Abstract
Human induced pluripotent stem cell (iPSC)-derived neurons are being increasingly used for high content imaging and screening. However, iPSC-derived neuronal differentiation and maturation is time-intensive, often requiring >8 weeks. Unfortunately, the differentiating and maturing iPSC-derived neuronal cultures also tend to migrate and coalesce into ganglion-like clusters making single-cell analysis challenging, especially in miniaturized formats. Using our defined extracellular matrix and low oxygen culturing conditions for the differentiation and maturation of human cortical neurons, we further modified neuronal progenitor cell seeding densities and feeder layer-free culturing conditions in miniaturized formats (i.e., 96 well) to decrease neuronal clustering, enhance single-cell identification and reduce edge effects usually observed after extended neuronal cell culture. Subsequent algorithm development refined capabilities to distinguish and identify single mature neurons, as identified by NeuN expression, from large cellular aggregates, which were excluded from image analysis. Incorporation of astrocyte conditioned medium during differentiation and maturation periods significantly increased the percentage (i.e., ~10% to ~30%) of mature neurons (i.e., NeuN+) detected at 4-weeks post-differentiation. Pilot, proof of concept studies using this optimized assay system yielded negligible edge effects and robust Z-factors in population-based as well as image-based neurotoxicity assay formats. Moreover, moxidectin, an FDA-approved drug with documented neurotoxic adverse effects, was identified as a hit using both screening formats. This miniaturized, feeder layer-free format and image analysis algorithm provides a foundational imaging and screening platform, which enables quantitative single-cell analysis of differentiated human neurons.
Keywords: Neuron, Feeder layer-free, Algorithm, NeuN, Edge effects, Miniaturization
Introduction
Neurological diseases comprise a vast array of disorders impacting the brain and nervous system. With aging worldwide human populations, neurological and neurodegenerative disorders are expected to increase in prevalence [1–3]. We are most familiar with non-communicable neurological and neurodegenerative diseases such as migraines, epilepsy, multiple sclerosis and Alzheimer’s Disease [1]; however, the COVID19 pandemic highlights the fact that infectious or viral diseases also may have detrimental neurological effects [4–6]. While some of the neurological effects of SARS-CoV-2 infection have been identified (e.g., anosmia, dysgeusia, seizures, confusion), their long term ramifications are not yet understood and SARS-CoV-2 infection may increase the risk of dementia [7], nerve damage [8] and stroke [9,10]. The COVID19 pandemic and the recent clinical failures of neurologically targeted drugs underscores the need for new model systems to study neurological disorders and to discover new therapeutics.
Human induced pluripotent stem cell-derived (iPSC) neurons are increasingly being used to establish models of neurological processes in healthy and disease states [11,12]. This is mainly because of their ability to recapitulate – at the cellular level – specific aspects of neurodevelopment and neurodegenerative/neurological disorders previously only detected in patient-derived samples and/or in vivo models. Hence, iPSC-derived neuron models, including organoids, “brain on a chip”, neurospheres, and various two/three dimensional systems are forming key components of neurophysiologically relevant model systems to understand, for example, neurite outgrowth, neurogenesis, synaptogenesis and neural network generation [13–16]. However, miniaturization of iPSC-derived neuron-based systems is challenging because of the required extended differentiation and maturation times [17,18], the tendency for the neurons to cluster into large groups and the asynchronous differentiation and maturation of the neuronal cultures. Moreover, full physiological maturation of iPSC-derived neurons most likely requires co-culturing (i.e., feeder layer) with astrocytes, or other cell populations, further complicating miniaturization and standardization efforts [17].
In the studies presented here, we sought to develop a miniaturized, feeder layer-free iPSC-derived human cortical neuron assay system, specifically designed to identify cortical neurons expressing the NeuN maturity marker (i.e., mature neurons) from an asynchronously matured “mixed” neuron culture. Using our imaging platform, which is compatible with high content applications, and feeder layer-free neuronal progenitor cells (NPCs) derived from healthy human patients, we have further refined iPSC-derived NPC seeding, handling and culturing conditions to minimize neuronal clustering associated with neuronal maturation. Moreover, we have developed an imaging-based algorithm to identify and quantify single iPSC-derived cortical neurons from a mixed neuron culture comprising large cellular aggregates and more dispersed neurons. Our miniaturized assay system may form the basis for neurological disease modeling and improve mature neuron-focused (i.e., NeuN+) screening-based studies.
Materials and methods
Derivation of neuronal progenitor cells (NPCs) from induced pluripotent stem cells (iPSCs) and NPC culturing procedures.
Original fibroblast lines for iPSC derivation were purchased from the NIGMS Human Genetic Cell Repository (Coriell Institute for Medical Research, Camden, NJ) by the University of Virginia (UVA) Stem Cell Core. Subsequently, iPSC-derived NPC lines were generated by and obtained from the UVA Stem Cell Core (Charlottesville, VA), as previously described [19]. Briefly, a UVA Stem Cell Core established iPSC line was used to generate CNS-associated neuronal progenitor cells (NPCs) using the STEMdiff™ SMADi inhibition-mediated Neural Induction Kit (StemCell Technologies, Cambridge, MA) and following the manufacturer’s monolayer culturing protocol. iPSCs were resuspended in STEMdiff Neural Induction Medium (NIM) supplemented with (SMADi) Neural Induction Supplement (NIS) and Y-27632 ROCK inhibitors. Cells were plated (2×106 cells/well) on a Matrigel™-coated 6-well plate with daily medium changes using STEMdiff NIM containing SMADi NIS. At passage 3, resulting NPCs were then switched to NPC medium (DMEM/F12 supplemented with Glutamax, N2 (2%), B27 without vitamin A (1%), mouse laminin (1 μg/mL), and human FGF (20 ng/mL)). Per standard UVA Stem Cell Core’s quality control criteria, cells displayed an NPC-like morphology and robustly expressed the NPC markers Pax6, Sox2, and Nestin and had no expression of the iPSC marker Oct4, and negligible expression of Doublecortin and Map2 as determined by immunofluorescence (IF).
Confirmed NPC populations were routinely maintained on Matrigel™-coated dishes in NPC medium [i.e., DMEM-F12 supplemented with Glutamax (Gibco, Gaithersburg, MD), 2% B27 without vitamin A (Gibco) and 20 ng/mL thermostable recombinant human Fibroblast Growth Factor (Gibco)]. NPC lines were maintained in 5% CO2 and environmental O2 (~21%) at 37°C. NPC lines were subcultured (~1:4) approximately once per week (up to passage 15) using Accutase (MilliporeSigma, St. Louis, MO).
Poly-L-ornithine/laminin coating of tissue culture vessels for iPSC-derived neuronal differentiation and maturation.
All tissue culture plates were coated with 50 μg/mL poly-L-ornithine (Sigma Aldrich) for 24 h at 4°C then rinsed four times with sterile water. Mouse laminin (25 μg/mL; Gibco) was then added and incubated at 37°C for 4 h. Laminin solution was removed prior to NPC seeding.
Differentiation and maturation into feeder layer-free iPSC-derived cortical neuron mixed culture.
iPSC-derived neuronal differentiation and maturation was performed under physiological O2 for neurons (5%) as previously described [19]. NPCs were seeded onto poly-L-ornithine/laminin-coated cyclic olefin co-polymer-based 96-well Cell-Carrier Ultra microtiter plates (PerkinElmer, Waltham, MA) at 8.5×102 cells/well in 50 μL of NPC medium unless otherwise stated. At 24 h post-seeding, the NPC maintenance medium was aspirated and replaced with 50 μL neuron differentiation medium [DMEM/F12+Glutamax, 2% B27 without insulin, 20 ng/mL brain derived neurotrophic factor (Shenandoah Biotechnology, Warwick, PA), 20 ng/mL glial cell derived neurotrophic factor (Shenandoah Biotechnology), 1 mM dibutyryl cyclic adenosine monophosphate (MilliporeSigma), 200 nM ascorbic acid (Millipore Sigma), 1 μg/mL mouse laminin (Invitrogen, Carlsbad), 100 ng/mL recombinant human insulin (Gibco) and penicillin (100 units/mL)-streptomycin (100 μg/mL) (Thermo Fisher Scientific, Waltham, MA)]. After 24 h, 50 μL of BrainPhys base medium supplemented with the above listed medium supplements was added to each well resulting in a 50:50 mixture (100 μL) of DMEM/F12 and BrainPhys base medium containing medium supplements. After an additional 24 h, the medium was replaced with 100 μL of 100% BrainPhys supplemented (as listed above) medium. iPSC-derived neuronal cultures received twice weekly 50% medium changes until experimentation. BrainPhys base medium [20] was obtained from StemCell Technologies (Cambridge, MA). Astrocyte conditioned medium was obtained from ScienCell Research Laboratories (Carlsbad, CA).
Antibodies and other reagents.
See Table 1 for primary and secondary antibody details. The nuclear dye, 4′6-diamidino-2-phenylindole (DAPI), was obtained from Life Technologies (Eugene, OR).
Table 1.
Antibodies used for immunofluorescence.
| Protein target | Vendor | Catalog number | Concentration (μg/mL) | Application |
|---|---|---|---|---|
| NeuN (RBFOX3) | Sigma | HPA030790 | 0.4 μg/mL | IF |
| Ki67-Alexa 488 | Cell Signaling | 11882 | 0.5 μg/mL | IF |
| Anti-Rabbit Alexa 647 | Cell Signaling | 4414 | 2 μg/mL | IF |
iPSC-derived neuron fixation, immunofluorescence (IF) and image acquisition.
Cells were fixed and processed for IF antibody staining as previously described [19]. The PerkinElmer Operetta CLS high content imaging system and its associated Harmony software package, located in the UVA Advanced Microscopy Core Facility, were used for image acquisition, algorithm development and analyses. All images were captured with a 20X water-immersion objective at a single focal plane in non-confocal mode. For whole plate analyses of nuclei counts per well, images were captured using Hoechst 33342 (channel 1) and 10 regions of interest (ROIs) per well. For analyses of neuronal markers, images were captured using Hoechst 33342 (channel 1), Alexa 488 (channel 2), and Alexa 647 (channel 3). Twenty-five ROIs per well were captured from 4-6 wells per group. The PerkinElmer Harmony 4.9 software was used to analyze all acquired images.
Drug library, compound dilution and pilot screening.
The Discovery probe-FDA approved drug library (2,100 compounds) was purchased from APExBIO (Boston, MA). Select compound plates were evaluated for pilot testing purposes. For drug dilution, 2 μL of 10 mM drug in 100% DMSO was diluted in 30 μL complete neuron culture medium, generating a 625 μM working concentration. An intermediate drug plate (i.e., 250 μM) was generated by transferring 16 μL of the 625 μM stock to 24 μL of complete neuron culture medium. The final drug screening concentration was 10 μM with a constant DMSO concentration of 0.1% in each assay well in 50 μL. Negative (vehicle) controls contained 0.1% DMSO, and positive neurotoxic controls contained 10% DMSO or 1% Triton X-100. Assay plates were allowed to incubate for 24 h at 37°C in the presence of 5% O2/5% CO2. 10 μL of CellTiter-Blue reagent (Promega, Madison, WI) were added to each assay plate well and incubated for 2 h. Data were captured on a Molecular Devices SpectraMax M5 (excitation560; emission590). Plates were then fixed and stained with DAPI as described above. Individual assay plate Z-factors were derived from the vehicle and positive controls. Primary actives were defined as compounds displaying ≥50% inhibition of signal readout.
Statistical Analysis.
Data were analyzed using GraphPad Prism 9.3 (La Jolla, CA). Data are presented as average (mean) ± standard deviation or standard error of the mean. p Values were calculated with Student’s t-test for comparisons involving 2 groups or one-way or two-way analysis of variance for comparisons involving >2 groups. p < 0.05 was considered statistically significant.
Results
NPC seeding optimization in miniaturized formats.
Differentiating and maturing iPSC-derived neuron cultures asynchronously proliferate, migrate and have the propensity to coalesce into ganglion-like clusters or neuronal cell bodies. This is not unexpected as neurons progress through multiple progenitor stages prior to maturation, and neuronal migration or locomotion is critical for the developing brain (reviewed in [21]). Unfortunately, this process makes single cell analysis challenging especially in miniaturized formats. Previously, using a 1,500-2,000 NPC seeding density in a 96 well format, we observed that neuronal coalescence was most noticeable after the rapid expansion of the NPC population post-differentiation (i.e., between weeks 2 and 3) (Fig. 1 and Supplemental Figure 1) [19]. Using our optimized extracellular matrix and low oxygen culturing conditions, we further modified NPC seeding densities and feeder layer-free culturing conditions in miniaturized formats to decrease cellular clustering and enhance the ability to distinguish individual nuclei. Thus, we reduced NPC seeding densities (i.e., 850, 1,000 and 1,500) in 96 well microtiter plates and initiated neuronal differentiation and maturation via medium changes as previously described [19,22]. Cellular clustering was most observable at the 1,000 and 1,500 NPC seeding densities at 4 weeks post-differentiation. By comparison the 850 NPC seeding density per well exhibited reduced cellular clustering (at 4 weeks post-differentiation) and enabled the identification of single cells for subsequent analysis (Fig. 1). Therefore, we focused on an 850 NPC per well seeding density for subsequent single cell imaging and quantification studies.
Fig. 1.

Reduction in neuronal clustering by lowering NPC seeding densities under feeder layer-free culturing conditions. Comparison of 850, 1000 and 1500 NPC seeding densities at 4 weeks post-differentiation. Global images based on DAPI+ staining from 9 fields of view are shown. Scale bar=500 μm.
Imaging algorithm development for single cell analysis.
Modifying microtiter plate handling and cell culturing conditions reduced, but did not eliminate, cellular clustering and unfortunately, fluorescence-based analyses are “intensity readout-biased” towards large cellular aggregates, increasing assay variability and hindering the ability to focus on single cells at lower fluorescent intensities. Unfortunately, available established algorithms were not adaptable to our study needs since they could not effectively remove large neuronal clusters from analyses. Thus, to identify single iPSC-derived neuronal cells, we created an image analysis algorithm (Supplemental Table 1) using the Perkin Elmer Operetta-associated Harmony software package that allowed us to computationally identify and remove large neuronal clusters from image analysis processes enabling single cell analysis (Fig. 2; Supplemental Table 1 for image processing steps). Cell nuclei were first identified using Hoechst 3334219 (ch 1) then well images were captured (Fig. 2, panel A). Within each well, a contiguous set of 25 individual fields of view were stitched together to create a global image for the well. The global input DAPI stained image was then inverted (cut-off quantile=100; Fig. 2, panel B) and a Gaussian smoothing filter (i.e., width 6 pixels) was applied to the inverted global DAPI image (Fig. 2, panel C). Regions comprising dense cell clusters, where individual nuclei could not be reliably distinguished and quantified, were then identified and excluded from further analysis. Using this new “filtered” image, the total population of dispersed, single DAPI+ nuclei was identified for downstream single cell analyses. DAPI+ nuclei at the image borders were excluded from analyses (Fig. 2, panel D). In addition, DAPI+ nuclei in nonviable cells were excluded from downstream analyses by setting the threshold of average intensity per nuclei at <45,000.
Fig. 2.

Differentiation of iPSC-derived neuronal clusters from single neurons by image inversion. A, Acquired image of DAPI+ nuclei. B, Inverted image. C, Gaussian smoothing of regions outside of cell clusters or groups. This “filtered” image (25 contiguous fields of view stitched into a single image) is used to identify and quantify individual nuclei via DAPI staining (then other cellular markers in downstream analyses). D, Single cell selection (One field of view, representative image). Grey=excluded cell clusters and assorted colors=single cells. After identifying the region of interest in the images for single cell analyses, nuclei were identified using the Hoechst 33342 channel and method M at a diameter of 12 μm, splitting sensitivity of 0.31 and common threshold of 0.4.
For neuronal marker identification and quantification, we then incorporated the intensity thresholds for other image acquisition channels (ch 2: Alexa 488, and ch 3: Alexa 647) for total individual DAPI+ nuclei (Supplemental Table 1). Once the thresholds for individual neuronal (i.e., NeuN+) populations were identified, we then quantified the number of objects (cells) within selected cell populations.
Uniform cellular distribution through pre-incubation at room temperature and amelioration of edge effects by modified culture handling procedures.
The prolonged incubation periods (i.e., >4 weeks in cell culture) required to differentiate and mature iPSC-derived neurons enhances the probability of uneven distribution of cells and edge effects. Usually variation in cellular distribution is introduced upon cell seeding and immediately after introduction into the tissue culture incubator environment [23]. In contrast, edge effects, resulting from medium evaporation and concentration due to temperature gradients [24], increases assay variability and are particularly detrimental to miniaturized cell-based assay formats, which are frequently used in high content imaging and screening activities.
To allow for even cellular distribution in our miniaturized neuronal cultures, we seeded NPCs into 96 well plates and allowed them to incubate at room temperature [24] from 0 to 60 minutes post-seeding. The iPSC-derived neuronal cultures then proceeded through differentiation and maturation as previously reported [19]. Moreover, after each biweekly half-volume medium change, plated neuronal cultures were rotated 18° upon return to the incubator and were positioned in different locations in the tissue culture incubator to control for positional effects. At 4-weeks post-differentiation, plates were fixed, stained with DAPI and full plate DAPI+ nuclei counts were quantified via the above-described algorithm (Supplemental Table 1). Acquired data showed that pre-incubation at room temperature for 30 and 60 min, after NPC seeding, significantly reduced the overall variability in DAPI+ nuclei counted per well (Fig. 3). The most variability in DAPI+ nuclei was observed in microtiter plates without pre-incubation (i.e., 0 min) post-NPC seeding. Heat maps and frequency distributions from two independent experiments (Supplemental Figures 2 and 3) confirm the amelioration of edge effects in DAPI + nuclei when microtiter plates were pre-incubated at room temperature prior to long term incubation at 37°C.
Fig. 3.
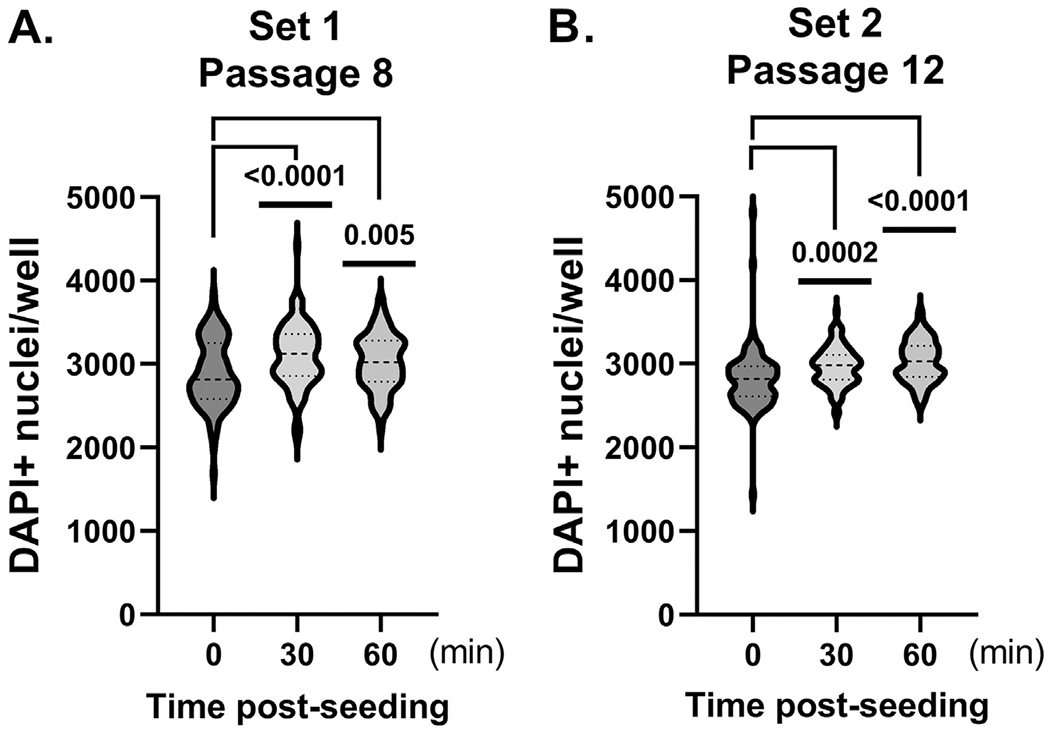
Pre-incubation at room temperature, environmental O2 and varied incubator positioning post-plating decreases well-to-well variability in miniaturized formats. NPCs were seeded at 850 cells/well in 96 well microtiter plates and subjected to 30 or 60 min pre-incubation on the benchtop prior to the initiation of differentiation and maturation procedures. At 4-weeks post-differentiation, plates were fixed, nuclei were stained with DAPI and DAPI+ nuclei quantified. A, set 1 (NPC passage 8). B, set 2 (passage 12). Significance of cell population data was determined using Kolmogorov–Smirnov (KS) [54] statistic. p-values are as indicated.
While pre-incubation and plate positioning during incubation ameliorated cellular distribution and DAPI-associated edge effects, quantification of DAPI + nuclei only reflected general cellular distribution within microtiter wells. To ascertain potential adverse miniaturization effects on neuronal maturation, we next concentrated on quantifying the expression of neuronal maturation markers. Although βIII tubulin has been used to identify neurons or cells with a neuronal identity, it is expressed early during neuron differentiation and, therefore, is detected in neuronal progenitor [25], immature and mature neurons [26,27]. Similarly, MAP2, another commonly used neuronal marker, is actually a family of three proteins (i.e., juvenile MAPC2, mature MAP2a/b), which are developmentally regulated with none being specific for mature neurons [28]. Thus, we focused on the expression of NeuN, which is a biomarker generally associated with post-mitotic mature neurons [29], within our DAPI + nuclei. NeuN is most commonly used to identify mature neurons in primary murine neuronal cultures [30,31], but has been less frequently used in the study of iPSC-derived neurons.
The data indicated there were no significant differences in the NeuN+ (i.e., ~15-20%) cell populations in inner and outer wells (Fig. 4). However, variation in the numbers of NeuN+ neurons within the outer wells was greater (versus inner wells) (Fig. 4). Nonetheless, these data suggested that iPSC-derived neuronal maturation occurred with equal efficiency and frequency regardless of the well location using our refined conditions. Using these NPC seeding and culturing conditions, we can expect to identify and analyze approximately ~1000 NeuN+ iPSC-derived neurons per well at 4 weeks post-differentiation. As expected, analysis of NeuN+ and Ki67+ cell populations also suggested that increasing NPC passage number decreases neuronal differentiation capability (Fig. 5) and selects for proliferative phenotypes as reflected by NeuN and Ki67 expression, respectively.
Fig. 4.

DAPI+ and NeuN+ cell numbers in inner and edge wells reveal minimal differences. A, Quantified total DAPI+ nuclei. B, Quantified NeuN+ cells (%). C, Images of DAPI+/NeuN + cells in inner versus edge wells. No significance detected.
Fig. 5.

Effect of NPC passage number on NeuN+ neuron populations 4 weeks post-differentiation. Quantification of- A, total DAPI+ nuclei, B, NeuN+ cells (%), C, Ki67+ cells (%), D, Ki67+/NeuN+ cells (%). N=12 per condition. Data presented as average ± standard deviation (percent). BP=BrainPhys.
Increased NeuN + neuronal populations in feeder layer-free cultures by the inclusion of astrocyte conditioned medium.
Although our miniaturized culturing system yielded a reasonable number of mature NeuN + iPSC-derived neurons for downstream applications at 4 weeks post-differentiation, predictably, prolonged passaging of NPCs resulted in populations biased toward cell proliferation with reduced differentiation potential (Fig. 5). To further maximize the quantity of mature iPSC-derived neurons in our cultures, we added astrocyte conditioned medium (ACM, 1%) to the neuronal differentiation and maturation medium for the full 4-week differentiation period or for 1-week, after the rapid expansion that occurs during week 2-3 post-differentiation [19]. ACM contains >180 defined soluble secreted factors, cytokines and extracellular matrix proteins [32] which are critical for neuronal differentiation and maturation [33,34]. The data showed that, in general, the inclusion of ACM to the neuronal culturing medium significantly increased the quantity of mature, NeuN+ (i.e., ~30%) neurons at 4-weeks post-differentiation (Fig. 6) without impacting Ki67 expression (Supplemental Figure 4). However, ACM supplementation for the full 4 weeks (versus 1 week) of neuronal differentiation may be more effective than short-term exposure (Fig. 6, Supplemental Figure 4).
Fig. 6.

Addition of astrocyte-conditioned medium to iPSC-derived neuronal differentiation and maturation medium increases the percentage of NeuN expressing cells. iPSC-derived NPCs were cultured in medium supplemented with 1% astrocyte conditioned medium (ACM) for 4 weeks or for one week (i.e., after the week 2-3 post-differentiation cell proliferation) [19]. Quantification of- A, total DAPI+ nuclei. B, NeuN+ cells (%). C, DAPI+/NeuN+ cells. BP, BrainPhys.
Pilot neurotoxicity studies identify limited edge effects.
To assess the suitability of our miniaturized, human feeder layer-free iPSC-derived neurons for screening-associated applications, we performed a small-scale pilot neurotoxicity study (i.e., 80 approved and experimental drugs) using a traditional metabolic assay format followed by analysis using our single cell image-based algorithm. We specifically were interested in identifying generalized edge effects under screening-associated conditions. DMSO tolerance assessments indicated both endpoints tolerated similar DMSO concentrations (Supplemental Figure 5); however, 0.1% DMSO was used during screening activities. iPSC-derived neurons were treated with 10 μM drug for 24 hr and effects on viability were evaluated. Results using the population-based metabolic endpoint (i.e., CellTiter-Blue) and a SpectraMax M5 plate reader revealed that edge effects were negligible (Fig. 7 panel A) and there was a clear differentiation between the positive (MAX) and negative (MIN) controls, which yielded a robust Z-factor (i.e., Z-factor=0.7). Comparable data collected using the imaging-based single neuron algorithm also confirmed the lack of edge effects after prolonged cell culture; however, there was more well-to-well variation within the plate (Fig. 7 panels B and D), including within the positive controls. This variation may be associated with post-processing of the microtiter plate for imaging, which will be unavoidable during because of the fix and read steps this antibody-based assay format will require. Nonetheless, the Z-factor (i.e. Z-factor=0.4) was acceptable for an imaging format. In our proof of concept studies, moxidectin was identified and confirmed as a “neurotoxic” hit (Fig. 7 panels C and D) using both assay formats. Moxidectin is an orally bioavailable anthelmintic drug approved in the US for onchocerciasis (i.e., river blindness) in 2018 [35]. Moxidectin crosses the blood brain barrier more easily than its structurally related drug, ivermectin, which is also approved (www.access.FDA.gov) as a systemic treatment for onchocerciasis. Common adverse effects of moxidectin are associated with neurotoxicity (e.g., loss of consciousness, confusion, hallucinations, headache, mood/mental changes, seizures) (www.access.FDA.gov). Data from additional compound evaluations (Supplemental Figure 6) reinforce the lack of edge effects and support the reproducibility of both endpoints (i.e., metabolic and toxicity). These data suggest that our miniaturized iPSC-derived neuron format will be amenable with screening applications.
Fig. 7.

Pilot studies using population- and single cell image-based screening formats reveal no generalized edge effects in 4 week neuronal cultures. Panels A and B, Heat maps of screening plate using CellTiter-Blue (RFU) and DAPI+ cells as readouts. Panels C and D, Screening plate data from CellTiter-Blue (RFU) and DAPI+ cells as readouts. MAX ( ); MIN (
); MIN ( ); Hits (
); Hits ( ); and Test compounds (
); and Test compounds ( ). RFU=Relative fluorescence units.
). RFU=Relative fluorescence units.
Discussion
Late-stage drug failures for neurological indications, which are expensive and require considerable patient participation, are well known and generally attributed to a disconnect between preclinical model systems and human patients [36]. The human brain is an extremely complex organ and it is challenging to replicate its complexity and its many pathologies in vitro. More representative and predictive in vitro models that are scalable and can be used in high throughput/high content assay formats are needed, especially those that can identify and focus upon specific neuronal subpopulations. iPSC-derived neuron model systems are particularly attractive as physiologically relevant neurological models, although there has been a surprising lack of standardized maturation and miniaturization procedures for the in vitro systems especially when using low oxygen culturing conditions. However, iPSC-derived neurons have been used in miniaturized formats (i.e., 384 well assay plates) but they focus on using commercially available pre-plated, pre-formed spheroids [37] matured 9.5 weeks, bulk neuronal differentiation [38] or co-culturing with astrocytes for extended culturing [39]. Moreover, as mentioned previously, NeuN has not been commonly used as a neuronal maturation marker in the assay miniaturization process. Unfortunately, iPSC-derived neuron populations are also known to exhibit asynchronous differentiation and maturation, which results in a mixed population of intermediate cellular populations (e.g., radial glial cells, intermediate progenitors) as well as immature and mature neurons. Many neuron-based screening studies use βIII tubulin or MAP2 to identify neurons [40], which do not conclusively identify mature neurons and their usage may merely reflect the robustness of the available antibody tools. Thus, it is critical to establish the maturity level of individual in vitro iPSC-derived neuronal subpopulations, if quantifying drug effects on mature neurons (e.g., NeuN expressing) is a primary goal.
One significant benefit of iPSC-derived neurons is that they allow one to study multiple stages of neuronal development, namely neurogenesis to fully mature neurons. Nonetheless, there are concerns regarding their epigenetic signature and the corresponding effects on gene expression [41]. When adult somatic cells are reprogrammed into iPSCs by enforced expression of transcription factors, they are reverted to an embryonic state [42, 43]. Previous work indicates that the epigenetic signatures of the iPSCs reflect a “fetal” or “prenatal” age [44] versus the actual chronological age of the “donor” somatic cells, which are typically adult or “aged”, and that fetal state endures during the subsequent conversion to NPCs and differentiation into neurons. This is understandably problematic when one is using iPSC-derived neurons in studies of neurodegenerative disorders typically associated with aged human populations. An interesting alternative is to use “iNeurons”, which, in contrast, are directly converted from adult fibroblasts and exhibit adult, rather than fetal epigenetic signatures [12,45], hence, their allure. It is notable, however, that iNeurons bypass the multifunctional neuronal progenitor stage limiting their usefulness for studies in neurodevelopment (e.g., neurogenesis) [46]. Moreover, NeuN expression in iNeurons is also not 100% in prevalence, indicative of a mixed versus “pure” population of iNeurons, and suggests that iNeuron differentiation and maturation is also asynchronous.
Recent evidence suggests that environmental cues, such as those derived from the extracellular matrix (ECM), act as mediators of stem cell function, lineage commitment and epigenetic status [47]. Moreover, material cues can actually influence and regulate the epigenome [47] underscoring that the issue may be more complex than originally surmised. Since the vast majority of iPSC-derived neuron studies are performed using minimal ECM components (i.e., poly-L-ornithine, laminin), we believe it may be possible to change their epigenome by increasing ECM biocomplexity (i.e., adding hyaluronan, brevican, versican, tenascin-R, vitronectin) [48]; however, further studies will need to be performed including studies using multi-component systems (e.g., co-cultures, three dimensional). Although our cell-based model might have the appearance of two dimensionality, the inclusion of laminin, poly-L-ornithine and astrocyte conditioned medium (ACM) enhances its biocomplexity by providing additional extracellular cues for neuronal differentiation, maturation, synapse formation, extracellular matrix remodeling and neuronal viability [32,49,50]. As a result, we were able to standardize and miniaturize the iPSC-derived cortical neuron culturing process and increase the percent of NeuN+ neurons in our cultures enhancing the pool of mature neurons available for single cell image-based analyses. Furthermore, our methodology can be adapted for other iPSC-derived neuronal subpopulations providing the appropriate growth, differentiation and maturation signals are present.
Confirming previous observations [51–53], we detected a decrease in the differentiation potential (i.e., NeuN+ expression) of the NPCs with increasing passage number, which also correlated with an increase in Ki67 expression, indicating a selection bias during NPC maintenance. This general loss of differentiation capability of the “parent” NPC population suggests that both two and three dimensional iPSC-derived neurons cultures will be affected, and that screening assays may be biased towards the identification of cell cycle- or proliferation-based inhibitors. Overall, we have developed a defined and novel iPSC-based human cortical neuron model system that is compatible with high throughput and high content screening platforms. This miniaturized assay format is adaptable and is compatible with population-based endpoints as well as single cell analysis. The developed companion image-based algorithm enables the identification and quantification of single neurons. This foundational, and straightforward, system can be used independently or adapted to include more complex ECM and additional cell populations (i.e., astrocytes, microglia) to enhance its three dimensional physiological relevancy.
Supplementary Material
Acknowledgments
This research was made possible through funding from the National Institutes of Health (R01 - AG063400, R01-AG063400-02S1, ERS), S10OD021723-01A1 (JSL), the Cure Alzheimer’s Fund (ERS, JSL, GSB), the Owens Family Fund (JSL, GB), the Rick Sharp Alzheimer’s Disease Foundation (ERS, JSL, GB) and the Fiske Drug Discovery Laboratory.
Footnotes
Declaration of interests
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.slasd.2022.10.002.
References
- [1].Bazargan-Hejazi S, Dehghan K, Edwards C, et al. The health burden of non-communicable neurological disorders in the USA between 1990 and 2017. Brain Commun 2020;2:fcaa097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Collaborators GUND, Feigin VL, Vos T, et al. Burden of neurological disorders across the US from 1990-2017: A global burden of disease study. JAMA Neurol 2021;78:165–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hou Y, Dan X, Babbar M, et al. Ageing as a risk factor for neurodegenerative disease. Nat Rev Neurol 2019;15:565–81. [DOI] [PubMed] [Google Scholar]
- [4].Taquet M, Geddes JR, Husain M, et al. 6-month neurological and psychiatric outcomes in 236 379 survivors of COVID-19: a retrospective cohort study using electronic health records. Lancet Psychiatry 2021;8:416–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Marshall M COVID and the brain: researchers zero in on how damage occurs. Nature 2021;595:484–5. [DOI] [PubMed] [Google Scholar]
- [6].Wouk J, Rechenchoski DZ, Rodrigues BCD, et al. Viral infections and their relationship to neurological disorders. Arch Virol 2021;166:733–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang Q, Davis PB, Gurney ME, et al. COVID-19 and dementia: Analyses of risk, disparity, and outcomes from electronic health records in the US. Alzheimers Dement 2021;17:1297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fernandez CE, Franz CK, Ko JH, et al. Imaging review of peripheral nerve injuries in patients with COVID-19. Radiology 2021;298:E117–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Qureshi AI, Baskett WI, Huang W, et al. Acute ischemic stroke and COVID-19: An analysis of 27, 676 Patients. Stroke 2021;52:905–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Marshall M How COVID-19 can damage the brain. Nature 2020;585:342–3. [DOI] [PubMed] [Google Scholar]
- [11].McKinney CE. Using induced pluripotent stem cells derived neurons to model brain diseases. Neural Regen Res 2017;12:1062–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mertens J, Reid D, Lau S, et al. Aging in a Dish: iPSC-derived and directly induced neurons for studying brain aging and age-related neurodegenerative diseases. Annu Rev Genet 2018;52:271–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Middelkamp HHT, Verboven AHA, De Sa Vivas AG;, et al. Cell type-specific changes in transcriptomic profiles of endothelial cells, iPSC-derived neurons and astrocytes cultured on microfluidic chips. Sci Rep 2021;11:2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Liu L, Koo Y, Russell T, et al. Three-dimensional brain-on-chip model using human iPSC-derived GABAergic neurons and astrocytes: Butyrylcholinesterase post-treatment for acute malathion exposure. PLoS One 2020;15:e0230335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Begum AN, Guoynes C, Cho J, et al. Rapid generation of sub-type, region-specific neurons and neural networks from human pluripotent stem cell-derived neurospheres. Stem Cell Res 2015;15:731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Koroleva A, Deiwick A, El-Tamer A, et al. In vitro development of human iPSC-derived functional neuronal networks on laser-fabricated 3D scaffolds. ACS Appl Mater Interfaces 2021;13:7839–53. [DOI] [PubMed] [Google Scholar]
- [17].Burke EE, Chenoweth JG, Shin JH, et al. Dissecting transcriptomic signatures of neuronal differentiation and maturation using iPSCs. Nat Commun 2020;11:462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shi Y, Kirwan P, Livesey FJ. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc 2012;7:1836–46. [DOI] [PubMed] [Google Scholar]
- [19].Sharlow ER, Llaneza DC, Mendelson AJ, et al. Extracellular cues accelerate neurogenesis of induced pluripotent stem cell derived neurons. bioRxiv 2021. 2021.08.17.456634. [Google Scholar]
- [20].Bardy C, van den Hurk M, Eames T, et al. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc Natl Acad Sci U S A 2015;112:E2725–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kawauchi T Cellullar insights into cerebral cortical development: focusing on the locomotion mode of neuronal migration. Front Cell Neurosci 2015;9:394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gunhanlar N, Shpak G, van der Kroeg M, et al. A simplified protocol for differentiation of electrophysiologically mature neuronal networks from human induced pluripotent stem cells. Mol Psychiatry 2018;23:1336–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Reynolds PM, Holzmann Rasmussen C, Hansson M, et al. Controlling fluid flow to improve cell seeding uniformity. PLoS One 2018;13:e0207211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lundholt BK, Scudder KM, Pagliaro L. A simple technique for reducing edge effect in cell-based assays. J Biomol Screen 2003;8:566–70. [DOI] [PubMed] [Google Scholar]
- [25].Rodriguez-Jimenez FJ, Vilches A, Perez-Arago MA, et al. Activation of neurogenesis in multipotent stem cells cultured in vitro and in the spinal cord tissue after severe injury by inhibition of glycogen synthase kinase-3. Neurotherapeutics 2021;18:515–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chacon J, Rogers CD. Early expression of Tubulin Beta-III in avian cranial neural crest cells. Gene Expr Patterns 2019;34:119067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mariani J, Coppola G, Zhang P, et al. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 2015;162:375–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Soltani MH, Pichardo R, Song Z, et al. Microtubule-associated protein 2, a marker of neuronal differentiation, induces mitotic defects, inhibits growth of melanoma cells, and predicts metastatic potential of cutaneous melanoma. Am J Pathol 2005;166:1841–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sarnat HB. Immunocytochemical markers of neuronal maturation in human diagnostic neuropathology. Cell Tissue Res 2015;359:279–94. [DOI] [PubMed] [Google Scholar]
- [30].Seward ME, Swanson E, Norambuena A, et al. Amyloid-beta signals through tau to drive ectopic neuronal cell cycle re-entry in Alzheimer’s disease. J Cell Sci 2013;126:1278–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kodis EJ, Choi S, Swanson E, et al. N-methyl-D-aspartate receptor-mediated calcium influx connects amyloid-beta oligomers to ectopic neuronal cell cycle reentry in Alzheimer’s disease. Alzheimers Dement 2018;14:1302–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dowell JA, Johnson JA, Li L. Identification of astrocyte secreted proteins with a combination of shotgun proteomics and bioinformatics. J Proteome Res 2009;8:4135–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Moore NH, Costa LG, Shaffer SA, et al. Shotgun proteomics implicates extracellular matrix proteins and protease systems in neuronal development induced by astrocyte cholinergic stimulation. J Neurochem 2009;108:891–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Keene SD, Greco TM, Parastatidis I, et al. Mass spectrometric and computational analysis of cytokine-induced alterations in the astrocyte secretome. Proteomics 2009;9:768–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ebied AM, Na J, Cooper-DeHoff RM. New drug approvals in 2018 - another record year!. Am J Med 2019;132:1038–43. [DOI] [PubMed] [Google Scholar]
- [36].Hwang TJ, Carpenter D, Lauffenburger JC, et al. Failure of investigational drugs in late-stage clinical development and publication of trial results. JAMA Intern Med 2016;176:1826–33. [DOI] [PubMed] [Google Scholar]
- [37].Boutin ME, Strong CE, Van Hese B, et al. A multiparametric calcium signal screening platform using iPSC-derived cortical neural spheroids. SLAS Discov 2022;27:209–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].MacMullen C, Davis RL. High-throughput phenotypic assay for compounds that influence mitochondrial health using iPSC-derived human neurons. SLAS Discov 2021;26:811–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Traub S, Stahl H, Rosenbrock H, et al. Upscaling of hiPS cell-derived neurons for high-throughput screening. SLAS Discov 2017;22:274–86. [DOI] [PubMed] [Google Scholar]
- [40].Oh HN, Park S, Lee S, et al. In vitro neurotoxicity evaluation of biocidal disinfectants in a human neuron-astrocyte co-culture model. Toxicol In Vitro 2022;84:105449. [DOI] [PubMed] [Google Scholar]
- [41].Steg LC, Shireby GL, Imm J, et al. Novel epigenetic clock for fetal brain development predicts prenatal age for cellular stem cell models and derived neurons. Mol Brain 2021;14:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Medvedev SP, Pokushalov EA, Zakian SM. Epigenetics of pluripotent cells. Acta Naturae 2012;4:28–46. [PMC free article] [PubMed] [Google Scholar]
- [43].Bilic J, Izpisua Belmonte JC. Concise review: Induced pluripotent stem cells versus embryonic stem cells: close enough or yet too far apart? Stem Cells 2012;30:33–41. [DOI] [PubMed] [Google Scholar]
- [44].Imm J, Pishva E, Ali M, et al. Characterization of DNA methylomic signatures in induced pluripotent stem cells during neuronal differentiation. Front Cell Dev Biol 2021;9:647981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mertens J, Paquola ACM, Ku M, et al. Directly reprogrammed human neurons retain aging-associated transcriptomic signatures and reveal age-related nucleocytoplasmic defects. Cell Stem Cell 2015;17:705–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Xu Z, Su S, Zhou S, et al. How to reprogram human fibroblasts to neurons. Cell Biosci 2020;10:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Crowder SW, Leonardo V, Whittaker T, et al. Material cues as potent regulators of epigenetics and stem cell function. Cell Stem Cell 2016;18:39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Cummings J, Lee G, Zhong K, et al. Alzheimer’s disease drug development pipeline: 2021. Alzheimers Dement (N Y) 2021;7:e12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Song C, Wu YS, Yang ZY, et al. Astrocyte-conditioned medium protects prefrontal cortical neurons from glutamate-induced cell death by inhibiting TNF-alpha expression. Neuroimmunomodulation 2019;26:33–42. [DOI] [PubMed] [Google Scholar]
- [50].Jha MK, Kim JH, Song GJ, et al. Functional dissection of astrocyte-secreted proteins: Implications in brain health and diseases. Prog Neurobiol 2018;162:37–69. [DOI] [PubMed] [Google Scholar]
- [51].Wilson PG, Stice SS. Development and differentiation of neural rosettes derived from human embryonic stem cells. Stem Cell Rev 2006;2:67–77. [DOI] [PubMed] [Google Scholar]
- [52].Paavilainen T, Pelkonen A, Makinen ME, et al. Effect of prolonged differentiation on functional maturation of human pluripotent stem cell-derived neuronal cultures. Stem Cell Res 2018;27:151–61. [DOI] [PubMed] [Google Scholar]
- [53].Muratore CR, Srikanth P, Callahan DG, et al. Comparison and optimization of hiPSC forebrain cortical differentiation protocols. PLoS One 2014;9:e105807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kochanek SJ, Close DA, Wang AX, et al. Confirmation of selected synergistic cancer drug combinations identified in an HTS campaign and exploration of drug efflux transporter contributions to the mode of synergy. SLAS Discov 2019;24:653–68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


