ABSTRACT
The pathogenesis of osteoporosis, a common disease with great morbidity and mortality, comprises environmental and genetic factors. As with other complex disorders, the genetic basis of osteoporosis has been difficult to identify. Nevertheless, several approaches have been undertaken in the past decades in order to identify candidate genes for bone fragility, including the study of rare monogenic syndromes with striking bone phenotypes (e.g. osteogenesis imperfecta and osteopetroses), the analysis of individuals or families with extreme osteoporotic phenotypes (e.g. idiopathic juvenile and pregnancy-related osteoporosis), and, chiefly, genome-wide association studies (GWAS) in large populations. Altogether, these efforts have greatly increased the understanding of molecular mechanisms behind bone remodelling, which has rapidly translated into the development of novel therapeutic strategies, exemplified by the tales of cathepsin K (CTSK) and sclerostin (SOST). Additional biological evidence of involvement in bone physiology still lacks for several candidate genes arisen from GWAS, opening an opportunity for the discovery of new mechanisms regulating bone strength, particularly with the advent of high-throughput genomic technologies. In this review, candidate genes for bone fragility will be presented in comprehensive tables and discussed with regard to how their association with osteoporosis emerged, highlighting key players such as LRP5, WNT1 and PLS3. Current limitations in our understanding of the genetic contribution to osteoporosis, such as yet unidentified genetic modifiers, may be overcome in the near future with better genotypic and phenotypic characterisation of large populations and the detailed study of candidate genes in informative individuals with marked phenotype.
Keywords: GWAS, mutation, fracture, low bone mass, bone remodeling
INTRODUCTION
Osteoporosis is a common disease characterized by low bone mineral density (BMD) and microarchitectural deterioration, leading to increased fracture risk with great morbidity and mortality, resulting in social and economic burden (1,2). Clinical diagnosis of osteoporosis is established by assessing BMD by dual-energy X-ray absorptiometry (DXA), a predictor of fracture risk, or by the occurrence of fragility fractures (3,4).
Osteoporosis is a complex disorder, influenced by both environmental and genetic factors. In the study of complex disorders, the genetic influence can be inferred from estimations of heritability, i.e., the portion of phenotypic variance attributable to cumulative genetic factors (5). In osteoporosis, BMD heritability has been estimated from 50 to 85% and, more variably, fracture heritability has ranged from 25 to 68% (6,7). Supporting the intuitive concept that the genetic influence should be more pronounced in cases of early or idiopathic osteoporosis, fracture heritability is higher for fractures occurring before 70 years of age (8).
The identification of human genes associated with bone fragility started around the 1990s through the study of monogenic syndromes with marked skeletal phenotypes such as osteogenesis imperfecta due to COL1A1 and COL1A2 defects (9) and osteopetrosis due to TCIRG1 defects (10). In 2001, the breakthrough discovery of the involvement of the Wnt signalling pathway on the regulation of bone remodelling was made possible by the study of rare conditions such as osteoporosis-pseudoglioma syndrome (OPPG) due to LRP5 mutations (11) and sclerosteosis due to SOST defects (12,13). More recently, the study of subjects with extreme phenotypes of osteoporosis, such as idiopathic juvenile osteoporosis and pregnancy-associated osteoporosis has yielded WNT1 and PLS3 as novel regulators of bone strength (14-16).
The advent of genome-wide association studies (GWAS) expanded the horizon of the genetic contribution to osteoporosis. Following a proof of concept study in 2007 (17), two pioneer GWAS for BMD were published in 2008 (18,19), identifying five significant loci associated with BMD, four of them near genes already known or suspected to be involved in the pathophysiology of osteoporosis (RANKL, OPG, ESR1, LRP5). Highlighting the potential of GWAS for gene discovery, the remaining locus mapped to novel candidate gene ZBTB40, later confirmed by subsequent analyses (20). Since then, more than twenty GWAS have been performed interrogating genetic association to BMD, quantitative ultrasound and/or fracture, implicating more than 90 candidate genes for osteoporosis. The function of some of these genes in bone metabolism was only recognized following their identification by GWAS (for example, AXIN1 and WLS), but for the majority of candidates a biological mechanism remains unknown (7).
The identification of molecular pathways in osteoporosis has important implications not only for the recognition of individuals in risk, aiming for a personalized medical approach, but also for the development of new therapeutic strategies, as exemplified by the advent of sclerostin inhibition as a potential treatment for osteoporosis roughly ten years after the identification of SOST defects (21). Considering the fast paced evolution in the field, it is important to gather genetic factors involved with osteoporosis from multiple experimental sources and revise them in light of their contribution to our pathophysiological insight. In this review, a thorough and up-to-date list of candidate genes for bone fragility will be presented and discussed according to how they emerged: from rare monogenic diseases with high impact on bone strength, from extreme phenotypes of osteoporosis and/or from GWAS.
LITERATURE SEARCH STRATEGY
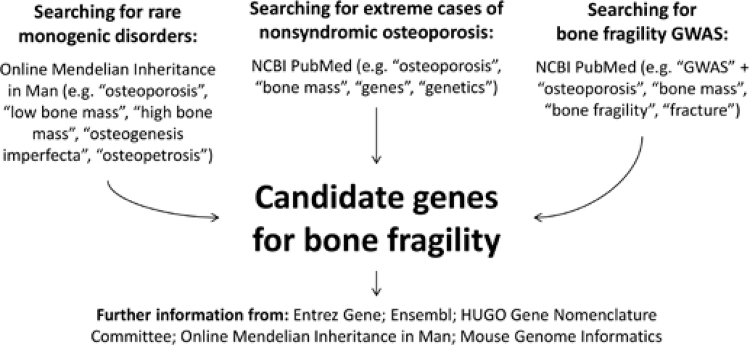
In order to identify genes associated with bone fragility, a broad literature search strategy was devised (Figure 1). A systematic review of original and review articles indexed on PubMed published until October 2015 using the descriptors “osteoporosis”, “genes”, “genetics”, and “bone mass” was undertaken. To retrieve all GWAS on bone fragility, search queries “GWAS and osteoporosis”, “GWAS and fractures”, “GWAS and bone fragility”, and “GWAS and BMD” were used. To enhance our discovery of monogenic disorders associated with altered bone mass or strength, the Online Mendelian Inheritance in Man® (OMIM®) database was also searched using standard descriptors. Mouse phenotypic data for identified candidate genes were obtained from the Mouse Genome Informatics (MGI) online database, and gene function information was searched on NCBI’s Entrez Gene database.
Figure 1. Scheme of the literature search strategy devised in order to identify candidate genes for bone fragility from rare monogenic phenotypes, extreme nonsyndromic cases of osteoporosis and genome wide association studies (GWAS) with bone fragility endpoints.
CANDIDATE GENES EMERGING FROM RARE MONOGENIC DISORDERS
The study of monogenic diseases with high impact on bone strength has enabled the identification of several pivotal mechanisms involved in bone physiology (22). For example, osteogenesis imperfecta has shown the importance of bone collagen matrix quality; Van Buchem disease, Hajdu-Cheney syndrome and autosomal recessive osteopetrosis have revealed important signalling pathways (namely Wnt, Notch and RANK-RANKL-OPG) that regulate bone remodelling; and pycnodysostosis has given insight into the pivotal action of cathepsin K in osteoclast function. On par with a recently proposed taxonomy of rare genetic disorders of bone metabolism (22), monogenic diseases will be presented according to how they affect bone strength. Candidate genes for bone fragility arising from these disorders are presented in Table 1.
Table 1. Genes associated with rare monogenic diseases with high impact on bone mass/strength.
| Gene | OMIM id | Protein function | Disease | Phenotype |
|---|---|---|---|---|
| COL1A1 | 120150 | Type 1 collagen | Osteogenesis imperfecta | Low BMD and increased fracture risk; severity varies from perinatal lethality to asymptomatic; extra-skeletal features include blue sclerae, dentinogenesis imperfecta and hearing loss |
| COL1A2 | 120160 | Type 1 collagen | ||
| BMP1 | 112264 | C-propeptide cleavage | ||
| CRTAP | 605497 | Collagen hydroxylation | ||
| FKBP10 | 607063 | Collagen processing | ||
| IFITM5 | 614757 | Mineralization | ||
| P3H1 | 610339 | Collagen hydroxylation | ||
| PLS3 | 300131 | Actin-binding | ||
| PPIB | 123841 | Collagen hydroxylation | ||
| SEC24D | 607186 | ER procollagen processing | ||
| SERPINF1 | 172860 | Collagen chaperoning | ||
| SERPINH1 | 600943 | Mineralization | ||
| SP7 | 606633 | Ob regulation | ||
| TMEM38B | 611236 | Cation channel | ||
| WNT1 | 164820 | Ob activation/Wnt signalling (ligand) | ||
| SEC24D | 607186 | ER procollagen processing | Cole-carpenter syndrome | Bone fragility; craniosynostosis; ocular proptosis; hydrocephalus; distinctive facial features |
| P4HB | 176790 | ER procollagen processing | ||
| FKBP10 | 607063 | Collagen processing | Bruck syndrome | Congenital contractures; early onset of fractures; short stature; severe limb deformity; progressive scoliosis |
| PLOD2 | 601865 | ER procollagen processing | ||
| TCIRG1 | 604592 | Oc function | Osteopetrosis | High BMD; skeletal deformities; compression of noble structures and occupation of bone marrow space; variable severity and age of onset |
| CLCN7 | 602727 | Oc function | ||
| OSTM1 | 607649 | Oc homeostasis | ||
| PLEKHM1 | 611466 | Oc function | ||
| CA2 | 611492 | Oc function | ||
| SNX10 | 614780 | Oc homeostasis | ||
| TNFRSF11A | 603499 | Oc activation (RANK) | ||
| TNFSF11 | 602642 | Oc activation (RANKL) | ||
| CTSK | 601105 | Oc function | Pycnodysostosis | Short stature; skull deformities; acroosteolysis; high BMD; increased fracture risk |
| SOST | 605740 | Ob activation/Wnt signalling (antagonist) | Sclerosteosis, van Buchem disease | High BMD; increased bone strength; increased head circumference; compression of noble structures; enlarged mandible; syndactyly; high stature |
| LRP5 | 603506 | Ob activation/Wnt signalling (receptor) | High bone mass syndrome | High BMD; increased bone strength; widened mandible; torus palatinus |
| Osteoporosis-pseudoglioma | Early-onset osteoporosis; ocular pseudoglioma or vitreoretinopathy | |||
| NOTCH2 | 600275 | Notch signalling | Hajdu-Cheney syndrome | Osteoporosis; short stature; acroosteolysis; distinctive facial features |
Proven or proposed protein functions are shown. Ob: osteoblast; Oc: osteoclast; OMIM id: online Mendelian inheritance in men identifier; ER: endoplasmic reticulum; RANK: receptor activator of nuclear factor kappa-β; RANKL: RANK ligand.
Monogenic diseases affecting the bone matrix
Osteogenesis imperfecta (OI) is a systemic disease characterized by high incidence of low-trauma fractures since birth or childhood due to defects in the bone matrix, chiefly in the quantity or quality of type I collagen (23,24). Clinical presentation is highly heterogeneous, with severity ranging from perinatal lethality to mostly asymptomatic. Extraskeletal features, such as blue sclerae, defective tooth development and hearing loss as well as family history may be present, allowing for an easier diagnosis. When none of these features are present, diagnosing OI can be challenging due to the overlap with idiopathic osteoporosis. Most commonly, OI is an autosomal dominant condition caused by mutations in COL1A1 and COL1A2 leading to clinical forms I to IV (25). Type V OI has recently been shown to be caused by mutations in IFITM5, also transmitted in an autosomal dominant pattern; the exact role of IFITM5 in determining bone strength remains elusive (26,27). Several rarer forms of OI with autosomal recessive inheritance exist, and the list of candidate genes for such phenotypes is ever increasing (Table 1). Most genes associated with recessive OI are directly or indirectly involved with type I collagen modification and/or assembly, but for some a mechanism is still unknown (28). Collectively, OI demonstrates how defects in bone material properties may have a substantial impact on bone strength.
More than 400 genetic skeletal disorders have been described, with around 360 genes implicated (29). A number of these skeletal dysplasias may also lead to bone fragility. In particular, Bruck syndrome and Cole-Carpenter syndrome have marked fragility, and their heterogeneous genetic bases overlap with OI (Table 1). Bruck syndrome, characterised by congenital joint contractures and early onset of fractures, can be caused by mutations in FKBP10 or PLOD2, and Cole-Carpenter syndrome, characterised by bone fragility, craniosynostosis and distinctive facies, has been associated with P4HB and SEC24D defects. Mutations in FKBP10 and SEC24D have also been implicated in OI, meaning that variants with variable biological impact may have different phenotypic expression and lead to isolated bone fragility (30,31).
Monogenic diseases affecting bone remodelling
Impairment of osteoclast-mediated bone resorption is known to lead to high bone mass syndromes such as osteopetrosis and pycnodysostosis (Table 1). In spite of the high bone mass, a high fracture risk is usually observed due to impaired bone renewal leading to poor quality.
Osteopetrosis is characterized by skeletal deformities, nerve compression and bone marrow occupation, and may present with variable degree of severity and inheritance patterns. Defects in the RANK-RANKL-OPG pathway, pivotal to osteoclast differentiation and activation, lead to autosomal recessive osteopetrosis due to a reduced number of osteoclasts (32). In contrast, defects in several genes involved in osteoclast function may lead to osteopetrosis with a normal or high number of osteoclasts. Of note, mutations in CLCN7, CA2 and TCIRG1, disrupting the regulation of organelle pH and acid secretion, may cause osteopetrosis by affecting the osteoclast ability to dissolve the bone matrix (32).
Pycnodysostosis, marked by high bone mass, short stature, skull deformities and acroosteolysis, is caused by mutations in CTSK encoding cathepsin K, an enzyme secreted by osteoclasts and crucial to bone resorption (33). The identification of CTSK defects as the cause of pycnodysostosis in 1996, and subsequent studies of its function in bone resorption, has led to the development of cathepsin K inhibition as a promising therapeutic approach for osteoporosis 20 years later, highlighting the importance of recognising molecular mechanisms in order to advance medical care and the fast pace of translation in this burgeoning field (34).
Disruption in bone formation may lead to either low BMD, and consequently decreased bone strength, or may inversely cause abnormally high BMD, with stronger bone and possibly decreased risk of fracture. Defects in members of the Wnt signalling pathway, key to osteoblast activation and function, illustrate how these opposite phenotypes might ensue (35). Activation of the Wnt receptor LRP5 ultimately leads to increased beta-catenin and osteoblast activity. Inactivating mutations in LRP5 lead to osteoporosis-pseudoglyoma syndrome, characterized by severe early-onset osteoporosis and ocular malformation, whereas gain-of-function LRP5 mutations (which abolish interaction with inhibitors Dkk-1 and sclerostin) lead to the high bone mass syndrome endosteal hyperostosis (Worth disease) (11,36). Accordingly, loss of the bone-specific Wnt inhibitor sclerostin (SOST) due to inactivating SOST mutations or deletion of its regulatory region lead to sclerosteosis and Van Buchem disease, marked by high BMD with skeletal deformities such as jaw and cranial enlargement (12,13,37). The painstaking study of these rare disorders led to recognition of sclerostin’s crucial repressive role in bone formation; its inhibition is currently being investigated in the treatment of osteoporosis in randomised clinical trials and may represent a paradigm shift in osteoporosis care in the near future (21).
Finally, Hajdu-Cheney syndrome, a rare form of syndromic osteoporosis accompanied by coarse and dysmorphic facies, short stature and acroosteolysis, is caused by NOTCH2 mutations disrupting Notch signalling (38,39). While the molecular physiology of Notch signalling in bone is still incompletely understood, the pronounced bone fragility in Hajdu-Cheney syndrome underlines the opportunity for novel therapeutic strategies targeting this pathway.
It should be noted that genetic defects associated with osteomalacia, primarily relating to bone mineralization, may also lead to osteoporosis-like milder phenotypes characterized by fragility fractures; within this vast group of disorders, attention is currently drawn to heterozygous ALPL mutations leading to adult hypophosphatasia, which has been proposed to be a potentially under-recognised cause of bone fragility (40).
CANDIDATE GENES IDENTIFIED THROUGH EXTREME CASES OF OSTEOPOROSIS
The quest for the genetic basis of a few extreme cases of nonsyndromic idiopathic osteoporosis has been reported in the literature. In general, a candidate gene approach has been applied, focussing on genes associated with OI and, more recently, Wnt signalling. Even though most studies have involved small cohorts and somewhat limited genetic approaches, the advent of massively parallel sequencing is rapidly boosting our capability for establishing a molecular diagnosis in these cases. Candidate genes identified in this manner are assembled in Table 2.
Table 2. Genes associated with idiopathic osteoporosis.
| Gene | OMIM id | Function | Phenotype | Study design | Reference |
|---|---|---|---|---|---|
| LRP5 | 603506 | Wnt signalling (receptor) | Juvenile osteoporosis | Candidate gene analysis (3 genes) | (43) |
| Vertebral fractures during pregnancy | Candidate gene analysis (3 genes) | (46) | |||
| Postpartum vertebral fractures | Candidate gene analysis (2 genes) | (47) | |||
| Idiopathic juvenile osteoporosis | WES, analysis focussed on candidate genes (14 genes) | (45) | |||
| DKK1 | 605189 | Wnt signalling (antagonist) | Juvenile osteoporosis | Candidate gene analysis (8 genes) | (44) |
| WNT3A | 606359 | Wnt signalling (ligand) | Juvenile osteoporosis | Candidate gene analysis (8 genes) | (44) |
| MTHFR | 607093 | Homocysteine metabolism | Postpartum vertebral fractures | Candidate gene analysis (2 genes) | (47) |
| PLS3 | 300131 | Actin-binding protein | X-linked osteoporosis | Massively parallel sequencing strategies | (16,48,49) |
| WNT1 | 164820 | Wnt signalling (ligand) | Early-onset autosomal dominant osteoporosis | Massively parallel sequencing strategies | (14,15) |
OMIM id: online Mendelian inheritance in men identifier; WES: whole-exome sequencing.
Initially, well known OI genes COL1A1 and COL1A2 posed as conspicuous candidates for mutational analysis in individuals with bone fragility. In 1991, Spotila and cols. investigated a 52-yo postmenopausal woman with low bone mass and a vertebral fracture, identifying a COL1A2 mutation (41). Of note, this patient had mildly blue sclerae and mild hearing loss, suggesting a mild presentation of OI. In 1994, the same group of authors undertook a mutational analysis of COL1A1 and COL1A2 in a cohort of 26 individuals with low bone density, identifying other two mutations in COL1A1 in association with milder phenotypes (42).
As novel molecular mechanisms in bone fragility were recognised, further genes became candidates for investigation. In 2005 and 2012, Hartikka and cols. and Korvala and cols. reported the mutational analysis of a cohort of children with idiopathic osteoporosis, examining a total of 11 candidate genes mainly associated with autosomal dominant OI or the Wnt signalling pathway (43,44). Initially, Hartikka and cols. studied COL1A1, COL1A2 and LRP5, identifying three distinct mutations in LRP5 in 3 children, with some evidence of familial segregation (43). Later, Korvala and cols. studied 8 new candidate genes, and identified rare sequence variants in two children (44). In one subject they found a heterozygous missense variant in WNT3A, which was also present in an affected sister, inherited from their mother who presented with post-menopausal osteoporosis. Nonetheless, the paternal family, who did not carry this variant, had a prominent history of adult osteoporosis and fractures, suggesting that other genetic factors might also be associated with the more severe/early-onset phenotype. In the other subject, a rare variant in DKK1, a well-known inhibitor of Wnt signalling, was identified, albeit with incomplete segregation (44).
Further studies have associated LRP5 variants with an array of extreme osteoporosis phenotypes (Table 2). Also using a candidate gene approach, Fahiminiya and cols., Campos-Obando and cols., and Cook and cols. have studied single cases and found three different LRP5 variants in two women with pregnancy-related osteoporosis and vertebral fractures, and one boy with idiopathic juvenile osteoporosis (45-47). Segregation analyses did not show clear relationships between variants and phenotype, again suggesting the association of additional genetic and/or environmental factors. One of the subjects with pregnancy-related osteoporosis was also homozygous for the MTHFR gene C677T polymorphism, which has been associated with several health outcomes including fracture risk and low BMD (47) MTHFR encodes for methylenetetrahydrofolate reductase, an enzyme involved in folate, homocysteine and amino acid metabolism.
The emergence of high throughput technologies allowed de novo discovery of candidate genes associated with familial idiopathic osteoporosis. In 2013, two groups independently identified WNT1 mutations in this context. Keupp and cols. performed whole exome sequencing in a four-generation family with early-onset autosomal dominant osteoporosis, identifying a heterozygous WNT1 mutation segregating with the phenotype (14). Laine and cols reported the genomewide linkage analysis followed by targeted parallel sequencing of another family with a similar presentation, also leading to the identification of a heterozygous WNT1 mutation (15). Notably, both groups found homozygous WNT1 mutations in families with severe recessive OI, suggesting a phenotypic spectrum of severity in relation to the molecular defects. Even though other Wnt family members were already well-known regulators of bone remodelling, these reports unravelled the importance of WNT1 in bone strength.
The discovery of entirely novel mechanisms in bone fragility has also been made possible by massively parallel sequencing. In 2013, Van Dijk and cols. performed X-linked whole exome sequencing in a family with X-linked osteoporosis, identifying a deleterious frameshift mutation in PLS3, a new factor in bone metabolism (16). Four additional PLS3 mutations were found in further four families. Notably, male individuals in these families carrying hemizygous PLS3 variants presented with overt osteoporotic fractures while female carriers had milder phenotypes with low bone mass. Additionally, a rare PLS3 variant (rs140121121) was found in 5 unrelated males with osteoporotic fractures and then studied in a large Dutch cohort, showing an association with increased fracture risk in elderly heterozygous female carriers, thus suggesting a role for this variant in common osteoporosis (16).
Further reports have supported a causative role for PLS3 mutations in the genesis of X-linked osteoporosis (48,49). While the biological role of PLS3 in bone is still largely unknown, a disturbance in osteocyte mechanosensing has been proposed as a putative mechanism based on animal model observations (16).
Taken together, these reports support a robust genetic contribution for extreme cases of osteoporosis, with potential translational implications for the care of common osteoporosis. Nevertheless, the individual impact of these variants on phenotype is still incompletely understood, and additional genetic factors may account for variable phenotypic expression in some cases.
CANDIDATE GENES IDENTIFIED THROUGH GENOME-WIDE ASSOCIATION STUDIES (GWAS)
As with other multifactorial diseases, common osteoporosis has long been hypothesized to be caused by multiple common variants each exerting a small influence on phenotype (7). Therefore, the technological breakthrough of GWAS was wholly embraced in the field, and at least twenty-nine low BMD and/or fractures GWAS have been published since 2008, including original studies and meta-analyses. As a result, most of the genes associated with bone fragility until now have been identified through such studies, totalling more than 70 loci and, respectively, more than 90 genes, which are listed on Table 3.
Table 3. Genes associated with bone mineral density or fracture risk in major genome-wide association studies.
| Candidate gene | BMD p-value (Fracture p-value) | SNP | References |
|---|---|---|---|
| ABCF2 | 7.3x10-9 | rs7812088 | GEFOS2 [Ref. (20)] |
| ABL1* | 3.4x10-22 | rs7851693 | GEFOS2 |
| ADAMTS18 | 2.1x10-8 | rs16945612 | Xiong 2009 [Ref. (54)] |
| ALDH7A1 | 6.4x10-6 (2.1x10-9) | rs13182402 | Guo 2010 [Ref. (55)] |
| ANAPC1 | 1.5x10-9 | rs17040773 | GEFOS2 |
| ARHGAP1 | 5.1x10-18 | rs7932354 | GEFOS1 [Ref. (51)], GEFOS2 |
| ATP6V1G1 | 3.0x10-9 | rs10817638 | Tan 2015 [Ref. (56)] |
| AXIN1* | 1.0x10-16 | rs9921222 | GEFOS2 |
| C12orf23 | 9.6x10-10 | rs1053051 | GEFOS2 |
| C7orf76 | 8.1x10-48 (5.9x10-11) | rs4727338 | GEFOS1, GEFOS2 |
| CCDC170 | 4.0x10-35 | rs4869742 | GEFOS1, GEFOS2, Styrkarsdottir 2008 [Ref. (19)] & 2009 [Ref. (50)] |
| CDC5L | 5.6x10-11 | rs163879 | GEFOS2 |
| CLCN7* | 1.5x10-16 | rs163879 | GEFOS2 |
| CLDN14 | 4.2x10-9 | rs170183 | Zhang 2014 [Ref. (57)] |
| COLEC10 | 3.2x10-39 | rs2062377 | GEFOS1, GEFOS2, Styrkarsdottir 2008, Richards 2008 [Ref. (18)] |
| CPED1 | 6.0x10-11 | rs13245690 | GEFOS2, Zheng 2012 [Ref. (58)] & 2015 [Ref. (53)] |
| CPN1 | 9.0x10-10 | rs7084921 | GEFOS2 |
| CREB3L1* | 5.1x10-18 | rs7932354 | GEFOS1, GEFOS2 |
| CRHR1 | 1.4x10-8 | rs9303521 | GEFOS1 |
| CTNNB1* | 4.4x10-25 | rs430727 | GEFOS1, GEFOS2 |
| CYLD | 1.9x10-22 | rs1566045 | GEFOS2 |
| DCDC1 | 2.2x10-11 | rs163879 | GEFOS1, GEFOS2 |
| DCDC5 | 2.2x10-11 | rs163879 | GEFOS1, GEFOS2 |
| DHH | 1.2x10-15 | rs12821008 | GEFOS2 |
| DKK1* | 1.6x10-12 (9.0x10-9) | rs1373004 | GEFOS2 |
| DMP1* | 1.2x10-27 (1.7x10-8) | rs6532023 | GEFOS2, Duncan 2011 [Ref. (52)] |
| DNM3 | 8.5x10-15 | rs479336 | GEFOS2 |
| EN1* | 2x10-14 (2x10-11) | rs11692564 | Zheng 2015 |
| ERC1 | 5.6x10-12 | rs2887571 | GEFOS2 |
| ESR1* | 4.0x10-35 | rs4869742 | GEFOS1, GEFOS2, Styrkarsdottir 2008 & 2009 |
| F2 | 5.1x10-18 | rs7932354 | GEFOS1, GEFOS2 |
| FAM210A | 4.9x10-8 (8.8x10-13) | rs4796995 | GEFOS2 |
| FAM3C | 1.0x10-11 | rs7776725 | Cho 2009 [Ref. (59)] |
| FAM9A | 1.2x10-8 | rs5934507 | GEFOS2 |
| FAM9B | 1.2x10-8 | rs5934507 | GEFOS2 |
| FKBP11* | 1.2x10-15 | rs12821008 | GEFOS2 |
| FMN2 | 1.9x10-9 | rs9287237 | Paternoster 2013 [Ref. (60)] |
| FOXC2* | 1.0x10-14 | rs10048146 | GEFOS1, GEFOS2 |
| FOXL1 | 1.0x10-14 | rs10048146 | GEFOS1, GEFOS2 |
| FUBP3 | 3.4x10-22 | rs7851693 | GEFOS2 |
| GALNT3* | 4.8x10-10 | rs6710518 | Duncan 2011 |
| GPATCH1 | 6.6x10-11 | rs10416218 | GEFOS2 |
| GPR68* | 2.0x10-15 | rs1286083 | GEFOS2 |
| GREM2* | 1.9x10-9 | rs9287237 | Paternoster 2013 |
| HDAC5 | 1.7x10-8 | rs228769 | GEFOS1 |
| IBSP* | 1.2x10-27 (1.7x10-8) | rs6532023 | Duncan 2011 |
| IDUA | 5.2x10-15 | rs3755955 | GEFOS2 |
| INSIG2 | 1.2x10-10 | rs1878526 | GEFOS2 |
| JAG1* | 3.1x10-19 | rs1878526 | GEFOS2, Kung 2010 [Ref. (61)] |
| KAL1 | 1.2x10-8 | rs5934507 | GEFOS2 |
| KCNMA1 | 5.0x10-19 | rs7071206 | GEFOS2 |
| KIAA2018 | 4.1x10-10 | rs1026364 | GEFOS2 |
| LACTB2 | 1.9x10-8 | rs7017914 | GEFOS2 |
| LEKR1 | 4.5x10-12 | rs344081 | GEFOS2 |
| LGR4 | 1.3x10-10 | rs587777005 | Styrkarsdottir 2013 [Ref. (62)] |
| LIN7C | 4.9x10-8 | rs10835187 | GEFOS2 |
| LRP4* | 5.1x10-18 | rs7932354 | GEFOS1, GEFOS2 |
| LRP5* | 2.1x10-26 (1.4x10-8) | rs3736228 | GEFOS1, GEFOS2, Kaufman 2008 [Ref. (63)] |
| MARK3 | 5.2x10-16 | rs11623869 | GEFOS1, GEFOS2, Sttyrkarsdottir 2009 |
| MECOM* | 3.6x10-8 | rs784288 | Hwang 2013 [Ref. (64)] |
| MEF2C | 4.5x10-61 | rs1366594 | GEFOS1, GEFOS2, Duncan 2011 |
| MEPE* | 1.2x10-27 (1.7x10-8) | rs6532023 | GEFOS2 |
| MPP7 | 2.4x10-16 | rs3905706 | GEFOS2 |
| NBR1* | 2.0x10-11 | rs4792909 | GEFOS, Styrkarsdottir 2009 |
| NTAN1 | 1.7x10-10 | rs4985155 | GEFOS2 |
| PDXDC1 | 1.7x10-10 | rs4985155 | GEFOS2 |
| PKDCC* | 1.3x10-9 | rs7584262 | GEFOS2 |
| PTHLH* | 1.9x10-12 | rs7953528 | GEFOS2 |
| RPS6KA5 | 2.0x10-15 | rs1286083 | GEFOS2 |
| RSPO3* | 3.0x10-8 | rs13204965 | Duncan 2011 |
| RUNX2* | 5.6x10-11 | rs11755164 | GEFOS2 |
| SALL1* | 1.9x10-22 | rs1566045 | GEFOS2 |
| SHFM1* | 8.1x10-48 (5.9x10-11) | rs4727338 | GEFOS1, GEFOS2 |
| SLC25A13 | 8.1x10-48 (5.9x10-11) | rs4727338 | GEFOS2 |
| SMG6 | 9.8x10-19 | rs4790881 | GEFOS2 |
| SMOC1* | 4.0x10-13 | rs227425 | Zhang 2014 |
| SOST* | 2.0x10-11 | rs4792909 | GEFOS2, Styrkarsdottir 2009 |
| SOX4* | 2.7x10-13 | rs9466056 | GEFOS2 |
| SOX6* | 1.1x10-32 | rs7108738 | GEFOS1, GEFOS2, Hsu 2010 [Ref. (65)] |
| SOX9* | 1.9x10-11 | rs7217932 | GEFOS2 |
| SP7* | 3.0x10-20 | rs2016266 | GEFOS1, GEFOS2, Styrkarsdottir 2009, Timpson 2009 [Ref. (66)] |
| SPP1* | 1.2x10-27 (1.7x10-8) | rs6532023 | GEFOS2 |
| SPTBN1 | 2.3x10-18 (2.6x10-8) | rs4233949 | GEFOS1, GEFOS2 |
| STARD3NL | 3.8x10-38 | rs6959212 | GEFOS1, GEFOS2 |
| SUCO* | 8.5x10-15 | rs479336 | GEFOS2 |
| SUPT3H | 5.6x10-11 | rs11755164 | GEFOS2 |
| TNFRSF11A (RANK)* | 1.6x10-17 | rs884205 | GEFOS1, GEFOS2, Styrkarsdottir 2009 |
| TNFRSF11B (OPG)* | 3.2x10-39 | rs2062377 | GEFOS1, GEFOS2, Styrkarsdottir 2008, Richards 2008 |
| TNFSF11 (RANKL)* | 5.4x10-25 | rs9533090 | GEFOS1, GEFOS2, Styrkarsdottir 2008 & 2009 |
| WLS* | 2.6x10-13 | rs1430742 | GEFOS1, Hsu 2010, Duncan 2011 |
| WNT16* | 3.2x10-51 | rs3801387 | GEFOS2, Zheng 2012 & 2015 |
| WNT5B* | 5.6x10-12 | rs2887571 | GEFOS2 |
| XKR9 | 1.9x10-8 | rs7017914 | GEFOS2 |
| ZBTB40 | 7.4x10-57 | rs6426749 | GEFOS1, GEFOS2, Duncan 2011, Styrkarsdottir 2008 |
| ZNF408 | 5.1x10-18 | rs7932354 | GEFOS1, GEFOS2 |
Strongest BMD/fracture p-values and corresponding single nucleotide polymorphisms (SNPs, identified according to dbSNP) are shown; only signals with a p-value less than 5x10-8 were included. * Indicates genes for which additional evidence of involvement in bone development and metabolism is available.
The first two major GWAS were published in 2008 by Styrkarsdottir and cols. and Richards and cols, interrogating genetic association to low BMD and low trauma fractures (18,19). Whole sample sizes comprised 13,786 and 8,557 individuals, respectively, and five major genes were identified: OPG, RANKL, LRP5, ESR1 and ZBTB40 (Table 3). As previously mentioned, OPG and RANKL regulate osteoclast differentiation and activation, and LRP5 is a crucial mediator of Wnt signalling in bone formation. ESR1, which encodes for the oestrogen receptor, has long been considered a candidate gene for osteoporosis, based on earlier linkage studies and oestrogens’ prominent physiological role in bone remodelling. A remaining locus identified by Styrkarsdottir and cols., rs7524102, was strongly associated with both spine and hip BMD but obvious candidate genes lacked in its vicinity. Subsequent GWAS have confirmed this locus on larger cohorts (20,50-52), with p-value reaching 7.4x10-57 for association with hip BMD (20). Since these signals map to an intergenic region, the association has been attributed to the closest gene, ZBTB40. Up to now, a biological role for ZBTB40 in human or animal health is largely unknown.
The largest published GWAS, GEFOS2, was published in 2012 comprising data from > 80,000 subjects for BMD and > 130,000 fracture cases and controls (20). This study alone was able to identify 56 loci associated with BMD and 14 loci related to fracture risk, but still could only explain 5.8% of the genetic contribution to femoral neck BMD. These striking numbers epitomize both the great strength of GWAS in identifying genes related to common diseases and their great limitations in explaining the total genetic variability of such diseases, a concept commonly referred to as the missing heritability (5,7).
In 2015, a breakthrough GWAS based on whole-genome sequencing was published by Zheng and cols., with enough power to detect the effects of low-frequency variants (minor allele frequency [MAF] between 1-5%), which are usually not contemplated by genotype-based GWAS (53). Using this approach, the novel candidate gene EN1 was identified, significantly related to both BMD and fracture risk. Animal models and in vitro studies indicate a possible role for EN1 in osteoblasts, offering an exciting opportunity for the discovery of new mechanisms in bone formation (53). Finally, this study also suggests that lower frequency variants may have higher impact on BMD and fractures, warranting further studies.
A full list of the major 95 genes identified by GWAS is presented on Table 3. Remarkably, evidence of involvement in bone physiology is currently available for only 41 genes (shown in table). The remaining 54 genes were selected based on their physical proximity to the GWAS signal, and therefore their biological association to bone fragility needs to be further scrutinized.
Future challenges
The genetics of osteoporosis have been increasingly unravelled during the past two decades. Gene defects underlying syndromic diseases with prominent skeletal phenotype have been identified, as well as genetic variants related to idiopathic and/or extreme osteoporosis. Technological advances have allowed unbiased de novo discovery of novel candidate genes and also of numerous loci associated to common osteoporosis. Through all these different strategies, several novel pathways regulating bone remodelling and matrix homeostasis have been recognised, pushing the boundaries of the therapeutic arsenal for bone fragility.
Concomitantly, however, gaps on our understanding of these processes have become apparent. For example, even with a great number of subjects and SNPs analysed, the largest GWAS to date can only explain 5.8% of the genetic contribution to BMD variability. Furthermore, most candidate genes or loci identified by high-throughput genomic analysis remain to have their role in bone metabolism fully elucidated. Altogether, these shortcomings pose as research challenges, warranting further exploration. In the foreseeable future, genomic analysis with enough power to detect the effects of low-frequency variants may lead to the discovery of missing heritability.
Gene defects so far identified in association with idiopathic osteoporosis are likely to have a major causative role in determining these phenotypes, but a clear genotype/phenotype correlation and precise co-segregation within families are still lacking in many cases, suggesting that a contribution of yet unfound genetic modifiers may exist. Further studies of idiopathic osteoporosis interrogating the role of candidate genes identified by GWAS for which a function in bone is still unknown might help identify such modifiers or even uncover major causative roles for some of these novel candidates. Additionally, animal models and in vitro studies may help to clarify their biological function in bone strength.
In conclusion, major advances in the genetics of bone fragility have allowed a deeper understanding of bone remodelling, with translational implications in many instances. Several experimental sources of candidate genes for osteoporosis have arisen, particularly due to the study of rarer informative individuals and families but also through the advent of genome-scale methods for genetic analysis. It is hoped that the continued and concerted effort of clinicians and researchers, and ongoing technological progress will further illuminate the genetic basis of osteoporosis and enable more precise treatment strategies in the near future.
Acknowledgements
M.G.M.R.-B. holds an institutional Capes scholarship (Program 33002010062P5), and B.F.-d.-S. holds a Sao Paulo Research Foundation (Fapesp) Young Investigator award (grant number 2011/12696-4).
REFERENCES
- 1.1. Consensus Development Conference. Prophylaxis and treatment of osteoporosis. Am J Med. 1991;90:107-10. [DOI] [PubMed]
- 2.2. Cooper C, Atkinson EJ, Jacobsen SJ, O’Fallon WM, Melton LJ, 3rd. Population-based study of survival after osteoporotic fractures. Am J Epidemiol. 1993;137:1001-5. [DOI] [PubMed]
- 3.3. Kanis JA. Assessment of fracture risk and its application to screening for postmenopausal osteoporosis: synopsis of a WHO report. WHO Study Group. Osteoporos Int. 1994;4:368-81. [DOI] [PubMed]
- 4.4. Siris ES, Adler R, Bilezikian J, Bolognese M, Dawson-Hughes B, Favus MJ, et al. The clinical diagnosis of osteoporosis: a position statement from the National Bone Health Alliance Working Group. Osteoporos Int. 2014;25:1439-43. [DOI] [PMC free article] [PubMed]
- 5.5. Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747-53. [DOI] [PMC free article] [PubMed]
- 6.6. Deng HW, Chen WM, Recker S, Stegman MR, Li JL, Davies KM, et al. Genetic determination of Colles’ fracture and differential bone mass in women with and without Colles’ fracture. J Bone Miner Res. 2000;15:1243-52. [DOI] [PubMed]
- 7.7. Richards JB, Zheng HF, Spector TD. Genetics of osteoporosis from genome-wide association studies: advances and challenges. Nat Rev Genet. 2012;13:576-88. [DOI] [PubMed]
- 8.8. Andrew T, Antioniades L, Scurrah KJ, Macgregor AJ, Spector TD. Risk of wrist fracture in women is heritable and is influenced by genes that are largely independent of those influencing BMD. J Bone Miner Res. 2005;20:67-74. [DOI] [PubMed]
- 9.9. Pope FM, Nicholls AC, McPheat J, Talmud P, Owen R. Collagen genes and proteins in osteogenesis imperfecta. J Med Genet. 1985;22:466-78. [DOI] [PMC free article] [PubMed]
- 10.10. Frattini A, Orchard PJ, Sobacchi C, Giliani S, Abinun M, Mattsson JP, et al. Defects in TCIRG1 subunit of the vacuolar proton pump are responsible for a subset of human autosomal recessive osteopetrosis. Nat Genet. 2000;25:343-6. [DOI] [PubMed]
- 11.11. Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513-23. [DOI] [PubMed]
- 12.12. Brunkow ME, Gardner JC, Van Ness J, Paeper BW, Kovacevich BR, Proll S, et al. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. Am J Hum Genet. 2001;68:577-89. [DOI] [PMC free article] [PubMed]
- 13.13. Balemans W, Ebeling M, Patel N, Van Hul E, Olson P, Dioszegi M, et al. Increased bone density in sclerosteosis is due to the deficiency of a novel secreted protein (SOST). Hum Mol Genet. 2001;10:537-43. [DOI] [PubMed]
- 14.14. Keupp K, Beleggia F, Kayserili H, Barnes AM, Steiner M, Semler O, et al. Mutations in WNT1 cause different forms of bone fragility. Am J Hum Genet. 2013;92:565-74. [DOI] [PMC free article] [PubMed]
- 15.15. Laine CM, Joeng KS, Campeau PM, Kiviranta R, Tarkkonen K, Grover M, et al. WNT1 mutations in early-onset osteoporosis and osteogenesis imperfecta. N Engl J Med. 2013;368:1809-16. [DOI] [PMC free article] [PubMed]
- 16.16. van Dijk FS, Zillikens MC, Micha D, Riessland M, Marcelis CL, de Die-Smulders CE, et al. PLS3 mutations in X-linked osteoporosis with fractures. N Engl J Med. 2013;369:1529-36. [DOI] [PubMed]
- 17.17. Kiel DP, Demissie S, Dupuis J, Lunetta KL, Murabito JM, Karasik D. Genome-wide association with bone mass and geometry in the Framingham Heart Study. BMC Med Genet. 2007;8 Suppl 1:S14. [DOI] [PMC free article] [PubMed]
- 18.18. Richards JB, Rivadeneira F, Inouye M, Pastinen TM, Soranzo N, Wilson SG, et al. Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet. 2008;371:1505-12. [DOI] [PMC free article] [PubMed]
- 19.19. Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, et al. Multiple genetic loci for bone mineral density and fractures. N Engl J Med. 2008;358:2355-65. [DOI] [PubMed]
- 20.20. Estrada K, Styrkarsdottir U, Evangelou E, Hsu YH, Duncan EL, Ntzani EE, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44:491-501. [DOI] [PMC free article] [PubMed]
- 21.21. McClung MR, Grauer A, Boonen S, Bolognese MA, Brown JP, Diez-Perez A et al. Romosozumab in postmenopausal women with low bone mineral density. N Engl J Med. 2014;370:412-20. [DOI] [PubMed]
- 22.22. Masi L, Agnusdei D, Bilezikian J, Chappard D, Chapurlat R, Cianferotti L, et al. Taxonomy of rare genetic metabolic bone disorders. Osteoporos Int. 2015;26:2529-58. [DOI] [PubMed]
- 23.23. Rauch F, Glorieux FH. Osteogenesis imperfecta. Lancet. 2004;363:1377-85. [DOI] [PubMed]
- 24.24. Forlino A, Cabral WA, Barnes AM, Marini JC. New perspectives on osteogenesis imperfecta. Nat Rev Endocrinol. 2011;7:540-57. [DOI] [PMC free article] [PubMed]
- 25.25. Sillence DO, Senn A, Danks DM. Genetic heterogeneity in osteogenesis imperfecta. J Med Genet. 1979;16:101-16. [DOI] [PMC free article] [PubMed]
- 26.26. Cho TJ, Lee KE, Lee SK, Song SJ, Kim KJ, Jeon D, et al. A single recurrent mutation in the 5’-UTR of IFITM5 causes osteogenesis imperfecta type V. Am J Hum Genet. 2012;91:343-8. [DOI] [PMC free article] [PubMed]
- 27.27. Semler O, Garbes L, Keupp K, Swan D, Zimmermann K, Becker J, et al. A mutation in the 5’-UTR of IFITM5 creates an in-frame start codon and causes autosomal-dominant osteogenesis imperfecta type V with hyperplastic callus. Am J Hum Genet. 2012;91:349-57. [DOI] [PMC free article] [PubMed]
- 28.28. Marini JC, Blissett AR. New genes in bone development: what’s new in osteogenesis imperfecta. J Clin Endocrinol Metab. 2013;98:3095-103. [DOI] [PMC free article] [PubMed]
- 29.29. Bonafe L, Cormier-Daire V, Hall C, Lachman R, Mortier G, Mundlos S, et al. Nosology and classification of genetic skeletal disorders: 2015 revision. Am J Med Genet A. 2015;167:2869-92. [DOI] [PubMed]
- 30.30. Kelley BP, Malfait F, Bonafe L, Baldridge D, Homan E, Symoens S, et al. Mutations in FKBP10 cause recessive osteogenesis imperfecta and Bruck syndrome. J Bone Miner Res. 2011;26:666-72. [DOI] [PMC free article] [PubMed]
- 31.31. Garbes L, Kim K, Riess A, Hoyer-Kuhn H, Beleggia F, Bevot A, et al. Mutations in SEC24D, encoding a component of the COPII machinery, cause a syndromic form of osteogenesis imperfecta. Am J Hum Genet. 2015;96:432-9. [DOI] [PMC free article] [PubMed]
- 32.32. Stark Z, Savarirayan R. Osteopetrosis. Orphanet J Rare Dis. 2009;4:5. [DOI] [PMC free article] [PubMed]
- 33.33. Gelb BD, Shi GP, Chapman HA, Desnick RJ. Pycnodysostosis, a lysosomal disease caused by cathepsin K deficiency. Science. 1996;273:1236-8. [DOI] [PubMed]
- 34.34. Bone HG, Dempster DW, Eisman JA, Greenspan SL, McClung MR, Nakamura T, et al. Odanacatib for the treatment of postmenopausal osteoporosis: development history and design and participant characteristics of LOFT, the Long-Term Odanacatib Fracture Trial. Osteoporos Int. 2015;26:699-712. [DOI] [PMC free article] [PubMed]
- 35.35. Baron R, Kneissel M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat Med. 2013;19:179-92. [DOI] [PubMed]
- 36.36. Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, et al. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. 2002;346:1513-21. [DOI] [PubMed]
- 37.37. Balemans W, Patel N, Ebeling M, Van Hul E, Wuyts W, Lacza C, et al. Identification of a 52 kb deletion downstream of the SOST gene in patients with van Buchem disease. J Med Genet. 2002;39:91-7. [DOI] [PMC free article] [PubMed]
- 38.38. Simpson MA, Irving MD, Asilmaz E, Gray MJ, Dafou D, Elmslie FV, et al. Mutations in NOTCH2 cause Hajdu-Cheney syndrome, a disorder of severe and progressive bone loss. Nat Genet. 2011;43:303-5. [DOI] [PubMed]
- 39.39. Isidor B, Lindenbaum P, Pichon O, Bezieau S, Dina C, Jacquemont S, et al. Truncating mutations in the last exon of NOTCH2 cause a rare skeletal disorder with osteoporosis. Nat Genet. 2011;43:306-8. [DOI] [PubMed]
- 40.40. Bianchi ML. Hypophosphatasia: an overview of the disease and its treatment. Osteoporos Int. 2015;26:2743-57. [DOI] [PubMed]
- 41.41. Spotila LD, Constantinou CD, Sereda L, Ganguly A, Riggs BL, Prockop DJ. Mutation in a gene for type I procollagen (COL1A2) in a woman with postmenopausal osteoporosis: evidence for phenotypic and genotypic overlap with mild osteogenesis imperfecta. Proc Natl Acad Sci U S A. 1991;88:5423-7. [DOI] [PMC free article] [PubMed]
- 42.42. Spotila LD, Colige A, Sereda L, Constantinou-Deltas CD, Whyte MP, Riggs BL, et al. Mutation analysis of coding sequences for type I procollagen in individuals with low bone density. J Bone Miner Res. 1994;9:923-32. [DOI] [PubMed]
- 43.43. Hartikka H, Makitie O, Mannikko M, Doria AS, Daneman A, Cole WG, et al. Heterozygous mutations in the LDL receptor-related protein 5 (LRP5) gene are associated with primary osteoporosis in children. J Bone Miner Res. 2005;20:783-9. [DOI] [PubMed]
- 44.44. Korvala J, Loija M, Makitie O, Sochett E, Juppner H, Schnabel D, et al. Rare variations in WNT3A and DKK1 may predispose carriers to primary osteoporosis. Eur J Med Genet. 2012;55:515-9. [DOI] [PubMed]
- 45.45. Fahiminiya S, Majewski J, Roughley P, Roschger P, Klaushofer K, Rauch F. Whole-exome sequencing reveals a heterozygous LRP5 mutation in a 6-year-old boy with vertebral compression fractures and low trabecular bone density. Bone. 2013;57:41-6. [DOI] [PubMed]
- 46.46. Campos-Obando N, Oei L, Hoefsloot LH, Kiewiet RM, Klaver CC, Simon ME, et al. Osteoporotic vertebral fractures during pregnancy: be aware of a potential underlying genetic cause. J Clin Endocrinol Metab. 2014;99:1107-11. [DOI] [PubMed]
- 47.47. Cook FJ, Mumm S, Whyte MP, Wenkert D. Pregnancy-associated osteoporosis with a heterozygous deactivating LDL receptor-related protein 5 (LRP5) mutation and a homozygous methylenetetrahydrofolate reductase (MTHFR) polymorphism. J Bone Miner Res. 2014;29:922-8. [DOI] [PubMed]
- 48.48. Fahiminiya S, Majewski J, Al-Jallad H, Moffatt P, Mort J, Glorieux FH, et al. Osteoporosis caused by mutations in PLS3: clinical and bone tissue characteristics. J Bone Miner Res. 2014;29:1805-14. [DOI] [PubMed]
- 49.49. Laine CM, Wessman M, Toiviainen-Salo S, Kaunisto MA, Mayranpaa MK, Laine T, et al. A novel splice mutation in PLS3 causes X-linked early onset low-turnover osteoporosis. J Bone Miner Res. 2015;30:510-8. [DOI] [PubMed]
- 50.50. Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, et al. New sequence variants associated with bone mineral density. Nat Genet. 2009;41:15-7. [DOI] [PubMed]
- 51.51. Rivadeneira F, Styrkarsdottir U, Estrada K, Halldorsson BV, Hsu YH, Richards JB, et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009;41:1199-206. [DOI] [PMC free article] [PubMed]
- 52.52. Duncan EL, Danoy P, Kemp JP, Leo PJ, McCloskey E, Nicholson GC, et al. Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genet. 2011;7:e1001372. [DOI] [PMC free article] [PubMed]
- 53.53. Zheng HF, Forgetta V, Hsu YH, Estrada K, Rosello-Diez A, Leo PJ, et al. Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature. 2015;526:112-7. [DOI] [PMC free article] [PubMed]
- 54.54. Xiong DH, Liu XG, Guo YF, Tan LJ, Wang L, Sha BY, et al. Genome-wide association and follow-up replication studies identified ADAMTS18 and TGFBR3 as bone mass candidate genes in different ethnic groups. Am J Hum Genet. 2009;84:388-98. [DOI] [PMC free article] [PubMed]
- 55.55. Guo Y, Tan LJ, Lei SF, Yang TL, Chen XD, Zhang F, et al. Genome-wide association study identifies ALDH7A1 as a novel susceptibility gene for osteoporosis. PLoS Genet. 2010;6:e1000806. [DOI] [PMC free article] [PubMed]
- 56.56. Tan LJ, Wang ZE, Wu KH, Chen XD, Zhu H, Lu S, et al. Bivariate Genome-Wide Association Study Implicates ATP6V1G1 as a Novel Pleiotropic Locus Underlying Osteoporosis and Age at Menarche. J Clin Endocrinol Metab. 2015;100:E1457-66. [DOI] [PMC free article] [PubMed]
- 57.57. Zhang L, Choi HJ, Estrada K, Leo PJ, Li J, Pei YF, et al. Multistage genome-wide association meta-analyses identified two new loci for bone mineral density. Hum Mol Genet. 2014;23:1923-33. [DOI] [PMC free article] [PubMed]
- 58.58. Zheng HF, Tobias JH, Duncan E, Evans DM, Eriksson J, Paternoster L, et al. WNT16 influences bone mineral density, cortical bone thickness, bone strength, and osteoporotic fracture risk. PLoS Genet. 2012;8:e1002745. [DOI] [PMC free article] [PubMed]
- 59.59. Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban HJ, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41:527-34. [DOI] [PubMed]
- 60.60. Paternoster L, Lorentzon M, Lehtimaki T, Eriksson J, Kahonen M, Raitakari O, et al. Genetic determinants of trabecular and cortical volumetric bone mineral densities and bone microstructure. PLoS Genet. 2013;9:e1003247. [DOI] [PMC free article] [PubMed]
- 61.61. Kung AW, Xiao SM, Cherny S, Li GH, Gao Y, Tso G, et al. Association of JAG1 with bone mineral density and osteoporotic fractures: a genome-wide association study and follow-up replication studies. Am J Hum Genet. 2010;86:229-39. [DOI] [PMC free article] [PubMed]
- 62.62. Styrkarsdottir U, Thorleifsson G, Sulem P, Gudbjartsson DF, Sigurdsson A, Jonasdottir A, et al. Nonsense mutation in the LGR4 gene is associated with several human diseases and other traits. Nature. 2013;497:517-20. [DOI] [PubMed]
- 63.63. Kaufman JM, Ostertag A, Saint-Pierre A, Cohen-Solal M, Boland A, Van Pottelbergh I, et al. Genome-wide linkage screen of bone mineral density (BMD) in European pedigrees ascertained through a male relative with low BMD values: evidence for quantitative trait loci on 17q21-23, 11q12-13, 13q12-14, and 22q11. J Clin Endocrinol Metab. 2008;93:3755-62. [DOI] [PubMed]
- 64.64. Hwang JY, Lee SH, Go MJ, Kim BJ, Kou I, Ikegawa S, et al. Meta-analysis identifies a MECOM gene as a novel predisposing factor of osteoporotic fracture. J Med Genet. 2013;50:212-9. [DOI] [PMC free article] [PubMed]
- 65.65. Hsu YH, Zillikens MC, Wilson SG, Farber CR, Demissie S, Soranzo N, et al. An integration of genome-wide association study and gene expression profiling to prioritize the discovery of novel susceptibility Loci for osteoporosis-related traits. PLoS Genet. 2010;6:e1000977. [DOI] [PMC free article] [PubMed]
- 66.66. Timpson NJ, Tobias JH, Richards JB, Soranzo N, Duncan EL, Sims AM, et al. Common variants in the region around Osterix are associated with bone mineral density and growth in childhood. Hum Mol Genet. 2009;18:1510-7. [DOI] [PMC free article] [PubMed]