Abstract
Medulloblastoma (MB) is one of the most frequent malignant brain tumors in children. The metastasis of MB outside the nervous system is associated with a poor prognosis. Our study aimed to explore the genes correlated with metastasis in MB. Using the data downloaded from the gene expression omnibus database, the differentially expressed genes were identified between the metastatic and nonmetastatic samples in MB, which were undergone functional enrichment. Prognosis related genes were identified using univariate Cox regression analysis. The gene set enrichment analysis was conducted to find MB metastasis related pathways. A total of 196 differentially expressed genes were identified between metastatic and nonmetastatic samples in MB patients, and these genes were significantly enriched in 483 gene ontology terms and 29 Kyoto encyclopedia of genes and genomes pathways. In addition, univariate Cox regression analysis screened the top 10 genes (CEMIP, GLCE, ART3, GABRA5, COLEC12, LIN28B, ZNF521, IL17RB, Fas apoptotic inhibitory molecule 2 (FAIM2), RCBTB2) that were significantly associated with survival of MB, among which FAIM2 was prominently expressed in cerebral cortex, cerebellum and hippocampus. The expression of FAIM2 was decreased in metastatic MB samples, and FAIM2 harbored missense mutations, amplifications and deep deletions in metastatic samples of MB. Moreover, a total of 25 pathways were significantly activated and 41 pathways were significantly inhibited in FAIM2 high expression group compared to FAIM2 low expression group in MB patients. FAIM2 was tightly correlated with metastasis in MB patients, and the low expression of FAIM2 was associated with poor prognosis.
Keywords: FAIM2, medulloblastoma, metastasis, prognosis
1. Introduction
Medulloblastoma (MB), one of the most common malignant brain tumors in children, accounts of ~63% of embryonal tumors in childhood intracranial.[1,2] The common diagnostic locations for pediatric MB are the cerebellum and brainstem, and standard treatment of MB including surgery, cytotoxic chemotherapy and cranio-spinal irradiation.[1,3] Unfortunately, these treatments can cause serious sequelae for the survivors, such as, hearing loss, vasculopathy, neurocognitive impairment, endocrine dysfunction and secondary cancers.[4–7] The metastasis of MB outside the nervous system is associated with a poor prognosis in MB patients, and the most common site of metastasis was bones (pelvis, femur, vertebrae), visceral organs and lymph glan.[8,9] It has been indicated that aberrant expression of certain genes has an important impact on the metastasis of MB. For example, the lower expression of ITRR1 and its coregulated genes (ATP1A2, MTTL7A, RGL1) are associated with processes in MB, and the lower expression of ANTXR1 and RGL1 are significantly correlated with worse overall survival (OS).[10] However, until now fewer clinical studies have identified novel genes directly against metastasis of MB. Accordingly, further investigation of metastasis related genes is helpful to better understand the molecular mechanisms of the occurrence of MB metastasis and provide more options for clinical treatment.
Fas apoptotic inhibitory molecule 2 (FAIM2) is a number of the transmembrane BAX inhibitor motif-containing family, and it is an antiapoptotic protein.[11,12] FAIM2 is primary expressed in nervous system related tissues, such as, brain (cortex, hippocampus, cerebellum) and spinal cord. The main biological function of FAIM2 is to inhibit Fas-induced cell apoptosis by direct interaction with Fas receptor and interaction with Bcl-xL to modulate calcium release at the endoplasmic reticulum.[12,13] Recently, several reports indicated that the expression of FAIM2 presents important role in metastasis and prognosis of cancer. In osteosarcoma samples, the closer to tumor blood vessels, the higher expression of FAIM2 is.[14] The higher expression of FAIM2 is associated with poor prognosis in non-small cell lung cancer.[15] In addition, the expression of FAIM2 has been reported in relation to differentiation of neuroblastoma (NBL).[16] The lower expression of FAIM2 reduces cell adhesion and promotes sphere growth and migration, thus increasing the metastatic capacity of NBL cells.[16] However, few reports have revealed the role of FAIM2 in metastasis and prognosis of MB.
In present study, we obtained a key MB metastasis associated gene FAIM2 by bioinformatic tools. Our research is expected to provide reference for the targeted therapy of MB metastasis in the future.
2. Materials and Methods
2.1. Data retrieval
The data was downloaded from the GSE124814, GSE202043, GSE10327, and GSE34355 datasets in the gene expression omnibus (https://www.ncbi.nlm.nih.gov/geo/) database. In GSE124814 dataset, there were a total of 1015 samples involving primary tumor, metastatic tumor and normal tissues, of which, a total of 652 samples had complete clinical information. GSE202043 and GSE10372 datasets are taken as validation sets, among which GSE202043 contains a total of 205 (129 samples had complete clinical information) samples, GSE10327 includes a total of 58 samples. GSE34355 is a methylation dataset including a total of 19 samples.
2.2. Differential gene analysis
The differential gene analysis between the metastatic and nonmetastatic groups was performed using limma package of R language (version 4.2.0).[17] The differentially expressed genes (DEGs) were screened using the |logFC| > 0.5 and P < .05.
2.3. Functional enrichment analysis
The DEGs were then subjected to (Gene ontology [GO], including biological process, molecular function, and cellular component) and Kyoto encyclopedia of genes and genomes (KEGG) enrichment analysis by the cluster Profiler function package in R language (version 4.2.0).[18] The adjusted P < .05 was considered significantly enriched.
2.4. Survival analysis
The OS of different groups was estimated with R language survival package and survminer package, based on the Kaplan–Meier method. The log-rank test was used to examine the significant difference between groups.
2.5. Gene set enrichment analysis (GSEA)
KEGG pathways were then explored using GSEA based on the cluster Profiler function package in R language (version 4.2.0). The |NES| > 1 and P < .05 was considered significantly enriched.
2.6. Gene mutations and copy number variation (CNV)
The mutations and CNV of genes in MB metastatic samples were analyzed using cBio Portal, which is an open access web analysis data resource for integrative exploration of genomics data across multiple cancers, including gene mutations, CNV and mRNA expression landscape of genes.[19]
2.7. Statistical analysis
All statistical analyses were performed using R software (Version 4.2.0; https://www.r-project.org/). The difference among various groups was determined by Wilcoxon-test. The OS was calculated using Kaplan–Meier method, and the survival between the 2 groups was performed with log-rank tests. All statistical tests were 2-sided, and P < .05 was considered significant.
3. Results
3.1. Metastasis related genes in MB
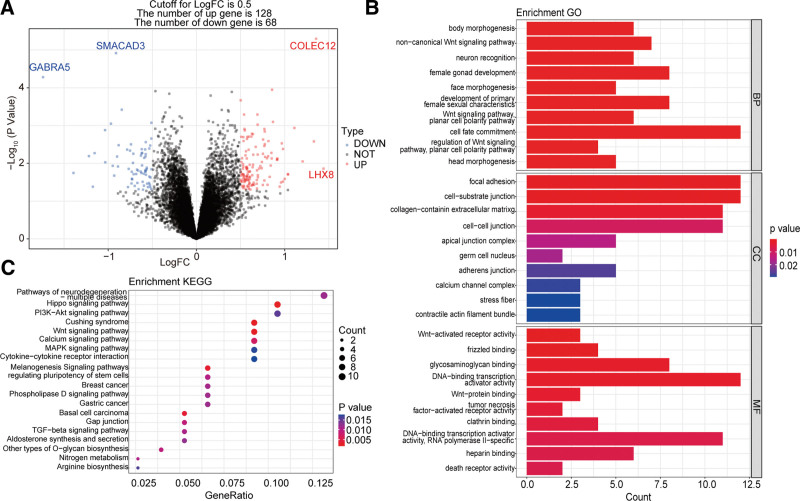
To explore the metastasis related genes in MB, we firstly analyzed the DEGs between metastatic and nonmetastatic groups in 88 MB from GSE124814 dataset. A total of 196 DEGs were identified in metastatic MB patients compared with nonmetastatic patients, including 128 downregulated genes and 68 upregulated genes (Fig. 1A). In addition, the GO enrichment analysis showed that these 196 genes were significantly enriched in body morphogenesis, focal adhesion, WNT-activated receptor activity and so on, and the top 10 GO terms were showed in Figure 1B. Moreover, these 196 genes were also significantly enriched in KEGG pathways, such as hippo signaling pathways, and the top 20 KEGG pathways were showed in Figure 1C. All enrichment results of GO and KEGG analysis were presented in (Table S1, Supplemental Digital Content, http://links.lww.com/MD/I841) and (Table S2, Supplemental Digital Content, http://links.lww.com/MD/I842), respectively.
Figure 1.
Metastasis related genes in MB. (A) DEGs between metastatic and nonmetastatic groups. (B) The top 10 significantly enriched GO terms. (C) The top twenty significantly enriched KEGG pathways. DEGs = differentially expressed genes, GO = gene ontology, KEGG = Kyoto encyclopedia of genes and genomes, MB = medulloblastoma.
3.2. FAIM2 was closely associated with the prognosis of MB
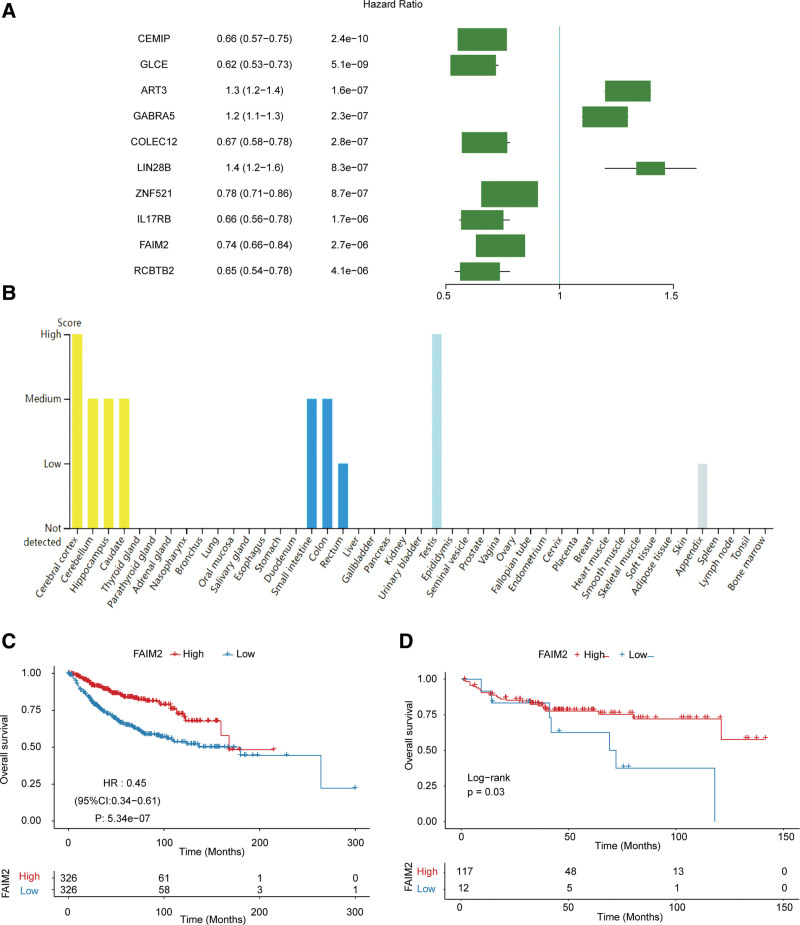
Metastasis of MB is an important prognostic factor for patients.[9] Therefore, we analyzed the potential association between 196 DEGs and prognosis of MB patients using univariate Cox regression, and screened top 10 genes (CEMIP, GLCE, ART3, GABRA5, COLEC12, LIN28B, ZNF521, IL17RB, FAIM2, RCBTB2) that were significantly associated with survival of MB (Fig. 2A). Among which FAIM2 was prominently expressed in cerebral cortex, cerebellum and hippocampus (Fig. 2B), which is consistent with the common metastatic location of MB in the brain.[9] Moreover, the MB patients with higher FAIM2 expression had better OS both in GSE124814 and GSE202043 datasets (Fig. 2C and D). These results suggested that higher expression of FAIM2 was associated with better prognosis in MB patients. Therefore, FAIM2 was selected as the target gene for subsequent study.
Figure 2.
FAIM2 was closely associated with the prognosis of MB. (A) The univariate Cox regression analysis results of DEGs. (B) Expression of FAIM2 in different tissues. The Kaplan–Meier curves of FAIM2 high and low expression groups in GSE124814. (C) And GSE202043. (D) Datasets. DEGs = differentially expressed genes. FAIM2 = Fas apoptotic inhibitory molecule 2, MB = medulloblastoma.
3.3. Repression of FAIM2 expression was associated with the metastasis of MB
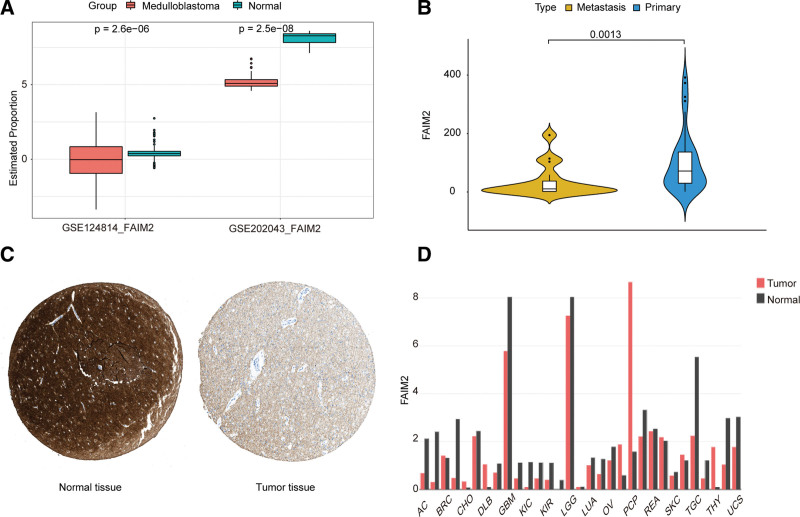
In addition, we found that the expression of FAIM2 was significantly lower in MB compared to the normal brain in GSE124814 dataset (Fig. 3A). To validate this result, we analyzed the expression of FAIM2 in MB and normal groups, metastatic and nonmetastatic groups in the validation set (GSE202043 and GSE10327), and found that the expression of FAIM2 in MB samples was also lower than that in normal brain samples (Fig. 3A) and lower in metastatic group than nonmetastatic group (Fig. 3B). Similarly, the expression of FAIM2 was significantly lower in MB samples compared to normal brain samples in HPA (https://www.proteinatlas.org/) database (Fig. 3C). Moreover, FAIM2 expression was significantly lower in tumor samples than that in normal brain samples in glioblastoma and low-grade gliomas (Fig. 3D). Collectively, our results indicated that the expression of FAIM2 was decreased in MB patients.
Figure 3.
Repression of FAIM2 expression was associated with the metastasis of MB. (A) Expression levels of FAIM2 in MB and normal samples. (B) Expression levels of FAIM2 in metastatic and nonmetastatic samples. (C) Expression of FAIM2 in MB samples and normal brain samples. (D) Expression of FAIM2 between tumors and normal samples in the different tumor types. FAIM2 = Fas apoptotic inhibitory molecule 2, MB = medulloblastoma.
3.4. Mutation of FAIM2 might contribute to its aberrant expression in metastatic MB
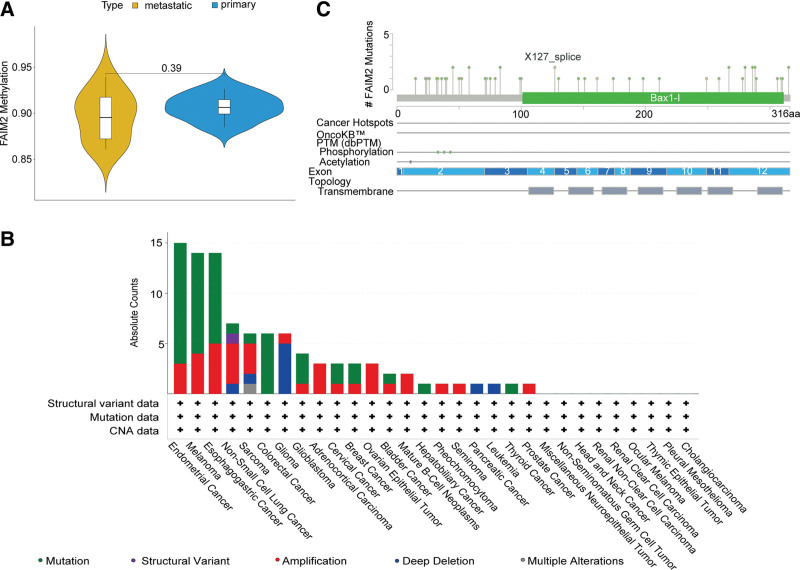
To explore why FAIM2 was aberrantly expressed in metastasis of MB, we analyzed its methylation and mutation status in MB. The methylation level of FAIM2 was not significantly different in metastatic and nonmetastatic samples (Fig. 4A). In addition, we analyzed the mutation status of FAIM2 in metastatic samples and found that FAIM2 harbored missense mutations, amplifications and deep deletions in 30 tumor datasets (Fig. 4B), and mainly focused on missense mutations (Fig. 4C). The results above demonstrated that FAIM2 might be aberrantly expressed in metastatic MB owing to its mutation.
Figure 4.
Methylation and mutation of FAIM2 in MB. (A) Methylation of FAIM2 in metastatic and nonmetastatic samples. (B) Mutation of FAIM2 in 30 tumor datasets. (C) Mutation information of FAIM2. FAIM2 = Fas apoptotic inhibitory molecule 2, MB = medulloblastoma.
3.5. Potential pathway of FAIM2 affecting MB metastasis
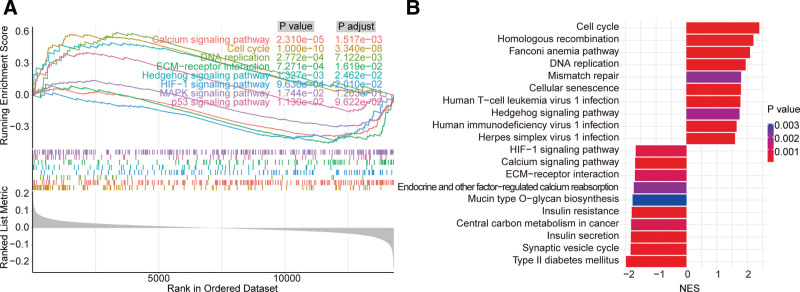
GSEA enrichment analysis found that a total of 25 pathways were significantly activated and 41 pathways were significantly inhibited in FAIM2 high expression group compared with FAIM2 low expression group in MB patients. Among which calcium signaling pathway, cell cycle, DNA replication, ECM-receptor interaction, hedgehog signaling pathway, HIF-1 signaling pathway, MAPK signaling pathway and p53 signaling pathway (Fig. 5A) were associated with cancer cell proliferation, angiogenesis and migration. The top 10 activated and inhibited pathways were showed in Figure 5B. All detailed results of pathways were showed in (Table S3, Supplemental Digital Content, http://links.lww.com/MD/I843).
Figure 5.
Potential pathway of FAIM2 affecting MB metastasis. (A) Signaling pathway associated with cancer cell proliferation, angiogenesis and migration. (B) Top 10 pathways of KEGG with significantly activated and inhibited. FAIM2 = Fas apoptotic inhibitory molecule 2, KEGG = Kyoto encyclopedia of genes and genomes, MB = medulloblastoma.
4. Discussion
Tumor metastasis is crucial factor involved in the prognosis of MB patients.[20] In our study, we collected and organized data in gene expression omnibus database to evaluate the genes related to metastasis in MB. We found that FAIM2 was tightly correlated with metastasis in MB patients, and the lower FAIM2 expression was associated with poor prognosis of MB patients.
Firstly, we identified that 196 DEGs between metastatic and nonmetastatic samples in MB patients, and these genes were significantly enriched in 483 GO terms and 29 KEGG pathways. In addition, univariate Cox regression analysis screened the top 10 genes (CEMIP, GLCE, ART3, GABRA5, COLEC12, LIN28B, ZNF521, IL17RB, FAIM2, RCBTB2) that were significantly associated with survival of MB, among which FAIM2 was prominently expressed in cerebral cortex, cerebellum and hippocampus, which is consistent with the common metastatic location of MB in the brain.[9] FAIM2 is an antiapoptotic protein, and the main biological function is to inhibit Fas-induced cell death.[12] In osteosarcoma samples, the closer to tumor blood vessels, the higher expression of FAIM2 is.[14] The higher expression of FAIM2 was associated with poor prognosis in Non-small cell lung cancer.[15] However, in NBL, the low level of FAIM2 has been reported to alter adhesion properties and increase metastatic potential of NBL cells, and the low level of FAIM2 was correlated with worse OS.[16] Hence, it is reasonable to observe the lower expression of FAIM2 was concerned with a poor prognosis in MB patients. These evidences suggest that FAIM2 plays distinct roles in different tumors due to the complexity of the tumor. Moreover, Liu et al[21] demonstrated that FAIM2 is a hub gene in MB, but the detailed role of FAIM2 in MB has not been investigated. Whereas, in our present work, we found that the expression of FAIM2 was decreased in MB patients, and the expression of FAIM2 in other brain tumors (glioblastoma and low-grade gliomas) were consistent with MB. These results indicated that the expression of FAIM2 was decreased in MB patients. We also found that FAIM2 harbored missense mutations, amplifications and deep deletions in 30 MB datasets. Therefore, FAIM2 might be aberrantly expressed in metastatic MB through mutation. We have firstly reported the impact of FAIM2 expression in metastasis of MB, and provide reference information for the treatment of MB metastasis in the future.
In addition, GSEA enrichment analysis found that a total of 25 pathways were significantly activated and 41 pathways were significantly inhibited in FAIM2 high expression group compared to FAIM2 low expression group in MB patients. Among which some signaling pathway were associated with cancer cell proliferation, angiogenesis and migration, such as cell cycle and calcium signaling pathway. FAIM2 was involved in calcium signaling regulation in the endoplasmic reticulum. FAIM2 reduces cytosolic Ca2+ by decreased calcium content in the endoplasmic reticulum,[22] and the overexpression of FAIM2 could reduce calcium release from the endoplasmic reticulum and mitochondrial cytochrome c release after FasL stimulation.[23] In addition, Ca2+ signaling plays a pivotal role in regulating cell cycle progression in tumor pathologies by controlling Ca2+ concentration in endoplasmic reticulum and mitochondria.[24,25] Moreover, previous studies have demonstrated that FAIM2 plays an important role in proliferation and metastasis of cancer cells. The low expression of FAIM2 could reduce cell adhesion and increase sphere growth and migration in NBL, and increased the metastatic capacity,[16] and the overexpression of FAIM2 can improve the proliferation migration and invasion of lung cancer cell.[26] These evidences suggested that FAIM2 might affect cancer cell proliferation, angiogenesis and migration by regulating calcium signaling pathway. Interestingly, we found that cell cycle was significantly activated and calcium signaling pathway was significantly inhibited in the FAIM2 high expression group compared to the low expression group. Therefore, we hypothesized that FAIM2 might affect MB metastasis through regulated calcium signaling pathway, which warrants further exploration in the future studies.
5. Conclusions
In present study, we have firstly reported the role of FAIM2 in MB patients. The results indicated that FAIM2 was tightly correlated with metastasis in MB patients, and the lower expression FAIM2 was associated with poor prognosis. Our results provide more reference information for targeted therapy of MB patients with metastasis in the future.
Author contributions
Conceptualization: Xiaojun Zhou.
Data curation: Xiaojun Zhou, Hao Zhao.
Formal analysis: Xiaojun Zhou.
Validation: Hao Zhao.
Writing – original draft: Xiaojun Zhou, Hao Zhao.
Supplementary Material
Abbreviations:
- CNV
- copy number variation
- DEGs
- differentially expressed genes
- FAIM2 =
- Fas apoptotic inhibitory molecule 2
- GO
- gene ontology
- GSEA
- gene set enrichment analysis
- KEGG
- Kyoto encyclopedia of genes and genomes
- MB
- medulloblastoma
- NBL
- neuroblastoma
- OS
- overall survival
Supplemental Digital Content is available for this article.
The datasets generated during and/or analyzed during the current study are publicly available.
The authors have no funding and conflicts of interest to disclose.
How to cite this article: Zhou X, Zhao H. FAIM2 is correlated with metastasis of medulloblastoma through bioinformatics analysis. Medicine 2023;102:16(e33591).
References
- [1].Northcott PA, Robinson GW, Kratz CP, et al. Medulloblastoma. Nat Rev Dis Primers. 2019;5:11. [DOI] [PubMed] [Google Scholar]
- [2].Ostrom QT, Gittleman H, Truitt G, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011-2015. Neuro Oncol. 2018;20(suppl_4):iv1–iv86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nasir A, Cardall A, Othman RT, et al. ABCB1 inhibition provides a novel therapeutic target to block TWIST1-induced migration in medulloblastoma. Neurooncol Adv. 2021;3:vdab030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Merchant TE, Kiehna EN, Li C, et al. Modeling radiation dosimetry to predict cognitive outcomes in pediatric patients with CNS embryonal tumors including medulloblastoma. Int J Radiat Oncol Biol Phys. 2006;65:210–21. [DOI] [PubMed] [Google Scholar]
- [5].Neglia JP, Robison LL, Stovall M, et al. New primary neoplasms of the central nervous system in survivors of childhood cancer: a report from the childhood cancer survivor study. J Natl Cancer Inst. 2006;98:1528–37. [DOI] [PubMed] [Google Scholar]
- [6].Van Ommeren R, Garzia L, Holgado BL, et al. The molecular biology of medulloblastoma metastasis. Brain Pathol. 2020;30:691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Blomstrand M, Brodin NP, Munck Af Rosenschold P, et al. Estimated clinical benefit of protecting neurogenesis in the developing brain during radiation therapy for pediatric medulloblastoma. Neuro Oncol. 2012;14:882–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kondoff SI, Milev MD, Laleva LN, et al. A case of early extraneural medulloblastoma metastases in a young adult. Asian J Neurosurg. 2015;10:331–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rickert CH. Extraneural metastases of paediatric brain tumours. Acta Neuropathol. 2003;105:309–27. [DOI] [PubMed] [Google Scholar]
- [10].das Chagas PF, de Sousa GR, Veronez LC, et al. Identification of ITPR1 as a hub gene of group 3 medulloblastoma and coregulated genes with potential prognostic values. J Mol Neurosci. 2022;72:633–41. [DOI] [PubMed] [Google Scholar]
- [11].Hong CJ, Yeon J, Yeo BK, et al. Fas-apoptotic inhibitory molecule 2 localizes to the lysosome and facilitates autophagosome-lysosome fusion through the LC3 interaction region motif-dependent interaction with LC3. FASEB J. 2020;34:161–79. [DOI] [PubMed] [Google Scholar]
- [12].Planells-Ferrer L, Urresti J, Coccia E, et al. Fas apoptosis inhibitory molecules: more than death-receptor antagonists in the nervous system. J Neurochem. 2016;139:11–21. [DOI] [PubMed] [Google Scholar]
- [13].Fernandez M, Segura MF, Sole C, et al. Lifeguard/neuronal membrane protein 35 regulates Fas ligand-mediated apoptosis in neurons via microdomain recruitment. J Neurochem. 2007;103:190–203. [DOI] [PubMed] [Google Scholar]
- [14].Pan Y, Zhang Y, Tang W, et al. Interstitial serum albumin empowers osteosarcoma cells with FAIM2 transcription to obtain viability via dedifferentiation. In Vitro Cell Dev Biol Anim. 2020;56:129–44. [DOI] [PubMed] [Google Scholar]
- [15].She K, Yang W, Li M, et al. FAIM2 promotes non-small cell lung cancer cell growth and bone metastasis by activating the WNT/beta-catenin pathway. Front Oncol. 2021;11:690142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Planells-Ferrer L, Urresti J, Soriano A, et al. MYCN repression of lifeguard/FAIM2 enhances neuroblastoma aggressiveness. Cell Death Dis. 2014;5:e1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Varan A, Sari N, Akalan N, et al. Extraneural metastasis in intracranial tumors in children: the experience of a single center. J Neurooncol. 2006;79:187–90. [DOI] [PubMed] [Google Scholar]
- [21].Liu Z, Zhang R, Sun Z, et al. Identification of hub genes and small-molecule compounds in medulloblastoma by integrated bioinformatic analyses. PeerJ. 2020;8:e8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lisak DA, Schacht T, Enders V, et al. The transmembrane bax inhibitor motif (TMBIM) containing protein family: tissue expression, intracellular localization and effects on the ER CA (2) (+)-filling state. Biochim Biophys Acta. 2015;1853:2104–14. [DOI] [PubMed] [Google Scholar]
- [23].Urresti J, Ruiz-Meana M, Coccia E, et al. Lifeguard inhibits fas ligand-mediated endoplasmic reticulum-calcium release mandatory for apoptosis in type II apoptotic cells. J Biol Chem. 2016;291:1221–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Marchi S, Giorgi C, Galluzzi L, et al. Ca (2+) fluxes and cancer. Mol Cell. 2020;78:1055–69. [DOI] [PubMed] [Google Scholar]
- [25].Patergnani S, Danese A, Bouhamida E, et al. Various aspects of calcium signaling in the regulation of apoptosis, autophagy, cell proliferation, and cancer. Int J Mol Sci. 2020;21:8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].She K, Yan H, Huang J, et al. miR-193b availability is antagonized by LncRNA-SNHG7 for FAIM2-induced tumour progression in non-small cell lung cancer. Cell Prolif. 2018;51:e12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.