Abstract
Multiple diseases, such as cancer and neural degeneration diseases, are related with the latent infection of DNA viruses. However, it is still difficult to clean up the latent DNA viruses and new anti-viral strategies are critical for disease treatment. Here, we screen a pool of small chemical molecules and identify UNC0379, an inhibitor for histone H4K20 methyltransferase SETD8, as an effective inhibitor for multiple DNA viruses. UNC0379 not only enhances the expression of anti-viral genes in THP-1 cells, but also repress DNA virus replication in multiple cell lines with defects in cGAS pathway. We prove that SETD8 promotes DNA virus replication in a manner dependent on its enzyme activity. Our results further indicated that SETD8 is required for PCNA stability, one factor critical for viral DNA replication. Viral infection stimulates the interaction between SETD8 and PCNA and thus enhances PCNA stability and viral DNA replication. Taken together, our study reveals a new mechanism for regulating viral DNA replication and provides a potential strategy for treatment of diseases related with DNA viruses.
Keywords: SETD8, UNC0379, PCNA, DNA virus, DNA replication
Graphical abstract
Highlights
-
•
UNC0379 represses DNA viruses in human cells and an animal model.
-
•
Histone methylase SETD8 promotes HSV-1 replication dependent on methylation.
-
•
UNC0379 enhances anti-viral gene expression in THP-1 cells.
-
•
SETD8 promotes viral DNA replication via PCNA methylation and stabilization.
1. Introduction
Virus infection is the cause for many diseases and one of the greatest threats for human health all over the world. Compared with RNA viruses, DNA viruses infect a large population of human beings in the world and are much more difficult to be cleaned due to its latent infection (Dong et al., 2021; Li et al., 2017; Whitley and Roizman, 2001). Innate immunity mediated by cGAS pathway has been considered the major pathway in human cells fighting against DNA viruses (Miller et al., 2021; Hu and Shu, 2020; Hertzog and Rehwinkel, 2020; Cai et al., 2014). The double strand DNA of viruses is recognized by cGAS, which synthesizes the second messenger cGAMP to activate the pathway. cGAMP then activates the important adaptor protein MITA (also known as STING), which recruits and promotes phosphorylation of TANK binding kinase 1 (TBK1) and the subsequent phosphorylation of interferon regulatory factor 3 (IRF3) (Zhang et al., 2016; Zhong et al., 2008). Phosphorylated IRF3 enters nucleus to turn on the expression of type I interferons. Then interferons further activate JAK/STAT pathway to express multiple anti-viral genes to eliminate viruses (Yang and Shu, 2020; Hu and Shu, 2020). Interestingly, in many cell types, especially the cultured cancer cell lines, cGAS pathway is defect upon virus infection. It is interesting to investigate whether other anti-viral pathways exist in these cells.
DNA viruses are usually much bigger than RNA viruses and encode more proteins, which can lead to much more complexed cell response. The transcriptomic studies have shown that DNA virus infection often causes the alteration of a great number of host genes, which indicates that the virus-host-cell interaction is very complicated (Gao et al., 2020). After infection, the genome of DNA viruses enters host cell nuclear immediately, and transcribes viral genes and undergoes fast DNA replication (Conn and Schang, 2013; Placek and Berger, 2010; Whitley and Roizman, 2001). The viruses use a combination of viral and host protein machineries to push forward its life cycle. For example, during HSV-1 DNA replication, several viral proteins such as ICP4 and ICP8, along with cellular PCNA and topoisomerases, were enriched on viral genomes (Dembowski and Deluca, 2015; Muylaert et al., 2011).
Recent studies have shown that many epigenetic factors, such as histones, bind to viral genome and play important roles and viral gene transcription and DNA replication (Conn and Schang, 2013; Placek and Berger, 2010). Different forms of histone modifications have been also detected on viral genome (Gao et al., 2020). They appear on viral genome quickly after infection and change dynamically during viral life cycle. It was reported that formation of heterochromatin marks on HSV-1 genome is critical for ordered gene expression (Cliffe et al., 2009). Inhibition of LSD1, a histone H3K4 demethylase, results in heterochromatic suppression of the HSV-1 genome and subsequently affects viral infection (Hill et al., 2014; Liang et al., 2009). An epigenomic study has mapped nucleosome positioning on HSV-1 genome via high throughput sequencing (Oh et al., 2015). The epigenomic study of HSV-1 genome has shown that histone H3K9me3 decreases and H3K27ac increases during viral life cycle, and an inhibitor of H3K27ac, C646, is able to repress HSV-1 in cultured cells (Gao et al., 2020). The landscapes of multiple histone modifications on KHSV genome also have been reported in vitro and in vivo (Sun et al., 2017; Toth et al., 2010, 2013; Hilton et al., 2013). It is interesting to know whether other modified histones also exist on viral genome and regulates viral activities.
In the current study, to identify potential epigenetics-related small molecular chemicals repressing DNA viruses, we performed a screen with a pool with inhibitors of epigenetic factors. Then UNC0379, an inhibitor for H4K20 methyltransferase SETD8, was identified to be capable of repressing multiple DNA viruses. Then our further study reveals that SETD8 promotes DNA virus replication through stabilizing the host factor PCNA and facilitating viral genome replications.
2. Results
2.1. UNC0379 represses DNA viruses in multiple cell lines
To identify novel drugs to repress DNA viruses, we first performed a screen with a library composed of small molecular chemicals of epigenetic inhibitors. The results showed that UNC0379, an inhibitor of histone H4K20 methyltransferase SETD8 (Ma et al., 2014), and EPZ5676, an inhibitor of H3K79 methyltransferase DOT1L (Daigle et al., 2013), significantly repressed HSV-1 replication in U2OS cells (Fig. 1A). In the following experiments, EPZ5676 did not show stable results in virus repression, so, we focused on UNC0379 for further study. We checked the toxicity of UNC0379 to U2OS cells, and the result showed that drug concentration lower than 5 μM was safe to cells (Fig. 1B). Then we examined the effect of UNC0379 at different doses and time points, and found that 5uM UNC0379 repressed HSV-1 replication by 70% in 10 h post-infection (Fig. 1C&D). We then checked the function of UNC0379 in multiple cell lines by checking viral DNA, RNA and plaque assay, and found that it successfully repressed HSV-1 replication in human foreskin fibroblast (HFF), human kidney cell lines HEK293 and 293T, mouse lung fibroblast (MLF), mouse embryonic fibroblast (MEF), human monocyte leukemia cell line THP-1, and human colon cancer cell line HCT116, indicating UNC0379 is able to repress HSV-1 in multiple mammalian cell lines (Fig. 1E–G, Sup. Fig. S1A-D). UNC0379 also repressed the DNA and RNA level of HSV-1, MCMV and HCMV in the indicated cells, indicating that it is able to repress different DNA viruses (Sup. Fig. S1E&F). Then we checked its effect on RNA virus, and found that UNC0379 did not repress the replication of SeV in 293T and THP-1 cells (Sup. Fig. S1G). All the above data demonstrated that UNC0379 is a potent inhibitor for DNA viruses in multiple cell lines.
Fig. 1.
UNC0379 inhibitsHSV-1replication. (A) HFF cells were infected with HSV-1 and treated with inhibitors of histone modification enzymes.12 h later, expression of viral US11 gene were measure by quantitative RT-PCR. (B) U2OS cells were treated with UNC0379 for 12 h at the indicated concentrations and cell viability was measured by MTT assay. (C) U2OS cells were infected with HSV-1 and treated with UNC0379 for 10 h at the indicated concentrations. The amount of viral DNA was measured by quantitative PCR. (D) U2OS cells were infected with HSV-1 for 10 h and treated with 5 μM UNC0379 for the indicated time. The amount of viral DNA was measured by quantitative PCR. (E–F) HFF cells were infected with HSV-1 and treated with 5 μM UNC0379 for 10 h. The amounts of viral DNA (F) and viral genes expression (E) were measure by quantitative PCR. (G) Virus plaque assay with the supernatants of Vero cells infected with HSV-1 (MOI = 0.5) for 10 h with or without UNC0379. The results in all experiments represent the means (±SD) of at least three independent experiments. Statistical analysis was performed using an unpaired Student's t-test. ∗ means p-value ≤ 0.05, ∗∗ for p-value ≤ 0.01, ∗∗∗ for p-value ≤ 0.001.
2.2. SETD8 promotes HSV-1 replication in mammalian cells
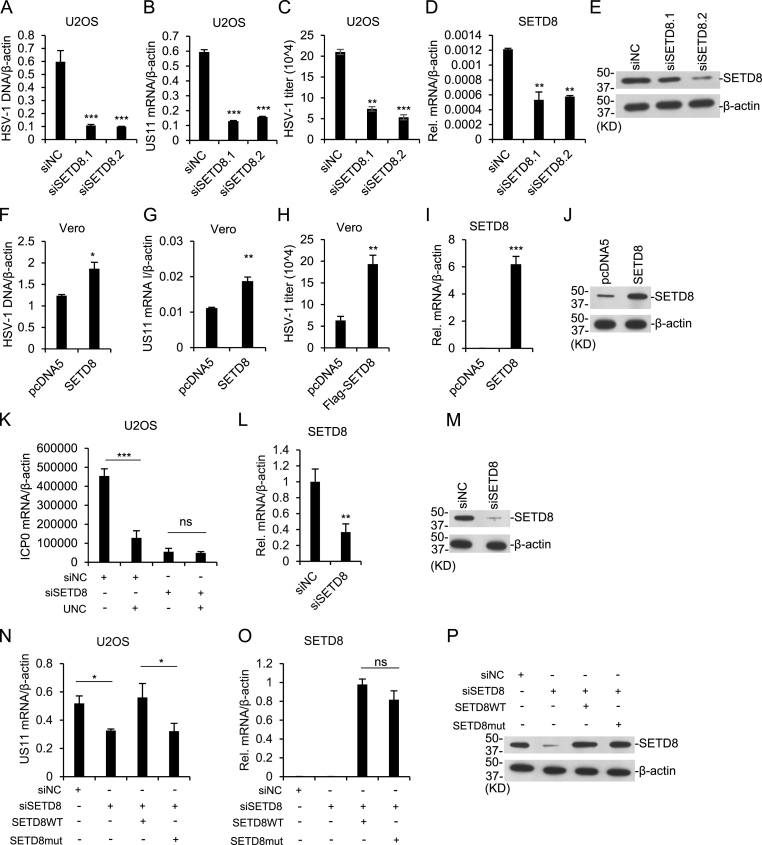
Since UNC0379 is an inhibitor for histone H4K20 methyltransferase SETD8, we further examined whether SETD8 regulates DNA virus replication. We knocked down SETD8 in U2OS cells with two different siRNAs, and found that SETD8 depletion significantly reduced the amount of HSV-1 DNA, mRNA level of viral US11 gene, and virus titer (Fig. 2A–E). To confirm the result, we exogenous expressed SETD8 in Vero cells and found that SETD8 expression increased HSV-1 DNA, US11 expression and virus titer (Fig. 2F–J). When we knocked down SETD8 in U2OS cells and treated with UNC0379, drug treatment could not decrease the expression of viral gene ICP0 (Fig. 2K–M), indicating that UNC0379 could not repress HSV-1 without SETD8. Moreover, we expressed SETD8 in SETD8 knockdown cells, and found that SETD8 expression rescued HSV-1 gene expression decreased by SETD8 siRNA, but a SETD8 catalytic mutant did not (Fig. 2N–P). These results demonstrated that SETD8 promotes HSV-1 replication in mammalian cells, which is dependent on its enzyme catalytic function.
Fig. 2.
Setd8 deficiency inhibitsHSV-1replication. (A–E) U2OS cells were transfected with SETD8 siRNAs and infected with HSV-1 for 10 h. The amounts of viral DNA (A), viral genes expression (B) and SETD8 expression (D&E) were measured by quantitative PCR and immunoblot. Virus plaque assay (C) was performed with the supernatants of U2OS cells infected with HSV-1 (MOI = 0.5) for 10 h with or without SETD8 knockdown. (F–J) Vero cells were transfected with Flag-SETD8 and infected with HSV-1 for 10 h. The viral DNA amount (F), viral genes expression (G), virus plaque assay (H) and SETD8 expression (I&J) were analyzed as above. (K–M) U2OS cells were transfected with SETD8 siRNA, treated with or without 5 μM UNC0379 and infected with HSV-1 for 10 h. Viral genes expression (K) and SETD8 expression (L&M) were measured. (N–P) SETD8 was knocked down in U2OS cells with siRNA and then wild type SETD8 or R265G mutant was expressed. Viral genes expression (N) and SETD8 expression (O&P) were measured after HSV-1 infection for 10 h. The results in all experiments represent the means (±SD) of at least three independent experiments. Statistical analysis was performed using an unpaired Student's t-test. ∗ means p-value ≤ 0.05, ∗∗ for p-value ≤ 0.01, ∗∗∗ for p-value ≤ 0.001.
2.3. UNC0379 enhances the expression of anti-viral genes in THP-1 cells
Since SETD8 is a mono-methyltransferase of histone H4K20 and known to be related with transcription regulation, we performed RNA-Seq analysis to study the potential target genes of UNC0379. We infected THP-1 cells with HSV-1 and then treated with UNC0379 for 12 h, and collected cells for RNA-Seq. We took all the differential expressed genes (DEGs) for analysis, clustered then to four groups and then performed gene function analysis (Fig. 3A, Sup. Fig. S3A-E, Sup. Table 1–3). Genes of cluster 3 was activated after virus infection and further elevated after UNC0379 treatment. The functions of these genes were enriched in defense response to virus and immune response, suggesting cluster 3 genes might play important roles in repressing HSV-1 (Fig. 3B). We selected some genes related with defend response to virus and validated in THP-1 cells. The results confirmed that the expression of IFN-α, IFN-β, IFN-γ, IL-6, ISG20, TLR7, OSA1, TNFRSF11A increased after UNC0379 treatment; meanwhile, US11 expression decreased as expected (Fig. 3C).
Fig. 3.
UNC0379 promotesanti-viralgene expression inTHP-1. (A) THP1 cells were infected with HSV-1 and treated with 5 μM UNC0379 for 10 h. RNA-seq was performed and the heat map shows the expression (FPKM) of all the DEGs compared with negative control. Four clusters of genes were identified. (B) Bar plot shows the function and pathway analysis of cluster 3, gradient color filled with p-value. (C&D) THP1 cells (C) and U2OS cells (D) were infected with HSV-1 and treated with or without 5 μM UNC0379 for 10 h. The immune genes expression was measured by quantitative RT-PCR. The results represent the means (±SD) of at least three independent experiments. Statistical analysis was performed using an unpaired Student's t-test. ∗ means p-value ≤ 0.05, ∗∗ for p-value ≤ 0.01, ∗∗∗ for p-value ≤ 0.001.
2.4. The virus-repressive function of UNC0379 is not dependent on H4K20me1
However, considering UNC0379 is able to repress DNA viruses in multiple cell line, and it is well known that anti-viral pathway is not activated after HSV-1 treatment in some cell lines, such as U2OS and HCT116, it is possible UNC0379 represses DNA viruses through other mechanisms. We examined the expression of anti-viral genes in U2OS cells, and no significant difference was observed. RNA –Seq analysis was also performed with U2OS cells. SETD8 was knocked down in U2OS cells with two different siRNAs and cells were infected with HSV-1. We did not find any pathways or biological processes related with viral defense were enriched in the DEGs (Sup. Fig. S4).
Our above study showed that the enzymatic activity of SETD8 is involved in its function of promoting virus, then we investigated whether H4K20 methylation is involved. ChIP-Seq analysis of H4K20me1 was performed in THP-1 cells, and H4K20me1 enrichment on the DEGs of the above four clusters was analyzed (Sup. Fig. S5A-D). We did not observe that H4K20me1 enrichment on cluster 3 genes decreased after UNC0379 treatment, on the contrary, the average level slightly increased (Sup. Fig. S5C). We also examined H4K20me1 level on some genes of cluster 3 with ChIP-PCR, and did not find any significant difference after UNC0379 treatment (Sup. Fig. S5E). Then we wondered whether H4K20me1 enriched on viral genome was involved in the process and analyzed H4K20me1 level on HSV-1 genome. The result showed that the average viral mRNA level decreased after UNC0379 treatment and H4K20me1 was significantly enriched on viral genome in THP-1 cells (Sup. Fig. S5F). However, UNC0379 treatment did not significantly affect H4K20me1 level on the viral genome (Sup. Fig. S5F).
Based on the above results, we speculate that the function of UNC0379 in repressing DNA viruses is probably not through H4K20me1.
2.5. PCNA is required for HSV-1 DNA replication
To further explore the underlying mechanism, we examined potential involved mechanisms one by one, such as autophagy process, p53 signaling pathway and viral DNA replication. However, UNC0379 is able to repress HSV-1 in ATG5−/− and p53−/− cells, suggesting that both autophagy and p53 are not involved (Sup. Fig. S6A-D). One report showed that SETD8 can methylate PCNA, and in the absence of SETD8, PCNA is degraded by ubiquitination-dependent proteasome pathway (Takawa et al., 2012; Oda et al., 2010). Another study showed that PCNA is potentially involved in the replication of DNA viruses (Sanders et al., 2015; Dembowski and Deluca, 2015). Then we speculated that SETD8 may be required for DNA virus replication through PCNA stabilization. Our results showed that HSV-1 infection did not affect PCNA level in cells significantly; and UNC0379 treatment in Vero cells decreased PCNA protein level in a dose-dependent manner (Fig. 4A). Knockdown of SETD8 by siRNAs in U2OS cells showed a similar effect on PCNA (Fig. 4B). Since PCNA is critical for DNA replication in host cells, knockdown of PCNA either by siRNAs or CRISPR/sgRNA greatly repressed cell growth. So, we examined the role of PCNA by expressing PCNA in cells. PCNA expression successfully increased HSV-1 titer, DNA amount and gene expression in Vero and U2OS cells (Fig. 4C–G, Sup. Fig. S7A-C). These indicate that SETD8 regulates PCNA stability and PCNA is required for HSV-1 replication.
Fig. 4.
PCNA is required forHSV-1replication. (A) Immunoblot assay of PCNA in Vero cells after HSV-1 infection and UNC0379 treatment for 10 h. (B) Immunoblot assay of PCNA and SETD8 in U2OS cells after SETD8 knockdown and HSV-1 infection. (C–G) Vero cells were transfected with Flag-PCNA plasmid for 48 h and infected with HSV-1 for 10 h. The amounts of viral DNA (E), viral genes expression (F), virus titer (G) and PCNA expression (H&I) were measured. The results in all experiments represent the means (±SD) of at least three independent experiments. Statistical analysis was performed using an unpaired Student's t-test. ∗ means p-value ≤ 0.05, ∗∗ for p-value ≤ 0.01, ∗∗∗ for p-value ≤ 0.001.
2.6. SETD8 promotes HSV-1 replication through stabilizing PCNA
To investigate whether SETD8 regulates HSV-1 replication through PCNA, we expressed PCNA in Vero cells, and then infected the cells with HSV-1 and treated with UNC0379. PCNA expression successfully rescued the viral DNA amount and gene expression inhibited by UNC0379 (Fig. 5A–D, Sup. Fig. S7D-F). We performed the similar experiments using siRNAs to knock down SETD8, instead of drug treatment. SETD8 knockdown did not affect PCNA mRNA level; while PCNA expression also rescued HSV-1 DNA amount and gene expression repressed by SETD8 knockdown (Fig. 5E–G, Sup. Fig. S7G). To investigate whether SETD8 promotes HSV-1 replication through PCNA modification, we constructed a PCNA mutant of the methylation site lysine 248 to alanine (Takawa et al., 2012). Expression of PCNA K248A mutant did not rescue HSV-1 repressed by UNC0379 treatment (Fig. 5H–K), indicating PCNA methylation by SETD8 is responsible for its function on HSV-1 replication.
Fig. 5.
PCNA rescuesHSV-1repression by SETD8 deficiency. (A–D) Vero cells were transfected with Flag-PCNA plasmids for 48 h and followed by HSV-1 infection and 5 μM UNC0379 treatment for 10 h. The amounts of viral DNA (A), viral genes expression (B) and PCNA expression (C&D) were assayed by quantitative PCR and immunoblotting. (E–G) U2OS cells were transfected SETD8 siRNA and PCNA plasmid. The amounts of viral DNA (E), viral genes expression (F) and PCNA expression (G) were measured after HSV-1 infection for 10 h. (H–K) Vero cells were transfected with PCNA K248A mutant plasmid for 48 h and followed by 5 μM UNC0379 treatment and HSV-1 infection for 10 h. The amounts of viral DNA (H), viral genes expression (I) and PCNA expression (J&K) were measured. (L) Immunoblot assay of PCNA and SETD8 in U2OS cells after SETD8 overexpression and HSV-1 infection. The number below the blots indicates the relative amount of Flag-Setd8/actin or PCNA/actin. (M) FLAG-SETD8 were expressed in 293T cells. After 48 h, cells were infected with or without HSV-1 for 10 h. Cells were IPed with anti-FLAG, and immunoprecipitants were blotted with anti-FLAG and anti-PCNA antibodies, respectively. The number below the blots indicates the relative amount of PCNA/Flag-Setd8. The results in all experiments represent the means (±SD) of at least three independent experiments. Statistical analysis was performed using an unpaired Student's t-test. ∗ means p-value ≤ 0.05, ∗∗ for p-value ≤ 0.01, ∗∗∗ for p-value ≤ 0.001.
2.7. HSV-1 infection increases PCNA amount
To investigate the effect of virus infection on SETD8 and PCNA, we checked their protein level after HSV-1 infection. As expected, SETD8 expression increased PCNA level; after HSV-1 infection, endogenous PCNA amount further increased in the cells (Fig. 5L). These indicate that HSV-1 infection stabilizes PCNA protein. To study whether interaction of PCNA and SETD8 is regulated by HSV-1 infection, we examined the interaction with or without HSV-1. The result of immunoprecipitation assay show that Flag-SETD8 successfully pulled down endogenous PCNA, and HSV-1 infection increased the amount of PCNA pulled down by SETD8 (Fig. 5M). However, considering the ratio between PCNA and SETD8, the affinity of PCNA-SETD8 interaction did not significantly change (Fig. 5M).
2.8. UNC0379 represses HSV-1 replication in vivo
To study whether UNC0379 is able to repress DNA virus in vivo, we infected the mice with HSV-1 and intraperitoneal injected 2 mg/kg UNC0379. In the preliminary experiments, UNC0379 did not cause any obvious effect at the used dosage. The survival analysis show that drug treatment significantly increased the animal survival rates (Fig. 6A). Drug treatment also significantly repressed HSV-1 titer and genomic DNA amount in the lung and brain tissues (Fig. 6B–C). We then isolated macrophage from mouse bone marrow (BMDM). Treatment with UNC0379 or SETD8 knockdown with siRNAs successfully repressed HSV-1 in BMDM (Fig. 6D–G). We then exogenous expressed PCNA in BMDM, which rescued HSV-1 replication inhibited by UNC0379 (Fig. 6H–J). These indicate that UNC0379 is able to repress HSV-1 in the primary cells and animal model.
Fig. 6.
UNC0379 treatment repressesHSV-1in vivo. (A) Survival (Kaplan-Meier curve) of mice intraperitoneal injected with HSV-1 (1 × 108 PFU per mouse) with (n = 15) or without UNC0379 treatment (2 mg/kg, intraperitoneal injection). Animal survival was monitored for 12 days. (B–C) The viral DNA amounts of lung (B) and viral titers of brain (C) from mice intravenously injected with HSV-1 (2.5 × 106) and UNC0379 (2 mg/kg) for 48 h (n = 5). (D) The primary BMDM cells isolated from the mouse bone were infected with HSV-1 and treated with UNC0379 for 10 h. The amount of viral DNA was measured. (E–G) SETD8 was knocked down in BMDM cells by siRNA and cells were infected with HSV-1 for 10 h. The amounts of viral DNA (E) and SETD8 expression (F&G) were measured. (H–J) The amounts of viral DNA (H) and PCNA expression (I&J) in BMDM cells transfected with PCNA plasmid for 48 h and treated with 5 μM UNC0379 and HSV-1 for 10 h. The results in all experiments represent the means (±SD) of at least three independent experiments. Statistical analysis was performed using an unpaired Student's t-test. ∗ means p-value ≤ 0.05, ∗∗ for p-value ≤ 0.01, ∗∗∗ for p-value ≤ 0.001.
3. Discussion
Viral infection has been one of the most critical threats to human health around the world. DNA viruses infect a large population all through the world and lead to multiple diseases, such as cancer and neural degeneration (Dong et al., 2021; Li et al., 2017; Krstanovic et al., 2021). It is always critical to discover the regulatory mechanisms of viral life cycle and develop new therapies. In our study, we identify an epigenetic enzyme, SETD8, is critical for DNA virus DNA replication. SETD8 is critical for maintain the stability of PCNA, an important host factor for DNA replication for both host cells and viruses. An enzymatic inhibitor for SETD8, UNC0379, is capable of repressing virus replication in vitro and in vivo. It suggests that it might be a useful strategy to repress DNA viruses through inhibiting PCNA and DNA replication.
PCNA is involved in the DNA replication machinery for both mammalian cells and DNA viruses. SETD8 inhibition by UNC0379 not only represses viruses, but also affects host cells. Based on our observation, we did not see any obvious effects on animals at the current dosage, which greatly inhibits virus replication. For the cultured cells, the long-term knockdown or knockout of SETD8 did cause growth defects. So, we speculate that a short-term treatment of UNC0379 is safe, which is perhaps good enough for virus repression. Future studies are still required to clarify whether it is safe to use the approach to treat the related diseases.
Besides its function in regulating DNA replication, we also found that UNC0379 treatment upregulates the expression of anti-viral genes in THP-1 cells. Among them, we identified multiple genes downstream of cGAS/STING anti-viral pathways, such as IFNs and ISGs. Unfortunately, we did not observe the result in the isolated BMDM cells. But, it is still possible that UNC0379 may be able to regulate anti-viral gene expression in certain cell types.
In our study, we find that SETD8 interacts with PCNA in the normal cells, and virus infection increases the total and SETD8-bound PCNA level. It indicates that PCNA stabilization by SETD8 occurs regularly in host cells, and it is probably that DNA viruses take the advantage to facilitate viral replication. However, it is still unknown how viruses stimulate the process, and it is possible that viruses increase PCNA through other approaches and the increased amount of SETD8-bound PCNA is due to the elevated total PCNA level. It will be interesting to further investigate the underlying mechanism.
Taken together, our study reveals a novel mechanism for regulation of viral DNA replication and provides a potential strategy to treat the related diseases.
4. Methods
4.1. Ethics statement and animal housing
Mice were maintained in the special pathogen-free facility of College of Life Sciences at Wuhan University. All the animal operations were following the laboratory animal guidelines of Wuhan University and approved by the Animal Experimentations Ethics Committee of Wuhan University (Protocol NO. 14110B). No patient study was involved and the consent to participate is not applicable.
All the mice were born and maintained under pathogen-free condition at 20–24 °C with a humidity of 40%–70% and a 12/12-h dark/light cycle (lights on at 7:00 a.m., lights off at 7:00 p.m.), with free access of water and food (Animal Center of College of Life Sciences, Wuhan University).
4.2. Cell lines and reagents
Antibodies recognizing H4K20me1 (Abcam, Ab9051), β-actin (ABclonal, AC026), Flag (Sigma, F2555), SETD8(ABclonal, A4316) and PCNA (ABclonal, a12427) were purchased from the indicated merchants. THP-1, U2OS cells and Vero cells were obtained from Cell Bank of Chinese Academy of Science. THP-1 and U2OS were cultured in RPMI-1640 medium (Gibco) and Vero in DMEM (Gibco), both supplemented with 10% FBS (BI).
4.3. Virus propagation
HSV-1 strain KOS was gifted by Dr. Hong-Bing Shu of Wuhan University. 2 × 106 Vero cells were seeded in a 10-cm dish. After 24 h, cells were infected with virus (MOI = 0.5). 48 h later, the supernatant was collected and sodium chloride solution was added to the supernatant to a final concentration of 0.5 M. Then isopyknic PEG was added and the supernatant was incubated at 4 °C overnight. The resulted mixture was centrifuged at 8000 rpm for 1 h and the sediment was collected as virus granule. The virus granule sediment was dissolved in PBS buffer and kept at −80 °C. We have added the above information in the method section.
4.4. Viral infection
Cells were seeded in six-well plates (1 × 106 cells per well). After 24 h, cells were infected with HSV-1 for 10 h (MOI = 0.5). The cells were collected for qPCR or immunoblot assays. For mice infection, 8-week old male C57 wildtype littermates were intraperitoneal injected with HSV-1 (1 × 108 PFU per mouse), with or without UNC0379 (2 mg/kg, n = 15 per treatment). The survival of animals was monitored every day. The brain and lungs from mice (n = 5) were collected for qPCR and virus titer detection at 48 h post-injection.
4.5. Sample preparation for RNA-Seq
Typically, 1 × 107 cells were used for RNA extraction using Ultrapure RNA Kit (CWBIO, CW0581M). Cells were lysed in 1 mL TRIzon and incubated at room temperature for 5 min. Then 200 μL chloroform was added and the cells were shaken drastically. After centrifugation of 12000 rpm at 4 °C for 10 min, the upper layer of supernatant was passed through an adsorption column, and RNA was eluted with 50 μL RNase-free water. The RNA-Seq library was constructed with NEBNext Poly (A) mRNA Magnetic Isolation Module (NEB E7490) and NEBNext Ultra II Non-Directional RNA Second Strand Synthesis Module (NEB E6111). Briefly, poly-A mRNA was purified with poly-T magnetic beads and first and second strand cDNA was prepared. The newly synthesized cDNA was purified with AMPure XP beads (1:1) and eluted in 50 μL nucleotide-free water. The subsequent procedures were the same as ChIP-seq library construction described previously. RNA-seq libraries were sequenced by Illumina Hiseq X Ten platform with pair-end of 150 bp.
4.6. Sample preparation for ChIP-Seq
Around 1 × 107 cells were fixed with 1% formaldehyde and quenched by glycine. The cells were washed three times with PBS and then harvested in ChIP lysis buffer (50 mM Tris–HCl, pH7.6, 1 mM CaCl2, 0.2% Triton X-100). DNA was digested to 150–300 bp by MNase (SIGMA) before extensive centrifugation. Four volume of ChIP dilution buffer (20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS) was added to the supernatant. The resulted lysate was then incubated with protein G beads and antibodies at 4 °C overnight. The beads were washed five times and DNA was eluted by Chip elution buffer (0.1 M NaHCO3, 1% SDS, 20 μg/mL proteinase K). The elution was incubated at 65 °C overnight and DNA was extracted with DNA purification kit (TIANGEN). ChIP-seq libraries were constructed by VATHS Universal DNA Library Prep Kit for Illumina (Vazyme ND604). Briefly, 50 μL purified ChIP DNA (8–10 ng) was end-reparied for dA tailing, followed by adaptor ligation. Each adaptor marked with a barcode of 6 bp which can be recognized after mixing different sample together. Adaptor-ligated ChIP DNA was purified by AMPure XP beads (1:1) and then amplifying by PCR of 11–13 cycles with the primer matching with adaptor universal part. Amplified ChIP DNA was purified again using AMPure XP beads (1:1) in 35 μL EB elution buffer. For multiplexing, libraries with different barcode were mixed together with equal molar quantities by considering appropriate sequencing depth (30–40 million reads per library). Libraries were sequenced by Illumina Hiseq X Ten platform with pair-end reads of 150 bp.
4.7. Viral transcriptomic analysis
The sequenced reads were first quality-controlled using FASTQC software (v0.10.1) and then the adapter sequences were trimmed with Cutadapt software (v1.16). The reads were then mapped to the human hg19 reference genome along with HSV-1 reference genome (GenBank: JN555585.1) using Bowtie2 (v2.1.0) (Langmead and Salzberg, 2012) with provided annotations of human and HSV-1. Because the HSV genome have a large number of repetitive elements which can be mapped to multiple loci in the HSV genome and bowtie2 only report one of the best alignments, Samtools (v1.4.1) was used to screen out the reads that were uniquely mapped to the HSV-1 genome, with the parameter samtools view –b –F4 (Li et al., 2009). Bedtools (v2.25.0) was used to quantify the mapped reads across the HSV-1 genome with the parameter bedtools intersect –wa –c (Quinlan and Hall, 2010). Then the data were normalized by the total number of mapped reads to host genomes. To evaluate the average virus gene expression level, the normalized data were further normalized by the virus copy number of each time point. Then the bdg2bw software was used to generate bigwig format files which were finally uploaded to IGV to visualize the locations of RNA-seq reads across the HSV-1 genome.
4.8. Data analysis of viral epigenome
The data quality of each sample was controlled using the FASTQC tool and then the adapter sequences and low-quality sequences in the reads were trimmed with Cutadapt tool. The resulted reads were then mapped to the HSV-1 reference genome (GenBank: JN555585.1) using the Bowtie2 read mapper with default parameters and only uniquely mapped reads were kept by Samtools for further analysis. Bedtools software was used to calculate histone modifications signals on HSV-1 genome. The histone modifications were normalized by the total mapped data to the host genome and further divided by the copy number at the corresponding time point. Then the bdg2bw software was used to generate bigwig format files and finally the bigwig files were uploaded to IGV to visualize the histone modifications enrichment level across the HSV-1 genome.
4.9. Data analysis of histone modifications on the host genome
The reads were mapped to hg19 genome using the Bowtie2 tool with default parameters and unique reads were further analyzed using Samtools. Then the RPKM values were generated in each 2 kb bin for the entire hg19 genome. Such enrichment was compared between different time points and histone modifications samples for correlation analysis. The DAVID web-tool (v6.8) was used to identify the GO terms of differential expressed genes (fold change >2) (Huang Da et al., 2009).
4.10. Plaque assay
The supernatants of treated Vero cultures were used to infect monolayers of Vero cells. One hour later, the supernatants were removed and the infected Vero cells were washed with pre-warmed PBS twice, followed by incubation for 48 h with DMEM containing 2% methylcellulose. The cells were fixed with 4% paraformaldehyde for 15 min and stained with 1% crystal violet for 30 min before counting the plaques.
4.11. MTT assay
5 × 103 cells/well Vero cells were seeded in 96-well plate and cultured for 12 h. After UNC0379 treatment, 5 μL of MTT (5 μg/mL) were added to every single well in the plate which was incubated for 4 h at 37 °C. Then 100 μL of solubilization buffer (50% DNF and 30% SDS in distilled water) were added to the wells in the plate which was incubated for 4h at 37 °C. After the incubation, the absorbance was measured at 570 nm in a microplate reader.
4.12. Co-immunoprecipitation and immunoblot assay
Cells were collected and lysed for 15 min with 400 μL lysis buffer (20 mM Tris·HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40) containing inhibitors for protease and phosphotases (Biotool). Cell lysates (300 μL) were incubated with IgG or specific antibodies together with protein G agarose for 2–4 h. The immunoprecipitates were washed for three times with 1 mL prelysis buffer and subjected to immunoblot analysis. The rest of lysates (100 μL) were subjected to immunoblot analysis for detecting desired protein expression.
4.13. Reverse transcription and quantitative PCR
Cells were scraped down and collected with centrifugation. Total RNA was extracted with RNA extraction kit (Aidlab) according to the manufacturer's manual. Approximately 1 μg of total RNA was used for reverse transcription with a first strand cDNA synthesis kit (Toyobo). The resulted cDNA was then assayed with quantitative PCR. β-actin was used for normalization. The sequences of primers are in Supplementary Table 4. Assays were repeated at least three times. Data were shown as average values ± SD of at least three representative experiments. P-value was calculated using student's t-test. The primer information is included in Sup. Table 4.
4.14. Preparation of BMDMs
Cells were isolated from the thigh bone marrow of eight weeks old male C57 wildtype mice. We striped the thighbones and washed down the bone marrow cells with PBS injection (1%FBS+1%PS). The cells were then cultured in 10% M-CSF-containing conditional medium from L929 cells for 3–5 days.
4.15. Statistical analysis
For all the experimental studies, the assays were repeated at least three times. At least two biological replicates were performed for NGS studies; and for all other experiments, at least three biological replicates were performed. Data were shown as average values ± SD and p value. Student's t-test was used for comparison between groups.
Data access
The original deep sequencing data were submitted to SRA database, which can be accessed by BioProjectID: PRJNA819114. The other data are available within the article, supplementary Information or available from the authors upon request.
Author contribution
CL, YC and TSB performed most of the experiments; LQY and CJD performed bioinformatics analyses; WM and LLY directed the project and provided funding; WM, CL and LLY wrote the manuscript.
Acknowledgement
We appreciate Dr. Hong-Bing Shu of Wuhan University for sharing reagents and project discussion. This work was supported by Ministry of Science and Technology of China (2016YFA0502100), National Natural Science Foundation of China to Lian-Yun Li (3217050383) and Min Wu (81972647 and 31771503), and the Fundamental Research Funds for the Central Universities.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.cellin.2022.100033.
Contributor Information
Min Wu, Email: wumin@whu.edu.cn.
Lian-Yun Li, Email: lilianyun@whu.edu.cn.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- Cai X., Chiu Y.H., Chen Z.J. The cgas-cgamp-Sting pathway of cytosolic Dna sensing and signaling. Mol. Cell. 2014;54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- Cliffe A.R., Garber D.A., Knipe D.M. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J. Virol. 2009;83:8182–8190. doi: 10.1128/JVI.00712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn K.L., Schang L.M. Chromatin dynamics during lytic infection with herpes simplex virus 1. Viruses. 2013;5:1758–1786. doi: 10.3390/v5071758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle S.R., Olhava E.J., Therkelsen C.A., Basavapathruni A., Jin L., Boriack-Sjodin P.A., Allain C.J., Klaus C.R., Raimondi A., Scott M.P., Waters N.J., Chesworth R., Moyer M.P., Copeland R.A., Richon V.M., Pollock R.M. Potent inhibition of Dot1l as treatment of Mll-fusion leukemia. Blood. 2013;122:1017–1025. doi: 10.1182/blood-2013-04-497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembowski J.A., Deluca N.A. Selective recruitment of nuclear factors to productively replicating herpes simplex virus genomes. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Dong J., Xiang M., Lei P., Li Z., Zhang F., Sun X., Niu D., Bai L., Lan K. Ndrg1 facilitates lytic replication of Kaposi's sarcoma-associated herpesvirus by maintaining the stability of the Kshv helicase. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C., Chen L., Tang S.B., Long Q.Y., He J.L., Zhang N.A., Shu H.B., Chen Z.X., Wu M., Li L.Y. The epigenetic landscapes of histone modifications on Hsv-1 genome in human Thp-1 cells. Antivir. Res. 2020;176 doi: 10.1016/j.antiviral.2020.104730. [DOI] [PubMed] [Google Scholar]
- Hertzog J., Rehwinkel J. Regulation and inhibition of the Dna sensor cgas. EMBO Rep. 2020;21 doi: 10.15252/embr.202051345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J.M., Quenelle D.C., Cardin R.D., Vogel J.L., Clement C., Bravo F.J., Foster T.P., Bosch-Marce M., Raja P., Lee J.S., Bernstein D.I., Krause P.R., Knipe D.M., Kristie T.M. Inhibition of Lsd1 reduces herpesvirus infection, shedding, and recurrence by promoting epigenetic suppression of viral genomes. Sci. Transl. Med. 2014;6 doi: 10.1126/scitranslmed.3010643. 265ra169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton I.B., Simon J.M., Lieb J.D., Davis I.J., Damania B., Dittmer D.P. The open chromatin landscape of Kaposi's sarcoma-associated herpesvirus. J. Virol. 2013;87:11831–11842. doi: 10.1128/JVI.01685-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M.M., Shu H.B. Innate immune response to Cytoplasmic Dna: mechanisms and diseases. Annu. Rev. Immunol. 2020;38:79–98. doi: 10.1146/annurev-immunol-070119-115052. [DOI] [PubMed] [Google Scholar]
- Huang Da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using David bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Krstanovic F., Britt W.J., Jonjic S., Brizic I. Cytomegalovirus infection and inflammation in developing brain. Viruses. 2021;13 doi: 10.3390/v13061078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing S. The sequence alignment/map format and samtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Bai L., Dong J., Sun R., Lan K. Kaposi's sarcoma-associated herpesvirus: epidemiology and molecular biology. Adv. Exp. Med. Biol. 2017;1018:91–127. doi: 10.1007/978-981-10-5765-6_7. [DOI] [PubMed] [Google Scholar]
- Liang Y., Vogel J.L., Narayanan A., Peng H., Kristie T.M. Inhibition of the histone demethylase Lsd1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat. Med. 2009;15:1312–1317. doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A., Yu W., Li F., Bleich R.M., Herold J.M., Butler K.V., Norris J.L., Korboukh V., Tripathy A., Janzen W.P., Arrowsmith C.H., Frye S.V., Vedadi M., Brown P.J., Jin J. Discovery of a selective, substrate-competitive inhibitor of the lysine methyltransferase Setd8. J. Med. Chem. 2014;57:6822–6833. doi: 10.1021/jm500871s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K.N., Victorelli S.G., Salmonowicz H., Dasgupta N., Liu T., Passos J.F., Adams P.D. Cytoplasmic Dna: sources, sensing, and role in aging and disease. Cell. 2021;184:5506–5526. doi: 10.1016/j.cell.2021.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muylaert I., Tang K.W., Elias P. Replication and recombination of herpes simplex virus Dna. J. Biol. Chem. 2011;286:15619–15624. doi: 10.1074/jbc.R111.233981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H., Hubner M.R., Beck D.B., Vermeulen M., Hurwitz J., Spector D.L., Reinberg D. Regulation of the histone H4 monomethylase Pr-Set7 by Crl4(Cdt2)-mediated Pcna-dependent degradation during Dna damage. Mol. Cell. 2010;40:364–376. doi: 10.1016/j.molcel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J., Sanders I.F., Chen E.Z., Li H., Tobias J.W., Isett R.B., Penubarthi S., Sun H., Baldwin D.A., Fraser N.W. Genome wide nucleosome mapping for Hsv-1 shows nucleosomes are deposited at preferred positions during lytic infection. PLoS One. 2015;10 doi: 10.1371/journal.pone.0117471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placek B.J., Berger S.L. Chromatin dynamics during herpes simplex virus-1 lytic infection. Biochim. Biophys. Acta. 2010;1799:223–227. doi: 10.1016/j.bbagrm.2010.01.012. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. Bedtools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders I., Boyer M., Fraser N.W. Early nucleosome deposition on, and replication of, Hsv Dna requires cell factor Pcna. J. Neurovirol. 2015;21:358–369. doi: 10.1007/s13365-015-0321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R., Tan X., Wang X., Wang X., Yang L., Robertson E.S., Lan K. Epigenetic landscape of Kaposi's sarcoma-associated herpesvirus genome in classic Kaposi's sarcoma tissues. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takawa M., Cho H.S., Hayami S., Toyokawa G., Kogure M., Yamane Y., Iwai Y., Maejima K., Ueda K., Masuda A., Dohmae N., Field H.I., Tsunoda T., Kobayashi T., Akasu T., Sugiyama M., Ohnuma S., Atomi Y., Ponder B.A., Nakamura Y., Hamamoto R. Histone lysine methyltransferase Setd8 promotes carcinogenesis by deregulating Pcna expression. Cancer Res. 2012;72:3217–3227. doi: 10.1158/0008-5472.CAN-11-3701. [DOI] [PubMed] [Google Scholar]
- Toth Z., Brulois K., Jung J.U. The chromatin landscape of Kaposi's sarcoma-associated herpesvirus. Viruses. 2013;5:1346–1373. doi: 10.3390/v5051346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth Z., Maglinte D.T., Lee S.H., Lee H.R., Wong L.Y., Brulois K.F., Lee S., Buckley J.D., Laird P.W., Marquez V.E., Jung J.U. Epigenetic analysis of Kshv latent and lytic genomes. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley R.J., Roizman B. Herpes simplex virus infections. Lancet. 2001;357:1513–1518. doi: 10.1016/S0140-6736(00)04638-9. [DOI] [PubMed] [Google Scholar]
- Yang Q., Shu H.B. Deciphering the pathways to antiviral innate immunity and inflammation. Adv. Immunol. 2020;145:1–36. doi: 10.1016/bs.ai.2019.11.001. [DOI] [PubMed] [Google Scholar]
- Zhang M., Zhang M.X., Zhang Q., Zhu G.F., Yuan L., Zhang D.E., Zhu Q., Yao J., Shu H.B., Zhong B. Usp18 recruits Usp20 to promote innate antiviral response through deubiquitinating Sting/Mita. Cell Res. 2016;26:1302–1319. doi: 10.1038/cr.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B., Yang Y., Li S., Wang Y.Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P., Shu H.B. The adaptor protein Mita links virus-sensing receptors to Irf3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.