Abstract
Patient registries are a very important and essential tool for investigating rare diseases, as most physicians only see a limited number of cases during their career. Diseases of multi-organ autoimmunity and autoinflammation are especially challenging, as they are characterized by diverse clinical phenotypes and highly variable expressivity. The GAIN consortium (German multi-organ Auto Immunity Network) developed a dataset addressing these challenges. ICD-11, HPO, and ATC codes were incorporated to document various clinical manifestations and medications with a defined terminology. The GAIN dataset comprises detailed information on genetics, phenotypes, medication, and laboratory values. Between November 2019 and July 2022, twelve centers from Europe have registered 419 patients with multi-organ autoimmunity or autoinflammation. The median age at onset of symptoms was 13 years (IQR 3–28) and the median delay from onset to diagnosis was 5 years (IQR 1–14). Of 354 (84.5%) patients who were genetically tested, 248 (59.2%) had a defined monogenetic cause. For 87 (20.8%) patients, no mutation was found and for 19 (4.5%), the result was pending. The most common gene affected was NFkB1 (48, 11.5%), and the second common was CTLA4 (40, 9.5%), both genetic patient groups being fostered by specific research projects within GAIN. The GAIN registry may serve as a valuable resource for research in the inborn error of immunity community by providing a platform for etiological and diagnostic research projects, as well as observational trials on treatment options.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10875-023-01472-0.
Keywords: Inborn error of immunity, Autoimmunity, Autoinflammation, Immune-dysregulation, Primary immunodeficiency, Epidemiology, Rare diseases
Introduction
The management of patients with rare diseases is challenging for treating physicians, as many questions of patients stay unanswered due to the lack of data and the sparse knowledge for making treatment decisions. In addition, research to address these questions is hindered by the very limited availability of research samples of these rare patients. Patients with immune dysregulation and multi-organ autoimmunity belong to a group of patients with diverse rare genetic causes presenting with partly life-threatening autoimmune or autoinflammatory diseases of several organs. We still do not know how prevalent these diseases are, and detailed clinical, genetic, and quality of life information from these patients is lacking. Our aim was to document this type of data in a structured and regular manner and share it with the research community. Before, there was no data structure which was capable of capturing the diverse phenotypes and genotypes of patients with multi-organ autoimmune or autoinflammatory diseases in sufficient detail. Here, we present the GAIN registry, capable of addressing these challenges using international classification systems. This registry was set up in the context of the GAIN consortium (German multi-organ AutoImmunity Network; www.g-a-i-n.de). The registry is its central project and interconnects all other research projects within the consortium. We integrated the GAIN registry on the online registry platform of the ESID registry (European Society for Immunodeficiency; https://esid.org/Working-Parties/Registry-Working-Party/ESID-Registry). The ESID registry is a well-established, European-wide registry in the field of primary immunodeficiencies. Every documenting center of the ESID registry can easily participate in the GAIN registry project. The main research question addresses the prevalence of patients with multi-organ autoimmunity or autoinflammation. Second, we aim at answering basic questions of patients such as, “What can I expect from life in the future?” To answer this, we collect whenever possible the timing of events to document the disease progression. Third, our detailed dataset aims to address the relationship between the underlying gene variant and the actual expressivity of the disease. In addition, we designed the registry to serve as a platform for more detailed research projects. Patients with specific genetic or phenotypic traits can be identified. Detailed information on infections and the causing pathogens, organ involvements, comorbidities, allergies, vaccination status, treatment regiments, and genetic information may be documented into the registry.
Methods
Organization
The GAIN registry is part of the German multi-organ AutoImmunity Network (GAIN) consortium (BMBF, funding code 01GM1910A) and complements wet-lab research projects, a clinical pharmacological trial (ABACHAI), and a joint consortial GAIN Biobank, all part of the first GAIN funding period from 2019 to 2022.
How to Participate
The GAIN registry was implemented on the platform of the European Society for Immunodeficiency (ESID) registry [1] and is hence open to all centers that wish to participate and conclude an agreement with the ESID registry and have the approval of their local ethics committees. The ESID registry includes a core dataset (level 1), which is also always documented for GAIN patients. The detailed GAIN dataset is therefore an ESID level 2 dataset. All participating centers can hand in research proposals to the GAIN consortium. Patients with written informed consent of the ESID registry may be included into the GAIN registry. No other patient consent is necessary. Identifying information of patients are not stored in the ESID database, but can be matched locally [2] depending on the users’ access rights. For more details and information of the technical background of the ESID registry, see Scheible et al. (2019) [1]. All GAIN patients are directly made available to the ESID registry for other secondary research level 2 studies.
Research Goals
First, the level 2 GAIN registry dataset in combination with the core ESID registry dataset (level 1) was designed to estimate the minimal prevalence and key parameters and characteristics of immune dysregulatory diseases as outlined in Table 1. Only the minimal prevalence can be estimated as the participation in the registry is voluntary.
Table 1.
Research interest of the GAIN registry
| 1 | Minimal prevalence of immune-dysregulatory diseases |
| 2 | Median age of disease onset |
| 3 | Median age at diagnosis and diagnostic delay |
| 4 | Median survival |
| 5 | Detailed description of phenotypic manifestations of the disease, including infections and comorbidities |
| 6 | Detailed description of treatment |
| 7 | Detailed description of genetic variants |
| 8 | Quality of life of patients |
Second, we want to investigate — by longitudinal follow-up — risk factors associated with the expressivity of the disease, especially disease initiation, progression, and treatment response. With detailed description of genetic variants, we hope to identify new genetic and molecular pathways associated with multi-organ autoimmunity/autoinflammation.
Third, our detailed dataset aims to address the relationship between the underlying gene variant and the actual expressivity of the disease. We want to offer a platform for more detailed research projects for various subsets of multi-organ autoimmune/autoinflammatory patient groups, which alone would never have the resources to build up their own registry.
Inclusion Criteria for Patients
Patients are eligible for documentation into the GAIN registry, when they show a multi-organ pathology with either an autoimmunity or an autoinflammation phenotype or have a proven pathogenic mutation in a gene known to cause multi-organ autoimmunity/autoinflammation.
A list of genes known to cause multi-organ autoimmunity/autoinflammation can be found in Table 2. Genes are being added to the list whenever new evidence emerges. However, we also include patients into the GAIN registry with an unknown genetic cause to be able to investigate unknown genetic determinants. In addition, patients need to fulfill the ESID registry criteria [3]. The treating physician decides whether their patients fulfill these inclusion criteria. Using these criteria, we aim at including patients in the longitudinal data collection who are carriers of a mutation but do not show a multi-organ autoimmune/autoinflammation phenotype yet. Patients with an acquired immunodeficiency are not included into the study.
Table 2.
Candidate genes known to cause multi-organ autoimmunity or autoinflammation
| Gene abbreviation | Gene name |
|---|---|
| AIRE | Autoimmune regulator |
| ADA2 | Adenosine deaminase 2 |
| BACH2 | BTB domain and CNC homolog 2 |
| CD25/IL2-RA | Interleukin 2 receptor subunit alpha |
| CTLA4 | Cytotoxic T-lymphocyte associated protein 4 |
| FOXP3 | Forkhead box P3 |
| ICOS | Inducible T cell costimulator |
| LAT | Linker for activation of T cells |
| LRBA | LPS responsive beige-like anchor protein |
| NFKB1 | Nuclear factor kappa B subunit 1 |
| NFKB2 | Nuclear factor kappa B subunit 2 |
| PIK3CD | Phosphoinositide 3 kinase catalytic subunit delta |
| PIK3R1 | Phosphoinositide 3 kinase regulatory subunit 1 |
| PRKCD | Protein kinase C delta |
| RELA | Nuclear factor kappa B subunit 3 |
| SOCS1 | Suppressor of cytokine signaling 1 |
| STAT1-GOF | Signal transducer and activator of transcription 1 |
| STAT3-GOF | Signal transducer and activator of transcription 3 |
| TNFRSF13B | Tumor necrosis factor receptor superfamily member 13B |
| TPP2 | Tripeptidyl peptidase 2 |
GOF, gain-of-function
Ongoing Projects and Their Research Goals
The detailed data structure of the registry offers to answer specific research questions in subgroups of patients. Ongoing projects within the registry are listed in Table 3 with their inclusion criteria and research goals. Most projects use the possibility to capture detailed information on the history of symptoms, organ manifestations, and infections for the patients to describe their clinical phenotype. Three projects use the detailed documentation of treatment such as the history of prescribed drugs with its dose, frequency, treatment duration, adverse events, and outcome for the patient. Other possible research questions which may be answered by the dataset are as follows:
How are titer responses to vaccinations related to the infection history?
Do certain patient subgroups suffer more often from allergies?
Which type of cancer is more prevalent in patients with a given genetic diagnosis?
Are these cancers associated with the persistence of specific viruses?
Table 3.
Ongoing projects within the GAIN registry
| Project name | CTLA4 insufficiency | NFKB1 insufficiency | NFKB2 insufficiency | ADA2 deficiency | CARD11-GOF disease | RELA insufficiency | STAT3-GOF disease |
|---|---|---|---|---|---|---|---|
| Responsible person | Bodo Grimbacher | Bodo Grimbacher | Bodo Grimbacher | Fabian Hauck | Fabian Hauck | Min-Ae Lee-Kirsch | Stephan Ehl |
| Inclusion criteria | All patients with pathogenic mutations in CTLA4 | All patients with pathogenic mutations in NFKB1 | All patients with pathogenic mutations in NFKB2 | All patients with pathogenic mutations in ADA2 | All patients with pathogenic gain-of-function mutations in CARD11 | All patients with pathogenic mutations in RELA | All patients with pathogenic gain-of-function mutations in STAT3 |
| Exclusion criteria | Not consented individuals | Not consented individuals | Not consented individuals | Not consented individuals | Not consented individuals | Not consented individuals | Not consented individuals |
| No. of patients | 40 | 48 | 18 | 10 | starting 01/2023 | starting 01/2023 | 12 |
| Main research goals | Which complications are common in CTLA4 insufficiency? Which drugs are successfully used to treat these? | Which complications are common in NFKB1 insufficiency? Which drugs are successfully used to treat these? | Which complications are common in NFKB2 insufficiency? Which drugs are successfully used to treat these? | Describe the extended clinical phenotype of patients with biallelic mutations in ADA2 | Describe the extended clinical phenotype of patients with gain-of-function mutations in CARD11 | Describe the extended clinical phenotype of patients with mutations in RELA | Describe the extended clinical phenotype of patients with gain-of-function mutations in STAT3 |
| Secondary research goals | Identify biomarkers for treatment response and treatment failure | Identify biomarkers for treatment response and treatment failure | Identify biomarkers for treatment response and treatment failure. Assess the importance of anti-interferon antibodies in patients with NFKB2 insufficiency | Identify biomarkers for treatment response and treatment failure | Identify biomarkers for treatment response and treatment failure | Identify biomarkers for treatment response and treatment failure | Identify biomarkers for treatment response and treatment failure |
| Starting time | 2019 | 2019 | 2019 | 2019 | 2023 | 2023 | 2019 |
| Planned project duration | 9 years | 9 years | 9 years | 9 years | 6 years | 6 years | 9 years |
Recruiting Measures
Currently, twelve centers are documenting into the GAIN registry, ten originating from various regions in Germany and one center in Italy and one in Portugal each (Supplementary Table 1). Any ESID documenting center may include GAIN patients prospectively (patient has not previously been entered into the ESID registry) or retrospectively (patient had been documented into ESID before and now the GAIN level 2 dataset is being filled). Moreover, with local ethics committee approval, deceased patients may also be included.
Data Sources
Information entered into the GAIN registry derives mainly from three sources: The first and most important are (electronic) patient records. Information needs to be manually retrieved from medical letters and reports, coded if necessary, and entered into the online forms of the registry. Second are local research databases, for example, with genetic information on patients. The third data sources are electronic and paper questionnaires, designed to support the collection of information, which is not regularly available from patient records, such as quality of life data. For quality assurance, the documentation specialists will check documented data for plausibility and completeness and resolve open issues with the treating physicians.
Statistics
The analyses of the collected dataset focused on simple descriptive summary statistics, performed with R version 4.2.2 and Microsoft Excel 2010. If exact dates were missing but necessary for calculation, the middle of the month (15th) or the middle of the year (1st of July) was assumed. For the summary of genes with genetic variants, information from the GAIN registry was supplemented with information from ESID level 1 entries.
Results
Design and Implementation of the GAIN Registry
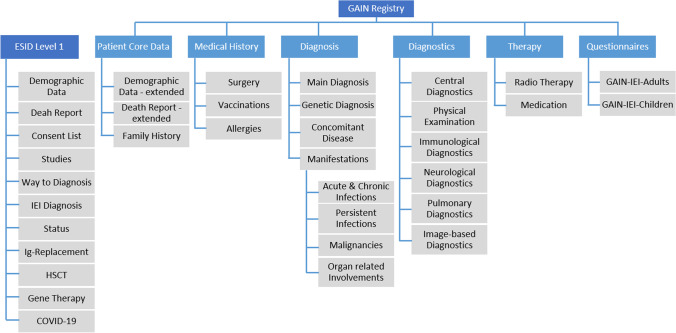
To overcome the need and expenses of creating a new registry for each new rare disease subgroup, we designed and implemented a dataset which is capable of capturing various manifestations of different forms of autoimmune/autoinflammatory diseases. In order to mirror a complex disease, a data structure with a complexity similar to an electronic patient record was developed (Fig. 1). The full GAIN entry forms can be accessed by the demonstration version of the ESID registry via https://cci-esid-reg-demo-app.uniklinik-freiburg.de/EERS with the username “demouser” and the password “Demo-2019” [1]. The GAIN dataset is designed to collect prospective information. Information which does not change, such as information on consanguinity, is only collected once. Information on laboratory and diagnostic values, as well as questionnaires, can be documented at each visit. A visit and documentation into the GAIN registry is aimed for at least once a year. In addition, therapies, infections, malignancies, and other organ-related symptoms and pathologies are captured in lists, including start and end dates. A GAIN-specific user guide that amends the general ESID registry user manual and answers frequently asked questions (in English) and a video tutorial (in German) were developed to support documentation and are available at the GAIN project website https://www.g-a-i-n.de/register/. From November 2019 on, patients have been registered into the GAIN registry and basic information was entered into the ESID level 1 dataset. Since the beginning of the year 2021, the GAIN dataset level 2 was also available for documentation.
Fig. 1.
Overview of GAIN registry dataset structure and the ESID basic level 1 dataset
Coding Systems for Increased Interoperability
Standardized terminology is a prerequisite to merge or to compare datasets. To foster the interoperability and sustainability of the GAIN dataset, it is based on international coding systems. The main clinical diagnosis of the patient is coded with the ORPHA nomenclature and ICD (International Classification of Diseases) codes [4]. The ORPHA nomenclature is maintained by the European Commission funded consortium “orphanet” and was developed especially for rare diseases [5].
The ICD codes are the global standard for diagnostic health information, developed and maintained by the WHO. We implemented the ICD-11 version, the more detailed and updated version of the commonly used ICD-10 code, as it was meant to replace ICD-10 in clinical information systems in the medium term. ICD-11 codes are being used to code all disease manifestations of immunodeficient patients, including infections, malignancies, and organ-related manifestations of autoimmunity or autoinflammation. In addition, the extension code of ICD-11 is used for the classification of pathogens, allergens, body sites, and tissues in the GAIN registry.
For the documentation of symptoms, we opted for the Human Phenotype Ontology (HPO) and implemented it as a REST-Application Programming Interface in the registry. This type of implementation assures that coding requests are sent directly to HPO servers; thereby, the evolving ontology is always up to date. The HPO is an international ontology, developed by the Monarch initiative, funded by the National Institute of Health of the USA [6].
Medications are coded by their ingredients using the ATC code (anatomical-therapeutic-chemical classification) [7]. In addition, if available, the full product name of the medication is documented based on a table requested from the European Medicines agency, comprising all drugs approved in Europe [8]. The documentation of drug allergies also uses this set of codes.
Classifications are performed by the treating physician or the documentation specialist. To document the classification process, also the original wording of the primary data source is recorded in original language.
Preselection of Common Diseases and Symptoms
Classification systems and ontologies enable a specific documentation of a large repertoire of disease and symptom entities. To facilitate coding, we included suggestions for common malignancies, organ involvements, and symptoms for patients with multi-organ autoimmunity/autoinflammation. These suggestions were developed by experienced clinicians of the GAIN consortium. When one suggestion is chosen, the corresponding ICD-11 or HPO code is complemented automatically.
Genetic Diagnosis
To enable phenotype-genotype correlations, the genetic information needs to be documented in detail. When collecting genetic data from different laboratories and different genetic tests, a common language and the specification of the reference genome are needed; otherwise, information might become ambiguous. To ensure standardized data entry of the genetic variant, users are asked to verify the HGVS (human genome variation society) conform format with the syntax checker developed by Lefter et al. [9]. In addition, the transcript ID can be selected either in Ensemble or RefSeq by searching the gene name to avoid typing errors [10, 11].
Capturing Incomplete Data
A good data quality of primary sources is a prerequisite for good data quality in the registry. Information is available in varying degrees of completeness in primary data sources (medical letters and records) and consequently in the GAIN registry. The GAIN dataset allows data entry of partly available information to use as much information as possible. If the exact date for an event is unknown, only the year or the year and the month can be specified. In addition, we offer the options, “Currently unknown,” “Truly unknown,” “Currently ongoing,” “Currently unknown if ongoing,” “Not ongoing but date currently unknown,” and “Not ongoing but date truly unknown” for diseases and symptoms to take into account the prospective nature of the documentation.
Cohort Description
Until the 27th of July 2022, 419 patients have been registered. 38.7% (162) of all patients were prospectively included, and 61.3% (257) retrospectively, as they had been already registered into the ESID registry. For 64.2% (269) of patients, the basic ESID level 1 dataset and two follow-ups or more had been documented. For 45.4% (132), the detailed GAIN dataset has been documented at least once. A total of 354 patients (86.5%) were genetically tested. For 59.2% (248), at least one gene variant was reported. In total, variants in 39 different genes were reported. Most registered patients had a defect in the NFKB1 gene (48 patients, 11.5%; see Table 4).
Table 4.
Characteristics of patients in the GAIN registry
| Characteristics | Total (n = 419) | No. | % |
|---|---|---|---|
| Gender | Male | 201 | 48.0% |
| Female | 217 | 51.8% | |
| Missing | 1 | 0.2% | |
| Agea | Mean age (SD), 40 (±19) | ||
| 0 – 9 | 25 | 6.0% | |
| 10 – 19 | 49 | 11.7% | |
| 20 – 29 | 67 | 16.0% | |
| 30 – 39 | 69 | 16.5% | |
| 40 – 49 | 61 | 14.6% | |
| 50 –59 | 84 | 20.0% | |
| 60 – 69 | 41 | 9.8% | |
| 70 – 79 | 17 | 4.1% | |
| 80+ | 6 | 1.4% | |
| Status | Alive | 399 | 95.2% |
| Deceased | 14 | 3.3% | |
| Discharged | 1 | 0.2% | |
| Lost to follow-up | 5 | 1.2% | |
| Mutated geneb | NFKB1 | 48 | 11.5% |
| CTLA4 | 40 | 9.5% | |
| TNFRSF13B | 39 | 9.3% | |
| NFKB2 | 18 | 4.3% | |
| LRBA | 13 | 3.1% | |
| STAT3 GOF | 12 | 2.9% | |
| ADA2 | 10 | 2.4% | |
| STAT1 | 9 | 2.1% | |
| STAT3 | 9 | 2.1% | |
| IKZF1 | 7 | 1.7% | |
| PIK3CD | 7 | 1.7% | |
| ICOS | 6 | 1.4% | |
| FAS | 5 | 1.2% | |
| UNC13D | 3 | 0.7% | |
| BACH2 | 2 | 0.5% | |
| CARMIL2 | 2 | 0.5% | |
| FLG | 2 | 0.5% | |
| MST1 | 2 | 0.5% | |
| SH2D1A | 2 | 0.5% | |
| Other | 22 | 5.3% | |
| Not tested | 43 | 10.3% | |
| No mutation found | 87 | 20.8% | |
| No information availablec | 41 | 9.8% | |
aIn case of death, lost to follow-up, and discharge, age at death or last information was used
bSeveral mutated genes per patient are possible. Therefore, percentages do not sum up to 100%
cPending result, or no information
We compared the gene variants of the GAIN registry with the main categories of the International Union of Immunological Societies (IUIS) tables [12], which categorized the genetic defects in different disease classes. Patients in the GAIN registry did not only fall into the “Table 4 Diseases of Immune Dysregulation” or “Table 7 Autoinflammatory Disorders” but also in more distant main categories as seen in Table 5. One patient inherited a gene variant in a gene not listed by the IUIS tables.
Table 5
| Table 1 Immunodeficiencies affecting cellular and humoral immunity | |
| DCLRE1C (ARTEMIS) | DNA Cross-Link Repair 1C |
| ICOS | Inducible T Cell Costimulator |
| IKBKB | Inhibitor Of Nuclear Factor Kappa B Kinase Subunit Beta |
| IKZF1 | IKAROS Family Zinc Finger 1 |
| STK4 (MST1) | Serine/Threonine Kinase 4 |
| Table 2 Combined immunodeficiencies with associated or syndromic features | |
| IKBKB | Inhibitor Of Nuclear Factor Kappa B Kinase Subunit Beta |
| SPINK5 | Serine Peptidase Inhibitor Kazal Type 5 |
| STAT3 | Signal Transducer and Activator Of Transcription 3 |
| WAS | WASP Actin Nucleation Promoting Factor |
| Table 3 Predominantly Antibody Deficiencies | |
| BTK | Bruton Tyrosine Kinase |
| IKZF1 | IKAROS Family Zinc Finger 1 |
| NFκB1 | Nuclear Factor Kappa B Subunit 1 |
| NFκB2 | Nuclear Factor Kappa B Subunit 2 |
| PIK3CD | Phosphoinositide 3 Kinase Catalytic Subunit Delta |
| PIK3R1 | Phosphoinositide 3 Kinase Regulatory Subunit 1 |
| TNFRSF13B | Tumor Necrosis Factor Receptor Superfamily Member 13B |
| TNFRSF13C | TNF Receptor Superfamily Member 13C |
| UNG | Uracil DNA Glycosylase |
| Table 4 Diseases of Immune Dysregulation | |
| AIRE | Autoimmune Regulator |
| BACH2 | BTB Domain and CNC Homolog 2 |
| CARMIL2 | Capping Protein Regulator And Myosin 1 Linker 2 |
| CD70 | CD70 Molecule |
| CTLA4 | Cytotoxic T-Lymphocyte Associated Protein 4 |
| FAS | Fas Cell Surface Death Receptor |
| FOXP3 | Forkhead Box P3 |
| LRBA | LPS Responsive Beige-Like Anchor Protein |
| RIPK1 | Receptor Interacting Serine/Threonine Kinase 1 |
| SH2D1A | SH2 Domain Containing 1A |
| SOCS1 | Suppressor of Cytokine Signalling 1 |
| STAT3 | Signal Transducer and Activator Of Transcription 3 |
| UNC13D | Unc-13 Homolog D |
| XIAP | X-Linked Inhibitor Of Apoptosis |
| Table 5 Congenital defects of phagocyte number or function | |
| CYBC1 | Cytochrome B-245 Chaperone 1 |
| JAGN1 | Jagunal Homolog 1 |
| WAS | WASP Actin Nucleation Promoting Factor |
| Table 6 Defects in intrinsic and innate immunity | |
| STAT1 | Signal Transducer and Activator Of Transcription 1 |
| Table 7 Autoinflammatory Disorders | |
| ADA2 | Adenosine Deaminase 2 |
| NLRP3 | NLR Family Pyrin Domain Containing 3 |
| PLCG2 | Phospholipase C Gamma 2 |
| SYK | Spleen Associated Tyrosine Kinase |
| TNFAIP3 | TNF Alpha Induced Protein 3 |
| TNFRSF1A | TNF Receptor Superfamily Member 1A |
| No IUIS Main category table | |
| FLG | Filaggrin |
The Patient’s Path to Diagnosis
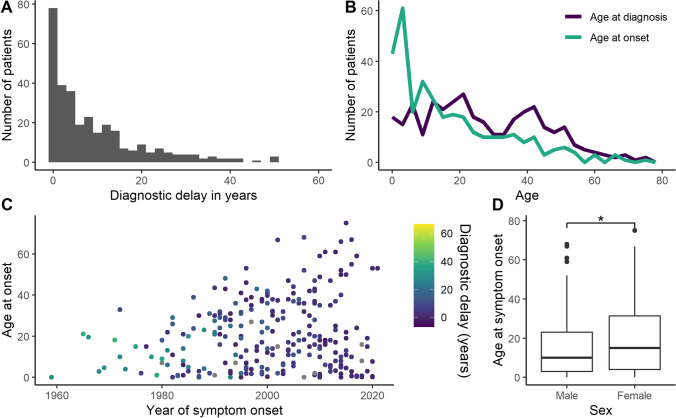
In any healthcare system, it typically takes a long time to diagnose patients with rare diseases. The resulting delay of an appropriate treatment leads to increased patient morbidity and considerable consumption of healthcare resources [13, 14]. As our patient group suffers from a wide range of symptoms, we were interested in the time span from symptom onset to clinical diagnosis, i.e., the diagnostic delay. Symptom onset was defined as first symptom suggestive of an inborn error of immunity (IEI) based on the retrospective physicians’ assessment. The diagnostic delay spans a large range (see Fig. 2A). After presenting with symptoms suggestive of IEI, 22.9% (88) of patients were diagnosed within 1 year. However, the median time of diagnostic delay was 5 years (IQR 1–14). A large proportion of patients lacked a clear diagnosis after symptom onset for several years and even decades.
Fig. 2.
Delayed diagnosis for patients with multi-organ autoimmunity/autoinflammation. A Histogram of diagnostic delay. B Frequency polygon for age at diagnosis and age at onset in years, 3 years grouped for plotting. C Age at onset and diagnostic delay for the calendar year of symptom onset. It is difficult to estimate calendar time effects with our cohort. Patients with symptom onset several decades ago and in later age are underrepresented in the cohort, due to the prospective inclusion of mostly living patients in the registry. D Male patients experience symptom onset earlier than females. Wilcoxon rank sum test p = 0.024, n = 352
Options for documenting first symptoms were infection, immune dysregulation, malignancy, syndromal manifestations, other, and unknown. For most patients, clinical symptoms suggested an IEI (385 patients, 91.9%). No symptom was apparent for 14 patients (3.3%) who were diagnosed by laboratory values (10 patients) or because of a known familial genetic defect (four patients). For 20 patients (4.8%), this information was unknown. Of the 385 patients with IEI-related symptoms, infection (285 patients, 74.0%) was the most prominent, followed by immune dysregulation (196, 50.9%). Immune dysregulation was defined as lymphoproliferation (splenomegaly, hepatomegaly, lymphadenopathy), granuloma formation, autoimmunity (e.g., cytopenia, thyroid disease, joint disease, hepatitis, vitiligo, alopecia, diabetes), inflammatory bowel disease, celiac disease, vasculitis, eczema, or autoinflammatory disease. Both infections and symptoms of immune dysregulation were apparent for 119 patients (28.4%) who were diagnosed based on symptoms. Less frequent as presenting symptom was a syndromal phenotype (18, 4.7%), malignancies (6, 1.6%), and others with unclear categorization (24, 6.2%).
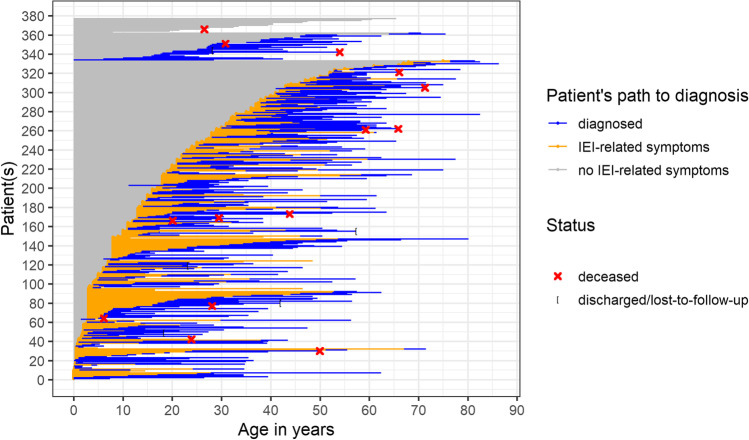
For some genetic causes of IEI with — or at risk for — rare multi-organ autoimmunity, incomplete penetrance and varying expressivity were reported [15, 16]. Therefore, we analyzed the age at onset of the first symptom which was suggestive for an IEI. Symptom onset spanned across a wide range of ages. The median age of symptom onset was 12 years (interquartile range (IQR) 3–26.3) (Fig. 2B; Fig. 3). Though male and female patients seem to be affected equally, male patients experienced onset of symptoms significantly earlier than female patients (Fig. 2D). An X-linked mutation could only be verified for six patients, suggesting other gender- and sex-related environmental factors and/or genetic contributions. The median age at diagnosis was 25 years (IQR 13–41). The comparison of the distribution of age at onset and age at diagnosis makes clear that even patients with early onset of symptoms may experience a considerable diagnostic delay (Fig. 2B; Fig. 3).
Fig. 3.
Patient’s path to diagnosis. Depicted are patients who reported symptoms related to inborn error of immunity (IEI) (n = 385). For patients at the top, date of symptom onset (n = 45) and/or date of diagnosis was unknown (n = 33) or they were genetically diagnosed before developing symptoms (n = 1). If only the partial information of the year was known, the middle of the year was used for the graph. In cases when the disease onset was given in a 5-year range, the middle of the range was used
The availability of genetic analysis has increased within the past decades and could affect the diagnostic delay over time. However, assessing this change with the current patient sample is difficult. This is due to a selection bias as most patients were included alive. Therefore, patients with a late onset of symptoms decades ago are underrepresented in our registry, as depicted in Fig. 2C by the nearly empty upper left corner of the graph. In addition, patients with a long diagnostic delay with symptom onset within the last years will still be diagnosed in the future. Therefore, our analysis of diagnostic delay can only constitute a snap shot of the patients already diagnosed.
Discussion
We developed the GAIN registry dataset, which is able to capture detailed clinical and genetic data for these diverse diseases. To our knowledge, no other dataset was available to accomplish this goal. Previously existing data sets did not provide the possibility to describe the autoimmune manifestations and therapies in this detail and resolution of time [1, 17]. Another ongoing ESID registry advancement, the immune deficiency and dysregulation activity (IDDA) score, focuses on estimated disease burden for an overlapping patient collective, which is also relevant, but not capable of describing clinical manifestations in detail [18]. Even the data set of the autoinflammatory disease alliance registry, which was published in the end of 2022 [19], does not provide the detail needed for the description clinical manifestations in a standardized manner. This was made possible by the utilization of coding systems and ontologies, such as the ICD-11 and the HPO. The international and interlinked nature of these coding systems enables interoperability and sustainability of the GAIN dataset. The GAIN registry enables the collection of information on rare patients with inborn errors of immunity in Europe by using the European platform of the ESID registry. The GAIN registry can serve as platform to identify subgroups of patients for researchers interested in special genes or aspects of immune dysregulation. Thereby, the GAIN registry may be used in the future as a basis for etiological and diagnostic studies, and may serve as a resource to recruit patients for clinical trials on treatment options. Due to the use of broad but also detailed ontologies, other classes of diseases outside the immunological field with or without heterogeneous phenotypes could make use of the GAIN dataset. Currently, the dataset focuses on clinical manifestations of the patients and is dependent on laborious manual data entry, as most registries in Germany. To address questions of patients as “What do I have to expect from life?” other aspects such as the quality of life should complement the collection of clinical information on these rare patients. As this information is not present in standard clinical information systems, other forms of data acquisition into the registry may complement the picture in the future. The manual data entry hampers the complete and up-to-date documentation in the registry. In addition, the detailed genetic documentation needs especially trained personal. Fortunately, the Medicine Informatics Initiative in Germany takes the first steps in the right direction, by providing infrastructure for automatic data import from clinical information systems. Since physicians themselves decide whether inclusion criteria are met, and since documentation will still be mostly manual, a structure for detailed quality control is needed.
As we only have a limited number of patients (n = 419) at risk for or with rare multi-organ autoimmunity/autoinflammation and a limited time of follow-up for most patients, a detailed analysis of patients will follow in the upcoming years. Due to the prospective nature of the registry, the evaluation of the diagnostic delay is biased by the selection of mostly living patients, already diagnosed. Nevertheless, a 5-year (IQR 1–14) diagnostic delay of patients presenting with clinical symptoms is comparable to the diagnostic delay of 6 years (IQR 2.5–10.5) for patients with autoinflammatory disorders, collected by the UK Primary Immune Deficiency registry via the same ESID registry platform [17]. This unfortunately highlights that patients with the diverse phenotype of multi-organ autoimmunity or multi-organ autoinflammation are diagnosed later than patients with other diseases such as common variable immunodeficiencies (CVID) [17, 20].
The investigation of the total prevalence of patients with immune dysregulation syndromes is hampered by the limited number of participating centers and the inhomogeneous distribution of specialized clinical centers mainly in Germany. Therefore, the GAIN initiative tries to include centers distributed all over Europe to investigate the minimal prevalence of disorders with multi-organ autoimmunity/autoinflammation with this registry as a first step to describe and understand these rare diseases.
The GAIN registry fosters the cooperation and clinical and scientific exchange in the field of genetically determined immune dysregulation in Germany and beyond and complements the research projects of the GAIN consortium. The GAIN registry may form the basis for future research to improve patient’s life and quality of life, and helps to answer urgent questions of patients and their families.
Supplementary Information
Acknowledgements
This study was performed by the GAIN registry in collaboration with the ESID registry and used the ESID Online Database for data acquisition. We want to thank the ESID Registry Working Party for their support and collaboration.
Abbreviations
- ABACHAI
clinical trial investigating the safety and efficacy of the drug abatacept (ABA) in patients with CTLA4 haploinsufficiency (CHAI) or LRBA deficiency
- ATC
Anatomical-Therapeutic-Chemical classification
- BMBF
Bundesministerium für Bildung und Forschung, German Federal Ministry of Education and Research
- COVID-19
corona virus disease 2019
- CVID
common variable immunodeficiency
- DFG
Deutsche ForschungsGemeinschaft, German Research Foundation
- ESID
European Society for Immunodeficiencies
- et al.
et alii, and others
- GAIN
German multi-organ Autoimmunity Network
- GOF
gain-of-function
- HGVS
Human Genome Variation Society
- HPO
Human Phenotype Ontology
- HSCT
hematopoietic stem cell transplantation
- ICD
International Classification of Diseases
- IEI
inborn error of immunity
- Ig
immunoglobulin
- IQR
interquartile range
- IUIS
international union of immunological societies
- n
number; group size
- ORPHA
standard nomenclature of rare diseases
- p
p-value
- RefSeq
Reference Sequence
- RESIST
DFG funded excellence cluster Resolving Infection Susceptibility
- REST
REpresentational State Transfer
- SD
standard deviation
- WHO
World Health Organization
Author Contribution
B.G., G.K., A.N., P.S., S.R., M.R.J.S., K.W., F.A., S.E., F.H., B.F.H., U.B., M.K., U. G, A.C.G.d.O., and G.M., S.E-H. contributed to the study conception and design of the registry. S.R. realized the registry technically. Testing was performed by S.R., P.S., S. E-H., and G.M. Data collection was performed by S.E-H., G.M., M.K., B.G., K.W., F.A., R.K., H.v.B., S.B., P.B., S.E., F.H., M.C., M.F., M.B., J.F.N., U.G., B.F.H, and U.B.. Data were analyzed by P.S. and she wrote the first draft of the manuscript. All authors commented on previous versions of the manuscript and read and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. The GAIN registry is supported by the Center for Chronic Immunodeficiency and since 2019 funded by the Federal Ministry of Education and Research (BMBF, funding code 01GM1910A). The ESID registry platform is provided by the European Society for Immunodeficiencies (www.esid.org) and supported by societies’ internal budget and industry cooperation. Currently, the ESID registry receives external funds from Pharming.
The documentation specialist S.E-H. was funded by the Federal Ministry of Education and Research (BMBF, Support Code: 01GM1517C), by the European Society for Immunodeficiencies (ESID), 2018, by the Care-for-Rare Foundation, by PROimmun e.V., and by a restricted grant from LFB, CSL Behring, and Grifols. For GAIN, she was funded by the BMBF (GAIN support code: 01GM1910A). For RESIST, she was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy - EXC 2155 - project number 390874280. M.K. is supported by the Deutsche Forschungsgemeinschaft (DFG) SFB1160_2 as clinician scientist associated to IMM-PACT-Program, Faculty of Medicine, University of Freiburg, Freiburg, Germany. The views expressed in the submitted article are those of the authors’ and not an official position of their institutions or funders.
Data Availability
All participating centers can hand in research proposals to the GAIN consortium to access data of the GAIN registry. The created data structure of the GAIN registry can be interactively explored via the publicly accessible demonstration version of the ESID registry [1].
Declarations
Ethics Approval
The ESID registry and herewith the GAIN registry follows the guidelines outlined by the Declaration of Helsinki and was approved by the ethics committee of the University of Freiburg and by the local ethics committees of the participating centers.
Consent to Participate
A written informed consent was obtained from all individual participants included in the registry.
Consent for Publication
The written informed consent to participate in the registry also includes the consent to publish the data. Except for Fig. 2C and Fig. 3, only aggregated information is presented. No individual information is presented here which could lead to an identification of an individual.
Conflict of Interest
The authors declare no competing interests.
Footnotes
Mikko R. J. Seppänen On behalf of the ESID registry working party https://esid.org/Working-Parties/Registry-Working-Party
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Gerhard Kindle and Bodo Grimbacher contributed equally to this work.
Contributor Information
Paulina Staus, Email: paulina.staus@uniklinik-freiburg.de.
Bodo Grimbacher, Email: bodo.grimbacher@uniklinik-freiburg.de.
References
- 1.Scheible R, Rusch S, Guzman D, Mahlaoui N, Ehl S, Kindle G. The NEW ESID online database network. Bioinformatics. 2019;35:5367–5369. doi: 10.1093/bioinformatics/btz525. [DOI] [PubMed] [Google Scholar]
- 2.Lablans M, Borg A, Ückert F. A RESTful interface to pseudonymization services in modern web applications. BMC Med Inform Decis Mak. 2015;15:2. doi: 10.1186/s12911-014-0123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seidel MG, Kindle G, Gathmann B, Quinti I, Buckland M, van Montfrans J, et al. The European Society for Immunodeficiencies (ESID) registry working definitions for the clinical diagnosis of inborn errors of immunity. J Allergy Clin Immunol Pract. 2019;7:1763–1770. doi: 10.1016/j.jaip.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 4.WHO World Health Organization . International classification of diseases for mortality and morbidity statistics (11th Revision) 2018. [Google Scholar]
- 5.Orphanet: the online rare disease and orphan drug data base. Orphanet nomenclature of rare diseases. 1999. http://www.orpha.net. Accessed 16 Nov 2020
- 6.Köhler S, Gargano M, Matentzoglu N, Carmody LC, Lewis-Smith D, Vasilevsky NA, et al. The Human Phenotype Ontology in 2021. Nucleic Acids Res. 2021;49:D1207–D1217. doi: 10.1093/nar/gkaa1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO Collaborating Centre for Drug Statistics Methodology. ATC classification index with DDDs. Oslo, Norway; 2018. 2019
- 8.EMA European Medicine Agency . Article 57 export of “Brand name” for EU authorised medicines. 2020. [Google Scholar]
- 9.Lefter M, Vis JK, Vermaat M, den Dunnen JT, Taschner PEM, Laros JFJ. Next generation HGVS nomenclature checker. Bioinformatics. 2021; 10.1093/bioinformatics/btab051. [DOI] [PMC free article] [PubMed]
- 10.Yates AD, Achuthan P, Akanni W, Allen J, Allen J, Alvarez-Jarreta J, et al. Ensembl 2020. Nucleic Acids Res. 2020;48:D682–D688. doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bethesda (MD), National Library of Medicine (US), National Center for Biotechnology Information. National Center for Biotechnology Information (NCBI). 1988. https://www.ncbi.nlm.nih.gov/. Accessed 8 Mar 2022
- 12.Tangye SG, Al-Herz W, Bousfiha A, Cunningham-Rundles C, Franco JL, Holland SM, et al. Human Inborn Errors of Immunity: 2022 Update on the Classification from the International Union of Immunological Societies Expert Committee. J Clin Immunol. 2022;42:1473–507. doi: 10.1007/s10875-022-01289-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elsink K, van Montfrans JM, van Gijn ME, Blom M, van Hagen PM, Kuijpers TW, Frederix GW. Cost and impact of early diagnosis in primary immunodeficiency disease: a literature review. Clin Immunol. 2020;213:108359. doi: 10.1016/j.clim.2020.108359. [DOI] [PubMed] [Google Scholar]
- 14.Joshi AY, Iyer VN, Hagan JB, St. Sauver JL, Boyce TG. Incidence and temporal trends of primary immunodeficiency: a population-based cohort study. Mayo Clin Proc. 2009;84:16–22. doi: 10.4065/84.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schubert D, Bode C, Kenefeck R, Hou TZ, Wing JB, Kennedy A, et al. Autosomal-dominant immune dysregulation syndrome in humans with CTLA4 mutations. Nat Med. 2014;20:1410–1416. doi: 10.1038/nm.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fliegauf M, Bryant VL, Frede N, Slade C, Woon S-T, Lehnert K, et al. Haploinsufficiency of the NF-κB1 subunit p50 in common variable immunodeficiency. Am J Hum Genet. 2015;97:389–403. doi: 10.1016/j.ajhg.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shillitoe B, Bangs C, Guzman D, Gennery AR, Longhurst HJ, Slatter M, et al. The United Kingdom Primary Immune Deficiency (UKPID) registry 2012 to 2017. Clin Exp Immunol. 2018;192:284–291. doi: 10.1111/cei.13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seidel MG, Tesch VK, Yang L, Hauck F, Horn AL, Smolle MA, et al. The Immune Deficiency and Dysregulation Activity (IDDA2.1 ‘Kaleidoscope’) score and other clinical measures in inborn errors of immunity. J Clin Immunol. 2022;42:484–498. doi: 10.1007/s10875-021-01177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaggiano C, Vitale A, Tufan A, Ragab G, Aragona E, Wiesik-Szewczyk E, et al. The Autoinflammatory Diseases Alliance Registry of monogenic autoinflammatory diseases. Front Med (Lausanne). 2022;9:980679. doi: 10.3389/fmed.2022.980679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Helou SM, Biegner A-K, Bode S, Ehl SR, Heeg M, Maccari ME, et al. The German National Registry of Primary Immunodeficiencies (2012–2017) Front Immunol. 2019;10:1272. doi: 10.3389/fimmu.2019.01272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All participating centers can hand in research proposals to the GAIN consortium to access data of the GAIN registry. The created data structure of the GAIN registry can be interactively explored via the publicly accessible demonstration version of the ESID registry [1].