Significance
Establishing coevolutionary associations among genes can clarify their function. A method to identify gene coevolution is the comparison of “phylogenetic profiles”, vectors describing presence/absence of genes in a genome set. However, coevolution can be hidden in poorly similar profiles when the history of the genes does not coincide with the history of their interaction. We have developed a procedure that can detect hidden coevolutionary interactions. We provide a proof-of-concept of this ability by validating the identification of the last gene of purine degradation in animals and other eukaryotes, which reveals a connection of glyoxylate cycle and purine catabolism. Software and datasets provided here can be used to uncover other significant associations in biological processes, cellular components, and metabolic pathways.
Keywords: coevolution, gene association, statistical significance, glyoxylate cycle, purine catabolism
Abstract
Coevolution at the gene level, as reflected by correlated events of gene loss or gain, can be revealed by phylogenetic profile analysis. The optimal method and metric for comparing phylogenetic profiles, especially in eukaryotic genomes, are not yet established. Here, we describe a procedure suitable for large-scale analysis, which can reveal coevolution based on the assessment of the statistical significance of correlated presence/absence transitions between gene pairs. This metric can identify coevolution in profiles with low overall similarities and is not affected by similarities lacking coevolutionary information. We applied the procedure to a large collection of 60,912 orthologous gene groups (orthogroups) in 1,264 eukaryotic genomes extracted from OrthoDB. We found significant cotransition scores for 7,825 orthogroups associated in 2,401 coevolving modules linking known and unknown genes in protein complexes and biological pathways. To demonstrate the ability of the method to predict hidden gene associations, we validated through experiments the involvement of vertebrate malate synthase-like genes in the conversion of (S)-ureidoglycolate into glyoxylate and urea, the last step of purine catabolism. This identification explains the presence of glyoxylate cycle genes in metazoa and suggests an anaplerotic role of purine degradation in early eukaryotes.
Coevolution, i.e., the reciprocal evolutionary change of interacting biological entities, can be observed at different molecular levels (1), ranging from individual amino acid sites (2, 3) to the genome scale (4, 5). An extreme example of coevolution at the gene level is when the existence of a gene in a genome is related to the existence of other genes (6–9). This is observed in genes coding for proteins interacting in macromolecular complexes (10, 11), signaling pathways (12), and metabolic pathways (13, 14).
A widely used technique to infer coevolution among genes is the comparison of their “phylogenetic profiles” (PPs), vectors describing the presence or absence of a gene (or protein) in a list of organisms (9). A central tenet of PP analysis is that functionally linked genes should have matching or similar profiles. However, there is no established optimal method to build and compare PPs; in more than 20 y of research, and particularly in recent years, various methods and procedures have been used (15–25). Variants of PP analysis include methods for orthologous gene identification and encoding presence/absence information in profiles, and methods and metrics to infer coevolution from profile comparisons. An enhanced PP method has been proposed which combines information on gene presence in extant/ancestral nodes of a taxonomic tree with information on gene duplication and loss and uses fast heuristics for profile comparison (20). Very recently, machine learning has been successfully applied to the identification of functionally related profiles, although limited to human genes (25).
Similarity in PPs can also be determined by shared phylogenetic inheritance (15, 19). This is a confounding factor in PP analysis especially for eukaryotic genomes, whose gene contents are dominated by shared inheritance and lack strong coevolutionary signals generated by horizontal transfer of functional units (i.e., operons). Unlike accurate phylogenetic methods (15, 16), approximate methods that take shared inheritance into account scale well with the expansion of biological databases and are suitable for big data analysis (18–20). These methods, however, have certain limitations. First, they lack a well-defined metric to assess the statistical significance of coevolutionary associations. Second, they maintain an evaluation of global similarity among profiles by penalizing mismatches between extant (18, 19) or extant/ancestral (20) states, according to a model in which the history of a gene partnership coincides with that of the genes. Conversely, gene coevolution can follow more complex dynamics, with interactions established or abolished in specific clades of the tree of life. It has been shown that local coevolution can be detected by performing PP analysis clade-wise (22–25), but this leaves the issue of which clades or clade combinations to choose to carry out the analysis.
To overcome these limitations, we developed a metric to score and assess the significance (i.e., P-value) of correlated presence/absence transitions across tree-ordered genomes, taken as proxies of shared gene losses or gains (18, 26). The use of this metric enables the identification of dynamic gene interactions in which partnerships can be established later in time, or later divorce due to, e.g., nonorthologous gene displacement (27). These interactions can be hidden in profiles with low overall similarity. We applied the metric to the analysis of a large dataset of eukaryotic orthologous genes as defined in OrthoDB (28). To provide a proof-of-concept of the ability of the method to detect hidden coevolutionary relationships in metabolic pathways, we experimentally validated the identification of the long-sought gene responsible for the last step of purine degradation in metazoans and other eukaryotes, namely the formation of glyoxylate and urea from ureidoglycolate (29). Interestingly, the gene shares homology and database annotation with malate synthase, the gene responsible for the formation of malate from glyoxylate and acetyl-CoA in the glyoxylate cycle (30).
Results
Pipeline and Metrics for Coevolutionary Analysis of Eukaryotic Genes.
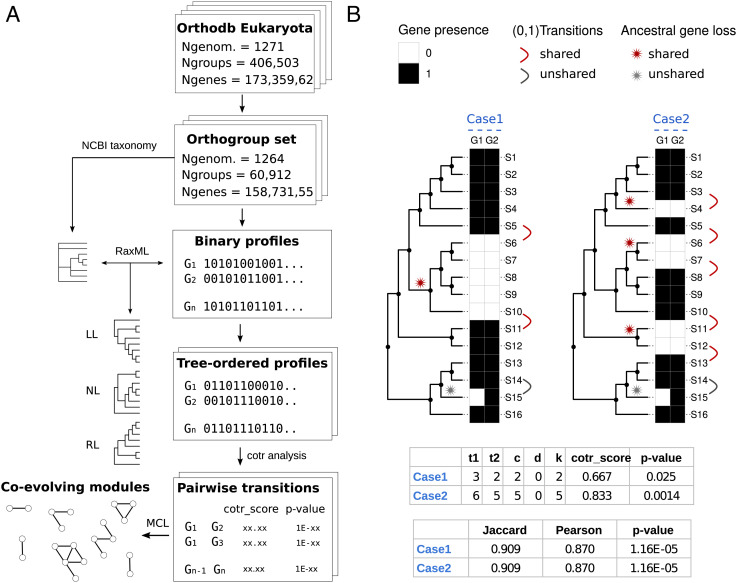
For the coevolutionary analysis (Fig. 1A), we leveraged the collection of eukaryotic orthologous groups (orthogroups) provided by OrthoDB (28), which was filtered to include only genomes of distinct species (n = 1,264) and orthogroups present in at least 1% of genomes. The selected dataset contained 60,912 of the 406,503 orthogroups while retaining the majority of genes (~159M/~173M). This dataset was used to build a large matrix (60,912 × 1,264) of binary profiles encoding the presence in each genome of one or more genes of each orthogroup as “1” and the absence as “0” (Fig. 1A). The matrix columns (genomes) were then ordered according to a taxonomy-constrained phylogenetic tree built using the transposed profile matrix to solve unresolved relationships of the ncbi phylogenetic tree. Different orientations of the same tree—right-ladderized (RL), left-ladderized (LL), and nonladderized (NL)—were used to generate tree-ordered profiles for subsequent analysis (Fig. 1A).
Fig. 1.
Pipeline and metrics for coevolutionary analysis. (A) Scheme of the workflow used for coevolutionary analysis. (B) Scheme illustrating the metrics used for measuring score and significance of coevolutionary associations. Two evolutionary scenarios (Case 1 and Case 2) with 16 species (S1 to S16) related by a phylogenetic tree and two orthogroups (G1, G2) with the same correspondence (15 matches, 1 mismatch) of presence (black squares) and absence (white squares) states are compared. For the two cases, reported are the score and significance computed from the enumeration of state transitions (1→0, 0→1) along the phylogenetic profile vectors. The cotr_score is a function of the total state transitions (t1, t2) and the number of concordant (c) and discordant (d) transitions (k = c − d; see also SI Appendix, Fig. S1). Unshared transitions (gray) are counted in the total number of transitions (t1 or t2), but they are considered neither concordant or discordant. Significance (P-value) is calculated from the transition table using Fisher’s exact test (Methods). Standard measures of profile similarities (Jaccard, Pearson) and Pearson P-values are reported for comparison. Although the associations between G1 and G2 receive the same scores and significance by global measures of similarity, the cotr_score is more significant in Case 2, consistent with the higher occurrence of shared gene losses, and the higher confidence of functional association.
For each pairwise combination of the tree-ordered profiles, we calculate the score and significance of coevolutionary transitions, a distinctive feature of our method (Fig. 1B). Instead of relying on similarity measures between profiles, we focus on state transitions (i.e., 1->0 or 0->1) shared among PPs (18). This measure relates to the number of correlated evolutionary events among orthogroups, enabling to distinguish between shared phylogenetic inheritance and gene coevolution through discrimination of presence/absence patterns with different coevolutionary information content (compare Case1 and Case2 in Fig. 1B). The pairwise cotransition score (cotr_score), calculated as a function of total transitions observed in each orthogroup (t1 and t2) and the number of concordant (c) or discordant (d) state transitions, quantifies the fraction of similarity in evolutionary transitions involving two orthogroups. Correlated profiles (Fig. 1B) are expected to have cotr_scores >0 and ≤1, while anticorrelated profiles (SI Appendix, Fig. S1) are expected to have cotr_scores ≥-1 and <0.
Since in the dataset few unshared transitions deriving from biological exceptions or erroneous gene calls are often observed in correlated profiles (see, e.g., S14/S15 in Fig. 1B), profiles with a higher number of shared transitions tend to have higher scores than profiles with a lower number (compare the cotr_scores in Fig. 1B). Nevertheless, in the presence of a low number of transitions, it is possible to obtain high cotr_scores with a relatively high probability of random occurrence. The probability of obtaining a particular cotr_score by chance (p-value) was estimated from the pairwise transition tables using Fisher’s exact test (see Methods for details).
Finally, pairwise relationships that reached a predetermined level of significance (adjusted P-value < 10−3) in all tree orientations were clustered using Markov clustering (MCL) (31) and the inverse of P-value as a similarity measure to identify coevolving modules, i.e., coevolutionary relationships involving two or more orthogroups (Fig. 1A).
Results of the Coevolutionary Analysis.
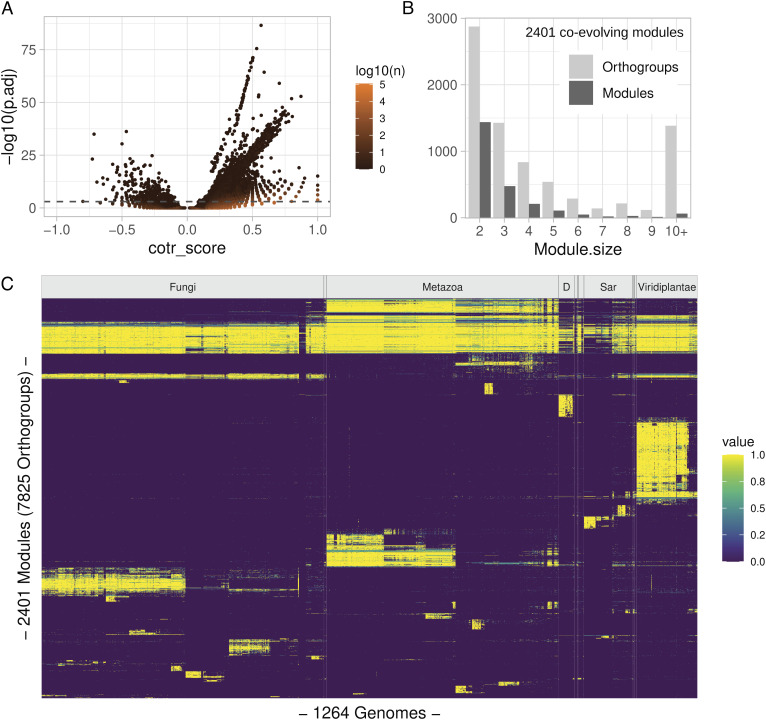
By applying the cotr analysis to the eukaryotic dataset with the species ordered according to a RL tree, we obtained 4,727,281 orthogroup pairs with unadjusted P-values < 10−3 (Fig. 2A). After P-value correction for multiple comparisons (SI Appendix, Fig. S2), we selected 57,716 pairs with significant adjusted P-values (P.adj < 10−3). Of these pairs, only a small part (530) had a negative score. In addition, at variance with positively scored pairs, negatively scored pairs shared sequence similarity (SI Appendix, Fig. S3), suggesting that they originated from problems in orthogroup construction (see next chapter). We focused on positively scored pairs for the rest of our analysis.
Fig. 2.
General results of the coevolutionary analysis. (A) Volcano plot showing the relation between score (cotr_score) and significance (adjusted P-values; P. adj) of 4,727,281 orthogroup pairs with unadjusted P-value <10−3; the numerosity of individual dots is indicated by the dot color as shown in the scale bar. The dashed gray line represents the p.adj cutoff (1e-3) used in subsequent analyses; transitions were calculated using an RL tree. (B) Size distribution of 2,401 coevolving modules obtained by applying the Markov cluster (MCL) algorithm to individual orthogroup pairs; 22,865 pairs with positive cotr_score and significant p.adj value in all fully resolved tree orientations were considered. (C) Organism distribution of coevolving modules. Colors indicate the fraction of orthogroups belonging to the same module present in individual genomes as shown in the scale bar. Vertical lines indicate the boundaries of taxonomic groups at the kingdom level. D=Discoba; groups with <20 members are unlabeled. Species are ordered according to an RL tree polarized with Viridiplantae as the starting node. Modules are ordered according to their Canberra distance in species distribution.
Quantitatively similar results were obtained with the species ordered by LL and NL trees. However, the set of significant orthogroup pairs obtained with different tree orientations was not completely overlapping, with about 15 to 16% of the significant pairs found in unique tree orientations (SI Appendix, Fig. S4A). Pairs with the most significant p-values were found in the subset shared by different tree orientations (SI Appendix, Fig. S4B). We selected this subset of 22,865 shared pairs for cluster analysis, obtaining 2,401 coevolving modules connecting a total of 7,825 orthogroups (Fig. 2B). The most represented modules involve association between two orthogroups. However, most orthogroups (63%) were found associated in modules with more than two members, and ~1,400 orthogroups were found in large modules with ≥10 members (Fig. 2B).
The distribution of the module presence across tree-ordered eukaryotic organisms (Fig. 2C) shows that a minor fraction of the 2,401 coevolving modules is composed by orthogroups that are found in the majority of eukaryotic genomes, whereas most of the modules comprise orthogroups present in specific taxonomic groups. At the kingdom level, the majority of coevolving modules are associated with Viridiplantae (37%), Metazoa (21%), and Discoba (14%). At lower taxonomic levels, groups with large fractions of associated modules include Oomycota, Mollusca, and Chordata among phyla, Mammals, Agaricomycetes, and Sordariomycetes among classes, Lepidoptera (i.e., butterflies) and Culicidae (i.e., mosquitos) among orders and families, respectively (SI Appendix, Fig. S5). Most of the coevolutionary signal, defined as the relative fraction of presence/absence transitions in coevolving modules across genomes, is found at the intersection of taxonomic groups both between and within kingdoms (Fig. 2C and SI Appendix, Fig. S6). The majority of such signals is not found in Opisthokonts (including Fungi and Metazoa), but in the remaining branches of eukaryote phylogeny, which are less represented by complete genomes (SI Appendix, Fig. S6).
Performance of the Method.
We evaluated the general performance of the method by constructing receiver operating characteristic (ROC) curves and measuring the area under the curve (AUC) parameter in curated datasets already used in previous coevolutionary studies (16), containing protein pairs known to interact (or known not to interact) in yeast (SI Appendix, Fig. S7) or humans (SI Appendix, Fig. S8). The use of a reference set of physically interacting proteins provides only a crude estimation of the true- and false-positive rates. In fact, interacting pairs could not have a coevolutionary signal in binary profiles (e.g., two universal components of the ribosome) as well as coevolving proteins could not have an experimentally detected interaction. However, the use of these or other similar reference sets for benchmarking a coevolutionary analysis is justified by the absence of independent evidence of gene coevolution, at variance with the availability of independent evidence for, e.g., sequence homology (32) and alignment (33).
We initially compared the results obtained by penalizing or not penalizing consecutive state transitions and found an appreciable improvement in AUC (0.67 vs. 0.62 in the yeast set) when consecutive state transitions were penalized (SI Appendix, Figs. S7A and S8A). We then compared the results obtained with different tree orders for both a fully resolved tree (“raxml”) and a partially resolved tree obtained with the ncbi taxonomy classification (“ncbi”) and found no appreciable differences in performance using different trees or different tree orientations. However, all trees provided largely better AUC values with respect to a random tree (SI Appendix, Figs. S7B and S8B). We also compared the performance of our method applied to a tree orientation of choice (“raxml.LR”) with other methods and obtained AUC values higher than those obtained with more conventional methods and similar to a recently published phylogeny-aware method (20) in the yeast dataset (SI Appendix, Fig. S7C) but lower in the human dataset (SI Appendix, Fig. S8C). Differences observed in this method comparison can depend on differences both in the scoring system and in the construction of orthologous groups (28, 34). However, we observed a very similar performance when the cotr analysis was conducted using a different orthology method (20, 34), both in the yeast and human datasets (SI Appendix, Figs. S7D and S8D). No improvement in the performance of different methods was observed when the analysis was restricted to specific clades including only fungal (for yeast) or metazoan (for humans) genomes (SI Appendix, Figs, S7E and S8E).
Various steps in the data generating process, including genome sequencing, gene calling, and orthogroup construction, can introduce bias in the analysis and produce false positive or negative results. An example is represented by orthogroup pairs with negative scores, which were shown to have high sequence similarities (SI Appendix, Fig. S3). Examination of such cases revealed that they mostly originate by the splitting of an orthologous group (SI Appendix, Fig. S9). Such problems in orthogroup construction are also a potential source of false negatives. We manually inspected the first one hundred largest modules (SI Appendix, Fig. S10) and noticed two modules with unusually high numbers of transitions, present particularly in plants (module #4) or scattered across eukaryotes (module #35). A search with the genes included in these modules established that they are of organellar origin, representing the almost complete collection of chloroplast- and mitochondria-encoded genes (SI Appendix, Fig. S11). Although these genes are certainly coevolving, they are considered false positives as their presence/absence pattern is due to their inclusion/exclusion from the genome source data.
Assessing the Functional Relationships of Coevolving Orthogroups.
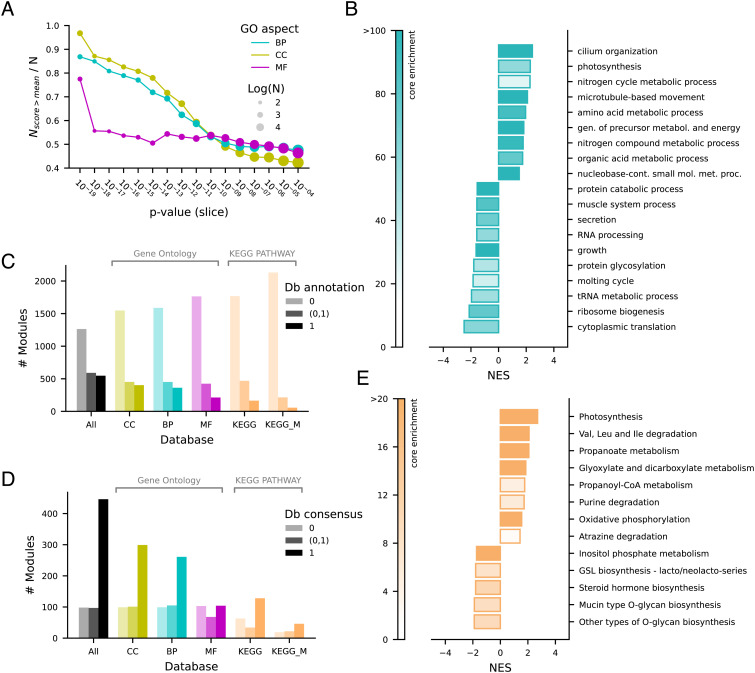
We used pathway annotation provided by different databases for the statistical assessment of the functional relationships of coevolving orthogroups (Fig. 3). We found that the ranking of orthogroup pairs established by significance is related to the degree of overlap in gene ontology (GO) experimental annotations for the cellular component (CC) and biological process (BP) aspects and to a lesser extent for the molecular function (MF) aspect (Fig. 3A). This suggests that the gene products of significant orthogroup pairs detected by our procedure often have the same locations relative to cellular structures (either cellular compartments, or stable macromolecular complexes) and participate in the same BP, while are less likely to share the specific molecular-level activity (for instance, if a member of a pair has a protein kinase or hydrolase activity, this does not imply the same activity for the coevolving orthogroup). Noteworthy, the curves showing the relation of annotation scores and cotr significance start to flatten at unadjusted P-values of ~10−10, approximately corresponding to our 10−3 cutoff for adjusted P-values (SI Appendix, Fig. S2).
Fig. 3.
Functional relationships of coevolving orthogroups. (A) Similarity of gene ontology (GO) experimental annotation in orthogroup pairs binned by unadjusted P-values. The fraction of orthogroup pairs with semantic similarity scores above the mean is reported for the GO aspects cellular component (CC), biological process (BP), and molecular function (MF). (B) Enrichment bar-plot of GO BP using an enrichment P-value cutoff of 0.001; NES = normalized enrichment score. (C) Module experimental annotation (fraction of orthogroups) in GO, KEGG, and KEGG metabolism (KEGG_M) databases. (D) Consensus of database annotation in modules. The database annotation consensus (Db consensus) indicates the fraction of annotated orthogroups included in the same GO-Slim term or KEGG map. (E) Enrichment bar-plot of KEGG metabolism using an enrichment P-value cutoff of 0.01; NES = normalized enrichment score.
To identify which particular GO terms involve coevolving proteins, we performed an orthogroup-level enrichment analysis (Fig. 3B and SI Appendix, Fig. S12). This analysis revealed enriched BP terms of the GO-slim subset (Fig. 3B), mostly in keeping with previous coevolutionary analyses such as, e.g., cilium organization (18, 35), photosynthesis (36), microtubule-based movement (37), amino acid metabolism (25). A large number of terms are found depleted of coevolving orthogroups. Understandably, these comprise universally conserved biological processes such as cytoplasmic translation and ribosome biogenesis. Also expected are terms related to the metabolism of proteins (protein catabolism) and nucleic acids (RNA metabolic process). Enriched CC terms confirmed the presence of ciliary structure, while only few, very general MF terms (such as oxidoreductase activity) were found enriched in coevolving genes.
We performed a module-level analysis using GO and KEGG databases to evaluate the fraction of modules that can be assigned to particular biological processes, CCs, and metabolic pathways, as well as the consensus annotation within modules. We found that about 55% of the modules are devoid of experimental annotation, about 20% have all annotated orthogroups, while 25% have only a fraction of annotated orthogroups, providing the possibility to predict the function of the unannotated ones through their associations (Fig. 3C). In modules with at least two annotated orthogroups, there is a general consensus with database annotations (Fig. 3D). However, in a substantial fraction of cases there is no or partial consensus. These modules can represent false-positive associations or still unknown connections between different pathways or processes.
Identification of Missing Genes in Metabolic Pathways through Cotr Analysis.
With the aim to provide an experimental validation of functional associations predicted by our method, we focused on modules mapping in KEGG metabolism. Only a minor fraction of the coevolving modules (263/2,401) in our dataset maps to KEGG metabolic pathways (Fig. 3 C–E). However, the identification of a significant association between a gene (orthogroup) not assigned to a metabolic pathway and known genes of the pathway provides the opportunity to make testable predictions about the function of unassigned genes. This opportunity is facilitated by knowledge of “pathway holes” (38), metabolic reactions for which no gene has been identified. In this case, unassigned genes identified by coevolution can be deemed as candidates for the unassigned reaction of the pathway.
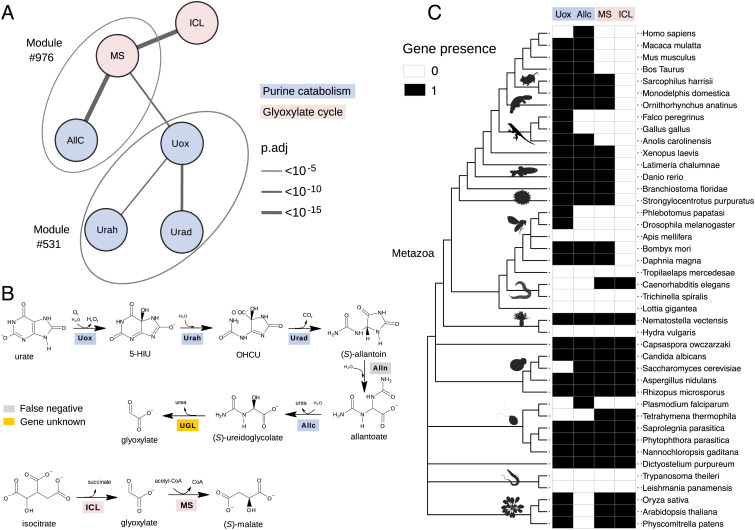
Among the metabolic pathways that were found to be enriched in coevolving genes is the purine degradation pathway (Fig. 3E), consistent with previous evidence (14, 39). Noteworthy, a gene responsible for the last step of the pathway, the formation of glyoxylate and urea from ureidoglycolate, has never been identified in metazoa, despite the demonstration of the existence of an ureidoglycolate lyase (UGL) activity in the tissues of some animals, including vertebrates (40). By inspecting the coevolutionary associations in our dataset, we observed a significant association between an orthogroup annotated as “malate synthase” (MS, 358540at2759) and orthogroups assigned to allantoicase (Allc, 563639at2759) and uricase (Uox, 906540at2759), responsible, respectively, for the penultimate and first steps of purine degradation (Fig. 4 A and B). MS is also significantly associated with Isocitrate lyase (ICL, 905115at2759), a gene involved with MS in the glyoxylate cycle (Fig. 4 A and B). Consistently, glyoxylate metabolism is another KEGG pathway enriched in coevolving genes (Fig. 3E). In our module collection, MS and Allc are found in the same module (#976), while Uox is found in a different module (#531) together with other purine degradation genes (Fig. 4A).
Fig. 4.
Identification of a missing gene in purine catabolism through cotr analysis. (A) Schematic network showing significant cotr associations of orthogroups representing urate oxidase (Uox) and allantoicase (Allc) with malate synthase (MS); other significant associations involve isocitrate lyase (ICL), HIU hydrolase (Urah), and OHCU decarboxylase (Urad). Modules defined by mcl clustering are encircled by ovals. (B) Scheme of the purine degradation and glyoxylate shunt reactions with the corresponding enzymes: True-positive genes retrieved by cotr analysis are shaded as in A; the false-negative allantoinase (Alln), member of the dihydropyrimidinase multigene family, is shaded gray; ureidoglycolate lyase (UGL) encoded by an unknown gene in Metazoa is shaded yellow. (C) Distribution map of Uox, Allc, MS, and ICL orthogroups across the ncbi phylogeny of selected eukaryotic species with emphasis on Metazoa. PhyloPic silhouettes (https://www.phylopic.org) are added to selected branches to aid species identification. A less detailed diagram of the gene distribution in 1,264 species is shown in SI Appendix, Fig. S13.
It should be noted that identification of such a coevolutionary association between MS and purine degradation genes would not have been possible with standard metrics of phylogenetic profile similarity due to the limited overlap of the MS profile with those of genes involved in purine catabolism (Fig. 4C and SI Appendix, Fig. S13). According to the Jaccard index, MS ranks over 300th and 500th positions with Allc and Uox, while it ranks first and third according to the cotr metrics. Both the Jaccard and cotr rankings are able to retrieve the known association between MS and ICL, while only the cotr ranking is able to retrieve the known association between Allc and Uox (SI Appendix, Fig. S13). A significant association between MS and purine degradation genes was also retrieved by cotr analysis using a different orthology dataset (34). This analysis confirmed the association of MS with AllC and Uox and found a significant score also with allantoinase (SI Appendix, Fig. S13), a gene not identified in the previous analysis (Fig. 4 A and B). Also with this dataset, most associations of purine degradation genes obtained a low Jaccard score.
Danio rerio MS-Like Encodes UGL, the Last Enzyme of Purine Degradation in Metazoa.
The known MS function is the conversion of glyoxylate and acetyl-CoA into the Krebs cycle intermediate (S)-malate, a reaction of the glyoxylate shunt (Fig. 4B). The observed association with purine degradation genes could be explained by the common metabolite (glyoxylate) of the two pathways. However, the fact that this association is particularly observed in metazoa (Fig. 4C), in which evidence of glyoxylate cycle activities is controversial (30), suggested the possibility that this gene in metazoa could instead or also be involved in the unassigned reaction (UGL) of purine catabolism.
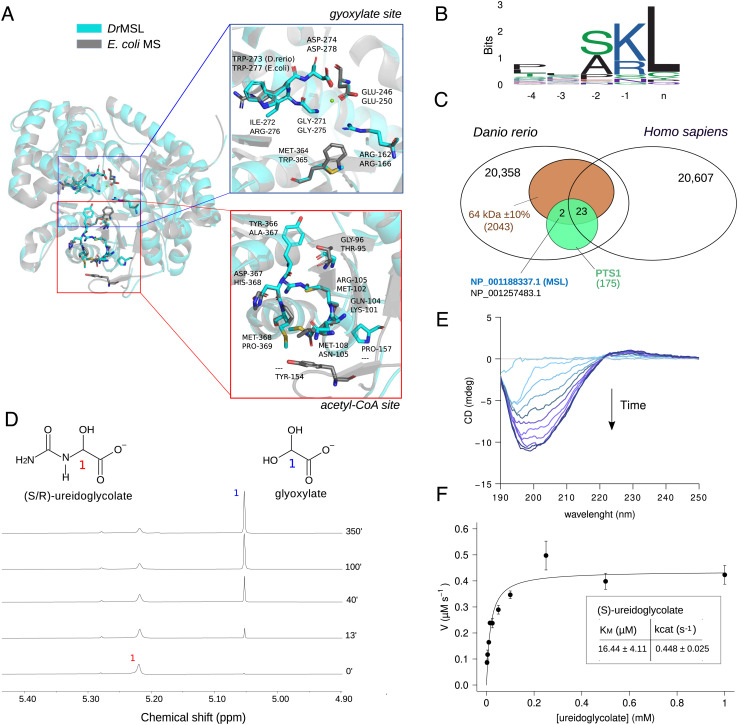
We examined the conservation of amino acid residues of proteins included in the MS orthogroup in multiple alignments and structural model comparisons (Fig. 5A and SI Appendix, Fig. S14). We found a large group of MS sequences in metazoa and other eukaryotes with distinct modifications at the active sites with respect to validated MS. In particular, these proteins, exemplified by the MS-like sequence of D. rerio [Danio rerio malate synthase-like (DrMSL), ncbi accession: NP_001188337] have a preserved glyoxylate-binding site except for the substitution of two conserved residues (Arg276Ile and Trp365Met) (Fig. 5 A, Upper). By contrast, no conservation of residue identities or chemical properties is observed at the site of acetyl-CoA binding (Fig. 5 A, Lower), consistent with the presence of a different activity. MS-like proteins are further distinguished by a 2-aa insertion at the interface of glyoxylate and acetyl-CoA-binding sites (SI Appendix, Fig. S14).
Fig. 5.
Danio rerio malate synthase-like (DrMSL) encodes ureidoglycolate lyase (UGL). (A) Cartoon representation of the DrMSL 3D homology model superimposed on the experimental structure of E.coli malate synthase A (PDB ID: 3CUZ) (41). Residues relevant for MS activity and the corresponding residues in DrMSL are drawn in sticks. The close-up panels show lack of conservation at the acetyl-CoA-binding site (red box) and conservation of residues involved in glyoxylate binding (blue box), except for two substitutions in DrMSL. (B) Sequence logo of MS and MS-like C-terminal sequences of selected eukaryotic species depicting the presence of a PTS1 motif. (C) Venn diagram of Homo sapiens and D. rerio proteomes with intersections defined by the expected features of the UGL enzyme (MW: 64 kDa ± 10%, PTS1 signal, present in D. rerio not in Homo sapiens). Accession numbers of the two proteins retrieved by the search (MSL and “protein brambleberry precursor”) are written in blue and in black. (D) Stacked plots of 1H NMR spectra of 52.5 mM ureidoglycolate in 95% D2O recorded at different time points after the addition of 2 µM DrMSL preincubated with 3 mM MgCl2. (E) Circular dichroism (CD) time-evolution spectra of 2.5 mM ureidoglycolate in the presence of 1 µM DrMSL, showing formation of the (R)-ureidoglycolate spectrum (42). (F) Fitting with the Michaelis–Menten equation of the initial velocity (V0) of 1 µM DrMSL with different substrate concentrations; the calculated kinetics constants are shown in the inset. Error bars represent SD of three independent experiments.
We checked the compatibility between DrMSL and UGL enzyme features described in purified fish liver extracts (40). We found a match between the experimentally determined and the calculated molecular mass (64 vs. 62.486 kDa), and between the peroxisomal localization of the activity and the presence of a peroxisome targeting signal (PTS1) in MSL proteins (Fig. 5B). Furthermore, absence of MSL genes in placentals (Fig. 4C) is consistent with the evidence that these organisms do not possess a true UGL enzyme (43). Interestingly, when a filter based on these expected features (molecular mass, PTS, in D. rerio not in Homo sapiens) was applied at the proteome level, only two compatible proteins were retrieved, one of which was DrMSL (Fig. 5C and SI Appendix, Fig. S15).
We validated the presence of UGL activity on the recombinant DrMSL protein overexpressed in Escherichia coli (Fig. 5 D–F and SI Appendix, Fig. S16) using chemically synthesized racemic ureidoglycolate. In the presence of purified DrMSL, we observed by 1H NMR the decrease of the ureidoglycolate peak at 5.22 ppm and the parallel increase of the glyoxylate peak at 5.05 ppm (Fig. 5D). The rate of the enzymatic reaction was clearly distinguishable from the rate of spontaneous hydrolysis of ureidoglycolate (SI Appendix, Fig. S16C). However, only half of the substrate appeared to be converted enzymatically. Circular dichroism spectroscopy provided evidence that the enzyme specifically converts the natural enantiomer (S) of ureidoglycolate (Fig. 5E) (42). Ammonia release was observed only in the presence of urease (SI Appendix, Fig. S16D), proving that the enzyme is a UGL (EC 4.3.2.3) as opposed to the ureidoglycolate amidohydrolase (EC 3.5.1.116), which releases glyoxylate and ammonia and is found in plants (39, 44). In a continuous coupled assay at various substrate concentrations, the enzyme exhibited Michaelis–Menten kinetics (Fig. 5F) with a catalytic efficiency (kcat/KM) of 2.7*104 s−1 M−1. These results indicate that the MS-like gene of D. rerio encodes the UGL enzyme responsible for the conversion of ureidoglycolate into gyoxylate and urea in the last step of purine degradation.
Evolutionary and Functional Divergence of MS and UGL in Eukaryotes.
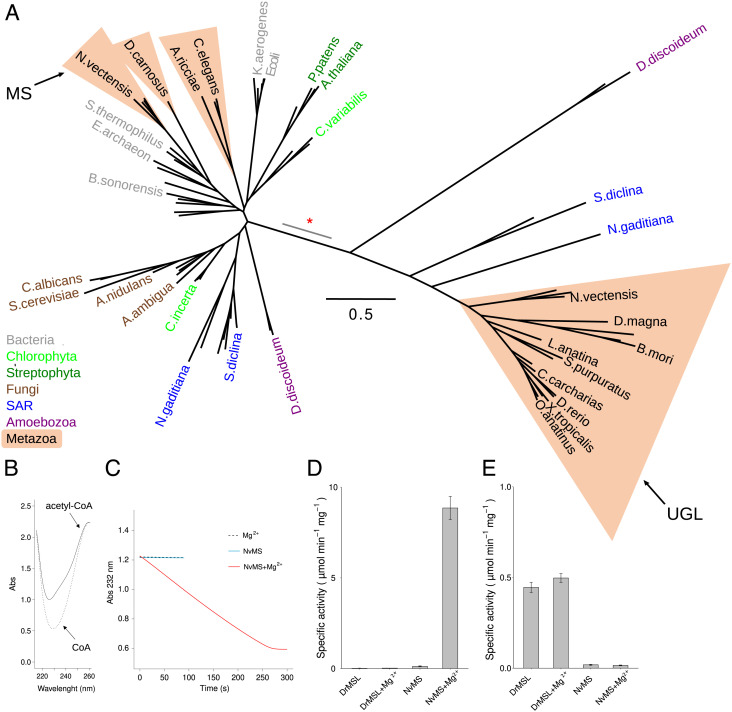
Phylogenetic analysis of proteins in the MS orthogroup, including bacterial homologs, provided evidence that the D. rerio protein characterized here belongs to a separated group of the MS family tree (Fig. 6A). This group (hereafter “UGL group”) includes most of the metazoan genes and genes found in Amoebozoa and the SAR clade. The other group (“MS group”) includes characterized glyoxylate cycle genes of plants (Arabidopsis thaliana) (45) and fungi (Saccharomyces cerevisiae) (46), but also homologs in some metazoan phyla such as Cnidaria (Nematostella vectensis), Rotifera (Adineta ricciae), and Nematoda (Caenorhabditis elegans) (47). A gene duplication event was inferred in the branch separating the two groups (red asterisk in Fig. 6A), as deduced by presence of both gene copies in species of Amoebozoa (e.g. Dictyostelium discoideum), SAR (e.g. Saprolegnia diclina), and Metazoa (e.g. N. vectensis). Eukaryotic sequences in the MS group are only in partial agreement with the organism phylogeny and are intermixed with bacterial sequences, suggesting the occurrence of horizontal gene transfer events (30). Conversely, the UGL group contains only eukaryotic sequences and, in consideration of the challenges in solving deep eukaryotic relationships with single-gene phylogenies (48), in approximate agreement with organism phylogeny (Fig. 6A), consistent with vertical transmission.
Fig. 6.
Evolutionary and functional divergence of malate synthase and ureidoglycolate lyase. (A) Unrooted maximum likelihood tree of MS and UGL sequences constructed using PhyML with the LG model (49). The scale bar corresponds to the number of calculated substitutions per site (0.5). Selected terminal nodes are labeled with the abbreviated species name; metazoan species are included in salmon triangles and other species are colored according to taxonomy. Proteins characterized in this work as malate synthase (MS) and ureidoglycolate lyase (UGL) are indicated by arrows. The branch of the inferred gene duplication separating MS and UGL is indicated by a red asterisk; the gray segment indicates uncertainty in the node position along the branch. (B) Superimposed spectra of acetyl-CoA (0.25 mM, solid line) and CoA (0.25 mM, dotted line). (C) Kinetics of the condensation of acetyl-CoA (0.25 mM) and glyoxylate (0.50 mM) catalyzed by NvMS, monitored at 232 nm. The assay was performed in the presence of 1 mM MgCl2 (dashed line), or 0.125 µM NvMS (light blue line), or both MgCl2 and NvMS (red line). (D) Malate synthase-specific activity of DrMSL and NvMS in the presence or in the absence of 1 mM MgCl2. (E) Ureidoglycolate lyase-specific activity of DrMSL and NvMS in the presence or in the absence of 1mM MgCl2.
When analyzed for residue conservation, the metazoan sequences of the MS group showed strict conservation of glyoxylate and acetyl-CoA-binding sites (SI Appendix, Figs. S14 and S17). We confirmed the presence of the MS activity on the recombinant protein of the sea anemone N. vectensis (NvMS, ncbi accession: XP_001639526.2) overexpressed in E. coli (Fig. 6 B–D and SI Appendix, Fig. S18). In the presence of glyoxylate and acetyl-CoA, NvMS caused a decrease in the UV signal at 232 nm, consistent with the release of CoA from acetyl-CoA. The reaction was strictly dependent on Mg2+ (Fig. 6 C and D) as already observed in MS enzymes (50). No CoA release was observed with DrMSL in the presence or absence of metal ions (Fig. 6D), suggesting that this protein is unable to catalyze the MS reaction. On the other hand, NvMS was unable to catalyze the UGL reaction as opposed to DrMSL, which catalyzed this reaction independently of the presence of Mg2+ or other metals (Fig. 6E and SI Appendix, Fig. S16E). These results provide evidence that genes enclosed in the MS and UGL groups encode functionally specialized enzymes.
Overall, our results support the malate synthase annotation for metazoan genes enclosed in the MS group and the presence of glyoxylate cycle in basal animal lineages, while sequences of metazoa and other eukaryotes enclosed in the UGL group and sharing the distinctive features of the D. rerio protein should be renamed UGL and assigned to purine catabolism. A proposal for a new name (UGL) and symbol (ugl) for the D. rerio gene has been submitted to the Zebrafish Information Network.
Discussion
Based on the statistical evaluation of the significance of coevolutionary gene transitions and the nonpenalization of nonmatching profile regions, the method described here enables the identification of local correlations in PPs. This orthogonal approach can reveal associations between orthogroups that originated at different times during evolution and “orthogroups” comprising genes with different functions as exemplified by the case studied here.
We are aware that our analysis has some limitations. The identification of shared transitions depends on the species order on the phylogenetic tree. As equivalent trees can have different leaf orders, the same tree can produce different cotransition scores and significance. Also, the method is rather sensitive to noise in genomic data: Since the number of shared transitions in coevolving genes is typically low (a median of 6 in our dataset at P.adj < 10−3), false transitions due to problems in genome sequencing, gene calls, or orthogroup construction, substantially affect the scores and sensitivity (Fig. 1B). The latter issue could be ameliorated in the future by the expansion and accuracy improvement of genomic data, while the first issue could be addressed by the identification of cotransitions on the ancestral rather than extant states of a phylogenetic tree. We note that the same metrics and statistics described here can be directly applied to cotransitions identified on tree edges.
Our coevolutionary analysis allowed us to recognize a gene family included in the malate synthase orthogroup as UGL through identification of a significant association with genes of purine catabolism, particularly Allc. Both UGL and Allc have complex evolutionary histories that do not coincide with the history of their association. UGL originated by gene duplication in early eukaryotes, but was lost in some organisms possessing Allc, such as fungi (Fig. 6A) in which the same function is fulfilled by a nonhomologous gene (51, 52) -a possible case of nonorthologous gene displacement. Allc genes with possibly a different function (53) have been retained in some organisms, such as reptiles and placentals (Fig. 4B), despite truncation of the pathway and loss of upstream (Alln) and downstream (UGL) genes—an example of how a gene can survive the loss of its partners. As a consequence of these evolutionary events, even after correction of the MS orthology into two separated MS and UGL groups, a very low similarity is observed between the UGL and Allc profiles (Jaccard score 0.18), although the association is still retrieved with high significance by cotr analysis (SI Appendix, Fig. S19).
This case of poorly similar profiles with highly significant cotr scores is not uncommon in our dataset. About 50% of the coevolving pairs and modules identified by our analysis have low or intermediate Jaccard scores (SI Appendix, Fig. S20 A and B) although with a high degree of consensus in database annotation (SI Appendix, Fig. S20C), and there is little overlap between rankings based on cotr significance or profile similarity (SI Appendix, Fig. S20D). An additional example in our module collection is nitrate assimilation (SI Appendix, Fig. S21), in which the known association between nitrate and nitrite reductase is retrieved with high significance in the presence of a low PP similarity due to reticulate evolution of analogous enzymes (54). The identification of highly significant associations in spite of evolutionary dynamics confounding phylogenetic profile similarity illustrates the ability of the procedure to detect complex gene interactions. Though effective with discontinuous patterns of coevolution, the cotr analysis has limitations in identifying genes associated with highly conserved biological processes, possibly explaining the observed performance in protein–protein interaction datasets.
Our results clarify the evolution of purine catabolism and glyoxylate cycle in eukaryotes. Glyoxylate cycle genes MS and ICL were probably present early in eukaryotes and also in metazoa as suggested by their presence in unicellular organisms of the Filozoan clade (i.e., Capsaspora owczarzaki, see Fig. 4C) and in basal animal lineages (i.e., Cnidaria, see Figs. 4C and 6A). However, they have been lost by most animal lineages. By contrast, UGL genes, which were similarly present at the origin of metazoans, have been retained in most lineages, although with many independent losses due to truncation of the purine degradation pathway (Fig. 4C and SI Appendix, Fig. S19). The distribution of UGL genes highlights differences in purine metabolism between placentals and other mammals (55), suggesting that marsupials and monotremes have a complete degradation pathway to glyoxylate (Fig. 4 B and C). In contrast, the pathway has been truncated to allantoin in the placental ancestor and further to urate in some mammals, including humans (56, 57).
Purine degradation and glyoxylate cycle are united by the fact that they are located, at least for some reactions, in the peroxisome, as evidenced by the presence of PTS1 or PTS2 signals in UGL and MS proteins (SI Appendix, Fig. S22). A functional connection is also suggested by instances in bacteria of MS genes included in purine degradation operons (e.g., the glcB gene of Paraglaciecola arctica), and the coordinated regulation of MS and purine catabolism observed in fungi (58). The degradation of allantoin locus (DAL) of Saccharomyces cerevisiae, a gene cluster for purine catabolism (59), includes a malate synthase gene (DAL7). Such cases, however, represent genuine MS genes as supported by the presence of dedicated UGLs of the bacterial/fungal type, such as the yeast DAL3.
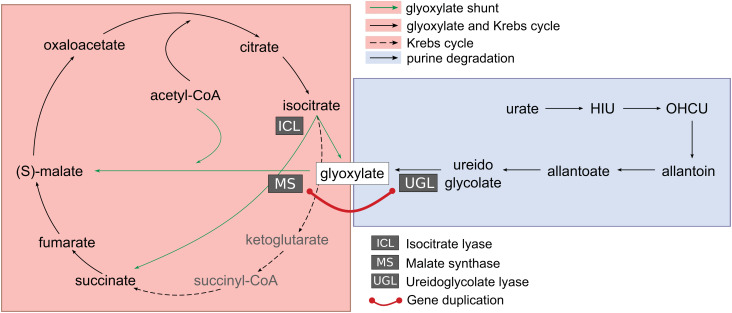
Our results now reveal the existence of an evolutionary link between genes involved in glyoxylate cycle and purine degradation (Fig. 7), as MS and UGL originated by duplication of an ancestral gene before separation of SAR and Metazoa (Fig. 6A), which is thought to have occurred about 1.5 Gya (60). One question concerns the metabolic function of the progenitor of the modern MS and UGL genes. According to a neofunctionalization scenario, the ancestral function could be assumed to be that of malate synthase, which could have been neofunctionalized into UGL through loss of function of the acetyl-CoA-binding domain (as opposed to the less likely gain of function from an ancestral UGL gene). According to a subfunctionalization scenario, a gene encoding a bifunctional protein with MS and UGL activities could be the ancestor of specialized genes with distinct activities. This scenario implies that in early eukaryotes, ureidoglycolate deriving from purine degradation was a significant source of Krebs cycle intermediate through the sequential UGL and MS reactions catalyzed by the same ancestral protein (Fig. 7). This could have provided early eukaryotes with a means to utilize the purine ring of nucleic acids as an energy source in the presence of available acetyl-CoA for the MS reaction.
Fig. 7.
Evolutionary connection of glyoxylate cycle and purine catabolism. The glyoxylate shunt (green arrows) bypasses two decarboxylation reactions of the Krebs cycle by converting isocitrate into succinate and glyoxylate through the enzyme isocitrate lyase (ICL). Glyoxylate is converted into (S)-malate, a Krebs cycle intermediate, by malate synthase (MS) through condensation with acetyl-CoA. Glyoxylate is also formed as the end product of purine degradation from ureidoglycolate in the reaction catalyzed by ureidoglycolate lyase (UGL). The evolutionary origin of MS and UGL by an ancient gene duplication in eukaryotes (red line) suggests a link between the two metabolic pathways and an anaplerotic role of purine catabolism in early eukaryotes.
Materials and Methods
Cotransition Analysis.
A memory-efficient algorithm for the enumeration of cotransitions was implemented in Python (https://github.com/lab83bio/Cotransitions). In a dataset containing the presence (“1”) or absence (“0”) of orthogroup genes (rows) in a list of species (columns) ordered according to phylogeny, the iterative difference is computed so that, e.g., the vector (1,1,0,0,1,1) representing a gene absent in the third and fourth columns, is encoded as (0,0,−1,0,1,0). Then, for each orthogroup, we determine the sets of column positions with present→absent (“−1”) and absent→present (“1”) transitions. Finally, set intersections are determined for each orthogroup pair to obtain the number of concordant (same sign) and discordant (different sign) transitions. These values are processed with an R script to compute the cotransition (cotr) score as follows:
where t1 and t2 are the total number of transitions for orthogroups 1 and 2, and k is the value concordant minus discordant. The cotr_score ranges from −1 to 1, as k can have positive (correlated transitions) and negative (anticorrelated transitions) values.
The probability to obtain by chance the observed cotr_score (significance) was calculated through the one-tailed Fisher’s exact test from the 2 × 2 contingency table:
| x’ | t1 − x’ | t1 |
| t2 − x’ | n − t1 − t2 + x’ | n − t1 |
| t2 | n − t2 | n |
where is the total number of positions in the transition vector (i.e. the number of genomes), and is . The use of the above contingency table is based on the following considerations. Given that the probability of observing by chance a number of concordant transitions ( ) equal or greater to that observed ( ) is:
and
The probability mass function is that of a hypergeometric distribution, in fact, Fisher’s exact test, which has been previously applied to the 2 × 2 table of presence/absence states of two genes (15, 61), can also be applied to the 2 × 2 table of presence/absence transitions. The P-value obtained by the test provides an exact measure of the significance if only concordant (or discordant) × transitions are counted and all transitions are counted equally. As in our case, consecutive transitions are considered only once, and opposite-sign transitions are penalized (Fig. 1B and SI Appendix, Fig. S1) and the value in the 2 × 2 table is x’ ≤ x, the test gives an upper bound estimation of the significance. P-values for individual tests were adjusted for multiple tests using the Holm correction. To obtain coevolving modules, orthogroup pairs with adjusted P-values <10−3 were clustered with mcl (v. 14-137) using the -log10(P.adj) as a similarity measure and an inflation parameter (“−I”) of 2.5.
Phylogenetic Profile Construction.
Orthogroups were downloaded from the OrthoDB database (v. 10.1) and parsed with R scripts to generate gene presence/absence tables. Data were filtered to retain only orthogroups at the eukaryota level according to the OrthoDB hierarchy and present in at least 1% of genomes. In the profile matrix, presence or absence of orthogroup genes (rows) in each genome (columns) were encoded as “1” and “0”, respectively. Genome columns were ordered according to the unresolved tree (684 internal nodes) of ncbi taxonomy (ver. Sept. 2022), or to a fully resolved tree (1263 internal node) obtained with RAxML (v. 8.2.12) (62) using the BINCAT model on the transposed profile matrix and the ncbi tree as constraint. Different tree orders were obtained through the ‘ladderize’ function of the ape package (63). In spite of the uncertainties in the root of the eukaryote phylogeny, we use Viridiplantae as the starting node to polarize unrooted trees and determine leaf orders. Casual polarization of the eukaryotic tree can produce otherwise incoherent leaf orders (e.g., with the splitting of Opisthokonta). For comparison, the same procedure was repeated using the hierarchical orthologous groups (HOGs) of the OMA database (Nov.2022 release), obtained by parsing data in the OrthoXML format with the PyHam library (64) and considering all and only eukaryotic genes at their most basal group level.
Pathway Analysis.
The orthogroup dataset resulting from our analysis was annotated with Uniprot-mapped GO terms only with experimental evidence codes (“EXP”, “IDA”, “IMP”, “IPI”, “IEP”, “IGI”, “HTP”, “HDA”, “HMP”, “HGI”, “HEP”, “IC”, “TAS”), as well as with KEGG maps and modules of the general and metabolism section of the KEGG pathway database. The semantic similarity scores of the GO terms of orthogroup pairs were calculated with the python package pygosemsim (https://github.com/mojaie/pygosemsim). The enrichment analysis was performed with the GSEA function of the clusterProfiler R library (65). Details of the procedure and of other bioinformatics analysis are reported in SI Appendix.
Experimental Validation.
MS and UGL gene functions predicted by the coevolutionary analysis were validated using biochemical assays on isolated proteins. DrMSL and NvMS proteins were overproduced in E. coli using synthetic clones purchased from GenScript and purified by affinity and size-exclusion chromatography with FPLC (SI Appendix, Figs. S16 and S18). Ureidoglycolate was synthesized according to a previously described procedure (66) with some modifications, as confirmed by 1H and 13C NMR spectroscopy (SI Appendix, Fig. S23). Details of the protein recombinant expression and purification, spectroscopic assays, and other experimental procedures are reported in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Yuval Tabach for valuable discussion at the ISMB2019. This work was supported by the Italian Ministry for Education, University and Research PRIN grant 2017483NH8 to R.P. and benefited from the equipment and framework of the COMP-HUB and COMP-R Initiatives, funded by the “Departments of Excellence” program of the Italian Ministry for University and Research (MIUR, 2018-2022 and MUR, 2023-2027), and from the High Performance Computing facility of the University of Parma, Italy.
Author contributions
R.P. designed research; E.D., M.M., D.C., A.S., and R.P. performed research; M.M., C.D.R., F.M., and R.P. contributed new reagents/analytic tools; E.D., M.M., C.D.R., and G.M. analyzed data; and R.P. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Software and notebooks to reproduce the analysis are available through GitHub (https://github.com/lab83bio/Cotransitions) (67). The datasets generated by the analysis are available through the Zenodo Open Data repository (https://doi.org/10.5281/zenodo.7578797) (68).
Supporting Information
References
- 1.de Juan D., Pazos F., Valencia A., Emerging methods in protein co-evolution. Nat. Rev. Genet. 14, 249–261 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Burger L., Van Nimwegen E., Disentangling direct from indirect co-evolution of residues in protein alignments. PLoS Comput. Biol. 6, e1000633 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamisetty H., Ovchinnikov S., Baker D., Assessing the utility of coevolution-based residue–residue contact predictions in a sequence-and structure-rich era. Proc. Natl. Acad. Sci. U.S.A. 110, 15674–15679 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ebert D., Fields P. D., Host–parasite co-evolution and its genomic signature. Nat. Rev. Genet. 21, 754–768 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Tatusov R. L., Koonin E. V., Lipman D. J., A genomic perspective on protein families. Science 278, 631–637 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Gaasterland T., Ragan M. A., Microbial genescapes: Phyletic and functional patterns of ORF distribution among prokaryotes. Microb. Comp. Genomics 3, 199–217 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Huynen M. A., Bork P., Measuring genome evolution. Proc. Natl. Acad. Sci. 95, 5849–5856 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pellegrini M., Marcotte E. M., Thompson M. J., Eisenberg D., Yeates T. O., Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc. Natl. Acad. Sci. U.S.A. 96, 4285–4288 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang J. Y., et al. , Dual sensing of physiologic pH and calcium by EFCAB9 regulates sperm motility. Cell 177, 1480–1494.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramazzina I., Folli C., Secchi A., Berni R., Percudani R., Completing the uric acid degradation pathway through phylogenetic comparison of whole genomes. Nat. Chem. Biol. 2, 144–148 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Sloan D. B., et al. , Cytonuclear integration and co-evolution. Nat. Rev. Genet. 19, 635–648 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabaldón T., Rainey D., Huynen M. A., Tracing the evolution of a large protein complex in the eukaryotes, NADH: Ubiquinone oxidoreductase (complex I). J. Mol. Biol. 348, 857–870 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Radhakrishnan G. V., et al. , An ancestral signalling pathway is conserved in intracellular symbioses-forming plant lineages. Nat. Plants 6, 280–289 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Cunningham F. X., Lafond T. P., Gantt E., Evidence of a role for LytB in the nonmevalonate pathway of isoprenoid biosynthesis. J. Bacteriol. 182, 5841–5848 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barker D., Pagel M., Predicting functional gene links from phylogenetic-statistical analyses of whole genomes. PLoS Comput. Biol. 1, e3 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ta H. X., Koskinen P., Holm L., A novel method for assigning functional linkages to proteins using enhanced phylogenetic trees. Bioinforma. Oxf. Engl. 27, 700–706 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Li Y., Calvo S. E., Gutman R., Liu J. S., Mootha V. K., Expansion of biological pathways based on evolutionary inference. Cell 158, 213–225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dey G., Jaimovich A., Collins S. R., Seki A., Meyer T., Systematic discovery of human gene function and principles of modular organization through phylogenetic profiling. Cell Rep. 10, 993–1006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cokus S., Mizutani S., Pellegrini M., An improved method for identifying functionally linked proteins using phylogenetic profiles. BMC Bioinformatics 8, S7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moi D., Kilchoer L., Aguilar P. S., Dessimoz C., Scalable phylogenetic profiling using MinHash uncovers likely eukaryotic sexual reproduction genes. PLOS Comput. Biol. 16, e1007553 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang Y., Li M., Li X., Yang Y., GFICLEE: Ultrafast tree-based phylogenetic profile method inferring gene function at the genomic-wide level. BMC Genomics 22, 774 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherill-Rofe D., et al. , Mapping global and local coevolution across 600 species to identify novel homologous recombination repair genes. Genome Res. 29, 439–448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin J., Lee I., Co-inheritance analysis within the domains of life substantially improves network inference by phylogenetic profiling. PLOS One 10, e0139006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsaban T., et al. , CladeOScope: Functional interactions through the prism of clade-wise co-evolution. NAR Genomics Bioinforma. 3, lqab024 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stupp D., et al. , Co-evolution based machine-learning for predicting functional interactions between human genes. Nat. Commun. 12, 6454 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dey G., Meyer T., Phylogenetic profiling for probing the modular architecture of the human genome. Cell Syst. 1, 106–115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zdobnov E. M., et al. , OrthoDB in 2020: Evolutionary and functional annotations of orthologs. Nucleic Acids Res. 49, D389–D393 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koonin E. V., Mushegian A. R., Bork P., Non-orthologous gene displacement. Trends Genet. TIG 12, 334–336 (1996). [PubMed] [Google Scholar]
- 29.Noguchi T., Takada Y., Fujiwara S., Degradation of uric acid to urea and glyoxylate in peroxisomes. J. Biol. Chem. 254, 5272–5275 (1979). [PubMed] [Google Scholar]
- 30.Kondrashov F. A., Koonin E. V., Morgunov I. G., Finogenova T. V., Kondrashova M. N., Evolution of glyoxylate cycle enzymes in Metazoa: Evidence of multiple horizontal transfer events and pseudogene formation. Biol. Direct 1, 31 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enright A. J., An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brenner S. E., Chothia C., Hubbard T. J. P., Assessing sequence comparison methods with reliable structurally identified distant evolutionary relationships. Proc. Natl. Acad. Sci. 95, 6073–6078 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson J., Plewniak F., Poch O., BAliBASE: A benchmark alignment database for the evaluation of multiple alignment programs. Bioinformatics 15, 87–88 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Altenhoff A. M., et al. , OMA orthology in 2021: Website overhaul, conserved isoforms, ancestral gene order and more. Nucleic Acids Res. 49, D373–D379 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nevers Y., et al. , Insights into ciliary genes and evolution from multi-level phylogenetic profiling. Mol. Biol. Evol. 34, 2016–2034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulkidjanian A. Y., et al. , The cyanobacterial genome core and the origin of photosynthesis. Proc. Natl. Acad. Sci. 103, 13126–13131 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hooff J. J., Tromer E., Wijk L. M., Snel B., Kops G. J., Evolutionary dynamics of the kinetochore network in eukaryotes as revealed by comparative genomics. EMBO Rep. 18, 1559–1571 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romero P., et al. , Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 6, R2 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Werner A. K., Romeis T., Witte C.-P., Ureide catabolism in Arabidopsis thaliana and Escherichia coli. Nat. Chem. Biol. 6, 19–21 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Takada Y., Noguchi T., Ureidoglycollate lyase, a new metalloenzyme of peroxisomal urate degradation in marine fish liver. Biochem. J. 235, 391–397 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lohman J. R., Olson A. C., Remington S. J., Atomic resolution structures of Escherichia coli and Bacillus anthracis malate synthase A: Comparison with isoform G and implications for structure-based drug discovery. Protein Sci. 17, 1935–1945 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.‘s-Gravenmade E. J., Vogels G. D., Van der Drift C., Hydrolysis, racemization and absolute configuration of ureidoglycolate, a substrate of allantoicase. Biochim. Biophys. Acta Enzymol. 198, 569–582 (1970). [DOI] [PubMed] [Google Scholar]
- 43.Fujiwara S., Noguchi T., Degradation of purines: Only ureidoglycollate lyase out of four allantoin-degrading enzymes is present in mammals. Biochem. J. 312, 315–318 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shin I., Han K., Rhee S., Structural insights into the substrate specificity of (S)-ureidoglycolate amidohydrolase and its comparison with allantoate amidohydrolase. J. Mol. Biol. 426, 3028–3040 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Cornah J. E., Germain V., Ward J. L., Beale M. H., Smith S. M., Lipid utilization, gluconeogenesis, and seedling growth in arabidopsis mutants lacking the glyoxylate cycle enzyme malate synthase. J. Biol. Chem. 279, 42916–42923 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Hartig A., et al. , Differentially regulated malate synthase genes participate in carbon and nitrogen metabolism of S. cerevisiae. Nucleic Acids Res. 20, 5677–5686 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu F., Thatcher J. D., Barral J. M., Epstein H. F., Bifunctional glyoxylate cycle protein of Caenorhabditis elegans: A developmentally regulated protein of intestine and muscle. Dev. Biol. 169, 399–414 (1995). [DOI] [PubMed] [Google Scholar]
- 48.Ren R., et al. , Phylogenetic resolution of deep eukaryotic and fungal relationships using highly conserved low-copy nuclear genes. Genome Biol. Evol. 8, 2683–2701 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guindon S., et al. , New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Schmid G., Durchschlag H., Biedermann G., Eggerer H., Jaenicke R., Molecular structure of malate synthase and structural changes upon ligand binding to the enzyme. Biochem. Biophys. Res. Commun. 58, 419–426 (1974). [DOI] [PubMed] [Google Scholar]
- 51.Percudani R., Carnevali D., Puggioni V., Ureidoglycolate hydrolase, amidohydrolase, lyase: How errors in biological databases are incorporated in scientific papers and vice versa. Database (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galanopoulou K., et al. , Purine utilization proteins in the Eurotiales: Cellular compartmentalization, phylogenetic conservation and divergence. Fungal Genet. Biol. 69, 96–108 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Vigetti D., Monetti C., Prati M., Gornati R., Bernardini G., Genomic organization and chromosome localization of the murine and human allantoicase gene. Gene 289, 13–17 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Ocaña-Pallarès E., Najle S. R., Scazzocchio C., Ruiz-Trillo I., Reticulate evolution in eukaryotes: Origin and evolution of the nitrate assimilation pathway. PLOS Genet. 15, e1007986 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keebaugh A. C., Thomas J. W., The genomes of the South American opossum (Monodelphis domestica) and platypus (Ornithorhynchus anatinus) encode a more complete purine catabolic pathway than placental mammals. Comp. Biochem. Physiol. Part D Genomics Proteomics 4, 174–178 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marchetti M., et al. , Catalysis and structure of Zebrafish urate oxidase provide insights into the origin of hyperuricemia in hominoids. Sci. Rep. 6, 38302 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharma V., Hiller M., Losses of human disease-associated genes in placental mammals. NAR Genomics Bioinforma. 2, lqz012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zambuzzi-Carvalho P. F., et al. , The malate synthase of Paracoccidioides brasiliensis Pb 01 is required in the glyoxylate cycle and in the allantoin degradation pathway. Med. Mycol. 47, 734–744 (2009). [DOI] [PubMed] [Google Scholar]
- 59.Wong S., Wolfe K. H., Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nat. Genet. 37, 777–782 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Hedges S. B., Marin J., Suleski M., Paymer M., Kumar S., TRee of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 32, 835–845 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barker D., Meade A., Pagel M., Constrained models of evolution lead to improved prediction of functional linkage from correlated gain and loss of genes. Bioinformatics 23, 14–20 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paradis E., K., Schliep, ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Train C.-M., Pignatelli M., Altenhoff A., Dessimoz C., iHam and pyHam: Visualizing and processing hierarchical orthologous groups. Bioinformatics 35, 2504–2506 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu T., et al. , clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation 2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winkler R. G., Blevins D. G., Randall D. D., Ureide catabolism in soybeans: III. Ureidoglycolate amidohydrolase and allantoate amidohydrolase are activities of an allantoate degrading enzyme complex. Plant Physiol. 86, 1084–1088 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Percudani R., Malatesta M., De C.. Rito., Statistical analysis of co-evolutionary transitions among genes. Github. https://github.com/lab83bio/Cotransitions. Deposited 3 February 2023
- 68.Dembech E., et al. , Identification of hidden associations among eukaryotic genes through statistical analysis of coevolutionary transitions. Zenodo. 10.5281/zenodo.7578797. Deposited 3 November 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Software and notebooks to reproduce the analysis are available through GitHub (https://github.com/lab83bio/Cotransitions) (67). The datasets generated by the analysis are available through the Zenodo Open Data repository (https://doi.org/10.5281/zenodo.7578797) (68).