ABSTRACT
The human gut microbiota continues to demonstrate its importance in human health and disease, largely owing to the countless number of studies investigating the fecal microbiota. Underrepresented in these studies, however, is the role played by microbial communities found in the small intestine, which, given the essential function of the small intestine in nutrient absorption, host metabolism, and immunity, is likely highly relevant. This review provides an overview of the methods used to study the microbiota composition and dynamics along different sections of the small intestine. Furthermore, it explores the role of the microbiota in facilitating the small intestine in its physiological functions and discusses how disruption of the microbial equilibrium can influence disease development. The evidence suggests that the small intestinal microbiota is an important regulator of human health and its characterization has the potential to greatly advance gut microbiome research and the development of novel disease diagnostics and therapeutics.
KEYWORDS: small Intestine, microbiota, microbiome, metabolism, immunity, human health
Introduction
Trillions of bacteria, fungi, viruses, and archaea take residence in our gastrointestinal (GI) tract. These gut-based microorganisms, better known as the gut microbiota, are crucial for human health and perturbations in their preferred assembly have been associated with an extensive range of pathologies.1 It is therefore of no surprise that the gut microbiota continues to command the attention of both scientific researchers and the health industry alike.
Although referred to broadly as the gut microbiota, the human intestine harbors distinct communities of microbes – microbial niches – along its length.2 With more and more research being conducted, it is becoming increasingly apparent that these microbial niches establish unique interactions with the host and perform different functions within the body. This is significant, considering that the majority of studies investigating the gut microbiota are based on fecal samples – which more accurately represent the distal intestinal (colonic) microbiota – leaving the functions performed by the microbes in the proximal parts of the intestinal tract largely unknown. Given that the small intestine (SI) is essential for nutrient absorption from the diet, as well as provides an extensive and favorable site for important immune, metabolic, neural, and endocrine responses to take place, it is highly plausible that its microbiota is equally fundamental.
Evidence is indeed beginning to emerge that the SI microbiota play a key role in human health and disease. This review discusses the challenges and current methods of studying the SI microbiota, what is known about its composition and temporal dynamics, and finally its potential role in host metabolism, immunity, and disease.
Studying the human SI microbiome
Studying the SI microbiome in humans is challenging due to its poor accessibility and high temporal dynamics. Many studies currently rely on routine endoscopies, intestinal resections, or sudden death victims, to obtain SI samples.3–9 These sampling methods, however, have a number of limitations (Table 1). Firstly, endoscopic procedures are invasive, can only be performed by a specialist, and are therefore not easily justified for individuals without GI-related symptoms. Secondly, the lavage treatment that precedes endoscopic procedures may disrupt the endogenous intestinal environment, introducing bias. Thirdly, these methods are prone to contamination from other parts of the intestinal tract during retrieval of the sample. Furthermore, longitudinal samples are difficult to acquire. Lastly, material obtained during these procedures is predominantly mucosal biopsies. Analysis of the mucosa-associated microbiota is limited by 16S rRNA sequencing, which cannot compete with the higher taxonomic and functional resolution achieved with metagenomic shotgun sequencing. Individuals with an ileostomy or ileal pouch-anal anastomosis provide a unique cohort in which to non-invasively and longitudinally sample the SI lumen, circumventing many of the aforementioned problems.10–12 However, the limited number of individuals and disease context pose alternative challenges. An altered anatomy and possible exposure of the luminal contents to the external environment are also suggested limitations; however, according to the findings in the recently published study of Yilmaz et al., the intestinal microbial content is not significantly perturbed as a result of an ileostomy.13 Additional methods of sampling the SI microbiota include the novel method detailed by Dreskin et al. for simultaneously sampling the proximal luminal and mucosal gut microbiome, without the risk of contamination that is usually associated with regular endoscopies.14 Sampling via this method, however, is limited to the proximal SI and is still invasive. Similar restrictions also apply to the 4-lumen catheter with multiple aspiration ports described in the study of Seekatz et al.15 Attempts are also being made to develop endoscopic capsules that allow noninvasive, no contamination sampling along the entire GI tract, with some promising results.16,17
Table 1.
Studies sampling the SI microbiota and their methods.
| Study cohort | Sampling method | Location in SI | Microbial analysis | Limitations | Main Species Identified | Reference |
|---|---|---|---|---|---|---|
| 54-y-old female, previous good health | Mucosal biopsies via colonoscopy and using a Watson intestinal biopsy capsule | Jejunum Distal ileum |
16S rRNA gene amplicon sequencing |
|
Jejunum: Streptococcus & Proteobacteria Distal ileum: Bacteroidetes & Clostridium clusters XIVa and IV |
Wang et al. 2005 [3] |
| 26 patients who underwent emergency surgery requiring colonic resection | Mucosal biopsies following colonic resection | Terminal ileum | 16S rRNA gene amplicon sequencing |
|
Bacteroides, Bifidobacteria, Lactobacilli & Enterobacteria | Ahmed et al. 2007 [4] |
| 3 deceased elderly individuals | Intestinal contents obtained during autopsy | Jejunum Ileum |
16S rRNA gene amplicon sequencing |
|
Streptococcus, Lactobacillus, ‘Gammaproteobacteria’, the Enterococcus group & the Bacteroides group | Hayashi et al. 2005 [5] |
| 5 individuals with an ileostomy & 4 healthy individuals |
Ileostomy effluent Intestinal fluid using an intraluminal naso-ileal catheter |
(Distal) SI Jejunum, ileum & terminal |
GI tract-specific phylogenetic microarray HITChip |
|
Streptococcus, E.coli, Clostridium, high G+C organisms | Zoetendal et al. 2012 [12] |
| 5 participants undergoing colonoscopy | Mucosal biopsies via colonoscopy | Terminal ileum | Metagenomic shotgun sequencing |
|
B. vulgatus, B. thetaiotaomicron, B. uniformis & B. caccae | Vaga et al. 2020 [6] |
| Liver disease patients undergoing esophagogastroduodenoscopy | Duodenal aspirate and biopsies during upper endoscopy – novel technique detailed by the authors | Duodenum | 16S rRNA gene amplicon sequencing |
|
Aspirate: Veillonella, Streptococcus, Prevotella Biopsy: Streptococcus, S24–7 (Muribaculaceae), Parabacteroides |
Dreskin et al. 2021 [14] |
| 49 individuals with an ileostomy 9 individuals with an Ileoanal pouch |
Ileostomy effluent Fecal sample |
SI | Metagenomic shotgun sequencing |
|
Streptococcus, Escherichia, Veillonella, Blautia, Actinomyces, Enterococcus, Lactobacillus | Ruigrok et al. 2021 [10] |
| 27 patients undergoing radical cystectomy | Lab cultivation of samples obtained by rubbing a swab against the ileal mucosa 25 cm from the ileocecal valve | Ileum | MALDI-TOF/Biotyper |
|
viridans streptococci, Candida, Actinomyces, Rothia & Lactobacillus species. | Villmones et al. 2021 [7] |
| 8 fasted healthy individuals | Orally intubation with a four-lumen catheter with multiple aspiration ports – multiple samples | Duodenum Jejunum |
16S rRNA gene amplicon sequencing |
|
Firmicutes, Proteobacteria & Bacteroidetes (duodenum) | Seekatz et al. 2019 [15] |
| Ex vivo study on lamb SI | A novel capsule robot designed by the authors: samples mucosa and intestinal contests | SI | / |
|
/ | Rehan et al. 2020 [16] |
| 27 patients undergoing radical cystectomy with urinary diversion | Sample obtained valve by rubbing a swab against the luminal wall about 25 cm proximal to the ileocecal during surgery | Distal ileum | 16S rRNA gene amplicon sequencing plus targeted sequencing of alternative genes when necessary for higher taxonomic resolution |
|
Streptococcus, Granulicatella, Actinomyces, Solobacterium, Rothia, Gemella & TM7(G-1) | Villmones et al. 2018 [8] |
| 6 patients undergoing gastric bypass surgery | Sample obtained valve by rubbing a swab against the luminal wall during surgery | Jejunum | 16S rRNA gene amplicon sequencing |
|
Streptococcus, Granulicatella, Schaalia odontolytica complex & Gemella | Villmones et al. 2022 [9] |
| 7 individuals with an ileostomy | Ileostomy effluent | SI | HITChip based on SSU rRNA gene sequences |
|
Streptococcus, Veillonella & Clostridium cluster I | Booijink et al. 2010 [11] |
| 30 individuals with an ileostomy | Ileostomy effluent | SI | 16S rRNA gene amplicon sequencing |
|
Lactobacillus, Clostridium, Streptococcus, Enterococcus, & Veillonella |
Yilmaz et al. 2022 [13] |
Abbreviations: SI, small intestine.
NB: Includes studies that also sampled the large intestine; however, only the methods used to study the SI and the corresponding results are detailed.
In addition to sampling, efforts have been made to model the SI both in vitro and in vivo. A potentially promising in vitro model for the human ileum and its microbiota is the single-stage fermenter developed by Stolaki et al.18 In vivo model studies have largely involved mice. Although differences between mouse and human GI tract and microbial composition are known – such as tract length in relation to the size of the species19 - these studies have allowed for functional exploration into the role of the SI microbiota in both healthy and disease-specific contexts. Future studies could also consider using piglet models given they have a more anatomically and physiologically similar GI tract to humans than mice.20
Overall, while the importance of the SI microbiota is gaining support and progress is being made to study its composition and function, there is still a need to develop uniform techniques that accurately model its dynamics.
The composition and temporal dynamics of the human SI microbiota
Despite the differences in study populations, sampling techniques, and analytic methods that complicate the process of characterizing the human SI microbiota, studies appear to agree on the main characteristics of this microbial ecosystem.
The microbial density in the SI microbiota is estimated at 103 - 108 cells/g, with an increasing gradient going from a low density in the duodenum (103) to a high density in the terminal ileum (108) which is still approximately 4-fold lower than in the colon.21 This increasing gradient reflects the more favorable conditions – lower acidity and lower concentrations of digestive enzymes and pancreatic, gastric, and bile acids – found in the terminal ileum compared with the duodenum and jejunum.2,22 The microbial ecosystem in the SI is also in general less diverse and more dynamic than in the colon11; to what extent, it depends on the location along the SI and whether one is describing the luminal content or mucosa-associated microbiota. The mucosal diversity in the ileum and colon, for example, do not differ significantly, whereas when comparing the luminal contents this difference is more pronounced.23 Spatial and temporal analysis of the SI microbiota has also shown that there is higher inter- and intra-individual variability in duodenal aspirates compared with those from the jejunum, suggesting that the duodenal microbial community is more dynamic.15
The SI microbiota is composed primarily of facultative anaerobic and aerobic bacteria, in particular, belonging to the phyla Firmicutes, Proteobacteria and Actinobacteria.3,5,8,10,12,14,15 This is in contrast to the obligate anaerobes that predominate in the colon.3,5,12 At the genus level, bacteria highly abundant in the SI include Lactobacillus, Veillonella, Streptococcus, Gemella, Actinomyces, and E. coli.3,5,8,10,12,14,15 Higher proportions of Bacteroidetes, namely Prevotella, and lower proportions of Firmicutes (Streptococcus, Veillonella, Gemella, and Lactobacillaceae) are found in the duodenum compared with the jejunum.15 Bacterial compositions in the ileum and jejunum appear to be comparable, despite a higher bacterial load in the ileum.2,8,9 Only select bacteria, including Clostridium, Lactobacillus, and Enterococcus species, are able to penetrate, or attach to, the mucus layer at the epithelium and use the mucus as an energy source, explaining some of the differences seen between the bacterial communities in the lumen versus the mucosa.24,25
The composition of the SI microbiota is also influenced by external factors. Two important factors are smoking and proton pump inhibitor (PPI) therapy.26,27 Smoking has been shown to disrupt both the duodenal mucosa-associated and luminal microbiota.26,27 In the lumen, the abundance of Prevotellaceae, Neisseriaceae, and Porphyromonadaceae is reduced in smokers, whereas Enterobacteriaceae and Lactobacillaceae abundances are increased. Smoking cessation eventually restores microbial composition to the extent that it more closely resembles that of a never-smoker. PPIs are a commonly prescribed drug for the prevention or treatment of gastric-acid related diseases, such as stomach ulcers and reflux complaints. PPIs inhibit gastric acid secretion, raising stomach pH and allowing certain microbes to thrive and colonize more distal sections of the GI tract. In the study of Lim et al., PPI use was shown to induce duodenal microbiota dysbiosis, characterized by an increase in Akkermansia muciniphila and Porphyromonas endodontalis and a decrease in Enterococcaceae, Coprococcus, Enterobacteriaceae, and Synergistes species.27
Rapid changes in the nutrient availability also acts as a factor shaping the microbiota composition along the small intestine. Using luminal stoma (ileo- and colostomy) contents from 114 patients with IBD or colorectal cancer, Yilmaz and colleagues demonstrated dynamic changes in the ileostomy-derived bacterial biomass and strain profiles following dietary intake,which was in contrast to the more stable microbiota of colostomy-derived samples.13
Lastly, it should be noted that belonging to SI microbiota are also viruses, archaea, and fungi. Although almost completely unexplored in the context of the SI, these microbes are increasingly being studied in the context of the fecal microbiome and are proving highly important for not only shaping the microbiota but also maintaining health and driving disease.28,29 Using ileostomy fluid and fresh colon resections from non-IBD and IBD patients to study the protective or pro-inflammatory potential of the viruses residing in the gut, the study of Adiliaghdam and colleagues gives us a first glimpse into the SI virome.30 The results revealed that eukaryotic viral families present in the SI include Anelloviridae, Papillomaviridae, Picornaviridae, and Virgaviridae, and bacteriophage families include Caudovirales, Siphoviridae, Myoviridae, and Podoviridae. Additional studies, however, are necessary to confirm these findings and define, if existing, a core SI virome. Given the paucity of studies describing the non-bacterial component of the SI microbiota, this review will henceforth continue to focus on the SI bacteriome.
The SI microbiota and host metabolic regulation
In addition to being the primary site of nutrient and energy absorption from the diet, the SI performs a number of other functions that are critical for host metabolism. In line with this, studies exploring the potential function of the SI microbiota using computational analyses have revealed an enrichment of bacterial pathways associated with energy and simple carbohydrate metabolism.10,12 In this section, we discuss the role of the SI in regulating host metabolism and the emerging evidence that places the SI microbiota in this role (Figure 1).
Figure 1.
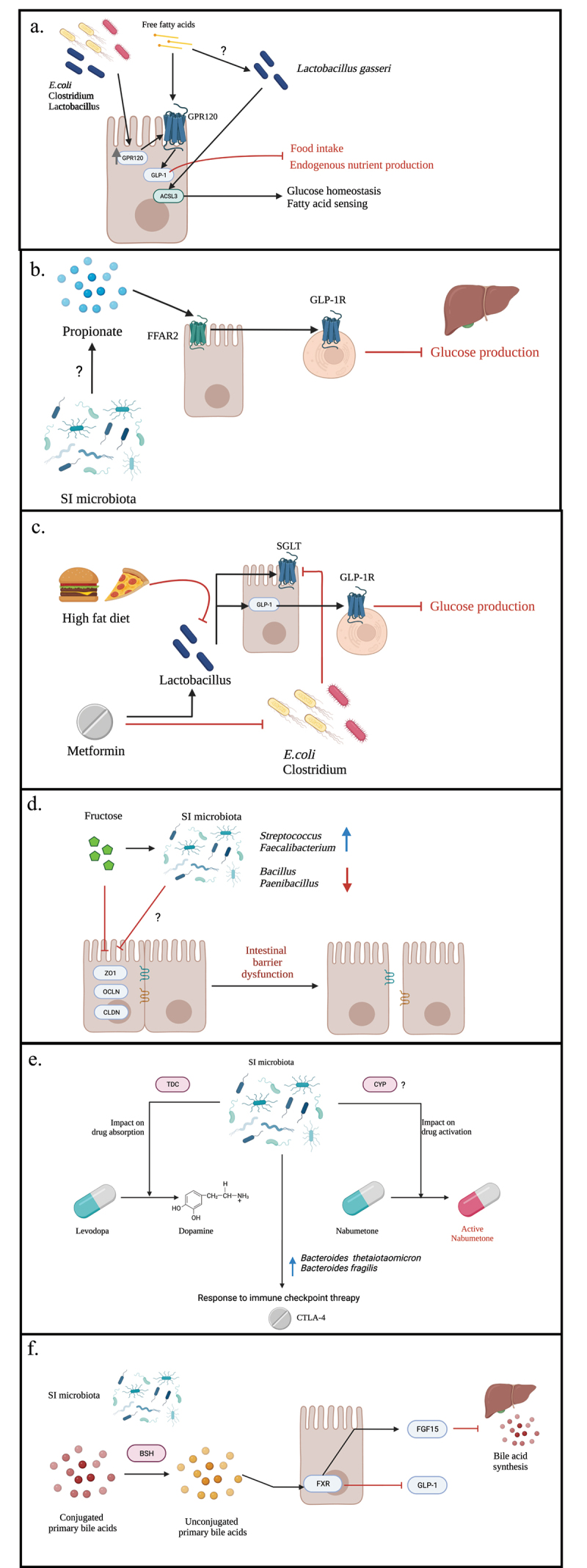
Interactions of small intestinal microbes and host metabolism.
Note: A schematic overview of the concepts discussed throughout this review regarding the role of the small intestinal microbiota in host metabolism. (a-c) nutrient sensing and energy homeostasis: a) SI bacteria can influence energy homeostasis and nutrient sensing through upregulation of GPR120 expression which, upon activation by free fatty acids, stimulates GLP-1 secretion. GLP-1 secretion in turn leads to decreased energy intake and endogenous nutrient production. SI bacteria may also facilitate fatty acid nutrient sensing and glucose homeostasis through increased ACSL3 protein expression and downstream signaling leading to decreased glucose production. b) It is proposed that SI bacteria can produce short-chain fatty acids (represented in the figure by propionate) which, via an FFAR2/GLP-1R dependent pathway, regulates glucose metabolism. c)A high fat diet and metformin modulate the SI microbiota in opposing ways which influence glucose metabolism. (d) fructose metabolism: excessive fructose intake is associated with reduced tight junction gene expression (ZO1, OLCN, CLDN) possibly mediated by (fructose-induced) SI microbial dysbiosis, leading to intestinal barrier dysfunction. (e) drug interactions:SI microbes are involved in the bioavailability of certain prescription drugs, via endogenous enzyme activity (TDC and CYP, respectively). Similarly, SI bacterial abundances are associated with the efficacy of anti-tumor treatments, namely CTLA-4 immunotherapy. (f) bile acid metabolism:SI microbes regulate bile acid pools in the small intestine via BSH enzyme activity, which in turn regulates bile acid synthesis and host/lipid metabolism. Figure created using BioRender (biorender.com)
Nutrient sensing and glucose/energy homeostasis
The SI has the ability to sense nutrients and trigger both local and systemic, hormonal, and neurological responses in the body that regulate energy and glucose homeostasis.31 Distributed throughout the intestinal epithelium – but of specific importance, here, in the SI – are hormone-secreting enteroendocrine cells (EECs)32 which, upon exposure to nutrients, secrete a variety of gut peptides including the hormones glucagon-like peptide 1 (GLP-1) and cholecystokinin (CCK) .33,34 These secretory products in turn act in an endocrine or paracrine fashion via the gut–brain metabolic axis and/or portal vagal-brain axis to prevent postprandial excess energy by suppressing food intake and endogenous nutrient production.31,35–37
Tahmasebi and colleagues were able to show that propionate infusion into the ileum of rats activates ileal mucosa-located free fatty acid receptor 2 (FFAR2).38 FFAR2 activation in turn triggers a negative feedback signal that, via a GLP-1 receptor-dependent neural network, results in reduced hepatic glucose production. Propionate is a short-chain fatty acid (SCFA) produced by bacterial dietary fiber fermentation. Although SCFAs are primarily synthesized by bacteria in the large intestine,39 these results suggest that ileal-derived fatty acids are important for GLP-1 release and glucose homeostasis. In line with this, supernatant from bacteria commonly found in the SI (Lactobacillus, Enterococcus, E. coli) have been demonstrated to increase G protein-coupled receptor 120 (GPR120) expression which, upon stimulation by free fatty acids, promotes secretion of GLP-1.40 Furthermore, changes to the upper SI microbiota composition in rats, as a result of a high-fat diet (HFD), were associated with disrupted upper SI fatty acid nutrient sensing and glucose homeostasis, driven by a decrease in long-chain fatty acyl-CoA synthase (ACSL3) expression.34 Direct transplantation of upper SI microbiota derived from rats fed a regular chow diet, as well as L. gasseri probiotic administration, reverses this phenotype, suggesting that SI bacteria are critical for the ACSL3-dependent regulation of glucoregulatory fatty acid-sensing pathways (Figure 1 a-b).
The role of microbes in SI nutrient sensing is further supported by the study of Bauer et al., investigating the glucose production reducing effect of diabetic medication, metformin.41 Metformin had previously been shown to increase upper SI GLP-1 secretion, stimulate upper SI sodium glucose cotransporter-1 (SGLT1) expression which regulates GLP-1 secretion, and lower glucose production through a GLP-1 R dependent neuronal axis, when infused into the upper SI.42–45 Furthermore, metformin intake was associated with alterations to the distal gut microbiota46, leading Bauer and colleagues to investigate whether changes to the upper SI microbiota also play a role in the glucoregulatory effects of metformin treatment.41 First, they confirmed that rats fed a HFD for as little as 3 days had disrupted upper SI glucose sensing, characterised by reduced SGLT1 expression and GLP-1 secretion. They then showed that an upper SI microbiota transplant from metformin-treated donor rats, leading to an increase in Lactobacillus salivarius and a decrease in Clostridium and E. coli in the upper SI, was able to restore the defective glucose sensing in HFD fed rats. Taken together, these results suggest that the glucoregulatory effects of metformin are partially driven by compositional changes in the upper SI microbiota, including reversal of the HFD-induced decrease in Lactobacillus species (Figure 1c).
Circadian rhythms and metabolism
Circadian rhythms are the physical, mental, and behavioral changes occurring within an organism that continuously cycles over a 24-hour period.47 Circadian rhythms are driven by a 24-hour internal clock – the circadian clock – found in almost all cells of the body, which is in turn driven by the light-regulated, ‘central’ circadian clock located in the brain.48 ‘Peripheral’ circadian clocks can also be influenced by factors other than the central clock, such as the time of food consumption, which regulates liver and intestinal circadian rhythms.49,50 Circadian rhythms are important for maintaining cellular and organ function, and dysregulation of the rhythms can lead to organ dysfunction. The link between circadian rhythms and host metabolism is well acknowledged47 and the SI and its microbiota appear to play an important role here.51 Previous studies had shown that diurnal fluctuations in intestinal microbial composition are necessary to the entrain peripheral circadian clocks that, in turn, maintain host metabolic homeostasis by inducing diurnal expression of hepatic and intestinal metabolic regulators of glucose, cholesterol, and fatty acid metabolism.48,52–54 However, these findings are based primarily on fecal samples, largely representing the large intestine. Dantas Machado and colleagues therefore focused on the process of peripheral circadian rhythm entrainment in the ileum.51 Here, they demonstrated that diurnal oscillations in ileal microbial composition and host ileal transcriptome exist, and both are disrupted in diet-induced obese mice, leading to perturbed peripheral circadian rhythms and dysmetabolism. Interestingly, time-restricted feeding was able to restore the ileal diurnal microbial dynamics, increase GLP-1 production and influence bile acid pools and signaling, suggesting that microbial dynamics in the SI are important for regulating host metabolism, and dietary/feeding patterns can significantly influence these dynamics.
Bile acid metabolism
The central role of the SI in bile acid metabolism is another important aspect of the SI’s regulatory function of host metabolism. Bile acids represent a key group of signaling molecules in the gut that primarily facilitate absorption of lipids and fat-soluble nutrients in the SI, but also regulate lipoprotein, glucose, and energy metabolism.55,56 Bile acids carry out their signaling functions by binding to nuclear receptor, farnesoid X receptor (FXR) and membrane-bound G protein-coupled bile acid receptor (TGR5). Both receptors are highly expressed in the ileum, where 90% of bile acids get reabsorbed. Constitutive bile acid-mediated activation of intestinal FXR has been shown to worsen obesity and increase insulin resistance and fatty liver.57–59 In the mouse study conducted by Jiang et al., FXR signaling inhibition reversed the metabolic dysfunctions associated with a high-fat diet and a genetic predisposition for metabolic related diseases, including obesity, insulin resistance, and fatty liver.60 Moreover, FXR signaling was enriched in the distal ileal mucosa of obese individuals, and mRNA levels of FXR signaling pathway molecules: fxr, small heterodimer partner (shp) and fibroblast growth factor 19 (fgf19), correlated with body mass index in these same individuals. FXR activation inhibits GLP-1 transcription and secretion by intestinal epithelial L-cells61, suggesting the beneficial metabolic effects of FXR inhibition involve increased intestinal GLP-1 production. In line with this, are the findings reported by Thomas and colleagues from their work in obese mice. TGR5 signaling was shown to stimulate intestinal GLP-1 release, which resulted in improved liver and pancreatic function with increased glucose tolerance in obese mice.62 These results emphasize the role of bile acid signaling in the intestinal regulation of energy and glucose homeostasis.
The gut microbiota is a key regulator of bile acid metabolism and signaling. Primary bile acids (PBAs), namely cholic acid (CA) and chenodeoxycholic acid (CDCA) in humans, are produced in the liver.56,63 PBAs are subsequently conjugated with glycine or taurine – making them water soluble – and secreted into the bile where they are either stored in the gallbladder or directly released into the SI for lipid and fat-soluble nutrient absorption. Once in the SI, the fate of PBAs is variable. Some PBAs are passively absorbed in the SI in their conjugated form, others are first deconjugated by SI bacteria before being passively reabsorbed, the majority are actively reabsorbed in the terminal ileum and approximately 5% continue on into the colon where they are converted by colonic bacteria into secondary bile acids (SBAs) and excreted in the feces. A small proportion of SBAs are also reabsorbed in the colon. Intestinal microbes are therefore critical for modulating host bile acid pools, which has an influence on host physiology. More detail on this microbial-dependent regulation of bile acid pools can be found in the review of Winston and Theriot63. The effect of SI bacteria on bile acid signaling and host metabolism is discussed further in this review in the context of metabolic diseases in the section ‘The SI microbiota and disease’.
Fructose metabolism
A more recently discovered function of the SI in fructose metabolism also warrants attention considering the strong correlation between high fructose intake and metabolic-related diseases such as obesity, type 2 diabetes, nonalcoholic fatty liver disease, kidney dysfunction, and cardiovascular disease.64 The conversion of fructose to glucose was previously assumed to takes place in the liver, on the basis that high levels of fructose catabolic enzymes are expressed in the liver, and the liver is highly sensitive to fructose leading to fatty liver disease.64,65 Jang and colleagues, however, revealed that the SI is in fact responsible for the conversion of fructose to glucose.66 This is hypothesized to play a protective role against fructose toxicity in the liver, provided fructose intake does not exceed the SI’s metabolic capacity. If fructose intake exceeds this threshold, fructose becomes available for intestinal bacteria, which, at least in the colon, can convert fructose into potentially hepatotoxic metabolites. Excess fructose intake may also drive intestinal barrier dysfunction, as a result of SI microbial dysbiosis.20 Fructose supplementation in piglets led to reduced ileal expression of three main tight junction genes, ZO1, OCLN, and CLDN, which correlated with decreased Bacillus and Paenibacillus (aerobes) and increased Streptococcus and Faecalibacterium (anaerobes) relative abundances in the ileum.20 Investigating the role of SI bacteria in fructose metabolism could therefore have significant implications for metabolic health (Figure 1d).
Drug bioavailability and response
The SI is also an important site for drug absorption, having a significant influence on drug efficacy. Bacteria in the gut can also metabolize drugs; however, studies have mainly focused on the metabolism occurring in the colon and, thus, the proportion of drugs that are not absorbed by the host.67 Bacteria in the SI, on the other hand, have the opportunity to influence the bioavailability of orally administered drugs by modulating the drug before the host has had a chance to absorb it. In the study performed by van Kessel and colleagues, fecal samples from patients with Parkinson’s disease (PD) and rats were used to investigate the possible modulation of levodopa by gut microbes.68 Levodopa, in combination with a decarboxylase inhibitor, is the primary treatment for PD – a degenerative disorder of the central nervous system. Treatment, however, is not always successful with a number of patients either not responding or having to increase their dosage regimen due to a decrease in treatment response over time. Given that the proximal SI (jejunum) is the primary site for levodopa absorption, and levodopa closely resembles the substrate of bacterial tyrosine decarboxylases (TDC),69–71 jejunal bacteria were hypothesized to mediate this reduced response to treatment. Van Kessel and colleagues observed that proximal SI bacteria, via tyrosine decarboxylase activity, were indeed able to efficiently convert levodopa to dopamine – the active form that is unable to cross the blood–brain barrier and exert its disease-modulating effects. The results implicate jejunal bacteria in the bioavailability of levodopa, providing a possible explanation as to why differences in the efficacy of levodopa are seen between patients with PD. Similarly, gut microbes are also linked to the metabolism of the nonsteroidal anti-inflammatory drug, nabumetone.72 Germ-free (GF) mice, compared with specific pathogen-free mice, exhibit increased small intestinal expression of drug metabolizing, cytochrome P450 (CYP) enzymes following oral administration of nabumetone. Cytochromes P450 convert nabumetone to its active form, highlighting the importance of gut microbes, plausibly specifically those in the SI, in regulating the effects of (nabumetone) drug treatment (Figure 1e).
In addition to drug bioavailability, the microbiota is related to drug response possibly via an interaction with the immune system. Antibodies targeting the immune checkpoint receptor protein and negative regulator of T cell activation, CTLA-4, have been successfully used to treat various cancers and improve survival outcomes, especially in the case of melanoma.73 The tumor fighting effect of CTLA-4 antibodies was studied in relation to gut microbes by Vétizou et al.74 Following injection of the CTLA-4 antibodies in wild-type mice, Vétizou and colleagues observed an increase in the relative abundance of Bacteroides thetaiotaomicron and Bacteroides fragilis at the SI mucosa, which was positively associated with CTLA-4 efficacy. These antitumor effects were absent or reduced in GF mice and mice treated with antibiotics, which were restored upon recolonization with B. thetaiotaomicron and B. fragilis. The role of SI microbes, therefore, also likely extends to the immunostimulatory effects of cancer immunotherapy, which if investigated further could tremendously enhance cancer therapy (Figure 1e).
The SI microbiota and immunity
The gut-associated lymphoid tissue (GALT) constitutes approximately 70% of the entire immune system in the human body, and the largest component is located in the SI.75 This immune tissue is not only critical for the maintenance of intestinal homeostasis but also protects the body against external factors that are able to penetrate the intestinal mucus barrier. Key to its role in maintaining intestinal immune homeostasis, GALT must sustain an equilibrium between, on the one hand, tolerance for diet-derived antigens and commensal microbes and, on the other hand, immunity toward pathogenic stimuli.76 Failure to maintain this equilibrium underlies the development of intestinal and extraintestinal immune-related disorders, such as type 1 diabetes, celiac disease, and inflammatory bowel disease (IBD). GALT has also more recently been linked to the development and organization of the enteric nervous system, which is closely linked to the central nervous system.77 Intestinal muscularis macrophages in particular have been shown to play a significant role here. Mice depleted of muscularis macrophages show gut dysmotility and a less-organized enteric nervous system.
GALT can be divided into three distinct components distributed within the different layers of the intestinal wall: diffuse lymphoid tissue within the lamina propria, isolated lymphocytes embedded within the epithelium (intraepithelial lymphocytes, IELs) and organized lymphoid follicles such as Peyer’s patches in the ileum.78 The diffuse lymphoid tissue is mainly composed of plasma cells, but also contains T lymphocytes (predominantly CD4+) and other innate immune cells. IELs mostly comprise CD8+ T cells and are almost exclusive to the intestinal immune system. Natural killer (NK) cells and NK-T cells are also significantly present in the epithelium. Peyer’s patches are abundant in CD4+ and CD8+ T cells and B cells, and have an overlying epithelium composed of M cells which allow transport of luminal antigens into the patches for an adaptive immune response.79 During inflammatory responses, these cells secrete increased levels of chemokines and IgA, which further perpetuates the inflammatory response.
Gut commensals are reliant on the intestinal immune system for their coexistence with the host, and increasing evidence shows that this dependence is mutual; the immune system is equally reliant on gut microbes for its own maturation and regulation.80 GF mice exhibit reduced gut lymphoid tissue expansion and present with several immunodeficiencies.78 Probiotic treatment with Lactobacillus kefiri in mice leads to increased fecal IgA abundances, reduced expression of proinflammatory mediators, and enhanced anti-inflammatory IL-10 release from Peyer’s patches and mesenteric lymph nodes81. Furthermore, bacteria-derived metabolites, such as SCFAs, tryptophan, and bile acid derivatives from bile salt hydrolase activity display immunoprotective properties.82–85 SCFAs, for example, increase antimicrobial peptide and mucus production and stimulate maturation and expansion of colonic regulatory T cells to reduce local inflammatory responses.83,85
Studies implicating SI microbes, specifically, are also emerging.86 This is not unexpected given the vast area of immune tissue in the SI alone and the fact that the SI mucus barrier is far less established than in the colon, allowing for closer contact between intestinal epithelial cells and luminal bacteria.87 One of the first steps in the intestinal defense cascade is the production and secretion of antimicrobial proteins (AMPs) by intestinal epithelial cells – a feature of innate immunity.78 AMP secretion, among other things, regulates bacterial colonization of the mucus, influencing an individual’s likelihood of infection. Expression of multiple AMPs, including regenerating islet-derived protein 3ɣ (REG3G) in the SI epithelium, in mice shows diurnal rhythmicity driven by an ICL3-STAT3 immune signaling pathway.86 Regulation of the ICL3-STAT3 immune signaling pathway is in turn dependent on host feeding-regulated rhythmic attachment of segmented filamentous bacteria (SFB) to the SI epithelium. These results indicate that, in addition to the diurnal microbial fluctuations in the SI that regulate host metabolism, rhythmic changes in the SI microbiota also influence diurnal rhythms in intestinal innate immunity that drive the variation in an individual’s susceptibility to infection across the day–night cycle.
Diurnal regulation of immune activity is also important for an adaptive response to repeated delivery of food into the SI. As mentioned earlier, the intestinal immune system must maintain a balance between immunity against pathogenic stimuli and tolerance for nonpathogenic antigens. The continual shifts in nutrient availability in the SI challenge this equilibrium. Resident epithelial cells and IELs must therefore be able to adapt to these changes. SI epithelial cells exhibit diurnal expression of the major histocompatibility complex (MHC) class II complex, which is dependent on host feeding-driven diurnal fluctuations in SI microbes.88 This diurnal expression of MHC class II establishes diurnal activation of intraepithelial T cell IL-10+ lymphocytes and thus diurnal secretion of IL-10, which leads to diurnal variation in SI barrier function, namely gut permeability. Such findings highlight the central role of SI microbes in maintaining intestinal tolerance for a highly variable, food-derived antigen burden.
Similarly, in the recent work of Grace Cao and colleagues, where mucosal immunity in the duodenum of mice was explored, Faecalibaculum rodentium along with other specific members of the duodenal microbiota was found to regulate duodenal epithelial homeostasis through increasing epithelial cell turnover rate, crypt proliferation, and MHC class II expression.89 This was further explained through microbial suppression of enterocyte retinoic acid production, leading to decreased eosinophil populations in the proximal SI. Decreased populations of eosinophils resulted in increased IEL-mediated production of interferon-γ, which stimulated the intra-epithelial cell turnover and MHCII expression.
The presence of immunomodulatory bacteria in the ileum has also been suggested in the context of allergy prevention. Following colonization of GF mice with feces from either healthy or cow’s milk allergy infants, Feehley et al. observed that mice colonized with the feces from healthy infants were protected against anaphylactic responses to cow’s milk allergen.90 Further correlation analyses between ileal bacteria and genes upregulated in the ileum identified the Clostridia species, Anaerostipes caccae, as a key player in this protective response. Additionally, antigen-specific Th2-dependent antibody and cytokine responses were reduced in mice monocolonised with A. caccae. It is therefore possible that Anaerostipes caccae induces a specific ileal epithelium gene expression profile, which prevents/attenuates an allergic response to dietary antigens.
Taking these results together, the SI is a key organ where interactions between the microbiota, environmental exposures (e.g., diet and infectious agents), and intestinal immune responses take place to maintain intestinal homeostasis and prevent immune-mediated disease development.
The SI microbiota and disease
The gut microbiota has been implicated in the pathogenesis of a myriad of diseases, and the mechanisms underlying these roles have been extensively studied and reviewed in relation to the large intestinal microbiota. In this section, the findings and potential mechanisms that explain the role of SI microbiota in the development and treatment of specific disorders will be discussed.
Functional GI disorders
Functional gastrointestinal disorders (FGIDs), which include functional dyspepsia (FD) and irritable bowel syndrome (IBS), have long been associated with a quantitative increase in bacteria in the SI, known as small intestinal bacterial overgrowth (SIBO).91 The mechanisms underlying this pathology, however, remain poorly understood and several studies have since failed to find a correlation between the presence of functional GI symptoms – such as diarrhea, abdominal pain and bloating – and SIBO, questioning the validity of this association.91–94 Discrepancies in the association observed between SIBO and the presence of dysbiosis are also present.91,95,96 Saffouri and colleagues, for example, failed to demonstrate a correlation between SIBO and duodenal luminal dysbiosis in patients with GI symptoms.91 Similarly, Yang et al. reported that SIBO does not correlate with duodenal luminal dysbiosis in patients with diarrhea-predominant subtype IBS (IBS-D); however, it does correlate with duodenal mucosal dysbiosis.95 In contrast, Bamba et al. – with the aim of identifying differences in the duodenal microbiome of symptomatic individuals with and without SIBO – found SIBO to be associated with duodenal luminal dysbiosis, characterized by significantly reduced α-diversity and compositional changes including an increase in Streptococcus spp. and Actinomyces spp. and a decrease in species belonging to the genera Bacteroides, Blautia, and Prevotella.96 These incongruities highlight the limitations of current SIBO diagnostic methods and challenge the significance of a SIBO-positive result, but also emphasize the need for deeper characterization of the SI microbiota in the context of FGIDs to elucidate its role in GI symptom development.
In the study of Zhong et al. involving nine patients with FD, quality of life (QoL), symptom responses to a standardized nutrient challenge and bacterial load – reported as the ratio between copies of bacterial 16S rRNA and human β-actin genes measured using quantitative PCR – were assessed.97 Interestingly, bacterial load negatively correlated with reported QoL and positively correlated with the total score of meal-related symptoms following the nutrient challenge, suggesting that bacterial load may be a better predictor of GI symptom development compared to SIBO. Similarly, higher absolute loads of disrupter taxa – bacteria identified as displacing common strict anaerobes in the duodenum – were associated with more severe GI symptoms.98 FD has been associated with an enrichment of Streptococcus, and a reduction in Prevotella, Veillonella, and Actinomyces species at the duodenal mucosa.97,99 Notably, a higher abundance of mucus-associated Streptococci, was also observed following PPI use in both patients with FD and healthy controls.100 The enrichment of Streptococcus bacteria observed at the mucosa in patients with FD may therefore reflect the effect of PPI use, rather than FD itself, on duodenal bacterial composition. The microbial composition at the duodenum mucosa, in the context of FD, is further influenced by the presence of an H. pylori infection101. Changes to the duodenal luminal microbiota in patients with common FGID symptoms include decreases in Porphyromonas, Prevotella, and Fusobacterium species.91 Investigating whether diet-associated increases in FGID symptom burden could be attributed to changes in the SI microbiota, Saffouri and colleagues also performed a short-term dietary intervention pilot study on 16 healthy individuals consuming a baseline high fiber diet.91 Participants were placed on a low fiber, high simple-sugar diet for 7 days, and stool and duodenal aspirates were obtained before and after the intervention. 80% of the participants developed GI symptoms during the course of the intervention, which was associated with a concomitant decrease in microbial diversity. Furthermore, there was an inverse relation between SI microbial diversity and SI permeability. A more recent study, conducted by Shanahan and colleagues, on the contrary, found no significant differences in long-term nutrient intake or quality of diet between individuals with FD (n = 56) and healthy controls (n = 30) nor did they find an association between habitual diet and duodenal mucosa-associated microbiota profiles.102 These results suggest that short-term diet-microbe-host interactions are important in driving the development of GI symptoms. How exactly microbes mediate the interaction between host and diet in the pathogenesis of FGIDs is still to be determined. However, in our recent study of the human SI microbiota we found that methane-producing bacterial pathways were significantly underrepresented in the SI when compared with the fecal microbiome.10 Methane has been linked to GI motility and constipation-predominant diseases, providing one possible theory implicating SI dysbiosis via increased methane production in GI symptom development.103,104
Obesity and metabolic disorders
As discussed earlier, the SI plays a significant role in maintaining host metabolic homeostasis. Disruption to this state of equilibrium forms the basis of metabolic disorder development, such as obesity, dyslipidemia and type 2 diabetes. In this section, we discuss dyslipidaemia and how the SI microbiota might be involved in driving a hyperlipidaemic state in humans.
Dyslipidemia, or hyperlipidemia, is the presence of excess fat or lipids such as cholesterol and triglycerides in the blood.105 Hyperlipidemia is an increasing health problem worldwide, and the complications associated with it, including cardiovascular disease, are devastating. Early treatment is paramount to complication prevention, in return also relieving the increasing pressures health care services are currently facing. GF mice have been shown to exhibit reduced lipid absorption and resistance to diet-induced obesity following a HFD. This phenotype is reversed upon colonization with microbes harvested from the SI of mice fed a HFD and irrespective of the diet consumed thereafter.106 Furthermore, proximal SI epithelial organoids grown in media conditioned by SI Clostridium bifermentans cultures show increased expression of lipid absorption genes such as Dgat2, suggesting that SI bacteria play a key role in facilitating digestive and absorptive responses in the SI to dietary lipids, important for host lipid regulation. Additional evidence linking microbes residing in the SI to hyperlipidemia involve bile acid (BA) metabolism.107 As detailed earlier, BAs are important components of lipid digestion and absorption,56 and regulation of BA pools is not only critical for their function but also contributes to the management of total blood serum cholesterol levels.107 BA synthesis is under the control of specific hepatic enzymes, depending on the pathway: classic (CYP7A1 & CYP8B1) or alternative (CYP27A1 & CYP7B1). Hepatic expression of BA-synthesizing enzymes is regulated by nuclear receptor FXR activity in the ileum, which mediates its effects through a fibroblast growth factor (FGF15)-dependent pathway.55 Increased FXR-FGF15 signaling leads to suppressed hepatic BA-synthesizing enzyme expression and thus reduced BA synthesis. Ileal FXR activity is dependent on the binding of specific BAs. Unconjugated BAs tend to increase ileal FXR activity, whilst conjugated BAs are mainly FXR antagonists or weak agonists.55,108 The proportion of conjugated to unconjugated BAs in the SI is modulated by resident bacteria - Lactobacillus, Bacillus, Streptococcus, and Lactococcus - which, via intrinsic bile salt hydrolase (BSH) activity, deconjugate conjugated BAs.55,109,110 The SI microbiota can therefore inhibit hepatic BA synthesis – and in return increase serum cholesterol levels – through BA pool modulation resulting in increased FXR-FGF15 signaling. This is further demonstrated in the study of Huang et al. investigating the lipid-reducing effects of a famous traditional Chinese tea, Pu-erh, using mice.108 Pu-erh tea, and more specifically its constituent theabrownin, reduces ileal abundances of BSH-producing microbes and attenuates ileal BSH activity, both in vivo and in cultured ileal microbes. This leads to tauro-conjugated BA accumulation in the ileum, FXR-FGF15 signaling inhibition and finally increased hepatic BA synthesis, with a subsequent reduction in serum and hepatic cholesterol and triglyceride levels (Figure 1f).
Environmental enteric dysfunction
Environmental enteric dysfunction (EED) is a disease of the SI characterized histopathologically by diminution in the number and height of intestinal villi, disruption of the epithelial barrier, and chronic inflammatory infiltrate. EED is a prevalent health problem, especially in developing countries. It is clinically associated with malabsorption and diarrhea and is thought to play a role in undernutrition, possibly mediated through the intestinal microbiota. Investigating this hypothesis using a collection of duodenal aspirate samples from 80 children with biopsy-confirmed EED, Chen and colleagues identified a strong correlation between the absolute levels of a group of 14 duodenal bacteria – among others Veillonella sp., Streptococcus sp., and Rothia mucilaginosa - and the degree of stunting.111 Furthermore, gnotobiotic mice colonized with duodenal strains cultured from children with EED developed enteropathy of the SI. Taken together, these results suggest a causal relationship between the SI microbiota, EED development, and growth stunting. Therapies targeting these EED-associated microbial changes may therefore prove vital for fighting undernutrition.
Liver disease (cirrhosis)
Cirrhosis describes scarring (fibrosis) of the liver as a result of long-term damage to the liver. Patients with cirrhosis, particularly when decompensated, have increased risk of developing a range of serious complications including systemic infections, spontaneous bacterial peritonitis, hepatic encephalopathy, and acute-on-chronic liver failure.112,113 Underlying this increased risk is thought to be increased intestinal permeability, specifically in the duodenum. 114 Investigating the relationship between mucosal bacteria and epithelial permeability in patients with compensated cirrhosis, Bloom et al., identified a distinct duodenal microbial community in the duodenum, characterized predominantly by increased Pseudomonadaceae (Proteobacteria) and decreased Lactobacillus, Bifidobacterium, and Clostridia species, that was associated with increased epithelial permeability.115 Although further studies are required to establish underlying mechanisms, previous reports have shown specific Lactobacillus and Bifidobacterium strains to decrease intestinal permeability,116,117 suggesting a role for the mucosa-associated duodenal microbiota in the development of complications associated with cirrhosis.
Inflammatory/Autoimmune disorders
A breakdown in the ability of the intestinal immune system to maintain a balance between immunity and tolerance underlies the development of several inflammatory-mediated disorders. As discussed previously, SI bacteria play an important role in this regulation. Consistent with this, studies have implicated the SI microbiota in a number of inflammatory disorders, with the underlying mechanisms slowly being unraveled. Here, we will focus on autoimmune/inflammatory diseases: IBD, pouchitis, type 1 diabetes and celiac disease.
Inflammatory bowel disease
IBD is a chronic relapsing and remitting inflammatory disorder of the GI tract, with the two most common clinical manifestations being Crohn’s disease (CD) and ulcerative colitis (UC). IBD has been extensively studied in the context of the gut microbiota118 and several bacteria common to the SI, such as Streptococci, Enterococci, Actinomyces, Veillonella spp., and Klebsiella pneumoniae have been repeatedly implicated in its pathogenesis.119–122 Colonization of the common SI resident Enterococcus faecalis was also shown to induce IBD in genetically susceptible mice.123 These observations have been linked to the idea of oralization – the ectopic colonization of typically oral bacteria in the colon – which is hypothesized to exacerbate an already dysbiotic and inflammatory state in patients with IBD.124 This is demonstrated in the study of Atarashi et al., whereby ectopic intestinal colonization of orally derived Klebsiella isolates induced TH1 cell activation and subsequent intestinal inflammation in genetically susceptible individuals.125 However, given that Klebsiella, and other common oral/IBD-associated bacteria, are abundant in the SI, as well as the close proximity of the SI to the colon, one could hypothesize that in fact the SI serves as a reservoir for pathobionts to translocate to the colon and cause damage. Furthermore, diurnal regulation of the diet-microbiota-MHC class II-IL10 axis is critical for maintaining SI immune response and barrier integrity. Using a Crohn-like enteritis mouse model, Tuganbaev and colleagues showed that concomitant disruption of the MHC class II-IL10 axis further exacerbates intestinal inflammation.88 These findings suggest that perturbations in the SI microbiota contribute to IBD pathogenesis, in part, through dysregulation of an MHC class II-dependent IL-10 signaling pathway.
Differences in disease manifestation between CD and UC may also be explained by SI bacteria. Using a cohort of 359 treatment-naive pediatric patients with CD or UC, as well as control individuals, Haberman and colleagues were able to identify ileal gene expression profiles and microbial communities specific to CD, irrespective of disease location (i.e., ileum or colon).126 These results hint at a central role for the ileum, and its microbiota, in the induction of IBD subtype CD. The association between intracellular pattern recognition receptor, NOD2, and CD supports this further. NOD2 is highly expressed in ileal Paneth cells where it regulates the ileal microbiota through secretion of antimicrobial molecules and controls pro-inflammatory immune responses to intestinal microbiota through upregulation of anti-inflammatory mediators and downregulation of pro-inflammatory cytokines.127,128 Mutations in NOD2 are the most strongly associated genetic risk factors for ileal CD,129–131 suggesting that dysregulation of the SI microbiota contributes to CD disease pathogenesis. Increased consumption of emulsifiers is also strongly linked to an increased risk of CD.132 Dietary emulsifiers polysorbate-80 and carboxymethyl cellulose were reported to induce compositional changes in the ileal microbiota of mice that lead to exacerbation of indomethacin-induced SI lesions via an interleukin-1β signaling pathway.133 Finally, extra-intestinal manifestation of creeping fat in individuals with CD is another IBD subtype-distinguishing feature that has recently been attributed to small intestinal bacteria.134 Creeping fat results from the migration of mesenteric adipose tissue to inflamed regions in the intestine, primarily in the SI whereby it undergoes hyperplasia and wraps around the intestinal wall. Ha and colleagues demonstrated, using CD ileal surgical resections containing creeping fat and gnotobiotic mice, that this restructuring of mesenteric adipose tissue is promoted by the translocation of a collection of ileal mucosa-associated bacteria, including Clostridium innocuum.134
UC patients, particularly those with right-sided presentation, are also at risk of developing primary sclerosing cholangitis (PCS), whereby the colitis is thought to be the first manifestation of the disease. PSC is characterized by inflammation and progressive fibrosis of the bile ducts with eventually liver disease.(135) Microbial dysbiosis in the upper GI and bile duct fluid, including an increase in abundance of Veillonella dispar and E. coli at the duodenal mucosa has been reported.(136)
Pouchitis
Although several different drug therapies are available to reduce symptoms and disease activity in individuals with IBD, an unfortunate proportion end up requiring a total colonic resection with an ileal pouch-anal anastomosis (pouch). This same surgical procedure is also carried out as a preventative treatment for individuals with familial adenomatous polyposis (FAP) due to a significantly increased risk of developing colorectal cancer.137 Notably, approximately 50% of individuals with a pouch due IBD go on to develop pouchitis – inflammation of the pouch – whereas this very rarely occurs in patients with FAP.138,139 Studies investigating the pathogenesis of pouchitis have found differences in pouch microbiomes between IBD and FAP pouches, as well as between UC pouches with and without pouchitis, indicating a role for the microbiota in driving pouchitis.140,141 This is further supported by the fact that pouchitis is often responsive to antibiotic treatment.142 In general, the microbiota of IBD pouches display a stronger shift toward a more colon-like composition, typified by a decrease in facultative anaerobes and an increase in obligate anaerobes, sulfate-reducing bacteria, and Clostridia species.143 In the study conducted by Sinha et al., metabolomic, transcriptomic, and metagenomic profiling in UC vs FAP pouches showed significantly reduced levels of SBAs, genes converting PBAs to SBAs and the family of SBA-producing bacteria, Ruminococcaceae141. Furthermore, SBA supplementation in mouse colitis models minimized the intestinal inflammation, in part via a TGR5 bile acid receptor-dependent pathway, suggesting that a previously existing dysbiosis in the newly created pouches of UC patients leads to SBA deficiency, which in turn drives a pro-inflammatory state.
Type 1 diabetes
The body’s progressive self-destruction of the insulin producing beta cells in the pancreas, causing type 1 diabetes (T1D), is increasingly being linked to intestinal abnormalities.144 These alterations include dysfunctional epithelial barrier function and abnormally active immune system. The role of the gut microbiota has therefore gained attention in this respect, including the SI microbiota given its close functional and spatial relationship with the pancreas, as well as shared blood supply. By means of inflammatory profiling and microbiome evaluation of duodenal biopsies from patients with T1D (n = 19), patients with celiac disease (included as controls for intestinal inflammatory disease; n = 19) and healthy controls (n = 16), Pellegrini et al., identified a T1D-specific microbial signature in the duodenal mucosa that correlated with a pro-inflammatory gene expression profile.144 The distinct microbial signature was characterized by an increase in Firmicutes and Firmicutes/Bacteroidetes ratio and a reduction in Proteobacteria and Bacteroidetes. Similar changes were also observed in the duodenal microbiota of diabetic rats.145 Notably, despite an overall increase in Firmicutes in duodenal mucosal biopsies from patients with T1D as compared to the healthy controls, Clostridia species abundances were reduced. Clostridia contribute significantly to butyrate production in the gut and are also key mucin-degrading bacteria, both of which are important for barrier integrity and possibly preventing autoantibody formation.146,147 Segmented filamentous bacterial strains in non-obese diabetic mice were also shown to induce autoimmune diabetes via T-helper 17 cell interaction in the SI lamina propria.148 Further evidence implicating the SI microbiota in T1D pathogenesis comes from the fecal microbiota transplantation (FMT) study of de Groot et al. in 2021.149 FMT preserved the residual beta cell function, halting the decline of endogenous insulin production. The mechanisms of which were linked to changes in both fecal and SI microbiota composition, SI gene expression, metabolite profiles, and T cell immunity.
Celiac disease
Celiac disease (CeD) is characterized by an intolerance for gluten, aberrantly activating the body’s immune system and causing damage, predominantly, to the SI.150 This loss of tolerance for gluten has been hypothesized to involve increased mucosal permeability and subsequent recruitment of T cells, brought about by changes in the intestinal microbial composition.151 Analysis of duodenal biopsies from patients with CeD have revealed a dominance of Proteobacteria such as Enterobacteriaceae bacteria, as well as a high prevalence of Bacteroidetes and Streptococcus species.151,152 Pseudomonas aeruginosa abundances are also increased in the duodenum of patients with CeD.153 P. aeruginosa is able to metabolize gluten, producing immunogenic peptides that more efficiently permeate the gut barrier and activate gluten-specific T cells. The bacteria also express a specific elastase that synergizes with gluten to amplify inflammation. Potential metabolic changes associated with the microbial changes in the duodenum of patients with CeD include an increase in alternative pathways for energy production such as D-glucarate, L-arabinose, D-galactarate, and biogenic amine degradation pathways and decreased SCFA production.152 A gluten-free diet is currently the only available and effective treatment for CeD.150 Complete remission, however, is not guaranteed, with some individuals still experiencing symptoms despite adhering to the strict diet. The role of duodenal microbiota in the persistence of symptoms following the removal of gluten from the diet was investigated by a group in Sweden.154 Patients on a gluten-free diet suffering from persistent symptoms, compared to without symptoms, exhibited lower microbial richness, higher relative abundance of Proteobacteria, and lower relative abundance of Bacteroidetes and Firmicutes. Although in its infancy, these results provide insight into possible future treatment alternatives for CeD patients.
Neurological disorders
The gut–brain axis describes the bidirectional interaction between the gut and brain that influences both neurological and intestinal functions.155 Increasingly more research is being conducted on the role played by gut microbes in effecting this two-way relationship, and a couple of studies focus specifically on the SI.
Already inside the womb, the influence of the SI microbiota on the brain is significant. Mice born to mothers exposed to a synthetic double-stranded RNA that mimics viral infection during pregnancy – eliciting an immune response termed maternal immune activation (MIA) – display cortical brain lesions and abnormal behavioral phenotypes.156 In contrast, offspring from antibiotic treated pregnant mice do not exhibit such abnormal phenotypes, suggesting that maternal intestinal microbes are required for the induction of MIA-associated behavioral and brain abnormalities in offspring. Ileal mucosa-associated segmented filamentous bacteria in pregnant mice are necessary to induce these neurodevelopmental disorders via TH17 cell expansion and activation.
Exploring the contribution of psychological stress toward disease exacerbation in patients with CD (i.e., increased intestinal inflammation), Shaler and colleagues observed a number of changes in the ileum of conventional specific-pathogen-free mice subjected to acute psychological stress.157 Firstly, psychological stress induced ileal nutritional immunity, evident by an upregulation of host genes associated with nutritional immunity and metal restriction, as well as glucocorticoid-mediated depletion of IL-22-producing immune cells that activate the protective mucosal antimicrobial defenses. This in turn led to ileal dysbiosis dominated by an expansion of, in particular, adherent-invasive Escherichia coli (AIEC) - a gut pathobiont that can evade SI defense mechanisms and is known to colonize ileal lesions in patients with CD.157,158 Deeper analysis showed that AIEC takes advantage of the increased nutritional immunity, in part through the expression of iron-scavenging siderophores, as well as the impaired host immunity, permitting its expansion and further enhancing the proinflammatory state in patients with CD. Other genera commonly found enriched in CD patients such as Enterococcus faecalis were also enriched following exposure to physiological stress, although this was not further explored. The results highlight the complex role of the SI microbiota within the gut–brain axis to influence intestinal health, which warrants further investigation.
The study of Miyauchi et al. highlights the potential of the SI microbiota to drive neurological disease, namely multiple sclerosis (MS).159 MS is an autoimmune-driven demyelinating disease of the brain and spinal cord. Miyauchi and colleagues demonstrated that mice treated orally with ampicillin were protected against demyelination of the spinal cord and infiltration of the spinal cord by inflammatory cells, hinting at a gut microbiota-mediated inflammatory response underlying MS.159 Further investigation using GF mice colonized with specific ampicillin-sensitive SI bacteria revealed that SI microorganisms act synergistically to activate myelin oligodendrocyte glycoprotein-specific T cells. These T cells are likely responsible for the pro-inflammatory and demyelination phenotype of experimental autoimmune encephalomyelitis mice. Given these results and previous reports of increased TH17 cells in the SI of patients with MS,160 future efforts should be made to explore the human SI microbiota in the context of MS.
Conclusion and future perspectives
The SI plays an essential role in host metabolism, immunity, and endocrine and neurological functions. Despite this, and the growing interest in its microbial communities, knowledge of the SI microbiota is still in its infancy and greatly lags behind that of the lower intestinal tract. In this review, we detailed the current methods used to sample and model the SI microbiota and their limitations. We also discussed the function of the SI in host metabolism and immunity and summarized the studies that provide evidence for, and insight into, the role played by the SI microbiota, including in the context of disease.
The SI microbiota is emerging as a critical component of human health by regulating fundamental processes in the SI that not only impact intestinal health but also influence extraintestinal physiological functions. In recent years, there has been a surge in fecal microbiota transplantation studies and a drive to develop clinical interventions, but with limited success. A potential reason for this could be the current bias toward fecal microbiome studies, upon which these trials are based, ignoring the important contribution of the SI microbiota. A better understanding of the metabolic capacity of the SI microbiota and its interactions with the host therefore has the potential to significantly advance gut microbiome research and its quest for novel diagnostic techniques and intervention strategies to improve human health. Similarly, dietary interventions have been designed following the conclusions derived from fecal microbiome studies. Considering that the small intestine is the main organ involved in nutrient absorption and metabolic regulation, future interventions should consider host–microbiota interactions within this organ.
One of the major factors restricting SI microbiota research, however, are the limitations and challenges of the current methods for sampling and modeling this poorly accessible and highly dynamic ecosystem. The field would therefore greatly benefit from more effort being made to develop uniform sampling techniques that provide accurate representations of the SI microbiota, are less invasive, and do not require specialists to perform. Additionally, the majority of our knowledge on the mechanisms underlying the SI microbiota–host interactions is derived from mouse/animal models. Developing in vitro or in vivo models that better recapitulate the human SI environment will be paramount to translating promising findings into human intervention studies and clinical trials.
Acknowledgements
We would like to acknowlegde A. Petrenko's involvement in the creative design of Figure 1.
Funding Statement
AVV and RKW are supported by Health~Holland grant LWV21.165/LSHM21077. RKW is supported by HORIZON-HLTH-2022-STAYHLTH-02-0 and the Seerave Foundation.
Disclosure statement
RKW acted as a consultant for Takeda and received unrestricted research grants from Takeda and Johnson and Johnson pharmaceuticals and speaker fees from AbbVie, MSD, Olympus and AstraZeneca. No potential conflict of interest were reported by other authors.
Data availability statement
Data sharing is not applicable to this article as no new data were created or analyzed in this review.
References
- 1.Clemente JC, Ursell LK, Parfrey LW, Knight R.. The impact of the gut microbiota on human health: an integrative view. Cell. 2012 Mar 16;148(6):1258–25. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dieterich W, Schink M, Zopf Y. Microbiota in the gastrointestinal tract. Med Sci (Basel). 2018 Dec 14;6(4):116. doi: 10.3390/medsci6040116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang M, Ahrné S, Jeppsson B, Molin G. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol Ecol. 2005 Oct 1;54(2):219–231. doi: 10.1016/j.femsec.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed S, Macfarlane GT, Fite A, McBain AJ, Gilbert P, Macfarlane S. Mucosa-associated bacterial diversity in relation to human terminal ileum and colonic biopsy samples. Appl Environ Microbiol. 2007. Nov;73(22):7435–7442. doi: 10.1128/AEM.01143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayashi H, Takahashi R, Nishi T, Sakamoto M, Benno Y. Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J Med Microbiol. 2005. Nov;54(11):1093–1101. doi: 10.1099/jmm.0.45935-0. [DOI] [PubMed] [Google Scholar]
- 6.Vaga S, Lee S, Ji B, Andreasson A, Talley NJ, Agréus L, Bidkhori, G, Kovatcheva-Datchary, P, Park, J, Lee, D, et al. Compositional and functional differences of the mucosal microbiota along the intestine of healthy individuals. Sci Rep 2020 Sep 11;10(1):14977–2. 10.1038/s41598-020-71939-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villmones HC, Halland A, Stenstad T, Ulvestad E, Weedon-Fekjær H, Kommedal Ø. The cultivable microbiota of the human distal ileum. Clin Microbiol Infect 2021. Jun;27(6):912.e7–912.e13. 10.1016/j.cmi.2020.08.021 [DOI] [PubMed] [Google Scholar]
- 8.Villmones HC, Haug ES, Ulvestad E, Grude N, Stenstad T, Halland A, Kommedal Ø. Species level description of the human ileal bacterial microbiota. Sci Rep 2018 Mar 16;8(1):4736–5. 10.1038/s41598-018-23198-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villmones HC, Svanevik M, Ulvestad E, Stenstad T, Anthonisen IL, Nygaard RM, Dyrhovden R, Kommedal Ø. Investigating the human jejunal microbiota. Sci Rep. 2022 Jan 31;12(1):1682–1689. doi: 10.1038/s41598-022-05723-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruigrok RAAA, Collij V, Sureda P, Klaassen MAY, Bolte LA, Jansen BH, Voskuil, M.D., Fu, J., Wijmenga, C.Zhernakova, A, et al. The composition and metabolic potential of the human small intestinal microbiota within the context of inflammatory bowel disease. J Crohns Colitis. 2021 Aug 2;15(8):1326–1338. doi: 10.1093/ecco-jcc/jjab020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Booijink CC, El-Aidy S, Rajilić-Stojanović M, Heilig HG, Troost FJ, Smidt H, Kleerebezem M, De Vos WM, Zoetendal EG. High temporal and inter-individual variation detected in the human ileal microbiota. Environ Microbiol. 2010. Dec;12(12):3213–3227. doi: 10.1111/j.1462-2920.2010.02294.x. [DOI] [PubMed] [Google Scholar]
- 12.Zoetendal EG, Raes J, van den Bogert B, Arumugam M, Booijink CC, Troost FJ, Bork P, Wels M, de Vos WM, Kleerebezem M. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. Isme J. 2012. Jul;6(7):1415–1426. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yilmaz B, Fuhrer T, Morgenthaler D, Krupka N, Wang D, Spari D, Candinas, D., Misselwitz, B., Beldi, G., Sauer, U., et al. Plasticity of the adult human small intestinal stoma microbiota. Cell Host & Microbe. 2022 Dec 14;30(12):1773–1787.e6. doi: 10.1016/j.chom.2022.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Dreskin BW, Luu K, Dong TS, Benhammou J, Lagishetty V, Vu J, Sanford, D., Durazo, F., Agopian, V.G., Jacobs, J.P., et al. Specimen collection and analysis of the Duodenal Microbiome. J Vis Exp. 2021 Jan 12;(167): 10.3791/61900. [DOI] [PubMed] [Google Scholar]
- 15.Seekatz AM, Schnizlein MK, Koenigsknecht MJ, Baker JR, Hasler WL, Bleske BE, Young, VB, Sun, D. SpAtial and temporal analysis of the stomach and small-intestinal microbiota in fasted healthy humans. mSphere 2019 Mar 13;4(2):e00126–19. 10.1128/mSphere.00126-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rehan M, Al-Bahadly I, Thomas DG, Avci E. Capsule robot for gut microbiota sampling using shape memory alloy spring. Int J Med Robot. 2020. Oct;16(5):1–14. doi: 10.1002/rcs.2140. [DOI] [PubMed] [Google Scholar]
- 17.Nimble science Simba Capsule. 2022; Available at: https://www.nimblesci.com/simbacapsule.
- 18.Stolaki M, Minekus M, Venema K, Lahti L, Smid EJ, Kleerebezem M, Zoetendal EG. Microbial communities in a dynamic in vitro model for the human ileum resemble the human ileal microbiota. FEMS Microbiol Ecol. 2019 Aug 1;95(8):fiz096. doi: 10.1093/femsec/fiz096. [DOI] [PubMed] [Google Scholar]
- 19.Hugenholtz F, de Vos WM. Mouse models for human intestinal microbiota research: a critical evaluation. Cell Mol Life Sci. 2018. Jan;75(1):149–160. doi: 10.1007/s00018-017-2693-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo P, Wang H, Ji L, Song P, Ma X. Impacts of Fructose on intestinal barrier function, inflammation and microbiota in a Piglet model. Nutrients. 2021 Oct 6;13(10):3515. doi: 10.3390/nu13103515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010. Jul;90(3):859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 22.Leser TD, Mølbak L. Better living through microbial action: the benefits of the mammalian gastrointestinal microbiota on the host. Environ Microbiol. 2009. Sep;11(9):2194–2206. doi: 10.1111/j.1462-2920.2009.01941.x. [DOI] [PubMed] [Google Scholar]
- 23.Hu S, Bourgonje AR, Gacesa R, Jansen BH, Björk JR, Bangma A, Hidding, I.J., van Dullemen, H.M., Visschedijk M.C, Faber, K.N., et al. Mucosal host–microbe interactions associate with clinical phenotypes in inflammatory bowel disease. bioRxiv. 2022; 2022.June.4. 494807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sommer F, Bäckhed F. The gut microbiota–masters of host development and physiology. Nat Rev Microbiol. 2013. Apr;11(4):227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 25.Juge N. Microbial adhesins to gastrointestinal mucus. Trends Microbiol. 2012. Jan;20(1):30–39. doi: 10.1016/j.tim.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Leite G, Barlow GM, Hosseini A, Parodi G, Pimentel ML, Wang J, Fiorentino A, Rezaie A, Pimentel M, Mathur R. Smoking has disruptive effects on the small bowel luminal microbiome. Sci Rep 2022 Apr 14;12(1):6231–z. 10.1038/s41598-022-10132-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim J, Shin J, Park J. Effect of a proton pump inhibitor on the Duodenum Microbiome of Gastric Ulcer patients. Life (Basel). 2022 Sep 27;12(10):1505. doi: 10.3390/life12101505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garmaeva S, Sinha T, Kurilshikov A, Fu J, Wijmenga C, Zhernakova A. Studying the gut virome in the metagenomic era: challenges and perspectives. BMC Biol 2019 Oct 28;17(1):84–y. 10.1186/s12915-019-0704-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Zhan H, Xu W, Yan S, Ng SC. The role of gut mycobiome in health and diseases. Therap Adv Gastroenterol. 2021 Sep 23;14:17562848211047130. doi: 10.1177/17562848211047130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adiliaghdam F, Amatullah H, Digumarthi S, Saunders TL, Rahman RU, Wong LP, Sadreyev, R, Droit, L, Paquette, J, Goyette, P, et al. Human enteric viruses autonomously shape inflammatory bowel disease phenotype through divergent innate immunomodulation. Sci Immunol. 2022 Apr 8;7(70):eabn6660. doi: 10.1126/sciimmunol.abn6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duca FA, Waise TMZ, Peppler WT, Lam TKT. The metabolic impact of small intestinal nutrient sensing. Nat Commun 2021 Feb 10;12(1):903–y. 10.1038/s41467-021-21235-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song Y, Koehler JA, Baggio LL, Powers AC, Sandoval DA, Drucker DJ. Gut-Proglucagon-Derived Peptides are essential for regulating Glucose Homeostasis in mice. Cell Metab. 2019 Nov 5;30(5):976–986.e3. doi: 10.1016/j.cmet.2019.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Covasa M, Stephens RW, Toderean R, Cobuz C. Intestinal sensing by gut Microbiota: targeting gut Peptides. Front Endocrinol (Lausanne). 2019 Feb 19;10:82. doi: 10.3389/fendo.2019.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bauer PV, Duca FA, Waise TMZ, Dranse HJ, Rasmussen BA, Puri A, Rasti M, O’Brien CA, Lam TKT. Lactobacillus gasseri in the upper small intestine impacts an ACSL3-Dependent Fatty Acid-SenSing pathway regulating whole-body Glucose Homeostasis. Cell Metab. 2018 Mar 6;27(3):572–587.e6. doi: 10.1016/j.cmet.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 35.Breen DM, Rasmussen BA, Kokorovic A, Wang R, Cheung GW, Lam TK. Jejunal nutrient sensing is required for duodenal-jejunal bypass surgery to rapidly lower glucose concentrations in uncontrolled diabetes. Nat Med. 2012. Jun;18(6):950–955. doi: 10.1038/nm.2745. [DOI] [PubMed] [Google Scholar]
- 36.Cheung GW, Kokorovic A, Lam CK, Chari M, Lam TK. Intestinal cholecystokinin controls glucose production through a neuronal network. Cell Metab. 2009. Aug;10(2):99–109. doi: 10.1016/j.cmet.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 37.Wang PY, Caspi L, Lam CK, Chari M, Li X, Light PE, Gutierrez-Juarez R, Ang M, Schwartz GJ, Lam TKT. Upper intestinal lipids trigger a gut–brain–liver axis to regulate glucose production. Nature. 2008 Apr 24;452(7190):1012–1016. doi: 10.1038/nature06852. [DOI] [PubMed] [Google Scholar]
- 38.Zadeh-Tahmasebi M, Duca FA, Rasmussen BA, Bauer PV, Côté CD, Filippi BM, Lam TKT. Activation of short and long chain fatty acid sensing machinery in the ileum lowers glucose production in vivo. J Biol Chem. 2016 Apr 15;291(16):8816–8824. doi: 10.1074/jbc.M116.718460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silva YP, Bernardi A, Frozza RL. THe role of short-chain fatty acids from gut microbiota in gut-brain communication. Front Endocrinol (Lausanne). 2020 Jan 31;11:25. doi: 10.3389/fendo.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fredborg M, Theil PK, Jensen BB, Purup S. G protein-coupled receptor 120 (GPR120) transcription in intestinal epithelial cells is significantly affected by bacteria belonging to the Bacteroides, Proteobacteria, and Firmicutes phyla. J Anim Sci. 2012. Dec;90(Suppl suppl_4):10–12. doi: 10.2527/jas.53792. [DOI] [PubMed] [Google Scholar]
- 41.Bauer PV, Duca FA, Waise TMZ, Rasmussen BA, Abraham MA, Dranse HJ, Puri A, O’Brien CA, Lam TKT. Metformin alters upper small intestinal microbiota that impact a Glucose-SGLT1-Sensing Glucoregulatory pathway. Cell Metab. 2018 Jan 9;27(1):101–117.e5. doi: 10.1016/j.cmet.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 42.Maida A, Lamont BJ, Cao X, Drucker DJ. Metformin regulates the incretin receptor axis via a pathway dependent on peroxisome proliferator-activated receptor-α in mice. Diabetologia. 2011. Feb;54(2):339–349. doi: 10.1007/s00125-010-1937-z. [DOI] [PubMed] [Google Scholar]
- 43.Vardarli I, Arndt E, Deacon CF, Holst JJ, Nauck MA. Effects of sitagliptin and metformin treatment on incretin hormone and insulin secretory responses to oral and “isoglycemic” intravenous glucose. Diabetes. 2014. Feb;63(2):663–674. doi: 10.2337/db13-0805. [DOI] [PubMed] [Google Scholar]
- 44.Lenzen S, Lortz S, Tiedge M. Effect of metformin on SGLT1, GLUT2, and GLUT5 hexose transporter gene expression in small intestine from rats. Biochem Pharmacol. 1996 Apr 12;51(7):893–896. doi: 10.1016/0006-2952(95)02243-0. [DOI] [PubMed] [Google Scholar]
- 45.Duca FA, Côté CD, Rasmussen BA, Zadeh-Tahmasebi M, Rutter GA, Filippi BM, Lam TKT. Metformin activates a duodenal Ampk–dependent pathway to lower hepatic glucose production in rats. Nat Med. 2015. May;21(5):506–511. doi: 10.1038/nm.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, Velásquez-Mejía EP, Carmona JA, Abad JM, de la Cuesta-Zuluaga J, Escobar JS. Metformin is associated with higher relative abundance of Mucin-Degrading Akkermansia muciniphilA and several short-chain fatty acid–producing microbiota in the gut. Diabetes Care. 2017. Jan;40(1):54–62. doi: 10.2337/dc16-1324. [DOI] [PubMed] [Google Scholar]
- 47.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A. 2014 Nov 11;111(45):16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bishehsari F, Voigt RM, Keshavarzian A. Circadian rhythms and the gut microbiota: from the metabolic syndrome to cancer. Nat Rev Endocrinol. 2020. Dec;16(12):731–739. doi: 10.1038/s41574-020-00427-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoogerwerf WA, Hellmich HL, Cornélissen G, Halberg F, Shahinian VB, Bostwick J, Savidge, TC, Cassone, VM. Clock gene expression in the murine gastrointestinal tract: endogenous rhythmicity and effects of a feeding regimen. Gastroenterology. 2007. Oct;133(4):1250–1260. doi: 10.1053/j.gastro.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 50.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001 Jan 19;291(5503):490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 51.Dantas Machado AC, Brown SD, Lingaraju A, Sivaganesh V, Martino C, Chaix A, Zhao, P., Pinto, A.F., Chang, M.W., Richter, R.A., et al. Diet and feeding pattern modulate diurnal dynamics of the ileal microbiome and transcriptome. Cell Rep. 2022 Jul 5;40(1):111008. doi: 10.1016/j.celrep.2022.111008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frazier K, Chang EB. Intersection of the gut microbiome and Circadian rhythms in metabolism. Trends Endocrinol Metab. 2020. Jan;31(1):25–36. doi: 10.1016/j.tem.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson, L., Katz, M.N., Korem, T., Zmora, N., et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014 Oct 23;159(3):514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 54.Zarrinpar A, Loomba R. Review article: the emerging interplay among the gastrointestinal tract, bile acids and incretins in the pathogenesis of diabetes and non-alcoholic fatty liver disease. Alimentary Pharmacology & Therapeutics. 2012. Nov;36(10):909–921. doi: 10.1111/apt.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013 Feb 5;17(2):225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 56.Yang J, Palmiotti A, Kuipers F. Emerging roles of bile acids in control of intestinal functions. Curr Opin Clin Nutr Metab Care. 2021 Mar 1;24(2):127–133. doi: 10.1097/MCO.0000000000000709. [DOI] [PubMed] [Google Scholar]
- 57.Ryan KK, Tremaroli V, Clemmensen C, Kovatcheva-Datchary P, Myronovych A, Karns R, Wilson-Pérez, H.E., Sandoval, D.A., Kohli, R., Bäckhed, F., et al. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature. 2014 May 8;509(7499):183–188. doi: 10.1038/nature13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watanabe M, Horai Y, Houten SM, Morimoto K, Sugizaki T, Arita E, Mataki, C., Sato, H., Tanigawara, Y., Schoonjans, K., et al. Lowering bile acid pool size with a synthetic farnesoid X receptor (FXR) agonist induces obesity and diabetes through reduced energy expenditure. J Biol Chem. 2011 Jul 29;286(30):26913–26920. doi: 10.1074/jbc.M111.248203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prawitt J, Abdelkarim M, Stroeve JH, Popescu I, Duez H, Velagapudi VR,Dumont, J., Bouchaert, E., Van Dijk, T.H., Lucas, A., et al. Farnesoid X receptor deficiency improves glucose homeostasis in mouse models of obesity. Diabetes. 2011. Jul;60(7):1861–1871. doi: 10.2337/db11-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang C, Xie C, Lv Y, Li J, Krausz KW, Shi J, Brocker, CN, Desai, D, Amin, SG, Bisson, WH, et al. Intestine-selective farnesoid X receptor inhibition improves obesity-related metabolic dysfunction. Nat Commun. 2015 Dec 15;6(1):10166. doi: 10.1038/ncomms10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trabelsi MS, Daoudi M, Prawitt J, Ducastel S, Touche V, Sayin SI,Perino A, Brighton CA, Sebti Y, Kluza J, et al. Farnesoid X receptor inhibits glucagon-like peptide-1 production by enteroendocrine L cells. Nat Commun. 2015 Jul 2;6(1):7629. doi: 10.1038/ncomms8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Macchiarulo, A., Yamamoto, H., Mataki, C., Pruzanski, M., et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009. Sep;10(3):167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winston JA, Theriot CM. Diversification of host bile acids by members of the gut microbiota. Gut Microbes. 2020;11(2):158–171. doi: 10.1080/19490976.2019.1674124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caliceti C, Calabria D, Roda A, Cicero AFG. Fructose intake, Serum Uric Acid, and Cardiometabolic disorders: a critical review. Nutrients. 2017 Apr 18;9(4):395. doi: 10.3390/nu9040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lanaspa MA, Ishimoto T, Li N, Cicerchi C, Orlicky DJ, Ruzycki P, Rivard, C, Inaba, S, Roncal-Jimenez, CA, Bales, ES, et al. Endogenous fructose production and metabolism in the liver contributes to the development of metabolic syndrome. Nat Commun. 2013;4(1):2434. doi: 10.1038/ncomms3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jang C, Hui S, Lu W, Cowan AJ, Morscher RJ, Lee G, Liu W, Tesz GJ, Birnbaum MJ, Rabinowitz JD. The small intestine converts dietary Fructose into Glucose and Organic Acids. Cell Metab. 2018 Feb 6;27(2):351–361.e3. doi: 10.1016/j.cmet.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vich Vila A, Collij V, Sanna S, Sinha T, Imhann F, Bourgonje AR, Mujagic, Z., Jonkers, D.M., Masclee, A.A., Fu, J. and Kurilshikov, A.. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat Commun 2020 Jan 17;11(1):362–z. 10.1038/s41467-019-14177-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Kessel SP, Frye AK, El-Gendy AO, Castejon M, Keshavarzian A, van Dijk G, van Kessel SP, van Dijk G, El Aidy S. Gut bacterial tyrosine decarboxylases restrict levels of levodopa in the treatment of Parkinson’s disease. Nat Commun 2019 Jan 18;10(1):310–y. 10.1038/s41467-019-08294-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gundert-Remy U, Hildebrandt R, Stiehl A, Weber E, Zürcher G, Da Prada M. Intestinal absorption of levodopa in man. Eur J Clin Pharmacol. 1983;25(1):69–72. doi: 10.1007/BF00544017. [DOI] [PubMed] [Google Scholar]
- 70.Perez M, Calles-Enríquez M, Nes I, Martin MC, Fernandez M, Ladero V, Alvarez MA. Tyramine biosynthesis is transcriptionally induced at low pH and improves the fitness of Enterococcus faecalis in acidic environments. Appl Microbiol Biotechnol. 2015. Apr;99(8):3547–3558. doi: 10.1007/s00253-014-6301-7. [DOI] [PubMed] [Google Scholar]
- 71.Zhu H, Xu G, Zhang K, Kong X, Han R, Zhou J, Ni Y. Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding. Sci Rep. 2016 Jun 13;6(1):27779. doi: 10.1038/srep27779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jourová L, Lišková B, Lněničková K, Zemanová N, Anzenbacher P, Hermanová P, Hudcovic T, Kozáková H, Anzenbacherová E. Presence or absence of microbiome modulates the response of mice organism to administered drug nabumetone. Physiol Res. 2020 Dec 31;69(Suppl 4):S583–594. doi: 10.33549/physiolres.934607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peggs KS, Quezada SA, Korman AJ, Allison JP. Principles and use of anti-CTLA4 antibody in human cancer immunotherapy. Curr Opin Immunol. 2006. Apr;18(2):206–213. doi: 10.1016/j.coi.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 74.Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz, S., Routy, B., Roberti, M.P., Duong, C.P., et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015 Nov 27;350(6264):1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vighi G, Marcucci F, Sensi L, Di Cara G, Frati F. Allergy and the gastrointestinal system. Clin Exp Immunol. 2008. Sep;153(Suppl Supplement_1):3–6. doi: 10.1111/j.1365-2249.2008.03713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tokuhara D, Kurashima Y, Kamioka M, Nakayama T, Ernst P, Kiyono H. A comprehensive understanding of the gut mucosal immune system in allergic inflammation. Allergol Int. 2019. Jan;68(1):17–25. doi: 10.1016/j.alit.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 77.Obata Y, Pachnis V. THe effect of microbiota and the immune system on the development and organization of the enteric nervous system. Gastroenterology. 2016. Nov;151(5):836–844. doi: 10.1053/j.gastro.2016.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shi N, Li N, Duan X, Niu H. Interaction between the gut microbiome and mucosal immune system. Mil Med Res. 2017 Apr 27;4(1):14–19. 10.1186/s40779-017-0122-9. eCollection 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ohno H. Intestinal M cells. J Biochem. 2016. Feb;159(2):151–160. doi: 10.1093/jb/mvv121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wiertsema SP, van Bergenhenegouwen J, Garssen J, Knippels LMJ. The interplay between the gut microbiome and the immune system in the context of infectious diseases throughout life and the role of nutrition in optimizing treatment strategies. Nutrients. 2021 Mar 9;13(3):886. doi: 10.3390/nu13030886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Carasi P, Racedo SM, Jacquot C, Romanin DE, Serradell MA, Urdaci MC. Impact of kefir derived Lactobacillus kefiri on the mucosal immune response and gut microbiota. J Immunol Res 2015. 2015;2015:361604. doi: 10.1155/2015/361604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schnupf P, Gaboriau-Routhiau V, Cerf-Bensussan N. Modulation of the gut microbiota to improve innate resistance. Curr Opin Immunol. 2018. Oct;54:137–144. doi: 10.1016/j.coi.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 83.Rannug A. How the AHR became Important in intestinal Homeostasis-A Diurnal FICZ/AHR/CYP1A1 feedback controls both immunity and immunopathology. Int J Mol Sci. 2020 Aug 8;21(16):5681. doi: 10.3390/ijms21165681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Labbé A, Ganopolsky JG, Martoni CJ, Prakash S, Jones ML. Bacterial bile metabolising gene abundance in Crohn’s, ulcerative colitis and type 2 diabetes metagenomes. PLoS One. 2014 Dec 17;9(12):e115175. doi: 10.1371/journal.pone.0115175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews, E, Ajami, NJ, Bonham, KS, Brislawn, CJ et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019. May;569(7758):655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brooks JF 2nd, Behrendt CL, Ruhn KA, Lee S, Raj P, Takahashi JS, Hooper LV. The microbiota coordinates diurnal rhythms in innate immunity with the circadian clock. Cell. 2021 Aug 5;184(16):4154–4167.e12. doi: 10.1016/j.cell.2021.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Johansson MEV, Sjövall H, Hansson GC. The gastrointestinal mucus system in health and disease. Nat Rev Gastroenterol Hepatol. 2013. Jun;10(6):352–361. doi: 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tuganbaev T, Mor U, Bashiardes S, Liwinski T, Nobs SP, Leshem A, Dori-Bachash, M, Thaiss, CA, Pinker, EY, Ratiner, K, et al. DIet diurnally regulates small intestinal Microbiome-Epithelial-Immune Homeostasis and Enteritis. Cell. 2020 Sep 17;182(6):1441–1459.e21. doi: 10.1016/j.cell.2020.08.027. [DOI] [PubMed] [Google Scholar]
- 89.Cao YG, Bae S, Villarreal J, Moy M, Chun E, Michaud M, Lang JK, Glickman JN, Lobel L, Garrett WS. Faecalibaculum rodentium remodels retinoic acid signaling to govern eosinophil-dependent intestinal epithelial homeostasis. Cell Host & Microbe. 2022 Aug 12;30(9):1295–1310.e8. doi: 10.1016/j.chom.2022.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Feehley T, Plunkett CH, Bao R, Choi Hong SM, Culleen E, Belda-Ferre P, Campbell, E, Aitoro, R, Nocerino, R, Paparo, L, et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat Med. 2019. Mar;25(3):448–453. doi: 10.1038/s41591-018-0324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Saffouri GB, Shields-Cutler RR, Chen J, Yang Y, Lekatz HR, Hale VL, Cho, JM, Battaglioli, EJ, Bhattarai, Y, Thompson, KJ, et al. Small intestinal microbial dysbiosis underlies symptoms associated with functional gastrointestinal disorders. Nat Commun. 2019 May 1;10(1):2012–2017. doi: 10.1038/s41467-019-09964-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Grover M, Kanazawa M, Palsson OS, Chitkara DK, Gangarosa LM, Drossman DA, Whitehead WE. Small intestinal bacterial overgrowth in irritable bowel syndrome: association with colon motility, bowel symptoms, and psychological distress. Neurogastroenterol Motil. 2008. Sep;20(9):998–1008. doi: 10.1111/j.1365-2982.2008.01142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ford AC, Spiegel BM, Talley NJ, Moayyedi P. Small intestinal bacterial overgrowth in irritable bowel syndrome: systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2009. Dec;7(12):1279–1286. doi: 10.1016/j.cgh.2009.06.031. [DOI] [PubMed] [Google Scholar]
- 94.Walters B, Vanner SJ. Detection of bacterial overgrowth in IBS using the lactulose H2 breath test: comparison with 14C-D-xylose and healthy controls. Am J Gastroenterol. 2005. Jul;100(7):1566–1570. doi: 10.1111/j.1572-0241.2005.40795.x. [DOI] [PubMed] [Google Scholar]
- 95.Yang M, Zhang L, Hong G, Li Y, Li G, Qian W, Xiong H, Bai T, Song J, Hou X. Duodenal and rectal mucosal microbiota related to small intestinal bacterial overgrowth in diarrhea-predominant irritable bowel syndrome. J Gastroenterol Hepatol. 2020. May;35(5):795–805. doi: 10.1111/jgh.14910. [DOI] [PubMed] [Google Scholar]
- 96.Bamba S, Imai T, Sasaki M, Ohno M, Yoshida S, Nishida A, Takahashi K, Inatomi O, Andoh A. Altered gut microbiota in patients with small intestinal bacterial overgrowth. J Gastroenterol Hepatol. 2022 Sep 30;38(1):61–69. doi: 10.1111/jgh.16013. [DOI] [PubMed] [Google Scholar]
- 97.Zhong L, Shanahan ER, Raj A, Koloski NA, Fletcher L, Morrison M, Walker MM, Talley NJ, Holtmann G. Dyspepsia and the microbiome: time to focus on the small intestine. Gut. 2017. Jun;66(6):1168–1169. doi: 10.1136/gutjnl-2016-312574. [DOI] [PubMed] [Google Scholar]
- 98.Barlow JT, Leite G, Romano AE, Sedighi R, Chang C, Celly S, Rezaie A, Mathur R, Pimentel M, Ismagilov RF. Quantitative sequencing clarifies the role of disruptor taxa, oral microbiota, and strict anaerobes in the human small-intestine microbiome. Microbiome 2021 Nov 2;9(1):214–2. 10.1186/s40168-021-01162-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fukui A, Takagi T, Naito Y, Inoue R, Kashiwagi S, Mizushima K, Inada, Y, Inoue, K, Harusato, A, Dohi, O, et al. Higher levels of streptococcus in upper gastrointestinal Mucosa associated with symptoms in patients with functional Dyspepsia. Digestion. 2020;101(1):38–45. doi: 10.1159/000504090. [DOI] [PubMed] [Google Scholar]
- 100.Wauters L, Tito RY, Ceulemans M, Lambaerts M, Accarie A, Rymenans L, Verspecht, C, Toth, J, Mols, R, Augustijns, P, et al. Duodenal Dysbiosis and relation to the efficacy of proton pump inhibitors in functional Dyspepsia. Int J Mol Sci. 2021 Dec 192224:13609. 10.3390/ijms222413609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Suárez-Jaramillo A, Baldeón ME, Prado B, Fornasini M, Cohen H, Flores N, Salvador I, Cargua O, Realpe J, Cárdenas PA. Duodenal microbiome in patients with or without Helicobacter pylori infection. Helicobacter. 2020. Dec;25(6):e12753. doi: 10.1111/hel.12753. [DOI] [PubMed] [Google Scholar]
- 102.Shanahan ER, Kang S, Staudacher H, Shah A, Do A, Burns G, Chachay, VS, Koloski, NA, Keely, S, Walker, MM, et al. Alterations to the duodenal microbiota are linked to gastric emptying and symptoms in functional dyspepsia. Gut. 2022 Sep 27;72(5):929–938. doi: 10.1136/gutjnl-2021-326158. [DOI] [PubMed] [Google Scholar]
- 103.Triantafyllou K, Chang C, Pimentel M. Methanogens, methane and gastrointestinal motility. J Neurogastroenterol Motil. 2014. Jan;20(1):31–40. doi: 10.5056/jnm.2014.20.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jahng J, Jung IS, Choi EJ, Conklin JL, Park H. The effects of methane and hydrogen gases produced by enteric bacteria on ileal motility and colonic transit time. Neurogastroenterol Motil 2012. Feb;24(2):185–90, e92. 10.1111/j.1365-2982.2011.01819.x [DOI] [PubMed] [Google Scholar]
- 105.Mitchell S, Roso S, Samuel M, Pladevall-Vila M. Unmet need in the hyperlipidaemia population with high risk of cardiovascular disease: a targeted literature review of observational studies. BMC Cardiovasc Disord 2016 Apr 26;16:74–3. 1 10.1186/s12872-016-0241-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Martinez-Guryn K, Hubert N, Frazier K, Urlass S, Musch MW, Ojeda P, Pierre, JF, Miyoshi, J, Sontag, TJ, Cham, CM, et al. SmAll intestine microbiota regulate host digestive and absorptive adaptive responses to dietary lipids. Cell Host & Microbe. 2018 Apr 11;23(4):458–469.e5. doi: 10.1016/j.chom.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jia X, Xu W, Zhang L, Li X, Wang R, Wu S. Impact of gut microbiota and microbiota-related metabolites on Hyperlipidemia. Front Cell Infect Microbiol. 2021 Aug 19;11:634780. doi: 10.3389/fcimb.2021.634780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huang F, Zheng X, Ma X, Jiang R, Zhou W, Zhou S, Zhang, Y, Lei, S, Wang, S, Kuang, J, et al. Theabrownin from Pu-erh tea attenuates hypercholesterolemia via modulation of gut microbiota and bile acid metabolism. Nat Commun 2019 Oct 31;10(1):4971–x. 10.1038/s41467-019-12896-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Degirolamo C, Rainaldi S, Bovenga F, Murzilli S, Moschetta A. Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the Fxr-Fgf15 axis in mice. Cell Rep. 2014 Apr 10;7(1):12–18. doi: 10.1016/j.celrep.2014.02.032. [DOI] [PubMed] [Google Scholar]
- 110.Rani RP, Anandharaj M, Ravindran AD. Characterization of bile salt Hydrolase from Lactobacillus gasseri FR4 anD demonstration of its substrate specificity and inhibitory mechanism using molecular docking analysis. Front Microbiol. 2017 May 31;8:1004. doi: 10.3389/fmicb.2017.01004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen RY, Kung VL, Das S, Hossain MS, Hibberd MC, Guruge J, Mahfuz, M, Begum, SKN, Rahman, MM, Fahim, SM, et al. Duodenal Microbiota in stunted undernourished children with Enteropathy. N Engl J Med. 2020 Jul 23;383(4):321–333. doi: 10.1056/NEJMoa1916004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rabiee A, Ximenes RO, Nikayin S, Hickner A, Juthani P, Rosen RH, Garcia‐tsao G. Factors associated with health-related quality of life in patients with cirrhosis: a systematic review. Liver Int. 2021. Jan;41(1):6–15. doi: 10.1111/liv.14680. [DOI] [PubMed] [Google Scholar]
- 113.D’Amico G, Garcia-Tsao G, Pagliaro L. Natural history and prognostic indicators of survival in cirrhosis: a systematic review of 118 studies. J Hepatol. 2006. Jan;44(1):217–231. doi: 10.1016/j.jhep.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 114.Du Plessis J, Vanheel H, Janssen CEI, Roos L, Slavik T, Stivaktas PI, Nieuwoudt, M, van Wyk, SG, Vieira, W, Pretorius, E, et al. Activated intestinal macrophages in patients with cirrhosis release NO and IL-6 that may disrupt intestinal barrier function. J Hepatol. 2013. Jun;58(6):1125–1132. doi: 10.1016/j.jhep.2013.01.038. [DOI] [PubMed] [Google Scholar]
- 115.Bloom PP, Rao K, Bassis CM, Zhou SY, Nojkov B, Owyang C, Young VB, Lok AS. DuodeNal permeability is associated with mucosal microbiota in compensated Cirrhosis. Clin Transl Gastroenterol. 2022 Oct 1;13(10):e00522. doi: 10.14309/ctg.0000000000000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hou Q, Huang Y, Wang Y, Liao L, Zhu Z, Zhang W, Liu Y, Li P, Chen X, Liu F. Lactobacillus casei LC01 regulates intestinal Epithelial permeability through miR-144 targeting of OCLN and ZO1. J Microbiol Biotechnol. 2020 Oct 28;30(10):1480–1487. doi: 10.4014/jmb.2002.02059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang W, Li H, Zhao N, Luo X, Liu S, Bao A, Chen Y, Wang H, Wang J, Wang J. Lactobacillus johnsonii BS15 combined with abdominal massage on intestinal permeability in rats with nonalcoholic fatty liver and cell biofilm repair. Bioengineered. 2021. Dec;12(1):6354–6363. doi: 10.1080/21655979.2021.1954134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. 2017. Oct;14(10):573–584. doi: 10.1038/nrgastro.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kojima A, Nakano K, Wada K, Takahashi H, Katayama K, Yoneda M, Higurashi, T, Nomura, R, Hokamura, K, Muranaka, Y, et al. Infection of specific strains of Streptococcus mutans, oral bacteria, confers a risk of ulcerative colitis. Sci Rep. 2012;2(1):332. doi: 10.1038/srep00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, Vatanen, T, Hall, AB, Mallick, H, McIver, LJ, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019. Feb;4(2):293–305. doi: 10.1038/s41564-018-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vich Vila A, Imhann F, Collij V, Jankipersadsing SA, Gurry T, Mujagic Z, Kurilshikov, A, Bonder, MJ, Jiang, X, Tigchelaar, EF, et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med. 2018 Dec 1910472:eaap8914. 10.1126/scitranslmed.aap8914. [DOI] [PubMed] [Google Scholar]
- 122.Seishima J, Iida N, Kitamura K, Yutani M, Wang Z, Seki A, Yamashita, T, Sakai, Y, Honda, M, Yamashita, T, et al. Gut-derived Enterococcus faecium from ulcerative colitis patients promotes colitis in a genetically susceptible mouse host. Genome Biol. 2019 Nov 25;20(1):252–259. doi: 10.1186/s13059-019-1879-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Balish E, Warner T. Enterococcus faecalis induces inflammatory bowel disease in interleukin-10 knockout mice. Am J Pathol. 2002. Jun;160(6):2253–2257. doi: 10.1016/S0002-9440(10)61172-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schmidt TS, Hayward MR, Coelho LP, Li SS, Costea PI, Voigt AY, Wirbel, J, Maistrenko, OM, Alves, RJ, Bergsten, E, et al. Extensive transmission of microbes along the gastrointestinal tract. Elife. 2019 Feb 12;8 10.7554/eLife.42693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, Kiguchi, Y, Yasuma, K, Watanabe, E, Tanoue, T, et al. Ectopic colonization of oral bacteria in the intestine drives T(H)1 cell induction and inflammation. Science. 2017 Oct 20;358(6361):359–365. doi: 10.1126/science.aan4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Haberman Y, Tickle TL, Dexheimer PJ, Kim MO, Tang D, Karns R, Baldassano, RN, Noe, JD, Rosh, J, Markowitz, J, et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014. Aug;124(8):3617–3633. doi: 10.1172/JCI75436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sidiq T, Yoshihama S, Downs I, Kobayashi KS. Nod2: a critical regulator of Ileal Microbiota and Crohn’s disease. Front Immunol. 2016 Sep 20;7:367. doi: 10.3389/fimmu.2016.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rochereau N, Roblin X, Michaud E, Gayet R, Chanut B, Jospin F, Corthésy B, Paul S. NOD2 deficiency increases retrograde transport of secretory IgA complexes in Crohn’s disease. Nat Commun 2021 Jan 11;12(1):261–0. 10.1038/s41467-020-20348-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dragasevic S, Stankovic B, Milosavljevic T, Sokic-Milutinovic A, Lukic S, Alempijevic T, Zukic, B, Kotur, N, Nikcevic, G, Pavlovic, S, et al. Genetic and environmental factors significant for the presentation and development of inflammatory bowel disease. European Journal of Gastroenterology & Hepatology. 2017. Aug;29(8):909–915. doi: 10.1097/MEG.0000000000000877. [DOI] [PubMed] [Google Scholar]
- 130.Goethel A, Turpin W, Rouquier S, Zanello G, Robertson SJ, Streutker CJ, Philpott DJ, Croitoru K. Nod2 influences microbial resilience and susceptibility to colitis following antibiotic exposure. Mucosal Immunol. 2019. May;12(3):720–732. doi: 10.1038/s41385-018-0128-y. [DOI] [PubMed] [Google Scholar]
- 131.Ramanan D, Tang MS, Bowcutt R, Loke P, Cadwell K. Bacterial sensor Nod2 prevents inflammation of the small intestine by restricting the expansion of the commensal Bacteroides vulgatus. Immunity. 2014 Aug 21;41(2):311–324. doi: 10.1016/j.immuni.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Roberts CL, Rushworth SL, Richman E, Rhodes JM. Hypothesis: increased consumption of emulsifiers as an explanation for the rising incidence of Crohn’s disease. J Crohns Colitis. 2013. May;7(4):338–341. doi: 10.1016/j.crohns.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 133.Furuhashi H, Higashiyama M, Okada Y, Kurihara C, Wada A, Horiuchi K, Hanawa, Y, Mizoguchi, A, Nishii, S, Inaba, K, et al. Dietary emulsifier polysorbate-80-induced small-intestinal vulnerability to indomethacin-induced lesions via dysbiosis. J Gastroenterol Hepatol. 2020. Jan;35(1):110–117. doi: 10.1111/jgh.14808. [DOI] [PubMed] [Google Scholar]
- 134.CWY H, Martin A, Sepich-Poore GD, Shi B, Wang Y, Gouin K, Humphrey, G, Sanders, K, Ratnayake, Y, Chan, KS, et al. Translocation of viable gut microbiota to Mesenteric AdiPose drives formation of creeping fat in humans. Cell. 2020 Oct 29;183(3):666–683.e17. doi: 10.1016/j.cell.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Karlsen TH, Folseraas T, Thorburn D, Vesterhus M. Primary sclerosing cholangitis - a comprehensive review. J Hepatol. 2017. Dec;67(6):1298–1323. doi: 10.1016/j.jhep.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 136.Liwinski T, Zenouzi R, John C, Ehlken H, Rühlemann MC, Bang C, Groth, S, Lieb, W, Kantowski, M, Andersen, N, et al. Alterations of the bile microbiome in primary sclerosing cholangitis. Gut. 2020. Apr;69(4):665–672. doi: 10.1136/gutjnl-2019-318416. [DOI] [PubMed] [Google Scholar]
- 137.Half E, Bercovich D, Rozen P. Familial adenomatous polyposis. Orphanet J Rare Dis 2009 Oct 12;4:22–22. 1 10.1186/1750-1172-4-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zella GC, Hait EJ, Glavan T, Gevers D, Ward DV, Kitts CL, Korzenik JR. Distinct microbiome in pouchitis compared to healthy pouches in ulcerative colitis and familial adenomatous polyposis. Inflamm Bowel Dis. 2011. May;17(5):1092–1100. doi: 10.1002/ibd.21460. [DOI] [PubMed] [Google Scholar]
- 139.Coffey JC, Rowan F, Burke J, Dochery NG, Kirwan WO, O’Connell PR. Pathogenesis of and unifying hypothesis for idiopathic pouchitis. Am J Gastroenterol. 2009. Apr;104(4):1013–1023. doi: 10.1038/ajg.2008.127. [DOI] [PubMed] [Google Scholar]
- 140.Turpin W, Kelly O, Borowski K, Boland K, Tyler A, Cohen Z, Croitoru K, Silverberg MS. Mucosa-associated microbiota in IleoaNal pouches may contribute to clinical symptoms, particularly stool frequency, independent of endoscopic disease activity. Clin Transl Gastroenterol. 2019 May 22;10(5):1–7. doi: 10.14309/ctg.0000000000000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sinha SR, Haileselassie Y, Nguyen LP, Tropini C, Wang M, Becker LS, Sim, D, Jarr, K, Spear, ET, Singh, G, et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host & Microbe. 2020 Apr 8;27(4):659–670.e5. doi: 10.1016/j.chom.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Navaneethan U, Shen B. Pros and cons of antibiotic therapy for pouchitis. Expert Rev Gastroenterol Hepatol. 2009. Oct;3(5):547–559. doi: 10.1586/egh.09.37. [DOI] [PubMed] [Google Scholar]
- 143.AJ K, Terry NA, Wu GD, Albenberg LG. THe structure and function of the human small intestinal microbiota: current understanding and future directions. Cell Mol Gastroenterol Hepatol. 2020;9(1):33–45. doi: 10.1016/j.jcmgh.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pellegrini S, Sordi V, Bolla AM, Saita D, Ferrarese R, Canducci F, Clementi, M, Invernizzi, F, Mariani, A, Bonfanti, R, et al. Duodenal Mucosa of patients with type 1 diabetes shows distinctive inflammatory profile and microbiota. J Clin Endocrinol Metab. 2017 May 1;102(5):1468–1477. doi: 10.1210/jc.2016-3222. [DOI] [PubMed] [Google Scholar]
- 145.Wirth R, Bódi N, Maróti G, Bagyánszki M, Talapka P, Fekete É, Bagi Z, Kovács KL. Regionally distinct alterations in the composition of the gut microbiota in rats with streptozotocin-induced diabetes. PLoS One. 2014 Dec 3;9(12):e110440. doi: 10.1371/journal.pone.0110440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, Casella, G, Drew, JC, Ilonen, J, Knip, M, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6(10):e25792. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dedrick S, Sundaresh B, Huang Q, Brady C, Yoo T, Cronin C, Rudnicki, C, Flood, M, Momeni, B, Ludvigsson, J, et al. The role of gut microbiota and environmental factors in type 1 diabetes Pathogenesis. Front Endocrinol (Lausanne). 2020 Feb 26;11:78. 10.3389/fendo.2020.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci U S A. 2011 Jul 12;108(28):11548–11553. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.de Groot P, Nikolic T, Pellegrini S, Sordi V, Imangaliyev S, Rampanelli E, Hanssen, N, Attaye, I, Bakker, G, Duinkerken, G, et al. Faecal microbiota transplantation halts progression of human new-onset type 1 diabetes in a randomised controlled trial. Gut. 2021. Jan;70(1):92–105. doi: 10.1136/gutjnl-2020-322630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kivelä L, Caminero A, Leffler DA, Pinto-Sanchez MI, Tye-Din JA, Lindfors K. Current and emerging therapies for coeliac disease. Nat Rev Gastroenterol Hepatol. 2021. Mar;18(3):181–195. doi: 10.1038/s41575-020-00378-1. [DOI] [PubMed] [Google Scholar]
- 151.Di Biase AR, Marasco G, Ravaioli F, Dajti E, Colecchia L, Righi B, D’Amico V, Festi D, Iughetti L, Colecchia A. Gut microbiota signatures and clinical manifestations in celiac disease children at onset: a pilot study. J Gastroenterol Hepatol. 2021. Feb;36(2):446–454. doi: 10.1111/jgh.15183. [DOI] [PubMed] [Google Scholar]
- 152.Arcila-Galvis JE, Loria-Kohen V, Ramírez de Molina A, Carrillo de Santa Pau E, Marcos-Zambrano LJ. A comprehensive map of microbial biomarkers along the gastrointestinal tract for celiac disease patients. Front Microbiol. 2022 Sep 13;13:956119. doi: 10.3389/fmicb.2022.956119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Caminero A, McCarville JL, Galipeau HJ, Deraison C, Bernier SP, Constante M, Rolland, C, Meisel, M, Murray, JA, Yu, XB, et al. Duodenal bacterial proteolytic activity determines sensitivity to dietary antigen through protease-activated receptor-2. Nat Commun. 2019 Mar 13;10(1):1198–1199. doi: 10.1038/s41467-019-09037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wacklin P, Laurikka P, Lindfors K, Collin P, Salmi T, Lähdeaho ML, Saavalainen, P, Mäki, M, Mättö, J, Kurppa, K, et al. Altered duodenal microbiota composition in celiac disease patients suffering from persistent symptoms on a long-term gluten-free diet. Am J Gastroenterol. 2014. Dec;109(12):1933–1941. doi: 10.1038/ajg.2014.355. [DOI] [PubMed] [Google Scholar]
- 155.Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol. 2012. Nov;10(11):735–742. doi: 10.1038/nrmicro2876. [DOI] [PubMed] [Google Scholar]
- 156.Kim S, Kim H, Yim YS, Ha S, Atarashi K, Tan TG, Longman, RS, Honda, K, Littman, DR, Choi, GB, et al. Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature. 2017 Sep 28;549(7673):528–532. doi: 10.1038/nature23910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Shaler CR, Parco AA, Elhenawy W, Dourka J, Jury J, Verdu EF, Coombes BK. Psychological stress impairs IL22-driven protective gut mucosal immunity against colonising pathobionts. Nat Commun 2021 Nov 18;12(1):6664–4. 10.1038/s41467-021-26992-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chervy M, Barnich N, Denizot J, Adherent-Invasive E. Coli: updAte on the lifestyle of a troublemaker in Crohn’s Disease. Int J Mol Sci. 2020 May 25;21(10):3734. doi: 10.3390/ijms21103734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Miyauchi E, Kim SW, Suda W, Kawasumi M, Onawa S, Taguchi-Atarashi N, Morita H, Taylor TD, Hattori M, Ohno H, et al. Gut microorganisms act together to exacerbate inflammation in spinal cords. Nature. 2020. Sep;585(7823):102–106. doi: 10.1038/s41586-020-2634-9. [DOI] [PubMed] [Google Scholar]
- 160.Cosorich I, Dalla-Costa G, Sorini C, Ferrarese R, Messina MJ, Dolpady J, Radice, E, Mariani, A, Testoni, PA, Canducci, F, et al. High frequency of intestinal T(H)17 cells correlates with microbiota alterations and disease activity in multiple sclerosis. Sci Adv. 2017 Jul 12;3(7):e1700492. doi: 10.1126/sciadv.1700492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this review.


