Key Points
Question
Are longitudinal changes in circulating tumor DNA (ctDNA) methylation effective in monitoring disease progression from molecular residual disease to recurrence?
Findings
In this cohort study of 299 patients with colorectal cancer, circulating tumor DNA status was evaluated with 6 DNA methylation markers. The presence of ctDNA was strongly associated with recurrence before and after surgery, after adjuvant chemotherapy, and during longitudinal monitoring.
Meaning
Findings suggest that the effectiveness of ctDNA methylation has potential in postoperative management; this simplified detection assay offers a pragmatic tool to aid residual disease detection, risk stratification, chemotherapy guidance, and relapse surveillance in clinical settings.
Abstract
Importance
Detection of molecular residual disease and risk stratification as early as possible may improve the treatment of patients with cancer. Efficient pragmatic tests are therefore required.
Objective
To measure circulating tumor DNA (ctDNA) with 6 DNA methylation markers in blood samples and to evaluate the association of the presence of ctDNA with colorectal cancer (CRC) recurrence throughout the disease course.
Design, Setting, and Participants
In this multicenter prospective longitudinal cohort study performed from December 12, 2019, to February 28, 2022, 350 patients with stage I to III CRC were recruited from 2 hospitals for collection of blood samples before and after surgery, during and after adjuvant chemotherapy, and every 3 months for up to 2 years. A multiplex, ctDNA methylation, quantitative polymerase chain reaction assay was used to detect ctDNA in plasma samples.
Results
A total of 299 patients with stage I to III CRC were evaluated. Of 296 patients with preoperative samples, 232 (78.4%) tested positive for any of the 6 ctDNA methylation markers. A total of 186 patients (62.2%) were male, and the mean (SD) age was 60.1 (10.3) years. At postoperative month 1, ctDNA-positive patients were 17.5 times more likely to relapse than were ctDNA-negative patients (hazard ratio [HR], 17.5; 95% CI, 8.9-34.4; P < .001). The integration of ctDNA and carcinoembryonic antigen tests showed risk stratification for recurrence with an HR of 19.0 (95% CI, 8.9-40.7; P < .001). Furthermore, ctDNA status at postoperative month 1 was strongly associated with prognosis in patients treated with adjuvant chemotherapy of different durations and intensities. After adjuvant chemotherapy, ctDNA-positive patients had a significantly shorter recurrence-free survival than did the ctDNA-negative patients (HR, 13.8; 95% CI, 5.9-32.1; P < .001). Longitudinal ctDNA analysis after the postdefinitive treatment showed a discriminating effect in that ctDNA-positive patients had poorer recurrence-free survival than did the ctDNA-negative patients (HR, 20.6; 95% CI, 9.5-44.9; P < .001). The discriminating effect was enhanced (HR, 68.8; 95% CI, 18.4-257.7; P < .001) when ctDNA status was maintained longitudinally. Postdefinitive treatment analysis detected CRC recurrence earlier than radiologically confirmed recurrence, with a median lead time of 3.3 months (IQR, 0.5-6.5 months).
Conclusions and Relevance
The findings of this cohort study suggest that longitudinal assessment of ctDNA methylation may enable the early detection of recurrence, potentially optimizing risk stratification and postoperative treatment of patients with CRC.
This cohort study investigates the effectiveness of circulating tumor DNA methylation in early detection of colorectal cancer recurrence.
Introduction
Colorectal cancer (CRC) is the second most common cause of cancer-related death worldwide.1 The 5-year survival rate remains as low as 60%,2,3 partially because of a relatively high recurrence rate after curative-intent therapies. Adjuvant chemotherapy after resection is routinely administered to patients with stage III or high-risk stage II disease. There is consensus that patients with stage III CRC benefit from adjuvant chemotherapy with sufficient evidence, but the advantage of adjuvant chemotherapy for patients with stage II CRC remains controversial.4,5,6,7,8,9 It is therefore crucial to stratify patients at high risk as early as possible to enhance decision-making for appropriate therapies and monitoring.
The standard of care for surveillance includes carcinoembryonic antigen blood tests, colonoscopy, and radiologic imaging.10,11,12 Owing to the rapid development in techniques, circulating tumor DNA (ctDNA) detected in patient blood samples is considered an effective indicator of disease progression from tumorigenesis to recurrence.13,14,15,16 Studies on postoperative ctDNA have been accumulating to explore its clinical utility in the detection of molecular residual disease, assessment of the effectiveness of adjuvant chemotherapy, and detection of recurrence.17,18,19,20,21 However, most of the research has focused on profiling genomic variations with ultradeep next-generation sequencing, which challenges implementation because of complex processes, long testing turnaround time, and high cost.21,22
Circulating tumor DNA methylation has been proposed as an alternative biomarker of recurrence in serial studies.23,24,25,26,27 A pilot study by our group demonstrated the feasibility of using 6 ctDNA methylation biomarkers for detecting early relapse in patients with CRC.28 Given the proposed use of ctDNA methylation in patient care, it is necessary to provide sufficient evidence to accelerate the integration of ctDNA into standard of care for patient surveillance.
We report the results of a prospective multicenter study of patients with stage I to III CRC. By using a simplified multiplex quantitative polymerase chain reaction method, we analyzed ctDNA methylation changes in patient plasma samples obtained preoperatively, postoperatively, during and after adjuvant chemotherapy treatment, and throughout the surveillance (every 3 months for up to 2 years).
Methods
Study Design
Patients with stage I to III CRC from December 12, 2019, to February 28, 2022, with R0 section and no evidence of any molecular residual disease on standard imaging were enrolled in this cohort study. Self-reported data on racial and ethnicity categories were collected; all patients were Asian. Blood samples were obtained preoperatively (approximately 7 days before surgery), approximately 1 month after surgery, and at follow-up time points (3-month intervals). Patients without any blood samples after surgery or nonrelapsed patients with less than 7 months of follow-up were excluded. Adjuvant chemotherapy was chosen by physicians and patients who were blinded to ctDNA results. Recurrence-free survival was used as the primary outcome, assessed with standard radiologic criteria, and calculated from the date of surgery to the date of verified radiologic recurrence. Details of the method are available in eAppendices 1 to 3 in Supplement 1. This study was approved by the ethics committee of Fudan University Shanghai Cancer Center and Shanghai Jiao Tong University Renji Hospital (NCT03737539). All participants provided written informed consent. The study followed the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guideline.
Measurement of ctDNA Methylation
Circulating tumor DNA methylation was measured with the ColonAiQ assay as previously described.28 The status of each marker was determined with its cutoff value. The ctDNA-positive sample was defined as having at least 1 positive marker; otherwise, it was defined as ctDNA negative. Details of the method are available in eAppendix 4 in Supplement 1.
Statistical Analysis
All data were analyzed and presented with descriptive statistics as the mean, range, and proportions. Data were analyzed from March 1 to June 30, 2022. Survival analyses were performed with the Kaplan-Meier method (log rank), and univariate and multivariable analyses were performed with Cox proportional hazards regression analysis. Statistical analyses were performed with R, version 4.1.3 (R Foundation for Statistical Computing). P values were based on 2-sided testing, and differences were considered statistically significant at P < .05. Details of the method are available in eAppendix 5 in Supplement 1.
Results
Patient Enrollment and Baseline ctDNA
A prospective longitudinal study was designed to trace the dynamic changes in ctDNA methylation in the blood samples of patients with CRC during treatment at 2 hospitals (Figure 1). From an initial cohort of 350 patients, a total of 299 eligible patients with stage I to III CRC were recruited, and 51 patients were excluded owing to insufficient follow-up or loss to follow-up (median, 21 months; range, 8-27 months). A total of 186 patients (62.2%) were male, and 113 (37.8%) were female. The mean (SD) age was 60.1 (10.3) years. During the study period, recurrence was confirmed by radiologic examination for 55 of 299 patients (18.4%) (eTable 1 and eTable 2 in Supplement 1). Altogether, 1228 blood samples were obtained from 299 patients multiple times (before and after surgery, during and after adjuvant chemotherapy, and every 3 months for up to 2 years), and ctDNA was measured with a multilocus, ctDNA methylation, quantitative polymerase chain reaction assay (eAppendix 4 and eTable 3 in Supplement 1).
Figure 1. Patient Enrollment and Definition of the Patient Subgroups.
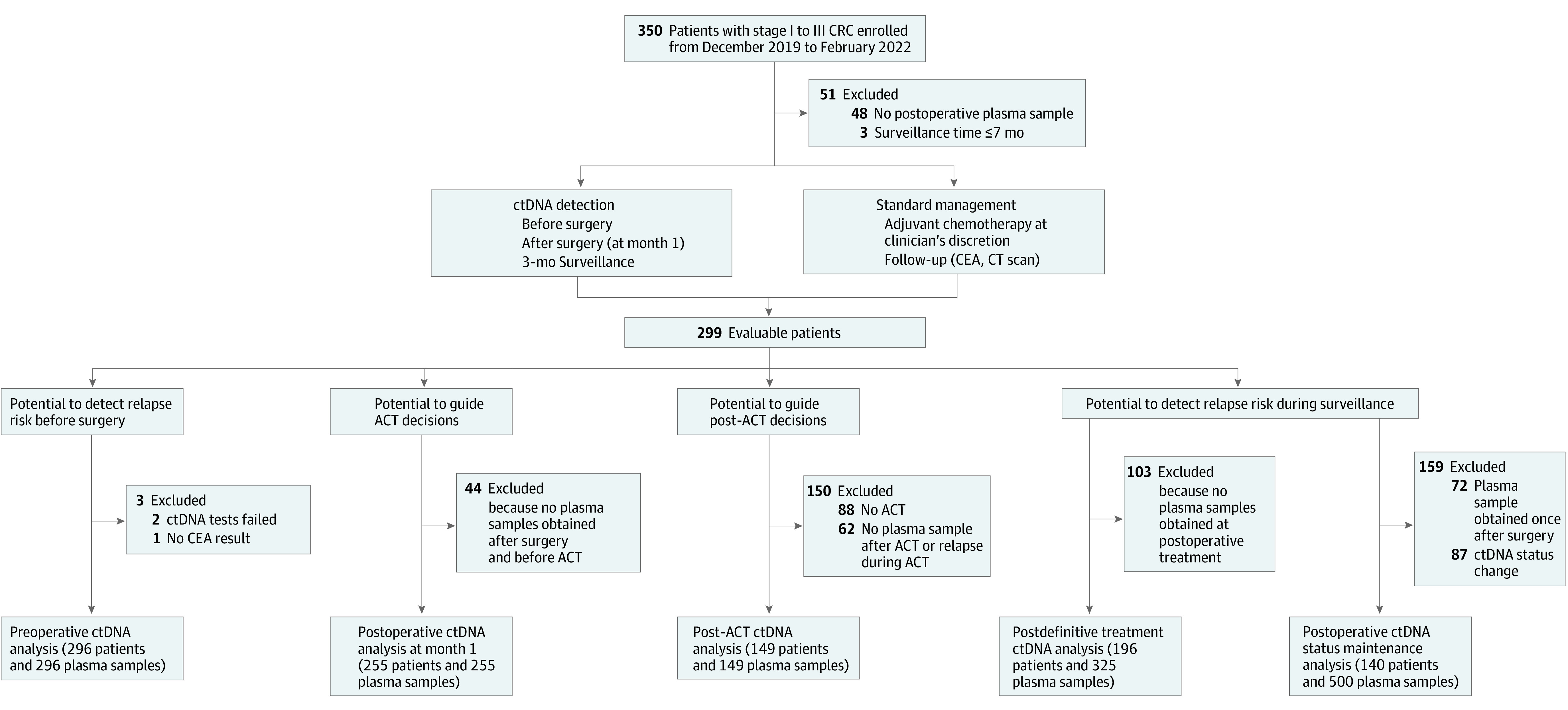
ACT indicates adjuvant chemotherapy; CEA, carcinoembryonic antigen; CRC, colorectal cancer; CT, computed tomography; and ctDNA, circulating tumor DNA.
Of 296 patients with preoperative samples, 232 (78.4%) tested positive for any of the 6 ctDNA methylation markers. As a baseline, blood ctDNA status was evaluated for the enrolled patients before surgery. The detection rates of CRC stages I, II, and III were 65.1% (41 of 63 patients), 82.7% (81 of 98 patients), and 81.5% (110 of 135 patients), respectively, which significantly outperformed carcinoembryonic antigen (eFigure 1 and eTable 4 in Supplement 1). A total of 51 of 54 patients with recurrence (94.4%) were ctDNA positive preoperatively, whereas 61 of 242 patients with nonrecurrence (25.2%) were ctDNA negative (eFigure 2 in Supplement 1). Of the 64 patients who were ctDNA negative preoperatively, only 3 (4.7%) relapsed and 61 (95.3%) remained relapse free.
Detection of Recurrence by ctDNA at Postoperative Month 1
To evaluate the residual molecular tumor after surgery and stratify patients’ risks of recurrence, we assessed the presence of ctDNA in blood samples obtained approximately 1 month after surgery (median, 3.2 weeks; range, 2.4-12.1 weeks). Of 255 patients with available plasma samples, 196 (76.9%) were ctDNA negative, and 59 (23.1%) were ctDNA positive (eTable 5 in Supplement 1). The recurrence risk of ctDNA-positive patients was significantly higher than that of ctDNA-negative patients (hazard ratio [HR], 17.5; 95% CI, 8.9-34.4; P < .001) (Figure 2A). At the end of follow-up, 185 of the 196 ctDNA-negative patients (94.4%) were recurrence free, whereas 39 of the 59 ctDNA-positive patients (66.1%) experienced recurrence. As early as postoperative month 1, the sensitivity to detect relapse was 78.0%, with 90.2% specificity, which was consistent across different stages (eFigure 3 in Supplement 1). Multivariable Cox regression analysis of clinicopathologic risk factors indicated that both ctDNA (HR, 17.0; 95% CI, 8.2-34.0; P < .001) and carcinoembryonic antigen (HR, 5.5; 95% CI, 2.3-13.0; P < .001) were significant prognostic factors associated with recurrence-free survival (Table and eTable 6 in Supplement 1).
Figure 2. Circulating Tumor DNA (ctDNA) Analysis of Plasma at Postoperative Month 1 for Detecting Colorectal Cancer Recurrence.
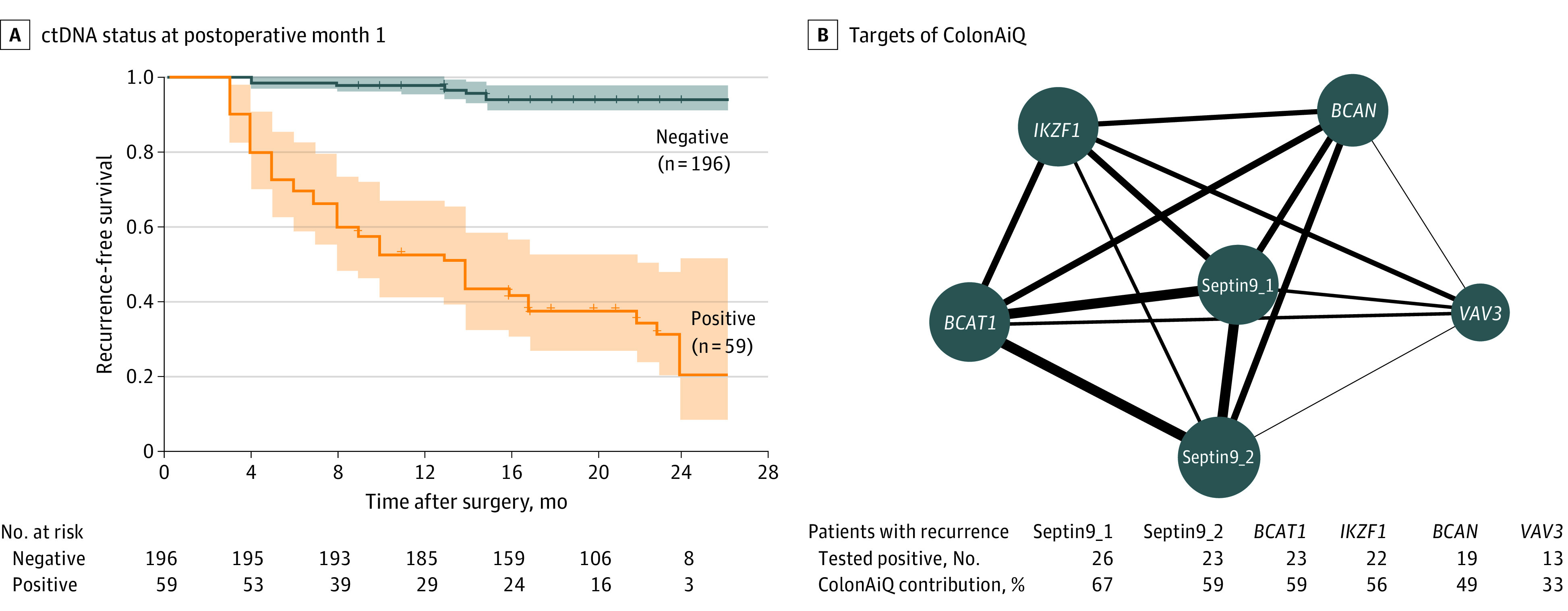
A, Kaplan-Meier estimates of recurrence-free survival, stratified by ctDNA status at postoperative month 1. The recurrence risk for ctDNA-positive patients was significantly higher than that of ctDNA-negative patients (hazard ratio, 17.5; 95% CI, 8.9-34.4; P < .001). P value was determined by the log-rank test. Shaded areas indicate 95% CIs. B, Network map of 6 targets of the ColonAiQ assay28 in patients with recurrence. The size of the nodes is proportional to the number of patients who tested positive, and the thickness of lines is proportional to the number of patients with recurrence who tested positive for 2 targets. Positive rates of 6 targets are shown. BCAN indicates brevican; BCAT1, branched-chain amino acid transaminase 1; IKZF1, ikaros family zinc finger 1; Septin9_1, septin 9 locus 1; Septin9_2, septin 9 locus 2; and VAV3, vav guanine nucleotide exchange factor 3.
Table. Univariate and Multivariable Cox Analysis of Recurrence-Free Survival by Clinicopathologic Variables and ctDNA Status and CEA Status at POM1.
| Variable | Univariate analysis | Multivariable analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Age (older than or equal to mean vs younger than mean) | 1.0 (0.6-1.7) | .96 | NA | NA |
| Sex (male vs female) | 1.0 (0.6-1.8) | .98 | NA | NA |
| Primary tumor site (left vs right) | 1.2 (0.6-2.3) | .64 | NA | NA |
| Histologic type (mucinous adenocarcinoma or SRCC vs adenocarcinoma) | 2.1 (1.1-4.1) | .02 | 1.6 (0.7-3.6) | .29 |
| Tumor differentiation (well moderated vs poor) | 0.5 (0.3-0.9) | .03 | 0.9 (0.4-1.7) | .71 |
| T stage (T4 vs T1, T2, or T3) | 3.1 (1.7-5.6) | <.001 | 1.3 (0.7-2.7) | .43 |
| Nodal involvement (N1-N2 vs N0) | 3.6 (1.9-7.0) | <.001 | 1.4 (0.6-3.1) | .43 |
| Invasion (yes vs no) | ||||
| Vascular | 2.7 (1.5-4.9) | .001 | 1.2 (0.6-2.6) | .60 |
| Perineural | 2.6 (1.5-4.6) | <.001 | 1.2 (0.6-2.3) | .62 |
| MMR status (proficient vs efficient) | 1.3 (0.3-5.4) | .71 | NA | NA |
| At POM1 (positive vs negative) | ||||
| CEA | 7.5 (3.9-14.0) | <.001 | 5.5 (2.3-13.0) | <.001 |
| ctDNA | 17.0 (8.7-34.0) | <.001 | 17.0 (8.2-34.0) | <.001 |
Abbreviations: CEA, carcinoembryonic antigen; ctDNA, circulating tumor DNA; HR, hazard ratio; MMR, mismatch repair; NA, not applicable; POM1, postoperative month 1; SRCC, signet-ring cell carcinoma.
To determine the association of methylation markers with recurrence, we individually investigated the positive rate of 6 markers, including septin 9, a well-studied ctDNA marker for CRC recurrence.29,30,31 The detection sensitivity of individual markers for recurrence varied from 26% to 52%, corresponding to a specificity of 95% to 98% (eTable 7 in Supplement 1). The contributions of the 6 loci were ranked as 67%, 59%, 59%, 56%, 49%, and 33%, respectively, indicating that these markers in combination capture a fuller spectrum of relapse than individual markers alone (Figure 2B).
Improved Recurrence Detection by Combination of ctDNA and Carcinoembryonic Antigen at Postoperative Month 1
Carcinoembryonic antigen routinely used in clinics32,33,34,35 was limited in assessing prognosis owing to the low detection rate preoperatively (101 of 296 patients with CRC [34.1%]); therefore, 195 patients (65.9%) could not be monitored postoperatively (eFigure 4 in Supplement 1). For patients who were carcinoembryonic antigen negative preoperatively, ctDNA positivity at postoperative month 1 was strongly associated with recurrence (HR, 35.2; 95% CI, 10.3-120.5; P < .001), suggesting an alternative aid for surveillance. Similarly, ctDNA at postoperative month 1 was also associated with recurrence for patients who were carcinoembryonic antigen positive preoperatively (HR, 9.1; 95% CI, 4.0-21.0; P < .001) (eFigure 5A and B in Supplement 1). We further combined the status of ctDNA and carcinoembryonic antigen at postoperative month 1 to assess their potential to improve detection of relapse (eAppendix 6 in Supplement 1). A patient with a positive test result in any of the 2 tests (ctDNA and carcinoembryonic antigen tests) was defined as positive. The integration of ctDNA and carcinoembryonic antigen reached 0.849 in the area under the curve compared with 0.839 and 0.623 for ctDNA and carcinoembryonic antigen, respectively (eFigure 5C in Supplement 1). In the combined strategy, the patients with positive test results had inferior prognoses compared with those who had negative results (HR, 19.0; 95% CI, 8.9-40.7; P < .001) (eFigure 5D in Supplement 1). Overall, the combined test detected recurrence with 83.3% sensitivity and 86.5% specificity (eTable 8 in Supplement 1).
Risk Stratification for Adjuvant Chemotherapy by Postoperative Month 1 ctDNA
To explore whether postoperative month 1 ctDNA can potentially guide the choice of adjuvant treatment, we performed univariate analysis for 100 patients with stage III CRC, and the subgroups were classified according to clinical risk evaluation for recurrence (high or low risk) and adjuvant treatment duration (3 to 6 months). Patients with stage III CRC were classified into clinically low-risk (T1-3N1) and high-risk (T4 or N2) groups with different probabilities of recurrence.7,36 Patients with stage III ctDNA-negative status at postoperative month 1 had significantly better outcomes regardless of the clinical high- or low-risk classification (eFigure 6A in Supplement 1). Similarly, ctDNA-negative patients had longer recurrence-free survival independent of adjuvant chemotherapy duration (eFigure 6B in Supplement 1). For patients with high-risk stage III CRC who were ctDNA positive at postoperative month 1, although the patient number was limited, we observed that 6-month adjuvant chemotherapy treatment might lead to a lower recurrence rate (eFigure 6C in Supplement 1), whereas 6-month adjuvant chemotherapy might not be superior to 3-month treatment in the low-risk subgroup (eFigure 6D in Supplement 1). Postoperative month 1 ctDNA provided prognostic indications for most categories of patients (eTable 9 and eTable 10 in Supplement 1). Four of the 5 ctDNA-positive patients relapsed, and all 5 ctDNA-negative patients were free of relapse in the subgroup of patients at high risk who were treated with 3 months of adjuvant chemotherapy. Similar results were observed for 62 patients with stage II disease treated with adjuvant chemotherapy (eFigure 7 in Supplement 1). Furthermore, ctDNA status at postoperative month 1 was strongly associated with prognosis in patients treated with adjuvant chemotherapy of different durations and intensities. For patients with stage I and low-risk stage II CRC with no adjuvant chemotherapy, all ctDNA-negative patients remained recurrence free, and 4 of 6 ctDNA-positive patients relapsed within 2 years (eFigure 8 in Supplement 1). Overall, ctDNA status at postoperative month 1 performed better than the current classifications in discriminating prognosis, potentially offering the best time point to guide and monitor adjuvant chemotherapy.
Association of Recurrence Risk With Post–Adjuvant Chemotherapy ctDNA
To evaluate ctDNA clearance after the completion of adjuvant chemotherapy, we analyzed the ctDNA status of the first blood sample obtained after chemotherapy. Of 149 patients with post–adjuvant chemotherapy blood samples, 22 experienced recurrence from 0.5 to 17.2 months after adjuvant chemotherapy, 12 of the 18 ctDNA-positive patients (66.7%) experienced recurrence, and 121 of the 131 ctDNA-negative patients (92.4%) remained recurrence free. The ctDNA-positive patients had a significantly shorter recurrence-free survival than ctDNA-negative patients (HR, 13.8; 95% CI, 5.9-32.1; P < .001) (eFigure 9 in Supplement 1). Limited by availability, 128 of the 211 adjuvant chemotherapy–treated patients provided both post–adjuvant chemotherapy and postoperative month 1 blood samples. As expected, ctDNA-positive results were associated with significantly reduced recurrence-free survival after adjuvant chemotherapy (HR, 10.9; 95% CI, 4.5-26.3; P < .001) and at postoperative month 1 (HR, 8.8; 95% CI, 3.5-22.3; P < .001) compared with recurrence-free survival for ctDNA-negative patients (eFigure 10A and B in Supplement 1). Regardless of the patients’ ctDNA status at postoperative month 1, the presence of ctDNA after adjuvant chemotherapy showed risk discrimination in recurrence-free survival (eFigure 10C-E in Supplement 1).
Longitudinal ctDNA Associated With Risk of Recurrence
To investigate the role of serial ctDNA analysis in surveillance, we assessed the time course of ctDNA positivity and its association with recurrence in each patient (eTable 3 and eFigure 11 in Supplement 1). Among 217 patients available with samples obtained at the first time point after definitive treatment, ctDNA positivity after curative treatment was associated with a reduced recurrence-free survival compared with ctDNA negativity (HR, 20.6; 95% CI, 9.5-44.9; P < .001) (Figure 3A; eAppendices 1-3 in Supplement 1). All patients with recurrence and ctDNA-positive test results had ctDNA detected earlier than radiologically confirmed recurrence, with a median lead time of 3.3 months (IQR, 0.5-6.5 months; Wilcoxon signed rank test) (Figure 3B). Unexpectedly, 10 of 12 adjuvant chemotherapy–treated patients showed ctDNA positivity before the start of adjuvant chemotherapy, indicating a high-risk subgroup. Next, we identified 226 patients with serial blood samples obtained after treatment and found that an inferior prognosis was associated with patients with ctDNA-positive results at any time point (HR, 13.5; 95% CI, 5.4-33.8; P < .001) (Figure 3C). A total of 140 patients exhibited a sustained ctDNA status longitudinally. Six of the 7 ctDNA-positive patients (85.7%) experienced recurrence within 12 months, whereas 129 of the 133 ctDNA-negative patients (97.0%) were relapse free. The ctDNA status maintenance of ctDNA-positive patients showed a significantly shorter recurrence-free survival than that of ctDNA-negative patients (HR, 68.8; 95% CI, 18.4-257.7; P < .001), indicating a worse prognosis when the residual ctDNA was retained (Figure 3D).
Figure 3. Longitudinal Circulating Tumor DNA (ctDNA) Analysis for Detecting Colorectal Cancer Recurrence.
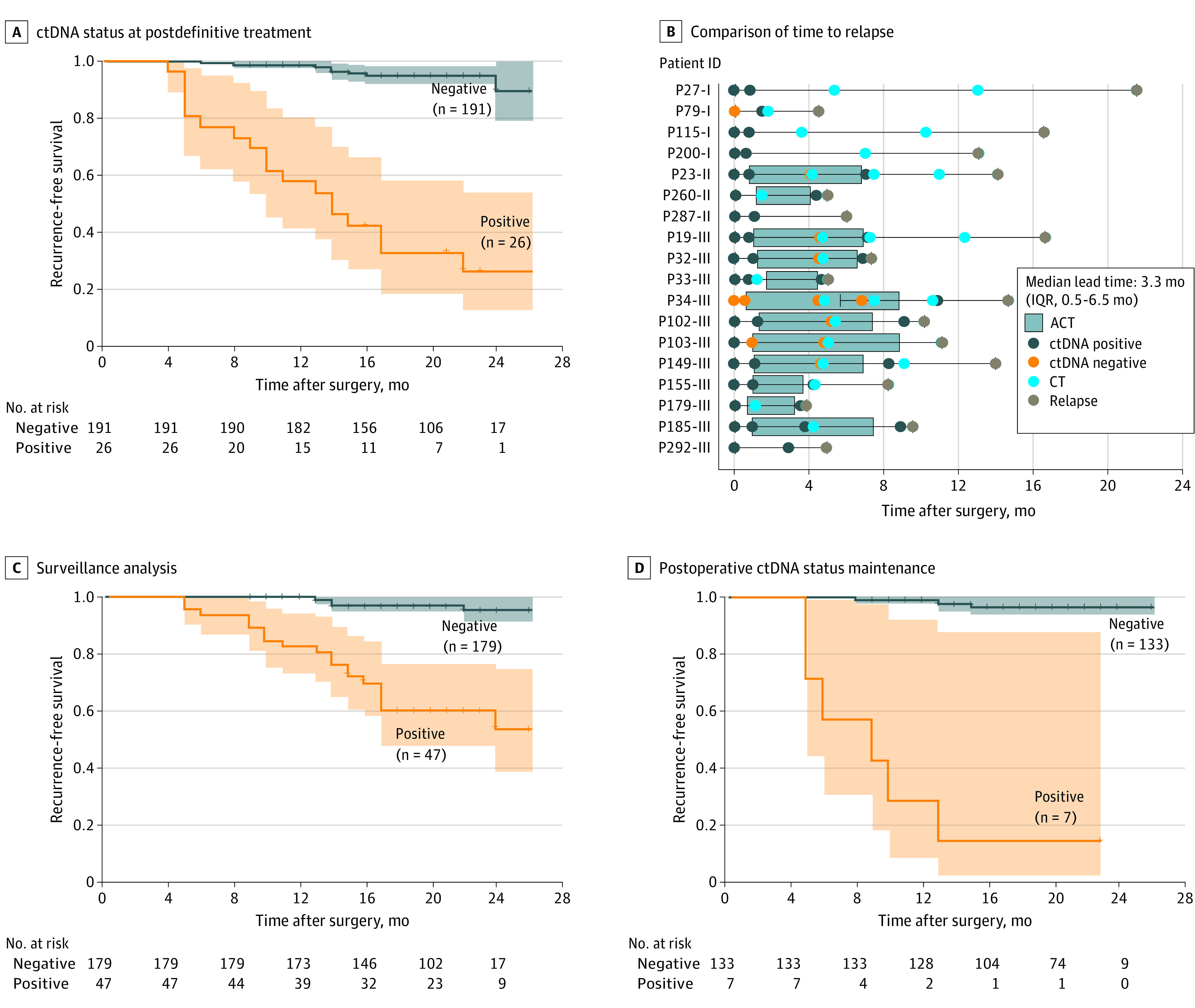
A, Kaplan-Meier estimates of recurrence-free survival (RFS) for patients who completed definitive treatment (surgery and adjuvant chemotherapy [ACT]), stratified by the first time point at postdefinitive treatment ctDNA status. After curative treatment, ctDNA positivity was associated with a strikingly reduced RFS compared with ctDNA negativity (hazard ratio [HR], 20.6; 95% CI, 9.5-44.9; P < .001). P value was determined by the log-rank test. B, Comparison of time to relapse by ctDNA status at postdefinitive-treatment and standard-of-care computed tomography (CT). ID indicates identification. C, Kaplan-Meier estimates of RFS for patients during surveillance, stratified by the longitudinal ctDNA status. A patient was classified as testing positive if there was at least 1 positive ctDNA test result. An inferior prognosis was associated with patients with ctDNA-positive results at any point (HR, 13.5; 95% CI, 5.4-33.8; P < .001). P value was determined by the log-rank test. D, Kaplan-Meier estimates of RFS stratified by ctDNA status in blood samples obtained after surgery. A patient was classified as testing positive (or negative) if all plasma samples were ctDNA positive (or negative). The ctDNA status maintenance indicated that ctDNA-positive patients had a significantly shorter RFS than ctDNA-negative patients (HR, 68.8; 95% CI, 18.4-257.7; P < .001), indicating a worse prognosis when the residual ctDNA was retained. P value was determined by the log-rank test. Shaded areas indicate 95% CIs.
Discussion
In this prospective longitudinal cohort study of 299 patients with CRC, we demonstrated effective ctDNA measurement for patients with CRC by using a noninvasive, multilocus, quantitative polymerase chain reaction assay targeting 6 DNA methylation markers. Our longitudinal analysis of the 299 patients suggested that the existence of molecular residual disease could be effectively detected by ctDNA methylation as early as postoperative month 1, which implies a high risk of recurrence. We found that ctDNA methylation was the most significant prognostic factor for recurrence-free survival among all clinicopathologic risk factors on multivariable analysis. The clinical utility of ctDNA methylation as a valuable biomarker was confirmed for postoperative risk stratification, adjuvant chemotherapy assessment, and recurrence warning throughout the disease course. This pragmatic test can largely simplify and facilitate the integration of ctDNA measurements in standard patient care.
A considerable proportion of patients with localized CRC21,22,37,38 can experience recurrence after radical surgery, which urgently demands a powerful tool for early detection of disease recurrence. An increasing number of studies in recent years have proposed ctDNA-guided postoperative management, suggesting that ctDNA represents changes in tumor burden throughout the clinical disease course. One of the most critical decisions is the selection of patients for adjuvant chemotherapy treatment after surgery based on risk stratification, which has a very restricted window and is currently determined by CRC stage and clinical risk factors. We found that compared with stage, carcinoembryonic antigen, and other clinical risk factors, ctDNA methylation was a significant prognostic factor associated with recurrence in multivariable analysis, which is in accordance with, and superior to, previous reports from ctDNA studies at postoperative month 1.18,19,20,22,25,39 In our study, we showed that at postoperative month 1, ctDNA-positive patients had a significantly higher risk of recurrence (HR, 17.5; 95% CI, 8.9-34.4; P < .001), indicating the reliability and effectiveness of ctDNA methylation in detecting molecular residual disease and recurrence. Furthermore, ctDNA methylation may greatly assist existing carcinoembryonic antigen tests that are routinely used in clinics. For patients who tested negative for preoperative carcinoembryonic antigen, ctDNA methylation at postoperative month 1 was associated with the detection of recurrence (HR, 35.2; 95% CI, 10.3-120.5; P < .001). The combination of carcinoembryonic antigen and ctDNA methylation tests was associated with improved efficiency of risk stratification, with an area under the curve of 0.849. The ctDNA status at postoperative month 1 can serve as a foundation for optimizing the clinical benefits of adjuvant chemotherapy and follow-up, implying a future paradigm change in ctDNA-guided treatment of patients with CRC.
The absence of ctDNA at earlier time points is a reliable indicator of a better prognosis, consistent with reports from other groups.40 We found that ctDNA-negative patients at postoperative month 1 had a low risk of relapse, with a 94.4% negative predictive value, regardless of adjuvant chemotherapy administration. Moreover, we assessed the persistent status of ctDNA in patients’ outcomes, in which ctDNA-positive patients with retained residual ctDNA were 68.8 times more likely to have recurrence than persistent ctDNA-negative patients. In other words, only 4 of 133 patients (3%) with serially negative ctDNA relapsed, similar to the rate reported for lung cancer.41 The absence of ctDNA before surgery is also indicative of prognosis, with 95.3% of ctDNA-negative patients remaining relapse free. Therefore, patients with ctDNA negativity may have an option to defer intensive prognostic treatment, which saves them from the toxic effects of chemotherapy and its complications. This patient group could benefit the most from surgery alone and from longitudinal ctDNA-based surveillance to monitor relapse risk.
In recent years, significant efforts in developing assays targeting ctDNA in blood have been made to monitor prognosis and reduce mortality due to CRC. Most ctDNA assays focus on genomic variations that lack the common patterns shared among a variety of patients. Special care must be taken to avoid the dropout of cancer signals if a fixed panel is applied. Therefore, we incorporated a ctDNA methylation assay to be used to the fullest extent, whereas epigenetic markers, such as methylated cytosines, have long been recognized as robust cancer biomarkers for patients with CRC.
The simultaneous detection of the methylation status of 6 genomes targeted specifically by CRC allows for robust detection and higher sensitivity. More important, the simplicity of the quantitative polymerase chain reaction assay workflow and the convenience of blood samples make it practical for clinical settings to detect molecular residual disease promptly, robustly, and cost-effectively.
According to the results of this study and other published studies,25,28,42 we propose the following potential clinical applications of ctDNA methylation: (1) for patients with positive ctDNA test results at postoperative month 1, adjuvant chemotherapy is recommended regardless of tumor stage; (2) for patients with negative ctDNA test results at postoperative month 1, there may be an opportunity for de-escalation of standard adjuvant chemotherapy; (3) escalation of adjuvant chemotherapy with extended duration may be considered if ctDNA test results are persistently positive after standard adjuvant chemotherapy; and (4) if a patient has ctDNA-positive test results during follow-up, more frequent radiologic examinations may be considered for early diagnosis of recurrence.
Strengths and Limitations
Our study has several potential limitations. First, we observed that adjuvant chemotherapy was associated with reduced recurrence risk for ctDNA-positive patients at postoperative month 1 (eFigure 10C in Supplement 1); however, the efficiency might be underestimated because the collected sample did not cover most adjuvant chemotherapy–treated patients (eTable 11 in Supplement 1). More than 2 centers for recruiting patients may be necessary to maximize the integrity of follow-up, sufficient samples for subgroup analysis, and generalization of conclusions. Second, longitudinal analysis after definitive treatment was limited in our study owing to patient compliance and sample availability at later follow-up time points. Improvements and adjustments will be required for patient follow-up treatment and sample collection in future studies. Third, these results suggest that the feasibility of using ctDNA methylation to measure ctDNA during postoperative monitoring was proven in our study. Future interventional clinical trials incorporating ctDNA methylation tests into standard of care are needed to validate the clinical utility and benefits of ctDNA methylation–guided management.
Conclusions
In conclusion, this cohort study demonstrated the robustness, reproducibility, and reliability of an easy-to-implement multilocus ctDNA methylation assay to accurately measure ctDNA changes throughout the disease course. We also demonstrated that ctDNA methylation is closely associated with tumor progression, potentially accelerating risk stratification, molecular residual disease assessment, and recurrence detection. The ctDNA status represents changes in tumor burden before and after surgery, during and after adjuvant chemotherapy, during surveillance, and eventually to recurrence. Our results may guide decision-making for selection of adjuvant chemotherapy for patients postoperatively, assessment of the duration and intensity of treatment, and prognostic patient care, which could benefit patients and reduce aggressive overtreatment.
eAppendix 1. Postdefinitive Treatment Group Analysis
eAppendix 2. Surveillance Analysis
eAppendix 3. Circulating Tumor DNA Maintenance Analysis
eAppendix 4. ColonAiQ Assay
eAppendix 5. Statistical Analysis
eAppendix 6. Carcinoembryonic Antigen (CEA) Test and Integration of CEA With Circulating Tumor DNA
eTable 1. Clinicopathologic Patient Characteristics
eTable 2. Characteristics of Enrolled Patients
eTable 3. Circulating Tumor DNA Results for All 1228 Plasma Samples
eTable 4. Preoperative and Postoperative (POM1) Carcinoembryonic Antigen (CEA) Results
eTable 5. Number of Recurrent Patients According to Circulating Tumor DNA Status at Postoperative Month 1 (POM1)
eTable 6. Multivariable Cox Analysis of Recurrence-Free Survival by Circulating Tumor DNA and Carcinoembryonic Antigen (CEA) Status at Postoperative Month 1
eTable 7. Performance of 6 Methylated DNA Markers of ColonAiQ in Detecting Colorectal Cancer Recurrence
eTable 8. Performance Estimates of Circulating Tumor DNA and Carcinoembryonic Antigen (CEA) Status at Postoperative Month 1 for Detecting Recurrence in Single and Combined Diagnoses
eTable 9. Number of Recurring Patients According to Circulating Tumor DNA Status at Postoperative Month 1 (POM1) in Stage II and III ACT-Treated Patients in the Subgroup Analysis
eTable 10. Univariate Cox Analysis of Recurrence-Free Survival by Postoperative Circulating Tumor DNA Status at Postoperative Month 1 in Patients With Stage III Cancer With Different Clinical Risk Stratification and Adjuvant Chemotherapy (ACT) Treatment Durations
eTable 11. Compliance Rates at the Key Times
eFigure 1. Preoperative Detection of Circulating Tumor DNA and Carcinoembryonic Antigen (CEA) in Patients With Stage I to III Colorectal Cancer
eFigure 2. Patients With Recurrence in Preoperative Circulating Tumor DNA (Carcinoembryonic Antigen [CEA])–positive and Circulating Tumor DNA (CEA)–Negative Patients
eFigure 3. Prognostic Value by Stages
eFigure 4. Kaplan-Meier Estimates of Recurrence-Free Survival (RFS) Stratified by Carcinoembryonic Antigen (CEA) Status at Postoperative Month 1 for Patients Whose Preoperative CEA Levels Were Positive or Negative
eFigure 5. Subgroup Analysis of Circulating Tumor DNA Status in Patients With Preoperative and Postoperative Carcinoembryonic Antigen (CEA) Test
eFigure 6. Circulating Tumor DNA Status Postoperatively as a Prognostic Factor for Recurrence-Free Survival (RFS) in Patients With Stage III Cancer With Different Clinical Risk Stratifications and Adjuvant Chemotherapy (ACT) Treatment Durations
eFigure 7. Circulating Tumor DNA Status Postoperatively as a Prognostic Factor for Recurrence-Free Survival (RFS) in Patients With Stage II Cancer With Different Clinical Risk Stratifications and Adjuvant Chemotherapy (ACT) Treatment Durations
eFigure 8. Kaplan-Meier Estimates of Recurrence-Free Survival (RFS) Stratified by Circulating Tumor DNA Status at Postoperative Month 1 in Low-Risk Patients Without Adjuvant Chemotherapy (ACT)
eFigure 9. Postadjuvant Chemotherapy (ACT) Analysis for Detecting Colorectal Cancer Recurrence
eFigure 10. Risk Stratification by Circulating Tumor DNA in Patients Whose Plasma Was Collected at Both Postadjuvant Chemotherapy (ACT) and Postoperative Month 1 (POM1)
eFigure 11. Longitudinal Plot of Circulating Tumor DNA Status During Treatment in 299 Patients
Data Sharing Statement
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209-249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Jiang Y, Yuan H, Li Z, et al. Global pattern and trends of colorectal cancer survival: a systematic review of population-based registration data. Cancer Biol Med. 2021;19(2):175-186. doi: 10.20892/j.issn.2095-3941.2020.0634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moghimi-Dehkordi B, Safaee A. An overview of colorectal cancer survival rates and prognosis in Asia. World J Gastrointest Oncol. 2012;4(4):71-75. doi: 10.4251/wjgo.v4.i4.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saltz LB, Kelsen DP. Adjuvant treatment of colorectal cancer. Annu Rev Med. 1997;48(1):191-202. doi: 10.1146/annurev.med.48.1.191 [DOI] [PubMed] [Google Scholar]
- 5.Shibutani M, Maeda K, Kashiwagi S, Hirakawa K, Ohira M. Effect of adjuvant chemotherapy on survival of elderly patients with stage III colorectal cancer. Anticancer Res. 2021;41(7):3615-3624. doi: 10.21873/anticanres.15150 [DOI] [PubMed] [Google Scholar]
- 6.Baxter NN, Kennedy EB, Bergsland E, et al. Adjuvant therapy for stage II colon cancer: ASCO guideline update. J Clin Oncol. 2022;40(8):892-910. doi: 10.1200/JCO.21.02538 [DOI] [PubMed] [Google Scholar]
- 7.Argilés G, Tabernero J, Labianca R, et al; ESMO Guidelines Committee. Localised colon cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2020;31(10):1291-1305. doi: 10.1016/j.annonc.2020.06.022 [DOI] [PubMed] [Google Scholar]
- 8.Rebuzzi SE, Pesola G, Martelli V, Sobrero AF. Adjuvant chemotherapy for stage II colon cancer. Cancers (Basel). 2020;12(9):2584. doi: 10.3390/cancers12092584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.André T, de Gramont A, Vernerey D, et al. Adjuvant fluorouracil, leucovorin, and oxaliplatin in stage II to III colon cancer: updated 10-year survival and outcomes according to BRAF mutation and mismatch repair status of the MOSAIC study. J Clin Oncol. 2015;33(35):4176-4187. doi: 10.1200/JCO.2015.63.4238 [DOI] [PubMed] [Google Scholar]
- 10.Lakemeyer L, Sander S, Wittau M, Henne-Bruns D, Kornmann M, Lemke J. Diagnostic and prognostic value of CEA and CA19-9 in colorectal cancer. Diseases. 2021;9(1):21. doi: 10.3390/diseases9010021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campos-da-Paz M, Dórea JG, Galdino AS, Lacava ZGM, de Fatima Menezes Almeida Santos M. Carcinoembryonic antigen (CEA) and hepatic metastasis in colorectal cancer: update on biomarker for clinical and biotechnological approaches. Recent Pat Biotechnol. 2018;12(4):269-279. doi: 10.2174/1872208312666180731104244 [DOI] [PubMed] [Google Scholar]
- 12.Lieberman D. Colorectal cancer screening with colonoscopy. JAMA Intern Med. 2016;176(7):903-904. doi: 10.1001/jamainternmed.2016.1333 [DOI] [PubMed] [Google Scholar]
- 13.Warton K, Samimi G. Methylation of cell-free circulating DNA in the diagnosis of cancer. Front Mol Biosci. 2015;2:13. doi: 10.3389/fmolb.2015.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warton K, Mahon KL, Samimi G. Methylated circulating tumor DNA in blood: power in cancer prognosis and response. Endocr Relat Cancer. 2016;23(3):R157-R171. doi: 10.1530/ERC-15-0369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baylin SB, Jones PA. A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer. 2011;11(10):726-734. doi: 10.1038/nrc3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985-990. doi: 10.1038/nm.1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang DS, Yang H, Liu XY, et al. Dynamic monitoring of circulating tumor DNA to predict prognosis and efficacy of adjuvant chemotherapy after resection of colorectal liver metastases. Theranostics. 2021;11(14):7018-7028. doi: 10.7150/thno.59644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8(346):346ra92. doi: 10.1126/scitranslmed.aaf6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henriksen TV, Tarazona N, Frydendahl A, et al. Circulating tumor DNA in stage III colorectal cancer, beyond minimal residual disease detection, toward assessment of adjuvant therapy efficacy and clinical behavior of recurrences. Clin Cancer Res. 2022;28(3):507-517. doi: 10.1158/1078-0432.CCR-21-2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen G, Peng J, Xiao Q, et al. Postoperative circulating tumor DNA as markers of recurrence risk in stages II to III colorectal cancer. J Hematol Oncol. 2021;14(1):80. doi: 10.1186/s13045-021-01089-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarazona N, Gimeno-Valiente F, Gambardella V, et al. Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized colon cancer. Ann Oncol. 2019;30(11):1804-1812. doi: 10.1093/annonc/mdz390 [DOI] [PubMed] [Google Scholar]
- 22.Reinert T, Henriksen TV, Christensen E, et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 2019;5(8):1124-1131. doi: 10.1001/jamaoncol.2019.0528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leon Arellano M, García-Arranz M, Guadalajara H, Olivera-Salazar R, Valdes-Sanchez T, García-Olmo D. Analysis of septin 9 gene hypermethylation as follow-up biomarker of colorectal cancer patients after curative surgery. Diagnostics (Basel). 2022;12(4):993. doi: 10.3390/diagnostics12040993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tham C, Chew M, Soong R, et al. Postoperative serum methylation levels of TAC1 and SEPT9 are independent predictors of recurrence and survival of patients with colorectal cancer. Cancer. 2014;120(20):3131-3141. doi: 10.1002/cncr.28802 [DOI] [PubMed] [Google Scholar]
- 25.Jin S, Zhu D, Shao F, et al. Efficient detection and post-surgical monitoring of colon cancer with a multi-marker DNA methylation liquid biopsy. Proc Natl Acad Sci U S A. 2021;118(5):e2017421118. doi: 10.1073/pnas.2017421118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young GP, Pedersen SK, Mansfield S, et al. A cross-sectional study comparing a blood test for methylated BCAT1 and IKZF1 tumor-derived DNA with CEA for detection of recurrent colorectal cancer. Cancer Med. 2016;5(10):2763-2772. doi: 10.1002/cam4.868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musher BL, Melson JE, Amato G, et al. Evaluation of circulating tumor DNA for methylated BCAT1 and IKZF1 to detect recurrence of stage II/stage III colorectal cancer (CRC). Cancer Epidemiol Biomarkers Prev. 2020;29(12):2702-2709. doi: 10.1158/1055-9965.EPI-20-0574 [DOI] [PubMed] [Google Scholar]
- 28.Cai G, Cai M, Feng Z, et al. ; ColonAiQ Group . A multilocus blood-based assay targeting circulating tumor DNA methylation enables early detection and early relapse prediction of colorectal cancer. Gastroenterology. 2021;161(6):2053-2056. doi: 10.1053/j.gastro.2021.08.054 [DOI] [PubMed] [Google Scholar]
- 29.Warren JD, Xiong W, Bunker AM, et al. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 2011;9:133. doi: 10.1186/1741-7015-9-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Church TR, Wandell M, Lofton-Day C, et al. ; PRESEPT Clinical Study Steering Committee, Investigators and Study Team . Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63(2):317-325. doi: 10.1136/gutjnl-2012-304149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.deVos T, Tetzner R, Model F, et al. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem. 2009;55(7):1337-1346. doi: 10.1373/clinchem.2008.115808 [DOI] [PubMed] [Google Scholar]
- 32.Yeh CY, Hsieh PS, Chiang JM, et al. Preoperative carcinoembryonic antigen elevation in colorectal cancer. Hepatogastroenterology. 2011;58(109):1171-1176. doi: 10.5754/hge10564 [DOI] [PubMed] [Google Scholar]
- 33.Maehara Y, Kusumoto T, Takahashi I, et al. Predictive value of preoperative carcinoembryonic antigen levels for the prognosis of patients with well-differentiated gastric cancer: a multivariate analysis. Oncology. 1994;51(3):234-237. doi: 10.1159/000227340 [DOI] [PubMed] [Google Scholar]
- 34.Takagawa R, Fujii S, Ohta M, et al. Preoperative serum carcinoembryonic antigen level as a predictive factor of recurrence after curative resection of colorectal cancer. Ann Surg Oncol. 2008;15(12):3433-3439. doi: 10.1245/s10434-008-0168-8 [DOI] [PubMed] [Google Scholar]
- 35.Allende T, García Muñiz JL, Vizoso F, et al. Preoperative serum levels of the carcinoembryonic antigen (CEA) and prognosis in colorectal cancer. Article in Spanish. Rev Esp Med Nucl. 2001;20(5):358-364. doi: 10.1016/S0212-6982(01)71974-X [DOI] [PubMed] [Google Scholar]
- 36.Grothey A, Sobrero AF, Shields AF, et al. Duration of adjuvant chemotherapy for stage III colon cancer. N Engl J Med. 2018;378(13):1177-1188. doi: 10.1056/NEJMoa1713709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Li L, Cohen JD, et al. Prognostic potential of circulating tumor DNA measurement in postoperative surveillance of nonmetastatic colorectal cancer. JAMA Oncol. 2019;5(8):1118-1123. doi: 10.1001/jamaoncol.2019.0512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobrero A, Lonardi S, Rosati G, et al. ; TOSCA Investigators . FOLFOX or CAPOX in stage II to III colon cancer: efficacy results of the Italian Three or Six Colon Adjuvant trial. J Clin Oncol. 2018;36(15):1478-1485. doi: 10.1200/JCO.2017.76.2187 [DOI] [PubMed] [Google Scholar]
- 39.Symonds EL, Pedersen SK, Murray DH, et al. Circulating tumour DNA for monitoring colorectal cancer—a prospective cohort study to assess relationship to tissue methylation, cancer characteristics and surgical resection. Clin Epigenetics. 2018;10:63. doi: 10.1186/s13148-018-0500-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kotaka M, Shirasu H, Watanabe J, et al. Association of circulating tumor DNA dynamics with clinical outcomes in the adjuvant setting for patients with colorectal cancer from an observational GALAXY study in CIRCULATE-Japan. Jpn J Clin Oncol. 2022;40(4)(suppl):9. doi: 10.1200/JCO.2022.40.4_suppl.009 [DOI] [Google Scholar]
- 41.Zhang JT, Liu SY, Gao W, et al. Longitudinal undetectable molecular residual disease defines potentially cured population in localized non–small cell lung cancer. Cancer Discov. 2022;12(7):1690-1701. doi: 10.1158/2159-8290.CD-21-1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mo S, Dai W, Wang H, et al. Early detection and prognosis prediction for colorectal cancer by circulating tumour DNA methylation haplotypes: a multicentre cohort study. EClinicalMedicine. 2023;55:101717. doi: 10.1016/j.eclinm.2022.101717 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eAppendix 1. Postdefinitive Treatment Group Analysis
eAppendix 2. Surveillance Analysis
eAppendix 3. Circulating Tumor DNA Maintenance Analysis
eAppendix 4. ColonAiQ Assay
eAppendix 5. Statistical Analysis
eAppendix 6. Carcinoembryonic Antigen (CEA) Test and Integration of CEA With Circulating Tumor DNA
eTable 1. Clinicopathologic Patient Characteristics
eTable 2. Characteristics of Enrolled Patients
eTable 3. Circulating Tumor DNA Results for All 1228 Plasma Samples
eTable 4. Preoperative and Postoperative (POM1) Carcinoembryonic Antigen (CEA) Results
eTable 5. Number of Recurrent Patients According to Circulating Tumor DNA Status at Postoperative Month 1 (POM1)
eTable 6. Multivariable Cox Analysis of Recurrence-Free Survival by Circulating Tumor DNA and Carcinoembryonic Antigen (CEA) Status at Postoperative Month 1
eTable 7. Performance of 6 Methylated DNA Markers of ColonAiQ in Detecting Colorectal Cancer Recurrence
eTable 8. Performance Estimates of Circulating Tumor DNA and Carcinoembryonic Antigen (CEA) Status at Postoperative Month 1 for Detecting Recurrence in Single and Combined Diagnoses
eTable 9. Number of Recurring Patients According to Circulating Tumor DNA Status at Postoperative Month 1 (POM1) in Stage II and III ACT-Treated Patients in the Subgroup Analysis
eTable 10. Univariate Cox Analysis of Recurrence-Free Survival by Postoperative Circulating Tumor DNA Status at Postoperative Month 1 in Patients With Stage III Cancer With Different Clinical Risk Stratification and Adjuvant Chemotherapy (ACT) Treatment Durations
eTable 11. Compliance Rates at the Key Times
eFigure 1. Preoperative Detection of Circulating Tumor DNA and Carcinoembryonic Antigen (CEA) in Patients With Stage I to III Colorectal Cancer
eFigure 2. Patients With Recurrence in Preoperative Circulating Tumor DNA (Carcinoembryonic Antigen [CEA])–positive and Circulating Tumor DNA (CEA)–Negative Patients
eFigure 3. Prognostic Value by Stages
eFigure 4. Kaplan-Meier Estimates of Recurrence-Free Survival (RFS) Stratified by Carcinoembryonic Antigen (CEA) Status at Postoperative Month 1 for Patients Whose Preoperative CEA Levels Were Positive or Negative
eFigure 5. Subgroup Analysis of Circulating Tumor DNA Status in Patients With Preoperative and Postoperative Carcinoembryonic Antigen (CEA) Test
eFigure 6. Circulating Tumor DNA Status Postoperatively as a Prognostic Factor for Recurrence-Free Survival (RFS) in Patients With Stage III Cancer With Different Clinical Risk Stratifications and Adjuvant Chemotherapy (ACT) Treatment Durations
eFigure 7. Circulating Tumor DNA Status Postoperatively as a Prognostic Factor for Recurrence-Free Survival (RFS) in Patients With Stage II Cancer With Different Clinical Risk Stratifications and Adjuvant Chemotherapy (ACT) Treatment Durations
eFigure 8. Kaplan-Meier Estimates of Recurrence-Free Survival (RFS) Stratified by Circulating Tumor DNA Status at Postoperative Month 1 in Low-Risk Patients Without Adjuvant Chemotherapy (ACT)
eFigure 9. Postadjuvant Chemotherapy (ACT) Analysis for Detecting Colorectal Cancer Recurrence
eFigure 10. Risk Stratification by Circulating Tumor DNA in Patients Whose Plasma Was Collected at Both Postadjuvant Chemotherapy (ACT) and Postoperative Month 1 (POM1)
eFigure 11. Longitudinal Plot of Circulating Tumor DNA Status During Treatment in 299 Patients
Data Sharing Statement


