RNA editing refers to the post-transcriptional modification of RNA sequences. Diverse eukaryotes are capable of mRNA editing, most commonly through enzymatic deamination of adenosine-to-inosine (A-to-I) (1–3). The resulting ‘I’ is read by the translational machinery as guanosine (‘G’) (4). Thus, A-to-I editing within coding sequences can increase proteome complexity by generating new protein variants with amino acid substitutions or recoded stop codons. These new protein variants may be functionally distinct from their genome-encoded counterparts, but only a handful of protein variants generated by A-to-I editing have demonstrated functions. Widespread application of DNA- and RNA-sequencing has revealed that A-to-I editing is prevalent in mammals, cephalopods, insects, and other organisms including some fungi (5–9). The idea that A-to-I editing provides an adaptive advantage by recoding mRNAs is attractive, but evidence for this is limited. In PNAS, Xin et al. explore the role of A-to-I editing during sexual development of Fusarium graminearum, a filamentous ascomycete (10). The authors demonstrate that conserved A-to-I editing sites are functionally important during fungal development and provide evidence that A-to-I editing provides an adaptive advantage. In addition, these findings provide new mechanistic insights into multicellular development in the fungal kingdom and have major implications for understanding the complete coding potential of eukaryotic genomes.
The authors demonstrate that conserved A-to-I editing sites are functionally important during fungal development and provide evidence that A-to-I editing provides an adaptive advantage.
A-to-I editing was first identified through biochemical studies (11). In animals, members of the adenosine deaminase acting on RNA (ADAR) family target double-stranded RNAs for unwinding and adenosine deamination (12). ADAR enzymes can modify endogenous RNA duplexes to prevent aberrant antiviral responses (13). More recently, genomic approaches have uncovered thousands of A-to-I editing sites in a variety of metazoan organisms, including mammals, flies, and cephalopods (5–7, 9). The extent to which editing sites alter coding sequences varies between organisms. In primates, most editing sites occur in repetitive Alu elements, though a small fraction of A-to-I editing events impacts coding regions (5, 14). In flies and cephalopods, editing sites that generate nonsynonymous amino acid substitutions make up a significant fraction of total sites. The number of A-to-I editing sites that alter coding potential of mRNAs ranges from thousands (>1,000 in flies and >3,000 in humans) to over 70,000 in cephalopods (12). Genes with neuronal functions appear to be common targets of the A-to-I editing machinery in metazoans, but only a handful of observed A-to-I editing sites have demonstrated functions (6, 15, 16). For example, editing of the Q/R site in mammalian glutamate receptor 2 is essential for postnatal survival in mice (17). It is reasonable to assume that mRNA recoding via A-to-I editing provides an evolutionary advantage, especially given the widespread occurrence in metazoans, yet studies to address this question have provided contradictory results. A-to-I editing has been linked to temperature adaptation in flies and cephalopods, whereas researchers studying A-to-I editing in humans concluded that editing was nonadaptive (18–20).
It is experimentally challenging to demonstrate functional importance of individual A-to-I editing sites. Editing often occurs at low frequency, making it difficult to distinguish bona fide A-to-I editing sites from DNA sequencing errors. Labor-intensive genetic approaches are required to demonstrate that individual editing sites are functionally relevant, but results of genetic studies can be difficult to interpret because phenotypes are often subtle or tissue specific. The recent discovery that certain ascomycete fungi perform extensive A-to-I editing during sexual development is exciting because it creates an opportunity to apply facile fungal genetics to investigate A-to-I editing (8, 21).
In the present study, Xin et al. carried out comparative genomic analyses and genetic studies to investigate the role of A-to-I editing during fruiting body development of F. graminearum. Previous studies with F. graminearum identified over 26,000 putative A-to-I editing sites, including 429 sites that result in nonsynonymous substitutions and are shared across three species of sordariomycete fungi, F. graminearum, Neurospora crassa, and Neurospora tetrasperma (8, 21). These genes were classified as conserved missense editing (CME) genes. In this study, 21 CME genes that are edited at high frequency in both F. graminearum and N. crassa were investigated.
The authors first constructed deletion alleles of selected CME genes and examined their phenotypes. This revealed seven CME genes required for normal sexual development of F. graminearum. Next, CME deletion strains were transformed with an “nonedited” allele, which only produces the genome-encoded protein variant. In parallel, CME deletion strains were transformed with an allele that mimics the edited transcript (i.e., an ‘A’ to ‘G’ missense allele). The ‘nonedited’ allele was able to rescue the phenotype for most CME deletion strains, demonstrating that editing of these sites is not required for proper dev elopment. It remains possible that these editing-deficient alleles give rise to subtle phenotypes that were not investigated in this study. Significantly, the ‘nonedited’ allele of CME5 failed to rescue developmental defects of a CME5 deletion mutant, whereas an ‘A’ to ‘G’ missense allele restored normal development. These data demonstrate that A-to-I editing of CME5 is essential for normal sexual development. CME5 encodes a predicted kinesin-8 motor. Editing is predicted to result in an arginine to glycine substitution at position 986. Remarkably, phylogenetic analysis of CME5 homologs across the fungal tree of life revealed that glycine is rarely hard-wired into the genome at this position, indicating the genome-encoded allele is conserved and likely ancestral. Together, these data indicate that the genome-encoded allele likely provides an important function during vegetative growth and that editing operates to produce a new protein variant with a critical function during sexual development.
Experiments with a second gene provided additional evidence that A-to-I editing is adaptive in F. graminearum. CME11 encodes a predicted chromatin remodeling enzyme. A-to-I editing of CME11 results in the substitution of a putative phosphorylated residue, threonine-304 to an alanine. In contrast to CME5, developmental defects of CME11 strains could not be rescued by the nonedited allele or a ‘G’ allele that mimics the editing product. A third experiment showed that introduction of both ‘nonedited’ and ‘G’ alleles into the CME11 deletion strain rescued the developmental phenotype. Thus, both edited and unedited CME11 transcripts are required for fruiting body development in F. graminearum. Together, these data show that nonsynonymous amino acid substitutions introduced by the A-to-I editing machinery are critical for fungal development and suggest that proteome diversification by A-to-I editing provides an adaptive advantage.
These findings have major implications for understanding regulatory mechanisms that control fungal physiology and development. In at least some Sordariomycetes, both genome-encoded mRNAs and novel mRNA isoforms generated by A-to-I editing are translated to produce functionally important proteins. This work provides strong motivation for future studies of A-to-I editing in other fungi. In addition to several Sordariomycetes, A-to-I editing operates during sexual development of Pyronema confluens, a member of the Pezizomycetes that last shared a common ancestor with the sordariomycete lineage over 400 Mya (22, 23). It seems likely that A-to-I editing is broadly important for fungal development. In both F. graminearum and N. crassa, A-to-I editing exhibits remarkable tissue specificity. Will studies of other fungi reveal A-to-I editing during additional growth conditions or cell types?
Identification of the A-to-I editing machinery and understanding how editing is regulated both spatially and temporally are important goals for future studies. Only a fraction of mRNA molecules is subject to A-to-I editing, enabling cells to express edited and unedited transcripts simultaneously, as observed here for CME11 (Fig. 1). Editing levels at individual sites may be controlled by accessory factors, perhaps in response to specific stimuli. The fact that A-to-I editing is a dynamic process in fungi and metazoans supports the idea that unknown regulators of A-to-I editing are yet to be discovered. More broadly, it seems likely that proteome diversification by A-to-I editing is important for additional cell type-specific functions in metazoans. As mentioned above, editing is common in neuronal genes in insects, cephalopods, and mammals, suggesting that A-to-I editing may regulate the establishment or maintenance of neuronal cell identity or at least recode key neuronal functions.
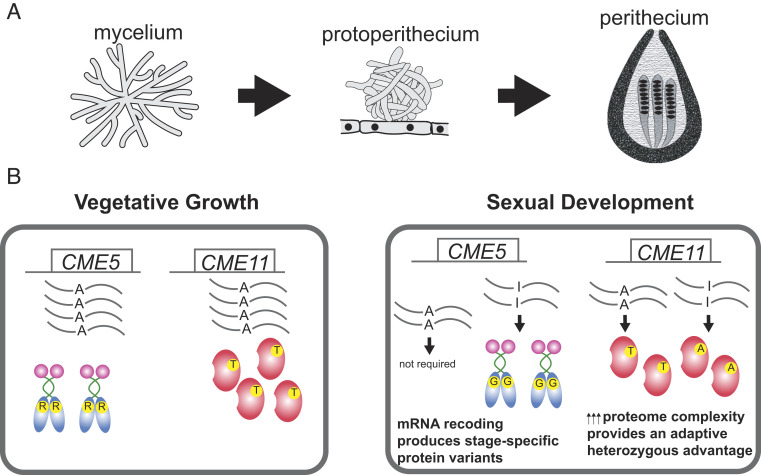
Fig. 1.
A-to-I mRNA editing expands proteome complexity to drive fungal development and provide an adaptive advantage. (A) Sordariomycete fungi grow vegetatively as mycelia or differentiate to produce multicellular structures during sexual development (protoperithecia and perithecia). A-to-I mRNA editing operates exclusively during sexual development. (B) Genetic studies of two conserved missense editing (CME) genes in F. graminearum demonstrate that A-to-I editing provides an adaptive advantage. Recoding of CME5 mRNA by A-to-I editing replaces the ancestral, genome-encoded arginine (R) with a glycine (G) to generate a stage-specific protein variant required for normal development of perithecia. In addition, expression of both edited and unedited CME11 transcripts is required for normal perithecial development, suggesting that editing confers a heterozygous advantage.
A-to-I editing expands the coding potential of the genome, giving rise to new protein variants in the population that can be selected upon. The findings of Xin et al. provide evidence that A-to-I editing in fungi is both functionally important and adaptive. These results have far-reaching implications for understanding eukaryotic proteome complexity, developmental biology, and human disease states.
Acknowledgments
I would like to thank Dr. Michael Freitag for comments on this manuscript. My research is supported by grants from the NIH/National Institute of General Medicine (R01GM132644) and the National Institute of Allergy and Infectious Disease (R01AI162989; co-PI with R.B. Meagher).
Author contributions
Z.A.L. wrote the paper.
Competing interests
The author declares no competing interest.
Footnotes
See companion article, “Experimental evidence for the functional importance and adaptive advantage of A-to-I RNA editing in fungi,” 10.1073/pnas.2219029120.
References
- 1.Eisenberg E., Proteome diversification by RNA editing. Methods Mol. Biol. 2181, 229–251 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Eisenberg E., Levanon E. Y., A-to-I RNA editing–Immune protector and transcriptome diversifier. Nat. Rev. Genet. 19, 473–490 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Nishikura K., A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 17, 83–96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Licht K., et al. , Inosine induces context-dependent recoding and translational stalling. Nucleic Acids Res. 47, 3–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bazak L., et al. , A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 24, 365–376 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graveley B. R., et al. , The developmental transcriptome of Drosophila melanogaster. Nature 471, 473–479 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albertin C. B., et al. , The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature 524, 220–224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H., et al. , Genome-wide A-to-I RNA editing in fungi independent of ADAR enzymes. Genome Res. 26, 499–509 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan M. H., et al. , Dynamic landscape and regulation of RNA editing in mammals. Nature 550, 249–254 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xin K., et al. , Experimental evidence for the functional importance and adaptive advantage of A-to-I RNA editing in fungi. Proc. Natl. Acad. Sci. U.S.A. 120, e2219029120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bass B. L., Weintraub H., An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55, 1089–1098 (1988). [DOI] [PubMed] [Google Scholar]
- 12.Yablonovitch A. L., Deng P., Jacobson D., Li J. B., The evolution and adaptation of A-to-I RNA editing. PLoS Genet. 13, e1007064 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mannion N. M., et al. , The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 9, 1482–1494 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniel C., Silberberg G., Behm M., Ohman M., Alu elements shape the primate transcriptome by cis-regulation of RNA editing. Genome Biol. 15, R28 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alon S., et al. , The majority of transcripts in the squid nervous system are extensively recoded by A-to-I RNA editing. Elife 4, e05198 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuddleston W. H., et al. , Spatiotemporal and genetic regulation of A-to-I editing throughout human brain development. Cell Rep. 41, 111585 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brusa R., et al. , Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science 270, 1677–1680 (1995). [DOI] [PubMed] [Google Scholar]
- 18.Buchumenski I., et al. , Dynamic hyper-editing underlies temperature adaptation in Drosophila. PLoS Genet. 13, e1006931 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garrett S., Rosenthal J. J., RNA editing underlies temperature adaptation in K+ channels from polar octopuses. Science 335, 848–851 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu G., Zhang J., Human coding RNA editing is generally nonadaptive. Proc. Natl. Acad. Sci. U.S.A. 111, 3769–3774 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H., et al. , A-to-I RNA editing is developmentally regulated and generally adaptive for sexual reproduction in Neurospora crassa. Proc. Natl. Acad. Sci. U.S.A. 114, E7756–E7765 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teichert I., Dahlmann T. A., Kuck U., Nowrousian M., RNA editing during sexual development occurs in distantly related filamentous ascomycetes. Genome Biol. Evol. 9, 855–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prieto M., Wedin M., Dating the diversification of the major lineages of Ascomycota (Fungi). PLoS One 8, e65576 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]