Abstract
Identifying key proteins from protein-protein interaction (PPI) networks is one of the most fundamental and important tasks for computational biologists. However, the protein interactions obtained by high-throughput technology are characterized by a high false positive rate, which severely hinders the prediction accuracy of the current computational methods. In this paper, we propose a novel strategy to identify key proteins by constructing reliable PPI networks. Five Gene Ontology (GO)-based semantic similarity measurements (Jiang, Lin, Rel, Resnik, and Wang) are used to calculate the confidence scores for protein pairs under three annotation terms (Molecular function (MF), Biological process (BP), and Cellular component (CC)). The protein pairs with low similarity values are assumed to be low-confidence links, and the refined PPI networks are constructed by filtering the low-confidence links. Six topology-based centrality methods (the BC, DC, EC, NC, SC, and aveNC) are applied to test the performance of the measurements under the original network and refined network. We systematically compare the performance of the five semantic similarity metrics with the three GO annotation terms on four benchmark datasets, and the simulation results show that the performance of these centrality methods under refined PPI networks is relatively better than that under the original networks. Resnik with a BP annotation term performs best among all five metrics with the three annotation terms. These findings suggest the importance of semantic similarity metrics in measuring the reliability of the links between proteins and highlight the Resnik metric with the BP annotation term as a favourable choice.
Introduction
Proteins are crucial components of cell and tissue structures and are cornerstones used by an organism to maintain normal life activities. Due to the different roles each protein plays in the life activities of organisms, proteins are divided into essential proteins and nonessential proteins. The deletion or elimination of essential proteins may result in normal cellular function disorders or diseases and may even affect the development and survival of organisms [1, 2]. Previous studies have shown that when a virus attacks the human body, it attacks essential proteins first [3]. For instance, when studying the novel coronavirus, the most important aspect is to determine several possible target proteins and then use super-large computer-aided drug screening to find effective antiviral drugs. Therefore, identifying key proteins has vital application prospects in disease diagnosis [4], drug discovery [5], and drug design [6].
Traditional biological experiments can only be carried out in a limited number of species and are expensive and time consuming [7]. Fortunately, with the rapid development of high-throughput technology, many PPI data have been accumulated, and these provide a convenient condition for identifying essential proteins with computational methods.
PPI networks provide a comprehensive view of the global interaction structure of an organism’s proteome. Initially, the key proteins were predicted by measuring topologic properties. In 2001, Jeong [8] pointed out that proteins involved in more interactions in PPI networks have higher possibilities of being key proteins; this is known as the centrality-lethality rule. Subsequently, a series of topological structure-based approaches were developed, such as the betweenness centrality (BC) [9], eigenvector centrality (EC) [10], neighborhood centrality (NC) [11], subgraph centrality (SC) [12], strength centrality (StrC) [13], average neighbor centrality (aveNC) [14], closeness centrality (CC) [15], information centrality (IC) [16], local average connectivity (LAC) [17], local interaction density (LID) [18], maximum neighborhood component (MNC), density of maximum neighborhood component (DMNC) [19], TP and TP-NC [20]. The performance of these centrality methods depends on the quality of the utilized PPI networks.
PPI networks retrieved from high-throughput techniques are incomplete and inherently noisy [21]. The reliability of yeast two-hybrid assays is approximately 50%, even for the well-studied Saccharomyces cerevisiae species; this impairs the prediction performance of the available topology-based methods.
To overcome the influence of false positive data in PPI networks, two categories of methods have been developed to improve the performance of identifying essential proteins. The first category identifies essential proteins by combining the topological properties of PPI networks with various biological data, such as Gene Ontology (GO) annotation data [22–27], gene expression profiles [23, 25, 27–31], subcellular localizations [24, 25, 31], the domain features of proteins [32], orthologous information [30, 33], and protein complex information [34, 35]. Previous studies have demonstrated that the efficient and effective integration of multiple sources of data could yield better results for identifying essential proteins. For example, Kim [22] proposed that adding gene-level annotation information, such as GO terms, to detect essential proteins would result in higher accuracy than that of existing methods. Li et al. [29] introduced a novel essential protein prediction algorithm named CPPK. CPPK predicts key proteins with a combination of network topology properties and gene expression data. Zhang et al. [23] developed a new method named TEO that combines PPI networks, and gene expression profiles with GO annotation terms, and it achieved higher accuracy in predicting key proteins than previously developed. Peng et al. [32] developed a method called UDoNC that utilize protein domain information and the topology of given PPI network. Lei et al. [24] introduced a novel strategy named RSG using RNA-Seq, GO information, and subcellular localization. Zhang et al. [25] developed TEGS, a new strategy to predict key proteins, which improved prediction accuracy by integrating network topology with subcellular localization information, gene expression profiles, and GO annotation datasets. Peng et al. [33] developed a novel measure to predict key proteins by adding orthologous data. Zhang et al. [30] designed OGN by using gene expressions, orthologies, and network topologies to identify key proteins. Li et al. [34] introduced a novel idea that combines protein complexes information with the topological properties of PPI networks.
The methods in the second category predict key proteins based on refined networks by filtering the false positive interactions in the original network. For instance, Kim et al. [26] designed a motif-based method named MCGO, which utilizes Gene Ontology annotation data to prune several uninformative edges from the given network. Li et al. [31] proposed a novel approach to reconstruct PPI networks by using gene expression information and subcellular localization information. Liu et al. [36] developed a new algorithm, EPPSO, to identify key proteins according to improved particle swarm optimization and reconstructed PPI networks by combining the topology information of the PPI networks with other biological information. Lei et al. [27] presented RWEP, which utilizes GO terms and gene expression data to construct a new weighted PPI network, and a random walk with the restart algorithm is applied to quantify the essentiality value of the protein. Simulation results show that RWEP dominates topology-based approaches in predicting key proteins. However, the performance of these approaches is still unsatisfactory, and many methods are complicated and involve many steps, which might hinder their wide application in biological research.
GO annotation is a system of uniform and normative descriptions of the genes and gene products of all species. A GO annotation collects information on the molecular function (MF), biological process (BP), and cellular component (CC) of different organisms. The GO-based semantic similarity metric (SSM) is a numerical measure that is used to estimate the semantic intimacy between two terms and is widely used for measuring the functional similarities between proteins [37–39]. Five widely used SSMs, Jiang [40], Lin [41], Rel [42], Resnik [43], and Wang [44], are applied to calculate the GO semantic similarity values at present. However, each of the SSMs focuses on characterizing particular aspects of GO annotation terms and has strengths as well as weaknesses. The advantages and disadvantages of these SSMs in evaluating GO semantic similarities are important for predicting key proteins.
In this paper, we comprehensively discuss the aforementioned five semantic similarity measurements in combination with three subontology (BP, CC, and MF) terms on the identification of essential proteins. Six centrality methods (the BC, DC, EC, NC, SC, and aveNC) are applied on refined GO-PPI networks and the results are compared with those of the same methods on the original PPI networks. Extensive comparisons have been conducted under different conditions, and the simulation results offer a reference to biologists when investigating the essential proteins of PPI networks.
Methods
In this part, six conventional centrality methods (the BC, DC, EC, NC, SC, and aveNC) are reviewed briefly. Then, refined PPI network construction methods are described in detail. Additionally, the utilized datasets and evaluation metrics are presented.
Centrality methods
PPI networks are abstracted into graph structures, which are denoted as PPI = (P, E), where P is composed of proteins and E represents the set of interactions between proteins. PPI networks are stored as adjacent matrices. The six centrality calculation methods are calculated as follows.
- BC
where Sp(i, j) represents the number of shortest paths between protein i and j that go through protein p and S(i, j) represents the number of shortest paths between protein i and protein j. Considering the global characteristics of PPI networks, this method can identify some nodes whose degrees are not high but play a vital role in the connection of the given network.(1) - DC
where deg(p) represents the number of proteins connected to p directly, which is called the degree of p. And ap,u ∈ A represents the interactions between proteins p and u.(2) - EC
where α is a eigenvector of the adjacency matrix A and αmax(p) is the pth component of the eigenvector belonging to the maximum eigenvalue λmax.(3) - NC
where Np and Nu represent the neighboring sets of proteins p and u, respectively. ECC is the edge clustering coefficient. This method characterizes the connection relationships between a node and its neighbors; that is, the similarity of the relationship between two proteins is described by calculating the number of common neighbor nodes.(4) - SC
where μl(p) represents the number of loops whose starting and ending proteins are p and the lengths of these loops are l. In complex networks, essential proteins tend to form dense subgraphs. The shorter the loop is, the more likely the protein is to be in a dense subgraph and to be a key protein.(5) - aveNC
where Np represents the set of protein p’s neighbors. The significance of a protein is measured by its neighbors.(6)
Constructing refined PPI network by applying GO-based SSMs
There are two kinds of measures used to record confidence scores for a PPI network. One relies on interaction data [45], and the other takes gene expression values [46], functional similarities [39, 47], and other information into consideration [37]. According to the basic idea that proteins interacting in the same cell have a higher possibility of being involved in a similar biological process than that do not interact, we assume that the protein pairs with smaller semantic similarity values are more likely to be false positive links.
Five widely used methods, Jiang, Lin, Rel, Resnik, and Wang, are applied to compute semantic similarities based on the GO terms between proteins, and these are denoted as confidence scores. Wang determines the confidence scores between two proteins according to the locations of their corresponding GO terms in the GO graph and their ancestor terms’ relationships. The other four methods are based on information content (IC), which depends on the probabilities of the two GO terms involved and their closest common ancestor terms in the corpus of the GO annotation information.
The details of the five SSMs (semantic similarity metric) are shown as follows:
-
Resnik
Resnik believes that information content (IC) is the most informative common ancestor (MICA) [48]. The similarity between protein pairs m and v in this method is denoted as
where C represents the set of common ancestors of m and v. The IC mentioned above is denoted as IC(t) = −lnp(t), where p(t) represents the probability of occurrence in the GO corpus and IC is used to express the specificity of a protein.(7) -
Lin and Jiang
It seems that the performance of Resnik is valid for calculating the similarity of two terms, but it cannot distinguish between terms that have the same MICA. To tackle this problem, Lin and Jiang developed new methods with comprehensive consideration of the ICs between protein pairs and their MICAs. The similarity of two proteins based on the Lin and Jiang methods is defined as(8) (9) -
Rel
Shortcomings still exist in the approaches developed by Lin and Jiang. The similarity between two terms is overestimated when a protein is an ancestor of another. In addition, these approaches ignore the specificities of the two terms. By combining Resnik and Lin, Rel presented a novel measure to capture the similarity between two terms. The similarity between two proteins is defined as(10) -
Wang
Wang is a hybrid method that combines the number of common ancestors with the locations of these ancestors in the GO graph when calculating the similarity between two terms. GO terms are presented as directed acyclic graphs (DAGs). Suppose that Gv = (Pv, Ev) is a GAG for a GO term v, where Pv contains the ancestor terms of v including itself, and Ev is composed of edges that connect the GO terms in Gv. Other terms closer to v in Gv contribute more to its semantics. The contribution of a protein u to the semantics of protein v in Gv is defined as the S-value of u and is calculated as
where ωe(0 < ωe < 1) is the semantic contribution factor for edge e ∈ Ev that links term u with its child term u′. And SV(v) is used to compare the semantics of two GO terms, and SV(v)is defined as(11) (12)
The semantic similarity between protein pairs m and v is denoted as
| (13) |
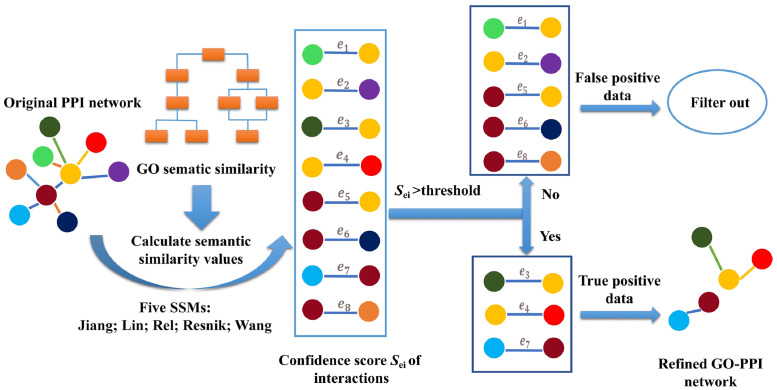
In this article, we apply five GO-based semantic similarity measurements to measure the reliability of protein pairs. For each SSM, we first compute the confidence scores for all of the protein pairs, and then construct refined PPI networks by filtering the interactions with low confidence scores. The refined PPI networks we obtain by measuring the GO semantic similarity are named GO-PPI for short, and the network refined by using the Resnik metric under the BP annotation term is named Resnik-BP GO-PPI for short. The main idea of constructing a refined GO-PPI network is shown in Fig 1.
Fig 1. The process of constructing a GO-PPI network.
Experimental data
To compare the performance of these centrality methods under different combinations of strategies, we choose the well-studied Saccharomyces cerevisiae PPI data for experiments, as they are widely applied for testing the performance of new methods. The datasets include the YDIP dataset composed of 5093 proteins and 24743 interactions, the new DIP dataset, which includes 4928 proteins and 17201 interactions, the Krogan dataset containing 7123 interactions among 2708 proteins, and the Krogan Extended dataset, which consists of 3672 proteins with 14317 interactions. A summary of these datasets is presented in Table 1.
Table 1. The detailed information of four PPI datasets.
| Dataset | Proteins | Interactions | Essential proteins | Density |
|---|---|---|---|---|
| YDIP | 5093 | 24743 | 1167 | 0.0019 |
| DIP PPI | 4928 | 17201 | 1150 | 0.0014 |
| Krogan | 2708 | 7123 | 786 | 0.0019 |
| Krogan Extended | 3672 | 14317 | 929 | 0.0021 |
The GO annotation information of each protein is downloaded from the Saccharomyces Genome Database, which was released on September 10th, 2020.
The benchmark of a known essential protein dataset including 1285 proteins is collected from four different databases (MIPS [49], SGD [50], DEG [51], and SGDP (http://www.sequence.stanford.edu/group/).
Evaluation metrics
To measure the efficiency of the proposed strategy, we calculate the numbers of key proteins predicted correctly among the top 600 ranked proteins, and the corresponding prediction precisions of the six topology-based methods are also calculated under the original PPI network and refined GO-PPI network. The prediction precision is denoted as
| (14) |
where TP describes the number of true positives, and FP describes the number of false positives.
Results and discussion
To evaluate whether the performance of the reconstructed GO-PPI network is better than that of the corresponding original PPI network in identifying key proteins, six topology structure-based methods (the BC, DC, EC, NC, SC, and aveNC) are applied in the experiments. We compare the numbers of key proteins identified properly and the prediction precisions under different types of strategies. The threshold for GO semantic similarity is set to 0.33 for filtering the unreliable links in the PPI networks.
Analysis of the original network and refined GO-PPI network
The number of interactions in a network influences the speed of calculation for identifying essential proteins. The lower the number of interactions, the less time is required for the calculation. Therefore, we compute the number of interactions and the portions of key proteins under the original PPI network and refined the GO-PPI network for the YDIP dataset. As shown in Table 2, the number of interactions declines dramatically after filtering the links with low-confidence scores, and more than half of the interactions are filtered, so the computational efficiency is greatly improved. Furthermore, the numbers of proteins and key proteins are reduced, but the portion of essential proteins is increased, which is more beneficial for identifying key proteins. For example, in networks with the application of the Resnik metric, the proportions of essential proteins under the three subontologies (the BP, CC, and MF) reach 39.83%, 41.55%, and 37.82%, respectively, while they are 22.91% in the original PPI network.
Table 2. The number of interactions and the portions of essential proteins under the original PPI network and GO-PPI network for the YDIP dataset.
| Ontology | SSMs | Network | No. interactions | No. proteins | Portion of essential proteins | ||
|---|---|---|---|---|---|---|---|
| Reserved | Filtered | Nonessential | Essential | ||||
| original PPI | 24743 | 0 | 5093 | 1167 | 22.91% | ||
| BP | Jiang | GO-PPI | 5456 | 19287 | 2313 | 785 | 33.94% |
| Lin | GO-PPI | 6323 | 18420 | 2522 | 833 | 33.03% | |
| Rel | GO-PPI | 5746 | 18997 | 2428 | 825 | 33.98% | |
| Resnik | GO-PPI | 3336 | 21407 | 1725 | 687 | 39.83% | |
| Wang | GO-PPI | 5418 | 19325 | 2644 | 864 | 32.68% | |
| CC | Jiang | GO-PPI | 11762 | 12981 | 3535 | 951 | 26.90% |
| Lin | GO-PPI | 8408 | 16335 | 3107 | 900 | 28.97% | |
| Rel | GO-PPI | 6140 | 18603 | 2760 | 849 | 30.76% | |
| Resnik | GO-PPI | 2018 | 22725 | 1160 | 482 | 41.55% | |
| Wang | GO-PPI | 18082 | 6661 | 4182 | 1069 | 25.56% | |
| MF | Jiang | GO-PPI | 3640 | 21103 | 1821 | 615 | 33.77% |
| Lin | GO-PPI | 3177 | 21566 | 1733 | 594 | 34.28% | |
| Rel | GO-PPI | 2821 | 21922 | 1667 | 585 | 35.09% | |
| Resnik | GO-PPI | 1331 | 23412 | 1092 | 413 | 37.82% | |
| Wang | GO-PPI | 4902 | 19841 | 2733 | 792 | 28.98% | |
In the meantime, we study the interactions that rank among the top 600. As the numbers of interactions are different for the original network and the reconstructed GO-PPI network, we compute the proportions of the interactions between essential protein pairs (Ess-ess), essential and nonessential protein pairs (Ess-noness), and nonessential pairs (Noness-noness), and the results for the YDIP dataset under the BP subontology are shown in Table 3. It can be seen that the portion of Ess-ess interactions is significantly improved under the five refined GO-PPI networks, and the portions of Ess-noness and Noness-noness interactions under the GO-PPI network are much lower than those under the original PPI network. We can also see that Wang achieves the best performance compared with those of the other four SSMs. For instance, the portion of essential pairs reaches 57.99% when using the NC method under Wang, which is the highest for the six different networks. And the interactions between the essential and nonessential pairs are only 12.87% of total interactions under Wang versus 37.27% under the original PPI network for the SC method.
Table 3. The portions of interactions under the original PPI network and GO-PPI network for the YDIP dataset (BP).
| SSMs | Network | Interactions | BC | DC | EC | NC | SC |
|---|---|---|---|---|---|---|---|
| original PPI | Ess-ess | 17.11% | 20.03% | 17.21% | 32.11% | 17.21% | |
| Ess-noness | 37.64% | 35.91% | 37.27% | 28.12% | 37.27% | ||
| Noness-noness | 45.25% | 44.06% | 45.52% | 39.77% | 45.52% | ||
| Jiang | GO-PPI | Ess-ess | 34.85% | 46.72% | 47.32% | 55.98% | 49.46% |
| Ess-noness | 28.34% | 20.12% | 17.41% | 16.87% | 15.94% | ||
| Noness-noness | 36.82% | 33.16% | 35.27% | 27.14% | 34.60% | ||
| Lin | GO-PPI | Ess-ess | 31.87% | 45.07% | 47.65% | 53.64% | 48.64% |
| Ess-noness | 28.85% | 20.15% | 18.05% | 17.38% | 17.42% | ||
| Noness-noness | 39.28% | 34.78% | 34.30% | 28.98% | 33.94% | ||
| Rel | GO-PPI | Ess-ess | 33.51% | 47.71% | 52.74% | 56.22% | 52.50% |
| Ess-noness | 28.95% | 20.18% | 15.10% | 17.12% | 15.59% | ||
| Noness-noness | 37.53% | 32.12% | 32.16% | 26.66% | 31.91% | ||
| Resnik | GO-PPI | Ess-ess | 34.85% | 46.72% | 47.32% | 55.98% | 49.46% |
| Ess-noness | 28.34% | 20.12% | 17.41% | 16.87% | 15.94% | ||
| Noness-noness | 36.82% | 33.16% | 35.27% | 27.14% | 34.60% | ||
| Wang | GO-PPI | Ess-ess | 35.57% | 51.87% | 55.39% | 57.99% | 55.99% |
| Ess-noness | 29.18% | 17.90% | 13.79% | 17.06% | 12.87% | ||
| Noness-noness | 35.24% | 30.23% | 30.81% | 24.95% | 31.14% |
Comparison of the numbers of true predictions under different strategies
In this part, we do a systematic evaluation of the performance of the newly constructed networks on the four test datasets. For each dataset, we adopt five SSMs to calculate confidence scores for the protein pairs in the PPI network under the three GO annotation terms (the BP, MF, and CC) and obtain fifteen kinds of refined GO-PPI networks. Six centrality methods are applied to predict the key proteins of the newly constructed GO-PPI network and the original PPI network.
Table 4 presents the numbers of essential proteins correctly identified from the top 600 candidate proteins of the original network and refined GO-PPI network with different SSMs under the three sub-ontologies (the BP, CC, and MF). As seen from Table 4, for the YDIP dataset, the numbers of essential proteins correctly identified under the six centrality methods on each of the newly constructed GO-PPI networks are consistently larger than those under the corresponding original PPI networks. For example, compared to the original network, the EC method yields an improvement of 57.01% on the Wang-BP (the Wang method under BP subontology) network, and the aveNC method provides an improvement of 300% on the Resnik-BP network. In terms of three subontologies, the performance of these methods under the refined GO-PPI network obtained with BP annotation term is significantly better than it under CC and MF annotation terms, especially for the Resnik and Wang methods.
Table 4. The numbers of essential proteins detected by the six centrality methods under different strategies for the YDIP dataset (top 600).
| Ontology | SSMs | Network | BC | DC | EC | NC | SC | aveNC |
|---|---|---|---|---|---|---|---|---|
| original PPI | 220 | 251 | 221 | 309 | 221 | 80 | ||
| BP | Jiang | GO-PPI | 266 | 327 | 310 | 351 | 324 | 221 |
| Lin | GO-PPI | 257 | 329 | 332 | 349 | 334 | 201 | |
| Rel | GO-PPI | 266 | 336 | 344 | 353 | 341 | 222 | |
| Resnik | GO-PPI | 311 | 363 | 340 | 351 | 349 | 320 | |
| Wang | GO-PPI | 265 | 344 | 347 | 356 | 346 | 259 | |
| CC | Jiang | GO-PPI | 222 | 253 | 217 | 325 | 214 | 100 |
| Lin | GO-PPI | 246 | 291 | 237 | 321 | 247 | 124 | |
| Rel | GO-PPI | 237 | 299 | 263 | 318 | 274 | 173 | |
| Resnik | GO-PPI | 301 | 317 | 280 | 315 | 291 | 295 | |
| Wang | GO-PPI | 229 | 269 | 228 | 328 | 228 | 92 | |
| MF | Jiang | GO-PPI | 239 | 272 | 246 | 276 | 249 | 184 |
| Lin | GO-PPI | 238 | 269 | 240 | 291 | 249 | 216 | |
| Rel | GO-PPI | 237 | 267 | 248 | 289 | 254 | 227 | |
| Resnik | GO-PPI | 237 | 261 | 238 | 242 | 243 | 234 | |
| Wang | GO-PPI | 230 | 278 | 259 | 289 | 232 | 145 |
To verify the superiority of the newly proposed strategy, we calculate the number of key proteins identified correctly by each method under three GO subontology terms for the reduced DIP PPI dataset, the Krogan dataset, and the Krogan Extended dataset. The calculation results are listed in Tables 5–7.
Table 5. The number of essential proteins detected by the six centrality methods under different strategies for the new DIP dataset (top 600).
| Ontology | SSMs | Network | BC | DC | EC | NC | SC | aveNC |
|---|---|---|---|---|---|---|---|---|
| original PPI | 239 | 274 | 160 | 318 | 163 | 72 | ||
| BP | Jiang | GO-PPI | 285 | 347 | 337 | 351 | 333 | 258 |
| Lin | GO-PPI | 275 | 350 | 332 | 346 | 329 | 228 | |
| Rel | GO-PPI | 282 | 358 | 345 | 349 | 345 | 261 | |
| Resnik | GO-PPI | 355 | 370 | 348 | 351 | 345 | 340 | |
| Wang | GO-PPI | 285 | 349 | 336 | 352 | 349 | 285 | |
| CC | Jiang | GO-PPI | 226 | 289 | 148 | 315 | 201 | 123 |
| Lin | GO-PPI | 253 | 309 | 203 | 311 | 293 | 171 | |
| Rel | GO-PPI | 261 | 319 | 279 | 303 | 294 | 212 | |
| Resnik | GO-PPI | 298 | 309 | 280 | 308 | 321 | 301 | |
| Wang | GO-PPI | 252 | 294 | 216 | 340 | 272 | 94 | |
| MF | Jiang | GO-PPI | 244 | 276 | 261 | 285 | 256 | 226 |
| Lin | GO-PPI | 260 | 276 | 254 | 288 | 260 | 230 | |
| Rel | GO-PPI | 265 | 286 | 278 | 286 | 279 | 241 | |
| Resnik | GO-PPI | 231 | 250 | 239 | 242 | 248 | 240 | |
| Wang | GO-PPI | 242 | 282 | 256 | 306 | 225 | 171 |
Table 7. The number of essential proteins detected by the six centrality methods under different strategies for the Krogan Extended dataset (top 600).
| Ontology | SSMs | Network | BC | DC | EC | NC | SC | aveNC |
|---|---|---|---|---|---|---|---|---|
| original PPI | 240 | 271 | 227 | 305 | 227 | 103 | ||
| BP | Jiang | GO-PPI | 264 | 324 | 277 | 326 | 280 | 253 |
| Lin | GO-PPI | 265 | 323 | 284 | 328 | 297 | 251 | |
| Rel | GO-PPI | 264 | 327 | 276 | 329 | 299 | 266 | |
| Resnik | GO-PPI | 317 | 327 | 306 | 326 | 307 | 305 | |
| Wang | GO-PPI | 275 | 332 | 317 | 327 | 316 | 288 | |
| CC | Jiang | GO-PPI | 215 | 259 | 215 | 313 | 217 | 149 |
| Lin | GO-PPI | 219 | 276 | 250 | 304 | 267 | 190 | |
| Rel | GO-PPI | 231 | 286 | 270 | 305 | 282 | 254 | |
| Resnik | GO-PPI | 296 | 301 | 288 | 303 | 283 | 308 | |
| Wang | GO-PPI | 253 | 275 | 242 | 309 | 241 | 147 | |
| MF | Jiang | GO-PPI | 253 | 273 | 249 | 267 | 251 | 229 |
| Lin | GO-PPI | 266 | 280 | 263 | 250 | 250 | 247 | |
| Rel | GO-PPI | 265 | 285 | 258 | 255 | 250 | 254 | |
| Resnik | GO-PPI | 258 | 259 | 250 | 251 | 254 | 257 | |
| Wang | GO-PPI | 227 | 281 | 235 | 276 | 241 | 183 |
For the new DIP PPI dataset, the comparison results are shown in Table 5. We can observe that the six centrality methods perform best under the refined GO-PPI network constructed by using the Resnik metric with the BP subontology, suggesting that this network is relatively more accurate and complete than it is under the MF and CC subontologies.
However, for the MF and CC subontologies, some of the centrality methods perform poorly under the refined GO-PPI network, such as the BC method under the Jiang-CC (the Jiang method under the CC subontology) PPI network and the DC method under the Resnik-MF (the Resnik method under the MF subontology) PPI network. The maximum number of essential proteins predicted by the NC method in all five newly constructed PPI networks under the MF subontology is 306, which is compared to the 318 correctly predicted essential proteins under the original PPI network. Considering the number of interactions under the refined GO-PPI network in the MF subontology (Table 2), this is might due to the GO annotation under MF is incomplete for the protein pairs in the new DIP dataset; therefore, the confidence scores of many true interacting protein pairs are assigned to 0, and the refined network constructed by using the five SSMs is relatively sparse, which hinders the performance of the NC centrality approach in identifying key proteins.
As seen from the results obtained using the Krogan dataset in Table 6, the performance of these six centrality methods under the refined GO-PPI networks constructed by using the five SSMs with the BP and CC annotation terms dominates the number of true key proteins predicted under the original networks. In particular, under the GO-PPI network filtered by the Wang method under the BP term, the numbers of correctly identified proteins achieved by the two centrality methods (the DC and NC) reach 336, which is significantly larger than that on the original PPI network. For the CC annotation term, the network filtered by using the Resnik metric is relatively more precise than other methods in predicting key proteins. Compared to the number of correct predictions obtained under the original PPI network, more than half of the centrality methods performed better under the newly constructed network with the MF sub-annotation term, except for the DC and NC methods.
Table 6. The number of essential proteins detected by the six centrality methods under different strategies for the Krogan dataset (top 600).
| Ontology | SSMs | Network | BC | DC | EC | NC | SC | aveNC |
|---|---|---|---|---|---|---|---|---|
| original PPI | 227 | 288 | 228 | 305 | 242 | 141 | ||
| BP | Jiang | GO-PPI | 302 | 317 | 268 | 325 | 278 | 297 |
| Lin | GO-PPI | 298 | 320 | 287 | 324 | 304 | 290 | |
| Rel | GO-PPI | 305 | 329 | 290 | 323 | 300 | 293 | |
| Resnik | GO-PPI | 311 | 328 | 299 | 312 | 305 | 325 | |
| Wang | GO-PPI | 308 | 336 | 309 | 336 | 307 | 301 | |
| CC | Jiang | GO-PPI | 217 | 279 | 245 | 309 | 255 | 236 |
| Lin | GO-PPI | 236 | 290 | 250 | 309 | 281 | 268 | |
| Rel | GO-PPI | 262 | 295 | 264 | 311 | 291 | 282 | |
| Resnik | GO-PPI | 300 | 309 | 289 | 305 | 305 | 304 | |
| Wang | GO-PPI | 235 | 292 | 243 | 300 | 263 | 170 | |
| MF | Jiang | GO-PPI | 266 | 270 | 254 | 266 | 254 | 269 |
| Lin | GO-PPI | 266 | 273 | 247 | 271 | 260 | 269 | |
| Rel | GO-PPI | 271 | 278 | 263 | 273 | 272 | 275 | |
| Resnik | GO-PPI | 252 | 254 | 256 | 254 | 260 | 257 | |
| Wang | GO-PPI | 264 | 276 | 229 | 264 | 240 | 251 |
Similar results are obtained on the Krogan Extended dataset and listed in Table 7. The number of key proteins truly predicted under the newly refined GO-PPI networks constructed with the BP subontology is consistently larger than that under the original PPI networks, and the refined network dominates the the network constructed with the CC and MF subontologies in terms of performance.
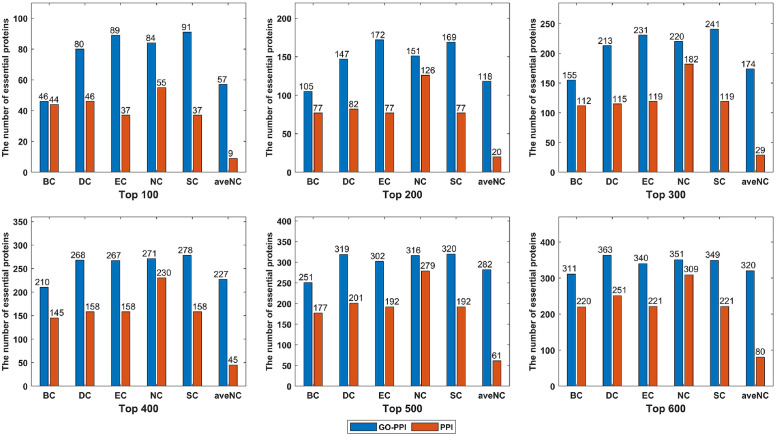
To further investigate the performance of the six centrality methods under the newly refined networks, we take the network constructed by using the Resnik metric with BP subontology for the YDIP dataset as an example. We calculate the numbers of key proteins predicted by these centrality approaches among the top 100, 200, 300, 400, 500, and 600 ranked candidates. As shown in Fig 2, the performance of these six topology-based methods is highly improved under the reconstructed GO-PPI network in terms of the number of key proteins identified correctly. Particularly, for the SC method, 91 out of 100 candidate predicted proteins are correctly identified, which is significantly more than those predicted by all of the other state-of-the-art approaches. When compared to the results of the original PPI network, 85.22% and 94.12% improvements are still achieved by the DC and EC methods under the GO-PPI networks for the top 300 candidates. For the SC and aveNC approaches, the improvements yielded are both greater than 100% with the application of the GO-PPI networks when predicting the top 300 candidate proteins.
Fig 2. The numbers of key proteins predicted correctly under the original PPI network and reconstructed GO-PPI network for the YDIP dataset.
Comparison of prediction precision for the six centrality methods
To validate the advantage of the reconstructed GO-PPI network in predicting key proteins intuitively, six centrality approaches (the BC, DC, EC, NC, SC, and aveNC) are taken to predict key proteins under the original PPI network and reconstructed GO-PPI network.
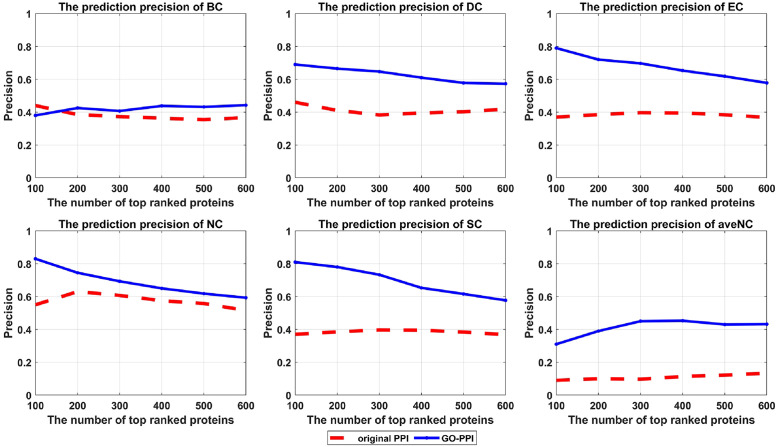
Fig 3 shows the prediction precision comparison for the six centrality approaches under the original PPI network and GO-PPI network reconstructed by using the Wang method with the BP subontology information for the YDIP dataset. Fig 3 shows that the prediction precisions of these six methods under the newly constructed GO-PPI network show significant improvements over those obtained with the original PPI network.
Fig 3. Prediction precision of six centrality methods under the original PPI network and reconstructed GO-PPI network for the YDIP dataset.
Comparison of ROC curves
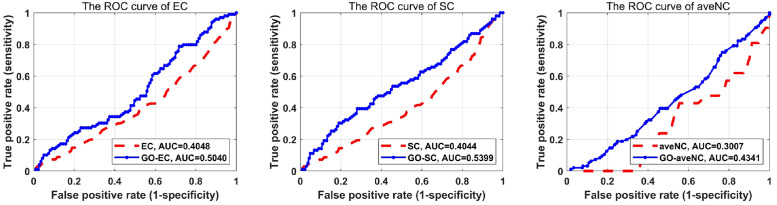
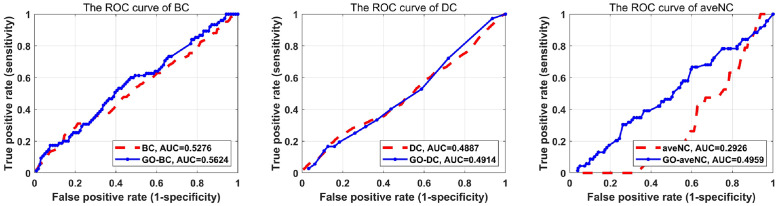
To further exhibit the performance of proposed strategy, we compared the ROC curves of different methods under original PPI networks and corresponding GO-PPI networks. The top 600 ranked proteins predicted by each method are assumed as essential, the rest proteins are non-essential. For the gold-standard essential proteins in GO-PPI is obtained from the original true essential protein sets and filtered the proteins that are not in GO-PPI network. The rank value of each protein in original PPI network and GO-PPI network are normalized, and the true positive rate as well as false positive rate is calculated by using the threshold value varies in [0, 1]. We draw the ROC curve by using the obtained true positive rate and false positive fate. AUC means the area under the ROC curve and calculated by using trapz function in Matlab. The comparison of ROC curves as well as AUC value under original new DIP PPI and YDIP PPI network are shown as following Figs 4 and 5. As shown in Figs 4 and 5, the ROC curves under GO-PPI network is higher than the corresponding original PPI network, suggesting that the GO-PPI network we constructed is reliable for predicting essential proteins.
Fig 4. The comparison of ROC curves for original new DIP PPI network and Wang-BP (the Wang method under the BP subontology) PPI network under (left) EC method, (middle) SC method and (right) aveNC method.
Fig 5. The comparison of ROC curves for original YDIP PPI network and Resink-BP (the Resink method under the BP subontology) PPI network under (left) BC method, (middle) DC method and (right) aveNC method.
Analysis of the effect of the threshold
Since the new GO-PPI network is constructed by filtering the unreliable links in the original PPI network, we need to choose an appropriate threshold to distinguish false positive data and real interactions. However, the threshold value is related to the SSMs and the quality of a given PPI network, and different thresholds should be set for different SSMs to achieve the best performance.
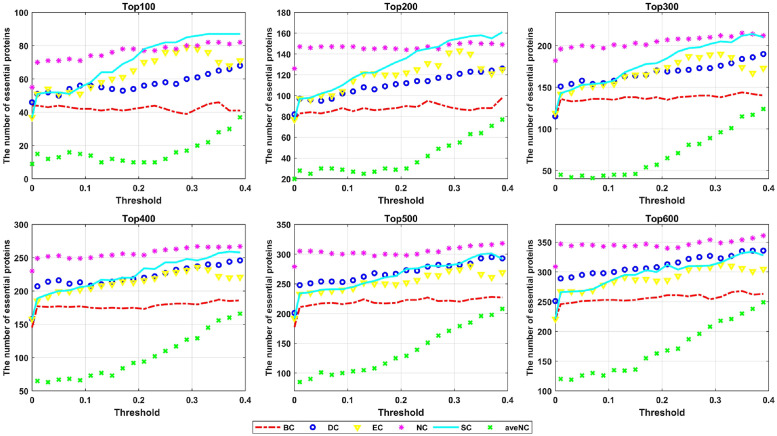
To investigate the effect of the threshold on the performance of the methods in identifying essential proteins, we plot the true numbers of key proteins identified among the top 100, 200, 300, 400, 500, and 600 candidates as functions of the threshold value for the YDIP Jiang-BP network in Fig 6. As shown in Fig 6, the numbers of correct predictions increase with increasing threshold value for all of the methods, especially the DC, SC, and aveNC methods. The results show that GO semantic similarity is efficient in filtering unreliable links in PPI networks, and almost all of the considered methods achieve the maximum number of correctly predicted essential proteins with a relatively large threshold value.
Fig 6. The numbers of correctly identified key proteins among the top 100, 200, 300, 400, 500, and 600 candidates for different method vs the threshold value.
Conclusions
Predicting essential proteins by developing computational methods from PPI networks has been a hot topic in recent years. However, the PPIs obtained by high-throughput technology at present have high false positive rates. False interactions in PPI networks have great effects on the performance of computational methods in terms of predicting key proteins. Semantic similarity measures have been shown to be useful for assessing the confidence scores between linked protein pairs. The best of the five current widely used semantic similarity measurements for selecting appropriate metrics to measure the reliability of interactions remains unclear.
This paper presents a comparison between GO-PPI networks newly constructed by five semantic similarity methods with three GO annotation terms and corresponding original PPI networks. The six topological-based centrality methods (the BC, DC, EC, NC, SC, and aveNC) are used to calculate the numbers of correct predictions and the precisions for the 600 top-ranked candidate proteins under the newly constructed GO-PPI networks and original networks. The comparison results suggest that the prediction accuracies under each of the newly constructed GO-PPI networks are consistently higher than those under the original PPI network. In particular, the networks constructed by using the semantic similarity metrics of Resnik and Wang under the BP annotation term are most reliable for predicting essential proteins among these topological-based centrality methods. These results suggest that constructing a new PPI network by using the Resnik and Wang metrics under the BP annotation term can filter out some false positive data effectively and improve the quality of the network, which is also the direction of future research.
Data Availability
The source simulation code and data used in this paper is available at https://github.com/wzhangwhu/EPI.
Funding Statement
This work is supported by National Natural Science Foundation of China Grant No.12161039 and 61802125, and Natural Science Foundation of Jiangxi Province Grant No.20212ACB211002,No.20224BAB201011 and No.20181BAB202006.
References
- 1. Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999; 285(5429): 901–906. doi: 10.1126/science.285.5429.901 [DOI] [PubMed] [Google Scholar]
- 2. Glass JI, Hutchison CA III, Smith HO. and Venter JC. A systems biology tour de force for a near-minimal bacterium. Molecular systems biology. 2009; 5(1): 330. doi: 10.1038/msb.2009.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Acencio ML and Lemke N. Towards the prediction of essential genes by integration of network topology, cellular localization and biological process information. BMC bioinformatics. 2009; 10(1):1–18. doi: 10.1186/1471-2105-10-290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Furney SJ, Albà M. and López-Bigas N. Differences in the evolutionary history of disease genes affected by dominant or recessive mutations. BMC genomics. 2006; 7(1): 1–11. doi: 10.1186/1471-2164-7-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Steinmetz LM, Scharfe C, Deutschbauer AM, Mokranjac D, Herman ZS, Jones T, et al. Systematic screen for human disease genes in yeast. Nature genetics. 2002; 31(4): 400–404. doi: 10.1038/ng929 [DOI] [PubMed] [Google Scholar]
- 6. Lu Y., Deng J, Rhodes JC, Lu H. and Lu LJ. Predicting essential genes for identifying potential drug targets in Aspergillus fumigatus. Computational biology and chemistry. 2014; 50: 29–40. doi: 10.1016/j.compbiolchem.2014.01.011 [DOI] [PubMed] [Google Scholar]
- 7. Tang X, Wang J, Zhong J. and Pan Y. Predicting essential proteins based on weighted degree centrality. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2014; 11(2): 407–418. doi: 10.1109/TCBB.2013.2295318 [DOI] [PubMed] [Google Scholar]
- 8. Jeong H, Mason SP, Barabási AL. and Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001; 411(6833): 41–42. doi: 10.1038/35075138 [DOI] [PubMed] [Google Scholar]
- 9. Joy MP, Brock A, Ingber DE. and Huang S. High-betweenness proteins in the yeast protein interaction network. Journal of Biomedicine and Biotechnology. 2005; 2005(2): 96–103. doi: 10.1155/JBB.2005.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bonacich P. Power and centrality: A family of measures. American journal of sociology. 1987; 92(5): 1170–1182. doi: 10.1086/228631 [DOI] [Google Scholar]
- 11. Wang J, Li M, Wang H. and Pan Y. Identification of essential proteins based on edge clustering coefficient. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2012; 9(4):1070–1080. doi: 10.1109/TCBB.2011.147 [DOI] [PubMed] [Google Scholar]
- 12. Estrada E. and Rodríguez-Velázquez JA. Subgraph centrality in complex networks. Physical Review E. 2005; 71(5): 056103. doi: 10.1103/PhysRevE.71.056103 [DOI] [PubMed] [Google Scholar]
- 13. Barrat A, Barthélemy M, Pastor-Satorras R. and Vespignani A. The architecture of complex weighted networks. Proceedings of the national academy of sciences. 2004; 101(11): 3747–3752. doi: 10.1073/pnas.0400087101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He DR, Liu ZH. and Wang BH. Complex Systems and Complex Networks. Higher Education Press, Beijing. 2009; pp.130–131 (Chinese). ISBN: 978-7-04-025627-7 [Google Scholar]
- 15. Wuchty S. and Stadler PF. Centers of complex network. Journal of Theoretical Biology. 2003; 223(1): 45–53. doi: 10.1016/S0022-5193(03)00071-7 [DOI] [PubMed] [Google Scholar]
- 16. Stephenson K. and Zelen M. Rethinking centrality: Methods and examples. Social networks. 1989; 11(1): 1–37. doi: 10.1016/0378-8733(89)90016-6 [DOI] [Google Scholar]
- 17. Li M, Wang J, Chen X, Wang H. and Pan Y. A local average connectivity-based method for identifying essential proteins from the network level. Computational biology and chemistry. 2011; 35(3): 143–150. doi: 10.1016/j.compbiolchem.2011.04.002 [DOI] [PubMed] [Google Scholar]
- 18. Qi Y. and Luo J. Prediction of essential proteins based on local interaction density. IEEE/ACM transactions on computational biology and bioinformatics. 2016; 13(6): 1170–1182. doi: 10.1109/TCBB.2015.2509989 [DOI] [PubMed] [Google Scholar]
- 19. Lin CY, Chin CH, Wu HH, Chen SH, Ho CW. and Ko MT. Hubba: hub objects analyzer-a framework of interactome hubs identification for network biology. Nucleic acids research. 2008; 36(suppl_2): W438–W443. doi: 10.1093/nar/gkn257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li M, Lu Y, Wang J, Wu FX. and Pan Y. A topology potential-based method for identifying essential proteins from PPI networks. IEEE/ACM transactions on computational biology and bioinformatics. 2015; 12(2): 372–383. doi: 10.1109/TCBB.2014.2361350 [DOI] [PubMed] [Google Scholar]
- 21. Sprinzak E, Sattath S. and Margalit H. How reliable are experimental protein-protein interaction data? Journal of molecular biology. 2003; 327(5): 919–923. [DOI] [PubMed] [Google Scholar]
- 22. Kim W. Prediction of essential proteins using topological properties in GO-pruned PPI network based on machine learning methods. Tsinghua Science and Technology. 2012; 17(6): 645–658. doi: 10.1109/TST.2012.6374366 [DOI] [Google Scholar]
- 23.Kim W, Li M, Wang J. and Pan Y. Essential protein discovery based on network motif and gene ontology. 2011 IEEE International Conference on Bioinformatics and Biomedicine. 2011; pp: 470–475.
- 24. Lei X, Yang X. and Fujita H. Random walk based method to identify essential proteins by integrating network topology and biological characteristics. Knowledge-Based Systems. 2019; 167: 53–67. doi: 10.1016/j.knosys.2019.01.012 [DOI] [Google Scholar]
- 25. Lei X, Zhao J, Fujita H. and Zhang A. Predicting essential proteins based on RNA-Seq, subcellular localization and GO annotation datasets. Knowledge-Based Systems. 2018; 151: 136–148. doi: 10.1016/j.knosys.2018.03.027 [DOI] [Google Scholar]
- 26. Zhang W, Xu J, Li Y. and Zou X. Detecting essential proteins based on network topology, gene expression data, and Gene Ontology information. IEEE/ACM transactions on computational biology and bioinformatics,2018; 15(1): 109–116. doi: 10.1109/TCBB.2016.2615931 [DOI] [PubMed] [Google Scholar]
- 27. Zhang W, Xu J. and Zou X. Predicting essential proteins by integrating network topology, subcellular localization information, gene expression profile and GO annotation data. IEEE/ACM transactions on computational biology and bioinformatics. 2020; 17(6): 2053–2061. doi: 10.1109/TCBB.2019.2916038 [DOI] [PubMed] [Google Scholar]
- 28. Li M, Ni P, Chen X, Wang J, Wu FX. and Pan Y.Construction of refined protein interaction network for predicting essential proteins. IEEE/ACM transactions on computational biology and bioinformatics. 2019; 16(4): 1386–1397. doi: 10.1109/TCBB.2017.2665482 [DOI] [PubMed] [Google Scholar]
- 29. Li M, Zhang H, Wang J. and Pan Y. A new essential protein discovery method based on the integration of protein-protein interaction and gene expression data. BMC systems biology. 2012; 6(1): 15. doi: 10.1186/1752-0509-6-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li M, Zheng R, Zhang H, Wang J. and Pan Y. Effective identification of essential proteins based on priori knowledge, network topology and gene expressions. Methods. 2014; 67(3):325–333. doi: 10.1016/j.ymeth.2014.02.016 [DOI] [PubMed] [Google Scholar]
- 31. Zhang X, Xiao W and Hu X. Predicting essential proteins by integrating orthology, gene expressions, and PPI networks. PloS one, 2018; 13(4): e0195410. doi: 10.1371/journal.pone.0195410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peng W, Wang J, Cheng Y, Lu Y, Wu F. and Pan Y. UDoNC: an algorithm for identifying essential proteins based on protein domains and protein-protein interaction networks. IEEE/ACM transactions on computational biology and bioinformatics. 2015; 12(2): 276–288. doi: 10.1109/TCBB.2014.2338317 [DOI] [PubMed] [Google Scholar]
- 33. Peng W, Wang J, Wang W, Liu Q, Wu FX. and Pan Y. Iteration method for predicting essential proteins based on orthology and protein-protein interaction networks. BMC systems biology. 2012; 6(1): 1–17. doi: 10.1186/1752-0509-6-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li M, Lu Y, Niu Z. and Wu FX. United complex centrality for identification of essential proteins from PPI networks. IEEE/ACM transactions on computational biology and bioinformatics. 2017; 14(2): 370–380. doi: 10.1109/TCBB.2015.2394487 [DOI] [PubMed] [Google Scholar]
- 35. Luo J. and Qi Y. Identification of essential proteins based on a new combination of local interaction density and protein complexes. PloS one. 2015; 10(6): e0131418. doi: 10.1371/journal.pone.0131418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu W, Wang J, Chen L. and Chen B. Prediction of protein essentiality by the improved particle swarm optimization. Soft Computing. 2018; 22(20): 6657–6669. doi: 10.1007/s00500-017-2964-1 [DOI] [Google Scholar]
- 37. Jain S. and Bader GD. An improved method for scoring protein-protein interactions using semantic similarity within the gene ontology. BMC bioinformatics. 2010; 11(1): 562. doi: 10.1186/1471-2105-11-562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Paul M. and Anand A. Impact of low-confidence interactions on computational identification of protein complexes. Journal of Bioinformatics and Computational Biology. 2020; 18(4):2050025. doi: 10.1142/S0219720020500250 [DOI] [PubMed] [Google Scholar]
- 39. Yu G. Gene Ontology semantic similarity analysis using GOSemSim. In:Kidder B.(eds)Stem Cell Transcriptional Networks. Methods in Molecular Biology. 2020; 2117: 207–215. doi: 10.1007/978-1-0716-0301-7_11 [DOI] [PubMed] [Google Scholar]
- 40.Jiang JJ. and Conrath DW. Semantic similarity based on corpus statistics and lexical taxonomy. Proceedings of 10th International Conference on Research in Computational Linguistics (ROCLING97). 1997.
- 41.Lin D. An information-theoretic definition of similarity. Proceedings of the 15th International Conference on Machine Learning. Morgan Kaufmann Publishers Inc., San Francisco, CA, USA. 1998; pp: 296–304.
- 42. Schlicker A, Domingues FS, Rahnenführer J. and Lengauer T. A new measure for functional similarity of gene products based on Gene Ontology. BMC Bioinformatics. 2006; 7(1): 1–16. doi: 10.1186/1471-2105-7-302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Resnik P. Using information content to evaluate semantic similarity in a taxonomy. Proceedings of the 14th international joint conference on Artificial intelligence, Morgan Kaufmann Publishers Inc., San Francisco, CA, USA. 1995; pp. 448–453.
- 44. Wang JZ, Du Z, Payattakool R, Yu PS. and Chen CF. A new method to measure the semantic similarity of GO terms. Bioinformatics. 2007; 23(10): 1274–1281. doi: 10.1093/bioinformatics/btm087 [DOI] [PubMed] [Google Scholar]
- 45. Gilchrist MA, Salter LA. and Wagner A. A statistical framework for combining and interpreting proteomic datasets. Bioinformatics. 2004; 20(5): 689–700. doi: 10.1093/bioinformatics/btg469 [DOI] [PubMed] [Google Scholar]
- 46. Deng M, Sun F. and Chen T. Assessment of the reliability of protein-protein interactions and protein function prediction. Biocomputing. 2003; 2002: 140–151. [PubMed] [Google Scholar]
- 47. Lin X, Liu M, and Chen XW. Assessing reliability of protein-protein interactions by integrative analysis of data in model organisms. BMC bioinformatics. 2009; 10(4): 1–14. doi: 10.1186/1471-2105-10-S4-S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Paul M. and Anand A. A new family of similarity measures for scoring confidence of protein interactions using Gene Ontology. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2018; pp: 459107. [DOI] [PubMed] [Google Scholar]
- 49. Mewes HW, Frishman D, Mayer KFX, Münsterkötter M, Noubibou O, Pagel P, et al. MIPS: Analysis and Annotation of Proteins from Whole Genomes in 2005. Nucleic Acids Research. 2006; 34(suppl_1): D169–D172. doi: 10.1093/nar/gkj148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, et al. SGD: Saccharomyces Genome Database. Nucleic Acids Research. 1998; 26(1): 73–79. doi: 10.1093/nar/26.1.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang R. and Lin Y. DEG 5.0, A Database of Essential genes in both Prokaryotes and Eukaryotes. Nucleic Acids Research, 2009; 37(suppl_1): D455–D458. doi: 10.1093/nar/gkn858 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The source simulation code and data used in this paper is available at https://github.com/wzhangwhu/EPI.