Abstract
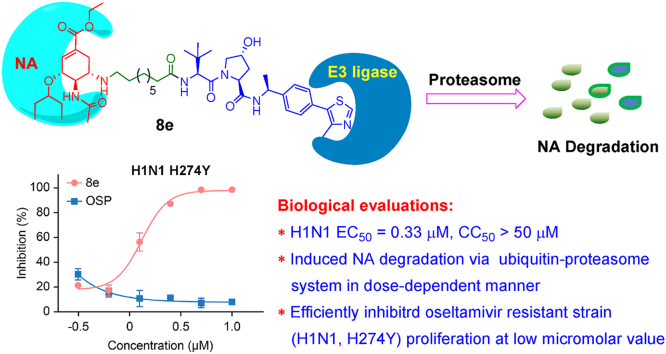
Annual and sporadic influenza outbreaks pose a great threat to human health and the economy worldwide. Moreover, the frequent mutation of influenza viruses caused by antigen drift complicates the application of antiviral therapeutics. As such, there is an urgent need for novel antiviral agents to tackle the problem of insufficient efficacy of licensed drugs. Inspired by the success of the newly emerged PROTACs (PROteolysis TArgeting Chimeras) strategy, we report herein the design and synthesis of novel PROTAC molecules based on an oseltamivir scaffold to combat severe annual influenza outbreaks. Among these, several compounds showed good anti-H1N1 activity and efficient influenza neuraminidase (NA) degradation activity. The best compound, 8e, effectively induced influenza NA degradation in a dose-dependent manner and relied on the ubiquitin–proteasome pathway. Moreover, Compound 8e exhibited potent antiviral activity toward both wild-type H1N1 virus and an oseltamivir-resistant strain (H1N1, H274Y). A molecular docking study demonstrated that Compound 8e had good hydrogen-bonding and hydrophobic interactions with both the active sites of NA and Von Hippel-Lindau (VHL) proteins, which could effectively drive the favorable interaction of these two proteins. Thus, as the first report of a successful anti-influenza PROTAC, this proof of concept will greatly widen the application range of the PROTAC technique to antiviral drug discovery.
Keywords: Influenza, PROTACs, Neuraminidase, Oseltamivir, Antiviral drug
Graphical abstract
Highlights
-
•
A series of oseltamivir derived PROTACs with diverse linkers and E3 ligands were synthesized.
-
•
The Compound 8e exhibited potent antiviral activity toward both wild-type H1N1 virus and an oseltamivir-resistant strain.
-
•
Mechanistic studies indicated that 8e degraded the NA protein through the ubiquitin-proteasome pathway.
-
•
This proof of concept work will greatly widen the application range of the PROTAC technique in antiviral drug discovery.
1. Introduction
Annual and sporadic influenza outbreaks have resulted in an enormous burden on human health and the economy worldwide (Grienke et al., 2010; Kadam and Wilson, 2017). The WHO has reported that the annual seasonal influenza epidemic involves approximately 3–5 million cases of severe illness, with 290–650 thousand deaths (Zhang et al., 2018b). Influenza-related clinical symptoms vary from mild diseases to severe lethal pneumonia and nervous system damage (Krammer et al., 2018; Lowen Anice et al., 2008; Sellers et al., 2017). Outbreaks from the 1918 Spanish flu to the 2009 swine flu have cost thousands of people's lives, and the frequent mutation of influenza viruses caused by antigen drift complicates the application of antiviral therapeutics (Hensley et al., 2009). Thus, the search for anti-influenza drugs with novel scaffolds or new mechanisms is urgently needed to tackle the problem of insufficient efficacy of licensed vaccines and drugs.
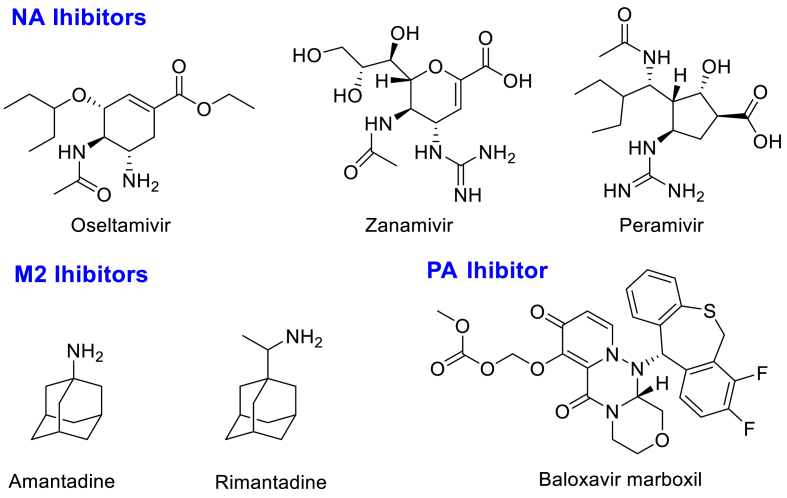
Influenza virus is a negative-stranded RNA virus belonging to the Orthomyxoviridae family (Schrauwen and Fouchier, 2014). The life cycle of influenza virus has rigorous and precise steps (Paules and Subbarao, 2017). In addition, there are many key proteins involved in influenza virus proliferation, including RNA-dependent RNA polymerase (RdRp) and neuraminidase (NA) (Christopher, 2007; Schrauwen et al., 2014). Therefore, targeting the critical protein involved in virus propagation to develop influenza inhibitors has garnered much attention. Amantadine (an M2 inhibitor, Fig. 1) was the first licensed anti-influenza drug in 1966 and has since been gradually withdrawn from clinical use due to severe nervous system side effects and drug resistance (Rey-Carrizo et al., 2013; Wang et al., 2013). Oseltamivir (Fig. 1), as the first orally available influenza NA inhibitor, is widely used to treat influenza infection as a first-line therapy (Zhang et al., 2018b). Other anti-influenza drugs targeting NA, such as zanamivir and peramivir (Fig. 1), are dosed intravenously or inhaled because of their high polarity (Anuwongcharoen et al., 2016). Despite the great success of the discovery of influenza NA inhibitors, various mutants have been identified with the frequent use of oseltamivir (Collins et al., 2008; Memoli et al., 2010). Although these mutants remain sensitive to zanamivir, they limit the clinical application of oseltamivir, with a better drug compliance. In addition, the search for robust anti-influenza agents has also focused on RdRp inhibitors, which have been confirmed to have antiviral activities ranging from good to excellent (Credille et al., 2016; Massari et al., 2015, Massari et al., 2021; McGowan et al., 2019). Recently, two RdRp inhibitors, favipiravir and baloxavir marboxil (PA inhibitor, Fig. 1), were approved by Japan and the FDA, respectively, for the treatment of influenza infections (Du et al., 2020; Takashita et al., 2016). However, due to the severe teratogenicity of favipiravir and the emerging drug resistance of baloxavir marboxil (Hirotsu et al., 2020; Zaraket and Saito, 2016), the therapeutic efficacy of these two drugs remains a huge obstacle. Thus, considering the enormous threats associated with influenza infections, the development of inhibitors with diverse scaffolds is urgently needed.
Fig. 1.
Chemical structures of representative approvedanti-influenzadrugs.
PROteolysis TArgeting Chimeras (PROTACs) are a newly emerging modality utilized for targeted protein depletion by recruiting the intracellular ubiquitin−proteasome system (Cromm and Crews, 2017; Neklesa et al., 2017). PROTACs are bifunctional molecules that combine the POI (protein of interest) ligand and E3 ligase ligand via diverse linkers (Lai and Crews, 2017; Naro et al., 2020; Ottis et al., 2017). Notoriously, drug resistance to viral infection has represented a huge obstacle for the discovery of novel antiviral agents. PROTACs, as a robust chemical biology approach, have drawn great attention from medicinal chemists for drug discovery due to their superior substoichiometric catalytic nature and ability to handle drug resistance compared with traditional occupancy-driven pharmacology (Lai and Crews, 2017; Zheng et al., 2021). To date, the milestones of PROTAC technology development, from the initially reported PROTAC targeting methionine aminopeptidase-2 (MetAp-2) by recruiting peptide E3 ligase ligands to small molecule-based PROTACs, have facilitated intense research in small-molecule drug discovery (Sakamoto et al., 2001, 2003; Schneekloth et al., 2004, 2008). Intriguingly, the PROTAC strategy has been extensively applied for the degradation of diverse targets, showing efficient activity toward both wild-type and mutants and serving as clinical candidates for the treatment of various cancers (Hu et al., 2019; Lai et al., 2016; Su et al., 2019; Yang et al., 2020; Zhou et al., 2018). Recently, the first reported virus-targeted PROTAC efficiently inhibited virus replication in both wild-type and mutant strains by degrading hepatitis C virus (HCV) NS3 protease based on the telaprevir structure, which shed light on the discovery of novel antiviral agents based on a degradation strategy (de Wispelaere et al., 2019). In addition, a few studies have identified degraders that have activity in SARS-CoV-2 infections (Desantis et al., 2021; Haniff et al., 2020).
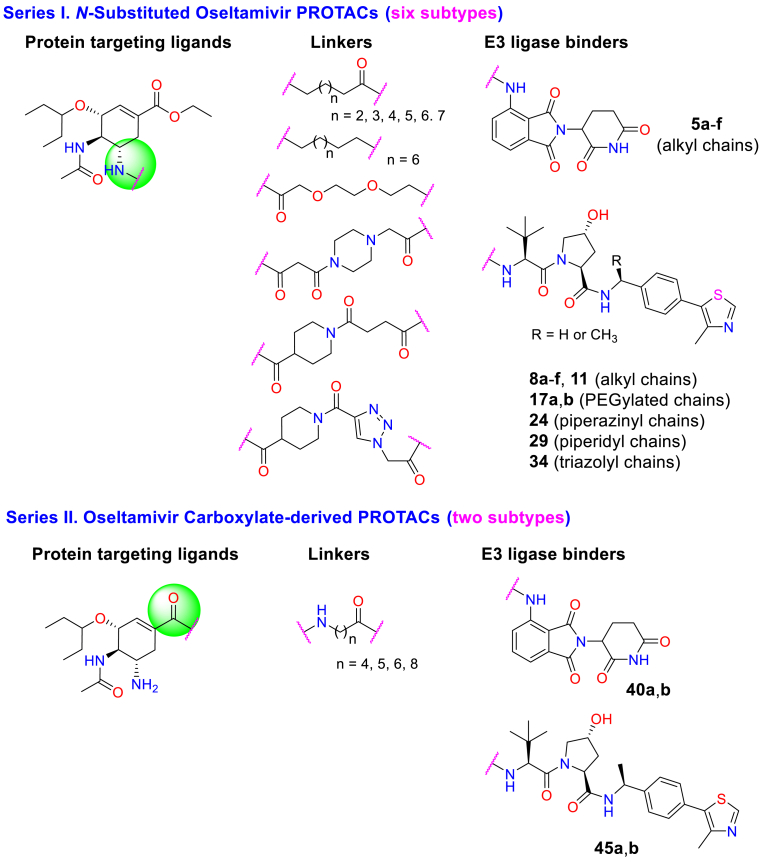
Inspired by these works and considering the continuing detriment of influenza epidemics, we report herein the design and synthesis of oseltamivir-based PROTACs aimed at combating influenza diseases. For the structural modification of oseltamivir, intense research has always focused on the amino- or carboxylate-moiety by targeting an additional 150-cavity or 430-cavity, respectively (Ju et al., 2020; Zhang et al., 2018a), which could improve the antiviral activity and overcome drug resistance. Hence, we designed and synthesized a variety of oseltamivir-based PROTACs by exploiting the widely used E3 ligase ligands VHL or CRBN via different linkers, including flexible or rigid linkages such as alkyl, PEGylated, piperidyl, piperazinyl and triazolyl chains (Fig. 2). The systematic structure-activity relationship (SAR) study indicated that several compounds showed high potency against H1N1 proliferation in vitro. We found that the best compound, 8e, exhibited good antiviral activity toward both the wild-type H1N1 virus and an oseltamivir-resistant strain (H1N1, H274Y). Mechanistic studies illustrated that Compound 8e degraded the NA protein in a dose-dependent manner and relied on the ubiquitin–proteasome pathway.
Fig. 2.
Design strategy ofoseltamivir-basedPROTACs with diverse linkers through use of different E3 ligase ligands.
2. Results
2.1. SAR study on anti-H1N1 activity of synthesized oseltamivir-based PROTACs
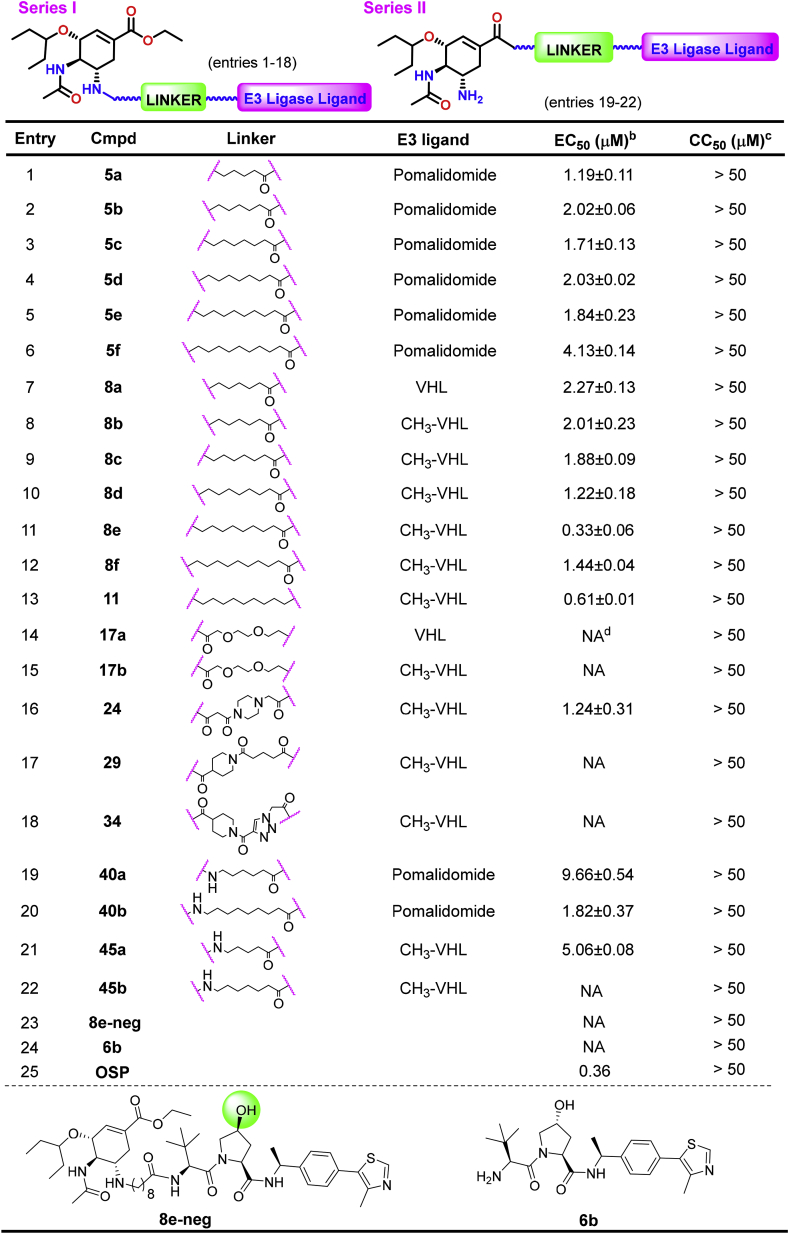
Because the structural modifications of oseltamivir have been mainly focused on the amino or carboxylate moieties, we designed and synthesized two series (eight subtypes) of PROTACs with diversified linkers by connecting E3 ligase ligands on these two binding sites of oseltamivir (Fig. 2, see Schemes S1–8 for the synthetic details). Then, the anti-H1N1 activity of these synthesized compounds was evaluated by plaque formation assays in MDCK cells, in which oseltamivir phosphate (OSP) was used as the positive control (Table 1). In general, the SAR study of this type of PROTAC revealed that most compounds showed good anti-H1N1 activity, in which N-substituted oseltamivir PROTACs (Series I) were usually more potent than carboxylate-derived PROTACs (Series II). Moreover, the type of linkers and the selectivity of the E3 ligands also impacted the anti-H1N1 efficacy of these PROTACs. First, for the N-substituted oseltamivir PROTACs with alkyl chains of varying lengths, when pomalidomide was selected as the E3 ligand, Compounds 5a-f (Scheme S1A, entries 1–6) exhibited good antiviral activity with low micromolar values. We found that the VHL ligand-derived PROTACs 8a-f (Scheme S1B, entries 7–12) showed comparable activity to those with a pomalidomide ligand. In particular, Compound 8e, with a nine-carbon alkyl chain, exerted the best antiviral activity (EC50 = 0.33 μM), which was better than the reference drug oseltamivir phosphate (OSP, Table 1, entries 11 vs. 25). Next, we synthesized Compound 11, a close analog of 8e with an aklyl amine linker at the binding site of CH3-VHL, which showed a slightly lower antiviral efficacy of 0.61 μM compared with the latter (entries 11 vs. 13). In addition, diverse structural modifications were conducted on linkers (Schemes S3–6, Table 1), including the PEGylated chain (17a,b), piperazine (24), piperidine (29) or triazole moiety (34), to investigate the impact of different linkers on antiviral activity. However, except for 24 (entry 16), most PROTACs were essentially inactive against H1N1 proliferation (Table 1, entries 16 vs. 14, 15, 17,18). To explore the mechanism of action of the antiviral activity of the best compound, 8e, we synthesized the negative control Compound 8e-neg (Scheme S8, Table 1), which could not recruit the E3 ligase due to the configuration inversion of the hydroxyl group in the pyrrolidinyl moiety. Subsequently, we evaluated the anti-H1N1 activity of Compounds 8e-neg and CH3-VHL ligand 6b; as expected, these two compounds exhibited no activity toward virus proliferation.
Table 1.
Anti-H1N1 activity of N-substituted oseltamivir PROTACsa.
The anti-H1N1 activity of all synthesized PROTACs was evaluated by plaque formation assays in MDCK cells. Oseltamivir phosphate (OSP) was used as the positive control. All data were acquired from at least three independent experiments. bEC50: effective concentration protecting 50% of cells from virus infection. cCC50: minimum concentration inducing 50% death of normal cells. dNA: no activity (EC50 > 50 μM).
In addition, structural modification of the oseltamivir scaffold has also focused on the carboxylate moiety, although it always displays inferior antiviral activity compared with amino-derived analogs (Ju et al., 2020; Zhang et al., 2018a). For comparison, oseltamivir carboxylate-derived PROTACs (40a, b and 45a, b, entries 19–22) were also designed and synthesized. However, these carboxylate-based PROTACs showed weaker antiviral activity than the amino-based PROTACs (Table 1, 40a vs. 5c, 45b vs. 8d). It is worth noting that all synthesized compounds exhibited no obvious cytotoxicity to normal cells, with a CC50 > 50 μM (Table 1).
2.2. Degradation activity of synthesized oseltamivir-based PROTACs
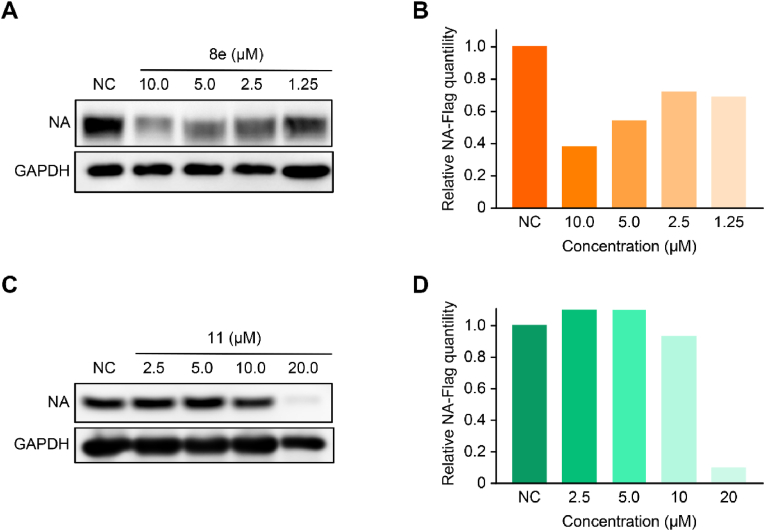
The influenza NA protein degradation activity of the synthesized compounds was further evaluated. The full-length fragment of NA was amplified from pPolI-WSN-NA and inserted into Flag-tagged vectors. 293T cells were transfected with the plasmids, and compounds were added after 8 h. Next, after incubation for 24 h, western blotting was utilized to evaluate the NA protein level. For the N-substituted oseltamivir PROTACs, Compounds 8e, 11 and 17a,b with alkyl or PEGylated chains at fixed lengths induced the degradation of the influenza NA protein at 20 μM, of which Compounds 8e and 11 could degrade NA with better efficacy in a dose-dependent manner (Fig. 3 and S1). As shown in Fig. S1, we observed that oseltamivir carboxylate-based PROTACs (40a,b and 45a,b) at 10 μM showed no degradation activity. Alternately, we speculated that the PROTACs cannot lead to the degradation of the NA protein, possibly due to the difference of linker subtypes or length. In addition, OSP or CH3-VHL-ligand (6b) exhibited no degradation activity in this experiment.
Fig. 3.
Degradation activity of NA protein in the presence of PROTACs 8e and 11 at different concentrations. (A, C) The NA degradation level was evaluated by western blot. (B, D) Western blot bands were quantified by ImageJ. The data were obtained from at least two independent assays.
2.3. Compound 8e efficiently inhibited H1N1 proliferation in vitro
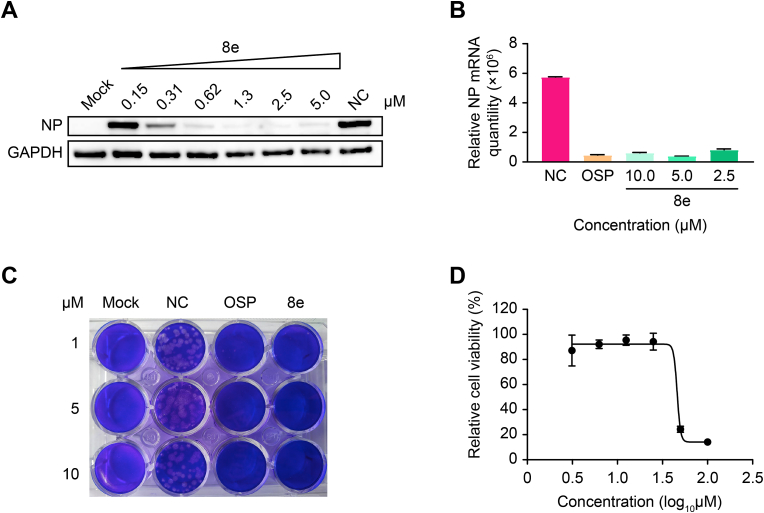
Due to the good NA degradation activity and antiviral activity of Compound 8e, we further verified the in vitro antiviral activity via western blot, plaque formation and qRT–PCR assays. As shown in Fig. 4A, Compound 8e significantly inhibited the expression level of viral nucleoprotein (NP) in a dose-dependent manner and completely suppressed the expression of NP at 1.3 μM. Even when the concentration of 8e was reduced to 0.31 μM, the virus inhibition rate was still greater than 50%. Alternatively, the qRT–PCR data demonstrated that Compound 8e could significantly reduce the viral NP mRNA level in the supernatant, with OSP used as a positive control (Fig. 4B). Additionally, we observed that Compound 8e exerted comparable anti-H1N1 activity to that of OSP via plaque assays (Fig. 4C). Additionally, this compound showed no obvious cytotoxicity in MDCK cells, as depicted in Fig. 4D.
Fig. 4.
In vitroefficacy against H1N1 replication of Compound 8e; OSP was used as the positive control. (A) Western blot assay, (B) qRT–PCR assay and (C) plaque formation assay were used to evaluate the antiviral activity of Compound 8e. The concentration of OSP in the qRT–PCR assay was 5 μM. Mock, blank control; NC, negative control (treated with DMSO). (D) The cytotoxicity of Compound 8e was determined with the CCK-8 reagent.
2.4. Mechanism of action studies of compound 8e
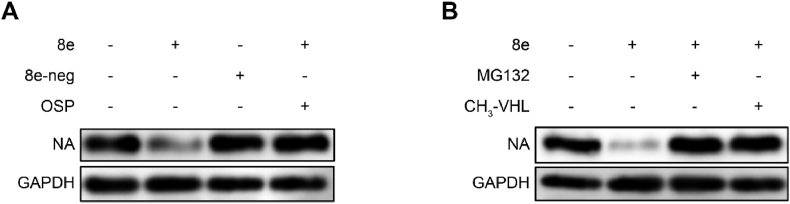
As mentioned above, Compound 8e could degrade the influenza NA protein and inhibit virus proliferation. Hence, we conducted mechanistic studies to further investigate the degradation mechanism of Compound 8e. First, we observed that 8e-neg exhibited no degradation activity toward NA protein because it lost the binding affinity of E3 ligase (Fig. 5B). Moreover, 8e-neg displayed no antiviral activity, which indicated that the antiviral activity of 8e relied on its degradation activity. When the NA inhibitor OSP was added, it disrupted the degradation efficacy of 8e (Fig. 5A), which indicated competitive binding with NA of both compounds. Furthermore, to verify whether this compound induced target protein degradation via the ubiquitin-proteasomal pathway, the effect of 8e was assayed by introducing the E3 ligand CH3-VHL and proteasome inhibitor MG132 (Fig. 5B). In 293T cells, after treatment with 5 μM MG132 (proteasome inhibitor) or CH3-VHL (VHL ligase ligand), the degradation of NA protein was rescued. These results proved that Compound 8e was an NA protein-targeted PROTAC and degraded NA through the ubiquitin−proteasome system.
Fig. 5.
Compound 8e induced NA degradation via the ubiquitin−proteasome system. (A) NA degradation activity of Compound 8e was determined after treatment with 8e-neg (10 μM) or OSP (10 μM). (B) NA degradation activity of Compound 8e (10 μM) after treatment with MG132 (10 μM) or CH3-VHL (10 μM).
2.5. Antiviral activity toward an oseltamivir-resistant strain (H1N1, H274Y)
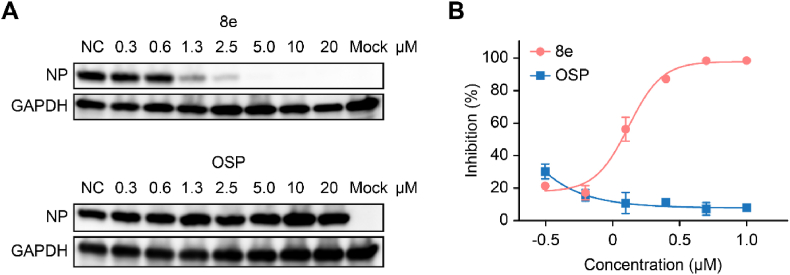
Due to drug resistance of the frequent use of oseltamivir and the ability of PROTACs to handle drug resistance, we evaluated the antiviral activity of Compound 8e against an oseltamivir-resistant strain (H1N1, H274Y). As shown in Fig. 6, Compound 8e significantly reduced the viral NP protein (A) and mRNA (B) levels of the resistant strain in a dose-dependent manner, while OSP displayed no antiviral activity in this assay.
Fig. 6.
Antiviral activity of Compound 8e against anoseltamivir-resistantstrain (H1N1, H274Y). Western blot (A) and qRT–PCR (B) were used to evaluate the NP protein or mRNA level, respectively, after treatment with Compound 8e or OSP.
2.6. Molecular docking
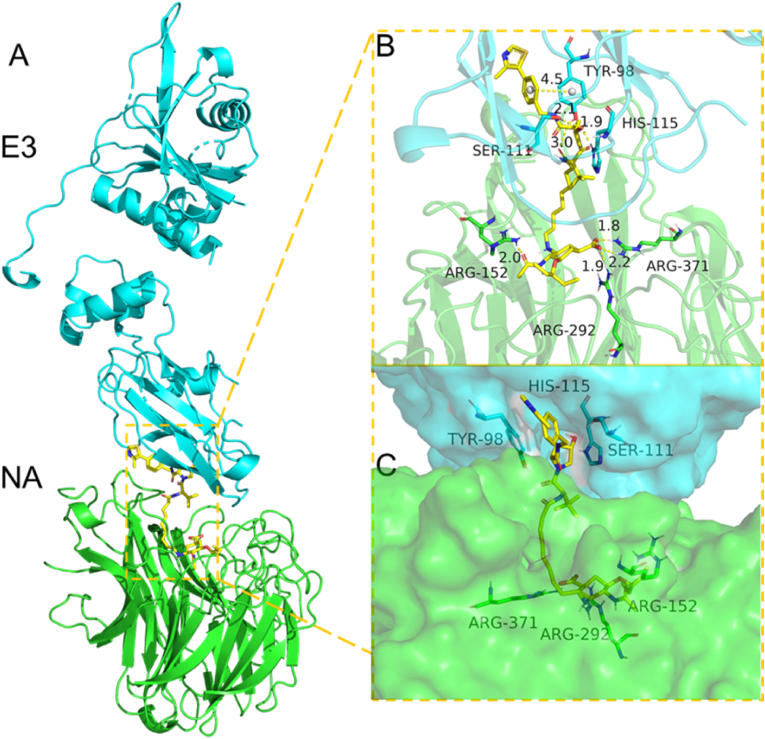
To further understand the interaction mode of PROTAC 8e with influenza NA protein and VHL ligase, we used molecular docking to mimic the formation of the NA-8e-VHL ternary complex. The docking experiment was performed by utilizing MOE2019.01 software, and the result was processed with PyMOL. The binding mode of this ternary complex indicated an extremely low energy of −15.69 kcal/mol (Fig. 7, Table S1). As observed, Compound 8e was located in the middle region between the NA-VHL protein–protein interaction surface. In addition, both ends of Compound 8e showed good hydrogen-bonding and hydrophobic interactions with both the active sites of NA and VHL proteins, which could effectively drive the interaction and binding affinity of the two proteins.
Fig. 7.
Predicted binding mode of the NA-8e-VHL ternary complex (NA protein PDB: 2HU0, VHL protein PDB: 5NW2). (A) The backbone of the protein was rendered in a tube and colored green (NA protein) and blue (E3 protein). (B, C) The binding mode of 8e with E3 and NA. Hydrogen bond distance and π-stacking distance are represented in yellow.
2.6.1. Pharmacokinetic (PK) studies of 8e in rat
Considering the good in vitro inhibitory activity of influenza virus proliferation and NA degradation activity of compound 8e, we further evaluated the PK properties of this compound, wherein the key PK parameters are summarized in Table 2. The compound 8e was administrated via single intravenous (iv) injection and achieved a maximum plasma concentration (Cmax) of 31.2 ng/mL at 4.8 min; at the same time, it possessed a half-life of 1.6 h and high clearance (1575.3 mL/(min·kg)). These modest PK parameters make it possible for further optimal modification of the lead compounds to improve bioavailability.
Table 2.
In vivo PK parameters of compound 8e in rat.
| Cmpda | Routeb | Speciesc | Cmax (ng/mL)d | T1/2 (h) | AUC0−t (h·ng/mL) | Vss (L/kg) | Cl (mL/(min·kg)) |
|---|---|---|---|---|---|---|---|
| 8e | iv | rat | 31.2 | 1.6 | 27.9 | 178.5 | 1575.3 |
The compound 8e was formulated as solution with 5% DMSO and 10% solutol in Saline.
Dosage: 3 mg/kg via single intravenous injection. Plasma samples were measured for drug exposure by LC-MS/MS.
Sprague-Dawley rat was used (n = 3).
The maximum drug concentration (Cmax) was observed at t = 4.8 min, the first sampling time point after iv administration.
3. Discussion
Annual influenza epidemics around the world have led to serious burdens on people's health. To date, FDA-approved anti-influenza drugs show an insufficient efficacy for the treatment of influenza infections due to their severe side effects and the development of drug resistance. In recent years, PROTAC technology has been widely used for drug discovery, including undruggable targets, which suggests the broad application prospects of this technique. Additionally, degraders of the HCV protease NS3/4A based on the PROTAC strategy with good antiviral activities against both wild-type and mutant strains have been identified. Thus, in this study, we designed and synthesized a broad panel of PROTACs based on an oseltamivir scaffold to exhibit potent antiviral activity against wild-type and oseltamivir-resistant strains and provide an alternative strategy for the discovery of anti-influenza agents.
We initially evaluated the anti-H1N1 activity of these synthesized oseltamivir PROTACs, and the results showed that most compounds inhibited virus proliferation at low-to sub-micromolar values, of which Compounds 8e and 11 were identified as having the better antiviral activity (EC50 = 0.33 or 0.61 μM). Based on the ability of PROTACS to induce the degradation of target proteins, we performed western blot assay to assess the degradation activity of these PROTACs. The results illustrated that Compounds 8e, 11 and 17a,b could degrade the NA protein significantly at 10 or 20 μg/mL. To confirm that the inhibitory activity of 8e was mainly dependent on its degradation activity, we synthesized Compound 8e-neg, which could not recruit the E3 ligase. Both the results of the anti-H1N1 activity assay and the degradation activity assays suggested that Compound 8e-neg showed no inhibitory activity or degradation activity, indicating that Compound 8e exhibited good anti-influenza activity by degrading the NA protein.
Since the formation of a ternary complex of PROTACs with the target protein and E3 ligase was identified to play a key role in degradation activity (Luh et al., 2020), we evaluated the degradation efficacy of Compound 8e by adding the NA inhibitor (OSP) and E3 ligase binder (CH3-VHL, 6b). As observed, both competitive inhibitors could disrupt the formation of the ternary complex, thus leading to loss of the degradation efficacy of the target protein NA. Furthermore, to confirm that the degradation activity of Compound 8e was dependent on the ubiquitin–proteasome system, we used a proteasome inhibitor (MG132). The results showed that the degradation efficacy was completely rescued. As stated above, Compound 8e induced NA degradation via the ubiquitin–proteasome system.
As reported, the PROTAC strategy can be utilized to tackle the problem of drug resistance. Herein, we tested the antiviral efficacy of Compound 8e toward an oseltamivir-resistant strain (H1N1, H274Y). The western blot results demonstrated that this compound could significantly inhibit virus proliferation at low micromolar values, while oseltamivir showed no antiviral activity in this assay.
Lastly, we evaluated the PK profiles of compound 8e in rat to assess its druggability. Preliminary PK studies indicated that compound 8e showed modest PK properties, while it's worthwhile to be further optimized (NA binder, linker and E3 ligase ligand optimizations) to identify a better orally bioavailable NA degrader to combat influenza infections.
In conclusion, we report herein the first discovery of influenza NA protein-targeting PROTAC degraders based on an oseltamivir scaffold to provide an alternative for potential treatments of influenza infections. However, one issue is that only a few PROTACs displayed moderate degradation activity toward the target protein, and the correlation between inhibitory activity and degradation activity remains unclear and needs to be further elucidated. We believe that this PROTAC technology will expand the horizons of discovery for novel antiviral agents for existing or newly emerging virus epidemics.
4. Materials and methods
4.1. Plaque formation assays
MDCK cells were seeded into 12-well plates and infected with 70 PFU H1N1 virus per well without fetal bovine serum, followed by incubation for 1 h at 37 °C. Then, the medium on the cells was replaced with 2 × MEM medium containing test compounds at the indicated concentrations in 0.5% agarose. After incubation at 37 °C for 72 h, the cells were fixed with 3% formaldehyde and stained with 1% crystal violet.
4.2. Western blot
Cells were lysed in RIPA reagent, and the cellular extracts were mixed with SDS gel loading buffer and then resolved by SDS–PAGE. The membranes were blocked in skim milk and incubated with the indicated primary antibodies. After incubation with the secondary antibodies, the membranes were soaked with ECL reagents (GE) for visualization.
4.2.1. qRT–PCR
Virion RNA in the supernatant was extracted with the Viral Nucleic Acid Purification Kit. Viral RNA was reverse transcribed into cDNA. qRT–PCR was performed with qPCR SYBR Green Master Mix using QuantStudio 6.
4.3. NA degradation assays
The full-length fragment of NA was amplified from pPolI-WSN-NA and inserted into Flag-tagged vectors. 293 T cells were transfected with the plasmids, and Compound 8e was added after 8 h. Next, Western blot was utilized to assess the NA-Flag protein level. For competitive inhibition of the proteasome, 5 μM MG132 or CH3-VHL or 10 μM OSP or CH3-VHL was added at the same time as the PROTAC after transfection.
4.4. Molecular docking
PROTAC 8e was processed with Chemdraw 3D to minimize the energy and then saved in pdb format. The PDB data for NA protein and VHL ligase were downloaded from the PDB archive and then pretreated with the protein preparation unit of MOE2019.01, which includes removal of H2O, supplementation of missing components and minimization of the energy. The docking experiment was conducted by MOE2019.01, and the pretreated compound was selected to dock with the two proteins 50 times. Then, the docking result and binding mode were determined with PyMOL 2.1 software.
4.5. Pharmacokinetic studies
In PK studies, compound 8e was administrated via intravenous injection (iv, 3 mg/kg), nine SD (Sprague-Dawley) rats were divided equally into 3 groups (n = 3). All animal study procedures followed the Guide for the Care and Use Committee at Wuhan University (permit no. S01320070 A, Wuhan, China). Firstly, testing compound 8e was dissolved in 5% DMSO, 10% solutol and 85% saline. After compound was administrated, plasma samples were acquired at 0.083, 0.25, 0.5, 1, 2, 4, 8 and 24 h respectively. The samples were anticoagulated with heparin sodium and centrifugated at 6800 g for 6 min at 2–8 °C. Finally, the sample was analyzed by LC-MS/MS and the PK parameters were calculated via phoenix WinNonlin 7.0.
Author contributions
C.D., H.-B.Z., K.L., and S.W. conceived the study. Z.X., X.L., X.M., W.Z., Q.C., F.C., X.D., J.L. synthesized the compounds and performed the western blots and cell viability assays. All authors have given approval to the final version of the manuscript.
Conflicts of interest
There are no conflicts to declare.
Acknowledgments
We thank Nanjing University of Chinese Medicine for providing support for the molecular docking experiment. This study was supported by the National Key R&D Program of China (2020YFA0908800), the NSFC (82073690, 81773557, 82173676, 81971976, 32188101), the Major Project of Technology Innovation Program of Hubei Province (2018ACA123), the Fundamental Research Funds for the Central Universities in China (2042021kf1033), and the Seed Funds for International Joint Research Platform of Wuhan University (KYPT-ZD-9).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cellin.2022.100030.
Contributor Information
Ke Lan, Email: klan@whu.edu.cn.
Shuwen Wu, Email: shuwenwu@whu.edu.cn.
Hai-Bing Zhou, Email: zhouhb@whu.edu.cn.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- Anuwongcharoen N., Shoombuatong W., Tantimongcolwat T., Prachayasittikul V., Nantasenamat C. Exploring the chemical space of influenza neuraminidase inhibitors. PeerJ. 2016;4 doi: 10.7717/peerj.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher F.B. Influenza viruses: basic biology and potential drug targets. Infect. Disord. - Drug Targets. 2007;7:282–293. doi: 10.2174/187152607783018745. [DOI] [PubMed] [Google Scholar]
- Collins P.J., Haire L.F., Lin Y.P., Liu J., Russell R.J., Walker P.A., Skehel J.J., Martin S.R., Hay A.J., Gamblin S.J. Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants. Nature. 2008;453:1258–1261. doi: 10.1038/nature06956. [DOI] [PubMed] [Google Scholar]
- Credille C.V., Chen Y., Cohen S.M. Fragment-based identification of influenza endonuclease inhibitors. J. Med. Chem. 2016;59:6444–6454. doi: 10.1021/acs.jmedchem.6b00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromm P.M., Crews C.M. Targeted protein degradation: from chemical biology to drug discovery. Cell Chem. Biol. 2017;24:1181–1190. doi: 10.1016/j.chembiol.2017.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wispelaere M., Du G., Donovan K.A., Zhang T., Eleuteri N.A., Yuan J.C., Gray N.S., et al. Small molecule degraders of the hepatitis C virus protease reduce susceptibility to resistance mutations. Nat. Commun. 2019;10:3468. doi: 10.1038/s41467-019-11429-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desantis J., Mercorelli B., Celegato M., Croci F., Bazzacco A., Baroni M., Goracci L. Indomethacin-based PROTACs as pan-coronavirus antiviral agents. Eur. J. Med. Chem. 2021;226:113814. doi: 10.1016/j.ejmech.2021.113814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Nugent C., Galvani A.P., Krug R.M., Meyers L.A. Modeling mitigation of influenza epidemics by baloxavir. Nat. Commun. 2020;11:2750. doi: 10.1038/s41467-020-16585-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienke U., Schmidtke M., Kirchmair J., Pfarr K., Wutzler P., Dürrwald R., Wolber G., Liedl K.R., Stuppner H., Rollinger J.M. Antiviral potential and molecular insight into neuraminidase inhibiting diarylheptanoids from alpinia katsumadai. J. Med. Chem. 2010;53:778–786. doi: 10.1021/jm901440f. [DOI] [PubMed] [Google Scholar]
- Haniff H.S., Tong Y., Liu X., Chen J.L., Suresh B.M., Andrews R.J., Peterson J.M., O'Leary C.A., Benhamou R.I., Moss W.N., et al. Targeting the SARS-CoV-2 RNA genome with small molecule binders and ribonuclease targeting chimera (RIBOTAC) degraders. ACS Cent. Sci. 2020;6:1713–1721. doi: 10.1021/acscentsci.0c00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley S.E., Das S.R., Bailey A.L., Schmidt L.M., Hickman H.D., Jayaraman A., Viswanathan K., Raman R., Sasisekharan R., Bennink J.R., et al. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science. 2009;326:734–736. doi: 10.1126/science.1178258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu N., Sakaguchi H., Sato C., Ishibashi T., Baba K., Omoto S., Shishido T., Tsuchiya K., Hayden F.G., Uehara T., et al. Baloxavir marboxil in Japanese pediatric patients with influenza: safety and clinical and virologic outcomes. Clin. Infect. Dis. 2020;71:971–981. doi: 10.1093/cid/ciz908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Hu B., Wang M., Xu F., Miao B., Yang C.-Y., Wang M., Liu Z., Hayes D.F., Chinnaswamy K., et al. Discovery of ERD-308 as a highly potent proteolysis targeting chimera (PROTAC) degrader of estrogen receptor (ER) J. Med. Chem. 2019;62:1420–1442. doi: 10.1021/acs.jmedchem.8b01572. [DOI] [PubMed] [Google Scholar]
- Ju H., Xiu S., Ding X., Shang M., Jia R., Huang B., Liu X. Discovery of novel 1,2,3-triazole oseltamivir derivatives as potent influenza neuraminidase inhibitors targeting the 430-cavity. Eur. J. Med. Chem. 2020;187:111940. doi: 10.1016/j.ejmech.2019.111940. [DOI] [PubMed] [Google Scholar]
- Kadam R.U., Wilson I.A. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc. Natl. Acad. Sci. U. S. A. 2017;114:206–214. doi: 10.1073/pnas.1617020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F., Smith G.J.D., Fouchier R.A.M., Peiris M., Kedzierska K., Doherty P.C., Webster R.G., et al. Influenza. Nat. Rev. Dis. 2018;4:3. doi: 10.1038/s41572-018-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A.C., Crews C.M. Induced protein degradation: an emerging drug discovery paradigm. Nat. Rev. Drug Discov. 2017;16:101–114. doi: 10.1038/nrd.2016.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A.C., Toure M., Hellerschmied D., Salami J., Jaime-Figueroa S., Ko E., Hines J., Crews C.M. Modular PROTAC design for the degradation of oncogenic BCR-ABL. Angew. Chem. Int. Ed. 2016;55:807–810. doi: 10.1002/anie.201507634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowen Anice C., Steel J., Mubareka S., Palese P. High temperature (30°C) blocks aerosol but not contact transmission of influenza virus. J. Virol. 2008;82:5650–5652. doi: 10.1128/JVI.00325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luh L.M., Scheib U., Juenemann K., Wortmann L., Brands M., Cromm P.M. Prey for the proteasome: targeted protein degradation-A medicinal chemist's perspective. Angew Chem. Int. Ed. Engl. 2020;59:15448–15466. doi: 10.1002/anie.202004310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari S., Bertagnin C., Pismataro M.C., Donnadio A., Nannetti G., Felicetti T., Manfroni G., et al. Synthesis and characterization of 1,2,4-triazolo[1,5-a]pyrimidine-2-carboxamide-based compounds targeting the PA-PB1 interface of influenza A virus polymerase. Eur. J. Med. Chem. 2021;209:112944. doi: 10.1016/j.ejmech.2020.112944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari S., Nannetti G., Desantis J., Muratore G., Sabatini S., Manfroni G., Mercorelli B., Cecchetti V., Palù G., Cruciani G., et al. A broad anti-influenza hybrid small molecule that potently disrupts the interaction of polymerase acidic protein–basic protein 1 (PA-PB1) subunits. J. Med. Chem. 2015;58:3830–3842. doi: 10.1021/acs.jmedchem.5b00012. [DOI] [PubMed] [Google Scholar]
- McGowan D.C., Balemans W., Embrechts W., Motte M., Keown J.R., Buyck C., Corbera J., Funes M., Moreno L., Cooymans L., et al. Design, synthesis, and biological evaluation of novel indoles targeting the influenza PB2 cap binding region. J. Med. Chem. 2019;62:9680–9690. doi: 10.1021/acs.jmedchem.9b01091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memoli M.J., Hrabal R.J., Hassantoufighi A., Eichelberger M.C., Taubenberger J.K. Rapid selection of oseltamivirand peramivir-resistant pandemic H1N1 virus during therapy in 2 immunocompromised hosts. Clin. Infect. Dis. 2010;50:1252–1255. doi: 10.1086/651605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naro Y., Darrah K., Deiters A. Optical control of small molecule-induced protein degradation. J. Am. Chem. Soc. 2020;142:2193–2197. doi: 10.1021/jacs.9b12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neklesa T.K., Winkler J.D., Crews C.M. Targeted protein degradation by PROTACs. Pharmacol. Ther. 2017;174:138–144. doi: 10.1016/j.pharmthera.2017.02.027. [DOI] [PubMed] [Google Scholar]
- Ottis P., Toure M., Cromm P.M., Ko E., Gustafson J.L., Crews C.M. Assessing different E3 ligases for small molecule induced protein ubiquitination and degradation. ACS Chem. Biol. 2017;12:2570–2578. doi: 10.1021/acschembio.7b00485. [DOI] [PubMed] [Google Scholar]
- Paules C., Subbarao K. Influenza. Lancet. 2017;390:697–708. doi: 10.1016/S0140-6736(17)30129-0. [DOI] [PubMed] [Google Scholar]
- Rey-Carrizo M., Torres E., Ma C., Barniol-Xicota M., Wang J., Wu Y., Naesens L., DeGrado W.F., Lamb R.A., Pinto L.H., et al. 3-Azatetracyclo[5.2.1.15,8.01,5]undecane derivatives: from wild-type inhibitors of the M2 ion channel of influenza A virus to derivatives with potent activity against the V27A mutant. J. Med. Chem. 2013;56:9265–9274. doi: 10.1021/jm401340p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K.M., Kim K.B., Kumagai A., Mercurio F., Crews C.M., Deshaies R.J. Protacs: chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. U. S. A. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K.M., Kim K.B., Verma R., Ransick A., Stein B., Crews C.M., Deshaies R.J. Development of protacs to target cancer-promoting proteins for ubiquitination and degradation. Mol. Cell. Proteomics. 2003;2:1350–1358. doi: 10.1074/mcp.T300009-MCP200. [DOI] [PubMed] [Google Scholar]
- Schneekloth A.R., Pucheault M., Tae H.S., Crews C.M. Targeted intracellular protein degradation induced by a small molecule: en route to chemical proteomics. Bioorg. Med. Chem. Lett. 2008;18:5904–5908. doi: 10.1016/j.bmcl.2008.07.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneekloth J.S., Fonseca F.N., Koldobskiy M., Mandal A., Deshaies R., Sakamoto K., Crews C.M. Chemical genetic control of protein levels: selective in vivo targeted degradation. J. Am. Chem. Soc. 2004;126:3748–3754. doi: 10.1021/ja039025z. [DOI] [PubMed] [Google Scholar]
- Schrauwen E.J.A., de Graaf M., Herfst S., Rimmelzwaan G.F., Osterhaus A.D.M.E., Fouchier R.A.M. Determinants of virulence of influenza A virus. Eur. J. Clin. Microbiol. Infect. Dis. 2014;33:479–490. doi: 10.1007/s10096-013-1984-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen E.J.A., Fouchier R.A.M. Host adaptation and transmission of influenza A viruses in mammals. Emerg. Microb. Infect. 2014;3:1–10. doi: 10.1038/emi.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers S.A., Hagan R.S., Hayden F.G., Fischer W.A., Ii The hidden burden of influenza: a review of the extra-pulmonary complications of influenza infection. Influenza Other Respir. Viruses. 2017;11:372–393. doi: 10.1111/irv.12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Yang Z., Gao H., Yang H., Zhu S., An Z., Wang J., Li Q., Chandarlapaty S., Deng H., et al. Potent and preferential degradation of CDK6 via proteolysis targeting chimera degraders. J. Med. Chem. 2019;62:7575–7582. doi: 10.1021/acs.jmedchem.9b00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashita E., Ejima M., Ogawa R., Fujisaki S., Neumann G., Furuta Y., Kawaoka Y., Tashiro M., Odagiri T. Antiviral susceptibility of influenza viruses isolated from patients pre- and post-administration of favipiravir. Antivir. Res. 2016;132:170–177. doi: 10.1016/j.antiviral.2016.06.007. [DOI] [PubMed] [Google Scholar]
- Wang J., Ma C., Wang J., Jo H., Canturk B., Fiorin G., Pinto L.H., Lamb R.A., Klein M.L., DeGrado W.F. Discovery of novel dual inhibitors of the wild-type and the most prevalent drug-resistant mutant, S31N, of the M2 proton channel from influenza A virus. J. Med. Chem. 2013;56:2804–2812. doi: 10.1021/jm301538e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Gao H., Sun X., Sun Y., Qiu Y., Weng Q., Rao Y. Global PROTAC toolbox for degrading BCR–ABL overcomes drug-resistant mutants and adverse effects. J. Med. Chem. 2020;63:8567–8583. doi: 10.1021/acs.jmedchem.0c00967. [DOI] [PubMed] [Google Scholar]
- Zaraket H., Saito R. Japanese surveillance systems and treatment for influenza. Curr. Treat. Options Infect. Dis. 2016;8:311–328. doi: 10.1007/s40506-016-0085-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Murugan N.A., Tian Y., Bertagnin C., Fang Z., Kang D., Kong X., Jia H., Sun Z., Jia R., et al. Structure-based optimization of N-substituted oseltamivir derivatives as potent anti-influenza A virus agents with significantly improved potency against oseltamivir-resistant N1-H274Y variant. J. Med. Chem. 2018;61:9976–9999. doi: 10.1021/acs.jmedchem.8b01065. [DOI] [PubMed] [Google Scholar]
- Zhang J., Poongavanam V., Kang D., Bertagnin C., Lu H., Kong X., Ju H., Lu X., Gao P., Tian Y., et al. Optimization of N-substituted oseltamivir derivatives as potent inhibitors of group-1 and -2 influenza A neuraminidases, including a drug-resistant variant. J. Med. Chem. 2018;61:6379–6397. doi: 10.1021/acs.jmedchem.8b00929. [DOI] [PubMed] [Google Scholar]
- Zheng M., Huo J., Gu X., Wang Y., Wu C., Zhang Q., Wang W., Liu Y., Liu Y., Zhou X., et al. Rational design and synthesis of novel dual PROTACs for simultaneous degradation of EGFR and PARP. J. Med. Chem. 2021;64:7839–7852. doi: 10.1021/acs.jmedchem.1c00649. [DOI] [PubMed] [Google Scholar]
- Zhou B., Hu J., Xu F., Chen Z., Bai L., Fernandez-Salas E., Lin M., Liu L., Yang C.-Y., Zhao Y., et al. Discovery of a small-molecule degrader of bromodomain and extra-terminal (BET) proteins with picomolar cellular potencies and capable of achieving tumor regression. J. Med. Chem. 2018;61:462–481. doi: 10.1021/acs.jmedchem.6b01816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.