Significance
There is limited understanding as to why individuals exposed to chronic psychosocial stress have a higher disease risk and lower survival. Research in animal models in this area is still limited. We report the largest study yet on the impact of lifelong social stress on healthspan, aging-associated diseases, epigenome, and lifespan in multiple mouse laboratory strains. Low social status was generally adverse for lifespan, although the cost of a given social rank varied across strains. These results were associated with corresponding epigenetic changes assessed via DNA methylation in the liver. Overall, our work provides a biological base and a preclinical model, to study the impact of social determinants of health disparities and accelerated aging.
Keywords: aging, epigenetic, social status, social determinants of health, strain differences
Abstract
Sustained life stress and low socioeconomic status are among the major causes of aging-related diseases and decreased life expectancy. Experimental rodent models can help to identify the underlying mechanisms, yet very few studies address the long-term consequences of social stress on aging. We conducted a randomized study involving more than 300 male mice of commonly used laboratory strains (C57BL/6J, CD1, and Sv129Ev) chosen for the spontaneous aggression gradient and stress-vulnerability. Mice were exposed to a lifelong chronic psychosocial stress protocol to model social gradients in aging and disease vulnerability. Low social rank, inferred based on a discretized aggression index, was found to negatively impact lifespan in our study population. However, social rank interacted with genetic background in that low-ranking C57BL/6J, high-ranking Sv129Ev, and middle-ranking CD1 mice had lower survival, respectively, implying a cost of maintaining a given social rank that varies across strains. Machine learning linear discriminant analysis identified baseline fat-free mass as the most important predictor of mouse genetic background and social rank in the present dataset. Finally, strain and social rank differences were significantly associated with epigenetic changes, most significantly in Sv129Ev mice and in high-ranking compared to lower ranking subjects. Overall, we identified genetic background and social rank as critical contextual modifiers of aging and lifespan in an ethologically relevant rodent model of social stress, thereby providing a preclinical experimental paradigm to study the impact of social determinants of health disparities and accelerated aging.
With the world population age trajectory shifting upward, chronological and biological aging has become a salient issue representing one of the strongest risk factors for morbidity and mortality (1). A gradient for health and mortality outcomes exists both among and within countries, with individuals occupying low socioeconomic status (SES) encountering higher incidence of disease and lower survival (2–4). Nevertheless, a full understanding of the interplay between psychosocial risk factors and noncommunicable diseases is yet to be accomplished. For example, the role played by failure of adaptation to stress and social determinants of health have been described as critical research areas (5, 6) and differential exposure to stress as mitigated by individual coping capabilities has been indicated as one of the pathways linking SES, health, and ultimately all-cause mortality (2, 7).
The process of aging is recapitulated across species giving rise and reason to animal models, ranging from worms to nonhuman primates (8, 9). Laboratory mice are one of the most commonly used model organisms in biomedical research, due to the potential to translate findings obtained in controlled randomized experiments in a small-sized and relatively short-lived social mammal to humans. Nevertheless, most of what we know of the biology of aging in mice has been obtained from one strain characterized by minimal genetic heterogeneity (10), the C57BL/6, and under baseline stress-free conditions, overall limiting the scope of the results. Diversifying mouse strains in aging research and investigating the role of environmental stressors could elucidate factors contributing to the known differences in lifespan and age-related traits among individuals.
The negative consequence of stress on health and aging are generally acknowledged and have been reproduced in several social mammals including humans (3, 4); nevertheless, individual differences are commonly observed between and among populations, and it is unclear whether genetic predisposition or the relative degree of stress experienced by an individual are critical determinants of disease accrual and lifespan. The present study was designed to address this fundamental gap in knowledge and to test the hypothesis that genetic background moderates the effect of social rank on health and aging. The design of the present study is centered on the applicability of lifelong chronic psychosocial stress (LCPS) (11) to identify contextual modifiers of healthspan and lifespan in male mice. This was achieved using a large randomized population-based design comprised of three different strains (C57BL/6J, CD1, and Sv129Ev), chosen among the most commonly employed in biomedical research and presenting a gradient of agonistic behavior (12–16). The inbred C57BL/6J strain was chosen due to its favored status especially in aging research because of its susceptibility to age-related metabolic disorders worsened by high-calorie diets, such as obesity, type-2 diabetes, and atherosclerosis as well as for its modest territorial aggression (11, 17). The CD1 strain is the most popular outbred strain employed in research and possesses demonstrated coping capability and high territorial aggression (18, 19). Finally, the inbred 129 strains present a high level of anxiety and stress vulnerability and predisposition to a passive coping strategy (13, 19–22).
Our study identified contextual modifiers of health and longevity. An index of contextual agonistic behavior was developed to capture individual coping capabilities to the socially stressful situation. This index allowed us to highlight a gradient of vulnerability linking social rank to survival probability, at the bottom of which mice acquiring a low social rank manifested the shorter survival in the study population. However, social rank differentially affected survival vulnerability in the three strains, indicating low-ranking C57BL/6J, high-ranking Sv129Ev, and middle-ranking CD1 mice as the more vulnerable groups. Strain and social rank differences were associated with corresponding alterations in the patterns of DNA methylation, a biological phenomenon increasingly implicated in aging mechanisms and a multitude of diseases (23–25). These results demonstrate that contextual factors such as genetic background and social status shape health trajectory as well as life expectancy in mice, offering a preclinical model for social determinants of health and implicating epigenetic mechanisms.
Results
Genetic Background Is a Critical Modifier of Healthspan and Lifespan under Chronic Psychosocial Stress (CPS).
Healthspan and lifespan of three mouse strains (C57BL/6J, CD1, and Sv129Ev) were characterized in the context of the exposure to a LCPS model consisting of three phases: a baseline phase of 5 d (during which all mice were singly housed), followed by a 4-wk CPS phase (during which mice were exposed to daily defeats and sensory contact housing), and an aging phase lasting until spontaneous death (or euthanasia for humane reasons). In the aging phase, mice were housed in sensory contact, thus experiencing a continued degree of threat. This study used 346 male mice that were 12-wk old male mice which were distributed as follows: Each C57BL/6J (N = 173) mouse was randomized to be transferred as an intruder to the home cage of either a CD1 (N = 86) or Sv129Ev resident mouse (N = 87. Note: 1 Sv129Ev mouse died before the completion of the CPS due to an accidental unintentional event, and was therefore excluded from the analysis). This group allocation was designed to elicit the full spectrum of aggression and stress coping strategies enabling the study of their impact on both lifespan and healthspan. At the same time, this approach avoided the well-established low intrastrain aggression manifested by inbred strains of mice (16, 26, 27). A trained observer quantified aggression exhibited and received (see Materials and Methods for further details) by each mouse using a continuous scan sampling scoring system during the daily social interactions occurring in the 4 wk of the CPS phase.
The distribution of aggression corresponded to the expected gradient of territorial aggression with CD1>C57BL/6J>Sv129Ev males (SI Appendix, Fig. S1 A–C). Sv129Ev mice showed the lowest level of aggression exhibited [F(2,342) = 19.98, P < 0.001, = 0.105, obs. power = 0.999 Sv129Ev-CD1, P < 0.001; Sv129Ev-C57BL/6J, P = 0.004], and the highest level of aggression received [F(2,342) = 13.20, P < 0.001, = 0.072, obs. power = 0.997; Sv129Ev-C57BL/6J, P = 0.043; Sv129Ev-CD1, P < 0.001].
Although the three strains used in this study are common in biomedical research, no previous work directly compared them, prompting us to perform an initial strain comparison analysis. Food intake, body weight, fat mass, fat-free mass, and plasma glucose were significantly correlated with strain and chronological age (SI Appendix, Fig. S1 D–H and Table S1). In general, C57BL/6J mice consumed less calories than CD1 and Sv129Ev mice (SI Appendix, Fig. S1D and Table S1). CD1 mice had an overall significantly higher body weight, fat and fat-free mass than the other two strains, which also differ significantly from each other (SI Appendix, Fig. S1 E–G and Table S1). Body composition was exactly opposite between C57BL/6J and Sv129Ev mice, in that Sv129Ev mice maintained over time significantly more fat and significantly less fat-free mass than C57BL/6J mice (SI Appendix, Fig. S1 F and G and Table S1). Finally, plasma glucose levels (measured after 4 h of fasting) were found to be higher in C57BL/6J compared to both CD1 and Sv129Ev mice (SI Appendix, Fig. S1H and Table S1).
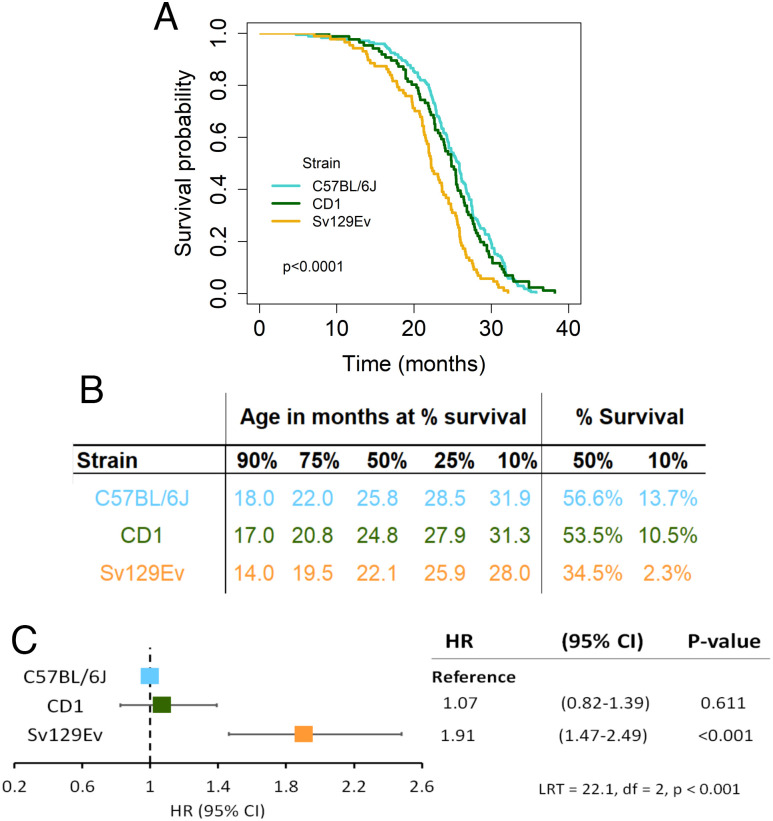
Survival was significantly associated with genetic background. Sv129Ev presented a significantly lower lifespan than either CD1 or C57BL/6J mice (HR = 1.91, P < 0.001), while CD1 mice did not differ compared to C57BL/6J as reference group (Fig. 1 A–C). Organ lesions detectable at necropsy differed by strain (SI Appendix, Fig. S2A and Table S2). Genital, hemorrhagic, and hepatic lesions differed significantly as far as the proportion of animals affected in each strain (SI Appendix, Fig. S2 B–J and Table S2). Across all strains, individuals presenting with a higher number of lesions had a longer lifespan, indicating an age-related accumulation of macroscopic nonlethal organ lesions (SI Appendix, Fig. S2 D, G, and J). Sv129Ev mice died at a younger age than other strains while having significantly fewer organ lesions detectable at necropsy (SI Appendix, Fig. S2A and Table S2).
Fig. 1.
Mouse genetic strain affects survival in the LPCS protocol. (A) Survival probability as a function of mouse strain is found to be significantly decreased in Sv129Ev (χ2 = 25.1 with 2 degree of freedom, P < 0.001, d = 0.560; Pairwise comparisons: log-rank test C57BL/6J vs. CD1, ns P > 0.999; C57BL/6J vs. Sv129Ev, P < 0.001; CD1 vs. Sv129Ev, P = 0.009, after Bonferroni adjustment for three comparisons). (B) Age (in months) when mice of the three strains reached respectively 90%, 75%, 50%, 25%, or 10% survivorship within each population (Left); % strain composition of overall surviving population both at median and maximum (10%) survival of the general population (Right). (C) Cox regression model examining the contribution of strain to the hazard of experiencing death (HR, hazard ratio; CI, confidence interval; LRT, likelihood ratio test).
Additionally, we used machine learning approaches to identify which of the measured variables could be the most informative determinants to discriminate between the three genetic backgrounds. The confusion matrix of the linear discriminant analysis (LDA) model was ~94% accurate in the test set reproducing mouse genetic background based on these predictors, thus matching almost exactly their annotation (P < 0.0001; SI Appendix, Table S3). The analysis of the contribution of the individual features to classifier performance evidenced that baseline fat-free mass and body weight, followed by baseline fat mass were the most important contributors to genetic background assignment (SI Appendix, Table S3).
The Aggression Index as an Indicator of Contextual Agonistic Behavior.
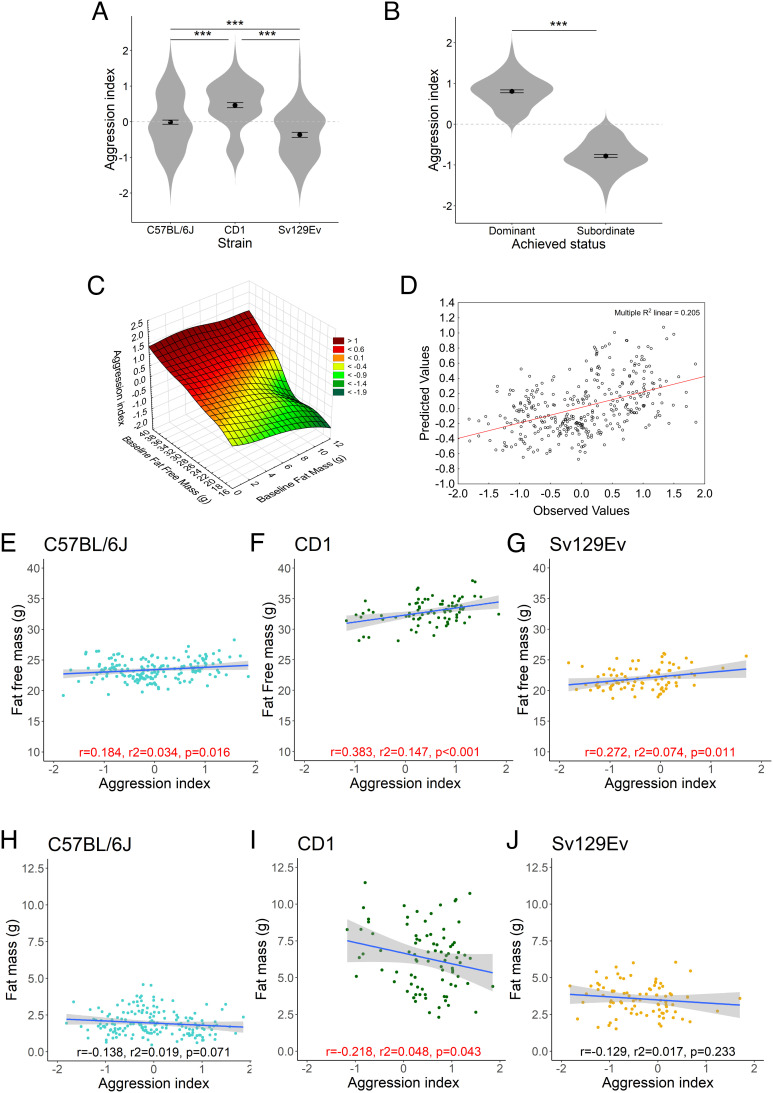
In chronic social stress protocols, mice are traditionally categorized as subordinate or dominant using an ethological criterion: dominant when exhibiting high levels and receiving low levels of aggression and subordinate in the opposite scenario (28). Albeit this is a convenient dichotomization, it suffers from several disadvantages in a population-based study, including uncertain categorization of individuals manifesting rank switch, low aggression, or open agonistic interactions without a clear social-rank definition. To address these limitations, we developed an approach to synthesize the level of contextual agonistic behavior and to capture the individual coping capability to the socially stressful situation designed after the Perceived Stress Scale (PSS) (29, 30). This index has been extensively validated against aging-related variables in humans (31, 32) and is based on two opposite-sign dimensions: the first dimension evaluates the level of perceived stress of an individual, while the second dimension evaluates the extent to which the individual’s ability to cope is perceived as surpassed. We adapted this construct to mice, developing an aggression index calculated upon the respective z-scores of either received or exhibited aggression, following a reductionist principle similar to the one of principal component analysis and that considered the consolidation of opposite-sign variables (see Materials and Methods for details). Confirming its biological relevance, the aggression index values were i) distributed similarly to the expected aggression gradient exhibited by the three strains CD1>C57BL/6J>Sv129Ev [F(2,342) = 29.22, P < 0.001, = 0.147, obs. power = 1.0] (Fig. 2A); and ii) clearly discriminated dominant and subordinate mice [F(1,231) = 1073, P < 0.001, = 0.821, obs. power = 1.0] (Fig. 2B). A multiple regression analysis with healthspan variables measured at baseline demonstrated that fat and fat-free mass are significant predictors of the aggression index outcome, albeit in opposite directions [R = 0.453, R2 = 0.205, F(2,342) = 44.15, P < 0.001]: fat mass being negatively associated [regression coefficient(SE): −10.078 (0.021)], and fat-free mass positively predicting the aggression index value [regression coefficient(SE): 0.092 (0.010)] (Fig. 2 C and D). Although only about 20% of the aggression index variance was explained by the multiple regression model including the two predictors, this result indicated a significant relationship between aggression index and body composition. The limited amount of the aggression index variance is plausibly explained in consideration of the complexity of the behaviors between the interacting animals.
Fig. 2.
Aggression index characterization. (A) Strains differed considerably for the level of aggression index with CD1 exhibiting higher levels than C57BL/6J and Sv129Ev; Sv129Ev expressed the lowest level [F(2,342) = 29.221, P < 0.001, = 0.105, obs. power = 0.999]. (B) Achieved social status was associated with significantly different aggression index values where dominant mouse had a significantly higher aggression index than subordinate mouse [F(1,231) = 1073, P < 0.001, = 0.072, obs. power = 0.997]. (C) 3D scatter plot of aggression index in relationship to initial values of fat and fat-free mass exhibited by mice. (D) Multiple regression scatterplot of observed and predicted aggression index values based on fat and fat-free mass predictors. (E–G) Scatterplot of the correlation between fat-free mass and aggression index in each strain. (H–J) Scatterplots of the correlation between fat mass and aggression index in each strain. Data represent group mean ± SEM in A and B. Asterisks represent significant differences from ANOVA with pairwise comparisons tested with Tukey’s honestly significant difference (HSD). In figures E through n, P < 0.05 are noted in red.
When strain was included as a factor, fat-free mass still remained significantly positively associated with aggression index within each strain [C57BL/6J: R = 0.206, R2 = 0.042, F(1,169) = 4.129, P < 0.05, regression coefficient (SE):0.076 −(0.04); CD1: R = 0.422, R2 = 0.178, F(1, 83) = 13.205, P < 0.001, regression coefficient (SE) 0.121 (0.03); Sv129Ev: R = 0.275, R2 = 0.076, F(1, 84) = 5.381, P < 0.05, regression coefficient (SE) 0.096 79(0.04)] (Fig. 2 E–J). This suggests that baseline fat-free mass might represent a more universal predictor for aggression index and social ranking, because it proved true both in smaller populations when analyzed within strain as well as across strains.
Social Status Described with the Discretized Aggression Index (DAI) Predicts Healthspan and Lifespan.
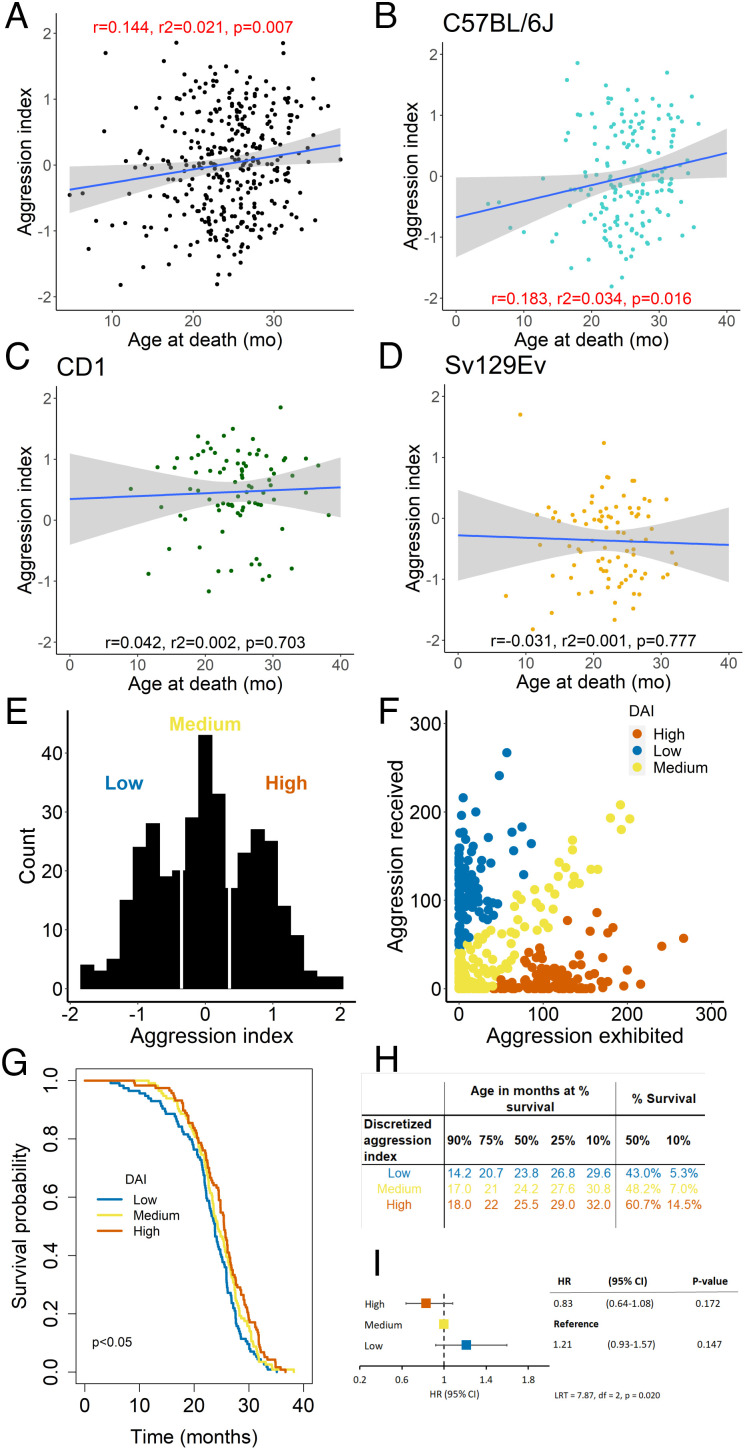
Next, we applied the aggression index to determine the role of the contextual aggression gradient and resulting social rank on health and aging. In the general population, the aggression index was significantly correlated with the age at death of the individuals (Fig. 3A), although this association was significant in C57BL/6J (Fig. 3B) but not in CD1 or Sv129Ev mice (Fig. 3 C and D). As a continuous variable, the aggression index related to a significantly lower hazard ratio (HR = 0.81, 95% CI: 0.70–0.94) and to a significantly higher number of lesions (SI Appendix, Fig. S3 A–D). Next, the aggression index was categorized into discrete groups, named low, medium, and high DAI, breaking the population on three groups based on tertiles (Fig. 3 E and F). The low-DAI group included all individuals that exhibited little and received high aggression (N = 117); the medium-DAI group included individuals that exhibited and received an intermediate level of aggression (N = 114); finally, the high-DAI group represented all individuals that exhibited high levels of aggression while receiving little (N = 114) (Fig. 3F).
Fig. 3.
Aggression index and mouse survival. Scatterplot of the correlation between aggression index and age at death for the general population (A), the C57BL/6J (B), CD1 (C), or Sv129Ev (D) strain. (E) Frequency distribution of aggression index with cutoffs identifying low, medium, and high groups. (F) Scatterplot of aggression exhibited and received identifying individuals within each DAI group. (G) Survival probability as a function of DAI is found to be impaired in the low-DAI group (Log-rank χ2 = 8.1 with 2 degrees of freedom, P = 0.020, d = 0.310; Pairwise comparisons for log-rank test low vs. medium, ns; low vs. high, P = 0.015 (significant at Bonferroni adjustment alpha 0.05/3 = 0.017); medium vs. high, ns). (H) Age (in months) when mice of within each of the three DAI categories reached respectively 90%, 75%, 50%, 25%, or 10% survivorship within each population (Left); % DAI composition of overall surviving population both at median and maximum (10%) survival of the general population (Right). (I) Cox regression model examining the contribution of DAI to the hazard of experiencing death (HR, hazard ratio; CI, confidence interval; LRT, likelihood ratio test).
The DAI had a significant association with mouse survival probability, with a small but significant shift to the left for the survival trajectory of mice belonging to the low-DAI category compared to the high-DAI category (Fig. 3G) corresponding to approximately 2 mo of lower median age at death and a lower proportion of mice reaching the median lifespan (Yates chi-squared = 7.66, df = 2, P = 0.0217, d = 0.301, obs. power = 0.999) (Fig. 3H). There was an overall significant association of the DAI with the hazard ratio that indicated ~20% increase of the risk of death in low compared to the medium DAI, while the high-DAI group was found to be protective with ~20% decreased hazard rate compared to the medium DAI (Fig. 3I). In line with the observed age-dependent accumulation of nonlethal organ lesions discussed above for the strain comparison, both medium- and high-DAI categories presented a heavier burden of lesions at necropsy (such as hepatic, genital, pulmonary, splenic) both for duration and representation in the sample population (see more details in SI Appendix, Fig. S3 E–J). Nevertheless, the disease burden for all three aggression index categories was inversely associated with lifespan (see more details in SI Appendix, Fig. S3 H–J).
Finally, machine learning approaches were performed to identify which of the continuous variables in the collected dataset were the most relevant determinants of DAI group classification. Our LDA model was able to discriminate between mice in the three groups (low, medium, and high DAI) with 60%, accuracy in the test set (P < 0.0001; SI Appendix, Table S3). ROC analyses of the combination of diagnostic variables revealed that the highest discriminative performance was achieved by baseline fat-free mass, followed by body weight in assigning DAI classification to our study subjects (SI Appendix, Table S3), and thus identified important predictors of achieved social rank during a dyadic social encounter.
Genetic Background and DAI.
The significant effect of genetic background on both healthspan and survival (Fig. 1 and SI Appendix, Fig. S1), somewhat limited the effect of the DAI categories when assessed within each strain (SI Appendix, Fig. S4 and Table S4). Food intake was the only healthspan parameter significantly affected by the DAI in the C57BL/6J strain, where both the medium- and the low-DAI groups consumed a higher amount of food than their high-DAI counterparts. In contrast, only fat mass was significantly increased in low compared to the high-DAI group in CD1 mice (SI Appendix, Table S4). Finally, in the Sv129Ev mice, the high-DAI group exhibited a unique phenotype characterized by increased food intake and fat mass when compared to the low-DAI group (SI Appendix, Table S4).
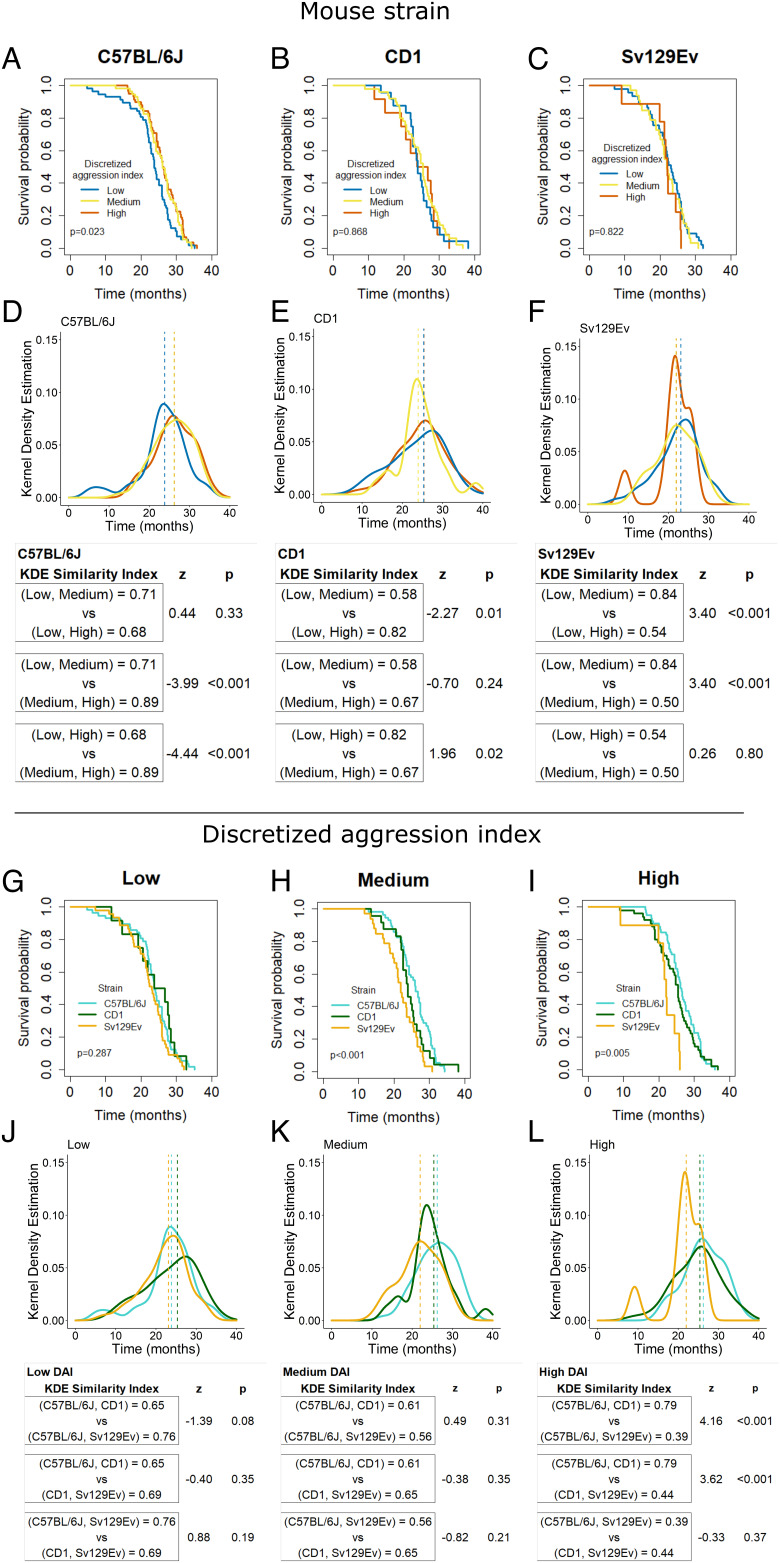
We next focused on the survival analysis. In addition to the Kaplan–Meier (KM) estimator and Cox proportional hazards models, we evaluated the probability of genetic background or DAI effects on survival by constructing the empirical distribution estimate of treatment group effects. This was done using a Gaussian kernel density estimation (KDE), based on smoothing histograms of survivorship frequencies with a predefined bandwidth (33). To quantify the similarity or difference among distributions thus described, we estimated the overlapping area intersected by two such KDE functions with an index η referred to as similarity index (34) having a range of 0 (complete separation) to 1 (complete identity). We conducted such analyses within each strain to estimate DAI effects (Fig. 4 A–F), and vice versa, within each DAI category to estimate strain effects (Fig. 4 G–L). In C57BL/6J mice, there was a significant overall effect on survival due to social rank, with a significantly lower median survival, higher hazard ratio, and lower similarity in the distribution of survival in the low compared to the other DAI groups (Fig. 4 A and D and SI Appendix, Tables S5 and S6). In CD1 and Sv129Ev mice, there was no significant difference due to DAI using the KM estimator. However, the KDE analysis of survival distributions with the similarity index identified the CD1 mice of the medium DAI category (Fig. 4 B and E and SI Appendix, Tables S5 and S6) and the Sv129Ev mice of the high DAI category (Fig. 4 C and F and SI Appendix, Tables S5 and S6) as the most dissimilar distributions and associated with lower survivorship compared to the other DAI groups within each strain.
Fig. 4.
(A–C) Survival probability as a function of DAI category within each of the three strains showing lower survivorship in low C57BL/6J DAI mice (A) (χ2 = 7.72 with 2 degrees of freedom, P = 0.023, d = 0.532), but no effect seen in either CD1 (B) or Sv129Ev mice (C). (D–F) distribution of death rates due to DAI smoothed through the KDE for each of the three strains; dashed lines indicate median values for each group and associated similarity index calculated for each of the three strains (see Materials and Methods and Results for details). (G–l) Survival probability as a function of mouse strain within each of the DAI categories showing that within low DAI (50% C57BL/6J, 10.5% CD1, 39.5% Sv129Ev) genetic background has no effect on survival (G), whereas the association with strain is still discernible in both medium (H) (χ2 = 13.6 with 2 degrees of freedom, P < 0.001, d = 0.736; 50% C57BL/6J, 21% cd1, 29% Sv129Ev) and High (I) (χ2 = 10.5 with 2 degrees of freedom, P = 0.005, d = 0.637; 50% C57BL/6J, 43% CD1, 7% Sv129Ev) DAI groups. (J–L) Distribution of death rates due to strain smoothed through the KDE for each of the three DAI groups; dashed lines indicate median values for each group and associated similarity indexes calculated for each of the three DAI groups comparing the three strains (see Materials and Methods and Results for details). * denotes significant differences after Bonferroni correction for multiple comparisons across groups (alpha 0.05/3 = 0.017 per test).
Interestingly, the significant effect of strain on mouse survival (Fig. 1) was absent within the category of low DAI (Fig. 4 G and J), while still being evident both in medium- and high-DAI categories (Fig. 4 H, I, K, and L and SI Appendix, Table S5). The incidence of deaths was similar across strains in both the low- and the medium-DAI groups (Fig. 4 J and K), while the high-DAI group showed a significantly lower similarity index when compared to other two strains (Fig. 4L). The result in the Sv129Ev strain is notable in that the high-DAI group within this strain exhibited lower aggression compared to the other two strains (SI Appendix, Fig. S1 A–C), yet they showed the lower survival, suggesting a high cost for maintaining dominance for this strain, which is not directly measured by the aggression index per se.
Overall, these data suggest that both strain and DAI have predictive power on survival: In particular, a higher cost of a low rank is observed in C57BL/6J, a greater cost of a high rank is observed in the Sv129 and, a higher cost of middle rank is observed for CD1 mice.
Strain and Social Rank Affect DNA Methylation Profiles.
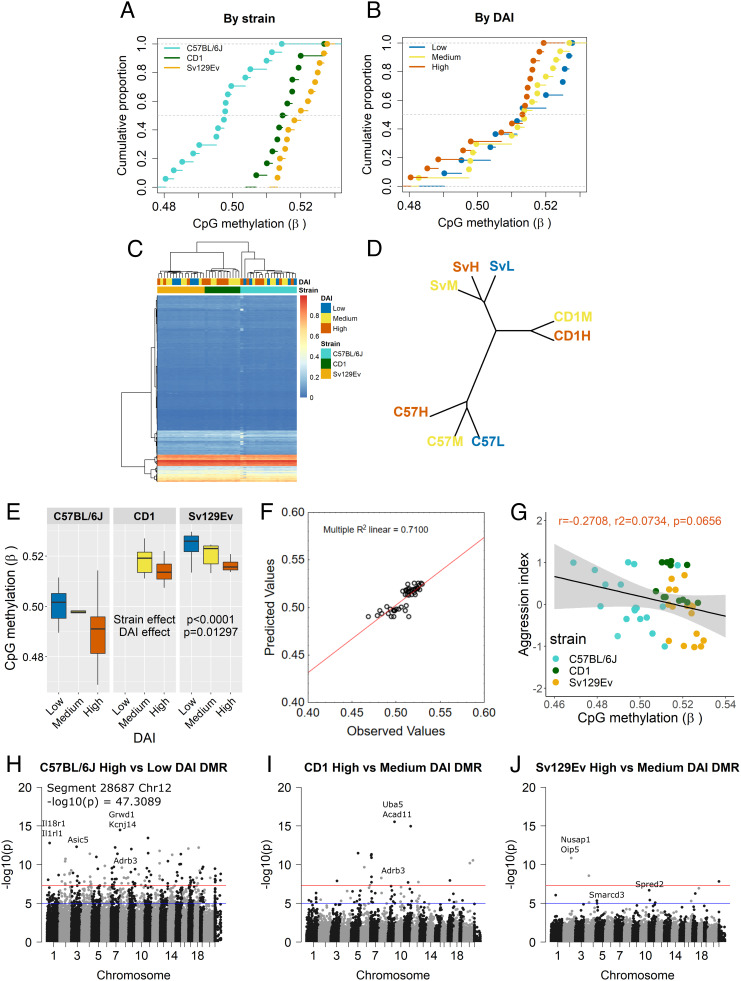
To characterize the possible source of the biological cost associated with strain and social rank, we tested DNA methylation changes, one of the hallmarks of cellular aging and an accepted biomarker of the aging process (35). Importantly, psychosocial stress has been linked to epigenetic aging and even aging acceleration in humans and other animals (2, 36, 37). Liver samples from a subset of the cohort killed at 17 mo were assayed with the recently released Illumina mouse methylation array [Infinium Mouse Methylation BeadChip, (38)] that profiles over 285,000 markers across diverse murine strains. Changes in the liver methylome have been shown to reflect changes in epigenetic aging due to lifespan-extending conditions in mice, with age-related methylation changes shared by mice and humans (39). Furthermore, our previous data showed the liver to be one of the organs mostly affected by lesions detectable at necropsy in mice under lifelong psychosocial stress as well as presenting social rank dependent changes in classic senescence makers such as p16 and p53 (11). The genome-wide DNA methylation profiling returned 287,050 targets from our samples, of which we retained 284,860 CpG probes. We excluded from further analysis any CpHs and single-nucleotide polymorphisms (SNPs) due to their low representation in our samples as well as CpG loci with missing data for >5% of individuals in the sample. The distribution of the average methylation level of CpG loci was found to be strongly dependent on strain (Fig. 5A), in agreement with previous data in rodents (40) and in humans, where shared genetic ancestry partially explains differential methylation between ethnic groups (41). In addition to genetic background, environmental factors contribute to variation in methylation both in rodents and humans (42). In the present study, we found that methylation varied as a function of social status as captured by the DAI (Fig. 5B). This result was confirmed by the unsupervised cluster analysis of the CpG probes and corresponding ~30,000 CpG islands, highlighting a segregation due to strain first and to DAI within strain (Fig. 5 C and D). Given that the experimental design was established long before organ collection and that the DAI was calculated a posteriori, on the entire population, no CD1 classified as low-DAI category was available in the cohort sampled at 17 mo for the DNA methylation assay. Nevertheless, these results demonstrate that, similar to other outcome variables, the power of DAI to predict the level of average methylation is strain dependent. In the C57BL/6J, low- and medium-DAI mice clustered closer to each other, while medium and high DAI segregated closer in the Sv129Ev strain (Fig. 5D). ANOVA showed a significant effect both of strain [F(2, 42) = 48.3, P < 0.001, = 0.707, obs. power = 1.0] and DAI [F(2, 42) = 4.4, P = 0.0179, = 0.187, obs. power = 0.769] on global DNA methylation levels, consisting of overall lower β values for C57BL/6J mice compared to CD1 and Sv129Ev mice (Tukey’s HSD tests: C57BL/6J vs. CD1, P = 0.0001; C57BL/6J vs. Sv129Ev, P = 0.0001; CD1 vs. Sv129Ev, P = 0.307), as well as lower methylation levels for the high DAI compared to both the low and medium groups (Tukey’s HSD tests: high vs. low, P = 0.039; high vs. medium, P = 0.025; low vs. medium, P = 0.996) (Fig. 5E).
Fig. 5.
(A and B) Empirical cumulative distribution function of the CpG loci methylation level (expressed as β value) in consideration of either strain (A) or DAI (B) in which horizontal lines represent that the same cumulative probability applies within the same β interval. (C and D) unsupervised cluster analysis of global DNA methylation shows grouping by strain and DAI: (C) Heatmap limited for clarity to 30370 CpG islands. Highlighted are strain and DAI; CpG islands were clustered in an unsupervised manner utilizing Euclidean distance and Ward.D2 clustering method. (D) Phyloepigenetic tree constructed from the methylation level of individual CpG loci, highlighting unsupervised clusters due to strain and to DAI within strain. (E) Tukey box-and-whisker plot of the average level of CpG loci methylation in consideration of the main effect of both strain and DAI. Due to the post hoc sample selection, no samples matched the low-DAI category of the CD1 harvested at 17 mo for the tissue bank. Box plot shows median, upper, and lower quartiles, minimum and maximum values. (F) Multiple regression scatterplot of observed and predicted average global CpG methylation levels based on strain and aggression index predictors. (G) Scatterplot of the correlation between global CpG methylation level and aggression index in the general population. (H–J) Features of DMRs. Each point in the Manhattan plot represents the location of a CpG region (x axis: autosomal chromosomes 1 to 19, chromosome X as 20 and chromosome Y as 21), defined by CpG loci grouped by Euclidean distance, and the association –log10p (y axis) for the effect of DAI within each of the three strains (H) C57BL/6J (segment 28687 was included in the plot not in scale for aesthetical reasons), (I) CD1, and (J) Sv129Ev. The genome-wide significant threshold is set at –log10(5e-08) (red line) and the suggestive line threshold at –log10(1e-05) (blue line).
A multiple regression analysis demonstrated that strain and DAI are significant predictors of the average methylation level [R = 0.8426, R2 = 0.7100, F(3, 43) = 35.0959, P < 0.0001]: aggression index as a continuous predictor was negatively associated [regression coefficient(SE): −0.0059 (0.0021)], and strain positively predicting the aggression index value with C57BL/6J representing the reference [CD1 regression coefficient(SE): 0.076 (0.0019); Sv129Ev regression coefficient(SE): 0.0079 (0.0018)] (Fig. 5F), explaining 72% of the dependent variable variance.
In the general study population, although aggression index had a statistically nonsignificant negative association with the average methylation level [R = 0.273, R2 = 0.075, F(1, 45) = 3.64, P = 0.063, regression coefficient (SE): −0.006 (0.003)] (Fig. 5G), a pattern emerged in the data distribution. A closer inspection identified a strain effect, which informed a follow-up analysis within each of the three strains: C57BL/6J aggression index was not significantly associated with methylation levels (SI Appendix, Fig. S5A), albeit showing the wider distribution in β values; CD1 strain β and aggression index values were narrowly distributed without a significant association between the two (SI Appendix, Fig. S5B); Sv129Ev mice had a significant negative association between the two variables, explaining approximately 30% of the sample variability, and their β values were narrowly distributed (SI Appendix, Fig. S5C). Taken together, the global methylation patterns demonstrated to be strain-specific but to be also reflective of individual social standing within each strain.
DNA methylation has given rise to several age predictors, including epigenetic clocks in humans (24, 35, 43, 44). Only recently, the methylation array used in this study has become available for mice, for which the available clocks are of limited transferability across datasets owing to analytical limitations (45). For the current dataset a significant effect of strain was revealed on the DNA methylation-based age predictor [F(2, 40) = 4.20, P = 0.022, = 0.173, obs. power = 0.705], while we could not identify a significant effect of DAI. In general, an older epigenetic age was identified for Sv129Ev compared to the C57BL/6J strain (Tukey’s HSD P = 0.0193) but not comparing Sv129Ev to CD1 (P = 0.5448) or C57BL/6J to CD1 strain (P = 0.2813) (SI Appendix, Fig. S5D). Future implementations of epigenetic clocks based on mouse DNA methylation array-derived data will likely allow the validity of this assessment to be improved.
Finally, using an epigenome-wide association study approach, we identified differentially methylated CpG loci (DMLs) and regions (DMRs) to investigate within each strain which features of the genome display altered methylation patterns due to the effect of DAI. The reference group for each strain was inversely based on their respective similarity index value (i.e., high vs. low DAI in C57BL/6J mice; high vs. medium DAI in both CD1 and Sv129Ev). Only a limited number of DMLs reached the multiple testing significance threshold for whole-epigenome data (P < 5e−08) or the commonly accepted suggestive threshold (P < 1e−05) (SI Appendix, Fig. S6 A, C, and E and Table S7). The highest significance was reached by cg38646594 on Chromosome 2 resulting hypermethylated in high- compared to low-DAI C57BL/6J mice, but no annotated gene corresponded to this probe yet, to the best of our knowledge. We subsequently conducted a regional analysis identifying several DMRs each containing ≥3 DMLs that were significantly associated with DAI groups (Fig. 5 H–J and SI Appendix, Table S8). Of these, ~65% were hypermethylated in high- compared to low-DAI C57BL/6J mice, ~21% in CD1 and ~70% in Sv129Ev where high was compared to medium DAI. Gene cluster methylation that was sensitive to DAI was found to be broadly related to immune system, metabolism of proteins, signal transduction, and gene expression (transcription) pathways identified using Reactome across all strains (SI Appendix, Fig. S6 B, D, and F and Table S8). In the C57BL/6J high-DAI group, we observed methylation changes when compared to the reference group reflecting an altered gene expression for Il18r1-Il1rl1 [implicated in the control of TH1/TH2 balance (46)], Adrb3 [a gene whose methylation levels have been reported to be associated with metabolic dysregulation (47)], and Kcnj14 [a gene implicated in contractile function (48)]. CD1 high-DAI group showed altered methylation at Uba5 [part of the UFMylation complex involved in protection against apoptosis (49)] and Nphp3 [implicated in organelle biogenesis and maintenance (50)], as well as Adrb3. Changes in the Sv129Ev high DAI group included: Smarcd3 [whose hypermethylation has been associated with accumulation of p21 in cells and DNA damage (51)], Spred2 (a key negative regulator of MAPK signaling, also linked to metabolic abnormalities), and Oip5 [implicated in cell cycle and chromatin organization (52)].
Overall, the results of the DNA methylation patterns reveal epigenetic changes in line with increased risk conferred by genetic background (strain) and social rank (DAI) on healthspan and lifespan.
Discussion
The development of experimental models recapitulating the negative effect of adverse social determinants of health and aging (SDoHA) in humans is of crucial importance. However, studies in mouse models of social stress are often neglected in aging studies, thus missing a critical opportunity in translational research. Here, we present the larger study conducted to date on the consequences of a lifetime of chronic social stress on healthspan and survival of males of multiple mouse strains commonly used in biomedical research, C57BL/6J, Sv129Ev, and CD1. We demonstrate that the link between chronic social stress and individual vulnerability to aging-relevant outcomes is moderated by genetic background and is modified by social rank, both of which can be predicted using machine learning algorithm by baseline fat-free mass. We also show an association between the negative effect of social factors on health and aging, and changes in DNA methylation patterns in the liver, suggesting the existence of epigenetic mechanisms linking negative social relationships and genetic background, possibly affecting individual biological age.
Genetic Background Is a Contextual Predictor of Healthspan and Lifespan under Chronic Social Stress.
Genetic background and environmental factors shape healthspan and lifespan trajectories. Specifically, it is well established that aging is associated with alterations in body weight and body composition. Similar to humans, aging in mice is linked to a biphasic trajectory due to accumulation of fat mass (at least up to the seventh decade in humans and ~24 mo in mice) followed by a decrease, and discrete glucose intolerance reflecting the development of insulin resistance (53). Different murine genetic backgrounds exhibit a wide range of adiposity throughout their lifespan suggesting a strong genetic contribution. From a metabolic vulnerability standpoint and under standard housing stress-free conditions, the three strains used in this study are distributed along a gradient, where C57BL/6J mice are considered obesity- and type 2 diabetes-prone, CD1 intermediate, and Sv129Ev obesity-resistant (18, 54–56). In our study, C57BL/6J mice developed and sustained higher levels of plasma glucose compared to the other strains independently from obesity (57–59), and could be at the origin of the heightened vulnerability of this strain to the effect of psychosocial stress on impaired glucose homeostasis (60).
Unexpectedly, Sv129Ev mice accumulated more fat mass than C57BL/6J, and lost fat-free mass over time in parallel to lower glucose levels, which can be considered an important risk factor. Indeed, low glucose in mice is associated with increased mortality (61), in contrast to primates where the opposite relationship is generally true. Excessive adiposity has also been hypothesized to depend on their lower autonomic stress responsivity compared to the C57BL/6J strain in which stress-induced sustained autonomic response can contribute to limit fat mass accrual (13). Furthermore, during the LCPS a sustained hyperphagic response was manifested by Sv129Ev mice – more pronounced than in C57BL/6J subjects—thus facilitating excessive fat accumulation. Obesity and excessive adiposity are consistently found related to higher all-cause mortality across human populations (3). The Sv129Ev strain proved to have the shortest lifespan of all strains concomitant with excess adiposity. This result is at odds with data published on the various Sv129 substrains in standard housing conditions that describe similar lifespan to C57BL/6 and CD1 male mice (SI Appendix, Table S9), overall, suggesting that stress vulnerability can play a critical role.
The end-of-life pathology of the three strains is well characterized in standard conditions (62–64). Our data describe a wide spectrum of abnormal organ findings with a strain-specific signature: i) biliary and pulmonary findings more common in CD1; ii). genital lesions more abundant in C57BL/6J; and iii) the Sv129Ev strain, manifested fewer lesions than the other strains possibly due to their shorter survival time that limited the accumulation of nonlethal organ lesions.
In consideration of the above, the lifespan shortening of Sv129Ev mice can be proposed to be the result of an overall stress vulnerability leading to hyperphagia-driven increased fat mass, commonly linked to sustained inflammation (65).
Social Status Is a Contextual Modifier of Healthspan and Lifespan.
Attention has recently been called to complement the classical biological pillars of aging with “social hallmarks of aging” (5) in order to explain the significant amount of variance in age-related diseases. Low SES, adverse life events, psychological states, and behaviors are introduced as interconnected elements comprising the contributors to social adversity that crucially affect health and aging (5, 66). Among them, low SES is proposed as the most fundamental because it is connected to multiple types of resources and is linked to damaging biological changes affecting both developmental capacity and age-related health outcomes (5, 66).
We aimed at assessing if social gradients could affect healthspan and lifespan in our mouse model. We developed an aggression index including both active (aggression exhibited) and passive (aggression received) behaviors. This index was modeled after one of the most widely used instruments to measure the perception of stress in humans, the PSS (29, 30), also shown to predict biological aging and survival (31, 32). Importantly, our aggression index, and thus social rank, was predicted by baseline fat-free mass both across strains and within each strain, thus adding to recent data of biological predictors of social rank in male mice (67) and altogether suggesting that genetic determinants of quantifiable physiological features can predispose to high or low social rank attainment.
We categorized the DAI using the tertiles of its distribution: The high-DAI group indicates social dominance; the low-DAI group corresponds to the expression of social subordination; the medium-DAI group reflects a continuum of undefined and/or unstable social rank. In the general population, mice belonging to the low-DAI category had a significantly shorter life expectancy compared to their high- and medium-DAI counterparts. However, the detrimental consequences on healthspan and lifespan due to social rank were modulated by the genetic background: a low social rank was associated with a higher cost in C57BL/6J; a high social rank was associated with a higher cost in Sv129Ev; and finally, in CD1 a higher cost of achieving an intermediate rank (characterized by various degrees of rank instability) emerged. Strain vulnerability in response to chronic subordination stress exposure has previously been established (68), and our work extends this characterization to a longevity study.
In humans, morbidity and mortality rates are higher for individuals of low SES compared to those of high SES, with people in lower SES groups reporting greater chronic stress (3, 4, 69). Seminal work by McEwen and others defined the concept of allostatic load, by theorizing that the subjugation to stress would lead individuals to suffer physiological consequences to a degree mitigated or exacerbated as a function of differential coping strategies (70). Heightened chronic stress in low-SES individuals may translate to different biologic risk factors for diseases (3, 71) because stress exposure can impact health directly and indirectly by various mechanisms including for example immune activation (72, 73). Consistently, chronic inflammation, splenomegaly, and increase in the levels of circulating proinflammatory cytokines have recently been shown in mice selectively bred for subordinate traits in the test tube challenge that manifest a significant survival deficit (74). Human longevity stratifies on the basis of race and SES among other factors (4). Overall, our model in a controlled experimental condition recapitulates critical variables linking SES to health and aging in a model organism where social factors can be randomized and mechanistic studies conducted. Furthermore, our data show that the extent to which the DAI modifies life trajectory in male mice is dependent on the genetic predisposition (the strain in this study) that defines individual stress susceptibility, coping capacity, and the trade-off of achieving and maintaining social status.
Genetic Background and Social Status Reflections on Epigenetic Changes.
Phenotypic outcomes are thought to be the product of the interplay between environmental and genetic factors resulting in epigenetic changes (23), with genome-wide CpG methylation representing one of the main mechanisms linked to both aging and cumulative life stress effects (23, 36). Consistent with this premise, evidence of genome-wide methylation changes, both due to strain and DAI, were found in the present study. In particular, the Sv129Ev strain had the higher degree of overall global methylation compared to both CD1 and C57BL/6J mice; on the other hand, a low social rank (reflected by low DAI) was found to be linked to higher global methylation levels. While the understanding of the implications of age-related DNA methylation changes in mice is a field still in its infancy (75, 76), available data reveal age-related changes in promoters and increased methylation in development and differentiation-related pathways (25). More importantly, the relevance of methylation patterns changes is underscored by the tendency of evolutionarily conserved elements between humans and mice to gain methylation during aging (75).
The epigenetics of aging has gained momentum in the last decade, due to the growing availability of epigenetic clock predictors for both mice and humans (24, 35, 39, 77, 78). We took advantage of an existing mouse age predictor (38) that highlighted a significant age acceleration in the Sv129Ev compared to the C57BL/6J strain, in line with their overall healthspan and lifespan characteristics but without differences with regard to CD1 mice. It must be pointed out that this age predictor has been developed mostly in the C57BL/6 and Sv129 strain, so it might not be as accurate for CD1 mice. Nevertheless, this epigenetic clock predictor did not support an age acceleration due to DAI, in contrast to the effect seen in global methylation levels. While previous and present data are in agreement with the methylome and DNA age-related variance due to a strain-level factor (40), the contribution of social standing on age acceleration warrants further testing and the development of dedicated algorithms that should be calibrated with social standing variables.
To gain insight on the plausible functional consequences of the global methylome remodeling, we conducted differential methylation analyses, highlighting distinct methylation signatures within each strain due to DAI and followed them with gene pathway analyses. Immune system, metabolism of proteins, signal transduction, and gene expression-related pathways were found to show methylation changes due to DAI across all strains (see Results for details). These changes include several genes implicated in the regulation of TH1/TH2 balance as well as metabolism, including: Adrb3 [a gene whose methylation levels have been reported to be associated with metabolic dysregulation (47)], which was found to be differentially regulated in high-DAI subjects of C57BL/6J and CD1 strains vs. respective reference groups; Spred2 [a key negative regulator of MAPK signaling, also linked to metabolic abnormalities (79)], which was increased high-DAI groups.
Accelerated biological aging is increasingly associated with changes in methylation patterns and cellular senescence (23–25), and recently a mechanistic link has been proposed between the epigenome and stress (80), uncovering one of the possible avenues through which social stress responses shaped the rate of aging (11) in the present study.
Limitations of the Study and Future Direction.
Studies in laboratory mice present several advantages to model SDoHA in humans: i) Numerous wild-derived, outbred, variable, and inbred strains exist, allowing to study complex interactions between gene variants as well as epigenetic and environmental factors (15, 81); ii) mice are one of the shortest-lived mammals (median lifespan ~2 y); and finally iii), they are amenable to conduct intent-to-treat randomization designs of social variables, which are unfeasible and unethical in humans. Despite these advantages, this field is still in its infancy, and our study represents the larger to date. Yet, this study has limitations. First, in our paradigm social adversities started at young-adult age. In human and nonhuman primates adverse childhood experience is one of the strongest predictors of health and aging (3, 82). Thus, in future studies the application of social stress could range from preconception to old age. Second, our study lacks a reference group. Partially addressing this limitation, our data show that the DAI group showing the lower adverse effect of LCPS manifested a lifespan similar to published studies in standard housing conditions (SI Appendix, Table S9). Furthermore, in a lifelong study focused on SDoHA it is difficult, if not impossible, to define a priori a single reference/control condition since common housing practice (e.g., individual housing, grouping with siblings, grouping with nonsiblings) can per se elicit chronic stress and impact aging (83). Thus, future studies are necessary to compare such social factors for their impact on healthspan and lifespan. Last, this study was limited to male mice. Although a social rank can be identified in female laboratory mice (84, 85), female laboratory mice do not manifest territorial behavior, and their aggression is significantly less than the one of males or wild female mice (86). Additionally, no study describes the effect of social stress or social rank on aging or lifespan in female mice. Thus, future studies should attempt to develop models of adverse social relationships including social isolation and social instability with the potential to affect aging of female mice.
Conclusion
Our study identified genetic and social factors determining the degree of vulnerability to the negative effect of chronic stress on survival and additionally shows how agonistic behavior and social rank may be significant modifiers of healthspan, DNA methylation pattern, and lifespan. Overall, our study represents an important contribution toward the development of animal models allowing the identification of the biological bases of social determinants of health.
Materials and Methods
Animals.
C57BL/6J (Jackson Labs), CD1 (Charles River Labs), and Sv129Ev (Taconic Farms) male mice were purchased at 10 wk of age and housed at 12:12-h light:dark cycle at 22 ± 2 °C. At 12 wk of age, mice were randomly allocated to the experimental conditions. The diets (Research diet) used in this study were a standard diet (D12405B), a high-fat diet (D12451), and a mature rodent maintenance diet (D10012M) (Further details in SI Appendix, Methods). Cages contained corn cob bedding that was changed every 7 d. Cotton nestlets were placed in the cages for enrichment. Animal experiments were approved by the IACUC, University of Minnesota.
LCPS Model.
This study was designed to analyze the survival and healthspan of the mice as a function of genetic background as well as of an unbiased segregation based on contextual aggression received/exhibited. The LCPS model consisted of three phases: a baseline phase of 5 d, during which all mice were singly housed, followed by a 4-wk CPS phase in which mice were exposed to daily defeats and sensory contact housing, and an aging phase lasting until spontaneous death or euthanasia for humane reasons. In the aging phase, mice were housed in sensory contact thus experiencing a continued degree of threat stress related to the previous encounters.
Each C57BL/6J (N = 173) male mouse was randomized using a simple randomization procedure of flipping a coin (87) to be transferred as an intruder to the home cage of either a CD1 (N = 86) or Sv129Ev (N = 87) resident mouse. The CD1 strain manifests high territorial aggression, while the Sv129Ev manifests a much lower level of territorial aggression and can thus be considered a weaker dominant (12–14).
Mortality (age at death) was the primary outcome. Animals were checked daily to determine vital status, and date of death was recorded to the nearest day. Mice found dead at each daily inspection were considered as censored deaths and were necropsied. Conditions considered cause for euthanasia, as approved by the IACUC, University of Minnesota included: prolonged respiratory distress, tumor growth that impedes activity (such as motion, eating, or drinking), severe deformities or self-mutilation, inability to eat or drink, or ataxia that prevents normal functions of eating and drinking.
Healthspan Outcomes.
Body weight (in g) and food intake (in kcal), body composition (fat mass and fat-free mass, both in g, Echo MRI 3-in-1, Echo Medical System), and 4-h fasting plasma glucose (in mg/dL, Accu-chek Aviva, Roche) were recorded regularly.
DAI.
Aggression was measured during the daily social interactions by direct observation. Both aggression exhibited and received by each mouse was recorded in the course of the full social interaction using a continuous scan sampling scoring system. Aggression exhibited consisted of initiating aggressive grooming, biting, kicking, boxing, and chasing; aggression received consisted of receiving the aforementioned behaviors while displaying either upright postures or flight behavior, and squeaking vocalizations (88). Based on the data recorder over the 4-wk CPS phase, total counts of aggression exhibited and received were calculated. An aggression index was obtained on the total values of aggression exhibited (xb) and received (rcvd) by the experimental subjects, using the z-scores z = (x−μ)/σ of the two parameters, where x is the observed aggression exhibited value for zxb, and x is the observed aggression received values for zrcvd, μ is the respective mean and σ is the SD: zxb = (x−μ)/σ; zrcvd = (x−μ)/σ. For each individual, zxb and zrcvd were averaged after changing the sign of zrcvd giving rise to the formula: Aggression index = [zxb + (−zrcvd)]/2. The obtained score was then discretized according to tertiles as well as by visual observation of the frequency distribution of values in three categories: low, medium, and high.
DNA Methylation Sample Preparation.
As previously described (11), in a second experiment 70 C57BL/6J, 35 CD1, and 35 Sv129Ev mice purchased from the respective vendor at 10 wk of age were exposed to LCPS. Mice were killed at 17 mo of age for the collection of tissue specimens. The dissection was rapidly executed after asphyxiation by CO2 by removing major organs, cut into appropriate size pieces, and either flash frozen in liquid nitrogen or placed in 4% paraformaldehyde (PFA) for preservation. Flash-frozen tissue samples were stored at −80 °C. A subset of 48 liver samples was subsequently selected for DNA methylation analysis (n = 18 C57BL/6J, n = 12 CD1, n = 16 Sv129Ev). DNA was purified from the liver tissue using standard approached described in SI Appendix, Methods.
DNA Methylation and Array Processing.
The DNA methylation assay was performed at the University of Minnesota Genomics Center (https://genomics.umn.edu/) according to the standard manufacturer’s protocol (http://illumina.com/) as detailed in SI Appendix, Methods.
Statistical Analysis.
The survival analyses were implemented in R Studio using the methodological references given below. Survival probability was assessed through the use of the nonparametric Log-rank (Mantel–Haenszel) test to compare the differences in KM survival curves using the “survival” package of the R language (version 3.2-11). When significant, pairwise comparisons were conducted using Bonferroni corrected log-rank test performed between two groups of interest at a time. The “coxme” survival package was used to fit the Cox proportional hazard model to actual lifespans to calculate the hazard ratio (HR) where “pair” was fitted as random factor. Median and maximum survival were assessed using chi-square tests to determine statistical significance. Maximum lifespans were calculated as the proportion of mice that were still alive when the total population reached 90% mortality (89). Median and maximum survival presented as percentages were analyzed using the Gao–Allison method (90) for testing differences at survival milestones with tau for median and maximum survival calculated per strain and DAI group. Multiple linear regression models were conducted to test for the predictive value of baseline healthspan parameters (i.e., food intake, body weight, fat mass, fat-free mass, and glucose) on the aggression index. All parameters that resulted nonsignificant contributors were excluded from further analysis (i.e. diet). The overall healthspan was analyzed by entering food intake, body weight, body composition (fat mass and fat-free mass), and glucose in mixed effects linear models as dependent parameter using the “nlme” package of the R language (version 3.1-152). The models for all parameters were tested and run using time, strain, or DAI as fixed factors; time and individual nested within pair for random factors; the models also included an autocorrelation function for individual repeated measures as well as an unconstrained optimization using the BFGS variable metric algorithm. Machine learning was applied to the entire dataset comprising the baseline healthspan variables, the aggression index, age, and lesions at death, to identify their relative importance as classifiers for genetic background and DAI (for the latter the aggression index variable was omitted from the model as a predictor). LDA models were validated by partitioning the dataset into two samples: the training set (80% of the dataset) for either genetic background or DAI (high, medium and low categories) selected at random, and the validation set with the remaining 20%. The models were fitted with the learning sample and validated by using the validation sample to assess their accuracy. LDA was conducted using the “caret” (version 6.0-91) package in R. The model classification accuracy was assessed in the validation set calculating the overall accuracy (%) and the κ, which provides an overall accuracy assessment for the classification based on commission and omission errors for all classes. The death distribution was visualized as density plots using a Gaussian kernel density estimate (KDE) technique to smooth the histogram of survivorship frequencies using a predefined bandwidth (33), thus constructing the probability density function of the variable death. The degree of similarity between distribution was analyzed using a similarity index calculated with the R package “overlapping” (version 1.7) (34). This similarity index estimates the overlapping of the area intersected by two density functions. It is considered a normalized measure (values 0 indicating complete separation, to 1 indicating identity) of association that provides a way to quantify the agreement between two real probability distributions in terms of their density function (34). Similarity indexes between different groups were statistically compared two at a time using Fisher’s transformation, assessing the observed z test statistic at a Bonferroni corrected alpha level for statistical significance using the R package “cocor” (version 1.1-3). A P-value of ≤0.05 was considered statistically significant for these outcomes when Bonferroni’s multiple comparison correction was not required or other significance criteria needed to be adopted (see below).
For DNA methylation data, heatmaps were generated for CpG islands (defined as regions >500 bp, >55% GC and expected/observed CpG ratio of >0.65; this restricted the sample for this analysis to ~30,000 CpG islands) using the R package “pheatmap” (version 1.0.12) to incorporate strain and DAI to cluster CpG islands by Euclidean distance and the Ward.D2 clustering method. Similarly, all CpG loci underwent hierarchical clustering using the R “ape” package (version 5.6) to verify the segregation due to DAI within each strain. Multiple linear regression models were conducted to test for the predictive value of baseline healthspan parameters (i.e., food intake, body weight, fat mass, fat-free mass, and glucose), strain and aggression index on the level of CpG loci methylation. The R package “SeSAMe” (version 1.2.4) was used following its default pipeline (91) for DNA age prediction as well as for differential methylation analyses. Mouse DNA age prediction uses a function looking for overlapping probes and estimates age using an aging multitissue model built from 321 MM285 probes (Illumina array 44) trained similarly to the human Horvath clock on C57BL/6 and Sv129 strains (absolute mean error = 1.2 mo). To evaluate differential methylation levels for each CpG locus, linear models were fitted using DAI as a covariate for each strain. These models were constructed with the appropriate reference group estimates (low DAI for C57BL/6J; medium DAI for both CD1 and Sv129Ev strain). The Benjamini–Hochberg (BH) correction was applied to control for false discovery rate (FDR). This analysis was followed by DMR analysis on the significantly differentially methylated probes using Euclidean distance to group CpG markers and then combined P-values for each segment.Sensible Step-wise Analysis of DNA Methylation (SeSAMe) annotates DMRs to University of California, Santa Cruz (UCSC) RefGene from the Illumina annotation file. Manhattan plots were then generated using the “qqman” R package (version 0.1.8) for DMLs and DMRs identified across the genome and significance thresholds were applied using the Bonferroni correction for multiple comparisons (P < 5e−08) as well as a suggestive threshold (P < 1e−05). Pathway analysis was performed using the ReactomePA pathway analysis tool [Pathway Browser 3.7, Database release 39, (92)] that uses hypergeometric distribution testing to determine enriched pathways in the analyzed dataset (17 to 24 genes per dataset), producing a probability score corrected for FDR with the BH method.
The following estimators of effect size are included. Conditional and Marginal R2 for linear mixed models, partial eta2 ( ) for ANOVAs, Cohen’s d for chi-square tests, η2[H] for Kruskal–Wallis tests and r for Wilcoxon rank sum tests. For survival analyses, KM survival analyses are complemented with Cox PH regressions, providing HR and its CI. For power calculation, we provide observed power values for ANOVA-related results. The least powerful design in our study are the three groups’ comparisons, where under alternate hypotheses referring to noncentral distributions with 2 and 341 degree of freedom, we have 80% power to explain approximately 17% of the variance. Wherever applicable, power calculations were executed with G*Power 3.1.9.7. For linear mixed models, due to their complexity, simulation-based power analyses were employed for the fixed effects with the “simr” R package (version 1.2.0). Additional details on the statistical design and software used can be found in SI Appendix, Methods.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
Supported by NIH/NIA R01AG043972, R61 AG078520, Minnesota partnership for Biotechnology and Molecular Genomic #18.4, Fesler-Lampert Chair in Aging Studies and Department of Integrative Biology and Physiology, University of Minnesota, Grant Accelerator Program, to A.B. We wish to thank J. Tung and R. Campbell for statistical advice on an earlier version of the manuscript, W. Zhou for invaluable advice on Sensible Step-wise Analysis of DNA Methylation (SeSAMe) use and methylation data handling, C. Erickson, J. McCallum, N. Spielman, R. Mansk and S. McGonigle for their help with the study, and the staff of the Research Animal Resources at the University of Minnesota for animal care. We also acknowledge the support of the University of Minnesota Genomics Center (SCR_012413) and the Minnesota Supercomputing Institute.
Author contributions
M.R. and A.B. designed research; M.R. and K.N.-D. performed research; B.H.C. contributed new reagents/analytic tools; M.R., B.H.C., and A.B. analyzed data; K.N.-D. and B.H.C revised/edited versions of the paper; and M.R. and A.B. wrote the paper.
Competing interests
B.H.C. is a full-time employee of FOXO Technologies Inc., which seeks to commercialize epigenetic technologies in the life insurance industry. B.H.C. owns stock in Illumina Inc., the manufacturer of the DNA methylation arrays used in this study. B.H.C. is listed as a co-inventor in filed patents on commercial applications of epigenetic prediction models. The other authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Maria Razzoli, Email: mrazzoli@umn.edu.
Alessandro Bartolomucci, Email: abartolo@umn.edu.
Data, Materials, and Software Availability
All data can be accessed at Mendeley Data (https://doi.org/10.17632/kfkhgw359t.1) (93). The DNAm data are deposited in NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE216631) (94, 95). R codes can be found in the Zenodo repository (https://doi.org/10.5281/zenodo.7737666) (96).
Supporting Information
References
- 1.Olshansky S. J., From lifespan to healthspan. JAMA – J. Am. Med. Assoc. 320, 1323–1324 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Epel E. S., The geroscience agenda: Toxic stress, hormetic stress, and the rate of aging. Ageing Res. Rev. 63, 101167 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kivimäki M., Bartolomucci A., Kawachi I., The multiple roles of life stress in metabolic disorders. Nat. Rev. Endocrinol. 2022, 1–18 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snyder-Mackler N., et al. , Social determinants of health and survival in humans and other animals. Science (1979) 368, eaax9553 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crimmins E. M., Social hallmarks of aging: Suggestions for geroscience research. Ageing Res. Rev. 63, 101136 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kennedy B. K., et al. , Geroscience: Linking aging to chronic disease. Cell 159, 709–713 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adler N. E., Stewart J., Health disparities across the lifespan: Meaning, methods, and mechanisms. Ann. N.Y. Acad. Sci. Ann. N.Y. Acad. Sci. 1186, 5–23 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Mitchell S. J., Scheibye-Knudsen M., Longo D. L., de Cabo R., Animal models of aging research: Implications for human aging and age-related diseases. Annu. Rev. Anim. Biosci. 3, 283–303 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Bronikowski A. M., et al. , Aging in the natural world: Comparative data reveal similar mortality patterns across primates. Science 1979, 1325–1328 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selman C., Swindell W. R., Putting a strain on diversity. EMBO J. 37, e100862 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Razzoli M., et al. , Social stress shortens lifespan in mice. Aging Cell 17, e12778 (2018), 10.1111/acel.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartolomucci A., et al. , Increased vulnerability to psychosocial stress in heterozygous serotonin transporter knockout mice. Dis. Model Mech. 3, 459–470 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Dadomo H., et al. , Vulnerability to chronic subordination stress-induced depression-like disorders in adult 129SvEv male mice. Prog. Neuropsychopharmacol Biol. Psychiatry 35, 1461–1471 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Lidster K., Owen K., Browne W. J., Prescott M. J., Cage aggression in group-housed laboratory male mice: An international data crowdsourcing project. Sci. Rep. 9, 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell S. J., et al. , Effects of sex, strain, and energy intake on hallmarks of aging in mice. Cell Metab. 23, 1093–1112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parmigiani S., Palanza P., Rodgers J., Ferrari P. F., Selection, evolution of behavior and animal models in behavioral neuroscience. Neurosci. Biobehav. Rev. 23, 957–970 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Sundberg J. P., et al. , The mouse as a model for understanding chronic diseases of aging: The histopathologic basis of aging in inbred mice. Pathobiol. Aging Age Relat. Dis. 1 (2011), 10.3402/pba.v1i0.7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boleij H., et al. , Chronic social stress does not affect behavioural habituation in male CD1 mice. Behav. Brain Res. 273, 34–44 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Sultana R., Ogundele O. M., Lee C. C., Contrasting characteristic behaviours among common laboratory mouse strains. R. Soc. Open Sci. 6, 190574 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Contet C., Rawlins J. N. P., Bannerman D. M., Faster is not surer - A comparison of C57BL/6J and 129S2/Sv mouse strains in the watermaze. Behav. Brain Res. 125, 261–267 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Võikar V., Kõks S., Vasar E., Rauvala H., Strain and gender differences in the behavior of mouse lines commonly used in transgenic studies. Physiol. Behav. 72, 271–281 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Narvik J., et al. , Metabolic profile associated with distinct behavioral coping strategies of 129Sv and Bl6 mice in repeated motility test. Sci. Rep. 8, 4–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benayoun B. A., Pollina E. A., Brunet A., Epigenetic regulation of ageing: Linking environmental inputs to genomic stability. Nat. Rev. Mol. Biol. 16, 593–610 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hannum G., et al. , Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49, 359–367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maegawa S., et al. , Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 20, 332–340 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miczek K. A., Maxson S. C., Fish E. W., Faccidomo S., Aggressive behavioral phenotypes in mice. Behav. Brain Res. 125, 167–181 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Hsieh L. S., Wen J. H., Miyares L., Lombroso P. J., Bordey A., Outbred CD1 mice are as suitable as inbred C57BL/6J mice in performing social tasks. Neurosci. Lett. 637, 142–147 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanchard R. J., Mckittrick C. R., Blanchard D. C., Animal models of social stress: Effects on behavior and brain neurochemical systems. Physiol. Behav. 73, 261–271 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Cohen S., Kamarck T., Mermelstein R., A global measure of perceived stress. J. Health Soc. Behav. 24, 385–396 (1983). [PubMed] [Google Scholar]
- 30.Golden-Kreutz D. M., Browne M. W., Frierson G. M., Andersen B. L., Assessing stress in cancer patients: A second-order factor analysis model for the perceived stress scale. Assessment 11, 216–223 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Epel E. S., et al. , Accelerated telomere shortening in response to life stress. Proc. Natl. Acad. Sci. U.S.A. 101, 17312–17315 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casaletto K. B., et al. , Perceived stress is associated with accelerated monocyte/macrophage aging trajectories in clinically normal adults. Am. J. Geriatric Psychiatry 26, 952–963 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silverman B. W., Density Estimation: For Statistics and Data Analysis 1–175 (Chapman and Hall, London, United Kingdom, 2018). [Google Scholar]
- 34.Pastore M., Calcagnì A., Measuring distribution similarities between samples: A distribution-free overlapping index. Front. Psychol. 10, 1089 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horvath S., DNA methylation age of human tissues and cell types. Genome Biol. 14, R115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson J. A., et al. , High social status males experience accelerated epigenetic aging in wild baboons. Elife 10, e66128 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palma-Gudiel H., Fañanás L., Horvath S., Zannas A. S., Psychosocial stress and epigenetic aging. Int. Rev. Neurobiol. 150, 107–128 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Zhou W., et al. , DNA methylation dynamics and dysregulation delineated by high-throughput profiling in the mouse. Cell Genom. 2, 100144 (2022) 10.1016/j.xgen.2022.100144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang T., et al. , Epigenetic aging signatures in mice livers are slowed by dwarfism, calorie restriction and rapamycin treatment. Genome Biol. 18, 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandoval-Sierra J. V., et al. , Body weight and high-fat diet are associated with epigenetic aging in female members of the BXD murine family. Aging Cell 19, e13207 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galanter J. M., et al. , Differential methylation between ethnic sub-groups reflects the effect of genetic ancestry and environmental exposures. Elife 6, e20532 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klengel T., Pape J., Binder E. B., Mehta D., The role of DNA methylation in stress-related psychiatric disorders. Neuropharmacology 80, 115–132 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Chen B. H., et al. , DNA methylation-based measures of biological age: Meta-analysis predicting time to death. Aging 8, 1844–1865 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levine M. E., et al. , An epigenetic biomarker of aging for lifespan and healthspan. Aging 10, 573–591 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simpson D. J., Chandra T., Epigenetic age prediction. Aging Cell 20, e13452 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hemmers S., Schizas M., Rudensky A. Y., T reg cell–intrinsic requirements for ST2 signaling in health and neuroinflammation. J. Exp. Med. 218, e20201234 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lima R. P. A., et al. , Methylation profile of the ADRB3 gene and its association with lipid profile and nutritional status in adults. Biol. Res. 52, 21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang X., Lee S. H., Lu H., Sanders K. M., Koh S. D., Molecular and functional characterization of inwardly rectifying K+ currents in murine proximal colon. J. Physiol. 596, 379 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fang B., Li Z., Qiu Y., Cho N., Yoo H. M., Inhibition of uba5 expression and induction of autophagy in breast cancer cells by usenamine a. Biomolecules 11, 1348 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olbrich H., et al. , Mutations in a novel gene, NPHP3, cause adolescent nephronophthisis, tapeto-retinal degeneration and hepatic fibrosis. Nat. Genetics 34, 455–459 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Tropée R., et al. , The SWI/SNF subunit SMARCD3 regulates cell cycle progression and predicts survival outcome in ER+ breast cancer. Breast Cancer Res. Treat 185, 601 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu M., et al. , Characterization of Opa interacting protein 5 as a new biomarker and therapeutic target for oral cancer. Int. J. Oncol. 60, 1–11 (2022). [DOI] [PubMed] [Google Scholar]
- 53.Pappas L. E., Nagy T. R., The translation of age-related body composition findings from rodents to humans. Eur. J. Clin. Nutr. 73, 172–178 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vitali A., et al. , The adipose organ of obesity-prone C57BL/6J mice is composed of mixed white and brown adipocytes. J. Lipid Res. 53, 619–629 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y., Bolze F., Fromme T., Klingenspor M., Intrinsic differences in BRITE adipogenesis of primary adipocytes from two different mouse strains. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1841, 1345–1352 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Sievert T., Laska M., Behavioral Responses of CD-1 Mice to Six Predator Odor Components. Chem. Senses 41, 399–406 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Kaku K., Fiedorek F. T., Province M., Permutt M. A., Genetic analysis of glucose tolerance in inbred mouse strains. Evidence for polygenic control. Diabetes 37, 707–713 (1988). [DOI] [PubMed] [Google Scholar]
- 58.Kooptiwut S., et al. , Comparison of insulin secretory function in two mouse models with different susceptibility to β-cell failure. Endocrinology 143, 2085–2092 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Toye A. A., et al. , A genetic and physiological study of impaired glucose homeostasis control in C57BL/6J mice. Diabetologia 48, 675–686 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Razzoli M., et al. , Social stress shortens lifespan in mice. Aging Cell 17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Palliyaguru D. L., et al. , Fasting blood glucose as a predictor of mortality: Lost in translation. Cell Metab. 33, 2189–2200.e3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Snyder J. M., Ward J. M., Treuting P. M., Cause-of-death analysis in rodent aging studies. Vet Pathol. 53, 233–243 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Son W. C., Factors contributory to early death of young CD-1 mice in carcinogenicity studies. Toxicol. Lett. 145, 88–98 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Hirano M., et al. , Mortality, major cause of moribundity, and spontaneous tumors in CD-1 mice *. Toxicol Pathol. 16, 340–349 (1988). [DOI] [PubMed] [Google Scholar]
- 65.Franceschi C., Garagnani P., Parini P., Giuliani C., Santoro A., Inflammaging: A new immune–metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 14, 576–590 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Braveman P., Egerter S., Williams D. R., The social determinants of health: Coming of age. Annu. Rev. Public Health 32, 381–398 (2011). [DOI] [PubMed] [Google Scholar]
- 67.Lee W., et al. , Distinct immune and transcriptomic profiles in dominant versus subordinate males in mouse social hierarchies. Brain Behav. Immun. 103, 130–144 (2022). [DOI] [PubMed] [Google Scholar]
- 68.Ebner K., Singewald N., Individual differences in stress susceptibility and stress inhibitory mechanisms. Curr. Opin. Behav. Sci. 14, 54–64 (2017). [Google Scholar]
- 69.Bassuk S. S., Berkman L. F., Amick B. C. III, Socioeconomic status and mortality among the elderly: Findings from four US communities. Am. J. Epidemiol. 155, 520–533 (2002). [DOI] [PubMed] [Google Scholar]
- 70.McEwen B. S., Protective and damaging effects of stress mediators. New England J. Med. 338, 171–179 (1998). [DOI] [PubMed] [Google Scholar]
- 71.Kivimäki M., Steptoe A., Effects of stress on the development and progression of cardiovascular disease. Nat. Rev. Cardiol. 15, 215–229 (2018). [DOI] [PubMed] [Google Scholar]
- 72.Bartolomucci A., Social stress, immune functions and disease in rodents. Front. Neuroendocrinol. 28, 28–49 (2007). [DOI] [PubMed] [Google Scholar]
- 73.Furman D., et al. , Chronic inflammation in the etiology of disease across the life span. Nat. Med. 25, 1822–1832 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bairachnaya M., Agranyoni O., Antoch M., Michaelevski I., Pinhasov A., Innate sensitivity to stress facilitates inflammation, alters metabolism and shortens lifespan in a mouse model of social hierarchy. Aging 11, 9901–9911 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sziráki A., Tyshkovskiy A., Gladyshev V. N., Global remodeling of the mouse DNA methylome during aging and in response to calorie restriction. Aging Cell 17, e12738 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Poganik J. R., et al. , Biological age is increased by stress and restored upon recovery. bioRxiv [Preprint] (2022). https://www.biorxiv.org/content/10.1101/2022.05.04.490686v1 (Accessed 17 May 2022).
- 77.Stubbs T. M., et al. , Multi-tissue DNA methylation age predictor in mouse. Genome Biol. 18, 68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thompson M. J., et al. , A multi-tissue full lifespan epigenetic clock for mice. Aging 10, 2832–2854 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ohkura T., et al. , Spred2 regulates high fat diet-induced adipose tissue inflammation, and metabolic abnormalities in mice. Front. Immunol. 10, 17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zannas A. S., et al. , Epigenetic upregulation of FKBP5 by aging and stress contributes to NF-κB-driven inflammation and cardiovascular risk. Proc. Natl. Acad. Sci. U.S.A. 166, 11370–11379 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bou Sleiman M., et al. , Sex- and age-dependent genetics of longevity in a heterogeneous mouse population. Science (1979) 377, eabo3191 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tung J., Archie E. A., Altmann J., Alberts S. C., Cumulative early life adversity predicts longevity in wild baboons. Nat. Commun. 7, 11181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bartolomucci A., et al. , Social factors and individual vulnerability to chronic stress exposure. Neurosci. Biobehav. Rev. 29, 67–81 (2005). [DOI] [PubMed] [Google Scholar]
- 84.Williamson C. M., et al. , Social hierarchy position in female mice is associated with plasma corticosterone levels and hypothalamic gene expression. Sci. Rep. 9, 7324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Karamihalev S., et al. , Social dominance mediates behavioral adaptation to chronic stress in a sex-specific manner. Elife 9, 1–18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dadomo H., et al. , What is stressful for females? Differential effects of unpredictable environmental or social stress in CD1 female mice. Horm Behav. 98, 22–32 (2018). [DOI] [PubMed] [Google Scholar]
- 87.Suresh K., An overview of randomization techniques: An unbiased assessment of outcome in clinical research. J. Hum. Reprod. Sci. 4, 8–11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 88.Bartolomucci A., et al. , Social status in mice: Behavioral, endocrine and immune changes are context dependent. Physiol. Behav. 73 (2001). [DOI] [PubMed] [Google Scholar]
- 89.Hofmann J. W., et al. , Reduced expression of MYC increases longevity and enhances healthspan. Cell 2912, 477–488 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gao G., Wan W., Zhang S., Redden D. T., Allison D. B., Testing for differences in distribution tails to test for differences in "maximum" lifespan. BMC Med. Res. Methodol. 8, 49 (2008), 10.1186/1471-2288-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou W., Triche T. J., Laird P. W., Shen H., SeSAMe: Reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucleic Acids Res. 46, 123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yu G., He Q. Y., ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 12, 477–479 (2016). [DOI] [PubMed] [Google Scholar]
- 93.Razzoli M., 2022-11755RR. Mendeley Data. https://data.mendeley.com/datasets/kfkhgw359t/1. Deposited 14 April 2021.
- 94.Edgar R., Domrachev M., Lash A. E., Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Razzoli M., Epigenome analysis of liver samples from C57BL/6J, CD1, and Sv129Ev mice in lifelong chronic psychosocial stress (LCPS). Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?%26acc=GSE216631. Deposited 26 October 2022.
- 96.Razzoli M., 2022-11755RR: codes. Zenodo. 10.5281/zenodo.7737666. Deposited 15 March 2023. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All data can be accessed at Mendeley Data (https://doi.org/10.17632/kfkhgw359t.1) (93). The DNAm data are deposited in NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE216631) (94, 95). R codes can be found in the Zenodo repository (https://doi.org/10.5281/zenodo.7737666) (96).