Abstract
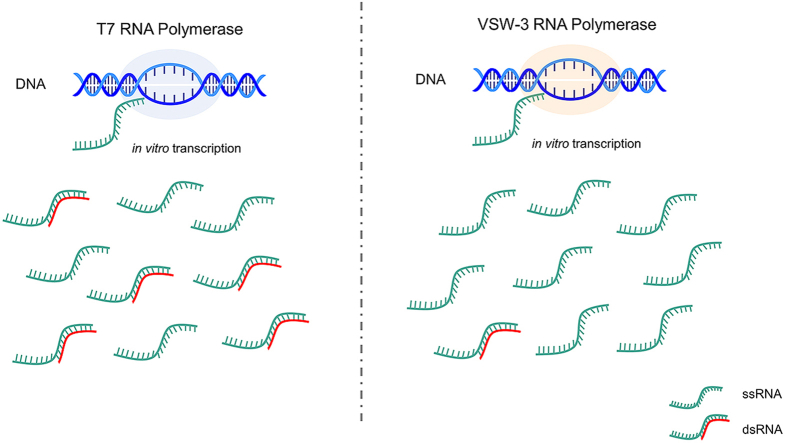
In vitro preparation of mRNA is a key step for mRNA therapeutics. The widely used T7 RNA polymerase (RNAP) was shown to have many by-products during in vitro transcription (IVT) process, among which double-stranded RNA (dsRNA) is the major by-product to activate the intracellular immune response. Here, we describe the use of a new VSW-3 RNAP that reduced dsRNA production during IVT and the resulting mRNA exhibited low inflammatory stimulation in cells. Compared to T7 RNAP transcripts, these mRNA exhibited superior protein expression levels, with an average of 14-fold increase in Hela cells and 5-fold increase in mice. In addition, we found that VSW-3 RNAP did not require modified nucleotides to improve protein production of IVT products. Our data suggest that VSW-3 RNAP could be a useful tool for mRNA therapeutics.
Graphical abstract
Highlights
-
•
Compared to T7 RNAP, VSW-3 RNAP reduces dsRNA byproducts.
-
•
mRNA by VSW-3 RNAP exhibits higher levels of protein expression than that by T7 RNAP.
-
•
VSW-3 RNAP does not need modified nucleotides for mRNA production.
1. Introduction
mRNA therapeutics has been developed for decades with a number of ongoing clinical trials (Beck et al., 2021). Recently, mRNA vaccines were developed to mitigate SARS-CoV-2 pandemic (Corbett et al., 2020; Jackson et al., 2020; Laczko et al., 2020). The design and production of clinical-scale vaccines in a short period of time has proved utility of mRNA therapeutics, which refers to the delivery of mRNA into cells and the expression of functional proteins for therapeutic purpose (Pardi et al., 2020; Sahin et al., 2014; Xu et al., 2020). At present, in vitro transcription (IVT) is the method to obtain large-scale mRNA, and T7 RNA polymerase (RNAP) is mostly used for IVT (Borkotoky and Murali, 2018; Sahin et al., 2014). When the unpurified mRNA is introduced into cells or animals, the presence of double-stranded RNA (dsRNA) as by-product of IVT activates cytoplasmic sensors including RIG-I and MDA5 (Luo et al., 2011; Mu et al., 2018; Pichlmair et al., 2009). These reactions limit the performance of transfected mRNA through translation suppression and RNA degradation (Chitrakar et al., 2021; Sahin et al., 2014). Although RNA purification methods including HPLC and Ribonuclease III digestion work well in small-scale preparations, these methods are not straightforward to scale-up for mass-production (Kariko et al., 2011; Weissman et al., 2013). The cellulose-based method is easy to scale up, but a large portion of mRNA could be lost during this process (Baiersdorfer et al., 2019; Pardi et al., 2020). Therefore, reducing the formation of dsRNA during transcription process is suitable to large-scale preparation of mRNA.
Recently, a new RNAP from the Pseudomonas fluorescens (P. fluorescens) bacteriophage VSW-3 was characterized (Zhang et al., 2017). The promoter of this VSW-3 RNAP was identified, and RNA can be efficiently produced by this RNAP at room temperature (25 °C) (Xia et al., 2020). Here, we describe that this VSW-3 RNAP significantly reduced the production of dsRNA without the need of incorporation of modified nucleotides during IVT, and the resulting mRNA achieved a higher expression level and lower innate immune response in cells and rodents than mRNA by T7 RNAP.
2. Results
2.1. The mRNA produced by VSW-3 RNAP has higher expression in cells than those by T7 RNAP
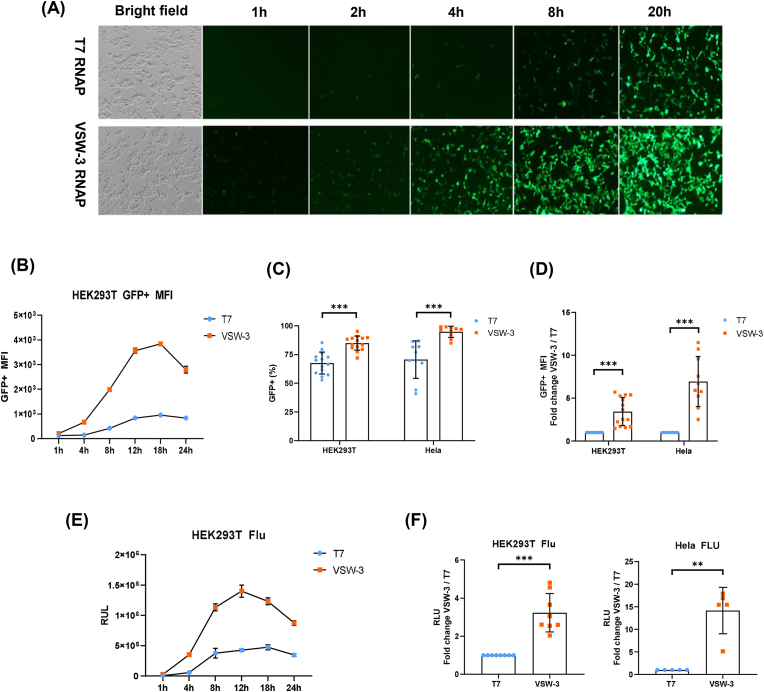
We found that GFP mRNA produced by IVT using VSW-3 RNAP had a higher expression level than that using T7 RNAP (Fig. 1A and B). The flow cytometry analysis and fluorescence microscopy showed that VSW-3 RNAP's transcription product had higher expression than that of T7 RNAP in the time course from 1 to 24 h in HEK293T cells (Fig. 1A and B). The VSW-3 group had more GFP-positive cells and higher median fluorescence intensity (MFI) than T7 group in both HEK293T cells and Hela cells (Fig. 1C and D). Strikingly, the expression level of GFP mRNA transcribed by VSW-3 was about 3 and 8 folds higher than that of T7 group in HEK293T cells and Hela cells, respectively (Fig. 1D). We observed similar trend of GFP expression in several other cell lines including N2A and RAW264.7 (data not shown). The firefly luciferase (Flu) mRNA had similar performance in HEK293T and Hela cells as GFP mRNA by VSW-3 or T7 RNAP (Fig. 1E and F).
Fig. 1.
Expression of GFP and Flu mRNA in HEK239T and Hela cells. (A) Green fluorescence signals were detected by fluorescence microscopy at various time points after transfection. (B) Flow cytometry was applied to detect the green fluorescence signals of HEK293T cells at various time points after transfection of mRNA, and median fluorescence intensity was determined. (C) Twenty hours after transfection of GFP mRNA into HEK293T and Hela cells, the ratios of GFP-positive cells were determined by flow cytometry. (D) Twenty hours after transfection of GFP mRNA into HEK293T (n = 14) and Hela (n = 10) cells, the MFI of GFP-positive cells was calculated. (E) The Flu mRNA produced by VSW-3 or T7 RNAP were transfected into HEK293T cells, and relative light unit (RLU) of luciferase was determined at different time points. (F) Flu mRNA transcribed by VSW-3 or T7 RNAP were transfected into HEK239T (n = 8) and Hela (n = 5) cells. The RLU were detected at 20 h after transfection of mRNA. Data are shown as mean ± SD. n referred to the numbers of batches of mRNA transcripts prepared by VSW-3 or T7 RNAP. Paired t-test was used. ∗∗p < 0.01, ∗∗∗p < 0.001.
2.2. The mRNA produced by VSW-3 RNAP has better expression in vivo than those by T7 RNAP
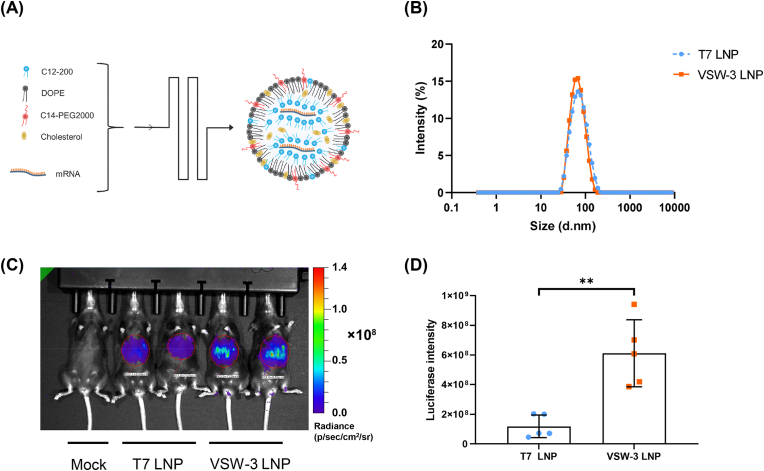
To compare mRNA transcribed by VSW-3 and T7 RNAP in vivo, we encapsulated each Flu mRNA in lipid nanoparticles (LNP). LNP encapsulated each mRNA showed similar particle size (VSW-3 LNP: Z-average = 66.7 nm, the polydispersity index (PDI) = 0.145; T7 LNP: Z-average = 63.0 nm, PDI = 0.129) and encapsulation rate (VSW-3 LNP:94.0%; T7 LNP:97.5%) (Fig. 2A and B). We determined expression of Flu in vivo 6 h after LNP injection into mice. The expression level of Flu mRNA in VSW-3 group was 5 folds higher than that in T7 group (Fig. 2C and D).
Fig. 2.
The expression levels of Flu mRNA produced by VSW-3 or T7 RNAP in mice. (A) Schematic diagram of Flu mRNA-LNP preparation. (B) Size distribution of Flu mRNA-LNP particles in VSW-3 group and T7 group. (C) Flu mRNA-LNP prepared by VSW-3 or T7 were injected into mice via tail vein. Animals in mock group were injected with normal saline. Six hours after injection, the mice were anesthetized for imaging, and regions of interest (ROI) were determined by amiview software. (D) The Luciferase intensity of the VSW-3 and the T7 group were compared. Data are shown as mean ± SD. Unpaired t-test was used. ∗∗p < 0.01, n = 5.
2.3. VSW-3 RNAP's mRNA products induce lower inflammation than those by T7 RNAP due to less dsRNA byproducts
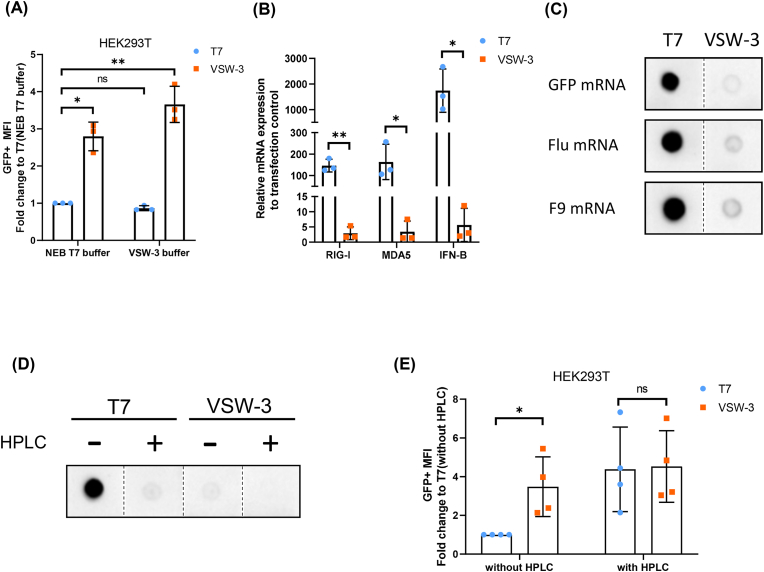
We compared GFP mRNA obtained by these two RNAP using different transcription buffers (the transcription buffer corresponding to VSW-3, named as VSW-3 buffer and the New England Biolabs RNAPol Reaction Buffer, #B9012S, named as NEB T7 buffer). Compared with NEB T7 buffer, VSW-3 buffer has higher MgCl2 content (16 mM vs 6 mM). In HEK293T cells, the mRNA transcribed by VSW-3 RNAP has a higher expression level using both buffers than T7 using respective ones during IVT (Fig. 3A).
Fig. 3.
The dsRNA byproducts in mRNA produced by the VSW-3 or T7 RNAP. (A) The IVT buffer of VSW-3 and NEB (#M0251S) were used to produce GFP mRNA which was transfected into HEK293T cells, and the MFI of GFP-positive cells was determined by flow cytometry. (B) Twenty hours after transfection, the expression levels of RIG-I, MDA5, and IFN-β in HEK293T cells were determined by qPCR. The negative control group was transfection agent treated only. (C) Dot blot analysis of dsRNA of GFP, Flu and F9 mRNA produced by VSW-3 or T7 RNAP. (D) Dot blot analysis of dsRNA in GFP mRNA with or without HPLC purification. For the dot blot analysis, 0.1 μg of RNA was loaded for each mRNA. (E) The expression of GFP mRNA with or without HPLC purification in HEK293T cells were determined by flow cytometry. n = 3 for A, B and n = 4 for E, n refers to the numbers of batches of mRNA transcripts prepared by VSW-3 or T7 RNAP. Data are shown as mean ± SD. Paired t-test was used. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
After HEK293T cells were transfected with each mRNA for 20 h, the expression levels of dsRNA related recognition receptors RIG-I and MDA5 were 87 and 48 folds higher in T7 group than those in the VSW-3 group, and IFN-β was more than 300 folds higher in T7 group (Fig. 3B, Supplementary Table 1). Therefore, we reasoned that dsRNA could be the reason for the difference in protein expression by transfected mRNA between these two groups. We used dot blot assay that applied J2 monoclonal antibody to recognize dsRNA. We found that GFP, Flu and coagulation factor IX (F9) mRNA transcribed by T7 RNAP contained more dsRNA than those by VSW-3 RNAP (Fig. 3C, Fig. S1A). After purification by HPLC, the dsRNA in GFP mRNA by T7 RNAP was substantially decreased (Fig. 3D. Fig. S1B). The T7 RNAP produced, HPLC purified GFP mRNA generate comparable MFI of GFP as that by VSW-3 RNAP (Fig. 3E).
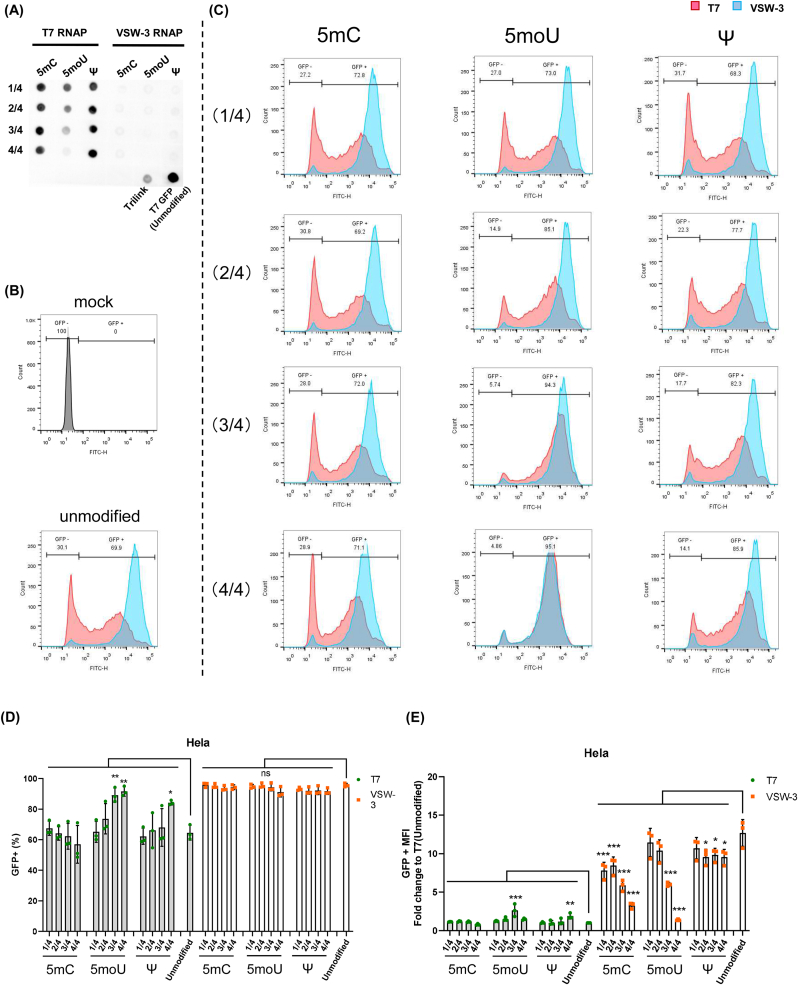
2.4. Effects of modified nucleotides on protein production
Incorporation of modified nucleotides into mRNA by T7 RNAP has been shown to reduce the activation of immune responses (Kariko et al., 2005; Richner et al., 2017). We investigated the effects of modified nucleotides on mRNA by VSW-3 RNAP. Three widely used modified nucleotides, 5-methylcytidine (5mC), 5-methoxyuridine (5moU), and pseudouridine (Ψ) were incorporated them into the transcription process in proportions of 1/4, 2/4, 3/4, and 4/4. The purity of GFP mRNA products synthesized by T7 RNAP was improved by incorporation of 5moU and Ψ in T7 group (Fig. S1C). We performed dot blot assay for these mRNAs. In T7 group, the imprinting after incorporation of 5moU but not 5mC or Ψ substantially decreased, and no imprinting was observed in the VSW-3 group with or without modified nucleotides (Fig. 4A). These GFP mRNAs were transfected into Hela cells and analyzed by flow cytometry. As the proportion of modified nucleotides increased in transcribed mRNA, the 5mU or Ψ modification could increase GFP expression in Hela cells of T7 RNAP group (Fig. 4B–E). Ψ modification has shown to increase mRNA production by reducing activation of RNA-dependent protein kinase (Anderson et al., 2010). Compared with unmodified nucleotides, the MFI of all of these modified GFP mRNA in VSW-3 group decreased (Fig. 4E). The similar results of were observed in HEK293T cells and using Flu mRNA (Figs. S1D–F). Recently, a thermostable T7 RNAP mutant named TsT7 (Hi-T7 RNA polymerase) was engineered, and this T7 mutant reduced dsRNA byproducts in IVT (Wu et al., 2020). We compared Flu mRNA transcribed by TsT7 and VSW-3 RNAP. Flu mRNA by VSW-3 RNAP exhibited near 2 folds higher protein production than that by TsT7 RNAP in HEK293T cells (Fig. S2A). Dot blot assay indicated that the dsRNA in Flu mRNA by VSW-3 RNAP was less than that by HsT7 RNAP (Figs. S2B–C).
Fig. 4.
Incorporation of modified nucleotides for mRNA by VSW-3 or T7 RNAP. (A) Dot-blot analysis of dsRNA signals in GFP mRNA synthesized by T7 or VSW-3 RNAP with modified nucleotides of different proportions (1/4, 2/4, 3/4, 4/4 indicated proportion of modified nucleotides used in IVT). GFP mRNA purchased from Trilink (L-7601) and unmodified GFP mRNA generated by T7 RNAP were served as controls. For the dot blot analysis, 0.1 μg of RNA was loaded for each mRNA. (B–E) The expression of mRNA with various modifications produced by T7 or VSW-3 RNAP in Hela cells was determined by flow cytometry. Flow cytometry charts were shown in B–C. The percentages of GFP-positive cells and MFI were shown in D-E. Data are shown as mean ± SD, n = 3. One-way ANOVA and Dunnett's multiple comparisons test were used. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
3. Discussion
In this study, we demonstrate that IVT using VSW-3 RNAP yields mRNA products with fewer dsRNA by-products than T7 RNAP, thereby causing higher protein expression and lower immunogenicity in cells than mRNA produced by T7 RNAP.
IVT-derived dsRNA can be removed by HPLC purification or cellulose column (Baiersdorfer et al., 2019; Kariko et al., 2011). During these processes, a large portion of the transcript is lost (Pardi et al., 2020). Moreover, in HPLC purification process, toxic reagents such as acetonitrile are used. The transcription products of VSW-3 have low level of dsRNA by-products. It might save the step of removing dsRNA.
Incorporation of modified nucleotides in IVT reduces immunogenicity of transcripts and improves mRNA translation capacity (Andries et al., 2015). However, incorporation of modified nucleotides reduced the expression of mRNA produced by VSW-3 RNAP (Fig. 4E, Figs. 1E–F). Using modified nucleotides in IVT could be a double-edged sword (Lu et al., 2020; Vaidyanathan et al., 2018). Incorporation of modified nucleotides in mRNA can assist to avoid activating innate immunity but interfere with the mRNA translation process to a certain extent (Thess et al., 2015). Because T7 RNAP's by-products activate a strong immune response, the consequence of compromising translation may be masked by effects of modified nucleotides on evading immunity (Kormann et al., 2011; Vaidyanathan et al., 2018). While modified nucleotides could not further reduce the immunogenicity of mRNA produced by VSW-3 RNAP, the effect of suppression translation by modified nucleotides stands out (Fig. 4 and Fig. S2).
In conclusion, mRNA produced by VSW-3 RNAP has less dsRNA and higher ability of protein expression than those by T7 RNAP. VSW-3 RNAP could be a convenient tool for both research and mRNA therapeutics.
4. Materials and methods
4.1. IVT mRNA synthesis
The transcription template was amplified by PCR from the plasmid encoding GFP, Flu and F9 (Supplementary Tables 1-2). T7 RNA polymerase (#M0251S), Hi-T7 RNA polymerase (#M0658S) and VSW-3 RNA polymerase were used for in vitro transcription to obtain the corresponding mRNA. DNase I was used to remove the DNA template, and the resulting mRNA was purified by Monarch RNA Cleanup Kit (New England Biolab). The mRNA was enzymatically capped (Vaccinia Capping System, New England Biolab), and the poly A tail was added using E. coli Poly(A) enzyme (New England Biolab). The HPLC purification was performed as previously described (Kariko et al., 2011). For synthesis of GFP mRNA bearing modified nucleosides, the transcription reaction was carried out with the replacement of rNTPs with the corresponding proportion of modified nucleotide. Each nucleotide including their modified ones were present at final concentration of 4 mM in IVT.
4.2. Cell transfection
Human embryonic kidney 293T cells (HEK293T) and Hela were obtained from ATCC. HEK293T cells were cultured in DMEM (Gibco) supplemented with L-glutamine and 10% fetal bovine serum. Hela cells were cultured in DMEM/F12 medium supplemented with 10% fetal bovine serum. The cells were tested using PCR to confirm free of mycoplasma contamination.
HEK293T or Hela cells were cultured in 24-well plates and transfected by lipofectamine 2000 (Thermo Fisher Scientific) (1.5 μL lipofectamine 2000 and 500 ng mRNA). Flow cytometry was performed at 20 h after transfection to determined GFP expression. The activity of Firefly luciferase was determined by Luciferase Assay Systems E1500 (Promega).
4.3. Formulation of lipid nanoparticles
The ionizable lipid c12-200, cholesterol, C14-PEG 2000, DOPE (1,2-allyl-sn-glycerol-3-phosphoethanolamine) were mixed at molecular ratio of 35:46.5:2.5:16. The lipid mixture and mRNA were formulated into LNP by microfluidic control. The particle sizes, mRNA concentration, and encapsulation efficiency were determined as previously described (Yin et al., 2016).
4.4. Animal experiments
All animal studies were approved by the Animal Care and Ethical Committee at Wuhan University. The prepared mRNA encapsulated in LNP was injected into 7–8 weeks old mice through the tail vein (1 mg/kg). D-luciferin potassium salt solution was intraperitoneally injected 6 h later LNP treatment (Ramaswamy et al., 2017). The mice were imaged after anesthesia and region of interest (ROI) were counted, and the ROI were determined by amiview software.
Funding
This work is kindly supported by National Key R&D Program of China (2019YFA0802801 and 2018YFA0801401 to H.Y.), the National Natural Science Foundation of China (31871345 and 32071442 to H.Y. 32150009 and 31870165 to B.Z.), Medical Science Advancement Program (Basic Medical Sciences) of Wuhan University (TFJC2018004), the Fundamental Research Funds for the Central Universities (to H.Y.), the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2020-PT320-004 to H.Y.), Applied Basic Frontier Program of Wuhan City (2020020601012216 to H.Y.), Hubei Health Commission Young Investigator award (to H.Y.), startup funding from Wuhan University (to H.Y), Science, Technology and Innovation Commission of Shenzhen Municipality (JCYJ 20210324115811032 to B.Z.) and Basic and Applied Basic Research Fund of Guangdong Province (2021A1515110376 to H.X.).
Author contribution statement
G.W. and H.X. conceived the project. G.W., H.X. and H.Y. designed the experiments. G.W. and H.X. carried out the experiments and wrote the manuscript, R.C., Q.C. and Y.X. participated the experiments. B.Z. provided materials support. H.Y. reviewed and edited the manuscript. All authors analyzed the data and contributed to the final manuscript.
Availability
Any methods and data are available online.
Conflict of interest
The authors declared that they have no conflicts of interest to this work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cellin.2022.100056.
Contributor Information
Hao Yin, Email: haoyin@whu.edu.cn.
Heng Xia, Email: xiaheng86@hotmail.com.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Kariko K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andries O., Mc Cafferty S., De Smedt S.C., Weiss R., Sanders N.N., Kitada T. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Contr. Release. 2015;217:337–344. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- Baiersdorfer M., Boros G., Muramatsu H., Mahiny A., Vlatkovic I., Sahin U., Kariko K. A facile method for the removal of dsRNA contaminant from in vitro-transcribed mRNA. Mol. Ther. Nucleic Acids. 2019;15:26–35. doi: 10.1016/j.omtn.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck J.D., Reidenbach D., Salomon N., Sahin U., Tureci O., Vormehr M., Kranz L.M. mRNA therapeutics in cancer immunotherapy. Mol. Cancer. 2021;20:69. doi: 10.1186/s12943-021-01348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkotoky S., Murali A. The highly efficient T7 RNA polymerase: a wonder macromolecule in biological realm. Int. J. Biol. Macromol. 2018;118:49–56. doi: 10.1016/j.ijbiomac.2018.05.198. [DOI] [PubMed] [Google Scholar]
- Chitrakar A., Solorio-Kirpichyan K., Prangley E., Rath S., Du J., Korennykh A. Introns encode dsRNAs undetected by RIG-I/MDA5/interferons and sensed via RNase L. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2102134118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schafer A., Ziwawo C.T., DiPiazza A.T., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariko K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Kariko K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kormann M.S., Hasenpusch G., Aneja M.K., Nica G., Flemmer A.W., Herber-Jonat S., Huppmann M., Mays L.E., Illenyi M., Schams A., et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- Laczko D., Hogan M.J., Toulmin S.A., Hicks P., Lederer K., Gaudette B.T., Castano D., Amanat F., Muramatsu H., Oguin T.H., 3rd, et al. A single immunization with nucleoside-modified mRNA vaccines elicits strong cellular and humoral immune responses against SARS-CoV-2 in mice. Immunity. 2020;53:724–732 e727. doi: 10.1016/j.immuni.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Lu G., Tan S., Xia J., Xiong H., Yu X., Qi Q., Yu X., Li L., Yu H., et al. A COVID-19 mRNA vaccine encoding SARS-CoV-2 virus-like particles induces a strong antiviral-like immune response in mice. Cell Res. 2020;30:936–939. doi: 10.1038/s41422-020-00392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., Ding S.C., Vela A., Kohlway A., Lindenbach B.D., Pyle A.M. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X., Greenwald E., Ahmad S., Hur S. An origin of the immunogenicity of in vitro transcribed RNA. Nucleic Acids Res. 2018;46:5239–5249. doi: 10.1093/nar/gky177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Weissman D. Recent advances in mRNA vaccine technology. Curr. Opin. Immunol. 2020;65:14–20. doi: 10.1016/j.coi.2020.01.008. [DOI] [PubMed] [Google Scholar]
- Pichlmair A., Schulz O., Tan C.P., Rehwinkel J., Kato H., Takeuchi O., Akira S., Way M., Schiavo G., Reis e Sousa C. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S., Tonnu N., Tachikawa K., Limphong P., Vega J.B., Karmali P.P., Chivukula P., Verma I.M. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E1941–E1950. doi: 10.1073/pnas.1619653114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C., et al. Modified mRNA vaccines protect against Zika virus infection. Cell. 2017;168:1114–1125 e1110. doi: 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U., Kariko K., Tureci O. mRNA-based therapeutics--developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- Thess A., Grund S., Mui B.L., Hope M.J., Baumhof P., Fotin-Mleczek M., Schlake T. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan S., Azizian K.T., Haque A., Henderson J.M., Hendel A., Shore S., Antony J.S., Hogrefe R.I., Kormann M.S.D., Porteus M.H., et al. Uridine depletion and chemical modification increase Cas9 mRNA activity and reduce immunogenicity without HPLC purification. Mol. Ther. Nucleic Acids. 2018;12:530–542. doi: 10.1016/j.omtn.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D., Pardi N., Muramatsu H., Kariko K. HPLC purification of in vitro transcribed long RNA. Methods Mol. Biol. 2013;969:43–54. doi: 10.1007/978-1-62703-260-5_3. [DOI] [PubMed] [Google Scholar]
- Wu M.Z., Asahara H., Tzertzinis G., Roy B. Synthesis of low immunogenicity RNA with high-temperature in vitro transcription. RNA. 2020;26:345–360. doi: 10.1261/rna.073858.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Jiang Y., Cheng R., Yu B., Lu X., Wu H., Zhu B. In vitro transcription using psychrophilic phage VSW-3 RNA polymerase. bioRxiv. 2020;(14):297226. doi: 10.1080/15476286.2022.2139113. 09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Yang K., Li R., Zhang L. mRNA vaccine era-mechanisms, drug platform and clinical prospection. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21186582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A., et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Zhang Z., Li J., Qin K., Wei Y., Zhang Q.…Ji X. Complete genome sequence of the lytic cold-active Pseudomonas fluorescens bacteriophage VSW-3 from Napahai plateau wetland. Virus Genes. 2017;53:146–150. doi: 10.1007/s11262-016-1403-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.