Abstract
Covalently closed, single-stranded circular RNAs can be produced from viral RNA genomes as well as from the processing of cellular housekeeping noncoding RNAs and precursor messenger RNAs. Recent transcriptomic studies have surprisingly uncovered that many protein-coding genes can be subjected to backsplicing, leading to widespread expression of a specific type of circular RNAs (circRNAs) in eukaryotic cells. Here, we discuss experimental strategies used to discover and characterize diverse circRNAs at both the genome and individual gene scales. We further highlight the current understanding of how circRNAs are generated and how the mature transcripts function. Some circRNAs act as noncoding RNAs to impact gene regulation by serving as decoys or competitors for microRNAs and proteins. Others form extensive networks of ribonucleoprotein complexes or encode functional peptides that are translated in response to certain cellular stresses. Overall, circRNAs have emerged as an important class of RNA molecules in gene expression regulation that impact many physiological processes, including early development, immune responses, neurogenesis, and tumorigenesis.
Keywords: backsplicing, circRNA, microRNA, noncoding RNA, pre-mRNA splicing, translation
INTRODUCTION
Unlike other well-known classes of RNAs that were originally named because of their function [e.g., messenger RNA (mRNA)], cellular location [e.g., small nucleolar RNA (snoRNA)], and/or size [e.g., long noncoding RNA (lncRNA)], circular RNAs were so designated because of their unique covalently closed structures. RNA circles without accessible 5’- and 3’-ends were first reported in plant viroids as rodlike structures that were observable on electron micrographs (Sanger et al. 1976). Since then, additional viroid circular RNA genomes have been identified as well as other distinct types of circular RNAs that are classified according to their mechanisms of production. These include circular transcripts that are generated from the processing of cellular noncoding sequences and those that are processed from eukaryotic precursor mRNAs (pre-mRNAs) in a spliceosome-dependent manner.
Pathogenic viroid circular RNA genomes have largely been identified in plants (Flores et al. 1999, Sanger et al. 1976), but hepatitis δ virus (HDV), a satellite virus of hepatitis B virus, is also a circular RNA (Kos et al. 1986) (Table 1a). These transcripts replicate using rolling-circle RNA polymerization followed by cleavage catalyzed by either a host enzyme or embedded ribozyme sequences. The intermediate sequences are then ligated by host enzymes or intramolecular self-ligation to form additional copies of mature circular RNA genomes (Côté & Perreault 1997; Flores et al. 1999, 2011). Beyond viroids, circular RNAs are processed from a variety of noncoding sequences in cells, including the noncoding regions of mitochondrial RNAs, ribosomal RNAs (rRNAs), and transfer RNAs (tRNAs) [see review by Lasda & Parker (2014)]. For example, groups I and II self-splicing introns (Cech 1990, Nielsen et al. 2003) are processed to form circular RNAs in both bacteria and eukaryotes (Inoue et al. 1986, Li-Pook-Than & Bonen 2006, Molina-Sánchez et al. 2006, Tabak et al. 1987) Table 1b). These include the introns of rRNA precursors in the protozoa Tetrahymena (Grabowski et al. 1981) and the archaebacterium Desulfurococcus mobilis (Kjems & Garrett 1988). Furthermore, the intron of tRNATrp in the euryarchaeote Haloferax volcanii forms a stable RNA circle after being cleaved from precursor RNAs and ligated by an RNA ligase (Clouet d’Orval et al. 2001, Salgia et al. 2003, Singh et al. 2004). Circular RNAs have also been found to be processed from some snoRNAs, ribonuclease (RNase) P RNA, and 7S RNA in archaea (Danan et al. 2012, Tang et al. 2002).
Table 1.
A chronological view of circular RNA discoveries
| Different types of RNA circles | Methods to study RNA circles | Reference(s) |
|---|---|---|
| a: Covalently closed viral circular RNA genomes | ||
| Circular viroid RNAs | Electrophoresis, thermal denaturation, and electron micrograph | Sanger et al. 1976 |
| Circular RNA genome of hepatitis delta virus | Electron micrograph and Northern blotting | Kos et al. 1986 |
| Self-cleavage and self-ligation of rolling viroid RNA circles in vitro | Primer extension, self-cleavage, and ligation | Côté & Perreault 1997 |
| b: Circular (intermediate) RNAs processed from noncoding sequences | ||
| Yeast mitochondrial RNA circles from cytochrome oxidase gene intron | Electrophoresis, electron micrograph, and RNA–DNA hybridization | Arnberg et al. 1980, Hensgens et al. 1983, Tabak et al. 1987 |
| Circular RNA from self-spliced introns of Tetrahymena rRNA precursors | Electrophoresis, electron micrograph, and endonuclease digestion | Grabowski et al. 1981, Inoue et al. 1986 |
| Circular RNA from introns of archaeal rRNA precursors | Northern blotting, sequencing, and in vitro splicing | Kjems & Garrett 1988 |
| Circular RNA from introns of archaeal tRNA precursors | Northern blotting, in vitro ligation or analysis, and electrophoresis | Clouet d’Orval et al. 2001, Salgia et al. 2003 |
| Various types of circular RNAs from archaeal noncoding transcriptomes | Northern blotting, RT-PCR, and electrophoresis | Tang et al. 2002, Danan et al. 2012 |
| c: Circular RNAs processed from RNA polymerase II–transcribed precursor RNAs (spliceosome dependent) | ||
| Circular RNA in HeLa cytoplasm with unknown origin and function | Electron micrograph | Hsu & Coca-Prados 1979 |
| Abnormally spliced (scrambled) transcripts at consensus splice sites | PCR cloning, sequencing, and nonpolyadenylated RNA isolation | Nigro et al. 1991 |
| Splicing of ets-1 exons in an aberrant order at consensus splice sites | PCR and RNase protection | Cocquerelle et al. 1992 |
| Circular transcripts of human ets-1 by nuclear pre-mRNA missplicing | PCR and poly(A)-RNA sucrose gradient | Cocquerelle et al. 1993 |
| Circular transcripts of mouse testis-determining gene Sry | RACE, RNase protection, and RNase H digestion | Capel et al. 1993 |
| Inverted repeats required for mouse Sry transcript circularization | Northern blotting, RT-PCR, and molecular cloning | Dubin et al. 1995 |
| Translation of circular RNAs by eukaryotic translational apparatus | In vitro circularization and translation | Chen & Sarnow 1995 |
| Spliceosome-dependent Sry circular RNA formation and recapitulation | Assays of RACE, RNase protection, and RNase H digestion | Pasman et al. 1996 |
Abbreviations: cDNA, complementary DNA; pre-mRNA, precursor messenger RNA; RACE, rapid amplification of cDNA ends; RNase, ribonuclease; rRNA, ribosomal RNA; RT-PCR, reverse transcription polymerase chain reaction; tRNA, transfer RNA.
Another major group of circular RNA (and the focus of this review, named circRNA hereafter) is eukaryotic specific and is produced by the splicing of pre-mRNAs in a spliceosome-dependent manner [for reviews, please see Chen (2016, 2020); Kristensen et al. (2019); Lasda & Parker (2014); Li et al. (2018); Wilusz (2018); and Xiao et al. (2020)]. Eukaryotic circular transcripts were first detected by electron microscopy in the late 1970s, but their origin and function were unknown at that time (Hsu & Coca-Prados 1979) (Table 1c). Not until the 1990s was it realized that some of these detected transcripts likely represented exons derived from protein-coding genes that were covalently joined at consensus splice sites to form mature circles (Capel et al. 1993; Cocquerelle et al. 1992, 1993; Dubin et al. 1995; Nigro et al. 1991; Pasman et al. 1996; Zaphiropoulos 1996). Due to their generally low levels of expression (~1/100th the level of the normally spliced transcript generated from the host gene) (Capel et al. 1993, Cocquerelle et al. 1993, Dubin et al. 1995, Pasman et al. 1996, Zhang et al. 2016a), whether these RNA circles represented biological noise of pre-mRNA splicing reactions was obscure. Nonetheless, Capel et al. (1993) found that more than 90% of the transcripts derived from the mouse Sry gene, which determines sex in mammals, were circular in adult mouse testis. This was the first indication that circRNAs can be the predominant outputs of some protein-coding genes, suggestive of functionality.
The advent of deep sequencing technologies coupled to methods that enrich for polyadeny-lated RNAs (mRNA-seq) uncovered pervasive transcription in eukaryotes (Cloonan et al. 2008, Graveley 2008, Mortazavi et al. 2008, Pan et al. 2008, Wang et al. 2008, Wilhelm et al. 2008). However, the lack of a poly(A) tail on circular RNAs caused the expression signals of these transcripts to be limited in mRNA-seq studies (Burd et al. 2010). This situation turned around when deep sequencing of total RNA (depleted of rRNAs) or nonpolyadenylated RNAs came to the fore (Jeck et al. 2013, Memczak et al. 2013, Salzman et al. 2012, Yang et al. 2011). These approaches enabled the genome-wide identification of two major classes of eukaryotic circular RNAs that can be generated when the spliceosome acts on RNA polymerase II (Pol II) transcripts. Circular intronic RNAs (ciRNAs), also known as stable intronic sequence RNAs (sisRNAs), are generated when a subset of intron lariats fails to be debranched (Talhouarne & Gall 2014, Zhang et al. 2013). In addition, it was determined that many exons can form circRNAs when pre-mRNAs are backspliced (Ashwal-Fluss et al. 2014, Hansen et al. 2013, Jeck et al. 2013, Liang & Wilusz 2014, Memczak et al. 2013, Salzman et al. 2012, X.-O. Zhang et al. 2014) (Figure 1a). Building off these initial observations, a plethora of studies have now characterized the expression patterns, biogenesis mechanisms, and function of these spliceosome-dependent circRNAs. Here, we summarize recent progress in this field that has revealed the tremendous regulatory potential of eukaryotic circRNAs produced from the backsplicing of pre-mRNA exons.
Figure 1.
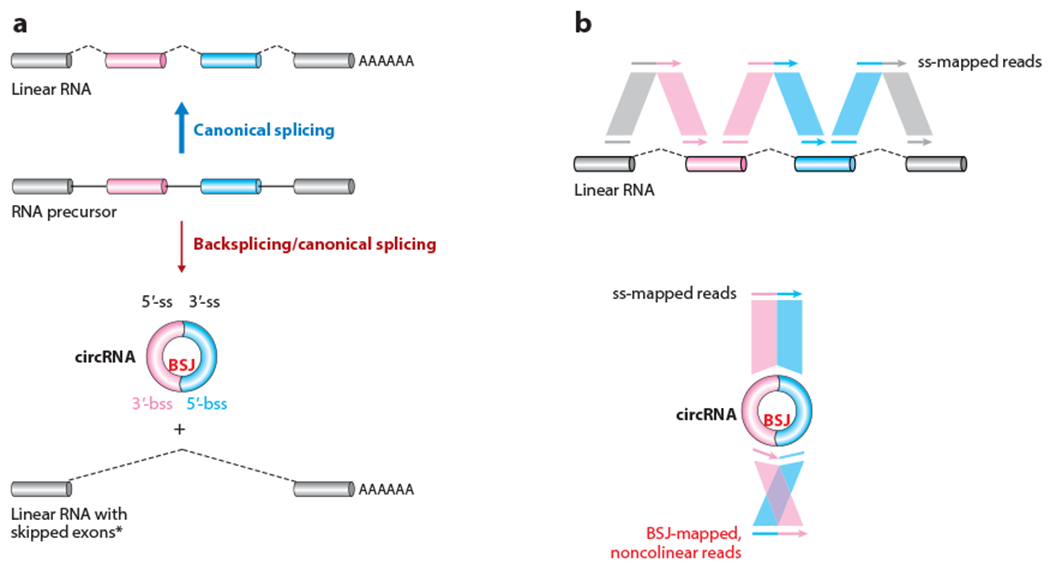
circRNAs can be produced from the backsplicing of precursor (m)RNAs. (a, top) An RNA precursor containing multiple exons can be subjected to canonical splicing, leading to the formation of a linear RNA with all exons included. (Bottom) In contrast, backsplicing and canonical splicing can lead to the formation of a circRNA with two exons, and an alternatively spliced linear RNA with skipped circle-forming exons. Note that the alternatively spliced linear RNA is only sometimes observed, as indicated by the asterisk. Panel a adapted with permission from X. Li et al. (2018); copyright 2018 Elsevier. (b) Short RNA-sequencing reads are mapped to (top) the splicing junction sites of a linear RNA and (bottom) the BSJ site of a circRNA. The BSJ-mapped, noncolinear reads are critical for identification of circRNAs from transcriptomic studies. Abbreviations: BSJ, backsplicing junction; bss, backsplice site; circRNA, circular RNA from exon backsplicing; mRNA, messenger RNA; ss, splice site.
GENOME-WIDE PROFILING OF CIRCRNAS
Because circRNAs are often coexpressed with their cognate linear (m)RNAs from their host gene loci, the primary sequences of the mature linear RNAs and circRNAs often fully overlap except for the unique backsplicing junction (BSJ) present in circRNAs (Figure 1a). To enable efficient circRNA profiling and characterization, it thus has been necessary to develop specific biochemical enrichment strategies for circRNAs as well as computational pipelines that identify sequencing reads that specifically map to BSJs (Chen 2016, 2020; Jeck & Sharpless 2014; Kristensen et al. 2019; Szabo & Salzman 2016) (Figure 1b).
circRNA Enrichment Strategies
circRNAs lack poly(A) tails and most are expressed at low levels, so different strategies have been applied to biochemically enrich circRNAs prior to performing short-read deep sequencing (Ma et al. 2021). At a minimum, abundant rRNAs must be depleted prior to sequencing, and this approach (ribo– RNA-seq) has revealed hundreds of circRNAs that are expressed at levels comparable to those of their cognate linear RNAs (Jeck et al. 2013, Memczak et al. 2013, Salzman et al. 2012). Ribo– RNA-seq (Jeck et al. 2013, Memczak et al. 2013, Salzman et al. 2012, X.-O. Zhang et al. 2016) allows a direct quantitative comparison between the expression levels of circRNAs and those of their cognate linear RNAs (Ma et al. 2019), as both nonpolyadenylated and polyadenylated RNAs are sequenced. To enrich for circRNAs, one can select for nonpolyadenylated RNAs by depleting both rRNAs and polyadenylated (m)RNAs (Yang et al. 2011). By comparing the data obtained from poly(A)– RNA-seq libraries to standard oligo(dT)-primed poly(A)+ RNA-seq libraries, many excised introns and exons can be detected that correspond to ciRNAs (Zhang et al. 2013) and circRNAs (Salzman et al. 2012; X.-O. Zhang et al. 2014, 2016), respectively, including some with complex alternative splicing patterns.
To go even further, ribonuclease R (RNase R), an exoribonuclease that degrades most [but not all; see Panda et al. (2017) and Xiao & Wilusz (2019)] linear RNAs (Suzuki et al. 2006), can be used to enrich circRNAs in ribo– or poly(A)– RNA fractions (Jeck et al. 2013, X.-O. Zhang et al. 2014). Indeed, many more individual circRNAs can be identified using the RNase R RNA-seq approach than with ribo– or poly(A)– RNA-seq (Jeck et al. 2013, Memczak et al. 2013, Salzman et al. 2012, X.-O. Zhang et al. 2014), confirming the robustness of RNase R for circRNA enrichment. RNase R treatment additionally serves as a useful means to detect or remove false positive linear RNAs. Trans-splicing events and artifacts of reverse transcriptase template switching can result in sequencing reads that resemble BSJs, but such reads should be depleted after RNase R treatment (Chuang et al. 2018, Jeck & Sharpless 2014, Szabo & Salzman 2016, Tang et al. 2018, Yu et al. 2014). RNase R has thus been widely used for circRNA validation (Burd et al. 2010, Hansen et al. 2013, Memczak et al. 2013, Salzman et al. 2012), but note that some circRNAs, such as cerebellar degeneration-related protein 1 antisense transcript (CDR1as, also known as circular RNA sponge for miR-7, ciRS-7) (Hansen et al. 2011, 2013; Memczak et al. 2013), have been observed to sometimes be partially depleted in RNase R RNA-seq experiments (Jeck et al. 2013). This is because circRNAs, especially long ones, can be partially degraded by prolonged RNase R incubation (Zhang et al. 2016b). Additional tips to ensure robust circRNA studies have been well summarized recently (Dodbele et al. 2021, Li et al. 2018).
Bioinformatic and Sequencing Approaches for circRNAs
To then identify reads or fragments mapping to BSJs of circRNAs in short-read sequencing data, multiple computational pipelines have been developed that employ distinct strategies (Gao & Zhao 2018; Hansen 2016, 2018; López-Jiménez et al. 2018; Szabo & Salzman 2016). These pipelines have variable efficiencies and accuracies for circRNA profiling, so the application of two or more pipelines has been recommended to reduce circRNA prediction biases (Hansen 2016, 2018; X. Zeng et al. 2017). Nonetheless, all of these pipelines are generally consistent on the identification of highly expressed circRNAs (Hansen 2016, 2018).
Due to the limited read lengths that can be achieved using short-read sequencing data, the exact internal sequences (away from the BSJ) of circRNAs are often ambiguous, especially for long circRNAs. There has thus recently been a significant effort to develop long-read sequencing platforms (Stark et al. 2019) and their corresponding bioinformatic pipelines for genome-wide circRNA profiling, mostly using nanopore technologies (Liu et al. 2021,Rahimi et al. 2021b, Xin et al. 2021, J. Zhang et al. 2021) [for a review, please see Rahimi et al. (2021a) and Zhang & Zhao (2021)]. Compared to short-read sequencing, long-read sequencing approaches have provided more accurate information on the alternative splicing landscape of circRNAs, including the identification of intron retention events, microexons, and exons that are circRNA specific (not observed in the associated cognate linear RNA) (Rahimi et al. 2021b). However, current long-read sequencing approaches can suffer from a number of disadvantages, including high cost, biased enrichment of circRNAs of different lengths, and high sequencing error rates (Rahimi et al. 2021a).
There is also much interest in characterizing the transcriptome, including circRNAs, not just in bulk cell populations but at single-cell or nucleus resolution (Slyper et al. 2020, Stark et al. 2019). Most of these current approaches require the complementary DNA to be primed and amplified with oligo(dT) primers and thus they largely miss circRNA expression. Nonetheless, using random primers, circRNA profiling was obtained at single-cell resolution from mouse preimplantation embryos (Fan et al. 2015). In the future, with improved technologies for circRNA enrichment, long-read sequencing, and advanced computational pipelines, even more precise circRNA expression profiles across cell lines and tissues should be obtained.
Genomic Expression Patterns and Features of circRNAs
circRNAs have now been profiled in a growing number of eukaryotic cell types and tissues, with several public databases summarizing some of the sequencing data (Dong et al. 2018, Glazar et al. 2014, Wu et al. 2020). These efforts have revealed that circRNAs are typically produced along with their cognate linear RNAs from the host gene loci (Figure 1a), but circRNAs accumulate to much lower levels (Jeck et al. 2013, Memczak et al. 2013, Salzman et al. 2012, X.-O. Zhang et al. 2014). A small portion of circRNAs can be detected at higher levels than their cognate linear RNAs (Salzman et al. 2012, 2013), partially due to their natural resistance to degradation by exonucleases. Across tissue types, circRNAs often accumulate to the highest levels in brains, where their levels increase during neuronal differentiation (Zhang et al. 2016a) and are associated with neuronal development and plasticity (Rybak-Wolf et al. 2015, You et al. 2015). These observations suggest that circRNAs can accumulate in slowly dividing cells. Consistent with this idea, decreased circRNA levels have been detected in rapidly dividing colorectal cancer cell lines (Bachmayr-Heyda et al. 2015) and prostate cancers (S. Chen et al. 2019, Vo et al. 2019).
Most circRNAs are derived from previously annotated exons (exons observed in linear RNAs) that are flanked by canonical splice sites, consistent with a critical role for the spliceosome in their biogenesis (Jeck et al. 2013, Liang & Wilusz 2014, Memczak et al. 2013, Salzman et al. 2012, Starke et al. 2015, X.-O. Zhang et al. 2014). circRNAs usually contain two to three exons that are derived from the middle of their host genes (X.-O. Zhang et al. 2014), although they can also sometimes be generated from readthrough transcripts (Liang et al. 2017, Vo et al. 2019). Strikingly, the introns flanking circRNA-forming exons are often much longer than those flanking non-circRNA-forming exons, and this has been observed in multiple species, including flies, pig, rice, and humans (Jeck et al. 2013, Kramer et al. 2015, Lu et al. 2015, Salzman et al. 2013, Veno et al. 2015, X.-O. Zhang et al. 2014). Repetitive elements, such as Alu in human and SINE in mouse, are often enriched in these long flanking introns that promote circRNA biogenesis (Jeck & Sharpless 2014, Jeck et al. 2013, Liang & Wilusz 2014, X.-O. Zhang et al. 2014).
Interestingly, multiple circRNAs can be generated from individual gene loci in a process known as alternative circularization (X.-O. Zhang et al. 2014). This includes alternative backsplicing events (the use of different splice sites to form the BSJ) as well as all four canonical types of alternative splicing (cassette exon, intron retention, and alternative 5′- and 3′-splice site selection) within circRNA-forming exons (Gao et al. 2016, X.-O. Zhang et al. 2016). For example, two major circRNA transcripts can be produced from the human CAMSAP1 locus, and they contain the same BSJ but different internal structures: with or without a retained intron (Salzman et al. 2013; X.-O. Zhang et al. 2014, 2016). Likewise, the human XPO1 locus produces two circRNA transcripts with the same BSJ that differ by the inclusion of a cassette exon (X.-O. Zhang et al. 2016).
REGULATION OF CIRCRNA PRODUCTION, CONFORMATION, AND TURNOVER
In agreement with the original reports that identified sporadic circRNAs in eukaryotes (Capel et al. 1993; Cocquerelle et al. 1992, 1993; Nigro et al. 1991), genome-wide profiling has now shown that most circRNAs are generated via backsplicing reactions when the spliceosome joins a downstream 5’-splice site to an upstream 3’-splice site (Jeck et al. 2013; Liang & Wilusz 2014; Memczak et al. 2013; Salzman et al. 2012, 2013; X.-O. Zhang et al. 2014). Backsplicing can thus be viewed as a type of alternative splicing (Gao et al. 2016, Jeck & Sharpless 2014, X.-O. Zhang et al. 2016), with circRNA steady-state levels reflecting the dynamic equilibrium of their production and degradation rates (Rabani et al. 2011). Here, we focus on the predominant mechanisms that regulate circRNA levels, but note that a few circRNAs may be generated by the U12 minor spliceosome (Szabo et al. 2015).
Coupling with Transcription
The processes of mRNA capping, splicing, and polyadenylation are closely coupled with Pol II transcription (Bentley 2014). Backsplicing is likewise associated with Pol II transcription, although there is evidence that circRNA biogenesis can occur either co- or posttranscriptionally (Ashwal-Fluss et al. 2014, Zhang et al. 2016a). On the one hand, chromatin-associated circRNAs have been identified from fly heads and mouse liver (Ashwal-Fluss et al. 2014), suggestive of cotranscriptional production. Mutants of the human Pol II large subunit that alter transcription elongation rates (TERs) (Fong et al. 2014) likewise suggest the cotranscriptional nature of circRNA formation, as fast Pol II produces more circRNAs and slow Pol II produces fewer (Ashwal-Fluss et al. 2014, Zhang et al. 2016a) (Figure 2a). The mean TER of circRNA-producing gene loci is higher than that of non-circRNA-producing ones (Zhang et al. 2016a), consistent with the notion that fast transcription favors backsplicing. Fast transcription may facilitate cross talk between distal splice sites (both donor and acceptor) as well as RNA pairing between distal complementary sequences in flanking introns (discussed further below in the section titled Regulation of Backsplicing by cis-Elements), but the detailed mechanisms remain poorly understood.
Figure 2.
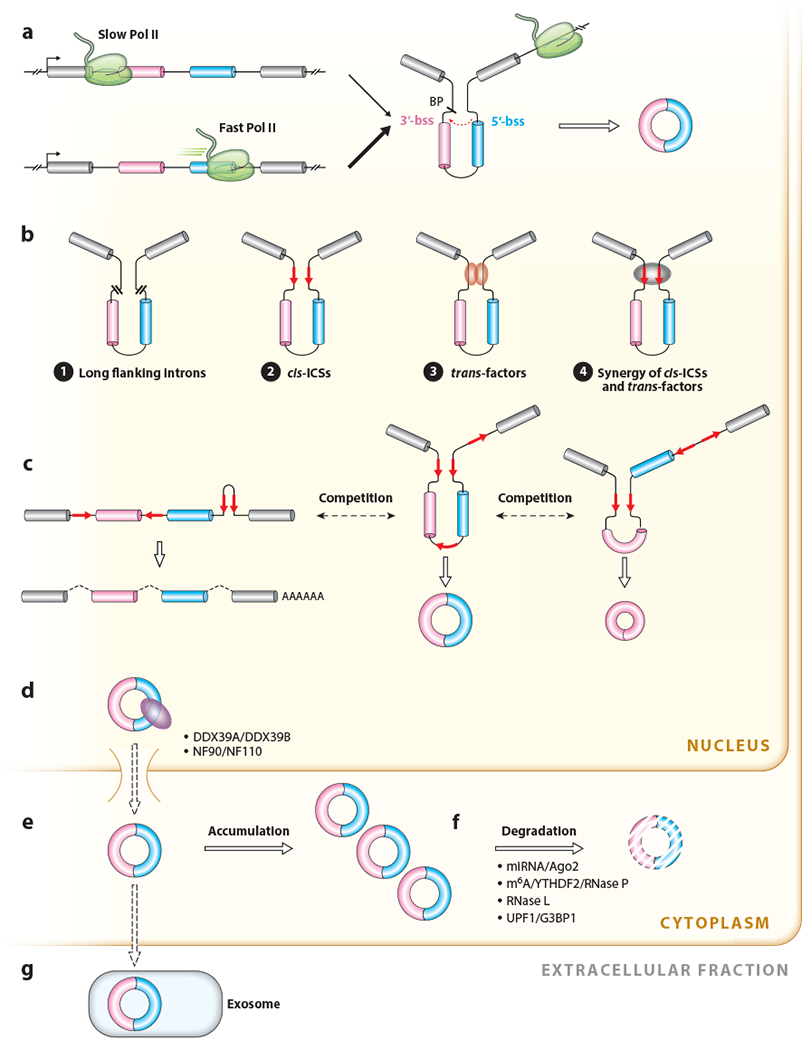
The life cycle of circRNAs in cells. (a) Backsplicing is coupled to Pol II transcription. Fast Pol II elongation can lead to enhanced efficiency of backsplicing catalyzed by the spliceosome. (b) Regulation of backsplicing. (❶) Long introns usually flank circRNA-forming exons. (❷) ICSs (red arrows) in flanking introns of circRNA-forming exons can facilitate backsplicing by forming transient intronic RNA duplexes that bring the splice sites into close proximity. (❸) trans-factors bind to intronic ICSs or other sequences in the pre-mRNA to directly bridge distal splice sites to promote backsplicing. (❹) cis- and trans-factors synergistically modulate exon backsplicing. (c) Competition of RNA pairing across introns or within an intron modulates splicing and backsplicing reactions. In this example, when repeats flanking exons 2 and 3 base pair to one another, (middle) a two-exon circRNA is produced. In contrast, when repeats flanking exon 2 base pair to one another, (right) a single exon circRNA is produced. (d) circRNA export is modulated by different proteins and occurs in a length-dependent manner. (e) circRNAs are stable in the cytoplasm and sometimes can accumulate to high levels. (f) Pathways of circRNA degradation in the cytoplasm. (g) Secretion of circRNAs from cells in exosomes. Abbreviations: BP, branch point; bss, backsplicing site; circRNA, circular RNA; ICS, intronic complementary sequence; m6A, N6-methyladenosine; miRNA, microRNA; Pol II, RNA polymerase II; pre-mRNA, precursor messenger RNA; RNase, ribonuclease.
On the other hand, analyzing the correlation between the 3’-end processing of pre-RNAs and the production of circRNAs from expression vectors revealed that backsplicing can occur posttranscriptionally (Liang et al. 2017, 2021; Liang & Wilusz 2014). Only when a downstream polyadenylation signal (or an alternative 3’-end processing signal) was present could circRNAs be efficiently generated from expression vectors (Liang & Wilusz 2014). Consistent with this observation, 4-thiouridine (4sU) pulse-chase labeling of nascent RNAs revealed that most BSJ-mapped reads could only be observed after prolonged 4sU treatments when the transcription of almost all circRNA-producing genes has been completed (Zhang et al. 2016a). This suggests most backsplicing events occur posttranscriptionally.
Interplay with Canonical Splicing
Given that the majority of backsplicing events are produced from exons with canonical splice sites that are also observed in linear RNAs (Ashwal-Fluss et al. 2014; X. Li et al. 2019; Liang & Wilusz 2014; Starke et al. 2015; Vo et al. 2019; Wang & Wang 2015; X.-O. Zhang et al. 2014, 2016), there is inevitably an interplay or competition between backsplicing and canonical splicing events. There are currently two known ways by which the spliceosome generates circRNAs that differ depending on the order of splicing events (Chen & Yang 2015, Jeck & Sharpless 2014, Jeck et al. 2013). In the direct backsplicing model, backsplicing preferentially happens before canonical splicing, resulting in production of a circRNA and a splicing intermediate that is sometimes further processed to a mature linear RNA that lacks the circRNA-forming exon(s) (Chen & Yang 2015, Jeck & Sharpless 2014, Jeck et al. 2013). In contrast, in the lariat intermediate model, a canonical splicing event happens first to produce a mature linear RNA as well as an intron lariat intermediate containing skipped exons. The latter is subsequently backspliced to produce a circRNA consisting of the exons that were originally skipped (Barrett et al. 2017, Chen & Yang 2015, Jeck & Sharpless 2014, Jeck et al. 2013). Stable alternatively spliced linear RNAs lacking the circRNA-forming exon(s) are produced at some circRNA-producing loci (Ashwal-Fluss et al. 2014, Kelly et al. 2015, Zaphiropoulos 1996, X.-O. Zhang et al. 2014), but how cells determine which mechanism is used to generate a given circRNA remains poorly understand.
Regulation of Backsplicing by cis-Elements
Numerous lines of evidence have shown that both cis-elements and trans-factors, acting in both independent and synergistic manners, can facilitate backsplicing [see reviews by Chen (2020) and Xiao et al. (2020)] (Figure 2b). So far, no specific exonic motifs have been found to be generally required for backsplicing. Instead, circRNA production is typically driven by the flanking intronic sequences. The presence of long introns flanking the circRNA-forming exons appears to be an intrinsic determinant for circRNA biogenesis in some species, including fly (Westholm et al. 2014) and rice (Lu et al. 2015) (Figure 2b). Within these long flanking introns are often orientation-opposite intronic complementary sequences (ICSs) (most often, but not always, from repetitive elements) that can base pair, especially in mammals (Barrett et al. 2015, Capel et al. 1993, Jeck et al. 2013, Kramer et al. 2015, Liang & Wilusz 2014, X.-O. Zhang et al. 2014) (Figure 2b). Consistent with ICSs having a critical function in promoting backsplicing, circRNA formation is impaired when ICSs are disrupted or deleted from endogenous gene loci (Xia et al. 2018, Zhang et al. 2016a, Zheng et al. 2016), and novel circRNAs can be produced in cancer cells when chromosomal translocations cause the formation of new ICS pairs (Guarnerio et al. 2016, Wu et al. 2019).
Cross-species comparisons have shown that a small portion of circRNAs are conserved between human and mouse, and the expression of these circRNAs is correlated with the co-occurrence of ICSs in both species (Dong et al. 2017). In primates, complementary inverted-repeat Alu elements in the flanking introns play the largest role in driving circRNA production, while B1/B2/B4 SINEs contribute the most in mice (Dong et al. 2017). It nonetheless has been shown that ICSs as short as 30–40 nucleotides are sufficient to promote circRNA production from expression vectors (Liang & Wilusz 2014, Starke et al. 2015).
Most introns contain many repetitive elements, which provides the opportunity for intronic pairing to occur between distinct sets of ICSs and the generation of complex patterns of circRNA expression (Figure 2c). For example, when ICSs in an individual intron (rather than in separate introns) base pair, a linear RNA tends to be produced, and circRNA formation is decreased (X.-O. Zhang et al. 2014) (Figure 2c). In contrast, the base pairing of different combinations of ICSs that bracket alternative backsplice sites can lead to alternative backsplice site selection to produce multiple circRNAs from a single gene locus (Gao et al. 2016; X.-O. Zhang et al. 2014, 2016) (Figure 2c).
Regulation of Backsplicing by trans-Factors
In addition to being regulated by cis-elements, backsplicing reactions are further controlled by trans-acting factors that often act synergistically with the associated intronic cis-elements (Kramer et al. 2015). Modulating components of the spliceosomal machinery can affect the efficiency of splicing reactions. Notably, the depletion of core spliceosome components, including those in the U1 and U2 small nuclear ribonucleoproteins (snRNPs), results in circRNA upregulation and concomitant downregulation of linear RNAs (Liang et al. 2017). Increased circRNA expression was likewise found in rat neurons treated with the splicing inhibitor isoginkgetin (Wang et al. 2019). Beyond core spliceosome components, many RNA-binding proteins (RBPs) have now been shown to associate with the introns flanking circRNA-producing exons to modulate circRNA expression levels (Figure 2b). This includes a number of serine-arginine proteins, heterogeneous nuclear ribonucleoproteins (hnRNPs) (Kramer et al. 2015) (e.g., hnRNP L) (Fei et al. 2017), and other known regulators of alternative splicing such as RNA-binding motif protein 20 (RBM20) (Khan et al. 2016) and FUS (Errichelli et al. 2017). In some cases, it is well understood how RBP binding can lead to physiologically relevant changes in gene outputs. For example, in fly, muscleblind (MBL, also known as MBNL1) directly interacts with the MBL pre-mRNA to facilitate circMBL biogenesis, thereby limiting linear MBL mRNA (and hence MBL protein) expression (Ashwal-Fluss et al. 2014). In humans, the splicing regulator Quaking associates with the introns flanking many circRNAs to promote their production during the epithelial-to-mesenchymal transition (Conn et al. 2015).
A significant number of RBPs directly bind to ICSs through their double-stranded RNA (dsRNA)-binding domains (dsRBDs) to modulate the efficiency of backsplicing reactions (Figure 2b). This includes adenosine deaminase acting on RNA 1 (ADAR1), which catalyzes adenosine-to-inosine editing to disrupt the pairing of ICSs and reduce circRNA production in worm and human (Ivanov et al. 2015, Rybak-Wolf et al. 2015). The dsRBD-containing nuclear RNA helicase DHX9 can likewise unwind paired intronic repeat elements that flank circRNA-forming exons to inhibit backsplicing reactions (Aktaş et al. 2017, Ottesen et al. 2019). However, some dsRBD-containing RBPs bind to ICSs to promote circRNA production, most notably nuclear factor 90 (NF90) and NF110 (X. Li et al. 2017) and the splicing factor SFPQ (Stagsted et al. 2021). Considering that all of these RBPs have overlapping binding capacities to bind to ICSs, there must be cross talk with one another, but key details of how they combinatorially dictate circRNA levels remain elusive. A genome-wide screen has further identified several dozen additional RBPs that also likely regulate circRNA production (X. Li et al. 2017). Further investigations are thus warranted to comprehensively understand the mechanisms of how different RBPs collectively modulate circRNA production and the choice between backsplicing versus canonical splicing.
Modification, Export, and Stability of circRNAs
Once produced, circRNAs can undergo nucleotide modifications, export to the cytoplasm, and ultimately degradation. Genome-wide analyses suggest that N6-methyladenosine (m6A) modifications occur in circRNAs at levels similar to those observed in linear RNAs (Y.G. Chen et al. 2019, Shi et al. 2019, Zhou et al. 2017), and this can have multiple potential functional consequences. For example, m6A modifications may affect circRNA nucleocytoplasmic export, possibly in a manner similar to how linear mRNA export can be promoted by the m6A-binding protein YTHDC1 (Roundtree et al. 2017). With the help of YTHDF3, another m6A-binding protein (A. Li et al. 2017), a noncanonical eIF4G protein (eIF4G2), can be recruited to m6A-circRNAs to initiate cap-independent translation from internal ribosome entry site (IRES) sequences (Yang et al. 2017) (discussed further below in the section titled Translatable circRNAs). It has further been shown that some m6A-circRNAs can be recognized by YTHDF2 and HRSP12 (also known as 2-iminobutanoate or 2-iminopropanoate deaminase) (Jarrous 2017), enabling recruitment of the ribonuclease complex RNase P/MRP for circRNA degradation (Park et al. 2019).
Similar to most linear RNAs, fully processed circRNAs are typically exported to the cytoplasm (Jeck et al. 2013, Salzman et al. 2012) (Figure 2d), although a small number of incompletely processed circRNAs, such as those with retained introns, remain in the nucleus (Conn et al. 2017, Z. Li et al. 2015, Veno et al. 2015). The detailed mechanism of circRNA nucleocytoplasmic export has yet to be fully characterized, although some key aspects have been revealed. A small scale RNA interference (RNAi) screen in fly cells showed that circRNA export occurs in a length-dependent manner and that the ATP-dependent RNA helicase Hel25E is required for the export of long (>800 nucleotides), but not short, circRNAs (Huang et al. 2018). Similar regulation was observed in human cells: UAP56 (also known as DDX39B), a homolog of Hel25E, controls the export of long (>1,298 nucleotides) circRNAs, whereas URH49 (also known as DDX39A), another homolog of Hel25E, controls the export of short (<356 nucleotides) circRNAs (Huang et al. 2018). Strikingly, the key amino acid motif responsible for the export of long circRNAs is highly conserved between fly Hel25E and human UAP56 (but is absent from URH49), and UAP56 could rescue the functional loss of Hel25E on long circRNA export in fly cells (Huang et al. 2018). Nonetheless, the factors that control the export of medium-length circRNAs (e.g., those between 400 and 1,300 nucleotides in human cells) remain unclear. NF90/NF110, which shuttle between the nucleus and cytoplasm, may mediate the export of some circRNAs, as these factors can interact with both flanking ICSs in the nucleus and fully processed circRNAs in the cytoplasm (X. Li et al. 2017). In addition, other factors, such as m6A modifications (Zhou et al. 2017) or RNA duplexes within circRNAs (Liu et al. 2019), may recruit additional proteins to modulate nucleocytoplasmic transport.
Given their lack of free ends, circRNAs are naturally resistant to degradation initiated by exonucleases and typically have much longer half-lives than do linear RNAs (Enuka et al. 2016, Liu et al. 2022, Zhang et al. 2016a). This enables some circRNAs to accumulate to high levels (Figure 2e), especially in cells with slow division rates, such as neurons and aging tissues (Rybak-Wolf et al. 2015, Westholm et al. 2014, You et al. 2015, Zhang et al. 2016a). Nevertheless, circRNAs can still undergo decay via several mechanisms that are initiated by endonucleases (Figure 2f). For example, the CDR1as/ciRS-7 circRNA contains a sequence with perfect complementarity to the microRNA (miRNA) miR-671, thereby enabling miR-671 to initiate its decay in an Argonaute 2 (Ago2)-dependent manner (Hansen et al. 2011, Kleaveland et al. 2018). This situation is unique, however, as miRNA target sites in other circRNAs are only partially complementary to miRNAs and are unable to trigger Ago2 endonucleolytic cleavage (Hansen et al. 2013, Jeck & Sharpless 2014, Memczak et al. 2013). Instead, several other endonucleases appear to mediate the degradation of other circRNAs. Upon certain viral infections, activated RNase L can rapidly (within an hour) degrade the vast majority of circRNAs (Liu et al. 2019). A subset of m6A-modified circRNAs additionally can be degraded by the ribonuclease complex RNase P/MRP (Park et al. 2019). Finally, a small portion of structured circRNAs can be specifically degraded by the endonuclease G3BP1 when it is in complex with the ATP-dependent RNA helicase upstream frameshift 1 (UPF1) (Fischer et al. 2020). How exactly these endonuclease activities are regulated under physiological conditions remain to be explored; there are likely additional enzymes that degrade circRNAs that await discovery. Nevertheless, the long stability of circRNAs endows these molecules with the potential to carry out important cellular functions (discussed in the section titled Regulatory Roles of circRNAs and Their Modes of Action) as well as be secreted to extra-cellular fractions, such as exosomes (Y. Li et al. 2015, Memczak et al. 2015) (Figure 2g).
REGULATORY ROLES OF CIRCRNAS AND THEIR MODES OF ACTION
Although most of the initial circRNAs identified in the 1990s were expressed at low levels and thought to have little functional potential, there were hints that these transcripts could be related to certain biological and pathological conditions. For example, circSRY, which contains 16 target sites for miR-138 (Hansen et al. 2013) and a long open reading frame (ORF) (Capel et al. 1993), was proposed to be required during mouse spermatogenesis (Capel et al. 1993). However, circSRY was barely loaded onto polysomes, so its translation potential was of unclear significance (Capel et al. 1993). In another example, an early study found that circANRIL is transcribed from a locus associated with atherosclerotic cardiovascular disease on chromosome 9p21, and that the expression of these circRNAs was correlated with atherosclerosis risk (Burd et al. 2010).
Physiological and Pathological Expression Dynamics of circRNAs
With the advent of large-scale profiling of circRNAs, a growing number of studies have revealed circRNA expression patterns in physiological and pathological contexts (Figure 3). In early stage mouse embryos, over 2,800 circRNAs are expressed during the zygote to blastocyst stages (Fan et al. 2015) (Figure 3), and CRISPR-Cas13 knockdown approaches have recently revealed that circMan1a2 is required during mouse embryo preimplantation development (S. Li et al. 2021). A large number of circRNAs are likewise increasingly expressed during murine spermatogenesis (Figure 3), specifically at the stage when late pachytene spermatocytes develop into round and then elongating spermatids (Tang et al. 2020). Interestingly, many such circRNAs appear to have translation potential, as they have ORFs with m6A-modified start codons (Tang et al. 2020). Although their physiological relevance during spermatogenesis remains to be examined, removal of a specific noncoding circRNA, circBoule, was suggested to dampen male fertility in fly and mouse (Gao et al. 2020).
Figure 3.
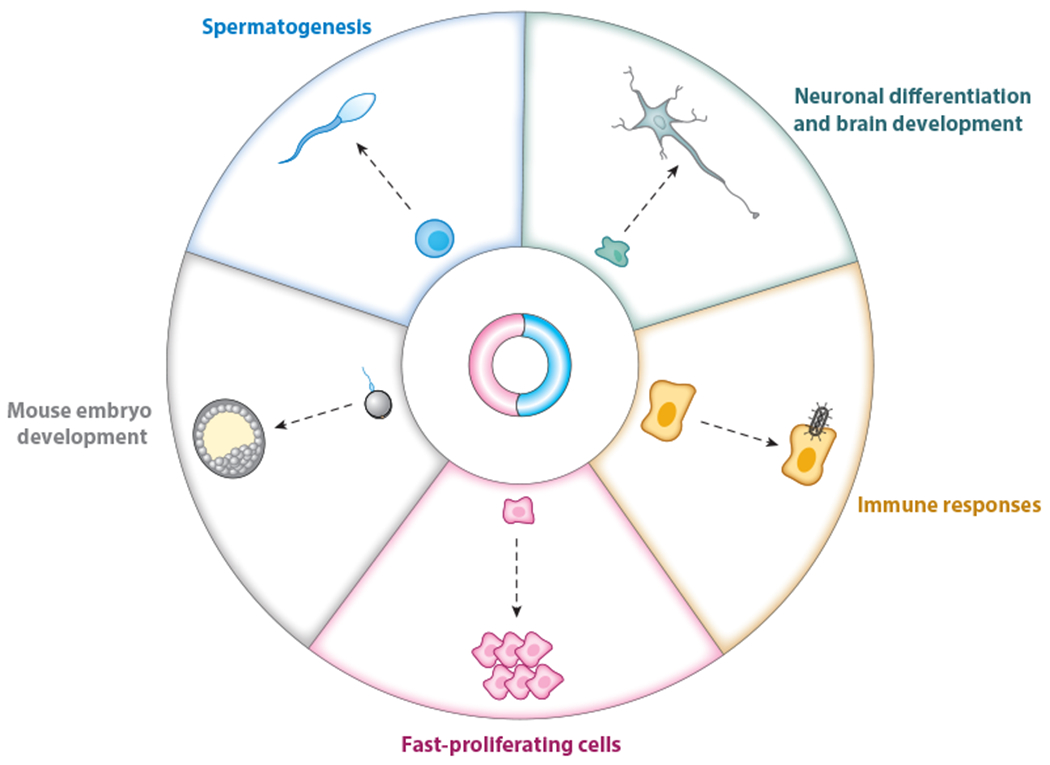
Altered cellular and physiological expression patterns of circular RNAs (circRNAs). Up- and downregulated circRNA expression has been observed in different biological and physiological contexts, including embryo and neuronal development, spermatogenesis, immune responses, and tumorigenesis. Although some specific circRNAs have been shown to participate in these different processes, the regulatory roles of most of these altered circRNAs await further investigation.
circRNAs also have been shown to generally accumulate with age (in a manner independent of expression of their linear cognate mRNAs) in C. elegans (Cortés-López et al. 2018), fly head (Westholm et al. 2014) and pig brain (Veno et al. 2015), as well as in mouse cortex and hippocampus (Rybak-Wolf et al. 2015, You et al. 2015) and human brain (Szabo et al. 2015). This appears to be because of the long half-lives of circRNAs. In support of this model, nascent RNA-seq has shown that the production rates of circRNAs are largely unchanged between human embryonic stem cells (hESCs) and hESC-derived forebrain neurons, yet the steady-state levels of circRNAs in forebrain neurons are significantly higher (Zhang et al. 2016a) (Figure 3). Note that global circRNA accumulation in aged flies can be affected by insulin signaling and that circSfl promotes lifespan of insulin mutant flies (Weigelt et al. 2020).
In contrast, a general decrease in circRNA abundance has been reported in several cancers (Bachmayr-Heyda et al. 2015, S. Chen et al. 2019, Vo et al. 2019) (Figure 3), likely because circRNAs can be diluted by rapid cell division. Indeed, there is a negative correlation between global circRNA abundance and proliferation (Bachmayr-Heyda et al. 2015). Although circRNAs are generally reduced in fast-proliferating cells, several oncogenic fusion circRNAs (f-circRNAs) have been identified in cells containing cancer-associated chromosomal translocations (Guarnerio et al. 2016, Wu et al. 2019). Such f-circRNAs are required for proliferation of these cancer cells, but the underlying molecular mechanisms remain unknown (Guarnerio et al. 2016, Wu et al. 2019).
Upon infection with some viruses, including the encephalomyocarditis virus, circRNA levels are globally decreased due to activation of the nonspecific RNase L endonuclease (Liu et al. 2019) (Figure 3). RNase L is also activated in the autoimmune disease systemic lupus erythematosus and, correspondingly, overall reduced circRNA levels have been observed (Liu et al. 2019). Similar trends have been reported in patients with chronic inflammatory skin diseases (Moldovan et al. 2019, 2021). Note that these altered circRNA expression patterns are independent of changes in cognate mRNA expression, suggesting that at least a subset of circRNAs may play regulatory roles in these diseases. We now summarize the modes of action of some well-studied circRNAs (Figure 4), which have largely been characterized using laboratory cultured cell lines.
Figure 4.
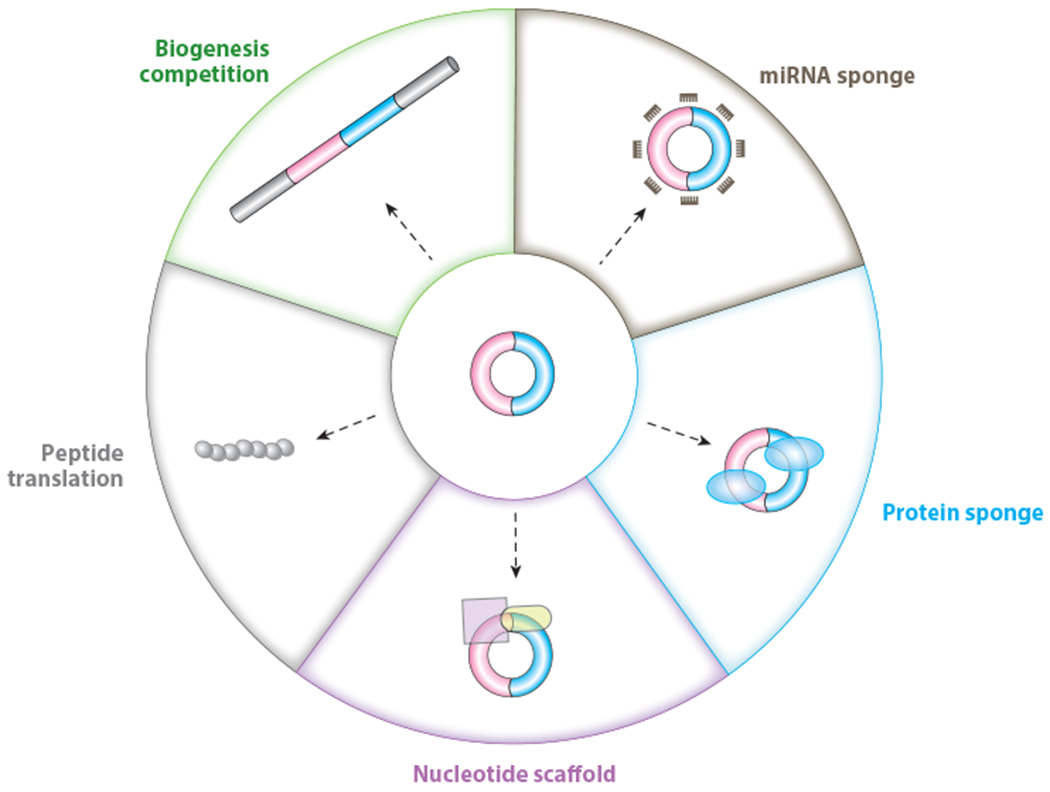
A schematic of different modes of action of circRNAs currently annotated in cells. In addition to the interplay between backsplicing and canonical splicing for the same splice site selection that can ultimately affect pre-mRNA splicing, mature circRNAs themselves can act as decoys or sponges for miRNAs and proteins, and as RNA scaffolds. A small subset of circRNAs can serve as templates for protein translation. Abbreviations: circRNA, circular RNA; miRNA, microRNA.
Modes of circRNA Action
Due to the unique life cycles of circRNAs, the act of their biogenesis, their conformation and binding partners, and their long stability can all contribute to their cellular regulatory potential. Backsplicing is in direct competition with canonical splicing events, and thus just the act of producing a circRNA can sometimes lead to the downregulation of cognate linear RNA levels and/or change the sets of linear isoforms expressed in cells. Alternatively, the mature circRNAs themselves can carry out biological functions, especially by binding to miRNAs, proteins, or other RNA species, sometimes in an inhibitory or competitive manner and other times in a scaffolding role that enables functional complexes to be formed. Other circRNAs appear to be translated to generate peptides with biological functions.
circRNAs as Decoys or Sponges for microRNAs
Given their largely cytoplasmic localization and cellular stability, many studies have pursued the idea that circRNAs might function as competing endogenous RNAs (ceRNAs) (Salmena et al. 2011) that modulate the bioavailability of miRNAs (Jeck et al. 2013) (Figure 4). The most prominent example is CDR1as/ciRS-7, a circRNA derived from an lncRNA (LINC00632) that is antisense to the CDR1 protein-coding gene (which is notably not expressed in most tissues) (Barrett et al. 2017; Hansen et al. 2011, 2013; Memczak et al. 2013). CDR1as/ciRS-7 is abundantly expressed in mammalian brains (Hansen et al. 2013, Memczak et al. 2013) with a particular enrichment in excitatory neurons (Piwecka et al. 2017). In addition to a perfect miR-671 target site that can enable cleavage of CDR1as/ciRS-7 by Ago2 (Hansen et al. 2011), CDR1as/ciRS-7 harbors 74 conserved binding sites for miR-7. Upon reducing CDR1s/ciRS-7 expression in human cell lines, the expression of mRNAs with miR-7 binding sites was likewise reduced, suggesting that this circRNA can act as a decoy or sponge for miR-7 (Hansen et al. 2013, Memczak et al. 2013). Interestingly, when the entire genomic region corresponding to Cdr1as/ciRS-7 was removed using CRISPR/Cas9, mice had phenotypes resembling neuropsychiatric disorders, including a dysfunction of excitatory synaptic transmission (Piwecka et al. 2017). This was not, however, only due to simple alterations in miR-7 sponging, as the expression of miR-7 was also reduced in Cdr1as/ciRS-7 knockout mice (Piwecka et al. 2017). Cdr1as/ciRS-7 is thus required for miR-7 stabilization in vivo and, indeed, a network of lncRNAs, including Cyrano, is now recognized to control miR-7 levels (Kleaveland et al. 2018). Cyrano can bind miR-7 and promote its destruction by inducing 3’ tailing and trimming (Kleaveland et al. 2018). Upon deletion of Cyrano in mice, increased levels of miR-7 are observed along with decreased Cdr1as/ciRS-7 expression, in part due to the slicing of Cdr1as/ciRS-7 by miR-671 (Kleaveland et al. 2018). Beyond a role in excitatory synaptic transmission in the brain, Cdr1as/ciRS-7 has also been implicated in insulin secretion (Xu et al. 2015) and in cancer (melanoma) progression (Hanniford et al. 2020). The latter notably occurs via an miR-7-independent mechanism.
Beyond Cdr1as/ciRS-7, additional abundant circRNAs have been proposed to serve as miRNA decoys or sponges. circHIPK2 modulates astrocyte activation by sponging miR124–2HG (Huang et al. 2017), circZNF1 plays a role in epidermal stem cell differentiation by sponging miR-23b-3p (Kristensen et al. 2018), and circBIRC6 controls hESC pluripotency and differentiation by sequestering both miR-34a and miR-145 (Yu et al. 2017). Nonetheless, most mammalian circRNAs are expressed at low levels, with relatively few miRNA binding sites (Guo et al. 2014). This makes most circRNAs poor candidates for acting as ceRNAs for miRNAs (Bosson et al. 2014, Denzler et al. 2014).
circRNAs as Decoys or Sponges for Proteins
In addition to binding RNAs, circRNAs can bind to proteins, and some act as decoys that prevent those protein factors from acting elsewhere (Figure 4). This mode of regulation is exemplified by Drosophila circMbl (Ashwal-Fluss et al. 2014). MBL, the protein encoded by the cognate gene, promotes circMbl production by binding to the flanking introns and then subsequently interacts with mature circMbl transcripts (Ashwal-Fluss et al. 2014). When MBL protein levels are high, circMbl backsplicing is promoted at the expense of canonical splicing, thereby preventing the expression of additional linear Mbl mRNAs and MBL proteins. The existing MBL proteins can be further sequestered by the mature circMbl transcripts, ensuring an efficient negative feedback loop (Ashwal-Fluss et al. 2014). In another example, circANRIL, which is associated with atherosclerotic cardiovascular disease (Burd et al. 2010), binds the C-terminal lysine-rich domain of pescadillo zebrafish homolog 1 (PES1) (Holdt et al. 2016). PES1 is a key component of the PES1-BOP1-WDR12 (PeBoW) complex that promotes precursor rRNA (pre-rRNA) processing to mature 28S and 5.8S rRNAs. The binding of PES1 to circANRIL can inhibit rRNA maturation, leading to impaired ribosome biogenesis, nucleolar stress, and cell death in vascular smooth muscle cells and macrophages related to atherosclerosis (Holdt et al. 2016).
Just as circRNAs need to be expressed at high levels to function as efficient ceRNAs, the stoichiometry between circRNAs and their binding proteins needs to be evaluated before proposing a decoy or sponge model. Rather than acting alone, recent work has suggested that circRNAs can function as a group to sequester dsRNA-binding proteins, such as NF90/NF110 (X. Li et al. 2017) and PKR (Liu et al. 2019), due to their tendency to form 16–26–base pair imperfect double-stranded (ds) regions. Indeed, a number of dsRNA-binding proteins with degenerate binding motifs, including oligoadenylate-synthetase and ADAR1–150, can bind in vitro–synthesized circRNAs (Liu et al. 2019). In another example, the circRNA cia-cGAS contains a dsRNA region that enables strong binding to the DNA-binding domain of the DNA sensor cyclic GMP–AMP synthase (cGAS) in the nucleus. This circRNA thus acts to competitively inhibit the binding of self-DNA to cGAS, thereby preventing inappropriate type I interferon activation and maintaining quiescent hematopoietic stem cells in the bone marrow (Xia et al. 2018).
circRNAs can simultaneously interact with miRNAs and proteins and, therefore, these modes of action are not mutually exclusive. Spatial localization, as well as the relative amounts of accessible circRNA and protein molecules, plays an important role in defining the exact function of the transcript. For example, circFAM120A is an abundant circRNA that is expressed at more than 20 copies per HeLa and HEK 293 cell (S. Li et al. 2021). This particular circRNA is enriched to monoribosomes, where it binds the translation inhibitor IGF2BP2, preventing it from interacting with the linear cognate FAM120A mRNA. Even though IGF2BP2 is present at a level of hundreds of thousands of copies per cell (Hein et al. 2015), this local niche of circFAM120A on monoribosomes enables this circRNA to carry out a critical function: It ensures robust FAM120A translation, which is required in the AKT pathway for cell proliferation (S. Li et al. 2021).
circRNAs as RNA Scaffolds
Some circRNA-protein interactions do not inhibit protein function but instead enable formation of complexes (circRNPs) that are involved in gene regulation. For example, circFOXO3 can promote cell cycle progression by interacting with cyclin-dependent kinase 2 (CDK2) and cyclin-dependent kinase inhibitor 1 (or p21) to impact CDK2 function (Du et al. 2016). In another example, circAmotl1 binds AKT1 and phosphoinositide-dependent kinase 1 (PDK1), leading to AKT1 phosphorylation and facilitating the cardioprotective nuclear translocation of pAKT in neonatal human cardiac tissue (Y. Zeng et al. 2017). Additionally, circACC1 acts as an RNA component of the AMP-activated protein kinase (AMPK) holoenzyme to stabilize and promote the enzymatic activity of AMPK by forming a ternary complex with the regulatory AMPK beta and gamma subunits (Q. Li et al. 2019). As a result, circACC1 modulates metabolic adaptation during serum deprivation by increasing glycolysis and beta-oxidation.
circRNAs can also act as important scaffolds in the nucleus and the mitochondrion. circPOK has been proposed to interact with and activate the ILF2/3 complex in the nucleus, promoting the transcription of ILF2/3-regulated proproliferative and pro-angiogenic factors in the context of mesenchymal tumor progression (Guarnerio et al. 2019). circKcnt2 inhibits Batf expression by recruiting the nucleosome-remodeling deacetylase complex onto the Batf promoter, which promotes colitis resolution (Liu et al. 2020). The mitochondrial DNA–encoded circSCAR transcript inhibits mitochondrial reactive oxygen species output by forming a complex with ATP synthase 5B (ATP5B) and shutting down the mitochondrial permeability transition pore (Zhao et al. 2020). In each of these examples, the stoichiometry between circRNA and its interacting partners still remains somewhat unclear, along with the mechanistic details of how each circRNP is formed, especially at the structural level.
Translatable circRNAs
In addition to the multiple noncoding roles that circRNAs can perform, recent work suggests that a subset of endogenous circRNAs may be translated into protein using cap-independent translation mechanisms. Engineered circRNAs that contain strong viral IRESs have long been known to be translated (Chen & Sarnow 1995). Such engineered transcripts can easily be generated in cells using expression vectors (Kramer et al. 2015, X. Li et al. 2017, Wang & Wang 2015) and in vitro using direct (or splint) ligation methods (Liu et al. 2022) or by using self-splicing introns (Chen et al. 2017; Ford & Ares 1994; Wesselhoeft et al. 2018, 2019). Nonetheless, there has been significant debate as to whether endogenous circRNAs are translated. Some ribosome-profiling data sets have provided little evidence for circRNA translation (Capel et al. 1993, Guo et al. 2014, Jeck et al. 2013, Stagsted et al. 2019), while others have suggested a subset of endogenous circRNAs [e.g., circZNF609 (Legnini et al. 2017), circMbl (Pamudurti et al. 2017), circSfl (Weigelt et al. 2020), and circFGFR1 (Chen et al. 2021)] are translatable. Such transcripts have been proposed to often contain regions that are complementary to 18S rRNA as well as a structured RNA element that facilitates IRES-dependent circRNA translation (Chen et al. 2021). Alternatively, m6A modifications have been reported to drive circRNA translation by recruiting the translation initiation factor eIF4G2 and the m6A reader YTHDF3 (Yang et al. 2017).
Cap-independent translation mechanisms generally have low efficiency, and thus protein products translated from circRNAs are limited under normal conditions (Legnini et al. 2017, Pamudurti et al. 2017, Yang et al. 2017). However, upon cellular stress, cap-dependent mRNA translation is generally suppressed, thereby allowing products of cap-independent circRNA translation to become elevated during starvation (Pamudurti et al. 2017) and heat shock (Chen et al. 2021, Yang et al. 2017). For example, the circFGFR1-encoded protein, which is in frame with full-length FGFR1, can function as an antagonist of the FGFR1 oncoprotein and can suppress FGFR1 signaling and cell growth during heat shock (Chen et al. 2021). In a somewhat analogous manner, circ-E-Cad can be translated into an oncogenic secretory E-cadherin protein variant that activates epidermal growth factor (EGF) receptor (EGFR) signaling independently of EGF, leading to glioma stem cell tumorigenicity (Gao et al. 2021). Emerging large-scale analyses of the translatome of human hearts (van Heesch et al. 2019), murine spermatogenesis (Tang et al. 2020), and additional cell lines and tissues (Chen et al. 2021) have suggested the existence of other circRNA-encoded proteins. However, these data need to be interpreted cautiously because standard quality control metrics that increase the robustness of ribosome-profiling data cannot be applied to circRNAs, and the robustness of false discovery rate metrics for mass spectrometry of circRNA-derived peptides has been called into question (Hansen 2021).
EMERGING ROLES OF OTHER TYPES OF CIRCULAR RNAS
This review has focused on the biogenesis and functions of circRNAs produced from backspliced exons, but other types of circular RNAs with distinct properties deserve mention. Hundreds of ciRNAs are generated in a splicing-dependent manner and are derived from intron lariats that fail to be debranched at their 2′,5′-phosphodiester bond. Once produced, these transcripts tend to localize in the nucleus where they can promote Pol II transcription (Zhang et al. 2013). For example, ci-ankrd52 maintains an open conformation that can form a stronger R-loop with its parental locus than can the cognate pre-mRNA. Upon ciRNA removal via RNase H1 cleavage, it has been proposed that this R-loop is resolved, and transcriptional elongation is promoted (X. Li et al. 2021). ciRNAs represent a subset of sisRNAs, which can be linear or circular and were first reported in Xenopus tropicalis oocytes (Gardner et al. 2012). Thousands of RNase R–resistant sisRNAs preferentially localize to the cytoplasm, and they can be transmitted to the fertilized egg and persist during early embryogenesis (Talhouarne & Gall 2014). For example, the maternally inherited sisRNA sisR-4 is produced from the deadpan gene, which is essential for X. tropicalis development. Female, but not male, mutants for sisR-4 produce embryos that fail to hatch. This appears to be because sisR-4 promotes the transcription of its host gene during embryogenesis via a positive feedback loop that activates an enhancer present in the intron where sisR-4 is encoded (Tay & Pek 2017).
DNA virus genomes can also produce circular RNAs that are likely involved in cell proliferation, but the underlying mechanisms are largely unknown (Huang et al. 2019, Tagawa et al. 2018, Toptan et al. 2018, Ungerleider et al. 2018, Zhao et al. 2019). In other kingdoms, such as Archaea, circular RNAs derived from noncoding sequences have also been suggested to have roles in gene expression regulation (Danan et al. 2012). For instance, the maturation of some tRNAs (Soma et al. 2007) and rRNAs (Tang et al. 2002) in Archaea requires the existence of their circular intermediates. Circular RNAs derived from archaeal tRNA and rRNA introns have additionally been suggested to contain ORFs that encode proteins (Burggraf et al. 1993, Dalgaard & Garrett 1992, Kjems & Garrett 1988) or contain C/D box snoRNAs that can guide RNA modifications (Clouet d’Orval et al. 2001, Singh et al. 2004, Starostina et al. 2004).
CONCLUSIONS
Recent technological advances have unveiled that circRNAs have diverse expression patterns, unique biogenesis mechanisms, and distinct modes of action. Many significant advances have been made, but the sequence overlap of circRNAs with their cognate linear mRNA sequences has made it somewhat complicated to precisely define their regulatory roles. Nonetheless, best experimental practices have now been provided to help discriminate these circles from linear cognate mRNAs [see reviews by Dodbele et al. (2021) and Li et al. (2018)]. Hence, we fully expect more regulatory functions and modes of action of this large class of RNA molecules to emerge, especially in physiological in vivo conditions. For example, loss-of-function circRNA studies can now be achieved not only by using RNAi-mediated approaches but also by using different CRISPR-Cas13 systems and guide RNAs targeting the BSJ sites in different contexts (Ai et al. 2022, S. Li et al. 2021, Y. Zhang et al. 2021). In addition, base editors can be used to target backsplice sites of predominantly circularized exons to specifically knock out some circRNAs (Gao et al. 2022). These emerging technologies open new doors to better appreciate circRNA functions in various contexts. In addition, they may enable disease-related circRNAs, such as the steatosis-to-NASH progression–associated circRNA SCAR (Zhao et al. 2020), to serve as potential therapeutic targets.
Beyond revealing how endogenous circRNAs are regulated and function, it is worthwhile to mention that the stability of these transcripts has endowed them with unique capabilities that can be very useful for biotechnological applications. circRNAs can serve as biomarkers (Bahn et al. 2015, Y. Li et al. 2015, Memczak et al. 2015) for cancers (Bachmayr-Heyda et al. 2015, Vo et al. 2019), virus infection (Ungerleider et al. 2018), and autoimmune diseases (Liu et al. 2019; Moldovan et al. 2019,2021). In addition, circRNAs represent a novel gene expression platform for generating abundant, stable RNAs that contain RNA aptamer sequences (Litke & Jaffrey 2019, Liu et al. 2022) or encode proteins (Chen et al. 2017, Wesselhoeft et al. 2019). In total, the last 10 years of circRNA research has laid a promising foundation for future studies to further understand how these transcripts control key biological processes as well as how circRNAs can be applied as useful technologies in biomedical research.
ACKNOWLEDGMENTS
We apologize to colleagues whose work could not be discussed owing to space limitations. Support for this work was provided by the National Key Research and Development Program of China (grant 2021YFA1300500), the CAS Project for Young Scientists in Basic Research (grant YSBR-009), the National Natural Science Foundation of China (NSFC) (grants 91940303 and 31725009), and the Howard Hughes Medical Institute International Program (grant 55008728) to L.-L.C.; by the National Institutes of Health (grants R35-GM119735 and R01-NS099371) to J.E.W.; and by the NSFC (grants 31730111, 31925011, and 91940306) to L.Y. L.-L.C. acknowledges support from the Xplorer Prize. J.E.W. is a Cancer Prevention and Research Institute of Texas (CPRIT) Scholar in Cancer Research.
Footnotes
DISCLOSURE STATEMENT
J.E.W. serves as a consultant for Laronde.
The Annual Review of Cell and Developmental Biology is online at cellbio.annualreviews.org
LITERATURE CITED
- Ai Y, Liang D, Wilusz JE. 2022. CRISPR/Cas13 effectors have differing extents of off-target effects that limit their utility in eukaryotic cells. Nucleic Acids Res. 50:e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktaş T, Avşar Ilik I, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, et al. 2017. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 544:115–19 [DOI] [PubMed] [Google Scholar]
- Arnberg AC, Van Ommen GJ, Grivell LA, Van Bruggen EF, Borst P. 1980. Some yeast mitochondrial RNAs are circular. Cell 19:313–19 [DOI] [PubMed] [Google Scholar]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, et al. 2014. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56:55–66 [DOI] [PubMed] [Google Scholar]
- Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, et al. 2015. Correlation of circular RNA abundance with proliferation – exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci. Rep 5:8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn JH, Zhang Q, Li F, Chan T-M, Lin X, et al. 2015. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem 61:221–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SP, Parker KR, Horn C, Mata M, Salzman J. 2017. ciRS-7 exonic sequence is embedded in a long non-coding RNA locus. PLOS Genet. 13:e1007114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SP, Wang PL, Salzman J. 2015. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. eLife 4:e07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. 2014. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet 15:163–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosson AD, Zamudio JR, Sharp PA. 2014. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol. Cell 56:347–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. 2010. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLOS Genet. 6:e1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burggraf S, Larsen N, Woese CR, Stetter KO. 1993. An intron within the 16S ribosomal RNA gene of the archaeon Pyrobaculum aerophilum. PNAS 90:2547–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel B, Swain A, Nicolis S, Hacker A, Walter M, et al. 1993. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73:1019–30 [DOI] [PubMed] [Google Scholar]
- Cech TR. 1990. Self-splicing of group I introns. Annu. Rev. Biochem 59:543–68 [DOI] [PubMed] [Google Scholar]
- Chen C-K, Cheng R, Demeter J, Chen J, Weingarten-Gabbay S, et al. 2021. Structured elements drive extensive circular RNA translation. Mol. Cell 81:4300–18.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-Y, Sarnow P. 1995. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268:415–17 [DOI] [PubMed] [Google Scholar]
- Chen L-L. 2016. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol 17:205–11 [DOI] [PubMed] [Google Scholar]
- Chen L-L. 2020. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol 21:475–90 [DOI] [PubMed] [Google Scholar]
- Chen L-L, Yang L. 2015. Regulation of circRNA biogenesis. RNA Biol. 12:381–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Huang V, Xu X, Livingstone J, Soares F, et al. 2019. Widespread and functional RNA circularization in localized prostate cancer. Cell 176:831–43.e22 [DOI] [PubMed] [Google Scholar]
- Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, et al. 2019. N6-methyladenosine modification controls circular RNA immunity. Mol. Cell 76:96–109.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, et al. 2017. Sensing self and foreign circular RNAs by intron identity. Mol. Cell 67:228–38.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang T-J, Chen Y-J, Chen C-Y, Mai T-L, Wang Y-D, et al. 2018. Integrative transcriptome sequencing reveals extensive alternative trans-splicing and cis-backsplicing in human cells. Nucleic Acids Res. 46:3671–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, et al. 2008. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat. Methods 5:613–19 [DOI] [PubMed] [Google Scholar]
- Clouet d’Orval B, Bortolin M-L, Gaspin C, Bachellerie J-P 2001. Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Res. 29:4518–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerelle C, Daubersies P, Majerus MA, Kerckaert JP, Bailleul B. 1992. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 11:1095–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. 1993. Mis-splicing yields circular RNA molecules. FASEB J. 7:155–60 [DOI] [PubMed] [Google Scholar]
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, et al. 2015. The RNA binding protein Quaking regulates formation of circRNAs. Cell 160:1125–34 [DOI] [PubMed] [Google Scholar]
- Conn VM, Hugouvieux V, Nayak A, Conos SA, Capovilla G, et al. 2017. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 3:17053. [DOI] [PubMed] [Google Scholar]
- Cortés-López M, Gruner MR, Cooper DA, Gruner HN, Voda AI, et al. 2018. Global accumulation of circRNAs during aging in Caenorhabditis elegans. BMC Genom. 19:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté F, Perreault J-P. 1997. Peach latent mosaic viroid is locked by a 2′,5′-phosphodiester bond produced by in vitro self-ligation. J. Mol. Biol 273:533–43 [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Garrett RA. 1992. Protein-coding introns from the 23S rRNA-encoding gene form stable circles in the hyperthermophilic archaeon Pyrobaculum organotrophum. Gene 121:103–10 [DOI] [PubMed] [Google Scholar]
- Danan M, Schwartz S, Edelheit S, Sorek R. 2012. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 40:3131–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. 2014. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell 54:766–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodbele S, Mutlu N, Wilusz JE. 2021. Best practices to ensure robust investigation of circular RNAs: pitfalls and tips. EMBO Rep. 22:e52072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R, Ma X-K, Chen L-L, Yang L. 2017. Increased complexity of circRNA expression during species evolution. RNA Biol 14:1064–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R, Ma X-K, Li G-W, Yang L. 2018. CIRCpedia v2: An updated database for comprehensive circular RNA annotation and expression comparison. Genom. Proteom. Bioinform 16:226–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. 2016. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 44:2846–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin RA, Kazmi MA, Ostrer H. 1995. Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene 167:245–48 [DOI] [PubMed] [Google Scholar]
- Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y.2016. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 44:1370–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, et al. 2017. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun 8:14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Zhang X, Wu X, Guo H, Hu Y, et al. 2015. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 16:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei T, Chen Y, Xiao T, Li W, Cato L, et al. 2017. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. PNAS 114:E5207–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer JW, Busa VF, Shao Y, Leung AKL. 2020. Structure-mediated RNA decay by UPF1 and G3BP1. Mol. Cell 78:70–84.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores R, Grubb D, Elleuch A, Nohales MA, Delgado S, Gago S. 2011. Rolling-circle replication of viroids, viroid-like satellite RNAs and hepatitis delta virus: variations on a theme. RNA Biol. 8:200–6 [DOI] [PubMed] [Google Scholar]
- Flores R, Navarro JA, de la Pena M, Navarro B, Ambros S, Vera A. 1999. Viroids with hammerhead ribozymes: some unique structural and functional aspects with respect to other members of the group. Biol. Chem 380:849–54 [DOI] [PubMed] [Google Scholar]
- Fong N, Kim H, Zhou Y, Ji X, Qiu J, et al. 2014. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev. 28:2663–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford E, Ares M Jr. 1994. Synthesis of circular RNA in bacteria and yeast using RNA cyclase ribozymes derived from a group I intron of phage T4. PNAS 91:3117–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Chang S, Xia W, Wang X, Zhang C, et al. 2020. Circular RNAs from BOULE play conserved roles in protection against stress-induced fertility decline. Sci. Adv 6(46):eabb7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Ma X-K, Li X, Li G-W, Liu C-X, et al. 2022. Knockout of circRNAs by base editing back-splice sites of circularized exons. Genome Biol. 23:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Xia X, Li F, Zhang M, Zhou H, et al. 2021. Circular RNA-encoded oncogenic E-cadherin variant promotes glioblastoma tumorigenicity through activation of EGFR-STAT3 signalling. Nat. Cell Biol 23:278–91 [DOI] [PubMed] [Google Scholar]
- Gao Y, Wang J, Zheng Y, Zhang J, Chen S, Zhao F. 2016. Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat. Commun 7:12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhao F 2018. Computational strategies for exploring circular RNAs. Trends Genet. 34:389–400 [DOI] [PubMed] [Google Scholar]
- Gardner EJ, Nizami ZF, Talbot CC Jr., Gall JG. 2012. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Genes Dev. 26:2550–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazar P, Papavasileiou P, Rajewsky N. 2014. circBase: a database for circular RNAs. RNA 20:1666–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski PJ, Zaug AJ, Cech TR. 1981. The intervening sequence of the ribosomal RNA precursor is converted to a circular RNA in isolated nuclei of tetrahymena. Cell 23:467–76 [DOI] [PubMed] [Google Scholar]
- Graveley BR. 2008. Molecular biology: power sequencing. Nature 453:1197–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, et al. 2016. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell 165:289–302 [DOI] [PubMed] [Google Scholar]
- Guarnerio J, Zhang Y, Cheloni G, Panella R, Katon JM, et al. 2019. Intragenic antagonistic roles of protein and circRNA in tumorigenesis. Cell Res. 29:628–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Agarwal V, Guo H, Bartel DP. 2014. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 15:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanniford D, Ulloa-Morales A, Karz A, Berzoti-Coelho MG, Moubarak RS, et al. 2020. Epigenetic silencing of CDR1as drives IGF2BP3-mediated melanoma invasion and metastasis. Cancer Cell 37:55–70.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB.2018. Improved circRNA identification by combining prediction algorithms. Front. Cell Dev. Biol 6:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB. 2021. Signal and noise in circRNA translation. Methods 196:68–73 [DOI] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, et al. 2013. Natural RNA circles function as efficient microRNA sponges. Nature 495:384–88 [DOI] [PubMed] [Google Scholar]
- Hansen TB, Veno MT, Damgaard CK, Kjems J. 2016. Comparison of circular RNA prediction tools. Nucleic Acids Res. 44:e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, et al. 2011. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 30:4414–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, et al. 2015. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163:712–23 [DOI] [PubMed] [Google Scholar]
- Hensgens LA, Arnberg AC, Roosendaal E, Van Der Horst G, Van Der Veen R, et al. 1983. Variation, transcription and circular RNAs of the mitochondrial gene for subunit I of cytochrome c oxidase. J. Mol. Biol 164:35–58 [DOI] [PubMed] [Google Scholar]
- Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, et al. 2016. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun 7:12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MT, Coca-Prados M. 1979. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280:339–40 [DOI] [PubMed] [Google Scholar]
- Huang C, Liang D, Tatomer DC, Wilusz JE. 2018. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 32:639–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J-T, Chen J-N, Gong L-P, Bi Y-H, Liang J, et al. 2019. Identification of virus-encoded circular RNA. Virology 529:144–51 [DOI] [PubMed] [Google Scholar]
- Huang R, Zhang Y, Han B, Bai Y, Zhou R, et al. 2017. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124–2HG. Autophagy 13:1722–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Sullivan FX, Cech TR. 1986. New reactions of the ribosomal RNA precursor of Tetrahymena and the mechanism of self-splicing. J. Mol. Biol 189:143–65 [DOI] [PubMed] [Google Scholar]
- Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, et al. 2015. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 10:170–77 [DOI] [PubMed] [Google Scholar]
- Jarrous N. 2017. Roles of RNase P and its subunits. Trends Genet. 33:594–603 [DOI] [PubMed] [Google Scholar]
- Jeck WR, Sharpless NE. 2014. Detecting and characterizing circular RNAs. Nat. Biotechnol 32:453–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, et al. 2013. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19:141–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S, Greenman C, Cook PR, Papantonis A. 2015. Exon skipping is correlated with exon circularization. J. Mol. Biol 427:2414–17 [DOI] [PubMed] [Google Scholar]
- Khan MA, Reckman YJ, Aufiero S, van den Hoogenhof MM, van der Made I, et al. 2016. RBM20 regulates circular RNA production from the Titin gene. Circ. Res 119:996–1003 [DOI] [PubMed] [Google Scholar]
- Kjems J, Garrett RA. 1988. Novel splicing mechanism for the ribosomal RNA intron in the archaebacterium Desulfurococcus mobilis. Cell 54:693–703 [DOI] [PubMed] [Google Scholar]
- Kleaveland B, Shi CY, Stefano J, Bartel DP. 2018. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell 174:350–62.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. 1986. The hepatitis delta (δ) virus possesses a circular RNA. Nature 323:558–60 [DOI] [PubMed] [Google Scholar]
- Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, et al. 2015. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 29:2168–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. 2019. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet 20:675–91 [DOI] [PubMed] [Google Scholar]
- Kristensen LS, Okholm TLH, Veno MT, Kjems J. 2018. Circular RNAs are abundantly expressed and upregulated during human epidermal stem cell differentiation. RNA Biol. 15:280–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasda E, Parker R. 2014. Circular RNAs: diversity of form and function. RNA 20:1829–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, et al. 2017. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 66:22–37.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Chen Y-S, Ping X-L, Yang X, Xiao W, et al. 2017. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res 27:444–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wang Y, Wu S, Zhou Z, Ding X, et al. 2019. CircACC1 regulates assembly and activation of AMPK complex under metabolic stress. Cell Metab. 30:157–73.e7 [DOI] [PubMed] [Google Scholar]
- Li S, Li X, Xue W, Zhang L, Yang LZ, et al.2021. Screening for functional circular RNAs using the CRISPR-Cas13 system. Nat. Methods 18:51–59 [DOI] [PubMed] [Google Scholar]
- Li X, Liu C-X, Xue W, Zhang Y, Jiang S, et al. 2017. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol. Cell 67:214–27.e7 [DOI] [PubMed] [Google Scholar]
- Li X, Liu S, Zhang L, Issaian A, Hill RC, et al. 2019. A unified mechanism for intron and exon definition and back-splicing. Nature 573:375–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yang L, Chen L-L. 2018. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell 71:428–42 [DOI] [PubMed] [Google Scholar]
- Li X, Zhang J-L, Lei Y-N, Liu X-Q, Xue W, et al. 2021. Linking circular intronic RNA degradation and function in transcription by RNase H1. Sci. China Life Sci 64:1795–809 [DOI] [PubMed] [Google Scholar]
- Li Y, Zheng Q, Bao C, Li S, Guo W, et al. 2015. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 25:981–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Huang C, Bao C, Chen L, Lin M, et al. 2015. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol 22:256–64 [DOI] [PubMed] [Google Scholar]
- Li-Pook-Than J, Bonen L. 2006. Multiple physical forms of excised group II intron RNAs in wheat mitochondria. Nucleic Acids Res. 34:2782–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Tatomer DC, Luo Z, Wu H, Yang L, et al. 2017. The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol. Cell 68:940–54.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Tatomer DC, Wilusz JE. 2021. Use of circular RNAs as markers of readthrough transcription to identify factors regulating cleavage/polyadenylation events. Methods 196:121–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Wilusz JE. 2014. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 28:2233–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litke JL, Jaffrey SR. 2019. Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat. Biotechnol 37:667–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Ye B, Zhu X, Yang L, Li H, et al. 2020. An inducible circular RNA circKcnt2 inhibits ILC3 activation to facilitate colitis resolution. Nat. Commun 11:4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C-X, Guo S-K, Nan F, Xu Y-F, Yang L, Chen L-L. 2022. RNA circles with minimized immunogenicity as potent PKR inhibitors. Mol. Cell 82:420–34.e6 [DOI] [PubMed] [Google Scholar]
- Liu C-X, Li X, Nan F, Jiang S, Gao X, et al. 2019. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177:865–80.e21 [DOI] [PubMed] [Google Scholar]
- Liu Z, Tao C, Li S, Du M, Bai Y, et al. 2021. circFL-seq reveals full-length circular RNAs with rolling circular reverse transcription and nanopore sequencing. eLife 10:e69457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Jiménez E, Rojas AM, Andrés-León E. 2018. RNA sequencing and prediction tools for circular RNAs analysis. Adv. Exp. Med. Biol 1087:17–33 [DOI] [PubMed] [Google Scholar]
- Lu T, Cui L, Zhou Y, Zhu C, Fan D, et al. 2015. Transcriptome-wide investigation of circular RNAs in rice. RNA 21:2076–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X-K, Wang M-R, Liu C-X, Dong R, Carmichael GG, et al. 2019. CIRCexplorer3: a CLEAR pipeline for direct comparison of circular and linear RNA expression. Genom. Proteom. Bioinform 17:511–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X-K, Xue W, Chen L-L, Yang L. 2021. CIRCexplorer pipelines for circRNA annotation and quantification from non-polyadenylated RNA-seq datasets. Methods 196:3–10 [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, et al. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–38 [DOI] [PubMed] [Google Scholar]
- Memczak S, Papavasileiou P, Peters O, Rajewsky N. 2015. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLOS ONE 10:e0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan LI, Hansen TB, Veno MT, Okholm TLH, Andersen TL, et al. 2019. High-throughput RNA sequencing from paired lesional- and non-lesional skin reveals major alterations in the psoriasis circRNAome. BMC Med. Genom 12:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan LI, Tsoi LC, Ranjitha U, Hager H, Weidinger S, et al. 2021. Characterization of circular RNA transcriptomes in psoriasis and atopic dermatitis reveals disease-specific expression profiles. Exp. Dermatol 30:1187–96 [DOI] [PubMed] [Google Scholar]
- Molina-Sánchez MD, Martinez-Abarca F, Toro N. 2006. Excision of the Sinorhizobium meliloti group II intron RmInt1 as circles in vivo. J. Biol. Chem 281:28737–44 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5:621–28 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Fiskaa T, Birgisdottir AB, Haugen P, Einvik C, Johansen S. 2003. The ability to form full-length intron RNA circles is a general property of nuclear group I introns. RNA 9:1464–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, et al. 1991. Scrambled exons. Cell 64:607–13 [DOI] [PubMed] [Google Scholar]
- Ottesen EW, Luo D, Seo J, Singh NN, Singh RN. 2019. Human Survival Motor Neuron genes generate a vast repertoire of circular RNAs. Nucleic Acids Res. 47:2884–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, et al. 2017. Translation of circRNAs. Mol. Cell 66:9–21.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet 40:1413–15 [DOI] [PubMed] [Google Scholar]
- Panda AC, De S, Grammatikakis I, Munk R, Yang X, et al. 2017. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res. 45:e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park OH, Ha H, Lee Y, Boo SH, Kwon DH, et al. 2019. Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Mol. Cell 74:494–507.e8 [DOI] [PubMed] [Google Scholar]
- Pasman Z, Been MD, Garcia-Blanco MA. 1996. Exon circularization in mammalian nuclear extracts. RNA 2:603–10 [PMC free article] [PubMed] [Google Scholar]
- Piwecka M, Glažar P, Hernandez-Miranda LR, Memczak S, Wolf SA,et al. 2017. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357:eaam8526. [DOI] [PubMed] [Google Scholar]
- Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, et al. 2011. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol 29:436–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi K, Faerch Nielsen A, Veno MT, Kjems J. 2021a. Nanopore long-read sequencing of circRNAs. Methods 196:23–29 [DOI] [PubMed] [Google Scholar]
- Rahimi K, Veno MT, Dupont DM, Kjems J. 2021b. Nanopore sequencing of brain-derived full-length circRNAs reveals circRNA-specific exon usage, intron retention and microexons. Nat. Commun 12:4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Luo G-Z, Zhang Z, Wang X, Zhou T, et al. 2017. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife 6:e31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, et al. 2015. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 58:870–85 [DOI] [PubMed] [Google Scholar]
- Salgia SR, Singh SK, Gurha P, Gupta R. 2003. Two reactions of Haloferax volcanii RNA splicing enzymes: joining of exons and circularization of introns. RNA 9:319–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. 2011. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146:353–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. 2013. Cell-type specific features of circular RNA expression. PLOS Genet. 9:e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. 2012. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLOS ONE 7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. 1976. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. PNAS 73:3852–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wei J, He C. 2019. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74:640–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Gurha P, Tran EJ, Maxwell ES, Gupta R. 2004. Sequential 2′-O-methylation of archaeal pre-tRNATrp nucleotides is guided by the intron-encoded but trans-acting box C/D ribonucleoprotein of pre-tRNA. J. Biol. Chem 279:47661–71 [DOI] [PubMed] [Google Scholar]
- Slyper M, Porter CBM, Ashenberg O, Waldman J, Drokhlyansky E, et al. 2020. A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat. Med 26:792–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma A, Onodera A, Sugahara J, Kanai A, Yachie N, et al. 2007. Permuted tRNA genes expressed via a circular RNA intermediate in Cyanidioschyzon merolae. Science 318:450–53 [DOI] [PubMed] [Google Scholar]
- Stagsted LVW, Nielsen KM, Daugaard I, Hansen TB. 2019. Noncoding AUG circRNAs constitute an abundant and conserved subclass of circles. Life Sci. Alliance 2:398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagsted LVW, O’Leary ET, Ebbesen KK, Hansen TB. 2021. The RNA-binding protein SFPQ preserves long-intron splicing and regulates circRNA biogenesis in mammals. eLife 10:e63088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark R, Grzelak M, Hadfield J. 2019. RNA sequencing: the teenage years. Nat. Rev. Genet 20:631–56 [DOI] [PubMed] [Google Scholar]
- Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, et al. 2015. Exon circularization requires canonical splice signals. Cell Rep 10:103–11 [DOI] [PubMed] [Google Scholar]
- Starostina NG, Marshburn S, Johnson LS, Eddy SR, Terns RM, Terns MP. 2004. Circular box C/D RNAs in Pyrococcus furiosus. PNAS 101:14097–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. 2006. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 34:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo L, Morey R, Palpant NJ, Wang PL, Afari N,et al. 2015. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 16:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo L, Salzman J. 2016. Detecting circular RNAs: bioinformatic and experimental challenges. Nat. Rev. Genet 17:679–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak HF, Van der Horst G, Kamps AM, Arnberg AC. 1987. Interlocked RNA circle formation by a self-splicing yeast mitochondrial group I intron. Cell 48:101–10 [DOI] [PubMed] [Google Scholar]
- Tagawa T, Gao S, Koparde VN, Gonzalez M, Spouge JL, et al. 2018. Discovery of Kaposi’s sarcoma herpesvirus-encoded circular RNAs and a human antiviral circular RNA. PNAS 115:12805–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talhouarne GJ, Gall JG. 2014. Lariat intronic RNAs in the cytoplasm of Xenopus tropicalis oocytes. RNA 20:1476–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Xie Y, Yu T, Liu N, Wang Z, et al. 2020. m6A-dependent biogenesis of circular RNAs in male germ cells. Cell Res. 30:211–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Yu T, Xie Y, Wang Z, McSwiggin H, et al. 2018. Template switching causes artificial junction formation and false identification of circular RNAs. bioRxiv 259556. 10.1101/259556 [DOI] [Google Scholar]
- Tang TH, Rozhdestvensky TS, d’Orval BC, Bortolin ML, Huber H, et al. 2002. RNomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res. 30:921–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay ML, Pek JW. 2017. Maternally inherited stable intronic sequence RNA triggers a self-reinforcing feedback loop during development. Curr. Biol 27:1062–67 [DOI] [PubMed] [Google Scholar]
- Toptan T, Abere B, Nalesnik MA, Swerdlow SH, Ranganathan S, et al. 2018. Circular DNA tumor viruses make circular RNAs. PNAS 115:E8737–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerleider N, Concha M, Lin Z, Roberts C, Wang X, et al. 2018. The Epstein Barr virus circRNAome. PLOS Pathog. 14:e1007206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heesch S, Witte F, Schneider-Lunitz V, Schulz JF, Adami E, et al. 2019. The translational landscape of the human heart. Cell 178:242–60.e29 [DOI] [PubMed] [Google Scholar]
- Veno MT, Hansen TB, Veno ST, Clausen BH, Grebing M, et al. 2015. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 16:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, et al. 2019. The landscape of circular RNA in cancer. Cell 176:869–81.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, et al. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456:470–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Hou J, Muller-McNicoll M, Chen W, Schuman EM. 2019. Long and repeat-rich intronic sequences favor circular RNA formation under conditions of reduced spliceosome activity. iScience 20:237–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Z. 2015. Efficient backsplicing produces translatable circular mRNAs. RNA 21:172–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt CM, Sehgal R, Tain LS, Cheng J, Esser J, et al. 2020. An insulin-sensitive circular RNA that regulates lifespan in Drosophila. Mol. Cell 79:268–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesselhoeft RA, Kowalski PS, Anderson DG. 2018. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun 9:2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesselhoeft RA, Kowalski PS, Parker-Hale FC, Huang Y, Bisaria N, Anderson DG. 2019. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol. Cell 74:508–20.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm JO, Miura P, Olson S, Shenker S, Joseph B, et al. 2014. Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 9:1966–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, et al. 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453:1239–43 [DOI] [PubMed] [Google Scholar]
- Wilusz JE. 2018. A 360° view of circular RNAs: from biogenesis to functions. Wiley Interdiscip. Rev. RNA 9:e1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Liao X, Gong Y, He J, Zhou J-K, et al. 2019. Circular RNA F-circSR derived from SLC34A2-ROS1 fusion gene promotes cell migration in non-small cell lung cancer. Mol. Cancer 18:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Ji P, Zhao F. 2020. CircAtlas: an integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol. 21:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P, Wang S, Ye B, Du Y, Li C, et al. 2018. A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity 48:688–701.e7 [DOI] [PubMed] [Google Scholar]
- Xiao M-S, Ai Y, Wilusz JE. 2020. Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol. 30:226–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M-S, Wilusz JE. 2019. An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3′ ends. Nucleic Acids Res. 47:8755–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin R, Gao Y, Gao Y, Wang R, Kadash-Edmondson KE, et al. 2021. isoCirc catalogs full-length circular RNA isoforms in human transcriptomes. Nat. Commun 12:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Guo S, Li W, Yu P. 2015. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci. Rep 5:12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Duff MO, Graveley BR, Carmichael GG, Chen L-L. 2011. Genomewide characterization of non-polyadenylated RNAs. Genome Biol 12:R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fan X, Mao M, Song X, Wu P, et al. 2017. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 27:626–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X, Vlatkovic I, Babic A, Will T, Epstein I, et al. 2015. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci 18:603–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C-Y, Li T-C, Wu Y-Y, Yeh C-H, Chiang W, et al. 2017. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat. Commun 8:1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C-Y, Liu H-J, Hung L-Y, Kuo H-C, Chuang T-J. 2014. Is an observed non-co-linear RNA product spliced in trans, in cis or just in vitro? Nucleic Acids Res. 42:9410–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaphiropoulos PG. 1996. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. PNAS 93:6536–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Lin W, Guo M, Zou Q. 2017. A comprehensive overview and evaluation of circular RNA detection tools. PLOS Comput. Biol 13:e1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Du WW, Wu Y, Yang Z, Awan FM. 2017. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics 16:3842–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Hou L, Zuo Z, Ji P, Zhang X, et al. 2021. Comprehensive profiling of circular RNAs with nanopore sequencing and CIRI-long. Nat. Biotechnol 39:836–45 [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhao F. 2021. Reconstruction of circular RNAs using Illumina and Nanopore RNA-seq datasets. Methods 196:17–22 [DOI] [PubMed] [Google Scholar]
- Zhang X-O, Dong R, Zhang Y, Zhang J-L, Luo Z, et al. 2016. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 26:1277–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X-O, Wang H-B, Zhang Y, Lu X, Chen L-L, Yang L. 2014. Complementary sequence-mediated exon circularization. Cell 159:134–47 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Nguyen TM, Zhang XO, Wang L, Phan T, et al. 2021. Optimized RNA-targeting CRISPR/Cas13d technology outperforms shRNA in identifying functional circRNAs. Genome Biol. 22:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xue W, Li X, Zhang J, Chen S, et al. 2016a. The biogenesis of nascent circular RNAs. Cell Rep. 15:611–24 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang L, Chen L-L. 2016b. Characterization of circular RNAs. Methods Mol. Biol 1402:215–27 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang X-O, Chen T, Xiang J-F, Yin Q-F, et al. 2013. Circular intronic long noncoding RNAs. Mol. Cell 51:792–806 [DOI] [PubMed] [Google Scholar]
- Zhao J, Lee EE, Kim J, Yang R, Chamseddin B, et al. 2019. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat. Commun 10:2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Liu J, Deng H, Ma R, Liao J-Y, et al. 2020. Targeting mitochondria-located circRNA SCAR alleviates NASH via reducing mROS output. Cell 183:76–93.e22 [DOI] [PubMed] [Google Scholar]
- Zheng Q, Bao C, Guo W, Li S, Chen J, et al. 2016. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun 7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, et al. 2017. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 20:2262–76 [DOI] [PMC free article] [PubMed] [Google Scholar]


