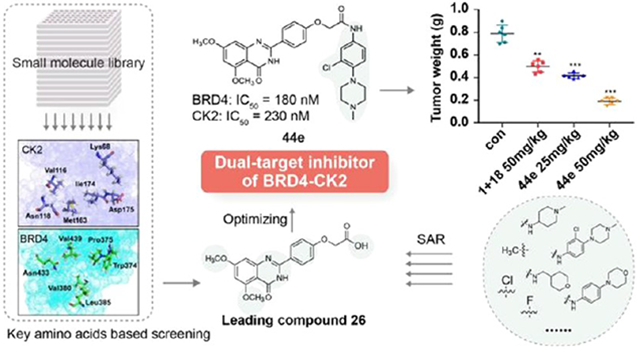
Abstract
Bromodomain-containing protein 4 (BRD4) is an attractive epigenetic target in human cancers. Inhibiting the phosphorylation of BRD4 by casein kinase 2 (CK2) is a potential strategy to overcome drug resistance in cancer therapy. The present study describes the synthesis of multiple BRD4–CK2 dual inhibitors based on rational drug design, structure–activity relationship, and in vitro and in vivo evaluations, and 44e was identified to possess potent and balanced activities against BRD4 (IC50 = 180 nM) and CK2 (IC50 = 230 nM). In vitro experiments show that 44e could inhibit the proliferation and induce apoptosis and autophagy-associated cell death of MDA-MB-231 and MDA-MB-468 cells. In two in vivo xenograft mouse models, 44e displays potent anticancer activity without obvious toxicities. Taken together, we successfully synthesized the first highly effective BRD4–CK2 dual inhibitor, which is expected to be an attractive therapeutic strategy for triple-negative breast cancer (TNBC).
Graphical Abstract
INTRODUCTION
Breast cancer (BC) is the most common female malignant tumor in the world, and more than 42 170 women died of BC in 2020.1,2 Triple-negative breast cancer (TNBC), which does not express estrogen receptor (ER), progesterone receptor (PR), or HER2/neu, significantly overlaps with the molecular subtype of basal-like breast cancer (BLBC).3 TNBC/BLBC cases account for 15% of all BC and mainly affect young women, people of African or Spanish origin, and those with BRCA1 mutations. It is highly aggressive and features high recurrence, metastasis, and heterogeneity. At present, surgery and conventional systemic cytotoxic chemotherapy are commonly used for TNBC treatment. However, its prognosis is far from satisfactory compared with advanced BC cases of other subtypes.4-6 Fortunately, targeted therapy drugs have improved cancer treatment for many people in a certain period of time.
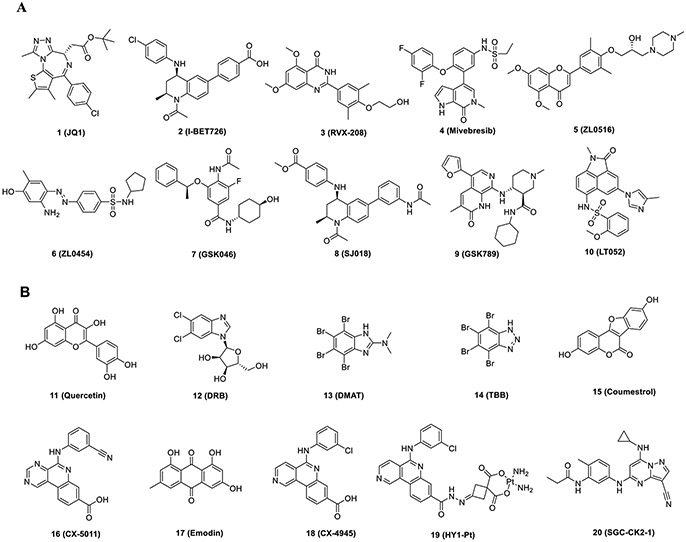
So far, the main targets and pathways of TNBC, including the bromodomain-containing protein 4 (BRD4),7 epidermal growth factor receptor (EGFR),8 mitogen-activated protein kinase (MAPK)–extracellular regulated protein kinases (ERK),9 phosphatidylinositol 3 kinase (PI3K)–protein kinase B (AKT),10 polyADP–ribose polymerase (PARP),11 vascular endothelial growth factor receptor 2 (VEGFR2),12 and human epidermal growth factor receptor 2 (HER2),12,13 have been well studied. However, drug resistance has become challenging with increased application of small molecule targeted drugs.14-16 How to prevent and reverse drug resistance in cancer patients has been a major concern. BRD4 is a member of the BET family, which serves as a vital transcriptional and epigenetic regulatory factor in cell cycle progression. It is also involved in cancer progression by promoting oncogene transcription through RNA polymerase II (Pol II) or regulating oncogene networks by inducing signal transduction after recognizing acetylated histones.17-19 BRD4 is of great significance in many hematopoietic and solid tumors expressing Myc and other oncogenes.20,21 It has been reported that dysregulated BRD4 was detected in breast cancer, colon cancer, and prostate cancer.22 The large-scale genome analysis of TNBC demonstrated that BRD4 is essential for the survival of TNBC cell populations.23 Knockdown of BRD4 in TNBC cells significantly inhibits the activity of Notch1, thus suppressing metastasis and invasion.24,25 Dysfunctional BRD4 is closely linked to the occurrence, development, invasion, metastasis, and prognosis of TNBC. Furthermore, inhibition of BRD4 could induce autophagy in breast cancer cells. BRD4 inhibitors are capable of inhibiting the transcription of oncogenes in TNBC, serving as promising therapeutic agents.26,27 BRD4 inhibitors have drawn significant attention in recent years. Multiple BET inhibitors have been reported, including JQ1 (1), I-BET726 (2), RVX-208 (3), mivebresib (4), ZL0516 (5), ZL0454 (6), GSK046 (7), SJ018 (8), GSK789 (9), and LT052 (10) (Figure 1A), and clinical trials with some BRD4 inhibitors evaluating the therapeutic efficacy against cancers are in progress.5,28,29 A growing number of BRD4 inhibitors have been discovered and applied to the treatment of TNBC, but the emergence of drug resistance was one of the reasons that greatly limited the clinical application.30,31 The latest study in 2021 suggested that the drug resistance mechanism of BET inhibitors varies with the cancer type, and inhibiting the phosphorylation of BRD4 by casein kinase 2 (CK2) is a potential strategy to overcome drug resistance in cancer patients.32
Figure 1.
Structures representing known BRD4 and CK2 inhibitors. (A) Representative published BRD4 inhibitors with diverse scaffolds. (B) Representative published CK2 inhibitors with diverse scaffolds.
CK2 is upregulated in various types of cancers, presenting a vital function in essential biological processes. It is considered as a potential therapeutic target for cancers.33 CK2 is composed of catalytic α/α′ and regulatory β subunits. During the past four decades, multiple CK2 inhibitors have been developed for human disease treatment, including quercetin (11), DRB (12), DMAT (13), TBB (14), coumestrol (15), CX-5011 (16) emodin (17), CX-4945 (18), HY1-Pt (19), and SGC-CK2-1 (20) (Figure 1B).34,35 Among them, 18 is the only small molecule CK2 inhibitor explored in phase I/II clinical trials. In addition, 18 effectively induces autophagy and apoptosis in pancreatic cancer cells. Studies have shown that CK2 could regulate autophagy through downstream targets of phosphorylation.36 CK2 has been validated as one of the proteins with the most genetic diversity in eukaryotic systems. It contains more than 300 phosphorylated substrates and is involved in various cellular processes, including the AKT/PI3K pathway, DNA damage repair, cell cycle progress, and cell growth. Among them, CK2-induced phosphorylation is essential for BRD4 binding to acetylated chromatin and occurs at the N-terminal cluster of phosphorylation sites (NPS), situated downstream of the second bromodomain (BD2), and the C-terminal cluster of phosphorylation sites (CPS), located downstream of the extraterminal (ET) domain, of BRD4.37 Both NPS and CPS are also found in other bromodomain and ET (BET) family proteins, including BRD2, BRD3, and BRDT.37 Shu et al.38 explored the inhibitory effect of BET in human breast cancer. It was found that MED1, a widely existing protein for Pol II-dependent transcription, binds to BRD4 in drug-resistant cells more tightly than in drug-sensitive cells, which results from BRD4-independent transcriptional activation of Myc gene caused by an increase of BRD4 phosphorylation due to decreased protein phosphatase 2A (PP2A) activity. In addition, CK2 inhibition leads to increased uptake of known drugs in multidrug-resistant cells.39-41 Currently, development of multitarget approaches targeting different proteins, especially dual target inhibitors with synergistic effects, has received increasing attention in anticancer drug development. Many dual BET/kinase inhibitors have been described.5 They can simultaneously regulate multiple targets in the disease network and have synergistic antitumor effects, providing a rationale for the development of dual inhibitors ofBRD4. Therefore, we speculate that inhibition of BRD4 and CK2 could both solve the problem of resistance of BRD4 inhibitors caused by phosphorylation and could further inhibit the proliferation and induce apoptosis and autophagy-associated cell death in TNBC. In conclusion, the design and synthesis of BRD4–CK2 dual inhibitors may provide a new therapeutic option for TNBC.
To achieve this goal, we first identified that BRD4 and CK2 could influence the progression and prognosis of TNBC through apoptosis and autophagy-associated cell death by networks and systems biology. A series of small molecule BRD4–CK2 dual inhibitors were then synthesized via virtual screening and rational drug design and assessed for their in vivo and in vitro biological activities. The obtained candidate compound 44e was found to inhibit colony formation of MDA-MB-231 and MDA-MB-468 TNBC cells, and dose-dependently induced cell apoptosis and autophagy-associated cell death. In vivo experiments further validated an acceptable anticancer effect of 44e in mice. Our findings demonstrated the therapeutic efficacy of 44e on TNBC through dual-inhibition of BRD4 and CK2, which is expected to be a promising candidate drug for TNBC.
RESULTS AND DISCUSSION
Bioinformatics Analysis.
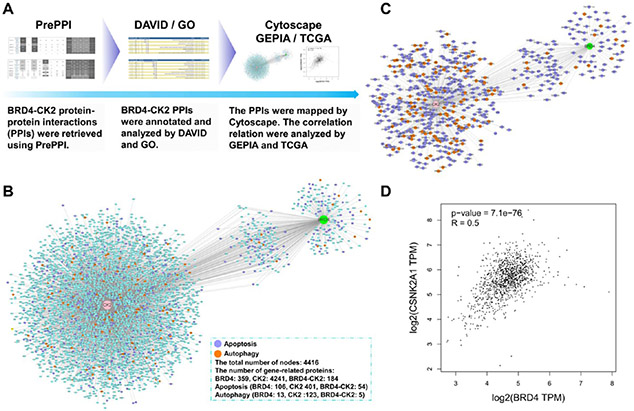
Bioinformatic analysis was performed to investigate the potential functions of BRD4 and CK2 in the development of breast cancer susceptibility genes (Figure 2A). First, the interacting proteins of BRD4 and CK2 were searched using the PrePPI database.42,43 It is shown that there are 359 and 4241 proteins interacting with BRD4 and CK2, respectively, with a regulatory relationship. These interacting proteins were subsequently annotated and clustered using the Gene Ontology (GO) and DAVID databases.44,45 Among them, 453 and 131 proteins were enriched in the apoptosis and autophagy pathways, respectively (Figure 2B,C). Finally, the GEPIA tool was used to analyze the breast cancer transcriptome data of BRD4 and CK2 obtained from the TCGA database; as shown in Figure 2D, a significantly positive correlation was identified between BRD4 and CK2 (R = 0.5, p = 7.1 × 10−76).46 This bioinformatic analysis correlates interaction between BRD4 and CK2 and underlines the progression and prognosis of TNBC through apoptosis and autophagy-associated cell death.
Figure 2.
Bioinformatics analysis. (A) Workflow of bioinformatics analysis of BRD4–CK2 association. (B) Clustering of BRD4 and CK2 interacting protein networks related to cell autophagy and apoptosis. (C) Predicted BRD4 and CK2 related proteins involved in the regulation of cell apoptosis and autophagy-associated cell death. (D) Correlation between BRD4 and CK2 in BC.
Design of Novel BRD4–CK2 Dual Inhibitors.
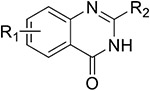
BET inhibitors are mainly categorized as pan-BET inhibitors, selective BRD4 inhibitors, and BET-BD1 or BET-BD2 inhibitors.5 BRD4 contains two conserved N-terminal bromodomains (BD1 and BD2) with high sequence similarity.18 There are many studies describing the unique functions of BD1 and BD2. However, in cancer research, the functional differences between BD1 and BD2 are not very clear.14 Currently, BRD4 inhibitors based on BD1 or BD2 selectivity have been reported.5
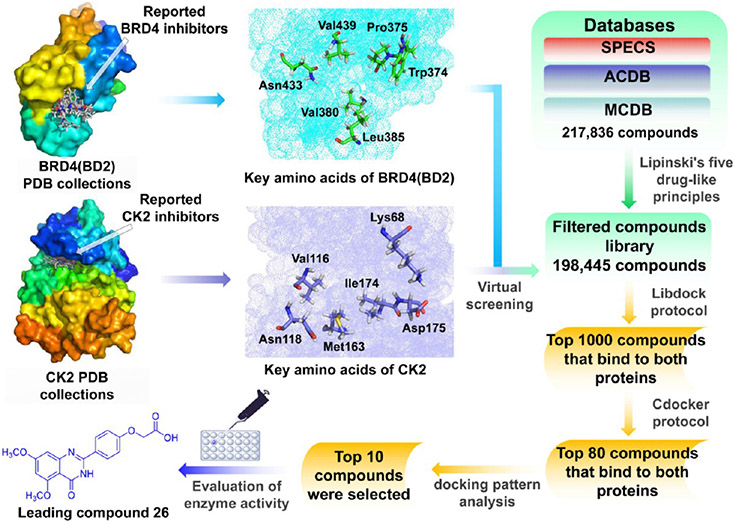
To design BRD4–CK2 dual inhibitors, we first analyzed the crystal structures of BRD4 (BD2) and CK2 for key inhibitor contact amino acid sites via molecular docking. BRD4 and CK2 small-molecule inhibitor complexes were downloaded from the Crystallography Open Database. Based on the reported interaction between BRD4 (BD2) or CK2 and small molecule inhibitors, key amino acid residues in different spatial positions in the active pocket were summarized. As shown in Figure 3, Trp374, Pro375, Val380, Leu385, Asn433, and Val439 were identified as key amino acid residues in BRD4-BD2 for interaction with small molecule inhibitors, while Lys68, Val116, Asn118, Met163, Ile174, and Asp175 were key amino acid residues for CK2 interaction with small molecule inhibitors. Subsequently, we screened BRD4–CK2 dual inhibitors through the multiple-docking strategy. A total of 217 836 compounds in the SPECS library of Topscience, ACDB, and MCDB were initially screened based on the Lipinski’s rule, and finally, 198 445 small molecules were obtained as a compound library for further analyses.2,47 Rapid molecular docking of compounds with BRD4 (BD2) (PDB ID 5UOO) and CK2 (PDB ID 6RFE) in the generated library was conducted using the LibDock module of Discovery Studio software (version 3.5), and the top 1000 matches that could bind to BRD4 and CK2 were selected. Molecular docking of the 1000 matches with BRD4 and CK2 was performed using the CDOCKER module. The obtained 80 matched compounds that could bind to BRD4 and CK2 were further analyzed for the docking mode, structural diversity, and conformational stability with key amino acids as important references, and finally, 10 compounds were obtained (Figure S1). Through assessing their kinase activity in vitro, we found that compound 26 at 10 μM presented an inhibitory effect on BRD4 and CK2 with an inhibitory ratio of 63% and 45%, respectively (Figure S2). Thus, 26 was selected as the lead compound for further design and structural optimization of BRD4–CK2 dual inhibitors.
Figure 3.
Workflow of designing BRD4–CK2 inhibitors based on extracted amino acid residues and virtual screening.
Synthetic Routes of BRD4–CK2 Dual Inhibitors.
The synthetic routes of all target compounds are shown in Schemes 1-3. Target compounds 33a–d and 34a–c were initially synthesized as shown in Scheme 1. Briefly, hydroxyacetophenone derivatives dissolved in pyridine were induced with benzoyl chloride derivatives under alkaline conditions for 1.5 h, thus obtaining the intermediates 31a–d. The intermediates were then dissolved in pyridine, heated at 50 °C for 1 h, and induced with KOH at 80 °C for 1 h. The obtained intermediates 32a–d resolved in HOAc were treated at 90 °C for 1 h, followed by cooling, vacuum distillation, and incubation with H2SO4 1% (v/v) in HOAc solution for 1 h, thus obtaining compounds 33a–d. They were then dissolved in toluene and subjected to heating reflux using the Lawesson’s reagent for 4 h. Finally, target compounds 34a–c were obtained.
Scheme 1a.
aReagents and conditions: (a) pyridine, DBU, 80 °C, 1.5 h; (b) pyridine, KOH, 50–80 °C, 2 h; (c) HOAc, 1% H2SO4, 90–110 °C, 2 h; (d) dry toluene, Lawesson’s reagent, 110 °C, 4 h.
Scheme 3a.
aReagents and conditions: (a) K2CO3, DMF, 80 °C, 3 h; (b) DCM, TFA, rt, 2 h; (c) DMAC, PTSA, NaHSO3, 120 °C, 4–8 h; (d) DMF, Et3N, HOBt, EDCI, rt, 24 h.
Target compounds 38a–s and the lead compound 26 were obtained as shown in Scheme 2. First, 3,5-dimethoxyaniline was converted into hydrochloride salt, which was then reacted with (COCl)2 at high temperature to obtain 4,6-dimethoxyindolinedione (35a). Compound 35a was subjected to 0.5 h heating reflux under alkaline conditions by adding the oxidant H2O2, and the intermediate 2-amino-4,6-dimethoxybenzamide (36a) was obtained after suction filtration. Compound 36a was induced with the carboxylic acid activator HOBt, the coupling agent EDCI, and N-methylmorpholine in THF solution. After condensation using ammonium hydroxide, an amide product, 2-amino-4,6-dimethoxybenzamide (37a), was obtained. The synthetic routes of intermediates 37b,c were similar to that of 37a. Finally, 37a–c were reacted with various aldehydes using PTSA as the catalyst and NaHSO3 as the additive, heated to reflux for 4–8 h, and purified by column chromatography, thus obtaining target compounds 38a–s and the lead compound 26.
Scheme 2a.
aReagents and conditions: (a) HCl/Et2O, rt, overnight; (b) (COCl)2, 160 °C, 2.5 h; (c) NaOH, H2O2, 80 °C, 0.5 h; (d) ammonium hydroxide, HOBt, EDCI, NMM, THF, overnight, rt; (e) benzaldehyde derivatives, DMAC, PTSA, NaHSO3, 120 °C, 4–8 h.
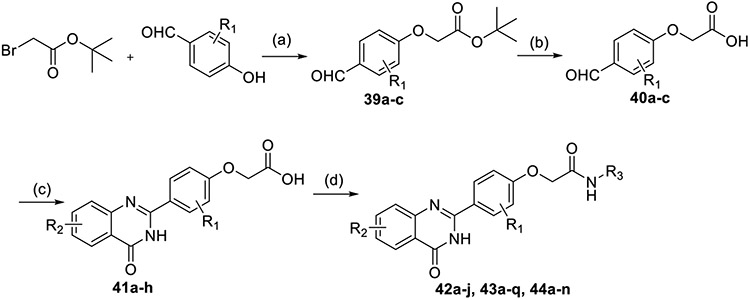
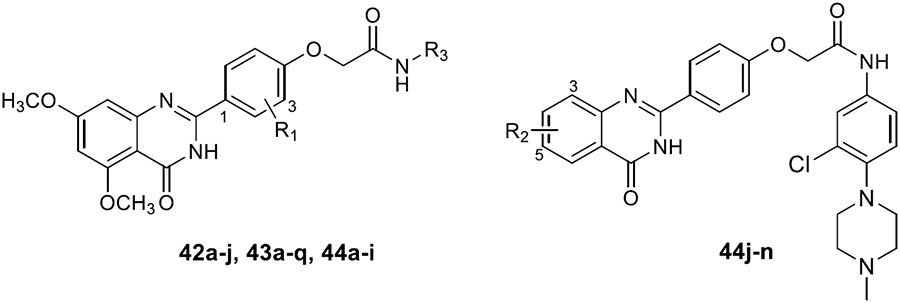
Target compounds 42a–j, 43a–q, and 44a–n were synthesized according to the preparation pathways depicted in Scheme 3. Under alkaline conditions, tert-butyl bromoacetate and p-hydroxybenzaldehyde derivatives in DMF developed substitution reaction, thus obtaining intermediates 39a–c. Later, intermediates 40a–c were obtained by hydrolysis of tert-butyl ester. Anthranilamide derivatives and 40a–c were dissolved in DMAc and reacted with NaHSO3 and the catalyst PTSA at high temperature for 4–8 h. The obtained intermediates 41a–h were incubated with amino substitution derivatives in DMF solution containing HOBt and EDCI. After 24 h stirring at room temperature, target compounds 42a–j, 43a–q, and 44a–n were separated by column chromatography.
Structural Optimization of the BRD4–CK2 Dual Inhibitors.
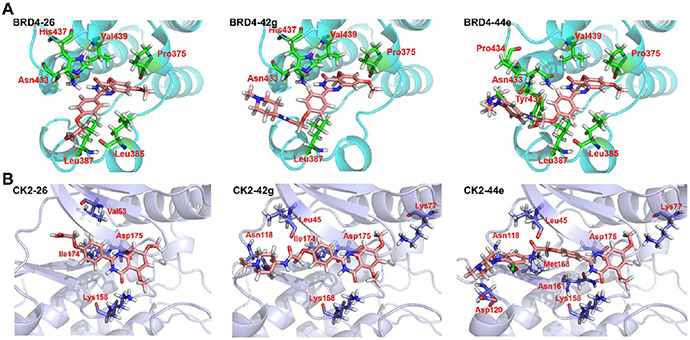
To optimize the structure of the lead compound, we analyzed the binding mode of compound 26 and the target proteins. Molecular docking analysis showed that the binding mode of compound 26 within BRD4 was similar to that of BRD4 inhibitor 3 (Figure 4A). As the binding mode of compound 26 within CK2 showed, the quinazolinone core scaffold was inserted deeply into the hydrophobic pocket, in which the O atom and the para-position N atom of amide groups formed two key hydrogen bonds with Lys158 and Asp175, respectively. Furthermore, the conjugation of the benzene ring of the quinazolinone core scaffold and Asp175 maintained the stable binding of the quinazolinone core scaffold and CK2, while the benzene ring on the other side formed hydrophobic interactions with Val53 and Ile174. However, the tail group of the compounds did not occupy the channel surrounding the hinge region. In addition, 26 did not have interactions with key amino acids like Asn118 and Met163 in the core group, which may be vital factors for influencing the binding stability and affinity with CK2 (Figure 4B).
Figure 4.
Binding mode analysis of 26, 42g, and 44e. (A, B) Docking poses show the interaction of 26, 42g, and 44e with BRD4 (BD2) (PDB ID 5UOO) and CK2 (PDB ID 6RFE), respectively. Oxygen atoms are colored in red and nitrogen atoms in blue.
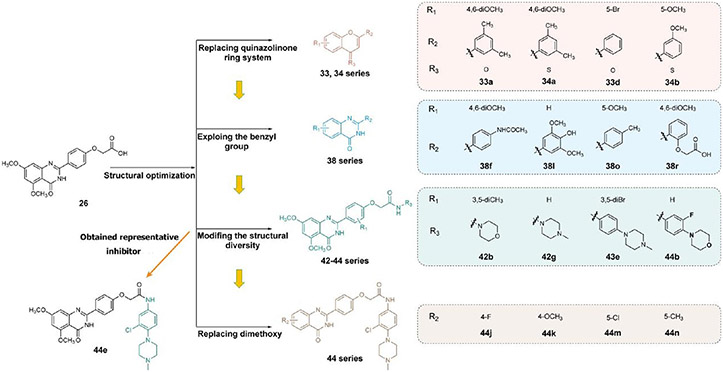
In the first round of modification, we verified the importance of the quinazolinone core scaffold in the lead compound with dual inhibition of BRD4 and CK2. The key N atom of quinazolinone was first replaced to synthesize compounds 33a–d. Their inhibitory rates against BRD4 and CK2 were determined at a concentration of 1 μM (Table 1), which, as expected, were lower than 15%. Compounds lost almost all inhibitory activity against TNBC cells. The inhibitory effects of 33a–d on target proteins were significantly reduced. Subsequently, compounds 34a–c were synthesized through introducing a S atom to replace the oxygen carbonyl, which barely had activity. Therefore, it is suggested that the quinazolinone core scaffold was the vital fragment for maintaining the compounds’ activities.
Table 1.
BRD4 and CK2 Inhibition Rates and In Vitro Anti-Proliferative Activities of Compounds 33a–d and 34a–c
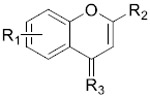
| |||||||
|---|---|---|---|---|---|---|---|
| NO. | R1 | R2 | R3 | Kinase inhibitory activity (1 μM, %)[a] |
Anti-proliferative active (IC50, μM)[b] |
||
| BRD4 | CK2 | MDA-MB-231 | MDA-MB-468 | ||||
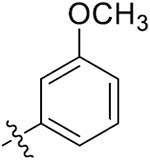
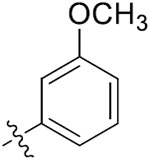
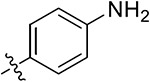
| 33a | 4,6-diOCH3 |
|
O | 13.55 ± 0.21 | 9.13 ± 1.96 | 47.75 ± 2.83 | 39.65 ± 2.15 |
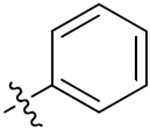
| 33b | 5-OCH3 |
|
O | 6.05 ± 1.29 | 12.56 ± 0.23 | 45.71 ± 3.04 | > 50 |
| 33c | 5-OCH3 |
|
O | 4.26 ± 0.71 | 10.37 ± 0.85 | > 50 | > 50 |
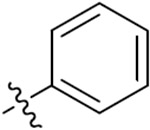
| 33d | 5-Br |
|
O | 5.92 ± 0.37 | 11.76 ± 0.48 | 33.48 ± 1.70 | 48.23 ± 3.61 |
| 34a | 4,6-diOCH3 |
|
S | 14.80 ± 0.53 | 7.32 ± 0.89 | > 50 | > 50 |
| 34b | 5-OCH3 |
|
S | 4.37 ± 0.60 | 14.19 ± 0.52 | > 50 | > 50 |
| 34c | 5-OCH3 |
|
S | 6.91 ± 0.34 | 13.95 ± 0.14 | > 50 | > 50 |
Reported compounds were tested in triplicate. Data are presented as mean ± SD.
IC50 values were determined from cell viability assay for 24 h.
In the second round of optimization, we retained the quinazolinone core scaffold and further optimized the tail group of 26. Target compounds 38a–s were synthesized by introducing various substituents (R1, R2) on the benzene ring. We speculated that these structures were able to form various interactions with channels surrounding the hinge region of CK2. Our findings showed that 38d, in which a 3,5-dimethyl-4-hydroxyphenyl group served as the R2 group, inhibited BRD4 by 21.25%. Compound 38p was synthesized by introducing an electron-withdrawing group, −COOH, and 38i introduced an electron-donating group, −OCH3. Compared with 26, 38p (16.81%) and 38i (16.83%) still presented a low inhibitory rate against CK2, and their inhibitory rate against BRD4 was not elevated (17.46% and 11.74%). In addition, compounds synthesized by substituting R1 as the H atom (38j–l) or a single methoxy (38m–o) presented a low inhibitory rate against BRD4, with no antiproliferative activity on MDA-MB-231 cells (IC50 > 50 μM). The dimethoxyphenyl group was found to be the key group responsible for inhibiting BRD4, which was similar to that of 3. Interestingly, inhibition was elevated to varying degrees in 38a, 38d, and 38f by substituting R1 with the dimethoxy group (Table 2).
Table 2.
BRD4 and CK2 Inhibition Rates and In Vitro Antiproliferative Activities of Compounds 38a–s and 26
| ||||||
|---|---|---|---|---|---|---|
| NO. | R1 | R2 | Kinase inhibitory activity (1 μM, %)[a] |
Anti-proliferative active (IC50, μM)[b] |
||
| BRD4 | CK2 | MDA-MB-231 | MDA-MB-468 | |||
| 38a | 4,6-diOCH3 |
|
16.26 ± 0.48 | 13.17 ± 2.23 | 45.61 ± 3.16 | 38.42 ± 0.47 |
| 38b | 4,6-diOCH3 |
|
18.18 ± 1.32 | 8.46 ± 0.41 | 25.21 ± 0.32 | 29.62 ± 1.29 |
| 38c | 4,6-diOCH3 |
|
10.65 ± 1.18 | 15.41 ± 0.20 | 36.52 ± 2.03 | > 50 |
| 38d | 4,6-diOCH3 |
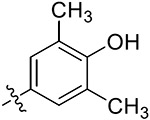
|
21.25 ± 1.65 | 16.13 ± 0.61 | 17.53 ± 3.23 | 19.26 ± 1.82 |
| 38e | 4,6-diOCH3 |
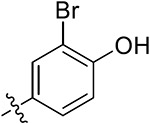
|
19.61 ± 2.71 | 10.22 ± 2.54 | 23.21 ± 2.61 | 22.32 ± 2.15 |
| 38f | 4,6-diOCH3 |
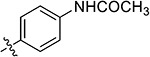
|
17.46 ± 0.38 | 14.14 ± 2.15 | 27.31 ± 0.19 | 24.71 ± 1.39 |
| 38g | 4,6-diOCH3 |
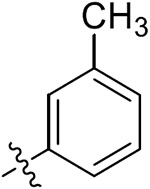
|
13.11 ± 1.24 | 9.46 ± 0.98 | 43.86 ± 0.62 | 39.41 ± 1.76 |
| 38h | 4,6-diOCH3 |
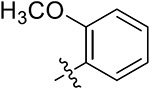
|
5.62 ± 2.28 | 17.57 ± 1.29 | >50 | 44.62 ± 2.11 |
| 38i | 4,6-diOCH3 |
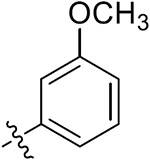
|
11.74 ± 2.42 | 16.83 ± 0.58 | 38.30 ± 1.54 | 29.34 ± 1.27 |
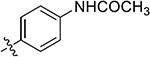
| 38j | H |
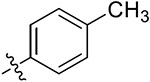
|
3.28 ± 1.45 | 7.72 ± 1.61 | >50 | >50 |
| 38k | H |
|
1.31 ± 2.22 | 14.59 ± 1.54 | >50 | >50 |
| 38l | H |
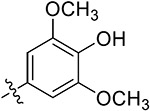
|
5.62 ± 0.41 | 11.83 ± 1.78 | >50 | >50 |
| 38m | 5-OCH3 |
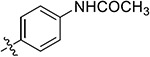
|
2.82 ± 0.02 | 14.40 ± 2.49 | >50 | 41.21 ± 3.25 |
| 38n | 5-OCH3 |
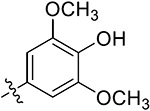
|
5.35 ± 0.42 | 14.33 ± 1.91 | >50 | 43.72 ± 2.08 |
| 38o | 5-OCH3 |

|
2.52 ± 1.03 | 13.63 ± 1.52 | >50 | 30.41 ± 1.63 |
| 38p | 4,6-diOCH3 |
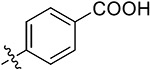
|
17.46 ± 2.30 | 16.81 ± 0.31 | 21.08 ± 0.16 | 8.33 ± 0.47 |
| 38q | 4,6-diOCH3 |
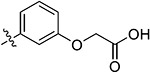
|
19.23 ± 0.31 | 21.63 ± 1.44 | 25.62 ± 2.36 | 22.73 ± 0.29 |
| 38r | 4,6-diOCH3 |
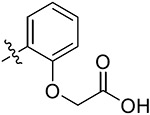
|
18.22 ± 1.53 | 20.43 ± 0.57 | 36.72 ± 1.08 | 18.56 ± 1.01 |
| 38s | 4,6-diOCH3 |
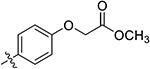
|
16.09 ± 2.64 | 17.17 ± 1.69 | 22.02 ± 2.05 | 13.82 ± 0.29 |
| 26 | 4,6-diOCH3 |
|
35.72 ± 1.53 | 26.44 ± 0.57 | 19.72 ± 1.08 | 18.56 ± 1.37 |
Reported compounds were tested in triplicate. Data are presented as mean ± SD.
IC50 values were determined from cell viability assay for 24 h.
Further, 38q and 38r were synthesized by moving the para-substituted acetic acid group on the benzene ring to the meta-position or ortho-position. Their inhibitory rate against CK2 was slightly reduced, but that against BRD4 was significantly reduced. Compounds 38q and 38r exhibited lower efficiency on TNBC cells compared with 26. Collectively, through altering the acetic acid chain at the end of the benzene group, the BRD4 and CK2 inhibition rates and in vitro antiproliferative activities of synthesized compounds were not elevated, suggesting that the terminal acetic acid group is an important group to maintain the inhibitory activity of the target compound.
According to the interaction between the lead compound 26 and CK2, we considered that extending the tail group of the molecule to strengthen the interactions, such as hydrophobic interactions, hydrogen bond interactions, and π–π stacking, in the hydrophobic channel surrounding the hinge region of CK2 could be a feasible approach. The quinazolinone core scaffold of 26 and the para-carboxyl substitution of the benzene ring are important structures for maintaining activity (Figure 4B). Therefore, the important pharmacophore was retained in the third round of modification, only optimizing the tail group.
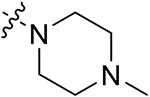
Through analyzing the structures of other BRD4 and CK2 inhibitors, it was found that the amide bond is responsible for the interaction formed in the hydrophobic channel. Therefore, it could be used to extend the molecular structure. Here, the amide bond was introduced to further boost the interaction to amino acid residues surrounding the hinge region of CK2. Later, compounds 42a–j were synthesized, retaining the dimethoxyphenyl group (Table 3) and the quinazolinone core scaffold. Through introduction of the morpholine derivatives (42a,b), tetrahydropyran derivatives (42c,d), N-methylpiperidine derivatives (42e,f), 1-methylpiperazine derivative (42g), and substituted benzene (42h,i), the extended length of substituents further strengthened the interaction between small molecules and the hinge region of CK2. BRD4 and CK2 activities of these compounds were elevated to a certain extent. Notably, the inhibitory rates of 42g against BRD4 and CK2 were 55.71% and 53.64% at the concentration of 1 μM. Meanwhile, 42g also showed acceptable cellular potency in MDA-MB-231 and MDA-MB-468 cells (IC50 = 12.82 and 13.65 μM, respectively). The introduction of the 1-methylpiperazine group was also conducive to the antiproliferative ability in TNBC cells.
Table 3.
BRD4 and CK2 Inhibition Rates and In Vitro Antiproliferative Activity of Compounds 3, 14, 42a–j, 43a–q, and 44a–n
| |||||||
|---|---|---|---|---|---|---|---|
| NO. | R1 | R2 | R3 | Kinase inhibitory activity (1 μM, %)[a] |
Anti-proliferative active (IC50, μM)[b] |
||
| BRIM | CK2 | MDA-MB-231 | MDA-MB-468 | ||||
| 42a | H | - |
|
45.29 ± 1.42 | 42.20 ± 1.25 | 14.71 ± 1.55 | 12.54 ± 0.87 |
| 42b | 3,5-diCH3 | - |
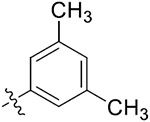
|
42.58 ± 0.82 | 40.31 ± 1.76 | 20.57 ± 0.62 | 18.50 ± 1.23 |
| 42c | H | - |
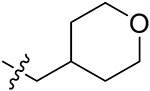
|
43.19 ± 3.47 | 43.43 ± 2.59 | 21.21 ± 2.80 | 26.61 ± 0.85 |
| 42d | 3,5-diCH3 | - |
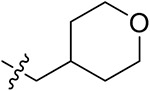
|
37.47 ± 1.42 | 33.70 ± 3.64 | 23.61 ± 0.58 | 25.56 ± 1.57 |
| 42e | 3,5-diBr | - |
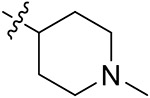
|
51.11 ± 3.47 | 47.43 ± 2.19 | 16.93 ± 0.87 | 14.81 ± 1.66 |
| 42f | 3,5-diCH3 | - |
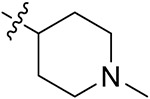
|
52.38 ± 1.15 | 49.23 ± 2.59 | 17.21 ± 2.80 | 15.55 ± 2.21 |
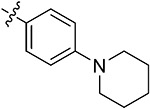
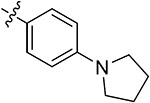
| 42g | H | - |
|
55.71 ± 2.02 | 53.64 ± 1.79 | 12.82 ± 1.72 | 13.65 ± 0.23 |
| 42h | H |
|
54.19 ± 1.28 | 47.43 ± 1.09 | 23.37 ±1.35 | 15.03 ± 0.26 | |
| 42i | 3,5-diCH3 |
|
53.50 ± 3.47 | 42.14 ± 0.33 | 24.41 ± 2.81 | 16.55 ± 1.80 | |
| 42j | 3,5-diCH3 | - |
|
56.14 ± 3.09 | 25.61 ± 6.57 | >50 | 30.31 ± 1.56 |
| 43a | H | - |
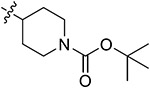
|
62.10 ± 0.52 | 55.68 ± 2.07 | 13.12 ± 1.67 | 16.62 ± 2.21 |
| 43b | 3,5-diCH3 | - |
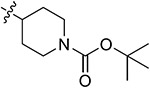
|
57.39 ± 2.06 | 51.38 ± 1.69 | 22.37 ± 2.30 | 17.24 ± 1.63 |
| 43c | 3,5-diCH3 | - |
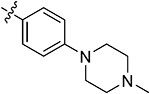
|
78.36 ± 4.07 | 69.31 ± 2.19 | 10.81 ± 1.85 | 12.59 ± 1.24 |
| 43d | 3,5-diCH3 | - |
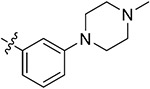
|
72.44 ± 2.87 | 57.08 ± 1.27 | 14.57 ± 1.64 | 15.74 ± 2.66 |
| 43e | 3,5-diBr | - |

|
62.58 ± 0.52 | 58.47 ± 1.23 | 13.64 ± 2.17 | 15.83 ± 1.11 |
| 43f | H | - |
|
82.30 ± 2.73 | 80.02 ± 4.22 | 6.46 ± 0.62 | 8.80 ± 1.15 |
| 43g | H | - |

|
74.33 ± 1.28 | 56.02 ± 1.90 | 13.22 ± 0.54 | 8.07 ± 1.68 |
| 43h | H | - |
|
69.01 ± 2.30 | 51.44 ± 0.65 | 17.32 ± 0.79 | 11.50 ± 0.54 |
| 43i | 3,5-diCH3 | - |
|
81.44 ± 1.15 | 63.61 ± 0.34 | 8.23 ± 1.39 | 9.49 ± 1.18 |
| 43j | 3,5-diCH3 | - |
|
73.71 ± 0.83 | 55.73 ± 1.86 | 11.26 ± 0.93 | 13.44 ± 0.77 |
| 43k | 3,5-diBr | - |
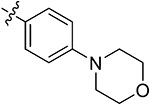
|
67.20 ± 1.52 | 58.29 ± 3.62 | 13.21 ± 2.21 | 15.11 ± 1.75 |
| 43l | H | - |
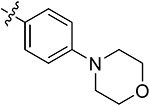
|
80.65 ± 3.98 | 75.39 ± 2.52 | 7.67 ± 1.03 | 9.54 ± 0.74 |
| 43m | H | - |

|
61.74 ± 2.03 | 60.02 ± 1.38 | 15.53 ± 0.91 | 17.80 ± 1.22 |
| 43n | 3,5-diCH3 | - |
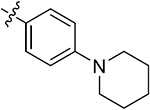
|
69.24 ± 3.67 | 70.51 ± 0.54 | 13.43 ± 2.53 | 9.43 ± 1.09 |
| 43o | 3,5-diBr | - |
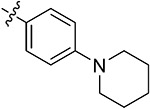
|
56.39 ± 0.46 | 62.47 ± 1.23 | 16.35 ± 2.02 | 15.63 ± 1.26 |
| 43p | H | - |
|
71.06 ± 2.96 | 75.20 ± 1.38 | 10.27 ± 2.03 | 11.63 ± 0.46 |
| 43q | H | - |
|
68.51 ± 1.72 | 67.52 ± 0.71 | 19.56 ± 1.63 | 7.82 ± 4.02 |
| 44a | 3,5-diCH3 | - |
|
83.29 ± 3.45 | 76.21 ± 2.53 | 10.30 ± 1.23 | 9.16 ± 0.93 |
| 44b | H | - |
|
85.43 ± 2.65 | 79.61 ± 3.65 | 7.09 ± 0.25 | 8.58 ± 1.51 |
| 44c | H |
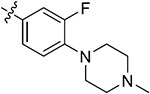
|
85.37 ± 1.58 | 82.46 ± 0.13 | 6.30 ± 2.37 | 7.55 ± 1.73 | |
| 44d | 3,5-diCH3 |
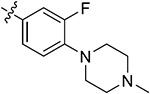
|
87.61 ± 2.73 | 80.23 ± 0.42 | 5.72 ± 1.45 | 8.33 ± 1.17 | |
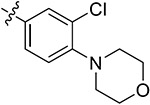
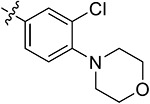
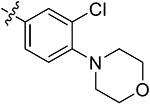
| 44e | H | - |
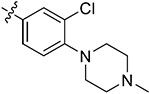
|
92.28 ± 0.66 | 90.65 ± 1.39 | 2.66 ± 0.81 | 3.52 ± 0.58 |
| 44f | 3,5-diCH3 | - |
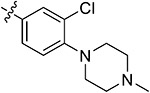
|
89.27 ± 1.47 | 85.40 ± 0.38 | 3.49 ± 1.23 | 4.58 ± 0.52 |
| 44g | 3,5-diBr | - |
|
79.30 ± 2.50 | 86.29 ± 1.47 | 4.02 ± 1.92 | 5.53 ± 0.28 |
| 44h | H | - |
|
83.52 ± 1.54 | 75.19 ± 0.73 | 4.52 ± 0.56 | 8.29 ± 1.92 |
| 44i | 3,5-diCH3 | - |
|
85.47 ± 0.28 | 70.13 ± 2.94 | 10.23 ± 0.95 | 6.31 ± 1.94 |
| 44j | - | 4-F | - | 45.26 ± 0.57 | 68.27 ± 1.39 | 6.94 ± 0.68 | 7.51 ± 1.13 |
| 44k | - | 4-OCH3 | - | 55.60 ± 1.62 | 72.03 ± 2.82 | 14.59 ± 0.77 | 16.36 ± 0.85 |
| 44l | - | 4-Cl | - | 51.49 ± 3.02 | 75.61 ± 1.12 | 14.40 ± 0.58 | 15.27 ± 0.72 |
| 44m | - | 5-Cl | - | 47.29 ± 1.16 | 65.93 ± 1.80 | 5.21 ± 0.82 | 6.23 ± 0.55 |
| 44n | - | 5-CH3 | - | 48.20 ± 0.47 | 74.70 ± 2.11 | 17.36 ± 0.56 | 16.22 ± 1.83 |
| 1 | - | - | - | 97.03 ± 1.32 | 6.60 ± 0.41 | 29.51 ± 1.70 | 30.19 ± 0.69 |
| 18 | - | - | - | 7.14 ± 1.51 | 98.53 ± 1.87 | 14.25 ± 0.62 | 20.92 ± 1.04 |
| 1 + | - | - | - | - | - | 7.37 ± 0.39 (CI = 0.78)d | 9.10 ± 0.14 (CI = 0.61d |
| 18 c | |||||||
Reported compounds were tested in triplicate. Data are presented as mean ± SD.
IC50 values were determined from cell viability assay for 24 h.
Combined at a mole ratio of 1:1.
Using the Chou–Talalay method.
The docking mode of 42g with BRD4 and CK2 revealed a similar key interaction site between the core scaffold structure of 42g and BRD4 to that of lead compounds (Figure 4). In particular, the introduction of the amide bond induced the formation of a hydrogen bond with Leu45 of CK2 and an intermolecular hydrogen bond between the 1-methylpiperazine group and Asn118. Since the hinge region of CK2 is relatively large, the length of the molecule was speculated to be prolonged here. We then introduced a phenyl group between the amide bond and the 1-methylpiperazine group, hoping to improve the interaction between this part and the hinge region of CK2, thus stabilizing the conformation of the 1-methylpiperazine group.
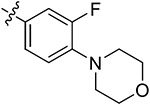
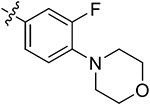
Subsequently, we further increased the length of the terminal substituents, while retaining the amide bond. As a result, the inhibitory activity of the newly synthesized compounds 43a,b against CK2 was not significantly improved. Considering the lack of correlation between the compounds and the wider hinge region of CK2, we introduced 1-methylpiperazine derivatives, morpholine derivatives, and five- and six-membered heterocyclic derivatives linking benzene rings to synthesize compounds 43c–q. Their inhibitory activities against BRD4 and CK2 have been greatly improved. Particularly, 43f, modified by introducing the 1-methylpiperazine group in the para-position of the benzene ring, presented pronounced inhibitory activity against BRD4 and CK2 with inhibitory rates of 82.30% and 80.02%, respectively, at a concentration of 1 μM, and its antiproliferative rate in TNBC cells was also improved (IC50 = 6.46 and 8.80 μM in individual MDA-MB-231 and MDA-MB-468 cells). Moving the 1-methylpiperazine group from the para-position of the benzene ring to the meta- (43g) and ortho-positions (43h) resulted in reduced inhibition against BDR4 and CK2. Later, we analyzed the structure–activity relationship (SAR) on the phenyl group, which directly bonded to the amide bond, and synthesized 44a–i. Compound 44e, modified by introducing a Cl atom at the meta-position, presented the optimal inhibitory activity against BRD4 (92.28%) and CK2 (90.65%), as well as the highest antiproliferative rates in MDA-MB-231 and MDA-MB-468 cells (IC50 = 2.66 and 3.52 μM, respectively), which was slightly better than that of 44c with substitution of the F atom at the meta-position. The introduction of the Cl atom at the meta-position of the benzene ring and of the 1-methylpiperazine group at the para-position were optimal groups for BRD4 and CK2. Next, the inhibitory activity of 44j–n with the Cl atom and the 1-methylpiperazine group against BRD4 was significantly reduced, further validating the importance of the dimethoxyphenyl group. However, in MDA-MB-231 and MDA-MB-468 cells, the antiproliferative effect of 44m was more pronounced than others, which may be explained by an off-target effect. The docking mode analysis revealed that the introduction of phenyl induced interaction between the benzene ring and Met163. The Cl atom on the benzene ring was of great significance in maintaining the conformation of the 1-methylpiperazine group and promoting the formation of intermolecular hydrogen bonds between H atoms of the 1-methylpiperazine group and Asn118 and Asp120 of CK2, thus triggering the stable combination with CK2.
Based on the above analysis, the dimethoxyphenyl group was of great significance to the inhibitory activity against BRD4. After being replaced with single methoxy or halogens (F, Cl) at different positions or H, the inhibitory activity against BRD4 was significantly affected (Figure 5). The quinazolinone core scaffold is a critical group leading to compounds’ activities. The inhibitory activity of target compounds with H substitution on the benzene ring connected to the quinazolinone core scaffold was superior to that of the compounds with methyl and Br substitutions. The introduction of the benzene ring linking the Cl atom at the meta-position and methylpiperazine at the para-position through an amide bond resulted in the best BRD4–CK2 dual inhibitory activity, as well as the antiproliferative ability of TNBC cells. During the process of evaluating the activity, 1 and 18 were used as positive control drugs (Table 3). In MDA-MB-231 and MDA-MB-468 cells, 1 and 18 might show a certain synergistic effect; the combination index (CI) calculated by the Chou–Talalay method was 0.78 and 0.61.48 In addition, we tested inhibitory activities of 44e against BET BDs and CK1 and CK2 because of the pronounced enzymatic activity and antiproliferative activity in vitro (Table 4). It is shown that 44e presented the most significant inhibitory activity against BRD4 (IC50 = 180 nM) compared with the other isoforms of BRD proteins. Moreover, compared with BRD4-BD1 (IC50 = 193 nM), 44e showed weak selectivity for BRD4-BD2 (IC50 = 98 nM). Compared with that of CK1, 44e presented higher selectivity to inhibition against CK2 (IC50 = 230 nM). Hence, 44e is an effective BRD4–CK2 dual inhibitor.
Figure 5.
Structural optimization and discovery of BRD4–CK2 dual-target inhibitors. Representative compounds are shown.
Table 4.
CK1, CK2, and BRD Inhibitory Profiles of Compounds 44e, 1, and 18 (nM)a
| target | 44e | 1 | 18 |
|---|---|---|---|
| BRD2 BD1 | >10000 | 185 ± 24 | b |
| BRD2 BD2 | 8700 ± 420 | 32 ± 4 | b |
| BRD3 BD1 | 9200 ± 670 | 76 ± 8 | b |
| BRD3 BD2 | >10000 | 61 ± 7 | b |
| BRD4 BD1 | 193 ± 60 | 92 ± 13 | b |
| BRD4 BD2 | 98 ± 35 | 44 ± 5 | b |
| BRD4 (BD1 + BD2) | 180 ± 20 | 84 ± 10 | b |
| BRDT BD1 | >10000 | 207 ± 23 | b |
| BRDT BD2 | >10000 | 185 ± 21 | b |
| CK2 | 230 ± 43 | b | 4 ± 1 |
| CK1 | >10000 | b | >10000 |
Reported compounds were tested in triplicate. Data are presented as mean ± SD.
Not determined.
Molecular docking analysis revealed the binding modes of 44e in the active pocket of BRD4(BD2) and CK2. The core scaffold of 44e was located inside the active pocket, forming intermolecular hydrogen bonds with amino acid residues Pro375 and Asn433 of BRD2 (BD2) and hydrophobic interactions with Pro375, Leu385, Leu387, and Val439, showing a similar molecular binding mode to that of 3. Compared to the lead compound 26, the hydrophobic interaction formed by the hydrogen on the side-chain benzene ring and Pro434 and intermolecular hydrogen bonds formed by the 1-methylpiperazine group and Tyr432 further promoted the binding of 44e and BRD4 (BD2). The core part of 44e was deeply inserted into the hydrophobic pocket of CK2, forming intermolecular hydrogen bonds with amino acid residues Lys77, Lys158, Asn161 of CK2 and a conjugation force with Asp175. An intermolecular hydrogen bond formed between the carbonyl group of the amide bond and the amino acid residue Leu45 was responsible for fixing the position and conformation of the compound in CK2. Compared with compound 42g, a conjugation force formed by the increased side-chain benzene ring and Met163, and intermolecular hydrogen bonds formed by the 1-methylpiperazine group and Asn118 and Asp120 in the hinge region enhanced the binding stability with CK2 as well (Figure 4B). Collectively, 44e is a novel chemical type for BRD4-CK2 dual inhibition, which might be helpful for subsequent exploration of novel BRD4-CK2 inhibitors.
Compound 44e Induces TNBC Cell Death by Targeting BRD4 and CK2.
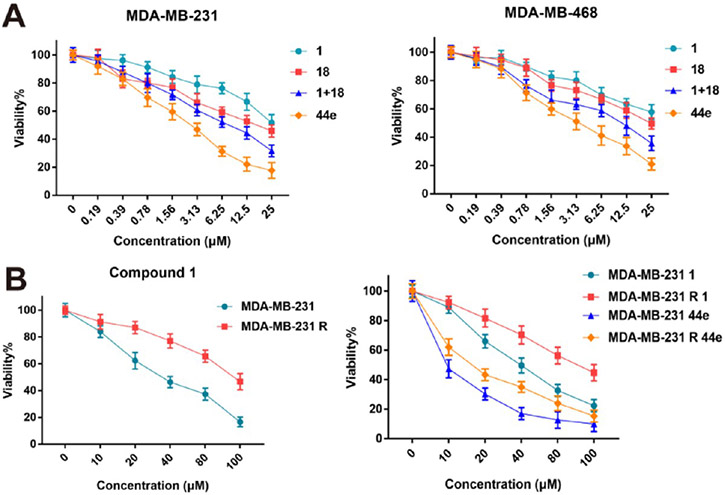
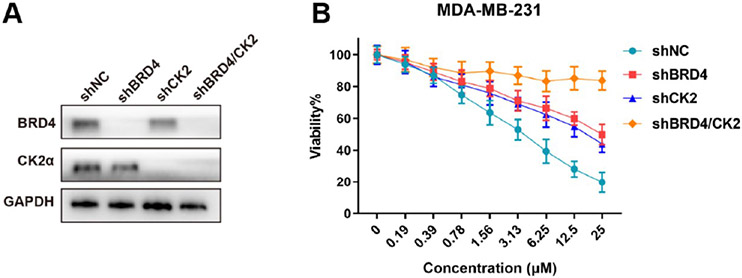
A combination administration of 1 and 18 (at a mole ratio of 1:1) in MDA-MB-231 and MDA-MB-468 cells resulted in a synergistic effect, while administration of the candidate compound 44e presented the most effective kinase inhibition and antiproliferation (Figure 6A and Table 3). Notably, it did not have obvious toxicity to the normal breast cell line MCF-10A (Figure S3). Considering the common concern of drug resistance with BRD4 inhibitors, we first assessed the anticancer activity of 44e against BRD4 inhibitor-resistant breast cancer cells. Briefly, compound 1-resistant MDA-MB-231 cell line (MDA-MB-231R) was generated by low-dose 1 selection at different time points. MDA-MB-231R cells and their parental cells were treated with different doses of 1, aiming to generate MDA-MB-231R.49 Later, MDA-MB-231R cells and their parental cells were treated with different doses of 1 or 44e. It was shown that 44e was able to alleviate drug resistance of 1 in MDA-MB-231R cells (p < 0.01), suggesting that 44e still presented an acceptable anticancer effect on 1-resistant breast cancers (Figure 6B).
Figure 6.
Antiproliferative activity of candidate compound 44e. (A) Antiproliferative activity of 1, 18, 1 + 18, and 44e toward MDA-MB-231 and MDA-MB-468 cells. (B) Antiproliferative activity of 44e toward compound 1-resistant MDA-MB-231R cells.
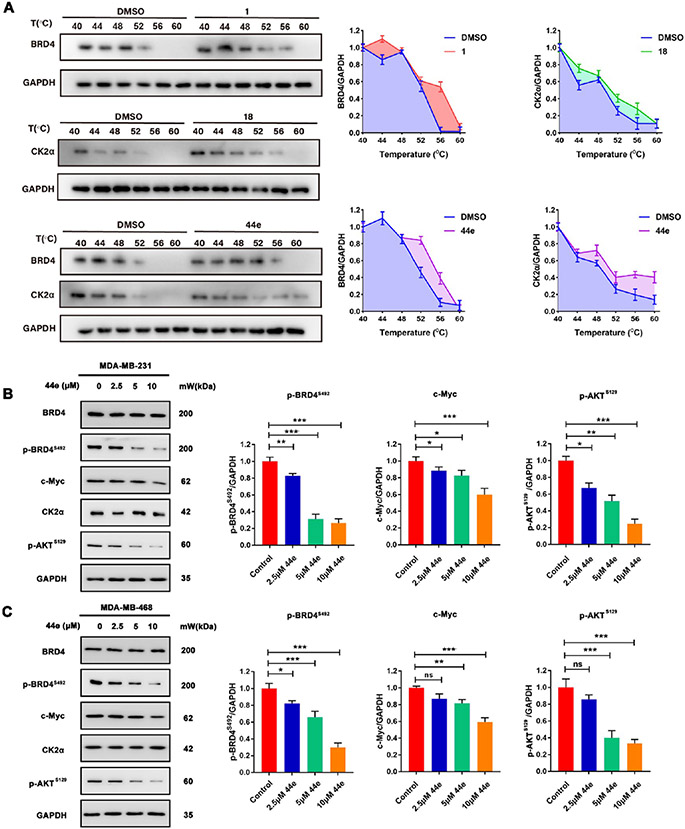
To further verify the BRD4 and CK2 inhibition efficacy of 44e treatment in TNBC cells, we silenced the expression of BRD4, CK2, and BRD4/CK2 in MDA-MB-231 cell lines with the BRD4 shRNA, CK2 shRNA, and BRD4/CK2 shRNA, respectively (Figure 7A). The expression of BRD4 and CK2 was significantly downregulated in MDA-MB-231 cells. Following a 24 h exposure to 44e, the results showed that the inhibitory effect of 44e was reduced to a certain extent after knockdown. This finding also verified the inhibition of BRD4 and CK2 by 44e in TNBC cell lines (Figure 7B). Next, to investigate whether 44e could bind to BRD4 and CK2α in cells, we performed CETSA assay in MDA-MB-231 cells. The results demonstrated that 44e improved the thermal stability of BRD4 and CK2α, indicating that 44e could directly combine with BRD4 and CK2α (Figure 8A). Subsequently, we determined whether 44e affects the key downstream proteins of BRD4 and CK2α. MDA-MB-231 and MDA-MB-468 cells were treated with 0, 2.5, 5, and 10 μM of 44e for 24 h, followed by detection of protein levels of BRD4, c-Myc, CK2α, and p-AKTS129 by Western blotting. Previously, the Chiang lab reported that the NPS in BRD4 is enriched in acidic residues, containing seven CK2 phosphorylation sites (S484, S488, S492, S494, S498, S499, S503).37 We verified that 44e dose-dependently downregulated the level of p-BRD4S492 (Figure 8B,C). Taken together, 44e could bind to BRD4 and CK2 in cells and inhibit the proliferation pathway regulated by BRD4 and CK2 but had no significant effect on the protein levels of BRD4 and CK2.
Figure 7.
Antiproliferative activity of candidate compound 44e relies on BRD4 and CK2. (A) BRD4, CK2, or BRD4/CK2 knockdown efficiency in MDA-MB-231 cells as determined by Western blots. GAPDH was used as a control. (B) MDA-MB-231 cell viability after 24 h at different concentrations of 44e.
Figure 8.
Western blotting assay. (A) CETSA assay detected the thermal stability of BRD4 and CK2 in MDA-MB-231 cells treated with 1, 18, and 44e. (B, C) Expression of BRD4, p-BRD4S492, c-Myc, CK2α, and p-AKTS129 in MDA-MB-231 and MDA-MB-468 cells treated with 0, 2.5, 5, and 10 μM of 44e for 24 h. Error bar shows SD; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 compared with the control groups.
Compound 44e Inhibits Colony Formation and Promotes Apoptosis in TNBC Cells.
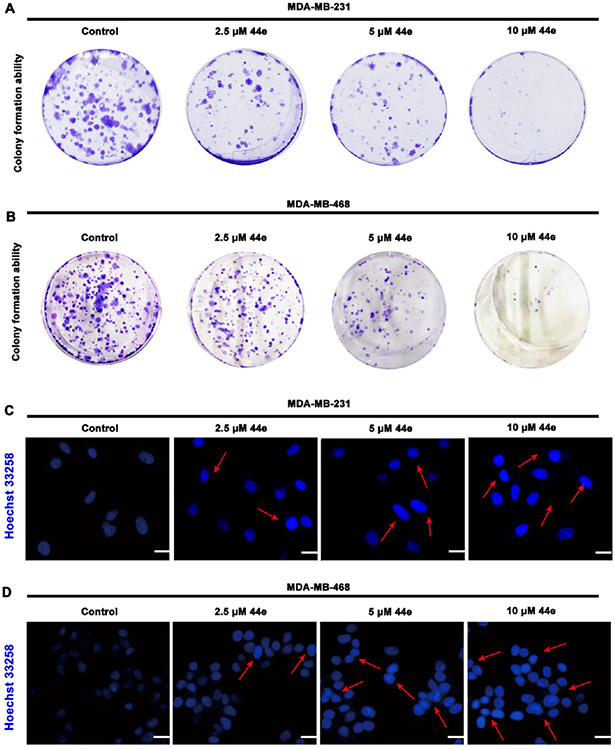
To further investigate the effect of 44e on TNBC cells, we first performed the colony formation assay to detect the long-term proliferation ability of TNBC cells after 44e treatment. The results showed that 44e dose-dependently suppressed growth and reduced the number of colonies in MDA-MB-231 and MDA-MB-468 cells (Figure 9A,B). In addition, MDA-MB-231 and MDA-MB-468 cells were treated with different doses of 44e for 24 h, followed by Hoechst 33258 staining to determine whether apoptosis was induced after 44e treatment. The results showed that the number of dead cells was dose-dependently elevated (Figure 9C,D). Collectively, these results revealed that apoptosis might be involved in 44e-induced cell death.
Figure 9.
Compound 44e inhibits colony formation and promotes apoptosis in TNBC cells. (A, B) Colony formation assay. MDA-MB-231 and MDA-MB-468 cells were treated with 0, 2.5, 5, and 10 μM of 44e for 7 days. (C, D) Hoechst 33258 staining assay. MDA-MB-231 and MDA-MB-468 cells treated with 0, 2.5, 5, and 10 μM of 44e for 24 h. Scale bar, 10 μm.
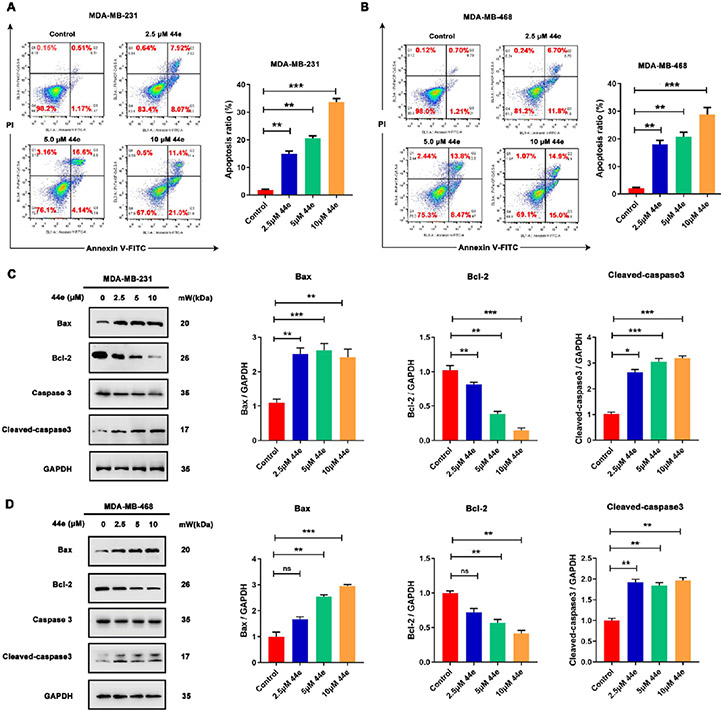
In order to confirm that 44e could induce apoptosis, flow cytometry was performed to measure cell apoptosis. Interestingly, 44e dose-dependently induced apoptosis of MDA-MB-231 and MDA-MB-468 cells (Figure 10A,B). As expected, 44e dose-dependently downregulated Bcl-2 but upregulated Bax and cleaved caspase-3 (Figure 10C,D). Taken together, 44e induces apoptosis in MDA-MB-231 and MDA-MB-468 cells.
Figure 10.
Apoptotic effect of 44e on TNBC cells. (A, B) MDA-MB-231 and MDA-MB-468 cells were treated with indicated concentrations of 44e for 24 h. Apoptosis ratios were determined by flow cytometry analysis of Annexin-V/PI double staining. A representative image and quantification of apoptosis are presented. (C, D) Cells were treated with 44e at different concentrations for 24 h. The levels of apoptosis-related proteins, including Bcl-2, Bax, caspase-3, and cleaved caspase-3, were analyzed by Western blotting. GAPDH was used as load control. Error bar shows SD; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 compared with the control groups.
Compound 44e Induces Autophagy in TNBC Cells.
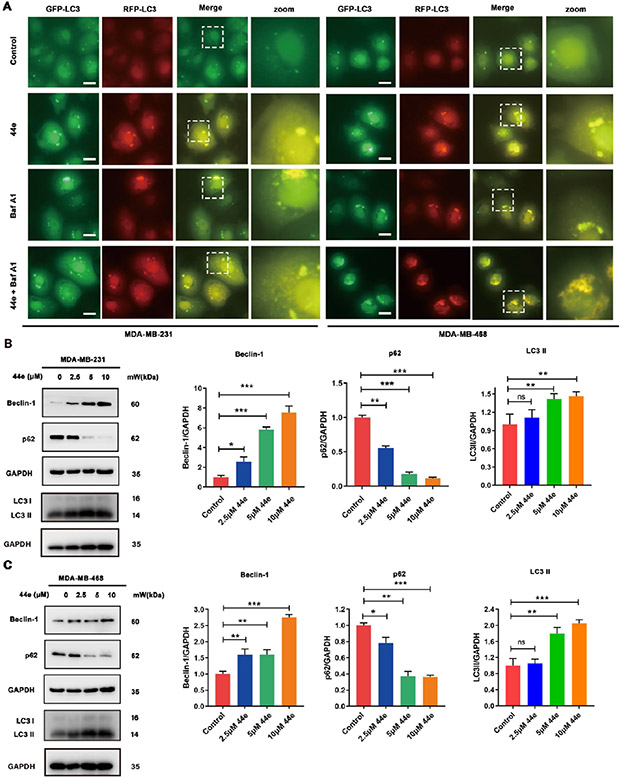
In our previous work, we found that BRD4 inhibition could induce autophagy in breast cancer cells.50 So we next checked whether 44e could induce autophagy in TNBC cells. MDA-MB-231 and MDA-MB-468 cells were transfected with the autophagy biosensor GFP-mRFP-LC3. With an inverted microscope, it was shown that 44e-induced autophagy flux was significantly increased (Figure 11A). Protein levels of autophagy markers were further detected by Western blotting. As expected, 44e significantly down-regulated the autophagy substrate p62 and up-regulated beclin-1 and LC3II (Figure 11B,C). Taken together, 44e significantly induced autophagy in MDA-MB-231 and MDA-MB-468 cells.
Figure 11.
Compound 44e induces regulated autophagy in TNBC cells. (A) MDA-MB-231 and MDA-MB-468 cells were transfected with GFP-mRFP-LC3 adenovirus, following co-incubation with 44e (5 μM) in the presence or absence of BafA1 (10 nM). Scale bar, 10 μm. (B, C) Expression levels of Beclin-1, LC3, and p62 in MDA-MB-231 and MDA-MB-468 cells treated with 5 μM 44e, analyzed by Western blotting. GAPDH was used as load control. Error bar shows SD; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 compared with the control groups.
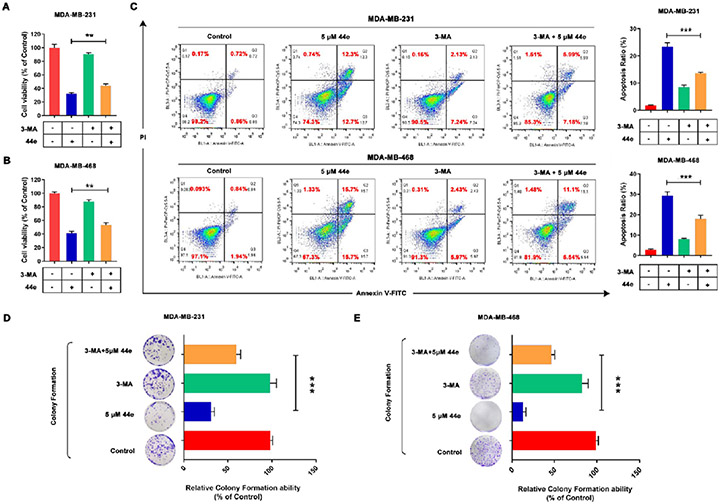
To investigate the role of autophagy in 44e-induced cell death, we employed the autophagy inhibitor 3-methyladenine (3-MA), which is commonly used to inhibit the autophagic process by blocking autophagosome formation.51 And according to previous findings, 3-MA was applied to show the relationship between cell death and autophagy.52 Upon the addition of 3-MA, the effect of 44e on cell death was significantly reduced, suggesting that autophagy plays a role in promoting TNBC cell death (Figure 12A,B). Similar results were also obtained from colony formation assay (Figure 12D,E), indicating that 44e induces autophagy-associated cell death in TNBC cells. In addition, a significantly lower apoptotic rate was detected in MDA-MB-231 and MDA-MB-468 cells treated with both 44e and 3-MA for 24 h compared to that of cells treated with 44e only (Figure 12C), suggesting that application of 3-MA was able to partially reverse 44e-induced apoptosis in TNBC cells. These data indicate that 44e-induced autophagy inhibited cell proliferation and promoted cell apoptosis.
Figure 12.
Mechanism of autophagy in 44e-induced cell death in TNBC cells. (A, B) MDA-MB-231 and MDA-MB-468 cell viability was measured by MTT assay. Cells were treated with 44e at 5 μM or combined with 3-MA at 1 mM treated for 24 h. 3-MA was added 1 h before treatment with 44e. (C) MDA-MB-231 and MDA-MB-468 cell apoptosis ratios were analyzed by flow cytometry with Annexin-V/PI double staining. Cells were treated with 44e at 5 μM or combined with 3-MA at 1 mM for 24 h. 3-MA was added 1 h before treatment with 44e. (D, E) Colony formation assay in MDA-MB-231 and MDA-MB-468 cells. Cells treated with 44e at 5 μM or combined with 3-MA at 1 mM. Error bar shows SD; *p < 0.05, **p < 0.01, ***p < 0.001 compared with the control groups.
Preliminary Assessment of Pharmacokinetics (PK) profile of 44e.
Based on the most potent BRD4–CK2 inhibitory activity of compound 44e and its antiproliferation and ability to induce apoptosis and autophagy-associated cell death in TNBC cells, we further studied the pharmacokinetic properties. As shown in Table 5, compound 44e was administered as a single dose at 1 mg/kg and 10 mg/kg by intravenous and oral routes, respectively. The results indicated that 44e could be absorbed rapidly from the intestine with the highest concentration observed at 5.14 ± 0.71 h and a maximum plasma concentration (Cmax) value of 206 ± 6 ng/mL. Furthermore, 44e had a reasonable area under the curve (AUC0–∞) value of 2079 ± 130 ng·h/mL and an acceptable oral bioavailability of 32.5%. Thus, it is ofgreat significance to further conduct in vivo tests of 44e on antitumor activity.
Table 5.
Pharmacokinetic Profile of 44ea
| parameter | iv (1 mg/kg) | po (10 mg/kg) |
|---|---|---|
| T1/2 (h) | 4.21 ± 0.57 | 5.14 ± 0.71 |
| Cmax (ng/mL) | 237 ± 11 | 206 ± 6 |
| AUC0–t (ng·h/mL) | 579 ± 49 | 2079 ± 130 |
| AUC0–∞ (ng·h/mL) | 588 ± 36 | 2090 ± 146 |
| Vz (L/kg) | 21.1 ± 2.6 | |
| CL ((mL/min)/kg) | 57.4 ± 1.3 | |
| F (%) | 32.5 |
All data were obtained by triple testing (mean ± SD) in SD rats.
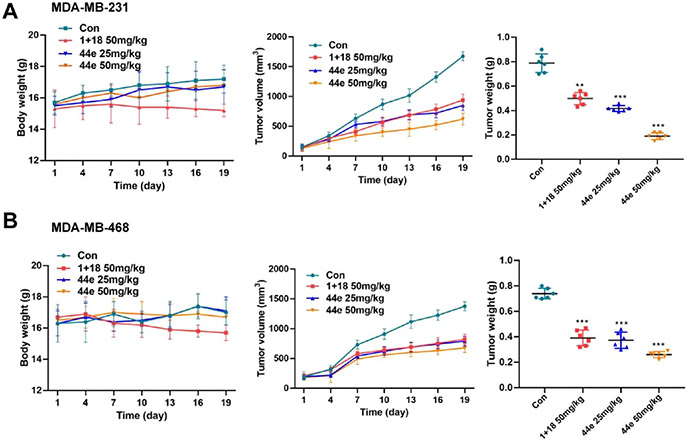
Compound 44e Inhibits Tumor Growth in TNBC Xenograft Models.
To further investigate the in vivo inhibitory effect of 44e on BRD4 and CK2, we generated MDA-MB-468 and MDA-MB-231 xenograft tumor models in mice. In each model, a total of 24 mice were randomly assigned into a control group (normal saline, n = 6), a combination group (1 and 18 combined at a mole ratio of 1:1, n = 6), a 44e 25 mg/kg group (n = 6), and a 44e 50 mg/kg group (n = 6). Intragastric administration of normal saline, 1 and 18, and 44e was given for consecutive 19 days. Compared with the control group, mice in the latter three groups all presented a dose-dependent tumor growth inhibition (TGI). In particular, 50 mg/kg intervention with 44e had the most pronounced TGI (63.8%) in the MDA-MB-231 xenograft tumor model (Figure 13A). No significant changes in mouse body weight and death were observed in both xenograft models treated with 44e. However, 1 + 18 demonstrated weak toxicity measured by body weight loss in treated mice (Figure 13A,B), which may be attributed to side effects of drug combinations.5 As expected, there was a significant difference between the 44e groups and the combination group. Compound 44e was more effective in these in vivo studies. Its safety was slightly better than that of the 1 + 18 combination. In addition, 44e’s skeleton is worthy of fUrther study and may have a positive effect on the development of BRD4–CK2 inhibitors. Moreover, the discovery of a potent inhibitor of BRD4–CK2 might provide chemical tools to help further research the physiological function of the two targets. Taken together, 44e is a promising potent anticancer candidate against TNBC without significant toxicities.
Figure 13.
Antitumor activity of 44e in the MDA-MB-231 and MDA-MB-468 xenograft models. (A) Body weights, tumor volumes, and tumor weights following treatment by oral administration with 25 or 50 mg/kg 44e or 50 mg/kg 1 + 18 in the MDA-MB-231 tumor xenograft model. (B) Body weights, tumor volumes, and tumor weights following the treatment by oral administration with 25 or 50 mg/kg 44e or 50 mg/kg 1 + 18 in the MDA-MB-468 tumor xenograft model. Error bar shows SD; *p < 0.05, **p < 0.01, ***p < 0.001 compared with the control groups.
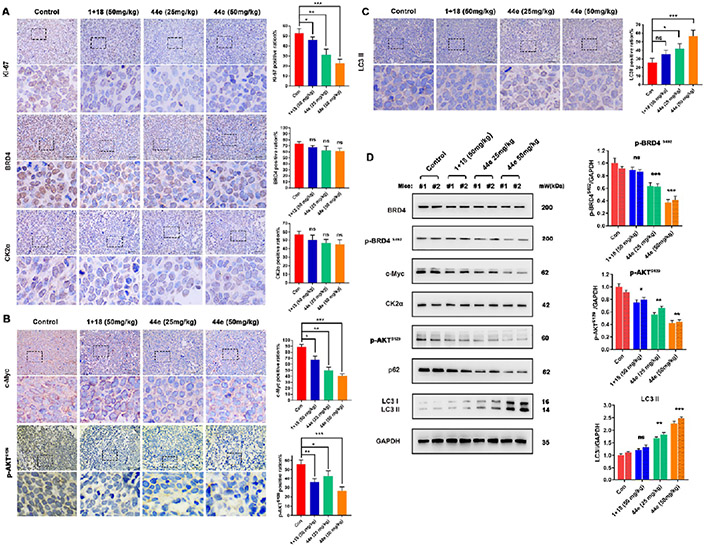
Next, the mouse heart, liver, spleen, lung, and kidney tissues were collected for HE staining. No significant morphologic or structural changes were observed (Figures S4 and S5). In addition, the immunohistochemical staining of Ki-67 in tumor tissue also indicated that 44e could significantly inhibit the proliferation of MDA-MB-231 xenograft tumor model (Figure 14A). To further clarify the therapeutic index of 44e in vivo, we detected positive expression of BRD4, CK2 (Figure 14A), and their downstream proteins c-Myc and p-AKTS129 (Figure 14B,C) in MDA-MB-231 xenograft tumor tissues by immunohistochemistry. Compared with those in the control group, the expression of c-Myc and p-AKTS129 was significantly reduced in 44e treated groups. Increased expression of LC3II in TNBC tissues of the 44e 25 mg/kg group and the 44e 50 mg/kg group further highlighted the occurrence of autophagy. Consistently, Western blot analysis showed similar results of c-Myc and p-AKTS129 protein levels in xenograft tissues of TNBC (Figure 14D). Altogether, 44e is a promising BRD4–CK2 dual inhibitor that exerts acceptable anticancer effect on TNBC.
Figure 14.
Potential therapeutic effects of 44e in vivo. (A–C) Representative images of IHC analysis of Ki-67, BRD4, c-Myc, CK2α, p-AKTS129, and LC3II markers in different groups from MDA-MB-231 xenograft tumor model. Scale bar, 40 μm. (D) Two individual tumor tissues (1 and 2) excised from the MDA-MB-231 xenograft tumor model were analyzed. The expression levels of BRD4, c-Myc, CK2α, p-AKTS129, p62, and LC3II were detected by Western blot analysis. Error bar shows SD; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 compared with the control groups.
CONCLUSION
In summary, this study aimed to identify small molecule BRD4–CK2 dual inhibitors that can be used as potential therapeutic for TNBC. An important interaction between BRD4 and CK2 in TNBC was validated through network analysis and systems biology. With available BRD4 (BD2) inhibitor and CK2 inhibitor cocrystal structures, key amino acids were first discovered. Following the virtual screening workflow, the lead compound 26 was first obtained. Then, a series of quinazolinone core scaffold derivatives was synthesized through rational drug design, synthesis, and in vivo and in vitro biological evaluations. We for the first time synthesized a selective inhibitor, 44e, that could target both BRD4 and CK2. Moreover, 44e was able to inhibit the proliferation of MDA-MB-231 and MDA-MB-468 cells and dose-dependently induce apoptotic and autophagy-associated cell death. Its dose-dependent TGI effect was also illustrated in mouse xenograft models of TNBC. Interestingly, 44e showed better anticancer activity, not only expanding the therapeutic index of BRD4 inhibitors but also reducing or preventing acquired resistance. Another advantage of 44e may be the reduction of toxicity against normal breast cell lines and tissues. In addition, 44e shows acceptable pharmacokinetic properties. Collectively, our findings support that the BRD4–CK2 dual inhibitor 44e used in the treatment of TNBC is worthy of further development and analysis.
EXPERIMENTAL SECTION
Chemistry Methods.
All chemicals including reagents and solvents were purchased as analytically pure forms from commercial sources and used without further purification. HRESIMS data were obtained on a Q-TOF micro-mass spectrometer (Waters, USA). The 1H and 13C nuclear magnetic resonance (NMR) spectra were recorded on a Bruker AV 400 spectrometer (Bruker Co. Ltd., Germany). Chemical shifts are given in parts per million (ppm), and TMS was used as an internal standard. High-performance liquid chromatography (HPLC) was performed on a Shimadzu LC-20 instrument (Kyoto, Japan) with Agilent ZORBAX Eclipse Plus C18 Column (3.5 μm, 150 mm × 4.6 mm) using MeOH/H2O as the mobile phase at a flow rate of 1.0 mL/min. The purity of the synthesized compounds was over 95% by HPLC. The HPLC trace of each compound is provided in the Supporting Information (SI). Melting points of the compounds were obtained on a Melting Point M-565 (Buchi, Switzerland).
Preparation of intermediate 31a.
1-(2-Hydroxy-4,6-dimethoxyphenyl)ethan-1-one (2.5 g, 12.7 mmol) dissolved in 20 mL of pyridine was incubated in 5 mL of the alkaline reagent DBU for 0.5 h. Then, 3,5-dimethylbenzoyl chloride (3.5 g, 20.8 mmol) was slowly dropped into the mixture, and the mixture was heated at 80 °C for 1 h. After cooling to room temperature, the pH of the mixture was adjusted to 5.0 by adding HCl. Then the mixture was extracted three times using ethyl acetate (EA), and the organic layers were combined. The extraction was dried using anhydrous Na2SO4, followed by vacuum distillation, and purified by column chromatography (PE/EA = 30:1–5:1), and finally the intermediate 31a (4 g, 95% yield) was obtained. 1H NMR (400 MHz, DMSO-d6), δ(ppm): 7.83 (2H, s), 7.58 (1H, s), 7.20 (1H, s), 6.56 (1H, s), 3.88 (3H, s), 3.82 (3H, s), 2.47 (3H, s), 2.33 (6H, s). HRMS (ESI)+ Calculated for C19H21O5 [M + H]+ m/z 329.1389, found 329.1387.
Preparation of Intermediate 32a.
The intermediate 31a (1 g, 3 mmol) was dissolved in 5 mL of pyridine at 50 °C for 1 h, and then KOH powder (0.4 g, 7 mmol) was added, and the mixture was heated at 80 °C for 1 h. After cooling to room temperature, the pH of the mixture was adjusted to 5.0 by adding HCl. Then the mixture was extracted three times using EA, and the organic layers were combined. The extraction was dried using anhydrous Na2SO4, followed by vacuum distillation, and intermediate 32a (0.5 g, 50% yield) was obtained. It was not necessary to purify 32a.
Preparation of 33a.
Intermediate 32a (200 mg, 0.6 mmol) was dissolved in 4 mL of HOAc at 90 °C for 1 h. After vacuum concentration, 5 mL of 1% H2SO4–HOAc (v/v) was added for reaction at 110 °C at 1 h. The reaction mixture was cooled in an ice–water mixture, and then the precipitant was filtered for purification by column chromatography (CH2O2/CH3OH = 80:1–1:1). Finally, the target compound 33a (120 mg, 76% yield) was obtained.
Synthesis routes of 33b–d were similar to that of 33a.
Preparation of 34a.
Compound 33a (100 mg, 0.3 mmol) dissolved in 4 mL of methylbenzene was incubated with Lawesson’s reagent (0.2 mmol) at 110 °C for 4 h. After concentration under vacuum conditions and purification by column chromatography (CH2Cl2/CH3OH = 100:1–1:1), the target compound 34a (55 mg, 56% yield) was obtained.
Synthesis routes of 34b,c were similar to that of 34a.
Preparation of 35a.
3,5-Dimethoxyaniline (8 g, 52 mmol) was dissolved in 100 mL of Et2O and stirred at 0 °C. After addition of 25 mL of saturated HCl–Et2O solution and reaction for 0.5 h, the mixture was then reacted at room temperature for 12 h. A total of 7 g of the hydrochloride was filtered and collected. Later, the hydrochloride (4 g, 21 mmol) was dissolved in 6 mL of (COCl)2 and refluxed at 160 °C for 2.5 h. After vacuum concentration, 20 mL of MeOH was added at 0 °C, followed by heating for 1 h, filtration, and washing. At last, intermediate 35a (5 g, 65% yield) was obtained. 1H NMR (400 MHz, CDCl3), δ(ppm): 11.06 (1H, s), 6.41 (2H, s), 5.81 (2H, s), 3.94 (3H, s), 3.76 (3H, s). HRMS (ESI)+ Calculated for C10H10NO4 [M + H]+ m/z 208.0611, found 208.0610.
Preparation of 36a.
Intermediate 35a was dissolved in 20 mL of 33% NaOH aqueous solution and reacted with 5 mL of 30% H2O2 aqueous solution. After refluxing at 80 °C for 1 h, the solution was cooled and then incubated in 20 mL of Na2S2O3 solution. The pH of the mixture was adjusted to 8.0 by addition of concentrated HCl, and then it was adjusted to 5.0 by addition of HOAc. The precipitant was washed using water and dried, and thus the intermediate 36a (1.3 g, 34% yield) was obtained. 1H NMR (400 MHz, CDCl3), δ(ppm): 11.06 (1H, s), 6.42 (2H, s), 5.81 (2H, s), 3.94 (3H, s), 3.76 (3H, s). HRMS (ESI)+ Calculated for C9H12NO4 [M + H] + m/z 198.0766, found 198.0760.
Preparation of 37a.
EDCI (0.75 g, 3.8 mmol), HOBt (0.50 g, 3.8 mmol), and NMM (0.38 g, 3.8 mmol) were added in 2-amino-4,6-dimethoxybenzoic acid (0.60 g, 3.0 mmol) dissolved in 20 mL of THF and reacted at room temperature for 15 min. Later, ammonium hydroxide 50% (v/v) aqueous solution was added and reacted for 12 h. After dilution by addition of 8 mL of H2O and extraction using CH2Cl2 (30 mL × 3), the extract was subjected to vacuum distillation and purified by column chromatography (DCM/MeOH = 100:1–60:1), and finally the intermediate 37a (0.46 g, 85% yield) was obtained. 1H NMR (400 MHz, DMSO-d6), δ(ppm): 7.46 (1H, s), 7.01 (1H, s), 6.88 (2H, s), 5.89 (1H, d, J = 2.4 Hz), 5.75 (1H, d, J = 2.4 Hz), 3.75 (3H, s), 3.69 (3H, s). HRMS (ESI)+ Calculated for C9H13N2O3 [M + H]+ m/z 197.0926, found 197.0924.
Compound 37c was synthesized based on a similar route, and 37b was commercially purchased.
General Procedures for Synthesizing 38a–s and the Lead Compound 26.
Intermediates 37a–c (0.9 mmol, 1 equiv) and benzaldehyde derivatives (0.9 mmol, 1 equiv) were dissolved in 15 mL of DMAc, in which NaHSO4 (1.05 mmol, 1.2 equiv) and PTSA (0.22 mmol, 0.24 equiv) were added. The reaction solution was stirred and refluxed at 120 °C for 4–8 h. Later, 100 mL of Ô was added, and the precipitant was filtered. The solid was purified by flash chromatography on silica gel (CH2O2/CH3OH = 80:1–10:1), and finally 38a–s and the lead compound 26 (41–65% synthetic yield) were obtained.
Preparation of Intermediate 39a.
tert-Butyl bromoacetate (3.0 mL, 1.2 equiv) and p-hydroxybenzaldehyde (2.5 g, 1 equiv) were dissolved in 10 mL of DMF, in which 5.2 g of K2O3 was added for 3 h reaction at 80 °C. Later, 80 mL of H2O was added, and the precipitant was filtered and dried. The solid was purified by flash chromatography on silica gel (CH2Cl2/CH3OH = 80:1–5:1), and finally intermediate 39a (2.6 g, 57% yield) was obtained. It was not necessary to purify 39a. 1H NMR (400 MHz, CD3OD), δ(ppm): 9.84 (1H, s), 7.86 (2H, d, J = 8.8 Hz), 7.06 (2H, d, J = 8.8 Hz), 4.71 (2H, s), 1.48 (9H, s). HRMS (ESI)+ Calculated for C13H17O3 [M + H]+ m/z 221.1178, found 221.1170.
Synthesis routes of 39b,c were similar to that of 39a.
Preparation of Intermediate 40a.
The intermediate 39a (2 g, 9 mmol) was dissolved in 5 mL of CH2Cl2 and reacted with 1.5 mL of TFA at room temperature for 2 h. The reaction solution was subjected to vacuum concentration, and the white solid 40a (1.2 g, 74% of synthesis yield) was obtained. It was not necessary to purify 40a. 1H NMR (400 MHz, CD3OD), δ(ppm): 12.28 (1H, s), 9.82 (1H, s), 7.84 (2H, d, J = 8.8 Hz), 7.04 (2H, d, J = 8.8 Hz), 3.68 (2H, s). HRMS (ESI)+ Calculated for C9H9O4 [M + H]+ m/z 181.0501, found 181.0507.
Synthesis routes of 40b,c were similar to that of 40a.
Preparation of 42a-j, 43a-q, and 44a-n.
Intermediates 40a–c (0.9 mmol, 1 equiv) and benzaldehyde derivatives (0.9 mmol, 1 equiv) were dissolved in 15 mL of DMAc, in which NaHSO4 (1.05 mmol, 1.2 equiv) and PTSA (0.22 mmol, 0.24 equiv) were added. The reaction solution was stirred and refluxed at 120 °C for 4–8 h. Later, 100 mL of H2O was added, and the precipitant was filtered. The solid was purified by flash chromatography on silica gel (CH2Cl2/CH3OH = 30:1–1:1), and finally 41a–h (42–66% synthetic yield) were obtained. Intermediates 41a–h (0.42 mmol, 1.2 equiv) were dissolved in 3 mL of DMF, in which HOBt (0.54 mmol, 1.3 equiv) was added and stirred at 0 °C for 10 min. Subsequently, EDCI (0.54 mmol, 1.3 equiv), reactants containing amino substitutions, and TEA (1.5 mmol, 3.5 equiv) were added and reacted at 0 °C for 1 h. The reaction mixture was further reacted at room temperature for 24 h. Later, 80 mL of H2O was added, and the precipitant was filtered. The solid was purified by flash chromatography on silica gel (CH2Cl2/CH3OH = 30:1–1:1), and finally end products 42a–j, 43a–q, and 44a–n (43–66% synthetic yield) were obtained.
2-(3,5-Dimethylphenyl)-5,7-dimethoxy-4H-chromen-4-one (33a).
White powder, mp 214–216 °C, yield 50.1%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 7.65 (2H, s), 7.19 (1H, s), 6.87 (1H, d, J = 2.4 Hz), 6.70 (1H,s), 6.49 (1H, d, J = 2.4 Hz), 3.90 (3H, s), 3.82 (3H, s), 2.35 (6H, s). 13C NMR (100 MHz, CF3COOD), δ (ppm): 178.6, 175.7, 173.3, 163.3, 162.1, 143.2(2), 140.1, 130.7, 127.8(2), 104.6, 103.4, 102.5, 96.8, 59.9, 58.8, 22.1(2). HRMS (ESI)+ Calculated for C19H19O4 [M + H]+ m/z 311.1283, found 311.1288.
2-(3,5-Dimethylphenyl)-6-methoxy-4H-chromen-4-one (33b).
White powder, mp 210–212 °C, yield 55.7%. 1H NMR (400 MHz, CDCl3), δ (ppm): 7.58 (1H, d, J = 2.4 Hz), 7.50–7.52 (3H, m), 7.28 (1H, dd, J = 8.0, 2.4 Hz), 7.16 (1H, s), 6.78 (1H, s), 3.90 (3H, s),2.40 (6H, s). 13C NMR (100 MHz, CDCl3), δ (ppm): 178.5, 163.8, 157.1, 151.3, 138.8(2), 133.4, 131.9, 124.7, 124.2(2), 123.8, 119.6, 106.9, 56.1, 21.5(2). HRMS (ESI)+ Calculated for C18H17O3 [M + H]+ m/z 281.1178, found 281.1184.
6-Methoxy-2-(2-methoxyphenyl)-4H-chromen-4-one (33c).
White powder, mp 180–183 °C, yield 58.4%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 7.90 (1H, dd, J = 8.4, 2.0 Hz), 7.68 (1H, dd, J = 8.4, 2.0 Hz), 7.56 (1H, td, J = 8.0, 2.4 Hz), 7.39–7.42 (1H, m), 7.25 (1H, d, J = 8.4 Hz), 7.15 (1H, td, J = 8.0, 2.4 Hz), 6.91 (1H, s), 3.92 (3H, s), 3.86 (3H, s). 13C NMR (100 MHz, CDCl3), δ (ppm): 178.9, 160.8, 158.1, 156.9, 151.5, 132.5, 129.4, 124.5, 123.8, 121.1, 120.9, 119.6, 112.0, 111.9, 104.8, 56.1, 55.8. HRMS (ESI)+ Calculated for C17H157O4 [M + H]+ m/z 283.0970, found 283.0963.
6-Methoxy-2-(2-methoxyphenyl)-4H-chromen-4-one (33d).
White powder, mp 192–195 °C, yield 51.2%. 1H NMR (400 MHz, CDCl3), δ (ppm): 8.36 (1H, d, J = 2.4 Hz), 7.91 (2H, dd, J = 8.4, 2.4 Hz), 7.78 (1H, dd, J = 8.4, 2.4 Hz), 7.51–7.56 (3H,m), 7.47 (1H, d, J = 8.4 Hz), 6.83 (1H, s). 13C NMR (100 MHz, CDCl3), δ (ppm): 177.2, 163.9 155.2, 136.9, 132.0, 131.5, 129.3(2), 128.5, 126.5, 125.4(2), 120.2, 118.8, 107.7, 104.8, 56.1, 55.8. HRMS (ESI)+ Calculated for C15H10BrO2 [M + H]+ m/z 300.9864, found 300.9866.
6-Methoxy-2-(2-methoxyphenyl)-4H-chromen-4-one (34a).
White powder, mp 211–213 °C, yield 53.4%. 1H NMR (400 MHz, CDCl3), δ (ppm): 7.54 (1H, s), 7.51 (2H, s), 7.13 (1H, s), 6.58 (1H, d, J = 2.4 Hz), 6.40 (1H, d, J = 2.4 Hz), 3.93 (3H, s), 3.91 (3H, s), 2.38 (6H, s). 13C NMR (100 MHz, CDCl3), δ (ppm): 200.3, 164.0, 161.6, 155.9, 150.3, 138.8(2), 133.2, 130.8, 124.1(2), 122.5, 117.9, 97.0, 92.9, 56.0(2), 21.5(2). HRMS (ESI)+ Calculated for C19H19O3S [M + H] + m/z 327.1055, found 327.1049.
6-Methoxy-2-(2-methoxyphenyl)-4H-chromen-4-one (34b).
White powder, mp 223–225 °C, yield 48.4%. 1H NMR (400 MHz, CDCl3), δ (ppm): 7.95–7.99 (3H, m), 7.81 (1H, s), 7.49–7.55 (4H, m), 7.32 (1H, dd, J = 8.4, 2.4 Hz), 3.93 (1H, s). 13C NMR (100 MHz, CDCl3), δ (ppm): 200.9, 158.0, 154.2, 146.5, 131.8, 131.3, 130.7, 129.3(2), 126.6(2), 124.6, 120.0, 119.9, 107.9, 56.1. HRMS (ESI)+ Calculated for C16H13O2S [M + H]+ m/z 269.0636, found 269.0628.
6-Methoxy-2-(2-methoxyphenyl)-4H-chromen-4-one (34c).
Light yellow solid, mp 193–195 °C, yield 55.1%. 1H NMR (400 MHz, CDCl3), δ (ppm): 8.15 (1H, s), 8.01 (1H, d, J = 3.2 Hz), 7.92 (1H, dd, J = 8.0, 2.4 Hz), 7.46–7.51 (2H, m), 7.30 (1H, dd, J = 8.0, 2.4 Hz), 7.10 (1H, td, J = 3.2 Hz), 7.03 (1H, d, J = 8.4 Hz), 3.95 (3H, s), 3.93 (3H, s). 13C NMR (100 MHz, CDCl3), δ (ppm): 201.0, 158.2, 157.9, 152.4, 146.8, 132.8, 130.6, 129.7, 124.9, 124.5, 121.1, 120.3, 119.9, 112.0, 107.9 56.0, 55.9. HRMS (ESI)+ Calculated for C17H15O3S [M + H] + m/z 299.0742, found 299.0750.
5,7-Dimethoxy-2-(p-tolyl)quinazolin-4(3H)-one (38a).
White powder, mp 254–256 °C, yield 56.1%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 11.95 (1H, s), 8.07 (2H, d, J = 8.0 Hz), 7.32 (2H, d, J = 8.0 Hz), 6.73 (1H, d, J = 2.0 Hz), 6.52 (1H, d, J = 2.0 Hz), 3.88 (3H, s), 3.84 (3H, s), 2.37 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 164.2, 160.9, 159.8, 153.0, 152.8, 141.4, 129.5, 129.1(2), 127.5(2), 104.7, 101.2, 97.6, 55.9, 55.6, 21.0. HRMS (ESI)+ Calculated for C17H17N2O3 [M + H]+ m/z 297.1239, found 297.1242.
2-(4-Bromo-3,5-dimethylphenyl)-5,7-dimethoxyquinazolin-4(3H)-one (38b).
White powder, mp 261–263 °C, yield 49.5%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 11.95 (1H, s), 7.99 (2H, s), 6.76 (1H, s), 6.54 (1H, s), 3.89 (3H, s), 3.85 (3H, s), 2.45 (6H, s). 13C NMR (100 MHz, CDCl3), δ (ppm): 164.8, 161.5, 160.1, 153.1, 152.7, 138.6(2), 131.2, 130.8, 127.9(2), 105.2, 101.6, 98.4, 56.5, 56.2, 23.9(2). HRMS (ESI)+ Calculated for C18H18BrN2O3 [M + H]+ m/z 391.0480, found 391.0472.
2-(4-Aminophenyl)-5,7-dimethoxyquinazolin-4(3H)-one (38c).
White powder, mp 244–246 °C, yield 55.2%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 7.94 (1H, s), 7.91 (1H, s), 6.66 (1H, d, J = 4.8 Hz), 6.63 (2H, d, J = 2.0 Hz), 6.45 (1H, d, J = 4.0 Hz), 3.88 (3H, s), 3.83 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 164.2, 161.0(2), 159.6, 153.3, 152.4, 129.3(2), 117.3, 113.0(2), 103.9, 99.9, 96.8, 55.9, 55.6. HRMS (ESI)+ Calculated for C16H16N3O3 [M + H]+ m/z 298.1192, found 298.1184.
2-(4-Hydroxy-3,5-dimethylphenyl)-5,7-dimethoxyquinazolin-4(3H)-one (38d).
Light yellow solid, mp 232–234 °C, yield 42.9%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 7.81 (2H, s), 6.75 (1H, d, J = 2.0 Hz), 6.54 (1H, d, J = 2.0 Hz), 3.89 (3H, s), 3.85 (3H, s), 2.24 (6H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 164.3, 161.0, 159.6, 156.9 153.2, 152.1, 128.2(2), 124.2(2), 121.9, 104.0, 100.3, 97.3, 56.0, 55.7, 16.7(2). HRMS (ESI)+ Calculated for C18H19N2O4 [M + H] + m/z 327.1345, found 327.1350.
2-(3-Bromo-4-hydroxyphenyl)-5,7-dimethoxyquinazolin-4(3H)-one (38e).
White powder, mp 246–249 °C, yield 56.0%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 11.1 (1H, s), 8.37 (1H, d, J = 2.4 Hz), 8.04 (1H, dd, J = 8.8, 2.4 Hz), 6.73 (1H, d, J = 2.4 Hz), 6.52 (1H, d, J = 2.4 Hz), 3.89 (3H, s), 3.84 (3H, s), 2.24 (6H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 164.4, 161.0, 159.5, 157.4, 153.9, 151.9, 132.7, 128.6, 123.6, 116.0, 109.5, 104.3, 100.5, 97.6, 56.0, 55.7. HRMS (ESI)+ Calculated for C16HMBrN2O4 [M + H]+ m/z 379.0116, found 379.0102.
N-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)phenyl)-acetamide (38f).
White powder, mp 245–247 °C, yield 64.3%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 11.91 (1H, s), 10.23 (1H, s), 8.12 (2H, d, J = 8.8 Hz), 7.71 (1H, d, J = 8.8 Hz), 6.71 (1H, d, J = 2.4 Hz), 6.51 (1H, d, J = 2.4 Hz), 3.88 (3H, s), 3.84 (3H, s), 2.08 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 168.8, 164.2, 161.0, 159.8, 153.1, 152.4, 142.2, 128.4(2), 126.4, 118.3(2), 104.6, 101.1, 97.5, 55.9, 55.6 24.2. HRMS (ESI)+ Calculated for C18H18N3O4 [M + H]+ m/z 340.1297, found 340.1292.
5,7-Dimethoxy-2-(m-tolyl)quinazolin-4(3H)-one (38g).
White powder, mp 237–239 °C, yield 52.9%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 11.94 (1H, s), 8.01 (1H, s), 7.93–7.95 (1H, m), 7.39–7.41 (2H, m), 6.76 (1H, d, J = 2.4 Hz), 6.53 (1H, d, J = 2.4 Hz), 3.89 (3H, s), 3.85 (3H, s), 2.39 (3H, s). 13C NMR (100 MHz, DMSO-d6 + CF3COOD), δ (ppm): 166.0, 162.1, 157.4(2), 143.8, 138.9, 135.0, 130.0, 129.2, 126.9, 126.7, 103.4, 99.0, 96.1, 56.7, 56.3, 20.9. HRMS (ESI)+ Calculated for C17H17N2O3 [M + H]+ m/z 297.1239, found 297.1237.
5,7-Dimethoxy-2-(2-methoxyphenyl)quinazolin-4(3H)-one (38h).
White powder, mp 257–260 °C, yield 48.2%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 11.51 (1H, s), 7.74 (1H, dd, J = 11.6, 2.4 Hz), 7.52 (1H, td, J = 11.6, 2.4 Hz), 7.18 (1H, d, J = 11.6 Hz), 7.08 (1H, t, J = 11.6 Hz), 6.71 (1H, d, J = 2.4 Hz), 6.54 (1H, d, J = 2.4 Hz), 3.87 (3H, s), 3.86 (3H, s), 3.84 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 164.1, 160.9, 158.8, 157.2, 153.3, 152.9, 132.3, 130.3, 122.1, 120.4, 111.9, 104.9, 101.2, 97.7, 56.0, 55.8, 55.6. HRMS (ESI)+ Calculated for C17H17N2O4 [M + H]+ m/z 313.1188, found 313.1186.
5,7-Dimethoxy-2-(3-methoxyphenyl)quinazolin-4(3H)-one (38i).
Light yellow solid, mp 225–227 °C, yield 41.0%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 12.03 (1H, s), 7.77 (1H, d, J = 8.0 Hz), 7.73 (1H, s), 7.43 (1H, t, J = 8.0 Hz), 7.12 (1H, dd, J = 8.0, 2.4 Hz), 6.76 (1H, d, J = 2.4 Hz), 6.56 (1H, d, J = 2.4 Hz), 3.89 (3H, s), 3.85 (6H, s). 13C NMR (100 MHz, DMSO-d6 + CF3COOD), δ (ppm): 166.0, 162.1, 159.6, 157.5, 157.0, 144.0, 130.2, 128.1, 121.8, 120.4, 114.6, 103.5, 99.1, 96.3, 56.8, 56.3, 55.8. HRMS (ESI)+ Calculated for C17H16NaN2O4 [M + Na]+ m/z 335.1008, found 335.0999.
2-(p-Tolyl)quinazolin-4(3H)-one (38j).
White powder, mp 244–246 °C, yield 51.7%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 12.44 (1H, s), 8.14 (1H, dd, J = 8.0, 2.0 Hz), 8.10 (2H, d, J = 8.0 Hz), 7.82 (1H, td, J = 8.0, 2.0 Hz), 7.12 (1H, dd, J = 8.0, 2.0 Hz), 7.50 (1H, t, J = 8.0 Hz), 7.35 (1H, d, J = 8.0 Hz), 2.39 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 162.2, 152.2, 148.8, 141.4, 134.5, 129.7, 129.2(2), 127.7(2), 127.4, 126.4, 125.8, 120.9, 21.0. HRMS (ESI)+ Calculated for C13H13N2O [M + H]+ m/z 237.1028, found 237.1029.
N-(4-(4-Oxo-1,4-dihydroquinazolin-2-yl)phenyl)acetamide (38k).
White powder, mp 231–234 °C, yield 47.0%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 12.39 (1H, s), 10.22 (1H, s), 8.12–8.16 (3H, m), 7.81 (1H, t, J = 8.0 Hz), 7.82 (1H, td, J = 8.0, 2.4 Hz), 7.69–7.75 (3H, m), 7.49 (1H, td, J = 8.0, 2.4 Hz), 2.09 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 168.8, 162.2, 158.8, 148.9, 142.2, 134.5, 128.5(2), 127.3, 126.8, 126.2, 125.8, 120.8, 118.4(2), 24.2. HRMS (ESI)+ Calculated for C16H14N3O2 [M + H]+ m/z 280.1086, found 280.1090.
2-(4-Hydroxy-3,5-dimethoxyphenyl)quinazolin-4(3H)-one (38l).
White powder, mp 246–248 °C, yield 60.2%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 12.37 (1H, s), 9.11 (1H, s), 8.12 (1H, dd, J = 8.0, Hz), 7.81 (1H, td, J = 8.0, 2.4 Hz), 7.71 (1H, d, J = 8.0 Hz), 7.57 (2H, s), 7.47 (1H, td, J = 8.0, 2.4 Hz), 3.88 (6H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 162.4, 152.0, 148.9, 147.8(2), 139.1, 134.5, 127.3, 126.0, 125.8, 122.1, 120.0, 105.3(2), 56.2(2). HRMS (ESI)+ Calculated for C16H15N2O4 [M + H]+ m/z 299.1032, found 299.1031.
N-(4-(6-Methoxy-4-oxo-1,4-dihydroquinazolin-2-yl)phenyl)-acetamide (38m).
White powder, mp 252–255 °C, yield 62.1%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 12.36 (1H, s), 10.20 (1H, s), 8.12 (2H, d, J = 8.4 Hz), 7.72 (2H, d, J = 8.4 Hz), 7.65 (1H, d, J = 8.4 Hz), 7.52 (1H, d, J = 2.4 Hz), 7.51 (1H, dd, J = 8.8, 2.4 Hz), 3.88 (3H, s), 2.09 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 168.6, 161.9, 157.4, 149.5, 143.2, 141.7, 128.9, 128.1(2), 126.8, 123.9, 121.4, 118.3(2), 105.7, 55.5, 24.0. HRMS (ESI)+ Calculated for C17H16N3O3 [M + H]+ m/z 310.1192, found 310.1193.
2-(4-Hydroxy-3,5-dimethoxyphenyl)-6-methoxyquinazolin-4(3H)-one (38n).
Light yellow solid, mp 261–263 °C, yield 65.0%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 7.68 (1H, d, J = 8.8 Hz), 7.53 (3H, s), 7.42 (1H, dd, J = 8.8, 2.8 Hz), 3.89 (9H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 162.0, 157.5, 150.3(2), 147.9, 142.5, 139.0, 128.4, 124.2, 121.7, 121.3, 106.0, 105.2(2), 56.2, 55.7, 55.6. HRMS (ESI)+ Calculated for C17H17N2O5 [M + H]+ m/z 329.1137, found 329.1135.
6-Methoxy-2-(p-tolyl)quinazolin-4(3H)-one (38o).
White powder, mp 249–251 °C, yield 64.8%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 8.07 (2H, d, J = 8.0 Hz), 7.68 (1H, d, J = 8.0 Hz), 7.54 (1H, d, J = 2.4 Hz), 7.43 (1H, dd, J = 8.8, 2.4 Hz), 7.34 (1H, d, J = 8.0 Hz), 3.89 (3H, s), 2.39 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 162.0, 157.7, 150.3, 142.8, 141.2, 129.7, 129.2(2), 128.8, 127.5(2), 124.1, 121.6, 105.9, 55.6, 21.0. HRMS (ESI)+ Calculated for C16H15N2O2 [m + h]+ m/z 267.1134, found 267.1137.
4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)benzoic Acid (38p).
White powder, mp 217–219 °C, yield 58.9%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 13.19 (1H, s), 12.17 (1H, s), 8.26 (2H, d, J = 8.8 Hz), 8.05 (2H, d, J = 8.8 Hz), 6.78 (1H, d, J = 2.4 Hz), 6.56 (1H, d, J = 2.4 Hz), 3.89 (3H, s), 3.86 (3H, s), 3.36 (4H, t, J = 6.4 Hz), 1.96–1.99 (4H, m). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 166.8, 164.3, 161.0, 159.7, 152.8, 152.2, 136.2, 133.0, 129.4(2), 127.9(2), 104.9, 101.5, 98.1, 56.0, 55.7. HRMS (ESI)+ Calculated for C17H15N2O5 [M + Na]+ m/z 349.0800, found 349.0782.
2-(3-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-phenoxy)acetic Acid (38q).
White powder, mp 209–211 °C, yield 45.2%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 12.04 (1H, s), 7.79 (1H, d, J = 8.0 Hz), 7.71 (1H, s), 7.11 (1H, dd, J = 8.0, 2.4 Hz), 6.75 (1H, d, J = 2.4 Hz), 6.54 (1H, d, J = 2.4 Hz), 4.77 (2H, s), 3.89 (3H, s), 3.84 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 164.7, 161.4, 160.2, 158.4, 153.2, 152.9, 149.3, 133.9, 130.1, 120.8, 118.7, 113.3, 105.2, 101.8, 98.3, 60.3, 56.4, 56.1. HRMS (ESI)+ Calculated for C18H17N2O6 [M + H]+ m/z 357.1087, found 357.1077.
2-(2-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-phenoxy)acetic Acid (38r).
Light yellow solid, mp 235–237 °C, yield 47.1%. 1HNMR(400 MHz, DMSO-d6), δ (ppm): 7.88 (1H, dd, J = 8.0, 2.4 Hz), 7.77 (1H, dt, J = 8.0, 2.4 Hz), 7.30–7.36 (2H, m), 6.89 (1H, d, J = 2.0 Hz), 6.76 (1H, d, J = 2.0 Hz), 4.96 (2H, s), 3.99 (3H, s), 3.97 (3H, s). 13C NMR (100 MHz, DMSO-d6 + CF3COOD), δ (ppm): 160.0 163.2, 158.4, 155.0, 154.8, 152.3, 151.9, 138.2, 132.2, 126.8, 118.4, 109.8, 98.7, 95.1, 90.7, 61.8, 51.8, 51.551.8, 51.5. HRMS (ESI)+ Calculated for C18H16NaN2O6 [M + Na]+ m/z 379.0906, found 379.0897.
Methyl 2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-phenoxy)acetate (38s).
White powder, mp 224–227 °C, yield 60.3%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 8.13 (2H, d, J = 8.8 Hz), 7.08 (2H, d, J = 8.8 Hz), 6.72 (1H, d, J = 2.0 Hz), 6.52 (1H, d, J = 2.0 Hz), 4.92 (2H, s), 3.89 (3H, s), 3.85 (3H, s), 3.71 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 168.9, 164.4, 161.1, 160.4, 159.6, 152.9, 129.6(2), 124.4, 114.6(2), 104.4, 100.4, 97.6, 64.6, 56.1, 55.7, 51.9. HRMS (ESI)+ Calculated for C19H18NaN2O6 [M + Na]+ m/z 393.1063, found 393.1058.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-phenoxy)acetic Acid (26).
White powder, mp 251–253 °C, yield 44.2%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 11.97 (1H, s), 8.13 (2H, d, J = 8.8 Hz), 7.04 (2H, d, J = 8.8 Hz), 6.72 (1H, d, J = 2.0 Hz), 6.51 (1H, d, J = 2.0 Hz), 4.79 (2H, s), 3.88 (3H, s), 3.84 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 169.9, 168.6, 164.3, 161.0, 160.4, 159.8, 152.6, 129.4(2), 124.7, 114.5(2), 104.5, 100.9, 97.5, 64.5, 56.0 55.6. HRMS (ESI)+ Calculated for C18H16NaN2O6 [M + Na]+ m/z 379.0906, found 379.0910.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-phenoxy)-N-morpholinoacetamide (42a).
Light yellow solid, mp 262–264 °C, yield 54.4%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 9.29 (1H, s), 8.90 (1H, s), 8.09–8.15 (2H, m), 7.08 (1H, d, J = 8.8 Hz), 7.02 (1H, d, J = 8.8 Hz), 6.71–6.73 (1H, m), 6.53 (1H, d, J = 2.4 Hz), 5.0 (1H, s), 4.56 (2H, s), 3.89 (3H, s), 3.85 (3H, s), 3.62 (4H, t, J = 4.4 Hz), 2.79 (4H, t, J = 4.4 Hz). 13C NMR (100 MHz, DMSO-d6 + CF3COOD), δ (ppm): 170.0, 167.2, 166.5, 164.1, 163.1, 157.8, 143.1, 133.0, 132.8, 118.9, 116.4, 116.3, 103.7, 99.8, 95.8, 67.2, 67.1, 65.6, 57.2, 57.0, 56.8, 56.0. HRMS (ESI)+ Calculated for C22H24NaN4O6 [M + Na]+ m/z 463.1594, found 463.1601.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-2,6-dimethylphenoxy)-N-morpholinoacetamide (42b)
White powder, mp 265–267 °C, yield 48.1%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 9.39 (1H, s), 8.81 (1H, s), 7.92 (2H, s), 6.69 (1H, s), 6.45 (1H, s), 4.60 (1H, s), 4.25 (2H, s), 3.87 (3H, s), 3.63 (3H, s), 3.64 (4H, t, J = 4.4 Hz), 2.82 (4H, t, J = 4.4Hz), 2.30 (6H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 169.3, 164.8, 163.6, 160.7, 158.1, 157.4, 153.7, 130.4(2), 128.3(2), 128.2, 105.0, 100.7, 97.0, 70.5, 65.9(2), 55.8, 55.5, 54.5(2), 16.1(2). HRMS (ESI)+ Calculated for C24H29N4O6 [M + H]+ m/z 469.2087, found 469.2103.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-phenoxy)-N-((tetrahydro-2H-pyran-4-yl)methyl)acetamide (42c).
White powder, mp 253–255 °C, yield 60.3%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 11.93 (1H, s), 8.16 (3H, d, J = 8.8 Hz), 7.07 (2H, d, J = 8.8 Hz), 6.71 (1H, d, J = 2.4 Hz), 6.52 (1H, d, J = 2.4 Hz), 4.59 (2H, s), 3.89 (3H, s), 3.84 (3H, s), 3.79–3.81 (2H, m), 3.23 (2H, t, J = 8.8 Hz), 3.03 (2H, t, J = 8.8 Hz), 1.49–1.52 (2H, m). 13C NMR (100 MHz, DMSO-d6 + CF3COOD), δ (ppm): 167.9, 166.8, 164.0, 162.8, 157.8, 157.6, 142.9, 132.6(2), 118.4, 116.1(2), 103.4, 99.5, 95.6, 67.8, 67.6(2), 57.1, 55.6, 44.9, 35.8, 31.2(2). HRMS (ESI)+ Calculated for C24H28N3O6 [M + H]+ m/z 454.1978, found 454.1978.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-2,6-dimethylphenoxy)-N-((tetrahydro-2H-pyran-4-yl)methyl)acetamide (42d).
Light yellow solid, mp 224–226 °C, yield 47.2%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 11.78 (1H, s), 8.24 (1H, t, J = 6.0 Hz), 7.90 (2H, s), 6.73 (1H, d, J = 2.4 Hz), 6.51 (1H, d, J = 2.4 Hz), 4.28 (2H, s), 3.88 (3H, s), 3.84 (3H, s), 3.26 (2H, t, J = 11.6 Hz), 3.09 (2H, t, J = 7.6 Hz), 2.31 (6H, s), 1.55 (2H, d, J = 8.8 Hz), 1.16–1.19 (2H, m). 13C NMR (100 MHz, DMSO-d6 + CF3COOD), δ (ppm): 168.1, 166.5, 162.5, 160.8, 157.4, 157.3, 143.0, 132.5(2), 131.0(2), 125.0, 103.4, 99.4, 95.7, 71.3, 67.4(2), 57.0, 56.5, 44.6, 35.5, 31.1(2), 16.5(2). HRMS (ESI)+ Calculated for C26H31NaN3O6 [M + Na]+ m/z 504.2111, found 504.2106.
2-(2,6-Dibromo-4-(5,7-dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)phenoxy)-N-(1-methylpiperidin-4-yl)acetamide (42e).
White powder, mp 267–269 °C, yield 45.5%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 9.50 (1H, s), 8.50 (2H, s), 6.80 (1H, s), 6.57 (1H, s), 4.48 (2H, s), 3.90 (3H, s), 3.86 (3H, s), 3.45–3.47 (2H, m), 3.07–3.08 (2H, m), 2.75–2.77 (2H, m), 2.97–2.99 (2H, m). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 166.0, 164.4, 160.9, 159.5, 154.1, 152.4, 149.7, 132.0(2), 131.2, 117.6(2), 104.8, 101.5, 98.2, 71.0, 56.0, 55.7, 52.7(2), 43.5, 42.6, 28.8(2). HRMS (ESI)+ Calculated for C24H27Br2N2O5 [M + H]+ m/z 611.0328, found 611.0320.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-2,6-dimethylphenoxy)-N-(1-methylpiperidin-4-yl)acetamide (42f).
White powder, mp 270–272 °C, yield 43.0%. 1H NMR (400 MHz, CDCl3), δ (ppm): 7.78 (1H, t, J = 6.0 Hz), 6.78 (1H, s), 6.41 (1H, s), 4.27 (2H, s), 3.91 (6H, s), 2.80 (2H, d, J = 4.8 Hz), 2.29 (6H, s), 2.14 (2H, t, J = 8.8 Hz), 2.01–2.03 (2H, m), 1.57–1.60 (2H, m). 13C NMR (100 MHz, CDCl3), δ (ppm): 167.0(2), 165.0, 164.8, 161.0, 157.2, 154.1, 131.1, 128.5(2), 125.1, 123.0, 105.0, 101.2, 98.2, 70.6, 56.2, 55.7, 54.5(2), 46.3, 45.8, 32.3(2), 16.5(2). HRMS (ESI)+ Calculated for C26H33N4O5 [M + H]+ m/z 481.2451, found 481.2443.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-phenoxy)-N-(4-methylpiperazin-1-yl)acetamide (42g).
Light yellow solid, mp 256–258 °C, yield 62.5%. 1H NMR (400 MHz, CDCl3 + CD3OD), δ (ppm): 8.25 (2H, d, J = 2.4 Hz), 7.35 (2H, d, J = 2.4 Hz), (1H, s), 6.72 (1H, s), 4.82 (2H, s), 4.16 (3H, s), 4.15 (3H, s), 3.09–3.12 (4H, m), 2.85–2.87 (4H, m), 2.57 (3H, s). 13C NMR (100 MHz, CDCl3 + CD3OD), δ (ppm): 166.0, 165.9, 165.2, 161.1, 159.9, 152.7, 129.2(2), 126.0, 114.8(2), 104.5, 100.8, 97.9, 66.7, 55.8, 55.5, 53.9(2), 49.2(2), 45.2. HRMS (ESI)+ Calculated for C23H27NaN5O5 [M + Na]+ m/z 476.1910, found 476.1904.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-phenoxy)-N-(3,4,5-trimethoxyphenyl)acetamide (42h).
White powder, mp 269–271 °C, yield 63.7%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 10.07 (1H, s), 8.15 (2H, d, d, J = 8.8 Hz), 7.13 (2H, d, J = 8.8 Hz), 7.06 (2H, s), 6.74 (1H, d, J = 2.4 Hz), 6.53 (1H, d, J = 2.4 Hz), 4.80 (2H, s), 3.89 (3H, s), 3.85 (3H, s), 3.74 (6H, s), 3.62 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 166.0, 164.4, 161.0, 160.7, 159.5, 153.0, 152.7(2), 151.7, 134.5, 133.7, 129.7(2), 124.2, 114.7(2), 104.3, 100.2, 97.6, 97.4(2), 67.1, 60.1(2), 56.0, 55.7(2). HRMS (ESI)+ Calculated for C27H27NaN3O8 [M + Na]+ m/z 544.1696, found 544.1694.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-2,6-dimethylphenoxy)-N-(3,4,5-trimethoxyphenyl)acetamide (42i).
Off-white solid, mp 240–242 °C, yield 54.9%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 9.98 (1H, s), 7.92 (2H, s), 7.18 (2H, s), 6.76 (1H, d, J = 2.4 Hz), 6.54 (1H, d, J = 2.4 Hz), 4.48 (2H, s), 3.89 (3H, s), 3.85 (3H, s), 3.75 (6H, s), 3.63 (3H, s), 2.36 (6H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 166.3, 164.4, 161.0, 159.5, 157.8, 152.8, 152.7(2), 152.0, 134.5, 133.8, 130.9(2), 128.6(2), 127.3, 104.5, 100.6, 97.7, 97.6(2), 71.0, 60.1, 56.0, 55.8(2), 55.7, 16.2(2). HRMS (ESI)+ Calculated for C29H31NaN3O8 [M + Na]+ m/z 572.2009, found 572.2007.
N-(1H-Benzo[d]imidazol-2-yl)-2-(4-(5,7-dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-2,6-dimethylphenoxy)acetamide (42j).
White powder, mp 257–260 °C, yield 57.2%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 7.93 (2H, s), 7.45–7.47 (2H, m), 7.10–7.11 (1H, m), 6.74 (1H, d, J = 2.4 Hz), 6.50 (1H, d, J = 2.4 Hz), 4.67 (2H, s), 3.89 (3H, s), 3.84 (3H, s), 2.37 (6H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 168.3, 164.2, 160.9, 160.1, 159.7, 159.1, 158.0, 153.1, 152.7, 146.6, 130.6(2), 130.4, 128.7, 128.4(2), 128.0, 121.2(2), 104.7, 101.1, 97.5, 70.9, 55.9, 55.6, 16.3(2). HRMS (ESl)+ Calculated for C27H26N5O5 [M + Na]+ m/z 522.1753, found 522.1752.
tert-Butyl-(2-(4-(5,7-dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)phenoxy)acetamido)piperidine-1-carboxylate (43a).
White powder, mp 244–246 °C, yield 64.4%. 1H NMR (400 MHz, CDCl3), δ (ppm): 8.26 (2H, d, J = 8.4 Hz), 7.06 (2H, d, J = 8.4 Hz), 6.46 (1H, d, J = 2.4 Hz), 6.42 (1H, d, J = 8.4 Hz), 4.56 (2H, s), 3.98 (3H, s), 3.93 (3H, s), 4.03–4.06 (4H, m), 2.87 (2H, t, J = 12.4 Hz), 1.94 (2H, t, J = 12.4 Hz), 1.45 (9H, s). 13C NMR (100 MHz, CDCl3), δ (ppm): 166.9, 161.9, 161.5, 159.9, 154.7, 152.2, 129.8(2), 115.0(2), 104.9, 98.5, 79.9, 67.5, 56.6, 55.9, 46.7, 42.8, 42.5, 32.1(2), 28.6(5). HRMS (ESI)+ Calculated for C28H35N4O7 [M + H]+ m/z 539.2506, found 539.2514.
tert-Butyl-(2-(4-(5,7-dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-2,6-dimethylphenoxy)acetamido)piperidine-1-carboxylate (43b).
White powder, mp 236–239 °C, yield 63.8%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 8.13 (1H, d, J = 8.4 Hz), 7.90 (2H, s), 6.72 (1H, d, J = 2.4 Hz), 6.48 (1H, d, J = 2.4 Hz), 4.26 (2H, s), 3.91–3.93 (2H, m), 3.89 (3H, s), 3.83 (3H, s), 3.31–3.33 (2H, m), 2.80–2.82 (2H, m), 2.29 (6H, s), 1.70–1.72 (2H, m), 1.39 (9H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 166.8, 164.0, 161.9, 160.9, 160.5, 153.2, 152.9, 130.6(2), 128.3(2), 104.8, 100.9, 97.4, 78.6, 70.8, 55.9, 55.5, 45.7, 42.7, 42.2, 31.2(2), 28.1(3), 16.1(2). HRMS (ESI)+ Calculated for C30H39N4O4 [M + Na]+ m/z 589.2638, found 589.2635.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-2,6-dimethylphenoxy)-N-(4-(4-methylpiperazin-1-yl)phenyl)acetamide (43c).
Off-white solid, mp 240–243 °C, yield 60.3%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 11.86 (1H, s), 9.86 (1H, s), 7.92 (2H, s), 7.55 (2H, d, J = 8.8 Hz), 6.90 (2H, d, J = 8.8 Hz), 6.74 (1H, d, J = 2.0 Hz), 6.51 (1H, d, J = 2.0 Hz), 4.47 (2H, s), 3.88 (3H, s), 3.84 (3H, s), (4H, t, J = 4.8 Hz), 2.44 (4H, t, J = 4.8 Hz), 2.35 (6H, s), 2.21 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 165.9, 164.3, 161.0, 159.8, 157.8, 153.1, 152.4, 147.6, 130.8(2), 130.2, 128.4(2), 127.9, 121.1(2), 115.6(2), 104.7, 101.2, 97.6, 71.2, 56.0, 55.7, 54.6(2), 48.5(2), 45.8, 16.8(2). HRMS (ESI)+ Calculated for C31H36N5O5 [M + H]+ m/z 558.2716, found 558.2719.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-2,6-dimethylphenoxy)-N-(3-(4-methylpiperazin-1-yl)phenyl)acetamide (43d).
White powder, mp 237–240 °C, yield 57.2%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 11.88 (1H, s), 10.01 (1H, s), 7.93 (2H, s), 7.42 (1H, s), 7.28 (1H, t, J = 8.0 Hz), 7.23 (1H, t, J = 8.0 Hz), 6.77 (1H, d, J = 2.4 Hz), 6.73 (1H, s), 6.51 (1H, d, J = 2.4 Hz), 4.49 (2H, s), 3.88 (3H, s), 3.84 (3H, s), 3.35–3.37 (4H, m), 3.17 (4H, s), 2.86 (3H, s), 2.36 (6H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 166.4, 165.6, 160.9, 159.7, 157.6, 153.0, 152.4, 149.9, 139.2, 130.7(2), 129.3, 128.3(2), 127.9, 111.7, 107.5, 104.6, 101.1, 97.6, 71.0, 57.8, 55.9, 55.6, 52.2(2), 45.7(2), 16.2(2). HRMS (ESI)+ Calculated for C31H35NaN5O5 [M + Na]+ m/z 580.2536, found 580.2534.
2-(2,6-Dibromo-4-(5,7-dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)phenoxy)-N-(4-(4-methylpiperazin-1-yl)phenyl)acetamide (43e).
Off-white solid, mp 270–272 °C, yield 55.7%. 1H NMR (400 MHz, acetic acid-d4), δ (ppm): 8.58 (2H, s), 7.58 (2H, d, J = 8.8 Hz), 7.22 (2H, d, J = 8.8 Hz), 7.05 (1H, d, J = 2.0 Hz), 6.71 (1H, d, J = 2.0 Hz), 5.01 (2H, s), 4.10 (3H, s), 4.11 (3H, s), 3.92 (4H, t, J = 4.8 Hz), 3.41 (4H, t, J = 4.8 Hz), 3.12 (3H, s). 13C NMR (100 MHz, acetic acid-d4), δ (ppm): 168.7, 167.8, 164.9.0, 163.1, 156.1, 154.8, 151.0, 148.8, 133.9(2), 132.9, 132.4, 123.9(2), 119.7(2), 119.2(2), 105.7, 103.0, 72.6, 57.2, 57.1, 54.7(2), 48.7(2), 44.3. HRMS (ESI)+ Calculated for C29H30Br2N5O5 [M + H]+ m/z 688.0593, found 688.0596.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-2,6-dimethylphenoxy)-N-(4-(4-methylpiperazin-1-yl)phenyl)acetamide (43f).
White powder, mp 267–269 °C, yield 58.2%. 1H NMR (400 MHz, acetic acid-d4), δ (ppm): 8.31 (2H, d, J = 8.0 Hz), 7.82 (2H, d, J = 8.0 Hz), 7.39 (2H, d, J = 8.0 Hz), 7.23 (2H, d, J = 8.0 Hz), 7.12 (1H, s), (1H, s), 5.00 (2H, s), 4.12 (3H, s), 4.09 (3H, s), 3.94 (4H, brs), 2.44 (4H, brs), 3.16 (3H, s). 13C NMR (100 MHz, acetic acid-d4), δ (ppm): 169.6, 167.9, 165.0, 163.2, 162.2, 154.9, 154.4, 148.8, 132.5, 131.6(2), 126.8, 124.2(2), 119.2(2), 116.8(2), 105.4, 102.3, 100.0, 68.8, 57.2, 57.1, 54.6(2), 48.7(2), 44.4. HRMS (ESI)+ Calculated for C29H32N5O5 [M + H]+ m/z 530.2403, found 530.2401.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-phenoxy)-N-(3-(4-methylpiperazin-1-yl)phenyl)acetamide (43g).
White powder, mp 256–258 °C, yield 66.0%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 11.95 (1H, s), 10.12 (1H, s), 8.17 (2H, d, J = 8.4 Hz), 7.38 (1H, s), 7.21 (1H, t, J = 8.0 Hz), 7.11–7.15 (2H, m), 6.77 (1H, dd, J = 8.0, 2.4 Hz), 6.72 (1H, d, J = 2.4 Hz), 6.52 (1H, d, J = 2.0 Hz), 4.82 (2H, s), 3.85 (3H, s), 3.82 (3H, s), 3.74–3.76 (2H, m), 3.51–3.53 (2H, m), 3.16 (2H, s), 2.96–2.98 (2H, m), 2.86 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 166.8, 166.6, 164.7, 161.5, 160.9, 153.5, 152.9, 150.5, 139.8, 129.9(2), 125.5, 115.1(2), 112.3, 112.0, 107.8, 105.0, 101.5, 97.9, 89.7, 67.5, 56.0, 55.7, 52.7(2), 46.2(2), 42.5. HRMS (ESI)+ Calculated for C29H31NaN5O5 [M + Na]+ m/z 552.2223, found 552.2222.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-phenoxy)-N-(2-(4-methylpiperazin-1-yl)phenyl)acetamide (43h).
Off-white solid, mp 252–255 °C, yield 58.6%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 9.96 (1H, s), 8.23 (2H, d, J = 2.4 Hz), 8.15 (1H, d, J = 8.4 Hz), 7.14–7.28 (4H, m), 6.71 (1H, d, J = 2.0 Hz), 6.52 (1H, d, J = 2.0 Hz), 4.90 (2H, s), 3.89 (3H, s), 3.85 (3H, s),3.51 −3.53 (4H, m), 2.98–3.03 (4H, m), 2.87 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 166.0, 164.3, 161.1, 160.0, 159.9, 158.7, 152.8, 152.5, 141.0, 132.0, 129.8(2), 125.5, 125.3, 124.8, 120.8, 114.8(2), 104.6, 100.9, 97.5, 67.4, 56.0, 55.6, 53.3(2), 48.5(2), 42.4. HRMS (ESI)+ Calculated for C29H31NaN5O5 [M + Na]+ m/z 552.2223, found 552.2215.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-phenoxy)-N-(4-morpholinophenyl)acetamide (43i).
White powder, mp 273–275 °C, yield 61.8%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 11.87 (1H, s), 9.89 (1H, s), 7.92 (2H, s), 7.57 (2H, d, J = 8.4 Hz), 6.92 (2H, d, J = 8.4 Hz), 6.74 (1H, d, J = 2.0 Hz), 6.52 (1H, d, J = 2.0 Hz), 4.45 (2H, s), 3.89 (3H, s), 3.84 (3H, s), 3.73 (4H, t, J = 4.8 Hz), 3.05 (4H, t, J = 4.8 Hz). 13C NMR (100 MHz, DMSO-d6 + CF3COOD), δ (ppm): 166.4, 165.3, 161.6, 159.5, 157.8, 155.4, 145.9, 142.0, 131.5(2), 129.9(2), 123.4(2), 121.1, 121.0(2), 119.5, 103.5, 98.5, 97.1, 71.2, 65.0(2), 56.4, 56.1, 52.0(2), 16.3(2). HRMS (ESI)+ Calculated for C30H33N4O4 [M + H]+ m/z 545.2400, found 545.2400.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-2,6-dimethylphenoxy)-N-(3-morpholinophenyl)acetamide (43j).
White powder, mp 264–267 °C, yield 48.3%. 1H NMR (400 MHz, DMSO-xyd6), δ (ppm): 11.87 (1H, s), 9.91 (1H, s), 7.92 (2H, s), 7.34 (1H, s), 7.15–7.23 (2H, m), 6.74 (1H, d, J = 2.4 Hz), 6.69 (1H, dd, J = 8.4, 2.4 Hz), 6.51 (1H, d, J = 2.8 Hz), 4.47 (2H, s), 3.88 (3H, s), 3.84 (3H, s), (4H, t, J = 4.8 Hz), 3.08 (4H, t, J = 4.8 Hz), 2.35 (6H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 167.7, 166.4, 164.3, 162.5, 161.0, 157.7, 152.4, 151.5, 139.1, 130.8(2), 129.1, 128.4(2), 127.9, 110.9(2), 106., 104.6, 101.2, 97.6, 71.0, 66.1(2), 56.0, 55.7, 48.5(2), 16.2(2). HRMS (ESI)+ Calculated for C30H32NaN4O6 [M + Na]+ m/z 567.2220, found 567.2211.
2-(2,6-Dibromo-4-(5,7-dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)phenoxy)-N-(4-morpholinophenyl)acetamide (43k).
White powder, mp 242–244 °C, yield 49.7%. 1H NMR (400 MHz, DMSO-d6 + CF3COOD), δ (ppm): 10.34 (1H, s), 8.39 (2H, s), 7.87 (2H, d, J = 8.4 Hz), 7.62 (2H, d, J = 8.4 Hz), 6.80 (1H, d, J = 2.0 Hz), 6.62 (1H, d, J = 2.0 Hz), 4.74 (2H, s), 3.95 (4H, t, J = 4.8 Hz), 3.86 (6H, s), 3.58 (4H, t, J = 4.8 Hz). 13C NMR (100 MHz, DMSO-d6 + CF3COOD), δ (ppm): 166.6, 166.4, 162.6, 156.9, 156.8, 153.8, 139.9, 138.6, 134.6(2), 122.5(2), 122.2(2), 118.6(2), 116.8, 115.0, 104.5, 99.8, 98.4, 72.4, 65.0(2), 57.0, 56.6, 55.4(2). HRMS (ESI)+ Calculated for C28H27Br2N4O6 [M + H]+ m/z 675.0277, found 675.0275.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-2,6-dimethylphenoxy)-N-(4-morpholinophenyl)acetamide (43l).
Off-white solid, mp 276–278 °C, yield 60.7%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 11.92 (1H, s), 9.93 (1H, s), 7.50 (2H, d, J = 8.8 Hz), 7.12 (2H, d, J = 8.8 Hz), 6.91 (2H, d, J = 8.8 Hz), 6.71 (1H, d, J = 2.4 Hz), 6.50 (1H, d, J = 2.4 Hz), 4.76 (2H, s), 3.88 (3H, s), 3.84 (3H, s), 3.72 (4H, t, J = 4.8 Hz), 3.05 (4H, t, J = 4.8 Hz). 13C NMR (100 MHz, CDCl3 + CF3COOD), δ (ppm): 169.0, 163.2, 162.0, 160.0, 156.4, 137.5, 131.3, 131.0, 123.4, 123.3, 121.8, 121.5, 119.0, 117.0, 116.9, 116.1, 116.0, 113.3, 101.0, 98.5, 94.8, 67.0, 64.5, 64.2, 56.9, 56.3, 56.1(2). HRMS (ESI)+ Calculated for C28H29N4O4 [M + H]+ m/z 517.2087, found 517.2086.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-phenoxy)-N-(3-morpholinophenyl)acetamide (43m).
White powder, mp 272–274 °C, yield 51.0%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 10.02 (1H, s), 8.16 (2H, d, J = 8.4 Hz), 7.28 (1H, t, J = 2.4 Hz), 7.09–7.16 (4H, m), 6.72 (1H, d, J = 2.4 Hz), 6.68 (1H, dd, J = 8.4, 2.4 Hz), 6.51 (1H, d, J = 2.4 Hz), 4.79 (2H, s), 3.88 (3H, s), 3.84 (3H, s), 3.72 (4H, t, J = 4.8 Hz), 3.06 (4H, t, J = 4.8 Hz). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 166.1(2), 164.3, 161.0, 160.5, 152.9, 152.5, 151.5, 139.2, 129.4(2), 129.2, 125.0, 114.6(2), 110.9, 110.7, 106.4, 104.5, 100.9, 97.5, 67.1, 66.1(2), 56.0, 55.6, 48.5(2). HRMS (ESI)+ Calculated for C28H29N4O6 [M + H]+ m/z 517.2087, found 517.2053.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-2,6-dimethylphenoxy)-N-(4-(piperidin-1-yl)phenyl)acetamide (43n).
Off-white solid, mp 265–267 °C, yield 56.1%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 11.87 (1H, s), 9.58 (1H, s), 7.92 (2H, s), 7.53 (2H, d, J = 8.8 Hz), 6.89 (2H, d, J = 8.8 Hz), 6.74 (1H, s), 6.52 (1H, s), 4.45 (2H, s), 3.89 (3H, s), 3.84 (3H, s), 3.06–3.08 (4H, m), 2.35 (6H, s), 1.58–1.63 (4H, m), 1.51–1.52 (2H, m). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 165.8, 164.2, 160.9, 157.8, 153.1, 152.4, 148.4, 130.8(2), 129.9, 128.7(2), 127.9, 121.1(2), 116.1(2), 104.7, 101.2, 97.6, 71.1, 56.0, 55.6, 50.0(2), 35.8, 25.3(2), 23.9, 16.2(2). HRMS (ESI)+ Calculated for C31H35N4O5 [M + H]+ m/z 543.2607, found 535.2607.
2-(2,6-Dibromo-4-(5,7-dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)phenoxy)-N-(4-(piperidin-1-yl)phenyl)acetamide (43o).
White powder, mp 271–273 °C, yield 62.0%. 1H NMR (400 MHz, DMSO-d6 + CF3COOD), δ (ppm): 10.48 (1H, s), 8.52 (2H, s), 7.94 (2H, d, J = 8.8 Hz), 7.73 (2H, d, J = 8.8 Hz), 6.85 (1H, d, J = 2.4 Hz), 6.65 (1H, d, J = 2.4 Hz), 4.78 (2H, s), 3.93 (3H, s), 3.91 (3H, s), 3.58–3.61 (4H, m), 1.94–1.97 (4H, m), 1.70 (2H, s). 13C NMR (100 MHz, DMSO-d6 + CF3COOD), δ (ppm): 166.0, 165.4, 165.3, 161.7, 155.2, 151.8, 142.1, 139.8, 138.2, 133.2(2), 122.4(2), 121.5, 121.4(2), 118.0(2), 104.6, 99.8, 98.9, 71.7, 56.8(2), 56.5, 56.1, 23.8(2), 21.0. HRMS (ESI)+ Calculated for C29H29Br2N4O5 [M + H]+ m/z 673.0484, found 673.0490.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-phenoxy)-N-(4-(piperidin-1-yl)phenyl)acetamide (43p).
White powder, mp 253–256 °C, yield 63.4%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 11.93 (1H, s), 9.89 (1H, s), 8.17 (2H, d, J = 8.8 Hz), 7.45 (1H, d, J = 8.8 Hz), 7.12 (2H, d, J = 8.8 Hz), 6.88 (1H, d, J = 8.8 Hz), 6.71 (1H, d, J = 2.4 Hz), 6.51 (1H, d, J = 2.4 Hz), 4.75 (2H, s), 3.88 (3H, s), (3H, s), 3.04–3.07 (4H, m), 1.57–1.62 (4H, m), 1.50–1.51 (2H, m). 13C NMR (100 MHz, DMSO-d6 + CF3COOD), δ (ppm): 166.6, 165.8, 162.8, 161.9, 157.6, 156.4, 144.2, 139.7, 137.9, 131.7(2), 122.3(2), 120.9, 120.8(2), 119.2, 115.3(2), 103.1, 98.7, 96.1, 67.2, 56.5(2), 56.2, 23.6(2), 20.8. HRMS (ESI)+ Calculated for C29H31N4O5 [M + H]+ m/z 515.2294, found 515.2300.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-phenoxy)-N-(4-(pyrrolidin-1-yl)phenyl)acetamide (43q).
Off-white solid, mp 243–245 °C, yield 50.7%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 9.80 (1H, s), 8.16 (2H, d, J = 9.2 Hz), 7.42 (2H, d, J = 9.2 Hz), 7.14 (2H, d, J = 8.8 Hz), 6.72 (1H, d, J = 2.4 Hz), 6.50–6.53 (2H, m), 4.74 (2H, s), 3.89 (3H, s), 3.85 (3H, s), 3.17–3.19 (4H, m), 1.92–1.95 (4H, m). 13C NMR (100 MHz, DMSO-d6 + CF3COOD), δ (ppm): 166.2, 163.2, 162.1(2), 157.3, 156.9, 143.3, 141.4, 132.0(2), 121.1(2), 119.9, 119.8(2), 118.6, 115.5(2), 103.0, 98.9, 95.6, 67.4, 56.8, 56.3, 55.3(2), 23.9(2). HRMS (ESI)+ Calculated for C28H29N4O5 [M + H]+ m/z 501.2138, found 501.2133.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-2,6-dimethylphenoxy)-N-(3-fluoro-4-morpholinophenyl)acetamide (44a).
White powder, mp 258–260 °C, yield 55.9%. 1H NMR (400 MHz, DMSO-d6 + CF3COOD), δ (ppm): 7.80 (1H, dd, J = 14.0, 2.4 Hz), 7.68 (2H, s), 7.47 (1H, dd, J = 9.2, 2.4 Hz), 7.36 (1H, t, J = 9.2 Hz), 6.83 (1H, d, J = 2.0 Hz), 6.55 (1H, d, J = 2.0 Hz), 4.46 (2H, s), 3.82–3.85, (4H, m), 3.81 (3H, s), 3.78 (3H, s), 3.34–3.36 (4H, m), 2.30 (6H, s). 13C NMR (100 MHz, DMSO-d6 + CF3COOD), δ (ppm): 169.2, 168.8, 164.3, 162.6, 159.3, 158.9, 155.8(250.2 Hz), 143.9, 134.7(2), 130.6(2), 132.6(2), 126.4, 126.2, 124.6, 122.9, 104.8, 101.2, 97.0, 72.7, 65.9, 65.5, 58.0, 57.7, 56.2(2), 17.7(2). HRMS (ESI)+ Calculated for C30H32FN4O6 [M + Na]+ m/z 585.2125, found 585.2122.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-phenoxy)-N-(3-fluoro-4-morpholinophenyl)acetamide (44b).
White powder, mp 236–238 °C, yield 58.0%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 10.1 (1H, s), 8.15 (2H, d, J = 8.8 Hz), 7.56 (1H, dd, J = 14.8, 2.4 Hz), 7.32 (1H, dd, J = 14.8, 2.4 Hz), 7.14 (2H, d, J = 14.8 Hz), 7.02, (1H, t, J = 9.6 Hz), 6.73 (1H, d, J = 2.4 Hz), 6.53 (1H, d, J = 2.4 Hz), 4.80 (2H, s), 3.89 (3H, s), 3.85 (3H, s),3.71–3.74 (4H, m), 2.94–2.96, (4H, m). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 166.1, 164.5, 161.1, 160.7, 159.6, 155.6, 153.2, 153.0, 151.8, 135.7(40.6 Hz), 133.5(36.0 Hz), 129.7(2), 124.3, 119.2, 115.8, 114.7, 108.2(100.8 Hz), 104.4, 100.3, 97.6, 67.1, 66.2(2), 56.1, 55.7, 50.8(2). HRMS (ESI)+ Calculated for C28H28FN4O6 [M + H]+ m/z 535.1993, found 535.1992.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-phenoxy)-N-(3-fluoro-4-(4-methylpiperazin-1-yl)phenyl)acetamide (44c).
Off-white solid, mp 245–247 °C, yield 59.9%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 11.94 (1H, s), 10.16 (1H, s), 8.17 (2H, d, J = 8.8 Hz), 7.55 (1H, dd, J = 14.8, 2.4 Hz), 7.30 (2H, dd, J = 8.8, 2.4 Hz), 7.13, (1H, d, J = 2.4 Hz), 7.00 (1H, t, J = 8.8 Hz), 6.71 (1H, d, J = 2.4 Hz), 6.51 (1H, d, J = 2.4 Hz), 4.79 (2H, s), 3.88 (3H, s), 3.84 (3H, s), 2.96, (4H, t, J = 4.8 Hz), 2.46 (4H, brs), 2.22 (3H, s). 13C NMR (100 MHz, DMSO-d6 + CF3COOD), δ (ppm): 166.2, 166.1, 163.3, 162.8, 162.2, 157.4, 157.0, 156.2, 153.7, 143.2, 134.7(42.8 Hz), 134.5(36.8 Hz), 120.2, 118.6, 116.0, 115.5(2), 108.5(100.8 Hz), 103.1, 98.9, 95.5, 67.4, 56.8, 56.7, 56.4, 56.3, 53.1(2), 47.9. HRMS (ESI)+ Calculated for C29H31FN5O5 [M + H]+ m/z 548.2309, found 548.2311.
2-(4-(5,7-Dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-2,6-dimethylphenoxy)-N-(3-fluoro-4-(4-methylpiperazin-1-yl)phenyl)-acetamide (44d).
White powder, mp 253–256 °C, yield 61.4%. 1H NMR (400 MHz, CDCl3), δ (ppm): 8.60 (1H, s), 7.85 (2H, s), 7.58 (1H, dd, J = 14.0, 2.4 Hz), 7.26–7.27 (1H, m), 6.94 (1H, d, J = 8.8 Hz), 6.81 (1H, s), 6.44 (1H, s), 4.40 (2H, s), 3.92 (6H, s), 3.10 (4H, t, J = 4.4 Hz), 2.61 (4H, t, J = 4.4 Hz), 2.36 (9H, s). 13C NMR (100 MHz, CDCl3), δ (ppm): 166.1, 165.2, 161.4, 157.0, 156.7, 154.2, 153.9, 152.3, 137.3(36.4 Hz), 131.9(36.8 Hz), 131.3(2), 129.1, 128.5(2), 119.1, 115.9, 109.2(56.4 Hz), 105.0, 101.4, 98.4, 70.5, 56.3, 55.8, 55.2(2), 50.7(2), 46.2, 16.5(2). HRMS (ESI)+ Calculated for C31H35FN5O5 [M + H]+ m/z 576.2622, found 576.2624.
N-(3-Chloro-4-(4-methylpiperazin-1-yl)phenyl)-2-(4-(5,7-dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)phenoxy)acetamide (44e).
White powder, mp 273–276 °C, yield 62.9%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 10.29 (1H, s), 8.17 (2H, d, J = 8.4 Hz), 7.85 (1H, s), 7.55 (1H, d, J = 8.8 Hz), 7.21 (1H, d, J = 8.8 Hz), 7.12 (2H, d, J = 8.8 Hz), 6.71 (1H, s), 6.50 (1H, s), 4.81 (2H, s), 3.88 (3H, s), 3.84 (3H, s), 3.30–3.37 (8H, m), 2.86 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 166.3, 164.2, 160.9, 160.3, 159.8, 153.1, 152.3, 143.1, 135.1, 129.3(2), 127.5, 125.1, 121.3(2), 119.3, 114.6(2), 104.5, 101.1, 97.4, 67.0, 55.9, 55.6, 52.9(2), 48.1(2), 42.2. HRMS (ESI)+ Calculated for C29H31ClN5O5 [M + H]+ m/z 564.2014, found 564.2011.
N-(3-Chloro-4-(4-methylpiperazin-1-yl)phenyl)-2-(4-(5,7-dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-2,6-dimethylphenoxy)-acetamide (44f).
White powder, mp 260–263 °C, yield 45.8%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 10.22 (1H, s), 7.93 (2H, s), 7.91 (1H, d, J = 2.4 Hz), 7.61 (1H, dd, J = 8.8, 2.4 Hz), 7.14 (1H, d, J = 8.8 Hz), 6.73 (1H, d, J = 2.4 Hz), 6.50 (1H, d, J = 2.4 Hz), 4.49 (2H, s), 3.88 (3H, s), 3.84 (3H, s), 3.17 (4H, brs), 2.93 (4H, brs), 2.35 (6H, s), 2.23 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 166.6, 164.1, 160.9, 157.6, 153.3, 153.0, 145.0, 134.3, 130.7(2), 128.4(2), 127.4, 121.7, 120.8, 119.6, 104.8, 101.0, 97.4, 79.2, 71.0, 56.0, 55.6, 54.9(2), 51.1, 48.6, 45.8, 16.3(2). HRMS (ESI)+ Calculated for C31H35ClN5O5 [M + H]+ m/z 614.2148, found 614.2174.
N-(3-Chloro-4-morpholinophenyl)-2-(2,6-dibromo-4-(5,7-dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)phenoxy)acetamide (44g).
White powder, mp 268–271 °C, yield 47.6%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 12.17 (1H, s), 10.21 (1H, s), 8.52 (2H, s), 7.88 (1H, d, J = 2.4 Hz), 7.56 (1H, dd, J = 8.8, 2.4 Hz), 7.16 (1H, d, J = 8.8 Hz), 6.81 (1H, s), 6.56 (1H, s), 4.67 (2H, s), 3.89 (3H, s), 3.85 (3H, s), 3.74, (4H, t, J = 4.4 Hz), 2.94 (4H, t, J = 4.0 Hz). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 165.1, 164.4, 160.9, 154.0, 152.4, 149.8, 144.8, 134.3, 132.0(2), 131.3, 127.4, 122.8, 121.7, 120.9, 119.7, 117.7(2), 104.8, 101.5, 98.2, 71.3, 66.4(2), 56.1, 55.7, 51.5(2). HRMS (ESI)+ Calculated for C28H25Br2ClNaN4O6 [M + Na]+ m/z 730.9707, found 730.9714.
N-(3-Chloro-4-morpholinophenyl)-2-(4-(5,7-dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)phenoxy)acetamide (44h).
White powder, mp 247–249 °C, yield 56.0%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 10.20 (1H, s), 8.15 (2H, d, J = 8.8 Hz), 7.52 (1H, dd, J = 8.8, 2.4 Hz), 7.13–7.16 (3H, m), 6.73 (1H, d, J = 2.4 Hz), 6.54 (1H, d, J = 2.4 Hz), 4.81 (2H, s), 3.89 (3H, s), 3.85 (3H, s), 3.73 (4H, t, J = 4.4 Hz), 2.92 (4H, t, J = 4.0 Hz). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 166.1, 164.4, 161.1, 160.6, 159.5, 153.0, 151.8, 144.6, 134.4, 129.7(2), 127.5, 124.3, 121.4, 120.9, 119.3, 114.7(2), 104.3, 100.3, 97.6, 67.0, 66.4(2), 56.0, 55.7, 51.5(2). HRMS (ESI)+ Calculated for C28H27ClNaN4O6 [M + Na]+ m/z 573.1517, found 573.1533.
N-(3-Chloro-4-morpholinophenyl)-2-(4-(5,7-dimethoxy-4-oxo-1,4-dihydroquinazolin-2-yl)-2,6-dimethylphenoxy)acetamide (44i).
Off-white solid, mp 258–261 °C, yield 51.5%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 10.17 (1H, s), 7.90 (3H, s), 7.62 (2H, dd, J = 8.8, Hz), 7.15 (1H, d, J = 8.8 Hz), 6.75 (1H, d, J = 2.4 Hz), 6.54(1H, d, J = 2.4 Hz), 4.49 (2H, s), 3.88 (3H, s), 3.85 (3H, s), 3.73 (4H, t, J = 4.4 Hz), 2.93 (4H, t, J = 4.0 Hz), 2.35 (6H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 165.5, 164.4, 161.0, 159.5, 157.9, 152.9, 151.9, 144.7, 134.4, 130.9(2), 128.6(2), 127.4, 127.2, 121.7, 120.8, 119.6, 104.5, 100.5, 97.7, 71.0, 66.4(2), 56.0, 55.7, 51.5(2), 16.2(2). HRMS (ESI)+ Calculated for C30H32ClN4O6 [M + H]+ m/z 579.2010, found 579.2018.
N-(3-Chloro-4-(4-methylpiperazin-1-yl)phenyl)-2-(4-(6-fluoro-4-oxo-1,4-dihydroquinazolin-2-yl)phenoxy)acetamide (44j).
White powder, mp 255–257 °C, yield 58.2%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 10.28 (1H, s), 8.18 (2H, d, J = 8.8 Hz), 7.87 (1H, d, J = 2.4 Hz), 7.76–7.80 (2H, m), 7.70 (1H, td, J = 8.8, 2.4 Hz), 7.56 (1H, dd, J = 8.8, 2.4, Hz), 7.22 (1H, d, J = 8.8 Hz), 7.15 (1H, d, J = 8.8 Hz), 3.52 (2H, d, J = 11.2 Hz), 3.38 (2H, d, J = 11.2 Hz), 3.19–3.22 (2H, m), 3.29 (2H, t, J = 13.2 Hz), 2.89 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 166.3, 161.8, 161.0, 158.6, 151.4, 145.7, 143.1, 135.1, 130.0(32 Hz), 129.4(2), 127.5, 125.3, 123.1(97.2 Hz), 121.9(33.2 Hz), 121.3(2), 119.3, 114.7(2), 110.6(93.2 Hz), 67.0, 52.8(2), 48.1(2), 42.5. HRMS (ESI)+ Calculated for C27H25ClFNaN5O3 [M + Na]+ m/z 544.1528, found 544.1526.
N-(3-Chloro-4-(4-methylpiperazin-1-yl)phenyl)-2-(4-(6-methoxy-4-oxo-1,4-dihydroquinazolin-2-yl)phenoxy)acetamide (44k).
White powder, mp 243–245 °C, yield 47.7%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 10.28 (1H, s), 8.03 (2H, d, J = 8.8 Hz), 7.85 (1H, d, J = 2.4 Hz), 7.80 (1H, d, J = 8.8 Hz), 7.60 (1H, d, J = 2.4 Hz), 7.51–7.56 (2H, m), 7.28 (2H, d, J = 8.8 Hz), 7.17 (1H, d, J = 8.8 Hz), 4.88 (2H, s), 3.90 (3H, s), 3.52 (2H, d, J = 12.0 Hz), 3.36 (2H, d, J = 12.0 Hz), 3.21 (2H, td, J = 12.0, 2.8 Hz), 2.97 (2H, td, J = 12.0, 2.8 Hz), 2.87 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 166.4, 162.1, 160.0(2), 157.5, 149.7, 143.1, 135.1, 129.1(2), 127.5, 125.7, 124.2, 124.1, 121.5. 121.4, 121.3, 119.3, 114.7(2), 105.9, 67.0, 55.7, 52.9(2), 48.1(2), 42.2. HRMS (ESI)+ Calculated for C28H29ClN5O4 [M + H]+ m/z 534.1908, found 534.1899.
N-(3-Chloro-4-(4-methylpiperazin-1-yl)phenyl)-2-(4-(6-chloro-4-oxo-1,4-dihydroquinazolin-2-yl)phenoxy)acetamide (44l).
White powder, mp 281–283 °C, yield 54.5%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 12.60 (1H, s), 10.30 (1H, s), 10.05 (1H, s), 8.20 (2H, d, J = 8.8 Hz), 8.06 (1H, d, J = 2.4 Hz), 7.86 (1H, d, J = 2.4 Hz), 7.83 (1H, dd, J = 8.8, 2.4 Hz), 7.72 (1H, d, J = 8.8 Hz), 7.55 (1H, dd, J = 8.8, 2.4 Hz), 7.22 (1H, d, J = 8.8 Hz), 7.15 (2H, d, J = 8.8 Hz), 4.83 (2H, s), 3.52–3.55 (2H, m), 3.38 (2H, d, J = 12.0 Hz), 3.18 (2H, td, J = 12.0, 2.8 Hz), 2.99 (2H, td, J = 12.0, 2.8 Hz), 2.89 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 166.3, 161.4, 160.5, 152.3, 147.6, 143.1, 135.1, 134.6, 130.3, 129.6(2), 129.5, 127.5, 125.2, 124.9, 121.9, 121.3(2), 119.3, 114.7(2), 67.0, 52.9(2), 48.1(2), 42.2. HRMS (ESI)+ Calculated for C27H26Cl2N5O3 [M + H]+ m/z 538.1413, found 538.1408.
N-(3-Chloro-4-(4-methylpiperazin-1-yl)phenyl)-2-(4-(7-chloro-4-oxo-1,4-dihydroquinazolin-2-yl)phenoxy)acetamide (44m).
Off-white solid, mp 280–282 °C, yield 46.2%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 12.51 (1H, s), 10.18 (1H, s), 8.19 (2H, d, J = 8.8 Hz), 8.12 (1H, d, J = 8.8 Hz), 7.80 (1H, d, J = 2.4 Hz), 7.45 (1H, d, J = 2.4 Hz), 7.51 (2H, dd, J = 8.8, 2.4 Hz), 7.13–7.17 (3H, m), 4.81 (2H, s), 3.33 (4H, brs), 2.91–2.93 (4H, m), 2.25 (3H, s). 13C NMR (100 MHz, DMSO-d6 + CF3COOD), δ (ppm): 166.3, 161.6, 160.9, 153.6, 149.4, 143.2, 139.4, 135.2, 129.9(2), 128.1, 127.7, 126.7, 125.9, 124.7, 121.5, 121.4, 119.6, 119.5, 114.9(2), 67.1, 53.0(2), 48.2(2), 42.2. HRMS (ESI)+ Calculated for C27H26Cl2N5O3 [M + H]+ m/z 538.1413, found 538.1415.
N-(3-Chloro-4-(4-methylpiperazin-1-yl)phenyl)-2-(4-(7-methyl-4-oxo-1,4-dihydroquinazolin-2-yl)phenoxy)acetamide (44n).
White powder, mp 279–282 °C, yield 55.5%. 1H NMR (400 MHz, DMSO-d6), δ (ppm): 12.33 (1H, s), 10.27 (1H, s), 9.91 (1H, s), 8.17 (1H, d, J = 8.8 Hz), 8.01 (1H, d, J = 8.4 Hz), 7.93 (2H, s), 7.85 (1H, d, J = 2.4 Hz), 7.56 (1H, dd, J = 8.8, 2.4 Hz), 7.52 (1H, s), 7.31 (1H, dd, J = 8.4, 2.4 Hz), 7.22 (1H, d, J = 8.8 Hz), 7.14 (1H, d, J = 8.8 Hz), 4.82 (2H, s), 3.52 (2H, d, J = 11.6 Hz), 3.38 (2H, d, J = 11.6 Hz), 3.20 (2H, t, J = 12.0 Hz), 2.96 (2H, t, J = 12.0 Hz), 2.88 (3H, s), 2.46 (3H, s). 13C NMR (100 MHz, DMSO-d6), δ (ppm): 166.3, 162.2, 160.3, 151.9, 148.8, 145.0, 143.1, 135.1, 129.4(2), 127.7, 127.5, 126.8, 125.7, 125.5, 121.3(2), 119.3, 118.3, 114.7(2), 67.0, 52.9(2), 48.1(2), 42.7, 21.4. HRMS (ESI)+ Calculated for C28H29ClN5O3 [M + H]+ m/z 518.1959, found 518.1953.
Bioinformatics Analysis.
PrePPI (https://honiglab.c2b2.columbia.edu/PrePPI/) was used to predict BRD4 and CK2 related PPI networks. Subsequently, these interacting proteins were annotated and clustered using the Gene Ontology (GO) and the DAVID databases. Finally, the GEPIA tool (http://gepia.cancer-pku.cn/) was used to analyze the breast cancer transcriptome data of BRD4 and CK2 obtained from the TCGA database, and a significantly positive correlations identified between BRD4 and CK2 PPI networks were produced by using Cytoscape (version 3.8.2)
Molecular Docking.
Molecular docking follows previous studies.14,51 Small molecules were constructed using the Accelrys Discovery Studio (version 3.5; Accelrys, SanDiego, CA, USA) using LibDock protocol. Small molecules were energy minimized with the CHARMm force field. Then CDOCKER protocol was performed to rerank the top 10 small molecule compounds. Other parameters were set as default values.
BRD4 and CK2 Enzymatic Assays.
The in vitro BRD and CK assays were performed by the drug discovery service of Huawei Pharmaceutical Company. The BRD enzymatic profiling assays were performed with TR-FRET technology.14 The CK2 and CK1 enzymatic profiling assays were performed by the methods reported in the literature.53
Cellular Thermal Shift Assay (CETSA).
CETSA was performed with intact cells to assay the interaction between 1, 18, 44e, and BRD4/CK2α. Briefly, MDA-MB-231 cells were incubated with DMSO (0.3% vol/vol), 1 (5 μM), 18 (5 μM), or 44e (5 μM). After 6 h treatment in the incubator, cells were collected and resuspended in PBS. Following heating at the indicated temperatures (48, 51, 54, 57, 60, and 63 °C) for 3 min, cell suspensions were lysed by three freeze–thaw cycles with liquid nitrogen, thawing at room temperature. The lysates were clarified by centrifugation at 17 000g for 20 min. The soluble proteins in the supernatant were collected and prepared for Western blotting to detect the indicated proteins.
Cell Culture, Antibodies, and Reagents.
MDA-MB-231, MDA-MB-468, and MCF-10A cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). MDA-MB-231 and MCF-10A cells were cultured in DMEM with 10% fetal bovine serum, while MDA-MB-468 cells were cultured in L-15 with 10% fetal bovine serum and incubated with 5% CO2. Antibodies used in this study were as follows: BRD4 (Cell Signaling Technology, no. 13440S), p-BRD4S492 (EMD Millipore, ABE no. 1451), c-Myc (Abcam, ab56), LC3B antibody (Abcam, ab51520), SQSTM1/p62 antibody (Abcam, ab56416), CK2α (Cell Signaling Technology, no. 2656S), p-AKTS129 (Abcam, 133458), GAPDH (Cell Signaling Technology, no. 5174), beclin-1 (Cell Signaling Technology, no. 3495), Bax (Cell Signaling Technology, no. 5023), Bcl-2 (Cell Signaling Technology, no. 2870), caspase-3 (Abcam, ab13847). MTT (M2128), 3-MA (M9281), and Hoechst 33258 were purchased from Solarbio. Bafilomycin A1 (ab120497) was purchased from Abcam.
Generation of JQ1-Resistant Cells.
To generate JQ1-resistant cells, MDA-MB-231 cells were subjected to gradual increases in JQ1 concentrations (72 h pulses every 2 weeks for 6 months).
Cell Viability Assay.
The MDA-MB-231, MDA-MB-468, or MCF-10A cells were dispersed in 96-well plates at a density of 6 × 104 cells/mL. After 24 h incubation, cells were treated with different concentrations of compounds for 24 h. Cell viability was measured by MTT assay. MTT (5 mg/mL) was added, and cells were incubated for an additional 4 h at 37 °C; then the absorbance value was determined with an enzyme-labeling instrument at 490 nm.
Colony Formation Assay.
Cells (500–800 per well) were seeded in 6-well plates and allowed to adhere overnight. Cells were treated with the indicated concentration of compounds or vehicle control for 7–10 days. Cells were fixed with formaldehyde (4%) and stained with crystal violet. The number of colonies was counted. Data represent the mean ± SD from 3 independent experiments performed in triplicate wells.
Annexin-V/PI Dual Staining.
Cells (1 × 105 per well) were cultured in six-well plates for 24 h. Cells were then cultured in the absence or presence of 44e (2.5, 5, or 10 μM) for 24 h and then washed twice with cold PBS. The early and late apoptosis cells were identified by flow cytometry (Becton Dickinson, Franklin Lakes, NJ) after applying an Annexin-V/PI Staining Kit (Roche, Germany). The detailed procedures were performed according to the corresponding manufacturer’s instructions.
Hoechst Staining.
Cells (2.5 × 104 per well) were plated in 24-well plates, left to adhere overnight, and then incubated with or without 44e (2.5, 5, or 10 μM) for 24 h. Then, the original growth medium was discarded, cells were washed twice with PBS, followed by addition of Hoechst 33258 for 5 min, and then observed under an Olympus fluorescence microscope.
Autophagy Assay.
A total of 2.5 × 104 cells/well were seeded in a 24-well plate. After transfection with GFP-mRFP-LC3 (HB-AP2100001, HANBIO, China) for 4 h, the original growth medium was replaced with fresh medium, and cells were incubated with or without compounds for 24 h at 37 °C, then analyzed under a fluorescence microscope.
Western Blot Analysis.
Cells were seeded in a 6-well plate (1.5 × 105 cells/well), grown at 37 °C for 24 h, and then treated with compounds and vehicle (DMSO) for 24 h, collected, centrifuged (3000 rpm for 5 min), dissociated, centrifuged again (12000g for 10 min), and quantified (the BCA kit, Beyotime). Western blot analysis was carried out by the following method: In brief, equal amounts of total protein in cell lysate were separated by 10–15% SDS-PAGE and then transferred to PVDF membranes. After being soaked in blocking buffer (5% skimmed milk), the membranes were incubated with corresponding primary antibodies at 4 °C overnight, followed by HRP-conjugated secondary antibody at room temperature for 2 h, and visualized using ECL as the HRP substrate.
Pharmacokinetic Studies.
The pharmacokinetic parameters of 44e were evaluated. Six Sprague–Dawley (SD) rats (body weight of 200–220 g) were randomly divided into two groups (3 rats in each group) for injection (iv) or oral administration (po). The saline solutions were formulated in 5% DMSO and 10% hydroxypropyl-β-cyclodextrin (HP-β-CD). Two groups of rats were administered a single concentration of 44e at 1 mg/kg (caudal vein injection) or 10 mg/kg (oral gavage). Blood samples were collected through the jugular vein at the specified time and centrifuged at 7000 rpm for 10 min to obtain the isolated plasma. LC-MS (5500QTRAP system, Applied Biosystems) was used to evaluate the plasma concentration of 44e.
Xenograft Tumor Models.
All experimental protocols used in this study were carried out in accordance with guidelines of the animal ethics committee (Sichuan University). Female nude mice (BALB/c, 6–8 weeks, 20–22 g, n = 48) were randomized into two groups (24 mice per group) and injected with MDA-MB-231 cells (5 × 106 cells/mouse) or MDA-MB-468 cells (5 × 106 cells/mouse). When the tumors of all mice reached 100 mm3 in volume (V = L × W2/2), the mice were randomly grouped by weight and coded according to the tumor types (n = 6 in each group) and treated via intragastric administration of 0.2 mL of saline, 1 + 18 (50 mg/kg, combined at a mole ratio of 1:1), or 44e (25 or 50 mg/kg) once a day for 19 days. Meanwhile, the tumor volume and body weight were measured every 3 days during treatment. At the end of treatment, all mice were sacrificed. The tumor tissues and main tissues were harvested and weighed. Then, the tumor tissues were frozen in liquid nitrogen or fixed in formalin immediately for immunohistochemistry analysis and HE staining.
HE Staining.
The fixed tissues were dehydrated and paraffin embedded. Then tissues were sliced into 4 mm thick slices, stained for 10–15 min with hematoxylin, washed for 2 min (tap water), differentiated for 30 s (different gradients hydrochloric acid and ethanol), washed for 5 min (warm water), and finally placed into the eosin solution for 2 min before regular dehydration and sealing with resinene. All the specimens were evaluated according to the pathological examination SOP procedure.
Immunohistochemistry Analysis (IHC).
Tumor sections were immersed in EDTA antigen retrieval buffer (pH 8.0) or citrate buffer (pH 6.0), and microwave was used for antigen retrieval. Then the slides were incubated with anti-Ki-67 antibody (1:500), anti-BRD4 antibody (1:500), anti-c-Myc antibody (1:500), anti-CK2α antibody (1:500), anti-p-AKTS129 antibody (1:400), or anti-LC3B antibody (1:400) for 30–40 min at 37 °C. Normal rabbit or mouse IgG was used as a negative control. The slides were then treated with HRP polymer conjugated secondary antibody for 30 min and developed with diamino-benzidine solution. Meyer’s hematoxylin was used as a counterstain.
Statistical Analysis.
All cell experiments were performed independently at least three times. The data are expressed as the mean ± SD and analyzed with GraphPad Prism 7.0. Statistical comparisons were made by one-way ANOVA and Student’s test. p < 0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (Grant Numbers 81922064, 81874290, 81903502, and 81803365), the Applied Basic Research Programs of Science and Technology Department of Sichuan Province (Grant Numbers 2020YJ0105, 2020YJ0094), the Fundamental Research Funds for the Central Universities (Grant Number 2021SCU12102), China Postdoctoral Science Foundation (Grant Number 2020M673268), and Health Commission of Sichuan Province (Grant Number 20PJ002). C.-M.C.’s research is sponsored by the U.S. National Institutes of Health (Grant 1RO1CA251698-01)and the Cancer Prevention and Research Institute of Texas (CPRIT; Grants RP180349 and RP190077).
ABBREVIATIONS
- AKT
protein kinase B
- BC
breast cancer
- BafA1
bafilomycin A1
- BLBC
basal-like breast cancer
- BRD4
bromodomain-containing protein 4
- CK2
casein kinase 2
- DBU
1,8-diazabicyclo[5.4.0]undec-7-ene
- DMAc
dimethylacetamide
- EDCI
1-ethyl-3-(3-dimethylaminopropyl)carbodiimide
- EGFR
epidermal growth factor receptor
- ER
estrogen receptor
- ERK
extracellular signal-regulated kinase
- GFP
green fluorescent protein
- HE
hematoxylin–eosin
- HER2
human epidermal growth factor receptor 2
- HOAc
acetic acid
- HOBt
hydroxybenzotriazole
- HRESIMS
high resolution electrospray ionization mass spectroscopy
- IHC
immunohistochemistry analysis
- IC50
half-maximal inhibitory concentration
- MAPK
mitogen-activated protein kinase
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- 3-MA
3-methyladenine
- NMM
N-methylmorpholine
- PE
polyethylene
- PARP
poly(ADP-ribose) polymerase
- PI3K
phosphoinositide 3-kinase
- PR
progesterone receptor
- PTSA
p-toluenesulfonic acid
- Q-TOF
quadrupole time of flight
- SAR
structure–activity relationship
- TCGA
The Cancer Genome Atlas
- TEA
triethylamine
- TFA
trifluoroacetic acid
- TNBC
triple-negative breast cancer
- TMS
tetramethylsilane
- TR-FRET
time resolved fluorescence resonance energy transfer
- VEGFR2
vascular endothelial growth factor 2
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.1c01382.
Molecular formula strings and the associated biochemical and biological data (CSV)
Top 10 candidate dual-target inhibitors of BRD4 and CK2, BRD4 and CK2 inhibition rates of top 10 candidate compounds, toxicity of candidate compound 44e to MCF-10A cells, representative images of main organ tissues after staining with hematoxylin–eosin (H&E) in MDA-MB-231 and MDA-MB-468 tumor xenograft models, molecular docking results, ATP competitive binding results, selectivity of 44e against other kinases, and MS and NMR data (PDF)
The authors declare no competing financial interest.
Contributor Information
Jifa Zhang, State Key Laboratory of Biotherapy and Cancer Center, Sichuan University-Oxford University Huaxi Gastrointestinal Cancer Centre, Innovation Center of Nursing Research, Nursing Key Laboratory of Sichuan Province, National Clinical Research Center for Geriatrics, West China Hospital of Sichuan University, Chengdu 610041 Sichuan, China.
Pan Tang, State Key Laboratory of Biotherapy and Cancer Center, Sichuan University-Oxford University Huaxi Gastrointestinal Cancer Centre, Innovation Center of Nursing Research, Nursing Key Laboratory of Sichuan Province, National Clinical Research Center for Geriatrics, West China Hospital of Sichuan University, Chengdu 610041 Sichuan, China.
Ling Zou, State Key Laboratory of Biotherapy and Cancer Center, Sichuan University-Oxford University Huaxi Gastrointestinal Cancer Centre, Innovation Center of Nursing Research, Nursing Key Laboratory of Sichuan Province, National Clinical Research Center for Geriatrics, West China Hospital of Sichuan University, Chengdu 610041 Sichuan, China.
Jin Zhang, State Key Laboratory of Biotherapy and Cancer Center, Sichuan University-Oxford University Huaxi Gastrointestinal Cancer Centre, Innovation Center of Nursing Research, Nursing Key Laboratory of Sichuan Province, National Clinical Research Center for Geriatrics, West China Hospital of Sichuan University, Chengdu 610041 Sichuan, China; School of Pharmaceutical Sciences, Health Science Center, Shenzhen University, Shenzhen 518060, China.
Juncheng Chen, State Key Laboratory of Biotherapy and Cancer Center, Sichuan University-Oxford University Huaxi Gastrointestinal Cancer Centre, Innovation Center of Nursing Research, Nursing Key Laboratory of Sichuan Province, National Clinical Research Center for Geriatrics, West China Hospital of Sichuan University, Chengdu 610041 Sichuan, China.
Chengcan Yang, State Key Laboratory of Biotherapy and Cancer Center, Sichuan University-Oxford University Huaxi Gastrointestinal Cancer Centre, Innovation Center of Nursing Research, Nursing Key Laboratory of Sichuan Province, National Clinical Research Center for Geriatrics, West China Hospital of Sichuan University, Chengdu 610041 Sichuan, China.
Gu He, State Key Laboratory of Biotherapy and Cancer Center, Sichuan University-Oxford University Huaxi Gastrointestinal Cancer Centre, Innovation Center of Nursing Research, Nursing Key Laboratory of Sichuan Province, National Clinical Research Center for Geriatrics, West China Hospital of Sichuan University, Chengdu 610041 Sichuan, China.
Bo Liu, State Key Laboratory of Biotherapy and Cancer Center, Sichuan University-Oxford University Huaxi Gastrointestinal Cancer Centre, Innovation Center of Nursing Research, Nursing Key Laboratory of Sichuan Province, National Clinical Research Center for Geriatrics, West China Hospital of Sichuan University, Chengdu 610041 Sichuan, China.
Jie Liu, State Key Laboratory of Biotherapy and Cancer Center, Sichuan University-Oxford University Huaxi Gastrointestinal Cancer Centre, Innovation Center of Nursing Research, Nursing Key Laboratory of Sichuan Province, National Clinical Research Center for Geriatrics, West China Hospital of Sichuan University, Chengdu 610041 Sichuan, China.
Cheng-Ming Chiang, Simmons Comprehensive Cancer Center, Department of Pharmacology, and Department of Biochemistry, University of Texas Southwestern Medical Center, Dallas, Texas 75390, United States.
Guan Wang, State Key Laboratory of Biotherapy and Cancer Center, Sichuan University-Oxford University Huaxi Gastrointestinal Cancer Centre, Innovation Center of Nursing Research, Nursing Key Laboratory of Sichuan Province, National Clinical Research Center for Geriatrics, West China Hospital of Sichuan University, Chengdu 610041 Sichuan, China.
Tinghong Ye, State Key Laboratory of Biotherapy and Cancer Center, Sichuan University-Oxford University Huaxi Gastrointestinal Cancer Centre, Innovation Center of Nursing Research, Nursing Key Laboratory of Sichuan Province, National Clinical Research Center for Geriatrics, West China Hospital of Sichuan University, Chengdu 610041 Sichuan, China.
Liang Ouyang, State Key Laboratory of Biotherapy and Cancer Center, Sichuan University-Oxford University Huaxi Gastrointestinal Cancer Centre, Innovation Center of Nursing Research, Nursing Key Laboratory of Sichuan Province, National Clinical Research Center for Geriatrics, West China Hospital of Sichuan University, Chengdu 610041 Sichuan, China.
REFERENCES
- (1).Siegel RL; Miller KD; Jemal A Cancer statistics, 2020. CA-Cancer. Ca-Cancer J. Clin 2020, 70, 7–30. [DOI] [PubMed] [Google Scholar]
- (2).Xu T; Zhang JF; Yang CC; Pluta R; Wang G; Ye TH; Ouyang L Identifification and optimization of 3-bromo-N’-(4-hydroxybenzylidene)-4-methylbenzohydrazide derivatives as mTOR inhibitors that induce autophagic cell death and apoptosis in triplenegative breast cancer. Eur. J. Med. Chem 2021, 219, 113424. [DOI] [PubMed] [Google Scholar]
- (3).Franzoi MA; Romano E; Piccart M Immunotherapy for early breast cancer: too soon, too superficial, or just right? Ann. Oncol 2021, 32, 323–336. [DOI] [PubMed] [Google Scholar]
- (4).Vagia E; Mahalingam D; Cristofanilli M The landscape of targeted therapies in TNBC. Cancers 2020, 12, 916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Tang P; Zhang JF; Liu J; Chiang CM; Ouyang L Targeting bromodomain and extraterminal proteins for drug discovery: from current progress to technological development. J. Med. Chem 2021, 64, 2419–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Zhang JF; Ouyang L Proteolysis-targeting chimeras in breast cancer therapy. Future Med. Chem 2020, 12, 2175–2177. [DOI] [PubMed] [Google Scholar]
- (7).Da Motta LL; Ledaki I; Purshouse K; Haider S; De Bastiani MA; Baban D; Morotti M; Steers G; Wigfield S; Bridges E; Li JL; Knapp S; Ebner D; Klamt F; Harris AL; McIntyre A The BET inhibitor JQ1 selectively impairs tumour response to hypoxia and downregulates CA9 and angiogenesis in triple negative breast cancer. Oncogene 2017, 36, 122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Verma N; Müller AK; Kothari C; Panayotopoulou E; Kedan A; Selitrennik M; Mills GB; Nguyen LK; Shin S; Karn T; Holtrich U; Lev S Targeting of PYK2 synergizes with EGFR antagonists in basal-like TNBC and circumvents HER3-associated resistance via the NEDD4-NDRG1 axis. Cancer Res. 2017, 77, 86–99. [DOI] [PubMed] [Google Scholar]
- (9).Loi S; Dushyanthen S; Beavis PA; Salgado R; Denkert C; Savas P; Combs S; Rimm DL; Giltnane JM; Estrada MV; Sánchez V; Sanders ME; Cook RS; Pilkinton MA; Mallal SA; Wang K; Miller VA; Stephens PJ; Yelensky R; Doimi FD; Gómez H; Ryzhov SV; Darcy PK; Arteaga CL; Balko JM RAS/MAPK activation is associated with reduced tumor-infiltrating lymphocytes in triple-negative breast cancer: therapeutic cooperation between MEK and PD-1/PD-L1 immune checkpoint inhibitors. Clin. Cancer Res 2016, 22, 1499–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Delaloge S; DeForceville L Targeting PI3K/AKT pathway in triple-negative breast cancer. Lancet Oncol. 2017, 18, 1293–1294. [DOI] [PubMed] [Google Scholar]
- (11).Curtin NJ; Szabo C Poly(ADP-ribose) polymerase inhibition: past, present and future. Nat. Rev. Drug Discovery 2020, 19, 711–736. [DOI] [PubMed] [Google Scholar]
- (12).Jhan JR; Andrechek ER Triple-negative breast cancer and the potential for targeted therapy. Pharmacogenomics 2017, 18, 1595–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Lord CJ; Ashworth A PARP inhibitors: synthetic lethality in the clinic. Science 2017, 355, 1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Chang XS; Sun DJ; Shi DF; Wang G; Chen Y; Zhang K; Tan HD; Liu J; Liu B; Ouyang L Design, synthesis, and biological evaluation of quinazolin-4(3H)-one derivatives co-targeting poly-(ADP-ribose) polymerase-1 and bromodomain containing protein 4 for breast cancer therapy. Acta Pharm. Sin. B 2021, 11, 156–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Gornstein EL; Sandefur S; Chung JH; Gay LM; Holmes O; Erlich RL; Soman S; Martin LK; Rose AV; Stephens PJ; Ross JS; Miller VA; Ali SM; Blau S BRCA2 reversion mutation associated with acquired resistance to olaparib in estrogen receptor-positive breast cancer detected by genomic profiling of tissue and liquid biopsy. Clin. Breast Cancer 2018, 18, 184–188. [DOI] [PubMed] [Google Scholar]
- (16).Feng L; Wang G; Chen Y; He G; Liu B; Liu J; Chiang CM; Ouyang L Dual-target inhibitors of bromodomain and extraterminal proteins in cancer: A review from medicinal chemistry perspectives. Med. Res. Rev 2021, DOI: 10.1002/med.21859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Filippakopoulos P; Knapp S Targeting bromodomains: epigenetic readers of lysine acetylation. Nat. Rev. Drug Discovery 2014, 13, 337–356. [DOI] [PubMed] [Google Scholar]
- (18).Wu SY; Chiang CM The double bromodomain-containing chromatin adaptor BRD4 and transcriptional regulation. J. Biol. Chem 2007, 282, 13141–13145. [DOI] [PubMed] [Google Scholar]
- (19).Wu SY; Nin DS; Lee AY; Simanski S; Kodadek T; Chiang CM BRD4 phosphorylation regulates HPV E2-mediated viral transcription, origin replication, and cellular MMP-9 expression. Cell Rep. 2016, 16, 1733–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Ramadoss M; Mahadevan V Targeting the cancer epigenome: synergistic therapy with bromodomain inhibitors. Drug Discovery Today 2018, 23, 76–89. [DOI] [PubMed] [Google Scholar]
- (21).Wu SY; Lee CF; Lai HT; Yu CT; Lee JE; Zuo H; Tsai SY; Tsai MJ; Ge K; Wan Y; Chiang CM Opposing functions of BRD4 isoforms in breast cancer. Mol. Cell 2020, 78, 1114–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Chiang CM Phospho-BRD4: transcription plasticity and drug targeting. Drug Discovery Today: Technol. 2016, 19, 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Marcotte R; Sayad A; Brown KR; Sanchez-Garcia F; Reimand J; Haider M; Virtanen C; Bradner JE; Bader GD; Mills GB; Pe’er D; Moffat J; Neel BG Functional genomic landscape of human breast cancer drivers, vulnerabilities, and resistance. Cell 2016, 164, 293–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Andrieu G; Tran AH; Strissel KJ; Denis GV BRD4 regulates breast cancer dissemination through Jagged1/Notch1 signaling. Cancer Res. 2016, 76, 6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zawistowski JS; Bevill SM; Goulet DR; Stuhlmiller TJ; Beltran AS; Olivares-Quintero JF; Singh D; Sciaky N; Parker JS; Rashid NU; Chen X; Duncan JS; Whittle MC; Angus SP; Velarde SH; Golitz BT; He X; Santos C; Darr DB; Gallagher K; Graves LM; Perou CM; Carey LA; Earp HS; Johnson GL Enhancer remodeling during adaptive bypass to MEK inhibition is attenuated by pharmacologic targeting of the p-TEFb complex. Cancer Discovery 2017, 7, 302–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Devaiah BN; Mu J; Akman B; Uppal S; Weissman JD; Cheng D; Baranello L; Nie Z; Levens D; Singer DS MYC protein stability is negatively regulated by BRD4. Proc. Natl. Acad. Sci. U. S. A 2020, 117, 13457–13467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Liu ZQ; Wang PY; Chen HY; Wold EA; Tian B; Brasier AR; Zhou J Drug discovery targeting bromodomain-containing protein 4. J. Med. Chem 2017, 60, 4533–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Pan ZP; Li X; Wang YJ; Jiang QL; Jiang L; Zhang M; Zhang N; Wu FB; Liu B; He G Discovery of thieno[2,3-d]pyrimidine-based hydroxamic acid derivatives as bromodomain-containing protein 4/histone deacetylase dual inhibitors induce autophagic cell death in colorectal carcinoma cells. J. Med. Chem 2020. 63, 3678–3700. [DOI] [PubMed] [Google Scholar]
- (29).Kulikowski E; Rakai BD; Wong N Inhibitors of bromodomain and extra-terminal proteins for treating multiple human diseases. Med. Res. Rev 2021, 41, 223–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Fong CY; Gilan O; Lam EY; Rubin AF; Ftouni S; Tyler D; Stanley K; Sinha D; Yeh P; Morison J; Giotopoulos G; Lugo D, Jeffrey P; Lee SC; Carpenter C; Gregory R; Ramsay RG; Lane SW; Abdel-Wahab O; Kouzarides T; Johnstone RW; Dawson SJ; Huntly BJ; Prinjha RK; Papenfuss AT; Dawson MA BET inhibitor resistance emerges from leukaemia stem cells. Nature 2015, 525, 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Rathert P; Roth M; Neumann T; Muerdter F; Roe JS; Muhar M; Deswal S; Cerny-Reiterer S; Peter B;Jude J; Hoffmann T; Boryn LM; Axelsson E; Schweifer N; Tontsch-Grunt U; Dow LE; Gianni D; Pearson M; Valent P; Stark A; Kraut N; Vakoc CR; Zuber J Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature 2015, 525, 543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Calder J; Nagelberg A; Luu J; Lu D; Lockwood WW Resistance to BET inhibitors in lung adenocarcinoma is mediated by casein kinase phosphorylation of BRD4. Oncogenesis. 2021, 10, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Licciardello MP; Workman P A new chemical probe challenges the broad cancer essentiality of CK2. Trends Pharmacol. Sci 2021, 42, 313–315. [DOI] [PubMed] [Google Scholar]
- (34).Wang YJ; Wang XY; Xu G; Gou SH Novel CK2-specifific Pt(II) compound reverses cisplatin-induced resistance by inhibiting cancer cell stemness and suppressing DNA damage repair in non-small cell lung cancer treatments. J. Med. Chem 2021, 64, 4163–4178. [DOI] [PubMed] [Google Scholar]
- (35).Wang YJ; Lv ZD; Chen FH; Wang X; Gou SH Discovery of 5-(3-Chlorophenylamino)benzo[c][2,6]naphthyridine derivatives as highly selective CK2 inhibitors with potent cancer cell stemness inhibition. J. Med. Chem 2021, 64, 5082–5098. [DOI] [PubMed] [Google Scholar]
- (36).Hwang DW; So KS; Kim SC; Park KM; Lee YJ; Kim SW; Choi CM; Rho JK; Choi YJ; Lee JC Autophagy induced by CX-4945, a casein kinase 2 inhibitor, enhances apoptosis in pancreatic cancer cell lines. Pancreas 2017, 46, 575–581. [DOI] [PubMed] [Google Scholar]
- (37).Wu SY; Lee AY; Lai HT; Zhang H; Chiang CM Phospho switch triggers BRD4 chromatin binding and activator recruitment for gene-specific targeting. Mol. Cell 2013, 49, 843–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Shu S; Lin CY; He HH; Witwicki RM; Tabassum DP; Roberts JM; Janiszewska M; Huh SJ; Liang Y; Ryan J; Doherty E; Mohammed H; Guo H; Stover DG; Ekram MB; Brown J; D’Santos C; Krop IE; Dillon D; McKeown M; Ott C; Qi J; Ni M; Rao PK; Duarte M; et al. Response and resistance to BET bromodomain inhibitors in triple negative breast cancer. Nature. 2016, 529, 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Becherel OJ; Jakob B; Cherry AL; Gueven N; Fusser M; Kijas AW; Peng C; Katyal S; McKinnon PJ; Chen J; Epe B; Smerdon SJ; Taucher-Scholz G; Lavin MF CK2 phosphorylation-dependent interaction between aprataxin and MDC1 in the DNA damage response. Nucleic Acids Res. 2010, 38, 1489–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Zhou B; Ritt DA; Morrison DK; Der CJ; Cox AD Protein kinase CK2α maintains extracellular signal-regulated kinase (ERK) activity in a CK2α kinase-independent manner to promote resistance to inhibitors of RAF and MEK but not ERK in BRAF mutant melanoma. J. Biol. Chem 2016, 291, 17804–17815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Loizou JI; El-Khamisy SF; Zlatanou A; Moore DJ; Chan DW; Qin J; Sarno S; Meggio F; Pinna LA; Caldecott KW The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell 2004, 117, 17–28. [DOI] [PubMed] [Google Scholar]
- (42).Zhang QC; Petrey D; Deng L; Qiang L; Shi Y; Thu CA; Bisikirska B; Lefebvre C; Accili D; Hunter T; Maniatis T; Califano A; Honig B Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature 2012, 490, 556–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Zhang QC; Petrey D; Garzón JI; Deng L; Honig B PrePPI: a structure-informed database of protein-protein interactions. Nucleic Acids Res. 2012, 41, D828–D833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Huang DW; Sherman BT; Lempicki RA Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Huang DW; Sherman BT; Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 2009, 4, 44–57. [DOI] [PubMed] [Google Scholar]
- (46).Tang ZF; Li CW; Kang BX; Gao G; Li C; Zhang ZM GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Zhang L; Zhang L; Guo Y; Xiao M; Feng L; Yang C; Wang G; Ouyang L MCDB: A comprehensive curated mitotic catastrophe database for retrieval, protein sequence alignment, and target prediction. Acta Pharm. Sin. B 2021, 11, 3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Wang G; Zhao YQ; Liu Y; Sun DJ; Zhen YQ; Liu J; Fu L; Zhang L; Ouyang L Discovery of a novel dual-target inhibitor of ERK1 and ERK5 that induces regulated cell death to overcome compensatory mechanism in specific tumor types. J. Med. Chem 2020, 63, 3976–3995. [DOI] [PubMed] [Google Scholar]
- (49).Noblejas-López MDM; Nieto-Jimenez C; Burgos M; GómezJuárez M; Montero JC; Esparís-Ogando A; Pandiella A; Galán-Moya EM; Ocaña A Activity of BET-proteolysis targeting chimeric (PROTAC) compounds in triple negative breast cancer. J. Exp. Clin. Cancer. Res 2019, 38, 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Ouyang L; Zhang L; Liu J; Fu L; Yao D; Zhao Y; Zhang S; Wang G;He G; Liu B Discovery of a small-molecule bromodomain-containing protein 4 (BRD4) inhibitor that induces AMP-activated protein kinase-modulated autophagy-associated cell death in breast cancer. J. Med. Chem 2017, 60, 9990–10012. [DOI] [PubMed] [Google Scholar]
- (51).Ouyang L; Zhang L; Zhang S; Yao D; Zhao Y; Wang G; Fu L; Lei P; Liu B Small-molecule activator of UNC-51-Like Kinase 1 (ULK1) that induces cytoprotective autophagy for parkinson’s disease treatment. J. Med. Chem 2018, 61, 2776–2792. [DOI] [PubMed] [Google Scholar]
- (52).Zhang L; Fu LL; Zhang SY; Zhang J; Zhao YQ; Zheng YX; He G; Yang SY; Ouyang L; Liu B Discovery of a small molecule targeting ULK1-modulated cell death of triple negative breast cancer in vitro and in vivo. Chem. Sci 2017, 8, 2687–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Kashem MA; Nelson RM; Yingling JD; Pullen SS; Prokopowicz AS; Jones JW; Wolak JP; Rogers GR; Morelock MM; Snow RJ; Homon CA; Jakes S Three mechanistically distinct kinase assays compared: Measurement of intrinsic ATPase activity identified the most comprehensive set of ITK inhibitors. J. Biomol Screening 2007, 12, 70–83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.