Abstract
Nasopharyngeal carcinoma (NPC) is a primary malignancy that originates from the nasopharyngeal region. It has been demonstrated that a decrease in the expression level of cell division cycle gene 25A (CDC25A) suppresses cell viability and induces apoptosis in a variety of different types of cancer. However, at present, the role of CDC25A in NPC has yet to be fully elucidated. Therefore, the aim of the present study was to investigate the role of CDC25A in NPC progression and to explore the potential underlying mechanism. Reverse transcription-quantitative PCR was performed to detect the relative mRNA levels of CDC25A and E2F transcription factor 1 (E2F1). Western blot analysis was subsequently used to determine the expression levels of CDC25A, Ki67, proliferating cell nuclear antigen (PCNA) and E2F1. CCK8 assay was employed to measure cell viability and flow cytometric analysis was employed to analyze the cell cycle. The binding sites between the CDC25A promoter and E2F1 were predicted using bioinformatics tools. Finally, luciferase reporter gene and chromatin immunoprecipitation assays were performed to verify the interaction between CDC25A and E2F1. The results obtained suggested that CDC25A is highly expressed in NPC cell lines and CDC25A silencing was found to inhibit cell proliferation, reduce the protein expression levels of Ki67 and PCNA and induce G1 arrest of NPC cells. Furthermore, E2F1 could bind CDC25A and positively regulate its expression at the transcriptional level. In addition, CDC25A silencing abolished the effects of E2F1 overexpression on cell proliferation and the cell cycle in NPC. Taken together, the findings of the present study showed that CDC25A silencing attenuated cell proliferation and induced cell cycle arrest in NPC and CDC25A was regulated by E2F1. Hence, CDC25A may be a promising therapeutic target for treatment of NPC.
Keywords: cell division cycle gene 25A, proliferation, cell cycle, E2F transcription factor 1, nasopharyngeal carcinoma
Introduction
Nasopharyngeal carcinoma (NPC) is a rare type of primary malignancy that originates from the nasopharyngeal region (1) and is predominant in southeast Asia and southern China (2,3). It is estimated that there were 129,079 new cases of NPC and 72,987 cancer mortalities caused by NPC worldwide in 2018 (4). In spite of the great progress that has been made in treating NPC with radiation therapy and combined chemotherapy, the prognosis of patients with NPC remains unsatisfactory, especially in patients at an advanced stage of the disease (5); ~30% of patients with NPC develop distant metastasis and/or recurrence (1,3), as a result of which the 5-year overall survival rate is reduced from ~90–70% (4). Owing to the annual increase of the NPC incidence rate, it is necessary to investigate the molecular mechanisms underlying the progression of NPC and to develop novel therapeutic strategies for NPC treatment.
Cell division cycle gene 25 (CDC25) family members have been demonstrated to function in several steps in multiple physiological processes, including cell cycle and mitosis (6). CDC25A is a core regulator of the cell cycle that has a dual-specific phosphatase activity. CDC25s cause the dephosphorylation of cyclin-dependent kinase (CDK)4, CDK6 and CDK2, stimulating G1 progression and accelerating into the S phase (6). CDC25A has been reported to be a promising biomarker for the diagnosis of non-small-cell lung cancer (NSCLC) and its upregulation has been shown to predict an unfavorable prognosis for patients with NSCLC (7). Moreover, CDC25A upregulation promotes the radioresistance of cervical cancer cells, whereas CDC25A knockdown reduces the cell survival rate and promotes apoptosis (8). A previous study demonstrated that reduced expression levels of CDC25A caused by the silencing of fibroblast growth factor receptor 2 contributed to the efficacy of cisplatin treatment, including attenuating cell viability and stimulating cell cycle arrest (9). Notably, it has been reported that radiation treatment can decrease CDC25A expression in human NPC cells, leading to cell cycle arrest and cell apoptosis (10). However, the specific mechanism underlying the role of CDC25A in NPC progression has yet to be fully elucidated. E2F transcription factor 1 (E2F1) belongs to the E2F family of transcription factors, participating in cellular differentiation and tissue development to maintain body metabolism and homeostasis as a metabolic regulator (11). An number of studies have highlighted the crucial role of E2F1 in various types of malignancies (12), including ovarian cancer (13), prostate cancer (14) and glioblastoma (15). It is noteworthy that the importance of E2F1 in NPC inflammation and tumorigenesis has also been identified in previously published studies (16,17). Nevertheless, whether CDC25A functions in NPC via transcriptional regulation of E2F1 remains unclear.
Therefore, the aim of the present study was to investigate the role of CDC25A in cell proliferation and cell cycle in NPC and to explore the potential underlying mechanism. The findings showed how E2F1 suppression-mediated CDC25A silencing attenuated cell proliferation and induced cell cycle arrest in NPC, thereby raising the possibility that CDC25A may be a promising therapeutic target for NPC treatment.
Materials and methods
Bioinformatic analysis
The Coexpedia database (http://www.coexpedia.org/) was used to analyze the co-expression between CDC25A and E2F1 (‘CDC25A’ entered in the Search panel and E2F1 search for in the Co-expressed Genes panel). The Cyclebase database (https://cyclebase.org/) was searched to identify E2F1 as a transcription factor that could regulate G1/S phase transition (‘E2F1’ entered in the in Search panel). In addition, the predicted binding sequences between E2F1 and the CDC25A promoter was analyzed using the JASPAR database (https://jaspar.genereg.net; relative profile score threshold: 80%).
Cell culture and transfection
A human nasopharyngeal epithelial cell line (NP69) and four NPC cell lines (C666-1, HNE-3, NPC-039 and HK1) were obtained from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences. The cells were cultured in RPMI-1640 medium (Thermo Fisher Scientific, Inc.). The NP69 cell line was used as the control group. Cells were supplied with 10% HyClone fetal bovine serum (HyClone; Cytiva) and cultured at 37°C with 5% CO2. Cells (5×103/well) at 80% confluence were transfected with 100 nM empty vector siRNA-negative control (NC; sequence, 5′-UUCUCCGAACGUGUCACGU-3′) or short interfering (si)RNA-CDC25A (siRNA-CDC25A-1, 5′-GCUUAGCUAGCAUUACUAACC-3′; and siRNA-CDC25A-2, 5′-GCGUGUCAUUGUUGUGUUUCA-3′) or siRNA-E2F1 (siRNA-E2F1-1, 5′-GAUGGUUAUGGUGAUCAAAGC-3′; and siRNA-E2F1-2, 5′-AGAUGGUUAUGGUGAUCAAAG-3′), all synthesized by Guangzhou RiboBio Co., Ltd. using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. In addition, the full-length sequences of E2F1 were inserted into pcDNA3.1 vector (Invitrogen; Thermo Fisher Scientific, Inc.) to construct E2F1 overexpression plasmid (OV-E2F1). The plasmids for pcDNA3.1 (400 ng/µl; OV-NC and OV-E2F1) were respectively transfected into HK1 cells using Lipofectamine 2000 for 48 h at 37°C according to the manufacturer's instructions. After 6 h, the medium was replaced with fresh DMEM (Thermo Fisher Scientific, Inc.) containing 10% FBS and maintained in a 5% CO2 incubator for a further 48 h.
Reverse transcription-quantitative (RT-q)PCR analysis
Total RNA was isolated from NPC cells (1×106 cells) using an RNA isolation kit (Tiangen Biotech Co., Ltd.), following the manufacturer's instructions. The RNA quality was examined using a NanoDrop™ 2000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc.). A RT kit (Takara Bio, Inc.) was used for RT to obtain the first-chain cDNAs, according to the manufacturer's protocol. A standard SYBR® Green PCR kit (Takara Bio, Inc.) was used to amplify the target fragments, according to the manufacturer's protocol. qPCR was performed at 50°C for 2 min and 95°C for 2 min, followed by 40 cycles at 95°C for 15 sec, 60°C for 1 min and extension at 72°C for 1 min, followed by a final extension step at 72°C for 10 min. The relative gene expression levels were calculated using the 2−ΔΔCq method (18). The cDNA fragment of the GAPDH gene was used as an internal control for analysis of the results. The primer sequences used were: CDC25A forward, 5′-TTCCTCTTTTTACACCCCAGTCA-3′ and reverse, 5′-TCGGTTGTCAAGGTTTGTAGTTC-3′; E2F1 forward, 5′-TGAGGGCATCCAGCTCATTG-3′ and reverse, 5′-AAACATCGATCGGGCCTTGT-3′; Cyclin D1 forward, 5′-CTGATTGGACAGGCATGGGT-3′ and reverse, 5′-GTGCCTGGAAGTCAACGGTA-3′; CDK4 forward, 5′-GTGTATGGGGCCGTAGGAAC-3′ and reverse, 5′-CAGTCGCCTCAGTAAAGCCA-3′; CDC6 forward, 5′-GCGAGGCCTGAGCTGTG-3′ and reverse, 5′-GCTGAGAGGCAGGGCTTTTA-3′; and GAPDH forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse, 5′-CACCCTGTTGCTGTAGCCAAA-3′. This experiment was repeated three times.
Western blot analysis
Following transfection, total proteins were extracted with RIPA lysis buffer (Wuhan Servicebio Technology Co., Ltd.) containing protease/phosphatase inhibitor cocktail. The total protein concentration was measured with a BCA protein assay kit (Beijing Solarbio Science and Technology Co., Ltd.). Equal amounts of protein per lane (25 µg) were separated by SDS-PAGE on 10% gels and were transferred onto PVDF membranes. Membranes were blocked with 5% skimmed milk for 1 h at room temperature and probed with primary antibodies at 4°C overnight. The primary antibodies included anti-CDC25A (cat. no. ab989; 1:1,000 dilution; Abcam), anti-Ki67 (cat. no. ab16667; 1:1,000; Abcam), anti-PCNA (cat. no. ab92552; 1:1,000; Abcam) and anti-E2F1 (cat. no. ab288369; 1:1,000; Abcam) antibodies. Subsequently, the membranes were incubated with HRP-conjugated goat anti-rabbit IgG (cat. no. ab6721; 1:2,000; Abcam) at room temperature for 2 h and images were acquired using a Tanon-5200 Chemiluminescence Imager (Tanon Science and Technology Co., Ltd.). Finally, the band density was analyzed using ImageJ software v1.8.0 (National Institutes of Health).
Cell counting kit-8 (CCK-8) assay
Following transfection, CCK-8 assay was performed to determine the cell viability. After incubating the HK1 cells for 0, 24, 48 and 72 h, CCK-8 solution (10 µl/well; Dojindo Laboratories, Inc.) was added and incubated with the cells for a further 2 h. Finally, the absorbance at 450 nm was measured using a microplate reader (Bio-Rad Laboratories, Inc.).
Flow cytometry
HK1 cells were planted in 6-well plates (1×105 cells/well) and transfected with or without OV-E2F1 and siRNA-CDC25A. At 24 h after transfection, the HK1 cells were collected. A Cell Cycle Assay kit (BD Biosciences) was used to analyze the cell cycle following the manufacturer's instructions. Briefly, HK1 cells were incubated with 200 µl liquid A for 10 min and 150 µl liquid B at 4°C for a further 10 min. Subsequently, HK1 cells were incubated with 120 µl liquid C in the dark at 4°C for 10 min prior to flow cytometry analysis on a BD FACSort system (BD Biosciences). Finally, the results were analyzed using ModFit software, version 3.2 (Verity Software House, Inc.).
Luciferase reporter gene assay
The full length of CDC25A promoter (FL group) and the full length of CDC25A promoter including mutational site 1 (CGTAATAT) (Site 1 group) and the full length of CDC25A promoter including mutational site 2 (GGCTATAT) (Site 2 group) were respectively cloned into the firefly luciferase reporter plasmid, pGL3-basic vector (Addgene, Inc.). HK1 cells were seeded in 24-well plates and cultured at 37°C. Subsequently, the OV-NC and OV-E2F1 overexpression plasmids were co-transfected into HK1 cells using Lipofectamine 2000 for 48 h. The cells were then lysed and the firefly luciferase activity was measured and normalized against Renilla luciferase activity using a Dual-Luciferase Reporter Assay System (Promega Corporation) according to the manufacturer's instructions. The relative activity of luciferase was determined through measuring the ratio of the two identified activities. The sequences of full length of CDC25A promoter are shown in Table SI.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed as previously described by using a Magna ChIP™ kit (MilliporeSigma). In brief, HK1 cells (1×106) were transfected with E2F1 overexpression plasmids for 48 h and subsequently DNA associated with specific immunoprecipitates or with mouse immunoglobulin G as a negative control was isolated and used as a template for PCR, amplifying the CDC25A promoter sequence containing the E2F1-binding site. qPCR was performed at 50°C for 2 min and 95°C for 2 min, followed by 40 cycles at 95°C for 15 sec and 60°C for 1 min as aforementioned using the SYBR Green PCR kit. The relative gene expression levels were calculated using the 2−ΔΔCq method (18). The primer sequence used in this study were as follows: CDC25A forward, 5′-CTTCTGAGAGCCGATGACCT-3′ and reverse, 5′-CACCTCTTACCCAGGCTGTC-3′ (19).
Statistical analysis
The statistical analysis was conducted using SPSS 22.0 software (IBM Corp.) and GraphPad Prism 6 software (GraphPad Software; Dotmatics). Data are expressed as the mean ± standard error of the mean of results derived from three independent experiments performed in triplicate. Statistical analysis was performed using one-way ANOVA followed by Tukey or Bonferroni post-hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
CDC25A is highly expressed in NPC cell lines
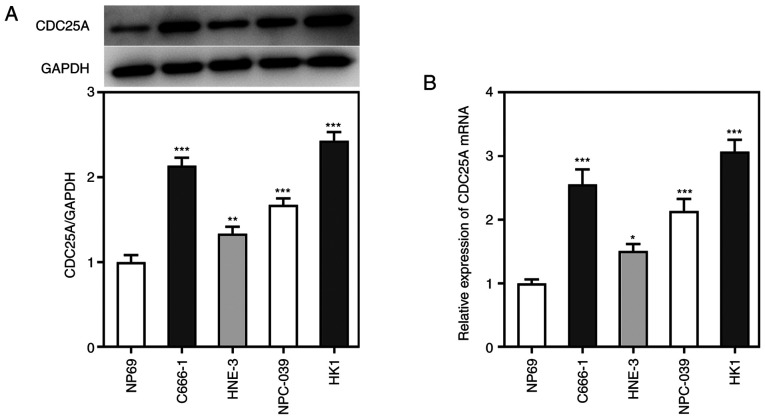
To explore the role of CDC25A in the progression of NPC, its expression levels were detected by western blot and RT-qPCR analyses. As shown in Fig. 1A and B, the results from the western blotting and RT-qPCR experiments showed that, compared with NP69 cells, the expression of CDC25A was significantly upregulated in the NPC cell lines. HK1 cells showed the highest mRNA and protein levels of CDC25A, therefore HK1 cells were selected as the cell line of choice in subsequent experiments. These results also suggested that CDC25A may be involved in NPC progression.
Figure 1.
CDC25A was highly expressed in NPC cell lines. (A) The protein expression of CDC25A was determined by western blotting. (B) The relative mRNA expression of CDC25A was quantified by reverse transcription-quantitative PCR. The data were expressed as mean ± standard error of the mean of three independent experiments. *P<0.05, **P<0.01, ***P<0.001 vs. NP69. CDC25A, cell division cycle gene 25A; NPC, nasopharyngeal carcinoma.
CDC25A silencing inhibits cell proliferation and induces G1 arrest in NPC
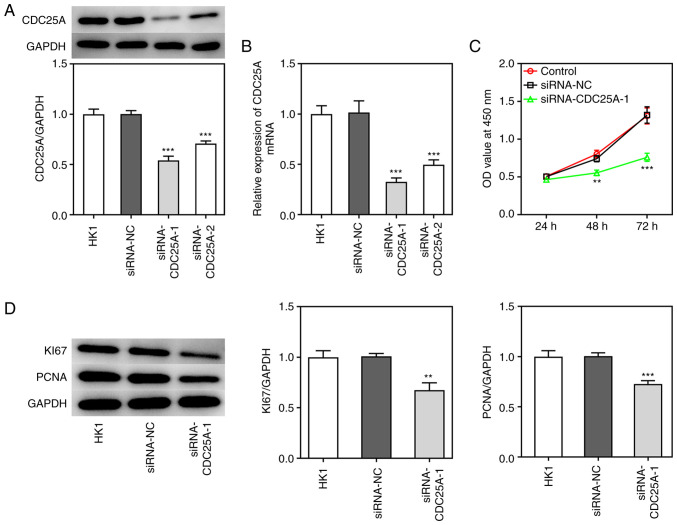
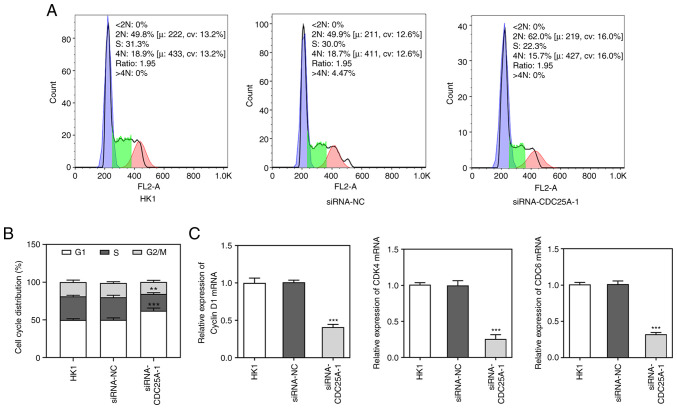
To investigate the role of CDC25A in NPC progression, the transfection of siRNA-CDC25A-1 and siRNA-CDC25A-2 was performed, first to achieve CDC25A silencing, as demonstrated by the results from western blotting and RT-qPCR analyses (Fig. 2A and B). Due to the lower expression level of CDC25A in the siRNA-CDC25A-1 group compared with that in the siRNA-CDC25A-2 group, siRNA-CDC25A-1 was used for further studies. Subsequently, the proliferation and cell cycle of HK1 cells were analyzed. The cell viability of HK1 cells transfected with siRNA-CDC25A was appraised using a CCK-8 assay. As shown in Fig. 2C, CDC25A silencing suppressed HK1 cell viability. Furthermore, CDC25A knockdown suppressed the expression of proliferation markers, including Ki67 and PCNA (Fig. 2D). Cell cycle was then analyzed by flow cytometry. As shown in Fig. 3A and B, it was observed that the numbers of HK1 cells transfected with siRNA-CDC25A in G1-phase were significantly larger compared with those of the siRNA-NC group. In addition, CDC25A knockdown reduced the levels of Cyclin D1, CDK4 and CDK6 (Fig. 3C). Therefore, it was possible to conclude that CDC25A silencing inhibited the proliferation and induced G1 arrest of HK1 cells.
Figure 2.
CDC25A silencing inhibited the proliferation of HK1 cells. (A) The protein expression of CDC25A was determined by western blotting. (B) The relative mRNA expression of CDC25A was quantified by reverse transcription-quantitative PCR. (C) HK1 cell viability was measured by CCK8 assay. (D) The expressions of Ki67 and PCNA were determined by western blotting. The data were expressed as mean ± standard error of the mean of three independent experiments. **P<0.01, ***P<0.001 vs. siRNA-NC. CDC25A, cell division cycle gene 25A; PCNA, proliferating cell nuclear antigen.
Figure 3.
CDC25A silencing induced G1 arrest of HK1 cells. (A) The cell cycle of HK1 cells was analyzed by flow cytometry and (B) quantified. The data were expressed as mean ± standard error of the mean with three independent times. (C) The protein expressions of Cyclin D1, CDK4 and CDK6 were determined by western blotting. **P<0.01, ***P<0.001 vs. siRNA-NC. CDC25A, cell division cycle gene 25A; CDK, cyclin-dependent kinase; si, short interfering; NC, negative control.
CDC25A is transcriptionally regulated by E2F1
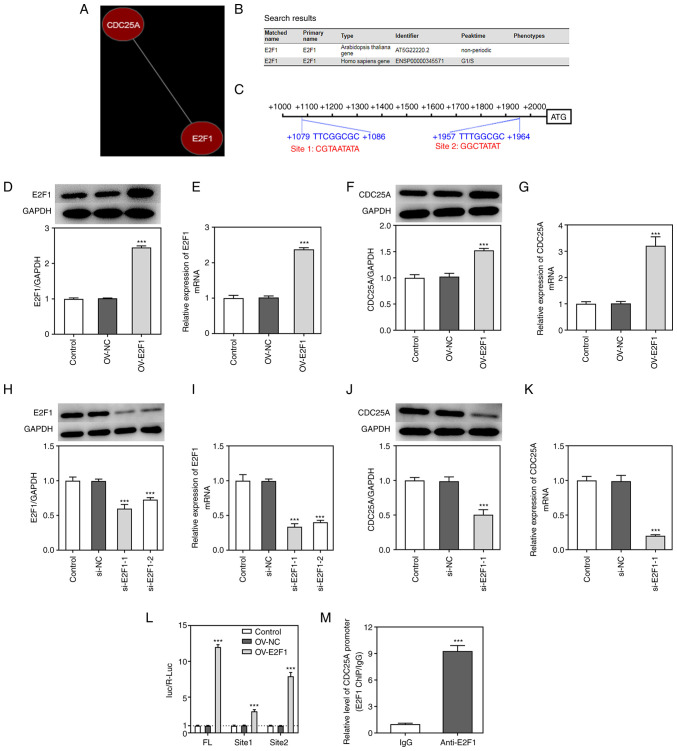
To investigate the potential mechanism underlying the role of CDC25A in NPC, co-expression between CDC25A and E2F1 was screened using the Coexpedia database (http://www.coexpedia.org/; Fig. 4A) and E2F1 was identified as a transcription factor that could regulate G1/S-phase transition using the Cyclebase database (https://cyclebase.org/; Fig. 4B), suggesting that E2F1 may exert a regulatory effect on CDC25A expression. The predicted binding sequences between E2F1 and the CDC25A promoter are shown in Fig. 4C. Subsequently, the OV-E2F1 plasmid was constructed to achieve E2F1 overexpression and the transfection efficacy was demonstrated through RT-qPCR and western blot analyses (Fig. 4D and E). Moreover, CDC25A upregulation resulted in E2F1 overexpression (Fig. 4F and G). The si-E2F1 plasmid was also constructed to achieve E2F1 knockdown. Due to the lower expression level of E2F1 that was observed, si-E2F1-1 was selected in subsequent experiments (Fig. 4H and I). Notably, E2F1 knockdown also caused downregulation of CDC25A expression (Fig. 4J and K). Furthermore, the results from the luciferase reporter assay showed that luciferase activity were reduced in Site 1 and Site 2 groups compared with the FL group (Fig. 4L). Moreover, ChIP assays corroborated that CDC25A could interact with E2F1 (Fig. 4M). These results indicated that CDC25A could both interact with E2F1 and was transcriptionally regulated by E2F1.
Figure 4.
CDC25A was transcriptionally regulated by E2F1 in HK1 cells. (A) The interaction between CDC25A and E2F1. (B) The search results from Cyclebase database. (C) The binding sites of CDC25A promoter and E2F1. The mutational fragments are in red. (D) The protein expression of E2F1 was determined by western blotting. (E) The relative mRNA expression of E2F1 was quantified by RT-qPCR. (F) The protein expression of CDC25A was determined by western blotting. (G) The relative mRNA expression of CDC25A was quantified by RT-qPCR. (H) The protein expression of E2F1 was determined by western blotting. (I) The relative mRNA expression of E2F1 was quantified by RT-qPCR. (J) The protein expression of CDC25A was determined by western blotting. (K) The relative mRNA expression of CDC25A was quantified by RT-qPCR. (L) The interaction between CDC25A and E2F1 was confirmed by luciferase reporter assay and ChIP assays. In luciferase reporter assay, FL group used the normal FL of CDC25A promoter while Site 1 and Site 2 groups used the FL of CDC25A promoter including the two mutational sequences of CDC25A promoter. (M) The interaction between CDC25A and E2F1 was confirmed by ChIP assay. The data were expressed as mean ± standard error of the mean of three independent experiments. ***P<0.001 vs. Control. CDC25A, cell division cycle gene 25A; E2F1, E2F transcription factor 1; RT-qPCR, reverse transcription-quantitative PCR; ChIP, chromatin immunoprecipitation; FL, full length.
CDC25A silencing abolished the effect of E2F1 overexpression on cell proliferation and cell cycle in NPC
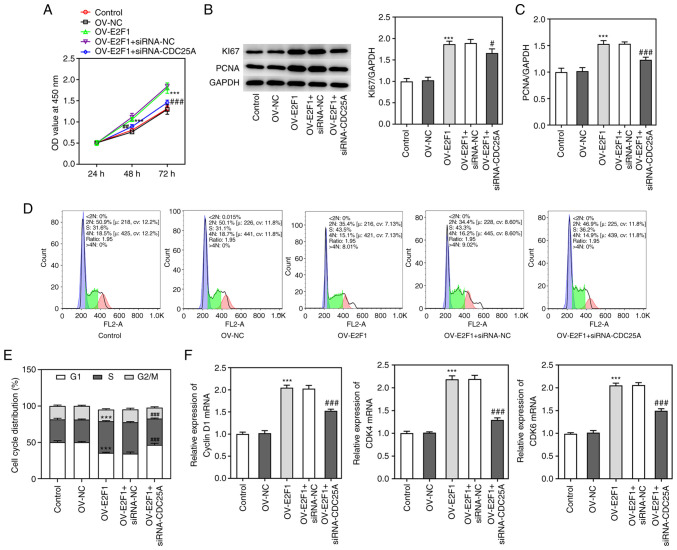
To study whether E2F1 could mediate the role of CDC25A in NPC progression, the proliferation of HK1 cells transfected with E2F1 overexpression plasmids and the cell cycle were both analyzed, as described above. As shown in Fig. 5A, E2F1 overexpression enhanced cell viability compared with the control group, whereas CDC25A silencing suppressed the viability of HK1 cells transfected with OV-E2F1. Additionally, the expression levels of Ki67 and PCNA were increased by E2F1 overexpression, whereas they were decreased through CDC25A silencing (Fig. 5B and C). More significantly, E2F1 overexpression promoted the transition of the cell cycle from G1-phase to S-phase in HK1 cells and this effect was partially reversed by CDC25A silencing (Fig. 5D and E). E2F1 overexpression increased the levels of Cyclin D1, CDK4 and CDK6 while CDC25A silencing reversed the effects of E2F1 overexpression on the levels of Cyclin D1, CDK4 and CDK6 (Fig. 5F). Taken together, these results implied that CDC25A silencing abolished the effect of E2F1 overexpression on the proliferation and cell cycle of HK1 cells.
Figure 5.
CDC25A silencing abolished the effect of E2F1 overexpression on cell proliferation and cell cycle in HK1 cells. (A) HK1 cell viability was measured by CCK8 assay. (B) The expression of Ki67 and PCNA were determined by western blot and (C) quantified. (D) The cell cycle of HK1 cells was analyzed by flow cytometry and (E) quantified. (F) The mRNA expressions of Cyclin D1, CDK4 and CDK6 were detected by qRT-PCR. The data were expressed as mean ± standard error of the mean of three independent experiments. ***P<0.001 vs. Control. ###P<0.001 vs. OV-E2F1. CDC25A, cell division cycle gene 25A; E2F1, E2F transcription factor 1; PCNA, proliferating cell nuclear antigen; OV, overexpression.
Discussion
Nasopharyngeal carcinoma (NPC) is a distinct head-and-neck cancer with high incidence of locoregional recurrence or metastasis (20). Broadly speaking, at present NPC is associated with an increasing number of new cases and mortality and it remains a major public health concern. However, the mechanisms underpinning the pathogenesis of NPC have yet to be fully elucidated. In the present study, the expression levels of CDC25A were found to be markedly elevated in all the NPC cell lines tested, including C666-1, HNE-3, NPC-039 and HK1 cells, compared with the normal nasopharyngeal epithelial cell line (NP69), suggesting that CDC25A may have an oncogenic role in NPC progression.
Accumulating evidence has identified CDC25A as an oncogene in multiple types of cancer, including ovarian cancer, liver cancer and breast cancer and it has been shown to be associated with the induction of chemoresistance (21,22). It has been reported that CDC25A knockdown inhibits cell proliferation, migration and invasion in colorectal cancer (23). CDC25A targeted by miR-34a-5p exacerbates cervical cancer growth and migration (24). Liu et al (25) also propose that CDC25A aggravates cell proliferation and impedes cell cycle in hepatocellular carcinoma. As far as is known at present, the latent mechanism underlying the carcinogenic role of CDC25A is complicated, primarily involving cell proliferation, apoptosis and the cell cycle (26,27). Collectively, the findings in the present study demonstrated that CDC25A silencing suppressed the viability of HK1 cells. Ki67 and PCNA are regarded as proliferation markers: Ki67, mainly expressed in the nucleus of proliferating cells, is closely associated with mitosis (28), whereas PCNA has also been documented to mediate cell proliferation via lipid phosphatase activity, protein phosphatase activity and various signaling pathways (29). The experimental data presented in this study have also shown that the expression levels of Ki67 and PCNA were suppressed by CDC25A knockdown, which further suggested that CDC25A interference could apparently suppress the proliferation of NPC cells. The accurate transition of the cell cycle from G1-phase to S-phase fulfills an important role in controlling cell proliferation and its dysregulation may contribute to oncogenesis (30). As a member of the CDC25 family of cell division cycle proteins, CDC25A promotes the transition of cells from G1-phase to S-phase by targeting cyclin A/CDK2 and cyclin E/CDK2 (26,31). For example, a previous study demonstrated that CDC25A has an important role in regulating cell cycle-mediated chemoresistance in colorectal cancer (32). CDC25A also aggravates the course of breast cancer via the cell-cycle pathway (21). In the present study, CDC25A silencing induced G1 arrest of HK1 cells, a finding that was consistent with the previous study. Hence, CDC25A may serve as an oncogene in NPC progression.
To investigate the potential mechanism underlying the role of CDC25A in NPC progression, additional studies were performed. E2F1 has been shown to function chiefly through transcriptional activation events in the E2F family of transcription factors. Forkhead box protein O1 halts cell proliferation by downregulating the E2F1-activated expression of NOD-, LRR- and pyrin domain-containing protein 3 (NLRP3) in prostate cancer (33). E2F1-mediated SEC61G promotes breast cancer tumor growth and metastasis (34). In a similar way, the co-expression of CDC25A and E2F1 was screened using the Coexpedia database and E2F1 was identified as a transcription factor that could regulate G1/S-phase transition through the Cyclebase database. E2F1 has been proposed to participate in myriad cell biological processes, including cell cycle, DNA repair, apoptosis, development, differentiation and metabolism (35,36). A previous study reported that the overexpression of E2F1 led to a decrease in the radiosensitivity of NPC cells (37). The findings from the present study suggested that E2F1, as a transcription factor, could bind to the promoter of CDC25A and thereby positively regulate its expression. Luciferase reporter assay showed that the luciferase activity were reduced in cells with mutational sites compared with the cells with the normal sequences of CDC25A promoter, which may be because the promoter of CDC25A with mutational sequences is harder to combined with E2F1. Notably, CDC25A silencing abolished the effect of E2F1 overexpression on the proliferation of HK1 cells and on the G1/S-phase cell cycle, indicating that E2F1 could mediate the role of CDC25A in NPC progression.
Taken together, the results of the present study have elucidated the specific role of CDC25A in cell proliferation and the cell cycle in NPC, increasing our understanding of the underlying regulatory mechanism by revealing the association between CDC25A and E2F1 transcription factor. Nevertheless, there are several limitations in the present study. It did not examine the expression of CDC25A in nasopharyngeal carcinoma patients or animals, so animal experiments in vivo and clinical trials in humans are required to augment the findings of the present study. Additionally, whether signaling pathways have any functional roles downstream of CDC25A also requires further investigation. The present study proved E2F1 targets to CDC25A in its transcription, but did not explore the mechanism the E2F1-CDC2A interaction in the cell cycle regulation and nor did it explore CyclinD, CyclinE, CDK2/CDC1 and other specific markers for cell cycle; these will be investigated in a future study. In addition, the present study only used the HK1 cell line to explore the functional role of CDC25A in NPC and the biological effects of CDC25A in other NPC cell lines will be explored in a future study. Moreover, the present study aimed to explore the effects of CDC25A on cell proliferation and cell cycle in nasopharyngeal carcinoma, thus it did not perform wound healing and invasion assays; the effects of CDC25A on cell migration and invasion will be explored in a future study.
In conclusion, the present study demonstrated for the first time, to the best of the authors' knowledge, that CDC25A silencing could attenuate cell proliferation and induce cell cycle arrest in NPC and the regulation of CDC25A were transcriptionally modulated by the E2F1 transcription factor. Therefore, the present study has provided valuable information that improves our understanding of the pathogenesis of NPC, also indicating that CDC25A may be a promising therapeutic target for NPC treatment.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by Medical Health Science and Technology Project of Zhejiang Provincial Health Commission (grant no. 2020KY695), and The Construction Fund of Key Medical Disciplines of Hangzhou (grant no. OO20200123).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
BW and QG designed the study, drafted and revised the manuscript. BW and FC analyzed the data and searched the literature. BW, QG and FC performed the experiments. BW and QG confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zhang P, He Q, Lei Y, Li Y, Wen X, Hong M, Zhang J, Ren X, Wang Y, Yang X, et al. m6A-mediated ZNF750 repression facilitates nasopharyngeal carcinoma progression. Cell Death Dis. 2018;9:1169. doi: 10.1038/s41419-018-1224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chow JC, Ngan RK, Cheung KM, Cho WC. Immunotherapeutic approaches in nasopharyngeal carcinoma. Expert Opin Biol Ther. 2019;19:1165–1172. doi: 10.1080/14712598.2019.1650910. [DOI] [PubMed] [Google Scholar]
- 3.Zheng ZQ, Li ZX, Zhou GQ, Lin L, Zhang LL, Lv JW, Huang XD, Liu RQ, Chen F, He XJ, et al. Long noncoding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and upregulate ITGB3. Cancer Res. 2019;79:4612–4626. doi: 10.1158/0008-5472.CAN-19-0799. [DOI] [PubMed] [Google Scholar]
- 4.Lee HM, Okuda KS, González FE, Patel V. Current perspectives on nasopharyngeal carcinoma. Adv Exp Med Biol. 2019;1164:11–34. doi: 10.1007/978-3-030-22254-3_2. [DOI] [PubMed] [Google Scholar]
- 5.Guo Y, Zhai J, Zhang J, Ni C, Zhou H. Improved radiotherapy sensitivity of nasopharyngeal carcinoma cells by miR-29-3p targeting COL1A1 3′-UTR. Med Sci Monit. 2019;25:3161–3169. doi: 10.12659/MSM.915624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenner AK, Reikvam H, Lavecchia A, Bruserud Ø. Therapeutic targeting the cell division cycle 25 (CDC25) phosphatases in human acute myeloid leukemia-the possibility to target several kinases through inhibition of the various CDC25 isoforms. Molecules. 2014;19:18414–18447. doi: 10.3390/molecules191118414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin TC, Lin PL, Cheng YW, Wu TC, Chou MC, Chen CY, Lee H. MicroRNA-184 deregulated by the MicroRNA-21 promotes tumor malignancy and poor outcomes in non-small cell lung cancer via targeting CDC25A and c-Myc. Ann Surg Oncol. 2015;22((Suppl 3)):S1532–S1539. doi: 10.1245/s10434-015-4595-z. [DOI] [PubMed] [Google Scholar]
- 8.Ding FN, Gao BH, Wu X, Gong CW, Wang WQ, Zhang SM. miR-122-5p modulates the radiosensitivity of cervical cancer cells by regulating cell division cycle 25A (CDC25A) FEBS Open Bio. 2019;9:1869–1879. doi: 10.1002/2211-5463.12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pu L, Su L, Kang X. The efficacy of cisplatin on nasopharyngeal carcinoma cells may be increased via the downregulation of fibroblast growth factor receptor 2. Int J Mol Med. 2019;44:57–66. doi: 10.3892/ijmm.2019.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li MY, Liu JQ, Chen DP, Li ZY, Qi B, He L, Yu Y, Yin WJ, Wang MY, Lin L. Radiotherapy induces cell cycle arrest and cell apoptosis in nasopharyngeal carcinoma via the ATM and Smad pathways. Cancer Biol Ther. 2017;18:681–693. doi: 10.1080/15384047.2017.1360442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denechaud PD, Fajas L, Giralt A. E2F1, a novel regulator of metabolism. Front Endocrinol (Lausanne) 2017;8:311. doi: 10.3389/fendo.2017.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen C, Li J, Chang S, Che G. Advancement of E2F1 in common tumors. Zhongguo Fei Ai Za Zhi. 2020;23:921–926. doi: 10.3779/j.issn.1009-3419.2020.101.32. (In Chinese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farra R, Dapas B, Grassi M, Benedetti F, Grassi G. E2F1 as a molecular drug target in ovarian cancer. Expert Opin Ther Targets. 2019;23:161–164. doi: 10.1080/14728222.2019.1579797. [DOI] [PubMed] [Google Scholar]
- 14.Bi XC, Pu XY, Liu JM, Huang S. Effect of transcription factor E2F1 expression on the invasion of prostate cancer. Zhonghua Yi Xue Za Zhi. 2017;97:2856–2859. doi: 10.3760/cma.j.issn.0376-2491.2017.36.016. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 15.Godoy PRDV, Donaires FS, Montaldi APL, Sakamoto-Hojo ET. Anti-proliferative effects of E2F1 suppression in glioblastoma cells. Cytogenet Genome Res. 2021;161:372–381. doi: 10.1159/000516997. [DOI] [PubMed] [Google Scholar]
- 16.Liu P, Zhang X, Li Z, Wei L, Peng Q, Liu C, Wu Y, Yan Q, Ma J. A significant role of transcription factors E2F in inflammation and tumorigenesis of nasopharyngeal carcinoma. Biochem Biophys Res Commun. 2020;524:816–824. doi: 10.1016/j.bbrc.2020.01.158. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Nie P, Zhu D. LncRNA HOXA10-AS activated by E2F1 facilitates proliferation and migration of nasopharyngeal carcinoma cells through sponging miR-582-3p to upregulate RAB31. Am J Rhinol Allergy. 2022;36:348–359. doi: 10.1177/19458924211064400. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Ru Lee W, Chen CC, Liu S, Safe S. 17beta-estradiol (E2) induces cdc25A gene expression in breast cancer cells by genomic and non-genomic pathways. J Cell Biochem. 2006;99:209–220. doi: 10.1002/jcb.20902. [DOI] [PubMed] [Google Scholar]
- 20.Xiao Z, Chen Z. Deciphering nasopharyngeal carcinoma pathogenesis via proteomics. Expert Rev Proteomics. 2019;16:475–485. doi: 10.1080/14789450.2019.1615891. [DOI] [PubMed] [Google Scholar]
- 21.Qin H, Liu W. MicroRNA-99a-5p suppresses breast cancer progression and cell-cycle pathway through downregulating CDC25A. J Cell Physiol. 2019;234:3526–3537. doi: 10.1002/jcp.26906. [DOI] [PubMed] [Google Scholar]
- 22.Sun Y, Li S, Yang L, Zhang D, Zhao Z, Gao J, Liu L. CDC25A facilitates chemo-resistance in ovarian cancer multicellular spheroids by promoting E-cadherin expression and arresting cell cycles. J Cancer. 2019;10:2874–2884. doi: 10.7150/jca.31329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin W, Xu J, Li C, Dai X, Wu T, Wen J. Circular RNA circ_0007142 facilitates colorectal cancer progression by modulating CDC25A expression via miR-122-5p. Onco Targets Ther. 2020;13:3689–3701. doi: 10.2147/OTT.S238338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang T, Cheng H. miR-34a-5p blocks cervical cancer growth and migration by downregulating CDC25A. J BUON. 2021;26:1768–1774. [PubMed] [Google Scholar]
- 25.Liu P, Xia P, Fu Q, Liu C, Luo Q, Cheng L, Yu P, Qin T, Zhang H. miR-199a-5p inhibits the proliferation of hepatocellular carcinoma cells by regulating CDC25A to induce cell cycle arrest. Biochem Biophys Res Commun. 2021;571:96–103. doi: 10.1016/j.bbrc.2021.07.035. [DOI] [PubMed] [Google Scholar]
- 26.Sadeghi H, Golalipour M, Yamchi A, Farazmandfar T, Shahbazi M. CDC25A pathway toward tumorigenesis: Molecular targets of CDC25A in cell-cycle regulation. J Cell Biochem. 2019;120:2919–2928. doi: 10.1002/jcb.26838. [DOI] [PubMed] [Google Scholar]
- 27.Shen T, Huang S. The role of Cdc25A in the regulation of cell proliferation and apoptosis. Anticancer Agents Med Chem. 2012;12:631–639. doi: 10.2174/187152012800617678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun X, Kaufman PD. Ki-67: More than a proliferation marker. Chromosoma. 2018;127:175–186. doi: 10.1007/s00412-018-0659-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Worby CA, Dixon JE. PTEN. Annu Rev Biochem. 2014;83:641–669. doi: 10.1146/annurev-biochem-082411-113907. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Wang Y, Wang S, Meng X, Song F, Huo W, Zhang S, Chang J, Li J, Zheng B, et al. Coxsackievirus A6 induces cell cycle arrest in G0/G1 phase for viral production. Front Cell Infect Microbiol. 2018;8:279. doi: 10.3389/fcimb.2018.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ditano JP, Sakurikar N, Eastman A. Activation of CDC25A phosphatase is limited by CDK2/cyclin A-mediated feedback inhibition. Cell Cycle. 2021;20:1308–1319. doi: 10.1080/15384101.2021.1938813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma Y, Wang R, Lu H, Li X, Zhang G, Fu F, Cao L, Zhan S, Wang Z, Deng Z, et al. B7-H3 promotes the cell cycle-mediated chemoresistance of colorectal cancer cells by regulating CDC25A. J Cancer. 2020;11:2158–2170. doi: 10.7150/jca.37255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang Y, Jiang L, Zhao X, Hu D, Zhao G, Luo S, Du X, Tang W. FOXO1 inhibits prostate cancer cell proliferation via suppressing E2F1 activated NPRL2 expression. Cell Biol Int. 2021;45:2510–2520. doi: 10.1002/cbin.11696. [DOI] [PubMed] [Google Scholar]
- 34.Ma J, He Z, Zhang H, Zhang W, Gao S, Ni X. SEC61G promotes breast cancer development and metastasis via modulating glycolysis and is transcriptionally regulated by E2F1. Cell Death Dis. 2021;12:550. doi: 10.1038/s41419-021-03797-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bramis J, Zacharatos P, Papaconstantinou I, Kotsinas A, Sigala F, Korkolis DP, Nikiteas N, Pazaiti A, Kittas C, Bastounis E, Gorgoulis VG. E2F-1 transcription factor immunoexpression is inversely associated with tumor growth in colon adenocarcinomas. Anticancer Res. 2004;24:3041–3047. [PubMed] [Google Scholar]
- 36.Chan AB, Huber AL, Lamia KA. Cryptochromes modulate E2F family transcription factors. Sci Rep. 2020;10:4077. doi: 10.1038/s41598-020-61087-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan Y, Wei X, Zhang W, Wang X, Wang K, Du B, Xiao J. Resveratrol enhances the radiosensitivity of nasopharyngeal carcinoma cells by downregulating E2F1. Oncol Rep. 2017;37:1833–1841. doi: 10.3892/or.2017.5413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.