Key Points
-
•
Crystallization studies map the binding of glenzocimab to the site of dimerization in the D2 domain of GPVI.
-
•
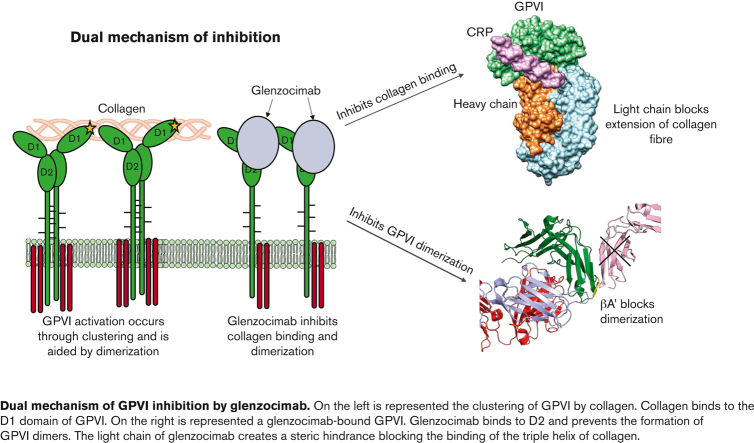
Glenzocimab inhibits GPVI interactions with CRP, collagen, and fibrin by loss of dimerization, conformational changes, and steric hindrance.
Visual Abstract
Abstract
Platelet glycoprotein VI (GPVI) is attracting interest as a potential target for the development of new antiplatelet molecules with a low bleeding risk. GPVI binding to vascular collagen initiates thrombus formation and GPVI interactions with fibrin promote the growth and stability of the thrombus. In this study, we show that glenzocimab, a clinical stage humanized antibody fragment (Fab) with a high affinity for GPVI, blocks the binding of both ligands through a combination of steric hindrance and structural change. A cocrystal of glenzocimab with an extracellular domain of monomeric GPVI was obtained and its structure determined to a resolution of 1.9 Å. The data revealed that (1) glenzocimab binds to the D2 domain of GPVI, GPVI dimerization was not observed in the crystal structure because glenzocimab prevented D2 homotypic interactions and the formation of dimers that have a high affinity for collagen and fibrin; and (2) the light variable domain of the GPVI-bound Fab causes steric hindrance that is predicted to prevent the collagen-related peptide (CRP)/collagen fibers from extending out of their binding site and preclude GPVI clustering and downstream signaling. Glenzocimab did not bind to a truncated GPVI missing loop residues 129 to 136, thus validating the epitope identified in the crystal structure. Overall, these findings demonstrate that the binding of glenzocimab to the D2 domain of GPVI induces steric hindrance and structural modifications that drive the inhibition of GPVI interactions with its major ligands.
Introduction
The finely tuned formation of a platelet plug at sites of vascular injury ensures hemostasis by preventing excessive blood loss. By contrast, uncontrolled platelet activation causes thrombotic events and acute ischemic events such as myocardial infarction or stroke. Moreover, platelets and immune cells act jointly in injured tissues, leading to thromboinflammation that contributes to cell death and organ dysfunction.1 Antiplatelet drugs are largely used for the treatment and prevention of arterial thrombosis, including coronary artery diseases and ischemic stroke. However, despite the considerable progress made in antiplatelet therapies, standard antiplatelet drugs, aspirin and P2Y12 antagonists, have nonoptimal efficacy and there is a major drawback in that they increase the risk of bleeding, which can be fatal.2 Thus, safe and efficient antiplatelet therapy remains an important unmet medical need.
Platelet glycoprotein VI (GPVI) is of interest because of its potential as a target for the development of new antiplatelet molecules involving a lower risk of bleeding.3 GPVI expression is restricted to megakaryocytes and platelets, and it is predicted that GPVI antagonists will not have offtarget effects.4 GPVI is expressed at a moderate level of 4000 to 6000 copies per platelet, which is favorable for pharmacological saturation.
GPVI was initially recognized as a receptor for collagen. Indeed, GPVI clustering by type I or type III collagen triggers platelet degranulation, aggregation, and exposure of procoagulant phospholipids at the platelet surface.5,6 Collagen is present in large quantities in atherosclerotic plaques and several studies have demonstrated that GPVI is a crucial factor in atherothrombosis.7, 8, 9, 10 The extracellular domain of GPVI consists of 2 immunoglobulin-like domains, D1 and D2.4 The crystal structures of the GPVI exodomain, with and without triple helical glycine-proline-hydroxyproline (GPO) motif (GPO)n-containing collagen-related peptide (CRP), have previously been solved revealing a dimer interface at D2 and the site of CRP binding at D1.11,12 A major advance in our understanding of the role of GPVI was the discovery that it binds to fibrin(ogen).13, 14, 15 Interactions between GPVI and fibrin and fibrinogen endow it with an important role in the growth and stability of a thrombus.16
GPVI-deficient animals have been shown to be protected in various models of thrombosis, such as lethal thromboembolism and atherothrombosis, myocardial ischemia reperfusion injury, and cerebral ischemia.17,18 However, GPVI is not strictly required for physiological hemostasis and individuals with a GPVI deficiency exhibit only a mild bleeding diathesis or even no bleeding at all.19,20 GPVI is therefore considered to have promise as a safe antithrombotic target.3,21
Glenzocimab, formerly designated ACT017, is a humanized Fab fragment derived from the mouse monoclonal antibody 9O12.22,23 Glenzocimab recapitulates the properties of the 9O12 Fab with the advantage of being administrable to humans. It retains nanomolar affinity for GPVI and inhibits GPVI interactions with collagen and fibrin(ogen) in vitro; in vivo, it protects humanized GPVI mouse models and nonhuman primates from thrombosis.15,23, 24, 25 In a first-in-human phase 1 trial, the safety outcome was favorable, and pharmacokinetic and pharmacodynamic data demonstrated the reversible binding of glenzocimab to platelets and inhibitory effect.26,27 A phase 2 clinical trial, ACTIMIS (NCT03803007), with glenzocimab in addition to thrombolysis or thrombectomy for patients with acute ischemic stroke has been completed, confirming the safety. Furthermore, despite the study being underpowered for efficacy, it provided promising results with a lower frequency of hemorrhagic transformation, a lower percentage of patients with poor outcome, and a reduction in the number of deaths in patients treated with glenzocimab compared with placebo.28
Here, our objective was to determine the mechanism of action of glenzocimab at the molecular level. For this purpose, we have analyzed the crystallographic structure of the binary GPVI/glenzocimab immunocomplex. We identified the area of interaction between the Fab and GPVI, which corresponds to a site that does not overlap with the collagen/CRP-binding site, and we show that the Fab hinders the homotypic interactions between GPVI at the cell surface. It creates steric hindrance blocking interactions with ligands and thus prevents subsequent platelet activation.
Materials and methods
Recombinant proteins
For affinity determination, the extracellular domain of GPVI residues 1 to 249 was expressed as either a monomer with an N-terminal His6-tag (GPVI-His) or as a dimer fused with the Fc (GPVI-Fc) domain of human immunoglobulin G1 (IgG1), as previously reported.23,29,30
For crystallography, the nonglycosylated human GPVI extracellular domain residues 1 to 183 (GPVIex) was produced by heterologous expression in Escherichia coli. Briefly, the protein was extracted from inclusion bodies, purified by affinity chromatography, and refolded as previously reported11 with some modifications described in supplemental Methods.
The GPVI variant, GPVI-Fc ΔC-C′ truncated of the hinge region (G129DPAPYKN136) was produced as previously described.31
Glenzocimab is a clinical grade IgG1,k humanized Fab fragment (ACT017) derived from the monoclonal mouse antibody 9O12. Glenzocimab was produced in Chinese hamster ovary cells using the GPEx protein expression technology (Catalent, US) and was obtained after protein L capture, followed by ion exchange chromatography and polishing steps (supplemental Figure 1). The product was concentrated at 10 g.L−1 in 20 mM citric acid with 130 mM sodium chloride, pH 5.0.23
Functional assays were performed mostly as previously described and details are reported in supplemental Methods.
Collagen-induced platelet activation was assessed by P-selectin exposure at the surface of platelets by flow cytometry.
Surface plasmon resonance analysis was performed with a BIACORE T200 apparatus (Cytiva, France). Briefly, glenzocimab was immobilized covalently on a CM5S sensor chip and GPVI-His at different concentrations was injected followed by dissociation and regeneration steps. For GPVI-Fc kinetic measurements, dimeric GPVI-Fc was captured and glenzocimab at different concentrations was injected.
GPVI dimerization and oligomerization were analyzed by fluorescence correlation spectroscopy (FCS), as previously described.32 Briefly, the molecular brightness of HEK293T cells transiently expressing monomeric CD86-eGFP, dimeric CD28-eGFP, or GPVI-eGFP was compared and the effect of glenzocimab on the molecular brightness of GPVI-eGFP determined.
For fibrin-induced platelet aggregation, solubilized fibrin was obtained as previously described.33 Washed human platelets were preincubated for 20 minutes at room temperature with vehicle, 9 μM eptifibatide, or glenzocimab 50 μg.mL−1 before aggregation was initiated by the addition of the fibrin monomers solution (200 μg.mL−1).
Solid-phase binding assays were performed according to previously described protocols.23,29 Data were analyzed using data Prism 9 software.
Crystallization and structure determination
To form the GPVIex and glenzocimab complex, a 1.3-fold molar excess of glenzocimab was mixed with GPVIex and incubated at 4°C for 1 hour. The complex was purified by size exclusion chromatography using a HiLoad 26/60 Superdex column (Cytiva) equilibrated with 20 mM Tris, 150 mM sodium chloride, pH 7.4. Two peaks were observed, 1 corresponding to the excess of Fab and the other to the binary complex GPVIex-Fab (supplemental Figure 1). The complex was isolated and concentrated to 4.5 mg.mL−1 for crystallography. Crystals were generated in 20% PEG 3350 with 0.1 M Bis-Tris propane (pH 8.5) and 0.2 M potassium thiocyanate, and diffraction data were collected at the station ×06DA of the Swiss Light Source, Paul Scherrer Institute, Villigen, Switzerland, (λ = 1.0000 Å) equipped with a Pilatus 2M-F detector. The data were processed using XDS and Aimless.34,35
The structure was determined using the Phaser molecular replacement software.36 The search models used for the heavy chain, light chain, and GPVI were Protein Data Bank (PDB) entries 5AZE, 3GKW, and 5OU7, respectively. One complex was found in the asymmetric unit, with a solvent content of 53%. The structure was initially refined using Refmac5 software37 and model building was carried out in Coot.38 Thereafter, the model was subject to refinement using Buster and model building was pursued in Coot.39 The Qt-PISA and NCONT programs were used to investigate the antigen-antibody interactions.40 The atomic coordinates and structure factors (PDB ID codes 7R58) have been deposited in the PDB (www.wwpdb.org). Structural models were generated from PDB coordinates using UCSF Chimera version 1.15.41
Results
Inhibitory properties of glenzocimab
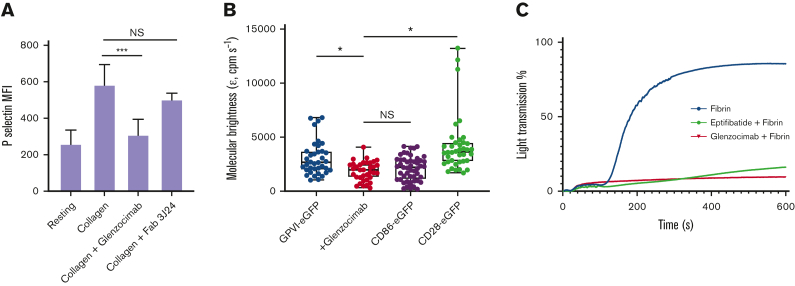
Glenzocimab reportedly inhibits GPVI binding to collagen and collagen-induced platelet aggregation.23 Here, we confirm that glenzocimab inhibits collagen-induced platelet activation as indicated by the inhibition of P-selectin exposure at the platelet surface (Figure 1A). The monoclonal Fab 3J24, which targets the D2 domain of GPVI29 had no inhibitory effect, indicating that the mechanism of inhibition by glenzocimab is specific.
Figure 1.
Inhibitory properties of glenzocimab. (A) Glenzocimab inhibits collagen-induced platelet activation. Washed human platelets were preincubated with glenzocimab or the 3J24 Fab (50 μg.mL−1) before activation was triggered by the addition of collagen (25 μg.mL−1) during 15 minutes at room temperature. Exposure of P-selectin was assessed by flow cytometry using a fluorescein isothiocyanate-coupled anti–P-selectin. Mean fluorescence intensities are shown. Data are the mean ± SD of 3 experiments made in triplicate. Statistical analysis was performed using one-way analysis of variance followed by a Tukey multiple comparisons test; ∗∗∗P < .001. (B) Glenzocimab inhibits GPVI dimerization and clustering. Box plot showing the effect of glenzocimab (50 μg.mL−1) on the molecular brightness (cpm s−1) of GPVI-eGFP alongside the molecular brightness (cpm s−1) of monomeric CD86-eGFP and dimeric CD28-eGFP control receptors in transfected HEK293T cells. For all box plots, center lines represent the median; box limits indicate the 25th and 75th percentiles and whiskers extend to minimum and maximum points. Significance was measured with Kruskal-Wallis with a Dunn post-hoc test in which P ≤ .05. FCS measurements were taken in 35 to 49 cells from 3 independent experiments. (C) Glenzocimab inhibits fibrin-induced platelet aggregation. Washed human platelets were preincubated with vehicle (red curve), 9μM eptifibatide (green curve), or glenzocimab (50 μg.mL−1) before aggregation was initiated by the addition of a solubilized fibrin (200 μg.mL−1). NS, not significant; SD, standard deviation.
GPVI is predominantly monomeric on resting platelets with a minor fraction present as a dimer.30, 31, 32 Dimers of recombinant soluble GPVI reportedly have a greater affinity for collagen than monomers.30,42 We have thus investigated whether glenzocimab could affect GPVI dimerization. We have analyzed the affinity of glenzocimab for soluble recombinant GPVI monomers and dimers by surface plasmon resonance (supplemental Figure 2). Glenzocimab bound to both monomeric and dimeric GPVI with a KD of 12.60 ± 0.04 nM and 1.310 ± 0.005 nM (mean ± standard deviation), respectively. We also examined whether glenzocimab could impact GPVI dimerization and clustering by FCS (Figure 1B). HEK293T cells expressing similar levels GPVI-eGFP, or monomeric CD86-eGFP, or dimeric CD28-eGFP (supplemental Figure 3) were compared for molecular brightness with the level of brightness governed by the degree of receptor dimerization/clustering. The molecular brightness of cells expressing GPVI-eGFP was intermediate between the brightness of cells expressing monomeric CD86-eGFP and cells expressing dimeric CD28-eGFP, confirming that GPVI is present at the cell surface as a mixture of monomers and dimers.32 In the presence of glenzocimab, the molecular brightness of GPVI-eGFP was lowered to the level of CD86-eGFP, indicating that glenzocimab inhibits GPVI dimerization and clustering.
GPVI interaction with fibrin has also been shown to promote platelet procoagulant activity and platelet spreading.13,14 Here, light transmission aggregometry showed that fibrin induces aggregation of washed platelets (Figure 1C). In these experiments, fibrin solubilized in acidic conditions repolymerizes when added to the platelet suspension. When the experiment was repeated in the presence of eptifibatide, a peptidic antagonist of αIIbβ3, no change in light transmission was observed, confirming that fibrin triggered αIIbβ3-dependent platelet aggregation. Glenzocimab completely inhibited fibrin-induced platelet aggregation.
Glenzocimab binds to the D2 domain of GPVI away from the collagen-binding site
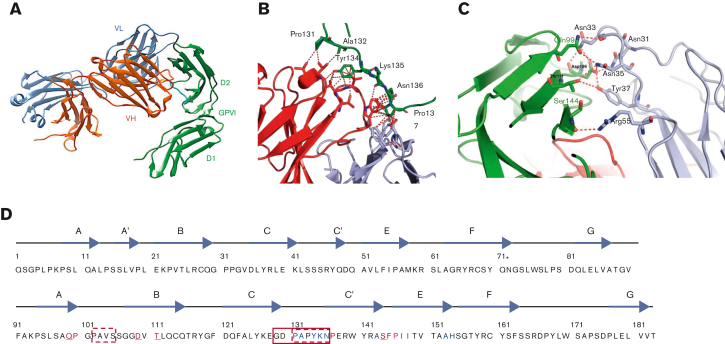
Studies were performed to solve the crystal structure of glenzocimab-bound GPVI. One complex of GPVIex-glenzocimab was found in the asymmetric unit (PDB ID: 7R58). The structure was determined to a resolution of 1.9 Å with the entire interface region between glenzocimab and GPVI well resolved. The data collected and refinement statistics are summarized in Table 1. No dimeric GPVI structures are present in the crystal (Figure 2A). The interaction area between GPVI and glenzocimab is 940 Å2 (503.6 Å2 for the heavy chain and 436.4 Å2 for the light chain). Glenzocimab bound largely to the D2 domain C-C′ loop region, with several residues in the C-C′ loop of GPVI situated within a distance of 4 Å from atoms of the Fab CDRs H1 (P131, A132, Y134) and H3 (Y134, K135, N136); the flanking amino acid P137 is in the vicinity of both CDRH3 and CDR L2. The multiple hydrophobic interactions between these residues and with both the light and heavy chain of glenzocimab are shown in Figure 2B. Epitope residues having atoms within a distance of 4 Å from atoms in glenzocimab are shown in Table 2. Ten potential hydrogen bonds were identified between GPVI and the VL domain of glenzocimab as defined by QtPISA35 (Figure 2C; Table 3). The discontinuous epitope of glenzocimab on GPVI is depicted in Figure 2D.
Table 1.
Crystallographicdata collection and refinement statistics.
| Data collection | Value |
|---|---|
| Resolution (Å) | 49.4-1.90 (1.95-1.90) |
| Wavelength (Å) | 10 000 |
| Space group | P21 21 21 |
| Unit cell a, b, c (Å) | 68.7, 93.2, 110.4 |
| Completeness (%) | 100 (99.5) |
| Redundancy | 13.2 (12.5) |
| No. of observations/unique reflections | 743 962/56 379 |
| <I/σ(I)> | 9.9 (1.6) |
| CC (1/2) (%) | 99.5 (59.8) |
| R merge (I) (%) | 25.5 (177.7) |
| Rcryst (F) (%) | 17.9 (28.6) |
| Rfree (F) (%) | 22.5 (21.4) |
| No. of nonhydrogen atoms | 5 539 |
| No. of water molecules | 779 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.09 |
| Mean B-factor, all atoms (Å2) | 24.2 |
| Mean B-factor, protein chain A, H, L (Å2) | 21.4, 21.8, 23.1 |
| Mean B-factor, solvent (Å2) | 36.2 |
| Mean B-factor, others (Å2) | 37.3 |
| Ramachandran plot quality ∗ | |
| Favored regions (%) | 98.1 |
| Allowed regions (%) | 1.9 |
| Outliers (%) | 0 |
Values in parentheses are for highest-resolution shell.
R.m.s., root mean square.
Calculated using a local Molprobity server.
Figure 2.
Delineation of the glenzocimab epitope based on GPVIex/glenzocimab crystal analysis. (A) The heavy chain (VH-CH1) of glenzocimab is depicted in red and the light chain (VL-CL) in blue. GPVI domain 2 (D2) bound to glenzocimab is represented in green as is GPVI domain 1 (D1). The D2 C-C′ loop (residues 131-137) is shown in cyan. (B) Hydrophobic contacts made between the D2 C-C′ loop (shown in green, and the heavy and light variable chains of glenzocimab in red and light blue, respectively). Hydrophobic contacts are shown as red dotted lines and represent residues that are within 4 Å or less from one another. (C) Hydrogen bonds between glenzocimab light chain CDRs (gray) and GPVI (green) according to Qt-PISA. (D) Representation of the glenzocimab epitope on the GPVIex sequence (top strand corresponds to D1 domain, bottom strand to D2 domain); the glenzocimab epitope is discontinuous. In red and in blue: residues having atoms within a distance of 4 Å from the VL and VH CDRs, respectively. In purple: residues with atoms within a distance of 4 Å from both VL and VH CDRs. Underlined are GPVI residues involved in hydrogen bonds with glenzocimab. The solid red box indicates the GPVI 129 to 136 truncation that results in the loss of glenzocimab binding and decreased binding to collagen. Red dashed boxes indicate GPVI residues truncated in the 5OU7, 5OU8, and 5OU9 crystals. Asterisk (∗) indicates N72.
Table 2.
GPVI-glenzocimabinteractions
| CDR | Glenzocimab | GPVI-CBD residues |
|---|---|---|
| CDR-H1 | Thr 28 | Pro 131, Ala 132 |
| Thr 30 | Pro 131 | |
| Ser 31 | Tyr 134 | |
| CDR-H2 | Asn 55 | Ala 153 |
| CDR-H3 | Thr 100 | Tyr 134 |
| Val 101 | Tyr 134, Ile147, His154 | |
| Val 102 | Leu 125, Tyr 134, Phe 145, Pro 146 | |
| Gly 103 | Pro 146 | |
| Asp 104 | Pro 146, Ile 148 | |
| Trp 105 | Lys 135, Asn 136, Pro 137 | |
| Tyr 106 | Ile 148 | |
| CDR-L1 | Asn 31 | Asp 109 |
| Asn 33 | Gln 99, Pro 100, Asp 109 | |
| Asn 35 | Gln 99, Asp 109, Thr 111 | |
| Tyr 37 | Asp 109, Thr 111, Pro 146, Ile 148 | |
| CDR-L2 | Tyr 54 | Pro 137 |
| Arg 55 | Ser 144, Pro 146 | |
| Phe 60 | Pro 137 | |
| Ser 61 | Pro 137 | |
| CDR-L3 | Leu 96 | Ile 148 |
Epitope residues were defined as GPVI residues having atoms within a distance of 4 Å from Fab atoms.
Table 3.
Hydrogen bondsinvolved in the GPVI-glenzocimab interface
| GPVI | Glenzocimab | Distance (Å) |
|---|---|---|
| Asp 109 (OD2) | Asn L31 (ND2) | 2.9 |
| Asp 109 (OD1) | Asn L33 (ND2) | 2.8 |
| Asp 109 (O) | Asn L33 (ND2) | 3.2 |
| Gln 99 (NE2) | Asn L33 (O) | 3.3 |
| Gln 99 (OE1) | Asn L35 (ND2) | 3.2 |
| Asp 109 (OD2) | Asn L35 (ND2) | 3.0 |
| Thr 111 (OG1) | Asn L35 (ND2) | 3.7∗ |
| Asp 109 (OD2) | Tyr L37 (OH) | 2.6 |
| Thr 111 (OG1) | Tyr L37 (OH) | 3.9∗ |
| Ser 144 (O) | Arg L55 (NH2) | 3.4∗ |
Distance was calculated using Qt-PISA.
Indicates a very long distance to being defined as a hydrogen bond.
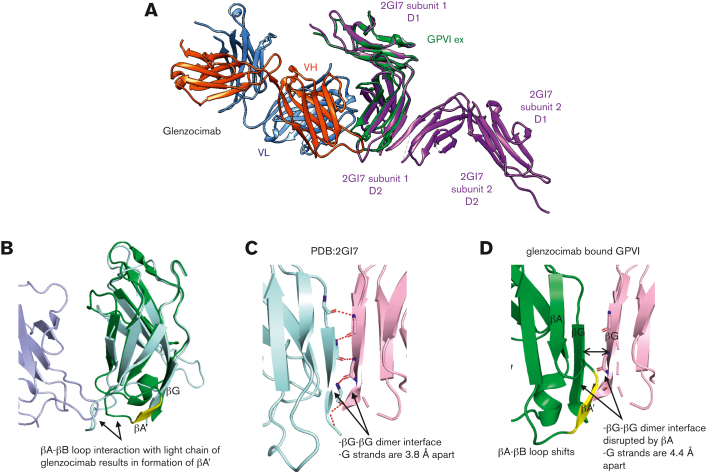
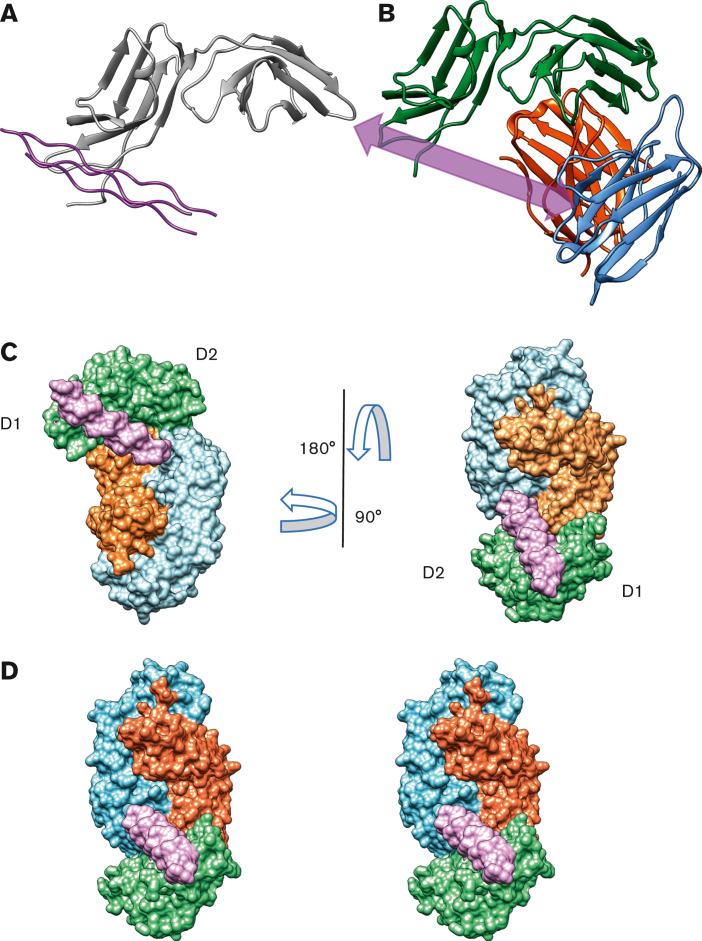
Superimposing the GPVIex-glenzocimab complex (PDB ID: 7R58) on the original crystal structure of the GPVI dimer (PDB ID: 2GI7) revealed a root mean square deviation of 1.4 Å2 for 161 aligned Cα atoms (Figure 3A). The 99 to 109 region, which forms a loop between the βA and βB strands of D2 in 2GI7 contains several residues involved in the interaction with glenzocimab CDR-L1 (N31, N33, N35, and Y37 are within hydrogen bonding distance), which results in a conformational change within this loop and the formation of a short β strand (A103VS105) named βA′ (Figure 3B) that interacts with the βG strand (residues 178-183). In 2GI7, the βG strands of the D2 domain from both subunits formed a continuous β sheet at the dimer interface (Figure 3C). In the GPVIex-glenzocimab complex, this β sheet was hampered by the newly formed βA′ strand, which makes polar contacts with the βG strand and prevents the interaction of βG with a second βG from an adjacent subunit (Figure 3D). As a consequence, no dimer formation was observed, consistent with the FCS data. As the C-C′ loop is part of the glenzocimab epitope and was shown to be involved in the formation of dimers of GPVI complexed to the Nb2 nanobody, the structures of 7R58 and 7NMU were compared (supplemental Figure 4A).31 This shows that the swapped domain structure cannot be formed in the presence of glenzocimab. The orientation of the C-C′ loop in the glenzocimab-bound structure also differs to that in the unbound nondomain swapped dimer (2GI7) (supplemental Figure 4B), highlighting that engaging glenzocimab causes a visible shift in this flexible loop resulting in minor changes within the D2 domain secondary structure.
Figure 3.
Structural comparison between GPVIex/glenzocimab and 2GI7 crystals. (A) Overview of the superimposed dimeric GPVIex (PDB ID: 2GI7) on the GPVIex/glenzocimab complex (PDB ID: 7R58). The heavy chain (VH-CH1) of glenzocimab is depicted in red and the light chain (VL-CL) in blue. GPVI domain 2 (D2) bound to glenzocimab is represented in green and GPVI without any ligand (PDB ID: 2GI7) in purple. (B) Close up view of the overlayed D2 domains of dimeric GPVI (PDB:2GI7) (pale cyan) and glenzocimab-bound GPVI (green) highlighting the rearrangements within D2 because of clashes between the βA-βB loop of GPVI and the light chain of glenzocimab (light blue). Contacts with the light chain of glenzocimab cause a conformational change βA-βB loop resulting in the formation of a new β-strand, βA′, (shown in yellow). (C) Zoomed-in view of the dimeric interface formed between 2 βG strands from 2 GPVI subunits in the dimeric crystal structure (PDB:2GI7). Subunits are colored pale cyan and light pink, respectively. Polar contacts between each strand are shown as red dashed lines with binding resides shown as sticks. (D) Zoomed-in view of the βG strand of glenzocimab-bound GPVI (green) superimposed with the alternate GPVI subunit found in the dimeric structure (light pink). The conformational changes in the glenzocimab-bound D2 domain result in the formation of a new βA′ strand, which blocks the interaction between the 2 βG strands. The formation of βA′ also results in a shift in the βG strand resulting in an increased gap of 0.6 Å between the 2 βG strands. Combined, these factors explain how GPVI dimerization is blocked by glenzocimab.
Glenzocimab creates steric hindrance preventing a CRP chain from binding to adjacent GPVIs
The GPVI-CRP crystal structures (PDB ID: 5OU8 and 5OU9) reveal that the CRP-binding motif of GPVI is present within a groove on D1 with an orientation that allows the binding of multiple GPVIs along a single triple helix.12 In the glenzocimab-GPVI cocrystal, the variable domain of the Fab light chain bound to D2 lies directly in the path of the D1-bound CRP chain, thus preventing the binding of longer CRP or collagen chains (Figure 4; supplemental Video).
Figure 4.
Glenzocimab inhibits CRP/collagen binding through steric hindrance. (A) D1 domain of GPVI (gray) in complex with CRP (magenta) (PDB ID: 5OU8). (B) D1 domain of GPVI (green) in complex with glenzocimab (PDB ID: 7R58). The VL domain of glenzocimab (light blue) at the front creates a barrier in the way of the CRP indicated by the arrow (magenta). (C) Surface representation of the glenzocimab-bound structure of GPVI (PDB ID: 7R58) with CRP binding superimposed from PDB ID: 5OU8. GPVI, CRP, and glenzocimab heavy and light chains are colored in green, magenta, orange, and blue, respectively. The binding of glenzocimab to the D2 domains means the light variable region of the Fab lies directly in the way of the bound CRP chain. Larger CRP and collagen chains would be prevented from binding owing to the obstruction by the light chain. (D) Stereo view of panel C.
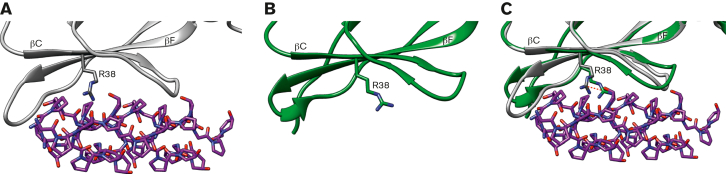
In addition, on binding to glenzocimab, the GPVIex D1 domain undergoes small structural changes. In the GPVI-CRP cocrystals, the CRP-binding groove within D1 is largely made up of βC and βF strands (Figure 5A). In the GPVI-glenzocimab complex, R38, a key CRP-binding residue, shifts 3.5 Å toward the CRP groove where it interacts with E40 and stacks against Y47 (Figure 5B-C). Combining the major steric hindrance caused by glenzocimab, the small conformational shifts, and inhibition of dimerization explains the strong inhibition caused by glenzocimab.
Figure 5.
The CRP-binding groove in the GPVIex/glenzocimab complex. (A) Zoomed-in view of the CRP-binding groove of the CRP-bound GPVI (PDB ID: 5OU8) (A) and glenzocimab-bound GPVI (PDB ID: 7R58) (B); structures shown in gray and green, respectively, with CRP shown in magenta. In (C) both structures are superimposed: the CRP-binding groove is largely made from the βC and βF strands within D1. In the glenzocimab-bound structure the R38 shifts 3.5 Å toward CRP and clashes with a CRP hydroxyproline residue. The side chain of R38 in both structures is shown as a stick, and the shift is shown using a red dotted line.
Truncating the C-C′ loop of the GPVI D2 domain alters the binding of glenzocimab
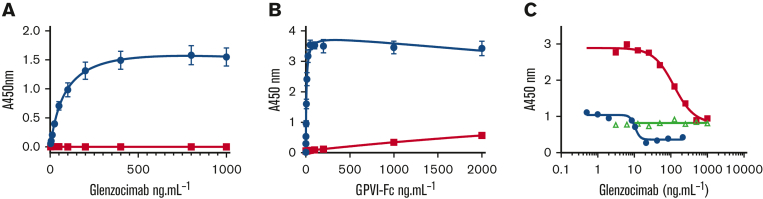
In order to validate the binding site for the antibody, we investigated whether truncating D2 C-C′ loop residues 129 to 136 affected the interaction of GPVI with glenzocimab. No binding of glenzocimab to a GPVI-Fc Δ129 to 136 coated surface was detected, even at concentrations up to 1 μg.mL−1 (20.5 nM), which exceed the 88 ng.mL−1 (1.8 nM) 50% effective concentration (EC50) observed for its binding to GPVI-Fc by an order of magnitude (Figure 6A). Similarly, the binding of GPVI-Fc Δ129 to 136 to immobilized glenzocimab was profoundly impaired, with very low levels of binding for concentrations higher than 1 μg.mL−1 (10.7 nM) whereas the EC50 observed with GPVI-Fc was 7.5 ng.mL−1 (0.05 nM) (Figure 6B). Taken together, these findings are consistent with the crystal structure of the complex and confirm the critical role of the D2 C-C′ loop in the glenzocimab epitope. The ability of glenzocimab to inhibit the binding of GPVI-Fc and GPVI-Fc Δ129 to 136 to collagen was then analyzed (Figure 6C). As previously reported, GPVI-Fc Δ129 to 136 bound to collagen but to a lesser degree than GPVI-Fc31 necessitating a higher concentration (20 μg.mL−1). No inhibition of GPVI-Fc Δ129 to 136 binding was observed in the presence of glenzocimab at levels up to 1 μg.mL−1 (20.5 nM). In contrast, glenzocimab efficiently inhibited the binding to GPVI-Fc at 2 and 20 μg.mL−1, with IC50 values of 10.52 and 121 ng.mL−1 (0.22, 2.52 nM ), respectively.
Figure 6.
Truncation of GPVI Δ129 to 136 disrupts the epitope of glenzocimab. GPVI-Fc Δ129 to 136 was compared with GPVI-Fc regarding interactions with glenzocimab in solid-phase assays. (A) Dose-dependent binding of glenzocimab to immobilized GPVI-Fc (blue circles) and GPVI-Fc Δ129 to 136 (red squares). The EC50 value of GPVI-Fc was of 85 ng.mL−1 (1.88 nM), in line with the KD value. No binding between glenzocimab and GPVI-Fc Δ129 to 136 could be observed even at 1 μg.mL−1, which is more than 10-fold the EC50 of GPVI-Fc. (B) Dose-dependent binding of GP-VI-Fc (blue circles) and GPVI-Fc Δ129 to 136 (red squares) to immobilized glenzocimab confirms defective interactions between glenzocimab and GPVI-Fc Δ129 to 136. Results are mean ± standard error of the mean of 3 experiments performed in triplicate. (C) Inhibition of GPVI-Fc binding to immobilized collagen by increasing concentrations of glenzocimab. Binding of GPVI-Fc at 2 μg.mL−1 (blue circles) or at 20 μg.mL−1 (red squares) to collagen was dose-dependently inhibited by glenzocimab with IC50 values of 10.25 and 113.7 μg.mL−1, respectively. In contrast, glenzocimab at 20 μg.mL−1 had no effect on the binding of GPVI-Fc Δ129 to 136 to collagen (green triangles).
Discussion
We have demonstrated that glenzocimab binds to an epitope located in the D2 domain of GPVI with a high affinity, and we propose a novel mechanism regarding the inhibition of GPVI interactions with its major ligands (Visual Abstract).
Consistent with previous data obtained with the Fab 9O12, we show that its humanized version (ACT017) of clinical grade (glenzocimab) inhibits platelet activation by collagen and by fibrin. Importantly, we show that glenzocimab inhibits GPVI dimerization and oligomerization.
Amino acid residues involved in the glenzocimab epitope were identified in the D2 domain of GPVI. Out of the 4 residues (Q99, D109, T111, and S144) involved in 10 hydrogen bonds at the GPVI-glenzocimab interface, all are well preserved in nonhuman primate GPVI whereas 2 are substituted for nonconservative residues in rodent GPVI (Q99>H; S144>N) leading to potential alteration of these hydrogen bonds. In addition, several residues involved in hydrophobic interactions are not conserved in rodent GPVI. Overall, these data provide direct evidence as to why glenzocimab does not cross-react with rodent GPVI.
Radiograph analysis provided a detailed view of the GPVI extracellular domain in complex with glenzocimab. The epitope is discontinuous. All of the GPVI residues involved in interactions with glenzocimab belong to the D2 domain of GPVI, as previously foreseen for the parental mouse IgG 9O12.29 Because the CRP/collagen-binding site on GPVI has been consistently identified within the D1 domain, as deduced from mutagenesis, docking, and the analysis of crystallographic structures, one can conclude that the collagen-binding site and the epitope targeted by glenzocimab do not overlap l.11,43,44 The mechanism by which glenzocimab inhibits GPVI activation results from 2 major effects: (1) glenzocimab prevents GPVI dimer formation through D2 and potentially promotes their dissociation, and (2) the light chain of glenzocimab provokes steric hindrance that prohibits the binding to adjacent GPVI-binding sites on CRP and collagen fibers. The loss of dimerization would also limit activation as it lowers the numbers of GPVI receptors in a cluster.45
Glenzocimab and the GPVI D2 domain
Several studies have established that the avidity of GPVI for collagen is increased by its dimerization. All these studies have been performed using the recombinant extracellular domain of GPVI fused to the Fc domain of human IgG.30,42 The ability of GPVI to form dimers was confirmed by different studies during which the GPVI ectodomain dimerized upon crystallization (PDB ID: 2GI7, 5NMU, 5OU7). In the first wild-type GPVI crystal described (PDB ID: 2GI7), 2 GPVI subunits formed homotypic interactions and were associated back-to-back via a β-sheet interaction between the parallel G strands (residues 178-183) of both D2 domains. The interaction between the 2 D2 domains no longer seems possible when glenzocimab binds to GPVI, and consistent with this, no GPVI dimers were observed in crystals from the GPVI-glenzocimab complex or when analyzed by FCS microscopy. This is in line with the observation that the deletion of D2 prevents the formation of dimers at the cell surface.32 Glenzocimab could even favor the dissociation of dimers by breaking the parallel β sheet made of 2 G strands.
With regard to GPVI-Fc, it has never been crystalized and the degree to which this construct mimics the structures of dimers in crystals remains unclear. In GPVI-Fc, both D2 domains are artificially maintained in close proximity via the Fc moiety and with some flexibility so that contacts between the G strands of the D2 domains are most likely not maintained. In this way, the binding of 1 Fab to GPVI-Fc should be sufficient to decrease the avidity for collagen. Furthermore, 2 Fabs could bind both D2 domains of GPVI-Fc.
The structure of the GPVIex-glenzocimab crystal indicates that the epitope includes 5 residues of the C-C′ loop (P131, A132, Y134, K135, and N136) and the flanking P137. The C-C′ loop (G129DPAPYKN136) has recently been shown to contribute to the formation of a domain swap between the D2 hinge regions of 2 GPVI subunits in the crystal of GPVI in complex with the nanobody Nb2 (PDB ID: 7NMU).31 Glenzocimab would prevent any access to, and structural rearrangement of, this loop. Slater et al truncated this region to test its functional significance. GPVI-Fc Δ129 to 136 bound with weaker affinity to collagen in DT-40 cells expressing full-length GPVI Δ129 to 136. Signaling in response to CRP or collagen was abolished.31 As expected, we observed that the deletion of residues 129 to 136 blocked the binding of glenzocimab, confirming the important contribution of the C-C′ loop to the epitope. Altogether, these observations argue for a significant contribution of the C-C′ loop in the GPVI function.
Glenzocimab and the collagen-binding domain
The GPVI-CRP complex has recently been solved, leading to a better understanding of the mechanism of GPVI activation by collagen.12 During these experiments a truncated GPVI, in which the 102 to 105 and 131 to 136 loops (ΔPAVS ΔPAPYKN) were deleted, was complexed to CRPs of different lengths (GPO5 or GPO3). Analysis of the crystals (PDB ID: 5OU8, 5OU9) indicated how the repeated GPVI-binding motifs on parallel triple helix can interact with adjacent GPVI. The most striking observation on the GPVI-glenzocimab complex is the position of the variable domain of the light chain that obstructs the passage of the triple helix. This steric hindrance prevents collagen binding to adjacent GPVI, impeding the clustering of GPVI that is critical for downstream signaling. Minor conformational changes also affected the collagen-binding groove, with the protrusion of the R38 side chain susceptible to clash with a hydroxyproline residue of CRP. This might contribute to alter the interactions between GPVI and fibrillar collagen in addition to steric clashes caused by glenzocimab VL domain.
It should also be noted that the glenzocimab epitope does not contain residues known to be involved in natural variants of human GPVI, which makes it possible to predict that the efficacy of glenzocimab will not be affected by GPVI polymorphisms.46 The GP6 sequence presents 5 nonsynonymous single nucleotide polymorphisms (rs1613662, rs1654416, rs2304167, rs1654413, and rs1671152), which are far distant from the residues involved in the glenzocimab epitope (supplemental Figure 5). This enables the prediction that glenzocimab binding to the 5 major GPVI haplotypes derived from the 1000 Genomes Project will be conserved.47 The clinical effect of glenzocimab should therefore not be dependent on the currently known GPVI polymorphisms.
Fibrin or fibrinogen have been identified as ligands for GPVI with important functional effects on thrombus growth and stability. Platelet GPVI is activated by fibrin, leading to the exposure of platelet procoagulant activity and the accretion of new platelets.13,14 GPVI has also been shown to allow the spreading of human platelets on immobilized fibrinogen and to trigger outside-in signaling after fibrinogen binding to integrin αIIbβ3.15 Recent studies are in favor of a greater avidity of dimeric GPVI for fibrin(ogen) compared with monomeric GPVI, and a mechanism involving GPVI clustering by fibrin fibers for platelet activation.48,49 The present observation that glenzocimab inhibits platelet aggregation by fibrin is in line with previously published data.14,25 Moreover, we observed that an αIIbβ3 integrin-specific inhibitor and glenzocimab block the aggregation to a similar level in agreement with the results of Perrella et al.50 This confirms that the integrin αIIbβ3 and GPVI play complementary roles in fibrin-induced platelet activation/aggregation. Both receptors are required and Syk is activated downstream of both. At the low concentration of fibrin used to prevent clotting in our experiments, its binding to integrin αIIbβ3 can trigger Syk activation but at a level insufficient to induce platelet aggregation. In turn, the interaction of fibrin with GPVI triggers a sufficient level of Syk activation for the full signaling cascade to occur but only when the integrin αIIbβ3 is functional. Together, functional and structural data indicate that glenzocimab alters the interaction between GPVI and fibrin and that the hinge region of the D2 domain may also regulate this interaction.
Conclusion
Resolving the crystal structure of the glenzocimab-GPVI complex has allowed us to identify a new mechanism by which the binding of glenzocimab to the hinge region of the D2 domain prevents interactions between GPVI and its main ligands, collagen and fibrin(ogen) and enables the inhibition of cell signaling downstream of GPVI and potentially the destabilization of platelet-platelet interactions. This original mechanism may be the basis of the powerful antithrombotic effect of glenzocimab in vitro and in vivo in animal models of thrombosis and supports its clinical development in thrombotic diseases such as acute ischemic stroke.
Conflict-of-interest disclosure: P.B. and M.J.-P. are founders and scientific advisers for Acticor-Biotech. D.F., K.L., and E.T. are employees at Acticor-Biotech. M.W. and N.R. are employees at SARomics Biostructures. The remaining authors declare no competing financial interests.
Acknowledgments
Surface plasmon resonance experiments were carried out using the facilities of the Montpellier Proteomics Platform (PPM/PP2I, BioCampus Montpellier). The authors acknowledge the Paul Scherrer Institut, Villigen, Switzerland, for provision of synchrotron radiation beamtime at beamline ×06DA and SLS and thank Tomizaki Takahashi for assistance. The authors also acknowledge the Imaging Suite at the University of Birmingham for support in imaging experiments. Imaging facilities used in this project were funded by the University of Birmingham, COMPARE, and the British Heart Foundation (IG/18/2/33544).
This research was funded, in part, by a Wellcome Trust Investigator Award (204951/B/16/Z). J.C.C. and A.S. are supported by a BHF Accelerator Grant (AA/18/2/34218). S.P.W. is a British Heart Foundation professor (CH03/003).
Authorship
Contribution: P.B. and M.J.-P. designed the study, interpreted the data, and wrote the manuscript; A.S. and S.P.W. provided GPVI-FcΔ129 to 136, interpreted the data, and edited the manuscript; M.W., J.C.C., and M.P. designed experiments, interpreted the data, and edited the manuscript; I.G.J. contributed to structural analysis; N.R., S.L., and D.F. performed experiments; E.T. edited the manuscript; and K.L. contributed to the design of the heterologous expression and purification of GPVIex.
Footnotes
Atomic coordinates and structure factors (PDB ID codes 7R58) have been deposited in the Protein Data Bank (www.wwpdb.org).
Data are available on request from the corresponding authors, Martine Jandrot-Perrus (martine.jandrot-perrus@inserm.fr) and Philippe Billiald (philippe.billiald@universite-paris-saclay.fr).
The full-text version of this article contains a data supplement.
Contributor Information
Philippe Billiald, Email: philippe.billiald@universite-paris-saclay.fr.
Martine Jandrot-Perrus, Email: martine.jandrot-perrus@inserm.fr.
Supplementary Material
References
- 1.Lebas H, Yahiaoui K, Martos R, Boulaftali Y. Platelets are at the nexus of vascular diseases. Front Cardiovasc Med. 2019;6:132. doi: 10.3389/fcvm.2019.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McFadyen JD, Schaff M, Peter K. Current and future antiplatelet therapies: emphasis on preserving haemostasis. Nat Rev Cardiol. 2018;15(3):181–191. doi: 10.1038/nrcardio.2017.206. [DOI] [PubMed] [Google Scholar]
- 3.Borst O, Gawaz M. Glycoprotein VI - novel target in antiplatelet medication. Pharmacol Ther. 2021;217:107630. doi: 10.1016/j.pharmthera.2020.107630. [DOI] [PubMed] [Google Scholar]
- 4.Jandrot-Perrus M, Busfield S, Lagrue AH, et al. Cloning, characterization, and functional studies of human and mouse glycoprotein VI: a platelet-specific collagen receptor from the immunoglobulin superfamily. Blood. 2000;96(5):1798–1807. [PubMed] [Google Scholar]
- 5.Nieswandt B, Watson SP. Platelet-collagen interaction: is GPVI the central receptor? Blood. 2003;102(2):449–461. doi: 10.1182/blood-2002-12-3882. [DOI] [PubMed] [Google Scholar]
- 6.Bergmeier W, Stefanini L. Platelet ITAM signaling. Curr Opin Hematol. 2013;20(5):445–450. doi: 10.1097/MOH.0b013e3283642267. [DOI] [PubMed] [Google Scholar]
- 7.Cosemans JM, Kuijpers MJ, Lecut C, et al. Contribution of platelet glycoprotein VI to the thrombogenic effect of collagens in fibrous atherosclerotic lesions. Atherosclerosis. 2005;181(1):19–27. doi: 10.1016/j.atherosclerosis.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 8.Penz S, Reininger AJ, Brandl R, et al. Human atheromatous plaques stimulate thrombus formation by activating platelet glycoprotein VI. FASEB J. 2005;19(8):898–909. doi: 10.1096/fj.04-2748com. [DOI] [PubMed] [Google Scholar]
- 9.Kuijpers MJ, Gilio K, Reitsma S, et al. Complementary roles of platelets and coagulation in thrombus formation on plaques acutely ruptured by targeted ultrasound treatment: a novel intravital model. J Thromb Haemost. 2009;7(1):152–161. doi: 10.1111/j.1538-7836.2008.03186.x. [DOI] [PubMed] [Google Scholar]
- 10.Hechler B, Gachet C. Comparison of two murine models of thrombosis induced by atherosclerotic plaque injury. Thromb Haemost. 2011;105(Suppl 1):S3–12. doi: 10.1160/THS10-11-0730. [DOI] [PubMed] [Google Scholar]
- 11.Horii K, Kahn ML, Herr AB. Structural basis for platelet collagen responses by the immune-type receptor glycoprotein VI. Blood. 2006;108(3):936–942. doi: 10.1182/blood-2006-01-010215. [DOI] [PubMed] [Google Scholar]
- 12.Feitsma LJ, Brondijk HC, Jarvis GE, et al. Structural insights into collagen binding by platelet receptor glycoprotein VI. Blood. 2022;139(20):3087–3098. doi: 10.1182/blood.2021013614. [DOI] [PubMed] [Google Scholar]
- 13.Alshehri OM, Hughes CE, Montague S, et al. Fibrin activates GPVI in human and mouse platelets. Blood. 2015;126(13):1601–1608. doi: 10.1182/blood-2015-04-641654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mammadova-Bach E, Ollivier V, Loyau S, et al. Platelet glycoprotein VI binds to polymerized fibrin and promotes thrombin generation. Blood. 2015;126(5):683–691. doi: 10.1182/blood-2015-02-629717. [DOI] [PubMed] [Google Scholar]
- 15.Mangin PH, Onselaer MB, Receveur N, et al. Immobilized fibrinogen activates human platelets through glycoprotein VI. Haematologica. 2018;103(5):898–907. doi: 10.3324/haematol.2017.182972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmed MU, Kaneva V, Loyau S, et al. Pharmacological blockade of glycoprotein VI promotes thrombus disaggregation in the absence of thrombin. Arterioscler Thromb Vasc Biol. 2020;40(9):2127–2142. doi: 10.1161/ATVBAHA.120.314301. [DOI] [PubMed] [Google Scholar]
- 17.Nieswandt B, Schulte V, Bergmeier W, et al. Long-term antithrombotic protection by in vivo depletion of platelet glycoprotein VI in mice. J Exp Med. 2001;193(4):459–469. doi: 10.1084/jem.193.4.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massberg S, Gawaz M, Gruner S, et al. A crucial role of glycoprotein VI for platelet recruitment to the injured arterial wall in vivo. J Exp Med. 2003;197(1):41–49. doi: 10.1084/jem.20020945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jandrot-Perrus M, Hermans C, Mezzano D. Platelet glycoprotein VI genetic quantitative and qualitative defects. Platelets. 2019;30(6):708–713. doi: 10.1080/09537104.2019.1610166. [DOI] [PubMed] [Google Scholar]
- 20.Nurden AT. Clinical significance of altered collagen-receptor functioning in platelets with emphasis on glycoprotein VI. Blood Rev. 2019;38:100592. doi: 10.1016/j.blre.2019.100592. [DOI] [PubMed] [Google Scholar]
- 21.Zahid M, Mangin P, Loyau S, et al. The future of glycoprotein VI as an antithrombotic target. J Thromb Haemost. 2012;10(12):2418–2427. doi: 10.1111/jth.12009. [DOI] [PubMed] [Google Scholar]
- 22.Mangin PH, Tang C, Bourdon C, et al. A humanized glycoprotein VI (GPVI) mouse model to assess the antithrombotic efficacies of anti-GPVI agents. J Pharmacol Exp Ther. 2012;341(1):156–163. doi: 10.1124/jpet.111.189050. [DOI] [PubMed] [Google Scholar]
- 23.Lebozec K, Jandrot-Perrus M, Avenard G, Favre-Bulle O, Billiald P. Design, development and characterization of ACT017, a humanized Fab that blocks platelet's glycoprotein VI function without causing bleeding risks. MAbs. 2017;9(6):945–958. doi: 10.1080/19420862.2017.1336592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lebozec K, Jandrot-Perrus M, Avenard G, Favre-Bulle O, Billiald P. Quality and cost assessment of a recombinant antibody fragment produced from mammalian, yeast and prokaryotic host cells: a case study prior to pharmaceutical development. N Biotechnol. 2018;44:31–40. doi: 10.1016/j.nbt.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 25.Lehmann M, Schoeman RM, Krohl PJ, et al. Platelets drive thrombus propagation in a hematocrit and glycoprotein VI-dependent manner in an in vitro venous thrombosis model. Arterioscler Thromb Vasc Biol. 2018;38(5):1052–1062. doi: 10.1161/ATVBAHA.118.310731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voors-Pette C, Lebozec K, Dogterom P, et al. Safety and tolerability, pharmacokinetics, and pharmacodynamics of ACT017, an antiplatelet GPVI (glycoprotein VI) Fab. Arterioscler Thromb Vasc Biol. 2019;39(5):956–964. doi: 10.1161/ATVBAHA.118.312314. [DOI] [PubMed] [Google Scholar]
- 27.Renaud L, Lebozec K, Voors-Pette C, et al. Population pharmacokinetic/pharmacodynamic modeling of glenzocimab (ACT017) a glycoprotein VI inhibitor of collagen-induced platelet aggregation. J Clin Pharmacol. 2020;60(9):1198–1208. doi: 10.1002/jcph.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazighi M, Richard S, Molina C, et al. Glenzocimab, an novel antithrombotic, is associated with reduced intracrnial hemorrhage and mortality rates when combined with standard-of-care reperfusion therapies: the ACTIMIS study. Eur Stroke J. 2022;7(suppl 1):574–1758. [Google Scholar]
- 29.Dumont B, Minullina I, Loyau S, et al. Chimeric Fc receptors identify ligand binding regions in human glycoprotein VI. J Mol Biol. 2006;361(5):877–887. doi: 10.1016/j.jmb.2006.06.053. [DOI] [PubMed] [Google Scholar]
- 30.Loyau S, Dumont B, Ollivier V, et al. Platelet glycoprotein VI dimerization, an active process inducing receptor competence, is an indicator of platelet reactivity. Arterioscler Thromb Vasc Biol. 2012;32(3):778–785. doi: 10.1161/ATVBAHA.111.241067. [DOI] [PubMed] [Google Scholar]
- 31.Slater A, Di Y, Clark JC, et al. Structural characterization of a novel GPVI-nanobody complex reveals a biologically active domain-swapped GPVI dimer. Blood. 2021;137(24):3443–3453. doi: 10.1182/blood.2020009440. [DOI] [PubMed] [Google Scholar]
- 32.Clark JC, Neagoe RAI, Zuidscherwoude M, et al. Evidence that GPVI is expressed as a mixture of monomers and dimers, and that the D2 domain is not essential for GPVI activation. Thromb Haemost. 2021;121(11):1435–1447. doi: 10.1055/a-1401-5014. [DOI] [PubMed] [Google Scholar]
- 33.Jandrot-Perrus M, Aurousseau MH, Rabiet MJ, Josso F. Fibrinogen Bondy: a new case of dysfibrinogenemia. Isolation of the abnormal fibrinogen molecules. Thromb Res. 1982;27(6):659–670. doi: 10.1016/0049-3848(82)90004-4. [DOI] [PubMed] [Google Scholar]
- 34.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69(pt 7):1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40(pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murshudov GN, Skubak P, Lebedev AA, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bricogne G, Blanc E, Brandl M, et al. Global Phasing Ltd; 2011. BUSTER version 2.11.7. [Google Scholar]
- 40.Winn MD, Ballard CC, Cowtan KD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 42.Miura Y, Takahashi T, Jung SM, Moroi M. Analysis of the interaction of platelet collagen receptor glycoprotein VI (GPVI) with collagen. A dimeric form of GPVI, but not the monomeric form, shows affinity to fibrous collagen. J Biol Chem. 2002;277(48):46197–46204. doi: 10.1074/jbc.M204029200. [DOI] [PubMed] [Google Scholar]
- 43.Smethurst PA, Joutsi-Korhonen L, O'Connor MN, et al. Identification of the primary collagen-binding surface on human glycoprotein VI by site-directed mutagenesis and by a blocking phage antibody. Blood. 2004;103(3):903–911. doi: 10.1182/blood-2003-01-0308. [DOI] [PubMed] [Google Scholar]
- 44.O'Connor MN, Smethurst PA, Farndale RW, Ouwehand WH. Gain- and loss-of-function mutants confirm the importance of apical residues to the primary interaction of human glycoprotein VI with collagen. J Thromb Haemost. 2006;4(4):869–873. doi: 10.1111/j.1538-7836.2005.01764.x. [DOI] [PubMed] [Google Scholar]
- 45.Slater A, Jandrot-Perrus M. GPVI and collagen: the final word? Blood. 2022;139(20):3005–3007. doi: 10.1182/blood.2022015962. [DOI] [PubMed] [Google Scholar]
- 46.Watkins NA, O'Connor MN, Rankin A, et al. Definition of novel GP6 polymorphisms and major difference in haplotype frequencies between populations by a combination of in-depth exon resequencing and genotyping with tag single nucleotide polymorphisms. J Thromb Haemost. 2006;4(6):1197–1205. doi: 10.1111/j.1538-7836.2006.01937.x. [DOI] [PubMed] [Google Scholar]
- 47.Genomes Project C, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moroi M, Induruwa I, Farndale RW, Jung SM. Dimers of the platelet collagen receptor glycoprotein VI bind specifically to fibrin fibers during clot formation, but not to intact fibrinogen. J Thromb Haemost. 2021;19(8):2056–2067. doi: 10.1111/jth.15399. [DOI] [PubMed] [Google Scholar]
- 49.Xu RG, Gauer JS, Baker SR, et al. GPVI (glycoprotein VI) interaction with fibrinogen is mediated by avidity and the fibrinogen alphaC-region. Arterioscler Thromb Vasc Biol. 2021;41(3):1092–1104. doi: 10.1161/ATVBAHA.120.315030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perrella G, Huang J, Provenzale I, et al. Nonredundant roles of platelet glycoprotein VI and Integrin alphaIIbbeta3 in fibrin-mediated microthrombus formation. Arterioscler Thromb Vasc Biol. 2021;41(2):e97–e111. doi: 10.1161/ATVBAHA.120.314641. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.