Summary
Exhausted T (TEX) cells are main immunotherapy targets in cancer, but it lacks a general identification method to characterize TEX cell in disease. To assess the characterization of TEX cell, we extract signature of TEX cell from large cancer and chronic infection cohorts. Based on single-cell transcriptomes, a systematic T cell exhaustion prediction (TEXP) model is designed to define TEX cell in cancer and chronic infection. We then prioritize 42 marker genes, including HAVCR2, PDCD1, TOX, TIGIT and LAG3, which are associated with T cell exhaustion. TEXP could identify high TEX and low TEX subtypes in pan-cancer of TCGA. The high TEX subtypes are characterized by high immune score, immune cell infiltration, high expression of TEX marker genes and poor prognosis. In summary, TEXP and marker genes provide a resource for understanding the function of TEX cell, with implications for immune prediction and immunotherapy in chronic infection and cancer.
Subject areas: Immunology, Bioinformatics, Cancer, Transcriptomics
Graphical abstract
Highlights
-
•
Exhausted T (TEX) marker genes are upregulated in exhaustion related datasets
-
•
TEX marker genes are enriched in a variety of immune functions
-
•
TEX marker genes recognize exhausted T cells in single-cell transcriptomes
-
•
High TEX subtype shows high immune infiltration and poor prognosis in pan-cancer
Immunology; Bioinformatics; Cancer; Transcriptomics
Introduction
When T cells are exposed to antigen for a long time, the expression of inhibitory receptors continues to increase. Then functional T cells are converted into exhausted T (TEX) cells, and their immune function, transcription and metabolism would be abnormal.1 This specific state of T cell is first investigated in a mouse model of chronic lymphocytic choriomeningitis virus (LCMV) infection, but recent studies have also discovered TEX cells in human chronic infection and cancers, which leads to the dysfunction of immune response for cancer patients.2,3 Immune checkpoint blockade therapy (PDCD1 and CTLA4) is subsequently developed to help restore T cell function, further demonstrating the importance of immune checkpoints in dysfunctional T cells. The immune checkpoint blocking therapy has shown to work well in few patients for melanoma, non-small-cell lung carcinoma, and kidney cancer, but not in most patients, and it is hard to provide an extensive and long-lasting effect for patients.4,5,6 This phenomenon may be caused by the diversification of inhibitory receptors in TEX cells during the tumor progress and the complicated tumor microenvironment.7 Moreover, some tumor-infiltrating T cells are exhausted status even without the high expression of PDCD1 or CTLA4.8
To characterize the features of TEX cells deeply, the gene expression patterns for TEX cells have been widely investigated across various cancer types or diseases with chronic infection. Different gene lists are identified as candidate marker genes based on their relatively high/low expression in TEX cells compared to other T cell types. Typically, the inhibitory receptors, such as PDCD1, HAVCR2 (TIM3) and TIGIT, are upregulated in TEX cells of cancer and chronic infection, which have been validated by different biological experimental methods.9,10 Besides, the transcription factor TOX which plays an important role during thymic development of CD4+ T lineage cells, natural killer and innate lymphoid cell is differentially expressed compared TEX cells with normal T cells in chronic infection model, melanoma, breast cancer, and lung cancer based on RNA-seq expression data.11TOX is also significantly overexpressed in melanoma and non-small cell lung cancer based on single-cell expression data.12 However, a large proportion of candidate marker genes are not always with consistent expression directions, such as KLRG1, CD160, LAG3, CTLA4, TBET, and EOMES.13,14 EOMES, a transcription factor, is highly expressed in chronic infection diseases but lowly expressed in liver cancer model.15 On the contrary, EOMES is overexpressed in lymphoma model and it has been demonstrated to exert bimodal functions in effector CD8+T cells and TEX cells in tumors.16 These results suggest that filtering stable marker genes of TEX cell is important, but many studies only focus on their interested direction and ignore the functional diversity. Moreover, these studies are mainly dependent on their own data, whose heterogeneity interferes with their reliability and prevents the identification of common markers. Therefore, it is essential to integrate currently available TEX transcriptomes across cancers and chronic infections to identify stable TEX markers.
With the development of single-cell technology, many studies have assayed the transcriptome profiles of immune cells in tumor microenvironment and identified immune cells including TEX cells. For example, Zhang et al. utilized single-cell expression profiles to identify different immune cell types in liver cancer, lung cancer, colorectal cancer and many other cancers, which used few marker genes to define immune cells such as LEF1 for naive T cell, CX3CR1 for effector T cell, LAYN for TEX cell and so on.17,18,19,20 Then we collected the exhaustion-related genes from Zhangs’ studies of liver cancer, lung cancer and colorectal cancer. The gene number was 82, 117, and 68 in liver cancer, lung cancer and colorectal cancer respectively, and the overlap of three sets was only 34. Marker genes in single-cell expression data were first identified by unsupervised clustering analysis, then they analyzed the differentially expressed genes as marker genes. This analysis process for single-cell data was also lack of commonality for marker genes. Therefore, marker genes identified by single-cell data needed to be further optimized, which could obtain robust information, accurately assess the influence of exhausted T cell state in tumor immune microenvironment and promote immune therapy.
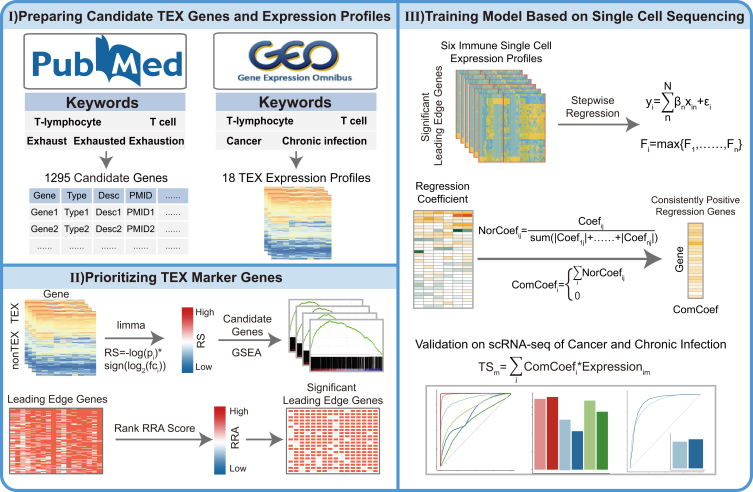
In this study, a systematic T cell exhaustion prediction (TEXP) model was developed to prioritize TEX marker genes and further explored TEX states in cancers (Figure 1). In the TEXP model, TEX-related gene list was obtained by searching PubMed. Then, Gene Set Enrichment Analysis (GSEA) and robust rank aggregation (RRA) were applied to filter genes based on multiple expression datasets from chronic infection in cancer, chronic hepatitis virus and chronic LCMV. Third, robust and stable gene signature was further refined by stepwise regression analysis based on single-cell transcriptomic data. Finally, the gene signature was used to predict the TEX tendency for TCGA transcriptomic data. The gene signature and framework contributed a novel resource that could serve future studies and improve the understanding of exhaustion in chronic infection and cancer.
Figure 1.
Schematic illustration of three steps of TEXP for the identification of exhausted pattern
Results
TEX candidate genes tend to be upregulated in chronic infection and cancer
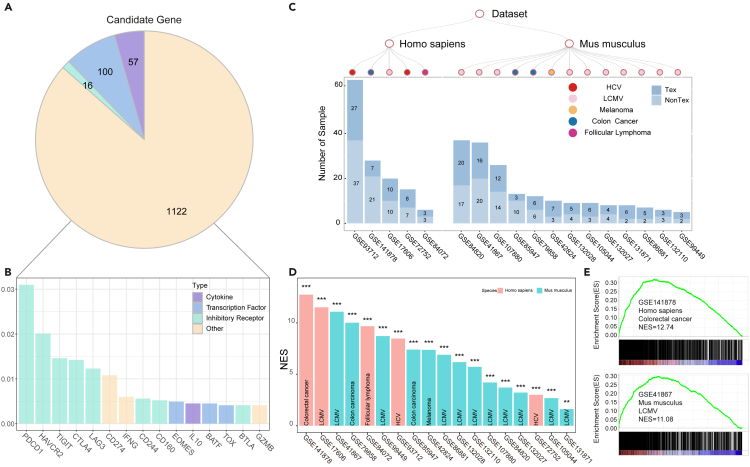
To extract the candidate genes associated with TEX cells, a combination of different key words of TEX and cell type was employed to search PubMed and 1,295 candidate genes were collected (Figure 2A). These candidate genes concluded 57 cytokines and their receptors, 16 inhibitory receptors and 100 transcription factors. After calculating the literature number, we found 1.16% candidates recurrently occurred in more than 10 literature, such as PDCD1, HAVCR2, TIGIT, CTLA4, LAG3, CD274, CD244, CD160, EOMES and IL10, indicating their key roles for TEX cells (Figure 2B). For example, inhibitory receptors PDCD1 and CTLA4 were the common targets for immune checkpoint blocking therapy, and LAG3, CD244, CD160 and HAVCR2 have also been regarded as inhibitory receptors to inhibit the immune process.21,22 These results suggested that inhibitory receptors played a major role in TEX cells. Besides, EOMES with different expression level in different datasets were also highly frequent in our gene lists.16 Therefore, it was necessary to integrate different datasets to explore the mechanism of TEX cells.
Figure 2.
The identification of TEX marker genes
(A) The distribution of candidate genes.
(B) The candidate genes with high frequency of literature mining.
(C) The distribution of bulk expression profiles. The dendrogram showed the species and infection type. The bar diagram showed the sample number.
(D) The NES value of 18 TEX expression profiles from GSEA.
(E) Enrichment of candidate genes in different types of data.
To characterize the consistent expression performance of candidate genes in TEX cells, 18 mRNA expression profiles for TEX cells in chronic infection and cancer were downloaded from GEO, which were composed of 5 human expression profiles and 13 mouse expression profiles23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39 (Figure 2C). First, the Pearson correlation coefficients among TEX samples were calculated through the candidate genes (Figure S1). The result showed that the datasets in different species and diseases had the similar expression pattern and could be used to explore the TEX phenotype. In addition, to measure whether candidate genes were upregulated in TEX cells, differential analysis between TEX and nonTEX samples were made for all datasets. The differential analysis was made through 18 mRNA expression profiles, respectively. Therefore, the different species and batches didn’t influence our result. The p value and fold-change were converted to an RS score for each gene in every dataset and genes in every dataset were ranked based on their own RS scores. Next, the activity of candidate genes was computed in all datasets through GSEA.40 The result showed that the candidate genes were significantly enriched in upregulated genes for all datasets (NES>0 and FDR<=0.05) (Figure 2D). For example, candidate genes were significantly enriched in upregulated genes in human colon cancer and mouse LCMV (Figures 2E and S2). This result also demonstrated the strong relationship between candidate genes and TEX cells. Leading edge analysis of GSEA could help determine the bona fide critical genes, but there were ranking differences for candidate genes in different datasets (FigureS3). Therefore, it was necessary to analyze their performance across these datasets comprehensively.
Prioritizing TEX marker genes
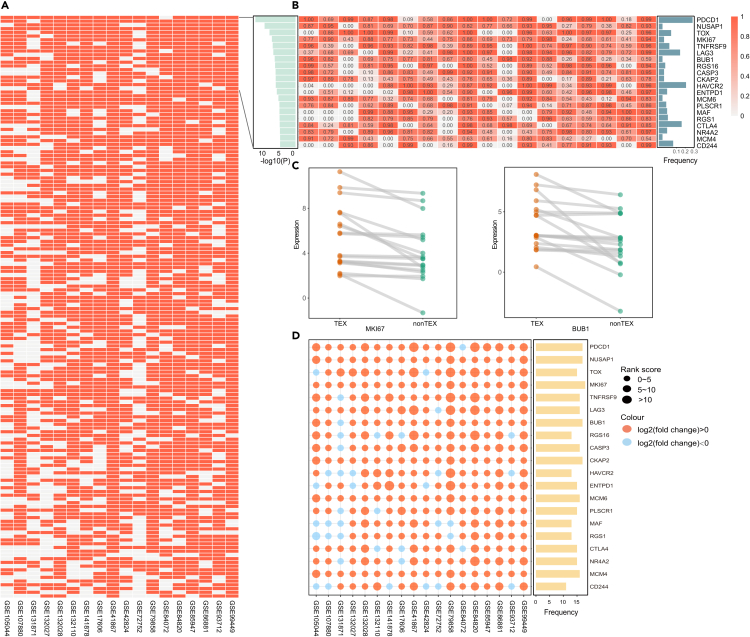
To filter the marker genes in TEX phenotype, it was hypothesized that marker genes should be stably activated in TEX state, so the RRA method was used to evaluate the consistently upregulated expression patterns.41 According to the RS score, we sorted the genes in descending order for 18 datasets and extracted genes in top-rank location of aggregated rank list. 156 genes were selected with the threshold value<=0.05 (Figures 3A and 3B). RRA assessed genes strongly contributing to the enrichment, including PDCD1, LAG3, CTLA4, TOX, CD38, TIGIT and HAVCR2, and their expressions were consistently upregulated in TEX samples (Figure 3D). Besides, some other genes rarely reported to be associated with immune response were top-ranked as well (Figures 3B and 3C). For example, MKI67 was highly expressed in 18 datasets, which was demonstrated to be upregulated in PD1 high TIM3+ CD8 T cells in the AT-3 tumor model and was associated with the immune tolerance signature in breast cancer.42,43 BUB1 was reported to be related to the tumor immune and immune infiltration.44,45 Therefore, some genes in TEX cells without highlighting in previous research were identified through our criteria.
Figure 3.
The optimization of candidate genes based on leading edge genes and RRA
(A) The global ranking result of candidate genes after RRA. The row and column represented gene and datasets, and the grid represented the leading edge genes in datasets.
(B) The ranking weight of top 20 genes from leading edge genes of GSEA. The left bar plot was the pvalue of RRA. The middle heatmap was the ranking weight of leading edge genes. The right bar plot represented the frequency of literature mining.
(C) The expression levels of MKI67 and BUB1 with low frequency of literature mining.
(D) The expression feature of top 20 genes for RRA. The left dot plot represented the expression levels between TEX and non-TEX samples. The size and color represented the RS score and log2(fold change). The right bar plot represented the overlap of leading edge genes in 18 expression profiles.
TEX marker genes involved in immune functions
To assess the function of 156 stable marker genes, we made functional enrichment analysis with Enrichr (https://maayanlab.cloud/Enrichr/) to validate their relevance with TEX functions (adjusted pvalue ≤0.05). These genes were significantly enriched in 67 biological processes, which were mainly involved in immune functions (Figure 4A), such as cytokine-mediated signaling pathway (p = 7.26E-07), regulation of B cell proliferation (p = 4.98E-05), positive regulation of immunoglobulin production (p = 0.004), regulation of regulatory T cell differentiation (p = 0.005), inflammatory response (p = 0.007), positive regulation of defense response (0.029) and so on. Then, to systematically characterize the effect of candidate genes on biological processes, the biological processes were categorized based on the upstream-downstream relationship of gene ontology, and cytokine regulation (23 subclasses), metabolic process regulation (12 subclasses) and T cell regulation (10 subclasses) were dominant in several broad categories (Figure 4A). Previous studies have demonstrated that various cytokines and immunomodulatory factors could be produced through the stimulation of chronic inflammation, which would stimulate other immunosuppressive cells, eliminate the metabolites related to T cell function, and produce enzymes to strengthen the immunosuppressive effect. Abnormalities in immune metabolic process could influence the fate of immune cells and regulate immunity, which was often used as a key to the cancer treatment. Besides, the candidate genes were also enriched in 33 KEGG pathways (Figure 4B), such as cell cycle (p = 7.66E-08), cytokine-cytokine receptor interaction (p = 5.62E-05), inflammatory bowel disease (p = 5.63E-05), Th17 cell differentiation (p = 9.96E-05), natural killer T cell regulation (p = 3.87E-04), TNF signaling pathway (p = 0.019), multiple cancer pathways (non-small-cell lung carcinoma, melanoma, Glioma, leukemia), and multiple viral infections (Kaposi sarcoma-associated herpes virus infection, Epstein-Barr virus infection and hepatitis C virus infection). The results showed that candidate genes not only played regulatory roles in helper T cell and effector T cell pathway, but also participated in cell cycle and cancer-related pathways. In addition, these genes were involved in a variety of viral infection-related pathways, suggesting that they were related to exhaustion of chronic infection and tumor development.46,47 Therefore, abnormalities of candidate genes could lead to dysregulation of immune response and inflammatory response, and might eventually lead to exhaustion of immune cells and escape of cancer cells.
Figure 4.
The result of functional enrichment analysis
(A) The heatmap was the result of biological process.
(B) The heatmap was the result of KEGG pathway. The row and column of heatmap represented the gene and functional name, respectively. The right bar plot was the p value of functional enrichment analysis. The bottom bar plot was the enriched gene number in each function. The Sankey diagram represented the upstream-downstream relationship of biological processes.
Validation of TEX marker genes in single-cell transcriptomes
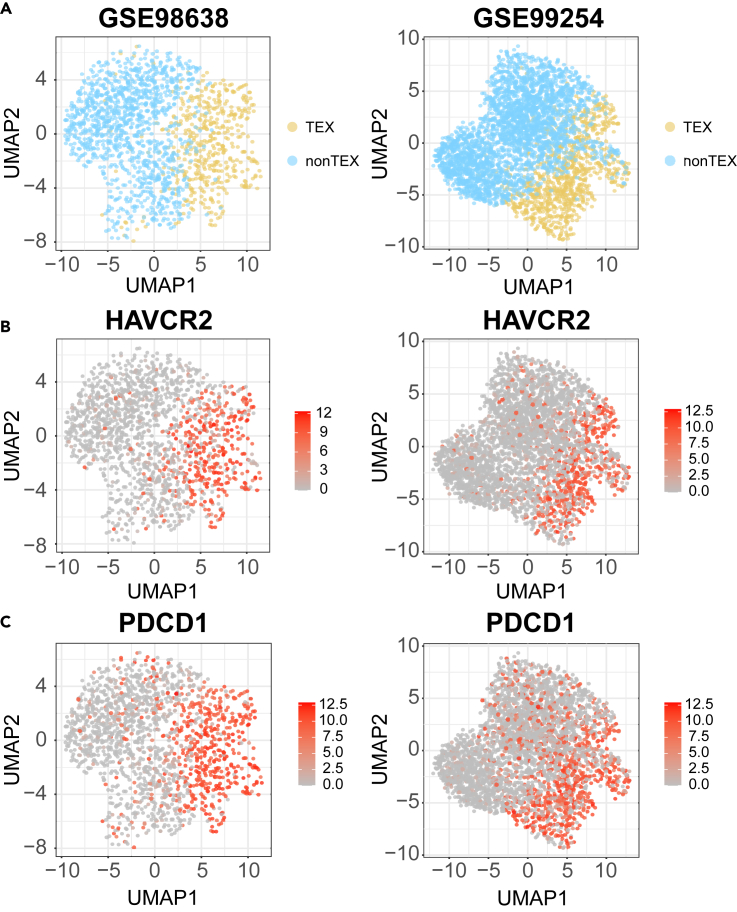
Single-cell transcriptome had more precise and reliable expression pattern than bulk expression data, so six human single-cell expression profiles were utilized to characterize the TEX cells.17,18,48,49,50 To visualize the complex phenotypes between TEX cells and nonTEX cells, a Uniform Mani-fold Approximation and Projection for Dimension Reduction (UMAP)-based dimensionality reduction approach was used to integrate the expression information of 156 marker genes based on single-cell expression profiles51 (Figure 5). The result showed that TEX and nonTEX cells could be divided into two groups and have distinct phenotype respectively (Figure 5A). Then the UAMP result displayed the expression pattern of marker genes. These genes were highly expressed in TEX cells (Figure 5B). Besides, cells locating in the intermediate position showed intermediate expression pattern and might play a role in transition between TEX cells and nonTEX cells.
Figure 5.
The UMAP result of single-cell expression profiles
(A) The cell distribution of TEX and nonTEX cells. The yellow and blue were the TEX and nonTEX cells.
(B) The expression distribution of HAVCR2 in GSE98638 and GSE99254.
(C) The expression distribution of PDCD1 in GSE98638 and GSE99254. The dark color represented high expression.
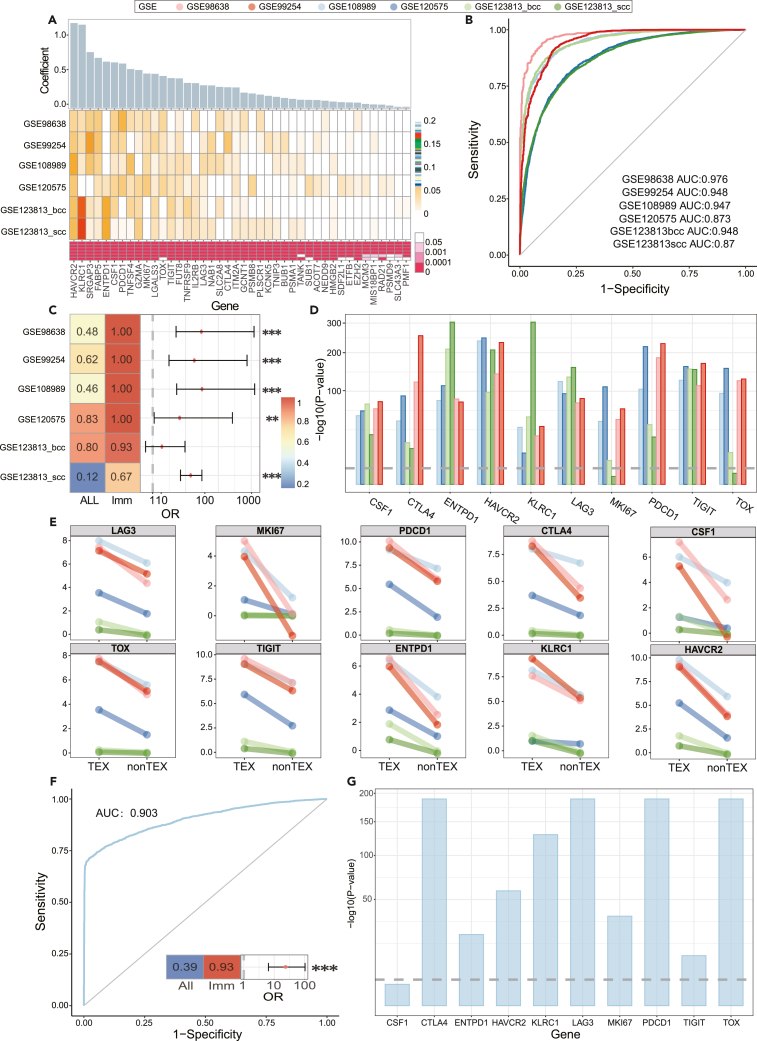
Marker genes have been demonstrated to play an important role in TEX cells and immune response. Because of the precision of single-cell transcriptome, six single-cell expression profiles were used to validate marker genes. First, 156 marker genes and 6 single-cell expression profiles were used to make multiple stepwise linear regression analysis (p value<=0.05). The marker genes were divided into three categories based on the regression coefficients in six expression profiles: (1) 42 consistently positive regression genes; (2) 26 consistently negative regression genes; and otherwise. Positive regression genes represented the positive relationship between gene expression and TEX level. Recent infiltration-related studies often used positive regression genes to analyze the tumor immune environment and demonstrated the reliability and stability of positive regression genes.52,53 Therefore, 42 consistently positive regression genes were selected to construct classifier model (Figure 6A and Table S1). The combined regression coefficient (ComCoef) was calculated based on coefficients in six single-cell expression profiles and was used to compute the TEX score (TS) for each cell. The classifier model was applied in six single-cell expression profiles. The result showed that the AUCs were greater than 0.85, which demonstrated the model efficiency on separating TEX and non-TEX cells (Figure 6B). In addition, most marker genes were significantly differential between TEX cells and nonTEX cells (P<=0.001, nonparametric rank-sum test), and more than 60% genes showed differentially expressed in six single-cell expression profiles respectively (Figures 6A and 6C). For example, LAG3, MKI67, PDCD1, CTLA4, CSF1, TOX, TIGHT, ENTPD1, KLRC1, and HAVCR2 were all highly expressed in TEX cells. Interestingly, the marker genes were more likely to show expression perturbation than other genes across most profiles (p< 0.01 in five out of six profiles, two-sided Fisher’s exact test) (Figures 6D and 6E). All marker genes were differential in GSE98638, GSE99254, GSE108989 and GSE120575, which revealed the perturbation of marker genes in TEX cells. In summary, marker genes could accurately assess TEX cells and provide diagnostic and therapeutic evidence for immunotherapy based on single-cell transcriptomes of human cancer.
Figure 6.
The characterization of single-cell expression data for human cancer and mouse chronic infection
(A) The regression coefficients of marker genes. The top bar plot represented the ComCoefs of marker genes. The middle heatmap represented the regression coefficients of marker genes in each single-cell expression profiles. The bottom heatmap represented the p-value of marker genes in stepwise regression analysis.
(B) The AUC value of six single-cell expression profiles.
(C) The left heatmap showed the proportion of differentially expressed genes in all genes and marker genes respectively. The right panel showed the odds ratios in six single-cell expression profiles. ∗∗∗ meant P<=0.001, ∗∗ meant 0.01≤P≤0.001, two-sided Fisher’s exact test.
(D) The significance of marker genes between TEX and nonTEX cells in six single-cell expression profiles. Threshold value P<=0.001, nonparametric rank-sum test.
(E) The expression levels of marker genes between TEX and non-TEX samples in six single-cell expression profiles. The xaxis shows the different cell types, and the yaxis shows the expression levels.
(F) The AUC value of mouse single-cell expression profile. The heatmap and panel represented the proportion of differentially expressed genes and odds ratios as well.
(G) The significance of marker genes between TEX and non-TEX cells in mouse single-cell expression profiles. Threshold value P<=0.001, nonparametric rank-sum test.
Studies have also found that chronic infection was often regarded as a precursor to cancer, and more and more research confirmed that patients with chronic inflammatory disorders would develop into cancer eventually. To explore the TEX mechanism in chronic infection, single-cell transcriptome (GSE131535) from mouse LCMV chronic infection was collected to validate the role of marker genes.54 The AUC value was 0.903 and demonstrated the role of marker genes in mouse chronic infection model (Figure 6F). Besides, the expression feature of marker genes was also estimated, which displayed a similar trend like human cancer model (Figures 6G and S4). For example, CTLA4, ENTPD1, HAVCR2, KLRC1, LAG3, MKI67, PDCD1, TIGIT, and TOX were highly and differentially expressed in TEX cells. The marker genes were also more disturbed than other genes in this dataset. Collectively, TEX model was a useful pipeline to identify the TEX feature in chronic infection and cancer.
TEX marker genes highlight immune subtypes in pan-cancer
To estimate the TEX application on bulk transcriptomes, 32 expression profiles were downloaded from TCGA and classifier was used to calculate the TSs for each patient. There was no pre-defined TEX classification for TCGA, therefore we utilized the TSs to define patient samples as follows: (1) High TEX subtype was defined as those with more than three-quarters of the quartile TS. (2) Low TEX subtype was defined as those with less than one-quarter of the quartile TS.
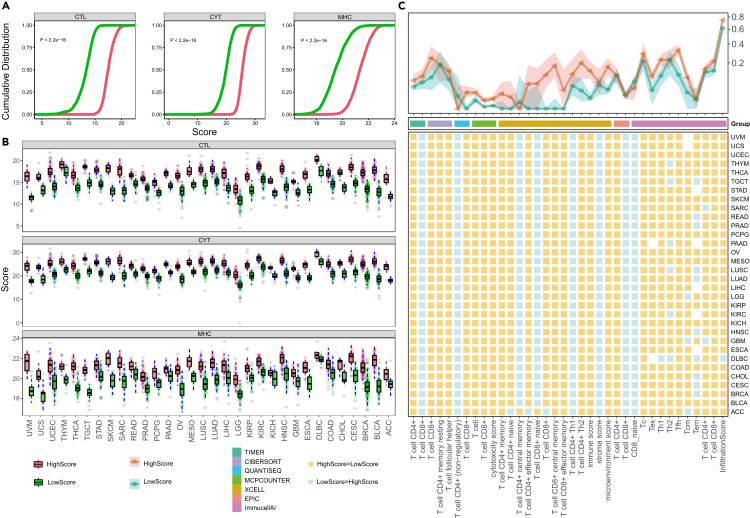
To assess the difference in immune effects between high TEX and low TEX subtypes, both subtypes were evaluated through multiple immune scores: MHC (major histocompatibility complex), CYT (cytotoxic activity), and CTL (cytotoxic T lymphocyte).55,56,57,58 These scores were defined as the average of marker genes as follows: HLA-A, HLA-B, HLA-C, TAP1, TAP2, NLRC5, PSMB9, PSMB8 and B2M for MHC; GZMA and PRF1 for CYT; GZMA, PRF1 and GZMB for CTL. The result showed that high TEX subtype had significantly higher MHC, CYT and CTL scores than low TEX subtype among pan-cancer samples (P<=2.2e-16, Kolmogorov-Smirnov Test) (Figure 7A). In addition, we used one-quarter quantile and three-quarter quantile to divide samples for individual cancer, and the immune score of high TEX subtype was higher than that of low TEX subtype. These results further suggested that the relationship between high exhaustion and immune score was uniform in different cancers (Figure 7B). Previous studies have demonstrated that CYT and CTL were positively correlated with the levels of TEX cells and inhibitory receptors (PDCD1, PDL1, CTLA4, LAG3, TIM3 and IDO1).55,59 In addition, the relationship between TEX subtypes and immune infiltration was examined. The immune infiltration tools and methods included TIMER2.0 and ImmuCellAI. TIMER2.0 has integrated xCell, MCP-counter, EPIC, QuanTIseq, CIBERSORT and TIMER.52,53,60,61,62,63,64,65 We next extracted T cell-related score and all kinds of T cells such as CD4+T cell, CD8+T cell, CD8+ Memory T cell, CD8+ Effector T cell, CD4+ naive T cell and exhausted T cell. In summary, 37 kinds of scores and immune infiltration levels were collected. Almost all results showed significantly higher infiltrating levels in high TEX subtype than low TEX subtypes for pan-cancer and individual cancer (Figure 7C). The stromal score demonstrated the positive correlation between TEX level and tumor malignant level.66,67 The T cell infiltration and immune-related score indicated that immune infiltration was accompanied by exhaustion of functional immune cells in cancer patients. And the infiltration score of TEX cell in ImmuCellAI was also consistent with our TEX score, which proved the reliability of our result. Previous studies have found that although cancer patients had a high level of immune infiltration, the functional immune cells were induced into dysfunctional status.68 The effect of immunotherapy did not achieve the desired result. Therefore, our hypothesis was that the T cell exhaustion would disturb therapeutic effect and prognosis for cancer patients.
Figure 7.
The immune score and infiltration of different TEX subtypes
(A) The immune score in different TEX subtypes for pan-cancer.
(B) The immune score in different TEX subtypes for individual cancer.
(C) The immune infiltration in different TEX subtypes. The top line plot represented the immune infiltration in different TEX subtypes for pan-cancer. The middle color bar represented the different tool sources of immune infiltration. The bottom heatmap represented the immune infiltration in different TEX subtypes for individual cancer.
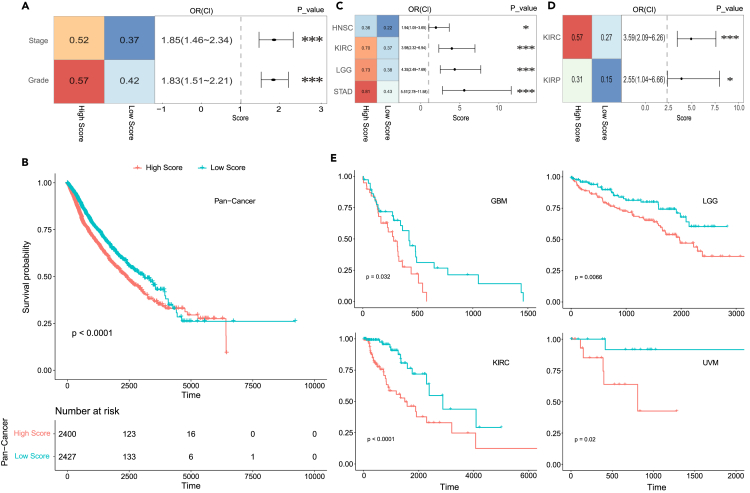
To explore the role of TEX marker genes in clinical application, the malignant level and patients’ prognosis were used to evaluate the exhausted score. There were two criteria for tumor malignant level: grade and stage. The tumors were classified into four levels: well differentiated G1, moderately differentiated G2, poorly differentiated G3, and poorly differentiated G4. Based on the TNM stage (T: Primary Tumor Range, N: Peripheral Lymph Node Metastasis, M: distal metastasis), the stages of TCGA tumors were divided into four levels: Stage I, stage II, stage III and stage IV. This tumor clinical information came from the Eighth Edition AJCC Cancer Staging Manual.69 10 cancers had grade information (CESC, ESCA, HNSC, KIRC, LGG, LIHC, OV, PAAD, STAD and UCEC), and 22 cancers had stage information (ACC, BLCA, BRCA, CESC, COAD, ESCA, HNSC, KIRC, KIRP, LIHC, LUAD, LUSC, MESO, OV, PAAD, READ, SKCM, STAD, TGCT, THCA, UCEC, and UVM). G1 and G2 were defined as low grade level, G3 and G4 as high grade level. Stage I and Stage II were defined as low stage level, and stage III and stage IV as high stage level. Based on the pan-cancer subtypes, high TEX subtype had more malignant patients than low TEX subtype (grade: 0.57 versus 0.42; 0.52 versus 0.37) (Figure 8A). Then survival analysis was applied to assess the relationship between survival prognosis and TEX subtypes, and high TEX subtype displayed poorer prognostic status than low TEX subtype (Figure 8B). Next, we estimated the malignant level and survival prognosis in individual cancer. Grade in HNSC, KIRC, LGG and STAD and stage in KIRC and KIRP had more malignant patients in high TEX subtype than in low TEX subtype (P<=0.05, two-sided Fisher’s exact test) (Figures 8C and 8D). Besides, high TEX subtype had poorer prognostic status than low TEX subtype in GBM, LGG, KIRC and UVM (P<=0.05, Logrank Test) (Figure 8E). Previous studies found that PD-1/PD-L1 checkpoint blockade therapy have not achieved breakthroughs in treating glioblastoma because of its low immunogenic response and immunosuppressive microenvironment. Moreover, the phenotypes of tumor infiltrating lymphocytes (TILs) in glioma specimens were found to be rich in CD95, PDCD1, PDL1, CTLA4, LAG3, and TIM3.70 Studies also showed a link between immunity and progression of kidney cancer. Kidney cancer might mediate immune dysfunction by inducing immunosuppressive cells to infiltrate the tumor microenvironment, and several possible mechanisms have also been identified to explain how these tumor-infiltrating cell types impeded the development of an anti-tumor immune response, including regulating the activity of effector T cell and antigen-presenting cell by inhibiting factors such as checkpoint molecules.71 Furthermore, inhibitory receptors CTLA4 and LAG3 have been demonstrated to be associated with poor prognosis in kidney cancer.72 Collectively, these results exhibited the negative role of TEX in clinical treatment and further provided new ideas to strengthen the efficacy of cancer immunotherapy.
Figure 8.
The clinical application of different TEX subtypes
(A) The left heatmap showed the proportion of malignant tumors in different TEX subtypes for pan-cancer. The right panel showed the odds ratios for pan-cancer. ∗∗∗ meant P<=0.001, ∗∗ meant 0.01≤P≤0.001, two-sided Fisher’s exact test.
(B) The survival curve between different TEX subtypes for pan-cancer.
(C) The left heatmap showed the proportion of malignant tumors based on grade in different TEX subtypes for individual cancer. The right panel showed the odds ratios for individual cancer.
(D) The left heatmap showed the proportion of malignant tumors based on stage in different TEX subtypes for individual cancer. The right panel showed the odds ratios for individual cancer.
(E) The survival curve between different TEX subtypes for individual cancer.
Discussion
Accumulating evidence suggests that TEX cells are important for immune response and immune therapy. However, the ability to identify TEX cells by existing methods has remained challenging because of the lack of distinct and stable marker genes of TEX cells. For example, many marker genes involved in exhaustion, such as PDCD1 and CTLA4, are also expressed in other activated cells.73,74 In this study, we first extracted TEX-related candidate genes through the literature mining, and explored expression feature of TEX cells using tumor and chronic infection transcriptomes, which revealed the transcriptional misregulation of TEX cells and highly biased to TEX phenotype. Then the stable marker genes were used to train a model through single-cell data and were able to identify TEX cells. It was also demonstrated that this model helped to identify TEX subtypes in 32 cancer types. The high TEX subtype was likely to display high immune score and high immune infiltration. Notably, high TEX subtype also led to high malignant level and poor prognosis for pan-cancer patients.
TEX cells are emerging as crucial regulators in immune system and immunotherapy. However, only a few stable TEX marker genes help to determine the identity of TEX cells. Therefore, TEXP model was constructed to prioritize stable marker genes and validate TEX role in pan-cancer. We found that TEX candidate genes tended to be upregulated in chronic infection and cancer. Besides, these genes were also associated with many immune-related functions such as cytokine regulation, T cell regulation, cytokine-cytokine receptor interaction, inflammatory bowel disease, natural killer T cell regulation and so on. Next, cancer and chronic infection single-cell transcriptomes were employed to validate the marker genes, which could help to recognize the identity of TEX cells in single-cell transcriptomes. Moreover, a significantly higher proportion of TEX marker genes were co-occurred with exhaustion in single-cell data.
Increasing studies suggest that TEX may interfere with immunotherapy. Therefore, TEXP model was used to determine a cutoff to classify tumors into high TEX subtype and low TEX subtype. The results showed that high TEX subtype had higher immune infiltration and immune score than low TEX subtype in pan-cancer. Furthermore, patients in high TEX subtype displayed higher tumor malignant level as well as worse survival prognosis than low TEX subtype, which went in the opposite direction from previous conclusions that higher immune infiltration and immune score indicated a better chance of survival. Many previous studies also demonstrated our finding that some tumors had a high infiltration level of cytotoxic T cells, but these T cells were induced as dysfunctional cells, which tended to keep a dysfunctional status and disturb the immune response in tumors.75,76,77 These results reminded that evaluating patients’ prognosis should consider of both immune infiltration and TEX level simultaneously.
In summary, TEXP provides some biological insights to analyze immune system. First, the enrichment results show that TEX candidate genes tend to be upregulated in TEX cells. These results indicate that highly expressed genes have more stable status in TEX cells. Second, TEXP helps prioritize TEX marker genes and its filtered genes are demonstrated to be associated with immune-related functions. Moreover, many reliable marker genes that are rarely analyzed before are identified. These results suggest that a proportion of marker genes could be detected at sufficiently robust expression levels to act as biomarkers. Third, based on single-cell transcriptomes, the characterization of TEX marker genes is validated. Finally, based on the bulk transcriptomes, it is demonstrated that TEXP could highlight different TEX subtypes (high TEX subtype, low TEX subtype), which show distinct responses to immune score, immune infiltration, malignancy and prognosis.
Limitations of the study
Identification of the marker genes is a critical step to investigate TEX function. However, limited and unstable marker genes restrict studies of TEX cells. In the present study, we prioritize the TEX marker genes and further investigate the function of TEX marker genes through immunity and clinical information. However, there are some limitations to our study. This study only focuses on the identification of TEX features, but does not provide the guidance to improve the efficacy for immunotherapy effectively. It is possible that this study could be used to further develop new direction to achieve a comprehensive prediction performance for clinical treatment.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| See Table S2 | This study | N/A |
| Software and algorithms | ||
| R version 4.0.2 or higher | Open source | https://www.r-project.org/ |
| GSEA | Open source | https:www.gsea-msigdb.org/ |
| Python (version 3.6) | Open source | https://www.python.org/ |
| randomForest (version 4.6.12 in R) | Open source | N/A |
| survival (version 3.1.12 in R) | Open source | N/A |
| ggplot2 (version 3.3.5 in R) | Open source | N/A |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Xia Li (lixia@hrbmu.edu.cn).
Materials availability
This study did not generate new materials.
Method details
Collection of exhaustion-related genes
In order to extract the candidate genes associated with TEX cells, PubMed was searched through the keywords (("t-lymphocytes" OR "T cell") AND (“exhaust” OR “exhausted” OR “exhaustion”)), and 2,529 exhaustion-related publications were available. Then, we carefully checked the abstracts or the full texts manually to obtain candidate genes. Since the candidate genes came from different versions of human or mouse reference genome, the homologous gene information of human and mouse as well as the Entrez ID information of human genes from NCBI were utilized to transform the candidate genes into standard human gene symbols. Finally, 1,295 candidate genes of exhausted T cells were collected.
Bulk and single-cell expression across different TEX types and cancer types
This analysis mainly consisted of three parts of data: bulk TEX expression data, single-cell TEX expression data and TCGA expression data. To verify the function of candidate genes in immune microenvironment, 18 sets of bulk mRNA expression profiles of tissues associated with TEX cells were downloaded from GEO database, which were composed of 5 human expression profiles and 13 mouse expression profiles. The data were mainly divided into two categories: cancer and chronic infection, which embraced melanoma, colon cancer, follicular lymphoma, normal tissues treated with lymphocyte meningitis virus (LCMV) or hepatitis virus (HBV, HCV), and tissues that were positive for common inhibitory receptors (PDCD1, TIM3, etc.). The detailed information for profiles were in Figure 1A. Next, six single-cell expression profiles of human cancers were downloaded from GEO, which included GSE98638, GSE99254, GSE108989, GSE120575 and GSE123813, and covered liver cancer, lung cancer, colorectal cancer, melanoma, basal cell carcinoma and squamous cell carcinoma. Besides, a single-cell expression profile of mouse treated with LCMV (GSE131535) was also obtained from GEO. For TCGA data, 32 mRNA expression profiles and their clinical information were downloaded. The clinical information of tumor patients, including the survival status, stage, grade and survival time were also obtained from the TCGA project. The sample information of different cancers was in Table S2.
According to the annotation information provided by each bulk dataset, the samples from GEO was divided into TEX and nonTEX. The exhaustion was mainly caused by continuous chronic inflammation stimulation to the functional T cells. TEX samples included cancer tissues, normal tissues treated with LCMV or hepatitis virus, and tissues that were positive for common inhibitory receptors (PDCD1, TIM3, etc.). NonTEX samples included tissues with immune function (initial T cells, effector T cells, memory T cells, etc.), tissues treated with acute virus, and tissues that were negative for common inhibitory receptors (PDCD1, TIM3, etc.). For single-cell data, the category of each cell was defined through the fluorescence-activated cell sorting analysis and annotation information of dataset.
The normalized microarray data and RNA-seq data did not need to be processed. For data without normalization, the microarray expression profiles were normalized via the R package “limma”, and the RNA-seq expression profiles were converted to CPM (Counts per million) with the R package “edgeR”.78,79 The single-cell expression profiles were normalized via the R package “Seurat” with the default parameter.80 All bulk and single-cell expression profiles were log-transformed. Based on the homology information and human gene annotation information in NCBI and GENCODE, all expression profiles were converted into the human Entrez ID. Finally, the expression profiles covered 23,048 human coding genes.
TEXP: Optimization of TEX marker genes
To identify the stable TEX marker genes, we proposed a computational method to prioritize candidate marker genes and explore their functions. Briefly, all genes were ranked based on their difference in expression compared TEX and nonTEX samples. The ranked gene list was subjected to candidate genes to explore whether the TEX genes were enriched in the top or bottom of the list. This process was repeated for all 18 bulk expression profiles of GEO. Based on the result of enrichment analysis, all genes with significantly higher RRA scores were identified in TEX samples.41
Identification of differential genes
We first used R package “limma” to make differential analysis for GEO bulk expression profiles.79 For each gene, we calculated the rank score (RS) as follows:
Where pi and fci was the p-value and fold-change value of differential analysis for gene i, respectively. The RS was calculated in 18 bulk expression profiles of GEO. All genes were ranked based on RS.
The prioritization of candidate genes
The enrichment analysis was made to identify exhaustion-related genes. Firstly, the GSEA was used to make enrichment analysis through the ranked gene list. Next, we collected the ranked leading edge genes of expression profiles whose FDR values were less than 0.05 and NES scores were more than 0. Finally, the robust rank aggregation method was applied to integrate the leading edge genes with the R package “RobustRankAggreg”, the genes with P-value less than 0.05 were regarded as marker genes associated with TEX stably.
The validation of TEX marker genes based on single-cell transcriptomes
To further prioritize TEX marker genes and validate the function of TEX marker genes, a TEX classifier was constructed through single-cell expression profiles and stepwise regression analysis. The stepwise regression analysis was carried out to further optimize the marker genes. Then the gene regression coefficients were integrated to construct the TEX classifier, and the efficacy of TEX classifier was validated in single-cell expression profiles.
Optimization of marker genes
For the single-cell expression profile, we first converted the TEX and non-TEX cell identity into 1 and 0, respectively. Then the cell identity and marker gene expression were regarded as dependent variable and independent variable to input into multiple stepwise linear regression analysis for training model. The multiple linear regression was as follow:
Where, y was the vector , and represented the cell identity in cell n; X was the matrix , and represented the expression level of gene p; was the vector , and represented the expression level of gene p in cell n; β was the vector , and represented the regression coefficient of gene p; ε was a constant term. The threshold value of multiple stepwise linear regression analysis was p ≤0.05, and the significant genes were used to make further analysis.
This multiple stepwise linear regression analysis was repeated for all six single-cell expression profiles. Next, the regression coefficient of significant genes from six single-cell expression profiles was obtained to construct TEX classifier.
The construction and validation of TEX classifier
We first built a regression coefficient matrix whose row was union set of significant genes in six single-cell expression profiles and column was the GEO ID (Table S1). The missing value in this matrix was filled with zero. For each gene, the regression coefficient was then normalized as follows:
Where i and j were the number of significant genes and single-cell expression profiles, Coefficientij was the regression coefficient of gene i in profile j. After normalization, the comprehensive regression coefficient was calculated to construct TEXP model classifier as follows:
Where i and j were the number of significant genes and single-cell expression profiles. In this section, we extracted the significant genes whose regression coefficients in six single-cell expression profiles were all no less than zero. Finally, the comprehensive coefficients were used to calculate to TEX score (TS) as follows:
Where i and m were the number of significant genes and cell number in each single-cell expression profiles. After calculating the TS for each cell, the efficacy of TEXP was validated in six single-cell expression profiles through the R package “pROC” respectively.
Quantification and statistical analysis
The detailed information of quantification and statistical analysis were described in method details.
Acknowledgments
This work was supported by the National Key R&D Program of China (2018YFC2000100), China Brain Project (2021ZD0202403), National Natural Science Foundation of China (62202094, 31970646, 32060152, 32070673, 32170676), HMU Marshal Initiative Funding (HMUMIF-21024), Natural Science Foundation for Distinguished Young Scholars of Heilongjiang Province of China (JQ2019C004), Heilongjiang Touyan Innovation Team Program, the Fundamental Research Funds for the Central Universities [2572022BH01] and the Postdoctoral Program of Heilongjiang Province of China [LBH-Z22065].
Author contributions
X.L., J.X., and Y.L. conceived of the project. C.Z., Q.S., and X.Z. designed and performed the research with contributions from K.X., M.Z., X.J., W.Z., D.L., and C.Y. C.Z. and X.Z. searched the candidate marker genes. C.Z. and Q.S. analyzed the single-cell data. J.X., Y.L., and X.L. supervised research and provided critical advice on the study. C.Z. and J.X. wrote the manuscript, with input from other co-authors.
Declaration of interests
The authors declare no competing interests.
Published: March 24, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106484.
Contributor Information
Yongsheng Li, Email: liyongsheng@hainmc.edu.cn.
Juan Xu, Email: xujuanbiocc@ems.hrbmu.edu.cn.
Xia Li, Email: lixia@hrbmu.edu.cn.
Supplemental information
Data and code availability
Data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Wei S.C., Levine J.H., Cogdill A.P., Zhao Y., Anang N.A.S., Andrews M.C., Sharma P., Wang J., Wargo J.A., Pe'er D., Allison J.P. Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell. 2017;170:1120–1133.e17. doi: 10.1016/j.cell.2017.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barber D.L., Wherry E.J., Masopust D., Zhu B., Allison J.P., Sharpe A.H., Freeman G.J., Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 3.Angelosanto J.M., Wherry E.J. Transcription factor regulation of CD8+ T-cell memory and exhaustion. Immunol. Rev. 2010;236:167–175. doi: 10.1111/j.1600-065X.2010.00927.x. [DOI] [PubMed] [Google Scholar]
- 4.Larkin J., Lao C.D., Urba W.J., McDermott D.F., Horak C., Jiang J., Wolchok J.D. Efficacy and safety of nivolumab in patients with BRAF V600 mutant and BRAFwild-type advanced melanoma: a pooled analysis of 4 clinical trials. JAMA Oncol. 2015;1:433–440. doi: 10.1001/jamaoncol.2015.1184. [DOI] [PubMed] [Google Scholar]
- 5.Borghaei H., Paz-Ares L., Horn L., Spigel D.R., Steins M., Ready N.E., Chow L.Q., Vokes E.E., Felip E., Holgado E., et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N. Engl. J. Med. 2015;373:1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quinn D.I., Lara P.N., Jr. Renal-cell cancer--targeting an immune checkpoint or multiple kinases. N. Engl. J. Med. 2015;373:1872–1874. doi: 10.1056/NEJMe1511252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wherry E.J. T cell exhaustion. Nat. Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 8.Odorizzi P.M., Pauken K.E., Paley M.A., Sharpe A., Wherry E.J. Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. J. Exp. Med. 2015;212:1125–1137. doi: 10.1084/jem.20142237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackburn S.D., Shin H., Haining W.N., Zou T., Workman C.J., Polley A., Betts M.R., Freeman G.J., Vignali D.A.A., Wherry E.J. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol. 2009;10:29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuchroo V.K., Anderson A.C., Petrovas C. Coinhibitory receptors and CD8 T cell exhaustion in chronic infections. Curr. Opin. HIV AIDS. 2014;9:439–445. doi: 10.1097/COH.0000000000000088. [DOI] [PubMed] [Google Scholar]
- 11.Scott A.C., Dündar F., Zumbo P., Chandran S.S., Klebanoff C.A., Shakiba M., Trivedi P., Menocal L., Appleby H., Camara S., et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature. 2019;571:270–274. doi: 10.1038/s41586-019-1324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim K., Park S., Park S.Y., Kim G., Park S.M., Cho J.W., Kim D.H., Park Y.M., Koh Y.W., Kim H.R., et al. Single-cell transcriptome analysis reveals TOX as a promoting factor for T cell exhaustion and a predictor for anti-PD-1 responses in human cancer. Genome Med. 2020;12:22. doi: 10.1186/s13073-020-00722-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baitsch L., Baumgaertner P., Devêvre E., Raghav S.K., Legat A., Barba L., Wieckowski S., Bouzourene H., Deplancke B., Romero P., et al. Exhaustion of tumor-specific CD8(+) T cells in metastases from melanoma patients. J. Clin. Invest. 2011;121:2350–2360. doi: 10.1172/JCI46102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baitsch L., Legat A., Barba L., Fuertes Marraco S.A., Rivals J.P., Baumgaertner P., Christiansen-Jucht C., Bouzourene H., Rimoldi D., Pircher H., et al. Extended co-expression of inhibitory receptors by human CD8 T-cells depending on differentiation, antigen-specificity and anatomical localization. PLoS One. 2012;7:e30852. doi: 10.1371/journal.pone.0030852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schietinger A., Philip M., Krisnawan V.E., Chiu E.Y., Delrow J.J., Basom R.S., Lauer P., Brockstedt D.G., Knoblaugh S.E., Hämmerling G.J., et al. Tumor-specific T cell dysfunction is a dynamic antigen-driven differentiation Program initiated early during tumorigenesis. Immunity. 2016;45:389–401. doi: 10.1016/j.immuni.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J., He Y., Hao J., Ni L., Dong C. High levels of eomes promote exhaustion of anti-tumor CD8(+) T cells. Front. Immunol. 2018;9:2981. doi: 10.3389/fimmu.2018.02981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng C., Zheng L., Yoo J.K., Guo H., Zhang Y., Guo X., Kang B., Hu R., Huang J.Y., Zhang Q., et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342–1356.e16. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 18.Guo X., Zhang Y., Zheng L., Zheng C., Song J., Zhang Q., Kang B., Liu Z., Jin L., Xing R., et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 2018;24:978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L., Yu X., Zheng L., Zhang Y., Li Y., Fang Q., Gao R., Kang B., Zhang Q., Huang J.Y., et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564:268–272. doi: 10.1038/s41586-018-0694-x. [DOI] [PubMed] [Google Scholar]
- 20.Zheng L., Qin S., Si W., Wang A., Xing B., Gao R., Ren X., Wang L., Wu X., Zhang J., et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science. 2021;374:abe6474. doi: 10.1126/science.abe6474. [DOI] [PubMed] [Google Scholar]
- 21.Esteva F.J., Hubbard-Lucey V.M., Tang J., Pusztai L. Immunotherapy and targeted therapy combinations in metastatic breast cancer. Lancet Oncol. 2019;20:e175–e186. doi: 10.1016/S1470-2045(19)30026-9. [DOI] [PubMed] [Google Scholar]
- 22.Chen D., Menon H., Verma V., Guo C., Ramapriyan R., Barsoumian H., Younes A., Hu Y., Wasley M., Cortez M.A., Welsh J. Response and outcomes after anti-CTLA4 versus anti-PD1 combined with stereotactic body radiation therapy for metastatic non-small cell lung cancer: retrospective analysis of two single-institution prospective trials. J. Immunother. Cancer. 2020;8:e000492. doi: 10.1136/jitc-2019-000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang R., Cheng S., Luo N., Gao R., Yu K., Kang B., Wang L., Zhang Q., Fang Q., Zhang L., et al. Distinct epigenetic features of tumor-reactive CD8+ T cells in colorectal cancer patients revealed by genome-wide DNA methylation analysis. Genome Biol. 2019;21:2. doi: 10.1186/s13059-019-1921-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolski D., Foote P.K., Chen D.Y., Lewis-Ximenez L.L., Fauvelle C., Aneja J., Walker A., Tonnerre P., Torres-Cornejo A., Kvistad D., et al. Early transcriptional divergence marks virus-specific primary human CD8(+) T cells in chronic versus acute infection. Immunity. 2017;47:648–663.e8. doi: 10.1016/j.immuni.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta P.K., Godec J., Wolski D., Adland E., Yates K., Pauken K.E., Cosgrove C., Ledderose C., Junger W.G., Robson S.C., et al. CD39 expression identifies terminally exhausted CD8+ T cells. PLoS Pathog. 2015;11:e1005177. doi: 10.1371/journal.ppat.1005177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gravelle P., Do C., Franchet C., Mueller S., Oberic L., Ysebaert L., Larocca L.M., Hohaus S., Calmels M.N., Frenois F.X., et al. Impaired functional responses in follicular lymphoma CD8(+)TIM-3(+) T lymphocytes following TCR engagement. OncoImmunology. 2016;5:e1224044. doi: 10.1080/2162402X.2016.1224044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Man K., Gabriel S.S., Liao Y., Gloury R., Preston S., Henstridge D.C., Pellegrini M., Zehn D., Berberich-Siebelt F., Febbraio M.A., et al. Transcription factor IRF4 promotes CD8(+) T cell exhaustion and limits the development of memory-like T cells during chronic infection. Immunity. 2017;47:1129–1141.e5. doi: 10.1016/j.immuni.2017.11.021. [DOI] [PubMed] [Google Scholar]
- 28.Doering T.A., Crawford A., Angelosanto J.M., Paley M.A., Ziegler C.G., Wherry E.J. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity. 2012;37:1130–1144. doi: 10.1016/j.immuni.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Argilaguet J., Pedragosa M., Esteve-Codina A., Riera G., Vidal E., Peligero-Cruz C., Casella V., Andreu D., Kaisho T., Bocharov G., et al. Systems analysis reveals complex biological processes during virus infection fate decisions. Genome Res. 2019;29:907–919. doi: 10.1101/gr.241372.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singer M., Wang C., Cong L., Marjanovic N.D., Kowalczyk M.S., Zhang H., Nyman J., Sakuishi K., Kurtulus S., Gennert D., et al. A distinct gene module for dysfunction uncoupled from activation in tumor-infiltrating T cells. Cell. 2016;166:1500–1511.e9. doi: 10.1016/j.cell.2016.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waugh K.A., Leach S.M., Moore B.L., Bruno T.C., Buhrman J.D., Slansky J.E. Molecular profile of tumor-specific CD8+ T cell hypofunction in a transplantable murine cancer model. J. Immunol. 2016;197:1477–1488. doi: 10.4049/jimmunol.1600589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giordano M., Henin C., Maurizio J., Imbratta C., Bourdely P., Buferne M., Baitsch L., Vanhille L., Sieweke M.H., Speiser D.E., et al. Molecular profiling of CD8 T cells in autochthonous melanoma identifies Maf as driver of exhaustion. EMBO J. 2015;34:2042–2058. doi: 10.15252/embj.201490786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alfei F., Kanev K., Hofmann M., Wu M., Ghoneim H.E., Roelli P., Utzschneider D.T., von Hoesslin M., Cullen J.G., Fan Y., et al. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature. 2019;571:265–269. doi: 10.1038/s41586-019-1326-9. [DOI] [PubMed] [Google Scholar]
- 34.Snell L.M., MacLeod B.L., Law J.C., Osokine I., Elsaesser H.J., Hezaveh K., Dickson R.J., Gavin M.A., Guidos C.J., McGaha T.L., Brooks D.G. CD8(+) T cell priming in established chronic viral infection preferentially directs differentiation of memory-like cells for sustained immunity. Immunity. 2018;49:678–694.e5. doi: 10.1016/j.immuni.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khan O., Giles J.R., McDonald S., Manne S., Ngiow S.F., Patel K.P., Werner M.T., Huang A.C., Alexander K.A., Wu J.E., et al. TOX transcriptionally and epigenetically programs CD8(+) T cell exhaustion. Nature. 2019;571:211–218. doi: 10.1038/s41586-019-1325-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pauken K.E., Sammons M.A., Odorizzi P.M., Manne S., Godec J., Khan O., Drake A.M., Chen Z., Sen D.R., Kurachi M., et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 2016;354:1160–1165. doi: 10.1126/science.aaf2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beyer M., Abdullah Z., Chemnitz J.M., Maisel D., Sander J., Lehmann C., Thabet Y., Shinde P.V., Schmidleithner L., Köhne M., et al. Tumor-necrosis factor impairs CD4(+) T cell-mediated immunological control in chronic viral infection. Nat. Immunol. 2016;17:593–603. doi: 10.1038/ni.3399. [DOI] [PubMed] [Google Scholar]
- 38.Jadhav R.R., Im S.J., Hu B., Hashimoto M., Li P., Lin J.X., Leonard W.J., Greenleaf W.J., Ahmed R., Goronzy J.J. Epigenetic signature of PD-1+ TCF1+ CD8 T cells that act as resource cells during chronic viral infection and respond to PD-1 blockade. Proc. Natl. Acad. Sci. USA. 2019;116:14113–14118. doi: 10.1073/pnas.1903520116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bengsch B., Ohtani T., Khan O., Setty M., Manne S., O'Brien S., Gherardini P.F., Herati R.S., Huang A.C., Chang K.M., et al. Epigenomic-guided mass cytometry profiling reveals disease-specific features of exhausted CD8 T cells. Immunity. 2018;48:1029–1045.e5. doi: 10.1016/j.immuni.2018.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 41.Kolde R., Laur S., Adler P., Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 2012;28:573–580. doi: 10.1093/bioinformatics/btr709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anurag M., Zhu M., Huang C., Vasaikar S., Wang J., Hoog J., Burugu S., Gao D., Suman V., Zhang X.H., et al. Immune checkpoint profiles in luminal B breast cancer (Alliance) J. Natl. Cancer Inst. 2020;112:737–746. doi: 10.1093/jnci/djz213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuertes Marraco S.A., Neubert N.J., Verdeil G., Speiser D.E. Inhibitory receptors beyond T cell exhaustion. Front. Immunol. 2015;6:310. doi: 10.3389/fimmu.2015.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian X., Xu W., Wang Y., Anwaier A., Wang H., Wan F., Zhu Y., Cao D., Shi G., Zhu Y., et al. Identification of tumor-infiltrating immune cells and prognostic validation of tumor-infiltrating mast cells in adrenocortical carcinoma: results from bioinformatics and real-world data. OncoImmunology. 2020;9:1784529. doi: 10.1080/2162402X.2020.1784529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu H., Zhou Q., Wei W., Qi B., Zeng F., Bao N., Li Q., Guo F., Xia S. The potential drug for treatment in pancreatic adenocarcinoma: a bioinformatical study based on distinct drug databases. Chin. Med. 2020;15:26. doi: 10.1186/s13020-020-00309-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Groth C., Hu X., Weber R., Fleming V., Altevogt P., Utikal J., Umansky V. Immunosuppression mediated by myeloid-derived suppressor cells (MDSCs) during tumour progression. Br. J. Cancer. 2019;120:16–25. doi: 10.1038/s41416-018-0333-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andrejeva G., Rathmell J.C. Similarities and distinctions of cancer and immune metabolism in inflammation and tumors. Cell Metab. 2017;26:49–70. doi: 10.1016/j.cmet.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y., Zheng L., Zhang L., Hu X., Ren X., Zhang Z. Deep single-cell RNA sequencing data of individual T cells from treatment-naive colorectal cancer patients. Sci. Data. 2019;6:131. doi: 10.1038/s41597-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sade-Feldman M., Yizhak K., Bjorgaard S.L., Ray J.P., de Boer C.G., Jenkins R.W., Lieb D.J., Chen J.H., Frederick D.T., Barzily-Rokni M., et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 2018;175:998–1013.e20. doi: 10.1016/j.cell.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yost K.E., Satpathy A.T., Wells D.K., Qi Y., Wang C., Kageyama R., McNamara K.L., Granja J.M., Sarin K.Y., Brown R.A., et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med. 2019;25:1251–1259. doi: 10.1038/s41591-019-0522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Becht E., McInnes L., Healy J., Dutertre C.A., Kwok I.W.H., Ng L.G., Ginhoux F., Newell E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 52.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aran D., Hu Z., Butte A.J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Z., Ji Z., Ngiow S.F., Manne S., Cai Z., Huang A.C., Johnson J., Staupe R.P., Bengsch B., Xu C., et al. TCF-1-Centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity. 2019;51:840–855.e5. doi: 10.1016/j.immuni.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rooney M.S., Shukla S.A., Wu C.J., Getz G., Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lauss M., Donia M., Harbst K., Andersen R., Mitra S., Rosengren F., Salim M., Vallon-Christersson J., Törngren T., Kvist A., et al. Mutational and putative neoantigen load predict clinical benefit of adoptive T cell therapy in melanoma. Nat. Commun. 2017;8:1738. doi: 10.1038/s41467-017-01460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weigelin B., Krause M., Friedl P. Cytotoxic T lymphocyte migration and effector function in the tumor microenvironment. Immunol. Lett. 2011;138:19–21. doi: 10.1016/j.imlet.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 58.Basu R., Whitlock B.M., Husson J., Le Floc'h A., Jin W., Oyler-Yaniv A., Dotiwala F., Giannone G., Hivroz C., Biais N., et al. Cytotoxic T cells use mechanical force to potentiate target cell killing. Cell. 2016;165:100–110. doi: 10.1016/j.cell.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Narayanan S., Kawaguchi T., Yan L., Peng X., Qi Q., Takabe K. Cytolytic activity score to assess anticancer immunity in colorectal cancer. Ann. Surg Oncol. 2018;25:2323–2331. doi: 10.1245/s10434-018-6506-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Becht E., Giraldo N.A., Lacroix L., Buttard B., Elarouci N., Petitprez F., Selves J., Laurent-Puig P., Sautès-Fridman C., Fridman W.H., de Reyniès A. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li B., Severson E., Pignon J.C., Zhao H., Li T., Novak J., Jiang P., Shen H., Aster J.C., Rodig S., et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17:174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Racle J., de Jonge K., Baumgaertner P., Speiser D.E., Gfeller D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife. 2017;6:e26476. doi: 10.7554/eLife.26476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Finotello F., Mayer C., Plattner C., Laschober G., Rieder D., Hackl H., Krogsdam A., Loncova Z., Posch W., Wilflingseder D., et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019;11:34. doi: 10.1186/s13073-019-0638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miao Y.R., Zhang Q., Lei Q., Luo M., Xie G.Y., Wang H., Guo A.Y. ImmuCellAI: a unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv. Sci. 2020;7:1902880. doi: 10.1002/advs.201902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li T., Fu J., Zeng Z., Cohen D., Li J., Chen Q., Li B., Liu X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 67.Kalluri R., Zeisberg M. Fibroblasts in cancer. Nat. Rev. Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 68.Jiang P., Gu S., Pan D., Fu J., Sahu A., Hu X., Li Z., Traugh N., Bu X., Li B., et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Amin M.B., Greene F.L., Edge S.B., Compton C.C., Gershenwald J.E., Brookland R.K., Meyer L., Gress D.M., Byrd D.R., Winchester D.P. The Eighth Edition AJCC Cancer Staging Manual: continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA. Cancer J. Clin. 2017;67:93–99. doi: 10.3322/caac.21388. [DOI] [PubMed] [Google Scholar]
- 70.Wang X., Guo G., Guan H., Yu Y., Lu J., Yu J. Challenges and potential of PD-1/PD-L1 checkpoint blockade immunotherapy for glioblastoma. J. Exp. Clin. Cancer Res. 2019;38:87. doi: 10.1186/s13046-019-1085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Díaz-Montero C.M., Rini B.I., Finke J.H. The immunology of renal cell carcinoma. Nat. Rev. Nephrol. 2020;16:721–735. doi: 10.1038/s41581-020-0316-3. [DOI] [PubMed] [Google Scholar]
- 72.Zhang S., Zhang E., Long J., Hu Z., Peng J., Liu L., Tang F., Li L., Ouyang Y., Zeng Z. Immune infiltration in renal cell carcinoma. Cancer Sci. 2019;110:1564–1572. doi: 10.1111/cas.13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Keir M.E., Butte M.J., Freeman G.J., Sharpe A.H. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mitsuiki N., Schwab C., Grimbacher B. What did we learn from CTLA-4 insufficiency on the human immune system? Immunol. Rev. 2019;287:33–49. doi: 10.1111/imr.12721. [DOI] [PubMed] [Google Scholar]
- 75.Gajewski T.F., Schreiber H., Fu Y.X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013;14:1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Joyce J.A., Fearon D.T. T cell exclusion, immune privilege, and the tumor microenvironment. Science. 2015;348:74–80. doi: 10.1126/science.aaa6204. [DOI] [PubMed] [Google Scholar]
- 77.Spranger S., Gajewski T.F. Tumor-intrinsic oncogene pathways mediating immune avoidance. OncoImmunology. 2016;5:e1086862. doi: 10.1080/2162402X.2015.1086862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.