Abstract
Background
Hybrid imaging became an instrumental part of medical imaging, particularly cancer imaging processes in clinical routine. To date, several radiomic and machine learning studies investigated the feasibility of in vivo tumor characterization with variable outcomes. This study aims to investigate the effect of recently proposed fuzzy radiomics and compare its predictive performance to conventional radiomics in cancer imaging cohorts. In addition, lesion vs. lesion+surrounding fuzzy and conventional radiomic analysis was conducted.
Methods
Previously published 11C Methionine (MET) positron emission tomography (PET) glioma, 18F-FDG PET/computed tomography (CT) lung, and 68GA-PSMA-11 PET/magneto-resonance imaging (MRI) prostate cancer retrospective cohorts were included in the analysis to predict their respective clinical endpoints. Four delineation methods including manually defined reference binary (Ref-B), its smoothed, fuzzified version (Ref-F), as well as extended binary (Ext-B) and its fuzzified version (Ext-F) were incorporated to extract imaging biomarker standardization initiative (IBSI)-conform radiomic features from each cohort. Machine learning for the four delineation approaches was performed utilizing a Monte Carlo cross-validation scheme to estimate the predictive performance of the four delineation methods.
Results
Reference fuzzy (Ref-F) delineation outperformed its binary delineation (Ref-B) counterpart in all cohorts within a volume range of 938–354987 mm3 with relative cross-validation area under the receiver operator characteristics curve (AUC) of +4.7–10.4. Compared to Ref-B, the highest AUC performance difference was observed by the Ref-F delineation in the glioma cohort (Ref-F: 0.74 vs. Ref-B: 0.70) and in the prostate cohort by Ref-F and Ext-F (Ref-F: 0.84, Ext-F: 0.86 vs. Ref-B: 0.80). In addition, fuzzy radiomics decreased feature redundancy by approx. 20%.
Conclusions
Fuzzy radiomics has the potential to increase predictive performance particularly in small lesion sizes compared to conventional binary radiomics in PET. We hypothesize that this effect is due to the ability of fuzzy radiomics to model partial volume effects and delineation uncertainties at small lesion boundaries. In addition, we consider that the lower redundancy of fuzzy radiomic features supports the identification of imaging biomarkers in future studies. Future studies shall consider systematically analyzing lesions and their surroundings with fuzzy and binary radiomics.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00259-023-06127-1.
Keywords: Machine learning, Radiomics, Fuzzy radiomics, PET, PET/CT, PET/MRI
Introduction
Cancer is one of the leading causes of death worldwide [1]. Medical imaging became an instrumental part of cancer detection, with positron emission tomography (PET) playing an important role in the imaging of metabolic activities, and the characterization of tumor heterogeneity in vivo [2]. Hybrid imaging systems relying on PET/computed tomography (CT) are considered the gold standard of cancer imaging, while PET/Magnetic resonance imaging (MRI) is in the process of wide-scale adoption worldwide [3, 4].
While imaging is routinely employed to detect tumors, to date, it is mainly used for visual inspection and for basic imaging biomarker measurements such as metabolic tumor volume [5]. In contrast, various studies proposed the utilization of radiomics—the approach to extract different imaging features from tumors—for analysis [6, 7]. Contrary to promising results, radiomic studies had been challenged by their poor reproducibility due to various factors such as biological and imaging differences, delineation, radiomic feature extraction equation variations as well as their parameters (e.g., resolution, bin size, binning method) [7, 8]. An important consolidation phase had been started with the proposal of the Imaging Biomarker Standardization Initiative (IBSI) [9]. However, IBSI does not cover all important aspects of radiomics such as delineation, which has a profound effect on radiomic features [2, 10–12] and it appears to be mainly affected by multi-observer variabilities and cohort, as well as imaging characteristics [10]. Consequently, delineation of tumor lesions had been extensively investigated in the corresponding literature [10, 13]. While various tumor delineation approaches had been proposed [14–16], certain studies looked into analyzing not only the lesions, but also their surroundings [17, 18]. Recently, deep learning (DL) has been demonstrated as a powerful technique to delineate suspicious lesions in PET for subsequent analysis with e.g., radiomics and machine learning (ML) [13, 19–22]. However, the common property of all the above approaches is that they result in a binary delineation mask or volume of interest (VOI) for radiomic analysis, meaning, that a particular PET voxel is either part of the analysis or not, regardless of how certain its membership in the given VOI is. This approach has various drawbacks. First, operating with binary masks renders the radiomic analysis sensitive to PET partial volume effects (PVE) especially at lesion boundaries [23]. Second, delineation errors may result in suboptimal radiomic analysis at lesion boundaries, regardless of PVE, and third, multi-observer variations result in different radiomic outcomes, which makes the repeatability of reported studies challenging [10, 13, 16]. Fuzzy radiomics had been presented as a potential approach to handle voxel membership uncertainties by relying on non-binary probability masks [24]. However, to date, it has not been utilized and evaluated in real clinical cancer settings.
In theory, fuzzy radiomics has numerous advantages. First, it can model and encompass PVE in the given mask as to the properties of the given imaging system. Second, it can also encode multiple observer’s delineations as a weighted mask. Third, it can consider not just the given lesion, but its surroundings with appropriate weights for the analysis. Last, it can directly handle DL delineation masks that are inherently probabilistic, but are routinely post-processed and dichotomized by a threshold to provide a binary mask for subsequent analysis [19, 20, 22, 25].
In light of the above, the aim of this study was to compare the effect of binary and fuzzy delineation masks in both lesions and their surroundings, through investigating the performance of ML prediction models built in various cancer cohorts to predict their clinical endpoints. Specifically, this study had the following objectives: (a) to collect various cancer imaging cohorts having different characteristics regarding the imaging systems and PET tracers involved; (b) to perform classic and fuzzy radiomic feature extraction relying on binary and fuzzy probability masks of lesions as well as their surroundings; and (c) to compare ML performance of predicting cohort-specific clinical endpoints relying on the above feature extraction approaches.
Methods
Cohorts
This study relied on already delineated lesions from three retrospectively available cancer imaging cohorts including 11C methionine (MET) PET glioma, 18F-FDG PET/CT lung and 68GA-PSMA-11 PET/MRI prostate cancer cases. For details of how these delineations were done, see Sec. Delineation. All cohorts had been previously investigated and presented in various ML studies [26–30] with follow-up up to 3 years. This study included lesions from the above cohort databases that fulfilled the minimum 64 voxel number constraint [31], resulting in 105, 543 and 121 delineated lesions in glioma, lung and prostate cases respectively.
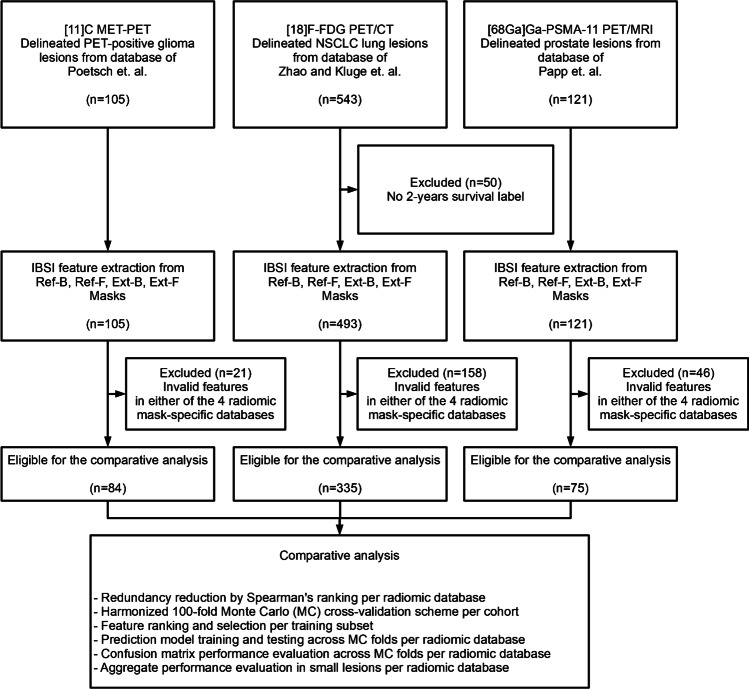
All cohorts had been approved for analysis by their respective institutional review boards and the need for informed consent was waived in retrospective studies. The clinical endpoints were 3-years survival, 2-years survival and low-vs-high risk in glioma, lung and prostate cohorts, respectively. See Supplemental: Patient Cohorts and Supplemental Table S1-S3 for patient and clinical characteristics of the utilized cohorts. See Fig. 1 for the CONSORT diagram of this study.
Fig. 1.
The CONSORT diagram of preparing and analyzing the cohorts of this study. Already delineated glioma, lung and prostate lesions (Ref-B) were collected from databases that were analyzed and published in prior studies [27–29]. Glioma and lung cohorts had one lesion per patient, while the prostate cohort contained multiple lesions per patient. Reference labels for glioma, lung and prostate lesions were 3-years survival, 2-years survival and low-vs-high risk, respectively. Three additional delineations were generated form Ref-B delineations: a fuzzy mask (Ref-F), an extended binary mask (Ext-B) and an extended fuzzy (Ext-F) mask. Fuzzy masks were generated by a three-dimensional Gaussian filtering, relying on the physical resolution of each cohort's PET imaging system. Samples having no variations or invalid radiomic values (e.g., by low-uptake lesions) were excluded from all delineation-specific databases to ensure a harmonized comparison among them. The resulted four databases per cohort underwent the comparative analysis (see the “Methods” section)
Delineation
This study did not intend to promote a new delineation approach, but intended to compare binary vs. fuzzy masks of lesions as well as their surroundings regarding their performance in predicting clinical endpoints. Therefore, this study included four delineation approaches to compare in each cohort, where the existing delineation of each cohort was serving as reference binary (Ref-B) mask provided by clinical experts in consensus. The number of clinicians involved in this step was cohort-specific (See Supplemental: Table S4).
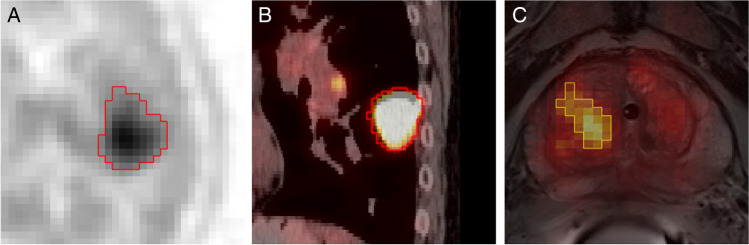
The delineation of cohort-specific lesions was originally performed in the Hermes Hybrid 3D software ver. 4.0.0 (Hermes Medical Solutions, Stockholm, Sweden) relying on standard three-dimensional (3D) iso-count VOIs [26, 27, 32]. Where needed, manual slice-by-slice modifications were performed to result in the final Ref-B VOI. See Fig. 2 for example screenshots of the delineation in each cohort. In addition to the above, a reference background region for tumor-to-background ratio (TBR) normalization of PET images was also available for each case (see Supplemental Table S4 for details).
Fig. 2.
Manual delineation (Ref-B) example views of the cohorts involved in this study. A Glioma tumor (axial view, inverted gray palette with range 0–7 SUV); B Lung tumor (sagittal view, PET hot metal palette with range 0–5 SUV, CT gray palette with range -100.200 HU); C Prostate tumor (oblique-axial view, PET hot iron palette with range 0–9 SUV, T2w MRI grey palette). PET—Positron Emission Tomography; SUV—Standard Uptake Value; T2w—T2-weighted magnetic resonance imaging (MRI)
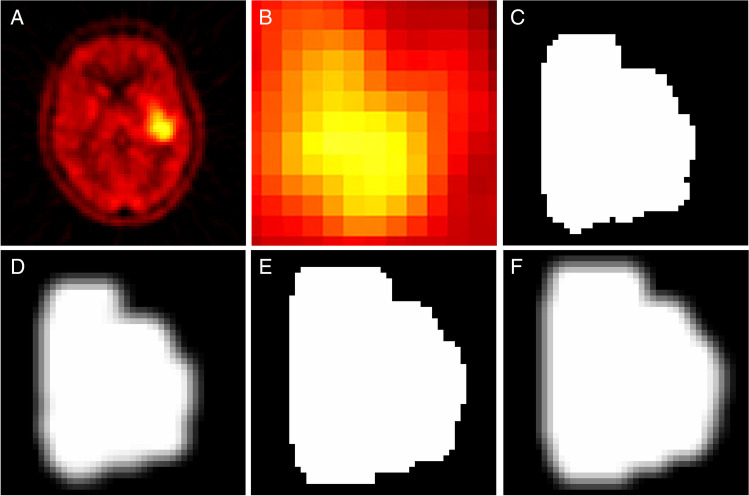
Based on the Ref-B VOI of each case, three additional delineation masks were generated: a reference fuzzy mask, by smoothing Ref-B by a 3D Gaussian filter [28] with a cohort-specific full-width half-maximum (FWHM), which corresponded to the physical resolution of the given imaging system in 3D (Ref-F); an extended binary mask by morphological dilating [33] Ref-B with 3 voxels in 3D (Ext-B), and an extended fuzzy mask by smoothing Ext-B mask with a 3D Gaussian kernel by applying the cohort-specific FWHM as smoothing parameter (Ext-F). The Gaussian FWHM for the 3D smoothing in case of both the reference (Ref-F) and extended (Ext-F) fuzzy mask generations were 5 mm, 4.7 mm, and 4.6 mm for glioma, lung and prostate cohorts respectively, according to the manufacturer-reported physical resolutions of their imaging systems (see Supplemental Table S4). For a visual comparison of an example lesion as well as the four masks, see Fig. 3.
Fig. 3.
A Axial view of an example lesion in the Glioma cohort (MET-PET, Hot iron palette with palette ranges 0–6 SUV). B Magnified axial view of the tumor C Reference binary delineation (Ref-B). D Reference fuzzy delineation (Ref-F). E Extended binary delineation (Ext-B). F Extended Fuzzy delineation (Ext-F). MET—methionine; PET—positron emission tomography; SUV—standard uptake value. Note that images C-F are delineated masks with 2 × 2 × 2 mm resampling for the radiomic analysis
Feature extraction and normalization
All extracted VOIs were resampled to 2 × 2 × 2 mm voxel resolution by using Kriging interpolation in 3D [26, 34] and the PET standardized uptake values (SUV) were normalized by the mean of their respective background regions (See Supplemental Table S4 for details). Each TBR PET lesion VOI was subject to radiomic feature extraction following the IBSI guidelines where only features of “very strong” and “strong” multi-centric consensus as of the IBSI [9] were extracted. The above steps were performed for each of the four delineation masks (Ref-B, Ref-F, Ext-B and Ext-F) and resulted in 153 features per sample. In case either of the four delineation-specific radiomics database had invalid numbers or numbers with no variations (e.g., due to too small uptake in lesions in relation to the bin width), the given sample was removed from all four databases. This was necessary to ensure that the harmonized cross-validation split configuration had identical train-test split samples for all four delineation processes for comparison. This step resulted in 84, 335, and 75 samples in glioma, lung and the prostate cohorts, respectively (see Fig. 1).
Due to the properties of fuzzy radiomics, its feature calculation equations are identical to those defined by IBSI. The only difference between fuzzy and binary radiomics is that fuzzy radiomics takes weights with any value between 0.0 and 1.0 into account when calculating intermediate metadata e.g., textural matrices [6]. In contrast, classic radiomics consider that the weight of a voxel is 1.0 if it is part of the given delineation mask, and 0.0 otherwise. Therefore, classic radiomics is a special variant of fuzzy radiomics in case the mask only has 0.0 and 1.0 values (See Supplemental: Fuzzy vs. Classic Radiomics for details). As of this relationship, this study only utilized the IBSI-validated fuzzy radiomics engine to calculate both binary (Ref-B and Ext-B) and fuzzy (Ref-F and Ext-F) radiomic values for further analysis (See Supplemental Table S4 for details).
Feature redundancy reduction
Since radiomic features are generally highly-redundant [35, 36], this study performed redundancy reduction (RR) in each of the four radiomic data tables corresponding to the four delineation methods of each cohort. The RR was performed by correlation matrix analysis with Spearman correlation coefficient 0.9 as threshold to identify redundant feature clusters [37]. From each redundant cluster, all features except the one with the highest variance were deleted per delineation type in each cohort.
Harmonized cross-validation scheme and data preprocessing
Hundred fold Monte Carlo (MC) cross validation with 90–10% train-test ratio was utilized to generate training-test subsets per cohort [32]. While the splitting was random, only unique folds were allowed to be generated, and no lesions from the same patient were allowed to be part of a train-test split at once, to avoid patient-level data leakage. This step was necessary for the prostate cohort, where patients had multiple lesions [27]. The fold split configuration was harmonized within the given cohort for all four delineation-specific radiomic datasets to avoid split-specific predictive performance variations during the performance evaluation step of the study. The 10% test ratio was calculated for the minority subgroup of each cohort, followed by selecting equal number of test samples according to this ratio in order to ensure that each prediction label subgroup had the same test subgroup count [32]. This way, each training subset remained imbalanced. To handle class imbalance, Synthetic Minority Oversampling Technique (SMOTE) was utilized to obtain equal numbers of subgroups in each training subset [38]. In order to remain in the original IBSI feature space for supporting imaging biomarker identification and analysis, this study did not employ dimensionality reduction [39], but feature selection. Feature selection in the training subset was performed by R-squared ranking [32] where the number of selected features f was calculated by Eq. 1:
where M represents the number of samples in the majority group within the given dataset. Since each cohort had a binary label to predict and the training subset ratio was 90%, 0.9 * 2 multiplier ensured that the number of features selected followed the curse of dimensionality rule [40] in relation to the number of samples in the preprocessed, SMOTE-extended training subset of each MC fold. See Table 1 for details of the collected cohorts in relation to sample counts and class imbalance ratios.
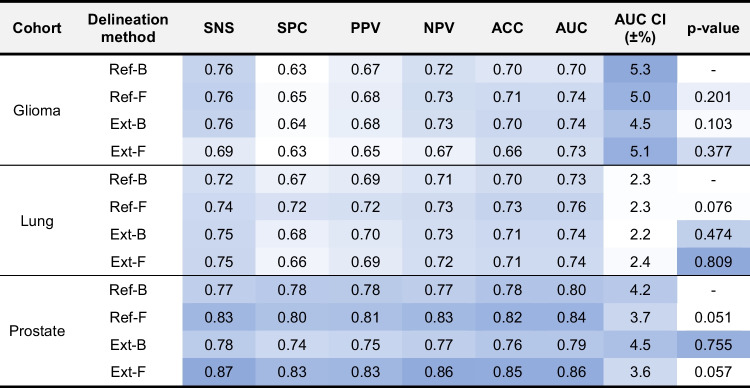
Table 1.
Imaging modalities, sample counts and clinical endpoints to predict in the collected cohorts of this study. For clinical and patient characteristics of each cohort, see Supplemental Table S1-S3. Original sample counts refer to the number of cases this study incorporated to its analysis. Harmonized sample counts refer to the number of samples that were mutually-present as valid across all four delineation-specific radiomic databases. Class imbalance ratio refers to the ratio of the minority subclass vs. the number of all samples in the harmonized radiomic datasets
| Cohort | Imaging | Sample count (original) | Sample count (harmonized) | Class imbalance ratio (harmonized) | To predict |
|---|---|---|---|---|---|
| Glioma | 11C-MET PET | 105 | 84 | 39% | 3-years survival |
| Lung | 18F-FDG PET/CT | 543 | 335 | 39% | 2-years survival |
| Prostate | 68GA-PSMA PET/MRI | 121 | 75 | 40% | Low-vs-high risk |
Prediction models
To minimize method-specific bias and the effect of bias-variance trade-off [41], mixed ensemble learning consisting of four different Random Forest (RF) [32, 42] and one multi-Gaussian (MG) [26] classifiers was built by analyzing each training subset of the given MC fold and for all four delineation-specific datasets (see Supplemental: Table S5 for parameters of the ML approaches). Each model predicted its cohort-specific binary label as of Table 1. The final prediction for a given input sample was provided as the majority vote of all RF and MG model instances. The above approach also allowed to eliminate the effects of hyperparameter variations when comparing the four delineation approaches across a harmonized cross-validation scheme, as the five model instances operated with a fixed hyperparameter set.
Performance evaluation
The number of true positive (TP), true negative (TN), false positive (FP), and false negative (FN) cases were calculated across the test subsets of each ML fold utilizing their respective ML models. Confusion matrix analytics including sensitivity (SNS), specificity (SPC), positive predictive value (PPV), negative predictive value (NPV), accuracy (ACC), and area under the receiver operator characteristics curve (AUC) were calculated for each of the four delineation-specific radiomic models in each cohort. The 100-fold cross-validation AUC values in-between Ref-B vs. Ref-F, Ext-B and Ext-F were correlated by ANOVA analysis where p < 0.05 was considered as significance threshold to reject the null hypothesis. In addition, cross-validation AUC confidence intervals (CI) with 95% confidence ranges were calculated for each ML prediction model.
Further to the above, aggregate performance increase analysis of the four delineation methods across the cohorts within a standardized volume range of 938–354987 mm3 (or approx. 117–4500 voxels per lesion respectively, with 2.0 mm uniform voxel resolution for IBSI analysis—See Supplemental Table S4 for details) was performed. This step categorized volumes into 10 percentile clusters and the number of correct classifications for each delineation-specific predictive model were calculated per percentile cluster.
Results
Effect of delineation on feature redundancy
A consistent pattern across the different delineation methods (Ref-B, Ref-F, Ext-B and Ext-F) was identified regarding their effect on feature redundancy. As such, fuzzy radiomics decreased feature redundancy after performing RR compared to binary radiomics (see Table 2). The highest non-redundant feature count was identified in the prostate group for Ext-F (n = 52) compared to Ref-B (n = 35) delineations. For the list of high-ranking features per delineation approach in each cohort, see Supplemental: Feature Ranking.
Table 2.
Number of IBSI radiomic features per cohort and per delineation method across feature extraction and data preprocessing steps including redundancy reduction and feature selection. Note that the number of IBSI features per-image extracted was 153 (See Supplemental Table S4); however, features having no variation or having invalid values (e.g., due to low uptake in the given lesion) were removed from the original IBSI features across all four delineation-specific datasets to ensure a unified comparison. IBSI imaging biomarker standardization initiative; Ref-B reference binary; Ref-F reference fuzzy; Ext-B extended binary; Ext-F extended fuzzy delineation
| Cohort | Delineation method | #Features (original) |
#Features (Redundancy reduced) |
#Features (Ranked and selected) |
|---|---|---|---|---|
| Glioma | Ref-B | 153 | 31 | 10 |
| Ref-F | 39 | 10 | ||
| Ext-B | 35 | 10 | ||
| Ext-F | 41 | 10 | ||
| Lung | Ref-B | 153 | 24 | 20 |
| Ref-F | 36 | 20 | ||
| Ext-B | 23 | 20 | ||
| Ext-F | 34 | 20 | ||
| Prostate | Ref-B | 153 | 35 | 9 |
| Ref-F | 42 | 9 | ||
| Ext-B | 35 | 9 | ||
| Ext-F | 52 | 9 |
Effect of delineation on feature ranking and selection
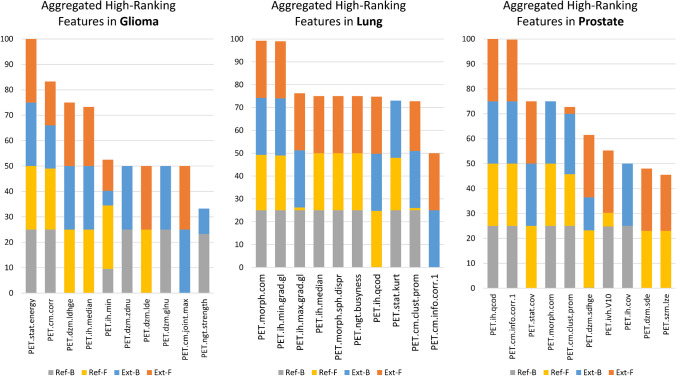
High-ranking feature distributions accumulated across the four delineation types per cohort are shown in Fig. 4. In all cohorts, the highest-ranking aggregate features were also selected as high-ranking across all four delineation types in all cohorts (Fig. 4). Across all cohorts, two features were selected as high-ranking across all four delineations. Three, seven and three additional features were present in three delineation types as high-ranking in glioma, lung and prostate cohorts, respectively.
Fig. 4.
Aggregated high-ranking features in glioma (A), lung (B), and prostate (C) cohorts across radiomics features extracted by four delineation approaches utilized. Feature names are composed of the modality (PET, CT) type, followed by the IBSI-standard feature identified [9]. Feature occurrences (y-axis) are in the range of 0–100 with units of %. Ext-B—extended binary; Ref-B—reference binary; Ext-F—extended fuzzy; Ref-F—reference fuzzy delineation
The per-delineation feature rankings (Supplemental: Feature Ranking), demonstrated a diverse distribution of feature importance across cohorts. Nevertheless, delineations that resulted in high predictive performance also tended to have a more-balanced feature rank distribution compared to those that had a skewed feature ranking distribution (Table 3).
Table 3.
Performance values for glioma, lung and prostate cohorts relying on four delineation approaches to evaluate test cases within the harmonized 100-fold Monte Carlo cross-validation scheme. P-values represent the ANOVA analysis results in-between the reference binary delineation (Ref-B) and the other three delineation methods across the cross-validation area under the receiver operator characteristics curves (AUC). SNS Sensitivity; SPC specificity; PPV positive predictive value; NPV negative predictive value; ACC accuracy; CI confidence interval; Ext-B extended binary; Ref-B reference binary; Ext-F extended fuzzy; Ref-F reference fuzzy delineation. Color scale is normalized between the lowest (white) and the highest (blue) values in each category, where SNS – AUC, AUC CI (±%) and p-value form their own categories. Note that since the test subsets in the cross-validation scheme were balanced and the training subsets underwent class imbalance correction, ACC values reflect on a balanced classifier and they are in line with balanced accuracy (a.k.a. the average SNS and SPC)
Performance evaluation
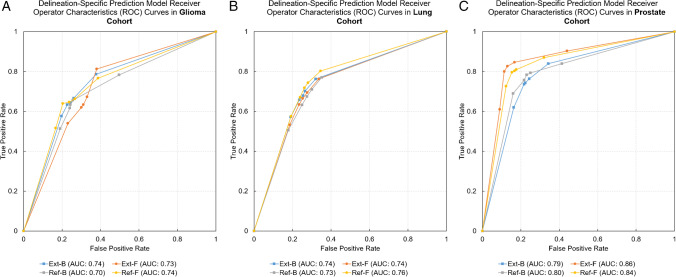
Predicting 3-year survival in the glioma cohort was highest with the Ref-F delineation (AUC: 0.74, ACC: 0.71) vs. all other approaches (AUC: 0.70–0.74, ACC: 0–66-0.70). Similarly, predicting 2-year survival in the lung cohort demonstrated the highest performance with the Ref-F delineation (AUC: 0.76, ACC: 0.73) compared to other approaches (AUC: 0.73–0.74, ACC: 0–70-0.71). Predicting low-vs-high risk in the prostate cohort yielded the highest performance by utilizing Ext-F (AUC: 0.86, ACC: 0.85) and Ref-F (AUC: 0.84, ACC: 0.82) compared to the other approaches (AUC: 0.79–0.80, ACC: 0.78–0.76). See Fig. 5 for the receiver operator characteristics (ROC) curve comparisons of each of the four delineation approaches in the cohorts. See Table 3 for the detailed test confusion matrix results of the four delineation methods in each cohort. ANOVA p-value analyses revealed that the null hypothesis of performance values having no differences cannot be rejected (p-value ranges 0.051–0.809). Nevertheless, in case of prostate, p = 0.051 between Ref-B vs. Ref-F and p = 0.057 between Ref-B vs. Ext-F were near the significance level. In these cases, the performance values were demonstrating the highest differences as well (see Table 3). A similar pattern was visible in case of lung with p = 0.076 between Ref-B vs. Ref-F.
Fig. 5.
Receiver operator characteristics (ROC) curves of cohort-specific prediction models as of their respective delineation methods. A Glioma cohort. B Lung cohort. C Prostate cohort. Ext-B—extended binary; Ref-B—reference binary; Ext-F—extended fuzzy; Ref-F—reference fuzzy delineation; AUC—area under the ROC curve
Aggregated performance increase across volume percentiles revealed that reference fuzzy (Ref-F) delineations outperformed reference binary (Ref-B) delineations within a common volume range across glioma, lung and prostate cohorts with an aggregate AUC of +4.7–10.4. (see Fig. 6). Nevertheless, the least significant differences among the four delineation approaches were present in the lung cohort, which was in line with the overall performance variations in this cohort when considering all its volume ranges (Table 3). The glioma cohort demonstrated that the advantage of fuzzy (Ref-F, Ext-F) delineations over binary ones (Ref-B, Ext-B) was already present in small lesions (~5000 mm3, ~625 voxel count) and further increased afterwards. The prostate cohort revealed that the advantage of fuzzy delineations – Ext-F in particular – became prominent at approx. 8000 mm3 or ~1000 voxel counts within lesions.
Fig. 6.
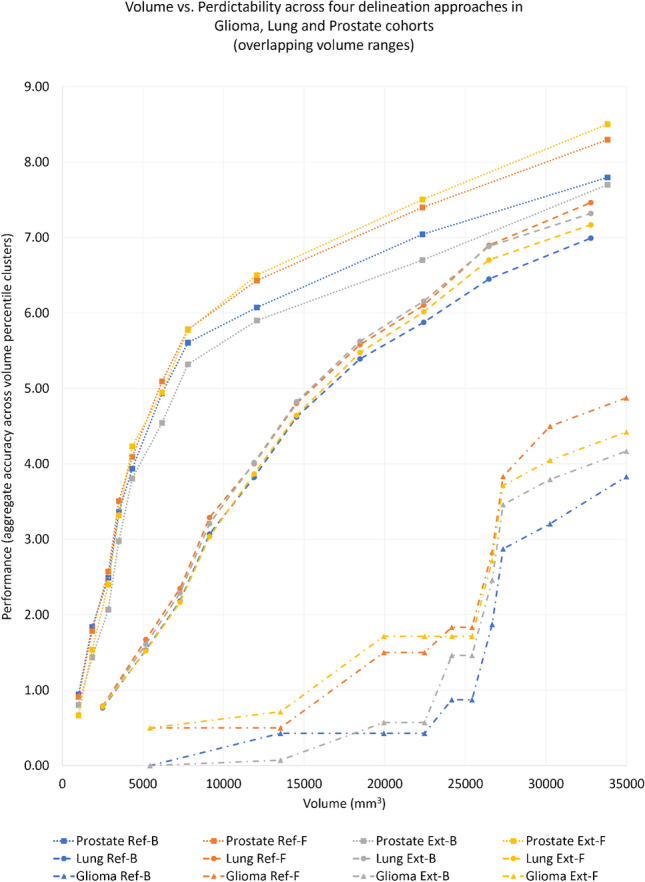
Aggregate performance increase of the four delineation types (Ref-B, Ref-F, Ext-B, Ext-F) across glioma, lung and prostate cohorts categorized to 10 percentile volume clusters within the common volume range of 938–354,987 mm3 or approx. 117–4500 voxels per lesion respectively. Ext-B—extended binary; Ref-B—reference binary; Ext-F—extended fuzzy; Ref-F—reference fuzzy delineation
Discussion
The effect of delineation on radiomic analyses in PET has been extensively investigated. To date, reports regarding reproducibility of predictive performance across various delineation approaches have been inconclusive and often cohort-specific [10, 43]. This study investigated the effect of conventional binary as well as fuzzy radiomics in both lesions and their surroundings by predicting clinically relevant endpoints in PET and hybrid imaging cancer cohorts. Across all cohorts, reference fuzzy (Ref-F) delineations outperformed reference binary (Ref-B) delineations with 3–4% AUC. We consider that this phenomenon is due to the fact that our fuzzy masks were specific to the given PET imaging system's physical resolutions, which allowed the modeling of partial volume effects (PVE) directly in the radiomics calculations. While the ANOVA p-value analysis could not reject the null hypothesis (a.k.a. performance values have no differences), cohort-specific predictive performance variations demonstrated a diverse pattern.
Specifically, the glioma MET-PET cohort had the highest AUC (0.74) and ACC (0.71) with the reference fuzzy (Ref-F) radiomics to predict 3-year survival. The second highest AUC (0.74) and ACC (0.70) was achieved by the and extended binary (Ext-B) delineation. We hypothesize that the relevance of Ext-B in this cohort is due to the infiltrating behavior of glioma [29, 44], which can be better characterized by employing extended binary radiomics.
In the lung 18F-FDG PET/CT cohort, Ext-B and fuzzy (Ext-F) delineations slightly increased predictive performance of 2-year survival, however, the highest performance increase was identifiable with the Ref-F delineation (Ref-F AUC: 0.76 vs. Ref-B AUC: 0.73). Since the PET acquisitions utilized no motion compensation, the reconstructed PET were subjects to motion artefacts [45–48]. Therefore, we argue that the reference delineations were already subjects to overestimation of the true metabolic tumor volumes in this cohort. Consistently, further extending the reference could not significantly increase predictive performance (p = 0.474–0.809). This implies that fuzzy radiomics may not be able to counter-balance motion artefact-related smoothing effects, which is logical, as motion may significantly alter the heterogeneity pattern within tumors, not only at the tumor boundaries [48].
Contrary to the above, the 68GA PSMA-11 PET/MRI study yielded the highest AUC of 0.86 with Ext-F, followed by the AUC of 0.84 with Ref-F delineation against binary delineations (Ext-B AUC: 0.79, Ref-B AUC: 0.80). While the generic superiority of reference fuzzy delineations was consistently demonstrated in this study, the highest performance of Ext-F delineation is considered to be due to the lowest feature redundancy achieved with this delineation. Specifically, the Ref-B delineation resulted in 35 non-redundant features before feature ranking and selection. In contrast, Ref-F and Ext-F resulted in 42 and 52 non-redundant features, respectively. Having a higher number of non-redundant features supports the identification of more high-ranking features, thus, may potentially yield high-performing models. Overall, the highest predictive performance was achieved in the prostate cohort. We consider the following reasons for this phenomenon: First, this cohort utilized a relatively new hybrid camera system and a high PET target resolution (2.08 × 2.08 × 2.03 mm) and here, reference binary delineations relied on full-mount histopathology slices [30, 49]. However, delineation was still performed on the PET images. This means, that in this cohort, the partial volume effect had the most-significant contribution to the delineation of prostate lesions [50]. Cohorts operating with relatively small lesions are more prone to delineation effects than, for example, binning [51]. This is logical, given, that small lesions are also more prone to the PVE [50, 52] or more sensitive to the absence of point-spread function (PSF) modelling [53]. The PVE was most prominent in our prostate cohort as it had the smallest lesions as well (average lesion volume in prostate: 10.9 cm3 vs. 113 cm3 in lung and 93 cm3 in glioma respectively), where a Ref-F delineation resulted in +4% cross-validation AUC. This finding was in line with those from Cysouw et al. [53] who investigated the predictive performance of various delineations in [18F]DCFPyL PET-CT prostate patients in combination with analyzing the effect of partial volume correction. The above findings imply that fuzzy radiomics can be ideal to not only handle delineation uncertainties at lesion edges, but to also model partial volume effects directly in the radiomic calculations themselves. Regardless of lesion size, following EARL guidelines and relying on imaging systems operating with FPS modelling has been proven to generally increase radiomic predictive performance in the context of delineation variations [13, 54, 55].
The aggregate performance analysis across the four delineation methods and cohorts within a common lesion volume range revealed that reference fuzzy (Ref-F) delineations in <35,000 mm3 lesions systematically outperformed the reference binary (Ref-B) delineations in all cohorts. While disease-specific imaging characteristics (e.g., infiltrating behavior) may influence these results, it is important to emphasize that all three cohorts were delineated by different clinicians, thus, our findings may also be subjects to interobserver variability bias. This implies that while fuzzy radiomics on its own has added value compared to conventional binary radiomics—especially in small lesions—future studies shall not exclude the analysis of extended fuzzy or binary regions around lesions within their investigations.
While fuzzy radiomics could naturally model a weighted average of multiple clinician-defined delineations, automated approaches have been repeatedly presented as more robust compared to manually-defined delineations that are prone to multi-observer variabilities [13, 14, 43, 56, 57]. In this regard, the study of Hatt et al. [10] investigated a wide-range of automated PET delineation approaches and concluded that while automated approaches have more accurate delineation's compared to simpler manual or semi-automated ones, the potential magnitude of advantage is mainly specific to the given cohort, the scanner and the imaging protocol. Recently, novel deep learning approaches have been reported to provide highly accurate and automated delineation in a wide range of lesion types [13, 58–62]. In the context of automated, especially DL approaches, we wish to emphasize that this study does not promote a particular fuzzy delineation approach, only the concept of incorporating probability weights into standard radiomics calculations. Deep learning is a naturally probabilistic approach; however, its output delineation is routinely post-processed and further dichotomized by a threshold to analyze the lesions by conventional radiomics afterwards [19, 20, 25, 61]. This step introduces an uncertainty into the dichotomized delineation mask [63–65], and overall, results in information loss. Dichotomization does not only influence analyzed lesion boundaries, but may also excludes lesions with relatively lower DL probabilities, that may otherwise be important for predicting the given clinical endpoint. Fuzzy radiomics on the other hand can organically fit the naturally probabilistic output of DL delineation approaches and can minimize the above uncertainties originated by utilizing thresholds.
Further to the above, fuzzy radiomics systematically decreased redundancy across radiomics features in all three involved cohorts by approximately 20%. Due to the naturally high redundancy of various radiomics features [66, 67], they need to undergo redundancy reduction prior to building machine learning models. Redundancy reduction approaches routinely select one from redundant clusters of features having the highest variance [2]. This, however, does not guarantee that the selected feature is the most predictive. Since fuzzy radiomics decreases redundancy, it may support the identification of precise imaging biomarkers in the future by better discriminating features that are otherwise prone to be redundant. Nevertheless, feature redundancy is a phenomenon which is not only affected by inherently similar radiomic calculations, but also by the volume effect [63, 68] which is feature-specific [17, 33]. In this regard, future studies shall investigate how fuzzy radiomics contributes to volume effects, given, that its contribution to decrease feature redundancy is significant.
When looking at the per-delineation feature ranking, a balanced feature rank distribution of high-ranking features was associated to a higher performance which is in line with prior reports [26, 69, 70]. Nevertheless, our aggregated feature ranking analysis suggests that features being high-ranking across multiple delineation types are able to characterize cohort-specific clinical endpoints, regardless of the chosen delineation type. Therefore, we consider such high-ranking features as robust properties of the given cohort to characterize the given clinical endpoint.
According to our findings, we consider that the advantages of fuzzy radiomics are the results of two phenomena: on the one hand, the ability to model imaging system-specific PVE in the radiomic models allows to handle delineation uncertainties, especially in small lesions. On the other hand, the higher number of non-redundant features increases the likelihood of identifying more high-ranking features for building prediction models when relying on fuzzy radiomics.
This study had limitations, namely, that it only utilized single-center cohorts. Nevertheless, the collected cohorts were from different camera systems and relied on various tracers. In addition, this study relied on a high, 100-fold Monte Carlo (MC) cross-validation scheme to estimate the predictive performance of its models built on its delineations and radiomics evaluations in order to minimize the chances of false discoveries. While we employed train-test splits across our MC folds, we relied on mixed ensemble learning to minimize the effects of bias-variance trade-off and we also avoided variations of hyperparameters that could have skewed differences among the four delineation variations.
Conclusions
Fuzzy radiomics can result in prediction models that outperform conventional binary radiomics-based models, especially in imaging cohorts operating with small lesion sizes. Nevertheless, cohort-specific investigations shall continue to investigate the impact of both fuzzy-vs-binary and lesion-vs-extended lesion volumes in future studies.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank Prof. I. Buvat’s valuable insights and suggestions to help shape the activities and outcomes of this work.
Author contribution
All authors were taking part in the conceptualization, internal review and approval of this paper. Specific contributions are as follows: MG: study design, cohort collection, ML evaluation; CPS: data preprocessing, statistical analysis; DK: data preprocessing, cross-validation; BE: fuzzy radiomics engine development; TTW, SR, KK, MZ, XL, MH, AH: cohort collection, curation, delineation and preparation; LP: study design, radiomics evaluation, supervision.
Funding
Open access funding provided by Medical University of Vienna. This study was funded by the EU grant ERACoSysMed, 4724-B HOLY 2020 (PI: A. Haug).
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
Consent to participate and for publication
Informed consent was obtained from all individual participants included in the study.
Competing interests
The authors declare no competing interests.
Footnotes
This article is part of the Topical Collection on Advanced Image Analyses (Radiomics and Artificial Intelligence)
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.IARC. Latest Global Cancer Data. Press Release N° 263. World Heal Organ [Internet]. 2018;(September):13–5. Available from: http://gco.iarc.fr/.
- 2.Papp L, Spielvogel CP, Rausch I, Hacker M, Beyer T. Personalizing Medicine Through Hybrid Imaging and Medical Big Data Analysis. Front Phys [Internet]. 2018 Jun 7;6. 10.3389/fphy.2018.00051/full.
- 3.Rosenkrantz AB, Friedman K, Chandarana H, Melsaether A, Moy L, Ding Y-S, et al. Current Status of Hybrid PET/MRI in Oncologic Imaging. Am J Roentgenol [Internet]. 2016 Jan;206(1):162–72. 10.2214/AJR.15.14968. [DOI] [PMC free article] [PubMed]
- 4.Kjaer A. Hybrid imaging with PET / CT and PET / MR. Cancer Imaging [Internet]. 2014;14(Suppl 1):O32. 10.1186/1470-7330-14-S1-O32.
- 5.Lee JW, Lee SM. Radiomics in oncological PET/CT: Clinical applications. Nucl Med Mol Imaging (2010) [Internet]. 2018 Oct 20;52:170–89. 10.1007/s13139-017-0500-y. [DOI] [PMC free article] [PubMed]
- 6.Hatt M, Tixier F, Visvikis D, Cheze Le Rest C. Radiomics in PET/CT: More Than Meets the Eye? J Nucl Med [Internet]. 2017 Mar;58(3):365–6. 10.2967/jnumed.116.184655. [DOI] [PubMed]
- 7.Yip SSF, Aerts HJWL. Applications and limitations of radiomics. Phys Med Biol [Internet]. 2016 Jul 7;61(13):R150–66. 10.1088/0031-9155/61/13/R150. [DOI] [PMC free article] [PubMed]
- 8.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology [Internet]. 2016 Feb;278(2):563–77. 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed]
- 9.Zwanenburg A, Leger S, Vallières M, Löck S, Initiative for the IBS. Image biomarker standardisation initiative. arXiv [Internet]. 2016;(November). Available from: http://arxiv.org/abs/1612.07003.
- 10.Hatt M, Lee JA, Schmidtlein CR, El Naqa I, Caldwell C, De Bernardi E, et al. Classification and evaluation strategies of auto-segmentation approaches for PET: Report of AAPM task group No. 211. Med Phys [Internet]. 2017 Jun;44(6):e1–42. 10.1002/mp.12124. [DOI] [PMC free article] [PubMed]
- 11.Carles M, Torres-Espallardo I, Alberich-Bayarri A, Olivas C, Bello P, Nestle U, et al. Evaluation of PET texture features with heterogeneous phantoms: Complementarity and effect of motion and segmentation method. Phys Med Biol [Internet]. 2017;62(2):652–68. 10.1088/1361-6560/62/2/652. [DOI] [PubMed]
- 12.Beichel RR, Smith BJ, Bauer C, Ulrich EJ, Ahmadvand P, Budzevich MM, et al. Multi-site quality and variability analysis of 3D FDG PET segmentations based on phantom and clinical image data: Med Phys [Internet]. 2017 Feb;44(2):479–96. 10.1002/mp.12041. [DOI] [PMC free article] [PubMed]
- 13.Hatt M, Krizsan AK, Rahmim A, Bradshaw TJ, Costa PF, Forgacs A, et al. Joint EANM/SNMMI guideline on radiomics in nuclear medicine. Eur J Nucl Med Mol Imaging [Internet]. 2023 Jan 3;50(2):352–75. 10.1007/s00259-022-06001-6. [DOI] [PMC free article] [PubMed]
- 14.Hatt M, Cheze le Rest C, Descourt P, Dekker A, De Ruysscher D, Oellers M, et al. Accurate Automatic Delineation of Heterogeneous Functional Volumes in Positron Emission Tomography for Oncology Applications. Int J Radiat Oncol Biol Phys [Internet]. 2010 May;77(1):301–8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20116934. [DOI] [PubMed]
- 15.Layer T, Blaickner M, Knäusl B, Georg D, Neuwirth J, Baum RP, et al. PET image segmentation using a Gaussian mixture model and Markov random fields. EJNMMI Phys [Internet]. 2015;2(1):1–15. 10.1186/s40658-015-0110-7. [DOI] [PMC free article] [PubMed]
- 16.Hatt M, Laurent B, Ouahabi A, Fayad H, Tan S, Li L, et al. The first MICCAI challenge on PET tumor segmentation. Med Image Anal [Internet]. 2018 Feb;44:177–95. 10.1016/j.media.2017.12.007. [DOI] [PMC free article] [PubMed]
- 17.Nyflot MJ, Yang F, Byrd D, Bowen SR, Sandison GA, Kinahan PE. Quantitative radiomics: impact of stochastic effects on textural feature analysis implies the need for standards. J Med imaging (Bellingham, Wash) [Internet]. 2015 Oct 5;2(4):041002. 10.1117/1.JMI.2.4.041002. [DOI] [PMC free article] [PubMed]
- 18.Pérez-Morales J, Tunali I, Stringfield O, Eschrich SA, Balagurunathan Y, Gillies RJ, et al. Peritumoral and intratumoral radiomic features predict survival outcomes among patients diagnosed in lung cancer screening. Sci Rep [Internet]. 2020 Dec 29;10(1):10528. 10.1038/s41598-020-67378-8. [DOI] [PMC free article] [PubMed]
- 19.Moe YM, Groendahl AR, Tomic O, Dale E, Malinen E, Futsaether CM. Deep learning-based auto-delineation of gross tumour volumes and involved nodes in PET/CT images of head and neck cancer patients. Eur J Nucl Med Mol Imaging [Internet]. 2021 Aug 9;48(9):2782–92. 10.1007/s00259-020-05125-x [DOI] [PMC free article] [PubMed]
- 20.Shiri I, Arabi H, Sanaat A, Jenabi E, Becker M, Zaidi H. Fully Automated Gross Tumor Volume Delineation From PET in Head and Neck Cancer Using Deep Learning Algorithms. Clin Nucl Med [Internet]. 2021 Nov;46(11):872–83. 10.1097/RLU.0000000000003789. [DOI] [PubMed]
- 21.Arabi H, Shiri I, Jenabi E, Becker M, Zaidi H. Deep Learning-based Automated Delineation of Head and Neck Malignant Lesions from PET Images. In: 2020 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC) [Internet]. IEEE; 2020. p. 1–3. 10.1109/NSS/MIC42677.2020.9507977.
- 22.Capobianco N, Sibille L, Chantadisai M, Gafita A, Langbein T, Platsch G, et al. Whole-body uptake classification and prostate cancer staging in 68Ga-PSMA-11 PET/CT using dual-tracer learning. Eur J Nucl Med Mol Imaging [Internet]. 2022 Jan 7;49(2):517–26. 10.1007/s00259-021-05473-2. [DOI] [PMC free article] [PubMed]
- 23.Hatt M, Tixier F, Cheze Le Rest C, Pradier O, Visvikis D. Robustness of intratumour18F-FDG PET uptake heterogeneity quantification for therapy response prediction in oesophageal carcinoma. Eur J Nucl Med Mol Imaging [Internet]. 2013 Oct;40(11):1662–71. 10.1007/s00259-013-2486-8. [DOI] [PubMed]
- 24.Papp L, Rausch I, Hacker M, Beyer T. Fuzzy Radiomics: A novel approach to minimize the effects of target delineation on radiomic models. In 2019. 10.1055/s-0039-1683478.
- 25.Andrearczyk V, Oreiller V, Boughdad S, Rest CC Le, Elhalawani H, Jreige M, et al. Overview of the HECKTOR Challenge at MICCAI 2021: Automatic Head and Neck Tumor Segmentation and Outcome Prediction in PET/CT Images. In 2022. p. 1–37. 10.1007/978-3-030-98253-9_1.
- 26.Papp L, Poetsch N, Grahovac M, Schmidbauer V, Woehrer A, Preusser M, et al. Glioma survival prediction with the combined analysis of in vivo 11C-MET-PET, ex vivo and patient features by supervised machine learning. J Nucl Med [Internet]. 2017;59(6):jnumed.117.202267. 10.2967/jnumed.117.202267. [DOI] [PubMed]
- 27.Papp L, Spielvogel CP, Grubmüller B, Grahovac M, Krajnc D, Ecsedi B, et al. Supervised machine learning enables non-invasive lesion characterization in primary prostate cancer with [68Ga]Ga-PSMA-11 PET/MRI. Eur J Nucl Med Mol Imaging [Internet]. 2020 Dec 19. 10.1007/s00259-020-05140-y. [DOI] [PMC free article] [PubMed]
- 28.Zhao M, Kluge K, Papp L, Grahovac M, Yang S, Jiang C, et al. Multi-lesion radiomics of PET/CT for non-invasive survival stratification and histologic tumor risk profiling in patients with lung adenocarcinoma. Eur Radiol [Internet]. 2022 Jul 28;32(10):7056–67. 10.1007/s00330-022-08999-7. [DOI] [PubMed]
- 29.Poetsch N, Woehrer A, Gesperger J, Furtner J, Haug AR, Wilhelm D, et al. Visual and semiquantitative 11C-methionine PET: an independent prognostic factor for survival of newly diagnosed and treatment-naïve gliomas. Neuro Oncol [Internet]. 2018 Feb 19;20(3):411–9. 10.1093/neuonc/nox177. [DOI] [PMC free article] [PubMed]
- 30.Hartenbach M, Hartenbach S, Bechtloff W, Danz B, Kraft K, Klemenz B, et al. Combined PET/MRI improves diagnostic accuracy in patients with prostate cancer: A prospective diagnostic trial. Clin Cancer Res [Internet]. 2014 Jun 15;20(12):3244–53. 10.1158/1078-0432.CCR-13-2653. [DOI] [PubMed]
- 31.Orlhac F, Nioche C, Klyuzhin I, Rahmim A, Buvat I. Radiomics in PET Imaging. PET Clin [Internet]. 2021 Oct;16(4):597–612. 10.1016/j.cpet.2021.06.007. [DOI] [PubMed]
- 32.Krajnc D, Papp L, Nakuz TS, Magometschnigg HF, Grahovac M, Spielvogel CP, et al. Breast Tumor Characterization Using [18F]FDG-PET/CT Imaging Combined with Data Preprocessing and Radiomics. Cancers (Basel) [Internet]. 2021;13(6). 10.3390/cancers13061249. [DOI] [PMC free article] [PubMed]
- 33.Papp L, Rausch I, Grahovac M, Hacker M, Beyer T. Optimized Feature Extraction for Radiomics Analysis of 18 F-FDG PET Imaging. J Nucl Med [Internet]. 2019 Jun;60(6):864–72. 10.2967/jnumed.118.217612. [DOI] [PubMed]
- 34.Stytz MR, Parrott RW. Using kriging for 3d medical imaging. Comput Med Imaging Graph. 1993;17(6):421–442. doi: 10.1016/0895-6111(93)90059-V. [DOI] [PubMed] [Google Scholar]
- 35.Parmar C, Leijenaar RTH, Grossmann P, Velazquez ER, Bussink J, Rietveld D, et al. Radiomic feature clusters and Prognostic Signatures specific for Lung and Head &neck cancer. Sci Rep [Internet]. 2015 Sep 5;5(1):11044. 10.1038/srep11044. [DOI] [PMC free article] [PubMed]
- 36.Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, et al. Radiomics: The process and the challenges. Magn Reson Imaging [Internet]. 2012;30(9):1234–48. 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed]
- 37.Pfaehler E, Mesotten L, Zhovannik I, Pieplenbosch S, Thomeer M, Vanhove K, et al. Plausibility and redundancy analysis to select FDG‐PET textural features in non‐small cell lung cancer. Med Phys [Internet]. 2021 Mar 6;48(3):1226–38. 10.1002/mp.14684. [DOI] [PMC free article] [PubMed]
- 38.Amin A, Anwar S, Adnan A, Nawaz M, Howard N, Qadir J, et al. Comparing Oversampling Techniques to Handle the Class Imbalance Problem: A Customer Churn Prediction Case Study. IEEE Access. 2016;4(October):7940–7957. doi: 10.1109/ACCESS.2016.2619719. [DOI] [Google Scholar]
- 39.Lambin P, Leijenaar RTH, Deist TM, Peerlings J, De Jong EEC, Van Timmeren J, et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–62. 10.1038/nrclinonc.2017.141. [DOI] [PubMed]
- 40.Ross KA, Jensen CS, Snodgrass R, Dyreson CE, Jensen CS, Snodgrass R, et al. Curse of Dimensionality. In: Encyclopedia of Database Systems [Internet]. Boston, MA: Springer US; 2009. p. 545–6. 10.1007/978-0-387-39940-9_133.
- 41.Belkin M, Hsu D, Ma S, Mandal S. Reconciling modern machine-learning practice and the classical bias–variance trade-off. Proc Natl Acad Sci [Internet]. 2019 Aug 6;116(32):15849–54. 10.1073/pnas.1903070116. [DOI] [PMC free article] [PubMed]
- 42.Breiman, L. Random Forests. Machine Learning. 2001;45:5–32. 10.1023/A:1010933404324.
- 43.Cook GJR, Azad G, Owczarczyk K, Siddique M, Goh V. Challenges and Promises of PET Radiomics. Int J Radiat Oncol Biol Phys [Internet]. 2018;102(4):1083–9. 10.1016/j.ijrobp.2017.12.268. [DOI] [PMC free article] [PubMed]
- 44.Vigneswaran K, Neill S, Hadjipanayis CG. Beyond the World Health Organization grading of infiltrating gliomas: advances in the molecular genetics of glioma classification. Ann Transl Med [Internet]. 2015 May;3(7):95. 10.3978/j.issn.2305-5839.2015.03.57. [DOI] [PMC free article] [PubMed]
- 45.Constanzo J, Wei L, Tseng H-H, El Naqa I. Radiomics in precision medicine for lung cancer. Transl Lung Cancer Res [Internet]. 2017 Dec;6(6):635–47. 10.21037/tlcr.2017.09.07. [DOI] [PMC free article] [PubMed]
- 46.Osman MM, Cohade C, Nakamoto Y, Wahl RL. Respiratory motion artifacts on PET emission images obtained using CT attenuation correction on PET-CT. Eur J Nucl Med Mol Imaging [Internet]. 2003 Apr 21;30(4):603–6. 10.1007/s00259-002-1024-x [DOI] [PubMed]
- 47.Du Q, Baine M, Bavitz K, McAllister J, Liang X, Yu H, et al. Radiomic feature stability across 4D respiratory phases and its impact on lung tumor prognosis prediction. Lee M-C, editor. PLoS One [Internet]. 2019 May 7;14(5):e0216480. 10.1371/journal.pone.0216480. [DOI] [PMC free article] [PubMed]
- 48.Ha S, Choi H, Paeng JC, Cheon GJ. Radiomics in Oncological PET/CT: a Methodological Overview. Nucl Med Mol Imaging (2010) [Internet]. 2019 Feb 15;53(1):14–29. 10.1007/s13139-019-00571-4. [DOI] [PMC free article] [PubMed]
- 49.Grubmüller B, Baltzer P, Hartenbach S, D’Andrea D, Helbich TH, Haug AR, et al. PSMA Ligand PET/MRI for Primary Prostate Cancer: Staging Performance and Clinical Impact. Clin Cancer Res [Internet]. 2018 Dec 15;24(24):6300–7. 10.1158/1078-0432.CCR-18-0768. [DOI] [PubMed]
- 50.Hatt M, Le Rest CC, Tixier F, Badic B, Schick U, Visvikis D. Radiomics: Data Are Also Images. J Nucl Med [Internet]. 2019 Sep 3;60(Supplement 2):38S-44S. 10.2967/jnumed.118.220582. [DOI] [PubMed]
- 51.van Velden FHP, Kramer GM, Frings V, Nissen IA, Mulder ER, de Langen AJ, et al. Repeatability of Radiomic Features in Non-Small-Cell Lung Cancer [18F]FDG-PET/CT Studies: Impact of Reconstruction and Delineation. Mol Imaging Biol [Internet]. 2016 Oct 26;18(5):788–95. 10.1007/s11307-016-0940-2. [DOI] [PMC free article] [PubMed]
- 52.Soret M, Bacharach SL, Buvat I. Partial-Volume Effect in PET Tumor Imaging. J Nucl Med [Internet]. 2007;48(6):932–45. 10.2967/jnumed.106.035774. [DOI] [PubMed]
- 53.Cysouw MCF, Jansen BHE, van de Brug T, Oprea-Lager DE, Pfaehler E, de Vries BM, et al. Machine learning-based analysis of [18F]DCFPyL PET radiomics for risk stratification in primary prostate cancer. Eur J Nucl Med Mol Imaging [Internet]. 2020 Jul 31. 10.1007/s00259-020-04971-z. [DOI] [PMC free article] [PubMed]
- 54.Pfaehler E, Beukinga RJ, Jong JR, Slart RHJA, Slump CH, Dierckx RAJO, et al. Repeatability of 18 F‐ FDG PET radiomic features: A phantom study to explore sensitivity to image reconstruction settings, noise, and delineation method. Med Phys [Internet]. 2019 Feb 28;46(2):665–78. 10.1002/mp.13322. [DOI] [PMC free article] [PubMed]
- 55.Lasnon C, Enilorac B, Popotte H, Aide N. Impact of the EARL harmonization program on automatic delineation of metabolic active tumour volumes (MATVs). EJNMMI Res [Internet]. 2017 Dec 31;7(1):30. 10.1186/s13550-017-0279-y. [DOI] [PMC free article] [PubMed]
- 56.Hatt M, Visvikis D, Albarghach NM, Tixier F, Pradier O, Cheze-Le RC. Prognostic value of18F-FDG PET image-based parameters in oesophageal cancer and impact of tumour delineation methodology. Eur J Nucl Med Mol Imaging. 2011;38(7):1191–1202. doi: 10.1007/s00259-011-1755-7. [DOI] [PubMed] [Google Scholar]
- 57.Bashir U, Azad G, Siddique MM, Dhillon S, Patel N, Bassett P, et al. The effects of segmentation algorithms on the measurement of 18F-FDG PET texture parameters in non-small cell lung cancer. EJNMMI Res [Internet]. 2017 Dec;7(1):60. 10.1186/s13550-017-0310-3. [DOI] [PMC free article] [PubMed]
- 58.Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods [Internet]. 2021 Feb 7;18(2):203–11. 10.1016/j.media.2020.101874. [DOI] [PubMed]
- 59.Cho J, Park K-S, Karki M, Lee E, Ko S, Kim JK, et al. Improving Sensitivity on Identification and Delineation of Intracranial Hemorrhage Lesion Using Cascaded Deep Learning Models. J Digit Imaging [Internet]. 2019 Jun 24;32(3):450–61. 10.1007/s10278-018-00172-1. [DOI] [PMC free article] [PubMed]
- 60.Mou L, Zhao Y, Fu H, Liu Y, Cheng J, Zheng Y, et al. CS2-Net: Deep learning segmentation of curvilinear structures in medical imaging. Med Image Anal [Internet]. 2021 Jan;67:101874. 10.1016/j.media.2020.101874. [DOI] [PubMed]
- 61.Capobianco N, Meignan MA, Cottereau A-S, Vercellino L, Sibille L, Spottiswoode B, et al. Deep learning FDG uptake classification enables total metabolic tumor volume estimation in diffuse large B-cell lymphoma. J Nucl Med [Internet]. 2020 Jun 12;jnumed.120.242412. 10.2967/jnumed.120.242412. [DOI] [PMC free article] [PubMed]
- 62.Papadimitroulas P, Brocki L, Christopher Chung N, Marchadour W, Vermet F, Gaubert L, et al. Artificial intelligence: Deep learning in oncological radiomics and challenges of interpretability and data harmonization. Phys Medica [Internet]. 2021 Mar;83:108–21. 10.1016/j.ejmp.2021.03.009. [DOI] [PubMed]
- 63.Boellaard R, Krak NC, Hoekstra OS, Lammertsma AA. Effects of Noise, Image Resolution, and ROI Definition on the Accuracy of Standard Uptake Values: A Simulation Study. J Nucl Med [Internet]. 2004;45(9):1519–27. Available from: http://jnm.snmjournals.org/cgi/content/abstract/45/9/1519. [PubMed]
- 64.Parmar C, Velazquez ER, Leijenaar R, Jermoumi M, Carvalho S, Mak RH, et al. Robust radiomics feature quantification using semiautomatic volumetric segmentation. Woloschak GE, editor. PLoS One [Internet]. 2014 Jul 15;9(7):e102107. 10.1371/journal.pone.0102107. [DOI] [PMC free article] [PubMed]
- 65.Galavis PE, Hollensen C, Jallow N, Paliwal B, Jeraj R. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncol (Madr) [Internet]. 2010 Oct 13;49(7):1012–6. 10.3109/0284186X.2010.498437. [DOI] [PMC free article] [PubMed]
- 66.Lu L, Ehmke RC, Schwartz LH, Zhao B. Assessing Agreement between Radiomic Features Computed for Multiple CT Imaging Settings. Tian J, editor. PLoS One [Internet]. 2016 Dec 29;11(12):e0166550. 10.1371/journal.pone.0166550. [DOI] [PMC free article] [PubMed]
- 67.Parmar C, Grossmann P, Rietveld D, Rietbergen MM, Lambin P, Aerts HJWL. Radiomic Machine-Learning Classifiers for Prognostic Biomarkers of Head and Neck Cancer. Front Oncol [Internet]. 2015;5. 10.3389/fonc.2015.00272/abstract. [DOI] [PMC free article] [PubMed]
- 68.Shah B, Srivastava N, Hirsch AE, Mercier G, Subramaniam RM. Intra-reader reliability of FDG PET volumetric tumor parameters: Effects of primary tumor size and segmentation methods. Ann Nucl Med. 2012;26(9):707–714. doi: 10.1007/s12149-012-0630-3. [DOI] [PubMed] [Google Scholar]
- 69.Bolón-Canedo V, Sechidis K, Sánchez-Maroño N, Alonso-Betanzos A, Brown G. Insights into distributed feature ranking. Inf Sci (Ny) [Internet]. 2019 Sep;496:378–98. 10.1016/j.ins.2018.09.045.
- 70.Dougherty E, Hua J, Sima C. Performance of Feature Selection Methods. Curr Genomics [Internet]. 2009 Sep 1;10(6):365–74. 10.2174/138920209789177629. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.