SUMMARY
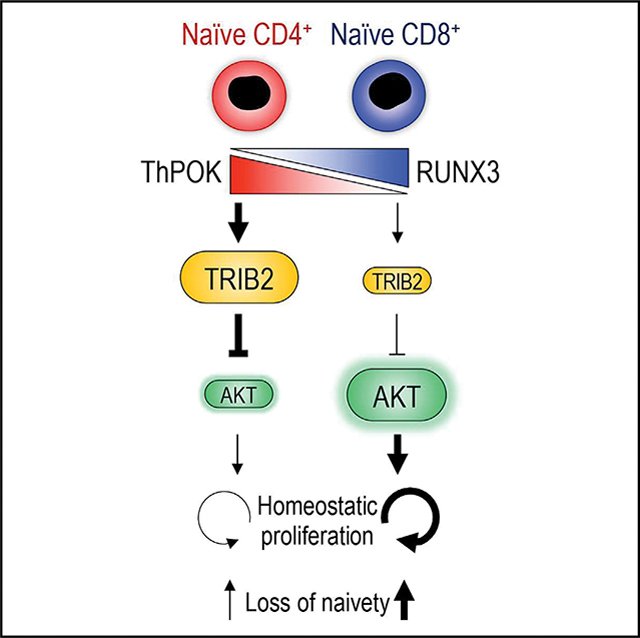
Naive CD4+ T cells are more resistant to age-related loss than naive CD8+ T cells, suggesting mechanisms that preferentially protect naive CD4+ T cells during aging. Here, we show that TRIB2 is more abundant in naive CD4+ than CD8+ T cells and counteracts quiescence exit by suppressing AKT activation. TRIB2 deficiency increases AKT activity and accelerates proliferation and differentiation in response to interleukin-7 (IL-7) in humans and during lymphopenia in mice. TRIB2 transcription is controlled by the lineage-determining transcription factors ThPOK and RUNX3. Ablation of Zbtb7b (encoding ThPOK) and Cbfb (obligatory RUNT cofactor) attenuates the difference in lymphopenia-induced proliferation between naive CD4+ and CD8+ cells. In older adults, ThPOK and TRIB2 expression wanes in naive CD4+ T cells, causing loss of naivety. These findings assign TRIB2 a key role in regulating T cell homeostasis and provide a model to explain the lesser resilience of CD8+ T cells to undergo changes with age.
In brief
Naive CD4+ and CD8+ T cells differ in homeostatic proliferation, which may explain the higher loss of naive CD8+ cells during human aging. Here, Cao et al. describe that TRIB2, regulated by ThPOK and RUNX3, protects naive CD4+ T cells from untimely quiescence exit and loss of naivety.
Graphical Abstract
INTRODUCTION
T cell aging is associated with an increased vulnerability to infections and malignant diseases.1 Although clearly multifactorial, a reduced number of T lymphocytes, and in particular naive cells, is a major determining factor. Mechanisms of T cell generation drastically change over a lifetime. While the thymus is the major source of T cells before puberty, it contributes to less than 20% of T cell production in early adulthood and further dwindles with older age. In humans, maintenance of the T cell compartment in adulthood mainly depends on homeostatic proliferation that occurs in the T cell zones of lymph nodes.2 Although homeostatic proliferation is relatively efficient up to older age, the frequency of naive T cells declines with age. The magnitude of this decline in humans is strikingly different for naive CD4+ and CD8+ T cells.3 Naive CD4+ T cells, although reduced, are still present in high frequencies in older adults, while naive CD8+ T cells are largely diminished. This difference is even larger if one considers that the phenotypically defined naive CD8+ T cell compartment includes stem-like memory cells.4 This is not the case for CD4+ T cells where stem-like memory T cells are contained in the central memory T cell subset.5,6 In biomarker studies, loss of naive CD8+ T cells is the strongest immunological correlate of age.7 These divergent response patterns of CD4+ and CD8+ T cells during aging cannot be attributed to thymic T cell generation but are likely caused by unequal peripheral homeostasis.
Homeostatic proliferation is less important for maintenance of the peripheral T cell compartment in mice.2,8 However, several lines of evidence suggest that homeostatic proliferation in the mouse is also T cell subset specific. In mice, naive cell differentiation into virtual memory cells driven by cytokines in the absence of antigens is largely restricted to CD8+ T cells.9 Such virtual memory cells accumulate with age and show evidence of dysfunction.10 Also, naive CD8+ T cells proliferate faster in response to homeostatic cytokines and largely outnumber CD4+ T cell after adoptive transfer into lymphopenic mice due to increased homeostatic proliferation.11,12
Peripheral naive T cell survival and homeostatic proliferation depend on interleukin-7 (IL-7) signaling and T cell receptor (TCR) recognition of self-antigen complexes/major histocompatibility complexes (MHCs).13 In secondary lymphoid organs, fibroblastic reticular cells support the maintenance of naive T cells by providing IL-7. The lymph node architecture undergoes changes with age, including accumulation of adipocytes and fibrosis. The latter has been shown to be accelerated by chronic or recurrent infections and has been shown to cause the decline in peripheral naive T cells.14 IL7R levels were shown to preferentially decline on CD8+ T cells with age, and a relatively CD8+ T cell-selective reduction in chromatin accessibility at the IL7R locus has been reported, suggesting that their survival may be impaired.15 However, the increased propensity of CD8+ T cells to proliferate and differentiate argues against such a simplified model. Turnover of CD8+ T cells under steady-state conditions is higher than that of CD4+ T cells, as evidenced by the frequency of Ki67+ cells.16 Many age-associated changes in chromatin accessibility and the transcriptome in naive CD4+ and CD8+ T cells resemble those of T cell differentiation. These changes are more pronounced in the CD8+ T cell compartment.15,17,18 Moreover, accumulation of effector memory T cells with age culminating in T effector memory CD45RA (TEMRA) cells is characteristic for CD8+ T cells. Collectively, these observations imply that CD8+ T cells are more prone to exit quiescence, leading to increased homeostatic proliferation and differentiation that may eventually lead to the depletion of naive cells. If this is a cell-intrinsic trait, it should be reflected by differential gene expression.
Here, we found that TRIB2, a regulator of AKT activity, is more expressed in naive CD4+ compared with naive CD8+ T cells. Differential expression was orchestrated by the CD4+ and CD8+ lineage-determining transcription factors ThPOK and RUNX3. ThPOK induced and RUNX3 repressed TRIB2 transcription. Low TRIB2 expression enabled elevated AKT activity in naive CD8+ T cells in response to homeostatic cytokines. Consequently, T cell activation and proliferation were greater in naive CD8+ T cells, and loss of naivety was more profound. Differential expression of Trib2 between naive CD4+ and CD8+ T cells was abolished in ThPOK/Cbfb (Cbfb encodes a mandatory RUNT cofactor) double knockout mice, almost equalizing lymphopenia-induced proliferation in these two subsets. Results were reproducible in Trib2 knockout mice.
RESULTS
Reduced TRIB2 expression enables higher turnover of naive CD8+ than CD4+ T cells
Circulating human naive (CD45RA+ CCR7+) CD8+ T cells are more severely depleted with increasing age than naive CD4+ T cells, indicating that homeostatic proliferation is less efficient to sustain their numbers (Figure 1A). IL-7 receptor (IL-7R) expression is slightly lower in human naive CD8+ than CD4+ T cells, raising the possibility that IL-7 signaling is more limiting for CD8+ than CD4+ T cells (Figure 1B). However, despite the reduced IL-7R expression, turnover rates under steady-state condition are higher for naive CD8+ T cells. Frequencies of Ki67+ T cells are higher in naive CD8+ T cells, raising the possibility that the preferential loss in naive CD8+ T cells is a consequence of a failure in maintaining quiescence (Figure 1C). To identify candidate genes that may be involved in differentially regulating T cell homeostasis, we compared gene expression of naive human CD4+ and CD8+ T cells in an available dataset (GEO: GSE75406).19 3,734 transcripts were differentially expressed at an adjusted significance level of p < 0.05 (Figure 1D). Among the top ten differentially expressed sequences were CD4 and CD8; CD40L, known as a CD4 help-related molecule20; CD248; TRIB2; and the pseudogene ADGRE4P. In our subsequent studies, we focused on TRIB2, which plays a role in several signaling pathways and restrains thymocyte proliferation in T cell development.21 Higher TRIB2 expression in purified naive CD4+ T cells was confirmed by qRT-PCR (Figures 1E and S1A) and immunoblot (Figures 1F and 1G). To investigate the role of TRIB2 in T cell homeostasis, we silenced TRIB2 in naive CD4+ T cells by culturing them with customized FANA-antisense oligonucleotides (Figure S1B). Low-level anti-CD28 antibody was added to improve silencing efficiency in non-activated cells. TRIB2 deficiency increased IL-7-induced proliferation to a level closer to the proliferation seen for naive CD8+ T cells (Figure 1H) as well as promoted TCR-induced proliferation (Figure 1I). Conversely, forced TRIB2 overexpression downregulated TCR-induced proliferation in naive CD4+ T cells (Figure 1J). Taken together, these data indicate that TRIB2 is involved in raising the threshold for CD4+ T cells to exit quiescence and to proliferate.
Figure 1. Increased expression of TRIB2 in human naive CD4+ T cells represses proliferation.
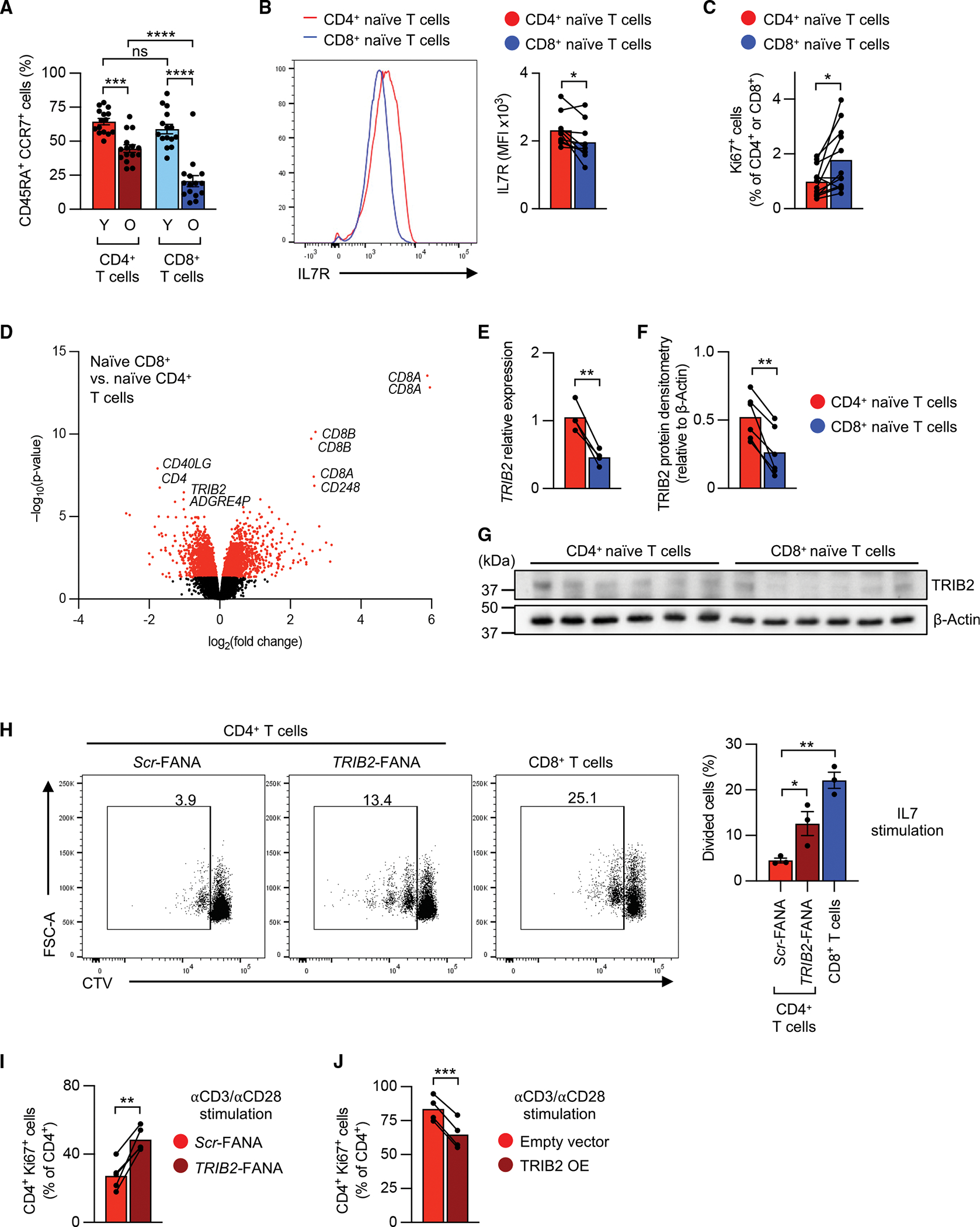
(A) Frequencies of human naive (CD45RA+ CCR7+) cells as percentage of total CD4+ or CD8+ T cells in young (Y; <35 years) and older (O; >65 years) individuals.
(B)Representative histograms and mean fluorescence intensities (MFIs) of IL-7 receptor (IL-7R) staining in naive CD4+ or CD8+ T cells as measured by flow cytometry.
(C) Frequencies of naive Ki67+ cells among CD4+ or CD8+ T cells.
(D) Volcano plot of differentially expressed (p < 0.05, red colored) transcripts in naive CD8+ vs. CD4+ T cells (analyzed from GEO: GSE75406). Top 10 differentially expressed transcripts are labeled.
(E–G) TRIB2 mRNA (E) and protein levels (F and G) in purified human naive CD4+ and CD8+ T cells.
(H) Representative flow cytometry plots and summary of frequencies of proliferating cells. Human naive CD4+ or CD8+ T cells were labeled with CellTrace Violet (CTV), then cultured on 2 μg/mL plate-bound anti-CD28 antibody with 50 ng/mL IL-7 for 7 days in the presence of TRIB2 or scrambled (Scr) FANA antisense oligonucleotides (FANA-ASOs).
(I) Proportion of Ki67+ cells after TRIB2 knockdown as determined by flow cytometry. Purified human naive CD4+ T cells were cultured on plate-bound anti-CD3/anti-CD28 antibodies (5 μg/mL) for 5 days in the presence of indicated FANA-ASOs.
(J) Proportion of Ki67+ cells after TRIB2 overexpression as determined by flow cytometry. Purified human naive CD4+ T cells were transduced with GFP-containing control (empty vector) or GFP-containing TRIB2-overexpressing (TRIB2-OE) lentivirus; after 5 days, Ki67+ cells within CD4+ GFP+ cells were determined. Results are shown as mean ± SEM in (A). Data points represent distinct biological replicates. Results are from 1 experiment (F) or pooled from 2 (C, E, and J), 3 (H and I), 5 (B), or 6 experiments (A). Data were compared by two-way ANOVA with post-hoc Tukey test (A), two-tailed, paired t tests (B, C, E, F, I, and J), or one-way ANOVA with post-hoc Dunnett test (H). *p < 0.05, **p % 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. ns, not significant.
See also Figure S1
TRIB2 inhibits AKT phosphorylation in T cells
AKT activation is required for T cell homeostatic proliferation, and higher AKT activation results in breaking T cell quiescence.22,23 TRIB2 has been reported to regulate AKT depending on the cell type, inhibiting it in liver cells and adipocytes while activating it in tumor cells.24–26 We hypothesized that TRIB2 prevents AKT activation in T cells and lowers the sensitivity of naive CD4+ T cells to homeostatic cytokine stimulation. Consistent with their higher homeostatic proliferation, naive CD8+ T cells had higher AKT phosphorylation than naive CD4+ T cells at both residues, Thr308 and Ser473, directly ex vivo (Figure 2A). This difference was even more pronounced after in vitro IL-7 stimulation. TRIB2-silencing in naive CD4+ T cells increased IL-7-induced AKT phosphorylation (Figure 2B), while not influencing classical IL-7 signaling, as determined by comparable STAT5 phosphorylation (Figure S1C). An increased number of TRIB2-silenced naive CD4+ T cells entered proliferation, which was completely abolished by AKT inhibition (Figure 2C). Conversely, overexpression of TRIB2 in naive CD8+ T cells reduced TCR-induced AKT phosphorylation at both sites (Figure 2D). To further explore TRIB2’s involvement in AKT signaling, we stimulated naive CD4+ T cells with low-dose anti-CD3/anti-CD28 antibodies in the presence of scrambled or TRIB2 FANA oligonucleotides (oligos) for 2 days; cells were rested for 2 additional days and then restimulated by TCR cross-linking. We found that TRIB2 silencing increased AKT and also ERK phosphorylation, but not proximal TCR signaling, as shown by equal phosphorylation of CD3ζ and ZAP70 (Figure 2E). As a consequence of the sustained AKT activation after TRIB2 silencing, S6 phosphorylation was higher and FOXO1 protein expression was lower on day 5 after stimulation (Figure 2F). Taken together, these data show that TRIB2 in naive CD4+ T cells restrains AKT activation to keep them quiescent.
Figure 2. TRIB2 inhibits AKT activation.
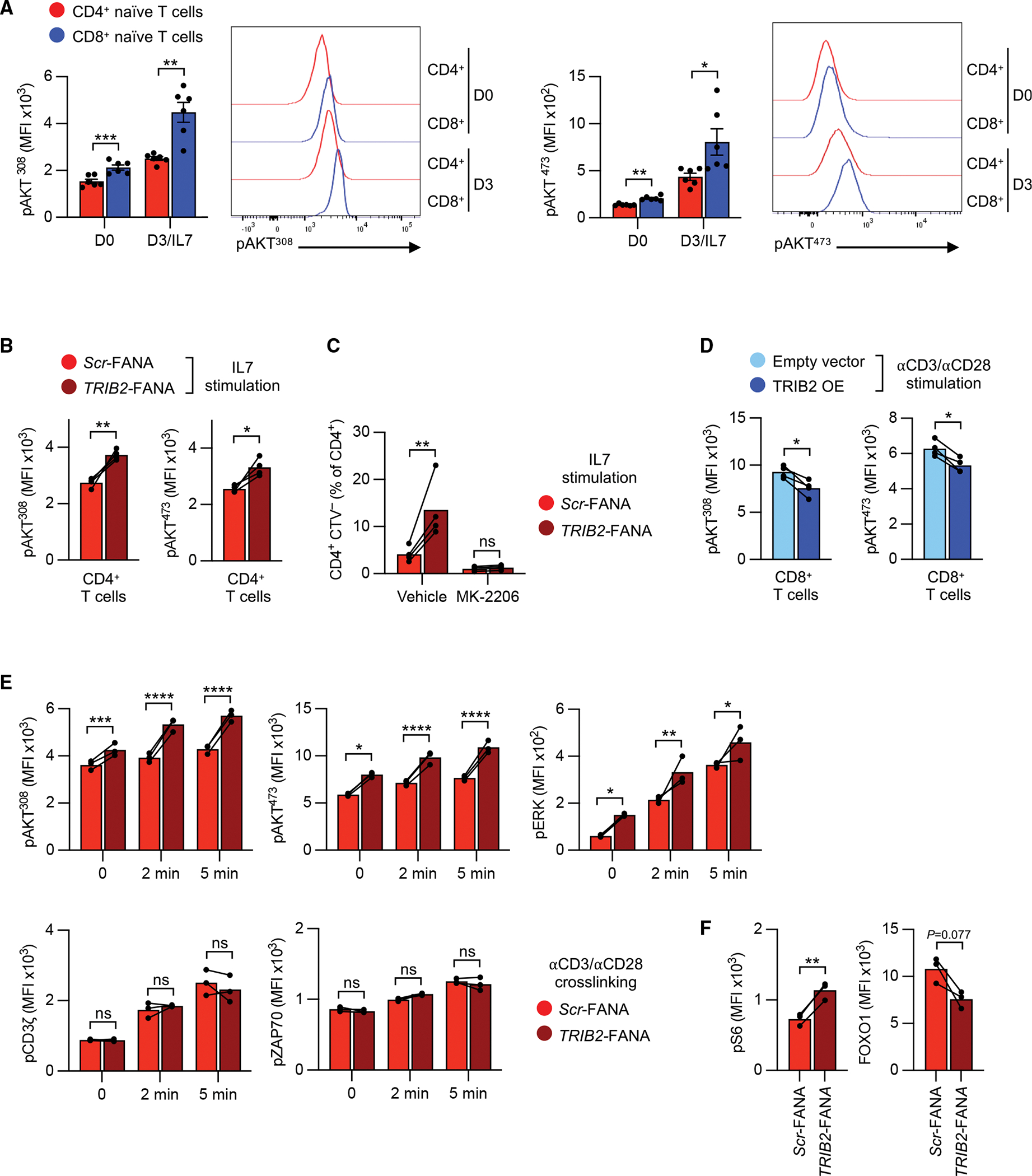
(A) Representative histograms and MFIs of phospho-AKT (Thr308 or Ser473) as measured by flow cytometry in purified human naive CD4+ or CD8+ T cells before and after stimulation with 50 ng/mL IL-7 for 3 days.
(B) Phospho-AKT (Thr308 or Ser473) in purified naive CD4+ T cells 3 days after stimulation with 50 ng/mL IL-7 and plate-bound anti-CD28 antibody (2 μg/mL) cultured with TRIB2-targeting or Scr FANA-ASOs.
(C) Proliferation as assessed by CTV dilution of purified naive CD4+ T cells that were treated with FANA-ASOs as in (B). Cells were cultured with IL-7, anti-CD28 antibody, and 5 μM AKT inhibitor MK-2206 or vehicle control for 7 days.
(D) Flow cytometry measurements of phospho-AKT (Thr308 or Ser473) in anti-CD3/anti-CD28-stimulated, purified, naive CD8+ T cells 3 days after transduction with GFP-containing empty vector or GFP-containing TRIB2-OE lentivirus.
(E) Kinetics of indicated phospho-proteins as measured by flow cytometry. Purified human naive CD4+ T cells were cultured on plate-bound anti-CD3/anti-CD28 antibody (2 μg/mL) in the presence of indicated FANA-ASOs for 2 days. Then, cells were rested for 2 additional days and restimulated by CD3/CD28 cross-linking.
(F) Flow cytometry measurements of phospho-S6 ribosomal protein (Ser235/236, pS6) and FOXO1 protein. Purified human naive CD4+ T cells were cultured on plate-bound anti-CD3/anti-CD28 antibodies (5 μg/mL) in the presence of indicated FANA-ASOs for 5 days.
Results are shown as mean ± SEM in (A). All datapoints represent distinct biological replicates. Results are pooled from 2 (B–D) or 3 experiments (A) or are representative of 2 experiments with 3 samples in each experiment (E and F). Data were compared by two-tailed, paired t tests (A, B, D, and F) or two-way ANOVA with post-hoc Šídák test (C and E). *p < 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. ns, not significant.
See also Figure S1
TRIB2 heightens the threshold for T cell activation and effector cell differentiation
To determine whether TRIB2 silencing promotes naive CD4+ T cells to exit quiescence and enter memory cell differentiation, we cultured naive CD4+ T cells with IL-7 and plate-bound anti-CD28 antibody with scramble or TRIB2 FANA oligos. After 7 days, the majority of control-transfected T cells had a naive CD45RA+ CCR7+ phenotype; in contrast, around 50% of TRIB2-silenced CD4+ cells had lost CD45RA expression, indicating that they had started to differentiate (Figure 3A). Also, TRIB2-silenced cells started to gain CD25 expression (Figure 3B). TRIB2 knockdown did not affect naive T cell survival (Figure S2A). Although less expressed in naive CD8+ T cells, TRIB2 silencing also promoted the loss of naive traits in naive CD8+ T cell (Figures S2B and S2C). To investigate the impact of TRIB2 on TCR-mediated activation, naive CD4+ T cells were stimulated with plate-bound anti-CD3/anti-CD28 antibodies with or without TRIB2 silencing FANA oligos for 5 days. Similar to the IL-7 culture, CD25 expression was higher in TRIB2-silenced CD4+ T cells (Figure 3C). Moreover, production of IL-2, tumor necrosis factor α (TNF-α), interferon γ (IFNγ), and GZMB were increased in TRIB2-silenced CD4+ T cells (Figures 3D and S2D). Silencing TRIB2 in naive CD8+ T cells also had similar effects (Figures S2E and S2F). In contrast, overexpression of TRIB2 in naive CD8+ T cells reduced GZMB levels and cytokine production after anti-CD3/anti-CD28 antibody stimulation (Figure 3E). Also, the inhibitory effects of TRIB2 in CD4+ T cells on cytokines, GZMB, and CD25 expression can be further enhanced by forced TRIB2 overexpression (Figures S2G and S2H). These findings support the notion that TRIB2 heightens the activation threshold of T cells and prevents their untimely differentiation into effector cells during homeostatic proliferation.
Figure 3. TRIB2 heightens the threshold for T cell activation and differentiation.
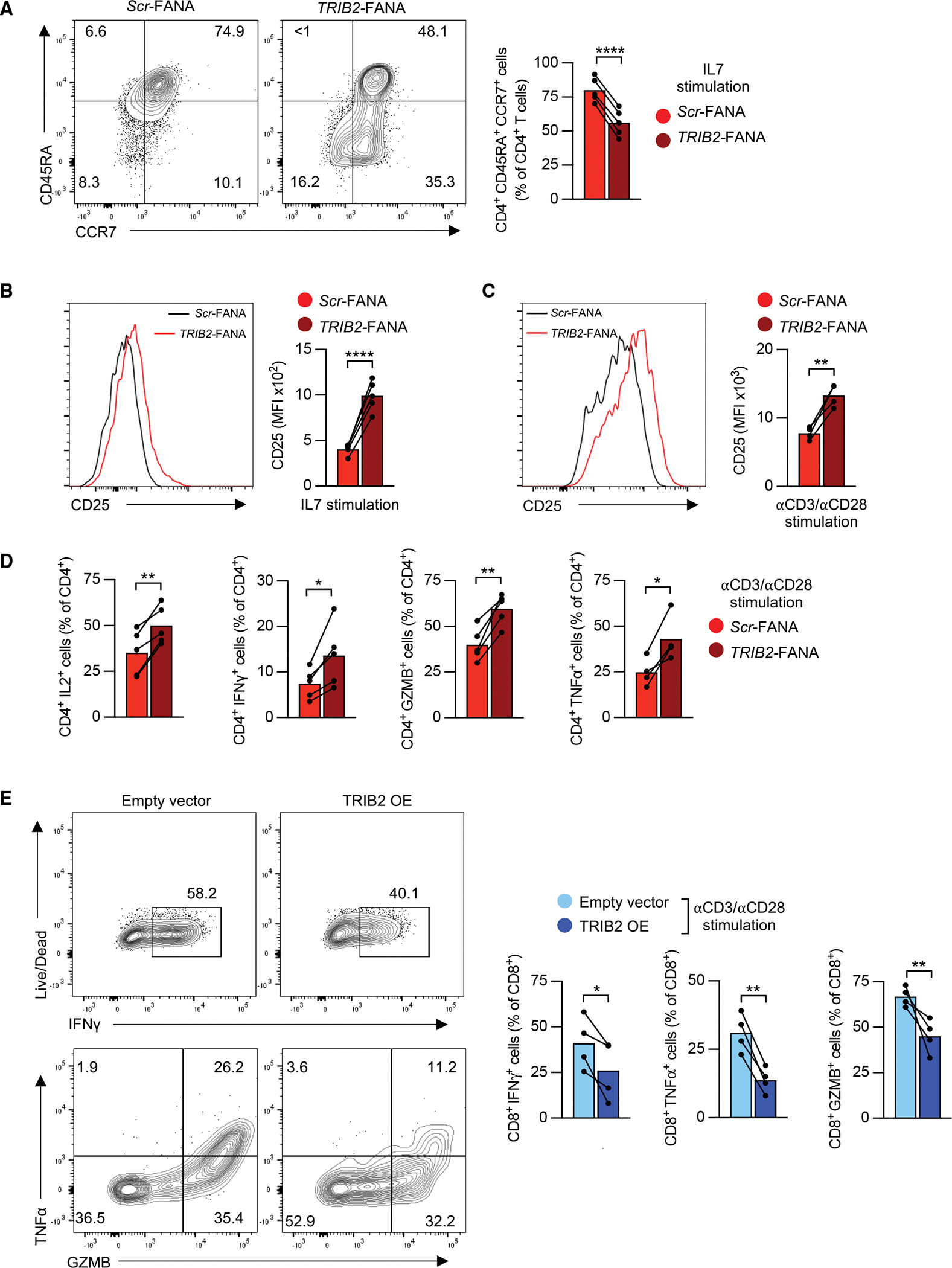
(A) Representative contour plots and frequencies of human naive CD45RA+ CCR7+ CD4+ T cells after FANA-ASO-mediated TRIB2 knockdown. Purified human naive CD4+ T cells were cultured with 50 ng/mL IL-7 and plate-bound anti-CD28 antibody (2 μg/mL) in the presence of indicated FANA-ASOs for 7 days.
(B) CD25 expression was determined in cells cultured as in (A). MFI, mean fluorescence intensity.
(C and D) CD25 expression (C) and cytokine production (D) in naive CD45RA+ CCR7+ CD4+ T cells after anti-CD3/anti-CD28 stimulation. Purified human naive CD4+ T cells were cultured with 5 μg/mL plate-bound anti-CD3/anti-CD28 antibodies for 5 days in the presence of indicated FANA-ASOs. For cytokine production, cells were restimulated with PMA and ionomycin for 4 h, and cytokine production within CD4+ cells was assessed.
(E) Human naive CD8+ T cells were stimulated with anti-CD3/anti-CD28 antibodies, transduced with GFP-containing, empty vector or GFP-containing TRIB2-OE lentivirus and cultured for 5 days. Cells were restimulated with PMA and ionomycin for 4 h, and cytokine production within CD8+ GFP+ cells was assessed. All datapoints represent distinct biological replicates.
Results are pooled from 2 (E) or 3 experiments (A–D). Data were compared by two-tailed, paired t tests (A–E). *p < 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
See also Figure S2
Lack of TRIB2 increases LIP of CD4+ T cells
Murine naive CD4+ and CD8+ T cells have similar differential Trib2 expression patterns to human T cells (Figure S3A). To examine TRIB2’s role in homeostatic proliferation in vivo, we used Trib2 knockout (KO) mice. TRIB2 deficiency was confirmed by immunoblot (Figure S3B). Consistent with a previous report,21 thymocyte numbers in Trib2 KO mice were increased due to elevated numbers of double-positive (DP) and single-positive (SP) cells (Figures S3C and S3D). In line with increased thymic output, the total number of splenocytes was increased in Trib2 KO mice (Figure S3E). However, spleens of Trib2 KO mice had similar frequencies of CD4+ and CD8+ T cell subsets and similar proportions of naive and memory CD4+ or CD8+ T cells (Figure S3F), suggesting that increased thymic output elevated T cell numbers in the periphery of Trib2 mice. Consistent with findings in TRIB2-silenced human naive CD4+ T cells, Trib2 deficiency rendered purified CD62Lhi CD44− naive CD4+ T cells more responsive to suboptimal stimulation with low doses of anti-CD3/anti-CD28 antibodies. They proliferated faster, expressed more activation markers, and produced more IFNγ (Figures S4A and S4B). To explore TRIB2’s impact on homeostatic proliferation, we used the model of lymphopenia-induced proliferation (LIP), in which accelerated proliferation is driven by self-MHC recognition and IL-7.27 CD62Lhi CD44− CD4+ and CD8+ T cells from wild-type (WT) or Trib2 KO mice (CD45.2) were labeled with CellTrace Violet (CTV) and adoptively transferred into irradiated CD45.1 WT mice; after 1 week, splenocytes were analyzed. Trib2-deficient naive T cells expanded more than WT naive T cells, mainly due to increased CD4+ T cell expansion (Figures 4A–4C). Analysis of CTV dilution also showed that Trib2 deficiency increased LIP of CD4+ T cells, thereby diminishing the difference between CD4+ and CD8+ T cells (Figure 4D). Accelerated LIP is associated with increased T cell differentiation, more so in CD8+ than CD4+ WT mice. Trib2 deficiency induced higher LIP-driven differentiation into CD122+ CD44+ memory cells for CD4+, but not CD8+, T cells (Figures 4E and 4F). Increased LIP in CD4+ T cells was not due to a change in IL-7R expression, consistent with the observation that TRIB2-silenced human naive CD4+ T cells had comparable phosphorylated (phospho)-STAT5 levels after IL-7 stimulation (Figures S4C and S1C). Taken together, TRIB2 restrains activation and proliferation of naive CD4+ T cells in lymphopenic mice, supporting a role of TRIB2 in maintaining homeostasis of CD4+ T cells.
Figure 4. Loss of Trib2 in CD4+ T cells increases lymphopenia-induced proliferation.
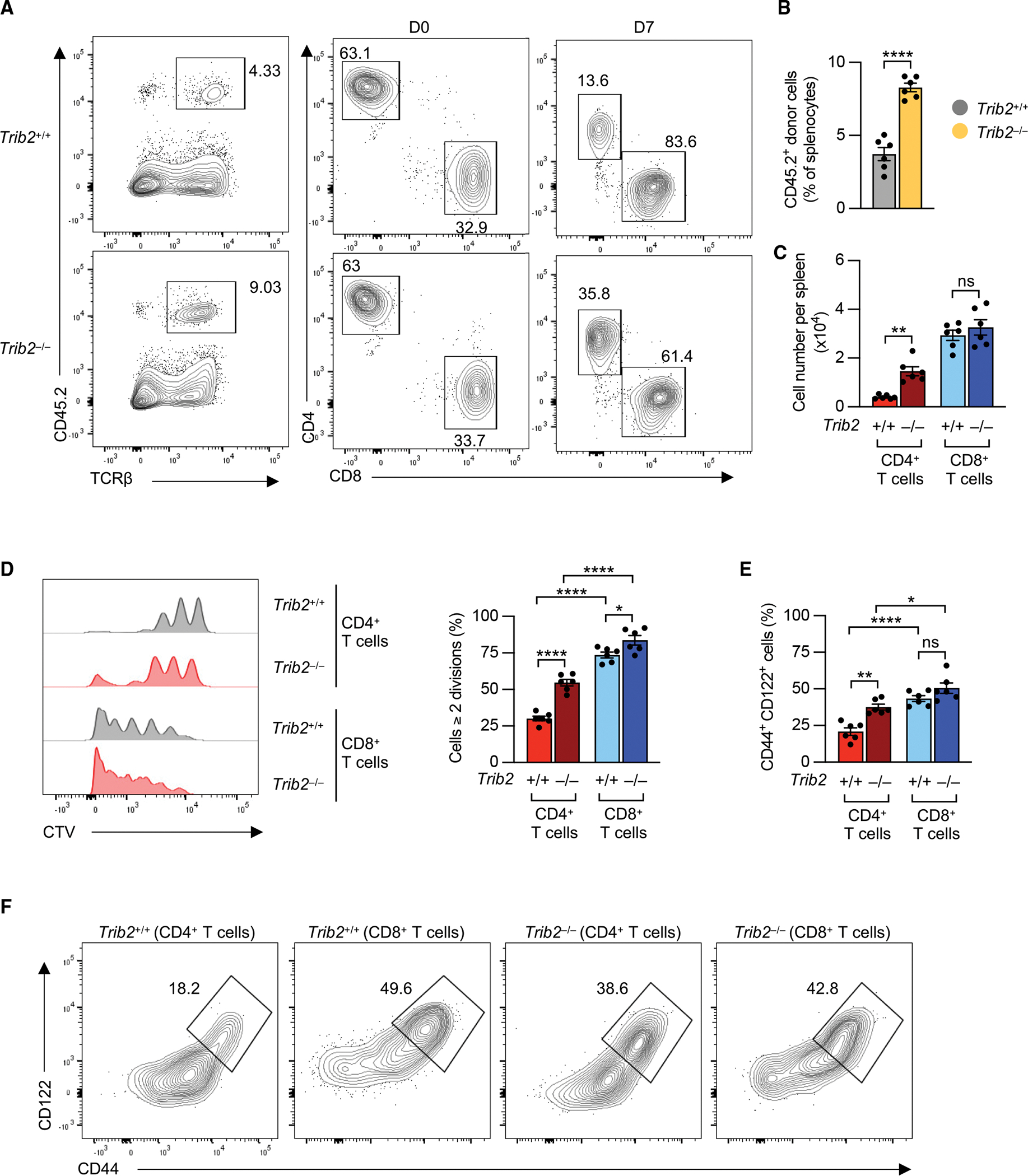
Fluorescence-activated cell sorting (FACS)-purified naive T cells from CD45.2 wild-type and Trib2 knockout mice were labeled with CTV and transferred into irradiated CD45.1 C57BL/6 mice. After 7 days, splenocytes were harvested, and frequencies of donor CD4+ and CD8+ T cells (A–C), CTV dilution (D), and differentiation state (E and F) were determined by flow cytometry. Results are shown as means ± SEM. Data are representative of 2 experiments with 4–6 mice per group in each experiment. Data were compared by two-tailed, unpaired t tests (B) or two-way ANOVA with post-hoc Tukey test (C–F), *p < 0.05, **p ≤ 0.01, ****p ≤ 0.0001. ns, not significant.
See also Figures S3 and S4.
ThPOK and RUNX3 regulate TRIB2 expression
ThPOK and RUNX3 are relatively cell lineage-specific transcription factors that cross-regulate each other and control the expression of the CD4 and CD8 genes. Expression of ThPOK is higher in human naive CD4+ and RUNX3 is higher in naive CD8+ T cells (Figure S5A). Analysis of ThPOK and RUNX3 chromatin immunoprecipitation sequencing (ChIP-seq) data of murine CD4+ or CD8+ T cells identified a ThPOK-binding peak at the Trib2 promoter region28 and several RUNX3 binding peaks across the Trib2 locus29 (Figure S5B). Indeed, ThPOK (ZBTB7B) silencing reduced, while RUNX3 silencing upregulated, TRIB2 expression (Figure 5A). ThPOK and RUNX3 silencing efficiency was confirmed by qRT-PCR and flow cytometry (Figures S5C–S5E). These data suggest that ThPOK and/or RUNX3 influence TRIB2 expression. Assay for transposase-accessible chromatin (ATAC)-seq of human naive CD4+ and CD8+ T cells revealed an open promoter region in both CD4+ and CD8+ T cells, which was less accessible in naive CD8+ T cells (Figure S5F). In reporter gene assays of the cloned TRIB2 promoter region, forced overexpression of ThPOK increased, while RUNX3 suppressed, the transcriptional activity (Figure 5B). Moreover, in ThPOK and RUNX3 ChIP assays, both transcription factors bound to this region (Figures 5C and 5D). To confirm that ThPOK and RUNX3 regulate TRIB2 expression independently and do not only cross-regulate each other, we checked Trib2 expression in purified naive CD4+ and CD8+ T cells from WT mice, mice with peripheral ThPOK (ThPOKpd) deficiency, peripheral Cbfb (an obligate RUNT cofactor) deficiency (Cbfbpd), and ThPOK Cbfb double peripheral deficiency (ThPOKpd Cbfbpd). Reminiscent of silencing in human cells, ThPOK deficiency reduced, while Cbfb deficiency increased, Trib2 expression (Figure 5E). The differential expression of Trib2 between naive CD4+ and CD8+ T cells was still present in the single-gene KO mice but was mostly abolished with ThPOK Cbfb double conditional KO (Figure 5E). These data suggest that ThPOK and RUNX3 regulate Trib2 independently and that either one contributes to Trib2’s differential expression in naive CD4+ and CD8+ T cells.
Figure 5. ThPOK and RUNX3 regulate TRIB2 expression and lymphopenia-induced T cell proliferation.
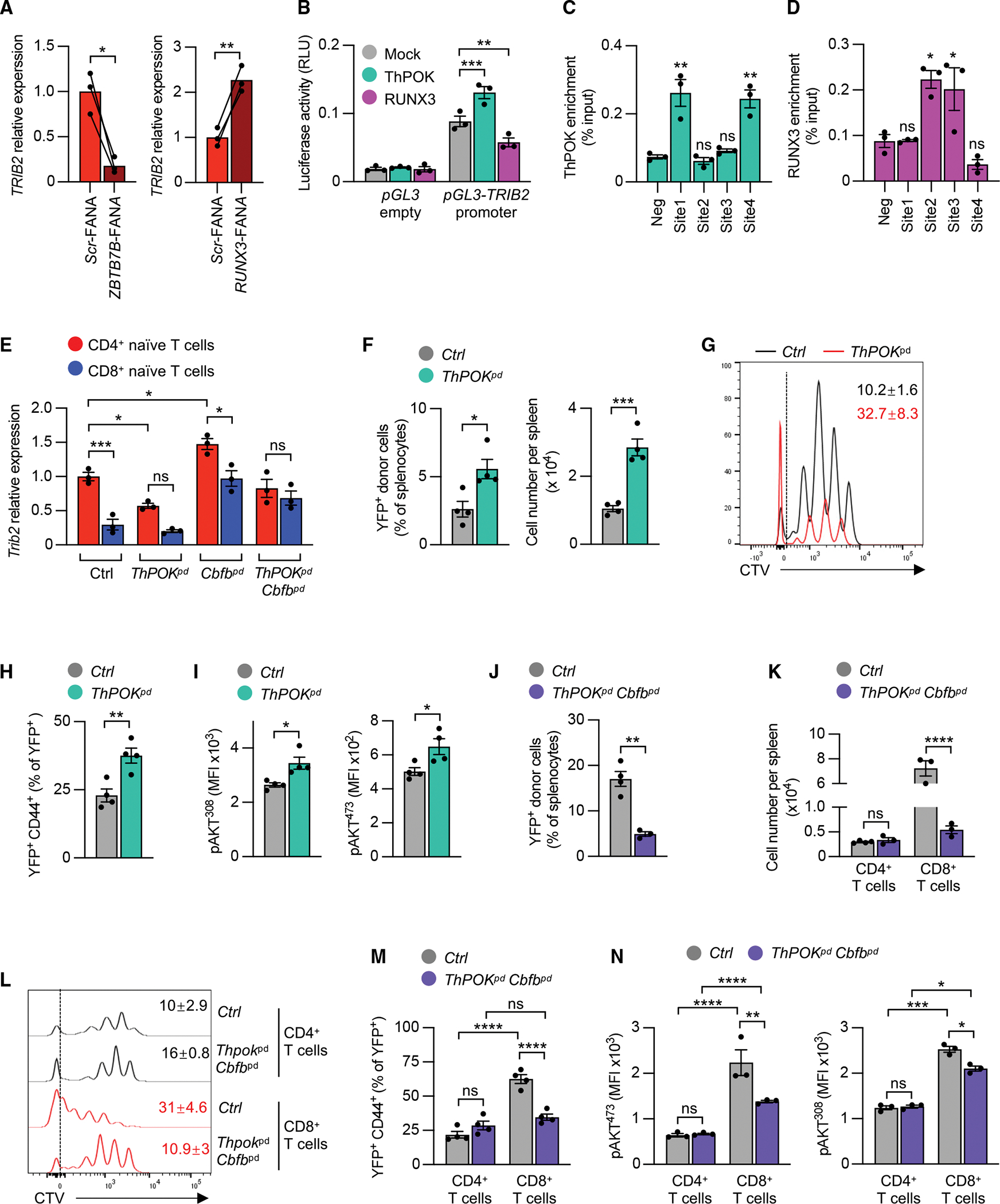
(A) qRT-PCR of TRIB2 transcripts after ZBTB7B (ThPOK) or RUNX3 knockdown. Purified human naive CD4+ T cells were cultured on plate-bound anti-CD3/anti-CD28 (5 μg/mL) antibodies in the presence of indicated FANA-ASOs for 5 days.
(B) Luciferase activity in HEK-293T cells 48 h after cotransfection with pGL3-basic or pGL3 reporter plasmid containing the TRIB2 promoter together with mock or ThPOK-or RUNX3-expressing plasmids.
(C and D) ChIP-qPCR of ThPOK (C) and RUNX3 binding (D) to TRIB2 locus sites (indicated in Figure S5F) in human naive unstimulated (C) and 48 h stimulated naive CD4+ T cells (D).
(E) Trib2 transcripts in purified naive CD4+ or CD8+ T cells from mice genetically modified via CD2-Cre as indicated. Control (Ctrl) mice were ThPOKfl/fl Cbfbfl/fl Rosa26-YFPfl/fl.
(F–I) Response of ThPOKpd T cells to lymphopenia. Sorted YFP+ CD4+ CD44– cells from CD2-Cre+ ThPOKfl/+ Rosa26-YFPfl/+ (Ctrl) and CD2-Cre+ ThPOKfl/fl Rosa26-YFPfl/fl (ThPOKpd) mice were labeled with CTV and adoptively transferred into irradiated wild-type mice. After 1 week, splenocytes were analyzed by flow cytometry for YFP+ donor cell abundance or total splenocyte numbers (F) and CTV dilution of YFP+ donor cells (G); numbers indicate mean proportion of cells in the highest CTV dilution (G, n = 4), CD44+ cell frequencies (H), and phospho-AKT in donor cells (I). Data are representative of 2 experiments with 4–5 mice in each group.
(J–N) Response of ThPOKpd Cbfbpd T cells to lymphopenia. Sorted YFP+ CD4+ CD44− and YFP+ CD8+ CD44− cells from CD2-Cre+ ThPOKfl/+ Cbfbfl/+ Rosa26-YFPfl/+ (Ctrl) or CD2-Cre+ ThpPOKfl/fl Cbfbfl/fl Rosa26-YFPfl/fl (ThPOKpd Cbfbpd) mice were labeled with CTV and adoptively transferred into irradiated wild-type mice. One week later, splenocytes were analyzed by flow cytometry for percentages and total numbers of YFP+ donor cells (J and K), CTV dilution of CD4+ and CD8+ donor cells (numbers indicate mean proportion of cells in the highest CTV dilution, n = 4) (L), frequencies of CD44-expressing cells (M), and phosphorylation of AKT in donor T cells (N). Data are representative of 3 experiments with 3–6 mice in each group.
Results are shown as means ± SEM (B–F, H–K, M, and N). All datapoints represent distinct biological replicates. Results are from 1 experiment (A, C, and D) or 3 experiments (B). Data were compared by two-tailed, paired t tests (A), one-way ANOVA with post-hoc Dunnett test (C and D), two-way ANOVA with post-hoc Tukey test (E, K, M, and N), or two-tailed, unpaired t tests (F and H–J), *p < 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. ns, not significant.</p/>See also Figures S5 and S6.
ThPOK and RUNX3 regulate LIP of naive T cells
Since TRIB2 transcription is regulated by ThPOK, ThPOK deficiency should reproduce the phenotype of Trib2-KO mice under lymphopenic conditions. We adoptively transferred naive CD4+ T cells alone into irradiated mice and found that similar to Trib2 KO, ThPOK-deficient naive CD4+ T cells expanded more than WT CD4+ T cells (Figures 5F, S6A, and S6B), driven by higher turnover rates of ThPOK-deficient naive CD4+ T cells, with 32% vs. 10% cells having divided at least 6 times (Figure 5G). Moreover, differentiation into memory cells was increased for ThPOK-deficient naive CD4+ T cells (Figures 5H and S6C). Mechanistically, ThPOK deficiency was associated with increased AKT activation in CD4+ T cells (Figure 5I). Conversely, LIP of naive CD4+ and CD8+ T cells from Cbfb conditional KO strains that had upregulated expression of Trib2 in both CD4+ and CD8+ T cells (Figure 5E) was reduced, mainly affecting CD8+ and, to a lesser degree, CD4+ T cells (Figures S6D–S6F).
Given that deletion of both ThPOK and Cbfb was needed to equalize expression of Trib2 in naive CD4+ and CD8+ T cells (Figure 5E), we compared the response of CD4+ and CD8+ T cells with these deficiencies during LIP. Indeed, expansion of naive T cells from ThPOK Cbfb double KO mice decreased significantly (Figures 5J, 5K, S6G, and S6H), mainly due to decreased proliferation of CD8+ T cells (Figure 5L). Similarly, memory differentiation of naive CD8+ T cells from ThPOK Cbfb double KO mice was reduced, and the difference between CD4+ and CD8+ cells was diminished (Figures 5M and S6I). AKT phosphorylation in ThPOK Cbfb double KO CD8+ T cells was lower compared with control T cells, while AKT phosphorylation in CD4+ T cells was unaffected by ThPOK Cbfb double KO (Figure 5N). Taken together, these findings show that both ThPOK and RUNX3 determine the differences in LIP of CD4+ and CD8+ T cells, probably by reciprocal regulation of Trib2.
ThPOK and RUNX3 set activation thresholds of human T cells
Since ThPOK and RUNX3 regulate LIP of murine T cells through TRIB2, we hypothesized that they play a role in maintaining T cell homeostasis in humans by setting activation thresholds. Silencing ThPOK in naive CD4+ T cells cultured with IL-7 decreased naive T cell frequencies (Figure 6A), accompanied by elevated CD25 expression (Figure 6B). Similar to TRIB2 silencing in naive CD4+ T cells, ThPOK silencing led to higher AKT phosphorylation (Figure 6C). Moreover, ThPOK-silenced T cells cultured with low-dose anti-CD3/anti-CD28 antibodies, followed by resting for 2 days, were more responsive to restimulation and responded to TCR cross-linking with increased AKT activation (Figure 6D), suggesting that ThPOK regulates CD4+ T cells quiescence through TRIB2-mediated inhibition of AKT. Consistent with RUNX3 repressing TRIB2 transcription, RUNX3 silencing in naive CD8+ T cells preserved the naive state, reduced CD25 expression, and decreased AKT phosphorylation after suboptimal anti-CD3/anti-CD28 stimulation (1 μg/mL anti-CD3/anti-CD28 plate-bound antibodies) (Figures 6E–6G). Downregulation of AKT phosphorylation by RUNX3 silencing can be reversed by TRIB2 silencing (Figure 6H). Taken together, the differential expression of TRIB2 controlled by ThPOK and RUNX3 accounts for different thresholds of AKT activation that may contribute to more sustainable T cell homeostasis of naive CD4+ than of naive CD8+ T cells with aging.
Figure 6. ThPOK and RUNX3 regulate human T cell responsiveness via TRIB2.
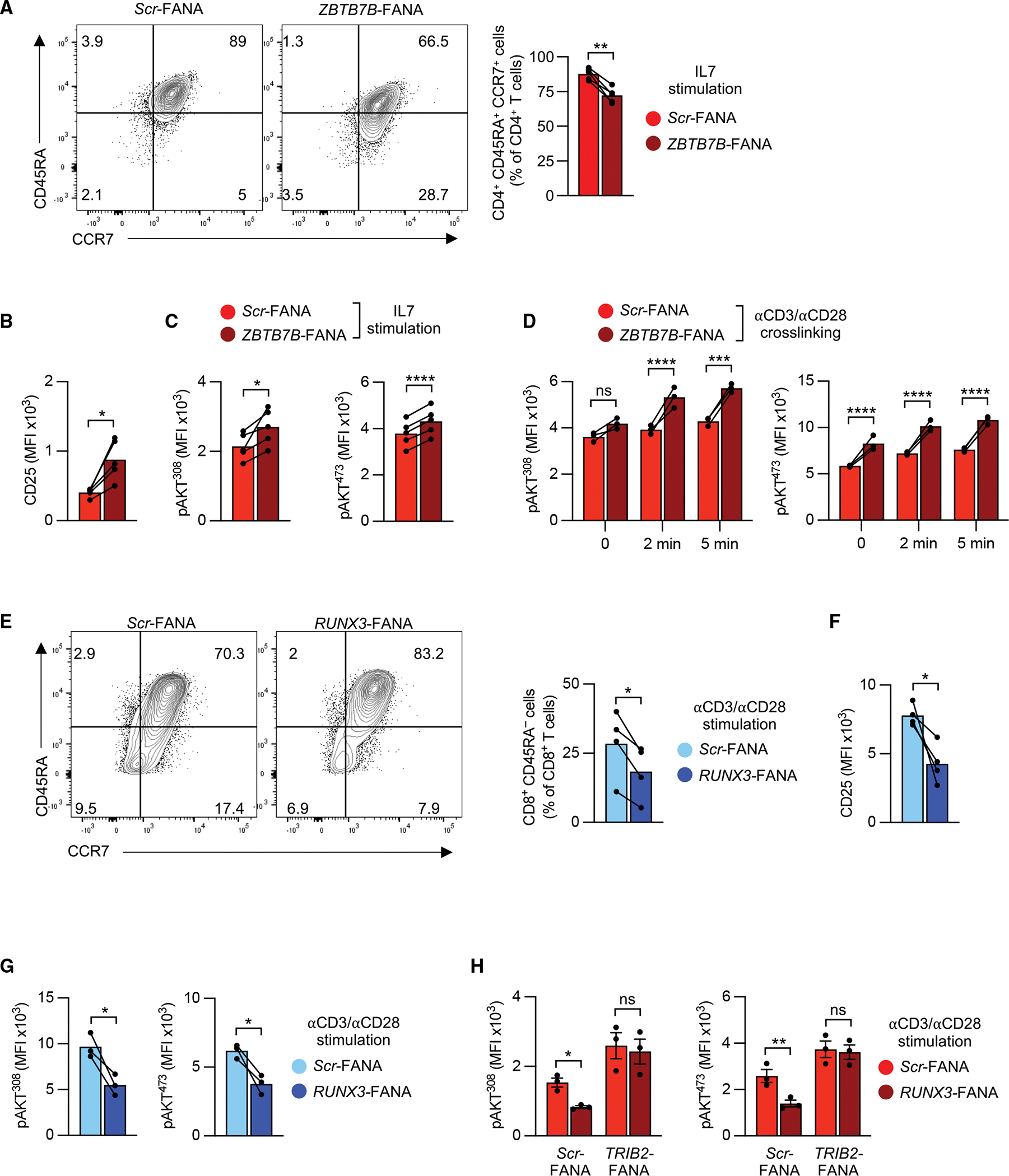
(A) Representative flow cytometry plots and quantification of human naive CD45RA+ CCR7+ CD4+ T cells after ZBTB7B (ThPOK) knockdown. Purified cells were cultured with IL-7 (50 ng/mL) and plate-bound anti-CD28 antibody (2 μg/mL) in the presence of indicated FANA-ASOs for 7 days.
(B) CD25 levels in cells from (A).
(C) Phospho-AKT (Thr308 or Ser473) was assessed 3 days post-knockdown in cells treated as in (A).
(D) Phospho-AKT (Thr308 or Ser473) kinetics after CD3/CD28 cross-linking. Purified human naive CD4+ T cells were cultured with plate-bound anti-CD3/anti-CD28 antibodies (2 μg/mL) in the presence of indicated FANA-ASOs for 2 days, rested for 2 additional days, and then restimulated by CD3/CD28 cross-linking. Results are representative of 3 experiments with 3 samples in each experiment.
(E–G) Representative flow cytometry plots and quantification of human naive CD45RA+ CCR7+ CD8+ T cells after RUNX3 knockdown. Purified human naive CD8+ T cells were cultured with plate-bound anti-CD3/anti-CD28 antibodies (1 μg/mL) in the presence of indicated FANA-ASOs. After 5 days, frequencies of CD45RA+ CCR7+ T cells (E) and CD25 MFI (F) were determined by flow cytometry. Phospho-AKT (Thr308 or Ser473) levels were measured by flow cytometry after 3 days (G).
(H) Phospho-AKT (Thr308 or Ser473) in purified human naive CD4+ T cells that were cultured with plate-bound anti-CD3/anti-CD28 antibodies (1 μg/mL) in the presence of indicated combinations of FANA-ASOs for 3 days. Results are shown as means ± SEM.
All datapoints represent distinct biological replicates. Results are from 1 experiment (H), pooled from 2 (A, B, E, and G) or 3 experiments (C), or are representative of 2 experiments with 3 samples in each experiment (D). Data were compared by two-tailed, paired t tests (A–C and E–G) or two-way ANOVA with post-hoc Šídák test (D and H). *p < 0.05, **p ≤ 0.01, ****p ≤ 0.0001. ns, not significant.
Decline in ThPOK and TRIB2 expression impairs CD4+ T cell quiescence in older adults
In older adults, homeostatic proliferation of naive T cells was increased, as documented by significantly higher frequencies of Ki67+ naive CD4+ T cells and a similar trend for Ki67+ naive CD8+ T cells (Figure 7A). Moreover, naive CD4+ T cells of older adults developed higher AKT phosphorylation when cultured with IL-7 (Figure 7B). TRIB2 expression declined with age, suggesting that the protective effect of TRIB2 on naive CD4+ T cell quiescence wanes at older age (Figures 7C and 7D). The loss in TRIB2 expression was due to a decline in ThPOK with age (Figure 7E), while RUNX3 expression remained stable (Figure 7F). Studies of ThPOK-deficient murine30 or human T cells (Figure S7A) have shown that ThPOK loss is associated with re-expression of CD8. Indeed, we found an increased frequency of CD4+ T cells with low expression of CD8 in older adults (Figure 7G), as previously reported.31 Almost all CD4+ CD8int T cells from older individuals were effector memory or TEMRA cells (Figure S7B). These cells expressed less ThPOK and TRIB2 and higher RUNX3 (Figure 7H). These data suggest that a decline in ThPOK with age causes reduced TRIB2 expression, resulting in increased turnover and emergence of a memory population that coexpresses CD4 and CD8.
Figure 7. Decline in ThPOK and TRIB2 expression in older adults.
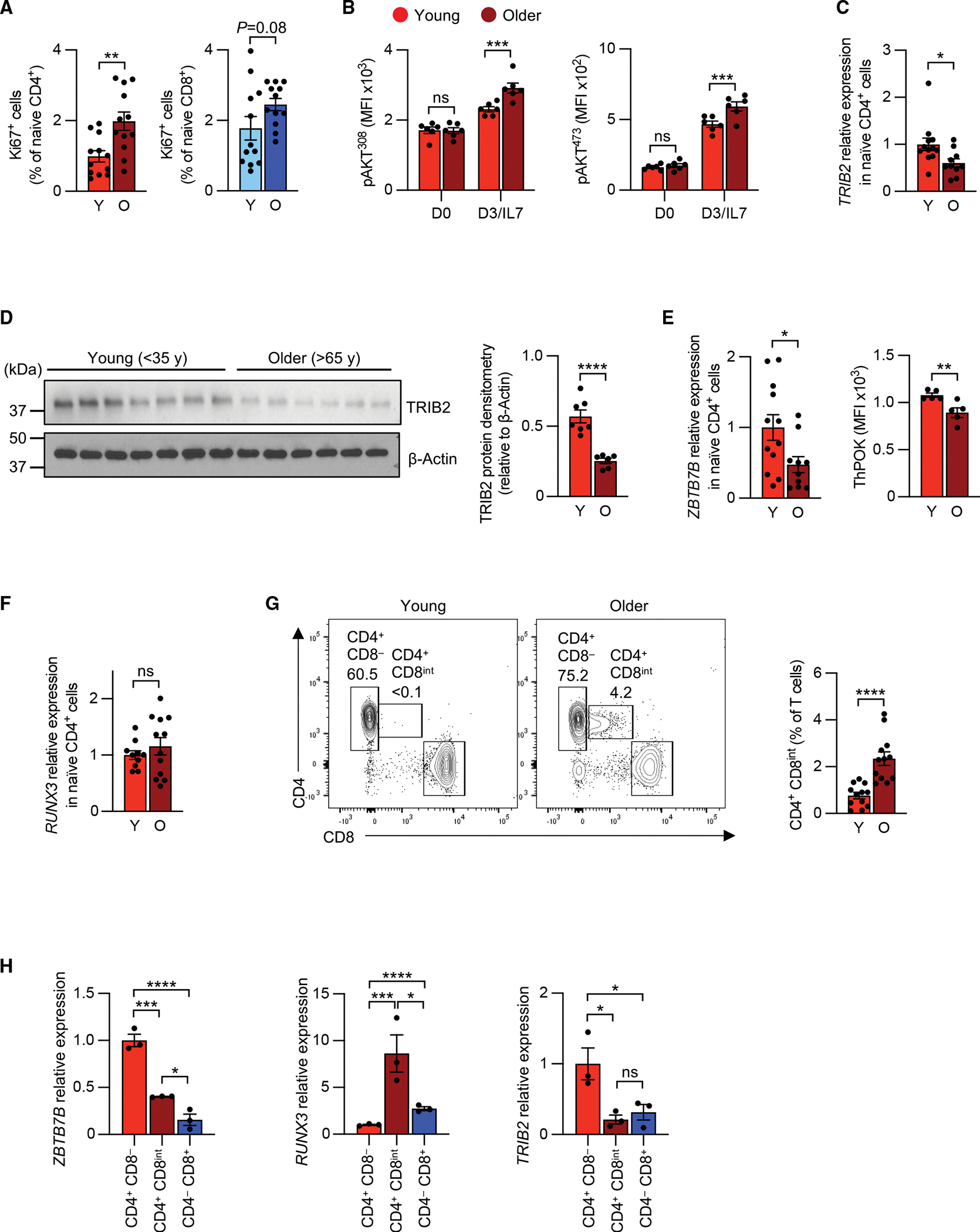
(A) Frequency of Ki67+ cells in naive CD4+ T cells from Y (<35 years) and O (>65 years) individuals. Results are from 2 experiments with 6 pairs in each experiment.
(B) IL-7-induced phospho-AKT (Thr308 or Ser473) in purified naive CD4+ T cells from Y and O individuals.
(C) qRT-PCR of TRIB2 expression in purified naive CD4+ T cells from 12 Y and 10 O individuals.
(D) Immunoblots and densitometric quantification of TRIB2 protein levels in naive CD4+ T cells from 7 Y and 6 O individuals. Membranes from Zhang et al. reblotted for TRIB2.32.
(E and F) ZBTB7B (ThPOK) transcript quantification, ThPOK MFI (E), and RUNX3 transcript quantification (F) as determined by qRT-PCR and/or flow cytometry.
(G) Flow cytometric quantification of CD4+ CD8int cell frequencies in peripheral blood CD3+ T cells from 12 Y and 12 O individuals.
(H) ZBTB7B (ThPOK), RUNX3, and TRIB2 expression in FACS-purified naive CD4+ CD8−, naive CD4− CD8+, and CD4+ CD8int T cells from O individuals as determined by qRT-PCR. Results are shown as mean ± SEM. All datapoints represent distinct biological replicates.
Results are from 1 experiment (C–F and H) pooled from 2 (A and B) or 4 experiments (G). Data were compared by two-tailed, unpaired t test (A and C–G), two-way ANOVA with post-hoc Šídák test (B), or one-way ANOVA with post-hoc Tukey test (H). *p < 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. ns, not significant.
See also Figure S7.
DISCUSSION
The T cell system constantly strives for balance between maintaining compartment size while avoiding inappropriate cell activation. At the same time, T cells have to be ready to rapidly enter and progress through the cell cycle when increased numbers of cells are needed to participate in an immune response. Naive T cells are quiescent, reside in G0 phase of the cell cycle, and have low metabolic, transcriptional, and translational activities.33 For self-renewal of naive T cells, cells undergo homeostatic proliferation that is driven by cytokines and low-grade TCR stimulation. Here, we describe that naive CD4+ and CD8+ T cells differ in the regulation of T cell maintenance due to higher expression of TRIB2 in CD4+ T cells. TRIB2 acts as an inhibitor of mTORC2- and PDK1-dependent phosphorylation of AKT. mTORC2 activation is induced by T cell growth factors that drive T cell homeostatic proliferation and survival. Reduced expression of TRIB2 in CD8+ T cells puts them at risk for sustained quiescence exit upon homeostatic signals and accounts for several characteristic features of naive CD8+ T cells, including the increased aging-associated acquisition of epigenetic differentiation signatures in naive CD8+ compared with CD4+ T cells in humans.15,17,18
The central regulator of quiescence exit is thought to be mTORC1, which induces the transcriptional and translational activity required for switching the cell to an anabolic state and subsequent cell-cycle progression and proliferation. Deficiencies in mTOR activators RAPTOR and, to a lesser extent, RHEB induce defects in quiescence exit due to inactivated mTORC1.34 Conversely, PI3K hyperactivity induces the proliferation of naive T cells and their spontaneous differentiation into different effector cell lineages, at least in part due to phosphorylating AKT and activating mTORC1.35,36 Repression of PI3K signaling via high expression of PTEN in naive T cells enforces quiescence until PTEN gets downregulated by TCR engagement.37 The mTORC2 pathway also contributes to quiescence exit by activating AKT and inducing the degradation of forkhead box protein O1 (FOXO1).
Our study suggests that naive CD8+ T cells have a higher propensity to be activated and to differentiate during homeostatic conditions, which becomes particularly critical during aging. Whether the more severe depletion of naive CD8+ T cells with age is primarily due to cell loss or differentiation into (virtual) memory cells remains to be determined. Naive CD8+ T cells continuously receive MHC class I-mediated TCR stimulation and may be depleted via activation-induce apoptosis. However, naive CD8+ T cells engage PILRα, a ligand for CD8α, as a safekeeping mechanism to counteract spontaneous activation and cell death.38
One of the enforcers of quiescence during IL-7 signaling is FOXP1.39 In vitro IL-7 stimulation of mouse naive T cells is effective in supporting survival but not in inducing cell division. In contrast, deletion of Foxp1 in peripheral T cells resulted in a modest increase in IL-7Ra that was remarkably effective in increasing IL-7-mediated proliferation of naive CD8+ T cells. Additionally, Foxp1-deficient T cells stimulated with IL-7 differentiated toward a memory-like phenotype.39 However, unlike TRIB2, FOXP1 is not differentially expressed in CD4+ and CD8+ T cells and, therefore, is unlikely to account for their distinct responsiveness.
TRIB2, a member of the Tribbles family, is a pseudo serine/threonine kinase and a binding partner of several key enzymes in major signaling pathways including, but not limited to, mitogen-activated protein kinases (MAPKs), nuclear factor κB (NF-κB), C/EBPα, and AKT.40 It is known as an oncogene that enhances cancer cell proliferation and confers tumor drug resistance through activating AKT or other pathways.26,41,42 In xenograft tumors, TRIB2 overexpression increased AKT Ser473 phosphorylation independently of PI3K or mTORC1 activity.26 Additionally, TRIB2 overexpression in hematopoietic stem cells perturbs myeloid development, promotes self-renewal, and causes acute myeloid leukemia (AML).42 In contrast, TRIB2 can also exhibit tumor-suppressive features, as TRIB2 overexpression in AML cells reduces cell proliferation and causes cell death.43 In lymphocytes, Trib2 deletion not only increased steady-state thymocyte proliferation but was also found to reduce T cell acute lymphoblastic leukemia latency.21 Consistent with these tumor-suppressive activities, TRIB2 acts as negative regulator of AKT in hepatocytes25 and adipocytes.24 TRIB325 and TRIB244 have been shown to preferentially bind to Thr308 unphosphorylated AKT, therefore possibly blocking Thr308 phosphorylation and subsequently reducing Ser473 phosphorylation in T cells. Given that MAPK signaling also participates in T cell homeostasis45 and ERK phosphorylation is elevated upon TRIB2 silencing (Figure 2E), TRIB2 may regulate naive T cell maintenance through pathways in addition to AKT.
Higher expression of TRIB2 in CD4+ T cells is a consequence of the expression of lineage-determining transcription factors. CD4+ and CD8+ T cells derive from common precursor cells, and lineage development is controlled by ThPOK and RUNX3 that cross-regulate each other in the expression of lineage-specific genes including CD4 and CD8A.46 Peripheral, mature CD4+ and CD8+ T cells continue to retain lineage-specific ratios of ThPOK/RUNX3 expression, making them master transcription factors to regulate CD4+ and CD8+ T cell-selective genes, including TRIB2.19,30,47 These molecules not only maintain CD4 or CD8 lineage integrity but also render CD4+ or CD8+ T cells distinctly susceptible to aging. Interestingly, RUNX3 and RUNX1 appear to have distinct functions in naive T cell homeostasis. RUNX3 promotes epigenetic reprogramming during naive CD8+ T cell differentiation,48 while RUNX1 is required for survival of murine CD4+ T cells via IL-7R levels.49 Given that CBFB is a general obligatory RUNT cofactor, the findings with Cbfb-deficient CD4+ T cells in the LIP host may be an underestimation.
Taken together, naive CD8+ T cells have less TRIB2, lowering the threshold to enter proliferation, activation, and differentiation—a direct consequence of lineage-determining transcription factors. This property may endow CD8+ T cells with the ability to respond faster when clonal expansion is warranted upon recognition of antigen from a pathogen. However, a low threshold to exit from quiescence comes with significant shortfalls in maintaining a functional CD8+ T cell repertoire with older age. Naive CD8+ T cells acquire memory T cell features through stimulation with homeostatic cytokines in the absence of antigen.9 Such virtual memory T cells are frequent in old mice10,50 and exhibit blunted TCR-mediated proliferation and increased apoptosis.10,51 Second, effector CD8+ T cells are expanded in older adults, where they may contribute to inflammaging and increased mortality.52,53 Finally, it is tempting to speculate that the CD8+ T cell-intrinsic trait of easier exit from quiescence compromises the maintenance of a functional naive CD8+ T cell compartment in humans and accounts for the disproportionate decline in naive CD8+ T cells, the single most striking feature of immune aging.54
To date, no therapeutic strategies exist to preserve the naive T cell compartment during aging. Identifying TRIB2 as a guardian of T cell homeostasis may open therapeutic opportunities since TRIB2-stabilizing small molecules have been recently discovered.44 Of note, one such compound, lapatinib, is FDA approved to treat breast and other cancers. Repurposing TRIB2-stabilizing compounds could be a therapeutically attractive avenue, either in isolation or by complementing current efforts investigating recombinant IL-7,55,56 to preserve the naive T cell compartment during aging.
Limitations of the study
Our study provides strong evidence that TRIB2 is differentially expressed in CD4+ and CD8+ T cells and that this differential expression is of functional importance for proliferation and differentiation of naive T cells in response to homeostatic cytokines and TCR activation. Given TRIB2’s role in regulating AKT activity and the importance of this pathway for IL-7-driven homeostatic proliferation, we mainly focused on AKT phosphorylation. Changes were consistent but subtle, and it cannot be excluded that TRIB2 acts through additional pathways. In our in vivo studies, we used a lymphopenia mouse model of accelerated homeostatic stress, but we have not directly assessed the role of TRIB2 on steady-state T cell homeostasis in humans or mice. We therefore can only conjecture what the implications are of reduced ThPOK and TRIB2 expression with advancing age. Most importantly, it is tempting to speculate, but it is ultimately unproven, that higher expression of TRIB2 accounts for the higher resilience of naive CD4+ T cells to age-associated depletion. Ultimate proof requires interventional studies, which, as with any study to prevent aging-related changes, are challenging.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Dr. Jörg J. Goronzy (goronzy.jorg@mayo.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This paper analyzes existing, publicly available data. The accession numbers for these datasets are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Anti-human CD3 (HIT3a) | BioLegend | Cat# 300318, RRID:AB_314054 |
| Anti-human CD3 (OKT3) | BioLegend | Cat# 317314, RRID:AB_571909 |
| Anti-human CD4 (RPA-T4) | BioLegend | Cat# 300506, RRID:AB_314074 |
| Anti-human CD8 (SK1) | BioLegend | Cat# 344723, RRID:AB_2562790 |
| Anti-human CCR7 (G043H7) | BioLegend | Cat# 353207, RRID:AB_11203894 |
| Anti-human CD25 (BC96) | BioLegend | Cat# 302635, RRID:AB_11219793 |
| Anti-human CD122 (TU27) | BioLegend | Cat# 339017, RRID:AB_2721347 |
| Anti-human IL2 (MQ1-17H12) | BioLegend | Cat# 500352, RRID:AB_2820083 |
| Anti-human TNFα (Mab11) | BioLegend | Cat# 502908, RRID:AB_315261 |
| Anti-human CD45RA (HI100) | BD Biosciences | Cat# 562886, RRID:AB_2737865 |
| Anti-human IFNγ (B27) | BD Biosciences | Cat# 562016, RRID:AB_10894955 |
| Anti-human Granzyme B (GB11) | BD Biosciences | Cat# 563388, RRID:AB_2738174 |
| Anti-human/mouse Ki67 (B56) | BD Biosciences | Cat# 561283, RRID:AB_10716060 |
| Anti-human/mouse pAKT-Ser473 (M89-61) | BD Biosciences | Cat# 560343, RRID:AB_1645397 |
| Anti-human/mouse pAKT-Thr308 (J1–223.371) | BD Biosciences | Cat# 558275, RRID:AB_2225329 |
| Anti-human pERK1/2-Thr202/Tyr204 (20A) | BD Biosciences | Cat# 612592, RRID:AB_399875 |
| Anti-human S6-pS235/pS236 (N7–548) | BD Biosciences | Cat# 560433, RRID:AB_2827879 |
| Anti-human FoxO1 (C29H4) | Cell Signaling Technologies | Cat# 58223S, RRID:AB_2799543 |
| Anti-human/mouse TRIB2 (D8P2X) | Cell Signaling Technologies | Cat# 13533S, RRID:AB_2798250 |
| Anti-human/mouse β-Actin (13E5) | Cell Signaling Technologies | Cat# 4970S, RRID:AB_2223172 |
| Anti-human ThPOK (D9V5T) | Cell Signaling Technologies | Cat# 13205, RRID:AB_2798147 |
| Anti-human RUNX3 (9F4A17) | BioLegend | Cat# 653603, RRID:AB_2687117 |
| Purified anti-human CD3 (UCHT1) | BioLegend | Cat# 300465, RRID:AB_11147760 |
| Purified anti-human CD28 (CD28.1) | BioLegend | Cat# 302943, RRID:AB_11150591 |
| Anti-mouse CD4 (RM4–5) | BioLegend | Cat# 100553, RRID:AB_2561388 |
| Anti-mouse CD4 (GK1.5) | BioLegend | Cat# 100449, RRID:AB_2564587 |
| Anti-mouse CD8α (53–6.7) | BioLegend | Cat# 100750, RRID:AB_2562610 |
| Anti-mouse TCRβ (H57–597) | BioLegend | Cat# 109219, RRID:AB_893624 |
| Anti-mouse CD44 (IM7) | BioLegend | Cat# 103040, RRID:AB_2616903 |
| Anti-mouse CD62L (MEL-14) | BioLegend | Cat# 104421, RRID:AB_493379 |
| Anti-mouse CD122 (TM-b1) | BioLegend | Cat# 123209, RRID:AB_940615 |
| Anti-mouse CD45.2 (104) | BioLegend | Cat# 109824, RRID:AB_830789 |
| Anti-mouse CD25 (PC61.5) | BioLegend | Cat# 102008, RRID:AB_312857 |
| Anti-mouse CD127 (IL7Ra) (A7R34) | BioLegend | Cat# 135041, RRID:AB_2572047 |
| Anti-mouse IFNγ (XMG1.2) | BioLegend | Cat# 505810, RRID:AB_315404 |
| Anti-mouse CD24 (M1/69) | BD Biosciences | Cat# 553261, RRID:AB_394740 |
| Purified anti-mouse CD3e (eBio500A2) | Invitrogen | Cat# 16–0033-85, RRID:AB_842785 |
| Purified anti-mouse CD28 (37.51) | Invitrogen | Cat# 16–0281-85, RRID:AB_468922 |
|
| ||
| Biological samples | ||
|
| ||
| Human peripheral blood (buffy coats) | Stanford University Blood bank | N/A |
| Human peripheral blood (leukoreduction system chambers) | Mayo Clinic Blood bank | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Human IL-7 | PeproTech | Cat# 200–07 |
| FANA antisense oligonucleotides | AUM BioTech | https://www.aumbiotech.com/ |
| MK-2206, AKT inhibitor | Selleckchem | Cat# S1078 |
|
| ||
| Critical commercial assays | ||
|
| ||
| EasySep Human Naive CD4+T cell isolation Kit II | StemCell Technologies | Cat# 19555 |
| EasySep Human Naive CD8+ T cell isolation Kit | StemCell Technologies | Cat# 19258 |
| Pan T cell isolation Kit II, mouse | Miltenyi Biotec | Cat# 130–095-130 |
| EasySep Mouse Naive CD4+ T cell Isolation Kit | StemCell Technologies | Cat# 19765 |
| EasySep Mouse Naive CD8+ T cell Isolation Kit | StemCell Technologies | Cat# 19858 |
| ChIP-IT High Sensitivity Kit | Active Motif | Cat# 53040 |
| Dual-Luciferase Reporter Assay System | Promega | Cat# E1910 |
| RNeasy Plus Mini Kit | Qiagen | Cat# 74134 |
| RNeasy Plus Micro Kit | Qiagen | Cat# 74034 |
| Maxima First Strand cDNA Synthesis Kit | Thermo Fisher Scientific | Cat# K1641 |
|
| ||
| Deposited data | ||
|
| ||
| Transcriptomic data of human naive CD4+ and CD8+ T cells | Serroukh et al.19 | GEO: GSE75406 |
| ThPOK ChIP-seq data of mouse CD4+ T cells | Ciucci et al.57 | GEO: GSE116506 |
| RUNX3 ChIP-seq data of mouse CD8+ T cells | Lotem et al.29 | GEO: GSE50130 |
| ATAC-seq data of human naive CD4+ T cells | Hu et al.17 | SRA: PRJNA478249 |
| ATAC-seq data of human naive CD8+ T cells | Hu et al.17 | dbGaP: phs001187.v1.p1 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: B6.Cg-Trib2tm1.1(crelERT2)Hze/J | The Jackson Laboratory | RRID:IMSR_JAX:022,865 |
| Mouse: CD2-Cre | Vacchio et al.30 | N/A |
| Mouse: conditional Zbtb7b/ThPOK knockout | Wang et al.47 | N/A |
| Mouse: conditional Cbfb knockout | Naoe et al.58 | N/A |
| Mouse: ROSA26-LoxP-STOP-LoxP-YFP | Srinivas et al.59 | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| see Table S1 for qPCR primers | N/A | N/A |
| see Table S1 for ChIP-PCR primers | N/A | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pHAGE-CMV-IRES-ZsGreen-W | Addgene, David Baltimore's unpublished plasmid | Addgene Plasmid #26532 |
| pGL3 basic | Promega | Cat# E1751 |
|
| ||
| Software and algorithms | ||
|
| ||
| FlowJo 10 | Tree Star | https://www.flowjo.com/ |
| Fiji | Schindelin et al.60 | https://imagej.net/ |
| Integrative Genomics Viewer (IGV) 2.10 | Broad Institute | https://software.broadinstitute.org/software/igv/ |
| Prism 9 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Study design
The objective of the study was to identify the mechanism(s) that account for differences of CD4+ and CD8+ T cells in their ability to maintain homeostasis and to thereby identify potential targets to improve naive T cell maintenance over lifetime. We used naive CD4+ or CD8+ T cells purified from young (20–35 years old) and older (65–85 years old) healthy individuals in which we silenced and overexpressed candidate genes that were differentially expressed in naive CD4+ and CD8+ T cells and evaluated the effect on in vitro signaling and function after stimulation with homeostatic cytokines and polyclonal activation. Sample sizes for population comparisons were chosen to provide 80% power with a level of significance of 5% when the difference in their means would be > 1.5 standard deviations. For intervention experiments or paired comparisons, we used a sample size of 3–6. Knockout or conditional knockout mice were used for validation of the effects seen in the in vitro studies of human T cells. We adoptively transferred naive T cells from these mice to irradiated, lymphopenic mice as a model of in vivo homeostatic proliferation. Recipient mice were randomly assigned to receive cells from wild-type versus indicated knockout mice. Data analysis was performed unblinded. Number of samples and experiments are specified in figure legends. No outliers were removed.
Human subject study population
Buffy coats or leukoreduction system (LRS) chambers from 86 blood or platelet donors were purchased from the Stanford University Blood Bank or Mayo Clinic Blood Bank. These samples were deidentified except for age and sex information. Samples from male and female individuals were used. Young adults were 35 years or younger, older adults were 65 years or older. In addition, PBMCs were obtained from 5 healthy volunteers who did not have an acute or active chronic disease and no history of cancer or autoimmune diseases. Chronic diseases such as hypertension were permitted if controlled by medication. Studies involving human subjects were approved by the Stanford University and Mayo Clinic Institutional Review Boards and participants gave informed written consent.
Mouse strains
Transgenic CD2-Cre mice30 were a gift from Dr. Paul E. Love (National Institutes of Health). Mice carrying loxP-flanked alleles for Zbtb7b/ThPOK,47 Cbfb58 and ROSA26-LoxP-STOP-LoxP-YFP59 were gifts from Dr. Rémy Bosselut (National Institutes of Health). CD45.1 and CD45.2 C57BL/6 mice and Trib2 knockout mice61 (Trib2-2A-CreERT2 knockin/knockout mice, #022865) were purchased from Jackson Laboratory. In Trib2 knockin/knockout mice, Exon 2 of the Trib2 gene was replaced with CreERT2 resulting in a Trib2 knockout allele and expression of CreERT2 (CreERT2 was not utilized in this study). Mice were used for experimentation at 6–14 weeks of age. Mice of both sexes were used. All mice were housed in the Stanford Research Animal Facility or Mayo Clinic Institutional Animal Facility. Protocols were approved by the Stanford University or Mayo Clinic Institutional Animal Care and Use Committee.
Primary human cell cultures
All data points referring to human samples represent distinct individuals. Human peripheral blood mononuclear cells (PBMCs) were isolated via density centrifugation using Lymphoprep (StemCell Technologies, #07861) according to the manufacturer’s instructions. Human naive CD4+ and naive CD8+ T cells were isolated from PBMCs using EasySep Human Naive CD4+ or CD8+ T cell isolation kits (StemCell Technologies, #19555 and #19258, respectively). The purity of purified human naive CD4+ or naive CD8+ T cells was routinely verified by flow cytometry (see Figure S1A). Cells were cultured in RPMI-1640 medium (Sigma Aldrich, R8758) supplemented with 10% FBS (GeminiBio, #900–108), 100 U/mL penicillin, and 100 U/mL streptomycin (Sigma Aldrich, P0781). For knock-down experiments, human naive CD4+ or naive CD8+ T cells were cultured with 2 μM of indicated FANA-ASOs (AUM BioTech). To improve silencing efficiency in non-activated cells, cells were cultured on plate-bound anti-CD28 antibody (2 μg/mL, BioLegend, #302943). To induce proliferation, cells were either stimulated with 50 ng/mL IL-7 (PeproTech, #200–07) for 7 days, or with plate-bound anti-CD3/anti-CD28 antibodies (both 5 μg/mL, BioLegend, #300465 and #302943) for 5 days. In AKT inhibition experiments, 5 μM MK-2206 (Selleckchem, S1078) was added at the beginning of culture.
Primary murine cell cultures
All data points referring to murine samples represent distinct mice. Murine splenocytes were extracted from spleens by mechanical dissociation and red blood cells were lysed with ACK lysis buffer (Gibco, A10492) for 5 min at room temperature. Naive CD4+ T cells were sorted on a BD FACSAria II or Melody cell sorter. The purity of sorted murine naive CD4+ and naive CD8+ T cells was routinely verified by flow cytometry (see Figure S4A). For stimulation experiments, cells were activated with plate-bound anti-CD3/anti-CD28 antibodies (both 2.5 μg/mL, Invitrogen, #16-0033-85 and #16-0281-85) for 4 days. For adoptive transfer experiments, total mouse T cells were first isolated from splenocytes by the Pan T cell isolation kit II (Miltenyi Biotec, #130-095-130). Then, naive CD4+ and naive CD8+ CD44− T cells were sorted on a BD FACSAria II or Melody cell sorter. Murine thymocytes were extracted from thymi by mechanical dissociation and red blood cells were lysed with ACK lysis buffer (Gibco, A10492) for 1 min at room temperature.
METHOD DETAILS
Adoptive transfer
Naive CD4+ and naive CD8+ CD44− T cells were sorted on a BD FACSAria II or Melody cell sorter. Sorted cells were labeled with CellTrace Violet (CTV, Thermo Fisher Scientific, #C34557) according to the manufacturer’s instructions. Sorted, stained naive CD4+ and CD8+ T cells were mixed at physiological ratios and the cell suspension (total of 1 million cells) was transferred into recipient mice via tail vein injection. For transfer experiments with ThPOK-deficient T cells, we transferred naive CD4+ CD44− T cells alone because ThPOK-deficient CD4+ T cells change their phenotype after adoptive transfer. They downregulated CD4 and expressed CD8 making them difficult to distinguish from true CD4− CD8+ T cells. CD45.1 and CD45.2 C57BL/6 recipient mice were irradiated with 6 Gy using a 137Cesium source one day before transfer. Cells from CD2-Cre+ ThPOKf/wt Cbfbf/wt YFPf/wt, ThPOKpd, Cbfbpd or ThPOKpd Cbfbpd were transferred into irradiated CD45.2 C57BL/6 mice and YFP was used to identify donor cells. Cells from WT or Trib2-KO mice were transferred into irradiated CD45.1 C57BL/6 mice and CD45.2 staining was used to identify donor cells. Mice were euthanized and splenocytes were analyzed 7 days post-transfer.
Lentivirus production and transduction
For overexpression studies, TRIB2 was subcloned into pHAGE-CMV-IRES-ZsGreen-W (Addgene, #26532). HEK-293T cells were transfected with this vector, along with psPAX2 (Addgene, #12260) and pMD2.G (Addgene, #12259) vectors using FuGENE (Promega, E2311). 72 h after transfection, the supernatant containing lentiviral particles was collected, filtered through a 0.45-μm syringe filter (Millipore) and concentrated using PEG-it solution (System Biosciences, LV810A-1). For lentiviral transduction, human naive CD4+ or CD8+ T cells were activated with anti-CD3/anti-CD28 Dynabeads (1:2) and transduced with the concentrated lentivirus in the presence of 8 μg/mL polybrene (Sigma Aldrich, TR-1003-G) and 10 U/mL human IL-2 (PeproTech, #200–02). After 1.5 days, lentivirus-containing medium was exchanged, and cells were transferred into new wells coated with anti-CD3/anti-CD28 antibodies (5 μg/mL).
T cell culture after gene silencing
Isolated human naive CD4+ T cells were cultured on plate-bound anti-CD3/anti-CD28 antibodies (both 2 μg/mL, BioLegend, #300465 and #392943) with indicated FANA-ASO (AUM BioTech) for 2 days, then transferred to empty wells to rest for 2 additional days. Rested cells were incubated with 5 μg/mL anti-CD3/anti-CD28 antibodies at 4°C for 30 min and washed with PBS. TCR cross-linking was performed by addition of 5 μg/ml F(ab’)2-goat anti-mouse IgG secondary antibodies (Invitrogen, #31166) in 100 μL PBS at 37°C. Alternatively, rested cells were stimulated with 50 μg/mL IL-7. At indicated time points after stimulation, cells were fixed with Lyse/Fix Buffer (BD Biosciences, #558049) at 37°C for 10 min and subjected to fluorescent antibody staining.
Flow cytometry
For flow cytometry cell surface staining, cells were stained with diluted antibodies (see key resources table) and a viability dye (Fixable Viability Dye eFluor 506 (1:1000, Invitrogen #65–0866-14) or LIVE/DEAD Fixable Blue Dead Cell Stain (1:1000, Invitrogen #L34962)). Staining was performed in PBS supplemented with 2% FBS for 30 min at 4°C. For intracellular cytokine assays, cells were stimulated with 50 ng/mL phorbol 12-myristate 13-acetate (PMA, Peprotech #1652981) and 500 ng/mL ionomycin (Peprotech, #5608212) in the presence of Brefeldin A (GolgiPlug, BD Biosciences #554724) for 4 h at 37°C. Intracellular cytokine staining was conducted using the Fixation/Permeabilization solution kit (BD Biosciences, #555028) according to the manufacturer’s instructions. For intracellular staining of phospho-proteins or Ki67, cells were fixed with BD Phosflow Lyse/Fix Buffer (BD Biosciences, #558049) and permeabilized with BD Perm III (BD Biosciences, #558050) according to the manufacturer’s instructions, followed by intracellular staining with indicated antibodies (see key resources table). Stained cells were analyzed using a BD LSRII, BD LSRFortessa flow cytometer (BD Biosciences) or 5-laser Cytek Aurora (Cytek Biosciences) and FlowJo software (Tree Star).
ATAC-seq analysis and ChIP-qPCR
Chromatin accessibilities of the TRIB2 locus in human naive CD4+ and CD8+ T cells were evaluated from previously published ATAC-seq datasets (SRA accession code PRJNA478249 and dbGaP accession code phs001187.v1.p1).17 Briefly, reads were aligned to the human reference genome (hg19) using Bowtie2, assigned to peaks using the Rsubread and read counts were normalized by Limma. Chromatin accessibility was visualized via the Integrative Genomics Viewer (IGV).
ChIP assays were perform using the ChIP-IT High Sensitivity kit (Active Motif, #53040) according to the manufacturer’s instructions. Briefly, cells were fixed with fresh 1 × fixation buffer and subjected to sonication on ice using an Active Motif sonicator to obtain DNA fragments sized 100 to 1000 bp. Sheared chromatin was immunoprecipitated with antibodies (4 μg for each reaction) specific for ThPOK (D9V5T, Cell Signaling Technology #13205) and RUNX3 (9F4A17, BioLegend #653603). ChIP-qPCR primers are listed in Table S1. Previously published ChIP-seq data of murine CD4+ and CD8+ T cells were obtained from GSE116506 (ThPOK ChIPseq)57 and GSE50130 (RUNX3 ChIP-seq).29
RNA isolation and quantitative RT-qPCR
For quantitative RT-qPCR, total RNA was extracted with the RNeasy Plus Mini or Plus Micro kit (Qiagen, #74134 and #74034) and cDNA was synthesized using the Maxima First Strand cDNA Synthesis Kit (Thermo Scientific, K1641). Quantitative RT-qPCR was performed on the ABI 7900HT system (Applied Biosystems) using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific, #4367659). RT-qPCR primers are listed in Table S1. Previously published transcriptomic data of human naive CD4+ and CD8+ T cells were obtained from GSE75406.19
Luciferase reporter assay
The TRIB2 promoter region (−511 to +1114 identified by ATAC-seq) was amplified with the following primers: Forward: 5′-AGAAA-TACAAGCCCAGGATG-3′; Reverse: 5′-TGAGTACCCGGGCTGCTAAG-3′ and was inserted into the pGL3 basic plasmid (Promega, #1751). ThPOK, or RUNX3 was subcloned into pHAGE-CMV-IRES-ZsGreen-W (Addgene, #26532) to generate overexpression plasmids. HEK-293T cells were co-transfected with the luciferase reporter plasmid, thymidine kinase promoter–Renilla luciferase reporter plasmid (Promega, E2271) and the ThPOK- or RUNX3-expressing plasmid or a control vector. After 48 h, luciferase activities were measured by the Dual-Luciferase Reporter Assay System (Promega, E1910) according to the manufacturer’s instructions.
Immunoblotting
Cells were lysed in RIPA buffer containing PMSF, protease and phosphatase inhibitors (Santa Cruz, sc-24948) for 30 min on ice. Protein lysates were separated on denaturing 4–15% gradient SDS-PAGE gels (BioRad, #4561086), transferred onto nitrocellulose membranes (BioRad, #170427) via the Trans-Blot Turbo system (Bio-Rad). Proteins were detected by antibodies against TRIB2 (D8P2X; 1:500, Cell Signaling Technology, #13533S) and β-Actin (13E5; 1:10,000, Cell Signaling Technology, #4970S). Membranes were developed using HRP-conjugated secondary antibodies (Cell Signaling Technology, #7074S) and SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific, #34096). Densitometry of protein bands was measured by Fiji (ImageJ) software.60
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using Prism software 9.0 (GraphPad). Unless stated otherwise, data are presented as mean with error bars indicating the standard error of the mean (SEM). Comparison of two groups was performed by paired or unpaired two-tailed Student’s t-tests. Two-way ANOVA with post-hoc Tukey, Šídák, or Dunnett test or one-way ANOVA with post-hoc Tukey or Dunnett test were used for multigroup comparisons, as appropriate. p-value less than 0.05 was considered statistically significant. Significance levels (*p < 0.05, **p ≤ 0.01, ***p ≤ 0.001 and ****p ≤ 0.0001) are indicated in figures and specific information is given in figure legends.
Supplementary Material
Highlights.
Expression of TRIB2 is higher in naive CD4+ than naive CD8+ T cells
TRIB2 transcription is upregulated by ThPOK and inhibited by RUNX3
TRIB2 in CD4+ T cells inhibits AKT activation in response to homeostatic cytokines
TRIB2 deficiency in CD8+ T cells enhances their proliferation and differentiation
ACKNOWLEDGMENTS
We thank R. Bosselut and P.E. Love from the National Institutes of Health for providing mouse strains and Xin Yang (Stanford University) for assistance in mouse studies. This work was supported by National Institutes of Health (NIH) R01AR042527, R01HL117913, R01AI108906, and R01HL142068 (all to C.M.W.); NIH R01AI108891, R01AG045779, R01AI129191, and U19AI057266 (all to J.J.G.); NIH T32AG049672 (to I.S.); and the National Natural Science Foundation of China 32270938 (to W.C.). This study was supported with resources and the use of facilities at the Palo Alto Veterans Administration Healthcare System and the Mayo Clinic Microscope and Cell Analysis Core. I.S. is a Glenn Foundation for Medical Research Postdoctoral Fellow. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112195.
REFERENCES
- 1.Goronzy JJ, and Weyand CM (2019). Mechanisms underlying T cell ageing. Nat. Rev. Immunol. 19, 573–583. 10.1038/s41577-019-0180-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.den Braber I, Mugwagwa T, Vrisekoop N, Westera L, Mögling R, de Boer AB, Willems N, Schrijver EHR, Spierenburg G, Gaiser K, et al. (2012). Maintenance of peripheral naive T cells is sustained by thymus output in mice but not humans. Immunity 36, 288–297. 10.1016/j.immuni.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Czesnikiewicz-Guzik M, Lee WW, Cui D, Hiruma Y, Lamar DL, Yang ZZ, Ouslander JG, Weyand CM, and Goronzy JJ (2008). T cell subset-specific susceptibility to aging. Clin. Immunol. 127, 107–118. 10.1016/j.clim.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MF, Almeida JR, Gostick E, Yu Z, Carpenito C, et al. (2011). A human memory T cell subset with stem cell-like properties. Nat. Med. 17, 1290–1297. 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akondy RS, Fitch M, Edupuganti S, Yang S, Kissick HT, Li KW, Youngblood BA, Abdelsamed HA, McGuire DJ, Cohen KW, et al. (2017). Origin and differentiation of human memory CD8 T cells after vaccination. Nature 552, 362–367. 10.1038/nature24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marraco SAF, Charlotte S, Laurène C, Philippe OG, Mathilde A, Samia AM, Nicole M, Nathalie R, Sophie W, Mauro D, et al. (2015). Long-lasting stem cell-like memory CD8+T cells with a naïve-like profile upon yellow fever vaccination. Sci. Transl. Med. 7. 10.1126/scitranslmed.aaa3700. [DOI] [PubMed] [Google Scholar]
- 7.Whiting CC, Siebert J, Newman AM, Du HW, Alizadeh AA, Goronzy J, Weyand CM, Krishnan E, Fathman CG, and Maecker HT (2015). Large-scale and comprehensive immune profiling and functional analysis of normal human aging. PLoS One 10, e0133627. 10.1371/journal.pone.0133627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsukamoto H, Clise-Dwyer K, Huston GE, Duso DK, Buck AL, Johnson LL, Haynes L, and Swain SL (2009). Age-associated increase in lifespan of naive CD4 T cells contributes to T-cell homeostasis but facilitates development of functional defects. Proc. Natl. Acad. Sci. USA 106, 18333–18338. 10.1073/pnas.0910139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haluszczak C, Akue AD, Hamilton SE, Johnson LDS, Pujanauski L, Teodorovic L, Jameson SC, and Kedl RM (2009). The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J. Exp. Med. 206, 435–448. 10.1084/jem.20081829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quinn KM, Fox A, Harland KL, Russ BE, Li J, Nguyen THO, Loh L, Olshanksy M, Naeem H, Tsyganov K, et al. (2018). Age-related decline in primary CD8(+) T cell responses is associated with the development of senescence in virtual memory CD8(+) T cells. Cell Rep. 23, 3512–3524. 10.1016/j.celrep.2018.05.057. [DOI] [PubMed] [Google Scholar]
- 11.Carrette F, and Surh CD (2012). IL-7 signaling and CD127 receptor regulation in the control of T cell homeostasis. Semin. Immunol. 24, 209–217. 10.1016/j.smim.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho J-H, Kim H-O, Surh CD, and Sprent J (2010). T cell receptor-dependent regulation of lipid rafts controls naive CD8+ T cell homeostasis. Immunity 32, 214–226. 10.1016/j.immuni.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Surh CD, and Sprent J (2008). Homeostasis of naive and memory T cells. Immunity 29, 848–862. 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Kityo C, Makamdop KN, Rothenberger M, Chipman JG, Hoskuldsson T, Beilman GJ, Grzywacz B, Mugyenyi P, Ssali F, Akondy RS, et al. (2018). Lymphoid tissue fibrosis is associated with impaired vaccine responses. J. Clin. Invest. 128, 2763–2773. 10.1172/JCI97377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ucar D, Márquez EJ, Chung CH, Marches R, Rossi RJ, Uyar A, Wu TC, George J, Stitzel ML, Palucka AK, et al. (2017). The chromatin accessibility signature of human immune aging stems from CD8+ T cells. J. Exp. Med. 214, 3123–3144. 10.1084/jem.20170416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naylor K, Li G, Vallejo AN, Lee WW, Koetz K, Bryl E, Witkowski J, Fulbright J, Weyand CM, and Goronzy JJ (2005). The influence of age on T cell generation and TCR diversity. J. Immunol. 174, 7446–7452. 10.4049/jimmunol.174.11.7446. [DOI] [PubMed] [Google Scholar]
- 17.Hu B, Jadhav RR, Gustafson CE, Le Saux S, Ye Z, Li X, Tian L, Weyand CM, and Goronzy JJ (2020). Distinct age-related epigenetic signatures in CD4 and CD8 T cells. Front. Immunol. 11, 585168. 10.3389/fimmu.2020.585168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moskowitz DM, Zhang DW, Hu B, Le Saux S, Yanes RE, Ye Z, Buenrostro JD, Weyand CM, Greenleaf WJ, and Goronzy JJ (2017). Epigenomics of human CD8 T cell differentiation and aging. Sci. Immunol. 2, eaag0192. 10.1126/sciimmunol.aag0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serroukh Y, Gu-Trantien C, Hooshiar Kashani B, Defrance M, Vu Manh TP, Azouz A, Detavernier A, Hoyois A, Das J, et al. (2018). The transcription factors Runx3 and ThPOK cross-regulate acquisition of cytotoxic function by human Th1 lymphocytes. Elife 7, e30496. 10.7554/eLife.30496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, and Noelle RJ (2009). Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol. Rev. 229, 152–172. 10.1111/j.1600-065X.2009.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang KL, O’Connor C, Veiga JP, McCarthy TV, and Keeshan K(2016). TRIB2 regulates normal and stress-induced thymocyte proliferation. Cell Discov. 2, 15050. 10.1038/celldisc.2015.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brzostek J, Gautam N, Zhao X, Chen EW, Mehta M, Tung DWH, Chua YL, Yap J, Cho SH, Sankaran S, et al. (2020). T cell receptor and cytokine signal integration in CD8+ T cells is mediated by the protein Themis. Nat. Immunol. 21, 186–198. 10.1038/s41590-019-0570-3. [DOI] [PubMed] [Google Scholar]
- 23.Liu M, Zhang J, Pinder BD, Liu Q, Wang D, Yao H, Gao Y, Toker A, Gao J, Peterson A, et al. (2021). WAVE2 suppresses mTOR activation to maintain T cell homeostasis and prevent autoimmunity. Science 371, eaaz4544. 10.1126/science.aaz4544. [DOI] [PubMed] [Google Scholar]
- 24.Naiki T, Saijou E, Miyaoka Y, Sekine K, and Miyajima A (2007). TRB2, a mouse Tribbles ortholog, suppresses adipocyte differentiation by inhibiting AKT and C/EBPβ. J. Biol. Chem. 282, 24075–24082. 10.1074/jbc.M701409200. [DOI] [PubMed] [Google Scholar]
- 25.Du K, Herzig S, Kulkarni RN, and Montminy M (2003). TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science 300, 1574–1577. 10.1126/science.1079817. [DOI] [PubMed] [Google Scholar]
- 26.Hill R, Madureira PA, Ferreira B, Baptista I, Machado S, Colac o, L., Dos Santos M, Liu N, Dopazo A, Ugurel S, et al. (2017). TRIB2 confers resistance to anti-cancer therapy by activating the serine/threonine protein kinase AKT. Nat. Commun. 8, 14687. 10.1038/ncomms14687. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Sprent J, and Surh CD (2011). Normal T cell homeostasis: the conversion of naive cells into memory-phenotype cells. Nat. Immunol. 12, 478–484. 10.1038/ni.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciucci T, Vacchio MS, Gao Y, Tomassoni Ardori F, Candia J, Mehta M, Zhao Y, Tran B, Pepper M, et al. (2019). The emergence and functional fitness of memory CD4+ T cells require the transcription factor thpok. Immunity 50, 91–105.e4. 10.1016/j.immuni.2018.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lotem J, Levanon D, Negreanu V, Leshkowitz D, Friedlander G, and Groner Y (2013). Runx3-mediated transcriptional program in cytotoxic lymphocytes. PLoS One 8, e80467. 10.1371/journal.pone.0080467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vacchio MS, Wang L, Bouladoux N, Carpenter AC, Xiong Y, Williams LC, Wohlfert E, Song KD, Belkaid Y, Love PE, and Bosselut R (2014). A ThPOK-LRF transcriptional node maintains the integrity and effector potential of post-thymic CD4+ T cells. Nat. Immunol. 15, 947–956. 10.1038/ni.2960 https://www.nature.com/articles/ni.2960#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghia P, Prato G, Stella S, Scielzo C, Geuna M, and Caligaris-Cap-pio F (2007). Age-dependent accumulation of monoclonal CD4+CD8+double positive T lymphocytes in the peripheral blood of the elderly. Br. J. Haematol. 139, 780–790. 10.1111/j.1365-2141.2007.06867.x. [DOI] [PubMed] [Google Scholar]
- 32.Zhang H, Jadhav RR, Cao W, Goronzy IN, Zhao TV, Jin J, Ohtsuki S, Hu Z, Morales J, Greenleaf WJ, et al. (2023). Aging-associated HELIOS deficiency in naive CD4+ T cells alters chromatin remodeling and promotes effector cell responses. Nat. Immunol. 24, 96–109. 10.1038/s41590-022-01369-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chapman NM, Boothby MR, and Chi H (2020). Metabolic coordination of T cell quiescence and activation. Nat. Rev. Immunol. 20, 55–70. 10.1038/s41577-019-0203-y. [DOI] [PubMed] [Google Scholar]
- 34.Yang K, Shrestha S, Zeng H, Karmaus PWF, Neale G, Vogel P, Guertin DA, Lamb RF, and Chi H (2013). T cell exit from quiescence and differentiation into Th2 cells depend on Raptor-mTORC1-mediated metabolic reprogramming. Immunity 39, 1043–1056. 10.1016/j.immuni.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki A, Yamaguchi MT, Ohteki T, Sasaki T, Kaisho T, Kimura Y, Yoshida R, Wakeham A, Higuchi T, Fukumoto M, et al. (2001). T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity 14, 523–534. 10.1016/s1074-7613(01)00134-0. [DOI] [PubMed] [Google Scholar]
- 36.Zeng H, Cohen S, Guy C, Shrestha S, Neale G, Brown SA, Cloer C, Kishton RJ, Gao X, Youngblood B, et al. (2016). mTORC1 and mTORC2 kinase signaling and glucose metabolism drive follicular helper T cell differentiation. Immunity 45, 540–554. 10.1016/j.immuni.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buckler JL, Walsh PT, Porrett PM, Choi Y, and Turka LA (2006). Cutting edge: T cell requirement for CD28 costimulation is due to negative regulation of TCR signals by PTEN. J. Immunol. 177, 4262–4266. 10.4049/jimmunol.177.7.4262. [DOI] [PubMed] [Google Scholar]
- 38.Zheng L, Han X, Yao S, Zhu Y, Klement J, Wu S, Ji L, Zhu G, Cheng X, Tobiasova Z, et al. (2022). The CD8alpha-PILRalpha interaction maintains CD8(+) T cell quiescence. Science 376, 996–1001. 10.1126/science.aaz8658. [DOI] [PubMed] [Google Scholar]
- 39.Feng X, Wang H, Takata H, Day TJ, Willen J, and Hu H (2011). Transcription factor Foxp1 exerts essential cell-intrinsic regulation of the quiescence of naive T cells. Nat. Immunol. 12, 544–550. 10.1038/ni.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richmond L, and Keeshan K (2020). Pseudokinases: a tribble-edged sword. FEBS J. 287, 4170–4182. 10.1111/febs.15096. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Park JS, Wei Y, Rajurkar M, Cotton JL, Fan Q, Lewis BC, Ji H, and Mao J (2013). TRIB2 acts downstream of Wnt/TCF in liver cancer cells to regulate YAP and C/EBPalpha function. Mol. Cell 51, 211–225. 10.1016/j.molcel.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keeshan K, He Y, Wouters BJ, Shestova O, Xu L, Sai H, Rodriguez CG, Maillard I, Tobias JW, Valk P, et al. (2006). Tribbles homolog 2 inactivates C/EBPalpha and causes acute myelogenous leukemia. Cancer Cell 10, 401–411. 10.1016/j.ccr.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gilby DC, Sung HY, Winship PR, Goodeve AC, Reilly JT, and Kiss-Toth E (2010). Tribbles-1 and -2 are tumour suppressors, downregulated in human acute myeloid leukaemia. Immunol. Lett. 130, 115–124. 10.1016/j.imlet.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 44.Foulkes DM, Byrne DP, Yeung W, Shrestha S, Bailey FP, Ferries S, Eyers CE, Keeshan K, Wells C, Drewry DH, et al. (2018). Covalent inhibitors of EGFR family protein kinases induce degradation of human Tribbles 2 (TRIB2) pseudokinase in cancer cells. Sci. Signal. 11, eaat7951. 10.1126/scisignal.aat7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X, Chang X, Facchinetti V, Zhuang Y, and Su B (2009). MEKK3 is essential for lymphopenia-induced T cell proliferation and Survival1. J. Immunol. 182, 3597–3608. 10.4049/jimmunol.0803738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carpenter AC, and Bosselut R (2010). Decision checkpoints in the thymus. Nat. Immunol. 11, 666–673. 10.1038/ni.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L, Wildt KF, Castro E, Xiong Y, Feigenbaum L, Tessarollo L, and Bosselut R (2008). The zinc finger transcription factor Zbtb7b represses CD8-lineage gene expression in peripheral CD4+ T cells. Immunity 29, 876–887. 10.1016/j.immuni.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang D, Diao H, Getzler AJ, Rogal W, Frederick MA, Milner J, Yu B, Crotty S, Goldrath AW, and Pipkin ME (2018). The transcription factor Runx3 establishes chromatin accessibility of cis-regulatory landscapes that drive memory cytotoxic T lymphocyte formation. Immunity 48, 659–674.e6. 10.1016/j.immuni.2018.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Egawa T, Tillman RE, Naoe Y, Taniuchi I, and Littman DR (2007). The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med. 204, 1945–1957. 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chiu BC, Martin BE, Stolberg VR, and Chensue SW (2013). Cutting edge: central memory CD8 T cells in aged mice are virtual memory cells. J. Immunol. 191, 5793–5796. 10.4049/jimmunol.1302509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Renkema KR, Li G, Wu A, Smithey MJ, and Nikolich-Žugich J (2014). Two separate defects affecting true naive or virtual memory T cell precursors combine to reduce naive T cell responses with aging. J. Immunol. 192, 151–159. 10.4049/jimmunol.1301453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wikby A, Ferguson F, Forsey R, Thompson J, Strindhall J, Löfgren S, Nilsson BO, Ernerudh J, Pawelec G, and Johansson B (2005). An immune risk phenotype, cognitive impairment, and survival in very late life: impact of allostatic load in Swedish octogenarian and nonagenarian humans. J. Gerontol. A Biol. Sci. Med. Sci. 60, 556–565. 10.1093/gerona/60.5.556. [DOI] [PubMed] [Google Scholar]
- 53.Olsson J, Wikby A, Johansson B, Löfgren S, Nilsson BO, and Ferguson FG (2001). Age-related change in peripheral blood T-lymphocyte subpopulations and cytomegalovirus infection in the very old: the Swedish longitudinal OCTO immune study. Mech. Ageing Dev. 121, 187–201. 10.1016/S0047-6374(00)00210-4. [DOI] [PubMed] [Google Scholar]
- 54.Zhang H, Weyand CM, and Goronzy JJ (2021). Hallmarks of the aging T-cell system. FEBS J. 288, 7123–7142. 10.1111/febs.15770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mackall CL, Fry TJ, and Gress RE (2011). Harnessing the biology of IL-7 for therapeutic application. Nat. Rev. Immunol. 11, 330–342. 10.1038/nri2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Velardi E, Tsai JJ, and van den Brink MRM (2021). T cell regeneration after immunological injury. Nat. Rev. Immunol. 21, 277–291. 10.1038/s41577-020-00457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ciucci T, Vacchio MS, Gao Y, Tomassoni Ardori F, Candia J, Mehta M, Zhao Y, Tran B, Pepper M, Tessarollo L, et al. (2019). The emergence and functional fitness of memory CD4(+) T cells require the transcription factor thpok. Immunity 50, 91–105.e4. 10.1016/j.immuni.2018.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Naoe Y, Setoguchi R, Akiyama K, Muroi S, Kuroda M, Hatam F, Littman DR, and Taniuchi I (2007). Repression of interleukin-4 in T helper type 1 cells by Runx/Cbfβ binding to the Il4 silencer. J. Exp. Med. 204, 1749–1755. 10.1084/jem.20062456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, and Costantini F (2001). Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4. 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M,Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harris JA, Hirokawa KE, Sorensen SA, Gu H, Mills M, Ng LL, Bohn P, Mortrud M, Ouellette B, Kidney J, et al. (2014). Anatomical characterization of Cre driver mice for neural circuit mapping and manipulation. Front. Neural Circ. 8, 76. 10.3389/fncir.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper analyzes existing, publicly available data. The accession numbers for these datasets are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Anti-human CD3 (HIT3a) | BioLegend | Cat# 300318, RRID:AB_314054 |
| Anti-human CD3 (OKT3) | BioLegend | Cat# 317314, RRID:AB_571909 |
| Anti-human CD4 (RPA-T4) | BioLegend | Cat# 300506, RRID:AB_314074 |
| Anti-human CD8 (SK1) | BioLegend | Cat# 344723, RRID:AB_2562790 |
| Anti-human CCR7 (G043H7) | BioLegend | Cat# 353207, RRID:AB_11203894 |
| Anti-human CD25 (BC96) | BioLegend | Cat# 302635, RRID:AB_11219793 |
| Anti-human CD122 (TU27) | BioLegend | Cat# 339017, RRID:AB_2721347 |
| Anti-human IL2 (MQ1-17H12) | BioLegend | Cat# 500352, RRID:AB_2820083 |
| Anti-human TNFα (Mab11) | BioLegend | Cat# 502908, RRID:AB_315261 |
| Anti-human CD45RA (HI100) | BD Biosciences | Cat# 562886, RRID:AB_2737865 |
| Anti-human IFNγ (B27) | BD Biosciences | Cat# 562016, RRID:AB_10894955 |
| Anti-human Granzyme B (GB11) | BD Biosciences | Cat# 563388, RRID:AB_2738174 |
| Anti-human/mouse Ki67 (B56) | BD Biosciences | Cat# 561283, RRID:AB_10716060 |
| Anti-human/mouse pAKT-Ser473 (M89-61) | BD Biosciences | Cat# 560343, RRID:AB_1645397 |
| Anti-human/mouse pAKT-Thr308 (J1–223.371) | BD Biosciences | Cat# 558275, RRID:AB_2225329 |
| Anti-human pERK1/2-Thr202/Tyr204 (20A) | BD Biosciences | Cat# 612592, RRID:AB_399875 |
| Anti-human S6-pS235/pS236 (N7–548) | BD Biosciences | Cat# 560433, RRID:AB_2827879 |
| Anti-human FoxO1 (C29H4) | Cell Signaling Technologies | Cat# 58223S, RRID:AB_2799543 |
| Anti-human/mouse TRIB2 (D8P2X) | Cell Signaling Technologies | Cat# 13533S, RRID:AB_2798250 |
| Anti-human/mouse β-Actin (13E5) | Cell Signaling Technologies | Cat# 4970S, RRID:AB_2223172 |
| Anti-human ThPOK (D9V5T) | Cell Signaling Technologies | Cat# 13205, RRID:AB_2798147 |
| Anti-human RUNX3 (9F4A17) | BioLegend | Cat# 653603, RRID:AB_2687117 |
| Purified anti-human CD3 (UCHT1) | BioLegend | Cat# 300465, RRID:AB_11147760 |
| Purified anti-human CD28 (CD28.1) | BioLegend | Cat# 302943, RRID:AB_11150591 |
| Anti-mouse CD4 (RM4–5) | BioLegend | Cat# 100553, RRID:AB_2561388 |
| Anti-mouse CD4 (GK1.5) | BioLegend | Cat# 100449, RRID:AB_2564587 |
| Anti-mouse CD8α (53–6.7) | BioLegend | Cat# 100750, RRID:AB_2562610 |
| Anti-mouse TCRβ (H57–597) | BioLegend | Cat# 109219, RRID:AB_893624 |
| Anti-mouse CD44 (IM7) | BioLegend | Cat# 103040, RRID:AB_2616903 |
| Anti-mouse CD62L (MEL-14) | BioLegend | Cat# 104421, RRID:AB_493379 |
| Anti-mouse CD122 (TM-b1) | BioLegend | Cat# 123209, RRID:AB_940615 |
| Anti-mouse CD45.2 (104) | BioLegend | Cat# 109824, RRID:AB_830789 |
| Anti-mouse CD25 (PC61.5) | BioLegend | Cat# 102008, RRID:AB_312857 |
| Anti-mouse CD127 (IL7Ra) (A7R34) | BioLegend | Cat# 135041, RRID:AB_2572047 |
| Anti-mouse IFNγ (XMG1.2) | BioLegend | Cat# 505810, RRID:AB_315404 |
| Anti-mouse CD24 (M1/69) | BD Biosciences | Cat# 553261, RRID:AB_394740 |
| Purified anti-mouse CD3e (eBio500A2) | Invitrogen | Cat# 16–0033-85, RRID:AB_842785 |
| Purified anti-mouse CD28 (37.51) | Invitrogen | Cat# 16–0281-85, RRID:AB_468922 |
|
| ||
| Biological samples | ||
|
| ||
| Human peripheral blood (buffy coats) | Stanford University Blood bank | N/A |
| Human peripheral blood (leukoreduction system chambers) | Mayo Clinic Blood bank | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Human IL-7 | PeproTech | Cat# 200–07 |
| FANA antisense oligonucleotides | AUM BioTech | https://www.aumbiotech.com/ |
| MK-2206, AKT inhibitor | Selleckchem | Cat# S1078 |
|
| ||
| Critical commercial assays | ||
|
| ||
| EasySep Human Naive CD4+T cell isolation Kit II | StemCell Technologies | Cat# 19555 |
| EasySep Human Naive CD8+ T cell isolation Kit | StemCell Technologies | Cat# 19258 |
| Pan T cell isolation Kit II, mouse | Miltenyi Biotec | Cat# 130–095-130 |
| EasySep Mouse Naive CD4+ T cell Isolation Kit | StemCell Technologies | Cat# 19765 |
| EasySep Mouse Naive CD8+ T cell Isolation Kit | StemCell Technologies | Cat# 19858 |
| ChIP-IT High Sensitivity Kit | Active Motif | Cat# 53040 |
| Dual-Luciferase Reporter Assay System | Promega | Cat# E1910 |
| RNeasy Plus Mini Kit | Qiagen | Cat# 74134 |
| RNeasy Plus Micro Kit | Qiagen | Cat# 74034 |
| Maxima First Strand cDNA Synthesis Kit | Thermo Fisher Scientific | Cat# K1641 |
|
| ||
| Deposited data | ||
|
| ||
| Transcriptomic data of human naive CD4+ and CD8+ T cells | Serroukh et al.19 | GEO: GSE75406 |
| ThPOK ChIP-seq data of mouse CD4+ T cells | Ciucci et al.57 | GEO: GSE116506 |
| RUNX3 ChIP-seq data of mouse CD8+ T cells | Lotem et al.29 | GEO: GSE50130 |
| ATAC-seq data of human naive CD4+ T cells | Hu et al.17 | SRA: PRJNA478249 |
| ATAC-seq data of human naive CD8+ T cells | Hu et al.17 | dbGaP: phs001187.v1.p1 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: B6.Cg-Trib2tm1.1(crelERT2)Hze/J | The Jackson Laboratory | RRID:IMSR_JAX:022,865 |
| Mouse: CD2-Cre | Vacchio et al.30 | N/A |
| Mouse: conditional Zbtb7b/ThPOK knockout | Wang et al.47 | N/A |
| Mouse: conditional Cbfb knockout | Naoe et al.58 | N/A |
| Mouse: ROSA26-LoxP-STOP-LoxP-YFP | Srinivas et al.59 | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| see Table S1 for qPCR primers | N/A | N/A |
| see Table S1 for ChIP-PCR primers | N/A | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pHAGE-CMV-IRES-ZsGreen-W | Addgene, David Baltimore's unpublished plasmid | Addgene Plasmid #26532 |
| pGL3 basic | Promega | Cat# E1751 |
|
| ||
| Software and algorithms | ||
|
| ||
| FlowJo 10 | Tree Star | https://www.flowjo.com/ |
| Fiji | Schindelin et al.60 | https://imagej.net/ |
| Integrative Genomics Viewer (IGV) 2.10 | Broad Institute | https://software.broadinstitute.org/software/igv/ |
| Prism 9 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |


