Significance
Growth rate, how quickly organisms grow and reproduce, is a key feature of biology. However, there are few measurements of microbial growth rates in soil, despite its crucial importance to terrestrial ecosystems and global environmental change. By measuring the uptake of isotopically labeled water, we can quantify microbial growth, even at exceedingly slow rates. We find that the growth rates of soil microorganisms are slower than those typically observed in culture as well as most previous estimates in soil. Surprisingly, we observe that lower-biomass soils exhibited faster growth than high-biomass soils and that more abundant microorganisms are not necessarily faster growing. Our results underscore the importance of considering slow and variable growth rates when studying microorganisms and their contributions to ecosystems.
Keywords: soil microbiology, growth rate, lipidomics, stable isotope probing
Abstract
The rate at which microorganisms grow and reproduce is fundamental to our understanding of microbial physiology and ecology. While soil microbiologists routinely quantify soil microbial biomass levels and the growth rates of individual taxa in culture, there is a limited understanding of how quickly microbes actually grow in soil. For this work, we posed the simple question: what are the growth rates of soil microorganisms? In this study, we measure these rates in three distinct soil environments using hydrogen-stable isotope probing of lipids with 2H-enriched water. This technique provides a taxa-agnostic quantification of in situ microbial growth from the degree of 2H enrichment of intact polar lipid compounds ascribed to bacteria and fungi. We find that growth rates in soil are quite slow and correspond to average generation times of 14 to 45 d but are also highly variable at the compound-specific level (4 to 402 d), suggesting differential growth rates among community subsets. We observe that low-biomass microbial communities exhibit more rapid growth rates than high-biomass communities, highlighting that biomass quantity alone does not predict microbial productivity in soil. Furthermore, within a given soil, the rates at which specific lipids are being synthesized do not relate to their quantity, suggesting a general decoupling of microbial abundance and growth in soil microbiomes. More generally, we demonstrate the utility of lipid-stable isotope probing for measuring microbial growth rates in soil and highlight the importance of measuring growth rates to complement more standard analyses of soil microbial communities.
The rate of microbial growth is a parameter commonly invoked in biogeochemical models of carbon flux, nutrient uptake, ecosystem productivity, and other soil assessments. However, growth rates of microorganisms are extremely variable, with estimated generation times ranging from minutes in laboratory-based culture to many months and years for organisms living in the Earth’s subsurface (1–3). Drastically different lifestyles and strategies characterize the microbial world, with oligotrophic systems sustaining slow-growing organisms on timescales far beyond those typically observed in more resource-rich environments that select for rapid growth and short generation times (4, 5).
Numerous studies have measured soil microbial biomass with the assumption that the size of the standing pool of microbial biomass is an important metric of soil microbial productivity (6–13). However, static biomass assessments are not necessarily a measure of the influence that microorganisms might have on ecosystem processes in real time, including nutrient and carbon cycling. Given that substantial numbers of microbial cells in soil may be slow-growing or dormant, it is important to assess the degree to which microbial activity is predicted by microbial abundance (14, 15) We cannot assume a priori that microbial communities with larger standing biomass are necessarily more productive. Just as net primary production, not standing biomass, is used as a growth parameter in plant systems, a similar metric is required to assess the productivity of soil microbial communities.
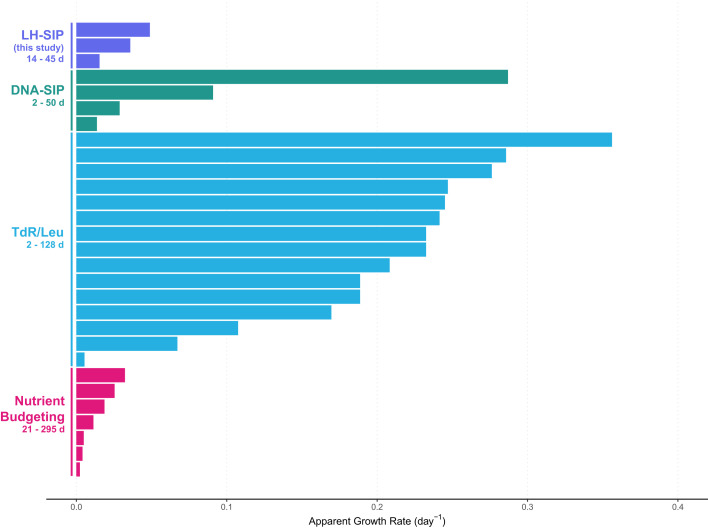
Growth rate is a critical parameter with which to assess ecosystem function, but there remains a lack of consensus around methods for measuring in situ microbial growth in soil systems. As a result, microbial growth rates in soil are poorly constrained, with previously reported community-level generation times varying by several orders of magnitude, variation that is strongly influenced by methodology (Fig. 3) (16–24). For a thorough overview of the preexisting methods for measuring soil microbial growth rates, see ref. 17.
Fig. 3.
Published estimates of soil microbial growth rates (16–24, 59–66) (collected as Dataset S3), including those inferred by LH-SIP (this study). We note that estimated growth rates with LH-SIP are much slower than those inferred by many DNA- and protein-based methods. Nutrient budgeting approaches, which typically measure the flux of carbon, nitrogen, or phosphorous through a soil to model microbial growth rate, generally lead to conservative estimates of microbial growth rates. Bar length corresponds to mean growth or turnover rate (day−1) reported (Materials and Methods). The ranges of corresponding generation times (days) are noted underneath the method label.
Stable isotope tracers (e.g., 13C, 2H, 15N, 18O) have been used to assess microbial activity in a wide variety of systems, from clinical samples to the deep biosphere (3, 16, 25–31). Stable isotope probing (SIP) relies on the addition of an isotopically labeled tracer followed by time-series measurements to calculate the rate of tracer incorporation into biomolecules by active microorganisms. SIP, unlike methods that rely on cell counting or biomass estimation, can provide information on biosynthetic or metabolic turnover independent of both the size of the standing biomass pool and the population dynamics of the community (i.e., population growth, steady state, or decline). Thus, SIP approaches are well suited to assess growth rates in soils, where standing biomass pools may vary widely, and population dynamics may be spatially or temporally variable.
Phospholipid fatty acids (PLFAs) have been routinely used to characterize soil microbial communities and provide two key advantages as biomarkers. First, PLFAs rapidly degrade upon cell death and are considered to represent only living organisms (32, 33). In contrast, other biomarkers such as soil DNA may survive for long time periods after cell death, potentially confounding efforts to study living cells (34). Second, despite their limited taxonomic information, quantification of PLFAs provides accurate estimates of microbial biomass and population size and can sensitively detect high-level shifts in the microbial community (35, 36). Here, we combine hydrogen SIP with traditional PLFA analysis in order to i) sensitively measure rates of microbial growth from living biomass and ii) determine the extent to which biomass abundance predicts microbial growth rates. To this end, we employ a dilute deuterated water (2H2O) tracer (2F = 5,000 ppm, δ2H = 31,357 ‰ vs. VSMOW) because all organisms incorporate water-derived H into their lipids during growth, and the addition of labeled water should not select for or against the growth of any organisms, thus rendering the tracer taxa-agnostic and nutritionally neutral. The incorporation of 2H from 2H2O into microbial membrane lipids can be measured by gas chromatography/pyrolysis/isotope ratio mass spectrometry (GC/P/IRMS) along a time series and converted into a growth rate, as previously demonstrated with clinical samples and marine sediments (26, 27, 37–40). This method, which we abbreviate as lipidomic hydrogen stable isotope probing (LH-SIP), offers several advantages for the estimation of microbial growth rates in situ. First, it allows us to use reasonably short incubation times (days) to measure the generation times of microorganisms, even if they are very long, as this method requires only a small fraction of lipids to be newly synthesized. Moreover, all microorganisms synthesize lipids regardless of their stage in the cell cycle or metabolic activity. Unlike nucleic acids and proteins, which can be resynthesized for cellular maintenance and repair even in the absence of growth, lipids are less likely to require repair and thus provide a more specific measurement of membrane and cell growth (41).
Isotope ratio mass spectrometers have a wide dynamic range that is well-suited for capturing the trace isotopic incorporation that is expected from exceptionally slow growth rates. The accuracy and dynamic range of IRMS instruments for measuring the isotopic ratio of 2H/1H allow for accurate quantification of 2H incorporation corresponding to a fraction of an organism’s generation time in the presence of less than 1% 2H2O (26) (SI Appendix, Supplementary Text).
For this study, we focused on three soils that represent different site and edaphic characteristics (Dataset S1), with these soils harboring distinct microbial communities (SI Appendix, Fig. S3). We directly measured growth rates in these soils with compound-specific LH-SIP, highlighting the utility of our approach, the relevance of measuring microbial growth rates in soil, and the broader implications of our results for understanding soil microbial dynamics.
Results and Discussion
Measuring the Growth Rates of Soil Microorganisms.
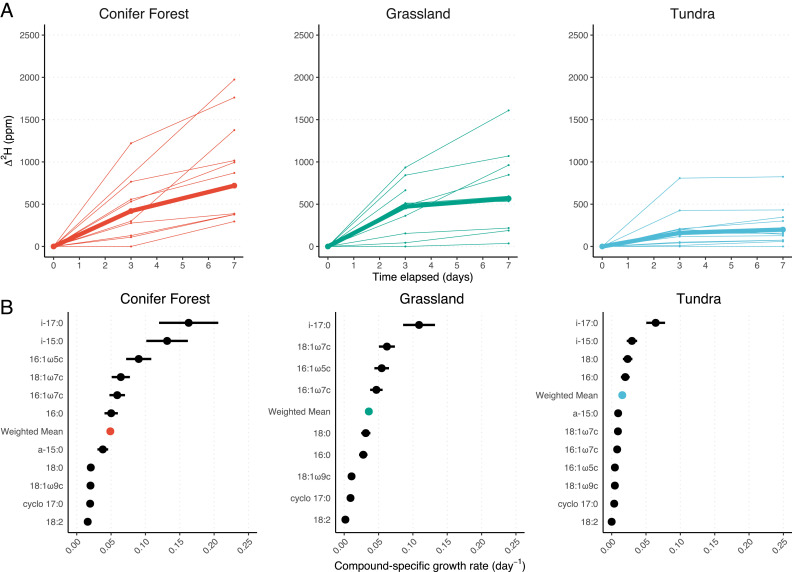
We measured 2H incorporation into soil microbial PLFA components at three time points during a 7-d incubation period in the presence of a dilute heavy water (2H2O) tracer. We detail these experiments in Materials and Methods. In brief, we incubated three soils (a sub-alpine conifer forest, prairie grassland, and an alpine tundra soil) in the presence of 5,000 ppm (δ2HVSMOW = 31,357 ‰) 2H2O, extracted intact PLFAs, and measured their abundances by gas chromatography flame ionization detection (GC-FID). The structures of PLFAs were determined by gas chromatography mass spectrometry (GC-MS) with PLFA isotopic compositions measured by gas chromatography isotope ratio mass spectrometry (GC-IRMS). We report isotopic values as fractional abundance (2F) in units of ppm where 2F = 2H/(2H + 1H). We observed 2H incorporation into fatty acids to range from 0 to 2,000 ppm in the presence of a 5,000-ppm 2H2O tracer (Fig. 1A). We inferred the growth rate and apparent generation times of microbial lipids using a previously derived relationship (26) (SI Appendix, Supplementary Text). In short, microbial growth (µ) is a logarithmic function of incubation time and the fractional hydrogen isotopic enrichment of new biomass relative to that of biomass at the start of the incubation. We report both lipid-specific growth rates as well as the abundance-weighted mean (i.e., community-level mean) growth rate for each soil. We report growth rates (day−1) at both the compound-specific level and as assemblage-level means weighted by compound-abundance. We also calculate generation times (days), a derived statistic estimating the time for complete reproduction of living biomass during clonal growth (Materials and Methods and SI Appendix, Supplementary Text). While the majority of the PLFAs detected are sourced from bacteria (42), PLFAs attributable to fungi (42, 43) are also reported.
Fig. 1.
∆2H enrichment, defined as change in 2F (ppm) relative to the start of the incubation, of microbial biomass increases over the course of the incubation (A), allowing the calculation of compound-specific and abundance-weighted-mean growth rates (B). The abundance-weighted mean ∆2H enrichment (A) is shown in bold, whereas compound-specific ∆2H are shown as thin lines. Growth rates (B) were calculated using the integrated ∆2H between 0 and 7 d. Error in growth rate corresponds to the propagated uncertainty of isotopic measurements and tracer assimilation (SI Appendix, Supplementary Text). Data used to generate this figure are available in Dataset S4.
Rates of Microbial Growth in Soil Are Slow.
The grassland and conifer forest soils both exhibited respective abundance-weighted mean microbial growth rates of 0.0358 d−1 and 0.0489 d−1 (corresponding to mean generation times of 19.3 d and 14.1 d respectively). Growth was far slower (0.0154 d−1) in the alpine tundra (mean generation time of 44.9 d). The alpine tundra site experiences the lowest mean annual temperature (MAT) of all the sites studied here, with a MAT of −3 °C (44), and is characterized by short (30 to 90 d) and cool growth seasons, where soil respiration begins immediately after soil begins to thaw underneath seasonal snowpack (45). The generation times we observed under conditions analogous to the warm season (20 °C) suggest that, even in the warm season, the majority of the microbial community at this site may not complete a single cell cycle.
Differences observed between specific compounds in all soils indicate that different taxonomic groups within a given soil may exhibit growth rates between 0.1629 and 0.0017 d−1, corresponding to generation times between 4.3 and 402.1 d (Dataset S4). This vast range indicates highly variable growth rates across different constituents of the soil microbiome (Fig. 1B). The majority of soil microorganisms in our study appear to be growing at extremely slow rates when compared to the maximum potential growth rates of many bacteria grown in culture (where generation times typically range from <1 to 100 h) (46). Although it is not surprising that the maximal growth of bacterial isolates in culture conditions does not represent in situ growth rates in soil, our finding that average, abundance-weighted generation times in soil microbial communities range from 14 to 45 d suggests that most soil microbes are oligotrophic, slow-growing, and/or dormant (15, 47), and studies of microbial growth in vitro may not be easily applicable to understanding microbial growth in situ.
Despite the slow growth rates inferred from our LH-SIP approach, these values should still be considered likely overestimates of ambient microbial growth rates. This is because the experimental conditions (conditions applied to many tracer approaches: water addition, sample homogenization, and stable temperatures) may provide more favorable growth conditions for many microbes compared to in situ conditions. The slow community-level and compound-specific growth rates of soil microbial communities measured here point to the importance and ubiquity of slow-growing life in soil systems.
Microbial Biomass Quantity Does Not Predict Growth.
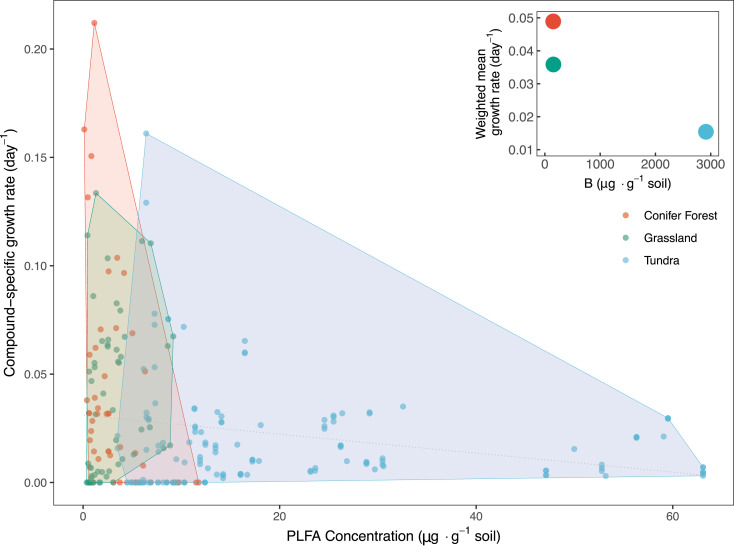
In all soils examined, we find no strong relationship between compound-specific abundance and inferred growth rate (Fig. 2). Compounds exhibiting more rapid rates of production were not necessarily more abundant than compounds exhibiting slower production rates. Overall, there exists a weak negative correlation across all samples between compound abundance and growth rate (r = −0.144, p = 0.016, Pearson’s). This indicates that rapidly growing taxa do not represent a large fraction of the soil microbial community at our field sites and, instead, most of the microbes found in bulk soil are relatively slow growing or dormant. Furthermore, on the time scale of our SIP incubation (0 to 7 d), growth of certain taxa did not clearly alter the bulk fatty acid profiles of the soils (SI Appendix, Fig. S4), contrary to what one might expect if a minority of taxa were overgrowing the community. The uniformity of PLFA profiles throughout the incubation supports the utility of 2H2O as a tracer of in situ microbial growth, as there was no apparent modification of microbial growth or population with the addition of the tracer.
Fig. 2.
Compound-specific PLFA abundance, measured via GC-FID (µg per g soil), plotted against estimated growth rates (day−1) in each of the three soils examined. There exists a very weak but statistically significant negative relationship between compound-specific growth rate and abundance (r = −0.14399, p = 0.016, Pearson’s). The thin dashed line notes the result of a linear model fit (y = −0.00025x + 0.02078, R2 = 0.02), further indicating a slightly negative trend with poor correlation. These results highlight the lack of a clear relationship between compound-specific pool sizes and growth rates in each soil. However, on the bulk scale, soils with lower total biomass, B, exhibited faster assemblage-level growth rates than the alpine tundra soil, which is relatively high in microbial biomass (Inset). This hints at a possible reciprocal relationship between total soil microbial biomass levels and microbiome turnover at the system level.
Because PLFA abundance is a measure of living microbial biomass (43), we next sought to examine whether the total quantity of microbial biomass predicted assemblage-level rates of microbial growth. In other words, do soils with more total microbial biomass also have higher rates of microbial growth? In the three soils examined here, we observed large differences between the sites in the total quantity of PLFAs, with the conifer forest and grassland soils having smaller quantities of intact lipids (150.7 and 154.3 µg g−1, respectively) than the tundra soil (2,913.2 µg g−1). These differences in PLFA abundances are mirrored by differences in organic matter loading for each of the soils (Dataset S1). At the same time, growth rates were slower in the tundra soil and faster in the grassland and conifer forest soils (Figs. 1 B and 2, Inset). Although microbial biomass has been considered a proxy for the microbial productivity of a given soil (6–13), here we find that total microbial biomass is inversely related to the rate at which this biomass is turning over. Our dataset delineates two distinct soil microbiome profiles: a comparatively fast growth but lower biomass soil (typified by the conifer forest and grassland soils) and a relatively slower growth but biomass-rich soil (the alpine tundra) (Fig. 2, Inset). We suggest that soil microbiomes should be assessed along independent axes of microbial biomass quantity and growth rate, parameters that do not necessarily covary. The location of any given soil along these axes is likely determined by biotic and abiotic conditions of the soil environment.
Lipidomic Data Provides Coarse Taxonomic Growth Signals.
Our 16S rRNA gene sequencing results (Materials and Methods) show that the microbial communities in the soils examined were distinct but composed of bacterial taxa that are typically dominant in soils (SI Appendix, Fig. S3): Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Proteobacteria, Planctomycetes, and Verrucomicrobia (48). To examine whether any of these phyla could be distinguished with our lipidomic data, we mined the fatty acid profiles of 4,959 taxa included in the Bacterial Diversity (BacDive) metadatabase (49). We observed that bacteria, at the phylum level, are broadly distinguishable based on their fatty acid profiles (SI Appendix, Figs. S1 and S2), a finding supported by previous characterization of microbial PLFAs (50, 51). We also note that all major PLFAs we predicted to be present and representative of these phyla, based on our analysis of BacDive profiles, were indeed represented in our extracted lipid pools. We find that growth rate patterns grouped by compound class are remarkably similar across all sample sites (Fig. 1B). For example, terminally branched bacterial iso-15:0 and iso-17:0 saturated fatty acids consistently exhibited some of the fastest rates of growth in each soil. These fatty acids are closely associated with the Acidobacteria and Bacteroidetes phyla (SI Appendix, Fig. S1). Conversely, the 18:1 and 18:2 unsaturated fatty acids exhibited slower growth rates. A large portion of the 18:1 and 18:2 unsaturated fatty acids likely represents the slower growth of saprotrophic fungi (51–57) whose lifestyles may differ markedly from those of their bacterial neighbors.
We note that the LH-SIP method is limited in its taxonomic specificity due to the conserved nature of many groups of lipids (i.e., multiple bacterial taxa produce the same membrane lipids) and the fact that the lipid profiles of some major soil bacterial taxa have not been well characterized (42). However, conservative inferences can be made regarding growth rates of bacteria and fungi at broad taxonomic levels based on the relative distributions of fatty acids, as demonstrated in previous studies (35, 36, 51, 58). Therefore, we provide the BacDive dataset (Dataset S2) for the benefit of researchers interested in using our approach in soil and other systems. However, we emphasize that lipidomic datasets like these benefit from sequencing-based approaches to couple taxonomic information on microbial communities to observed growth signatures.
Comparing Estimates of Soil Microbial Growth.
To assess how LH-SIP-derived estimates of microbial growth relate to previous estimates, we collected and compared 26 reports from prior studies that describe microbial growth rates in soil. These published estimates demonstrate disparate ranges that appear highly dependent on the methodology applied (16–24, 59–66) (Fig. 3). Previously published assemblage-level growth rate/turnover estimates vary between 0.002 and 0.356 d−1 (apparent generation times corresponding to 1.94 to 294 d). Much of this prior work has measured the incorporation of isotopically labeled thymidine (TdR) or leucine (Leu) (18–21, 59) and reported community-level growth rates typically between 0.08 and 0.35 d−1 (generation times of 1.95 to 10 d), aside from a single study reporting doubling times of 107 to 170 d (64) (Fig. 3 and Datasets S3 and S4). A potential source of difference in results between the LH-SIP method and these approaches is that DNA- and protein-amendment methods may likely bias toward faster-growing organisms because cells undergoing substantial translation or genomic replication are necessarily going to be captured at higher frequency and fidelity than organisms growing slowly. TdR and Leu approaches specifically could also stimulate microbial growth through the provision of carbon and nitrogen in the tracer solution. Perhaps most importantly, the short incubation times typically used with these approaches (usually <48 h) likely lead to only those taxa with generation times shorter than or approaching the incubation time incorporating sufficient quantities of the isotopic tracer for detection. Microbial taxa with longer generation times may not produce enough new DNA or protein to be detectable by these methods.
Following our literature survey, we sought to understand the utility and sensitivity of LH-SIP as a measurement of slow microbial growth (for more details, see SI Appendix, Supplemental Text). In brief, we calculated the propagated uncertainty in growth rates by accounting for errors inherent in measuring enriched isotopic abundances, soil moisture dilution of the tracer solution, microbial assimilation of hydrogen from water, and IRMS instrument precision. We determined that LH-SIP can accurately distinguish (with confidence ≥ 2σ) microbial generation times in the range of 5 to 700 d during a 7-d incubation period. IRMS instruments are highly sensitive to trace changes in 2H composition and, with isotopically enriched samples, are sensitive to growth rate differences corresponding to 0.0069 d−1 with the incubation parameters of our experiments (Materials and Methods). Interestingly, we found that hydrogen assimilation efficiency, the proportion of lipid hydrogen sourced from water as opposed to other sources (e.g., carbon sources), is the main control on uncertainty in LH-SIP measurements (SI Appendix, Figs. S7 and S10). In contrast, IRMS instrument precision and analytical corrections contribute minor components of the total uncertainty. In samples as complex as soil, it is difficult to parameterize hydrogen assimilation efficiency for an entire community of organisms, even assuming heterotrophy for the bulk of the community. We propose that LH-SIP measurements provide a more conservative estimate of soil microbial growth that captures the slow-growing majority of soil microbes. Because IRMS measurements can capture trace incorporation of 2H into the alkyl chains of membrane lipids, LH-SIP is uniquely suited to capturing slow growth rates both at the compound-specific and bulk scale.
Future Directions for LH-SIP.
The growth rates inferred by LH-SIP of living cells sharing lipid membrane constituents are aggregated. Therefore, growth rate heterogeneity at the cell-to-cell or species-to-species level cannot be resolved with LH-SIP. Single-cell methods including Raman spectroscopy or nanoSIMS (3, 28, 67) can elucidate cell-specific variation in growth rates while also measuring local mineralogical, elemental, or isotopic features. However, single-cell methods can be resource intensive and often require separation of intact cells from environmental matrices, which can be problematic in soil (68). A benefit of the LH-SIP approach is that it can provide community-level insights into anabolic growth activity that may be missed at the single-cell level, with the possibility for coarse taxonomic resolution that is standard for PLFA analysis (33, 35, 36, 51, 69). A two-pronged approach that pairs bulk-scale LH-SIP measurements with single-cell metrics of growth heterogeneity could be powerful.
As noted above, LH-SIP is limited in its taxonomic specificity due to the conserved nature of many PLFA classes (51). Because LH-SIP yields compound-specific growth rates, this method could be used in a highly targeted manner with systems containing less complex microbial communities or with strongly defined relationships between taxonomy and constituent lipid classes. Coupling LH-SIP with additional SIP (e.g., DNA or protein) or advanced lipidomic analyses has further potential to expand our understanding of the microbial physiology of slow growth in natural systems. For instance, future LH-SIP studies could take advantage of liquid chromatographic systems to elucidate relationships between intact polar lipid (IPL) head groups and associated growth rates. The use of signature biomarkers (e.g., glycerol dialkyl glycerol tetraether lipids (GDGT), bacteriohopanepolyols (BHPs), sterols, etc.) for an organism or group of organisms in a given environment would allow for growth rate assignments with greater taxonomic specificity, potentially down to the species and strain. For example, LH-SIP of phospholipid ether lipids could specifically target the growth rates of archaea in natural samples (58). The SIP of PLFAs with 13C (31) has been used with great utility in identifying microbial community members involved in the degradation of distinct substrates (36, 69) or identifying the use of specific metabolic pathways (70): dual stable-isotope (2H + 13C) probing of PLFAs holds great potential for the study of catabolic and anabolic microbial physiology in soils. Finally, LH-SIP could be coupled with already-established 18O-DNA qSIP methods to correlate highly sensitive compound-specific growth rates with direct sequencing of the active fractions of the microbial community.
Relevance of the Slow Growth of Soil Microorganisms.
The in vitro cultivation and isolation of microorganisms from natural systems are notoriously difficult (17, 71, 72). There are numerous proposed reasons for this phenomenon, including the fact that media formulations are imperfect or select for certain taxa, but it is plausible that many “wild” microorganisms are not adapted for the rapid growth that is required for isolation and enrichment using standard cultivation approaches. Slow growth in soil may imply severe limitations on maximum growth rates even under ideal laboratory conditions, given that biochemical adaptations to slow growth may not be easily overcome in a lab environment. Many soil microorganisms in culture are observed to grow slowly, even in “ideal” conditions and, in fact, may be inhibited by high substrate concentrations (73–76). Our observed growth rates (Fig. 1B) suggest that many soil microorganisms may be fundamentally difficult to cultivate due to time constraints on culturing experiments, as the amount of time for an organism to become visible on solid or in liquid media increases exponentially as doubling time increases (SI Appendix, Fig. S6). We also emphasize that maximum potential growth rates measured in vitro likely do not reflect actual growth rates in situ as culture conditions may not adequately replicate the availability or paucity of carbon sources, electron donors/acceptors, and interactions with other organisms. Additionally, maximum potential growth rates [estimated via genomic analyses or culture-based experiments (46, 77)] are fundamentally different metrics than a direct measurement of growth in an environmental setting: a microbe capable of rapid growth will not necessarily exhibit this behavior under environmental conditions. This is highlighted by the observation that a wide array of microbial groups (including those with high maximum potential growth rates in vitro) persist in a dormant or nearly dormant state in soil (14, 15).
Conclusions
Here, we present evidence that slow microbial growth is widespread in soil systems. Across the soils analyzed, PLFA abundances and growth rates were not strongly correlated at the compound-specific level (Fig. 2), indicating that the most abundant taxa are not necessarily the fastest growing. In addition, we observed that soils with lower total biomass exhibited higher rates of microbial growth, and vice versa. These results challenge the idea, often implicit in many studies documenting microbial biomass variation across soils, that higher biomass necessarily equates to higher soil microbial productivity. Instead, our results suggest that soil microbiomes operate on a continuum of growth rate and biomass quantity, with the largest proportion of standing microbial biomass representing oligotrophic or dormant taxa adapted to slow growth. Growth rates presented here occur on the order of weeks to months, comparable to estimates generated by carbon- and nutrient-budgeting models (Fig. 3). Our conclusion that slow-growing microorganisms appear to dominate the soil microbiome is in line with recent evidence that spatial variability in the composition of soil microbial communities typically exceeds the temporal variability observed at a given location (78, 79). Slow growth rates would be expected to attenuate short-term changes in overall microbial community composition, especially in soils with longer observed generation times. As microbial growth is a key regulator of a wide array of soil biogeochemical processes, our findings warrant additional studies that take advantage of the LH-SIP method described here to quantify variation in microbial growth rates across a broader array of soil types and conditions.
Materials and Methods
Soil Sampling and Incubation.
Soils were collected from three locations in central Colorado: a conifer forest located at Gordon Gulch Critical Zone Observatory, Boulder County, CO (40.01, −105.46); a prairie grassland located at Marshall Mesa, Boulder County, CO (39.95, −105.22); and an alpine tundra located at Niwot Ridge, Niwot Long-term Ecological Research Program (LTER) (40.05, −105.58) near Ward, CO. The top 10 cm of soil was excavated with a surface-sterilized trowel, soils were sieved to 2 mm to remove rocks and plant material and homogenized. Soils were stored in the dark at 4 °C before incubations were started. For the SIP incubations, a 10-g subsample of each soil was weighed into a centrifuge tube and combined with 10 mL of filter-sterilized water with ~5,000 ppm 2H2O (0.5 at% 2H, δ2HVSMOW ≈ 31,000 s ‰) and incubated at 20 °C for 0, 3, or 7 d. Isotopic composition of the incubation water was measured at the end of the time series experiment to account for the isotopic contributions of soil water. Samples were periodically shaken over the course of the incubation period to ensure uniform distribution of the tracer solution. At the end of the incubation period, excess incubation water was separated from the soil by centrifugation, decanted, and frozen for later isotopic analysis. Soil pellets were immediately flash-frozen by submerging in a dry ice ethanol bath and stored at −20 °C until lipid extraction.
Water Hydrogen Isotope Analysis.
The labeled incubation waters were analyzed for their H isotope composition (FL) after gravimetric dilution with water of known isotopic composition (1:1000 w/w) to get into the analytical range of available in-house standards previously calibrated to Vienna Standard Mean Ocean Water (VSMOW) and Standard Light Antarctic Precipitation (SLAP). Then, 1 µL of each sample was measured on a dual inlet Thermo Delta Plus XL isotope ratio mass spectrometer connected to an H-Device for water reduction by chromium powder at 850 °C (80). Measured isotope values in δ notation on the VSMOW-SLAP scale were converted to fractional abundances using the isotopic composition of VSMOW [RVSMOW = 2H/1H = 0.00015576, (81)] and the relation F = R/(1+R) = (δ + 1)/(1/RVSMOW + δ + 1) and corrected for the isotope dilution by mass-balance. The resulting isotopic composition of the tracer water was F = 5,015 ppm 2H (0.5015 at%; 31,357 ‰ vs. VSMOW). The isotopic composition of the labeled incubation water was diluted from this value after homogenization with the water in the soil depending on soil type and resulted in 4,363 ± 251 ppm 2H for conifer forest, 4,483 ± 49 ppm 2H for grassland, and 3,492 ± 43 ppm 2H for tundra soils. The latter values were considered to be what cells encountered during tracer incubation (a combination of both the tracer and water present in the soil) and were used for all growth rate calculations.
Lipid Extraction.
Frozen soil pellets were lyophilized for 24 h. Intact polar lipids were extracted from the dry pellets using a modified MTBE-based lipid extraction method (82, 83). In brief, 3.0 g of freeze-dried soil sample was added to a PTFE centrifuge tube. Then, 3 mL of methanol was added to the sample and vortexed. In addition, 10 mL of MTBE was added to the sample and incubated at 1 h at room temperature while shaking. To induce phase separation, 2.5 mL of MS-Grade water was added, and the mixture was centrifuged for 10 min at 1,000G at room temperature. The organic phase was carefully extracted and transferred to an organic-clean glass vial. This process was repeated three times in total. Total lipid extract (TLE) was dried down under a stream of N2 gas and the sample was stored dry at −20 °C until solid phase chromatography. Prior to MTBE extraction, 100 μg of 23-phosphatidylcholine (23-PC) was added to all soils as an internal extraction standard. All glassware was combusted at 450 °C for 8 h prior to use. All Teflon vessels were solvent washed by sonication in a 9:1 mixture of DCM:MeOH for two sets of 30 min. Empty vessels were extracted alongside samples to monitor for contamination. No contamination was detected in the extraction blanks.
Phospholipid Separation and Derivatization.
Phospholipid extract (PLE) was purified from TLE using silica gel chromatography (83) to focus isotopic analyses on lipids derived from intact cells [free phospholipids outside of cellular membranes degrade relatively rapidly with half-life estimates of 39 h at 15 °C (32)]. Combusted silica solid-phase extraction (SPE) columns containing 500 mg SiO2 were conditioned by the addition of 5 mL acetone, then two additions of 5 mL dichloromethane (DCM). TLE was redissolved in 0.5 mL DCM and transferred to the SPE column. Neutral lipids and glycolipids were eluted by the addition of 5 mL of DCM or acetone, respectively, and then dried down under a stream of N2 and stored dry at −20 °C. A PLE was eluted by the addition of 5 mL methanol to the column. PLE was similarly dried under a stream of N2 and stored with an N2-purged headspace at −20 °C.
The PLE was derivatized to fatty acid methyl esters (FAMEs) via base-catalyzed transesterification using methanolic base (84, 85). Transesterification was initiated by the addition of a mixture of 2 mL hexane and 1 mL 0.5 M NaOH in anhydrous methanol to dry PLE. The reaction mixture was allowed to proceed for 10 min at room temperature before being quenched by the addition of 140 μL of glacial (~17 M) acetic acid and 1 mL water. The organic phase was extracted three times with 4 mL hexane and dried down under a stream of N2. A recovery standard of 10 μg 21-phosphatidylcholine (21:0 PC) was added to each PLE before derivatization to assess reaction yield. Then, 10 μg of isobutyl palmitate (PAIBE) was added after derivatization to all samples as a quantification standard prior to analysis.
FAME Quantification and Identification.
A Thermo Scientific Trace 1310 Gas-Chromatograph equipped with a DB-5HT column (30 m × 0.250 mm, 0.10 µm) coupled to a flame-ionization detector (GC-FID) was used to quantify FAME concentrations (µg/g soil) and total amounts (µg extracted) based on peak area relative to the 23-PC extraction and PAIBE quantification standards, respectively. FAMEs were suspended in 100 μL n-hexane, and 1 uL was injected using a split-splitless injector run in splitless mode at 325 °C; split flow was 12.0 mL per min; splitless time was 0.80 min; purge flow was 5.00 mL/min; column flow rate was constant at 1.2 mL/min. The GC ramped according to the following program: 80 °C for 2 min, ramp at 20 °C/min for 5 min (to 140 °C), and ramp at 5 °C/min for 35 min (to 290 °C). The FID was held at 350 °C for the duration of the run. Major peaks were identified by retention time relative to a Bacterial Acid Methyl Ester standard (Millipore-Sigma) and a 37 FAME standard (Supelco). Peak identities were confirmed using a Thermo Scientific Trace 1310 Gas-Chromatograph coupled to a single quadrupole mass spectrometer (ISQ) using identical injection and chromatography conditions with mass scans from 50 to 550 amu and a scan time of 0.2 s in positive ion mode (electron impact). Due to the ambiguity associated with identifying double-bond position and stereochemistry for FAMEs containing multiple bonds, unsaturated compounds are identified only tentatively. FAMEs are referred to using the nomenclature z-x:y, where x is the total number of carbons in the fatty acid skeleton and y is the number of double bonds and their position (if known), while z-is a prefix describing additional structural features of the compound such as methylation and cyclization.
FAME Hydrogen Isotope Analysis.
The isotopic composition of FAMEs was measured on a Thermo Scientific 253 Plus stable isotope ratio mass spectrometer coupled to a Trace 1310 GC via Isolink II pyrolysis/combustion interface (GC/P/IRMS). Chromatographic conditions were identical to those from the GC-FID and GC-MS stated above except for extension of the temperature program to baseline separate all major analytes (40 °C hold for 2 min, 20 °C/min to 120 °C, then 2 °C/min ramp to 240 °C; 30 °C/min ramp to 330 °C, 4-min hold) and injection via programmable temperature vaporization inlet (ramped from 40 to 400 °C) to ensure quantitative transfer from the inlet to the column. Peaks were identified based on retention order and relative height based on coregistration with GC-FID and GC-MS chromatograms.
Measured isotope ratios were corrected for scale compression, linearity, and memory effects using natural abundance and isotopically enriched fatty acid esters of known isotopic composition ranging from –231.2 ‰ to +3,972 ‰ vs. VSMOW (SI Appendix, Supplementary Text). Memory (peak-to-peak carryover) effects are important to correct for given the wide range of peak areas and isotopic values encountered in this study and the known impact of memory effects on H isotope measurements (86, 87). Full memory effect corrections for standard mixtures of natural abundance and enriched fatty acid esters lead to a >44% improvement in residual SE (SI Appendix, Table S1 and Fig. S8). The multivariate linear regression calibration for the enriched samples (SI Appendix, Eqs. S1–S3 and Table S1) included standards ranging in mass 2 areas from 1.74 Vs (~25 ng) to 69.67 Vs (~975 ng) and lead to an overall RMSE of the calibration of 60.4‰ stemming from the substantial dynamic range of the isotope standards and believed to accurately reflect the elevated uncertainty that should be expected in the isotopically diverse samples. The conservative analytical SEs ranged from 46.5 to 269 ‰ depending on peak area with larger error estimates for smaller peaks that were more affected by memory effects (SI Appendix, Supplementary Text). The hydrogen isotope calibration was performed in R using the packages isoreader [v 1.3.0 (88)] and isoprocessor (v 0.6.11) available at github.com/isoverse.
Calibrated isotope ratios measured via GC/P/IRMS were further corrected for H added during derivatization to FAMEs, as well as for analytical and replicate error as follows: First, the 2F of methanol used for base-catalyzed transesterification was measured by taking an aliquot of anhydrous methanol reagent (the exact stock used for transesterification) and derivatizing a phthalic acid with a known H isotopic composition (Arndt Schimmelmann, Indiana University) via acid catalysis. The resulting phthalic methyl ester was analyzed by GC/P/IRMS and a correction was applied to all values (SI Appendix, Supplemental Text).
Growth Rate Calculations.
Calculations of biosynthetic activity focus on the isotopic composition of FAMEs because the hydrocarbon skeleton of fatty acids consists only of C–H bonds that are nonexchangeable on biological time scales (89), unlike the readily exchangeable H bound to O, N, P, and S in parts of lipid headgroups, proteins, and nucleic acids. The H tracer is thus stably incorporated into fatty acids tails during biological activity. The resulting isotopic enrichment of fatty acids in intact cellular lipids is described by the following equation (26):
where r is the specific biosynthesis rate (1/days); t is the duration of tracer exposure (days); a is the assimilation efficiency and fractionation of water hydrogen during lipid biosynthesis (see ref. 26 and SI Appendix, Fig. S7); and F0, Ft, and FL are the fractional abundances of 2H in fatty acids before tracer incorporation, in fatty acids at time t, and in the isotopically labeled sample water. Solving this equation for r makes it possible to infer from the incubation time and isotopic measurements how quickly cellular fatty acids turn over. With lipid biosynthesis reflecting a combination of growth and repair, this provides an upper/lower bound for the specific growth rate µ and apparent generation time TG of the microbial producers of a given lipid:
These calculations yield compound-specific growth rate and generation time estimates that can be viewed by themselves or aggregated into assemblage-level estimates of community growth by calculating an abundance-weighted mean:
where xi is the isotopic fractional abundance of compound i and wi is the relative abundance (weighting) of compound i. As discussed, these estimates aggregate cell-to-cell variations in growth. Uncertainty in each set of measurements was propagated through our calculations of µ by SE propagation (SI Appendix, Supplementary Text).
16S Ribosomal RNA Gene Sequencing.
To characterize the microbial community composition of each soil type before SIP incubation, DNA was extracted from soil subsamples in triplicate and with negative control using the DNeasy PowerSoil DNA isolation kit (Qiagen), according to the manufacturer’s instructions with one minor modification; samples were heated with Solution C1 for 10 min at 65 °C in a dry heat block prior to bead beating. Extracted DNA samples were amplified in duplicate using Platinum II Hot-Start PCR Master Mix (Thermo Fisher Scientific) and the 16S rRNA gene primers 515F and 806R with Illumina sequencing adapters and unique 12-bp barcodes. The PCR program was 94 °C for 2 min followed by 35 cycles of 94 °C (15 s), 60 °C (15 s), 68 °C (1 min), and a final extension at 72 °C for 10 min. Amplification was verified via gel electrophoresis. Amplicons were cleaned and normalized with the SequalPrep Normalization Plate (Thermo Fisher Scientific) following the manufacturer’s instructions and then pooled together. Sequencing was performed on an Illumina MiSeq using a v2 300 cycle kit with paired-end reads at the University of Colorado BioFrontiers Institute Next-Gen Sequencing Core Facility.
To prepare samples for analysis with the DADA2 (version 1.10.1) bioinformatic pipeline (90), reads were demultiplexed with adapters and primers were removed using standard settings for cutadapt (version 1.8.1, Martin 2011). We used standard filtering parameters with slight modifications for 2 × 150 bp chemistry where forward reads were not trimmed and reverse reads were trimmed (truncLen) to 140 base pairs. In addition, we truncated reads at the first nucleotide with a quality score (truncQ) below 11 and a maximum allowed error rate (maxEE) of 1. These filtering parameters resulted in a mean of 95.7% of reads retained, and this was visually assessed with quality profiles for each sample. Reads were dereplicated, paired ends were merged, amplicon sequence variants (ASVs) were assigned, and chimeras were removed (98.23% of reads were not chimeric). Finally, taxonomy was assigned to each ASV against the SILVA (v132) reference database (91). We removed all chloroplast, mitochondria, and eukaryotic reads from the ASV table, which resulted in an average of 36,029 reads per sample (range 27,074 to 58,528 reads), with the ASV table subsequently rarefied to 27,000 reads per sample. Blank samples had far fewer reads than actual samples (mean of 193 reads compared to 35,455 reads per sample), and the four genera detected in blanks (Thermus, Geobacillus, Deinococcus, and Pseudomonas) were not consistently detected and were below the 1% relative abundance threshold for inclusion in sample analyses. Taxonomic composition of the samples was compared across soil types (SI Appendix, Fig. S3).
BacDive Database Analysis and Literature Survey.
To infer relationships between lipid profiles observed across our SIP incubations and high-level taxonomy, we queried the Bacterial Metadiversity Database (BacDive) (49) for all available fatty acid profiles using the BacDive API client implemented by the BacDiveR package (92). We generated a table of 4,959 fatty acid profiles indexed by the taxonomy reported in the database and used principal component analysis to generate SI Appendix, Fig. S2, grouped at the phylum level. We appended 24 manually curated fungal fatty acid profiles to this dataset, and this combined table is available as Dataset S2. We looked at relationships between fatty acid profiles at the phylum level (SI Appendix, Fig. S1) and conducted a principal component analysis of fatty acid composition and taxonomy (SI Appendix, Fig. S2).
To compare LH-SIP-measured rates of growth to other methods, we surveyed 26 previously reported estimates of microbial growth and compared them to the abundance-weighted mean estimates of our study (Datasets S3 and S4). We collected specific passages, sentences, and tables and manually digitized these reported values, noting the specific mention of microbial growth. Studies use various terminology and units including growth rate (day−1), generation time (days), turnover rate (day−1), turnover time (days), or doubling times (days). Turnover time was converted to turnover rate by taking the reciprocal value. We include both “growth rates” and “turnover rates” as the distinction between the two is that turnover explicitly assumes a steady state of biomass (loss rates equal production rates). Generation time/doubling time estimates are converted to growth rates as described in this study. We plot the mean value from each study (Fig. 3), either reported in the manuscript or calculated from the upper- and lower-bounds reported in the manuscript.
Soil Geochemistry.
Soils were analyzed at the Colorado State University Soil, Water, and Plant Testing Laboratory for routine determination of soil characteristics. In short, a KCl extract was used to quantify soil nitrate (93). An AB-DTPA extract was used to quantify soil P, Zn, Fe, Mn, Cu, and S (94). Organic matter percentages were calculated by determining the weight loss of samples after ignition. These data are available in Dataset S1.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Acknowledgments
We are grateful to Julio Sepúlveda, Nadia Dildar, and Jonathan Raberg of the CU Boulder Organic Geochemistry Lab for valuable methodology input and Jessica Henley for assistance with 16S sequencing. We thank Jack Gugel, Toby Halamka, and Claire Karban for assistance with soil sampling. We thank the editor, as well as an anonymous reviewer and Alex Sessions for constructive feedback that substantially improved the manuscript. GC-FID, GC-MS, and GC-IRMS analyses were performed at the University of Colorado, Boulder Earth Systems Stable Isotope Lab core facility, RRID:SCR_019300. Isotopic determination of heavy water tracer solutions was conducted by the Feng Lab at Dartmouth College. This project was supported by a CU Boulder Grand Challenge Seed Grant to S.K. and N.F., a grant from the U.S. NSF (#2131837) to N.F., and a research grant from the Army Research Office (#78484-LS) to S.K. T.A.C. was supported by a NSF Graduate Research Fellowship and through the IQ Biology Program of the BioFrontiers Institute at the University of Colorado, Boulder. Logistical support for Niwot Ridge access was provided by the Niwot Ridge LTER program (NSF DEB – 1637686) with thanks to William D. Bowman for assistance with site access. S.J. was supported by the Canyonlands Research Center Graduate Research Scholars Program. The Canyonlands Research Station is supported by The Nature Conservancy.
Author contributions
T.A.C., N.F., and S.K. designed research; T.A.C., J.M., and S.J. performed research; T.A.C. and S.K. analyzed data; and T.A.C. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Analysis Scripts data have been deposited in GitHub (https://github.com/KopfLab/Caro-et-al.-Soil-Turnover) (95). All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Weinstock M. T., Hesek E. D., Wilson C. M., Gibson D. G., Vibrio natriegens as a fast-growing host for molecular biology. Nat. Methods 13, 849–851 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Hoehler T. M., Jørgensen B. B., Microbial life under extreme energy limitation. Nat. Rev. Microbiol. 11, 83–94 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Trembath-Reichert E., et al. , Methyl-compound use and slow growth characterize microbial life in 2-km-deep subseafloor coal and shale beds. Proc. Natl. Acad. Sci. U.S.A. 114, E9206–E9215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poindexter J. S., "Oligotrophy" in Advances in Microbial Ecology, Alexander M., Ed. (Springer, 1981), pp. 63–89. [Google Scholar]
- 5.Lauro F. M., et al. , The genomic basis of trophic strategy in marine bacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 15527–15533 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foote J. A., Boutton T. W., Scott D. A., Soil C and N storage and microbial biomass in US southern pine forests: Influence of forest management. For. Ecol. Manage. 355, 48–57 (2015). [Google Scholar]
- 7.Kaschuk G., Alberton O., Hungria M., Three decades of soil microbial biomass studies in Brazilian ecosystems: Lessons learned about soil quality and indications for improving sustainability. Soil Biol. Biochem. 42, 1–13 (2010). [Google Scholar]
- 8.Dalal R. C., Soil microbial biomass—What do the numbers really mean? Aust. J. Exp. Agric. 38, 649–665 (1998). [Google Scholar]
- 9.Chander K., Goyal S., Mundra M. C., Kapoor K. K., Organic matter, microbial biomass and enzyme activity of soils under different crop rotations in the tropics. Biol. Fertil. Soils 24, 306–310 (1997). [Google Scholar]
- 10.Bolton H., Elliott L. F., Papendick R. I., Bezdicek D. F., Soil microbial biomass and selected soil enzyme activities: Effect of fertilization and cropping practices. Soil Biol. Biochem. 17, 297–302 (1985). [Google Scholar]
- 11.Jenkinson D. S., Powlson D. S., The effects of biocidal treatments on metabolism in soil—V: A method for measuring soil biomass. Soil Biol. Biochem. 8, 209–213 (1976). [Google Scholar]
- 12.Wardle D. A., A comparative assessment of factors which influence microbial biomass carbon and nitrogen levels in soil. Biol. Rev. 67, 321–358 (1992). [Google Scholar]
- 13.Allen D. E., Singh B. P., Dalal R. C., "Soil health indicators under climate change: A review of current knowledge" in Soil Health and Climate Change, Soil Biology, Singh B. P., Cowie A. L., Chan K. Y., Eds. (Springer, 2011), pp. 25–45. [Google Scholar]
- 14.Lennon J. T., Jones S. E., Microbial seed banks: The ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 9, 119–130 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Blagodatskaya E., Kuzyakov Y., Active microorganisms in soil: Critical review of estimation criteria and approaches. Soil Biol. Biochem. 67, 192–211 (2013). [Google Scholar]
- 16.Koch B. J., et al. , Estimating taxon-specific population dynamics in diverse microbial communities. Ecosphere 9, e02090 (2018). [Google Scholar]
- 17.Rousk J., Bååth E., Growth of saprotrophic fungi and bacteria in soil: Growth of saprotrophic fungi and bacteria in soil. FEMS Microbiol. Ecol. 78, 17–30 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Bååth E., Growth rates of bacterial communities in soils at varying pH: A comparison of the thymidine and leucine incorporation techniques. Microb. Ecol. 36, 316–327 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Bååth E., Measurement of protein synthesis by soil bacterial assemblages with the leucine incorporation technique. Biol. Fertil. Soils 17, 147–153 (1994). [Google Scholar]
- 20.Bååth E., Thymidine and leucine incorporation in soil bacteria with different cell size. Microb. Ecol. 27, 267–278 (1994). [DOI] [PubMed] [Google Scholar]
- 21.Tibbles B. J., Harris J. M., Use of radiolabelled thymidine and leucine to estimate bacterial production in soils from continental Antarctica. Appl. Environ. Microbiol. 62, 694–701 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng W., Rhizosphere priming effect: Its functional relationships with microbial turnover, evapotranspiration, and C-N budgets. Soil Biol. Biochem. 41, 1795–1801 (2009). [Google Scholar]
- 23.Spohn M., Klaus K., Wanek W., Richter A., Microbial carbon use efficiency and biomass turnover times depending on soil depth—Implications for carbon cycling. Soil Biol. Biochem. 96, 74–81 (2016). [Google Scholar]
- 24.Hunt H. W., et al. , The detrital food web in a shortgrass prairie. Biol. Fert. Soils 3, 57–68 (1987). [Google Scholar]
- 25.Hungate B. A., et al. , Quantitative microbial ecology through stable isotope probing. Appl. Environ. Microbiol. 81, 7570–7581 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kopf S. H., et al. , Trace incorporation of heavy water reveals slow and heterogeneous pathogen growth rates in cystic fibrosis sputum. Proc. Natl. Acad. Sci. U.S.A. 113, E110–E116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wegener G., et al. , Assessing sub-seafloor microbial activity by combined stable isotope probing with deuterated water and 13C-bicarbonate. Environ. Microbiol. 14, 1517–1527 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Berry D., et al. , Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc. Natl. Acad. Sci. U.S.A. 112, E194–E203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer C. R., Bowen B. P., Pan C., Northen T. R., Banfield J. F., Stable-isotope probing reveals that hydrogen isotope fractionation in proteins and lipids in a microbial community are different and species-specific. ACS Chem. Biol. 8, 1755–1763 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Schwartz E., Characterization of growing microorganisms in soil by stable isotope probing with H218O. Appl. Environ. Microbiol. 73, 2541–2546 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evershed R. P., et al. , 13C-Labelling of lipids to investigate microbial communities in the environment. Current Opinion in Biotechnology 17, 72–82 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y., et al. , High turnover rate of free phospholipids in soil confirms the classic hypothesis of PLFA methodology. Soil Biol. Biochem. 135, 323–330 (2019). [Google Scholar]
- 33.Frostegård Å., Tunlid A., Bååth E., Use and misuse of PLFA measurements in soils. Soil Biol. Biochem. 43, 1621–1625 (2011). [Google Scholar]
- 34.Carini P., et al. , Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat. Microbiol. 2, 1–6 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Grayston S. J., et al. , Assessing shifts in microbial community structure across a range of grasslands of differing management intensity using CLPP, PLFA and community DNA techniques. Appl. Soil Ecol. 25, 63–84 (2004). [Google Scholar]
- 36.Ramsey P. W., Rillig M. C., Feris K. P., Holben W. E., Gannon J. E., Choice of methods for soil microbial community analysis: PLFA maximizes power compared to CLPP and PCR-based approaches. Pedobiologia 50, 275–280 (2006). [Google Scholar]
- 37.Neubauer C., et al. , Towards measuring growth rates of pathogens during infections by D2O-labeling lipidomics. Rapid Commun. Mass Spectrom. 32, 2129–2140 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Neubauer C., et al. , Refining the application of microbial lipids as tracers of Staphylococcus aureus growth rates in cystic fibrosis sputum. J. Bacteriol. 200, e00365-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wegener G., Kellermann M. Y., Elvert M., Tracking activity and function of microorganisms by stable isotope probing of membrane lipids. Curr. Opin. Biotechnol. 41, 43–52 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Kellermann M. Y., et al. , Autotrophy as a predominant mode of carbon fixation in anaerobic methane-oxidizing microbial communities. Proc. Natl. Acad. Sci. U.S.A. 109, 19321–19326 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kempes C. P., et al. , Drivers of bacterial maintenance and minimal energy requirements. Front. Microbiol. 8, 31 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joergensen R. G., Phospholipid fatty acids in soil—Drawbacks and future prospects. Biol. Fertil. Soils 58, 1–6 (2022). [Google Scholar]
- 43.Frostegård A., Bååth E., The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol. Fert. Soils 22, 59–65 (1996). [Google Scholar]
- 44.Greenland D., The climate of Niwot Ridge, Front Range, Colorado, U.S.A. Arctic Alpine Res. 21, 380–391 (1989). [Google Scholar]
- 45.Brooks P. D., Williams M. W., Schmidt S. K., Microbial activity under alpine snowpacks, Niwot Ridge, Colorado. Biogeochemistry 32, 93–113 (1996). [Google Scholar]
- 46.Weissman J. L., Hou S., Fuhrman J. A., Estimating maximal microbial growth rates from cultures, metagenomes, and single cells via codon usage patterns. Proc. Natl. Acad. Sci. U.S.A. 118, e2016810118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stone B. W. G., et al. , Life history strategies among soil bacteria—Dichotomy for few, continuum for many. ISME J. 17, 611–619 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delgado-Baquerizo M., et al. , A global atlas of the dominant bacteria found in soil. Science 359, 320–325 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Reimer L. C., et al. , BacDive in 2019: Bacterial phenotypic data for High-throughput biodiversity analysis. Nucl. Acids Res. 47, D631–D636 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willers C., Jansen van Rensburg P. J., Claassens S., Microbial signature lipid biomarker analysis—An approach that is still preferred, even amid various method modifications. J. Appl. Microbiol. 118, 1251–1263 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Willers C., Jansen van Rensburg P. J., Claassens S., Phospholipid fatty acid profiling of microbial communities–a review of interpretations and recent applications. J. Appl. Microbiol. 119, 1207–1218 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Dong H.-Y., Kong C.-H., Wang P., Huang Q.-L., Temporal variation of soil friedelin and microbial community under different land uses in a long-term agroecosystem. Soil Biol. Biochem. 69, 275–281 (2014). [Google Scholar]
- 53.Fichtner A., von Oheimb G., Härdtle W., Wilken C., Gutknecht J. L. M., Effects of anthropogenic disturbances on soil microbial communities in oak forests persist for more than 100 years. Soil Biol. Biochem. 70, 79–87 (2014). [Google Scholar]
- 54.Reinsch S., et al. , Short-term utilization of carbon by the soil microbial community under future climatic conditions in a temperate heathland. Soil Biol. Biochem. 68, 9–19 (2014). [Google Scholar]
- 55.Buckeridge K. M., Banerjee S., Siciliano S. D., Grogan P., The seasonal pattern of soil microbial community structure in mesic low arctic tundra. Soil Biol. Biochem. 65, 338–347 (2013). [Google Scholar]
- 56.Högberg M. N., Högbom L., Kleja D. B., Soil microbial community indices as predictors of soil solution chemistry and N leaching in Picea abies (L.). Karst. forests in S. Sweden. Plant Soil 372, 507–522 (2013). [Google Scholar]
- 57.García-Orenes F., Caravaca F., Morugán-Coronado A., Roldán A., Prolonged irrigation with municipal wastewater promotes a persistent and active soil microbial community in a semiarid agroecosystem. Agric. Water Manage. 149, 115–122 (2015). [Google Scholar]
- 58.Gattinger A., Günthner A., Schloter M., Munch J. C., Characterisation of Archaea in Soils by Polar Lipid Analysis. Acta Biotechnol. 23, 21–28 (2003). [Google Scholar]
- 59.Bååth E., Thymidine incorporation into macromolecules of bacteria extracted from soil by homogenization-centrifugation. Soil Biol. Biochem. 24, 1157–1165 (1992). [Google Scholar]
- 60.Uhlıřová E., Šantrůčková H., Growth rate of bacteria is affected by soil texture and extraction procedure. Soil Biol. Biochem. 35, 217–224 (2003). [Google Scholar]
- 61.Blazewicz S. J., Schwartz E., Firestone M. K., Growth and death of bacteria and fungi underlie rainfall-induced carbon dioxide pulses from seasonally dried soil. Ecology 95, 1162–1172 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Kouno K., Wu J., Brookes P. C., Turnover of biomass C and P in soil following incorporation of glucose or ryegrass. Soil Biol. Biochem. 34, 617–622 (2002). [Google Scholar]
- 63.Li J., et al. , Predictive genomic traits for bacterial growth in culture versus actual growth in soil. ISME J. 13, 2162–2172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harris D., Paul E. A., Measurement of bacterial growth rates in soil. Appl. Soil Ecol. 1, 277–290 (1994). [Google Scholar]
- 65.Perelo L. W., Munch J. C., Microbial immobilisation and turnover of 13C labelled substrates in two arable soils under field and laboratory conditions. Soil Biol. Biochem. 37, 2263–2272 (2005). [Google Scholar]
- 66.Bååth E., Thymidine incorporation into soil bacteria. Soil Biol. Biochem. 22, 803–810 (1990). [Google Scholar]
- 67.Eichorst S. A., et al. , Advancements in the application of NanoSIMS and Raman microspectroscopy to investigate the activity of microbial cells in soils. FEMS Microbiol. Ecol. 91, fiv106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Holmsgaard P. N., et al. , Bias in bacterial diversity as a result of Nycodenz extraction from bulk soil. Soil Biol. Biochem. 43, 2152–2159 (2011). [Google Scholar]
- 69.Yao H., Chapman S. J., Thornton B., Paterson E., 13C PLFAs: A key to open the soil microbial black box? Plant Soil 392, 3–15 (2015). [Google Scholar]
- 70.Wu W., Dijkstra P., Hungate B. A., Shi L., Dippold M. A., In situ diversity of metabolism and carbon use efficiency among soil bacteria. Sci. Adv. 8, eabq3958 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Staley J. T., Konopka A., Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39, 321–346 (1985). [DOI] [PubMed] [Google Scholar]
- 72.Prosser J. I., "Molecular and functional diversity in soil micro-organisms" in Diversity and Integration in Mycorrhizas, Smith S. E., Smith F. A., Eds. (Springer, Netherlands, 2002), pp. 9–17. [Google Scholar]
- 73.Pham V. H. T., Kim J., Cultivation of unculturable soil bacteria. Trends Biotechnol. 30, 475–484 (2012). [DOI] [PubMed] [Google Scholar]
- 74.Vartoukian S. R., Palmer R. M., Wade W. G., Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol. Lett. 309, 1–7 (2010). [DOI] [PubMed] [Google Scholar]
- 75.Davis K. E. R., Joseph S. J., Janssen P. H., Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 71, 826–834 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Overmann J., Abt B., Sikorski J., Present and future of culturing bacteria. Annu. Rev. Microbiol. 71, 711–730 (2017). [DOI] [PubMed] [Google Scholar]
- 77.Long A. M., Hou S., Ignacio-Espinoza J. C., Fuhrman J. A., Benchmarking microbial growth rate predictions from metagenomes. ISME J. 15, 183–195 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carini P., et al. , Effects of spatial variability and relic DNA removal on the detection of temporal dynamics in soil microbial communities. mBio 11, e02776-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lauber C. L., Ramirez K. S., Aanderud Z., Lennon J., Fierer N., Temporal variability in soil microbial communities across land-use types. ISME J. 7, 1641–1650 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Putman A. L., Feng X., Sonder L. J., Posmentier E. S., Annual variation in event-scale precipitation δ2H at Barrow, AK, reflects vapor source region. Atmos. Chem. Phys. 17, 4627–4639 (2017). [Google Scholar]
- 81.Meija J., et al. , Isotopic compositions of the elements 2013 (IUPAC Technical Report). Pure Appl. Chem. 88, 293–306 (2016). [Google Scholar]
- 82.Matyash V., Liebisch G., Kurzchalia T. V., Shevchenko A., Schwudke D., Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 49, 1137–1146 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Quideau S. A., et al. , Extraction and analysis of microbial phospholipid fatty acids in soils. J. Vis. Exp. 114, 54360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rodríguez-Ruiz J., Belarbi E.-H., Sánchez J. L. G., Alonso D. L., Rapid simultaneous lipid extraction and transesterification for fatty acid analyses. Biotechnol. Tech. 12, 689–691 (1998). [Google Scholar]
- 85.Griffiths M. J., van Hille R. P., Harrison S. T. L., Selection of direct transesterification as the preferred method for assay of fatty acid content of microalgae. Lipids 45, 1053–1060 (2010). [DOI] [PubMed] [Google Scholar]
- 86.Wang Y., Sessions A. L., Memory effects in compound-specific D/H analysis by gas chromatography/pyrolysis/isotope-ratio mass spectrometry. Anal. Chem. 80, 9162–9170 (2008). [DOI] [PubMed] [Google Scholar]
- 87.Liu H., Cao Y., Hu J., Liu Z., Liu W., Substantial peak size effect on compound-specific δD values analyzed on isotope ratio mass spectrometry. Chem. Geol. 590, 120721 (2022). [Google Scholar]
- 88.Kopf S., Davidheiser-Kroll B., Kocken I., Isoreader: An R package to read stable isotope data files for reproducible research. J. Open Source Softw. 6, 2878 (2021). [Google Scholar]
- 89.Schimmelmann A., Sessions A. L., Mastalerz M., Hydrogen isotopic (d/H) composition of organic matter during diagenesis and thermal maturation. Annu. Rev. Earth Planet. Sci. 34, 501–533 (2006). [Google Scholar]
- 90.Callahan B. J., et al. , DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Quast C., et al. , The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucl. Acids Res. 41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goeker M., BacDive: BacDive API Client (R Package Version 0.6.0/r885, R-Forge, 2021), https://r-forge.r-project.org/.
- 93.Keeney D. R., Nelson D. W., “Nitrogen—Inorganic forms” in Methods of Soil Analysis (John Wiley & Sons Ltd, 1983), pp. 643–698. [Google Scholar]
- 94.Rodriguez J. B., Self J. R., Peterson G. A., Westfall D. G., Sodium bicarbonate-DTPA test for macro- and micronutrient elements in soils. Commun. Soil Sci. Plant Anal. 30, 957–970 (1999). [Google Scholar]
- 95.Caro T. A., Kopf S., KopfLab/Caro-et-al.-Soil-Turnover. Github. https://github.com/KopfLab/Caro-et-al.-Soil-Turnover. Deposited 5 January 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Data Availability Statement
Analysis Scripts data have been deposited in GitHub (https://github.com/KopfLab/Caro-et-al.-Soil-Turnover) (95). All study data are included in the article and/or SI Appendix.