Abstract
Almost all the cholesterol in cellular membranes is associated with phospholipids in simple stoichiometric complexes. This limits the binding of sterol ligands such as filipin and perfringolysin O (PFO) to a small fraction of the total. We offer a simple mathematical model that characterizes this complexity. It posits that the cholesterol accessible to ligands has two forms: active cholesterol, which is that not complexed with phospholipids; and extractable cholesterol, that which ligands can capture competitively from the phospholipid complexes. Simulations based on the model match published data for the association of PFO oligomers with liposomes, plasma membranes, and the isolated endoplasmic reticulum. The model shows how the binding of a probe greatly underestimates cholesterol abundance when its affinity for the sterol is so weak that it competes poorly with the membrane phospholipids. Two examples are the understaining of plasma membranes by filipin and the failure of domain D4 of PFO to label their cytoplasmic leaflets. Conversely, the exaggerated staining of endolysosomes suggests that their cholesterol, being uncomplexed, is readily available. The model is also applicable to the association of cholesterol with intrinsic membrane proteins. For example, it supports the hypothesis that the sharp threshold in the regulation of homeostatic endoplasmic reticulum proteins by cholesterol derives from the cooperativity of their binding to the sterol weakly held by the phospholipids. Thus, the model explicates the complexity inherent in the binding of ligands like PFO and filipin to the small accessible fraction of membrane cholesterol.
Supplementary key words: accessible, active, complex, filipin, perfringolysin O, phospholipid, simulation, threshold
Sterols such as cholesterol are major constituents of eukaryotic plasma membrane bilayers and their association with endogenous proteins and exogenous ligands has been studied extensively; for example, references (1, 2, 3). It is not always appreciated, however, that such ligands do not bind all of the sterol in a given membrane; rather, as argued below, they only associate with a small fraction thereof, which we call accessible cholesterol. Consequently, probe binding data can be misleading. This complication in the binding of exogenous probes and integral proteins to membrane sterols has yet to be explored systematically. The present study initiates this inquiry using a simple model to simulate experimental data.
The central concept of the model is that the unesterified sterol associates with the polar lipids in biological membranes (4, 5). While presumably weak and short-lived, these interactions have simple stoichiometries and affinities characteristic of each phospholipid (4, 6, 7, 8). Absent a better term, these associations are called complexes. Sterol molecules in excess of the stoichiometric capacity of the polar lipids remain dispersed in the bilayer. This uncomplexed sterol has a high chemical activity (fugacity), as manifested by its relatively rapid transfer to other membranes as well as its greatly increased interaction with ligands (4, 7, 9, 10, 11, 12). This fraction has therefore been called active cholesterol. This chemically active sterol circulates throughout the cell where it binds to and cues a manifold of regulatory proteins that mediate sterol homeostasis (5, 12, 13, 14, 15). Small changes in the level of uncomplexed plasma membrane cholesterol at its physiologic threshold (namely, its stoichiometric equivalence with the phospholipids) elicit large feedback responses in the endoplasmic reticulum (ER) and mitochondria. This mechanism maintains the cholesterol in the plasma membrane at its resting level and keeps the endomembranes well below their capacity (5).
Not only can ligands bind the active cholesterol in excess of the complexation capacity of the phospholipids but they can also compete with the phospholipids for association with the sterol. We refer to the complexed sterol accessible to a ligand as extractable cholesterol. The active excess and extractable fractions together constitute the accessible cholesterol available to ligands (16, 17, 18, 19).
Of interest here are filipin and perfringolysin O (PFO), two water-soluble probes that bind sterols selectively and are frequently used to track their cellular disposition (1, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30). Filipin is a fluorescent polyene macrolide secreted by Streptomyces filipinensis (1, 31, 32). PFO is a protein secreted by Clostridium perfringens (33, 34). Upon binding to its sterol target, PFO reorients and oligomerizes within the bilayer where it forms cytolytic pores composed of rings of 30 or more monomers (34, 35, 36). The sterol binding domain of PFO, called D4, mimics the intact protein but is not lytic; it is therefore a popular probe (17, 25, 30, 34, 37). A related toxin, anthrolysin O, its nonlytic domain (ALOD4), and a mushroom protein (maistero-2) have been put to similar use (14, 17, 19, 38, 39). The binding of PFO to membranes has a hyperbolic dependence on the aqueous concentration of the protein and a sigmoidal dependence on the concentration of the membrane sterol (16, 18, 35). The threshold of this cholesterol dependence varies with the type and relative abundance of the sterol, the type of phospholipid and the ligand, as well as with the pH and other variables (16, 18, 32, 35, 40, 41, 42).
We offer a simple general model that describes the binding of a ligand to membrane sterols. Simulations closely matched the published data describing the interaction of PFO with cholesterol in two synthetic and two biological membranes using literature values for the relevant parameters. The model is not intended to provide realistic association constants for the ligands. Rather, it provides a conceptual framework that characterizes ligand binding to membrane sterols, clarifies puzzling findings in the literature, and suggests future experiments.
Materials and methods
A mathematical model for the binding of ligands to membrane sterols
We assume that a ligand (L) associates with cholesterol (C) in competition with membrane phospholipids (P). The relevant parameters are the sterol association constants and stoichiometries, both of the phospholipids and the ligand, as well as the possible oligomerization of the latter, as is the characteristic of cytolysins. We postulate an association equilibrium:
| (1) |
where C and P form CPr complexes with a stoichiometry r = 1 or 2 (5, 8). Defining Cu and Pu as the uncomplexed forms of the two lipids, we express the chemical activities of the membrane constituents as dimensionless mole fractions; namely, Cu/α, Pu/α, and (CPr)/α where α = (total C + total P). The association constant for complexation is then,
| (2) |
Two ligands (L) of interest, filipin and PFO, bind the sterol with 1:1 stoichiometry (34, 43, 44). Hence,
| (3) |
where CL represents this association. The ligand is not in the membrane; therefore, the chemical activity of the unbound form, designated as Lu, is taken as its abundance. The association constant for the binding of cholesterol to ligand is
| (4) |
We also assume that the binding of the ligand to the membrane sterol can result in its oligomerization (35). That is,
| (5) |
where n is the order of the oligomerization reaction. The equilibrium expression is
| (6) |
We modeled the binding of monomeric and oligomeric ligands to membrane sterols using Equations (1), (2), (3), (4), (5), (6). The computer code is presented in the Supplemental Information. Simulations are numerical solutions computed in MATLAB. We matched simulations to published cholesterol-dependent isotherms for the binding of PFO to various membranes. (See, for example, Fig. 1.) The simulations also yielded values for the fraction of a ligand associated with cholesterol as a function of the mole fraction of the sterol in the membrane, the fractional saturation of the ligand, the distribution of the sterol-bound ligand between its monomeric and oligomeric forms, the fraction of cholesterol that is uncomplexed, and the fraction of the phospholipid species associated with the sterol (shown in the figures where relevant).
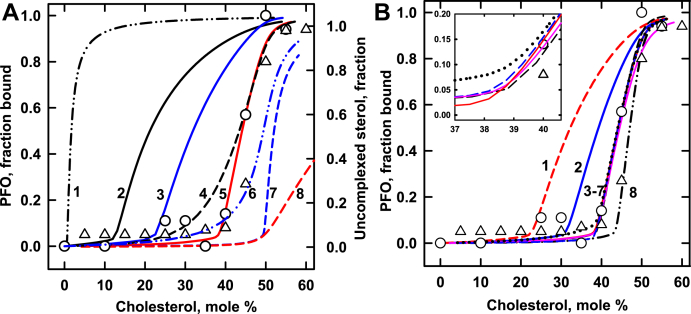
Fig. 1.
Modeling the binding of PFO to cholesterol in POPC membranes. Simulations (curves) are compared to experimental data (symbols) from two studies (41, 45). Calculations used the values in Table 1 and the published values, C:P = 1:1 and KP = 480 (8). The best match is curve 5 in Panel A, repeated as curve 5 in Panel B and in its inset. For reference, curve A8 shows uncomplexed cholesterol in the absence of a ligand. The inset in panel B shows a portion of the calculated curves with an expanded scale. Mole % cholesterol = 100 × moles cholesterol/(moles cholesterol + moles phospholipid). The phospholipid was set at unity in all simulations. PFO, perfringolysin O.
Simulated binding isotherms set the abundance of the phospholipid to unity and varied the sterol across its physiologic range. The abundance of PFO was set at 5 × 10−3. This is 1/200 of the phospholipid and typical of many experiments (41, 45, 46). Values for the sterol affinities and stoichiometries of the different membrane phospholipids (KP and CPr in Equation 2) were taken from published estimates (8). The sterol-loaded ligand was assumed to be either monomeric, dimeric [i.e., (CL)2], or oligomeric [i.e., (CL)30] (17, 41, 44, 47). We let the oligomerization constant for PFO 30-mers be KN = 1 × 10111. For each set of these values, the affinity of the probe for the sterol, KL, was varied to match simulations to experimental PFO binding curves by eye. Four attributes were optimized to match a simulated sigmoidal cholesterol-dependent isotherm to the corresponding published data set: its initial slope; the position of its threshold; its midpoint (i.e., 50% rise); and its plateau. (See, for example, Fig. 1.)
Each set of values that provided a good match to the experimental data occupied a tight parameter space. That is, as illustrated below, a modest change in any of the parameters typically undermined the fit to the data. Nevertheless, more than one set of values sometimes yielded isotherms closely congruent with experimental data (illustrated by curves 3–7 in Fig. 1B). Consequently, our estimates of KL and KN are illustrative and not definitive. Furthermore, simulated KL values cannot be directly related to the Kd values reported for PFO binding (17, 48). Thus, since realistic association constants were not sought or delivered, the use of a statistical best-fit analysis to obtain quantitative values was not appropriate. Rather, our goals were qualitative and conceptual: to see if a simple model sufficed to capture the form of ligand binding to membrane cholesterol and to use the model to rationalize puzzling data in the literature.
Results
We used a simple model to test the hypothesis that probes such as PFO do not accurately quantitate biological membrane cholesterol because they only bind a small fraction thereof (see above). We first simulated the published data for the association of PFO with liposomes composed of the sterol and different phospholipids; then, plasma membranes and the ER were explored. We not only matched simulations to experimental data but, to illustrate the properties of the model, we also tested the influence of the relevant parameters.
Binding of PFO to the cholesterol in POPC membranes
Figure 1 considers a membrane composed of POPC and varied cholesterol. The association of the sterol with the phospholipid in the absence of a ligand is simulated in curve 8 ( ) of Fig. 1A. The cholesterol is almost entirely complexed with the phospholipid up to a membrane concentration of 50 mol %, the stoichiometric equivalence point of their 1:1 association. Uncomplexed sterol rises rapidly beyond this threshold.
) of Fig. 1A. The cholesterol is almost entirely complexed with the phospholipid up to a membrane concentration of 50 mol %, the stoichiometric equivalence point of their 1:1 association. Uncomplexed sterol rises rapidly beyond this threshold.
The symbols in Fig. 1A give experimental data for the binding of PFO to cholesterol in POPC membranes. These data show a threshold that is well below the published equivalence point for POPC of 50 mol % (4, 8). The model suggests that this is because PFO is avid enough to bind the membrane by extracting a portion of the sterol from the phospholipid complexes. The data are simulated well by curve 5 in Fig. 1A, using plausible values for the relevant parameters and an oligomerization constant of 30 (Table 1). Not shown, the ligand in simulated curve 5 remains essentially monomeric up to an acute turning point at ∼39 mol % beyond which 30-mers become increasingly dominant.
Table 1.
Values used in Fig. 1
| Curve | KP | (CL)n | KL | KN | Code |
|---|---|---|---|---|---|
| Panel A | |||||
| A1 | 1 | 30 | 8 | 1 × 10111 | Black dash dot dot |
| A2 | 48 | 30 | 8 | 1 × 10111 | Black line |
| A3 | 480 | 30 | 32 | 1 × 10111 | Blue line |
| A4 | 480 | 2 | 8 | 4 × 104 | Black dash |
| A5 | 480 | 30 | 8 | 1 × 10111 | Red line |
| A6 | 480 | 1 | 20 | 0 | Blue dash dot dot |
| A7 | 480 | 30 | 1 | 1 × 10111 | Blue dash |
| A8 | 480 | Uncomplexed cholesterol, right axis | Red dash | ||
| Panel B | |||||
| B1 | 120 | 30 | 8 | 1 × 10111 | Red dash |
| B2 | 240 | 30 | 8 | 1 × 10111 | Blue line |
| B3 | 240 | 30 | 4 | 1 × 10111 | Black dots |
| B4 | 480 | 30 | 16 | 1 × 10102 | Blue dash |
| B5 | 480 | 30 | 8 | 1 × 10111 | Red line |
| B6 | 480 | 30 | 4 | 1 × 10120 | Pink line |
| B7 | 960 | 30 | 16 | 1 × 10111 | Black dash |
| B8 | 960 | 30 | 8 | 1 × 10111 | Black dash dot dot |
The behavior of the model is further explored in Fig. 1 through alternative simulations in which one parameter was changed at a time. Curve A1 simulates the binding of PFO 30-mers to the cholesterol in a membrane containing a hypothetical phospholipid with negligible sterol affinity; namely, KP = 1. The absence of significant competition by the phospholipid allows strong ligand binding at a very low cholesterol concentration. The curve has very slight sigmoidicity even though ∼90% of the PFO is oligomeric (not shown). Curve A2 shows that a higher sterol affinity for the phospholipid shifts the isotherm for ligand binding to the right. At the other extreme, ligands with very weak sterol affinity bind poorly until uncomplexed cholesterol becomes available at the stoichiometric equivalence point of the lipids, 50 mol % (curve A7). Increasing the sterol affinity of the probe shifts the isotherm to the left (curves A3 and A5 vs. curve A7). Curves A4 and A6 illustrate that monomeric and dimeric ligands bind by extracting cholesterol from phospholipid complexes but their isotherms are less sigmoidal than those of high oligomers (curve A5). As is the case for 30-mers, monomers, and dimers of high sterol affinity yield increasingly hyperbolic isotherms, while those with very low affinity resemble curve A7 (not shown).
Point mutations in the D4 domain of PFO have been shown to displace its binding curve, presumably by changing the strength of its association with the sterol (16, 18). Figure 1A illustrates this effect. A 4-fold increase in KL shifts the midpoint of curve A3 to the left of the best matched curve, A5, by 12.2 mol % cholesterol. An 8-fold reduction in KL moves the midpoint of curve A7 to the right of curve A5 by 7.9 mol % cholesterol.
Panel B in Fig. 1 shows the effect of varying other parameters on the calculated binding of PFO to cholesterol in POPC membranes. Curves B1, B2, B5, and B8 illustrate how competition from increasingly avid phospholipids shifts the isotherm to the right when other parameters are held constant. Curves B3–B7 show that various combinations of KP, KL, and KN can offset one another to produce nearly congruent matches to the data. Thus, simulated values for KL and KN are not uniquely determined.
Binding of PFO to cholesterol in DOPC membranes
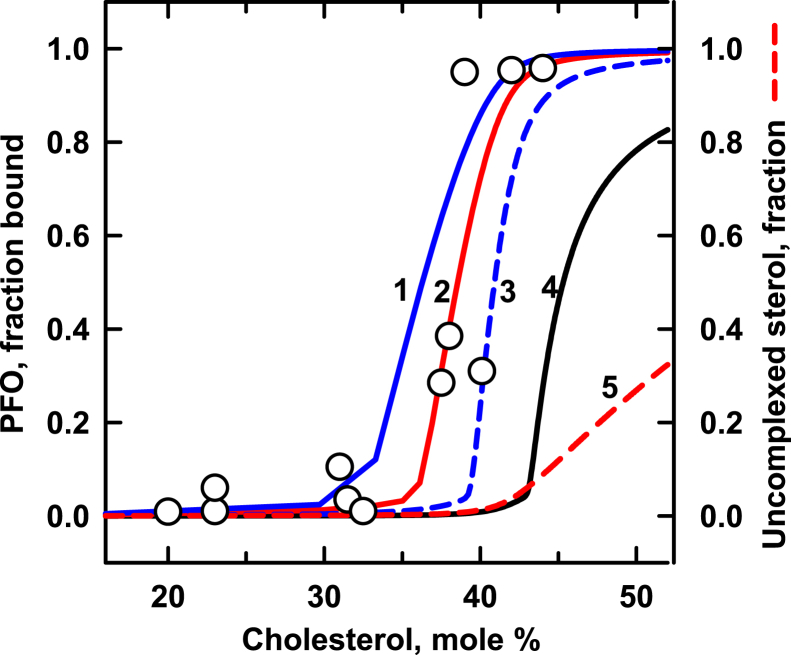
Figure 2 compares simulations with experimental data for liposomes composed of cholesterol and DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine) (41, 45). The best match for PFO dimers (curve 1) is weakly sigmoidal. Curve 2 shows a better match using (CL)30 and a published cholesterol stoichiometry for DOPC; namely, C:P = 1:2 (4, 8). A stoichiometry of C:P = 1:1 also approximates the data (curve 4). An even better match is obtained by assuming that DOPC forms cholesterol complexes with two stoichiometries, C:P = 1:1 and C:P = 1:2, in equal proportions (curve 3). Other sterols also form 1:1 complexes with DOPC as well as a mixed stoichiometry of 1:1 and 1:2 (40).
Fig. 2.
Modeling the binding of PFO to DOPC membranes. The procedure was as described for Fig. 1. The symbols depict experimental data from two studies (41, 45). Curves were calculated using the values in Table 2. A literature value for the sterol-phospholipid affinity constant, KP = 930, was assumed (8). The stoichiometric equivalence point for C:P = 1:2 is C/P = 0.5 mol/mol or 33 mol %; that for a membrane containing equal proportions of C:P = 1:1 and C:P = 1:2 is C/P = 0.75 mol/mole or 43 mol %. For reference, curve 5 shows uncomplexed cholesterol in the absence of a ligand, assuming C:P = 1:2, and curve 6 shows uncomplexed cholesterol in the absence of a ligand assuming an equimolar mixture of C:P = 1:1 and C:P = 1:2. DOPC, 1,2-dioleoyl-sn-glycero-3-phosphocholine; PFO, perfringolysin O.
Binding of PFO to plasma membrane cholesterol
Figure 3 shows calculated curves matched to experimental data for the binding of PFO to fibroblast plasma membranes (37). As in Figs. 1 and 2, PFO monomers and dimers do not provide satisfactory fits to the data (not shown). However, 30-mers work well (curve 2). Doubling KL shifts the midpoint of the isotherm of curve 1 to the left of curve 2 by 1.9 mol % cholesterol. Reducing the sterol affinity of the ligand by two-third moves the midpoint of curve 3 rightward of curve 2 by 2.5 mol % cholesterol. A ligand with very low affinity (curve 4) binds significantly only when uncomplexed cholesterol becomes available (curve 5).
Fig. 3.
Modeling perfringolysin O (PFO) binding to fibroblast plasma membranes. The procedure was as described for Fig. 1. The symbols depict experimental data from a published study (37). Curves were calculated using the values for KL in Table 3. A PFO: phospholipid mole ratio was not provided, so we used the value 5 × 10−3 to 1, as above. We assumed that the membranes contain ∼43 mol % cholesterol; hence, C/P ∼0.75 (49, 99). This corresponds to an equimolar mixture of phospholipids that bind cholesterol with stoichiometries of C:P = 1:1 and 1:2 (5). We assigned KP = 5,000 to the phospholipid with a stoichiometry of C:P = 1:1 and KP = 930 to the species with C:P = 1:2 (5, 8). We assumed the values (CL)30 and KN = 1 × 10111 as before. For reference, curve 5 shows the fraction of uncomplexed cholesterol in the absence of ligand.
The simulation in curve 2 in Fig. 3 gave values of ∼7.5% for the uncomplexed fraction of cholesterol and ∼0.6% for the fraction of the sterol bound to PFO at the physiologic set point of the plasma membrane, 43 mol % cholesterol (not shown). The value for the uncomplexed sterol fraction, ∼7.5%, accords with published values (5, 8). In contrast, it has been inferred from a PFO binding study that ∼37% of plasma membrane cholesterol is normally uncomplexed at its resting concentration (49). This is five times greater than the present result and the earlier estimates. The published value of 37% was based on the assumption that the fraction of the sterol accessible to PFO was that missing from cells starved of the sterol, since this treatment abrogated the binding of the probe (49). However, there is no reason to infer that the missing cholesterol was mostly uncomplexed. Indeed, if a third of plasma membrane cholesterol were free in resting cells, it would be readily available to cholesterol oxidase and methyl-β-cyclodextrin, which is not the case (8, 50, 51, 52). Our model suggests that the rise in PFO binding at ∼38 mol % cholesterol in Fig. 3 does not reflect the appearance of super-threshold uncomplexed sterol but, rather, extraction of complexed sterol by the ligand, as illustrated in Figs. 1 and 2. We conclude that the value of ∼37% is a large overestimate of the fraction of uncomplexed cholesterol in the resting plasma membrane (49). Indeed, homeostatic feedback mechanisms serve to minimize uncomplexed plasma membrane cholesterol (5).
Binding of PFO to cholesterol in ER membranes
The very low threshold value reported for ER cholesterol is of particular interest (41). Uncomplexed cholesterol circulates throughout the cell, signaling multiple homeostatic proteins therein (5, 12). The ER is the hub of this feedback system. Effectors such as acyl-coenzyme A (CoA):cholesterol acyltransferase (ACAT), 3-hydroxy-3-methyl-glutaryl-coenzyme A reductase and SREBP-2 are sharply modulated by small changes in ER cholesterol (12). It was reported that the threshold for the SREBP-2 response has a surprisingly low midpoint of ∼5 mol % cholesterol (41, 53). Two nonexclusive mechanisms for this striking finding were proposed in those studies. One is that the phospholipids in the ER, unlike those discussed above, have a stoichiometric equivalence point near 5 mol % cholesterol and that small changes in the uncomplexed ER sterol at that low threshold regulate the homeostatic proteins. If this were the case, however, the ER phospholipids would form C:P complexes of ∼1:20, which is difficult to imagine. The second hypothesis is that cooperativity in the binding of avid multimeric homeostatic proteins to ER cholesterol imposes the observed sharp threshold at ∼5 mol %.
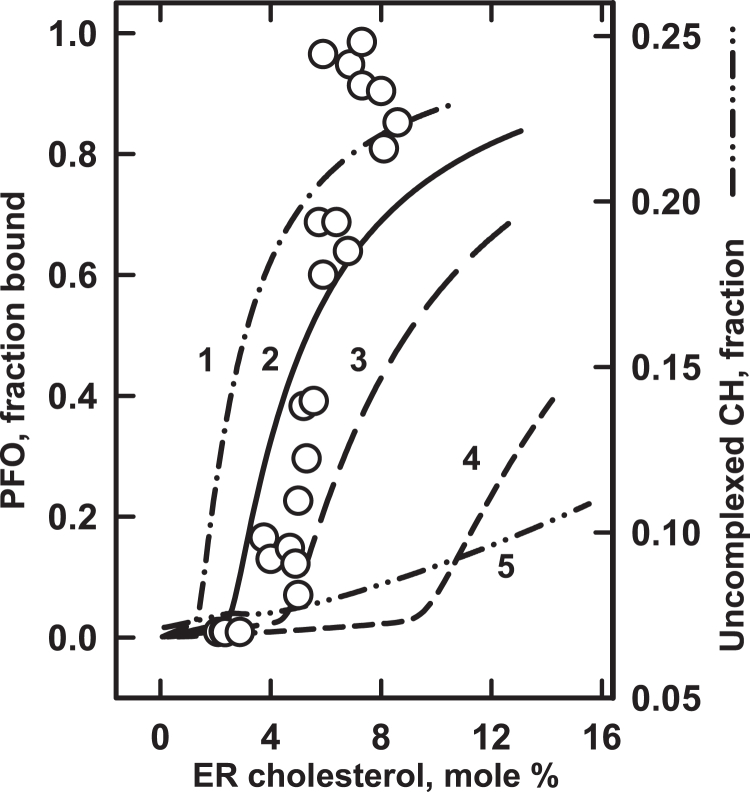
Sokolov et al. (41) went on to show that isolated ER membranes as well as their isolated lipids associate with PFO at ∼5 mol % cholesterol, precisely the threshold observed for SREBP-2 in vivo. Figure 4 shows our simulation of these data as a test of the two rival hypotheses. We found that very low values for the ER sterol–phospholipid affinity constant, KP, are required to place the threshold for the binding of PFO 30-mers near 5 mol % cholesterol (Table 4). (More than 90% of the PFO is oligomeric at that point; not shown.) In addition, curve 5 in Fig. 4 shows that the uncomplexed form of the cholesterol does not have a threshold in this region, contrary to the prediction of the first hypothesis.
Fig. 4.
Modeling the binding of PFO to ER membranes. The procedure was as described for Fig. 1. The symbols depict experimental data from a published study (41). Curves were calculated using the values for KA and KB in Table 4. The values KL = 14, (CL)30 and KN = 1 × 10111 were taken from the best match of PFO binding to DOPC membranes (curve 3 in Fig. 2 and Table 2). ER membranes contain diverse glycerophospholipids (100). It was assumed for simplicity that they had equal amounts of phospholipids A and B with stoichiometries of C:P = 1:1 and C:P = 1:2, respectively. For curve 2, we used previously estimated values for KA and KB (5). This curve gave the closest match to the data; the other three curves permuted its values (Table 4). Curve 5 charts the uncomplexed fraction of ER cholesterol (CH) on an expanded scale. DOPC, 1,2-dioleoyl-sn-glycero-3-phosphocholine; ER, endoplasmic reticulum; PFO, perfringolysin O.
Table 4.
Values used in Fig. 4
| Curve | KA | KB | Code |
|---|---|---|---|
| 1 | 10 | 5 | Dash dot dash |
| 2 | 21 | 10 | Solid line |
| 3 | 42 | 20 | Long dashes |
| 4 | 105 | 50 | Short dashes |
| 5 | Uncomplexed cholesterol, right axis | Dash dot dot dash | |
ER phospholipids are not known to be unusual (54). However, the calculated sterol affinity of the ER phospholipids in Table 4 is orders of magnitude lower than the values in Table 1, Table 2, Table 3 and those generally reported for biological membranes (5, 8). Other kinds of experiments have suggested similarly low affinities (5, 55). One explanation for the surprisingly low apparent sterol affinity of the ER phospholipids is that an undetected competitor reduces sterol binding (56). (Such an extraneous factor might be the reason that the experimental data for the ER could not be simulated as closely as those for the membranes shown in Fig. 1, Fig. 2, Fig. 3.) In any case, more experiments are needed to explicate the apparently low KP value for the ER.
Table 2.
Values used in Fig. 2
| Curve | C:P | (CL)n | KL | KN | Code |
|---|---|---|---|---|---|
| 1 | 1:2 | 2 | 10 | 1 × 105 | Blue dash |
| 2 | 1:2 | 30 | 1.1 | 1 × 10111 | Blue line |
| 3 | 1:1+1:2 | 30 | 14 | 1 × 10111 | Red line |
| 4 | 1:1 | 30 | 35 | 1 × 10111 | Black line |
| 5 | 1:2 | Uncomplexed cholesterol, right axis | Red dash | ||
| 6 | 1:1+1:2 | Uncomplexed cholesterol, right axis | Black dash | ||
Table 3.
Values used in Fig. 3
| Curve | KL | Code |
|---|---|---|
| 1 | 24 | Blue line |
| 2 | 12 | Red line |
| 3 | 4 | Blue dash |
| 4 | 1 | Black line |
| 5 | Uncomplexed cholesterol, right axis | Red dash |
Thresholds as low as ∼6 mol % have been reported for the binding of PFO to liposomes composed of equal mixtures of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (36). Figure 4 suggests that these low thresholds, like those for the ER, reflect the highly cooperative binding of PFO to very weakly held cholesterol rather than an unusually low lipid stoichiometry. This could also be the case for oligomeric homeostatic proteins such as SCAP (5). Future studies might distinguish between the two mechanisms originally postulated by determining the ER sterol threshold using a noncooperative probe such as cholesterol oxidase or a low affinity PFO variant such as L491S (16). One mechanism predicts a threshold at ∼5 mol % and the other predicts a much higher one.
Discussion
Applications of the model to probes for plasma membrane cholesterol
Our model encourages a reinterpretation of some puzzling findings in the literature. Consider filipin. The plasma membrane is particularly rich in cholesterol (5). Nevertheless, filipin and a similar macrolide, amphotericin B, label cell surfaces weakly compared to their robust staining of cytoplasmic organelles (16, 20, 24, 29, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67). The model suggests an explanation: ligands such as filipin compete poorly with the avid plasma membrane phospholipids for the sterol. Curve 4 in Fig. 3 illustrates this premise: a probe with KL = 1 barely binds to plasma membranes at their physiologic setpoint, ∼43 mol % cholesterol. This curve also explains why an enrichment of plasma membrane cholesterol of even 10% promotes strong filipin staining (24, 39, 68). Similarly, curve 4 accords with findings that probes that do not bind membranes strongly (namely, cholesterol oxidase and methyl-β-cyclodextrin), hardly access the complexed cholesterol in the unperturbed cell surface membrane but interact well when a small excess of uncomplexed cholesterol is introduced (8, 11, 50, 51, 52, 69). Likewise, a marginal increase in accessible cholesterol could be the reason that filipin stains cells overexpressing the sterol transporter, ABCG1, and sometimes untreated cells (22, 70). In one striking study, filipin failed to light up the surface of proliferating cells but did so strongly when they became quiescent (21). This large jump in filipin binding is more likely to reflect an increase in the accessibility of a small pool of uncomplexed cholesterol than a large change in total plasma membrane cholesterol. For the same reason, the total abolition of filipin staining of plasma membranes depleted by cyclodextrin extraction probably reflects a reduction in the small uncomplexed fraction rather than wholesale removal of the sterol (71).
Curve 2 in Fig. 3 models the robust binding of PFO to plasma membranes (16, 24, 72). This simulation suggests why the staining of membranes by this ligand is sometimes weak (23, 27, 39). That is, a marginal reduction of the plasma membrane sterol can significantly reduce the binding of the probe. The same mechanism applies to the decrease in cell surface staining by PFO D4 when delivery of intracellular cholesterol to the plasma membrane is diverted (27, 28, 73, 74). Similarly, the 2-fold increase in PFO staining observed in cells overexpressing the cholesterol transporter, ABCA1, is more likely to reflect an increment in the accessible fraction of their plasma membrane cholesterol than a doubling of the total for which there is no room (75, 76, 77). This reasoning can also be applied to the preferential labeling of microvilli by cytolysins (39, 72).
Flow cytometry shows a broad profile in the labeling of red blood cell populations by cytolysins (38, 78, 79). As suggested in one of those studies, some of this dispersion in stain intensity could reflect variation in the accessibility of the cholesterol across the sharp plasma membrane threshold rather than substantial heterogeneity in red cell sterol content (38).
The fluorescent D4 fragment of PFO reliably lights up cell surfaces (22, 24, 76, 77). However, it typically fails to stain the cytoplasmic side of the plasma membrane (24, 29). This has led to the inference that the inner leaflet of this bilayer is profoundly deficient in cholesterol (26, 80). Alternative hypotheses have been proposed (46, 81). Our analysis offers an additional explanation: D4 is less avid for the inner leaflet than for the outer. This is modeled in Fig. 3. Suppose that curve 2 with KL = 12 depicts the robust binding of D4 to the external surface of plasma membranes while curve 4 with KL = 1 represents its weak binding to the cytoplasmic side. The probe would then associate maximally with one leaflet and minimally with the other. Support for this prediction comes from the report that incrementing plasma membrane cholesterol promotes D4 binding to the inner leaflet (24, 46). Curve 4 accounts for this finding: a small increase in plasma membrane cholesterol above its set point at 43 mol % would foster robust binding. That D4 occasionally stains the cytoplasmic leaflet of the plasma membrane can then be explained by such an incidental enrichment (68). Here, as elsewhere, obtaining a complete isotherm for the sterol dependence of ligand binding would provide a good test of this interpretation.
That D4 has a lower affinity for the cytoplasmic than for the exofacial surface of the plasma membrane might reflect electrostatic repulsion between the negatively charged contact surface of the protein and the anionic head groups of the phospholipids enriched in the inner leaflet (82). The finding that PFO and its D4 domain bind significantly less well to anionic than to neutral liposomes supports this hypothesis (36, 46). Additional evidence is that the association of D4 with the cytoplasmic leaflet was strengthened by removing an acidic residue; namely, the point mutations D434S and D434A (18, 24, 26, 46, 66). These findings support the general premise that the apparent affinity of a probe for membrane cholesterol is conditioned by the environment of the sterol.
The D434S substitution transforms D4 into D4H, a strong ligand for the cholesterol in both leaflets of the plasma membrane (24, 25). However, even D4H sometimes fails to stain one surface or the other (63, 73, 83, 84). As above, Fig. 3 predicts that this can occur when the accessible plasma membrane cholesterol is marginally reduced by an experimental or physiological alteration. A case in point: cytoplasmic mCherry-D4H decorated the cytoplasmic leaflet poorly in control cells but strongly in those overexpressing ORP2, the intermembrane phosphatidylinositol 4,5-bisphosphate/sterol exchanger (66). ORP2 moves intracellular cholesterol to the plasma membrane; thus, in those experiments, it could have enriched the sterol beyond the threshold needed for probe binding. While the authors inferred that ORP2 regulates total plasma membrane sterol, our model suggests that it adjusts the level of accessible cholesterol at the physiologic threshold, leaving the major fraction essentially unchanged. This hypothesis could be tested by tracking plasma membrane cholesterol chemically and gauging active cholesterol with a reporter such as cholesterol oxidase.
Recently, D4H was modified with two additional point substitutions to create GST-D4H∗-mCherry (65, 67). This variant did not label the plasma membrane well but lit up endolysosomes. In contrast to the authors’ interpretation, our model predicts that the extra modification reduces the sterol affinity of the probe which would therefore stain the plasma membrane less well (curve 4 in Fig. 3). Prediction: marginal increments in plasma membrane cholesterol will significantly boost staining by GST-D4H∗-mCherry.
Application of the model to the binding of probes to endolysosomes
Filipin and cytolysins light up late endolysosomes whether or not they are engorged with ingested sterol. But why do these probes stain the organelles strongly compared to the cholesterol-rich plasma membrane (20, 23, 24, 26, 27, 29, 58, 59, 63, 64, 65, 66, 67, 68, 80, 84)? One reason might be that the sterol in the endolysosomes has been released from its association with ingested lipoproteins and membranes. As illustrated by curve 1 in Fig. 1, sterol molecules free of phospholipid complexes are very accessible to ligands. Indeed, PFO binds membrane-free cholesterol ∼100-times better than that in DOPC liposomes (17).
Conclusions
The model allows us to conceptualize the association of ligands with membrane sterols, interpret published results, and suggest new experiments. The basic premise is that almost all the sterol in the membrane is complexed with phospholipids so that ligands can bind only a small fraction thereof. Accessible cholesterol has two components: that which exceeds the complexation capacity of the phospholipids at their stoichiometric equivalence point (termed active cholesterol) and that which ligands can extract from sterol-phospholipid complexes as they bind (termed extractable cholesterol). Simulations show that PFO associates with the four membranes studied by extracting cholesterol from phospholipid complexes. In contrast, weakly bound probes such as cholesterol oxidase and methyl-β-cyclodextrin require the active sterol in excess of its stoichiometric equivalence with the phospholipids (8, 50, 51, 52, 69). The complementary roles played by these two forms of accessible cholesterol in the binding of ligands have not previously been clarified.
As seen in the figures, the binding of ligands to membrane cholesterol always has a sigmoidal isotherm. There are three sources of this sigmoidicity. First, titrations generally show a characteristic break at their stoichiometric equivalence point (i.e., at saturation). This threshold is illustrated by curve 8 in Fig. 1A and curve 5 in Fig. 3. Second, sigmoidicity arises from the cooperative formation of ligand oligomers. This, for example, sharpens the acute rise of curve 5 in Fig. 1A. Third, competition by the phospholipids for the sterol inevitably imposes sigmoidicity on a ligand binding isotherm, even when the ligand is monomeric (e.g., curve 6 in Fig. 1A) (85). Each of these three types of sigmoidicity plays a role in the regulation of cholesterol homeostasis by the ER. Specifically: (a) The level of ER cholesterol is set by its passive equilibration with the uncomplexed fraction of the plasma membrane sterol exceeding its stoichiometric equivalence point with the phospholipids (5). (b) SCAP tetramers regulate SREBP-2 activity with a highly cooperative dependence on ER sterol concentration (53, 86). Similarly, acyl-CoA:cholesterol acyltransferase and 3-hydroxy-3-methylglutaryl coenzyme A reductase have a high order dependence on the ER membrane sterol concentration (52, 87). (c) The model predicts that the activity of some monomeric ER proteins will have a weakly sigmoidal cholesterol dependence below the stoichiometric equivalence point of the phospholipids (see curve 6 in Fig. 1A). This hypothesis could be tested by determining the cholesterol dependence of the downregulation of squalene monooxygenase (13, 88).
While the simulated association constants for PFO binding and oligomerization lack absolute significance, there is a notable similarity among the KL values best matched to the data for the four membranes (Table 5). This agreement could signify that the probe experiences the cholesterol in diverse membranes in much the same way, differences in the sterol affinities and stoichiometries of the phospholipids notwithstanding. However, this is probably not always the case: the model predicts that the cholesterol affinity of the D4 fragment for the inner leaflet of the plasma membrane is significantly weaker than that for the outer leaflet (see the foregoing discussion). It could be that the predominant phosphorylcholine head groups of the outer surface phospholipids of the membranes examined in Fig. 1, Fig. 2, Fig. 3, Fig. 4 provide a similar environment for the sterol, leading to comparable KL values. In contrast, the anionic phospholipids prevalent on the cytoplasmic surfaces of plasma membranes would discourage D4 binding and reduce its apparent KL.
Table 5.
Summary of values for parameters
| Membrane | C:P | KP | (CL)n | KL | KN |
|---|---|---|---|---|---|
| POPC | 1:1 | 480 | 30 | 8 | 1 × 10111 |
| DOPC | 1:1 + 1:2 | 930 | 30 | 14 | 1 × 10111 |
| Plasma membrane | 1:1 + 1:2 | 5,000 + 930 | 30 | 12 | 1 × 10111 |
| Endoplasmic reticulum | 1:1 + 1:2 | 21 + 10 | 30 | 14 | 1 × 10111 |
DOPC, 1,2-dioleoyl-sn-glycero-3-phosphocholine; POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine.
The model suggests why the binding of even an avid ligand can be limited when the membrane cholesterol concentration is far below the stoichiometric equivalence point of the phospholipids and why the binding of a weak ligand can be robust when uncomplexed sterol is available. It also suggests that the relatively poor staining of plasma membranes by filipin reflects its weak affinity for the cholesterol held in tight phospholipid complexes. The conditional failure of D4 to bind to the cytoplasmic surface of the plasma membrane could be another instance of weak competition with the phospholipids while the strong labeling of endolysosomes by filipin is consistent with the availability of uncomplexed cholesterol therein. Despite these complexities, filipin and cytolysins have proved to be highly valuable (12, 28, 39, 66, 68, 72, 73, 74, 83, 89, 90, 91, 92). Going forward, the utility of these probes will increase with the recognition that their binding is limited to the small and dynamic accessible fraction of the membrane sterol, as evident in recent studies (28, 93, 94, 95).
A wide variety of membrane-intercalated amphiphiles also associate stoichiometrically with phospholipids, as reflected in their one-for-one displacement of cholesterol from its complexes and their ability to substitute for the sterol in bilayers (56, 96). The model predicts that, as is the case for sterols, proteins that bind membrane-intercalated bio-active lipids such as ceramides and arachidonic acid will also have to compete for them with the phospholipids.
The model is generally applicable to the binding of any ligand to membrane sterols. It shows how cholesterol modulates the activity of a variety of integral plasma membrane proteins (97). For instance, morphogenetic signaling by Hedgehog is mediated by plasma membrane cholesterol at its physiologic threshold (73, 92, 98). In the case of the ER, homeostatic proteins bind cholesterol with high affinity and cooperativity so that small changes in its sterol concentration drive large feedback responses (5). This too is treated by our model.
Data availability
All data are contained within the article.
Supplemental data
This article contains supplemental data.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors are grateful to Aaron Dinner (University of Chicago), Irwin London (Stony Brook University), and Arun Radhakrishnan (University of Texas Southwestern Medical Center) for their valuable feedback on the manuscript. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author contributions
Y. L. and T. L. S. designed the project; Y. L. and T. L. S. executed the simulations; Y. L. and T. L. S. wrote the manuscript; S. M. A. T. and T. L. S. formulated the model; S. M. A. T. wrote the code.
Supplemental data
References
- 1.Gimpl G. Cholesterol-protein interaction: methods and cholesterol reporter molecules. Subcell. Biochem. 2010;51:1–45. doi: 10.1007/978-90-481-8622-8_1. [DOI] [PubMed] [Google Scholar]
- 2.Wüstner D., Solanko K. How cholesterol interacts with proteins and lipids during its intracellular transport. Biochim. Biophys. Acta Biomembr. 2015;1848:1908–1926. doi: 10.1016/j.bbamem.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Fantini J., Epand R.M., Barrantes F.J. In: Direct Mechanisms in Cholesterol Modulation of Protein Function. RosenhouseDantsker A., Bukiya A.N., editors. 2019. Cholesterol-recognition motifs in membrane proteins; pp. 3–25. [DOI] [PubMed] [Google Scholar]
- 4.Litz J.P., Thakkar N., Portet T., Keller S.L. Depletion with cyclodextrin reveals two populations of cholesterol in model lipid membranes. Biophys. J. 2016;110:635–645. doi: 10.1016/j.bpj.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steck T.L., Tabei S.M.A., Lange Y. A basic model for cell cholesterol homeostasis. Traffic. 2021;22:471–481. doi: 10.1111/tra.12816. [DOI] [PubMed] [Google Scholar]
- 6.Niu S.L., Litman B.J. Determination of membrane cholesterol partition coefficient using a lipid vesicle-cyclodextrin binary system: effect of phospholipid acyl chain unsaturation and headgroup composition. Biophys. J. 2002;83:3408–3415. doi: 10.1016/S0006-3495(02)75340-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lange Y., Steck T.L. Cholesterol homeostasis and the escape tendency (activity) of plasma membrane cholesterol. Prog. Lipid Res. 2008;47:319–332. doi: 10.1016/j.plipres.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lange Y., Tabei S.M.A., Ye J., Steck T.L. Stability and stoichiometry of bilayer phospholipid–cholesterol complexes: relationship to cellular sterol distribution and homeostasis. Biochemistry. 2013;52:6950–6959. doi: 10.1021/bi400862q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radhakrishnan A., McConnell H.M. Chemical activity of cholesterol in membranes. Biochemistry. 2000;39:8119–8124. doi: 10.1021/bi0005097. [DOI] [PubMed] [Google Scholar]
- 10.Steck T.L., Lange Y. Cell cholesterol homeostasis: mediation by active cholesterol. Trends Cell Biol. 2010;20:680–687. doi: 10.1016/j.tcb.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ayuyan A.G., Cohen F.S. The chemical potential of plasma membrane cholesterol: implications for cell biology. Biophys. J. 2018;114:904–918. doi: 10.1016/j.bpj.2017.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lange Y., Steck T.L. Active cholesterol 20 years on. Traffic. 2020;21:662–674. doi: 10.1111/tra.12762. [DOI] [PubMed] [Google Scholar]
- 13.Howe V., Sharpe L.J., Alexopoulos S.J., Kunze S.V., Chua N.K., Li D., et al. Cholesterol homeostasis: how do cells sense sterol excess? Chem. Phys. Lipids. 2016;199:170–178. doi: 10.1016/j.chemphyslip.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 14.Infante R.E., Radhakrishnan A. Continuous transport of a small fraction of plasma membrane cholesterol to endoplasmic reticulum regulates total cellular cholesterol. Elife. 2017;6 doi: 10.7554/eLife.25466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Juhl A.D., Wüstner D. Pathways and mechanisms of cellular cholesterol efflux—insight from imaging. Front. Cell Dev. Biol. 2022;10:834408. doi: 10.3389/fcell.2022.834408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson B.B., Moe P.C., Wang D., Rossi K., Trigatti B.L., Heuck A.P. Modifications in perfringolysin O domain 4 alter the cholesterol concentration threshold required for binding. Biochemistry. 2012;51:3373–3382. doi: 10.1021/bi3003132. [DOI] [PubMed] [Google Scholar]
- 17.Gay A., Rye D., Radhakrishnan A. Switch-like responses of two cholesterol sensors do not require protein oligomerization in membranes. Biophys. J. 2015;108:1459–1469. doi: 10.1016/j.bpj.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson B.B., Brena M., Anguita J., Heuck A.P. Mechanistic insights into the cholesterol-dependent binding of perfringolysin O-based probes and cell membranes. Sci. Rep. 2017;7:13793. doi: 10.1038/s41598-017-14002-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson K.A., Radhakrishnan A. The use of anthrolysin O and ostreolysin A to study cholesterol in cell membranes. Methods Enzymol. 2021;649:543–566. doi: 10.1016/bs.mie.2021.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porn M.I., Slotte J.P. Localization of cholesterol in sphingomyelinase-treated fibroblasts. Biochem. J. 1995;308:269–274. doi: 10.1042/bj3080269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaz G., Batetta B., Sanna F., Uda S., Reali C., Angius F., et al. Lipid droplet changes in proliferating and quiescent 3T3 fibroblasts. Histochem. Cell Biol. 2008;129:611–621. doi: 10.1007/s00418-008-0402-2. [DOI] [PubMed] [Google Scholar]
- 22.Ishitsuka R., Saito T., Osada H., Ohno-Iwashita Y., Kobayashi T. Fluorescence image screening for chemical compounds modifying cholesterol metabolism and distribution. J. Lipid Res. 2011;52:2084–2094. doi: 10.1194/jlr.D018184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwiatkowska K., Marszalek-Sadowska E., Traczyk G., Koprowski P., Musielak M., Lugowska A., et al. Visualization of cholesterol deposits in lysosomes of Niemann-Pick type C fibroblasts using recombinant perfringolysin O. Orphanet J. Rare Dis. 2014;9:64. doi: 10.1186/1750-1172-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maekawa M., Fairn G.D. Complementary probes reveal that phosphatidylserine is required for the proper transbilayer distribution of cholesterol. J. Cell Sci. 2015;128:1422–1433. doi: 10.1242/jcs.164715. [DOI] [PubMed] [Google Scholar]
- 25.Maekawa M., Yang Y., Fairn G.D. Perfringolysin O theta toxin as a tool to monitor the distribution and inhomogeneity of cholesterol in cellular membranes. Toxins (Basel) 2016;8:67. doi: 10.3390/toxins8030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu S.L., Sheng R., Jung J.H., Wang L., Stec E., O'Connor M.J., et al. Orthogonal lipid sensors identify transbilayer asymmetry of plasma membrane cholesterol. Nat. Chem. Biol. 2017;13:268–274. doi: 10.1038/nchembio.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilhelm L.P., Wendling C., Védie B., Kobayashi T., Chenard M.-P., Tomasetto C., et al. STARD3 mediates endoplasmic reticulum-to-endosome cholesterol transport at membrane contact sites. EMBO J. 2017;36:1412–1433. doi: 10.15252/embj.201695917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naito T., Ercan B., Krshnan L., Triebl A., Koh D.H.Z., Wei F.Y., et al. Movement of accessible plasma membrane cholesterol by the GRAMD1 lipid transfer protein complex. Elife. 2019;8 doi: 10.7554/eLife.51401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilhelm L.P., Voilquin L., Kobayashi T., Tomasetto C., Alpy F. Intracellular and plasma membrane cholesterol labeling and quantification using Filipin and GFP-D4. Methods Mol. Biol. 2019;1949:137–152. doi: 10.1007/978-1-4939-9136-5_11. [DOI] [PubMed] [Google Scholar]
- 30.Schoop V., Martello A., Eden E.R., Hoglinger D. Cellular cholesterol and how to find it. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2021;1866:158989. doi: 10.1016/j.bbalip.2021.158989. [DOI] [PubMed] [Google Scholar]
- 31.Norman A.W., Demel R.A., de Kruyff B., van Deenen L.L. Studies on the biological properties of polyene antibiotics. Evidence for the direct interaction of filipin with cholesterol. J. Biol. Chem. 1972;247:1918–1929. [PubMed] [Google Scholar]
- 32.Ohno-Iwashita Y., Shimada Y., Waheed A.A., Hayashi M., Inomata M., Nakamura M., et al. Perfringolysin O, a cholesterol-binding cytolysin, as a probe for lipid rafts. Anaerobe. 2004;10:125–134. doi: 10.1016/j.anaerobe.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Heuck A.P., Moe P.C., Johnson B.B. The cholesterol-dependent cytolysin family of gram-positive bacterial toxins. Subcell. Biochem. 2010;51:551–577. doi: 10.1007/978-90-481-8622-8_20. [DOI] [PubMed] [Google Scholar]
- 34.Wade K.R., Hotze E.M., Tweten R.K. Comprehensive Sourcebook of Bacterial Protein Toxins, 4th Edition. Elsevier; Amsterdam, Netherlands: 2015. Perfringolysin O and related cholesterol-dependent cytolysins: mechanism of pore formation; pp. 719–738. [Google Scholar]
- 35.Flanagan J.J., Tweten R.K., Johnson A.E., Heuck A.P. Cholesterol exposure at the membrane surface is necessary and sufficient to trigger perfringolysin O binding. Biochemistry. 2009;48:3977–3987. doi: 10.1021/bi9002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Q., London E. The influence of natural lipid asymmetry upon the conformation of a membrane-inserted protein (perfringolysin O) J. Biol. Chem. 2014;289:5467–5478. doi: 10.1074/jbc.M113.533943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das A., Goldstein J.L., Anderson D.D., Brown M.S., Radhakrishnan A. Use of mutant 125I-perfringolysin O to probe transport and organization of cholesterol in membranes of animal cells. Proc. Natl. Acad. Sci. U. S. A. 2013;110:10580–10585. doi: 10.1073/pnas.1309273110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chakrabarti R.S., Ingham S.A., Kozlitina J., Gay A., Cohen J.C., Radhakrishnan A., et al. Variability of cholesterol accessibility in human red blood cells measured using a bacterial cholesterol-binding toxin. Elife. 2017;6 doi: 10.7554/eLife.23355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamaji-Hasegawa A., Murate M., Inaba T., Dohmae N., Sato M., Fujimori F., et al. A novel sterol-binding protein reveals heterogeneous cholesterol distribution in neurite outgrowth and in late endosomes/lysosomes. Cell. Mol. Life Sci. 2022;79:324. doi: 10.1007/s00018-022-04339-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson L.D., Johnson A.E., London E. How interaction of perfringolysin O with membranes is controlled by sterol structure, lipid structure, and physiological low pH: insights into the origin of perfringolysin O-lipid raft interaction. J. Biol. Chem. 2008;283:4632–4642. doi: 10.1074/jbc.M709483200. [DOI] [PubMed] [Google Scholar]
- 41.Sokolov A., Radhakrishnan A. Accessibility of cholesterol in endoplasmic reticulum membranes and activation of SREBP-2 switch abruptly at a common cholesterol threshold. J. Biol. Chem. 2010;285:29480–29490. doi: 10.1074/jbc.M110.148254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin Q., London E. Transmembrane protein (perfringolysin O) association with ordered membrane domains (rafts) depends upon the raft-associating properties of protein-bound sterol. Biophys. J. 2013;105:2733–2742. doi: 10.1016/j.bpj.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patterson J., Holland J., Bieber L.L. Studies on the competition of polyene antibiotics for sterols. J. Antibiot. (Tokyo) 1979;32:646–653. doi: 10.7164/antibiotics.32.646. [DOI] [PubMed] [Google Scholar]
- 44.Farrand A.J., Hotze E.M., Sato T.K., Wade K.R., Wimley W.C., Johnson A.E., et al. The cholesterol-dependent cytolysin membrane-binding interface discriminates lipid environments of cholesterol to support β-barrel pore insertion. J. Biol. Chem. 2015;290:17733–17744. doi: 10.1074/jbc.M115.656769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bielska A.A., Olsen B.N., Gale S.E., Mydock-McGrane L., Krishnan K., Baker N.A., et al. Side-chain oxysterols modulate cholesterol accessibility through membrane remodeling. Biochemistry. 2014;53:3042–3051. doi: 10.1021/bi5000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Courtney K.C., Fung K.Y., Maxfield F.R., Fairn G.D., Zha X. Comment on 'Orthogonal lipid sensors identify transbilayer asymmetry of plasma membrane cholesterol'. Elife. 2018;7 doi: 10.7554/eLife.38493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Solovyova A.S., Nollmann M., Mitchell T.J., Byron O. The solution structure and oligomerization behavior of two bacterial toxins: pneumolysin and perfringolysin O. Biophys. J. 2004;87:540–552. doi: 10.1529/biophysj.104.039974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimada Y., Maruya M., Iwashita S., Ohno-Iwashita Y. The C-terminal domain of perfringolysin O is an essential cholesterol-binding unit targeting to cholesterol-rich microdomains. Eur. J. Biochem. 2002;269:6195–6203. doi: 10.1046/j.1432-1033.2002.03338.x. [DOI] [PubMed] [Google Scholar]
- 49.Das A., Brown M.S., Anderson D.D., Goldstein J.L., Radhakrishnan A. Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. Elife. 2014;3 doi: 10.7554/eLife.02882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patzer E.J., Wagner R.R., Barenholz Y. Cholesterol oxidase as a probe for studying membrane organisation. Nature. 1978;274:394–395. doi: 10.1038/274394a0. [DOI] [PubMed] [Google Scholar]
- 51.Lange Y., Cutler H.B., Steck T.L. The effect of cholesterol and other intercalated amphipaths on the contour and stability of the isolated red cell membrane. J. Biol. Chem. 1980;255:9331–9337. [PubMed] [Google Scholar]
- 52.Lange Y., Ye J., Steck T.L. How cholesterol homeostasis is regulated by plasma membrane cholesterol in excess of phospholipids. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11664–11667. doi: 10.1073/pnas.0404766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Radhakrishnan A., Goldstein J.L., McDonald J.G., Brown M.S. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 2008;8:512–521. doi: 10.1016/j.cmet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andreyev A.Y., Fahy E., Guan Z., Kelly S., Li X., McDonald J.G., et al. Subcellular organelle lipidomics in TLR-4-activated macrophages. J. Lipid Res. 2010;51:2785–2797. doi: 10.1194/jlr.M008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shaw T.R., Wisser K., Shaffner T.A., Gaffney A.D., Machta B.B., Veatch S.L. Chemical potential measurements constrain models of cholesterol-phosphatidylcholine interactions. Biophys. 2023 doi: 10.1016/j.bpj.2023.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lange Y., Ye J., Duban M.E., Steck T.L. Activation of membrane cholesterol by 63 amphipaths. Biochemistry. 2009;48:8505–8515. doi: 10.1021/bi900951r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Severs N.J., Simons H.L. Failure of filipin to detect cholesterol-rich domains in smooth muscle plasma membrane. Nature. 1983;303:637–638. doi: 10.1038/303637a0. [DOI] [PubMed] [Google Scholar]
- 58.Blanchette-Mackie E.J., Dwyer N.K., Amende L.M., Kruth H.S., Butler J.D., Sokol J., et al. Type-C Niemann-Pick disease: low density lipoprotein uptake is associated with premature cholesterol accumulation in the Golgi complex and excessive cholesterol storage in lysosomes. Proc. Natl. Acad. Sci. U. S. A. 1988;85:8022–8026. doi: 10.1073/pnas.85.21.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kobayashi T., Beuchat M.-H., Lindsay M., Frias S., Palmiter R.D., Sakuraba H., et al. Late endosomal membranes rich in lysobisphosphatidic acid regulate cholesterol transport. Nat. Cell Biol. 1999;1:113–118. doi: 10.1038/10084. [DOI] [PubMed] [Google Scholar]
- 60.Lange Y., Ye J., Rigney M., Steck T.L. Dynamics of lysosomal cholesterol in Niemann-Pick type C and normal human fibroblasts. J. Lipid Res. 2002;43:198–204. [PubMed] [Google Scholar]
- 61.Pipalia N.H., Huang A., Ralph H., Rujoi M., Maxfield F.R. Automated microscopy screening for compounds that partially revert cholesterol accumulation in Niemann-Pick C cells. J. Lipid Res. 2006;47:284–301. doi: 10.1194/jlr.M500388-JLR200. [DOI] [PubMed] [Google Scholar]
- 62.Castellano B.M., Thelen A.M., Moldavski O., Feltes M., van der Welle R.E., Mydock-McGrane L., et al. Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science. 2017;355:1306–1311. doi: 10.1126/science.aag1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chernov K.G., Neuvonen M., Brock I., Ikonen E., Verkhusha V.V. Introducing inducible fluorescent split cholesterol oxidase to mammalian cells. J. Biol. Chem. 2017;292:8811–8822. doi: 10.1074/jbc.M116.761718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koponen A., Arora A., Takahashi K., Kentala H., Kivela A.M., Jaaskelainen E., et al. ORP2 interacts with phosphoinositides and controls the subcellular distribution of cholesterol. Biochimie. 2019;158:90–101. doi: 10.1016/j.biochi.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 65.Lim C.-Y., Davis O.B., Shin H.R., Zhang J., Berdan C.A., Jiang X., et al. ER–lysosome contacts enable cholesterol sensing by mTORC1 and drive aberrant growth signalling in Niemann–Pick type C. Nat. Cell Biol. 2019;21:1206–1218. doi: 10.1038/s41556-019-0391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang H., Ma Q., Qi Y., Dong J., Du X., Rae J., et al. ORP2 delivers cholesterol to the plasma membrane in exchange for phosphatidylinositol 4, 5-bisphosphate (PI(4,5)P2) Mol. Cell. 2019;73:458–473.e7. doi: 10.1016/j.molcel.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 67.Roney J.C., Li S., Farfel-Becker T., Huang N., Sun T., Xie Y., et al. Lipid-mediated motor-adaptor sequestration impairs axonal lysosome delivery leading to autophagic stress and dystrophy in Niemann-Pick type C. Dev. Cell. 2021;56:1452–1468.e8. doi: 10.1016/j.devcel.2021.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abe M., Makino A., Hullin-Matsuda F., Kamijo K., Ohno-Iwashita Y., Hanada K., et al. A role for sphingomyelin-rich lipid domains in the accumulation of phosphatidylinositol-4,5-bisphosphate to the cleavage furrow during cytokinesis. Mol. Cell. Biol. 2012;32:1396–1407. doi: 10.1128/MCB.06113-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yancey P.G., Rodrigueza W.V., Kilsdonk E.P., Stoudt G.W., Johnson W.J., Phillips M.C., et al. Cellular cholesterol efflux mediated by cyclodextrins. Demonstration of kinetic pools and mechanism of efflux. J. Biol. Chem. 1996;271:16026–16034. doi: 10.1074/jbc.271.27.16026. [DOI] [PubMed] [Google Scholar]
- 70.Neufeld E.B., O'Brien K., Walts A.D., Stonik J.A., Demosky S.J., Malide D., et al. Cellular localization and trafficking of the human ABCG1 transporter. Biology (Basel) 2014;3:781–800. doi: 10.3390/biology3040781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ge L., Wang J., Qi W., Miao H.H., Cao J., Qu Y.X., et al. The cholesterol absorption inhibitor ezetimibe acts by blocking the sterol-induced internalization of NPC1L1. Cell Metab. 2008;7:508–519. doi: 10.1016/j.cmet.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 72.He C., Hu X., Jung R.S., Weston T.A., Sandoval N.P., Tontonoz P., et al. High-resolution imaging and quantification of plasma membrane cholesterol by NanoSIMS. Proc. Natl. Acad. Sci. U. S. A. 2017;114:2000–2005. doi: 10.1073/pnas.1621432114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kinnebrew M., Luchetti G., Sircar R., Frigui S., Viti L.V., Naito T., et al. Patched 1 reduces the accessibility of cholesterol in the outer leaflet of membranes. Elife. 2021;10 doi: 10.7554/eLife.70504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ercan B., Naito T., Koh D.H.Z., Dharmawan D., Saheki Y. Molecular basis of accessible plasma membrane cholesterol recognition by the GRAM domain of GRAMD1b. EMBO J. 2021;40 doi: 10.15252/embj.2020106524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lange Y., Ye J., Steck T.L. Essentially all excess fibroblast cholesterol moves from plasma membranes to intracellular compartments. PLoS One. 2014;9 doi: 10.1371/journal.pone.0098482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Okamoto Y., Tomioka M., Ogasawara F., Nagaiwa K., Kimura Y., Kioka N., et al. C-terminal of ABCA1 separately regulates cholesterol floppase activity and cholesterol efflux activity. Biosci. Biotechnol. Biochem. 2020;84:764–773. doi: 10.1080/09168451.2019.1700775. [DOI] [PubMed] [Google Scholar]
- 77.Ogasawara F., Kano F., Murata M., Kimura Y., Kioka N., Ueda K. Changes in the asymmetric distribution of cholesterol in the plasma membrane influence streptolysin O pore formation. Sci. Rep. 2019;9:4548. doi: 10.1038/s41598-019-39973-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li J., Lee P.L., Pfeffer S.R. Quantitative measurement of cholesterol in cell populations using flow cytometry and fluorescent perfringolysin O. Methods Mol. Biol. 2017;1583:85–95. doi: 10.1007/978-1-4939-6875-6_8. [DOI] [PubMed] [Google Scholar]
- 79.Gaigalas A.K., Zhang Y.Z., Tian L., Wang L. Sources of variability in the response of labeled microspheres and B cells during the analysis by a flow cytometer. Int. J. Mol. Sci. 2021;22:8256. doi: 10.3390/ijms22158256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Buwaneka P., Ralko A., Liu S.L., Cho W.H. Evaluation of the available cholesterol concentration in the inner leaflet of the plasma membrane of mammalian cells. J. Lipid Res. 2021;62:18. doi: 10.1016/j.jlr.2021.100084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Steck T.L., Lange Y. Transverse distribution of plasma membrane bilayer cholesterol: picking sides. Traffic. 2018;19:750–760. doi: 10.1111/tra.12586. [DOI] [PubMed] [Google Scholar]
- 82.Lorent J.H., Levental K.R., Ganesan L., Rivera-Longsworth G., Sezgin E., Doktorova M., et al. Plasma membranes are asymmetric in lipid unsaturation, packing and protein shape. Nat. Chem. Biol. 2020;16:644–652. doi: 10.1038/s41589-020-0529-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Truong D., Boddy K.C., Canadien V., Brabant D., Fairn G.D., D'Costa V.M., et al. Salmonella exploits host Rho GTPase signalling pathways through the phosphatase activity of SopB. Cell. Microbiol. 2018;20 doi: 10.1111/cmi.12938. [DOI] [PubMed] [Google Scholar]
- 84.Koponen A., Pan G., Kivelä A.M., Ralko A., Taskinen J.H., Arora A., et al. ORP2, a cholesterol transporter, regulates angiogenic signaling in endothelial cells. FASEB J. 2020;34:14671–14694. doi: 10.1096/fj.202000202R. [DOI] [PubMed] [Google Scholar]
- 85.London W.P., Steck T.L. Kinetics of enzyme reactions with interaction between a substrate and a (metal) modifier. Biochemistry. 1969;8:1767–1779. doi: 10.1021/bi00832a061. [DOI] [PubMed] [Google Scholar]
- 86.Brown M.S., Radhakrishnan A., Goldstein J.L. Retrospective on cholesterol homeostasis: the central role of Scap. Annu. Rev. Biochem. 2018;87:783–807. doi: 10.1146/annurev-biochem-062917-011852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rogers M.A., Liu J., Song B.-L., Li B.-L., Chang C.C.Y., Chang T.-Y. Acyl-CoA:cholesterol acyltransferases (ACATs/SOATs): enzymes with multiple sterols as substrates and as activators. J. Steroid Biochem. Mol. Biol. 2015;151:102–107. doi: 10.1016/j.jsbmb.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chua N.K., Brown A.J. The degron architecture of squalene monooxygenase and how specific lipids calibrate levels of this key cholesterol synthesis enzyme. Adv. Exp. Med. Biol. 2021;21:1–12. doi: 10.1007/5584_2020_583. [DOI] [PubMed] [Google Scholar]
- 89.Abrams M.E., Johnson K.A., Perelman S.S., Zhang L.S., Endapally S., Mar K.B., et al. Oxysterols provide innate immunity to bacterial infection by mobilizing cell surface accessible cholesterol. Nat. Microbiol. 2020;5:929–942. doi: 10.1038/s41564-020-0701-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Radhakrishnan A., Rohatgi R., Siebold C. Cholesterol access in cellular membranes controls Hedgehog signaling. Nat. Chem. Biol. 2020;16:1303–1313. doi: 10.1038/s41589-020-00678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Griffiths W.J., Wang Y. Sterols, oxysterols, and accessible cholesterol: signalling for homeostasis, in immunity and during development. Front. Physiol. 2021;12:723224. doi: 10.3389/fphys.2021.723224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kinnebrew M., Woolley Rachel E., Ansell T.B., Byrne Eamon F.X., Frigui S., Luchetti G., et al. Patched 1 regulates smoothened by controlling sterol binding to its extracellular cysteine-rich domain. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abm5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Takahashi K., Kanerva K., Vanharanta L., Almeida-Souza L., Lietha D., Olkkonen V.M., et al. ORP2 couples LDL-cholesterol transport to FAK activation by endosomal cholesterol/PI(4,5)P2 exchange. EMBO J. 2021;40 doi: 10.15252/embj.2020106871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gao G., Guo S., Zhang Q., Zhang H., Zhang C., Peng G. Kiaa1024L/Minar2 is essential for hearing by regulating cholesterol distribution in hair bundles. Elife. 2022;11 doi: 10.7554/eLife.80865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Palladino E.N.D., Bernas T., Green C.D., Weigel C., Singh S.K., Senkal C.E., et al. Sphingosine kinases regulate ER contacts with late endocytic organelles and cholesterol trafficking. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2204396119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lange Y., Ye J., Steck T.L. Activation of membrane cholesterol by displacement from phospholipids. J. Biol. Chem. 2005;280:36126–36131. doi: 10.1074/jbc.M507149200. [DOI] [PubMed] [Google Scholar]
- 97.Lange Y., Steck T.L. Active membrane cholesterol as a physiological effector. Chem. Phys. Lipids. 2016;199:74–93. doi: 10.1016/j.chemphyslip.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 98.Cooper M.K., Wassif C.A., Krakowiak P.A., Taipale J., Gong R., Kelley R.I., et al. A defective response to Hedgehog signaling in disorders of cholesterol biosynthesis. Nat. Genet. 2003;33:508–513. doi: 10.1038/ng1134. [DOI] [PubMed] [Google Scholar]
- 99.Endapally S., Frias D., Grzemska M., Gay A., Tomchick D.R., Radhakrishnan A. Molecular discrimination between two conformations of sphingomyelin in plasma membranes. Cell. 2019;176:1040–1053.e17. doi: 10.1016/j.cell.2018.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell. Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.