Abstract
Vision-and-language (V&L) models take image and text as input and learn to capture the associations between them. These models can potentially deal with the tasks that involve understanding medical images along with their associated text. However, applying V&L models in the medical domain is challenging due to the expensiveness of data annotations and the requirements of domain knowledge. In this paper, we identify that the visual representation in general V&L models is not suitable for processing medical data. To overcome this limitation, we propose BERTHop, a transformer-based model based on PixelHop++ and VisualBERT for better capturing the associations between clinical notes and medical images.
Experiments on the OpenI dataset, a commonly used thoracic disease diagnosis benchmark, show that BERTHop achieves an average Area Under the Curve (AUC) of 98.12% which is 1.62% higher than state-of-the-art while it is trained on a 9× smaller dataset.1
Keywords: Computer-aided diagnosis, Transfer learning, Vision & language model
1. Introduction
Computer-Aided Diagnosis (CADx) [10] systems provide valuable benefits for disease diagnosis including but not limited to improving the quality and consistency of the diagnosis and providing a second option to reduce medical mistakes. Although most existing studies focus on diagnosis based on medical images such as chest X-ray (CXR) images [2, 1], the radiology reports often contain substantial information in the text(e.g. patient history and previous studies) that are difficult to be detected from the image alone. Besides, the diagnosis from both image and text is more closely aligned with disease diagnosis by human experts. Therefore, V&L models that take both images and text as input can be potentially more accurate for CADx.
However, the shortage of annotated data in the medical domain makes utilizing V&L models challenging. Annotating medical data is an expensive process as it requires human experts. Although a couple of recent large-scale auto-labeled datasets have been provided for some medical tasks, e.g., chest X-ray [26, 12], they are often noisy (low-quality) and degrade the performance of models. Besides, such datasets are not available for most medical tasks. Therefore, training V&L models with limited annotated data remains a key challenge.
Recently, pre-trained V&L models have been proposed to reduce the amount of labeled data required for training an accurate downstream model [14, 25, 6] in the general domain (transfer learning). These models are first trained on large-scale image caption data with self-supervision signals (e.g., using masked language model loss 2) to learn the association between objects and text tokens. Then, the parameters of the pre-trained V&L models are used to initialize the downstream models and fine-tuned on the target tasks. In most V&L tasks, it has been reported that V&L pre-training is a major source of performance improvement However, we identify a key problem in applying common pre-trained V&L models for the medical domain: the large domain gap between the medical (target) and the general domain (source) makes such pre-train and fine-tune paradigm considerably less effective in the medical domain. Therefore, domain-specific designs need to be applied.
Notably, V&L models mainly leverage object-centric feature extraction methods such as Faster R-CNN [20] which is pre-trained on general domain to detect everyday objects, e.g., cats, and dogs. However, the abnormalities in the X-ray images do not resemble everyday objects and will likely be ignored by a general-domain object detector.
To overcome this challenge, we propose BERTHop, a transformer-based V&L model designed for medical applications. In BERTHop, the visual encoder of the V&L architecture is redesigned leveraging PixelHop++ [7] and is fully unsupervised which significantly reduces the need for labeled data [21]. PixelHop++ can extract image representations at different frequency levels. This is significantly beneficial for highlighting abnormalities in different levels to be captured by the transformer in relation to the input text.
Furthermore, BERTHop resolves the domain gap issue by leveraging a pre-trained language encoder, BlueBERT [18], a BERT [9] variant that has been trained on biomedical and clinical datasets.
2. Related Work
Transformer-based V&L models
Inspired by the success of BERT for NLP tasks, various transformer-based V&L models have been proposed [14, 6, 25]. They generally use an object detector pre-trained on Visual Genome [13] to extract visual features from an input image and then use a transformer to model visual features and input sentences. They are pre-trained on a massive amount of paired image-text data with a mask-and-predict objective similar to BERT.
Such models have been applied to many V&L applications. [28, 17, 8]. However, for transferring the knowledge from these pre-trained models, the data distribution of source and target should be close enough or otherwise we need enough data for the target domain to properly transfer the knowledge.
V&L models in the medical domain
Various V&L architectures have been proposed for disease diagnosis on CXR.
TieNet is a CNN-RNN-based model for V&L embedding integrating multi-level attention layers into an end-to-end CNN-RNN framework for disease diagnosisTieNet uses a ResNet-50 pre-trained for general-domain visual feature extraction and an RNN for V&L fusion. As a result, it requires a large amount of in-domain training data (ChestX-ray14) for adapting to the medical domain, limiting its practical usage.
Recently, Li et al. [15] evaluated the transferability of well-known pre-trained V&L models by fine-tuning them on MIMIC-CXR [12] and OpenI. However, the pre-trained models are designed and pre-trained for general-domain, and directly fine-tuning them with limited in-domain data leads to suboptimal performance. We refer to this method as VB w/BUTD (section 4.2).
PixelHop++ for visual feature learning
PixelHop++ is originally proposed as an alternative to deep convolutional neural networks for feature extraction from images and video frames in resource-constrained environments. It is a multi-level model which generates output channels representing an image at different frequencies. PixelHop++ is used in various applications and shown to be highly effective on small datasets. These applications include face gender classification [22], face recognition [23], deep fake detection [5], and medical application [16]. To the best of our knowledge, this is the first study which integrates PixelHop++ and DNN models. Although using The PixelHop++ features alone as input to the transformer (no input text) underperform other vision-only models i.e. ChexNet [19], Our proposed model takes advantage of the attention mechanism to combine visual features extracted from PixelHop++ and the language embedding to better find the association between both modalities.
3. Approach
Inspired by the architecture of VisualBERT, our framework uses a single transformer to integrate visual features and language embeddings. The overall framework of our proposed approach is shown in Fig. 1. We first utilize PixelHop++ to extract visual features from the X-ray image; then the text (a radiology report) is encoded into subword embeddings; a joint transformer is applied on top to model the relationship between two modalities and capture implicit alignments.
Fig. 1.
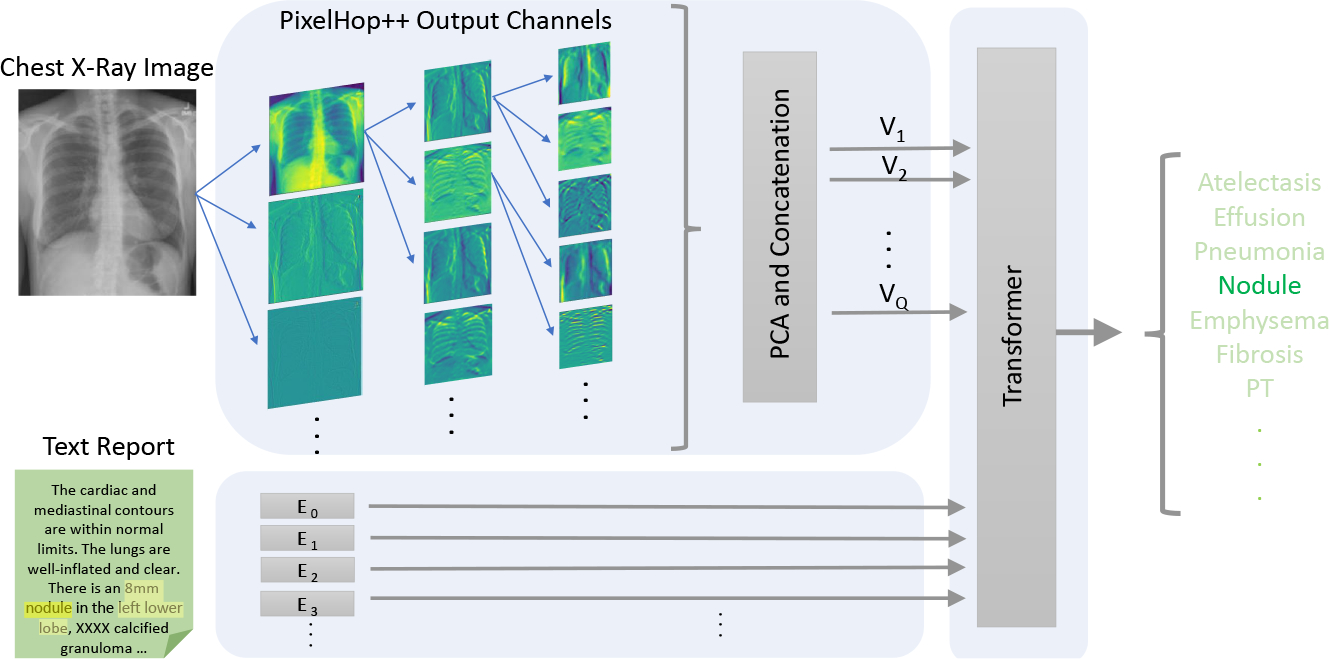
The proposed BERTHop framework for CXR disease diagnosis. A PixelHop++ model followed by a “PCA and concatenation” block is used to generate Q feature vectors. These features along with language embedding are fed to the transformer that is initialized with BlueBERT.
3.1. Visual encoder
We argue that extracting visual features from a general-domain object detector, i.e. the BUTD [3] approach that is dominant in most V&L tasks, is not suitable for the medical domain. BUTD3 takes an image and employs a ResNet-based Faster-RCNN [20] for object detection and feature extraction from each object. The detector is pre-trained on Visual Genome [13] to detect objects in everyday scenes. Such an approach fails to detect medical abnormalities when applied to X-ray images. The reason is that the abnormalities in the image, which are of high importance for facilitating diagnosis, usually do not resemble the normal notion of an “object” and will likely be ignored by a general-domain object detector. Further, there exists no large-scale annotated dataset for disease abnormality detection from which to train a reliable detector [24].
We propose to adopt PixelHop++ [7] for unsupervised visual feature learning in the medical domain, which has been shown to be highly effective when trained on small-scale datasets. The key idea of PixelHop++ is computing the parameters of its model by a closed-form expression without using back-propagation [21]. As PixelHop++ leverages PCA for computing parameters, the model is able to extract image representations at various frequencies in an unsupervised manner. Inspired by the architecture of DNN models, PixelHop++ is a multi-level model in which each level consists of one or several PixelHop++ units followed by a max-pooling layer. When training a PixelHop++ model, parameters of PixelHop++ units (kernels and biases) are computed, and during the inference, they are used for feature extraction from pixel blocks. Given the visual features from PixelHop++ and the radiology report, we then only train/fine-tune the transformer on the given task. PixelHop++ is unsupervised by nature meaning that it is not learned to extract specific features for a specific task. Therefore, the transformer has better access to raw features and has more flexibility to find the optimum alignments between input data (text and image) given less data.
3.2. In-domain text pre-training
In BERTHop, the text report plays an important role in guiding the transformer to pay more attention to the right visual features in the attention mechanism. The report is written by an expert radiologist, who lists the normal and abnormal observations in the “finding” section and other important patient information including patient history, body parts, and previous studies in the “impression” section of the report. The text style of the report is drastically different from that of the pre-training corpora of BERT (Wikipedia and BookCorpus) or V&L models (MSCOCO and Conceptual Captions). Therefore, we propose to use BlueBERT [18] as the backbone in BERTHop to better capture the text report information. Pre-training with text-only corpora has been reported to how only marginal or no benefit [25]. In the medical domain, however, we find that using a transformer pre-trained on in-domain text corpora as our initialized backbone serves as a simpler yet stronger approach. Previous methods [15] do not take such a significant domain gap into consideration. Rather, they initialize the transformer with a model trained on general-domain image-text corpora, as in most V&L tasks.
4. Experiments
In this section, we evaluate BERTHop on the OpenI dataset and compare it with other existing models. To understand the effectiveness of the model designs, we also conduct detailed studies to verify the value of the visual encoder and the transformer initialization.
4.1. Experiment setup
Dataset
We focus on the OpenI dataset comprising 3,996 reports and 8,121 associated images from 3,996 unique patients. Its labels include 14 commonly occurring thoracic chest diseases, OpenI is a reliable choice for both training and evaluating V&L models as it is annotated by experts (labels are not generated or learned from text reports or images). The disadvantage of using OpenI for training is that it contains a small amount of training data which is a challenge for DNN models. We apply the same pre-processing as TieNet and obtain 3,684 image-text pairs.
Model and training parameters
We first resize all images of OpenI to 206 × 206 and apply a three-level PixelHop++ for unsupervised feature learning from them. Then, we apply PCA to PixelHop++ output channels and concatenate the generated vectors to form a set of visual features of dimension D, i.e., . In BERTHop, is set to be 2048. In our experiments setup, is equal to 15 but may vary depending on the size of the output channels of the PixelHop++ model and also the number of PCA components.
As for the transformer backbone, we use BlueBERT-Base (Uncased, PubMed + MIMIC-III) from Huggingface [27], a transformer library. Having the visual features from the visual encoder and text embedding, we train the transformer on the training set of OpenI with 2,912 image-text pairs. We use batch size = 18, learning rate = 1e − 5, max-seq-length = 128, and Stochastic Gradient Descent (SGD) as the optimizer with momentum = 0.9 and train it for 240 epochs.
Evaluation metric
All mentioned datasets are highly imbalanced and mostly contain normal cases. Therefore, evaluating models using metrics such as accuracy does not reflect model performance. Instead, we follow prior studies to evaluate models based on Receiver Operating Characteristic (ROC) and Area Under the ROC Curve (AUC) score.
4.2. Main results
We train BERTHop on the OpenI training dataset containing 2,912 image-text pairs and evaluate it on the corresponding test set comprising 772 image-text pairs. The ROC curve for each disease is plotted in Fig. 2 b). We evaluate all the models using the same AUC implementation in scikit-learn [4]. Fig. 2 a) summarizes the performance of BERTHop compared with existing methods. The results demonstrate that BERTHop outperforms TieNet, which is the current best model, in 11 out of 14 thoracic disease diagnoses and achieves an average AUC of 98.23% which is 14.37%, 12.83%, and 1.73% higher than VB w/BUTD, TNNT, and TieNet, respectively. Note that TieNet has been trained on a much larger annotated dataset, i.e., the ChestX-ray14 dataset containing 108,948 training data while BERTHop is trained on only 2,912 case examples.
Fig. 2.
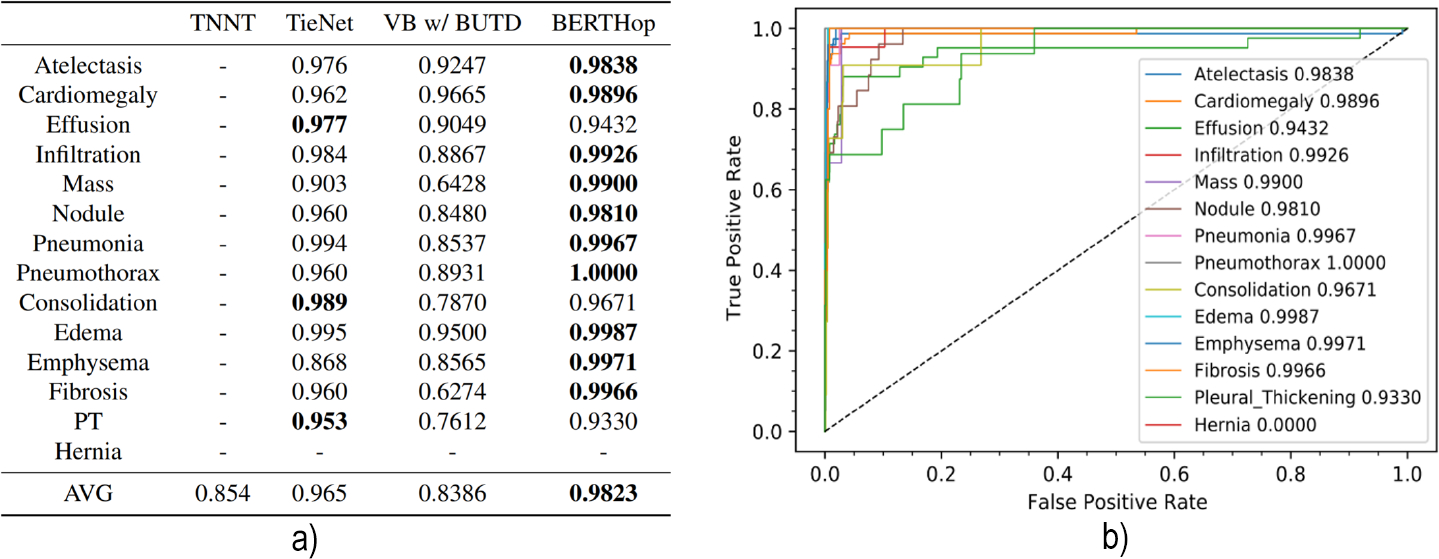
a) The AUC thoracic diseases diagnosis comparison of our model with other three methods on OpenI. BERTHop significantly outperforms models trained with a similar amount of data (e.g. VB w/BUTD). b) The ROC curve of BERTHop for all 14 thoracic diseases.
Regarding the VB w/BUTD results, we reevaluate the results based on the released code4 from the original authors. However, we cannot reproduce the results reported in the paper even after contacting the authors.
4.3. In-domain text pre-training
We further investigate the influence of different transformer backbone initialization on model performance by pairing it with different visual encoders. The results are listed in Table 1. First, we find that the proposed initialization with a model pre-trained on in-domain text corpora (BlueBERT) brings significant performance boosts when paired with PixelHop++. Initializing with BlueBERT gives a 6.46% performance increase compared to initializing with BERT. Second, when using BUTD, the model is less sensitive to the transformer initialization and the performance is generally low (from 83.09% to 85.64%). In contrast to other V&L tasks [14], general-domain V&L pre-training is not instrumental. The above findings suggest that for medical V&L applications, in-domain single modality pre-training can bring larger performance improvement than using pre-trained V&L models from the general domain, even though the latter is trained on a larger corpus. The relation and trade-off between single-modality pre-training and cross-modality pre-training are overlooked by previous works [14] and we advocate for future research on this.
Table 1.
Effect of the transformer backbones when paired with different visual encoders. When using BUTD features, the model becomes insensitive to the transformer initialization and the expensive V&L pre-training brings little benefit compared to BERT. When using PixelHop++, the model benefits significantly from BlueBERT, which is pre-trained on in-domain text corpora.
| Visual Encoder Transformer Backbone |
BUTD | PixelHop++ | |||
|---|---|---|---|---|---|
| VB | BERT | BlueBERT | BERT | BlueBERT | |
|
| |||||
| Atelectasis | 0.9247 | 0.8677 | 0.8866 | 0.9890 | 0.9838 |
| Cardiomegaly | 0.9665 | 0.8877 | 0.8875 | 0.9772 | 0.9896 |
| Effusion | 0.9049 | 0.8940 | 0.9120 | 0.9013 | 0.9432 |
| Mass | 0.6428 | 0.7365 | 0.7373 | 0.8886 | 0.9900 |
| Consolidation | 0.7870 | 0.8766 | 0.8906 | 0.8949 | 0.9671 |
| Emphysema | 0.8565 | 0.7313 | 0.8261 | 0.9641 | 0.9971 |
|
| |||||
| AVG | 0.8386 | 0.8309 | 0.8564 | 0.9177 | 0.9823 |
4.4. Visual encoder
To better understanding what visual encoder is suitable for medical applications, we compare three visual feature extraction methods (BUTD, ChexNet [19], and PixelHop++). In particular, we replace the visual encoder of BERTHop with different visual encoders and report their performance. BUTD extracts visual features from a Faster R-CNN pre-trained on Visual Genome, which is prevailing in recent V&L models.
ChexNet is a CNN-based method that is proposed for pneumonia disease detection. It is a 121-layer DenseNet [11] trained on the ChestX-ray14 dataset for pneumonia detection having all pneumonia cases labeled as positive examples and all other cases as negative examples. By modifying the loss function, it is also trained to classify all 14 thoracic diseases and achieved state-of-the-art among existing vision-only models, e.g., [26]. To augment the data, it extracts 10 crops from the image (4 corners and one center and horizontally flipped version of them) and feeds it into the network to generate a feature vector of dimension 1024 for each of them. In order to make it compatible with our transformer framework, we apply a linear transformation that maps feature vectors of size 1,024, generated by ChexNet, to 2,048. We fine-tune ChexNet and train the parameters of the linear transformation on the OpenI dataset. The results in Table 2 show that the visual encoder of BERTHop, PixelHop++, can provide more raw features of the CXR images as it uses a data-efficient method in an unsupervised manner and is capable of extracting task-agnostic image representations at different frequencies.
Table 2.
Comparison between different visual encoders (BUTD, ChexNet, and PixelHop++) under the same transformer backbone of BlueBERT. PixelHop++ outperforms BUTD and even ChexNet, which is pre-trained on a large in-domain disease diagnosis dataset.
| BUTD | ChexNet | PixelHop++ | |
|---|---|---|---|
|
| |||
| Atelectasis | 0.8866 | 0.9787 | 0.9838 |
| Cardiomegaly | 0.8875 | 0.9797 | 0.9896 |
| Effusion | 0.9120 | 0.8894 | 0.9432 |
| Mass | 0.7373 | 0.7529 | 0.9900 |
| Consolidation | 0.8906 | 0.9000 | 0.9671 |
| Emphysema | 0.8261 | 0.9067 | 0.9971 |
|
| |||
| AVG | 0.8564 | 0.8798 | 0.9823 |
5. Discussion and Conclusion
We proposed a high-performance data-efficient multimodal neural network model that jointly models X-ray images and clinical notes. In contrast with general V&L models which use an object detector to extract visual representations, our approach uses a parameter-effective visual encoder, PixelHop++, in an unsupervised setting. Our studies verify the effectiveness of the visual feature extractor PixelHop++ and the transformer backbone initialization BlueBERT. we illustrate that properly pre-training the transformer is of significance, which would provide valuable insight for designing future models. We urge our community to explore leveraging this method for other medical tasks suffering lack of annotated data. We believe that BERTHop is highly beneficial for reducing medical mistakes in disease diagnosis.
Footnotes
We will make our code public
part of the input is masked and the objective is to predict the masked words or image regions based on the remaining contexts
In the following, we use the term “BUTD” to refer to extracting visual features from a pre-trained object detector rather than the full model from [3].
References
- 1.Abiyev Rahib H and Ma’aitah Mohammad Khaleel Sallam. Deep convolutional neural networks for chest diseases detection. Journal of healthcare engineering, 2018, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allaouzi Imane and Ahmed Mohamed Ben. A novel approach for multi-label chest x-ray classification of common thorax diseases. IEEE Access, 7:64279–64288, 2019. [Google Scholar]
- 3.Anderson Peter, He Xiaodong, Buehler Chris, Teney Damien, Johnson Mark, Gould Stephen, and Zhang Lei. Bottom-up and top-down attention for image captioning and visual question answering. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 6077–6086, 2018. [Google Scholar]
- 4.Buitinck Lars, Louppe Gilles, Blondel Mathieu, Pedregosa Fabian, Mueller Andreas, Grisel Olivier, Niculae Vlad, Prettenhofer Peter, Gramfort Alexandre, Grobler Jaques, Layton Robert, VanderPlas Jake, Joly Arnaud, Holt Brian, and Varoquaux Gaël. API design for machine learning software: experiences from the scikit-learn project. In ECML PKDD Workshop: Languages for Data Mining and Machine Learning, pages 108–122, 2013. [Google Scholar]
- 5.Chen Hong-Shuo, Rouhsedaghat Mozhdeh, Ghani Hamza, Hu Shuowen, You Suya, and Kuo C. C. Jay. Defakehop: A light-weight high-performance deepfake detector, 2021.
- 6.Chen Yen-Chun, Li Linjie, Yu Licheng, El Kholy Ahmed, Ahmed Faisal, Gan Zhe, Cheng Yu, and Liu Jingjing. Uniter: Universal image-text representation learning. In European Conference on Computer Vision, pages 104–120. Springer, 2020. [Google Scholar]
- 7.Chen Yueru, Rouhsedaghat Mozhdeh, You Suya, Rao Raghuveer, and Kuo C-C Jay. Pixelhop++: A small successive-subspace-learning-based (ssl-based) model for image classification. In 2020 IEEE International Conference on Image Processing (ICIP), pages 3294–3298. IEEE, 2020. [Google Scholar]
- 8.Chou Shih-Han, Chao Wei-Lun, Lai Wei-Sheng, Sun Min, and Yang Ming-Hsuan. Visual question answering on 360deg images. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pages 1607–1616, 2020. [Google Scholar]
- 9.Devlin Jacob, Chang Ming-Wei, Lee Kenton, and Toutanova Kristina. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805, 2018. [Google Scholar]
- 10.Giger Maryellen L and Suzuki Kenji. Computer-aided diagnosis. In Biomedical information technology, pages 359–XXII. Elsevier, 2008. [Google Scholar]
- 11.Huang Gao, Liu Zhuang, Van Der Maaten Laurens, and Weinberger Kilian Q. Densely connected convolutional networks. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 4700–4708, 2017. [Google Scholar]
- 12.Johnson Alistair EW, Pollard Tom J, Greenbaum Nathaniel R, Lungren Matthew P, Deng Chih-ying, Peng Yifan, Lu Zhiyong, Mark Roger G, Berkowitz Seth J, and Horng Steven. Mimic-cxr-jpg, a large publicly available database of labeled chest radiographs. arXiv preprint arXiv:1901.07042, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krishna Ranjay, Zhu Yuke, Groth Oliver, Johnson Justin, Hata Kenji, Kravitz Joshua, Chen Stephanie, Kalantidis Yannis, Li Li-Jia, Shamma David A, et al. Visual genome: Connecting language and vision using crowdsourced dense image annotations. International journal of computer vision, 123(1):32–73, 2017. [Google Scholar]
- 14.Li Liunian Harold, Yatskar Mark, Yin Da, Hsieh Cho-Jui, and Chang Kai-Wei. Visualbert: A simple and performant baseline for vision and language. arXiv preprint arXiv:1908.03557, 2019. [Google Scholar]
- 15.Li Yikuan, Wang Hanyin, and Luo Yuan. A comparison of pre-trained vision-and-language models for multimodal representation learning across medical images and reports. In 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pages 1999–2004. IEEE, 2020. [Google Scholar]
- 16.Liu Xiaofeng, Xing Fangxu, Yang Chao, C-C Jay Kuo Suma Babu, El Fakhri Georges, Jenkins Thomas, and Woo Jonghye. Voxelhop: Successive subspace learning for als disease classification using structural mri. arXiv preprint arXiv:2101.05131, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Jiasen, Goswami Vedanuj, Rohrbach Marcus, Parikh Devi, and Lee Stefan. 12-in-1: Multi-task vision and language representation learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pages 10437–10446, 2020. [Google Scholar]
- 18.Peng Yifan, Yan Shankai, and Lu Zhiyong. Transfer learning in biomedical natural language processing: an evaluation of bert and elmo on ten benchmarking datasets. arXiv preprint arXiv:1906.05474, 2019. [Google Scholar]
- 19.Rajpurkar Pranav, Irvin Jeremy, Zhu Kaylie, Yang Brandon, Mehta Hershel, Duan Tony, Ding Daisy, Bagul Aarti, Langlotz Curtis, Shpanskaya Katie, et al. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv preprint arXiv:1711.05225, 2017. [Google Scholar]
- 20.Ren Shaoqing, He Kaiming, Girshick Ross, and Sun Jian. Faster r-cnn: towards real-time object detection with region proposal networks. IEEE transactions on pattern analysis and machine intelligence, 39(6):1137–1149, 2016. [DOI] [PubMed] [Google Scholar]
- 21.Rouhsedaghat Mozhdeh, Monajatipoor Masoud, Azizi Zohreh, and Kuo C-C Jay. Successive subspace learning: An overview. arXiv preprint arXiv:2103.00121, 2021. [Google Scholar]
- 22.Rouhsedaghat Mozhdeh, Wang Yifan, Ge Xiou, Hu Shuowen, You Suya, and Kuo C-C Jay. Facehop: A light-weight low-resolution face gender classification method. arXiv preprint arXiv:2007.09510, 2020. [Google Scholar]
- 23.Rouhsedaghat Mozhdeh, Wang Yifan, Hu Shuowen, You Suya, and Kuo C-C Jay. Low-resolution face recognition in resource-constrained environments. arXiv preprint arXiv:2011.11674, 2020. [Google Scholar]
- 24.Shin Hoo-Chang, Roth Holger R, Gao Mingchen, Lu Le, Xu Ziyue, Nogues Isabella, Yao Jianhua, Mollura Daniel, and Summers Ronald M. Deep convolutional neural networks for computer-aided detection: Cnn architectures, dataset characteristics and transfer learning. IEEE transactions on medical imaging, 35(5):1285–1298, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan Hao and Bansal Mohit. Lxmert: Learning cross-modality encoder representations from transformers. arXiv preprint arXiv:1908.07490, 2019. [Google Scholar]
- 26.Wang Xiaosong, Peng Yifan, Lu Le, Lu Zhiyong, Bagheri Mohammadhadi, and Summers Ronald M. Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 2097–2106, 2017. [Google Scholar]
- 27.Wolf Thomas, Debut Lysandre, Sanh Victor, Chaumond Julien, Delangue Clement, Moi Anthony, Cistac Pierric, Rault Tim, Louf Rémi, Funtowicz Morgan, et al. Huggingface’s transformers: State-of-the-art natural language processing. arXiv preprint arXiv:1910.03771, 2019. [Google Scholar]
- 28.Zhou Luowei, Palangi Hamid, Zhang Lei, Hu Houdong, Corso Jason, and Gao Jianfeng. Unified vision-language pre-training for image captioning and vqa. In Proceedings of the AAAI Conference on Artificial Intelligence, volume 34, pages 13041–13049, 2020. [Google Scholar]


