1 ∣. INTRODUCTION
The general pathway of iron sulfur (Fe-S) cluster biogenesis has been gradually elucidated over the years by a combination of genetic and biochemical studies. The components of the assembly machinery were found to be broadly distributed in all kingdoms of life and to share common features, such as the use of a cysteine desulfurase to provide inorganic sulfur, the central role of scaffold proteins for Fe-S cluster assembly, and a highly conserved transfer system, which consists of an HSP70 chaperone and a J-protein co-chaperone, that interacts with scaffold proteins to facilitate Fe-S cluster delivery to recipients. In this chapter, we provide an overview of our understanding of iron–sulfur cluster biogenesis in mammalian cells, discuss recent studies that shed light on the molecular interactions that govern iron–sulfur cluster assembly and introduce advances in the understanding of how the transfer complex efficiently engages recipient Fe-S target proteins to deliver a cluster.
2 ∣. BIOLOGICAL ROLES OF FE-S PROTEINS
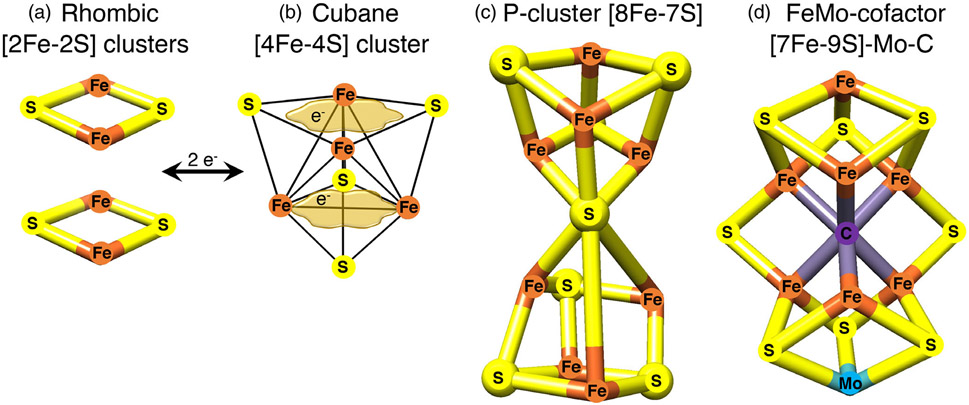
Fe-S clusters, among the oldest cofactors known in biology, may have played a critical role in the emergence of life on Earth more than 3 billion years ago, when they were incorporated into early metabolic pathways by primitive organisms.1 Their versatile chemical properties have fostered their pervasive use in virtually all organisms to execute an impressive number of reactions involved in fundamental cellular processes such as respiration, photosynthesis, metabolism, and nitrogen fixation. Fe-S clusters consist of iron and inorganic sulfide that are typically, but not exclusively, ligated to cysteinyl sulfur in proteins2 (Figure 1). The most common types are the tetranuclear [4Fe-4S]2+ clusters which can readily accept or donate single electrons. However, [2Fe-2S] clusters are also very abundant and are present in enzymes such as mammalian ferrochelatase, mitochondrial respiratory complexes I and II, ferredoxins, and Rieske proteins (Figure 1).
FIGURE 1.
Examples of Fe-S clusters found in proteins. (a) The common rhombic [2Fe-2S] clusters are initially assembled from inorganic iron and sulfur upon the main scaffold ISCU, and they can be utilized to generate much more complex Fe-S cofactors. (b) The tetranuclear or cubane clusters are formed by reductive coupling of two [2Fe-2S] clusters, in a process that requires two electrons (2e−). [4Fe-4S] clusters have the capability to delocalize electrons between iron sites (as depicted by the tan cloud-like frames, which illustrate that the iron-associated electrons are highly delocalized between neighboring iron atoms). (c) Larger Fe-S cofactors can attain much higher complexity in the P-cluster of nitrogenase or in the iron-molybdenum cofactor of nitrogenase depicted in (d), where an interstitial carbon atom occupies the core of the cofactor. Adopted from139
Fe-S clusters commonly mediate redox reactions in mitochondrial respiration, photosynthesis and nitrogen fixation, where they act as electron relay chains. A redox switch in the oxidation state of [4Fe-4S]2+ clusters was recently found to modulate the DNA-binding affinity of DNA-processing enzymes, including glycosylases, helicases and primases.3 Fe-S enzymes play critical roles in translation, as in the case of the ATP-binding cassette sub-family E member 1 (ABCE1), also known as RNase L inhibitor (Rli) in Saccharomyces cerevisiae.4
Besides their involvement in redox reactions, Fe-S enzymes are known to play essential roles in several metabolic pathways, with one of the most notable examples being mitochondrial aconitase of the TCA cycle, which ligates a solvent exposed [4Fe-4S] cluster that directly participates in substrate binding and conversion of citrate to isocitrate through acid-base catalysis.5 Cytosolic aconitase is a bifunctional enzyme (iron regulatory protein 1, IRP1/cytosolic aconitase, ACO1), endowed with a crucial role in the regulation of cellular iron homeostasis.6 Several bacterial dehydratases share with aconitase the ability to bind and activate substrates at the uniquely open iron site of their [4Fe-4S] clusters, in which the other three iron atoms are ligated to cysteines of the peptide backbone; these include the isopropylmalate isomerase involved in leucine biosynthesis, the Escherichia coli fumarase A that converts fumarate to malate, the dihydroxyacid dehydratase that is involved in the biosynthesis of branched chain amino acids, and the L-serine dehydratase that catalyzes the irreversible deamination of serine to pyruvate.2 Fe-S clusters are essential cofactors of the radical S-adenosylmethionine (SAM) superfamily of enzymes, eight of which have so far been reported to occur in pathways that directly affect human health and disease (reviewed in Reference 7), including molybdenum and lipoic acid cofactor biosynthesis,8,9 and the generation of specific tRNA modifications that affect codon-anticodon recognition and translation fidelity.10-12 Fe-S enzymes are also responsible for DNA replication and the maintenance/repair of the nuclear genome,13,14 as well as for cell division.15 Inactivating mutations in CISD2 (also known as NAF1), initially annotated as a zinc-finger protein16 and later characterized as a member of the NEET family of proteins that coordinates a labile [2Fe-2S] cluster,17 were recently reported to cause Wolfram syndrome 2,18 an autosomal recessive neurodegenerative disorder characterized by diabetes mellitus, sensorineural hearing loss, optic atrophy and neuropathy.
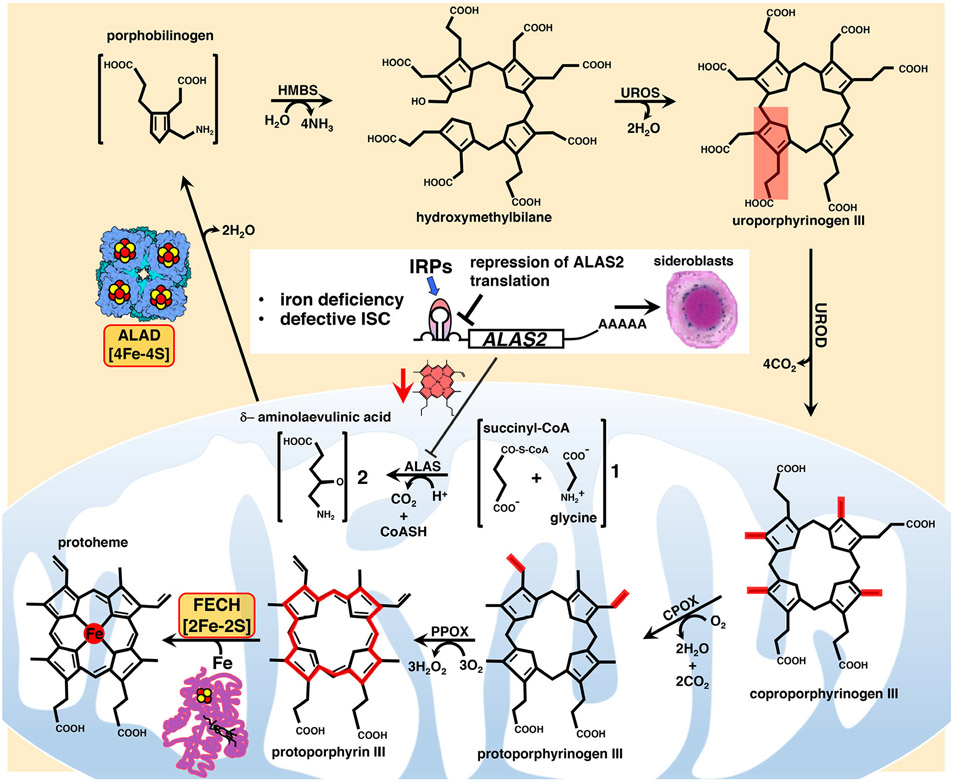
Fe-S clusters are also indispensable for heme biosynthesis (discussed in depth in ref. [19]). Heme consists of the protoporphyrin IX ring complexed with ferrous iron and it is necessary for the function of several proteins, including hemoglobin and myoglobin, cytochromes of the electron transport chain, catalase, and nitric oxide synthase. The mRNA of ALAS2 that encodes the erythroid specific 5-aminolevulinate synthase that catalyzes the first rate-limiting step of heme biosynthesis in mitochondria has an IRE in its 5′-UTR (untranslated region),20 which permits translational regulation by Iron Regulatory Proteins (IRPs) (Figure 2). Interestingly, the reaction catalyzed by ALAS2 represents one of four major points at which the heme and Fe-S cluster biosynthetic pathways intersect. Since IRP1 loses its IRE-binding activity when it ligates an Fe-S cluster, ALAS2 protein levels are indirectly regulated by Fe-S cluster biogenesis, because in conditions of iron deficiency or defective Fe-S cluster biogenesis, IRP1 is converted into its apo form, which binds to the IRE present in the 5′-UTR of the ALAS2 mRNA and represses its translation, thereby preventing accumulation of toxic porphyrin intermediates when heme biosynthesis cannot be sustained due to limited availability of iron. An additional point of intersection between Fe-S biogenesis and heme biosynthesis emerged for the first time from studies in zebrafish deficient in the Fe-S cluster biogenesis protein glutaredoxin 5 (GLRX5, in human).21 Genetic experiments demonstrated that in the shiraz zebrafish mutants, characterized by hypochromic anemia, loss of glutaredoxin 5 led to translational repression of ALAS2 mRNA due to activation of IRP1.21 Subsequent studies in fibroblasts derived from a patient affected by sideroblastic anemia caused by an inactivating mutation in GLRX5 and in cells depleted of GLRX5 by short interfering RNAs (siRNAs) confirmed that hyperactivation of IRP1 disrupted cellular iron homeostasis and repressed ALAS2 translation upon loss of function of GLRX5.22,23 A recent study revealed an additional point of intersection between heme biosynthesis and Fe-S cluster biogenesis that hinges on the presence of an oxidation-sensitive [2Fe-2S] cluster in the C-terminal domain of the F-box and leucine-rich repeats protein FBXL524 that recognizes IRP2 and promotes its iron- and oxygen-dependent degradation. Interestingly, studies for over a decade reported increased IRE-binding activities of IRPs upon loss of components of the Fe-S cluster biogenesis machinery.22,25-28 While loss of the cubane cluster in IRP1 in cells with defective Fe-S cluster biogenesis accounted for its increased IRE-binding activity, the mechanism that led to stabilization of IRP2 upon loss of Fe-S cluster biogenesis components had previously remained elusive until the discovery of a Fe-S cluster in FBXL5.24,29 By impairing Fe-S cluster incorporation into FBXL5, defective Fe-S cluster biogenesis hampers the interaction of FBXL5 with IRP2, leading to stabilization of IRP2 and consequently to the increase in its IRE-binding activity that represses ALAS2 translation. Once synthesized in mitochondria, ALA is exported to the cytosol, where ALA dehydratase (ALAD) catalyzes the second step of heme biosynthesis by condensing two ALA molecules into porphobilinogen (Figure 2). A recent study shed light on a previously unrecognized connection between heme biosynthesis and Fe-S cluster biogenesis with the discovery that human ALAD is a Fe-S protein.30 Finally, insertion of ferrous iron into protoporphyrin IX, which follows three additional enzymatic reactions in the cytosol and two in mitochondria, is catalyzed by ferrochelatase (FECH) (Figure 2), which, in higher eukaryotes, but not yeast, contains a [2Fe-2S] cluster31 that was found to contribute to the structural stabilization of the enzyme,32,33 thereby representing yet another point of convergence between the heme and Fe-S cluster biosynthetic pathways. Thus, defects that interfere with Fe-S cluster biogenesis impair heme biosynthesis by repressing ALAS2 synthesis in erythroid cells, and by inactivating the second and the last steps of heme biosynthesis catalyzed by the Fe-S enzymes ALAD and FECH, respectively.
FIGURE 2.
Main steps of heme biosynthesis in which the dependence of the pathway on the Fe-S enzymes FECH and ALAD and the main regulatory nodes are highlighted. Heme is synthesized in eight sequential steps that take place in the mitochondrial matrix and in the cytosol of mammalian cells. Four major points of intersection have been characterized between Fe-S cluster biogenesis and heme biosynthesis in mammalian cells (see the main text for detailed description). Mitochondrial dysfunction due to defects in heme biosynthesis or in Fe-S cluster biogenesis is the leading cause of a heterogenous group of inherited diseases, known as congenital sideroblastic anemias (see main text for details), characterized by ineffective heme biosynthesis and mitochondrial iron overload in erythroid progenitors, in which iron accumulation detected by Prussian blue staining in the mitochondria surrounding the nucleus gives erythroblasts their characteristic appearance of ringed sideroblasts
In the following sections, we will discuss de novo assembly of [2Fe-2S] clusters on the main scaffold protein, ISCU, with a focus on recent insights on the role of frataxin (FXN) that have emerged from the solution of the cryo-electron microscopy structures of the initial core biogenesis complex. We will then explore in-depth how Fe-S clusters are delivered to recipient proteins in both the mitochondrial matrix and cytosolic compartments of mammalian cells.
3 ∣. FE-S CLUSTER BIOGENESIS ON THE MAIN SCAFFOLD ISCU IN MAMMALIAN CELLS
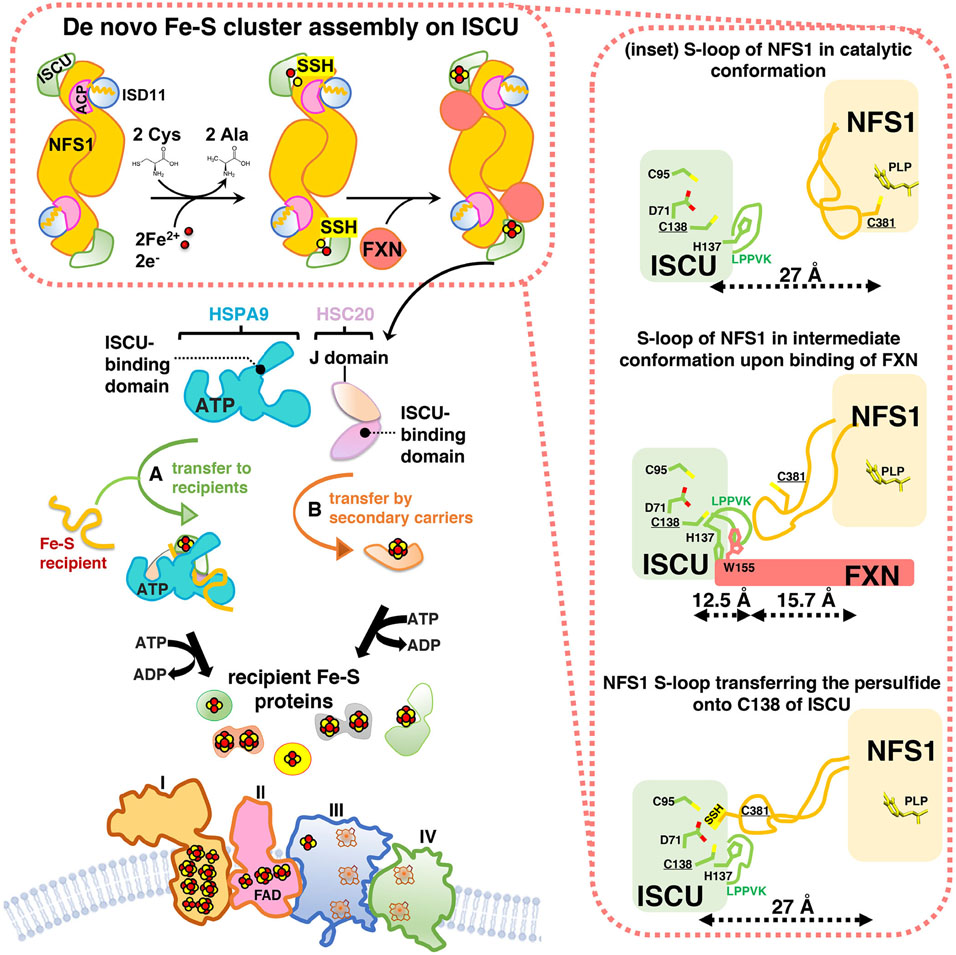
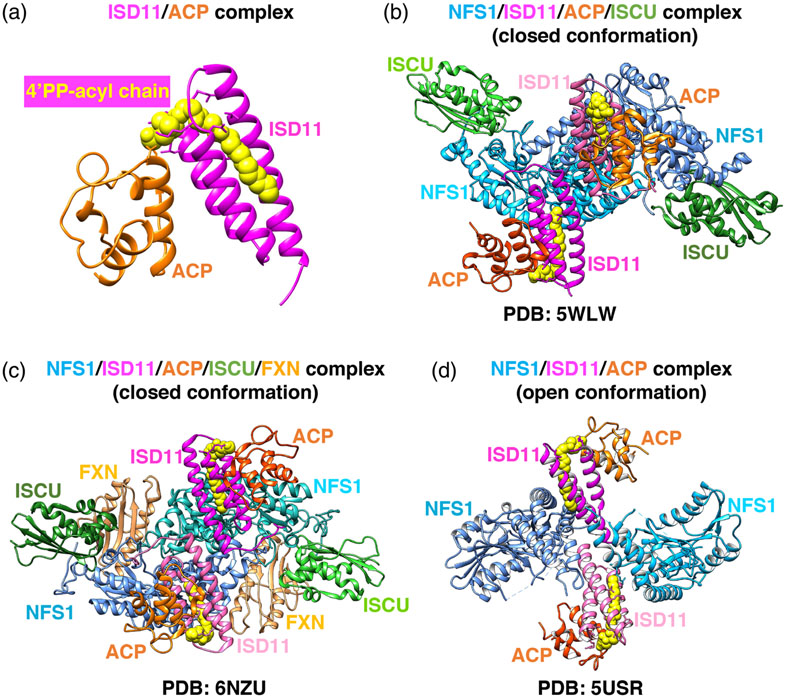
Fe-S cluster biogenesis is an evolutionarily highly conserved process. The first insights into the proteins that contributed to the pathway were obtained through analysis of a bacterial operon needed for Fe-S cluster biogenesis in nitrogen-fixing bacteria.34 De novo Fe-S cluster assembly, the pathway that assembles [2Fe-2S] clusters starting from inorganic sulfur and iron, is a complex, multi-protein-mediated process that involves an initial critical step catalyzed by a pyridoxal-phosphate (PLP) dependent homodimeric transaminase, the cysteine desulfurase NFS1. NFS1 converts cysteine into alanine, donating sulfur to generate a persulfide intermediate on a conserved cysteine residue (Cys381 of human NFS1) of a mobile S-transfer loop of the enzyme35 (Figure 3). The persulfide sulfur is then transferred to a conserved cysteine of the main scaffold protein ISCU (Cys138 of human mitochondrial ISCU) through a conformational change that is enhanced by frataxin (FXN).36,37 NFS1 is stabilized by the binding of the accessory protein ISD11, also known as LYRM4. The acyl-carrier protein ACP (NDUFAB1 in human) was also recently found to be part of the initial Fe-S cluster core biosynthetic complex and to bind ISD11.38 Combined structural and biochemical analyses showed that a critical aspect of the ISD11-ACP interaction involved the insertion of the entire acyl-chain and the 4′-phosphopantotheine (4′-PP) group of ACP into a hydrophobic barrel created by the triple-helical structure of ISD11 (Figure 4A), through a proposed mechanism known as chain-flipping.39-42 Mitochondrial ACP is a bacterial-type acyl-carrier protein involved in mitochondrial fatty acid biosynthesis (mFAS), a pathway that provides octanoic acid, the precursor of lipoic acid, through elongation of nascent acyl-chains bound to the 4′-PP cofactor attached to Ser112 of human ACP. The role of ACP in Fe-S cluster biogenesis remains unclear. It has been proposed that ACP may link fatty acid metabolism, through its role in mFAS, to Fe-S cluster biogenesis.38 Interestingly, an abnormal abundance of lipid droplets was detected in the myocardium of the muscle-specific frataxin-knockout mouse model,43 suggesting that one of the many steps of lipid metabolism was defective in cells with impaired Fe-S cluster assembly due to loss of FXN. In accordance, acute disruption of Fe-S cluster biogenesis was reported to cause a dramatic increase in the intracellular levels of glucose-derived citrate, which was diverted into fatty acid biosynthesis, leading to cytosolic lipid droplet accumulation.28 Mammalian ACP has also been crystallized as part of mitochondrial complex I, through its binding to the subunits known as LYR proteins NDUFA6 (aka LYRM6) and NDUFB9 (aka LYRM3).44
FIGURE 3.
Fe-S cluster biogenesis in mammalian mitochondria: an overview of the main steps. Nascent Fe-S clusters are assembled de novo on the main scaffold protein ISCU. A cysteine desulfurase, NFS1, forms a dimer to which monomers of the primary scaffold ISCU bind at either end. ISD11 (also known as LYRM4) and ACP with its bound acyl chain are structural components of the core complex in eukaryotes. NFS1, aided by its cofactor pyridoxal phosphate, provides inorganic sulfur, abstracted from cysteine, to the nascent cluster. Transient binding of frataxin (FXN) in a pocket-like region between NFS1 and ISCU promotes sulfur transfer from NFS1 to ISCU. The cluster assembles upon ISCU when iron is provided together with the reducing equivalents needed to generate the final electronic configuration of the cluster. A chaperone/co-chaperone complex binds to the LPPVK motif of ISCU and either facilitates direct transfer of Fe-S clusters to recipient proteins (pathway A) or mediates transfer to secondary carriers (e. g. NFU1, GLRX5, ISCA1, ISCA2, BOLA3; pathway B), which then donate Fe-S clusters to specific recipients (e.g., lipoic acid synthase and subunits of the respiratory chain complexes). The inset on the right shows the three different conformations of the mobile S-transfer catalytic loop of NFS1 during de novo Fe-S cluster assembly upon ISCU in its trajectory from the catalytic conformation, in which Cys381 of NFS1 is close to PLP and the substrate cysteine, to the intermediate conformation, recently structurally characterized41 produced by FXN-binding, to the final Fe-S cluster assembly conformation, in which the catalytic Cys381 of NFS1 donates sulfur to Cys138 of ISCU.39 Adopted from139
FIGURE 4.
Comparison of the three recently solved crystal structures of the human Fe-S cluster biogenesis core complex. (a) Structure of the ISD11/ACP complex. The acyl chain covalently attached to the 4'-PP group of ACP is shown in yellow and fits within the three-helical structure of ISD11. (b) Structure of the homodimeric (NFS1/ISD11/ACP/ISCU-Zn2+)2 complex39 (PDB ID: 5WLW). NFS1 protomers are in shades of blue, ISD11 in magenta, ACP in orange, and ISCU in green. (c) Structure of the homodimeric (NFS1/ISD11/ACP/ISCU-Zn2+-FXN)2 complex41 (PDB ID: 6NZU). FXN binds in a pocket-like region between ISCU and NFS1 protomers and it is colored in tan. (d) Structure of the (NFS1/ISD11/ACP)2 homodimeric complex in the “open” conformation40 (PDB ID: 5USR). Adopted from139
The LYR-motif family of proteins was initially defined through bioinformatics analysis as a class of small proteins that contained the Leu-Tyr-Arg motif near the N-terminus.45 The ACP/LYR protein interactions are mainly stabilized through the insertion of the acyl chain of ACP into the hydrophobic core of the helical structures of ISD11, NDUFA6, and NDUFB939,44 (Figure 4A).
Consistently, the deacylated ACPSer112Ala variant was unable to interact with the majority of the partners of wild type ACP, including ISD11 and the complex I sub-units.42 How acyl-ACP links Fe-S cluster biogenesis through its interaction with ISD11 to lipid metabolism remains unknown. Twelve Fe-S clusters are required for the assembly of a functional mitochondrial respiratory chain that utilizes the energy molecules (NADH and FADH2) generated from acetyl-CoA in the citric-acid cycle as primary substrates for oxidative phosphorylation (OXPHOS). Acetyl-CoA is also the precursor for the synthesis of the acyl chain of ACP. It has therefore been proposed that acylation of ACP may serve as a functional link between mitochondrial acetyl-CoA abundance and Fe-S cluster biogenesis.38
The ferredoxin (FDX)/ferredoxin reductase (FDXR) system assists Fe-S cluster assembly,46-48 by reducing the persulfide transferred from Cys381 of NFS1 to Cys138 of human ISCU.48 Combination of sulfane sulfur with Fe2+ on ISCU generates a [2Fe-2S]2+ cluster (Figure 3) that is subsequently transferred to recipient Fe-S apo-proteins or to secondary carriers by a chaperone/co-chaperone system that consists of HSPA9 and HSC20 (aka HSCB) in mammalian cells.49
4 ∣. ARCHITECTURE OF THE HUMAN MITOCHONDRIAL FE-S CLUSTER ASSEMBLY MACHINERY: THE COMPLEX FORMED BY NFS1, ISD11, ACP, ISCU AND FRATAXIN
The NFS1/ISD11/ACP/ISCU complex is a symmetric hetero-octamer, comprising two copies of each of the four constituent proteins39,41 (Figure 4B). NFS1, ISD11, and ACP form a homo-hexameric core, with ISCU bound to each end of the complex. Frataxin (FXN), which acts as an allosteric regulator that enhances sulfur transfer from Cys381 of the mobile S-loop of NFS1 to Cys138 of ISCU (Figure 3), has recently been co-crystallized with the NFS1/ISD11/ACP/ISCU complex41 (Figure 4C). FXN was found to occupy a cavity at the interface between NFS1 and ISCU, where it simultaneously interacted with both NFS1 protomers of the complex without directly contacting ISD1141 (Figure 4C), consistent with the structure of the closed conformation of the NFS1/ISD11/ACP/ISCU complex39 (Figure 4B), but not compatible with a previously reported open conformation of the NFS1/ISD11/ACP complex40 (Figure 4D), in which the two NFS1 protomers made minimal contacts with each other. The unusual arrangement of the NFS1/ISD11/ACP open complex40 revealed an incomplete substrate-binding pocket, a solvent-exposed PLP cofactor, a disordered mobile S-transfer loop, and an overall “open” architecture in which the ISD11 protomers mediated the interactions between NFS1 subunits40 (Figure 4D). To reconcile the two structures, it has been proposed that the NFS1/ISD11/ACP complex may exist in vivo as a mixture of the open and closed conformations in equilibrium. As eukaryotic NFS1 has a turnover rate that is lower than 10% of the prokaryotic cysteine desulfurase, IscS, it is conceivable that FXN stimulates the activity and the catalytic turnover rate of NFS135 by inducing an open-to-closed architectural rearrangement of the complex that repositions the second NFS1 protomer to complete formation of the active site and force a conformational change of the S-transfer loop. In contrast, prokaryotic IscS is proposed to mainly adopt the highly active “closed” conformation that is not further activated by the bacterial ortholog of FXN, CyaY.
5 ∣. INSIGHTS INTO THE FRATAXIN-MEDIATED ACTIVATION OF THE CORE FE-S CLUSTER BIOGENESIS COMPLEX REVEALED BY A NOVEL CRYO-ELECTRON MICROSCOPY STRUCTURE
In the closed conformation of the NFS1/ISD11/ACP/ISCU/FXN complex captured in a cryo-EM structure41 (Figure 4C), FXN occupied the interface between NFSI and ISCU, contacting one of the two NFS1 protomers at the catalytic S-loop, supporting that FXN functions as an allosteric modulator of Fe-S cluster biogenesis36 (Figures 2 and 4C). FXN also bound to two key regions of ISCU, one of which is the conserved ISCU-Alanine-loop (Ala66-Asp71), containing the Fe-S ligating residues Cys69 and Asp7128,50 (Figure 3), and the other is the ISCU-LPPVKLHC138SM140 motif, which not only contains the LPPVK motif recognized by the chaperone HSPA9 for downstream delivery of the cluster to recipient proteins, but also encompasses Cys138, which is the proposed sulfur acceptor from NFS136 (Figure 3), and Met140, a residue reported to confer FXN-dependence of Fe-S cluster biogenesis in S. cerevisiae,51 or FXN-independence in prokaryotes.52 The fact that the LPPVK motif of ISCU, required for the binding of the HSP70 chaperone, is buried at the FXN interface suggests that FXN must be displaced prior to binding of the HSPA9/HSC20 transfer complex to the main scaffold ISCU.
The source of iron for Fe-S cluster biogenesis in mitochondria remains elusive. The labile iron pool of the mitochondrial matrix, which has been calculated to be approximately 170 μM (15–20% of total mitochondrial iron content), may serve as a source of iron for Fe-S cluster biogenesis.53 It remains to be established whether a designated iron chaperone is required to donate iron to ISCU during initial cluster assembly.
The Fe-S cluster biogenesis core complex would therefore cycle between sulfur donation (accelerated by FXN) from NFS1 to iron loaded-ISCU, and electron donation (mediated by FDX/FDXR), followed by binding of the chaperone/co-chaperone complex (HSPA9/HSC20) that mediates transfer of de novo assembled clusters to recipient proteins. Since only reduced FDX was found to interact with the complex,54 it is possible that once electron donation is complete, oxidized FDX exits the complex allowing replacement by another reduced FDX or FXN.
6 ∣. A SPECIALIZED CHAPERONE-CO-CHAPERONE SYSTEM ENSURES EFFICIENT FE-S CLUSTER DELIVERY
Transfer of newly assembled clusters downstream of the main scaffold protein ISCU in mammalian cells relies on the activity of a highly conserved chaperone/co-chaperone system, consisting of HSPA9 and HSC20, analogous to the yeast Ssq1/Jac1 and the bacterial HscA/HscB complexes.
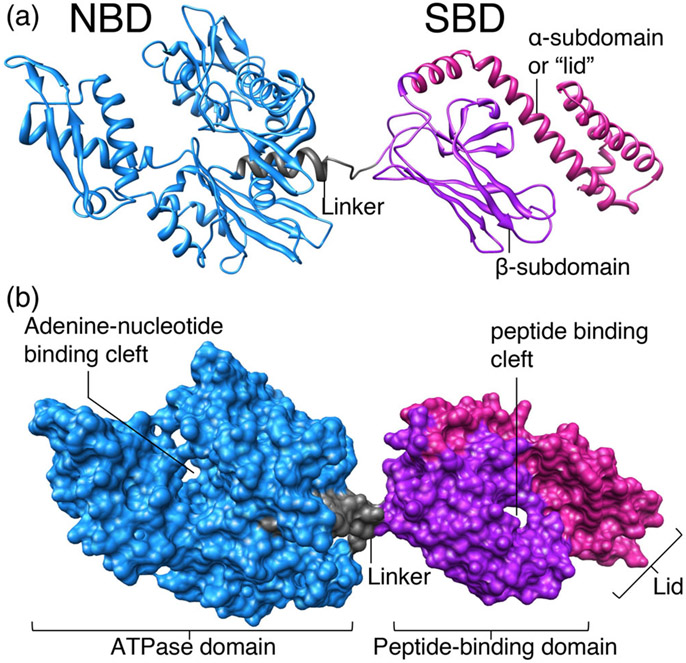
Heat shock 70 KDa proteins (HSP70s) are ubiquitous molecular chaperones that function in all major cellular compartments, participating in diverse cellular processes, including protein folding, degradation, translocation across membranes, and oligomeric assembly.55,56 All these activities depend on the ability of HSP70s to bind reversibly to short, unfolded segments of polypeptide chains. Most HSP70s are able to interact with a broad range of protein substrates, and recognition of these clients is determined by the intrinsic specificity of their binding partners, the HSP40 (Heat shock 40 KDa proteins) molecular chaperones, also known as co-chaper-ones.57 Co-chaperones or J-proteins invariably contain the so-called J-domain, which was named after the initially characterized DnaJ protein of Escherichia coli. The J-domain consists of a conserved ~70 amino acid signature region, which contains a histidine, proline and aspartic acid tripeptide (HPD) in a loop between the two main helices (II and III) that stimulates the ATPase activity of HSP70 chaperones.57 In fact, despite their remarkable functional diversity, HSP70s have high sequence identity and common fundamental structural features.58,59 All HSP70s possess a highly conserved N-terminal nucleotide binding domain (NBD), which displays weak ATPase activity, and a C-terminal domain, which forms the substrate binding domain (SBD), where client proteins bind (Figure 5).60
FIGURE 5.
Structure of a typical HSP70 chaperone, DnaK, the housekeeping HSP70 of E. coli, with ADP bound to the nucleotide-binding domain (PDB ID: 2KHO). The ATPase domain of DnaK, which is also known as nucleotide-binding domain (NBD), and the peptide-binding domain, which is also known as substrate-binding domain (SBD), are connected by a short, flexible, hydrophobic linker. The NBD and the SBD dock when the chaperone is bound to ATP, which is the conformation that exhibits low affinity for the substrate, due to the shortening of the linker, which causes displacement of the lid (also known as α-subdomain) in the SBD and allows easy access and egress of client proteins from the cleft. (a) and (b) are the ribbon and surface representation modes, respectively, of DnaK of E. coli (created from PDB structures using the UCSF ChimeraX program)
Molecular chaperones engage in a transient interaction with client proteins, which is regulated by the ATP binding and hydrolysis activities of the NBD. A conformational change generated upon hydrolysis of ATP stabilizes the HSP70's interaction with the substrate protein, while facilitating the multiple cellular processes in which HSP70s are involved. Exchange of ADP for ATP promotes client release and completes the binding cycle (Figure 6). HSP70s require a J-protein and, almost always, a nucleotide exchange factor (NEF) as key components that regulate their binding to client proteins and control the interaction with nucleotides (ATP or ADP). J-proteins or co-chaperones, in addition to assisting in substrate delivery, stimulate the HSP70 ATPase activity thereby stabilizing formation of the HSP70-substrate complex and facilitating selective substrate trapping.60
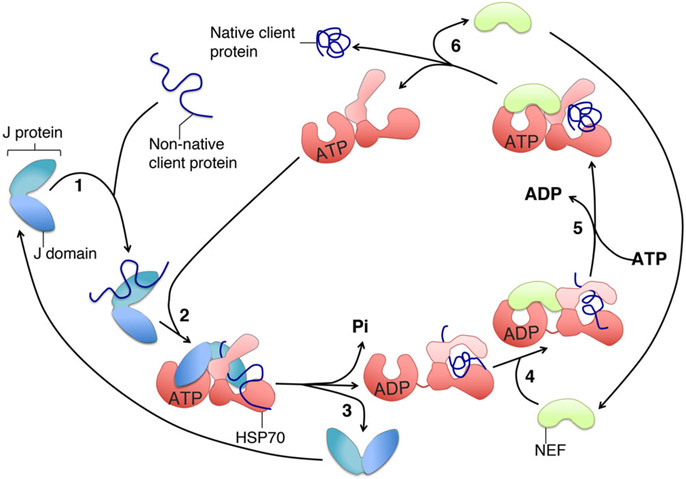
FIGURE 6.
ATPase cycle of the chaperone-co-chaperone core machinery. (1) The J-protein, also known as co-chaperone, binds to a subset of unfolded (non-native) substrate proteins (clients), and interacts with the HSP70 in the ATP-bound state through its J-domain (2). The client can then interact with the peptide-binding cleft, which is in the open conformation (2). ATP hydrolysis is simultaneously stimulated by both the J-domain of the co-chaperone and the client and causes a conformational change in the HSP70 that closes the lid of the SBD over the cleft and stabilizes the interaction with the client. (3) The J-protein leaves the complex. A nucleotide exchange factor (NEF), which exhibits high affinity for the ADP-bound state of the chaperone, binds to the HSP70-client complex (4), and exchanges ADP with ATP in the NBD (5). The client, after folding into the native conformation driven by the energy provided by hydrolysis of ATP, is ultimately released (6)
Most HSP70s display broad substrate specificity proportional to the variety of proteins that they have to recognize, and prokaryotes were initially thought to have a single HSP70, named DnaK, that served both stress-related and housekeeping functions.61 However, in 1994, a gene encoding a novel HSP70 was identified in E. coli and designated HscA (Heat shock cognate A).62,63 The gene was located within the isc (Iron–Sulfur Cluster) operon, immediately upstream of the fdx gene encoding a [2Fe-2S]-ferredoxin64 and downstream of a gene, named HscB, encoding a novel J-protein.63 The predicted HscA protein (also known as Hsc66 because it had a molecular weight of ~66KDa) exhibited 40% sequence identity to DnaK. Initial studies on the general biochemical properties of E. coli HscA and HscB revealed that the basal ATPase activity of HscA was stimulated four to six-fold by HscB,65,66 but not from DnaJ, the co-chaperone for DnaK, indicating a lack of cross-talk between the two chaperone systems. Despite the evidence that HscA exhibited general chaperone activity in vitro, due to its ability to suppress protein aggregation,66 the endogenous physiological substrate for the HscA/HscB system remained unknown until completion of the genome sequencing of E. coli revealed that the chaperone/co-chaperone system was encoded by a region of the genome, now known as isc operon, that exhibited similarity to the nif operon involved in biogenesis of Fe-S clusters of nitrogenase.67,68 The isc genes were found to be essential for cell growth. Inactivation of HscA or HscB reduced activities of Fe-S enzymes, such as glutamate synthase and succinate dehydrogenase, and significantly decreased the overall cell proliferation rates.69 Parallel studies in the yeast Saccharomyces cerevisiae revealed the presence of isc- and hsc-like genes,70 suggesting that the Fe-S biogenesis machinery was highly evolutionarily conserved. Experimental evidence that these genes encoded components of the Fe-S cluster assembly pathway came from the characterization of variants that suppressed oxidative damage in yeast strains in which superoxide dismutase 1 was deleted.71 Suppressor mutations were identified in two genes encoding a mitochondrial HSP70 and a J-type co-chaperone, respectively. The HSP70 protein was named Ssq1, and the co-chaperone, which was predicted to be targeted to the mitochondrial compartment, was named Jac1 (J-type accessory chaperone). Both Jac1 and Ssq1 mutant strains exhibited impaired mitochondrial respiration and reduced activities of succinate dehydrogenase and aconitase, a metabolic defect that had previously been reported for Nfs1 mutants, which were unable to mobilize sulfur for the initial assembly of the cluster. Subsequent work by several groups confirmed that Ssq1 and Jac1 were essential for Fe-S cluster biogenesis in yeast.72-76
The first experimental evidence for a specific molecular interaction of the chaperone/co-chaperone system with a critical component of the Fe-S assembly pathway, the main scaffold protein IscU, came in 2000 from studies on the effects of bacterial isc components on the ATPase activity of HscA.77 IscU was found to stimulate HscA's ATPase activity approximately eight-fold above the basal level. The co-chaperone HscB synergistically activated HscA's activity up to 400-fold and enhanced IscU binding to the ATP-bound form of HscA.77 Interestingly, IscU and/or HscB were unable to stimulate DnaK's ATPase activity, and DnaJ did not increase binding of IscU to HscA, suggesting that HscA formed a complex with HscB, and that the main scaffold protein IscU might serve as a substrate for this chaperone complex. The substrate specificity of HscA and the selective nature of the interaction between HscA and IscU were studied in more detail by screening a library of peptides for their ability to bind HscA, and, subsequently, by testing peptides that covered the entire IscU sequence.78,79 HscA was found to bind the residues between amino acids 99 and 103 of IscU, which contained the sequence Leu-Pro-Pro-Val-Lys. The LPPVK sequence is conserved among IscU proteins from bacteria to humans, suggesting that the mechanism of chaperone recognition and binding has been conserved during evolution. Importantly, the co-chaperone HscB was found to enhance the binding affinity of IscU for HscA,79 consistent with its role in targeting IscU to the HscA-ATP complex.
Studies of the yeast chaperone/co-chaperone/scaffold complex (Ssq1/Jac1/Isu1) provided results similar to those of the bacterial HscA/HscB/IscU system,80 and offered additional insights into the mechanism of chaperone-scaffold interactions. Ssq1 and Jac1 were found to act as partners in binding the scaffold protein Isu.80 More importantly, Isu was targeted to Ssq1 in a Jac1-dependent manner, and both Jac1 and Isu were required for efficient stimulation of Ssq1 ATPase activity.80 Interestingly, mitochondria of most eukaryotes, including the yeast Schizosaccharomyces pombe, contain a single multifunctional HSP70,57,81,82 which is homologous to mitochondrial Ssc1 of S. cerevisiae,83 that performs the dual task of housekeeping molecular chaperone (participating in protein translocation across the inner mitochondrial membrane and protein folding) and of specialized HSP70 for Fe-S cluster biogenesis. Only a subset of fungi, including S. cerevisiae and Candida albicans, contains a highly specialized mitochondrial HSP70, named Ssq1, that arose through gene duplication about 300 million years ago84 and that is unique in that it exhibits binding to a single protein substrate Isu1.85
7 ∣. TRANSFER OF FE-S CLUSTERS TO RECIPIENT PROTEINS: THE ATPASE CYCLE
Genetic and biochemical studies have clearly demonstrated that hydrolysis of ATP is essential for the chaperone activity of all the HSP70 proteins tested so far, and the rates of hydrolysis are very low under basal conditions.60 ATP hydrolysis triggers the closing of the α-sub-domain, also known as lid, in the SDB upon the substrate, which locks the substrate into position. In a thermodynamically coupled fashion, substrates activate the HSP70 ATPase activity typically by 2–10-fold, a low level of stimulation that is insufficient to drive the functional cycle of HSP70 chaperones. Instead, co-chaperones are required for the productive coupling of ATP hydrolysis with substrate association.57,86,87 On the basis of the data collected in bacteria and yeast, a model for the mode of action of the chaperone/co-chaperone system in Fe-S cluster biogenesis has been proposed86,88,89 (see Figure 7 for a schematic of the main steps). Accordingly, the co-chaperone (HscB in bacteria and Jac1 in yeast) starts the functional cycle of the cognate system by rapidly and transiently associating with the main substrate, the scaffold protein IscU, and with a recipient Fe-S apo-protein. IscU binds to the HSP70 protein (HscA or Ssq1, in bacteria or yeast, respectively) in a two-step process, which involves the transient interaction of the J-domain of the co-chaperone with the nucleotide binding domain (NBD) of the ATP-bound chaperone, and the interaction of IscU with the substrate binding domain (SBD). The chaperone in the ATP-bound state is in an open conformation, exhibiting the substrate-binding cavity to the interaction with IscU. Through a short interdomain communication between the SBD and the NBD in the chaperone, the simultaneous association of the substrate and the interaction of the NBD with the J-domain of the co-chaperone lowers the activation energy for the hydrolysis of ATP by the chaperone. Hydrolysis of ATP and the coupled conformational change in the SBD of HSP70 leads to a tighter interaction with the scaffold protein and represents the rate-limiting step in the overall cycle, which is usually more than 103-fold slower than release of the products ADP and phosphate.59 Nevertheless, under physiological conditions, nucleotide dissociation is a crucial step for substrate release. Therefore, exchange of ADP with ATP, which requires the opening of the nucleotide binding cleft, is a limiting step in the cycle, and it is accordingly highly regulated and subject to strong evolutionary variation. For HscA the dissociation rates for ADP ± Pi are 700-fold higher than those of the housekeeping chaperone DnaK.90 Therefore, in contrast to other HSP70 proteins, which are regulated at both ATP hydrolysis and ADP/ATP exchange rate, the HscA reaction cycle is regulated primarily at the hydrolysis step. Accordingly, HscA does not associate with nucleotide exchange factors to complete its cycle.91 In contradistinction, Ssq1 of S. cerevisiae has a higher nucleotide binding affinity than HscA and a slower ADP/ATP exchange rate.80 Therefore, Ssq1 must interact with the nucleotide exchange factor Mge1 to maintain high steady state rates of ATP hydrolysis.80 The structural basis for these strong kinetic differences in nucleotide dissociation between HSP70 homologs has been proposed to depend on the length of an exposed loop close to the NBD, and on the character of the amino acid residues that line the interface of the interaction of the SBD with the client protein.59,90 HscA, which exhibits the fastest turnover rate, has the shortest loop and lacks a hydrophobic patch (which consists of Leu257-Val59 in DnaK) and two putative salt bridges (Glu264-Arg56, and Glu267-Lys55 in DnaK) at the interface of the NBD.59 The physiological significance of the differences in maximal ATPase activity rates between HscA and Ssq1 determined in vitro with respect to the role of the chaperones in Fe-S cluster biogenesis in vivo is not known.
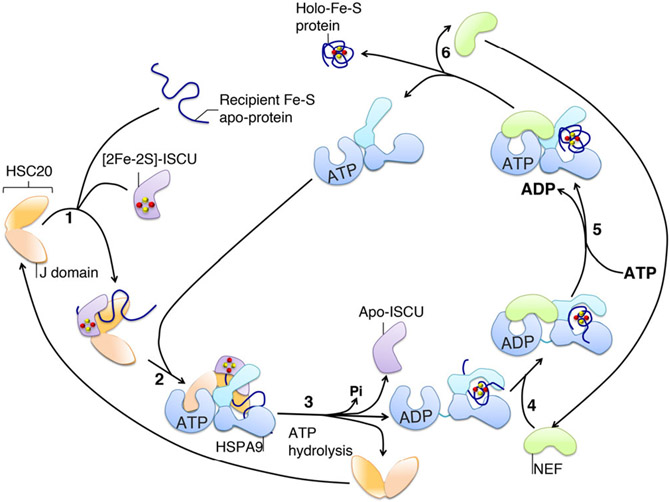
FIGURE 7.
Proposed schematic of the mode of action of the chaperone/co-chaperone system in Fe-S cluster delivery to recipient proteins. The co-chaperone HSC20 starts the functional cycle of the cognate system by associating with the scaffold protein ISCU, which is loaded with an Fe-S cluster, and with a recipient Fe-S apo-protein (1). (2) ISCU transiently interacts with the SBD of the HSP70 chaperone (HSPA9 in mammalian cells), whereas the J-domain of HSC20 contacts the NBD of the ATP-bound state of HSPA9. ATP-bound HSPA9 is initially in the open conformation, which exhibits the substrate-binding cavity to allow the interaction with ISCU. (3) The simultaneous association of ISCU and the interaction of the NBD of HSPA9 with the J-domain of HSC20 lowers the activation energy required for the hydrolysis of ATP. Hydrolysis of ATP and the coupled conformational change in the SBD of HSPA9 is proposed to facilitate cluster release from ISCU and transfer to the recipient protein. (4) A nucleotide exchange factor (NEF), which exhibits high affinity for the ADP-bound state of the HSP70 chaperone, binds to the HSP70-client complex and exchanges ADP with ATP in the NBD (5). The client, which has folded into the native conformation driven by the energy provided by hydrolysis of ATP, and has acquired its Fe-S cluster, is then released (6)
The mechanism by which energy release by the chaperone system is coupled to Fe-S cluster transfer to recipient proteins remains undefined. A full understanding of the transfer step requires detailed structural information of the components of the transfer machinery. Moreover, structures of different conformational states of the Fe-S transfer complex (chaperone/co-chaperone/scaffold protein) will be necessary to reveal the effects the chaperone/co-chaperone system has on the scaffold protein and its Fe-S cluster and how the process of Fe-S cluster transfer is regulated. The crystal structure of a region of the SBD of E. coli HscA (amino acid residues 390–543) bound to an IscU peptide (98ELPPVKIHC106) was determined to a resolution of 1.95 Å (Figure 8).92 The IscU peptide extends into a hydrophobic cleft within the β-subdomain (Figure 8), and a combination of nonpolar and hydrogen-bonding interactions appears to contribute to the binding affinity. The α-subdomain lies immediately above the binding cleft without making direct contacts with the peptide (Figure 8). The central proline residue of the IscU peptide is completely buried within a hydrophobic pocket in the middle of the cleft. The tight interaction of this proline with the residues lining the substrate-binding cleft explains the crucial requirement of the highly conserved PVK motif in all ISCU scaffold proteins for binding to the chaperone.79,93 The peptide in the HscA(SBD)-ELPPVIHC complex appears to be trapped in place by the lid-like structure of the α-subdomain of HscA (Figure 8).
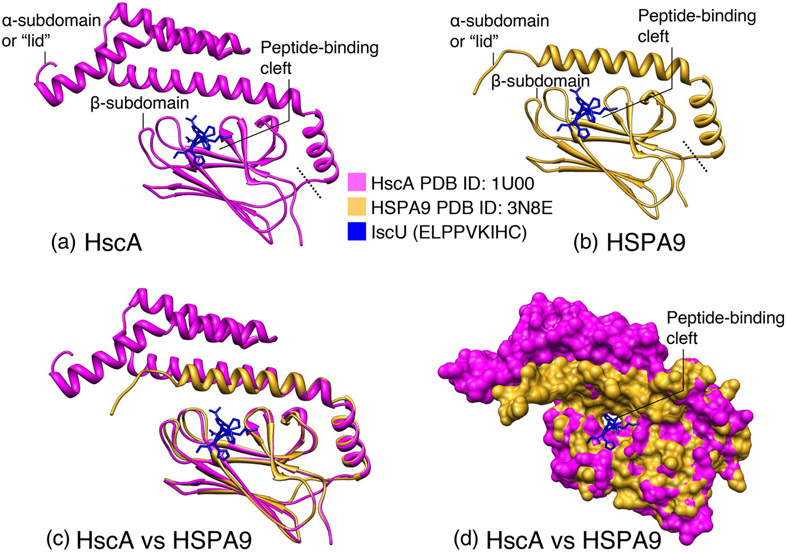
FIGURE 8.
Comparison of the crystal structures of the substrate-binding domains of HscA and HSPA9 in complex with the ELPPVKIHC peptide of IscU. (a) Structure of the E. coli HscA (SBD) in complex with ELPPVKIHC peptide of IscU (PDB ID: 1U00). The α-subdomain or “lid”, which comprises residues 390–498 of HscA, and the β-subdomain (residues 506–609) constitute the NBD; the dotted line delineates the separation between the two subdomains. The IscU peptide is depicted in blue. (b) Structure of the SDB of HSPA9 (PDB ID: 3N8E). The ELPPVKIHC peptide of IscU was docked to the peptide-binding cleft of the human chaperone. (c) Superimposition of the crystal structures of the substrate-binding domains of HscA and HSPA9. The α- and β-subdomains of the SBDs of HscA and HSPA9 were aligned, and the ribbon diagrams of the two structures were superimposed. Color coding: HscA in magenta, HSPA9 in gold. (d) Superimposition of the surface diagrams of the SBDs of HscA and HSPA9 in complex with the ELPPVKIHC peptide of IscU (created from PDB structures using the UCSF ChimeraX program)
Transfer of Fe-S clusters from holo-ISCU to recipient proteins depends on conformational changes in both the scaffold and the recipient protein. Studies using solution NMR spectroscopy have provided insights into the mechanism of nucleotide dependent interactions between the chaperone/co-chaperone system and the scaffold protein IscU.94,95 IscU was found to interconvert between two alternative conformations, a more structured state (known as the S-conformation) which resembled the holo-protein IscU-[2Fe-2S] and bound preferentially to the co-chaperone HscB, and a dynamically disordered form, which did not bind any metal and exhibited high affinity for HscA.94,95 Based on studies carried out by using purified components of the bacterial Fe-S transfer complex (HscA, HscB, and IscU),96 a model has been proposed to describe the molecular mechanism by which modulation of the ATPase activity of the chaperone by the co-chaperone is coupled to Fe-S cluster transfer from the scaffold protein IscU to recipient Fe-S apo-proteins. The co-chaperone HscB binds to and delivers [2Fe-2S]-IscU (IscU in the S-conformation) to the SBD of HscA; together, IscU and HscB synergistically enhance HscA's ATPase activity nearly 1000-fold.77,91 The substrate-binding domain of HscA undergoes conformational change upon hydrolysis of ATP and encloses the PVK peptide of IscU, which is located in close proximity to the cluster. The resulting conformational change may promote release of the [2Fe-2S] cluster from holo-IscU and transfer to the recipient protein.95-97
8 ∣. THE MAMMALIAN FE-S TRANSFER SYSTEM
Transfer of newly assembled Fe-S clusters downstream of the main scaffold protein ISCU in mammalian cells relies on the activity of a chaperone/co-chaperone system analogous to the bacterial and yeast complexes, which we have previously described.91,98 Mammalian cells lack an HSP70 chaperone specifically dedicated to Fe-S cluster biogenesis, but rather, like most eukaryotes, employ a multifunctional mitochondrial HSP70, recently identified as HSPA9 in mammalian cells (also known as mortalin/PBP74/GRP75).25,99 HSPA9 has been implicated in several different processes, including the facilitation of protein import within mitochondria, together with the co-chaperone DNAJC19 and the nucleotide exchange factor (NEF) BAP (also known as SIL1),100 intracellular trafficking, stress response, hematopoiesis, control of cell proliferation and tumor progression (reviewed in References 101-104). The absence of a chaperone that is solely dedicated to Fe-S cluster biogenesis in higher eukaryotes appears to warrant even greater participation of a specialized co-chaperone to specify the function of the cognate HSP70. On the basis of its high sequence similarity to the bacterial and yeast co-chaperones dedicated to Fe-S cluster biogenesis, the human DnaJ type III protein, HSC20, also referred to as DNAJC20 or HSCB, was predicted to be involved in the transfer of Fe-S clusters downstream of ISCU.105 Further characterization of the human co-chaperone established its role as an integral component of the Fe-S cluster transfer machinery.106 HSC20 shares 34% and 29% overall identity with the bacterial and fungal orthologs, respectively.105,106 The high degree of sequence homology translates into a remarkable structural conservation of the J- and C-terminal domains of bacterial and human co-chaperones dedicated to Fe-S cluster biogenesis107 (Figure 9). The crystal structures of HscB from E. coli,108 of Jac1 from S. cerevisiae,89 and of human HSC20107 revealed the presence of a conserved structural core that consists of two domains arranged in a L-shaped fold (Figure 9).
FIGURE 9.
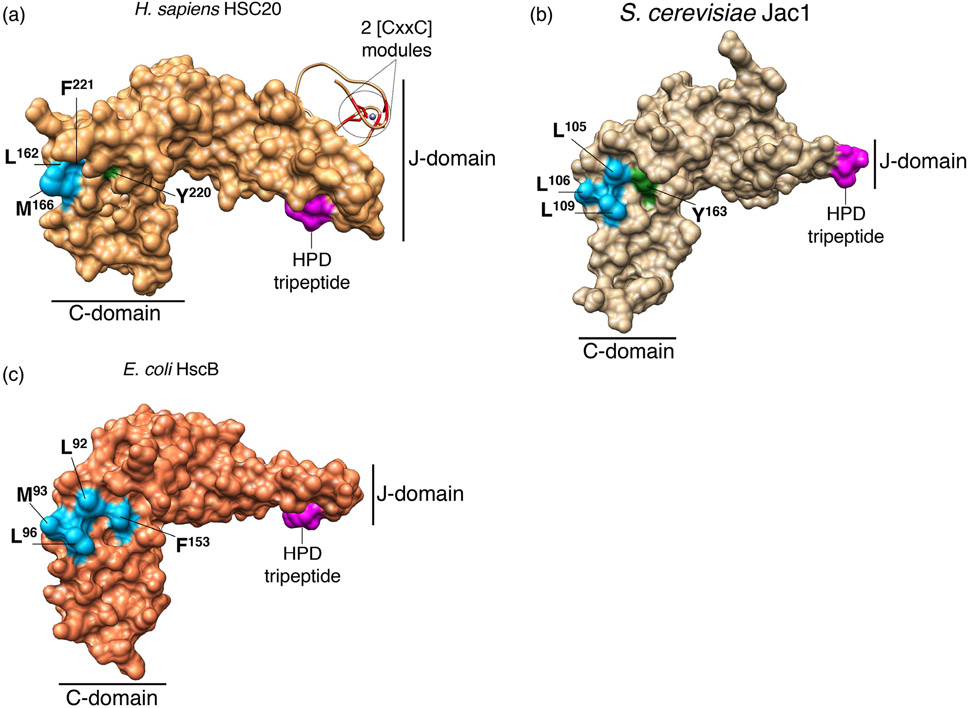
3D- structures of co-chaperones dedicated to Fe-S cluster biogenesis in H. sapiens (HSC20, panel A, PDB ID: 3BVO), S. cerevisiae (Jac1, panel B, PDB ID: 3UO2), and E. coli (HscB, panel C, 1FPO). Conserved amino acid residues that interact with the main scaffold ISCU on the surface of the C-terminal domains of the co-chaperones dedicated to Fe-S cluster biogenesis in human (a), yeast (b), and bacteria (c) are shown: in light blue are the hydrophobic amino acid residues (Leu, Met, and Phe), and in green the polar residues (Tyr). The His (H), Pro (P), Asp (d) tripeptide in the N-terminal domains (J-domains) of the co-chaperones is colored in magenta. Two CxxC modules, which coordinate zinc in the crystal structure of human HSC20, are shown in red
The N-terminal J-domain, which contains an invariant histidine, proline, aspartate (HPD) motif, is responsible for stimulating the ATPase activity of its HSP70 cognate chaperone,25,76,108,109 whereas the C terminus forms a three-helical bundle and is directly involved in binding the scaffold protein ISCU, with three highly conserved non-contiguous hydrophobic residues being of crucial importance for the HSC20-ISCU interaction.25,89,110 The N terminus of the human co-chaperone shows distinctive features compared to the specialized DnaJ type III proteins of bacteria and fungi, in that it contains, downstream of the mitochondrial targeting sequence (residues 1–2699), an additional domain, which harbors two CxxC modules (C41/C44 and C58/C61) that were found to coordinate a zinc ion in vitro107 (Figure 9). The physiological relevance of the unique N-terminal domain of HSC20 remains to be elucidated, though it may facilitate dimerization of the co-chaperone. The C-terminal domains of co-chaperones can selectively bind target substrates,111,112 facilitate refolding of denatured proteins, and enhance cell viability.113,114
As previously discussed, co-chaperones serve a dual function in Fe-S cluster biogenesis: they guide ISCU to the substrate binding domain of the HSP70 cognate chaperone, and they also activate the ATPase activity of the chaperone, thereby driving a conformational change that likely facilitates cluster release from ISCU and delivery to the final acceptor apo-protein or to intermediate carriers, which ultimately donate their clusters to specific recipients.91,97
9 ∣. DETERMINING SPECIFICITY OF FE-S CLUSTER DELIVERY: MOLECULAR FEATURES THAT GUIDE SELECTION OF RECIPIENT FE-S PROTEINS
HSC20 is the component of the Fe-S transfer machinery that was found to guide Fe-S cluster transfer from ISCU to recipient proteins.6,25,30,115-117 A stringent yeast two-hybrid (Y2H) screen was used to identify protein-targets bound by HSC20 as well as peptide motifs that might function as molecular signatures that guided the recruitment of the HSC20/HSPA9-transfer complex through direct binding to HSC20.25 The tripeptide Leu-Tyr-Arg and/or analogous sequences containing an hydrophobic amino acid in position 1, an aromatic residue (tyrosine or phenylalanine) in position 2, and arginine or lysine in position 3, were found to mediate direct binding of the Fe-S cluster transfer complex, consisting of HSC20, HSPA9 and [2Fe-2S]-ISCU, to recipient Fe-S apo-proteins to promote cluster acquisition.25,30,115-117 The initial identification of an LYR-like motif in the primary sequence of the heme biosynthetic enzyme aminolaevulinic acid dehydratase guided experimental work that led to the identification of a [4Fe-4S] cluster in the active site of the enzyme30 that had previously been thought to bind zinc, confirming the validity of the bioinformatic approach as a powerful tool to enable identification of thus far unrecognized Fe-S proteins.
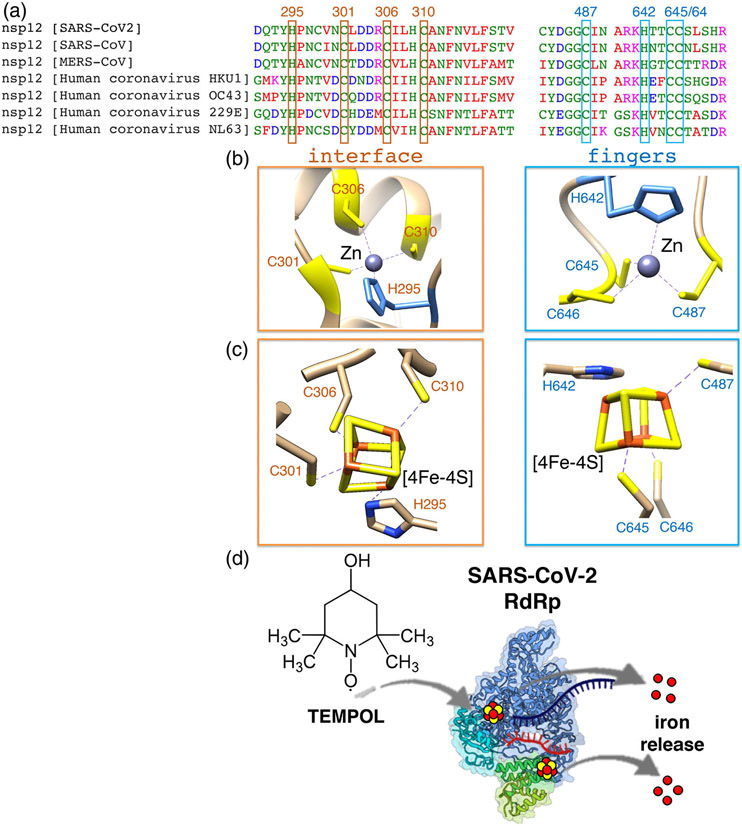
We recently reported that the RNA-dependent RNA polymerase (RdRp) required for replication of the viral genome and transcription of the genes of SARS-CoV-2, the causal agent of COVID-19, ligates two Fe-S metal cofactors in sites that were initially modeled as zinc centers in the available cryo-electron microscopy structures of the RdRp complex.117 Our discovery stemmed from the initial inspection of the amino acid sequence of the catalytic subunit, nsp12, of the RdRp complex that revealed the presence of two highly conserved LYR motifs, which were experimentally found to mediate transient interactions with the components of the de novo Fe-S cluster biogenesis machinery, HSC20, HSPA9 and ISCU, and with the late-acting cytoplasmic Fe-S assembly (CIA) factors, MMS19, CIAO1 and FAM96B. The two Fe-S clusters in the SARS-CoV-2 RdRp were found to be essential for replication and for the interaction with the viral helicase, nsp13, a component of the replication/transcription complex of SARS-CoV-2.117 Interestingly the residues that coordinate the two cubane clusters in nsp12 (three cysteines and one histidine in each of the two metal-ligating centers) are highly conserved among coronaviruses that infect humans (Figure 10A-C), including SARS-CoV, MERS-CoV and four coronaviruses that are the second most common cause of the seasonal cold after rhinoviruses, suggesting that other coronaviruses may rely on Fe-S cofactors for their replication.
FIGURE 10.
The RNA-dependent RNA polymerase (RdRp) of SARS-CoV-2 binds two oxidation-sensitive [4Fe-4S] clusters. (a) Multiple sequence alignments of the residues (three cysteines and one histidine in each site) ligating two [4Fe-4S] clusters in the catalytic subunit, nsp12, of the RdRp of SARS-CoV2 and other human coronaviruses (HCoVs). SARS-CoV (Severe Acute Respiratory Syndrome coronavirus), MERS-CoV (Middle East Respiratory Syndrome Coronavirus). HCoVs HKU1, OC43, 229E and NL63 are the second most frequent cause of the common cold after rhinoviruses. HCoVs may also be associated with acute otitis media or exacerbations of asthma. Less frequently, these viruses are associated with lower respiratory tract infections, including bronchiolitis and pneumonia, primarily in infants and immunocompromised children and adults. (b) Metal-ligating sites in the cryo-EM structure of the RdRp of SARS-CoV-2 (PDB ID: 6YYT) that were modeled as zinc centers are located at the interface between the nidovirus specific N-terminal domain of nsp12 (aka NiRAN domain) and the catalytic domain, and in the fingers subdomain within the catalytic domain. (c) Modeling of two [4Fe-4S] clusters in the metal ligating sites of nsp12 was performed by superimposing the cysteine resides ligating a [4Fe-4S] cluster in the Rhodothermus marinus HiPIP (High Potential Iron–Sulfur Protein) at 1.0 Å resolution (PDB: 3H31) to the 3Cys/His residues of nsp12 (PDB: 6YYT). (d). Schematic representation of the oxidative degradation of the two cubane Fe-S clusters in the SARS-CoV-2 RdRp by TEMPOL
We then attempted to exploit the sensitivity of Fe-S clusters to oxidative degradation29,118 to prevent coronavirus replication in cell culture models. Previous studies had shown that a stable nitroxide, TEMPOL (4-hydroxy-2,2,6,6-tetramethylpiperidin-1-oxyl), was beneficial in two different animal models of human conditions through its ability to oxidize and disassemble the Fe-S cluster of cytosolic aconitase (IRP1), leading to its conversion into the IRE-binding apo-form.119,120 Spectroscopic studies demonstrated that TEMPOL caused the disassembly of the Fe-S clusters in the RdRp, which led to the inactivation of the enzyme that failed to replicate a short RNA template in vitro (Figure 10D).117 The TEMPOL treatment of cells did not impact the activities of several mitochondrial Fe-S enzymes, including the respiratory complexes and mitochondrial aconitase, and the cytosolic enzyme dihydropirimidine dehydrogenase (DPYD), nor did it cause any cytotoxicity at doses up to 5 mM.117 Mitochondrial Fe-S enzymes are protected from the oxidative action of TEMPOL because they are within the mitochondrial matrix compartment that cannot be accessed by the drug. TEMPOL does not cross the mitochondrial double membrane, whereas a derivative of TEMPOL, called mito-TEMPOL, that contains a triphenyl phosphonium group, has been used for the delivery of the drug to mitochondria.121 On a short-term treatment, the turnover rates of endogenous Fe-S subunits of multi-protein complexes at physiological oxygen tensions determine whether these cofactors will be susceptible to TEMPOL. In fact, when the clusters are buried within the multi-protein complexes, they are protected and stable. The stage at which newly assembled Fe-S cofactors need to be inserted into a recipient protein endangers the cofactors because they may become exposed to TEMPOL and/or to other physiological oxidants prior to being fully ligated and integrated into the mature protein structure. During an active infection, viral proteins are synthesized at extremely high rates and their Fe-S cofactors may become the most abundant targets of TEMPOL, which may partially explain the apparent selective impairment of the RdRp of SARS-CoV-2 upon TEMPOL treatment. Finally, the redox potentials of the clusters affect their susceptibility to the oxidative action of TEMPOL. The two Fe-S cofactors in the RdRp of SARS-CoV-2 were found to have relatively low redox potentials (lower than −600 mV), which translated into their very low affinities for electrons, ineffective reduction upon treatment with dithionite, and susceptibility to donate an electron and undergo oxidation upon treatment with a mild oxidant, such as TEMPOL. With its low cytotoxicity and known access to tissues relevant to SARS-CoV-2 infection,122,123 TEMPOL and other related stable nitroxides may represent potent anti-SARS-CoV-2 therapies during active viral infection.
10 ∣. LONG-STANDING CONTROVERSY OVER THE EXISTENCE OF A PARALLEL SYSTEM FOR FE-S BIOGENESIS IN THE CYTOSOLIC/NUCLEAR COMPARTMENT OF MAMMALIAN CELLS
Fe-S clusters have emerged as essential cofactors in enzymes involved in all aspects of DNA processing.3,13,14 The cytoplasmic Fe-S cluster assembly (CIA) machinery is defined by having a primary location in the cytoplasm and by its function in the biogenesis of cytoplasmic and nuclear Fe-S proteins. The Fe-S cluster biogenesis components, on the other hand, are required for both mitochondrial and cytosolic Fe-S cluster biogenesis. In S. cerevisiae, Nfs1 is present in mitochondria and in the nucleus,124 whereas other Fe-S cluster biogenesis components had previously been detected exclusively in mitochondria, leading to the proposal that de novo Fe-S cluster assembly occurred only in the mitochondrial matrix compartment, whereas assembly of extra-mitochondrial Fe-S cofactors required export of a sulfur-containing compound (named X-S) from the mitochondrial matrix to the cytosol125-127 (Figure 11, left panel). In mammalian cells, a full complement of Fe-S biogenesis proteins is synthesized and targeted to the mitochondrial matrix, but almost all of these proteins have also been detected in the cytosolic/nuclear compartments of cells, where they localize as a result of alternative splicing of the transcript that also encodes the mitochondrial isoform,27 a weak mitochondrial targeting signal,128 and alternative utilization of initiator AUGs that retained or skipped the mitochondrial targeting sequence.129,130
FIGURE 11.
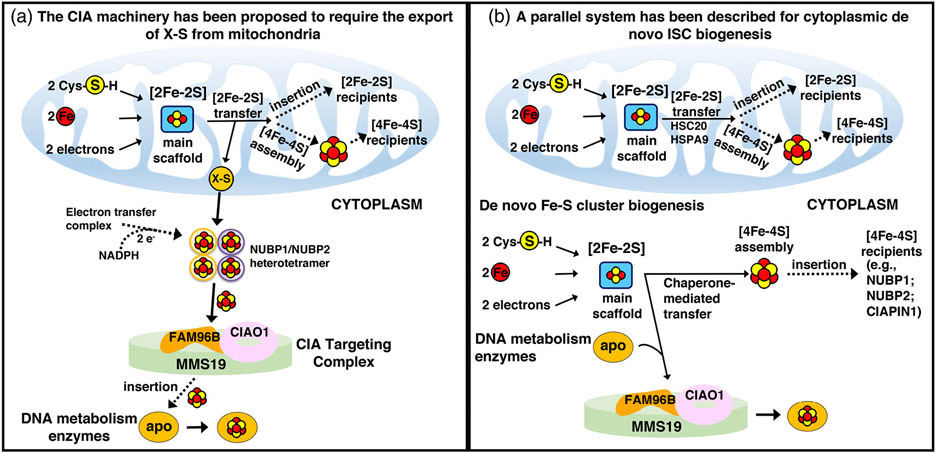
Alternative proposed models of cytoplasmic Fe-S cluster biogenesis. In the schematic on the left, assembly of cytoplasmic Fe-S clusters begins in mitochondria with components of the early Fe-S cluster biogenesis machinery synthesizing a sulfur-containing precursor (X-S) that is subsequently exported to the cytosol by the ABC transporter Atm1 (ABCB7 in human) and utilized by the CIA machinery for the biosynthesis of [4Fe-4S] clusters upon the main hetero-tetrameric complex composed of NUBP1 and NUBP2.131 In the model on the right, alternative isoforms of the core early Fe-S cluster biogenesis factors are present in the cytosol of mammalian cells where they initiate de novo assembly of [2Fe-2S] clusters on the main cytosolic scaffold protein ISCU1. A dedicated chaperone/co-chaperone system, consisting of HSPA9 and HSC20 (aka HSCB), either facilitates direct Fe-S cluster transfer to a subset of recipient cytosolic proteins (e.g., CIAPIN1, NUBP1, and NUBP2), or mediates transfer of Fe-S clusters to enzymes involved in DNA metabolism through direct binding of HSC20 to the LYR motif of the CIAO1 component of the CIA-targeting complex.128 Adopted from139
Therefore, two contrasting models have emerged to describe cytoplasmic Fe-S biogenesis127,131 (Figure 11).
One model, largely based on studies in yeast, proposed that assembly of extra-mitochondrial Fe-S proteins relied on the mitochondrial Fe-S cluster biogenesis machinery for the synthesis of a sulfur-containing compound (named X-S) that was exported to the cytoplasm by the ABC transporter Atm1 (ABCB7 in human) and utilized for the assembly of a [4Fe-4S] cluster transiently bound to the hetero-tetrameric NUBP1/NUBP2 complex (Nbp35/Cfd1, in yeast, respectively. Figure 11, left panel).131,132 The NADPH-dependent electron transfer system composed of the diflavin reductase Tah18 (NDOR1 in human) and the Fe-S protein Dre2 (CIAPIN1 in human) was proposed to provide electrons necessary for the assembly of [4Fe-4S] clusters on the NUBP1/NUBP2 hetero-tetramer, which was considered to function as the primary scaffold complex from which all cytoplasmic and nuclear Fe-S proteins acquired their cofactors.131 Nuclear Fe-S enzymes acquired their clusters from the CIA-targeting complex, consisting of CIAO1, FAM96B, and MMS19 (Figure 11, left panel).
The second proposed model for cytoplasmic Fe-S biogenesis (Figure 11, right panel) is based on studies that for more than two decades have demonstrated the existence of a de novo cytosolic Fe-S cluster biogenesis pathway in mammalian cells, which can supply the elemental components (i.e., iron and sulfur) and the biogenesis proteins required to build the cluster in the cytosol of mammalian cells.127,128 Consistent with this model, the pool of NFS1 that localizes to the cytosol129 is a functional enzyme that mobilizes sulfur from cysteine,133,134 a notion that questions the need for the export of a sulfur-containing compound out of mitochondria. Moreover, alternative isoforms of the core Fe-S cluster biogenesis components have been detected in the cytosol of mammalian cells,25-27,106,129,135-137 suggesting that Fe-S cluster biogenesis machineries may independently operate in parallel to generate nascent Fe-S clusters in multiple subcellular compartments of multicellular eukaryotes.
11 ∣. CONCLUDING REMARKS AND FUTURE PERSPECTIVES
It is possible that chaperone/co-chaperone complexes other than HSC20/HSPA9 may function in later steps of Fe-S biogenesis, by promoting cluster transfer from secondary carriers to Fe-S clients, while assisting the folding of the recipient proteins to accommodate the cluster and assume their final functional conformations. In fact, co-chaperones other than HSC20 have been identified in mass spectrometry analyses of interacting partners of components of the CIA machinery, CIAO1, FAM96B, and MMS19.14,138 Also, it is possible that other motifs, analogous to LYR, will be involved in guiding Fe-S cofactors to appropriate downstream recipients, preventing this highly complex machinery from participating in unproductive side reactions.
Finally, despite retention of a high degree of sequence homology of the Fe-S cluster biogenesis components from bacteria to human, and common basic molecular pathways for Fe-S biogenesis, the increased biological complexity in multicellular organisms evolved with a corresponding demand for more elaborate mechanisms of transcriptional/translational control, along with a greater diversity in the multi-subunit complexes that facilitate Fe-S cluster assembly in different compartments. This increased complexity may explain why a parallel pathway for de novo Fe-S cluster biogenesis is needed in the cytoplasm of mammalian cells, where alternative isoforms of the core Fe-S biogenesis components localize and operate.
Much progress has been made, but many basic questions remain unanswered in the complex process of mammalian Fe-S biogenesis (See Outstanding Questions).
12 ∣. OUTSTANDING QUESTIONS.
What is the source of iron for Fe-S cluster biogenesis? If low molecular mass iron complexes supply the iron for Fe-S cluster biogenesis, is a protein chaperone involved in donating iron to the main scaffold ISCU?
What is the role of the acyl carrier protein as a component of the initial Fe-S cluster biogenesis core complex?
Are the two different conformations of the NFS1/ISD11/ACP complexes, “open” and “closed” (PDB: 5USR and 5WLW, respectively) both present and functional in vivo?
How are specific Fe-S recipient apo-proteins such as LIAS targeted by secondary carriers to acquire their Fe-S cofactors?
What is the energy requirement, in terms of ATP molecules hydrolyzed by the chaperone/co-chaperone complex, required to facilitate cluster release from ISCU, transfer, and folding of the recipient protein?
Are chaperone/co-chaperone complexes other than HSPA9/HSC20 involved in Fe-S cluster delivery to specific subsets of Fe-S recipients?
Are most Fe-S clusters co-translationally inserted or can some, such as aconitase, acquire the Fe-S cluster after the protein has fully folded?
Are the intersections between Fe-S cluster biogenesis and lipid synthesis important in regulating respiration and supporting cellular energy supplies?
ACKNOWLEDGMENT
The authors would like to acknowledge support from the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Funding information
Intramural Research Program of the National Institutes of Health; Eunice Kennedy Shriver National Institute of Child Health and Human Development
Abbreviations:
- ABC
ATP-binding cassette
- ABCB7
ABC transporter of the inner mitochondrial membrane, subfamily B, member 7
- ACO1
cytosolic aconitase
- ACP
acyl carrier protein
- ALA
aminolaevulinic acid
- ALAD
ALA dehydratase
- eALAS/ALAS2
erythroid specific 5-aminolevulinate synthase
- FBXL5
F-box and leucine-rich repeat protein 5
- FDX
ferredoxin
- FDXR
ferredoxin reductase
- FECH
ferrochelatase
- Fe-S
iron-sulfur
- FRDA
Friedreich ataxia
- FXN
frataxin
- GLRX5
glutaredoxin 5
- HSP70
70-kDa heat shock protein
- HSP40
40-kD heat shock protein
- IRE
Iron Response Element
- IRP1/2
Iron Regulatory Protein 1/2
- NBD
Nucleotide Binding Domain
- NEF
Nucleotide Exchange Factor
- PLP
pyridoxal-phosphate
- RdRp
RNA-dependent RNA polymerase
- SAM
S-adenosylmethionine
- SBD
Substrate Binding Domain
- TCA
tricarboxylic acid
- UTR
untranslated region
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Martin W, Baross J, Kelley D, Russell MJ. Hydrothermal vents and the origin of life. Nat Rev Microbiol. 2008;6:805–814. [DOI] [PubMed] [Google Scholar]
- 2.Beinert H, Holm RH, Munck E. Iron-sulfur clusters: nature's modular, multipurpose structures. Science. 1997;277:653–659. [DOI] [PubMed] [Google Scholar]
- 3.Barton JK, Silva RMB, O'Brien E. Redox chemistry in the genome: Emergence of the [4Fe4S] cofactor in repair and replication. Annu Rev Biochem. 2019;88:163–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mancera-Martinez E, Brito Querido J, Valasek LS, Simonetti A, Hashem Y. ABCE1: A special factor that orchestrates translation at the crossroad between recycling and initiation. RNA Biol. 2017;14:1279–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beinert H, Kennedy MC, Stout CD. Aconitase as Ironminus signSulfur protein, enzyme, and iron-regulatory protein. Chem Rev. 1996;96:2335–2374. [DOI] [PubMed] [Google Scholar]
- 6.Rouault TA, Maio N. Biogenesis and functions of mammalian iron-sulfur proteins in the regulation of iron homeostasis and pivotal metabolic pathways. J Biol Chem. 2017;292:12744–12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landgraf BJ, McCarthy EL, Booker SJ. Radical S-adenosylmethionine enzymes in human health and disease. Annu Rev Biochem. 2016;85:485–514. [DOI] [PubMed] [Google Scholar]
- 8.Hanzelmann P, Schindelin H. Binding of 5'-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism. Proc Natl Acad Sci USA. 2006;103:6829–6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarthy EL, Booker SJ. Destruction and reformation of an iron-sulfur cluster during catalysis by lipoyl synthase. Science. 2017;358:373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei FY, Suzuki T, Watanabe S, et al. Deficit of tRNA(Lys) modification by Cdkal1 causes the development of type 2 diabetes in mice. J Clin Invest. 2011;121:3598–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei FY, Zhou B, Suzuki T, et al. Cdk5rap1-mediated 2-methylthio modification of mitochondrial tRNAs governs protein translation and contributes to myopathy in mice and humans. Cell Metab. 2015;21:428–442. [DOI] [PubMed] [Google Scholar]
- 12.Dauden MI, Jaciuk M, Weis F, et al. Molecular basis of tRNA recognition by the Elongator complex. Sci Adv. 2019;5:eaaw2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gari K, Leon Ortiz AM, Borel V, Flynn H, Skehel JM, Boulton SJ. MMS19 links cytoplasmic iron-sulfur cluster assembly to DNA metabolism. Science. 2012;337:243–245. [DOI] [PubMed] [Google Scholar]
- 14.Stehling O, Vashisht AA, Mascarenhas J, et al. MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science. 2012;337:195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ben-Shimon L, Paul VD, David-Kadoch G, et al. Fe-S cluster coordination of the chromokinesin KIF4A alters its subcellular localization during mitosis. J Cell Sci. 2018;131(12):jcs211433. [DOI] [PubMed] [Google Scholar]
- 16.Conlan AR, Axelrod HL, Cohen AE, et al. Crystal structure of Miner1: The redox-active 2Fe-2S protein causative in Wolfram syndrome 2. J Mol Biol. 2009;392:143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karmi O, Marjault HB, Pesce L, et al. The unique fold and lability of the [2Fe-2S] clusters of NEET proteins mediate their key functions in health and disease. J Biol Inorg Chem. 2018;23:599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mozzillo E, Delvecchio M, Carella M, et al. A novel CISD2 intragenic deletion, optic neuropathy and platelet aggregation defect in Wolfram syndrome type 2. BMC Med Genet. 2014;15:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maio N, Zhang DL, Ghosh MC, Jain A, SantaMaria AM, Rouault TA. Mechanisms of cellular iron sensing, regulation of erythropoiesis and mitochondrial iron utilization. Semin Hematol. 2021;58:161–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dandekar T, Stripecke R, Gray NK, et al. Identification of a novel iron-responsive element in murine and human erythroid delta-aminolevulinic acid synthase mRNA. EMBO J. 1991;10:1903–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wingert RA, Galloway JL, Barut B, et al. Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature. 2005;436:1035–1039. [DOI] [PubMed] [Google Scholar]
- 22.Ye H, Jeong SY, Ghosh MC, et al. Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts. J Clin Invest. 2010;120:1749–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camaschella C, Campanella A, De Falco L, et al. The human counterpart of zebrafish shiraz shows sideroblastic-like microcytic anemia and iron overload. Blood. 2007;110:1353–1358. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Shi H, Rajan M, Canarie ER, Hong S, Simoneschi D, Pagano M, Bush MF, Stoll S, Leibold EA, Zheng N (2020). FBXL5 regulates IRP2 stability in iron homeostasis via an oxygen-responsive [2Fe2S] cluster. Mol Cell 78, 31–41 e35, 31, 41.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maio N, Singh A, Uhrigshardt H, Saxena N, Tong WH, Rouault TA. Cochaperone binding to LYR motifs confers specificity of iron sulfur cluster delivery. Cell Metab. 2014;19:445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi Y, Ghosh MC, Tong WH, Rouault TA. Human ISD11 is essential for both iron-sulfur cluster assembly and maintenance of normal cellular iron homeostasis. Hum Mol Genet. 2009;18:3014–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong WH, Rouault TA. Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metab. 2006;3:199–210. [DOI] [PubMed] [Google Scholar]
- 28.Crooks DR, Maio N, Lane AN, et al. Acute loss of iron-sulfur clusters results in metabolic reprogramming and generation of lipid droplets in mammalian cells. J Biol Chem. 2018;293:8297–8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rouault TA, Maio N. How oxidation of a unique iron-sulfur cluster in FBXL5 regulates IRP2 levels and promotes regulation of iron metabolism proteins. Mol Cell. 2020;78:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu G, Sil D, Maio N, et al. Heme biosynthesis depends on previously unrecognized acquisition of iron-sulfur cofactors in human amino-levulinic acid dehydratase. Nat Commun. 2020;11:6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dailey HA, Finnegan MG, Johnson MK. Human ferrochelatase is an iron-sulfur protein. Biochemistry. 1994;33:403–407. [DOI] [PubMed] [Google Scholar]
- 32.Wu CK, Dailey HA, Rose JP, Burden A, Sellers VM, Wang BC. The 2.0 Å structure of human ferrochelatase, the terminal enzyme of heme biosynthesis. Nat Struct Biol. 2001;8:156–160. [DOI] [PubMed] [Google Scholar]
- 33.Crooks DR, Ghosh MC, Haller RG, Tong WH, Rouault TA. Posttranslational stability of the heme biosynthetic enzyme ferrochelatase is dependent on iron availability and intact iron-sulfur cluster assembly machinery. Blood. 2010;115:860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. [DOI] [PubMed] [Google Scholar]
- 35.Patra S, Barondeau DP. Mechanism of activation of the human cysteine desulfurase complex by frataxin. Proc Natl Acad Sci U S A. 2019;116:19421–19430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bridwell-Rabb J, Fox NG, Tsai CL, Winn AM, Barondeau DP. Human frataxin activates Fe-S cluster biosynthesis by facilitating sulfur transfer chemistry. Biochemistry. 2014;53:4904–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parent A, Elduque X, Cornu D, et al. Mammalian frataxin directly enhances sulfur transfer of NFS1 persulfide to both ISCU and free thiols. Nat Commun. 2015;6:5686. [DOI] [PubMed] [Google Scholar]
- 38.van Vranken JG, Jeong MY, Wei P, et al. The mitochondrial acyl carrier protein (ACP) coordinates mitochondrial fatty acid synthesis with iron sulfur cluster biogenesis. Elife. 2016;5:e17828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boniecki MT, Freibert SA, Muhlenhoff U, Lill R, Cygler M. Structure and functional dynamics of the mitochondrial Fe/S cluster synthesis complex. Nat Commun. 2017;8:1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cory SA, van Vranken JG, Brignole EJ, et al. Structure of human Fe-S assembly subcomplex reveals unexpected cysteine desulfurase architecture and acyl-ACP-ISD11 interactions. Proc Natl Acad Sci U S A. 2017;114:E5325–E5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fox NG, Yu X, Feng X, et al. Structure of the human frataxin-bound iron-sulfur cluster assembly complex provides insight into its activation mechanism. Nat Commun. 2019;10:2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Majmudar JD, Feng X, Fox NG, et al. 4'-Phosphopantetheine and long acyl chain-dependent interactions are integral to human mitochondrial acyl carrier protein function. Med-ChemComm. 2019;10:209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puccio H, Simon D, Cossee M, et al. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat Genet. 2001;27:181–186. [DOI] [PubMed] [Google Scholar]
- 44.Fiedorczuk K, Letts JA, Degliesposti G, Kaszuba K, Skehel M, Sazanov LA. Atomic structure of the entire mammalian mitochondrial complex I. Nature. 2016;538:406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Angerer H. Eukaryotic LYR proteins interact with mitochondrial protein complexes. Biology (Basel). 2015;4:133–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi Y, Ghosh M, Kovtunovych G, Crooks DR, Rouault TA. Both human ferredoxins 1 and 2 and ferredoxin reductase are important for iron-sulfur cluster biogenesis. Biochim Biophys Acta. 2012;1823:484–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai K, Tonelli M, Frederick RO, Markley JL. Human mitochondrial ferredoxin 1 (FDX1) and ferredoxin 2 (FDX2) both bind cysteine desulfurase and donate electrons for iron-sulfur cluster biosynthesis. Biochemistry. 2017;56:487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gervason S, Larkem D, Mansour AB, et al. Physiologically relevant reconstitution of iron-sulfur cluster biosynthesis uncovers persulfide-processing functions of ferredoxin-2 and frataxin. Nat Commun. 2019;10:3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maio N, Rouault TA. Iron-sulfur cluster biogenesis in mammalian cells: New insights into the molecular mechanisms of cluster delivery. Biochim Biophys Acta. 2015;1853:1493–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marinoni EN, de Oliveira JS, Nicolet Y, et al. (IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer. Angew Chem Int Ed Engl. 2012;51:5439–5442. [DOI] [PubMed] [Google Scholar]
- 51.Yoon H, Knight SA, Pandey A, et al. Turning Saccharomyces cerevisiae into a frataxin-independent organism. PLoS Genet. 2015;11:e1005135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roche B, Agrebi R, Huguenot A, Ollagnier de Choudens S, Barras F, Py B. Turning Escherichia coli into a frataxin-dependent organism. PLoS Genet. 2015;11:e1005134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindahl PA, Moore MJ. Labile low-molecular-mass metal complexes in mitochondria: Trials and tribulations of a burgeoning field. Biochemistry. 2016;55:4140–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cai K, Frederick RO, Dashti H, and Markley JL (2018). Architectural features of human mitochondrial cysteine desulfurase complexes from crosslinking mass spectrometry and small-angle X-ray scattering. Structure 26, 1127–1136 e1124, 1127, 1136.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alderson TR, Kim JH, Markley JL. Dynamical structures of Hsp70 and Hsp70-Hsp40 complexes. Structure. 2016;24:1014–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Balchin D, Hayer-Hartl M, Hartl FU. In vivo aspects of protein folding and quality control. Science. 2016;353:aac4354. [DOI] [PubMed] [Google Scholar]
- 57.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kominek J, Marszalek J, Neuveglise C, Craig EA, Williams BL. The complex evolutionary dynamics of Hsp70s: A genomic and functional perspective. Genome Biol Evol. 2013;5:2460–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mayer MP. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem Sci. 2013;38:507–514. [DOI] [PubMed] [Google Scholar]
- 60.Mayer MP, Bukau B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McKay DB. Structure and mechanism of 70-kDa heat-shock-related proteins. Adv Protein Chem. 1993;44:67–98. [DOI] [PubMed] [Google Scholar]
- 62.Seaton BL, Vickery LE. A gene encoding a DnaK/hsp70 homolog in Escherichia coli. Proc Natl Acad Sci U S A. 1994;91:2066–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kawula TH, Lelivelt MJ. Mutations in a gene encoding a new Hsp70 suppress rapid DNA inversion and bgl activation, but not proU derepression, in hns-1 mutant Escherichia coli. J Bacteriol. 1994;176:610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ta DT, Seaton BL, Vickery LE. Localization of the ferredoxin (fdx) gene on the physical map of the Escherichia coli chromosome. J Bacteriol. 1992;174:5760–5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vickery LE, Silberg JJ, Ta DT. Hsc66 and Hsc20, a new heat shock cognate molecular chaperone system from Escherichia coli. Protein Sci. 1997;6:1047–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Silberg JJ, Hoff KG, Vickery LE. The Hsc66-Hsc20 chaperone system in Escherichia coli: Chaperone activity and interactions with the DnaK-DnaJ-grpE system. J Bacteriol. 1998;180:6617–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blattner FR, Plunkett G 3rd, Bloch CA, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. [DOI] [PubMed] [Google Scholar]
- 68.Zheng L, Cash VL, Flint DH, Dean DR. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem. 1998;273:13264–13272. [DOI] [PubMed] [Google Scholar]
- 69.Tokumoto U, Takahashi Y. Genetic analysis of the isc operon in Escherichia coli involved in the biogenesis of cellular iron-sulfur proteins. J Biochem. 2001;130:63–71. [DOI] [PubMed] [Google Scholar]
- 70.Goffeau A, Barrell BG, Bussey H, et al. Life with 6000 genes. Science. 1996;274(546):563–547. [DOI] [PubMed] [Google Scholar]
- 71.Strain J, Lorenz CR, Bode J, et al. Suppressors of superoxide dismutase (SOD1) deficiency in Saccharomyces cerevisiae. Identification of proteins predicted to mediate iron-sulfur cluster assembly. J Biol Chem. 1998;273:31138–31144. [DOI] [PubMed] [Google Scholar]
- 72.Schilke B, Voisine C, Beinert H, Craig E. Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1999;96:10206–10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garland SA, Hoff K, Vickery LE, Culotta VC. Saccharomyces cerevisiae ISU1 and ISU2: Members of a well-conserved gene family for iron-sulfur cluster assembly. J Mol Biol. 1999;294:897–907. [DOI] [PubMed] [Google Scholar]
- 74.Kim R, Saxena S, Gordon DM, Pain D, Dancis A. J-domain protein, Jac1p, of yeast mitochondria required for iron homeostasis and activity of Fe-S cluster proteins. J Biol Chem. 2001;276:17524–17532. [DOI] [PubMed] [Google Scholar]
- 75.Lutz T, Westermann B, Neupert W, Herrmann JM. The mitochondrial proteins Ssq1 and Jac1 are required for the assembly of iron sulfur clusters in mitochondria. J Mol Biol. 2001;307:815–825. [DOI] [PubMed] [Google Scholar]
- 76.Voisine C, Cheng YC, Ohlson M, et al. Jac1, a mitochondrial J-type chaperone, is involved in the biogenesis of Fe/S clusters in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2001;98:1483–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hoff KG, Silberg JJ, Vickery LE. Interaction of the iron-sulfur cluster assembly protein IscU with the Hsc66/Hsc20 molecular chaperone system of Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:7790–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hoff KG, Ta DT, Tapley TL, Silberg JJ, Vickery LE. Hsc66 substrate specificity is directed toward a discrete region of the iron-sulfur cluster template protein IscU. J Biol Chem. 2002;277:27353–27359. [DOI] [PubMed] [Google Scholar]
- 79.Hoff KG, Cupp-Vickery JR, Vickery LE. Contributions of the LPPVK motif of the iron-sulfur template protein IscU to interactions with the Hsc66-Hsc20 chaperone system. J Biol Chem. 2003;278:37582–37589. [DOI] [PubMed] [Google Scholar]
- 80.Dutkiewicz R, Schilke B, Knieszner H, Walter W, Craig EA, Marszalek J. Ssq1, a mitochondrial Hsp70 involved in iron-sulfur (Fe/S) center biogenesis. Similarities to and differences from its bacterial counterpart. J Biol Chem. 2003;278:29719–29727. [DOI] [PubMed] [Google Scholar]
- 81.Leustek T, Dalie B, Amir-Shapira D, Brot N, Weissbach H. A member of the Hsp70 family is localized in mitochondria and resembles Escherichia coli DnaK. Proc Natl Acad Sci U S A. 1989;86:7805–7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mizzen LA, Chang C, Garrels JI, Welch WJ. Identification, characterization, and purification of two mammalian stress proteins present in mitochondria, grp 75, a member of the hsp 70 family and hsp 58, a homolog of the bacterial groEL protein. J Biol Chem. 1989;264:20664–20675. [PubMed] [Google Scholar]
- 83.Craig EA, Kramer J, Shilling J, et al. SSC1, an essential member of the yeast HSP70 multigene family, encodes a mitochondrial protein. Mol Cell Biol. 1989;9:3000–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Taylor JW, Berbee ML. Dating divergences in the fungal tree of life: Review and new analyses. Mycologia. 2006;98:838–849. [DOI] [PubMed] [Google Scholar]
- 85.Schilke B, Williams B, Knieszner H, et al. Evolution of mitochondrial chaperones utilized in Fe-S cluster biogenesis. Curr Biol. 2006;16:1660–1665. [DOI] [PubMed] [Google Scholar]
- 86.Karzai AW, McMacken R. A bipartite signaling mechanism involved in DnaJ-mediated activation of the Escherichia coli DnaK protein. J Biol Chem. 1996;271:11236–11246. [DOI] [PubMed] [Google Scholar]
- 87.Laufen T, Mayer MP, Beisel C, et al. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc Natl Acad Sci U S A. 1999;96:5452–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Silberg JJ, Tapley TL, Hoff KG, Vickery LE. Regulation of the HscA ATPase reaction cycle by the co-chaperone HscB and the iron-sulfur cluster assembly protein IscU. J Biol Chem. 2004;279:53924–53931. [DOI] [PubMed] [Google Scholar]
- 89.Ciesielski SJ, Schilke BA, Osipiuk J, et al. Interaction of J-protein co-chaperone Jac1 with Fe-S scaffold Isu is indispensable in vivo and conserved in evolution. J Mol Biol. 2012;417:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brehmer D, Rudiger S, Gassler CS, et al. Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat Struct Biol. 2001;8:427–432. [DOI] [PubMed] [Google Scholar]
- 91.Vickery LE, Cupp-Vickery JR. Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron-sulfur protein maturation. Crit Rev Biochem Mol Biol. 2007;42:95–111. [DOI] [PubMed] [Google Scholar]
- 92.Cupp-Vickery JR, Peterson JC, Ta DT, Vickery LE. Crystal structure of the molecular chaperone HscA substrate binding domain complexed with the IscU recognition peptide ELPPVKIHC. J Mol Biol. 2004;342:1265–1278. [DOI] [PubMed] [Google Scholar]
- 93.Dutkiewicz R, Schilke B, Cheng S, Knieszner H, Craig EA, Marszalek J. Sequence-specific interaction between mitochondrial Fe-S scaffold protein Isu and Hsp70 Ssq1 is essential for their in vivo function. J Biol Chem. 2004;279:29167–29174. [DOI] [PubMed] [Google Scholar]
- 94.Kim JH, Tonelli M, Markley JL. Disordered form of the scaffold protein IscU is the substrate for iron-sulfur cluster assembly on cysteine desulfurase. Proc Natl Acad Sci U S A. 2012;109:454–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim JH, Tonelli M, Frederick RO, Chow DC, Markley JL. Specialized Hsp70 chaperone (HscA) binds preferentially to the disordered form, whereas J-protein (HscB) binds preferentially to the structured form of the iron-sulfur cluster scaffold protein (IscU). J Biol Chem. 2012;287:31406–31413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alderson TR, Kim JH, Cai K, Frederick RO, Tonelli M, Markley JL. The specialized Hsp70 (HscA) interdomain linker binds to its nucleotide-binding domain and stimulates ATP hydrolysis in both cis and trans configurations. Biochemistry. 2014;53:7148–7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bonomi F, Iametti S, Morleo A, Ta D, Vickery LE. Facilitated transfer of IscU-[2Fe2S] clusters by chaperone-mediated ligand exchange. Biochemistry. 2011;50:9641–9650. [DOI] [PubMed] [Google Scholar]
- 98.Craig EA, Marszalek J. A specialized mitochondrial molecular chaperone system: A role in formation of Fe/S centers. Cell Mol Life Sci. 2002;59:1658–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shan Y, Cortopassi G. HSC20 interacts with frataxin and is involved in iron-sulfur cluster biogenesis and iron homeostasis. Hum Mol Genet. 2012;21:1457–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: Machineries and mechanisms. Cell. 2009;138:628–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wadhwa R, Taira K, Kaul SC. An Hsp70 family chaperone, mortalin/mthsp70/PBP74/Grp75: What, when, and where? Cell Stress Chaperones. 2002;7:309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kaul SC, Deocaris CC, Wadhwa R. Three faces of mortalin: A housekeeper, guardian and killer. Exp Gerontol. 2007;42:263–274. [DOI] [PubMed] [Google Scholar]
- 103.Deocaris CC, Lu WJ, Kaul SC, Wadhwa R. Druggability of mortalin for cancer and neuro-degenerative disorders. Curr Pharm Des. 2013;19:418–429. [PubMed] [Google Scholar]
- 104.Flachbartova Z, Kovacech B. Mortalin: A multipotent chaperone regulating cellular processes ranging from viral infection to neurodegeneration. Acta Virol. 2013;57:3–15. [DOI] [PubMed] [Google Scholar]
- 105.Sun G, Gargus JJ, Ta DT, Vickery LE. Identification of a novel candidate gene in the iron-sulfur pathway implicated in ataxia-susceptibility: Human gene encoding HscB, a J-type co-chaperone. J Hum Genet. 2003;48:415–419. [DOI] [PubMed] [Google Scholar]
- 106.Uhrigshardt H, Singh A, Kovtunovych G, Ghosh M, Rouault TA. Characterization of the human HSC20, an unusual DnaJ type III protein, involved in iron-sulfur cluster biogenesis. Hum Mol Genet. 2010;19:3816–3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bitto E, Bingman CA, Bittova L, et al. Structure of human J-type co-chaperone HscB reveals a tetracysteine metal-binding domain. J Biol Chem. 2008;283:30184–30192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cupp-Vickery JR, Vickery LE. Crystal structure of Hsc20, a J-type Co-chaperone from Escherichia coli. J Mol Biol. 2000;304:835–845. [DOI] [PubMed] [Google Scholar]
- 109.Knieszner H, Schilke B, Dutkiewicz R, et al. Compensation for a defective interaction of the hsp70 ssq1 with the mitochondrial Fe-S cluster scaffold isu. J Biol Chem. 2005;280: 28966–28972. [DOI] [PubMed] [Google Scholar]
- 110.Fuzery AK, Oh JJ, Ta DT, Vickery LE, Markley JL. Three hydrophobic amino acids in Escherichia coli HscB make the greatest contribution to the stability of the HscB-IscU complex. BMC Biochem. 2011;12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Perales-Calvo J, Muga A, Moro F. Role of DnaJ G/F-rich domain in conformational recognition and binding of protein substrates. J Biol Chem. 2010;285:34231–34239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Szabo A, Korszun R, Hartl FU, Flanagan J. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J. 1996;15:408–417. [PMC free article] [PubMed] [Google Scholar]
- 113.Lee S, Fan CY, Younger JM, Ren H, Cyr DM. Identification of essential residues in the type II Hsp40 Sis1 that function in polypeptide binding. J Biol Chem. 2002;277:21675–21682. [DOI] [PubMed] [Google Scholar]
- 114.Li J, Sha B. Structure-based mutagenesis studies of the peptide substrate binding fragment of type I heat-shock protein 40. Biochem J. 2005;386:453–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Maio N, Kim KS, Singh A, Rouault TA. A single adaptable cochaperone-scaffold complex delivers nascent iron-sulfur clusters to mammalian respiratory chain complexes I-III. Cell Metab. 2017;25:945–953.e6. [DOI] [PubMed] [Google Scholar]
- 116.Maio N, Ghezzi D, Verrigni D, et al. Disease-causing SDHAF1 mutations impair transfer of Fe-S clusters to SDHB. Cell Metab. 2016;23:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Maio N, Lafont BAP, Sil D, et al. Fe-S cofactors in the SARS-CoV-2 RNA-dependent RNA polymerase are potential antiviral targets. Science. 2021;373:236–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Imlay JA. Iron-Sulphur clusters and the problem with oxygen. Mol Microbiol. 2006;59:1073–1082. [DOI] [PubMed] [Google Scholar]
- 119.Ghosh MC, Tong WH, Zhang D, et al. Tempol-mediated activation of latent iron regulatory protein activity prevents symptoms of neurodegenerative disease in IRP2 knockout mice. Proc Natl Acad Sci U S A. 2008;105:12028–12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ghosh MC, Zhang DL, Ollivierre H, Eckhaus MA, Rouault TA. Translational repression of HIF2alpha expression in mice with Chuvash polycythemia reverses polycythemia. J Clin Invest. 2018;128:1317–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cunniff B, Benson K, Stumpff J, et al. Mitochondrial-targeted nitroxides disrupt mitochondrial architecture and inhibit expression of peroxiredoxin 3 and FOXM1 in malignant mesothelioma cells. J Cell Physiol. 2013;228:835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cotrim AP, Hyodo F, Matsumoto K, et al. Differential radiation protection of salivary glands versus tumor by Tempol with accompanying tissue assessment of Tempol by magnetic resonance imaging. Clin Cancer Res. 2007;13:4928–4933. [DOI] [PubMed] [Google Scholar]
- 123.Wang Y, Hai B, Ai L, et al. Tempol relieves lung injury in a rat model of chronic intermittent hypoxia via suppression of inflammation and oxidative stress. Iran J Basic Med Sci. 2018;21:1238–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nakai Y, Nakai M, Hayashi H, Kagamiyama H. Nuclear localization of yeast Nfs1p is required for cell survival. J Biol Chem. 2001;276:8314–8320. [DOI] [PubMed] [Google Scholar]
- 125.Lill R, Dutkiewicz R, Freibert SA, et al. The role of mitochondria and the CIA machinery in the maturation of cytosolic and nuclear iron-sulfur proteins. Eur J Cell Biol. 2015;94:280–291. [DOI] [PubMed] [Google Scholar]
- 126.Pandey AK, Pain J, Dancis A, Pain D. Mitochondria export iron-sulfur and sulfur intermediates to the cytoplasm for iron-sulfur cluster assembly and tRNA thiolation in yeast. J Biol Chem. 2019;294:9489–9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rouault TA. The indispensable role of mammalian iron sulfur proteins in function and regulation of multiple diverse metabolic pathways. Biometals. 2019;32:343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kim KS, Maio N, Singh A, Rouault TA. Cytosolic HSC20 integrates de novo iron-sulfur cluster biogenesis with the CIAO1-mediated transfer to recipients. Hum Mol Genet. 2018;27:837–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Land T, Rouault TA. Targeting of a human iron-sulfur cluster assembly enzyme, nifs, to different subcellular compartments is regulated through alternative AUG utilization. Mol Cell. 1998;2:807–815. [DOI] [PubMed] [Google Scholar]
- 130.Condo I, Malisan F, Guccini I, Serio D, Rufini A, Testi R. Molecular control of the cytosolic aconitase/IRP1 switch by extramitochondrial frataxin. Hum Mol Genet. 2010;19:1221–1229. [DOI] [PubMed] [Google Scholar]
- 131.Braymer JJ, Lill R. Iron-sulfur cluster biogenesis and trafficking in mitochondria. J Biol Chem. 2017;292:12754–12763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kispal G, Csere P, Prohl C, Lill R. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 1999;18:3981–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Marelja Z, Mullick Chowdhury M, Dosche C, et al. The L-cysteine desulfurase NFS1 is localized in the cytosol where it provides the sulfur for molybdenum cofactor biosynthesis in humans. PLoS One. 2013;8:e60869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Marelja Z, Stocklein W, Nimtz M, Leimkuhler S. A novel role for human Nfs1 in the cytoplasm: Nfs1 acts as a sulfur donor for MOCS3, a protein involved in molybdenum cofactor biosynthesis. J Biol Chem. 2008;283:25178–25185. [DOI] [PubMed] [Google Scholar]
- 135.Biederbick A, Stehling O, Rosser R, et al. Role of human mitochondrial Nfs1 in cytosolic iron-sulfur protein biogenesis and iron regulation. Mol Cell Biol. 2006;26:5675–5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tong WH, Jameson GN, Huynh BH, Rouault TA. Subcellular compartmentalization of human Nfu, an iron-sulfur cluster scaffold protein, and its ability to assemble a [4Fe-4S] cluster. Proc Natl Acad Sci U S A. 2003;100:9762–9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Thul PJ, Akesson L, Wiking M, et al. A subcellular map of the human proteome. Science. 2017;356(6340):eaal3321. [DOI] [PubMed] [Google Scholar]
- 138.Stehling O, Mascarenhas J, Vashisht AA, et al. Human CIA2A-FAM96A and CIA2B-FAM96B integrate iron homeostasis and maturation of different subsets of cytosolic-nuclear iron-sulfur proteins. Cell Metab. 2013;18:187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Maio N, Rouault TA. Outlining the complex pathway of mammalian Fe-S cluster biogenesis. Trends Biochem Sci. 2020;45:411–426. [DOI] [PMC free article] [PubMed] [Google Scholar]