Abstract
The androgen receptor is a key regulator of prostate cancer and the principal target of current prostate cancer therapies collectively termed androgen deprivation therapies. Insensitivity to these drugs is a hallmark of progression to a terminal disease state termed castration-resistant prostate cancer. Therefore, novel therapeutic options that slow progression of castration-resistant prostate cancer and combine effectively with existing agents are in urgent need. We show that JG-98, an allosteric inhibitor of HSP70, re-sensitizes castration-resistant prostate cancer to androgen deprivation drugs by targeting mitochondrial HSP70 (HSPA9) to suppress aerobic respiration. Rather than impacting androgen receptor stability as previously described, JG-98's primary effect is inhibition of mitochondrial translation, leading to disruption of electron transport chain activity. Although functionally distinct from HSPA9 inhibition, direct inhibition of the electron transport chain with a complex I or II inhibitor creates a similar physiological state capable of re-sensitizing castration-resistant prostate cancer to androgen deprivation therapies. These data identify a significant role for HspA9 in mitochondrial ribosome function and highlight an actionable metabolic vulnerability of castration-resistant prostate cancer.
Significance Statement.
Prostate cancer (PCa) is the second leading cause of cancer-related deaths among American men due to developing resistance against androgen receptor (AR) targeting therapies. PCa sensitivity to HSP70 inhibitors is attributed to AR dependence on HSP70. Here, we show low dose HSP70 inhibition in the presence of AR re-sensitizes castration-resistant prostate cancer (CRPC) to androgen signaling inhibitors. PCa vulnerability to HSP70 inhibition is due to unique mitoribosome dependence on mitochondrial HSP70 (HSPA9). Suppression of mitochondrial translation impacts aerobic respiration as mitoribosomes synthesize 13 integral electron transport chain (ETC) subunits; specific inhibition of ETC complex I or II also re-sensitizes CRPC to AR inhibitors. Our work uncovers a significant HSPA9 role in mitoribosome stability and highlights an actionable metabolic CRPC vulnerability.
Introduction
Maintenance of cellular proteostasis is an energetically costly process that is intimately associated with metabolism. Collapse of redox balance or ATP homeostasis activates pathways that suppress protein synthesis and promote protein turnover by autophagy and proteasomal degradation (1). Not surprisingly, the altered proliferation rate that accompanies oncogenic transformation often places a burden on both proteostasis and metabolism (2). Cancer cells therefore alter their metabolism to accommodate these changes and become increasingly dependent on chaperones that stabilize their stressed proteome (3). For this reason, therapeutic strategies that target these changes have become attractive avenues for discovery of new drugs that combat cancer progression.
Prostate cancer (PCa) is an androgen-dependent disease that requires the nuclear receptor androgen receptor (AR) to promote progression. Therefore, suppressing both circulating testosterone or AR activity (collectively referred to androgen-deprivation therapy—ADT) is the primary treatment strategy for both early and advanced stages of PCa (4). While initially effective, PCa often progresses to a state termed castration-resistant prostate cancer (CRPC) by bypassing the natural androgen transcriptional activities inhibited by ADT. This commonly occurs due to gene duplication of full-length (FL) AR or the production of constitutively active alternate splice variants of AR that no longer rely on the hormone binding domain to regulate AR transcription (5). Once the cancer has progressed to this state, PCa rapidly becomes a terminal disease resistant to ADT.
Alternative strategies that target AR stability have become a popular avenue of exploration in CRPC. Because AR signaling often remains important in CRPC, many studies have targeted HSP70 and HSP90 due to the well-documented dependence of AR on these molecular chaperones (6–8). Recent work from our group has shown that the allosteric HSP70 inhibitor JG-98 potently suppresses growth of CRPC cells in vitro and in vivo (7). This is accomplished through HSP70 binding to the AR unstructured N-terminal domain which is retained in constitutively active AR splice variants (9). Micromolar concentrations of JG-98 are therefore capable of decreasing the protein stability of all versions of AR, providing a promising therapeutic strategy to treat CRPC (7).
Rapid cell division, subsequent nutrient depletion, and proliferation in foreign sites of the body all put substantial stress on a tumor's natural metabolic program. Therefore, oncogenic transformation is often accompanied by a fundamental re-wiring of metabolism. In the case of PCa, this results in removal of the Zn2+ block on the TCA cycle required for citrate secretion into the seminal fluid in normal prostatic epithelium and subsequent transition from glycolysis to oxidative phosphorylation (OXPHOS) as the preferred energetic pathway (3). This metabolic shift likely puts substantial pressure on mitochondrial proteostasis as PCa metabolism relies more heavily on the electron transport chain (ETC) and OXPHOS compared with the normal prostate.
Mitochondrial proteostasis is complicated by many unique aspects of this organelle's structure and biology. First, mitochondria contain a vestigial genome that encodes ribosomal RNAs, tRNAs, and 13 protein-coding genes that generate integral subunits of the ETC. Thus, the overwhelming majority of the >1,000 mitochondrial proteins are encoded by the nucleus, translated in the cytosol, and translocated into mitochondria. Furthermore, imported proteins are transported across the outer and inner membranes in a largely unfolded state and require refolding upon arrival at their destination (10). This presents a particularly unique problem for the ordered assembly of large complexes such as the mitochondrial ribosome (mitoribosome) which contain approximately 80 subunits. Recent studies have shown that human mitochondrial ribosomal proteins (MRPs) are translocated into the mitochondrial matrix at levels that exceed what is required for mitoribosome assembly and are assembled in distinct modules in an orderly fashion (11). Therefore, any physiological or pharmacological condition that disrupts MRP translocation, folding, or assembly has a significant possibility of disrupting mitochondrial translation, respiration, and proteostasis.
In this study, we show that the mitoribosome is hypersensitive to the HSP70 inhibitor JG-98 due to the inhibitor's ability to disrupt mitochondrial HSP70 (HSPA9) activity. Our data show that suppression of mitochondrial translation by JG-98 disrupts the ETC in multiple cell types. Importantly, this activity is observed at concentrations far lower than the doses required to disrupt AR protein levels which, in the special case of CRPC, allows for secondary targeting of full-length AR activities with ADT drugs (7). Furthermore, specific inhibition of ETC complexes I and II, although mechanistically distinct from JG-98, provides an orthogonal confirmation of the impact HSP70 inhibition has on CRPC both in vitro and in vivo.
Results
Inhibition of HSP70 displays potent single-agent activity in CRPC while sensitizing cells to ADT
Treatment of 22Rv1 cells with high doses of HSP70 (JG-98) or HSP40 (C86) inhibitors promotes proteasome-mediated clearance of both full length and truncated forms of AR (7). Given the ability of JG-98 and C86 to selectively decrease variants of AR known to promote resistance to ADT, we asked whether treatment of the CRPC cell line 22Rv1 with JG-98 or C86 can synergize with otherwise ineffective concentrations of the ADT drugs enzalutamide or abiraterone (Fig. 1A and B and Fig. S1B-E). We used the HSP90 inhibitor ganetespib (STA-9090) as a control to distinguish effects due to loss of full-length androgen receptor (AR-FL) while retaining AR-v7 (a constitutively active splice variant lacking the ligand binding domain), as HSP90 inhibition is only able to promote degradation of AR-FL protein (7, 8). Only JG-98 was capable of re-sensitizing 22Rv1 to enzalutamide or abiraterone while the inactive analog JG-258 had minimal impact when combined with ADT drugs (Fig. 1B, Fig. S1A and B).
Fig. 1.
Inhibition of Hsp70 by JG-98 re-sensitizes CRPC cells to ADT drugs in vitro and in vivo. A) Incucyte growth analysis of 22Rv1 CRPC cells in the presence of JG-98. B) Incucyte growth analysis of 22Rv1 CRPC cells in the presence of 150 nM JG-98 +/− 10 µM enzalutamide or abiraterone. C) Mean tumor volume (± SEM) of 22Rv1 xenografts in mice treated with vehicle (n = 7), JG-231 (6 mg/kg; n = 7), enzalutamide (ENZ) (25 mg/kg; n = 7), or combo (n = 7). D) Mean tumor volume (±SD) in mice treated with JG-231 or combo at day 37 (post-22Rv1 injection). Mice were removed from the study when tumor volume surpassed 1,800 mm3. Statistical significance determined by one-tailed unpaired t test.
To confirm the in vitro cell growth data obtained with JG-98, we tested whether HSP70 inhibition with JG-231 (a JG-98 analog with improved in vivo properties) can sensitize 22Rv1 xenografts to treatment with enzalutamide. Treatment with JG-231 alone had a modest impact on growth in vivo, similar to published results (Figs. 1C and D and S1F) (7). However, the addition of enzalutamide to the JG-231 treatment cycle was able to further impede 22Rv1 xenograft growth, despite its having no impact when administered as a single agent (Fig. 1C and D, and Fig. S1F). These findings highlight the potential benefit of targeting HSP70 in vivo to re-sensitize CRPC to ADT drugs.
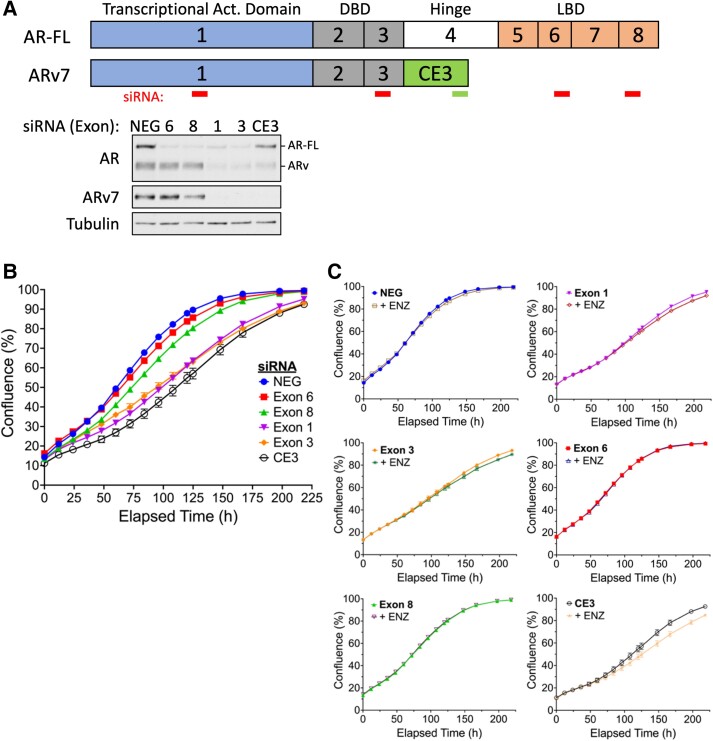
Enzalutamide sensitivity requires an intact, hormone-responsive AR signaling network. The emergence of alternate splice variants that are no longer hormone regulated is predicted to promote ADT resistance. Our initial hypothesis was that JG-98 and C86 would be able to re-sensitize CRPC cells to ADT by decreasing the impact ADT-resistant AR splice variants have on CRPC growth. However, numerous lines of evidence began to suggest that manipulation of AR protein levels was not related to JG-98's ability to re-sensitize CRPC to ADT. First, concentrations of JG-98 necessary to promote ADT-sensitivity had no impact on AR protein levels over a 4-day period (Fig. S1G). Second, C86 has a comparable effect on AR-FL and AR splice variant stability but was unable to promote ADT sensitivity (Fig. S1C, right panel) (7). And finally, C4-2 cells that lack AR splice variants also displayed ADT sensitivity when combined with low concentrations of JG-98 (Fig. S1E). To verify that manipulation of AR protein levels was not capable of re-sensitizing 22Rv1 to ADT observed with low concentrations of JG-98, we used an siRNA knockdown approach to eliminate specific populations of AR proteins (Fig. 2A). Depletion of AR-FL and/or AR splice variants with RNAi all resulted in a reduced growth rate but reached a similar confluence within 9 days (Fig. 2B). Importantly, no AR KD condition (especially ARv7) was capable of re-sensitizing 22Rv1 to enzalutamide, suggesting that manipulation of AR splice variant protein levels is not likely to be an effective strategy to promote ADT sensitivity in CRPC (Fig. 2C). These data collectively indicate that JG-98 re-sensitization to ADT is unrelated to its ability to decrease AR protein stability which occurs at much higher doses than those used in our Incucyte growth experiments.
Fig. 2.
siRNA-mediated knock down of AR-FL and/or AR splice variants in 22Rv1 cells does not support ADT re-sensitization. A) Map of siRNA complementarity (top); western blot analysis of 22Rv1 lysate transfected with respective siRNAs 24 h posttransfection (bottom). B) Incucyte growth analysis of 22Rv1 transfected with respective AR-specific siRNAs. C) Incucyte growth analysis of 22Rv1 measuring AR-KD impact on enzalutamide sensitivity.
JG-98 associates with mitochondria and suppresses aerobic respiration of CRPC cells
MKT-077, the HSP70 inhibitor from which the JG-series of allosteric inhibitors were developed, has been observed to specifically localize to mitochondria due to its cationic properties (7, 12). We tested whether JG-98 might also localize to mitochondria. Using HEK-293 cells, we assessed JG-98 physical association with isolated mitochondria by exploiting the compound's fluorescent characteristics. To distinguish between JG-98 binding to cytosolic HSC70 proteins associated with the outer mitochondrial membrane, we used proteinase K (PK) to strip mitochondrial surface proteins and to assess membrane-protected association of JG-98. Treatment of isolated mitochondria with PK resulted in loss of the outer mitochondrial membrane protein TOM20 without affecting matrix-associated HSP60, indicating our treatment stripped the outer mitochondrial membrane surface exposed proteins without disrupting mitochondrial integrity (Fig. S6B). PK treatment was only able to decrease JG-98 fluorescence by 30–40% indicating that a significant portion of mitochondrial-associated JG-98 resides within the matrix or intermembrane space (Fig. 3A).
Fig. 3.
JG-98 localizes to mitochondria and shifts metabolic energy preference in 22Rv1 cells from mitochondrial respiration to glycolysis in vitro and in vivo. A) JG-98 association with mitochondria isolated from HEK293 cells. Cells were treated with 250 nM or 500 nM JG-98 for 24 h prior to isolation of mitochondria, and isolated mitochondria were then treated with 0.01 U proteinase K prior to assessment of JG-98 association. Statistical significance determined by Tukey's multiple comparisons test. B) Oxygen consumption rate (OCR) of 22Rv1 cells treated with JG-98 (top) or inactive analog JG-258 (bottom) for 24 h. C) Energy plot of 22Rv1 cells after 24 h treatment with 100 nM or 150 nM JG-98 or JG-258. D) TMRE assay measuring the impact of JG-98 or JG-258 (24 h treatment) on 22Rv1 mitochondrial membrane potential. Ten micromolar FCCP was used as a positive control. All data are normalized to untreated 22Rv1 and statistical significance determined by Dunnett's multiple comparisons test. E) Quantitative in vivo imaging of 22Rv1 xenografts by determining [1-13C] lactate:pyruvate ratio (top) or [1-13C] bicarbonate:pyruvate ratio (bottom) in mice treated as described (inset) with 8 mg/kg JG-231. Statistical significance determined by two-tailed paired t test. ns, not significant; *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Given the strong mitochondrial association of JG-98, we next asked if this HSP70 inhibitor could impact mitochondrial respiration. Pretreatment of 22Rv1 CRPC cells with either 100 nM or 150 nM JG-98 for 24 h significantly decreased both the basal and maximal oxygen consumption rates (OCR) and was accompanied by an increase in glycolytic activity measured by the extracellular acidification rate (ECAR) (Fig. 3B and C, and Fig. S2A). Notably, this impact was not observed at comparable concentrations of JG-258 (Fig. 3B and C, and Fig. S2A). Finally, JG-98 treatment at concentrations comparable to those effective in our growth assays (compare JG-98 concentrations in Figs. 1A and 3B) resulted in a 25% decline in mitochondrial membrane potential, indicating that the HSP70 inhibitor moderately impacts the activity of the proton-pumping components of the ETC (Fig. 3D). Importantly, impact on membrane potential was not observed with JG-258. The lack of effect of other chaperone inhibitors highlighted that this property was unique to inhibition of HSP70, as treatment with inhibitors of HSP40 (C86) or HSP90 (STA-9090) at concentrations much higher than necessary for growth suppression in CRPC models failed to affect 22Rv1 OCR (Fig. S2B). These data strongly suggest that JG-98 growth suppression of 22Rv1 cells is likely related to inhibition of mitochondrial respiration.
We next tested whether JG-98 might also suppress mitochondrial respiration in vivo. To assess this possibility, we monitored the metabolic fate of hyperpolarized (HP) [1-13C] pyruvate in 22Rv1 xenografts using hyperpolarized magnetic resonance spectroscopic imaging (HP-MRSI). This technique allows noninvasive monitoring of the conversion of [1-13C] pyruvate to lactate (glycolysis) or bicarbonate (TCA cycle/aerobic respiration) (see Fig. S2C for detectable pyruvate metabolism schema) in the same xenograft before and 24 h after treatment with JG-231 (13). Comparable to our in vitro results, treatment with JG-231 resulted in an increased [1-13C] lactate signal, indicating up-regulation of glycolysis (Fig. 3E and Fig. S2D). We regularly observed decreased conversion of pyruvate to bicarbonate in response to JG-231 treatment, consistent with JG-98-mediated decrease in mitochondrial OXPHOS observed in vitro (Fig. 3B, C and E). However, this change in [1-C13] pyruvate metabolism was not statistically significant, partly due to the well-known difficulty in detecting bicarbonate conversion in these assays (Fig. 3E, bottom graph and Fig. S2D). Collectively, our data strongly support the hypothesis that a primary consequence of HSP70 inhibition in vitro and in vivo with JG-98/231 is suppression of mitochondrial respiration in CRPC.
ETC inhibition with complex I or complex II selective inhibitors suppress CRPC growth and restore sensitivity to ADT
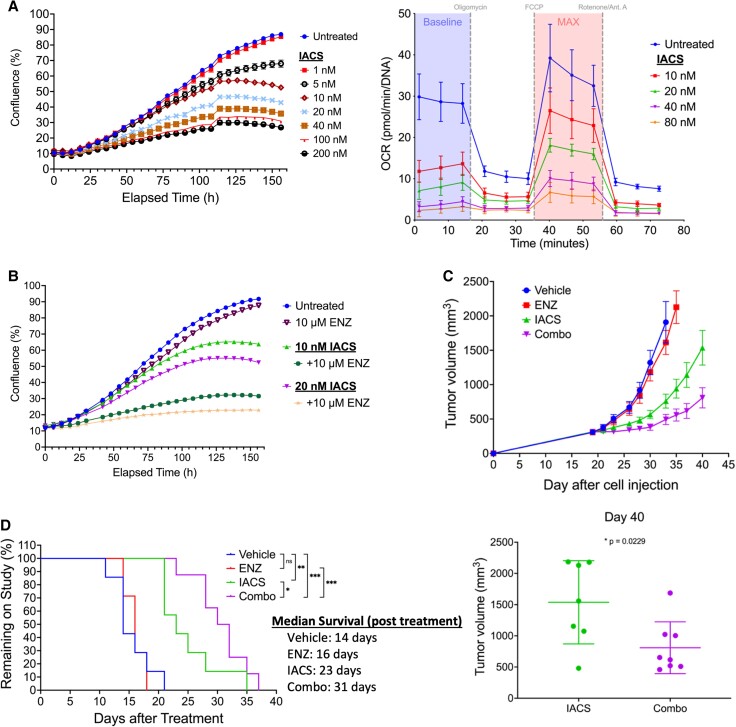
The impact of HSP70 inhibition on mitochondrial respiration led us to test whether directly targeting components of the ETC could phenocopy the metabolic effects of JG-98/231 in vitro and in vivo. To do this, we targeted the two electron entry points of the ETC (complex I and II) with therapeutically relevant inhibitors (14, 15). Treatment of 22Rv1 cells with complex I inhibitor (IACS-010759) (Fig. 4A and Fig. S3D) or complex II inhibitor (CNC-332) (Fig. S4A, B) each displayed strong growth suppression that correlated with decreased mitochondrial respiration and increased glycolysis. We repeated this experiment in C4-2 cells to test the reproducibility of this drug combination in another CRPC model over-expressing AR-FL only. While IACS alone had only a modest growth suppressive activity in C4-2 cells, combination with enzalutamide displayed a stronger effect (Fig. S3C). Both IACS-010759 and CNC-332 sensitized 22Rv1 in vitro to treatment with either enzalutamide or abiraterone (Fig. 4B, Fig. S3A-C, S4C, D), consistent with the hypothesis that the mechanistic basis of JG-98 re-sensitization to ADT drugs involves inhibition of ETC function.
Fig. 4.
ETC complex I inhibition sensitizes CRPC cells to ADT drugs. A) Incucyte growth analysis (left) or OCR response (right—24 h treatment) of 22Rv1 cells in the presence of increasing concentrations of IACS-010759. B) Incucyte growth analysis of 22Rv1 cells grown in the presence of 10 nM or 20 nM IACS-010759 +/− 10 µM enzalutamide. C).22Rv1 xenograft tumor growth represented as mean tumor volume ± SEM in mice treated with vehicle (n = 7), enzalutamide (25 mg/kg; n = 7), IACS-010759 (7.5 mg/kg; n = 7), or combo (n = 8). Mean tumor volume +/− SD at day 40 post-22Rv1 cell injection (right) in mice treated with IACS-010759 or combo. D) Durability of response of 22Rv1 xenografts in mice treated with vehicle, enzalutamide only, IACS-010759, or combo. Mice were removed from the study when tumor volume surpassed 1800 mm3; data presented as days after 22Rv1 cell injection. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001 using Gehan Breslow–Wilcoxon test or two-tailed unpaired t test.
To confirm the in vivo efficacy of IACS-010759/ENZ co-treatment, we tested their impact on growth suppression of 22Rv1 xenografts. IACS-010759 treatment alone was effective at slowing tumor growth by nearly 65% compared with vehicle-treated mice (Fig. 4C and D). Addition of enzalutamide to IACS-010759 increased the durability of the response, slowing 22Rv1 xenograft growth by an additional ∼35% (8 days) compared with IACS-010759 alone and by an additional 120% (17 days) compared with vehicle treated mice (Fig. 4C and D). Importantly, this treatment regimen was well-tolerated and caused no significant change in body weight (Fig. S5). These data are in general agreement with a recent in vitro study showing that circulating tumor cells (CTCs) isolated from 6 of 20 PCa patients responsive to enzalutamide displayed higher OXPHOS compared with the CTCs isolated from the remaining 14 patients. Furthermore, the six patients whose CTCs had higher OXPHOS developed resistance to enzalutamide less than 6 months after CTC collection (16). Comparison of these results to the efficacy of JG-98/ENZ co-treatment highlights the therapeutic benefit of adding complex I (or other ETC) inhibitors to extend the efficacy of ADT in ADT-resistant CRPC.
Acute inhibition of HSPA9 with JG-98 suppresses mitochondrial translation by targeting the mitoribosome
HSP70s promote the general folding of a significant portion of nascent and unfolded cytosolic peptides. Therefore, we sought to identify the JG-98-hypersensitive portion of the 22Rv1 proteome to uncover the most proximal JG-98 impact responsible for induction of mitochondrial disfunction. To do this, we employed tandem mass tag mass spectrometry (TMT-MS) which provides quantitative comparisons of protein samples from differentially treated cells. We treated 22Rv1 cells with 5 µM JG-98 for 4 h to maximize early detection of the proteomic impact of JG-98 while minimizing secondary cellular responses to HSP70 inhibition which may occur with prolonged exposure to the drug.
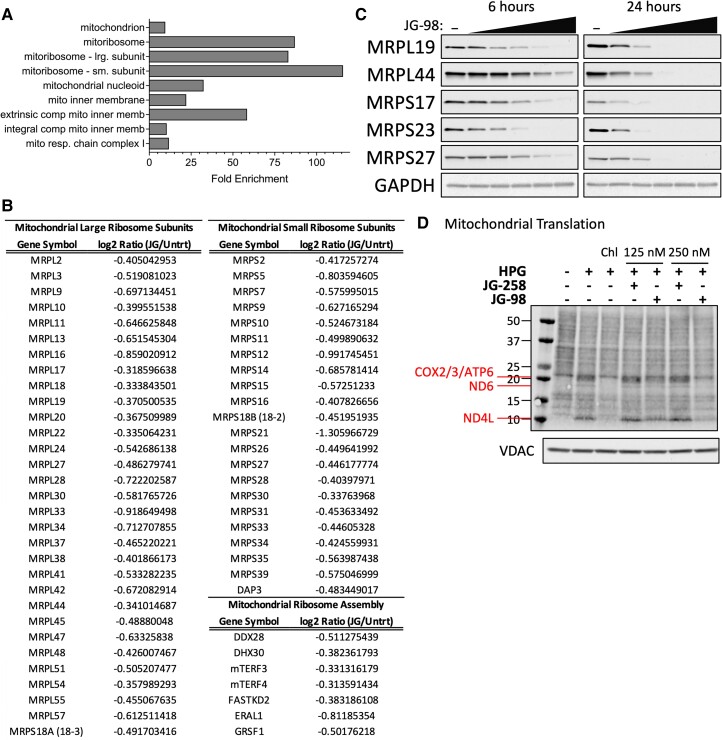
Collectively, 179 proteins were differentially affected (change ≥ 25%) compared with untreated cells. Approximately 80% (141/179) of the differentially detected proteins were negatively impacted by short-term JG-98 treatment indicating that we successfully captured the acute proteomic response to HSP70 inhibition (Table S1). Gene Ontology (GO) analysis of proteins that decrease in response to acute JG-98 showed that this data set was significantly enriched in mitochondrial proteins (Fig. 5A). Additionally, the JG-98 sensitive proteome was heavily enriched in MRPs comprising both large and small mitoribosome subunits including numerous proteins involved in mitoribosome assembly (Fig. 5B). Western blot analysis of 22Rv1 cells treated with JG-98 validated the TMT-MS results, confirming that acute JG-98 exposure had a strong destabilizing effect on all tested MRPs regardless of their presence in our TMT-MS data, indicating that HSP70 promotes the stability of mitoribosomal subunits (Fig. 5C).
Fig. 5.
JG-98 selectively decreases protein levels of mitochondrial ribosomal subunits. A) GO Cell Component enrichment analysis of most strongly down-regulated proteins in 22Rv1 cells after 4 h treatment with 5 µM JG-98. B) Table of mitochondrial ribosomal subunits (MRPs) and assembly factors down-regulated (≥ 25%) due to JG-98 treatment. C) Western blot analysis of select MRPs from 22Rv1 whole cell extract following treatment of intact cells with increasing concentrations of JG-98 (0, 1, 2.5, 5, 7.5, or 10 µM) for 6 h (left) or 24 h (right). D) Mitochondrial protein translation assay using mitochondria isolated from 22Rv1 cells treated with 125 nM or 250 nM JG-98 or inactive analog (JG-258) for 24 h. Detection of HPG incorporation into newly synthesized mitochondrial proteins (top panel); western blot detection of VDAC (bottom panel, load control).
Mitochondria are unique organelles that contain their own genome which encodes for only 13 protein-coding genes that are each essential subunits of ETC complexes (seven of which are subunits of complex I). We hypothesized that JG-98 impact on assembly of the highly complex multi-subunit mitoribosome would negatively affect the translation of mitochondrially synthesized proteins with a subsequent deleterious effect on aerobic respiration. To test this, we monitored mitochondrial translation by incorporation of the methionine analog, L-homopropargylglycine (HPG) which contains an alkyne moiety that allows for detection by click chemistry (17). In all mitochondrial translation assays, cytosolic translation was inhibited with harringtonine and mitochondrial translation was inhibited as indicated with chloramphenicol (17). Twenty-four-hour treatment with 150 or 250 nM JG-98 strongly decreased detection of mitochondrially encoded proteins, an activity that was absent following equivalent treatment with JG-258 (Fig. 5D).
JG-98 impacts MRP stability independent of cytosolic HSP70's impact on TOMs/TIMs
Cytosolic HSP70s promote the translocation of proteins across the endoplasmic reticulum and mitochondrial membranes. In the case of mitochondria, they are known to promote translocation of integral membrane proteins such as components of the inner and outer membrane pore complexes (10). To ensure MRP instability was not due to inhibition of their mitochondrial translocation, we isolated mitochondria from cells treated with JG-98 for 0–48 h and compared the impact on MRPs to that of components of the outer membrane (TOMs) and inner membrane (TIMs) pore complexes. We also tested the stability of TIM44, a member of the PAM complex which cooperates with HSPA9 to import translocating proteins into the matrix. JG-98 impact on MRP levels preceded any observable changes in TIM44 or the TOM/TIM complex proteins, indicating changes in MRP stability are independent of defects in mitochondrial import of cytosolically translated proteins (Fig. S6A).
JG-98 impacts MRP solubility by targeting HSPA9
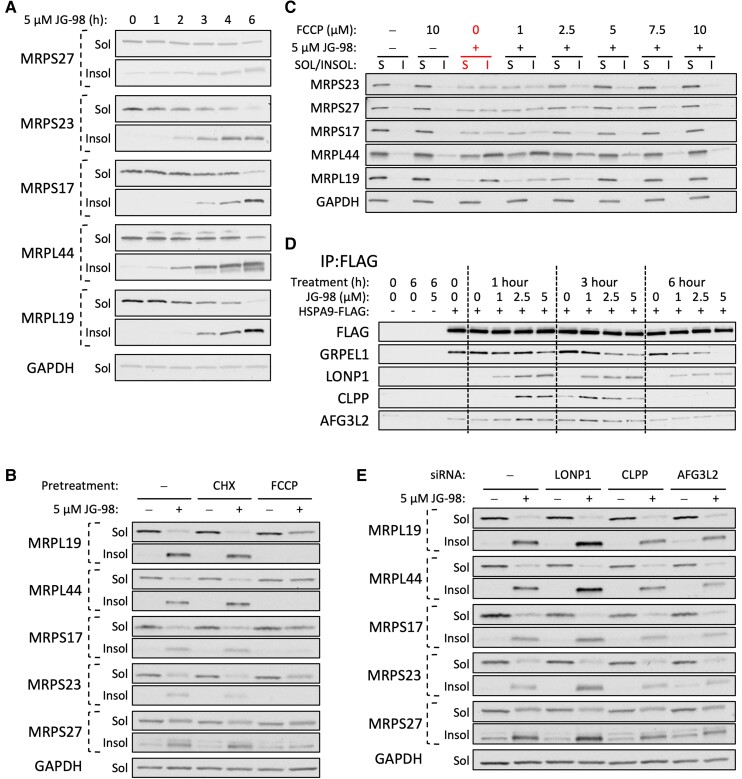
Mitoribosome assembly is a complicated process reliant on the coordinated cytosolic synthesis of over 80 subunits, translocation across the outer and inner membranes, and subsequent folding and assembly in the mitochondrial matrix (11, 18). Therefore, we monitored JG-98 impact on MRP aggregation as a measure of disruption of folding and/or protein stability. Treatment of 22Rv1 cells with 5 µM JG-98 for 1–6 h resulted in a time-dependent increase in insolubility of all MRPs tested that correlated with a loss of MRPs in the soluble fraction, indicating that MRP misfolding likely plays a significant role in JG-98 impact on the mitoribosome (Fig. 6A).
Fig. 6.
JG-98 targets mitochondrial HSP70 (HSPA9) to impact MRP stability and aggregation. A) Western blot of MRP protein solubility/insolubility in 22Rv1 cells treated with 5 µM JG-98 for 0, 1, 2, 3, 4, or 6 h. B) Western blot analysis of MRP solubility/insolubility in 22Rv1 cells pretreated with 10 µg/mL cycloheximide (CHX) or 10 µM FCCP +/− 5 µM JG-98 (3 h). C) FCCP impact on JG-98 induced MRP aggregation in 22Rv1 cells treated for 3 h. D) Anti-FLAG immunoprecipitation (IP) from HEK293 cells transfected with empty vector (EV) or HSPA9-FLAG. Cells were treated for 1, 3, or 6 h with 0, 1, 2.5, or 5 µM JG-98 prior to lysis and IP. GRPEL1 is a mitochondrial BAG/NEF; LONP1, CLPP, and AFG3L2 are mitochondrial matrix proteases. E) Impact of LONP1-, CLPP-, or AFG3L2-KD on JG-98-induced aggregation of MRP subunits (3 h treatment).
Mitochondrial translocation of matrix proteins commonly relies on positively charged N-terminal presequences that depend on an intact mitochondrial membrane potential to drive proteins through the TIM23 complex (19, 20). Furthermore, presequence translocation can be experimentally suppressed with FCCP treatment which collapses the mitochondrial proton gradient (18). We hypothesized that decreasing the influx of unfolded proteins translocated into the mitochondrial matrix might buffer against the JG-98 induced aggregation of MRP subunits. To test this, we inhibited cytosolic translation with cycloheximide (CHX) or disrupted presequence translocation with FCCP and monitored changes in MRP aggregation. While CHX had no impact on JG-98-dependent MRP aggregation, FCCP treatment substantially decreased JG-98's influence on both soluble and insoluble MRP fractions (Fig. 6B). We also monitored the impact of TIM23-KD as an additional test of the role mitochondrial import in MRP aggregation. Loss of TIM23 only offered mild protection from JG-98-dependent MRP aggregation (Fig. S7A).
Since FCCP treatment was consistently the most effective means of reversing JG-98 impact on MRPs, we sought to better understand the mechanistic basis of this phenomenon. Mitochondrial localization of the cationic HSP70 inhibitor, MKT-077, is partially reliant on the weak negative charge within the matrix created by the mitochondrial membrane potential (12). Molecules like FCCP that disrupt the mitochondrial membrane potential by collapsing the proton gradient across the inner membrane may be able to reduce mitochondrial association of JG-98 (which retains the cationic characteristics of MKT-077). To distinguish the impact of FCCP on translocation of presequence proteins from its effect on JG-98 localization, we titrated FCCP into JG-98 treated 22Rv1 cells and monitored MRP solubility and mitochondrial retention of JG-98. Our results show that 3 h treatment of 5 µM FCCP (half the concentration used in Fig. 6B) could reverse the impact of JG-98 on MRP solubility (Fig. 6C). However, FCCP did not disrupt mitochondrial association of JG-98 under these conditions (Fig. S6C), indicating that FCCP-mediated reversal of MRP aggregation is likely due to translocation defects rather than altered mitochondrial localization of JG-98. Because disruption of mitochondrial translocation alone had no impact on MRP stability, we conclude JG-98 potently disrupts the folding capacity of the mitochondrial matrix.
JG-98 targets a conserved allosteric site adjacent to the HSP70 nucleotide binding domain and disrupts binding of nucleotide exchange factors (NEFs), collectively stabilizing the ADP-bound state of HSP70, preventing the release of client proteins, and halting the conformational cycle of the chaperone (6, 21, 22). To monitor JG-98 impact on the HSPA9 conformational cycle, we tested its ability to disrupt HSPA9 interaction with the mitochondria-specific NEF GRPEL1. Treatment of HEK-293 cells expressing FLAG-tagged HSPA9 with JG-98 resulted in a dose- and time-dependent decrease in GRPEL1 association with HSPA9 without affecting GRPEL1 protein stability (Fig. 6D and Fig. S7B). JG-98 also induced transient association with numerous mitochondrial matrix proteases, including LONP1, CLPP, and AFG3L2 (Fig. 6D). Both LONP1 and CLPP association with HSPA9 increased substantially within 1–3 h of JG-98 treatment but dissipated by 6 h. Our results indicate that JG-98 has a significant impact on mitochondrial proteostasis, suggesting that mitochondrial matrix proteases are recruited to HSPA9 to help clear damaged or misfolded proteins. To test this possibility, we monitored the impact of LONP1-, CLPP-, or AFG3L2-KD on JG-98 induced MRP aggregation. Loss of LONP1 increased the level of MRP aggregation in response to JG-98 treatment while KD of CLPP and AFG3L2 slightly decreased the level of MRP aggregation (Fig. 6E). The impact of LONP1-KD on JG-98-mediated aggregation combined with its recruitment to HSPA9 in response to JG-98 treatment indicates that LONP1 likely senses a buildup of unfolded proteins and associates with HSPA9 to promote proteasomal clearance.
Mitochondrial DNAJA3 promotes the solubility of MRPs and expands the MRP-interaction network of HSPA9
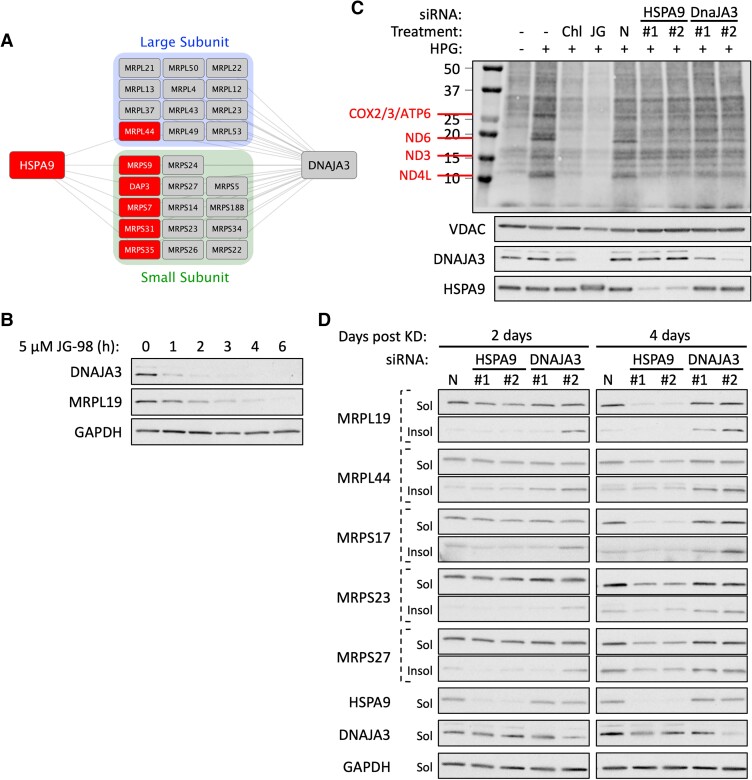
HSP40s (DNAJ proteins) are integral members of the HSP70 system capable of delivering proteins to HSP70 and stimulating HSP70 ATP hydrolysis, thereby converting HSP70 from a low to high affinity client binding state (23). DNAJ prevalence has also significantly expanded over evolutionary time to accommodate the increased complexity of HSP70 clientele in higher-order organisms (24). In human mitochondria, there are eight identified J-proteins. Our results have shown that JG-98 inhibition of HSPA9 destabilizes MRPs yet we were surprised to find that only 6 of the 80+ MRPs are known to interact with HSPA9 (25). We wondered if this discrepancy might be remedied by J-protein-mediated recruitment to HSPA9 and found that mitochondrial DNAJs interacted with 80% of the 78 MRPs cataloged in BioGrid, significantly expanding the network of mitoribosome proteins served by the HSPA9 system (Fig. S8) (25). Because DNAJA3 is well known to assist HSPA9 in folding of the mitochondrial matrix proteins, we analyzed its interaction network to better understand its contribution to MRP folding (20). We found that DNAJA3 interacts with 26 MRPs including the six known to interact with HspA9 (Fig. 7A) (25).
Fig. 7.
DNAJA3 expands HSPA9 MRP interactions and promotes the stability and functionality of the mitoribosome. A) Interaction network of HSPA9 and DNAJA3 with MRPs. B) Western blot analysis of DNAJA3 solubility/insolubility following treatment of 22Rv1 cells with 5 µM JG-98 (0–6 h). C) Mitochondrial protein translation assay using 22Rv1 cells 4 days post-transfection with two independent siRNAs directed towards HSPA9 or DNAJA3. Chloramphenicol (Chl) (mitochondria-specific translation inhibitor, 150 µg/ml) is administered 1 h prior to initiation of translation. Cells were pretreated with 500 nM JG-98 (24 h). Detection of HPG incorporation into newly synthesized mitochondrial proteins (top panel); western blot detection of VDAC (load control), and efficiency of DNAJA3/HSPA9 KD as shown (lower panels). D) Impact of siRNA-KD of HSPA9 or DNAJA3 on MRP solubility/insolubility in 22Rv1 cells 2 or 4 days posttransfection. N is negative control siRNA.
To better understand the role DNAJs might play in mitoribosome stability, we investigated DNAJA3 function in MRP stability and mitochondrial translation. As an initial test, we probed DNAJA3 protein levels in JG-98 treated 22Rv1 cells and observed a rapid decrease in the J protein that preceded the impact on MRPL19 (Fig. 7B). We therefore asked how KD of DNAJA3 or HSPA9 individually might contribute to JG-98 suppression of mitochondrial translation and MRP solubility. Surprisingly, neither RNAi treatment resulted in rapid reduction of mitoribosome translation efficiency or MRP stability and instead required between 2 and 4 days to observe significant defects in either process (Fig. 7C and D). Furthermore, DNAJA3 and HSPA9 KD each mimicked distinct activities of JG-98 treatment. While DNAJA3-KD had a minor effect on mitochondrial translation and soluble MRP levels, it had a greater impact on aggregation of all MRP subunits tested (Fig. 7C and D). HSPA9-KD, in contrast, had no observable effect on MRP aggregation but reduced MRP levels and mitochondrial translation after 4 days (Fig. 7C and D). Thus, comparing JG-98 impact on mitoribosome stability to the individual KDs of DNAJA3 or HSPA9 further underscores how the pleiotropic effects of JG-98 on HSPA9 can converge to result in a more potent and rapid biological effect.
Discussion
Inhibition of HSP70 by JG-98 has been well documented to suppress the growth of a wide range of cancer cells in vitro (6). In fact, previous work from our group and others showed its effectiveness in destabilizing both full-length and cancer-specific splice variants of AR via an HSP70 binding site in the highly conserved N-terminal domain (7, 9). Our previous study used JG-98 as a single agent to identify an alternate strategy that can deplete all forms of AR in these AR-dependent cells. The current study was initiated to determine whether variable JG-98 dosing might re-sensitize CRPC to ADT by depleting AR splice variants that contribute to ADT resistance. While JG-98 was capable of re-sensitizing CRPC to ADT, we found that doses necessary for this phenomenon spared AR protein levels and were related to a previously unrecognized dependence of the mitochondrial ribosome on the HSPA9 system. Our work suggests that mitoribosome sensitivity to JG-98 treatment explains the broad-spectrum sensitivity of cancer cells to JG-98, given that many cancers are not strictly dependent on steroid hormone receptors (26). In the unique context of CRPC, ADT re-sensitization by JG-98 spares the nuclear receptor necessary for ADT targeting.
Mitochondrial ribosome biogenesis has recently come into focus with numerous MS studies characterizing both the translocation and assembly of this large complex (11, 18). Assembly is a slow, complicated process entailing the cytosolic translation, translocation, and ordered association of 82 proteins onto the rRNA cores of the large and small mitoribosome subunits (27). Therefore, disruption of translocation rates or folding of any MRP or assembly factor is likely to have a significant impact on the functional complex. Our study finds JG-98 treatment disrupts the activity of the HSPA9 system and results in substantial loss of MRP stability that correlates with increased aggregation of individual MRPs. This is in general agreement with a recent study of multiple myeloma that also uncovered JG-98 sensitivity of the mitoribosome but failed to directly identify HSPA9 as the target of this response (28). Previous work has shed light on the importance of HSPA9 in both mitochondrial import and folding and our study has begun to unravel the relative impact each role plays in JG-98 induced MRP instability (29–31).
The presequence import pathway serves a substantial portion of mitochondrial matrix proteins (∼60% in yeast) (19, 20). The positively charged N-terminal presequence relies on an intact membrane potential to promote translocation into the mitochondria and has been experimentally manipulated to disrupt presequence translocation (18). Here we used FCCP to disrupt mitochondrial import and we show that it potently suppresses JG-98 influence on MRP aggregation, a likely result of a decrease in the unfolded protein burden within the mitochondrial matrix. Furthermore, it provides strong evidence for a model in which the primary impact of JG-98 on MRP stability relates to a buildup of unfolded proteins due to suppression of HSPA9 folding capacity. In contradiction to this is the surprising level of specificity JG-98 treatment has on mitoribosome stability since ∼40% of the acutely down-regulated proteins are MRPs or assembly factors. If JG-98 acted generally to block mitochondrial translocation or matrix folding capacity, we would expect a wider range of matrix targeted proteins to be affected by JG-98 in our TMT-MS results but this is not supported by our data. Future work will focus more specifically on HSPA9-mediated contributions to assembly/stability of the mitoribosome. The mitoribosome-specificity of JG-98 treatment is further confounded by the modest number of observed MRP interactions for HSPA9. However, we uncovered compelling evidence that mitochondrial DNAJs expand the HSPA9 network by associating with 57 additional MRPs and that DNAJA3 alone interacts with 26 MRPs (25). We found that JG-98 rapidly decreased DNAJA3 protein levels, leading us to attempt to mimic specific aspects of JG-98 treatment by assessing the impact of individual KD of HSPA9 or DNAJA3. While both KDs impacted MRP stability to some extent, neither was able to fully phenocopy JG-98 activity. HSPA9-KD resulted in a slow loss of MRP proteins but no apparent buildup of MRP aggregates while DNAJA3-KD promoted MRP aggregation. This result highlights the pleiotropic benefits of targeting HSPA9 with JG-98 and suggests that this pharmacological approach can achieve a far more potent impact by inhibiting the HSPA9 conformational cycle to simultaneously disrupt the natural activities of NEFs (GRPEL1 in mitochondria) and DNAJs (23).
JG-98 treatment also stimulated rapid but transient recruitment of the mitochondrial proteases LONP1, CLPP, and AFG3L2 to HSPA9 further validating JG-98 as having a significant impact on the re-wiring of mitochondrial proteostasis. We used RNAi-KD to decrease the levels of LONP1, CLPP, and AFG3L2 to better understand their role in JG-98-induced MRP aggregation. While loss of CLPP and AFG3L2 had minor impacts on MRP aggregation, KD of LONP1 in cells treated with JG-98 resulted in a significant buildup of MRP aggregates. We conclude that LONP1 recognizes an abundance of unfolded proteins and associates with HSPA9 to clear the unfolded or damaged proteins. Given the dual nature of both LONP1 and CLPP systems as chaperones and proteases, it is possible that loss of the proteases further handicaps the matrix chaperone network (32). However, this is not supported by our data since loss of LONP1 alone did not result in increased aggregation of MRPs, an observation commonly associated with LONP1 chaperone activity, again suggesting that single KO experiments do not mimic the pleiotropic pharmacological inhibition of HSPA9.
The mitoribosome is responsible for the translation of 13 integral membrane components of the ETC, seven of which encode for subunits of complex I. Thus, not surprisingly, JG-98's potent suppression of mitochondrial translation extends to disruption of OXPHOS. However, its impact on mitochondrial respiration is very likely to be an indirect result of decreased mitochondrial translation. This is supported by our data showing that JG-98 effects on respiration are delayed in relation to the impact on the mitoribosome, consistent with a diminished capacity to replenish damaged subunits of the ETC. It is this impact on mitochondrial respiration that not only demonstrates single-agent activity but also re-sensitizes CRPC cells to ADT drugs. Supporting this model, treatment with complex I (IACS-010759) or complex II (CNC-332) inhibitors phenocopied the effects of JG-98, although these agents are mechanistically distinct from each other. Our findings also indicate that rapid suppression of aerobic respiration with ETC-specific inhibitors results in a more durable combination with enzalutamide in vivo. Taken together, our data are consistent with the hypothesis that inhibition of OXPHOS is poorly tolerated by CRPC and is capable of re-sensitizing CRPC to ADT if AR is present. Unraveling the complex impact of HSPA9 and the mitochondrial cohort of DNAJ proteins on assembly/stability of the mitoribosome will be the subject of a future study.
Methods
Cell lines and cell culture
All cell lines (HEK-293, 22Rv1, and C4-2) were purchased from ATCC and grown as instructed by ATCC. HEK-293 was grown in DMEM (Corning 10-013-CV) supplemented with 10% (v/v) fetal bovine serum (FBS) (Gibco). 22Rv1 was cultured in RPMI-1640 (Corning 10-041-CV) supplemented with 10% (v/v) FBS. C4-2 cells were cultured in 4:1 DMEM/F12 K (ATCC, 30–2004) + 10% FBS supplemented with 5.7 mL T-media per 500 mL media. All cells were grown at 37˚C, 5% CO2 in a humidified incubator. All cell lines used in this study were tested for Mycoplasma prior to use and were authenticated by ATCC using short tandem repeat analysis.
Compounds used in this study
JG-98 (22) and its inactive analog JG-258 were synthesized by O. Johnson and provided by J. Gestwicki. CNC-332 (15) was synthesized and provided by J. Neuzil. IACS-010759 (14) was synthesized by J. Schrimp (NIH-NCATS). STA-9090 was provided by Synta Pharmaceuticals. C86 (3-NITRO-2′,4′,6′-TRIMETHOXYCHALCONE) was purchased from Indofine Chemical.
Western blots/antibody information
Cell lysate was prepared by aspirating media, rinsing with ice-cold PBS, and lysed with RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% DOC, and 0.1% SDS) containing fresh protease and phosphatase inhibitor (Roche) transferred to chilled Eppendorf tubes and briefly sonicated to disrupt genomic DNA. Lysate was clarified at 16,000×g for 15 min at 4˚C. When assessing aggregation, samples were lysed in Triton lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, and 1% Triton X-100) and clarified at 20,817×g for 10 min at 4°C. Soluble fractions were transferred to fresh, prechilled Eppendorf tubes and pellets were washed with 200 µL ice-cold PBS and resuspended in 2X SDS sample buffer. All SDS-PAGE and western blots were carried out on Criterion TGX 4–20% precast gradient gels (Bio-Rad) and transferred to nitrocellulose using the Trans-Blot Turbo System (Bio-Rad) following manufacturer's recommendations. Blots were blocked for 1 h in 5% nonfat milk/TBS-T (50 mM Tris pH 7.5, 300 mM NaCl, and 0.5% Tween 20), incubated with primary antibody (generally diluted in 5% BSA/TBS-T solutions) overnight at 4°C, and species-specific secondary antibody in milk/TBS-T was added for 1 h at room temperature. Protein was detected using the SuperSignal chemiluminescence detection system (ThermoFisher Scientific) and signals were visualized by autoradiography. The same blot was probed multiple times and, if necessary, sliced horizontally for better exposure of antibodies of the same species. Figure quality images were created by scanning relevant films; if brightness/contrast was adjusted, it was equivalently adjusted for the entire image. All antibodies used in this study are listed in table format in supplemental information. Western blots were generated from a minimum of two separate experiments.
JG-98 mitochondrial association
For JG-98 mitochondrial association experiments, ∼2–4×107 cells were treated with respective JG-98 concentrations for 24 h prior to isolation of mitochondria as previously described (33). Ten to 20 µg of mitochondria were then diluted into 400 µL 1X cold MS buffer (5 mM Tris-HCl pH 7.5, 210 mM mannitol, 70 mM sucrose, and 1 mM EDTA) +/− 0.01 U proteinase K (PK) (NEB Cat # P8107S) and incubated for 20 min on ice. All PK reactions were quenched with 4 mM PMSF and mitochondria were pelleted at 8,000×g for 5 min at 4°C and washed once with 400 µL 1X MS buffer. Mitochondrial pellets were gently resuspended in 1X MS buffer containing 500 nM MitoTracker Red CMXRos (Invitrogen) and incubated at 37°C for 30 min. Mitochondria were then analyzed using a Synergy H1 plate reader (Biotek) at ex 488/em 520 for JG-98 fluorescence and ex 579/em 610 for Mitotracker fluorescence. Mitotracker was used to normalize mitochondria associated JG-98 signal and ensure PK treatment did not compromise mitochondrial integrity. Data are presented as raw ratio or normalized to JG-98 (no PK) control. PK stripping of surface mitochondrial proteins was validated using TOM20 and HSP60 as mitochondrial outer membrane and for matrix controls, respectively. For experiments containing FCCP, FCCP was added at indicated concentrations at the time of JG-98 addition and incubated for 3–6 h prior to mitochondrial isolation.
Immunoprecipitations
HEK-293 cells were transfected with pcDNA3.1 HSPA9-FLAG (this study) using recommended DNA amounts and X-tremeGENE 9 (Roche) at 3:1 ratio. For negative control reactions, empty vector (EV, pcDNA3.1 C-FLAG) was used and transfected like HSPA9-FLAG. Cells were incubated for 2 days prior to harvesting for immunoprecipitations. Cells were lysed in TNES + 1% NP-40 [50 mM Tris (pH 7.5), 2 mM EDTA, 100 mM NaCl, and 1% NP-40] and equivalent concentrations of EV or HSPA9-FLAG lysate in equivalent volumes were incubated with FLAG-conjugated agarose beads for 1 h (Sigma). Beads were washed 3X in TNES + 1% NP-40 and eluted in 50 µL TNES + 1% NP-40 + 100 ng/µL 3XFLAG peptide (Sigma). Samples were analyzed by SDS-PAGE and western blot.
siRNA mediated knockdown
siRNA KD of genes was completed as recommended by RNAiMAX (Invitrogen) using siRNAs described in Table S2. siRNA silencing was allowed to proceed for 2 or 4 days, as described in figure legends.
Incucyte growth experiments
Incucyte growth experiments were carried out in 96-well plate formats seeding with 10,000 cells/well (22Rv1) or 2,000 cells/well (C4-2). For C4-2 experiments, wells were pretreated with 0.4 µg/well fibronectin (Sigma) diluted in PBS as instructed by manufacturer. Cells were allowed to adhere overnight prior to treatment with respective inhibitor concentrations and scanned using an Incucyte S3 (Sartorius) placed in a humidified incubator at 37°C with 5% CO2. Phase contrast images were taken at 10 × magnification every 6–24 h for 6–10 days. Cell-specific analysis definitions were created for C4-2 and 22Rv1 and used to calculate percent confluency at each time point. Standard error of the mean (34) analysis was generated for each condition using a minimum of four wells per condition (4–5 images/well). Experiments were then exported to excel files and analyzed by Prism 9 GraphPad. All figures are the result of a minimum of three separate experiments.
TMT-MS sample preparation and analysis
Six 60 mm dishes were seeded with 2×106 22Rv1 cells per dish. Forty-eight hours later, three dishes were treated with 5 µM JG-98 or an equivalent concentration of DMSO for 4 h. After treatment, media was aspirated, washed twice with PBS, and lysed with 150 µL RIPA buffer + protease and phosphatase inhibitors. Cells were transferred to prechilled 1.7 mL Eppendorf tubes, sonicated at 10% intensity to disrupt genomic DNA, and clarified at 16,100×g for 30 min at 4°C. Soluble extract was transferred to fresh prechilled tubes, flash froze, and stored at −85°C until analysis.
For TMT-MS, 50 µg of each sample was digested with trypsin and the tryptic peptides were then TMT-multiplex labeled. Equal amounts of the six trypsin-digested samples were combined and labeled with separate TMT-label for internal reference. All samples were then mixed and loaded onto a UHPLC column. Twenty-four separate fractions were collected and sequentially analyzed by 24×100 min LC/MS/MS runs. Raw MS data files were analyzed by Proteome Discoverer 2.4 for protein ID and TMT-tag based quantification. Changes ≥ 25% with P < 0.05, n = 3 were used as cutoffs for significance.
Gene Ontology analysis
Statistically significant decreases in protein abundance due to JG-98 treatment were analyzed by Gene Ontology analysis using the PANTHER Classification System (http://www.pantherdb.org/). Data were organized by GO Cell Component terms and presented as fold enrichment over expected occurrence within a data set.
Seahorse experiments
22Rv1 were seeded in an XFe96 cell plate and allowed to adhere overnight. For 24 h pretreatments with JG-98 or IACS-010759, 1.25–1.5×104 cells were seeded per well; 2.0–2.5×104 cells/well were used for Seahorse assays carried out the next day. On the day of the assay, the media was replaced with XF DMEM pH 7.4 supplemented with fresh 10 mM glucose, 1 mM sodium pyruvate, and 2 mM glutamine. Oxygen consumption and extracellular acidification rates (OCR and ECAR, respectively) were measured with a Seahorse XFe96 analyzer (Agilent). Mitochondrial stress test assays (Agilent) were conducted with the following final concentrations of inhibitors: 1 µM oligomycin, 1 µM FCCP, and 0.5 µM rotenone/antimycin A. Samples were normalized by CyQUANT (ThermoFisher). Analysis was conducted on Wave 2.6.3 (Agilent) and data presented are the result of at least four wells per condition and at least three individual experiments. OCR vs ECAR plots are the result of one of three separate baseline measurements. Data are presented as the mean +/− the standard deviation of rates.
In vivo xenograft mouse experiments
All animal experiments were approved by the NCI-Bethesda Animal Care and Use Committee (approved animal protocol number UOB-013-1-A). Four-week-old male athymic mice (Charles River) were allowed to mature and acclimate for 2 weeks. 1×106 low passage 22Rv1 cells were injected into the right thigh of each mouse. Once tumors reached a detectable volume (approximately 21 days postinoculation), mice were randomized and administered drug or vehicle [10% DMSO, 18% Cremophor RH 40 (Sigma), 3.6% dextrose, and 68.4% 1 M HEPES]. In vivo dosing was as follows: 6 mg/kg JG-231 by intraperitoneal injection (JG-231 dose was adjusted from 8 to 6 mg/kg after first administration) and 25 mg/kg enzalutamide (Selleckchem) by oral administration, or combination. JG-231 was administered 3 days per week (M/W/F) and enzalutamide was given 5 days per week (M-F) for 3 weeks. Animals were removed from the study once tumor dimension reached 20 mm in any direction.
For IACS-010759 studies, all conditions were similar to those stated above. Mice were administered vehicle (0.5% methyl cellulose) or 7.5 mg/kg IACS-010759 by oral gavage (M-F), 25 mg/kg enzalutamide by oral gavage (M-F), or a combination of the two drugs for 3 weeks.
Hyperpolarized [1-13C] magnetic resonance spectroscopic imaging
Hyperpolarized 13C-MRSI was performed as described previously (13). [1-13C] pyruvate (30 µL) containing 15 mmol/L OXO63 and 2.5 mmol/L gadolinium chelate was hyperpolarized using the Hypersense DNP polarizer (Oxford Instruments, Abingdon, UK). The hyperpolarized sample was rapidly dissolved in 4.5 mL of a superheated alkaline buffer and intravenously injected (12 µL/g body weight). Hyperpolarized 13C MRSI was performed on a 3 T scanner (MRSolutions, Guildford, Surrey, UK) using a 17 mm home-built 13C solenoid coil placed inside a saddle coil for detection of 1H (hydrogen atom). Anatomical images were taken with a fast spin-echo imaging sequence with a repetition time of 2,500 msec, an effective TE of 68 msec, one signal, an echo-train length of 8 echoes, a field of view of 28 mm × 28 mm with a 256×256 matrix, and a 2-mm section thickness with a 2-mm intersection gap. The 13C MR spectra were acquired every second with a spectral width of 3330 Hz, a repetition time of 1000 ms and a flip angle of 10°. 13C two-dimensional spectroscopic images were acquired 25 s after the start of the pyruvate injection, with a 28×28 mm field of view in an 8 mm axial slice, yielding a matrix size of 14×14 mm. The chemical shift values for each signal used in this study were—[1-13C] pyruvate: 170.6 ppm, [1-13C] lactate: 182.7 ppm, [1-13C] alanine: 176 ppm and [1-13C] bicarbonate: 163.5 ppm, respectively. Each mouse harboring xenografted tumors was imaged 1 h before (‘‘prestudy’’) and at 24 h after (‘‘poststudy’’) drug treatment, thus serving as its own control.
TMRE assay
Mitochondrial membrane potential was monitored by tetramethylrhodamine, ethyl ester, and perchlorate (TMRE) using a Synergy H1 plate reader (Biotek) and optimized as described in (35). 22Rv1 (0.5×106 cells/well) were used to seed 12-well plates sufficient for triplicate of each condition. Cells were incubated for ∼2 days (sufficient time for cultures to reach ∼80% confluency) prior to treating with 150 or 250 nM JG-98 or inactive analog JG-258 for 24 h. As a control for mitochondrial membrane depolarization, 10 µM FCCP was added to control wells for 1 h. All cells were then treated with 30 nM TMRE (Invitrogen) for 30 min. Media was then aspirated and washed with PBS, and cells were trypsinized with 0.25% Trypsin EDTA (Gibco). One ml of Fluorobrite DMEM (Gibco) + 10% FBS was added to each well to quench trypsin, cells were transferred to 1.7 ml Eppendorf tubes, and pelleted at 500×g for 5 min at 4°C. Cell pellets were resuspended in 320 µL fluorobrite DMEM + FBS. Two hundred microliters was used for measuring TMRE fluorescence (Ex 550 nm/Em 590 nm). One hundred microliters of remaining volume was then used to normalize TMRE fluorescence by protein concentration measured using the BCA reagent (Pierce).
Mitochondrial translation assay
The mitochondrial translation assay was adapted from Yousefi et al. (17). Briefly, 7.5×106 22Rv1 were used to seed each 10 cm dish in complete RPMI-1640 + 10% FBS and cells were incubated for 2 days to allow cells to reach ∼80% confluency. Cells were then treated with DMSO or 250 nM JG-98 or inactive analog, JG-258, for 24 h. After completion of JG treatment, all cells were washed with 10 mL PBS and replaced with RPMI-1640 (without methionine) + 10% dialyzed FBS. As a control for mitochondrial translation, one dish was pretreated where shown with 150 µg/mL chloramphenicol for 1 h. All cells were then treated with 500 µM Harringtonine (Cayman) for 20 min to block cytosolic translation prior to addition of 500 µM L-Homopropargylglycine hydrochloride (HPG) (Sigma) for 4 h. To monitor background, one dish was left without the addition of HPG. After completion of mitochondrial translation, cells were washed with PBS, trypsinized, transferred to prechilled 5 mL tubes, and pelleted at 200×g for 5 min. After aspirating media, cell pellets were washed with PBS and resuspended in RSB hypotonic buffer (10 mM Tris pH 7.5, 10 mM NaCl, 1.5 mM MgCl2). Mitochondria were isolated from each condition as described (33). Mitochondria were quantified by Bradford assay and 100 µg of mitochondrial protein from each condition was resuspended in 50 mM Tris pH 8.0, 1 mM PMSF, and 1% SDS. IRDye® 680RD Azide Infrared Dye (LI-COR Biosciences) was used to detect incorporated HPG mitochondrial translation by click-chemistry using the Click-iT kit (Thermo Fisher) per manufacturer's instructions. Once completed, each sample was resuspended in 50 µL 2X SDS-Load Buffer + 2% β-Mercaptoethanol, boiled, and stored at −80°C. Twenty-five microliters of each sample was loaded onto a Criterion TGX precast 4–20% gradient gel and transferred to nitrocellulose. IRDye® 680RD incorporation was detected by Odyssey Fc Imaging System (LI-COR Biosciences). Mitochondrial translation experiments were run in triplicate.
Supplementary Material
Acknowledgments
We gratefully acknowledge the training on HP-MRSI provided by Shun Kishimoto, M.D. We thank Dr. Matthew D. Hall, PhD, Director, Early Translation Branch, at National Center for Advancing Translational Sciences (NCATS) for synthesis of IACS-010759. We would also like to acknowledge Poochon Scientific for their work analyzing our mass spectrometry samples.
Contributor Information
Frank J Echtenkamp, Urologic Oncology Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA.
Ryo Ishida, Urologic Oncology Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA.
Genesis M Rivera-Marquez, Urologic Oncology Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA.
Marisa Maisiak, Urologic Oncology Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA.
Oleta T Johnson, Department of Pharmaceutical Chemistry and the Institute for Neurodegenerative Disease, University of California, San Francisco, San Francisco, CA 94158, USA.
Jonathan H Shrimp, Chemical Genomics Center, National Center for Advancing Translational Sciences, National Institutes of Health, Rockville, MD 20850, USA.
Arnav Sinha, Urologic Oncology Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA.
Stephen John Ralph, Cancure Ltd, Broadbeach, Queensland 4218, Australia.
Ian Nisbet, Cancure Ltd, Broadbeach, Queensland 4218, Australia.
Murali Krishna Cherukuri, Biophysics Section, Radiation Biology Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA.
Jason E Gestwicki, Department of Pharmaceutical Chemistry and the Institute for Neurodegenerative Disease, University of California, San Francisco, San Francisco, CA 94158, USA.
Leonard M Neckers, Urologic Oncology Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA.
Supplementary material
Supplementary material is available at PNAS Nexus online.
Funding
Funding for these studies was provided by the Intramural Program of the National Cancer Institute (L.M.N) and by a Department of Defense Award to L.M.N. (award # W81XWH-19-1-0716).
Author contributions
F.J.E. designed research, performed research, analyzed data, and wrote the paper; L.M.N. designed research and wrote the paper; R.I. performed research and analyzed data; G.M.R.M., M.M., and A.S. performed research; O.T.J., J.E.G., S.J.R., and I.N. contributed new reagents; M.K.C. contributed new techniques and analyzed the data.
Data availability
The datasets generated and analyzed during the current study are included in this manuscript and supplementary materials. Raw mass spectrometry data has been submitted to jPOSTrepo (https://repository.jpostdb.org) and ProteomeXchange (https://www.proteomexchange.org) accession #: PXD038684 and will be made public at time of acceptance.
References
- 1. Ottens F, Franz A, Hoppe T. 2021. Build-UPS and break-downs: metabolism impacts on proteostasis and aging. Cell Death Differ. 28:505–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dai C, Whitesell L, Rogers AB, Lindquist S. 2007. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 130:1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bader DA, McGuire SE. 2020. Tumour metabolism and its unique properties in prostate adenocarcinoma. Nat Rev Urol. 17:214–231. [DOI] [PubMed] [Google Scholar]
- 4. American Cancer Society . 2019. Hormone therapy for prostate cancer. Washington (DC): American Cancer Society. [Google Scholar]
- 5. Teo MY, Rathkopf DE, Kantoff P. 2019. Treatment of advanced prostate cancer. Annu Rev Med. 70:479–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li X, et al. 2015. Validation of the Hsp70-Bag3 protein-protein interaction as a potential therapeutic target in cancer. Mol Cancer Ther. 14:642–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moses MA, et al. 2018. Targeting the Hsp40/Hsp70 chaperone axis as a novel strategy to treat castration-resistant prostate cancer. Cancer Res. 78:4022–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shafi AA, Cox MB, Weigel NL. 2013. Androgen receptor splice variants are resistant to inhibitors of Hsp90 and FKBP52, which alter androgen receptor activity and expression. Steroids. 78:548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eftekharzadeh B, et al. 2019. Hsp70 and Hsp40 inhibit an inter-domain interaction necessary for transcriptional activity in the androgen receptor. Nat Commun. 10:3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Craig EA. 2018. Hsp70 at the membrane: driving protein translocation. BMC Biol. 16:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bogenhagen DF, Ostermeyer-Fay AG, Haley JD, Garcia-Diaz M. 2018. Kinetics and mechanism of mammalian mitochondrial ribosome assembly. Cell Rep. 22:1935–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koya K, et al. 1996. MKT-077, a novel rhodacyanine dye in clinical trials, exhibits anticarcinoma activity in preclinical studies based on selective mitochondrial accumulation. Cancer Res. 56:538–543. [PubMed] [Google Scholar]
- 13. Oshima N, et al. 2020. Dynamic imaging of LDH inhibition in tumors reveals rapid in vivo metabolic rewiring and vulnerability to combination therapy. Cell Rep. 30:1798–1810 e1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Molina JR, et al. 2018. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat Med. 24:1036–1046. [DOI] [PubMed] [Google Scholar]
- 15. Dong LF, et al. 2011. Mitochondrial targeting of vitamin E succinate enhances its pro-apoptotic and anti-cancer activity via mitochondrial complex II. J Biol Chem. 286:3717–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Basu HS, et al. 2021. Prostate cancer cells survive anti-androgen and mitochondrial metabolic inhibitors by modulating glycolysis and mitochondrial metabolic activities. Prostate. 81:799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yousefi R, et al. 2021. Monitoring mitochondrial translation in living cells. EMBO Rep. 22:e51635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schafer JA, Bozkurt S, Michaelis JB, Klann K, Munch C. 2022. Global mitochondrial protein import proteomics reveal distinct regulation by translation and translocation machinery. Mol Cell. 82:435–446 e437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vogtle FN, et al. 2009. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 139:428–439. [DOI] [PubMed] [Google Scholar]
- 20. Wiedemann N, Pfanner N. 2017. Mitochondrial machineries for protein import and assembly. Annu Rev Biochem. 86:685–714. [DOI] [PubMed] [Google Scholar]
- 21. Taylor IR, et al. 2018. High-throughput screen for inhibitors of protein-protein interactions in a reconstituted heat shock protein 70 (Hsp70) complex. J Biol Chem. 293:4014–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li X, et al. 2013. Analogs of the allosteric heat shock protein 70 (Hsp70) inhibitor, MKT-077, as anti-cancer agents. ACS Med Chem Lett. 4:1042–1047 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Faust O, Rosenzweig R. 2020. Structural and biochemical properties of Hsp40/Hsp70 chaperone system. Adv Exp Med Biol. 1243:3–20. [DOI] [PubMed] [Google Scholar]
- 24. Kampinga HH, Craig EA. 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 11:579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Piette BL, et al. 2021. Comprehensive interactome profiling of the human Hsp70 network highlights functional differentiation of J domains. Mol Cell. 81:2549–2565 e8. [DOI] [PubMed] [Google Scholar]
- 26. Ganguly S, Naik D, Muskara A, Mian OY. 2021. The nexus of endocrine signaling and cancer: how steroid hormones influence genomic stability. Endocrinology. 162:bqaa177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. De Silva D, Tu YT, Amunts A, Fontanesi F, Barrientos A. 2015. Mitochondrial ribosome assembly in health and disease. Cell Cycle. 14:2226–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ferguson ID, et al. 2022. Allosteric HSP70 inhibitors perturb mitochondrial proteostasis and overcome proteasome inhibitor resistance in multiple myeloma. Cell Chem Biol. 29:1288–1302 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Horst M, et al. 1997. Sequential action of two hsp70 complexes during protein import into mitochondria. EMBO J. 16:1842–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matouschek A, Pfanner N, Voos W. 2000. Protein unfolding by mitochondria. The Hsp70 import motor. EMBO Rep. 1:404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kang PJ, et al. 1990. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature. 348:137–143. [DOI] [PubMed] [Google Scholar]
- 32. Shin CS, et al. 2021. LONP1 And mtHSP70 cooperate to promote mitochondrial protein folding. Nat Commun. 12:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Clayton DA, Shadel GS. 2014. Isolation of mitochondria from tissue culture cells. Cold Spring Harb Protoc. 10:1109–1111. 10.1101/pdb.top074542. [DOI] [PubMed] [Google Scholar]
- 34. Dang EV, et al. 2011. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 146:772–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Perry SW, Norman JP, Barbieri J, Brown EB, Gelbard HA. 2011. Mitochondrial membrane potential probes and the proton gradient: a practical usage guide. Biotechniques. 50:98–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are included in this manuscript and supplementary materials. Raw mass spectrometry data has been submitted to jPOSTrepo (https://repository.jpostdb.org) and ProteomeXchange (https://www.proteomexchange.org) accession #: PXD038684 and will be made public at time of acceptance.