Abstract
Alpha7 nicotinic acetylcholine receptor (α7 nAChR), a hub of the cholinergic anti-inflammatory pathway (CAP), is required for the treatment of inflammatory diseases. HIV-1 infection can upregulate the expression of α7 nAChR in T lymphocytes and affect the role of CAP. However, whether α7 nAChR regulates HIV-1 infection in CD4+ T cells is unclear. In this study, we first found that activation of α7 nAChR by GTS-21 (an α7 nAChR agonist) can promote the transcription of HIV-1 proviral DNA. Then, through transcriptome sequencing analysis, we found that p38 MAPK signaling was enriched in GTS-21 treated HIV-latent T cells. Mechanistically, activation of α7 nAChR could increase reactive oxygen species (ROS), reduce DUSP1 and DUSP6, and consequently enhance the phosphorylation of p38 MAPK. By co-immunoprecipitation and liquid chromatography tandem mass spectrometry, we found that p-p38 MAPK interacted with Lamin B1 (LMNB1). Activation of α7 nAChR increased the binding between p-p38 MAPK and LMNB1. We confirmed that knockdown of MAPK14 significantly downregulated NFATC4, a key activator of HIV-1 transcription. Taken together, activation of the α7 nAChR could trigger ROS/p-p38 MAPK/LMNB1/NFATC4 signaling pathway enhancing HIV-1 transcription. We have revealed an unrecognized mechanism of α7 nAChR-mediated neuroimmune regulation of HIV infection.
Keywords: HIV-1, α7 nAChR, ROS, p38 MAPK, LMNB1
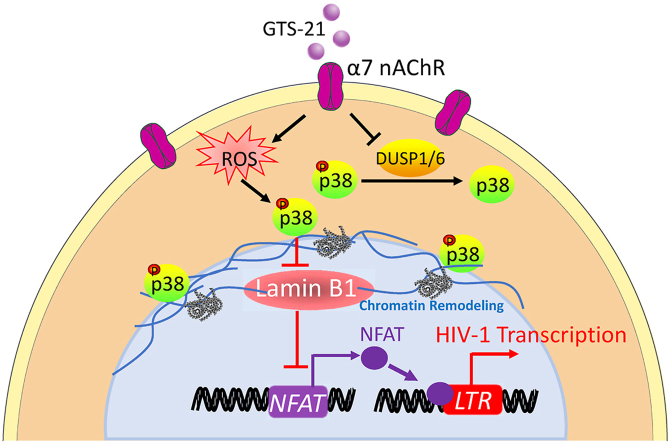
Graphical abstract
Whether α7 nAChR regulates HIV-1 infection in CD4+ T cells is unclear. Wen et al. reveal that activation of the α7 nAChR could trigger ROS/p38 MAPK/LMNB1/NFATC4 signaling pathway enhancing HIV-1 transcription.
Highlights
-
•
Activation of α7 nAChR promotes HIV-1 transcriptional initiation.
-
•
Activation of α7 nAChR reduces phosphorylation of p38 MAPK.
-
•
Phosphorylation of p38 MAPK is required for α7 nAChR-mediated HIV transcription.
-
•
Activation of α7 nAChR promotes HIV-1 transcription depending on interaction between p-p38 MAPK and LMNB1.
1. Introduction
Infection with the human immunodeficiency virus (HIV) is a threat to global health. Although the combination antiretroviral therapy (cART) has changed acquired immunodeficiency syndrome (AIDS) from fatal disease into chronic disease, current therapy can't eradicate the viral latent reservoirs. The persistence of a latent reservoir of replication-competent provirus remains a major obstacle for AIDS cure (Churchill et al., 2016; Margolis et al., 2016). Recent studies have identified that the maintenance of HIV-1 latency is regulated by multiple mechanisms, including epigenetic silencing, transcriptional interference, sequestration of transcription factors, and sequestration of the positive transcription elongation factor B complex, etc. (Ma et al., 2020; Ruelas and Greene, 2013). Deeply understanding the mechanisms of viral latency is urgently needed to uncover novel methods for eradicating the latent reservoir.
To study whether nervous system affects HIV latency, we approach the cholinergic anti-inflammatory pathway (CAP) that is mediated by vagus nerve-α7 nAChR signaling (Andersson and Tracey, 2012; Navarro et al., 2015; Woo et al., 2012). Stimulation of efferent vagus nerve initiates release of acetylcholine (ACh) by which stimulating the α7 nAChR expressed in macrophages to suppress pro-inflammatory cytokine production (Wang et al., 2003; Xie et al., 2020). Moreover, CHAT (choline acetyltransferase for ACh synthesis)-expressing memory T cell population in the mouse spleen builds up the neural circuits that implement the function of the CAP (Yang et al., 2017). Importantly, CHAT-expressing CD4+ T cells are implicated in the lymphocytic choriomeningitis virus infection in an IL-21-dependent manner (Yang et al., 2017).
The HIV-infected patients often suffer from chronic inflammation. In the macrophages isolated from HIV-infected patients, HIV-1 glycoprotein gp120 could upregulate the α7 nAChR in a CCR5-dependent way (Callahan et al., 2013). In neuronal cells, HIV-1-gp120 can interact with α7 nAChR, and this process may be involved in the development of HIV-associated neurocognitive disorder. Activation of α7 nAChR in neuronal cells reduced HIV-1-gp120-induced neurotoxicity (Ballester et al., 2012; Capo-Velez et al., 2018; Zhao et al., 2021). As we know, CD4+ T cells are the main HIV-1 infection target cells (Simon and Ho, 2003). HIV-1 infection can upregulate α7 nAChR expression in T lymphocytes and disrupt the effect of the CAP (Ballester et al., 2012; Delgado-Velez et al., 2015). Whether α7 nAChR signaling pathway plays a role in the HIV transcription or latency is little studied.
Oxidative stress might be associated with HIV-1 infection (Huang et al., 2020; Ivanov et al., 2016). A recent study has demonstrated that reactive oxygen species (ROS, including O2.-, OH., and H2O2) can regulate the replication of HIV-1 and vanadium pentoxide nanosheets could functionally mimic glutathione peroxidase to mitigate ROS and inhibit HIV-1 infection (Singh et al., 2021). Other studies also showed that HIV-1 protein Tat, Env and Nef can induce ROS production and antioxidants can inhibit viral replication (Daussy et al., 2021; Ivanov et al., 2016; Singh et al., 2021). ROS has also been regarded as a sensor in various signaling pathways, such as MAPK, NF-κB, and PI3K signaling pathways (Zhang et al., 2016). HIV-1 infection can trigger phosphorylation of MAPK by which affects apoptosis in host cells (Gupta et al., 2010; Kantner et al., 2013). In ACH2 cells (chronically infected with HIV-1), MAPK p38α can activate HIV-1 replication (Guo et al., 2014). Whether activation of α7 nAChR affects ROS production and phosphorylation of p38 MAPK and therefore alters HIV-1 transcription is unknown.
Nuclear peripheral chromatin-lamin B1 (LMNB1) interaction is required for global integrity of chromatin architecture and dynamics in human cells (Chang et al., 2020). Repressive chromatin is formed and modulated during HIV-1 infection. Lamin A/C tethered SUN2 to the nucleosomes 1 and 2 of the HIV-1 5′-LTR to block the initiation and elongation of HIV-1 transcription (Gupta et al., 2010). So far, whether and how activation of α7 nAChR influences interaction between p38 MAPK and LMNB1, and therefore affects NFAT-mediated HIV -1 transcription warrants further study.
Therefore, by means of RNAseq, co-immunoprecipitation, liquid chromatography tandem mass spectrometry, and other advanced methods, we have examined whether activation of α7 nAChR could initiate ROS/p-p38 MAPK/LMNB1/NFATC4 signaling by which augments HIV-1 transcription. We have identified that activation of α7 nAChR significantly regulated HIV-1 infectivity in a dose-dependent manner in ACH2 cells. Activation of α7 nAChR can promote phosphorylation of p38 MAPK by downregulating DUSP1 and DUSP6 expression and promoting ROS production. Knockdown of MAPK14 attenuated α7 nAChR activation-dependent HIV-1 infectivity. Mutant of phosphorylation site of p38 MAPK also reduced HIV-1 transcription. Interestingly, activation of α7 nAChR could specifically increase binding between p-p38 MAPK and LMNB1. Knockdown of LMNB1 increased NFATC4 and HIV-1 transcription. Thus, our findings have first clarified an unrecognized neuroregulatory mechanism that α7 nAChR activation promotes HIV-1 transcription via ROS/p-p38 MAPK/LMNB1/NFATC4 signaling, which will provide us novel strategies for AIDS therapy.
2. Results
2.1. Activation of α7 nAChR by its agonist GTS-21 promotes HIV-1 transcription in CD4+ T cells
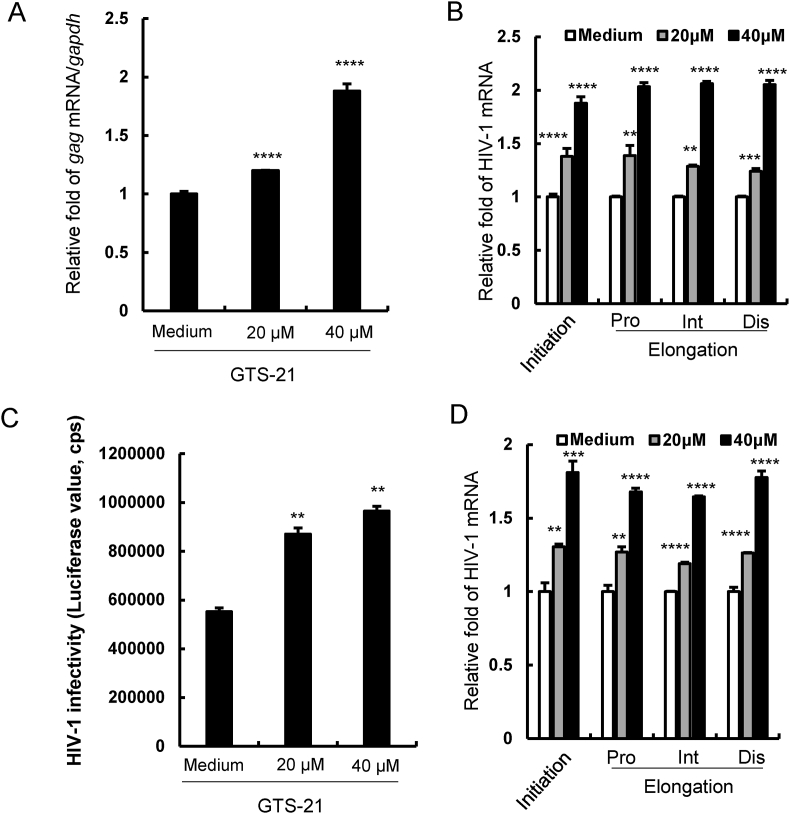
To investigate the activation of α7 nAChR whether modulates HIV-1 infection, we treated the HIV-1 latently infected CD4+ T cell ACH2 with different concentration of α7 nAChR agonist GTS-21 for 48 h and then detected the HIV-1 gag mRNA expression by RT-qPCR. We found that GTS-21 significantly increased the HIV-1 gag mRNA expression in a dose-dependent manner (Fig. 1A). HIV-1 long terminal repeat (LTR) promoter is required for viral transcription and production. Many transcription factors binding to LTR contribute to HIV-1 transcription, such as NF-κB, NFAT and TCF1 (Mukerjee et al., 2007; Pereira et al., 2000; Wen et al., 2022). To determine the role of α7 nAChR in HIV-1 transcription, HIV-1 transcription initiation and elongation were assessed by RT-qPCR with specific primers (Zhu et al., 2012). We found that activation of α7 nAChR significantly enhanced HIV-1 initial transcription (Fig. 1B). For confirmation, Jurkat T cells were acutely infected with single-cycle infectious HIV-Luc/VSV-G virus for 24 h, and then incubated with GTS-21 for 48 h. We found that viral transcription level was also increased after GTS-21 treatment (Fig. 1 C and D). These findings suggest that activation of α7 nAChR can promote HIV-1 transcription.
Fig. 1.
GTS-21 promotesHIV-1transcription in CD4+T cells. ACH2 cells were treated with GTS-21 at different concentration for 48 h, (A) the expression level of gag mRNA was detected by RT-qPCR, (B) and specific primers were used to quantify the initiation and elongation of HIV-1 transcription. (C, D) Jurkat T cells were acutely infected with pseudovirus HIV-Luc/NL4-3 (5 ng p24) for 24 h and treated with GTS-21 for 48 h, gag mRNA and the initiation and elongation of HIV-1 transcription were detected. Result is one representative from three independent repeats. Data are presented as mean ± SD. ∗∗P < 0.01, ∗∗∗P < 0.001 and ∗∗∗∗P < 0.0001 denote significant difference.
2.2. Activation of α7 nAChR enhances phosphorylation of p38 MAPK by downregulating DUSP1 and DUSP6, coinciding with an increase of HIV-1 transcription
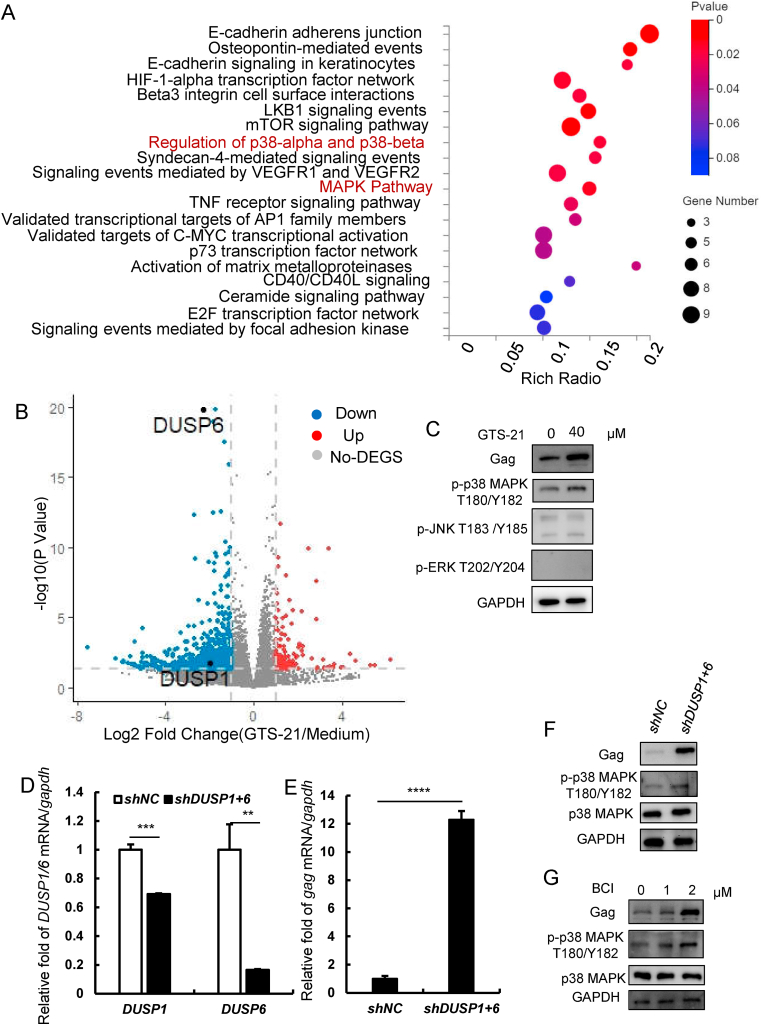
To address why activation of α7 nAChR could augment HIV-1 transcription, we assessed to RNA sequencing in ACH2 cells with or without GTS-21 stimulation. Via KEGG analysis, we found that genes regarding MAPK signaling pathway were significantly enriched in GTS-21-treated ACH2 cells (Fig. 2A). Through volcano plot analysis, we found that 126 upregulated and 840 downregulated genes were significantly different between medium-treated and GTS-21-treated ACH2 cells. Particularly, DUSP6 and DUSP1 were downregulated in GTS-21-treated group (Fig. 2B). Considering that DUSPs are able to inactivate the MAPK signaling pathway through dephosphorylating both threonine/serine and tyrosine residues of MAPK family (ERK, JNK, and p38 MAPK) (Chen et al., 2019; Ramkissoon et al., 2019), we attempted to examine whether activation of α7 nAChR would affect phosphorylation of MAPKs in GTS-21-treated ACH2 cells. By western blotting, we found that activation of α7 nAChR promoted phosphorylation of p38 MAPK in T180/Y182 site, but did not affect p-JNK T183/Y185 and p-ERK T202/Y204 (Fig. 2C). To test whether activation of α7 nAChR-mediated downregulation of DUSP1 and DUSP6 is contributed to phosphorylation of p38 MAPK, we used specific short hairpin RNA (shRNA) lentivirus to knock down DUSP1 and DUSP6 in ACH2 cells (Fig. 2D) and found that the level of p-p38 MAPK and HIV-1 gag expression were significantly increased in DUSP1 and DUSP6 knockdown group compared to the scrambled group (Fig. 2E and F). We confirmed that inhibition of DUSP1 and DUSP6 by their inhibitor BCI also enhanced the level of p-p38 MAPK and HIV-1 gag expression (Fig. 2G). These findings demonstrate that activation of α7 nAChR can decrease DUSP1 and DUSP6 expression, facilitate p38 MAPK phosphorylation, and drive HIV-1 transcription.
Fig. 2.
Activation of α7 nAChR enhances phosphorylation of p38 MAPK by downregulatingDUSP1andDUSP6. (A) MSigDB canonical pathways enrichment analysis of differentially expressed genes (DEGs) (P < 0.05) after GTS-21 treatment, bubble size indicates the absolute gene counts enriched in a term. (B) Volcano plot of DEGs comparing GTS-21-treated versus untreated (medium) cells. Three samples in either medium or GTS-21 treated ACH2 groups were used for RNAseq analysis. (C) ACH2 were treated with GTS-21 (40 μM) and detect the phosphorylation of MAPKs family members p38 MAPK, JNK and ERK, and gag protein level by western blotting. (D) The endogenous DUSP1/6 was knocked-down with lentiviruses containing specific shRNAs. (E) The gag mRNA level of negative control and DUSP1+6 knockdown groups were identified by RT-qPCR. (F) The levels of gag and p-p38 MAPK in two groups were detected by western blotting. (G) The inhibitor of DUSP1/6 were used to inhibit DUSP1 and DUSP6 for 8 h, then the gag and p-p38 MAPK levels were detected by western blotting. Result is one representative from three independent repeats. Data are presented as mean ± SD. ∗∗P < 0.01, ∗∗∗P < 0.001 and ∗∗∗∗P < 0.0001 denote significant difference.
2.3. Activation of α7 nAChR induces ROS production to promote the phosphorylation of p38 MAPK and HIV-1 transcription
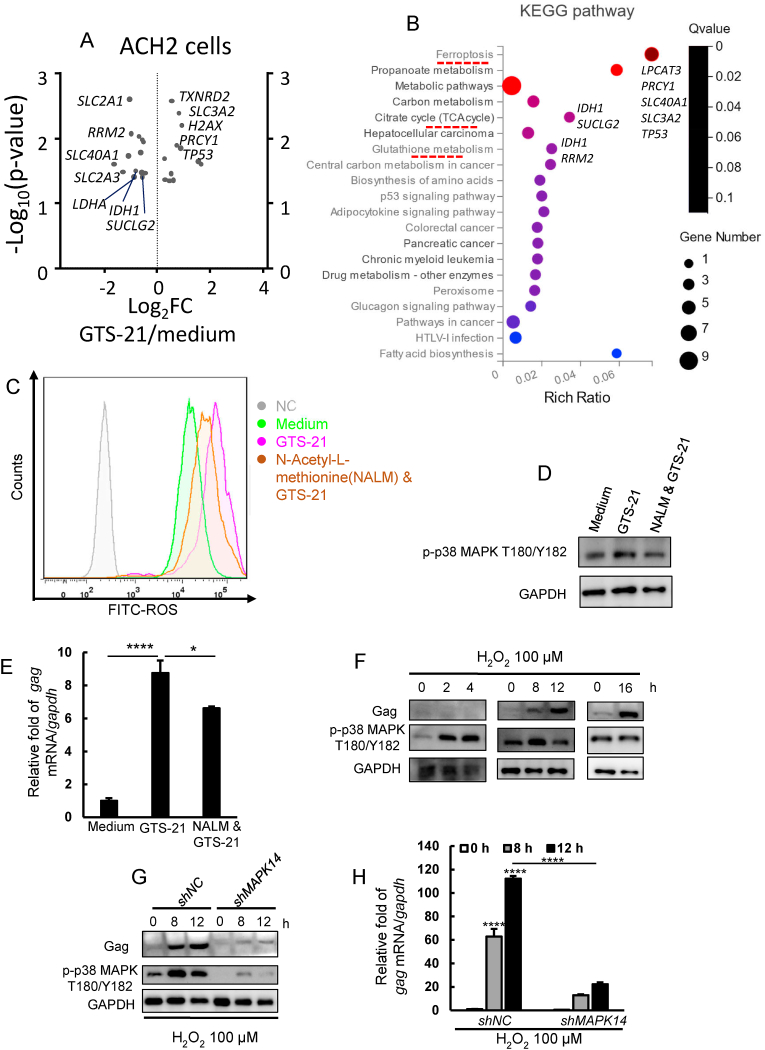
We performed gene enrichment regarding ferroptosis and oxidative stress by Wiki and KEGG pathway analysis. 25 differentially expressed genes were enriched in GTS-21 versus medium-treated ACH2 cells (Fig. 3A). KEGG pathway analysis showed that activation of α7 nAChR significantly affected genes related to TCA cycle (IDH1, SUCLG2), glutathione metabolism (IDH1, RRM2), ferroptosis (TP53, PRCY1), and DNA damage (TP53, H2AX) (Fig. 3B). Especially, genes regarding glucose transport (SLC2A1, SLC2A3), TCA cycle (IDH1, SUCLG2), and glutathione metabolism (IDH1, RRM2) were markedly downregulated in GTS-21-treated ACH2 cells (Fig. 3A), suggesting that activation of α7 nAChR might induce production of ROS by disrupting glutathione metabolism and oxidative phosphorylation. To prove this, we detected intracellular ROS level by staining ACH2 with fluorescent probes DCFH-DA and observed by flow cytometry. We found that treatment with GTS-21 obviously increased ROS production and the ROS scavenger N-Acetyl-L-methionine (NALM) could inhibit this effect (Fig. 3C). Next, we tested whether GTS-21 could increase phosphorylation of p38 MAPK by inducing ROS production (Liu et al., 2020; Wang et al., 2011; Xu et al., 2015). We detected the p-p38 MAPK and gag expression after GTS-21 or GTS-21 plus NALM treatment by western blotting and RT-qPCR. We found that GTS-21-induced increase of p-p38 MAPK and gag expression were markedly mitigated by NALM (Fig. 3D and E). To confirm the role of ROS in HIV-1 regulating transcriptional expression, we used hydrogen peroxide (H2O2, a kind of ROS) to stimulate ACH2 cells at different times. We found that phosphorylation of p38 MAPK and gag expression levels were significantly increased after 8 h H2O2 stimulation (Fig. 3F). We treated scrambled and MAPK14 knockdown cells with H2O2 for 8 and 12 h, and then detected the p-p38 MAPK and gag expression levels. We found that MAPK14 knockdown attenuated the boosting effect of H2O2 on HIV-1 transcription (Fig. 3G and H). These findings support that activation of α7 nAChR enhances ROS production and ROS-mediated HIV-1 transcription depends on phosphorylation of p38 MAPK.
Fig. 3.
Activation of α7 nAChR induces ROS production to promoteHIV-1transcription. (A) Volcano plot of upregulated and downregulated genes related to oxidative stress in the medium and GTS-21-treated ACH2 cells. (B) KEGG pathway analysis of upregulated and downregulated genes related to oxidative stress in the medium and GTS-21-treated ACH2 cells. Three samples in either medium or GTS-21 treated ACH2 groups were used for RNAseq analysis. (C) DCFH-DA fluorescent probes were used to label intracellular ROS, intracellular ROS level in untreated (Medium), GTS-21 (40 μM) treated and GTS-21(40 μM) &ROS scavenger N-Acetyl-L-methionine (NALM) (200 μM) treated groups by flow cytometry, cells without labeled with DCFH-DA probe were used as negative control (NC). (D, E) Detect the levels of p-p38 MAPK and gag transcription level in medium, GTS-21, GTS-21& NALM (200 μM) groups. (F) ACH2 cells were cultured with H2O2 (100 μM) for different times, and the level of p-p38 MAPK and gag were detected by western blotting. (G, H) Negative control and MAPK14 knockdown cells were cultured with H2O2 (100 μM) for 8, 12 h, (G) the levels of p-p38 MAPK and gag were detected by western blotting, (H) the expression level of gag mRNA was detected by RT-qPCR. Result is one representative from three independent repeats. Data are presented as mean ± SD. ∗P < 0.05, ∗∗∗∗P < 0.0001 denote significant difference.
2.4. The Thr180/Tyr182 site phosphorylation of p38 MAPK is requisite for activation of α7 nAChR-mediated HIV-1 transcription
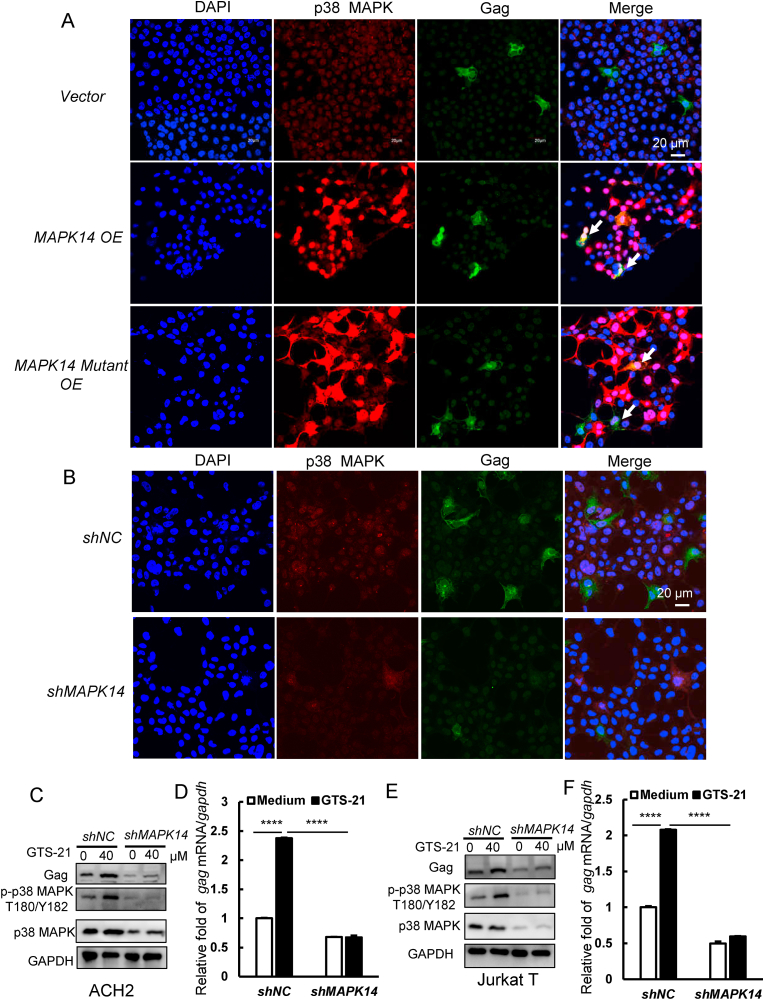
To explore whether the phosphorylation of p38 MAPK is required for promoting HIV-1 transcription, we constructed scrambled, MAPK14 overexpression, and MAPK14-Thr180/Tyr182 mutant plasmid. Plasmids were transfected into HEK293T cells for 24 h and then acutely infected with HIV-Luc/VSV-G virus for 24 h. The expression of p38 MAPK and HIV-1 gag protein were identified by immunofluorescence. We observed a significantly increased expression of gag in MAPK14-overexpressed cells but not in MAPK14-Thr180/Tyr182 mutant overexpressed cells. We also observed p38 MAPK and gag protein co-localized in the same cells (Fig. 4A). The expression of p38 MAPK and p-p38 MAPK were detected by western blotting (Fig. S1A). The HIV-Luc/VSV-G virus infection was increased in MAPK14-overexpressed cells but not in the MAPK14-Thr180/Tyr182 mutant overexpressed cells (Fig. S1B). Next, we knocked down MAPK14 in HEK293T cells by using shRNA lentivirus and observed the gag expression level by immunofluorescence. We found that gag expression level was significantly decreased in MAPK14 knockdown cells (Fig. 4B). This result was also confirmed by western blotting and RT-qPCR (Fig. S1 C and D). To further verify the phosphorylation of p38 MAPK is required for α7 nAChR activation-mediated HIV-1 transcription, we knocked down endogenous MAPK14 in ACH2 and Jurkat T cells and treated both negative control cells and MAPK14 knockdown cells with GTS-21 for 48 h and then detected the change of gag expression at both mRNA and protein levels. We found that MAPK14 knockdown markedly reduced α7 nAChR activation-mediated Thr180/Tyr182 site phosphorylation of p38 MAPK and gag expression at both mRNA and protein levels (Fig. 4C–F). These findings strongly support that Thr180/Tyr182 site phosphorylation of p38 MAPK is requisite for activation of α7 nAChR-mediated HIV-1 transcription.
Fig. 4.
Phosphorylation of p38 MAPK is required for activation of α7nAChR-mediated HIV-1 transcription. (A) HEK293T cells were transfected with vector, MAPK14 and MAPK14-Thr180/Tyr182 mutant (MAPK14 Mutant) plasmids and infected with HIV-Luc/NL4-3 pseudovirus (2 ng) for 48 h, then detected intracellular p38 MAPK and gag levels by immunofluorescence. (B) The endogenous MAPK14 was knocked-down with lentiviruses containing specific shRNA, the expression of gag was detected by immunofluorescence in negative control and MAPK14 knockdown groups. (C–F) In ACH2 and Jurkat T cells, the endogenous MAPK14 was knocked-down with lentiviruses containing specific shRNA, negative control and MAPK14 knockdown groups cells were treated with GTS-21 (40 μM) for 48 h, and then analyzed the transcription level of virus. Result is one representative from three independent repeats. Data are presented as mean ± SD. ∗∗∗P < 0.001 and ∗∗∗∗P < 0.0001 denote significant difference.
2.5. Activation of α7 nAChR increases binding between Thr180/Tyr182p-p38 MAPK and LMNB1 and contributes to structural abnormality of LMNB1
To directly view the Thr180/Tyr182p-p38 MAPK and gag expression, A549 cells were infected with HIV-Luc/VSV-G virus for 24 h and followed by GTS-21 treatment for 48 h. By immunofluorescence, we observed that both gag expression and p-p38 MAPK were upregulated in GTS-21 treated group. Most of p-p38 MAPK and gag were localized in the cytoplasm (Fig. S2A). By western blot, we confirmed that p-p38 MAPK and gag proteins were also accumulated in the cytoplasm in GTS-21-treated ACH2 cells (Fig. S2B). These findings suggest that promotion of HIV transcription by activation of α7 nAChR might be independent of nuclear translocation of p-p38 MAPK.
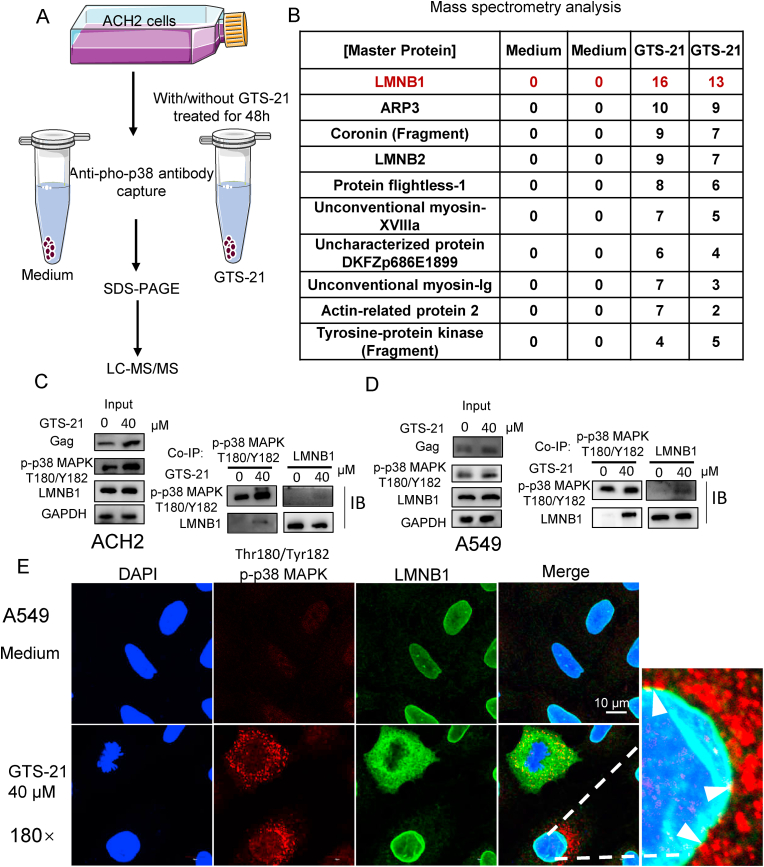
To probe p-p38 MAPK binding proteins, we performed anti-p-p38 MAPK pull down. The protein complex was analyzed by Liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Fig. 5A). We listed top 10 of p-p38 MAPK specifically-binding proteins enriched under GTS-21 treatment, LMNB1 was ranked in the 1st place (Fig. 5B). To confirm the interaction between LMNB1 and p-p38 MAPK, ACH2 cells treated with or without GTS-21 were lysed. The cell lysates were immunoprecipitated with anti-p-p38 MAPK or anti-LMNB1 antibodies. We found that GTS-21 treatment increased the binding between p-p38 MAPK and LMNB1 (Fig. 5C). We repeated the above experiments in A549 cells, the same phenomena were found (Fig. 5D). By immunofluorescence, we found that LMNB1 could spread in the cytosol when nucleus deformed and colocalization of p-p38 MAPK and LMNB1 occurred in the cytosol and surface of nucleus under GTS-21 stimulation (Fig. 5E). These findings strongly support that activation of α7 nAChR augments the interaction between LMNB1 and Thr180/Tyr182p-p38 MAPK. Phosphorylation of p38 MAPK is associated with structural abnormality of LMNB1.
Fig. 5.
Activation of α7 nAChR increases binding betweenThr180/Tyr182p-p38 MAPK and LMNB1. (A) ACH2 cells were treated with or without GTS-21 (40 μM) for 48 h, anti-p-p38 MAPK was used to capture the interaction proteins of p-p38 MAPK. (B) Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyze potential proteins interacting with p-p38 MAPK. The values in the table represent the number of non-specific peptides enriched. (C-E) The interaction between p-p38 MAPK and LMNB1 were confirmed in ACH2 and A549 cells. (C) Untreated or GTS-21 (40 μM) treated ACH2 cells were collected and pulled down interaction proteins by anti-p-p38 MAPK and anti- LMNB1 antibodies. The p-p38 MAPK and anti-LMNB1 were detected by western blotting. HIV-Luc/VSV-G pseudovirus infected A549 cells were untreated or treated with GTS-21 (40 μM) for 48 h, (D) cells were lysed and pulled down interaction proteins by anti-p-p38 MAPK8 and anti-LMNB1 antibodies, the p-p38 MAPK and anti-LMNB1 were detected by western blotting. (E) Cells were fixed and analyzed the intracellular co-localization of p-p38 MAPK and LMNB1 by immunofluorescence. Arrow heads indicate co-localization of p-p38 MAPK and LMNB1.
2.6. Presence of LMNB1 restricts NFAT family gene expression and HIV-1 transcription
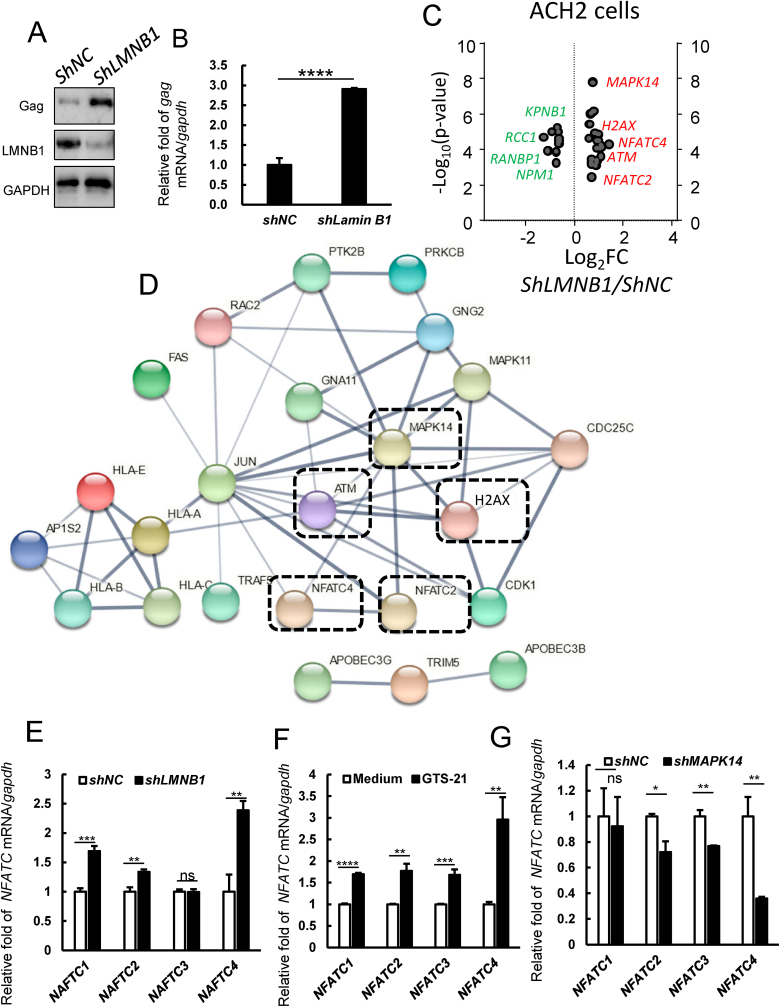
Considering that LMNB1 (Lamin B1) is a downstream factor of the activated α7 nAChR pathway, we knocked down LMNB1 in ACH2 cells and detected the expression of gag by RT-qPCR and western blot. We found that LMNB1 knockdown increased viral transcriptional expression at both protein and mRNA levels (Fig. 6A and B). To further explore the LMNB1 targeted genes, we performed transcriptome analysis on negative control and LMNB1 knockdown ACH2 cells. We uploaded 795 upregulated genes with FPKM>10 in the ShLMNB1 group to STRING database (https://cn.string-db.org/cgi/input?sessionId=bZw1NBphrlSh&input_page_active_form=multiple_identifiers). Through protein interaction network and KEGG pathway analysis, we found that 24 genes were related to HIV infection (Fig. S3). Using the same strategy, we uploaded 433 downregulated genes with FPKM>10 in the ShLMNB1 group to STRING platform, 37 genes associated with viral process were enriched by GO analysis (Fig. S4). Among these genes, 6 genes (KPNB1, NPM1, RCC1, SEH1L, RANBP1, and RAN) contributed to interactions of Rev with host cellular proteins (Fig. S5). We showed 24 upregulated and 10 downregulated genes in the LMNB1 knockdown group by Volcano plot (Fig. 6C). Among these genes, MAPK14, NFATC4, NFATC2, ATM, and H2AX were increased with LMNB1 knockdown, indicating that presence of LMNB1 restricts MAPK14 and NFAT gene expression and protects chromatin and DNA damage. To predict protein interaction, we inputted 24 upregulated genes into the STRING database and found that MAPK14 could interact with NFATC4 and NFATC2 (Fig. 6D), suggesting that LMNB1, p38 MAPK, NFAT family, ATM, and H2AX are in a protein interaction network. P-p38 MAPK engaging with LMNB1 might impair stability of chromatin and DNA, reflected by higher expression of ATM and H2AX.
Fig. 6.
LMNB1 restricts NFAT family gene expression andHIV-1transcription. (A, B) The endogenous LMNB1 was knocked-down with lentiviruses containing specific shRNA, the expression of LMNB1 and gag were detected by western blotting, the gag mRNA expression level was detected by RT-qPCR. (C) Volcano plot of enriched genes related to chromatin remodeling and transcription binding in the LMNB1 knockdown versus ShNC cells. (D) Protein interaction network in the upregulated genes regarding chromatin remodeling and transcription binding in the LMNB1 knockdown versus ShNC cells. (E–G) The effect of LMNB1 knockdown, GTS-21 treatment and MAPK14 knockdown on NFATC1, NFATC2, NFATC3 and NFATC4 gene expression was detected in ACH2 cells. Result is one representative from three independent repeats. Data are presented as mean ± SD. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 and ∗∗∗∗P < 0.0001 denote significant difference.
Given that LMNB1 is required for integrity of chromatin and transcription factor binding, we collected 500 genes regarding chromatin (GeneCards, https://www.genecards.org) and uploaded these genes into BGI BIG DATABASE (https://biosys.bgi.com/), where our RNAseq data were stored. We found that 139 genes were increased by 1.2-fold in the ShLMNB1 group. Using STRING database, GO enrichment was performed. We found that 45 were chromatin binding and 38 were transcription binding genes (Fig. S6). In 38 transcription binding genes, CDK9, NFATC1, NFATC2, NFATC4, SMARCA1, and SMARCD3 were upregulated when LMNB1 was knockdown (Fig. S7). These findings indicate that LMNB1 negatively regulates chromatin and transcription factor binding. We collected the p-p38 MAPK binding proteins from our Mass Spectrum database, and uploaded their genes to BGI BIG DATABASE (https://biosys.bgi.com/) of our LMNB1 knockdown experiments. 22 genes were upregulated in the LMNB1 knockdown group. GO analysis showed that these genes were related to cellular component organization (Fig. S8). Especially, H2AX and SMARCA1 contribute to chromatin remodeling, DNA damage, and transcription. This finding indicates that LMNB1 knockdown disrupts chromatin and causes DNA damage.
NFAT family members are key factors for initiation of transcription at the HIV-1 LTR (https://www.wikipathways.org/index.php/Pathway:WP3414). Therefore, we confirmed the expression levels of NFAT family genes in negative control and LMNB1 knockdown cells by RT-qPCR. The expressions of NFATC1, NFATC2 and NFATC4 were significantly increased in the LMNB1 knockdown group (Fig. 6E). Next, we examined the effect of GTS-21 treatment and MAPK14 knockdown on NFAT family gene expression. We found that α7 nAChR activation could increase NFAT family gene expression (Fig. 6F), while MAPK14 knockdown could reduce NFATC2, NFATC3, and NFATC4 expression (Fig. 6G). Collectively, these findings support that α7 nAChR activation could increase p-p38 MAPK, disassemble LMNB1, upregulate NFAT, and thereby promote HIV-1 transcription (Graphical Abstract).
3. Discussion
In this study, we demonstrate that α7 nAChR activation enhances ROS production and phosphorylation of p38 MAPK, which results in binding between p-p38 MAPK and LMNB1, disassembly of LMNB1, upregulation of NFAT, and promotion of HIV-1 transcription.
In the past decades, many studies have been focused on the regulatory effect of CAP on immune responses and bacterial infection, but less on the viral infection and replication. In our research group, we have studied the effect of activation of α7 nAChR on the replication of different viruses, including influenza, ZIKV, HIV, and SARS-CoV-2. We have reported the vagus nerve through α7 nAChR can regulate influenza virus infection (Gao et al., 2021). Here, we treated latent cell line ACH2 cells and pseudovirus acutely infected Jurkat T cells with α7 nAChR agonist GTS-21 and found that α7 nAChR activation can boost HIV-1 mRNA transcription.
It is well recognized that α7 nAChR activation could downregulate proinflammatory cytokines (Yang et al., 2017), ROS production (Navarro et al., 2015), and p38 MAPK phosphorylation (Uwada et al., 2020). In this study, we found that α7 nAChR activation increased ROS production and p38 MAPK phosphorylation. By RNAseq analysis, we detected that IDH1 and SUCLG1 were significantly downregulated in the GTS-21-treated ACH2 cells. It is known that IDH1 (coding isocitrate dehydrogenase-1) is an antioxidant related gene by generating NADPH(Sun et al., 2021). SUCLG1 (coding succinyl CoA ligase 1) is an enzyme in the Krebs cycle that converts succinyl-CoA to succinate and free coenzyme A, and converts ADP to ATP. Downregulation of IDH1 and SUCLG by α7 nAChR activation would disrupt Krebs cycle and NADPH production, and therefore reduce ROS removal capacity of glutathione (GSH)(Golub et al., 2019). SLC2A1 and SLC2A3 are main glucose transporters. Glucose carbons can be used for both the glycolytic and pentose phosphate (PPP) pathways to supply ATP and precursors for lipid and nucleotide synthesis, as well as for glutathione production (Seyfried et al., 2017). Thus, downregulation of SLC2A1 and SLC2A3 by α7 nAChR activation also contributes to less glutathione and higher ROS production. Under oxidative stress, TP53 was increased, which might consequently downregulate the antioxidant genes and increase ROS (Jung et al., 2013). HIV-1 viral infectivity factor interacts with TP53 to induce G2 cell cycle arrest and positively regulate viral replication (Izumi et al., 2010). H2AX is a well-known DNA damage marker. ROS and DNA damage reciprocally influence each other (Kang et al., 2012). Therefore, elevation of H2AX and TP53 also indicates that activation of α7 nAChR induces oxidative stress which facilitates HIV-1 transcription. This phenomenon has been recapitulated in the H2O2-treated ACH2 cells.
We have demonstrated that activation of α7 nAChR in macrophages and monocytes dampens proinflammatory responses (Su et al., 2007, 2010; Yang et al., 2017). However, in our unpublished studies, activation of α7 nAChR in bronchial epithelial cells and megakaryocytes promotes proinflammatory responses, which is similar to the findings in this study. Concurrently, our unpublished data suggest that activation of α7 nAChR could promote influenza and HIV-1 infection, but suppress replication of ZIKV and SARS-CoV-2. Thus, effect of activation of α7 nAChR on viral infection and proinflammatory responses depends on different cell types and pathological conditions.
The HIV-1 proviral DNA transcription occurred in the nucleus, while phosphorylation of p38 MAPK by activating α7 nAChR was mainly found in the cytoplasm. Therefore, the promotion effect of p-p38 MAPK on HIV-1 transcription might account for downstream nuclear factors. Lamins are important components of the nucleus, include Lamin A/C, Lamin B1 and Lamin B2 (de Leeuw et al., 2018; Karoutas and Akhtar, 2021). Lamin A/C tethered SUN2 to HIV-1 promoter and maintained the repressive chromatin to inhibit viral transcription and maintain viral latency (Sun et al., 2018). It has been noted that ERK and p38 kinase are important mediators of phosphorylation of histone leading to early gene expression, chromatin remodeling, and chromosome condensation (Zhong et al., 2000). p38 kinases could be recruited to the SWI-SNF chromatin-remodeling complex to regulate muscle-specific loci (Simone et al., 2004). p38 MAPK is able to regulate chromatin remodeling as well as DDB2 degradation for chromatin relaxation (Zhao et al., 2008). In the current study, activation of α7 nAChR increased the binding between p-p38 MAPK and LMNB1 causing nuclear disassembly and release of LMNB1 to the cytosol. The above process transmitted the α7 nAChR activation signaling to the nucleus for HIV-1 promotion. The LMNB1 knockdown transcriptome sequencing analysis proved this notion by increasing p38 MAPK signaling (MAPK14), DNA and chromatin damage (ATM and H2AX), and HIV-LTR binding (NFATC4, NFATC2) (Farrow et al., 2011; Rao et al., 1997) genes. Thus, p38 MAPK negatively regulates function of LMNB1 by which boosts HIV-1 transcription by NAFT family genes. The phosphorylated site of LMNB1 by p38 MAPK is worthy of further study.
We have to note that host cellular genes, KPNB1, NPM1, RCC1, SEH1L, RANBP1, and RAN, which could encode proteins interacting with Rev, were reduced in the ShLMNB1 group. Rev can mediate the export of unprocessed HIV-1 RNAs to the cytoplasm. Rev also participates HIV-1 RNA translation, stabilization, splicing and packaging (Truman et al., 2020). These findings may explain that α7 nAChR could enhance HIV-1 transcription during initiation, but did not affect the elongation (Fig. 1B).
The shortcoming of this study is that findings have not been confirmed by primary HIV-1-infected CD4+ T cells and animal models. The corresponding experiments will be conducted in the coming schedule. We have to mention that more than hundreds of papers used latent ACH2 cells in the HIV-1 study listed in the PubMed, which demonstrating that ACH2 cells are commonly recognized. In addition, our data could be produced in the Jurkat T cells as showed in Fig. 1 C-D. Two sets of RNAaeq data done in the ACH2 cells also reciprocally support our findings in Fig. 2, Fig. 3, Fig. 6.
Taken together, for the first time, we have elucidated that activation of α7 nAChR could enhance HIV-1 transcription through ROS/p-p38 MAPK/LMNB1/NFAT signaling pathway. In terms that electrical stimulation of vagus nerve can activate the CAP and inhibit inflammatory responses (Andersson and Tracey, 2012; Xie et al., 2020), our findings suggest that activation of α7 nAChR or electrical stimulation of vagus nerve might be useful for treating AIDS.
4. Materials and methods
4.1. Antibodies and compounds
Anti-phospho-p38 MAPK (Thr180/Tyr182) (9211S, Cell Signaling Technology), anti-p38α (sc-535, Santa Cruz), anti-Lamin B1(66095-1-Ig, Proteintech), anti-phospho-ERK(sc-81492, Santa Cruz), anti-phospho-JNK(sc-6254, Santa Cruz), anti-GAPDH (M20006; Abmart), anti-gag (ab63917, Abcam), anti-p24 (mouse ascites antibody), Alexa Fluor® 488 AffiniPure Goat Anti-Mouse IgG (Jackson Immunoresearch), Rhodamine(TRITC) AffiniPure Goat Anti-Rabbit IgG (Jackson Immunoresearch). α7 nicotinic acetylcholine receptor agonist: GTS-21 (ab120560, Abcam), DUSP1 and DUSP6 inhibitor: BCI (T10486, Topscience), ROS scavenger: N-Acetyl-L-methionine (T8059, Topscience).
4.2. Cells
The HIV-1 latently infected CD4+ CEM cell ACH2 was provided by Dr. Shi-Bo Jiang (Fudan University, Shanghai, China). Jurkat T cells were provided by Huan-Zhang Zhu (Fudan University, Shanghai, China). The HEK293T cells were kindly provided by Dr. Li Wu (The Ohio State University, USA). Jurkat T cells and ACH2 cells were cultured in RPMI 1640 medium (Gibco) supplemented with 10% fetal bovine serum (Gibco), 100 U/ml penicillin, and 100 μg/ml of streptomycin (Invitrogen) at 37 °C under 5% CO2. A549 (ATCC CCL-185) cells and HEK293T cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (Gibco), 100 U/ml penicillin, and 100 μg/ml of streptomycin (Invitrogen) at 37 °C under 5% CO2.
4.3. HIV-1 pseudovirus
Pseudotyped single-cycle infectious HIV-Luc/NL-3 was harvested from the co-transfection of HEK293T cells with the luciferase reporter HIV-1 proviral plasmid pLAI-Δ-env-Luc and the expression plasmid HIV-1-NL4-3 Env (CXCR4 tropic).
4.4. Real-time (RT-) qPCR
Total cellular RNA was extracted by TRIzol reagent (Invitrogen), and reverse transcribed into cDNA using a reverse transcriptase kit (Tiangen, Beijing, China). Real-time PCR was performed using the Thunderbird SYBR qPCR Mix (11203ES08; Yeasen) on the ABI QuantStudio 6 flex Real-Time PCR system. The primers were used as follows.
GAPDH: forward, 5′-ATC CCA TCA CCA TCT TCC AGG-3′ and reverse, 5′-CCT TCT CCA TGG TGG TGA AGA C-3′;
Gag: forward, 5′-GTG TGG AAA ATC TCT AGC AGT GG -3′ and reverse, 5′-CGC TCT CGC ACC CAT CTC-3′;
Initial primers targeted base pair 10–59 of the HIV-1 transcript, forward, 5′- GTT AGA CCA GAT CTG AGC CT-3′ and reverse, 5′- GTG GGT TCC CTA GTT AGC CA-3′; Proximal (Pro) primers targeted base pairs 29–180 of the HIV-1 transcript, forward, 5′- TGG GAG CTC TCT GGC TAA CT-3′ and reverse, 5′- TGC TAG AGA TTT TCC ACA CTG A-3′;
Intermediate (Int) primers targeted base pair 836–1015 of the HIV-1 transcript, forward, 5′- GTA ATA CCC ATG TTT TCA GCA TTA TC-3′ and reverse, 5′- TCTGGCCTG GTG CAA TAGG-3′;
Distal (Dis) primers targeted base pair 2341–2433 of HIV-1 transcript, forward, 5′- GAG AAC TCA AGA TTT CTG GGA AG-3′ and reverse, 5′- AAA ATA TGC ATC GCC CAC AT-3′.
4.5. RNA-Seq and RNA-Seq data analysis
The total RNA was extracted using TRIzol reagent (Invitrogen). After quality test, the total RNA of each sample was sequenced using the DNBSEQ platform (BGI BIG DATABASE (https://biosys.bgi.com/#/report/mrna/expression). Data were analyzed in online analysis software Dr. Tom. GeneCards (the Human Gene Database) (https://www.genecards.org) was used to search genes with known functions. STRING Database (https://string-db.org) was assessed to enrich the targeted genes which were differentially expressed in our study. GraphPad Prism 8.0.1 (San Diego, CA) was used to analyze difference of genes by Volcano plot.
4.6. Reactive oxygen species (ROS) assay
ROS were detected by using ROS Assay Kit-Highly Sensitive DCFH-DA kit (DOJINDO LABORATORIES). ACH2 cells were treated with compounds for 24 h and then washed twice with HBSS buffer. Add DCFH-DA Dye probe to the cells and mixed completely, cells were incubated at 37 °C for 30 min. Then wash cells for twice with HBSS buffer and detect ROS by Fortessa flow cytometer (BD Pharmingen) and analyzed with the assistance of FlowJo 7.6.1 software.
4.7. shRNAs
shRNA was cloned into the PLKO.1-puro shRNA expression vector. The packaging of shRNA lentiviruses was performed according to the PLKO.1 protocol (Addgene). Calcium phosphate-mediated transfection of HEK293T cells was used to generate shRNA lentiviruses. The targeted sequences of shRNAs:
negative control, 5′-TTCTCCGAACGTGTCACGTAT-3′;
MAPK14 shRNA, 5′-CCATGTTCAGTTCCTTATCTA-3′;
DUSP1 shRNA,5′-GAGGGTCACTACCAGTACAAG-3′
DUSP6 shRNA, 5′-CTGTGGTGTCTTGGTACATTG-3′
LMNB1 shRNA, 5′-GCTCAAAGAAGTACAGTCTTT-3′;
4.8. MAPK14 overexpression and MAPK14-Thr180/Tyr182 mutant plasmids
The full-length MAPK14 gene was cloned form HEK293T cDNA by PCR (2 × Phanta® Flash Master Mix, Vazyme), then cloned into the pCDNA3.1 plasmid by homologous recombination (ClonExpress Ultra One Step Cloning Kit, Vazyme). The MAPK14-Thr180/Tyr182 mutant plasmid was obtained by PCR point mutations (Mut Express MultiS Fast Mutagenesis Kit V2, Vazyme).
4.9. Confocal microscopy
Cells were fixed in 4% formaldehyde and permeabilized with 0.1% Triton-X 100. Then, the cells were blocked with PBS containing 5% BSA for 30 min at room temperature. Further, the cells were stained with p38 MAPK, p-p38 MAPK, LMNB1, and gag antibody, followed by secondary Alexa 488-labeled anti-mouse or Rhodamine (TRITC)-labeled anti-rabbit IgG antibodies (Jackson Immunoresearch) at room temperature. The nucleus was labeled with DAPI. Slides were imaged on a laser-scanning confocal microscope (Olympus FV-1200). Images were quantified by Image J Pro.
4.10. Co-immunoprecipitation and immunoblotting
For co-immunoprecipitation (co-IP), cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (1% protease and phosphatase inhibitor cocktail (P1049, Beyotime)) for 30 min on ice. After centrifugation for 10 min at 12,000×g, the lysates were incubated with the indicated antibody at 4 °C overnight. Protein G/A-labeled Dynabeads (Thermo Scientific) were added into each sample at 4 °C for 4 h for immunoprecipitation. Dynabeads were washed 3 times with lysis buffer. Proteins were eluted by boiling 10 min with loading buffer. The immunoprecipitates were separated by SDS-PAGE and analyzed by immunoblotting, five percent of total lysates was used as the input.
For immunoblotting, cells were lysed for 30 min at 4 °C in RIPA buffer. After centrifugation for 10 min at 12,000×g, supernatant was boiled in loading buffer and analyzed by SDS-PAGE. Specific primary antibodies were used, followed by horseradish peroxidase-conjugated goat anti-mouse IgG or goat anti-rabbit IgG (Jackson Immunoresearch) as the secondary antibodies.
4.11. Immunoprecipitation-Mass Spectrum
ACH2 cells treated with or without 40 μM GTS-21 for 48 h, cells were collected and lysed in RIPA buffer (1% protease and phosphatase inhibitor cocktail (P1049, Beyotime)) for 30 min on ice. After centrifugation for 10 min at 12,000×g, the lysates were incubated with 4 μg anti-phospho-p38 MAPK (Thr180/Tyr182) (9211S, Cell Signaling Technology) antibody at 4 °C overnight. 25 μl Protein A-labeled Dynabeads (Thermo Scientific) were added into each sample at 4 °C for 6 h for immunoprecipitation. Dynabeads were washed 3 times with lysis buffer. Proteins were eluted by boiling 10 min with loading buffer. The immunoprecipitates were separated by SDS-PAGE, each pull-down sample was run into the separation gel. We cut off the whole band as one sample and subjected it to in-gel trypsin digestion and Mass Spectrometry analysis (Performed by Orbitrap Velos Pro, Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences).
4.12. Statistical analysis
Statistics were calculated using GraphPad Prism software (GraphPad, San Diego, CA). An unpaired t-test was used unless there were multiple comparisons, in which case we used one-way ANOVA with a post hoc Bonferroni test (with a significance level of p < 0.05). The results are shown as mean ± SD.
Author contributions
J.W., J-H.W., and X.S. conceived, designed, and supervised the study and wrote the manuscript. J.W. and X.S. processed data analysis. C-Q.Z., J.C., S-T. S., Z-K.L., S-T., X. and H-X.Q. offered help for resources, validation, and discussion. All authors read and approved the contents of the manuscript.
Declaration of competing interest
The authors have declared that no competing interests exist.
Acknowledgments
This work is supported by grants from National Natural Science Foundation of China (81730001; 91942305; 81970075), Science and Technology Commission of Shanghai Municipality (20DZ2261200), Shanghai high-level local college innovation team supporting plan.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.cellin.2022.100028.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Andersson U., Tracey K.J. Reflex principles of immunological homeostasis. Annu. Rev. Immunol. 2012;30:313–335. doi: 10.1146/annurev-immunol-020711-075015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester L.Y., Capo-Velez C.M., Garcia-Beltran W.F., Ramos F.M., Vazquez-Rosa E., Rios R., Mercado J.R., Melendez R.I., Lasalde-Dominicci J.A. Up-regulation of the neuronal nicotinic receptor alpha7 by HIV glycoprotein 120: potential implications for HIV-associated neurocognitive disorder. J. Biol. Chem. 2012;287:3079–3086. doi: 10.1074/jbc.M111.262543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan P.M., Hutchings E.J., Kille N.J., Chapman J.M., Terry A.V., Jr. Positive allosteric modulator of alpha7 nicotinic-acetylcholine receptors, PNU-120596 augments the effects of donepezil on learning and memory in aged rodents and non-human primates. Neuropharmacology. 2013;67:201–212. doi: 10.1016/j.neuropharm.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capo-Velez C.M., Delgado-Velez M., Baez-Pagan C.A., Lasalde-Dominicci J.A. Nicotinic acetylcholine receptors in HIV: possible roles during HAND and inflammation. Cell. Mol. Neurobiol. 2018;38:1335–1348. doi: 10.1007/s10571-018-0603-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Li M., Shao S., Li C., Ai S., Xue B., Hou Y., Zhang Y., Li R., Fan X., et al. Nuclear peripheral chromatin-lamin B1 interaction is required for global integrity of chromatin architecture and dynamics in human cells. Protein Cell. 2020 doi: 10.1007/s13238-020-00794-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.F., Chuang H.C., Tan T.H. Regulation of dual-specificity phosphatase (DUSP) ubiquitination and protein stability. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20112668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill M.J., Deeks S.G., Margolis D.M., Siliciano R.F., Swanstrom R. HIV reservoirs: what, where and how to target them. Nat. Rev. Microbiol. 2016;14:55–60. doi: 10.1038/nrmicro.2015.5. [DOI] [PubMed] [Google Scholar]
- Daussy C.F., Galais M., Pradel B., Robert-Hebmann V., Sagnier S., Pattingre S., Biard-Piechaczyk M., Espert L. HIV-1 Env induces pexophagy and an oxidative stress leading to uninfected CD4(+) T cell death. Autophagy. 2021;17:2465–2474. doi: 10.1080/15548627.2020.1831814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw R., Gruenbaum Y., Medalia O. Nuclear lamins: thin filaments with major functions. Trends Cell Biol. 2018;28:34–45. doi: 10.1016/j.tcb.2017.08.004. [DOI] [PubMed] [Google Scholar]
- Delgado-Velez M., Baez-Pagan C.A., Gerena Y., Quesada O., Santiago-Perez L.I., Capo-Velez C.M., Wojna V., Melendez L., Leon-Rivera R., Silva W., et al. The alpha7-nicotinic receptor is upregulated in immune cells from HIV-seropositive women: consequences to the cholinergic anti-inflammatory response. Clin. Transl. Immunol. 2015;4:e53. doi: 10.1038/cti.2015.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow M.A., Kim E.Y., Wolinsky S.M., Sheehy A.M. NFAT and IRF proteins regulate transcription of the anti-HIV gene, APOBEC3G. J. Biol. Chem. 2011;286:2567–2577. doi: 10.1074/jbc.M110.154377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z.W., Li L., Huang Y.Y., Zhao C.Q., Xue S.J., Chen J., Yang Z.Z., Xu J.F., Su X. Vagal-alpha7nAChR signaling is required for lung anti-inflammatory responses and arginase 1 expression during an influenza infection. Acta Pharmacol. Sin. 2021;42:1642–1652. doi: 10.1038/s41401-020-00579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub D., Iyengar N., Dogra S., Wong T., Bready D., Tang K., Modrek A.S., Placantonakis D.G. Mutant isocitrate dehydrogenase inhibitors as targeted cancer therapeutics. Front. Oncol. 2019;9:417. doi: 10.3389/fonc.2019.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Zhao W., Hao W., Ren G., Lu J., Chen X. Cucurbitacin B induces DNA damage, G2/M phase arrest, and apoptosis mediated by reactive oxygen species (ROS) in leukemia K562 cells. Anti Cancer Agents Med. Chem. 2014;14:1146–1153. doi: 10.2174/1871520614666140601220915. [DOI] [PubMed] [Google Scholar]
- Gupta S.C., Mishra M., Sharma A., Deepak Balaji T.G., Kumar R., Mishra R.K., Chowdhuri D.K. Chlorpyrifos induces apoptosis and DNA damage in Drosophila through generation of reactive oxygen species. Ecotoxicol. Environ. Saf. 2010;73:1415–1423. doi: 10.1016/j.ecoenv.2010.05.013. [DOI] [PubMed] [Google Scholar]
- Huang B., Chen Q., Wang L., Gao X., Zhu W., Mu P., Deng Y. Aflatoxin B1 induces neurotoxicity through reactive oxygen species generation, DNA damage, apoptosis, and S-phase cell cycle arrest. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21186517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A.V., Valuev-Elliston V.T., Ivanova O.N., Kochetkov S.N., Starodubova E.S., Bartosch B., Isaguliants M.G. Oxidative stress during HIV infection: mechanisms and consequences. Oxid. Med. Cell. Longev. 2016;2016 doi: 10.1155/2016/8910396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi T., Io K., Matsui M., Shirakawa K., Shinohara M., Nagai Y., Kawahara M., Kobayashi M., Kondoh H., Misawa N., et al. HIV-1 viral infectivity factor interacts with TP53 to induce G2 cell cycle arrest and positively regulate viral replication. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20798–20803. doi: 10.1073/pnas.1008076107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H., Kim M.J., Kim D.O., Kim W.S., Yoon S.J., Park Y.J., Yoon S.R., Kim T.D., Suh H.W., Yun S., et al. TXNIP maintains the hematopoietic cell pool by switching the function of p53 under oxidative stress. Cell Metabol. 2013;18:75–85. doi: 10.1016/j.cmet.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Kang M.A., So E.Y., Simons A.L., Spitz D.R., Ouchi T. DNA damage induces reactive oxygen species generation through the H2AX-Nox1/Rac1 pathway. Cell Death Dis. 2012;3:e249. doi: 10.1038/cddis.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantner H.P., Warsch W., Delogu A., Bauer E., Esterbauer H., Casanova E., Sexl V., Stoiber D. ETV6/RUNX1 induces reactive oxygen species and drives the accumulation of DNA damage in B cells. Neoplasia. 2013;15:1292–1300. doi: 10.1593/neo.131310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karoutas A., Akhtar A. Functional mechanisms and abnormalities of the nuclear lamina. Nat. Cell Biol. 2021;23:116–126. doi: 10.1038/s41556-020-00630-5. [DOI] [PubMed] [Google Scholar]
- Liu B., Deng X., Jiang Q., Li G., Zhang J., Zhang N., Xin S., Xu K. Scoparone improves hepatic inflammation and autophagy in mice with nonalcoholic steatohepatitis by regulating the ROS/P38/Nrf2 axis and PI3K/AKT/mTOR pathway in macrophages. Biomed. Pharmacother. 2020;125 doi: 10.1016/j.biopha.2020.109895. [DOI] [PubMed] [Google Scholar]
- Ma L., Jiang Q.A., Sun L., Yang X., Huang H., Jin X., Zhang C., Wang J.H. X-linked RNA-binding motif protein modulates HIV-1 infection of CD4(+) T cells by maintaining the trimethylation of histone H3 lysine 9 at the downstream region of the 5' long terminal repeat of HIV proviral DNA. mBio. 2020;11 doi: 10.1128/mBio.03424-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis D.M., Garcia J.V., Hazuda D.J., Haynes B.F. Latency reversal and viral clearance to cure HIV-1. Science. 2016;353:aaf6517. doi: 10.1126/science.aaf6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukerjee R., Sawaya B.E., Khalili K., Amini S. Association of p65 and C/EBPbeta with HIV-1 LTR modulates transcription of the viral promoter. J. Cell. Biochem. 2007;100:1210–1216. doi: 10.1002/jcb.21109. [DOI] [PubMed] [Google Scholar]
- Navarro E., Buendia I., Parada E., Leon R., Jansen-Duerr P., Pircher H., Egea J., Lopez M.G. Alpha7 nicotinic receptor activation protects against oxidative stress via heme-oxygenase I induction. Biochem. Pharmacol. 2015;97:473–481. doi: 10.1016/j.bcp.2015.07.022. [DOI] [PubMed] [Google Scholar]
- Pereira L.A., Bentley K., Peeters A., Churchill M.J., Deacon N.J. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 2000;28:663–668. doi: 10.1093/nar/28.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramkissoon A., Chaney K.E., Milewski D., Williams K.B., Williams R.L., Choi K., Miller A., Kalin T.V., Pressey J.G., Szabo S., et al. Targeted inhibition of the dual specificity phosphatases DUSP1 and DUSP6 suppress MPNST growth via JNK. Clin. Cancer Res. 2019;25:4117–4127. doi: 10.1158/1078-0432.CCR-18-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A., Luo C., Hogan P.G. Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- Ruelas D.S., Greene W.C. An integrated overview of HIV-1 latency. Cell. 2013;155:519–529. doi: 10.1016/j.cell.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfried T.N., Yu G., Maroon J.C., D'Agostino D.P. Press-pulse: a novel therapeutic strategy for the metabolic management of cancer. Nutr. Metabol. 2017;14:19. doi: 10.1186/s12986-017-0178-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon V., Ho D.D. HIV-1 dynamics in vivo: implications for therapy. Nat. Rev. Microbiol. 2003;1:181–190. doi: 10.1038/nrmicro772. [DOI] [PubMed] [Google Scholar]
- Simone C., Forcales S.V., Hill D.A., Imbalzano A.N., Latella L., Puri P.L. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat. Genet. 2004;36:738–743. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- Singh S., Ghosh S., Pal V.K., Munshi M., Shekar P., Narasimha Murthy D.T., Mugesh G., Singh A. Antioxidant nanozyme counteracts HIV-1 by modulating intracellular redox potential. EMBO Mol. Med. 2021;13 doi: 10.15252/emmm.202013314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Lee J.W., Matthay Z.A., Mednick G., Uchida T., Fang X., Gupta N., Matthay M.A. Activation of the alpha7 nAChR reduces acid-induced acute lung injury in mice and rats. Am. J. Respir. Cell Mol. Biol. 2007;37:186–192. doi: 10.1165/rcmb.2006-0240OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Matthay M.A., Malik A.B. Requisite role of the cholinergic alpha7 nicotinic acetylcholine receptor pathway in suppressing Gram-negative sepsis-induced acute lung inflammatory injury. J. Immunol. 2010;184:401–410. doi: 10.4049/jimmunol.0901808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Guo Y., Fan Y., Wang Q., Zhang Q., Lai D. Decreased expression of IDH1 by chronic unpredictable stress suppresses proliferation and accelerates senescence of granulosa cells through ROS activated MAPK signaling pathways. Free Radic. Biol. Med. 2021;169:122–136. doi: 10.1016/j.freeradbiomed.2021.04.016. [DOI] [PubMed] [Google Scholar]
- Sun W.W., Jiao S., Sun L., Zhou Z., Jin X., Wang J.H. SUN2 modulates HIV-1 infection and latency through association with lamin A/C to maintain the repressive chromatin. mBio. 2018;9 doi: 10.1128/mBio.02408-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman C.T., Jarvelin A., Davis I., Castello A. HIV rev-isited. Open Biol. 2020;10 doi: 10.1098/rsob.200320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwada J., Nakazawa H., Mikami D., Islam M.S., Muramatsu I., Taniguchi T., Yazawa T. PNU-120596, a positive allosteric modulator of alpha7 nicotinic acetylcholine receptor, directly inhibits p38 MAPK. Biochem. Pharmacol. 2020;182 doi: 10.1016/j.bcp.2020.114297. [DOI] [PubMed] [Google Scholar]
- Wang H., Yu M., Ochani M., Amella C.A., Tanovic M., Susarla S., Li J.H., Wang H., Yang H., Ulloa L., et al. Nicotinic acetylcholine receptor alpha7 subunit is an essential regulator of inflammation. Nature. 2003;421:384–388. doi: 10.1038/nature01339. [DOI] [PubMed] [Google Scholar]
- Wang X., Liu J.Z., Hu J.X., Wu H., Li Y.L., Chen H.L., Bai H., Hai C.X. ROS-activated p38 MAPK/ERK-Akt cascade plays a central role in palmitic acid-stimulated hepatocyte proliferation. Free Radic. Biol. Med. 2011;51:539–551. doi: 10.1016/j.freeradbiomed.2011.04.019. [DOI] [PubMed] [Google Scholar]
- Wen J., Li X., Zhao Q.X., Yang X.F., Wu M.L., Yan Q., Chang J., Wang H., Jin X., Su X., et al. Pharmacological suppression of glycogen synthase kinase-3 reactivates HIV-1 from latency via activating Wnt/beta-catenin/TCF1 axis in CD4(+) T cells. Emerg. Microb. Infect. 2022;11:391–405. doi: 10.1080/22221751.2022.2026198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J.M., Shin D.Y., Lee S.J., Joe Y., Zheng M., Yim J.H., Callaway Z., Chung H.T. Curcumin protects retinal pigment epithelial cells against oxidative stress via induction of heme oxygenase-1 expression and reduction of reactive oxygen. Mol. Vis. 2012;18:901–908. [PMC free article] [PubMed] [Google Scholar]
- Xie H., Yepuri N., Meng Q., Dhawan R., Leech C.A., Chepurny O.G., Holz G.G., Cooney R.N. Therapeutic potential of alpha7 nicotinic acetylcholine receptor agonists to combat obesity, diabetes, and inflammation. Rev. Endocr. Metab. Disord. 2020;21:431–447. doi: 10.1007/s11154-020-09584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.G., Zhai Y.X., Chen J., Lu Y., Wang J.W., Quan C.S., Zhao R.X., Xiao X., He Q., Werle K.D., et al. LKB1 reduces ROS-mediated cell damage via activation of p38. Oncogene. 2015;34:3848–3859. doi: 10.1038/onc.2014.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Zhao C., Chen X., Jiang L., Su X. Monocytes primed with GTS-21/alpha7 nAChR (nicotinic acetylcholine receptor) agonist develop anti-inflammatory memory. QJM. 2017;110:437–445. doi: 10.1093/qjmed/hcx014. monthly journal of the Association of Physicians. [DOI] [PubMed] [Google Scholar]
- Zhang N., Huang L., Tian J., Chen X., Ke F., Zheng M., Xu J., Wu L. A novel synthetic novobiocin analog, FM-Nov17, induces DNA damage in CML cells through generation of reactive oxygen species. Pharmacol. Rep. : PR. 2016;68:423–428. doi: 10.1016/j.pharep.2015.11.002. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Barakat B.M., Qin S., Ray A., El-Mahdy M.A., Wani G., Arafa el S., Mir S.N., Wang Q.E., Wani A.A. The p38 mitogen-activated protein kinase augments nucleotide excision repair by mediating DDB2 degradation and chromatin relaxation. J. Biol. Chem. 2008;283:32553–32561. doi: 10.1074/jbc.M803963200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Wilson K., Uteshev V., He J.J. Activation of alpha7 nicotinic acetylcholine receptor ameliorates HIV-associated neurology and neuropathology. Brain. 2021;144:3355–3370. doi: 10.1093/brain/awab251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S.P., Ma W.Y., Dong Z. ERKs and p38 kinases mediate ultraviolet B-induced phosphorylation of histone H3 at serine 10. J. Biol. Chem. 2000;275:20980–20984. doi: 10.1074/jbc.M909934199. [DOI] [PubMed] [Google Scholar]
- Zhu J., Gaiha G.D., John S.P., Pertel T., Chin C.R., Gao G., Qu H., Walker B.D., Elledge S.J., Brass A.L. Reactivation of latent HIV-1 by inhibition of BRD4. Cell Rep. 2012;2:807–816. doi: 10.1016/j.celrep.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.