, we will analyze how auxin regulated pathways, RdDM and other pathways affect and integrate into the GRN reported here.
Introduction
Germline specification occurs during early embryo development in sexually reproducing animals. In contrast, germline cells are established from somatic cells in floral reproductive organs of adult seed plants (Hater et al. 2020). Because only one cell differentiates to become a female germline cell in most ovules, differentiation of this megaspore mother cell (MMC) is critical for plant sexual reproduction. MMC specification has been shown to be regulated at different levels involving signaling by hormones, small RNA pathways, cell cycle regulation, and other factors (Olmedo-Monfil et al. 2010; Schmidt et al. 2011; Tucker et al. 2012; She et al. 2013; Hernandez-Lagana et al. 2016; Cai et al. 2017; Li et al. 2017; Ferreira et al. 2018; Zhao et al. 2018).
In Arabidopsis (Arabidopsis thaliana), several epigenetic regulators involved in MMC differentiation have been revealed. The RNA-directed DNA methylation (RdDM) pathway component ARGONAUTE9 (AGO9) controls MMC specification through a noncell-autonomous small interfering RNA (siRNA)-mediated gene silencing mechanism (Olmedo-Monfil et al. 2010). Loss-of-function mutants of RdDM components, including AGO4 and AGO6, produce excess MMC-like cells. Additionally, some can bypass meiosis and develop into unreduced functional megaspores and embryo sacs (Hernandez-Lagana et al. 2016).
The SPOROCYTELESS/NOZZLE (SPL/NZZ) transcription factor, which is inhibited by RdDM, is necessary to promote MMC formation and limits female germline cell fate to a single cell (Mendes et al. 2020). Besides, the THO/TREX (transcription/export) complex inhibits MMC differentiation by affecting the biogenesis of a trans-acting siRNAs targeting AUXIN-RESPONSIVE FACTORs (tasiR-ARFs) (Cai et al. 2017). tasiR-ARFs are generated in the distal nucellar epidermis and move into the subepidermal cells to repress AUXIN RESPONSE FACTOR3 and thus suppress ectopic MMC fate in these cells (Su et al. 2020). Another epigenetics factor, SWITCH 2/SUCROSE NONFERMENTABLE 2-Related 1, which mediates the incorporation of histone H2A.Z at the WRKY28 locus, limits multiple MMC formation by promoting the expression of nucellar-specific WRKY28 leading to suppression of MMC cell fate (Zhao et al. 2018).
The INHIBITOR OF CYCLIN-DEPENDENT KINASE/KIP-RELATED PROTEIN (KRP) genes KRP4, KRP6, and KRP7 play a redundant role in restricting meiotic competence in a single cell through inhibiting A-type cyclin-dependent kinase (CDKA; 1)-dependent inactivation of RETINOBLASTOMA-RELATED 1 (RBR1). RBR1 represses cell proliferation of MMCs by regulating the degradation of KRPs and directly inhibiting activity of the stem cell factor WUSCHEL (WUS), thus repressing the entry of multiple cells into meiosis (Zhao et al. 2017). WUS also acts downstream of SPL in MMC development. Overexpression of SPL with the promoter of SEEDSTICK, which is specifically expressed in sporophyte ovule tissue, results in multiple MMCs during early ovule development (Mendes et al. 2020).
Brassinosteroids (BRs) are important hormones that regulate several processes during plant growth, development, and stress adaptation (Mitchell et al. 1970; Kutschera and Wang 2012; Nolan et al. 2017), and were recently shown to regulate MMC formation (Cai et al. 2022). BRs are sensed by cell surface receptor kinase complexes, including BRASSINOSTEROID INSENSITIVE 1 (BRI1) and its co-receptor BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1) (Li and Chory 1997; Li et al. 2002; Nam and Li 2002). Once BRI1 and BAK1 are activated (Hothorn et al. 2011; He et al. 2013), an intracellular signaling cascade is initiated (Kim et al. 2009), leading to the release of the negative regulator BRI1 KINASE INHIBITOR 1 (BKI1) from the plasma membrane localized BRI1–BAK1 complex (Wang and Chory 2006) to inactivating the second negative regulator BRASSINOSTEROID INSENSITIVE 2 (BIN2) by a phosphatase, BRI1 SUPPRESSOR 1 (Li et al. 2001; Yan et al. 2009). Inactivation of BIN2 can significantly induce the accumulation of nonphosphorylated forms of the two transcription factors BRI1 EMS SUPPRESSOR 1 (BES1) and BRASSINAZOLE RESISTANT 1 (BZR1) in the nucleus (Wang et al. 2002; Yin et al. 2002). Activated BES1 and BZR1 thereafter mediate expression of numerous downstream responsive genes by directly binding to the promoter of target genes or by interacting with other types of transcription factors (Sun et al. 2010).
In a recent study, we showed that a BR gradient is established in the distal nucellus of ovule primordia with a maximum in epidermal L1 layer cells. This BR-generated microenvironment is required to restrict female germline cell fate to a single L2 layer nucellus cell by activating the BRI1–BZR1–WRKY23 pathway (Cai et al. 2022). Whether BZR1 family members could be activated by BR-independent signaling and could trigger other downstream components in MMC specification is not known.
The ERECTA family (ERf) signaling pathway was initially linked to the growth and development of plant organs via cell proliferation (Shpak et al. 2004). In Arabidopsis, ERf consists of three genes: ERECTA (ER), ERECTA-LIKE1 (ERL1), and ERL2. ERf plays a redundant role in shoot apical meristem regulation, stomatal patterning, inflorescence architecture, and ovule development (Shpak et al. 2004). Ligands for ERf receptors belong to the evolutionarily conserved EPIDERMAL PATTERNING FACTOR (EPF)/EPF-like (EPFL)-family of cysteine-rich secreted peptides with 11 members in Arabidopsis. Some EPF/EPFL peptides act antagonistically in stomata development by competing for interaction with receptor complexes and consequently trigger different signaling readouts in the stomata cell lineage (Torii 2012). Moreover, while EPFL1, EPFL2, EPFL4, and EPFL6 function redundantly in the shoot apical meristem (Kosentka et al. 2019), EPFL4 and EPFL6 stimulate aboveground organ elongation (Uchida et al. 2012; Kosentka et al. 2019). Our recent study revealed that Arabidopsis ERf is enriched in epidermal nucellus cells of ovule primordia and loss-of-function of ERf induced supernumerary MMC-like cell formation (Hou et al. 2021). This observation suggested the involvement of ERf in MMC differentiation. However, ligands for ERf receptors during ovule development have not yet been identified and it was unclear how activated ERf complexes trigger downstream gene expression to restrict MMC formation.
We reported previously that quintuple mutants of the BZR1 transcription factor family (named qui-1) produced multiple MMCs in about 70% of ovules. This number exceeded by far the 20% multiple MMCs in BR receptor mutant ovules and indicated that other ligand–receptor complexes function upstream of the BZR1 family. Here, we reveal that EPFL-ERf ligand–receptor pairs act upstream of the BZR1 family and coordinate regulation of female germline specification by activating a nucleolar GTP-binding protein, NUCLEOSTEMIN-LIKE 1 (NSN1). In summary, this study elucidated a gene regulatory network that provides new insights for understanding how two different signaling pathways are interconnected to control female germline specification in concert.
Results
EPFL1, EPFL2, EPFL4, and EPFL6 are involved in MMC specification
We have recently shown that ERf members play an important role in MMC differentiation as ovules of the er erl1 erl2 triple mutant contained supernumerary MMC-like cells (Hou et al. 2021). To investigate whether ERf signaling regulates MMC differentiation in a dosage-dependent manner, we obtained single and double mutants of ERfs. Single and erl1 erl2 double mutants displayed normal fertility (Supplemental Fig. S1, A and B). er erl1 and er erl2 double as well as er erl1+ erl2 triple mutants showed reduced fertility compared with wild-type plants. er erl1 erl2+ and er erl1 erl2 triple mutants exhibited complete sterility (Supplemental Fig. S1, A and B and Fig. 1F). Nuclear localization of AGO9 and KNUCKLES (KNU) serve as excellent MMC markers (Rodriguez-Leal et al. 2015). Immunolocalization of AGO9 and pKNU:KNU-VENUS marker analysis revealed that ∼22% er erl1 erl2+ ovules and ∼40% er erl1 erl2 ovules displayed supernumerary enlarged MMC-like cells. This frequency is significantly higher than that in wild-type plants and double mutants (Supplemental Fig. S1, C–H and Fig. 1, G–L).
Figure 1.
Disruption of ERf signaling leads to the formation of multiple MMCs. MMC formation was compared in wild-type, epfl1,2,4,6 quadruple, and er erl1 erl2 triple mutants, respectively. A–E) Confocal microscopic analysis of indicated constructs in ovules from stage 1-III to stage 2-II. F) Siliques and quantification of seed-set percentage of indicated lines. Data represent means ± SD (n = 10 siliques from five independent plants). Seed-set percentage was calculated corresponding to the percentage of seeds/ovules. G) DIC observation of wild-type and mutants of premeiotic ovules (stage 2-I). Ovules in epfl1,2,4,6 and er erl1 erl2 mutants displayed supernumerary enlarged MMC-like cells (left panel) and no MMC formation (right panel). Arrows point toward nuclei of enlarged MMC-like cells. H) Statistical analysis of ovules showing multiple MMC-like cells in (G). Error bars indicate ± SD (n = 5 biological replicates, each replicate containing more than 100 ovules). I) AGO9 immunolocalization (green) in wild-type and mutants of premeiotic ovules (stage 2-I). Ovules in epfl1,2,4,6 and er erl1 erl2 mutants displayed supernumerary enlarged MMC-like cells (left panel) and no MMC formation (right panel). J) Statistical analysis of ovules showing AGO9 protein in multiple cells in (H). Error bars indicate ± SD (n = 5 biological replicates, each replicate containing more than 100 ovules). K) Signal corresponding to pKNU:KNU-VENUS (green) in wild-type and mutants of premeiotic ovules (stage 2-I). Ovules in epfl1,2,4,6 and er erl1 erl2 mutants displayed supernumerary enlarged MMC-like cells (left panel) and no MMC formation (right panel). L) Statistical analysis of ovules showing pKNU:KNU-VENUS in multiple cells in (I). Error bars indicate ± SD (n = 5 biological replicates, each replicate containing more than 100 ovules). Significance evaluations between wild-type and mutants were performed by student's t-test (**P < 0.01). Bars = 2 mm in (E). Bars = 10 µm in (A)–(D), and (G), (I) and (K).
To determine whether MMC-like cells present in er erl1 erl2 undergo meiosis, we performed immunostaining using antibodies against DMC1 (DISRUPTED MEIOTIC cDNA1), a recombinase expressed specifically in the MMC of wild-type ovules during meiosis (Supplemental Fig. S2). Expression of DMC1 was only detected in one MMC and not in the other enlarged MMC-like cells in the er erl1 erl2 triple mutant (Supplemental Fig. S2). This indicates that the excess MMC-like cells fail to enter meiosis. These results together suggested that ERf redundantly regulates plant fertility and restricts germline identity to a single MMC but does not control further entry into meiosis. Consistent with previous findings (Hou et al. 2021), we also found that about 18% of er erl1 erl2 ovules do not contain an MMC (Fig. 1, G–L), which was not observed in wild-type ovules. This suggests that ERf is also required for promoting MMC formation.
To narrow down the group of ligands that ERfs might perceive to regulate MMC differentiation, we investigated the expression patterns of EPFL1, EPFL2, EPFL4, and EPFL6, which function redundantly in the shoot apical meristem (Kosentka et al. 2019), by first using the VENUS transcriptional reporter assays. We found that EPFL1, EPFL4, and EPFL6 are predominately expressed in nucellar epidermal cells in ovule primordia from stage 1-III to stage 2-II (Fig. 1, A–D). This pattern is similar to the gene expression pattern of ERfs (Hou et al. 2021). We further generated ER-YFP protein fusion driven by the endogenous ER promoter and detected the receptor in the plasma membrane of both L1 and L2 layer cells of ovule primordia from stage 1-III to stage 2-II (Fig. 1E). To further test if EPFL1, EPFL2, EPFL4, and EPFL6 peptide ligands function in fertility regulation, we generated a series of mutants. Single and double mutants of EPFL1, EPFL2, EPFL4, and EPFL6 displayed normal fertility (Supplemental Fig. S3, A and B); however, triple mutants showed reduced fertility, and quadruple mutants (epfl1+,2,4,6 and epfl1,2,4,6) showed further reduced fertility (Fig. 1F and Supplemental Fig. S3, A and B). epfl1,2,4,6 displayed completely sterility (Fig. 1F). The sterile phenotype in epfl1,2,4,6 could be partially rescued by using EPFL1, EPFL2, EPFL4, and EPFL6 genes driven by their own promoters (Supplemental Fig. S4, A and C). These findings indicated that fertility regulation requires EPFL1, EPFL2, EPFL4, and EPFL6-mediated signaling in a dosage-dependent manner.
To determine whether EPFL1, EPFL2, EPFL4, and EPFL6 are involved in MMC differentiation, we analyzed the MMC specification phenotype in the epfl1,2,4,6 quadruple mutant. 86.9% (n = 189) of epfl1,2,4,6 mutant ovules displayed supernumerary enlarged MMC-like cells, which is significantly higher than the observed 0.86% (n = 177) in wild-type ovules (Fig. 1, G and H). The multiple MMC-like cells phenotype in epfl1,2,4,6 was partially rescued by pEPFL:EPFL constructs for EPFL1, EPFL2, EPFL4, and EPFL6, respectively (Supplemental Fig. S4, B and D).
To determine whether enlarged MMC-like cells also acquire MMC identity, we carried out whole-mount immunolocalization using anti-AGO9 antibodies. AGO9 was only detected in the nucleus of a single MMC in 96.8% (n = 155) wild-type ovules (Fig. 1, I and J). In contrast, in 86.5% (n = 163) of epfl1,2,4,6 ovules, AGO9 accumulated in the nuclei of more than one cell (Fig. 1, I and J). We further introduced the pKNU:KNU-VENUS marker into epfl1,2,4,6. KNU-VENUS was detected in the single MMC of wild-type ovule primordia with a frequency of 96.98% (n = 163) (Fig. 1, K and L). In epfl1,2,4,6, KNU-VENUS was observed in more than one cell in 84.2% (n = 196) ovules (Fig. 1, K and L). In contrast, the meiosis marker DMC1 was only detected in one MMC and not in the other enlarged MMC-like cells in epfl1,2,4,6 ovules (Supplemental Fig. S2). These data indicated that the excessive enlarged cells in epfl1,2,4,6 ovules obtained cellular identity of an MMC, but did not develop into fully functional MMCs undergoing meiosis. Triple mutants and epfl1+,2,4,6 quadruple mutants of EPFLs also developed supernumerary enlarged MMC-like cells that exhibited AGO9 and KNU positive signals (Supplemental Fig. S3). But similarly, DMC1 expression was only detected in a single MMC of the triple mutants and epfl1+,2,4,6 quadruple mutants (Supplemental Fig. S2). Moreover, similar to er erl1 erl2 ovules, epfl1,2,4,6 ovules lack an MMC at a frequency of 13.1% (n = 189) and also lack detectable AGO9 and KNU-VENUS signals at a similar frequency (13.5%; n = 163% and 15.8%; n = 196), respectively (Fig. 1, G–L). These data suggested that like ERfs, EPFL1, EPFL2, EPFL4, and EPFL6 also play a role in promoting MMC initiation, in addition to restricting MMC fate to a single cell.
Recent reports further uncovered the role of distal nucellar epidermal ovule cells in MMC specification (Su et al. 2020). To explore whether the supernumerary MMC-like cells phenotypes might be caused by defects in the distal nucellar epidermis, we specifically activated the expression of ER in the distal epidermal cells of er erl1 erl2 ovules by using SPL and WUS promoters. We found that specific activation of ER in epidermal cells is unable to rescue the MMC specification defects in er erl1 erl2 (Supplemental Fig. S5). However, ER driven by its own promoter can ultimately rescue the mutant phenotype in er erl1 erl2 (Supplemental Fig. S5). Altogether these findings indicated that EPFL1, EPFL2, EPFL4, and EPFL6 redundantly restrict germline cell fate to a single MMC, but do not regulate further progression into meiosis.
BZR1 family members act downstream of EPFL-ER ligand–receptor pairs in regulating MMC specification
To investigate whether known MMC specification factors are regulated by EPFL-ER signaling, we compared phenotypes of available mutants with epfl1,2,4,6 and er erl1 erl2 mutants. We found that the multiple MMCs phenotype of bri1-116 and qui-1 (quintuple bes1-1, bzr1-1, beh1-1, beh3-1, beh4-1 mutant) (Cai et al. 2022) appeared similar to that of epfl1,2,4,6 and er erl1 erl2 ovular mutants. The BRI1 receptor and the BZR1 transcription factor family, which consists with BZR1, BES1, BZR1/BES1 HOMOLOG1 (BEH1), BEH2, BEH3, and BEH4 of six members, serve as key components of the BR-signaling pathway (Chen et al. 2019).
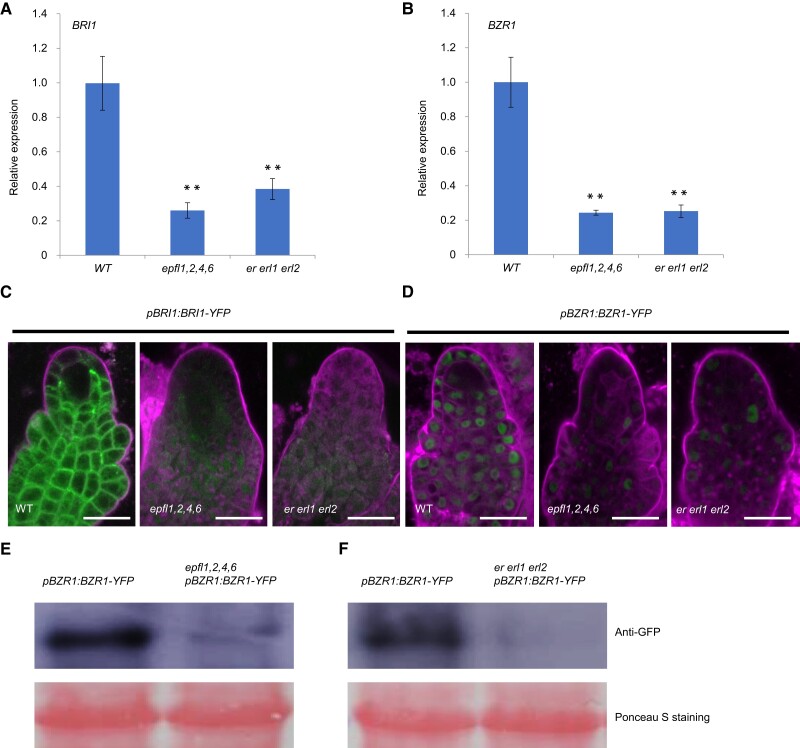
To investigate whether BR signaling is associated with EPFL-ER signaling, we first analyzed the expression level of BRI1 and BZR1 in epfl1,2,4 ,6 and er erl1 erl2 mutants and found that expression levels were strongly reduced compared to wild-type ovules (Fig. 2, A and B). Moreover, we also introduced pBRI1:BRI1-YFP and pBZR1:BZR1-YFP into epfl1,2,4,6 and er erl1 erl2 mutants, respectively, and found that in comparison to wild-type ovule primordia protein levels were very low (Fig. 2, C and D). Immunoblot analysis with antibody against GFP confirmed that BZR1-YFP protein concentration was low in epfl1,2,4,6 and er erl1 erl2 mutants (Fig. 2, E and F). To investigate whether BRI1-mediated signaling can alter ER expression via feedback regulation, we detected the expression level of ER and found that the expression of ER was unaltered in the bri1-116 mutant compared to wild-type ovules (Supplemental Fig. S6A). Expression level of pER:gER-YFP in the bri1-116 mutant was also comparable to that of wild-type ovules (Supplemental Fig. S6B). These results indicated that BRI1-BZR1 signaling likely acts downstream of EPFL-ER.
Figure 2.
BRI1 and BZR1 are activated by the EPFL-ERf signaling pathway. A) Relative expression analysis of BRI1 in wild-type, epfl1,2,4,6, and er erl1 erl2 mutants by RT-qPCR. Values are means ± SD from three biological replicates. B) Relative expression analysis of BZR1 in wild-type, epfl1,2,4,6, and er erl1 erl2 mutants by RT-qPCR. Values are means ± SD from three biological replicates. C) Confocal microscopic analysis of pBRI1:BRI1-YFP in wild-type, epfl1,2,4,6, and er erl1 erl2 ovules at stage 2-II. D) Confocal microscopic analysis of pBZR1:BZR1-YFP in wild-type, epfl1,2,4,6, and er erl1 erl2 ovules at stage 2-II. E) BZR1-YFP immunoblotting with anti-GFP antibody in wild-type and epfl1,2,4,6 background. Ponceau S staining is used as a loading control. F) BZR1-YFP immunoblotting with anti-GFP antibody in wild-type and er erl1 erl2 background. Ponceau S staining is used as a loading control. Significance evaluations between wild-type and mutants were performed by student's t-test (**P < 0.01). Bars = 10 µm.
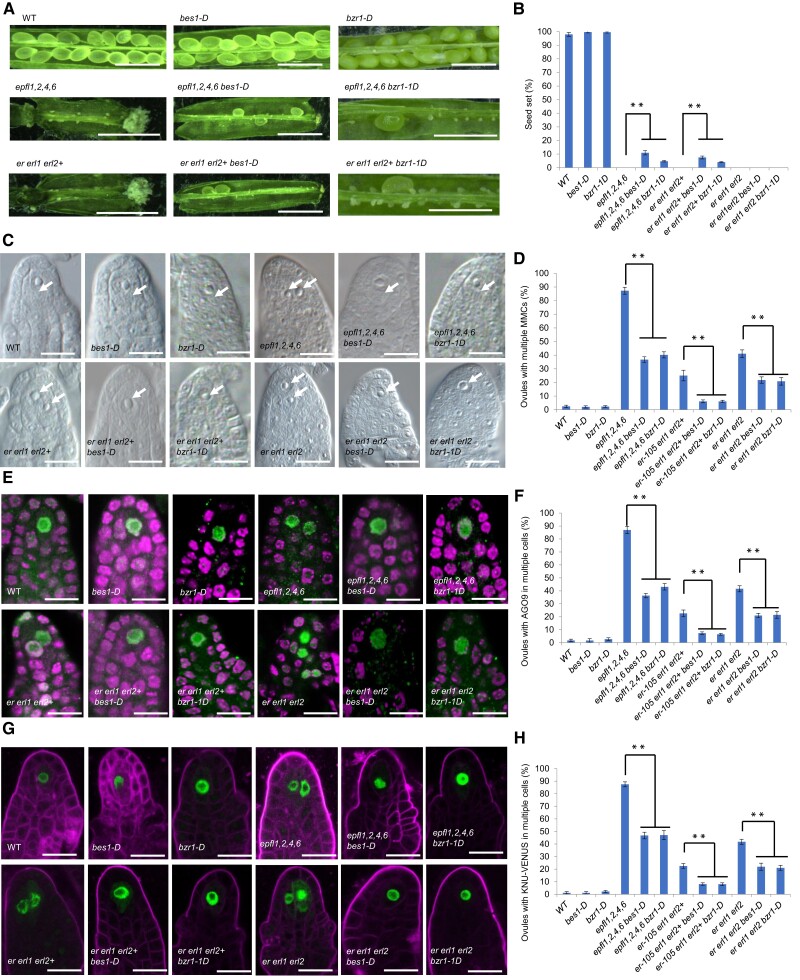
To further test this hypothesis, we introduced gain-of-function mutations of BZR1 and BES1 (bzr1-1D and bes1-D), respectively, into epfl1,2,4,6 and er erl1 erl2+ independently by genetic crossing. Genotypic and phenotypic analyses showed that bzr1-1D and bes1-D could partially restore the fertility of epfl1,2,4,6, er erl1 erl2+, but not that of er erl1 erl2 (Fig. 3, A and B). Observation by differential interference contrast (DIC) microscopy, AGO9 immunolocalization and pKNU:KNU-VENUS marker analysis showed that bzr1-1D and bes1-D could partially complement the multiple MMC-like cells defects in epfl1,2,4,6, er erl1 erl2+ and er erl1 erl2 (Fig. 3, C–H). These data suggest that the sterility of er erl1 erl2 is not only attributed to the defects in MMC specification.
Figure 3.
BES1 and BZR1 gain-of-function mutants partially rescue multiple MMC formation in EPFL-ERf signaling mutants. bes1-D and bzr1-1D gain-of-function mutants were introduced in the epfl1,2,4,6 quadruple mutant, the er erl1 erl2+ triple mutant, and the er erl1 erl2 triple mutant. A) Siliques and B) quantification of seed-set percentage of indicated mutant lines. Data represent means ± SD (n = 10 siliques from five independent plants). Seed-set percentage was calculated corresponding to the percentage of seeds/ovules. C) DIC observation of wild-type and indicated mutants of premeiotic ovules (stage 2-I). Arrows point toward nuclei of enlarged MMC-like cells. D) Statistical analysis of ovules showing multiple MMC-like cells in (C). Error bars indicate ± SD (n = 5 biological replicates, each replicate containing more than 100 ovules). E) AGO9 immunolocalization (green) in wild-type and indicated mutants of premeiotic ovules (stage 2-I). F) Statistical analysis of ovules showing AGO9 protein in multiple cells in (E). Error bars indicate ± SD (n = 5 biological replicates, each replicate containing more than 100 ovules). (G) Signal corresponding to pKNU:KNU-VENUS (green) in wild-type and mutants premeiotic ovules (stage 2-I). H) Statistical analysis of ovules showing pKNU:KNU-VENUS in multiple cells in (I). Error bars indicate ± SD (n = 5 biological replicates, each replicate containing more than 100 ovules). Significance evaluations between wild-type and mutants were performed by student's t-test (**P < 0.01). Bars = 2 mm in (A). Bars = 10 µm in (C), (E), and (G).
It was previously reported that BR treatment can increase the level of nonphosphorylated BZR1 and BES1 (Chen et al. 2019). To further determine whether BZR1 acts downstream of EFPL-ERf, we tested whether BZR1 phosphorylation levels change upon EPFL treatment. Immunoblotting analysis indicated that without treatment by EPFL1 peptide (mock experiment), wild-type inflorescences showed very low levels of nonphosphorylated BZR1 (Fig. 4A). Notably, treatment of EPFL1 peptide significantly induced accumulation of nonphosphorylated BZR1 (Fig. 4A).
Figure 4.
EPFL1 peptide treatment increases the level of nonphosphorylated BZR1. A) Immunoblotting analyses showing accumulation of nonphosphorylated BZR1 after EPFL1 peptide treatment. B) DIC observation of premeiotic ovules (stage 2-I) of epfl1+,2,4,6 and er erl1 erl2+ mutants, respectively, after EPFL1 peptide treatment. MMC-like cells are indicated by white arrows. C) Statistical analysis of ovules showing multiple MMC-like cells in (B). Error bars indicate ± SD (n = 5 biological replicates, each replicate containing more than 100 ovules).
We next examined whether EPFL peptide treatment can rescue the multiple MMCs phenotype in epfls and erf mutants, respectively. We applied EPFL1 peptide to inflorescences of epfl1+,2,4,6 and er erl1 erl2+ mutants, and found that exogenous treatment of EPFL1 peptide indeed repressed formation of multiple MMC-like cells in the epfl1+,2,4,6 mutant, but not in the er erl1 erl2+ mutant (Fig. 4, B and C). In conclusion, these findings indicated that BZR1 family members act downstream of the EPFL-ER ligand–receptor pair in MMC specification.
ER and BRI1 pathways genetically interact to prevent differentiation of multiple MMC-like cells
In a previous study, we showed that ER and BRI1 coordinately regulate inflorescence architecture (Cai et al. 2021a). To investigate whether ER and BRI1 signaling also coordinates female germline specification, we generated the er-105 bri1-119 double mutant. The er-105 bri1-119 double mutant is completely sterile showing a much more severe phenotype compared with er-105 and bri1-119 mutants, which display partial fertility (Supplemental Fig. S7, A and B). Moreover, 36.5% (n = 166) of er-105 bri1-119 ovules displayed supernumerary enlarged MMC-like cells, which was significantly higher than the observed 2.6% (n = 153) in wild-type ovules and each 2.0% in er-105 (n = 162) and bri1-119 (n = 150) single-mutant ovules, respectively (Supplemental Fig. S7, C and F).
To determine whether the enlarged cells acquired MMC identity, we looked for the presence of AGO9 and detected it in the nucleus of MMCs in 98.3% (n = 121) of wild-type ovules (Supplemental Fig. S7, D and G), but only in 38.1% (n = 147) of er-105 bri1-119 ovules where it accumulated in the nuclei of more than one cell (Supplemental Fig. S7, D and G). We also introduced pKNU:KNU-VENUS into the er-105 bri1-119 mutant by crossing. KNU-Venus was detected in the single MMC of 96.98% (n = 163) wild-type ovule primordia (Supplemental Fig. S7, E and H) but occurred in more than one cell in 36.7% (n = 185) of ovules in the er-105 bri1-119 mutant (Supplemental Fig. S7, E and H). DMC1 immunolocalization analysis indicated that only one of the enlarged MMC-like cells in er-105 bri1-119 ovules entered meiosis (Supplemental Fig. S2).
In addition, we also introduced gain-of-function mutants of BZR1 and BES1 (bzr1-1D and bes1-D, respectively) into er-105 bri1-119 by genetic crossing. Genotypic and phenotypic analyses indicated that bzr1-1D and bes1-D can partially restore the fertility of er-105 bri1-119 (Supplemental Fig. S7, A and B). Immunolocalization of AGO9 and the pKNU:KNU-VENUS marker showed that bzr1-1D and bes1-D can also partially rescue the multiple MMC-like phenotype in er-105 bri1-119 (Supplemental Fig. S7, C–H). These data together indicated that ER and BRI1 genetically interact to prevent formation of multiple MMC-like cells and that BZR1 family members act downstream of ER and BRI1 in MMC specification.
BZR1 directly targets to NSN1 depending on EPFL-ERf signaling
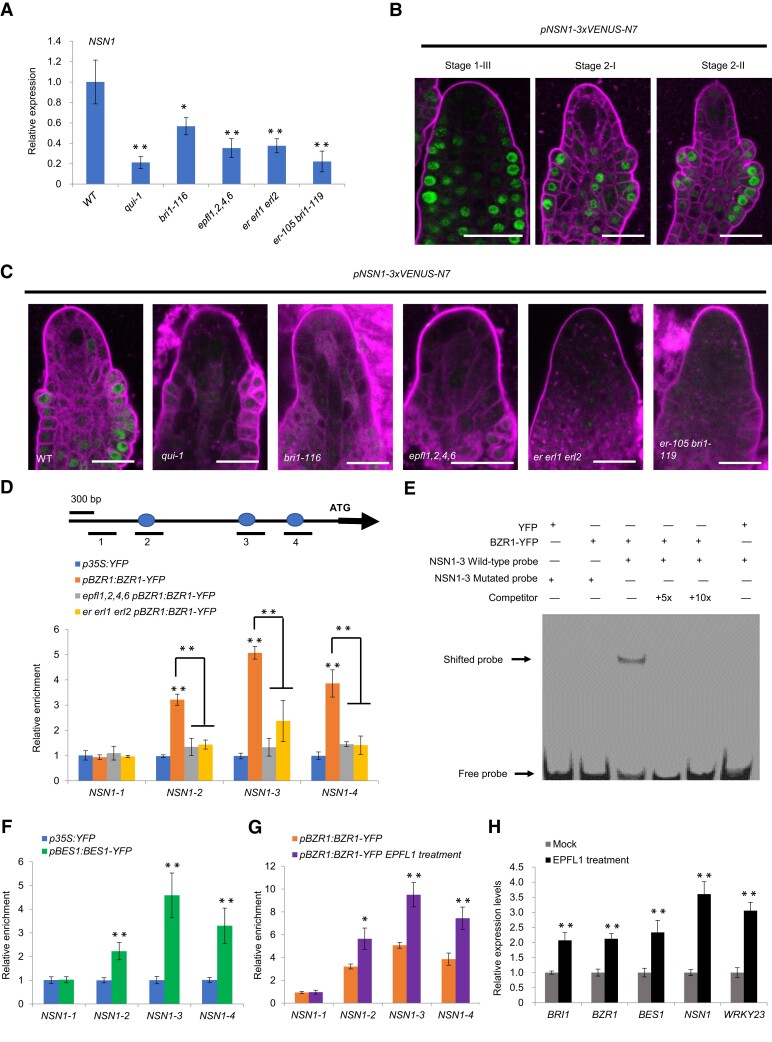
It has been reported that more than 7,000 genes (including high-confidence and low-confidence genes) are directly targeted by BZR1, which can either activate or repress their expression (Sun et al. 2010). To gain a deeper insight into the mechanisms of BZR1 family members regulating female germline initiation, we screened these 7,000 BZR1 targets genes and identified NSN1, which encodes a nucleolar GTP-binding protein, as a direct target gene. In our previous study, a homolog of NSN1 was identified as an MMC-stage preferentially expressed gene in the pineapple ovule (Zhao et al. 2021). RT-qPCR analysis showed that the transcript level of NSN1 was significantly reduced in qui-1 and bri1-116 mutants compared with that of wild-type ovules (Fig. 5A). This indicates that BZR1 and BR signaling activate the expression of NSN1. We also generated a promoter–reporter fusion construct (pNSN1:GUS) and found GUS expression in young seedling, mature leaf, inflorescence, sepal, petal, and pistil (Supplemental Fig. S8, A–E).
Figure 5.
BZR1 binds to E-boxes in the promoter of NSN1. A) RT-qPCR quantification of relative expression levels of NSN1 encoding a nucleolar GTP-binding protein in wild-type, epfl1,2,4,6, er erl1 erl2, bri1-116, bri1-116, and qui-1 (quintuple bes1-1, bzr1-1, beh1-1, beh3-1, beh4-1 mutant) ovule primordia (from stage 1-III to stage 2-III) attached to the septum. Error bars indicate SD (n = 3, biological repeats). Significance evaluations between wild-type and mutants were performed by Student's t-test (*P < 0.05; **P < 0.01). B) Confocal microscopic analysis of pNSN1-3xVENUS-N7 expression (green) in wild-type ovules from stage 1-III to stage 3-I. Bars = 10 µm. C) Confocal microscopic analysis of pNSN1-3xVENUS-N7 expression (green) in wild-type and mutants ovules from stage 1-III to stage 3-I. Bars = 10 µm. D) In vivo binding of BZR1 to the NSN1 promoter. ChIP-qPCR assays were performed with p35S:YFP, pBZR1:BZR1-YFP, epfl1,2,4,6 pBZR1:BZR1-YFP and er erl1 erl2 pBZR1:BZR1-YFP transgenic plants using an anti-GFP antibody. Error bars indicate SD (n = 3, biological repeats). Top panel shows a diagram of the NSN1 promoter. Blue ovals represent E-boxes and black bars below the diagram represent fragments amplified by ChIP-qPCR. Significance evaluations between wild type and mutants were performed by Student's t-test (**P < 0.01). E) EMSA of in vitro binding of BZR1-YFP to the NSN1 promoter fragment containing the E-box cis-element. Arrows indicate binding of BZR1 to Cy5 labeled probes (∼40 bp; −962 bp to −922 bp upstream of the TSS of NSN1). Unlabeled probe (5 times and 10 times of concentration, respectively) was added as a competitor. In the NSN1 mutated probe of the E-box CACATG was changed to AAAAAA. F) In vivo binding of BES1 to the NSN1 promoter. ChIP-qPCR assays were performed using p35S:YFP and pBES1:BES1-YFP transgenic plants with an anti-GFP antibody. ChIP-qPCR primers were the same as in (D). Error bars indicate SD (n = 3, biological repeats). Significance evaluations were performed by Student's t-test (**P < 0.01). G) In vivo binding of BZR1 to the NSN1 promoter. ChIP-qPCR assays were performed using pBES1:BES1-YFP transgenic after EPFL1 peptide treatment and an anti-GFP antibody. ChIP-qPCR primers were the same as in Fig. 5D. Error bars indicate SD (n = 3, biological repeats). Significance evaluations were performed by Student's t-test (*P < 0.05; **P < 0.01). H) RT-qPCR quantification of relative expression levels of BRI1, BZR1, BES1, NSN1, and WRKY23 after EPFL1 peptide treatment. Error bars indicate SD (n = 3, biological repeats). Significance evaluations were performed by Student's t-test (**P < 0.01).
To examine the expression pattern of NSN1 in developing ovules, we generated another promoter–reporter fusion construct (pNSN1:3xVENUS-N7) and found that it was broadly expressed in the proximal part of ovules at stage 1-III (Fig. 5B). At stage 2-I, NSN1 was predominately expressed in the epidermis of ovule primordia and its central region of the nucellus. At stage 2-II, NSN1 expression accumulated in the epidermis of the future inner and outer integuments (Fig. 5B). NSN1 expression was also observed in sporophytic ovule tissue in subsequent female gametophyte developmental stages of ovules (Supplemental Fig. S8F).
To investigate whether BZR1 and BRI1 signaling regulates the expression of NSN1, we introduced pNSN1:3xVENUS into qui-1 and bri1-116 mutants independently by genetic crossing. Expression of pNSN1:3xVENUS was downregulated in qui-1 and bri1-116 mutants compared with wild-type ovules (Fig. 5C). To determine whether BZR1 directly regulates NSN1 expression, we performed chromatin immunoprecipitation (ChIP) using a GFP antibody with floral buds expressing pBZR1:BZR1-YFP and p35S:YFP, respectively. BZR1 was enriched at E-boxes (DNA locations with the consensus sequence CANNTG) in promoter regions at ∼900 and ∼600 base pairs (bp) upstream of the NSN1 transcriptional start site (TSS), respectively (Fig. 5D). To further confirm the binding of BZR1 to E-boxes of the NSN1 promoter, we performed an electrophoretic mobility shift assay (EMSA). These experiments revealed that BZR1 binds to the E-box within the C region (∼900 bp upstream of the TSS) of the NSN1 promoter, but not to a mutated E-box in the same region (Fig. 5E). To determine whether BES1 can also directly bind to the promoter of NSN1, we performed ChIP-qPCR using a GFP antibody with pBES1:BES1-YFP and p35S:YFP floral buds. We found that BES1 was enriched at E-boxes in promoter regions at ∼900 and ∼600 bp upstream of the NSN1 promoter (Fig. 5F). These findings further indicated that NSN1 is a direct target of BZR1 family members.
The aforementioned results demonstrated that the BZR1 family members act downstream of the EPFL-ERf ligand–receptor complex. We thus thought to test whether the expression of NSN1 is regulated by EPFL-ERf. RT-qPCR analysis revealed that transcript levels of NSN1 were significantly reduced in epfl1,2,4,6 and er erl1 erl2 mutants compared with wild-type ovules (Fig. 5A). In addition, expression of pNSN1:3xVENUS was also downregulated in epfl1,2,4,6, er erl1 erl2 and er-105 bri1-119 mutants (Fig. 5C).
To further investigate whether BZR1 binding to NSN1 is dependent on EPFL-ERf ligand–receptor signaling, we expressed pBZR1:BZR1-YFP in epfl1,2,4,6 and er erl1 erl2 as well as in wild-type plants and performed ChIP-qPCR experiments using an anti-GFP antibody. We found that enrichment of BZR1 in the promoter regions of NSN1 was significantly reduced in epfl1,2,4,6 and er erl1 erl2 mutants compared to pBZR1:BZR1-YFP (Fig. 5D). We further performed ChIP-qPCR experiments using an anti-GFP antibody in pBZR1:BZR1-YFP floral buds after EPFL1 peptide treatment. We found that enrichment of BZR1 in the promoter regions of NSN1 was significantly increased after EPFL1 peptide application compared to the mock control (Fig. 5G). In addition, expression levels of BRI1, BZR1, BES1, NSN1, and WRKY23 were increased after EPFL1 peptide treatment compared to the control (Fig. 5H). These results further support the hypothesis that EPFL-ERf signaling regulates the expression of NSN1 and that binding of BZR1 to NSN1 depends on EPFL-ERf.
NSN1 is involved in preventing the formation of multiple MMC-like cells
To elucidate the function of NSN1, we analyzed a nsn1 T-DNA insertional mutant and a crispr-cas9 mutant (nsn1-cr) (Supplemental Fig. S8G). We found that the two independent nsn1 mutant lines flowered significantly later compared with wild-type plants (Supplemental Fig. S8H), and plant height and leaf size of nsn1 mutants were dramatically smaller (Supplemental Fig. S8I). The two independent nsn1 mutants showed also reduced fertility (Fig. 6, A and B). DIC microscopy observation revealed that nsn1 and nsn1-cr displayed multiple enlarged MMC-like cells in ovule primordia at a frequency of ∼39.9% (n = 185) and 35.6% (n = 164), respectively (Fig. 6, C and F). This frequency is significantly higher than that in wild-type ovules (2.48%, n = 166) (Fig. 6, C and F). Examination of AGO9 and KNU markers expression indicated that the multiple enlarged MMC-like cells in these lines obtained the identity of MMCs (Fig. 6, D, E, and G). The DMC1 signal was detected in only one MMC and not in the other enlarged MMC-like cells (Supplemental Fig. S9A). This indicates that the excess MMC-like cells failed to enter meiosis. In agreement with this finding, callose deposition was only visible in a single MMC but not in the other enlarged MMC-like cells in nsn1 ovules (Supplemental Fig. S9B). This phenotype is similar to that of epfl1,2,4,6, and er erl1 erl2 described above. pNSN1:NSN1-3Xvenus-N7, pBZR1:NSN1-3Xvenus-N7, and pER:NSN1-3xVENUS-N7 constructs were sufficient to rescue the low fertility rate and defective MMC specification phenotypes in nsn1, suggesting that NSN1 is responsible as a downstream factor for the mutant phenotype (Fig. 6, A–D, F, and G).
Figure 6.
NSN1 is required for preventing multiple MMC-like cells formation. A) Siliques and B) quantification of seed-set percentage of lines indicated. Data represent means ± SD (n = 10, siliques from five independent plants and two independent siliques from each plant; **P < 0.01 by t-test). Seed-set percentage was calculated corresponding to the percentage of seeds/ovules. C) DIC observation of wild-type, nsn1 mutants, and complementation lines showing premeiotic ovules (stage 2-I). Arrows point toward nuclei of enlarged MMC-like cells. D) AGO9 immunolocalization (green) in wild-type, nsn1 mutant, and premeiotic ovules of complementation lines (stage 2-I). E) Signal corresponding to pKNU:KNU-VENUS (green) in premeiotic ovules (stage 2-I) of wild-type and nsn1 mutant lines. F) Statistical analysis of ovules showing multiple MMC-like cells in (C). Error bars indicate ± SD (n = 5 biological replicates, each replicate containing more than 100 ovules). G) Statistical analysis of ovules showing AGO9 protein in multiple cells in (D). Error bars indicate ± SD (n = 5 biological replicates, each replicate containing more than 100 ovules). H) Expression pattern of pBZR1:NSN1-3xVENUS-N7 in ovules at stage 2-II. I) DIC observation of qui-1 and qui-1 pBZR1:NSN1-3xVENUS-N7 premeiotic ovules (stage 2-I). MMC-like cells are indicated by white arrows. J) Statistical analysis of ovules showing multiple MMC-like cells in (I). Error bars indicate ± SD (n = 5 biological replicates, each replicate containing more than 100 ovules). Significance evaluations between wild type and mutants were performed by student's t-test (**P < 0.01). Numbers in each panel denote the frequencies of the phenotypes shown. Bars = 1 mm in (A) and (N). Bars = 10 µm in (C)–(E), (H), (I), and (K–M).
To further confirm whether the observed phenotypes might be due primarily to defects in the distal nucellar epidermis, we specifically activated NSN1 in distal nucellar epidermal cells using SPL and WUS promoters and found that activation of NSN1 in the distal epidermal cell could not rescue the mutant phenotype in nsn1 (Supplemental Fig. S10, A and B). However, we specifically activated NSN1 in the inner integument primordia using the promoter of KLU, a cytochrome P450 gene (CYP78A5) and found that activation of NSN1 in the inner integument primordia almost completely rescued the multiple MMC phenotype in nsn1 (Supplemental Fig. S10, A and B). In addition, activation of NSN1 in the distal hypodermal cells surrounding the MMC by using the WRKY28 promoter can partially rescue the multiple MMC-like cells phenotype in nsn1 (Supplemental Fig. S10, A and B). This implies that NSN1 executes its role in restricting MMC fate to a single cell in multiple cell layers near the MMC.
NSN1 functions downstream of BZR1 in regulating MMC specification
To determine whether NSN1 indeed functions downstream of BZR1 to inhibit the formation of multiple MMCs in ovule primordia, we overexpressed NSN1 in sporophytic cells of qui-1 ovules by introducing a full-length NSN1 genomic fragment–reporter fusion construct with a nuclear localization signal under the control of the BZR1 promoter. In the T1 generation, 15 independent qui-1 transformants carrying the pBZR1:NSN1-3xVENUS-N7 transgene were obtained. These lines showed a significantly reduced frequency of multiple MMC-like cells compared to qui-1 ovules, but still a higher number than those in wild-type ovules (Fig. 6, H–J). This partial complementation indicates that NSN1 is a major but not the sole molecular player functioning downstream of BZR1 to inhibit excessive MMC formation in ovule primordia. Moreover, we also performed ectopic activation of NSN1 in distal epidermal cells by using SPL and WUS promoters and found that this approach could not rescue the mutant phenotype in qui-1 (Supplemental Fig. S11, A and B). This indicates that NSN1 functions in the nondistal epidermis of ovules.
To further confirm that BRI1-BZR1-NSN1 signaling functions with EPFL-ERf in a common pathway, we overexpressed BRI1, BZR1, and NSN1 by using the ER promoter in epfl1,2,4,6 and er erl1 erl2 mutants, respectively, and found that this partially rescued the MMC defects in epfl1,2,4,6 and er erl1 erl2 mutants, respectively (Supplemental Fig. S12). These results further suggested that the BRI1-BZR1-NSN1 signaling pathway acts downstream of EPFL-ERf in MMC specification.
NSN1 and WRKY23 genetically interact to prevent differentiation of multiple MMC-like cells
In a recent report, we showed that WRKY23 is a direct target gene of BZR1 (Cai et al. 2022). To determine whether expression of WRKY23 is also regulated by EPFL-ERf, we performed RT-qPCR analysis and revealed that transcript levels of WRKY23 were also significantly reduced in epfl1,2,4,6, er erl1 erl2, and er-105 bri1-119 mutants compared with wild-type ovules (Supplemental Fig. S13A). We further observed that expression of pWRKY23:GFP was also significantly reduced (Supplemental Fig. S13B). We next generated a nsn1 wrky23 double mutant and found that double mutant ovules contained multiple MMC-like cells at a frequency of 70.8% (n = 213), which was significantly higher than that in wild type (∼2%), nsn1 (∼40%), and wrky23 (∼20%) mutants (Supplemental Fig. S14, A and C). KNU marker expression analysis confirmed that the multiple enlarged MMC-like cells in the nsn1 wrky23 double mutant contained the molecular characteristics of MMCs (Supplemental Fig. S14, B and D). These results together suggested that NSN1 and WRKY23 may function together in MMC specification.
Discussion
The differentiation of a single L2-layer nucellus cells at the distal part of ovule primordia leading to the formation of the MMC as the initial cell of the female germline appears to be highly complex and tightly regulated by multiple interacting pathways. Usually, the centermost cell among the group of subepidermal nucellus cells will acquire MMC identity (Bower and Schnittger 2021; Hernandez-Lagana et al. 2021). The key events in MMC formation include female germline precursor cell selection, germline precursor cell expansion, germline commitment, and germline transition (Pinto et al. 2019). This is regulated by hormone-signaling pathways (especially by auxin and BR), the epigenetic RdDM pathway, cell cycle factors, and others (Bower and Schnittger 2021).
The ERf-signaling pathway first appeared in early terrestrial plants that evolved many innovations required to survive, e.g. at dryer environments (Shpak 2013). Each plant species usually contains only two or three ERf receptors, while its postulated EPF/EPFL peptide ligand family is relatively large, with 10 or more genes (Shpak 2013). Individual EPF/EPFLs are expressed in a unique spatio-temporal pattern and usually control a specific developmental process. EPF1, e.g. regulates stomata development in rice (Oryza sativa) (Bessho-Uehara et al. 2016). There are 11 EPF/EPFLs in Arabidopsis (Shimada et al. 2011). Mutant analyses showed that four genes, EPFL1, EPFL2, EPFL4, and EPFL6, are essential for the establishment of the shoot apical meristem (Kosentka et al. 2019). Here, we reported that the epfl1,2,4,6 quadruple mutant also exhibits a multiple-MMC phenotype suggesting that these genes function redundantly in MMC specification. EPFL1 and EPFL2 as well as EPFL4 and EPFL6 belong to two closely related clades. EPFL4 and EPFL6 are verified ERf ligands as they bind directly to ERfs. EPFL2 has been shown to bind to ERfs to regulate leaf margin morphogenesis (Takata et al. 2013).
Because all four EPFL genes have the potential to suppress stomata development when expressed in the epidermal tissue layer, they are considered as agonists of ERf receptors (Abrash et al. 2011). It has been reported that in the absence of ER and ERL1, ERL2 is haplo-insufficient for female fertility, whereas ERL1 is haplo-sufficient in the absence of ER and ERL2 (Pillitteri et al. 2007). This appears to occur also in MMC development and specification. In this study, double mutants of ER family members and er erl1+ erl2 showed only one MMC, while about 22% er erl1 erl2+ ovules and about 40% er erl1 erl2 ovules displayed supernumerary enlarged MMC-like cells. Taken together, our data suggest that the expression of EPFLs and ERfs overlap in sporophytic cells in ovule primordia and they act in concert to control MMC differentiation.
Moreover, about 87% of epfl1,2,4,6 ovules generate enlarged MMC-like cells, while only about 40% of er erl1 erl2 ovules contained enlarged MMC-like cells, suggesting that other receptors that work together with ERfs in higher order complexes in MMC specialization. A previous study indicated that the ERf and BRI1-signaling pathways coordinately regulate inflorescence architecture (Cai et al. 2021a). In this study, we found that bri1-119 and er-105 single mutant ovules differentiated only a single MMC; however, the er-105 bri-119 double mutant exhibited multiple MMCs, suggesting that ERf and BRI1 signaling also act together during MMC differentiation. Since the differentiated MMC is the first female germline cell in plants, plants evolved multiple ways to jointly ensure the accuracy of MMC specialization and thus reproductive success. It will now be important to elucidate higher-order receptor complexes or other factors involving in MMC specification. In addition to regulating MMC development, EPFL1/2/4/6 and ERf may also function in other stages of female gametophyte development, as the epfl1,2,4,6 and er erl1 erl2 mutants are completely sterile. In line with this finding, it was recently reported that EPFL1/2/4/6 and ERf play critical roles in regulating integument development in Arabidopsis (Li et al. 2022). The complete sterility in epfl1,2,4,6 and er erl1 erl2 mutants may be caused by defective megasporogenesis and megagametogenesis.
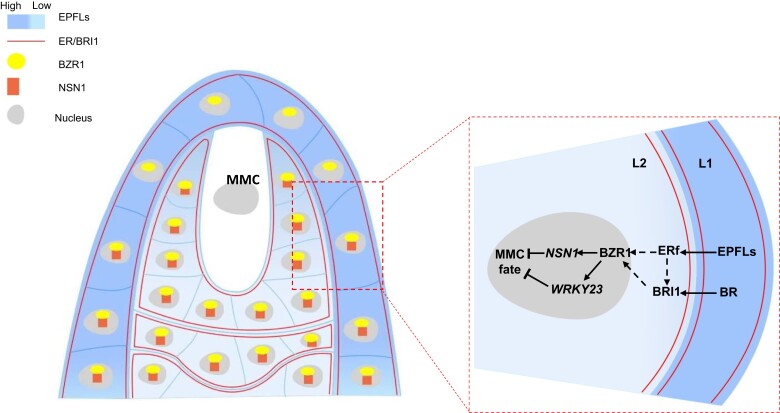
In another study, we demonstrated that BZR1 family transcription factors play redundant and critical roles in MMC differentiation (Cai et al. 2022). The BZR1 family quintuple mutant (qui-1) showed the same multiple MMC phenotypes similar to the epfl1,2,4,6 quadruple and er erl1 erl2 triple mutants, which implies that EPFL-ER and BZR1 family transcription factors may belong to the same gene regulatory network (GRN). This hypothesis was confirmed as bes1-D or bzr1-1D gain-of-function mutants could partially restore multiple MMC-like cells defects in epfl1,2,4,6, in er erl1 erl2 and in er-105 bri1-119. These findings together suggest that BZR1 family transcription factors function as downstream components of the EPFL-ER signaling complex. A recent study further indicated that BES1 family members serve also as downstream components of other non-BR-dependent pathways. It was shown that they act in the TAPETUM DETERMINANT 1 (TPD1)–EXTRA MICROSPOROCYTES1 (EMS1)–SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES 1 and 2 (SERK1/2) signaling pathway to regulate tapetum development (Chen et al. 2019). Additional signaling pathways may also use BZR1 family transcription factors as their downstream signaling components. Our study supports the observation that BZR1 family transcription factors can be shared by at least two distinctive signaling pathways in the regulation of MMC specification (Fig. 7).
Figure 7.
Model of EPFL-ER-BZR1-NSN1 signaling module regulating MMC specification. The model shows that activity of the BZR1 family regulates the repression of MMC fate in sporophytic cells via activating the expression of NSN1. This is shared by EPFL-ERf and BR-BRI1 signaling pathways. Expression of BRI1 is activated by EPFL-ERf, but does not feedback to the regulation of ERf signaling.
BZR1 family transcription factors can thus be considered as important hubs for crosstalk between BR and other signaling pathways. Many genes have been identified as downstream genes of BZR1 family transcription factors, such as DYSFUNCTIONAL TAPETUM1 in pollen development (Chen et al. 2019), PREs and DWF4 in whole plant growth (Zhang et al. 2009; Chen et al. 2019), and WRKY23 in MMC specification (Cai et al. 2022). Here, we added with NSN1 another direct downstream gene. The orthologue of NSN1 was previously reported to be expressed in pineapple (Ananas comosus) ovules at the MMC stage (Zhao et al. 2021). NSN1 encodes a GTP-binding protein, which contains multiple functional domains. These include nuclear localization signals, a GTP-binding domain and an RNA-binding domain. The nsn1 mutant was previously reported to display disrupted leaf polarity and meristem-like outgrowths in the adaxial leaf epidermis, which were accompanied by altered expression patterns of the stem cell marker gene CLAVATA3 (Wang et al. 2012a, b). NSN1 was further shown to genetically interact with the homeotic genes AGAMOUS and APETALA2 to regulate floral identity and maintenance of inflorescence meristem (Wang et al. 2012a, b). Here, we showed that NSN1 also genetically interacts with other transcription factors like WRKY23 for MMC differentiation. It will now be important to elucidate how it coordinates with WRKY23 to suppress MMC formation in ovule primordia.
Although this study significantly advanced our knowledge about the GRN controlling and restricting the initiation of female germline development to a single subepidermal nucellus cell, it is still unclear how expression of components of BR and EPFL-ERf receptor complexes and their shared downstream transcriptional regulators are suppressed once MMC differentiation is initiated. To complete the GRN, it will be important to analyze how auxin regulated pathways, RdDM, and other pathways affect and integrate into the GRN reported here.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia (Col) was used as wild-type plants. All mutants were in the Col background. Mutants and alleles used in this study were bri1-116, bri1-119, er-105, epfl1,2,4,6 (Kosentka et al. 2019), er-105 erl1-2 erl2-1 (simplified as er erl1 erl2), bes1-1 (SALK_098634), bzr1-1 (GK_857E04), beh1-1 (GK_432C09), beh3-1 (SALK_017577), beh4-1 (SALK_055421), nsn1 (SALK_ 029201), wrky23 (SALK_003943C), and bes1-1 bzr1-1 beh1-1 beh3-1 beh4-1 (qui-1) (Chen et al. 2019). Plants were grown under 16 h light (50 μmol m−2 s−1 white light)/8 h dark at 22 °C.
DIC observation of ovule structures
Ovules from wild-type, mutant, and transgenic plants were dissected from pistils at stage 8 to 13 flowers in a drop of chloral-hydrate solution (chloral-hydrate:H2O:glycerol = 8:2:1) (Zhao et al. 2014). Cleared ovules were observed and imaged using a ZEISS (Imager.A2) microscope with DIC optics.
Plasmid construction
To generate the pSPL:ER construct, a 2 kbp genomic fragment containing the putative SPL promoter was amplified with primers pSPL-F1 and pSPL-R1 (Supplemental Table S1). The same methods were used for generating pER:BRI1, pER:BZR1, pER:NSN1, and pEPFLs:EPFLs constructs. The full-length NSN1 cDNA was amplified with primers NSN1-F1 and NSN1-R1. The two PCR products were cloned into the pSPL4 3xVENUS-N7 vector with BamHI and XbaI restriction sites (Heisler et al. 2005). The same methods were used for generating pWUS:ER, pSPL:NSN1, pWUS:NSN1, pKLU:NSN1, and pWRKY28:NSN1 constructs. Promoter:3xVenus-N7 or Promoter:gene-3xVenus-N7 (for EPFL1, EPFL2, EPFL4, EPFL6, ERL1, and ERL2) were generated by the same method using the primers listed in Supplemental Table S1. PCR fragments were cloned into the pSPL4 3xVENUS-N7 vector with BamHI and XbaI restriction sites. The pNSN1:GUS constructs were generated by amplifying a 2 kbp sequence upstream of the NSN1 gene from WT genomic DNA using the primers listed in Supplemental Table S1. The PCR product was cloned into the pENTR/DTOPO vector (Invitrogen). pENTR/D-TOPO clones were recombined into the destination vectors pGWB533 (Nakagawa et al. 2007) using LR Clonase II (Invitrogen).
RT-qPCR
RNA samples were extracted from wild-type and mutant ovules at stage 2-I to 2-III (Cai et al. 2022) attached to the septum. To determine the relative transcript levels of selected genes, quantitative PCR was performed with specific primers (Supplemental Table S1) according to the manufacturer's instructions using the Bio-Rad real-time PCR system and the SYBR Premix Ex Taq II system (TaKaRa, Kyoto, Japan) as described previously (Cai et al. 2017). Relative transcript levels of analyzed genes were normalized using transcript levels of the reference gene HK2 (AT4G26410) (Cai et al. 2021c; Liu et al. 2021). Three biological replicates and two technical replicates for each sample were performed in quantitative PCR experiments.
ChIP-qPCR
ChIP was performed as previously described using inflorescences containing floral buds younger than stage 9. Polyclonal antigreen fluorescent protein (anti-GFP) antibody (ab290; Abcam) was used. ChIP experiment results were calculated by the Fold Enrichment Method (Cai et al. 2022). Fold enrichment of ChIP experiments was calculated using the HK2 gene as a reference. Primers used in ChIP-qPCR are listed in Supplemental Table S1.
Callose detection
Aniline blue staining and callose detection in megaspores were performed as previously described with minor modifications (Siddiqi et al. 2000). These include fixation of inflorescences in formaldehyde/alcohol/acetic acid for 16 h and incubation in 0.1% (w/v) aniline blue in 50 mM phosphate buffer (pH 11) for 8 to 12 h. Stained pistils were mounted in 30% (v/v) glycerol and observed with a BX63 microscope (OLYMPUS, Tokyo, Japan) at an excitation wavelength of 365 nm and by using an emission long-pass filter of 420 nm.
Whole-mount immunolocalization in ovules
Ovules were dissected from 30 to 40 pistils of stage 9 to 11 flowers (Robinson-Beers et al. 1992), fixed and processed according to a previously published protocol (Escobar-Guzman et al. 2015). AGO9 and DMC1 primary antibodies (Agrisera AS10673 and ABclonal Technology, respectively) were used at a dilution of 1:100. An Alexa Fluor 488 secondary antibody (Molecular Probes) was used at a dilution of 1:300. Samples were stained with propidium iodide (500 mg/mL) before mounting, and images were captured using a confocal microscope (Leica TCS SP8). For propidium iodide detection, excitation and emission wavelengths were 568 nm and 575 to 615 nm, respectively. For Alexa Fluor 488 detection, excitation and emission wavelengths were 488 nm and 500 to 550 nm, respectively. Laser intensity and gain were set to the same levels for all analyzed genotypes. For confocal fluorescence microscopy, ovules were prepared according to a previously reported method and mounted in 30% glycerol with 5 μM FM4-64 dye (Christensen et al. 1997). Ovules were stained for 5 min and analyzed using a Leica TCS SP8 microscope.
Electrophoretic mobility shift assay
EMSA was carried out as previously described (Heravi and Altenbuchner 2014). Labeled probes of wild-type and mutated DNA regions were synthesized using Cy5-labeled oligonucleotides (Supplemental Table S1). Unlabeled primer was used as a competitor (Supplemental Table S1). Recombinant proteins of BZR1-GFP or GFP alone were incubated either with wild-type, mutated or competitor probes as described (Heravi and Altenbuchner 2014) by using 5× binding buffer (1 M Tris–HCl pH 7.5, 5 M NaCl, 1 M potassium chloride, 1 M magnesium chloride, 0.5 M EDTA, 10 mg mL−1 BSA at pH 8.0) (Cai et al. 2021b). Reaction mixtures were separated by polyacrylamide gel electrophoresis and scanned by an Odyssey CLx instrument (Li-Cor).
Immunoblot
Proteins were separated by electrophoresis in 15% SDS-polyacrylamide gels and transferred to PVDF membranes in CAPS/methanol buffer (Liu et al. 2021). Primary antibodies specific for anti-GFP (ab290; Abcam; 1:200) were used.
EPFL1 peptide expression, purification, and in vitro treatment
For recombinant protein expression, the sequence of the C-terminal predicted mature EPFL1 peptide domain (residues 54–128) (Lee et al. 2012; Lin et al. 2017) was introduced into the pET32a-modified vector, which carries the 6xHis tag. The EPFL1-6xHis vector was transformed into Escherichia coli (BL21-DE3) and recombinant protein induced at 16 °C with 0.2 mM isopropyl β-D-1 thiogalactopyranoside overnight. Column buffer (20 mM Tris-HCl pH 7.4, 200 mM NaCl, 0.5 M EDTA, 1 mM PMSF, 1 mM DTT) was used to release recombinant protein. Supernatant was incubated with His Beads for 2 h at 4 °C. After enrichment, His beads were washed three times with column buffer and recombinant protein eluted for 1 h at 4 °C using column buffer with 10 mM maltose. Flowers of epfl1+,2,4,6, and er erl1 erl2+ were treated with and without 100 nM EPFL1 peptide, respectively, for 10 d and then MMC phenotypes observed with DIC microscopy. Inflorescences of pBZR1:BZR1-YFP were treated with and without 100 nM EPFL1 peptide, respectively, for 24 h. Total proteins were extracted with 2× SDS buffer containing 125 mM Tris (pH 6.8), 4% (w/v) SDS, 20% (v/v) glycerol, 20 mM DTT, and 0.02% (w/v) bromophenol blue. Proteins were separated by electrophoresis in 15% SDS-polyacrylamide gels and transferred to PVDF membranes in CAPS/methanol buffer (Liu et al. 2021). Primary antibodies specific for anti-GFP (ab290; Abcam; 1:200) were used for detecting the level of unphosphorylated BZR1.
Statistical analysis
All t-test analysis was conducted using Excel. A value of P < 0.05 was considered to be statistically significant. The results of statistical analyses are shown in Supplemental Data Set S1.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: NSN1 (AT3G07050), EPFL1 (AT5G10310), EPFL2 (AT4G37810), EPFL4 (AT4G14723), EPFL6 (AT2G30370), ERECTA (AT2G26330), ERL1 (AT5G62230), ERL2 (AT5G07180), BRI1 (AT4G39400), BZR1 (AT1G75080), BES1 (AT1G19350), WRKY23 (AT2G47260).
Supplementary Material
Acknowledgments
We thank Jia Li and Elena D. Shpak for providing seeds.
Contributor Information
Hanyang Cai, College of Life Sciences, Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Youmei Huang, College of Life Sciences, Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Liping Liu, Cell Biology and Plant Biochemistry, University of Regensburg, 93053 Regensburg, Germany.
Man Zhang, College of Life Sciences, Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fujian Agriculture and Forestry University, Fuzhou 350002, China; Tea Research Institute, Guangdong Academy of Agricultural Sciences & Guangdong Provincial Key Laboratory of Tea Plant Resources Innovation and Utilization, Dafeng Road 6, Tianhe District, Guangzhou 510640, China.
Mengnan Chai, College of Life Sciences, Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Xinpeng Xi, College of Life Sciences, Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Mohammad Aslam, College of Life Sciences, Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Lulu Wang, State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, Guangxi Key Lab of Sugarcane Biology, College of Agriculture, Guangxi University, Nanning 530004, China.
Suzhuo Ma, College of Life Sciences, Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Han Su, College of Life Sciences, Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Kaichuang Liu, College of Life Sciences, Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Yaru Tian, College of Life Sciences, Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Wenhui Zhu, College of Life Sciences, Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Jingang Qi, College of Life Sciences, Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Thomas Dresselhaus, Cell Biology and Plant Biochemistry, University of Regensburg, 93053 Regensburg, Germany.
Yuan Qin, College of Life Sciences, Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fujian Agriculture and Forestry University, Fuzhou 350002, China; State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, Guangxi Key Lab of Sugarcane Biology, College of Agriculture, Guangxi University, Nanning 530004, China.
Author contributions
H.C., Y.H., L.L. cloned the genes, generated transgenic lines and performed phenotypic as well as genetic analysis. M.Z., M.C., X.X., M.A., L.W., and S.Z. conducted ChIP analyses. H.S., K.L., and Y.T., performed quantitative PCR experiments. W.Z. and J.Q. analyzed the data. T.D. assisted with data interpretation. Y.Q. and H.C. designed the research and wrote the manuscript with input from all the authors.
Funding
This work was supported by the German Research Foundation (DFG) via Collaborative Research Center SFB960 to T.D., the National Natural Science Foundation of China (31970333, 32270366 and 32170352), the Science and Technology Program of Fujian Province (2019N5008), the Science and Technology Major Project of Guangxi (Gui Ke 2018-266-Z01) and a Guangxi Distinguished Experts Fellowship to Y.Q. The Excellent Youth Foundation of Fujian Province to H.C. (2022J06014) and Excellent Youth Foundation of Fujian Agriculture and Forestry University to H.C. (xjq202108) are also acknowledged.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. MMC formation in wild-type and ERf mutants.
Supplemental Figure S2. DMC1 immunolocalization in wild-type and ERf as well as EPFL mutants.
Supplemental Figure S3. MMC formation in wild-type and epfl mutants.
Supplemental Figure S4. EPFLs were sufficient to rescue the low fertility and multiple MMC phenotypes in the epfl1,2,4,6 mutant.
Supplemental Figure S5. ERf mutant is only complemented if ER is expressed in L2 layer cells.
Supplemental Figure S6. Lack of BRI1 does not alter ER expression in ovule primordia.
Supplemental Figure S7. BES1 and BZR1 gain-of-function mutants partially rescue the multiple MMC phenotype in the er-105 bri1-119 double mutant.
Supplemental Figure S8. Organ- and tissue-level expression pattern of NSN1.
Supplemental Figure S9. DMC1 immunolocalization and callose deposition in wild-type and nsn1 mutant ovules.
Supplemental Figure S10. Cell type specific expression of NSN1 in nsn1 mutant ovules.
Supplemental Figure S11. Ectopic expression of NSN1 in the L1 layer does not complement the multiple MMC phenotype in qui-1 mutant ovules.
Supplemental Figure S12. Overexpression of BRI1, BZR1, and NSN1 by using the ER promoter partially rescues the multiple MMC phenotype in epfl1,2,4,6 and er erl1 erl2 mutants.
Supplemental Figure S13. Relative expression level of WRKY23 is strongly reduced in epfl1,2,4,6, er erl1 erl2, and er-105 bri1-119 mutant ovules.
Supplemental Figure S14. NSN1 and WRKY23 genetically interact to prevent differentiation of multiple MMC-like cells.
Supplemental Table S1. Primers used in this paper.
Supplemental Data Set 1. Statistical data.
References
- Abrash EB, Davies KA, Bergmann DC. Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand-receptor interactions. Plant Cell. 2011:23(8):2864–2879. 10.1105/tpc.111.086637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho-Uehara K, Wang DR, Furuta T, Minami A, Nagai K, Gamuyao R, Asano K, Angeles-Shim RB, Shimizu Y, Ayano M, et al. Loss of function at RAE2, a previously unidentified EPFL, is required for awnlessness in cultivated Asian rice. Proc Natl Acad Sci U S A. 2016:113(32):8969–8974. 10.1073/pnas.1604849113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böwer F, Schnittger A. How to switch from mitosis to meiosis: regulation of germline entry in plants. Annu Rev Genet. 2021:55(1):427–452. 10.1146/annurev-genet-112618-043553 [DOI] [PubMed] [Google Scholar]
- Cai H, Chai M, Chen F, Huang Y, Zhang M, He Q, Liu L, Yan M, Qin Y. HBI1 acts downstream of ERECTA and SWR1 in regulating inflorescence architecture through the activation of the brassinosteroid and auxin signaling pathways. New Phytol. 2021a:229(1):414–428. 10.1111/nph.16840 [DOI] [PubMed] [Google Scholar]
- Cai H, Huang Y, Chen F, Liu L, Chai M, Zhang M, Yan M, Aslam M, He Q, Qin Y. ERECTA signaling regulates plant immune responses via chromatin-mediated promotion of WRKY33 binding to target genes. New Phytol. 2021b:230(2):737–756. 10.1111/nph.17200 [DOI] [PubMed] [Google Scholar]
- Cai H, Liu L, Huang Y, Zhu W, Qi J, Xi X, Aslam M, Dresselhaus T, Qin Y. Brassinosteroid signaling regulates female germline specification in Arabidopsis. Curr Biol. 2022:32(5):1102–1114.e1105. 10.1016/j.cub.2022.01.022 [DOI] [PubMed] [Google Scholar]
- Cai H, Liu L, Zhang M, Chai M, Huang Y, Chen F, Yan M, Su Z, Henderson I, Palanivelu R, et al. Spatiotemporal control of miR398 biogenesis, via chromatin remodeling and kinase signaling, ensures proper ovule development. Plant Cell. 2021c:33(5):1530–1553. 10.1093/plcell/koab056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Zhao L, Wang L, Zhang M, Su Z, Cheng Y, Zhao H, Qin Y. ERECTA Signaling controls Arabidopsis inflorescence architecture through chromatin-mediated activation of PRE1 expression. New Phytol. 2017:214(4): 1579–1596. 10.1111/nph.14521 [DOI] [PubMed] [Google Scholar]
- Chen W, Lv M, Wang Y, Wang PA, Cui Y, Li M, Wang R, Gou X, Li J. BES1 is activated by EMS1-TPD1-SERK1/2-mediated signaling to control tapetum development in Arabidopsis thaliana. Nat Commun. 2019:10(1):4164. 10.1038/s41467-019-12118-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen CA, King EJ, Jordan JR, Drews GN. Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex Plant Reprod. 1997:10(1):49–64. 10.1007/s004970050067 [DOI] [Google Scholar]
- Escobar-Guzmán R, Rodríguez-Leal D, Vielle-Calzada JP, Ronceret A. Whole-mount immunolocalization to study female meiosis in Arabidopsis. Nat Protoc. 2015:10(10):1535–1542. 10.1038/nprot.2015.098 [DOI] [PubMed] [Google Scholar]
- Ferreira LG, de Alencar Dusi DM, Irsigler AST, Gomes A, Mendes MA, Colombo L, de Campos Carneiro VT. GID1 expression is associated with ovule development of sexual and apomictic plants. Plant Cell Rep. 2018:37(2):293–306. 10.1007/s00299-017-2230-0 [DOI] [PubMed] [Google Scholar]
- Hater F, Nakel T, Groß-Hardt R. Reproductive multitasking: the female gametophyte. Annu Rev Plant Biol. 2020:71(1):517–546. 10.1146/annurev-arplant-081519-035943 [DOI] [PubMed] [Google Scholar]
- He K, Xu S, Li J. BAK1 directly regulates brassinosteroid perception and BRI1 activation. J Integr Plant Biol. 2013:55(12):1264–1270. 10.1111/jipb.12122 [DOI] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol. 2005:15(21):1899–1911. 10.1016/j.cub.2005.09.052 [DOI] [PubMed] [Google Scholar]
- Heravi KM, Altenbuchner J. Regulation of the Bacillus subtilis mannitol utilization genes: promoter structure and transcriptional activation by the wild-type regulator (MtlR) and its mutants. Microbiology (Reading). 2014:160(1):91–101. 10.1099/mic.0.071233-0 [DOI] [PubMed] [Google Scholar]
- Hernandez-Lagana E, Mosca G, Mendocilla-Sato E, Pires N, Frey A, Giraldo-Fonseca A, Michaud C, Grossniklaus U, Hamant O, Godin C, et al. Organ geometry channels reproductive cell fate in the Arabidopsis ovule primordium. Elife. 2021:10:e66031. 10.7554/eLife.66031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Lagana E, Rodriguez-Leal D, Lúa J, Vielle-Calzada JP. A multigenic network of ARGONAUTE4 clade members controls early megaspore formation in Arabidopsis. Genetics. 2016:204(3):1045–1056. 10.1534/genetics.116.188151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M, Belkhadir Y, Dreux M, Dabi T, Noel JP, Wilson IA, Chory J. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature. 2011:474(7352): 467–471. 10.1038/nature10153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Liu Y, Zhang M, Zhao L, Jin X, Liu L, Su Z, Cai H, Qin Y. High-throughput single-cell transcriptomics reveals the female germline differentiation trajectory in Arabidopsis thaliana. Commun Biol. 2021:4(1):1149. 10.1038/s42003-021-02676-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Guan S, Sun Y, Deng Z, Tang W, Shang JX, Sun Y, Burlingame AL, Wang ZY. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol. 2009:11(10):1254–1260. 10.1038/ncb1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosentka PZ, Overholt A, Maradiaga R, Mitoubsi O, Shpak ED. EPFL signals in the boundary region of the SAM restrict its size and promote leaf initiation. Plant Physiol. 2019:179(1):265–279. 10.1104/pp.18.00714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschera U, Wang ZY. Brassinosteroid action in flowering plants: a Darwinian perspective. J Exp Bot. 2012:63(10):3511–3522. 10.1093/jxb/ers065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Kuroha T, Hnilova M, Khatayevich D, Kanaoka MM, McAbee JM, Sarikaya M, Tamerler C, Torii KU. Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev. 2012:26(2):126–136. 10.1101/gad.179895.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997:90(5):929–938. 10.1016/S0092-8674(00)80357-8 [DOI] [PubMed] [Google Scholar]
- Li M, Lv M, Wang X, Cai Z, Yao H, Zhang D, Li H, Zhu M, Du W, Wang R, et al. The EPFL-ERf-SERK signaling controls integument development in Arabidopsis. New Phytol. 2022. 10.1111/nph.18701. Online ahead of print [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001:127(1):14–22. 10.1104/pp.127.1.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002:110(2):213–222. 10.1016/S0092-8674(02)00812-7 [DOI] [PubMed] [Google Scholar]
- Li L, Wu W, Zhao Y, Zheng B. A reciprocal inhibition between ARID1 and MET1 in male and female gametes in Arabidopsis. J Integr Plant Biol. 2017:59(9):657–668. 10.1111/jipb.12573 [DOI] [PubMed] [Google Scholar]
- Lin G, Zhang L, Han Z, Yang X, Liu W, Li E, Chang J, Qi Y, Shpak ED, Chai J. A receptor-like protein acts as a specificity switch for the regulation of stomatal development. Genes Dev. 2017:31(9):927–938. 10.1101/gad.297580.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Chai M, Huang Y, Qi J, Zhu W, Xi X, Chen F, Qin Y, Cai H. SDG2 regulates Arabidopsis inflorescence architecture through SWR1-ERECTA signaling pathway. iScience. 2021:24(11):103236. 10.1016/j.isci.2021.103236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes MA, Petrella R, Cucinotta M, Vignati E, Gatti S, Pinto SC, Bird DC, Gregis V, Dickinson H, Tucker MR, et al. The RNA-dependent DNA methylation pathway is required to restrict SPOROCYTELESS/NOZZLE expression to specify a single female germ cell precursor in Arabidopsis. Development. 2020:147(23):dev194274. 10.1242/dev.194274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JW, Mandava N, Worley JF, Plimmer JR, Smith MV. Brassins – a new family of plant hormones from rape pollen. Nature. 1970:225(5237):1065–1066. 10.1038/2251065a0 [DOI] [PubMed] [Google Scholar]
- doi: 10.1271/bbb.70216. Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem. 2007:71(8):2095–2100. [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002:110(2): 203–212. 10.1016/S0092-8674(02)00814-0 [DOI] [PubMed] [Google Scholar]
- Nolan T, Chen J, Yin Y. Cross-talk of brassinosteroid signaling in controlling growth and stress responses. Biochem J. 2017:474(16):2641–2661. 10.1042/BCJ20160633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo-Monfil V, Durán-Figueroa N, Arteaga-Vázquez M, Demesa-Arévalo E, Autran D, Grimanelli D, Slotkin RK, Martienssen RA, Vielle-Calzada JP. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 2010:464(7288):628–632. 10.1038/nature08828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri LJ, Bemis SM, Shpak ED, Torii KU. Haploinsufficiency after successive loss of signaling reveals a role for ERECTA-family genes in Arabidopsis ovule development. Development. 2007:134(17):3099–3109. 10.1242/dev.004788 [DOI] [PubMed] [Google Scholar]
- Pinto SC, Mendes MA, Coimbra S, Tucker MR. Revisiting the female germline and its expanding toolbox. Trends Plant Sci. 2019:24(5):455–467. 10.1016/j.tplants.2019.02.003 [DOI] [PubMed] [Google Scholar]
- Robinson-Beers K, Pruitt RE, Gasser CS. Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell. 1992:4(10):1237–1249. 10.2307/3869410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Leal D, Leon-Martinez G, Abad-Vivero U, Vielle-Calzada JP. Natural variation in epigenetic pathways affects the specification of female gamete precursors in Arabidopsis. Plant Cell. 2015:27(4):1034–1045. 10.1105/tpc.114.133009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Wuest SE, Vijverberg K, Baroux C, Kleen D, Grossniklaus U. Transcriptome analysis of the Arabidopsis megaspore mother cell uncovers the importance of RNA helicases for plant germline development. PLoS Biol. 2011:9(9):e1001155. 10.1371/journal.pbio.1001155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She W, Grimanelli D, Rutowicz K, Whitehead MW, Puzio M, Kotliński M, Jerzmanowski A, Baroux C. Chromatin reprogramming during the somatic-to-reproductive cell fate transition in plants. Development. 2013:140(19):4008–4019. 10.1242/dev.095034 [DOI] [PubMed] [Google Scholar]
- Shimada T, Sugano SS, Hara-Nishimura I. Positive and negative peptide signals control stomatal density. Cell Mol Life Sci. 2011:68(12):2081–2088. 10.1007/s00018-011-0685-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED. Diverse roles of ERECTA family genes in plant development. J Integr Plant Biol. 2013:55(12):1238–1250. 10.1111/jipb.12108 [DOI] [PubMed] [Google Scholar]
- Shpak ED, Berthiaume CT, Hill EJ, Torii KU. Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development. 2004:131(7):1491–1501. 10.1242/dev.01028 [DOI] [PubMed] [Google Scholar]
- Siddiqi I, Ganesh G, Grossniklaus U, Subbiah V. The dyad gene is required for progression through female meiosis in Arabidopsis. Development. 2000:127(1):197–207. 10.1242/dev.127.1.197 [DOI] [PubMed] [Google Scholar]
- Su Z, Wang N, Hou Z, Li B, Li D, Liu Y, Cai H, Qin Y, Chen X. Regulation of female germline specification via small RNA mobility in Arabidopsis. Plant Cell. 2020:32(9):2842–2854. 10.1105/tpc.20.00126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell. 2010:19(5):765–777. 10.1016/j.devcel.2010.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata N, Yokota K, Ohki S, Mori M, Taniguchi T, Kurita M. Evolutionary relationship and structural characterization of the EPF/EPFL gene family. PLoS One. 2013:8(6):e65183. 10.1371/journal.pone.0065183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU. Mix-and-match: ligand-receptor pairs in stomatal development and beyond. Trends Plant Sci. 2012:17(12):711–719. 10.1016/j.tplants.2012.06.013 [DOI] [PubMed] [Google Scholar]
- Tucker MR, Okada T, Hu Y, Scholefield A, Taylor JM, Koltunow AM. Somatic small RNA pathways promote the mitotic events of megagametogenesis during female reproductive development in Arabidopsis. Development. 2012:139(8):1399–1404. 10.1242/dev.075390 [DOI] [PubMed] [Google Scholar]
- Uchida N, Lee JS, Horst RJ, Lai HH, Kajita R, Kakimoto T, Tasaka M, Torii KU. Regulation of inflorescence architecture by intertissue layer ligand-receptor communication between endodermis and phloem. Proc Natl Acad Sci U S A. 2012:109(16):6337–6342. 10.1073/pnas.1117537109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006:313(5790):1118–1122. 10.1126/science.1127593 [DOI] [PubMed] [Google Scholar]
- Wang X, Gingrich DK, Deng Y, Hong Z. A nucleostemin-like GTPase required for normal apical and floral meristem development in Arabidopsis. Mol Biol Cell. 2012a:23(8):1446–1456. 10.1091/mbc.e11-09-0797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002:2(4):505–513. 10.1016/S1534-5807(02)00153-3 [DOI] [PubMed] [Google Scholar]
- Wang X, Xie B, Zhu M, Zhang Z, Hong Z. Nucleostemin-like 1 is required for embryogenesis and leaf development in Arabidopsis. Plant Mol Biol. 2012b:78(1–2):31–44. 10.1007/s11103-011-9840-7 [DOI] [PubMed] [Google Scholar]
- Yan Z, Zhao J, Peng P, Chihara RK, Li J. BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 2009:150(2): 710–721. 10.1104/pp.109.138099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002:109(2):181–191. 10.1016/S0092-8674(02)00721-3 [DOI] [PubMed] [Google Scholar]
- Zhang LY, Bai MY, Wu J, Zhu JY, Wang H, Zhang Z, Wang W, Sun Y, Zhao J, Sun X, et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell. 2009:21(12):3767–3780. 10.1105/tpc.109.070441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Bramsiepe J, Van Durme M, Komaki S, Prusicki MA, Maruyama D, Forner J, Medzihradszky A, Wijnker E, Harashima H, et al. RETINOBLASTOMA RELATED1 mediates germline entry in Arabidopsis. Science. 2017:356(6336):eaaf6532. 10.1126/science.aaf6532 [DOI] [PubMed] [Google Scholar]
- Zhao L, Cai H, Su Z, Wang L, Huang X, Zhang M, Chen P, Dai X, Zhao H, Palanivelu R, et al. KLU suppresses megasporocyte cell fate through SWR1-mediated activation of WRKY28 expression in Arabidopsis. Proc Natl Acad Sci U S A. 2018:115(3):E526–E535. 10.1073/pnas.1716054115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, He J, Cai H, Lin H, Li Y, Liu R, Yang Z, Qin Y. Comparative expression profiling reveals gene functions in female meiosis and gametophyte development in Arabidopsis. Plant J. 2014:80(4):615–628. 10.1111/tpj.12657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Liu L, Liu Y, Dou X, Cai H, Aslam M, Hou Z, Jin X, Li Y, Wang L, et al. Characterization of germline development and identification of genes associated with germline specification in pineapple. Hortic Res. 2021:8(1):239. 10.1038/s41438-021-00669-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.