Abstract
Background and Purpose:
The G-protein-coupled receptor GPR75 (Gq) and its ligand, the cytochrome P450-derived vasoactive eicosanoid 20-hydroxyeicosatetraenoic acid (20-HETE), are involved in the activation of pro-inflammatory and hypertensive signalling cascades contributing to diabetes, obesity, vascular dysfunction/remodelling, hypertension and cardiovascular disease. Little is known as to how, where and with what affinity 20-HETE interacts with GPR75.
Experimental Approach:
To better understand the pairing of 20-HETE and its receptor (GPR75), we used surface plasmon resonance (SPR) to determine binding affinity/kinetics. The PRESTO-Tango receptor-ome methodology for GPR75 overexpression was coupled with FLIPR Calcium 6 assays, homogeneous time-resolved fluorescence (HTRF) IP-1 and β-arrestin recruitment assays to determine receptor activation and downstream signalling events.
Key Results:
SPR confirmed 20-HETE binding to GPR75 with an estimated KD of 1.56 × 10−10 M. In GPR75-transfected HTLA cells, 20-HETE stimulated intracellular Ca2+ levels, IP-1 accumulation and β-arrestin recruitment, all of which were negated by known 20-HETE functional antagonists. Computational modelling of the putative ligand-binding pocket and mutation of Thr212 within the putative 20-HETE binding site abolished 20-HETE’s ability to stimulate GPR75 activation. Knockdown of GPR75 in human endothelial cells nullified 20-HETE-stimulated intracellular Ca2+.The chemokine CCL5, a suggested GPR75 ligand, binds to GPR75 (KD of 5.85 × 10−10 M) yet fails to activate GPR75; however, it inhibited 20-HETE’s ability to activate GPR75 signalling.
Conclusions and Implications:
We have identified 20-HETE as a high-affinity ligand for GPR75 and CCL5 as a low-affinity negative regulator of GPR75, providing additional evidence for the deorphanization of GPR75 as a 20-HETE receptor.
Keywords: 20-HETE, CCL5, cognate pairing, GPR75, receptor biology
1 |. INTRODUCTION
The recent COVID-19 pandemic has underscored the need for understanding the underlying aetiologies governing conditions and disease states that increase severity of illness when exposed to the virus, including obesity, hypertension, heart failure and chronic kidney disease (Lighter et al., 2020; Richardson et al., 2020; Schiffrin et al., 2020; Williamson et al., 2020). We have recently proposed the GPCR, GPR75, as the receptor for 20-hydroxy-5,8,11,14-eicosatetraenoic acid (20-HETE) (Garcia et al., 2017), an arachidonic acid metabolite and a vasoactive lipid whose formation is catalysed by enzymes of the cytochrome P450 (CYP) 4A/F family (Hoopes et al., 2015; Rocic & Schwartzman, 2018). The biological actions of 20-HETE have been linked to the pathogenesis of the diseases mentioned above (Fan et al., 2015, 2016; Gilani et al., 2021; Shekhar et al., 2019). GPR75 was first identified by Tarttelin et al. (1999) as a novel human GPCR and the corresponding gene maps to human chromosome 2p16, encoding a 540 amino acid protein. Initially, it was predominantly expressed in the retina with significant expression in cells surrounding retinal arterioles and in other areas of the brain (Sauer et al., 2001; Tarttelin et al., 1999). GPR75 is structurally unique among GPCRs, with about 200 amino acids, a putative third intracellular loop of 92 residues and an unusually long C-terminal tail of 169 residues.
Studies examining polymorphic associations of its five variants with retinal pathologies found no link between GPR75 and retinal-associated diseases (Sauer et al., 2001). Padmanabhan et al. (2006) identified GPR75 in a locus on chromosome 2p that shows significant linkage to anti-hypertensive responses in the British Genetics of Hypertension Study. However, the presence of numerous potential candidate genes such as PPP3R2 for calcineurin B and KCNK12 for the potassium two pore domain channel subfamily K member 12, questions GPR75 as a candidate gene. Ignatov et al. (2006) cloned the mouse orthologue of human GPR75 and documented its expression in brain and heart (Ignatov et al., 2006). They showed that the chemokine CCL5 increased IP3 and intracellular calcium (iCa2+) in CHO or HEK cells overexpressing the mouse Gpr75 via a Gq protein-coupled PLC-mediated signal transduction pathway. No direct binding studies of CCL5 to GPR75 were documented in that study. Recent studies reported that pancreatic islets as well as beta cell lines express GPR75 and that CCL5 stimulates insulin secretion via PLC-activated Ca2+ influx in a GPR75-dependent manner; however, they also did not document direct binding or interaction between GPR75 and CCL5 (Gencoglu et al., 2019; Liu et al., 2013). Importantly, the pairing of CCL5 and GPR75 could not be repeated in a β-arrestin assay (Davenport et al., 2013; Dedoni et al., 2018; Southern et al., 2013). The Nomenclature and Standards Committee of the International Union of Basic and Clinical Pharmacology (NC-IUPHAR) (Davenport et al., 2013) continues to classify GPR75 as an orphan receptor.
In a recent study, we showed that 20-HETE binds to and signals through GPR75 to affect vascular endothelial and smooth muscle cell function, endothelial-dependent vasodilation, and vascular contractility and remodelling (Garcia et al., 2017). These findings formed a logical basis for proposing 20-HETE as a ligand for GPR75. The NC-IUPHAR (Davenport et al., 2013) calls for establishing reproducibility of the pairing by demonstrating activity of the ligand at the receptor with potency that is consistent with a physiological function prior to the deorphanization of a GPCR through a battery of experiments incorporating binding studies, antagonists, receptor overexpression, second messenger signals and receptor gene deletion/knockdown. Using these NC-IUPHAR guidelines, the present study was undertaken to establish the structural and functional properties associated with the 20-HETE-GPR75 pairing and to provide substantial evidence supporting the deorphanization of GPR75 as a 20-HETE receptor. Moreover, these studies seek to identify the role of CCL5 as a putative regulator of GPR75 function.
2 |. METHODS
2.1 |. Surface plasmon resonance
Surface plasmon resonance (SPR) studies were conducted using the 2-channel OpenSPR Rev 4 instrument (Nicoya Lifesciences, Canada). In brief, human full-length recombinant GPR75 (H00010936-G01, Abnova, Taiwan) (isoelectric point [pI] = 8.3) was immobilized onto high sensitivity carboxyl sensors (SEN-HS-8-COOH) using Nicoya’s amine coupling kit (AMINESM-10) (4:1 EDC/NHS) at the desired ~8500–9300 response unit (RU) to detect small molecule binding to GPR75. For CCL5 (278-RN-010, Tocris/Bio-Techne, Minneapolis, MN) (IP = 8.49) binding studies, ~6000 RUs were immobilized across the SPR sensor surface. In all experiments, HBSS (14025134, Life Technologies) (pH 7.4) was used as a running buffer. Lipid injections including 20-HETE, arachidonic acid, arachidonoyl ethanolamide, 12(S)-HETE, 19(S)-HETE and 19(R)-HETE and the respective vehicle (HBSS containing 1% ethanol) were conducted using a Gastight Hamilton syringe (81165, Hamilton Company Inc, Reno, NV) and depending on the experiments, sample injections were conducted at a 20 or 50 μl min−1 flow rate across the sensor surface. CCL5 injections and treatments were conducted in HBSS pH 4.3 in the absence of BSA and run at a 20 μl min−1 flow rate across the GPR75 immobilized SPR sensor surface. NaOH (10 mM) was used as a regeneration solution. Data are presented as representative traces across multiple sensors. All binding kinetics and affinities were analysed using the TraceDrawer software (V 1.8.1, Ridgeview Instruments AB).
2.2 |. Cell culture
HTLA cells (an HEK293 cell line stably expressing a tTA-dependent luciferase reporter and a β-arrestin2-TEV fusion gene) when coupled with the PRESTO-Tango GPCR-ome screening assay are used as a novel tool to validate GPCR-ligand interactions as previously reported (Kroeze et al., 2015). HTLA cells were grown in DMEM (11965092, Thermo Fisher Scientific, Grand Island, NY) supplemented with 20% FBS (S11150H, Bio-Techne, Minneapolis, MN), penicillin (100 U ml−1)/streptomycin (0.1 mg ml−1) (15140122, Thermo Fisher Scientific, Grand Island, NY), 2-mM L-glutamine (25030081, Thermo Fisher Scientific, Grand Island, NY) and 100 μg ml−1 Hygromycin B (10687010, Thermo Fisher Scientific, Grand Island, NY) (Dogra et al., 2016; Kroeze et al., 2015). HTLA cells were plated onto their respective poly-l-lysine (102691, MP Biomedicals, LLC) coated plates prior to treatment. Human umbilical vein endothelial cells (HUVECs) were plated on 0.1% gelatin-coated plates and grown in EBM-2 basal medium (CC-3156, Lonza, Walkersville, MD) supplemented with EGM2 MV SingleQuots (CC-4147, Lonza, Walkersville, MD) using 10% FBS and used between passages 2 and 4.
2.3 |. Plasmids and cell transfection
HTLA cells underwent transfection using the calcium phosphate method as previously described (Dogra et al., 2016; Kroeze et al., 2015). In short, cells were transfected with the GPR75-Tango (66372 [RRID:Addgene_66372], Addgene, Watertown, MA) or respective GPR75 mutant constructs (S205A, T212N and S219A) to achieve a final receptor plasmid concentration of 0.1 μg per well. The receptor plasmid was mixed with 2-M calcium chloride (CaCl2) solution in DiH2O. This solution was then mixed with 2× HEPES buffered saline (HBS) and left to incubate at room temperature for 10 min (Kroeze et al., 2015). After 10 min of incubation, the solution was mixed and distributed evenly as to achieve the desired final 0.1 μg per well concentration followed by an overnight transfection at 37°C, 5% CO2. Upon the completion of transfection, samples underwent their specific assay protocol and treatment arm.
2.4 |. FLIPR Calcium 6 assay
Intracellular calcium measurements in HTLA cells and HUVECs were conducted using the FLIPR Calcium 6 Assay Kit (R8190, Molecular Devices, San Jose, CA) according to the manufacturer’s protocol. In brief, cells (HTLA cells or HUVECs) were plated onto clear bottom 96-well plates and underwent their respective interventions (i.e., transfection or siRNA). Upon 80%–90% confluency, cells were washed and starved (HTLA cells [serum free DMEM for 2 h], HUVECs [HBSS for 30 min]) prior to the addition of the reconstituted FLIPR Calcium 6 dye (dye was reconstituted in HBSS containing 20-mM HEPES). After the addition of the dye, cells were incubated (HTLA cells [2 h], HUVECs [30 min]) at 37°C, 5% CO2 prior to treatment. Finally, the 96-well plates were loaded into the SpectraMax iD5 multimode microplate reader (Molecular Devices, San Jose, CA). Respective treatments were injected using the iD5’s SmartInject injector system (50 μl), and fluorescence (Ex wavelength 484 nm, Em wavelength 525 nm) readings were conducted for 2 min (1 read per second) post-injection/treatment. Data are expressed as relative fluorescence units (RFU) (max–min). The respective compounds or treatments used during this assay include the following: Vehicle ((ethanol) HBSS containing 1.66% ethanol), 20-hydroxyeicosatetraenoic acid (20-HETE) (1 nM), N-disodiumsuccinate-20-hydroxyeicosa-6(Z),15(Z)-diencarboxamide (AAA) (1 nM), Vehicle (PBS), CCL5 (RANTES) in PBS (no BSA) (0.1–10 nM), BX471 (25 nM), SB328437 (80 nM) and DAPTA (20 nM). The ethanol content injected by the iD5’s SmartInject injector system when injecting 20-HETE, AAA or the combination of both etc. is a final 1.66% ethanol concentration of exposure to cells and therefore the Vehicle (ethanol) treatment arm in the most accurate control that captures the expected ethanol-dependent changes in intracellular calcium (Elamin et al., 2014). CCL5 was reconstituted only in PBS, as the recommended PBS containing 0.1% human or BSA by the manufacturer’s datasheet markedly affected calcium signalling as previously reported (Xia & Ren, 2009). Experiments were conducted concurrently and separated for presentation clarity and accessibility.
2.5 |. IP-1 assay
Inositol phosphate (IP-1) accumulation was assessed using the IP-One Gq kit (62IPAPEB, Cisbio, Bedford, MA) and following the manufacturer’s protocol as previously described (Garcia et al., 2017). This assay employs a competitive format wherein native IP1 produced by cells competes with d2-labelled IP-1 (acceptor) for binding to monoclonal anti-IP1 Tb2 + cryptate (donor). Briefly, HTLA cells were plated on a 96-well clear bottom plate and were transfected as previously described. On the treatment day, HTLA cells were starved in serum free DMEM for 2 h. Cells were then washed and treated with the appropriate reagents and their respective controls for 10 min. The IP-1 d2 reagent (acceptor) was added to each well followed by the IP-1 Tb Cryptate antibody (donor), and the plate was sealed and incubated for 1 h. Samples were then read on the SpectraMax iD5 multimode microplate reader (Molecular Devices, San Jose, CA) using a homogeneous time-resolved fluorescence (HTRF) protocol (Ex 350 nm, Em 665/620 nm HTRF). Additionally, a standard curve was generated using the provided IP-One Gq standard to determine the linear dynamic range of the assay per the manufacturer’s protocol. All calculations of the HTRF ratio ((665 nm/620 nm) × 104) were normalized to the sample/well’s protein concentration, and the final IP-1 concentration was interpolated from the IP-One Gq standard curve using GraphPad Prism (v 8.1.2). The data are expressed as IP-1 accumulation (fold change) relative to Vehicle (ethanol). Treatments/reagents used to evaluate IP-1 accumulation included the following: Vehicle ((ethanol) HBSS containing 0.01% ethanol), 20-hydroxyeicosatetraenoic acid (20-HETE) (1 nM), N-disodiumsuccinate-20-hydroxyeicosa-6(Z),15(Z)-diencarboxamide (AAA) (1 nM), Vehicle (PBS), CCL5 (RANTES) in PBS (no BSA) (0.1 nM), BX471 (25 nM), SB328437 (80 nM) and DAPTA (20 nM). Experiments were conducted concurrently and separated for presentation clarity.
2.6 |. β-Arrestin recruitment assay (PRESTO-Tango)
The PRESTO-Tango methodology was used for the screening of ligand–receptor activation using the readout of β-Arrestin recruitment as a measure. Transfected HTLA cells were starved in serum free DMEM for 2 h. Following this starvation period, cells were washed and treated with various reagents and left to incubate at 37°C, 5% CO2 for 2 h. After this incubation period, the treatment was removed and cells were fed with fresh complete media (DMEM, supplemented with 20% FBS) and left to incubate overnight at 37°C, 5% CO2. The following day, plates were retrieved and the growth media was replaced by the Bright-Glo™ Luciferase Assay System reagent (E2610, Promega, Madison WI) and left to incubate for 15 min at 37°C, 5% CO2. Luminescence was then assessed using the SpectraMax iD5 multimode microplate reader (Molecular Devices, San Jose, CA). Data are represented as fold change in luminescence using the retrieved relative fluorescence units from each sample. Treatments/reagents used to evaluate β-arrestin recruitment included the following: Vehicle ((ethanol) HBSS containing 0.01% ethanol) 20-hydroxyeicosatetraenoic acid (20-HETE) (0.0001–10 nM), N-disodiumsuccinate-20-hydroxyeicosa-6(Z),15(Z)-diencarboxamide (AAA) (1 nM), arachidonic acid (0.0001–10 nM), arachidonoyl ethanolamide (0.0001–10 nM), 12(S)-HETE (0.0001–10 nM), Vehicle (PBS), CCL5 (RANTES) in PBS (no BSA) (0.1–10 nM), BX471 (25 nM), SB328437 (80 nM) and DAPTA (20 nM). Experiments were conducted concurrently and separated for presentation clarity. The Log EC50 for all dose–response curves was generated using GraphPad Prism (ver. 8.3.0).
2.7 |. Molecular modeling and in silico molecular docking
For molecular modelling studies, the Schrödinger Software suite (Prime, Glide, LigPrep, etc.) was used alongside the GPR75 homology model (Protein Class: Class A (Rhodopsin), Species: Homosapiens, State: Active (Δ distance [Å]: 8.69), version 2019-09-03) (https://gpcrdb.org/structure/homology_models/gpr75_human_active/download_pdb). This model was composed using 5-HT2B receptor (sequence similarity: 35%) PDB: 5TUD as the primary template alongside various other templates including: 5ZBH (Y1 receptor), 5TUD (5-HT2B receptor), 4IB4 (5-HT2B receptor), 5TVN (5-HT2B receptor), 6CM4 (D2 receptor), 4PXZ (P2Y12 receptor), 5DYS (Rhodopsin), 5ZHP (M3 receptor), 4XT1 (US28), 3V2Y (S1P1 receptor), 3SN6 (β2-adrenoceptor) and 3VG9 (A2A receptor) (12 backbone templates/47 rotamer templates; source: https://gpcrdb.org/structure/homology_models/gpr75_human_active). The GPR75 receptor grid was generated using Glide, and the top ranked binding pocket/site was generated through SiteMap. In addition, Glide was used for all ligand-docking studies using the 20-HETE (PubChem CID: 5283157), 19(S)-HETE (PubChem CID: 9548883) and 19(R)-HETE (PubChem CID: 11244126) lipid models. The CCL5/RANTES model (PDB: 5DNF CC chemokine 5 [CCL5] oligomer in complex with heparin) was used and split to reflect its endogenous monomeric form for protein–protein interaction using the Biologics Protein–Protein Docking workflow. All models were visualized using Maestro (ver. 12.0).
2.8 |. Site-directed mutagenesis
GPR75 mutants were generated using PCR based site-directed mutagenesis. PCR was performed using the GPR75-Tango (66372 [RRID: Addgene_66372], Addgene, Watertown, MA) vector as a template, and primers for S205A (Forward-AAGGGTAAAGCTATTCTGGCTCTT and Reverse-CTACAACGTAAAGAGCCAGAATAG), T212N (Forward-TACGTTGTAGATTTTAACTTTTGCG and Reverse-ACGGCCACGCAA AAGTTAAAAT) and S219A (Forward-TGGCCGTCGTCGCAGTGTC and Reverse-CATGATGTAAGACACTGCGACGA). The PCR reactions were incubated with DpnI restriction enzyme (#R0176, New England Biolabs, Ipswich, MA) and transformed into DH5α to amplify mutated plasmids. The sequences of resulting mutant constructs were verified by direct DNA sequencing.
2.9 |. Immunofluorescence
For imaging studies, HTLA cells grown on poly-l-lysine-coated coverslips were fixed with 4% paraformaldehyde (PFA) for 30 min at room temperature. Cells were then washed three times with PBS and incubated with the Anti-FLAG M2 monoclonal antibody (Sigma-Aldrich Cat# F1804, [RRID:AB_262044], Sigma, St Louis, MO) at room temperature for 1 h (1: 500 dilution) as previously described. After the 1 h incubation, coverslips were transferred to 4°C and left to incubate further overnight. After washing, the Alexa Fluor 488 (anti-mouse) (Molecular Probes Cat# A-11029, [RRID:AB_138404], Life Technologies, Thermo Fisher Scientific, Grand Island, NY) secondary antibody was used as required for 1 h. HTLA cells were washed three times with PBS followed by mounting the coverslips onto slides using Vectashield Hardset Antifade Mounting Media (Vector Laboratories Cat# H-1400, [RRID:AB_2336787], Vector Laboratories Inc., Burlingame, CA). Images were acquired using the Leica PAULA Smart Cell Imager (Leica Microsystems, Buffalo Grove, IL).
2.10 |. Real-time PCR
For real-time PCR (RT-PCR), HTLA and HUVECs underwent the following protocol. RNA isolation was conducted using the RNeasy Mini Kit (74104, Qiagen, Hilden, Germany), followed by the quantification of the isolated product using the NanoDrop One/OneC Microvolume UV–Vis Spectrophotometer (Thermo Fisher Scientific, Grand Island, NY). cDNA was generated using the High-Capacity cDNA Reverse Transciption Kit (4368814, Applied Biosystems, Grand Island, NY) using the Miniamp Plus Thermal Cycler (Applied Biosystems). RT-PCR was performed using Fast SYBR Green Master Mix (4385610, Applied Biosystems) and the QuantStudio 3 Real-Time PCR System (Applied Biosystems) using the following primers and conditions:
QuantStudio 3 Real-Time PCR Conditions:
| Step # | Run temperature | Run time | # of repeats |
|---|---|---|---|
| 1 | 95°C | 20 s | 1× |
| 2 | 95°C | 3 s | 40× |
| 3 | 60°C | 60 s |
RT-PCR Primers:
| Primer name | Primer sequence |
|---|---|
| CCR1 Fwd | GGA CAA AGT CCC TTG GAA CC |
| CCR1 Rev | ACC AGG ATG TTT CCA ACC AG |
| CCR3 Fwd | AAT GAC TGT GAG CGG AGC AA |
| CCR3 Rev | CCT CTC TCC AAC AAA GGC G |
| CCR5 Fwd | TTT TCC AGC AAG AGG CTC C |
| CCR5 Rev | ATG TGC ACA ACT CTG ACT GG |
| GPR75 Fwd | GCC ACC CGG CAG GCT TAT CT |
| GPR75 Rev | TTC GGA GAG AAA TGT CTC CTT C |
| GAPDH Fwd | GAA GGT GAA GGT CGG AGT CA |
| GAPDH Rev | AAT GAA GGG GTC ATT GAT GG |
2.11 |. Transfection with siRNA
As previously described (Garcia et al., 2020), HUVECs between passages 2 and 4 were transfected with 40 nM siRNA targeting GPR75 (HSS116879, Thermo Fisher Scientific, Grand Island, NY), or a control siRNA (4390843, Ambion, Austin, TX) using Lipofectamine RNAiMAX (13778075, Thermo Fisher Scientific, Grand Island, NY) in Opti-MEM medium (31985070, Thermo Fisher Scientific, Grand Island, NY). After 5 h, the media was supplemented with media containing 2× growth factors overnight (Garcia et al., 2020). Cells were allowed to grow for 36 h and used for intracellular calcium measurements using the FLIPR Calcium 6 Assay system (R8190, Molecular Devices, San Jose, CA).
2.12 |. Western blotting
Western blotting was conducted as previously described (Garcia et al., 2020). RIPA was used as the cell lysis buffer, 20 μg of protein were loaded per sample, and the following primary antibodies were used: GPR75 (rabbit polyclonal, Abcam Cat# ab75581, [RRID:AB_1523717], Abcam, Cambridge, MA) and HSP90 (Santa Cruz Biotechnology Cat# sc-13119, [RRID:AB_675659], Santa Cruz Biotechnology, Dallas, TX). The appropriate LI-COR secondary IRDye antibodies (anti-rabbit (LI-COR Biosciences Cat# 926–32211, [RRID:AB_621843]) and anti-mouse (LI-COR Biosciences Cat# 926–32210, [RRID:AB_621842]) and LI-COR Odyssey Infrared Imaging System (LI-COR, Lincoln, NE) were used to detect and quantify immunoblots. The experimental detail complies with BJP guidelines for immunoblotting (Alexander et al., 2018).
2.13 |. Schematics
Schematics were created with BioRender.com.
2.14 |. Data, analysis and statistics
In compliance with the BJP’s declaration of scientific transparency and rigour, an n of at least 5 per group was used and statistics were not highlighted on data sets with n lower than 5 (P values provided). Sample and group sizes were chosen based the appropriate power analysis and observed variability across their respective assays. Randomization and blinding were not conducted or necessary for the studies performed throughout this manuscript. Group size is noted as the number of independent values and the appropriate Y axis denotes normalization of value to respective vehicle/control groups. No outliers were excluded and the manuscript complies with the BJP’s requirements on experimental design and analysis (Curtis et al., 2018). Results are presented as means ± SEM. Significance of differences between group means (P ≤ 0.05) was assessed using unpaired Student’s t test, one and two-way analysis of variance (ANOVA), followed by Tukey’s post hoc where applicable.
2.15 |. Materials
The following compounds were supplied by Dr. Falck: 20-HETE and N-disodium succinate-20-hydroxyeicosa-6(Z),15(Z)-diencarboxamide (AAA). Tocris/Bio-Techne, (Minneapolis, MN) supplied the following: BX471 (Cat no. 3496), CCL5 (RANTES) (Cat no. 278-RN-010), SB328437 (Cat no.3650) and DAPTA (Cat no.2423). Cayman Chemical (Ann Arbor, MI) supplied the following: 12(S)-HETE (Cat no. 34570), 19(S)-HETE (Cat no. 10007766), 19(R)-HETE (Cat no. 10007767), arachidonic acid (Cat no. 90010) and arachidonoyl ethanolamide (Cat no. 90050).
2.16 |. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in the IUPHAR/BPS Guide to PHARMACOLOGY (http://www.guidetopharmacology.org) (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/20 (Alexander et al., 2019).
3 |. RESULTS
3.1 |. 20-HETE binds to GPR75
To gain further insights and uncover the dynamics by which 20-HETE interacts with GPR75, we employed the use of SPR technology. As seen in Figure 1a, 20-HETE bound to immobilized recombinant GPR75 on the SPR sensor chip surface in a dose-dependent manner (0.0625–10 nM), exhibiting a binding affinity KD of 1.56 × 10−10 ± 2.43 × 10−13 M. To assess the specificity of this response, structurally related lipids including arachidonic acid, arachidonoyl ethanolamide and 12(S)-HETE, an eicosanoid recently identified as the ligand for GPR31 (Guo et al., 2011) (Figure 1b), were exposed to the GPR75 SPR sensor chip surface to determine receptor binding. As seen in Figure 1c and Figure S1A–C, all of the control lipids failed to exhibit any GPR75 binding, confirming the specificity of the 20-HETE/GPR75 pairing. These measures clearly identify 20-HETE as a high-affinity ligand for GPR75.
FIGURE 1.
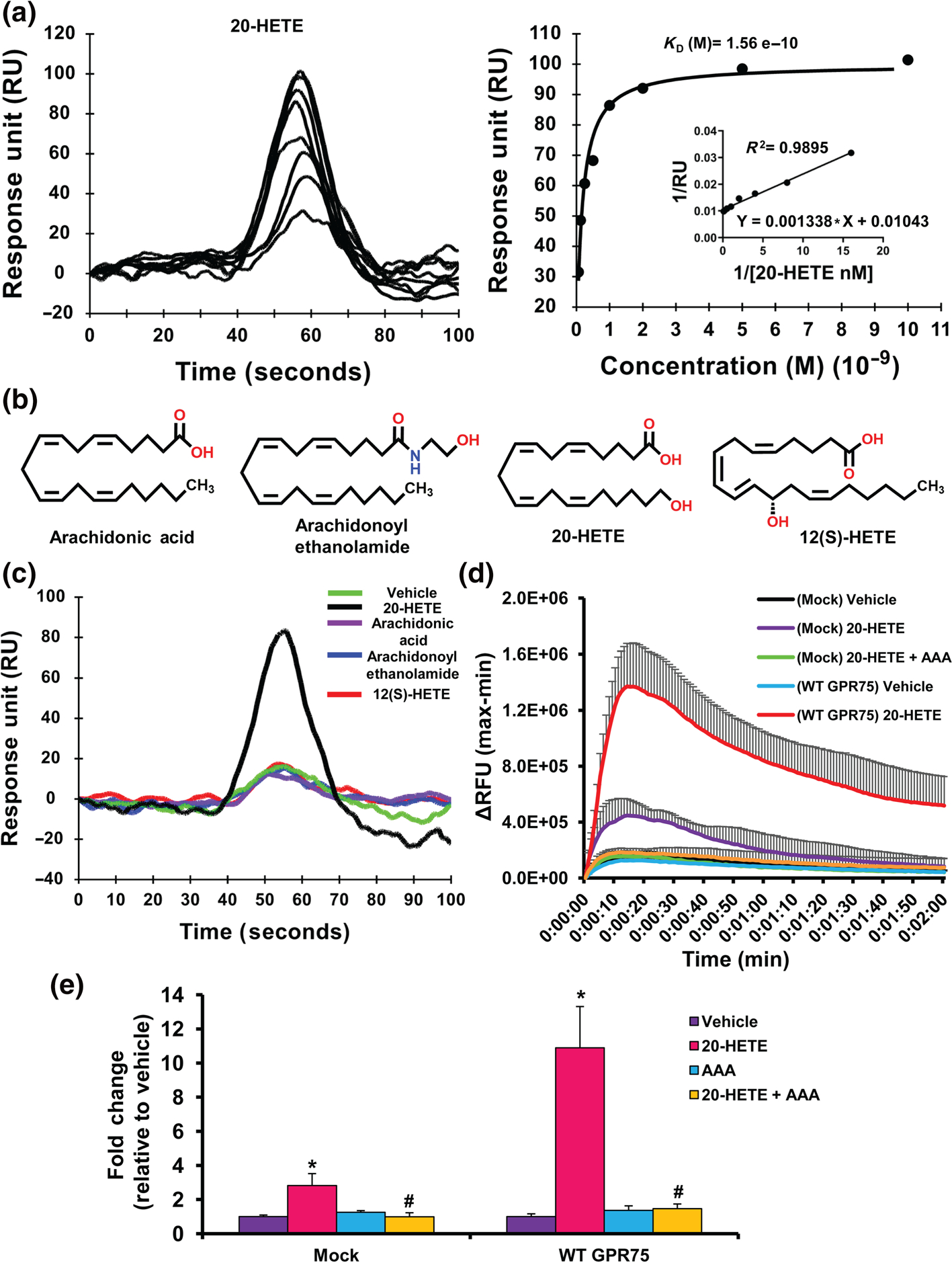
20-HETE binds to GPR75.(a) Surface plasmon resonance (SPR) analysis of 20-HETE-GPR75 binding, depicting the sensogram (normalized to vehicle (HBSS containing 1% ethanol) (left) of human recombinant GPR75 immobilized to the SPR sensor surface followed by 20-HETE injections (0.0625, 0.125, 0.25, 0.5, 1, 2, 5 and 10 nM) injected at a flow rate of 20 μl min−1 and affinity analysis (right). (b) Structures of arachidonic acid, arachidonoyl ethanolamide, 20-HETE and 12(S)-HETE. (c) SPR sensogram of human recombinant GPR75 immobilized to the SPR sensor surface followed by injections of vehicle (HBSS containing 1% ethanol), arachidonic acid, arachidonoyl ethanolamide, 12(S)-HETE and 20-HETE (1 nM) injected at a flow rate of 20 μl min−1; 20-HETE increases intracellular calcium (iCa2+) via GPR75. (d) FLIPR Calcium 6 assays of mock- or GPR75-transfected HTLA cells treated with vehicle (ethanol), 20-HETE (1 nM) and co-treatment of 20-HETE (1 nM) with AAA (1 nM), a 20-HETE receptor antagonist, showing post-treatment live calcium traces (2 min) or (e) peak treatment response expressed as fold change relative to vehicle-treated cells.Data shown are means ± SEM; n = 6.*P < 0.05, significantly different from vehicle (ethanol), #P < 0.05, significantly different from 20-HETE
3.2 |. 20-HETE increases intracellular Ca2+ (iCa2+) via GPR75
We employed the PRESTO-tango methodology (Dogra et al., 2016; Kroeze et al., 2015) (Figure S2) using HTLA cells transfected with a GPR75-tango construct to assess changes in G-protein-dependent and G-protein-independent signals upon receptor activation including iCa2+ and the accumulation of inositol phosphate (IP-1), both indicative of Gq-driven signalling, and the recruitment of β-arrestin. Administration of 20-HETE (1 nM) increased iCa2+ in both mock- and GPR75-transfected HTLA cells (Figure 1d,e). Moreover, 20-HETE-stimulated changes in iCa2+ were blocked by the 20-HETE functional antagonist N-disodium succinate-20-hydroxyeicosa-6(Z),15(Z)-diencarboxamide (AAA) (Gawrys et al., 2020; Sedlakova et al., 2018) (Figure 1e). The overexpression of GPR75 did not alter the response to the vehicle or the positive control ionomycin (20 nM) (Figure S1D,E). In addition, AAA bound to GPR75 but did not exert any significant increase in iCa2+ (Figure S3A,B). Taken together, these results indicate that the 20-HETE-mediated changes in iCa2+ are markedly amplified following transfection with GPR75 and inhibited by the functional antagonist of 20-HETE.
3.3 |. CCL5 does not promote iCa2+ increases through GPR75
Previous reports have implied a relationship between CCL5 (RANTES) and GPR75, going as far as to suggest that CCL5 can promote changes in iCa2+ through the activation of GPR75 (Ignatov et al., 2006; Liu et al., 2013). In order to confirm these claims, SPR experiments were conducted on GPR75 immobilized sensors to reveal a specific interaction between CCL5 and GPR75 (Figure 2a). CCL5 did not bind 20-HETE or similarly structured eicosanoids (12(S)-HETE) (Figure S4A). To determine the effects of CCL5 on iCa2+, mock- and GPR75-transfected HTLA cells were treated with CCL5 in the absence of BSA, as BSA alone can promote significant changes to iCa2+ (Figure S4B). Mock-transfected cells exhibited an almost two-fold increase in iCa2+ in response to CCL5 (0.1 nM) (Figure 2b,d). This increase was prevented in cells pretreated with a mixture of CCL5 receptor (CCR1/3/5) antagonists (CRA), which include the CCR1 antagonist BX471 (25 nM), the CCR3 antagonist SB328437 (80 nM) and the CCR5 antagonist DAPTA (20 nM), all of which are expressed in HTLA cells (Figures 2b,d and S3D). This finding suggests that the canonical CCRs are primarily responsible for the CCL5-mediated fluctuations in iCa2+. CRA-alone did not alter the response to vehicle or ionomycin (Figure S4C–E). Surprisingly, overexpression of GPR75 prevented the twofold CCL5-mediated changes in Ca2+ observed under mock-transfected conditions (Figure 2b). In addition, co-treatment of cells with CCL5 and the CRAs did not alter this response (Figure S4E). Interestingly, CCL5’s inability to elevate Ca2+ in the presence of overexpressed GPR75 could be overcome and unmasked by increasing CCL5 concentrations (1 and 10 nM) uncovering a leftward shift in iCa2+ (Figure S4F). The inability of CCL5 to promote increases in iCa2+, in the presence of abundant GPR75, suggests that the protein–protein interaction between CCL5 and GPR75 potentially sequesters CCL5 and renders it incapable of interacting with its canonically associated CCRs and therefore changes in Ca2+ signalling cannot be observed.
FIGURE 2.
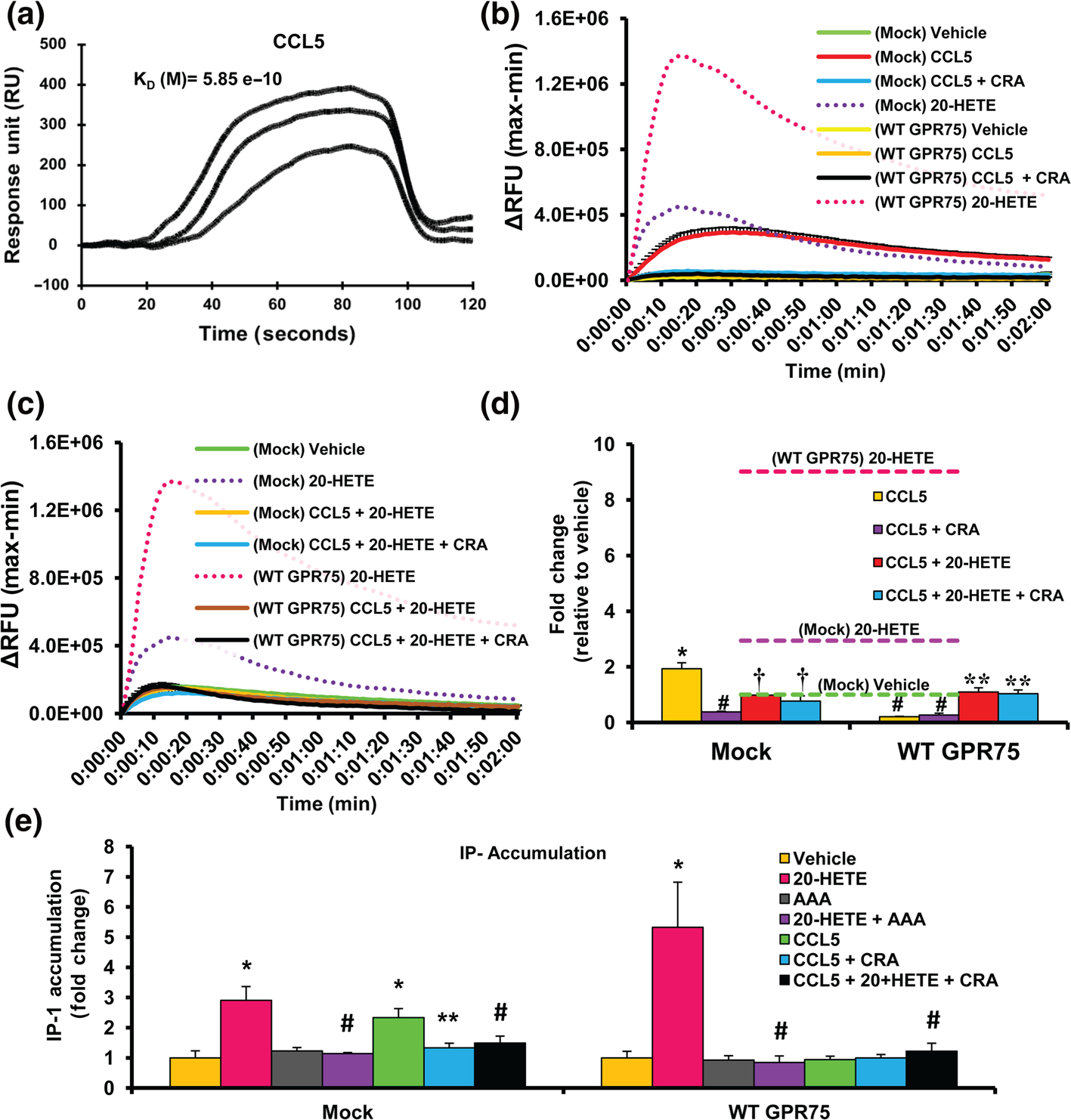
CCL5 increases intracellular calcium (iCa2+) and inositol phosphate-1 (IP-1) via its chemokine receptors and blocks 20-HETE-mediated increases in iCa2+ and IP-1. (a) Surface plasmon resonance (SPR) sensogram of human recombinant GPR75 immobilized to the SPR sensor surface followed by injections of CCL5/RANTES (0.01 nM) injected at a flow rate of 20 μl min−1. (b) FLIPR Calcium 6 assays showing mock- or GPR75-transfected HTLA cells treated with vehicle (PBS), 20-HETE (1 nM) and CCL5 (0.1 nM) in the presence and absence of CCL5 receptor antagonists (CRA) (BX471 targeting CCR1 (25 nM), SB328437 targeting CCR3 (80 nM) and DAPTA targeting CCR5 (20 nM)). (c) FLIPR Calcium 6 assays showing mock or GPR75 transfected HTLA cells treated with vehicle (ethanol), 20-HETE (1 nM), and cells pretreated with CCL5 (0.1 nM) followed by 20-HETE (1 nM) in the presence and absence of CRAs. (d) Peak treatment response expressed as fold change relative to mock-transfected, vehicle (ethanol)-treated cells. Dashed lines represent the maximal response of mock and GPR75-transfected cells to 20-HETE (1 nM). (e) Effect of 20-HETE (1 nM), AAA (1 nM) and CCL5 (0.1 nM) in the presence and absence of CRAs on IP-1 accumulation in HTLA cells transfected with or without WT GPR75-tango overexpression constructs. Data shown are means ± SEM; n = 6. *P < 0.05, significantly different from mock vehicle (PBS), #P < 0.05, significantly different from Mock CCL5, †P < 0.05, significantly different from Mock 20-HETE, **P < 0.05, significantly different from WT GPR75 20-HETE)
3.4 |. CCL5 blocks 20-HETE’s ability to increase iCa2+ and IP-1
To further characterize the relationship between 20-HETE, GPR75 and CCL5, mock- and GPR75-transfected HTLA cells were pretreated with CCL5 prior to the addition of 20-HETE. Surprisingly, CCL5 prevented the 20-HETE-mediated increases in iCa2+ (Figure 2c,d). Moreover, treatment of HTLA cells with CRAs prior to CCL5 addition did not alter the inhibition by CCL5 of 20-HETE-mediated changes in iCa2+, suggesting that the chemokine receptors conventionally associated with CCL5; CCR1, 3 and 5, are not responsible for this effect. We have previously shown that in endothelial cells, 20-HETE promotes the accumulation of inositol phosphate (IP-1) through GPR75 (Garcia et al., 2017). As seen in Figure 2e, the addition of 20-HETE to GPR75-transfected HTLA cells increased IP-1 accumulation, compared to mock-transfected cells. This 20-HETE-stimulated accumulation of IP-1 was also prevented by AAA (Figure 2e). Moreover, co-treatment of CCL5 and 20-HETE prevented the 20-HETE-mediated increases in IP-1 accumulation (Figure 2e).
3.5 |. 20-HETE-GPR75 pairing: β-arrestin recruitment
One of the primary benefits of the PRESTO-tango constructs is the ability to detect β-arrestin recruitment that may occur dependent or independent of G-protein activation (Kroeze et al., 2015); 20-HETE-treated HTLA cells expressing the GPR75-tango construct exhibited significant increases in β-arrestin recruitment in a concentration-dependent manner with an estimated LogEC50 of −12.15 M (Figure 3a). As seen in the SPR studies, all of the control lipids failed to promote any significant increase in β-arrestin recruitment to GPR75 in HTLA cells (Figure 3a). In subsequent studies, co-treatment of GPR75-transfected HTLA cells with equimolar concentrations of 20-HETE and AAA prevented the 20-HETE-mediated increase in β-arrestin recruitment to GPR75 (Figure 3b).
FIGURE 3.
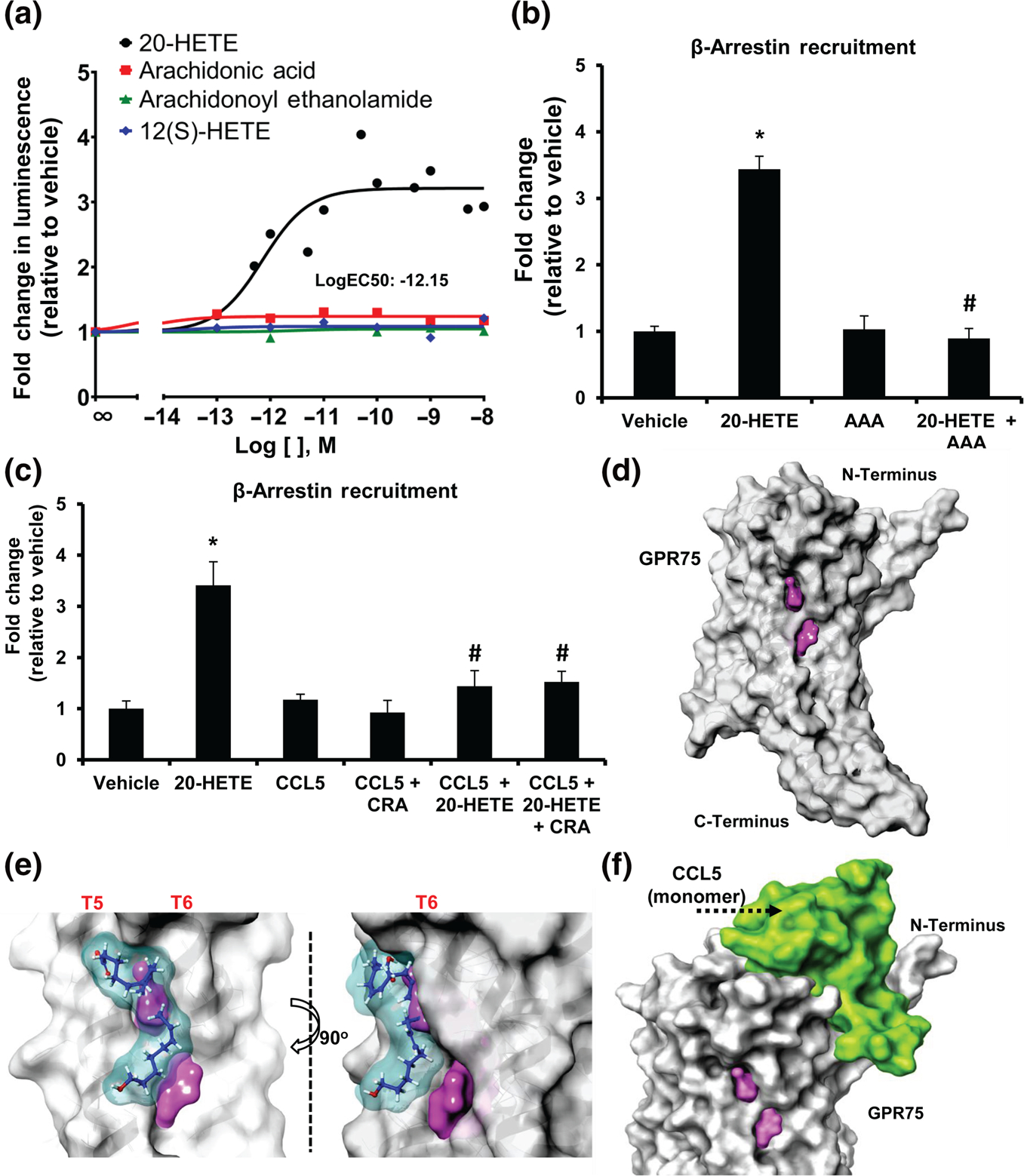
20-HETE, but not CCL5,increases β-arrestin recruitment to GPR75. (a) PRESTO-tango β-arrestin recruitment assay in HTLA cells overexpressing GPR75-tango constructs treated with increasing concentrations of 20-HETE, arachidonic acid, arachidonoyl ethanolamide and 12(S)-HETE; (b) fold change in β-arrestin recruitment in response to 20-HETE (1 nM), AAA (1 nM) and co-treatment of equimolar concentrations of 20-HETE and AAA (1 nM). (c) CCL5 blocks 20-HETE-mediated β-arrestin recruitment to GPR75. Fold change in β-arrestin recruitment in response to 20-HETE (1 nM), CCL5 (0.1 nM) and the co-treatment of CCL5 (0.1 nM) with 20-HETE (1 nM) in the presence and absence of CCL5 receptor antagonists (CRA) (BX471 targeting CCR1 (25 nM), SB328437 targeting CCR3 (80 nM) and DAPTA targeting CCR5 (20 nM)). (d–f) Molecular modelling of GPR75. GPR75 (grey) alongside the (d) ligand binding site (pink), (e) 20-HETE-GPR75 ligand–receptor binding model (20-HETE in blue) and (f) the CCL5-GPR75 protein–protein interaction model. Data shown are means ± SEM; n = 8–12). *P < 0.05, significantly different from vehicle (ethanol), #P < 0.05, significantly different from 20-HETE)
3.6 |. CCL5 prevents the 20-HETE-GPR75 pairing
As shown in Figures 3c and S8C, CCL5 (0.1–10 nM) alone or in the presence of CRAs failed to increase β-arrestin recruitment to GPR75, confirming previous reports (Davenport et al., 2013; Southern et al., 2013). However, co-treatment of 20-HETE and CCL5 in the presence and absence of CRAs prevented the 20-HETE-mediated increase in β-arrestin recruitment (Figure 3c). These results support our previous observations suggesting that CCL5 binding to GPR75 prevents 20-HETE from activating the receptor. Moreover, the calcium, IP-1 and β-arrestin data do not suggest or support the notion that CCL5 behaves as a biased agonist, preferentially activating one signalling event or cascade over another.
3.7 |. Distinct putative GPR75 binding sites for 20-HETE and CCL5
To better understand the pairing of 20-HETE and GPR75, we sought to identify the putative 20-HETE binding site located across the receptor. Molecular modelling of GPR75 uncovered a putative binding pocket within TM5 and TM6 (Site Score: 1.182, Dscore: 1.303) (Figure 3d). Our model further suggests that 20-HETE could interact within this binding pocket through several hydrogen-bond interactions (Docking Score: 3.229, Glide Emodel: −29.045) (Figure 3e). Interestingly, the CCL5-GPR75 protein–protein interaction model revealed the potential for CCL5 monomers to bind to the N-terminal cavities generated by GPR75’s transmembrane loops (Figure 3f). These models suggest that 20-HETE and CCL5 interact with GPR75 and potentially influence the receptor by binding to two distinct pockets (Figure S5).
Additional exploration of our 20-HETE-GPR75 binding model further revealed that the endogenous 20-HETE functional antagonist 19(R)-HETE (Alonso-Galicia et al., 1999; Zhang et al., 2005), which bound to GPR75 in our SPR studies (Figure S6A), interacts with the putative 20-HETE binding pocket within GPR75 TM5 and TM6 distinctly from its 19(S)-HETE stereoisomer antipode, which lacks the ability to functionally bind to GPR75 and antagonize 20-HETE (Figure S6A,C). Likewise, the AAA-GPR75 binding model exhibited similar interactions across the putative binding pocket (Figure S3C). In fact, experiments using 19(R)-HETE yielded similar results to experiments using AAA. Hence, co-treatment of HTLA GPR75-transfected cells with equimolar concentrations of 20-HETE and 19(R)-HETE also prevented the 20-HETE-mediated increases in β-arrestin recruitment to GPR75 (Figure S6B). As expected, based on our modelling (Figure S6C), the 20-HETE-mediated increase in β-arrestin recruitment was similar in the absence or presence of 19(S)-HETE (Figure S6B). These results confirm previous reports and point to the specificity of 20-HETE-GPR75 pairing.
3.8 |. Mutations in 20-HETE binding site alter its pairing with GPR75
Analysis of the ligand receptor docking model suggests the presence of several key residues involved in the formation of hydrogen bonds between 20-HETE and GPR75, including Serine205 (S205), Threonine212 (T212) and Serine219 (S219) (Figure 4a). Mutagenesis studies of HTLA cells transfected with the S205A, T212N and S219A GPR75-tango mutant constructs (Figure S7A,B) revealed that these mutants failed to display the same changes in iCa2+ observed upon addition of 20-HETE to WT GPR75-transfected cells (Figure 4b). Assessment of β-arrestin recruitment further uncovered striking differences between each GPR75 mutant when exposed to 20-HETE. As seen in Figure 4c, the S205A and S219A mutants exhibited significant rightward shift in 20-HETE-mediated β-arrestin recruitment compared to WT GPR75 (LogEC50(M) −11.13 and −11.01, respectively, compared to −12.15 for WT GPR75). The maximal response of S205A and S219A mutants to 20-HETE was not changed (Figure 4c). In contrast, the T212N GPR75 mutant failed to promote any significant changes to β-arrestin recruitment in response to 20-HETE (Figure 4c). Moreover, all three mutants failed to elicit the same changes in IP-1 accumulation observed upon addition of 20-HETE to WT GPR75-transfected cells (Figures 4d and S8B). Strikingly, the data on the T212N GPR75 mutant suggest that this location is vital for 20-HETE-mediated GPR75 activation, while the data on the S205A and S219A mutations uncovered the favouring of biased signalling towards a β-arrestin-specific pathway (Figure 4e). Importantly, all GPR75 mutants responded similarly to WT GPR75, when exposed to CCL5, suggesting that these mutants can still retain CCL5 from interacting with chemokine receptors (Figure S8A–C).
FIGURE 4.
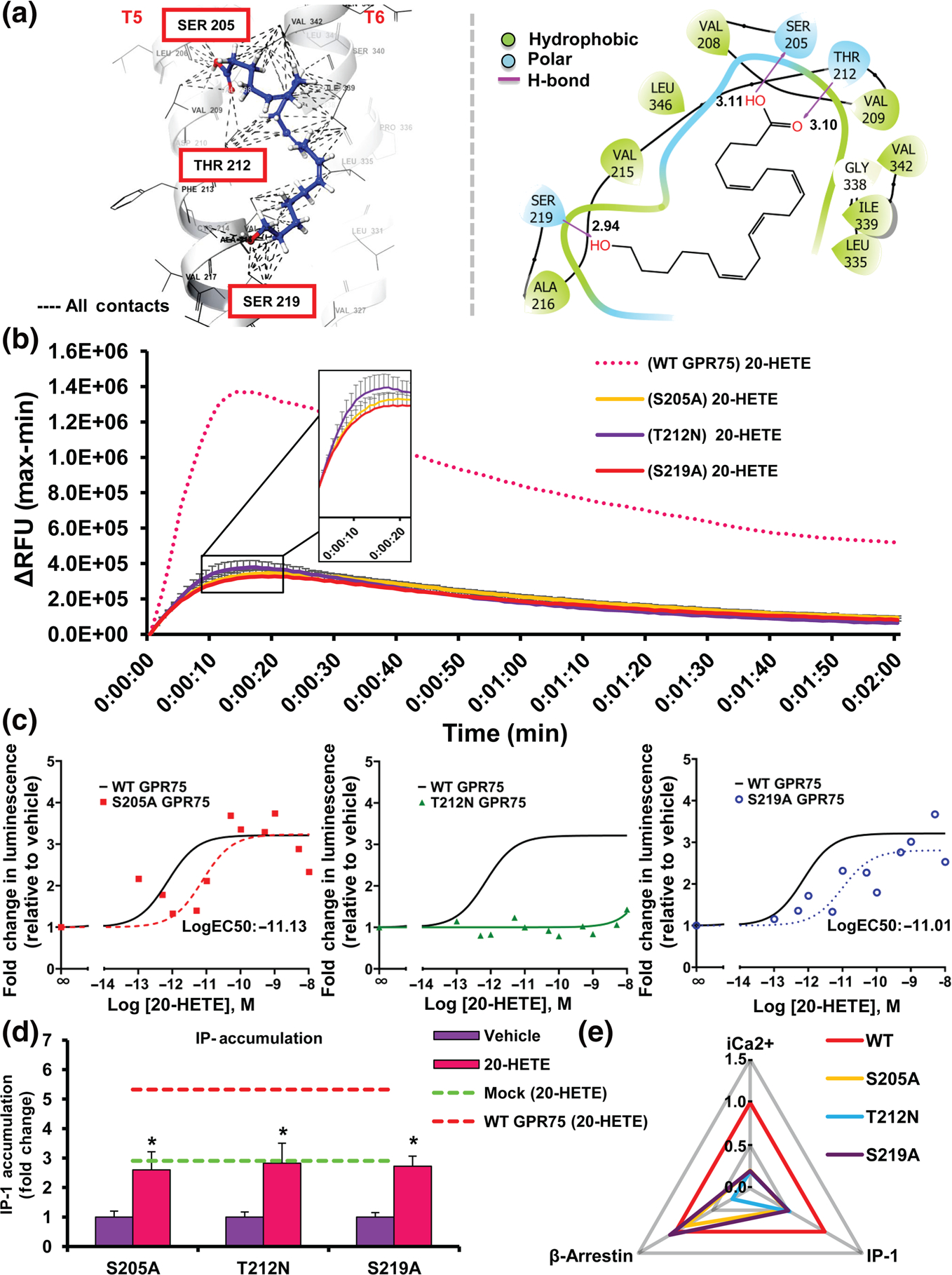
Mutations across the putative 20-HETE-GPR75 binding site alter the 20-HETE-mediated changes in intracellular calcium (iCa2+), IP-1 and β-arrestin. (a) Molecular modelling depicting the putative 20-HETE-GPR75 binding site (3D left, 2D right). (b) FLIPR Calcium 6 assays showing WT and mutant (S205A, T212N, S219A) GPR75-transfected HTLA cells treated with vehicle 20-HETE (1 nM) or CCL5 (0.1 nM). (c) PRESTO-tango β-arrestin recruitment assays in HTLA cells overexpressing WT and mutant (S205A, T212N, S219A) GPR75-tango constructs treated with increasing concentrations of 20-HETE (n = 8). (d) Effect of 20-HETE (1 nM) on IP-1 accumulation in HTLA cells transfected with mutant (S205A, T212N, S219A) GPR75-tango overexpression constructs. Dotted lines represent the maximal response observed in mock and GPR75-transfected cells to 20-HETE (1 nM). Data shown are means ± SEM; n = 6. *P < 0.05, significantly different from 20-HETE). (e) Web of bias plot illustrating the distinctions between the WT GPR75 and GPR75 mutants (S205A, T212N, S219A) responses to 20-HETE across three signalling pathways (iCa2+, IP-1 and β-arrestin). The signalling responses are normalized to WT (set to 1) and the plot distinguishes each mutant’s degree of deviation from the WT GPR75 transfected cells treated with 20-HETE (1 nM)
3.9 |. 20-HETE increases iCa2+ in HUVECs via GPR75
The vascular endothelium is a primary target for 20-HETE bioactivities leading to endothelial dysfunction and activation (Garcia et al., 2016, 2017). Here, we show that addition of 20-HETE to HUVECs promoted a robust increase in iCa2+ (Figures 5a and S9A). As observed in HTLA cells, HUVECs express GPR75 and the corresponding CCRs of interest (Figure S8D). In accordance with our observations in HTLA cells, co-treatment of HUVECs with equimolar concentrations of 20-HETE and the 20-HETE receptor antagonist, AAA, nullified 20-HETE’s ability to increase iCa2+ (Figures 5a and S9A). These studies clearly replicated our observations in HTLA cells, showing 20-HETE’s ability to promote changes in iCa2+ that can be blocked by its functional antagonist. Moreover, knockdown of GPR75 (Figure S9C,D) abolished the ability of 20-HETE to stimulate iCa2+ in HUVECs (Figure 5b), further indicating that 20-HETE signals through GPR75.
FIGURE 5.
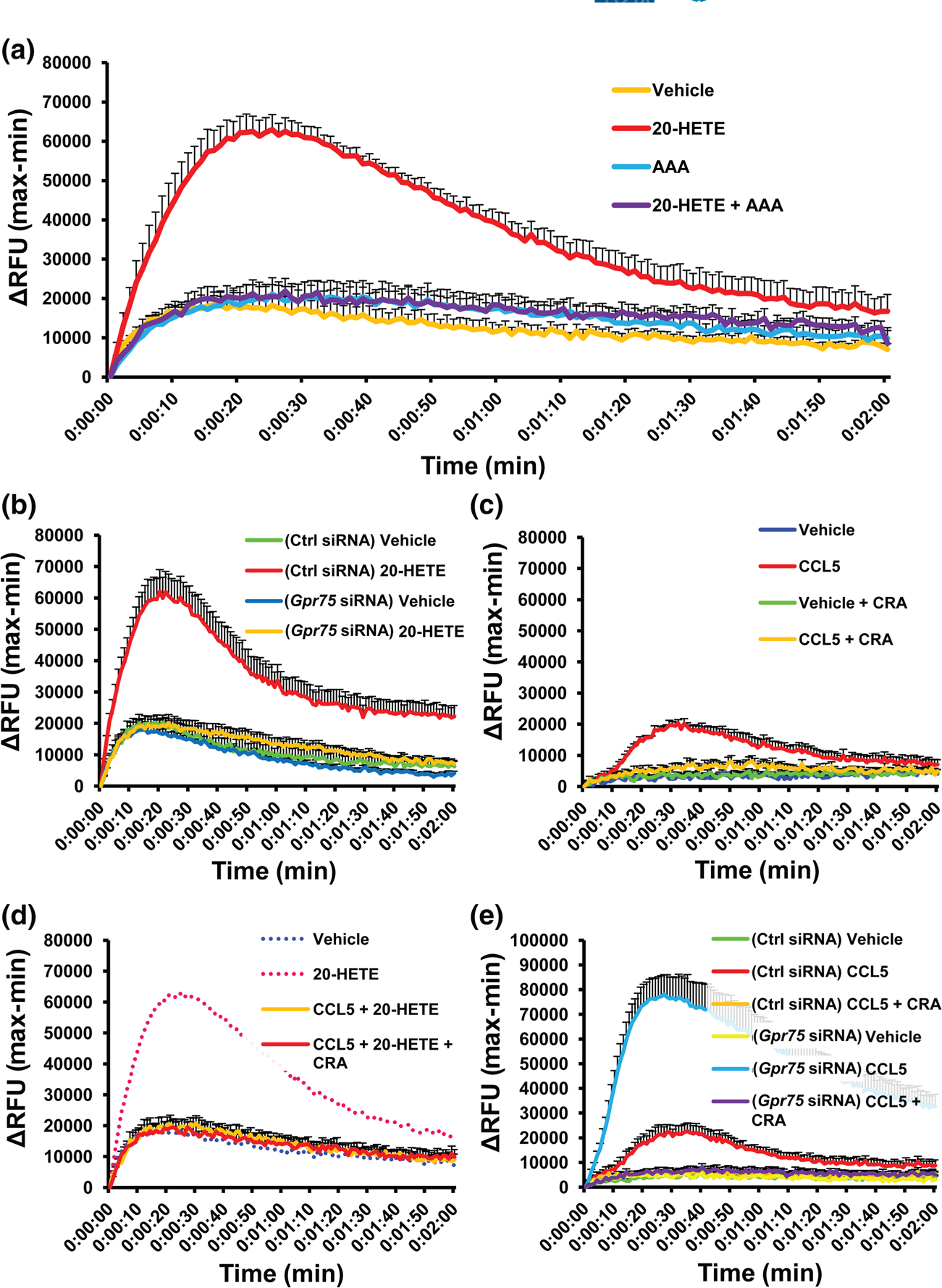
20-HETE, but not CCL5, increases intracellular calcium (iCa2+) via GPR75 in human umbilical vein endothelial cells (HUVECs). FLIPR Calcium 6 assays of HUVECs treated with; (a) vehicle (ethanol), 20-HETE (1 nM) and co-treatment of 20-HETE (1 nM) with AAA (1 nM), a 20-HETE receptor antagonist; (b) GPR75 knockdown in HUVECs abolishes 20-HETE-mediated increases in iCa2+. Effect of 20-HETE (1 nM) on intracellular calcium in control (Ctrl) and GPR75 siRNA-treated HUVECs. (c) CCL5 increases iCa2+ via its chemokine receptors. Effects of vehicle (PBS) and CCL5 (0.1 nM) in the presence and absence of CCL5 receptor antagonists (CRA) (BX471 targeting CCR1 [25 nM], SB328437 targeting CCR3 [80 nM] and DAPTA targeting CCR5 [20 nM]); (d) CCL5 blocks 20-HETE-stimulated increases in iCa2+. Effects of pretreatment of CCL5 (0.1 nM) ± CRAs followed by 20-HETE (1 nM). (e) Gpr75 knockdown intensifies CCL5 (RANTES)-mediated changes in iCa2+ in HUVECs. Effect of CCL5 (0.1 nM) on intracellular calcium in control (Ctrl) or GPR75 siRNA-treated HUVECs. Data shown are means ± SEM; n = 6
3.10 |. CCL5 prevents 20-HETE-mediated GPR75 signalling in HUVECs
Several reports have attempted to link CCL5-mediated changes in iCa2+ to GPR75 in different cell types including murine and human neuronal cell lines, transfected HEK and CHO cells as well as mouse and human islets (Dedoni et al., 2018; Ignatov et al., 2006; Liu et al., 2013). As seen in Figures 5c and S9B, addition of CCL5 to HUVECs increased iCa2+. As expected, pretreatment of the cells with CRAs abolished CCL5’s ability to increase intracellular calcium (Figure S9B). These results substantiate the notion that the CCL5-mediated changes in Ca2+ occur in these cells through a chemokine receptor-dependent mechanism and not through GPR75.
The ability of CCL5 to interfere with 20-HETE-GPR75 pairing was also observed in endothelial cells. As seen in Figure 5d, pretreatment of HUVECs with CCL5 prevented 20-HETE-mediated changes in iCa2+ (Figure S9A). However, knockdown of GPR75 in HUVECs (Figures 5e and S9D,E) markedly amplified CCL5-stimulated iCa2+ with a threefold increase in signal intensity, reaching upwards of 13-fold. These effects were completely nullified in the presence of the chemokine receptor antagonist cocktail (CRA) (Figures 5e and S9D,E), once again suggesting that CCL5-mediated changes in Ca2+ are not conducted through GPR75. Taken together, our data support the notion that the presence of GPR75 in HUVECs facilitates an association with CCL5 that diverts the chemokine from interacting with its cognate chemokine receptors.
4 |. DISCUSSION
The NC-IUPHAR receptor deorphanization criteria require the use of various pharmacological assays in order to accurately determine the nature of a ligand receptor pairing through the use of agonists, antagonists, binding studies, receptor gene amplification and receptor gene deletion experiments (Davenport et al., 2013). Here, we set out with a clear goal to definitively establish the intimate pairing of 20-HETE and its receptor GPR75. For the first time, direct and specific high-affinity binding between 20-HETE and GPR75 was observed using the SPR technology platform, further substantiating our previous report and providing sensitive measures of binding affinity between the ligand–receptor pair. This high-affinity binding observed between 20-HETE and GPR75 differs greatly from the reported low-affinity binding detected between 20-HETE and GPR40/FFAR1, a non-selective receptor binding medium and long-chain fatty acids, expressed primarily in β-cells of pancreatic islets (Park et al., 2018; Tunaru et al., 2018). Moreover, receptor overexpression studies revealed 20-HETE’s ability to promote rapid and significant increases in intracellular calcium, IP-1 accumulation and β-arrestin recruitment; three key cellular responses that occur specifically through GPR75 and rely on 20-HETE as its cognate ligand. Importantly, 20-HETE binding to and activation of GPR75 (Gq and β-arrestin) was achieved at comparable concentrations known to elicit 20-HETE’s bioactions (Cheng et al., 2012; Garcia et al., 2016). The dependency of 20-HETE on the presence of GPR75 was illustrated by experiments in which knockdown of GPR75 prevented 20-HETE from increasing iCa2+ in primary vascular endothelial cells. Moreover, all of these molecular mechanisms driven by 20-HETE via GPR75 were ablated in the presence of functional antagonists, highlighting the specificity of these interactions. Coupled together with our previous radioligand binding studies and animal experiments (Garcia et al., 2017), the studies presented here establish the close and specific pairing of 20-HETE and GPR75 and call for the classification of GPR75 as a receptor for 20-HETE.
Our current study also sheds light into a paradox that has proposed CCL5 as a GPR75 ligand, making it the fourth receptor alongside CCR1, CCR3 and CCR5 that this promiscuous chemokine influences or interacts with (Dedoni et al., 2018). Early studies by Ignatov et al. (2006) and Liu et al. (2013) stated that CCL5 influences iCa2+ through GPR75. In our hands, CCL5 was able to increase iCa2+ in HTLA and human endothelial cells, but these changes were clearly shown to not be dependent on GPR75. The differential expression patterns of CCL5 receptors across cell lines (e.g., endothelial vs. neuronal) together with sequence differences in GPR75 among species (e.g., mouse vs. human) and the potential existence of multiple GPR75 variants may account for these differences. In addition, our studies show that CCL5’s ability to promote changes in calcium and IP-1 accumulation is actually diminished when there is an over-expression of GPR75. Furthermore, in line with previous reports (Southern et al., 2013), CCL5 was unable to increase recruitment of β-arrestin to GPR75, providing additional evidence that although CCL5 can bind to GPR75, it does not induce any conventional or biased agonist signal through the receptor. Interestingly, for the first time, our data show that CCL5, through its interactions with GPR75, interferes with 20-HETE’s bioactions (Figure 6). Based on our models, we hypothesize that the antagonism by CCL5 of 20-HETE’s ability to activate GPR75 occurs at a distinct site located across the extracellular receptor region. Further studies are necessary to identify the nature of this protein–protein interaction and how it influences 20-HETE binding and GPR75 function. Moreover, special emphasis should be paid to how receptor binding, activation, coupling and downstream signalling cascades are altered across unique cell and tissue types to best uncover new spatiotemporal relationships between these mediators.
FIGURE 6.
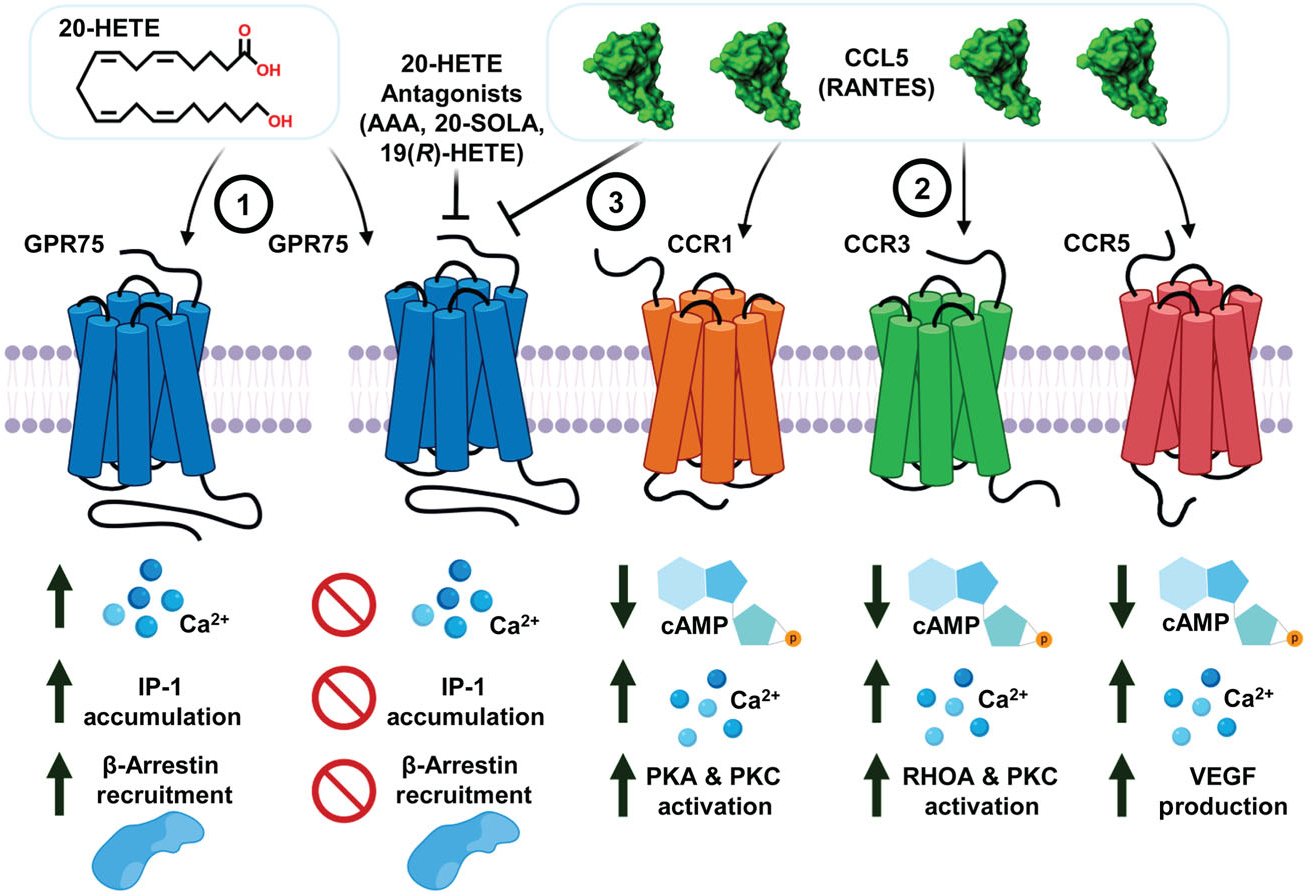
20-HETE promotes Ca2+ signalling, IP-1 accumulation and β-arrestin recruitment through its receptor GPR75. (1) 20-HETE binds to and activates GPR75, setting in motion a signalling program that promotes Gq-coupled changes to calcium, the accumulation of IP-1 and the recruitment of β-arrestin to GPR75. These events precede 20-HETE-mediated changes to pro-inflammatory signals such as the activation of NF-kB, uncoupling of endothelial nitric oxide synthase (eNOS) and the upregulation of angiotensin converting enzyme (ACE). These effects are prevented by 20-HETE receptor antagonists (e.g., AAA, 2,5,8,11,14,17-hexaoxanonadecan-19-yl 20-hydroxyeicosa-6(Z),15(Z)-dienoate (20-SOLA) and 19(R)-HETE). (2) The chemokine CCL5 predominantly interacts with the chemokine receptors CCR1, CCR3 and CCR5 to elicit Gi-coupled changes to cAMP and secondary changes to intracellular calcium. (3) CCL5, through its interaction with GPR75, can prevent and block the 20-HETE-GPR75 pairing and the ensuing changes in both calcium and PLC-mediated signalling cascades
The structure of the 20-HETE-GPR75 ligand–receptor interface revealed a putative ligand-binding site located across TM5 and TM6. Interestingly, recent studies have identified that it is not uncommon for ligands, particularly small lipids, to mediate receptor activation via binding pockets or sites located across the transmembrane domain (Szlenk et al., 2019; Yuan & Xu, 2018). Mutagenesis studies targeting possible hydrogen bond forming residues within the 20-HETE binding site revealed several key attributes of the binding pocket and identified residues critical for lipid-receptor signalling. The S205A and S219A GPR75 mutants exhibit a biased ligand–receptor signal cascade favouring β-arrestin recruitment while the T212N mutant completely disrupted 20-HETE’s ability to activate GPR75. The importance of residues involved in the formation of the hydrophobic pocket for 20-HETE binding to GPR75 merits further investigation. Currently, this binding site is comprised of several residues including Valine209 (V209), Valine215 (V215), Alanine 216 (A216), Leucine335 (L335), Isoleucine339 (I339), Valine 342 (V342) and Leucine246 (L246). An extensive investigation cataloguing this newfound region of interest has the potential to provide immense insight into the properties governing 20-HETE-receptor binding and more importantly, the information gained can be used to design and develop novel 20-HETE receptor blockers and /antagonists.
The studies presented here illustrate the intimate relationship between 20-HETE and GPR75. The dynamic interaction between 20-HETE, CCL5 and GPR75 is striking and speaks to the complexity and intricate nature of all receptor interactions and their respective signalling outcomes. Currently, GPR75 is identified as an understudied protein and listed under the National Institute of Health (NIH) as a candidate under the Illuminating the Druggable Genome (IDG) program as part of the druggable human GPCRome; 20-HETE, a pro-inflammatory and vasoactive lipid (Hoopes et al., 2015), has been proposed as a potent contributor and critical player in various pathologies including hypertension (Garcia et al., 2017; Shekhar et al., 2019), chronic kidney disease (CKD) (Zoccali et al., 2015), ischemic stroke (Crago et al., 2011; Liao et al., 2016; Poloyac et al., 2006), vascular dementia (Shabir et al., 2018), myocardial infarction-driven heart failure (Rocic & Schwartzman, 2018) and obesity-mediated insulin resistance (Gilani et al., 2018, 2021; Issan et al., 2013; Tsai et al., 2009). Most recently, these pathologies have come to the forefront as these underlying medical conditions are high risk factors and can contribute to the severity of COVID-19 outcomes in patients (Schiffrin et al., 2020). Further understanding of the molecular interactions occurring between 20-HETE and the 20-HETE receptor, GPR75, will enable the development of novel blockers, antibodies and biased agonists/ligands that have the potential to provide a potent therapeutic benefit across a vast array of conditions.
20-HETE and CCL5, in their own right, contribute to the development and progression of cardiovascular disease (Noels et al., 2019; Rocic & Schwartzman, 2018). Therefore, additional studies examining the potential for synergism or antagonism between these two mediators particularly in the context of atherosclerosis and myocardial infarction (MI) would benefit the field greatly. With respect to ligand–receptor interactions, context is key. It is still unclear how the 20-HETE-GPR75 pairing influences the renal tubular effects associated with 20-HETE. Moreover, uncovering the presence of splice variants, mutations and how they alter the aetiology of pathologies associated with 20-HETE (i.e., hypertension, diabetes, renal injury and obesity) remain unique avenues left to be explored. The 20-HETE-GPR75 pairing and its role across the brain will also be an interesting topic to explore as a recent study identified GPR75’s expression across cerebral endothelial cells, pericytes, astrocytes and neurons in rats (Gonzalez-Fernandez et al., 2020). The brain has high levels of GPR75 expression (Garcia et al., 2017). Lastly, determining the significance of hydrophobic interactions, identification of novel properties that favour agonists/antagonist binding and assessing the affinity of new agonists and antagonists of this 20-HETE receptor are also just a few of the ever-growing number of topics left to be realized. Nevertheless, the work presented here provides a wealth of new information and a new set of implications with regard to how lipids, chemokines and receptors interact and facilitate intracellular signals.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health grants PO1034300 and HL139793 (MLS), diversity supplement to Victor Garcia HL139793-1S, American Heart Association Scientist Development Grant (16SDG27090040) (EP) and by the Robert A. Welch Foundation (I-0011) (JRF). We acknowledge Dr. William C. Sessa from Yale University for his guidance throughout this project.
Abbreviations:
- 20-HETE
20-hydroxy-5,8,11,14-eicosatetraenoic acid
- AAA
N-disodium succinate-20-hydroxyeicosa-6(Z),15(Z)-diencarboxamide
- CDK
chronic kidney disease
- CRA
chemokine receptor antagonist
- iCa2+
intracellular Ca2+
- IDG
Illuminating the Druggable Genome
- IP-1
inositol phosphate
- SPR
surface plasmon resonance
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
DECLARATION OF TRANSPARENCY AND SCIENTIFIC RIGOUR
This Declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research as stated in the BJP guidelines for Design & Analysis, and Immunoblotting and Immunochemistry, and as recommended by funding agencies, publishers and other organizations engaged with supporting research.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request. Some data may not be made available because of privacy or ethical restrictions.
REFERENCES
- Alexander SPH, Christopoulos A, Davenport AP, Kelly E, Mathie A, Peters JA, Veale EL, Armstrong JF, Faccenda E, Harding SD, Pawson AJ, Sharman JL, Southan C, Davies JA, & CGTP Collaborators. (2019). The concise guide to PHARMACOLOGY 2019/20: G protein-coupled receptors. British Journal of Pharmacology, 176(Suppl 1), S21–S141. 10.1111/bph.14748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Roberts RE, Broughton BRS, Sobey CG, George CH, Stanford SC, Cirino G, Docherty JR, Giembycz MA, Hoyer D, Insel PA, Izzo AA, Ji Y, MacEwan DJ, Mangum J, Wonnacott S, & Ahluwalia A (2018). Goals and practicalities of immunoblotting and immunohistochemistry: A guide for submission to the British Journal of Pharmacology. British Journal of Pharmacology, 175(3), 407–411. 10.1111/bph.14112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Galicia M, Falck JR, Reddy KM, & Roman RJ (1999). 20-HETE agonists and antagonists in the renal circulation. The American Journal of Physiology, 277(5), F790–F796. 10.1152/ajprenal.1999.277.5.F790 [DOI] [PubMed] [Google Scholar]
- Cheng J, Garcia V, Ding Y, Wu CC, Thakar K, Falck JR, Ramu E, & Schwartzman ML (2012). Induction of angiotensin-converting enzyme and activation of the renin-angiotensin system contribute to 20-hydroxyeicosatetraenoic acid-mediated endothelial dysfunction. Arteriosclerosis, Thrombosis, and Vascular Biology, 32(8), 1917–1924. 10.1161/ATVBAHA.112.248344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crago EA, Thampatty BP, Sherwood PR, Kuo CW, Bender C, Balzer J, Horowitz M, & Poloyac SM (2011). Cerebrospinal fluid 20-HETE is associated with delayed cerebral ischemia and poor outcomes after aneurysmal subarachnoid hemorrhage. Stroke, 42(7), 1872–1877. 10.1161/STROKEAHA.110.605816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MJ, Alexander S, Cirino G, Docherty JR, George CH, Giembycz MA, Hoyer D, Insel PA, Izzo AA, Ji Y, MacEwan DJ, Sobey CG, Stanford SC, Teixeira MM, Wonnacott S, & Ahluwalia A (2018). Experimental design and analysis and their reporting II: Updated and simplified guidance for authors and peer reviewers. British Journal of Pharmacology, 175(7), 987–993. 10.1111/bph.14153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport AP, Alexander SP, Sharman JL, Pawson AJ, Benson HE, Monaghan AE, Liew WC, Mpamhanga CP, Bonner TI, Neubig RR, Pin JP, Spedding M, & Harmar AJ (2013). International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list: Recommendations for new pairings with cognate ligands. Pharmacological Reviews, 65(3), 967–986. 10.1124/pr.112.007179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedoni S, Campbell LA, Harvey BK, Avdoshina V, & Mocchetti I (2018). The orphan G-protein-coupled receptor 75 signaling is activated by the chemokine CCL5. Journal of Neurochemistry, 146(5), 526–539. 10.1111/jnc.14463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra S, Sona C, Kumar A, & Yadav PN (2016). Tango assay for ligand-induced GPCR-β-arrestin2 interaction: Application in drug discovery. Methods in Cell Biology, 132, 233–254. 10.1016/bs.mcb.2015.11.001 [DOI] [PubMed] [Google Scholar]
- Elamin E, Masclee A, Dekker J, & Jonkers D (2014). Ethanol disrupts intestinal epithelial tight junction integrity through intracellular calcium-mediated rho/ROCK activation. American Journal of Physiology. Gastrointestinal and Liver Physiology, 306(8), G677–G685. 10.1152/ajpgi.00236.2013 [DOI] [PubMed] [Google Scholar]
- Fan F, Ge Y, Lv W, Elliott MR, Muroya Y, Hirata T, Booz GW, & Roman RJ (2016). Molecular mechanisms and cell signaling of 20-hydroxyeicosatetraenoic acid in vascular pathophysiology. Front Biosci (Landmark Ed), 21, 1427–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan F, Muroya Y, & Roman RJ (2015). Cytochrome P450 eicosanoids in hypertension and renal disease. Current Opinion in Nephrology and Hypertension, 24(1), 37–46. 10.1097/MNH.0000000000000088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Gilani A, Shkolnik B, Pandey V, Zhang FF, Dakarapu R, Gandham SK, Reddy NR, Graves JP, Gruzdev A, Zeldin DC, Capdevila JH, Falck JR, & Schwartzman ML (2017). 20-HETE signals through G-protein-coupled receptor GPR75 (Gq) to affect vascular function and trigger hypertension. Circulation Research, 120(11), 1776–1788. 10.1161/CIRCRESAHA.116.310525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Park EJ, Siragusa M, Frohlich F, Mahfuzul Haque M, Pascale JV, Heberlein KR, Isakson BE, Stuehr DJ, & Sessa WC (2020). Unbiased proteomics identifies plasminogen activator inhibitor-1 as a negative regulator of endothelial nitric oxide synthase. Proceedings of the National Academy of Sciences of the United States of America, 117(17), 9497–9507. 10.1073/pnas.1918761117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Shkolnik B, Milhau L, Falck JR, & Schwartzman ML (2016). 20-HETE activates the transcription of angiotensin-converting enzyme via nuclear factor-κB translocation and promoter binding. The Journal of Pharmacology and Experimental Therapeutics, 356(3), 525–533. 10.1124/jpet.115.229377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawrys O, Huskova Z, Baranowska I, Walkowska A, Sadowski J, Kikerlova S, Vanourkova Z, Honetschlägerova Z, Škaroupkova P, Cervenka L, & Falck JR (2020). Combined treatment with epoxyeicosatrienoic acid analog and 20-hydroxyeicosatetraenoic acid antagonist provides substantial hypotensive effect in spontaneously hypertensive rats. Journal of Hypertension, 38, 1802–1810. 10.1097/HJH.0000000000002462 [DOI] [PubMed] [Google Scholar]
- Gencoglu H, Sahin K, & Jones PM (2019). Determining the insulin secretion potential for certain specific G-protein coupled receptors in MIN6 pancreatic beta cells. Turkish Journal of Medical Sciences, 49(1), 403–411. 10.3906/sag-1712-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilani A, Agostinucci K, Hossain S, Pascale JV, Garcia V, Adebesin AM, Falck JR, & Schwartzman ML (2021). 20-HETE interferes with insulin signaling and contributes to obesity-driven insulin resistance. Prostaglandins & Other Lipid Mediators, 152, 106485. 10.1016/j.prostaglandins.2020.106485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilani A, Pandey V, Garcia V, Agostinucci K, Singh SP, Schragenheim J, Bellner L, Falck JR, Paudyal MP, Capdevila JH, Abraham NG, & Laniado Schwartzman M (2018). High-fat diet-induced obesity and insulin resistance in CYP4a14(/) mice is mediated by 20-HETE. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology, 315(5), R934–R944. 10.1152/ajpregu.00125.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Fernandez E, Staursky D, Lucas K, Nguyen BV, Li M, Liu Y, Washington C, Coolen LM, Fan F, & Roman RJ (2020). 20-HETE enzymes and receptors in the neurovascular unit: Implications in cerebrovascular disease. Frontiers in Neurology, 11, 983. 10.3389/fneur.2020.00983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Zhang W, Giroux C, Cai Y, Ekambaram P, Dilly AK, Hsu A, Zhou S, Maddipati KR, Liu J, Joshi S, Tucker SC, Lee MJ, & Honn KV (2011). Identification of the orphan G protein-coupled receptor GPR31 as a receptor for 12-(S)-hydroxyeicosatetraenoic acid. The Journal of Biological Chemistry, 286(39), 33832–33840. 10.1074/jbc.M110.216564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding SD, Sharman JL, Faccenda E, Southan C, Pawson AJ, Ireland S, Gray AJG, Bruce L, Alexander SPH, Anderton S, Bryant C, Davenport AP, Doerig C, Fabbro D, Levi-Schaffer F, Spedding M, Davies JA, & NC-IUPHAR. (2018). The IUPHAR/BPS guide to PHARMACOLOGY in 2018: Updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Research, 46(D1), D1091–D1106. 10.1093/nar/gkx1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopes SL, Garcia V, Edin ML, Schwartzman ML, & Zeldin DC (2015). Vascular actions of 20-HETE. Prostaglandins & Other Lipid Mediators, 120, 9–16. 10.1016/j.prostaglandins.2015.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatov A, Robert J, Gregory-Evans C, & Schaller HC (2006). RANTES stimulates Ca2+ mobilization and inositol trisphosphate (IP3) formation in cells transfected with G protein-coupled receptor 75. British Journal of Pharmacology, 149(5), 490–497. 10.1038/sj.bjp.0706909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issan Y, Hochhauser E, Guo A, Gotlinger KH, Kornowski R, Leshem-Lev D, Lev E, Porat E, Snir E, Thompson CI, Abraham NG, & Laniado-Schwartzman M (2013). Elevated level of pro-inflammatory eicosanoids and EPC dysfunction in diabetic patients with cardiac ischemia. Prostaglandins & Other Lipid Mediators, 100–101, 15–21. 10.1016/j.prostaglandins.2012.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeze WK, Sassano MF, Huang XP, Lansu K, McCorvy JD, Giguère PM, Sciaky N, & Roth BL (2015). PRESTO-tango as an open-source resource for interrogation of the druggable human GPCRome. Nature Structural & Molecular Biology, 22(5), 362–369. 10.1038/nsmb.3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D, Yi X, Zhang B, Zhou Q, & Lin J (2016). Interaction between CYP4F2 rs2108622 and CPY4A11 rs9333025 variants is significantly correlated with susceptibility to ischemic stroke and 20-Hydroxyeicosatetraenoic acid level. Genetic Testing and Molecular Biomarkers, 20(5), 223–228. 10.1089/gtmb.2015.0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lighter J, Phillips M, Hochman S, Sterling S, Johnson D, Francois F, & Stachel A (2020). Obesity in patients younger than 60 years is a risk factor for COVID-19 hospital admission. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America, 71(15), 896–897. 10.1093/cid/ciaa415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Hassan Z, Amisten S, King AJ, Bowe JE, Huang GC, Jones PM, & Persaud SJ (2013). The novel chemokine receptor, G-protein-coupled receptor 75, is expressed by islets and is coupled to stimulation of insulin secretion and improved glucose homeostasis. Diabetologia, 56(11), 2467–2476. 10.1007/s00125-013-3022-x [DOI] [PubMed] [Google Scholar]
- Noels H, Weber C, & Koenen RR (2019). Chemokines as therapeutic targets in cardiovascular disease. Arteriosclerosis, Thrombosis, and Vascular Biology, 39(4), 583–592. 10.1161/ATVBAHA.118.312037 [DOI] [PubMed] [Google Scholar]
- Padmanabhan S, Wallace C, Munroe PB, Dobson R, Brown M, Samani N, Clayton D, Farrall M, Webster J, Lathrop M, Caulfield M, Dominiczak AF, & Connell JM (2006). Chromosome 2p shows significant linkage to antihypertensive response in the British Genetics of Hypertension Study. Hypertension, 47(3), 603–608. 10.1161/01.HYP.0000197947.62601.9d [DOI] [PubMed] [Google Scholar]
- Park SK, Herrnreiter A, Pfister SL, Gauthier KM, Falck BA, Falck JR, & Campbell WB (2018). GPR40 is a low-affinity epoxyeicosatrienoic acid receptor in vascular cells. The Journal of Biological Chemistry, 293(27), 10675–10691. 10.1074/jbc.RA117.001297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poloyac SM, Zhang Y, Bies RR, Kochanek PM, & Graham SH (2006). Protective effect of the 20-HETE inhibitor HET0016 on brain damage after temporary focal ischemia. Journal of Cerebral Blood Flow and Metabolism: Official Journal of the International Society of Cerebral Blood Flow and Metabolism, 26(12), 1551–1561. 10.1038/sj.jcbfm.9600309 [DOI] [PubMed] [Google Scholar]
- Richardson S, Hirsch JS, Narasimhan M, Crawford JM, McGinn T, Davidson KW, and the Northwell COVID-19 Research Consortium, Barnaby DP, Becker LB, Chelico JD, Cohen SL, Cookingham J, Coppa K, Diefenbach MA, Dominello AJ, Duer-Hefele J, Falzon L, Gitlin J, Hajizadeh N, … Zanos TP (2020). Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA, 323(20), 2052–2059. 10.1001/jama.2020.6775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocic P, & Schwartzman ML (2018). 20-HETE in the regulation of vascular and cardiac function. Pharmacology & Therapeutics, 192, 74–87. 10.1016/j.pharmthera.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer CG, White K, Stohr H, Grimm T, Hutchinson A, Bernstein PS, Lewis RA, Simonelli F, Pauleikhoff D, Allikmets R, & Weber BH (2001). Evaluation of the G protein coupled receptor-75 (GPR75) in age related macular degeneration. The British Journal of Ophthalmology, 85(8), 969–975. 10.1136/bjo.85.8.969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffrin EL, Flack JM, Ito S, Muntner P, & Webb RC (2020). Hypertension and COVID-19. American Journal of Hypertension, 33(5), 373–374. 10.1093/ajh/hpaa057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlakova L, Kikerlova S, Huskova Z, Cervenkova L, Chabova VC, Zicha J, Falck JR, Imig JD, Kompanowska-Jezierska E, Sadowski J, & Kratký V (2018). 20-Hydroxyeicosatetraenoic acid antagonist attenuates the development of malignant hypertension and reverses it once established: A study in Cyp1a1-Ren-2 transgenic rats. Bioscience Reports, 38(5), BSR20171496. 10.1042/BSR20171496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabir O, Berwick J, & Francis SE (2018). Neurovascular dysfunction in vascular dementia, Alzheimer’s and atherosclerosis. BMC Neuroscience, 19(1), 62. 10.1186/s12868-018-0465-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar S, Varghese K, Li M, Fan L, Booz GW, Roman RJ, & Fan F (2019). Conflicting roles of 20-HETE in hypertension and stroke. International Journal of Molecular Sciences, 20(18), 4500. 10.3390/ijms20184500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern C, Cook JM, Neetoo-Isseljee Z, Taylor DL, Kettleborough CA, Merritt A, Bassoni DL, Raab WJ, Quinn E, Wehrman TS, Davenport AP, Brown AJ, Green A, Wigglesworth MJ, & Rees S (2013). Screening beta-arrestin recruitment for the identification of natural ligands for orphan G-protein-coupled receptors. Journal of Biomolecular Screening, 18(5), 599–609. 10.1177/1087057113475480 [DOI] [PubMed] [Google Scholar]
- Szlenk CT, Gc JB, & Natesan S (2019). Does the lipid bilayer orchestrate access and binding of ligands to transmembrane Orthosteric/allosteric sites of G protein-coupled receptors? Molecular Pharmacology, 96(5), 527–541. 10.1124/mol.118.115113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarttelin EE, Kirschner LS, Bellingham J, Baffi J, Taymans SE, Gregory-Evans K, Csaky K, Stratakis CA, & Gregory-Evans CY (1999). Cloning and characterization of a novel orphan G-protein-coupled receptor localized to human chromosome 2p16. Biochemical and Biophysical Research Communications, 260(1), 174–180. 10.1006/bbrc.1999.0753 [DOI] [PubMed] [Google Scholar]
- Tsai IJ, Croft KD, Mori TA, Falck JR, Beilin LJ, Puddey IB, & Barden AE (2009). 20-HETE and F2-isoprostanes in the metabolic syndrome: The effect of weight reduction. Free Radical Biology & Medicine, 46(2), 263–270. 10.1016/j.freeradbiomed.2008.10.028 [DOI] [PubMed] [Google Scholar]
- Tunaru S, Bonnavion R, Brandenburger I, Preussner J, Thomas D, Scholich K, & Offermanns S (2018). 20-HETE promotes glucose-stimulated insulin secretion in an autocrine manner through FFAR1. Nature Communications, 9(1), 177. 10.1038/s41467-017-02539-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson EJ, Walker AJ, Bhaskaran K, Bacon S, Bates C, Morton CE, Curtis HJ, Mehrkar A, Evans D, Inglesby P, [Google Scholar]
- Cockburn J, McDonald HI, MacKenna B, Tomlinson L, Douglas IJ, Rentsch CT, Mathur R, Wong AYS, Grieve R, … Goldacre B (2020). Factors associated with COVID-19-related death using OpenSAFELY. Nature, 584(7821), 430–436. 10.1038/s41586-020-2521-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, & Ren D (2009). The BSA-induced Ca2+ influx during sperm capacitation is CATSPER channel-dependent. Reproductive Biology and Endocrinology: RB&E, 7, 119. 10.1186/1477-7827-7-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, & Xu Y (2018). Recent trends and applications of molecular modeling in GPCR-ligand recognition and structure-based drug design. International Journal of Molecular Sciences, 19(7), 2105. 10.3390/ijms19072105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Deng H, Kemp R, Singh H, Gopal VR, Falck JR, Laniado-Schwartzman M, & Nasjletti A (2005). Decreased levels of cytochrome P450 2E1-derived eicosanoids sensitize renal arteries to constrictor agonists in spontaneously hypertensive rats. Hypertension, 45(1), 103–108. 10.1161/01.HYP.0000150782.28485.91 [DOI] [PubMed] [Google Scholar]
- Zoccali C, Mallamaci F, & Grassi G (2015). 20-Hydroxyeicosatetraenoic acid, a far-reaching autacoid in chronic kidney disease: Hypertension and beyond. Journal of Hypertension, 33(9), 1764–1766. 10.1097/HJH.0000000000000678 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. Some data may not be made available because of privacy or ethical restrictions.


