Abstract
The neuron-glia cross-talk is critical to brain homeostasis and is particularly affected by neurodegenerative diseases. How neurons manipulate the neuron-astrocyte interaction under pathological conditions, such as hyperphosphorylated tau, a pathological hallmark in Alzheimer’s disease (AD), remains elusive. In this study, we identified excessively elevated neuronal expression of adenosine receptor 1 (Adora1 or A1R) in 3×Tg mice, MAPT P301L (rTg4510) mice, patients with AD, and patient-derived neurons. The up-regulation of A1R was found to be tau pathology dependent and posttranscriptionally regulated by Mef2c via miR-133a-3p. Rebuilding the miR-133a-3p/A1R signal effectively rescued synaptic and memory impairments in AD mice. Furthermore, neuronal A1R promoted the release of lipocalin 2 (Lcn2) and resulted in astrocyte activation. Last, silencing neuronal Lcn2 in AD mice ameliorated astrocyte activation and restored synaptic plasticity and learning/memory. Our findings reveal that the tau pathology remodels neuron-glial cross-talk and promotes neurodegenerative progression. Approaches targeting A1R and modulating this signaling pathway might be a potential therapeutic strategy for AD.
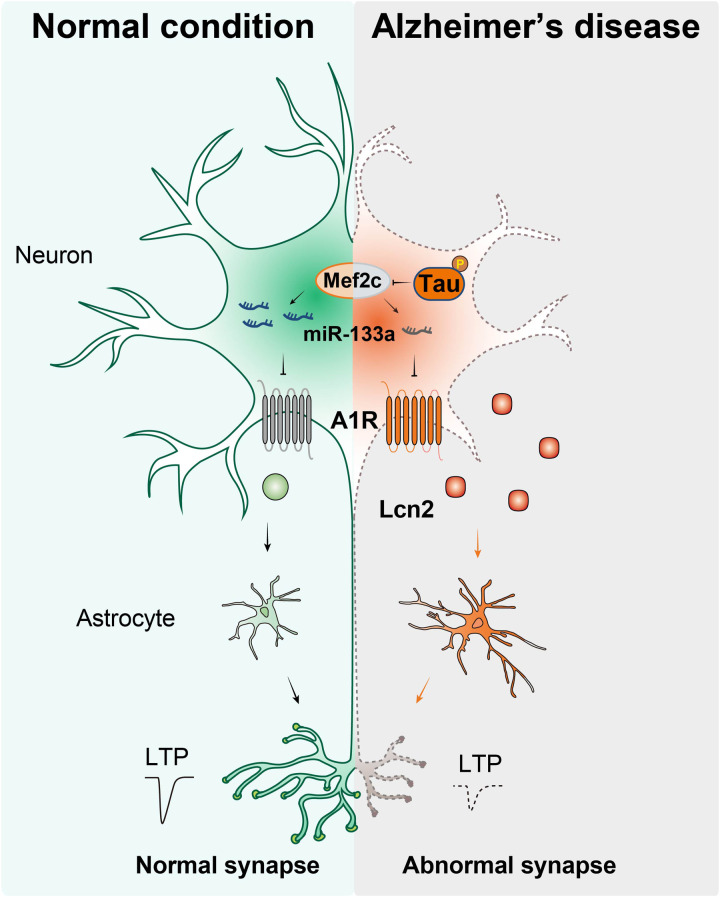
Tau pathology triggers Mef2c/miR-133a/A1R signal and promotes neuronal Lcn2 releasing to remodel the neuron-glial cross-talk.
INTRODUCTION
Alzheimer’s disease (AD) is one of most common neurodegenerative disorders in the elderly (1). The pathological hallmarks of AD are the accumulation of extracellular β-amyloid (Aβ) and the formation of intracellular aggregated phosphorylated tau (tangles) in the brain accompanied by synaptic dysfunction and neuron loss (2). Clinically, the most important manifestation of AD is progressive cognitive decline. To date, the detailed mechanisms underlying cognitive impairment remain unclear, although numerous experimental studies have suggested critical roles for Aβ and phosphorylated tau in the memory decline observed in individuals with AD (3). Thus, a better understanding of the mechanism underlying the relationship between AD pathological hallmarks such as tau tangles and the cognitive decline that manifests in the clinic and a search for potential approaches to target these underlying molecular pathways might be beneficial for AD therapy.
In the central nervous system, neurons are surrounded by different types of nonneuronal cells, such as astrocytes, microglia, and oligodendrocytes (4). The cross-talk between neurons and nonneuronal cells plays crucial roles in orchestrating physiological brain functions (4) and participates in the pathogenesis of multiple neurological diseases, such as AD. In response to tauopathy, reactive astrocytes lose their neurosupportive functions and participate in the impairments of the neuronal network and induce AD-like symptoms (5, 6). Aβ augments glucose uptake and glutamate release from astrocytes and then induces oxidative stress and overactivation of N-methyl-d-aspartate receptors, leading to synaptic disorders (7, 8). Meanwhile, Aβ stimulation increases the release of proinflammatory agents from astrocytes and subsequently initiates neuroinflammation and neuronal death (9). In addition, a recent study reported that astrocyte-derived apolipoprotein E particles specifically inhibit cholesterol biosynthesis in neurons by vectoring a set of microRNAs (miRNAs) (10). All of these evidences suggest a critical role of astrocytes in the cellular and functional degeneration of AD through alterations of neuron-glial cross-talk. However, whether neuronal malfunction, such as tau pathology, could disturb astrocytic homeostasis and participate in neurodegeneration in AD remains unclear.
Recently, several studies have revealed a critical role for adenosine and its receptors in neuromodulatory and neuroprotective effects on a variety of neurological diseases (11–14). Four G protein–coupled receptors (A1, A2A, A2B, and A3) for adenosine have been identified, and they are widely distributed in the brain and are highly involved in many aspects of brain function, such as cognition and memory, as well as neuronal injury and degeneration (15). Adenosine receptor A1 (A1R) is an inhibitory adenosine receptor that is expressed at high levels in the brain (16). Under physiological conditions, A1R actively maintains synaptic homeostasis (17, 18) and regulates neuronal activity through neuro-microglial cross-talk (11). Activation of A1R not only diminishes synaptic plasticity in the hippocampus (19) but also impairs learning and memory in rodents (20). In the AD brain, increased A1R immunoreactivity is detected in neurons and accompanied by AD-specific pathological changes (21, 22). The up-regulation of A1R has also been observed in the hippocampus of 5×FAD mice with impaired fear memory (23). A1R activation triggers neuronal death (24), and an A1R antagonist alleviates axonopathy in tau ΔK280 mice (25), implying its critical role in mediating AD pathology and cognitive decline. Although A1R activation is proposed to neutralize the hyperexcitability and excitotoxic network occurring in the AD parenchyma (26), administration of caffeine, a nonselective A1R antagonist, exerts a protective effect on AD, and higher coffee intake may improve the cognitive dysfunction of patients with AD by reducing pathological amyloid deposition (27). Abrogating A1R signaling ameliorates the inflammatory response in aged mice (28), suggesting a crucial role for A1R activation/up-regulation in neuron-glial cross-talk in individuals with AD.
In this study, we found that the A1R protein level was up-regulated in the hippocampus of 3×Tg mice, MAPT P301L (rTg4510) mice, brains of patients with AD, and induced pluripotent stem cell (iPSC)–derived AD neurons. A1R is posttranscriptionally regulated by tau via miR-133a-3p inhibition. We found that up-regulated A1R in individuals AD was mainly localized in neurons with a disrupted synaptic morphology, which were surrounded by reactive astrocytes and neuroinflammation. The medium from cultured neurons with A1R overexpression or miR-133a-3p inhibition stimulated astrocytes and the inflammatory response, which are widely observed in AD brains. Transferring the media of these reactive astrocytes to normal neuronal culture results in synaptic dysfunction in neurons. Genetic deletion or silencing of neuronal A1R with a short hairpin RNA (shRNA) notably reduced glial activation, neuroinflammation, synaptic dysfunction, and memory impairments in 3×Tg mice. By performing RNA sequencing (RNA-seq) of hippocampal CA3 neurons dissociated by laser capture microdissection, we identified that A1R knockout (AKO) reduced the neuronal expression of lipocalin 2 (Lcn2), which led to the stimulation of glial cells and reciprocally impaired dendritic morphology and spine maturation in vitro. Last, knockdown of Lcn2 in neurons effectively suppressed astrocyte activation, inflammation, synaptic disorders, and cognitive dysfunction in 3×Tg mice.
RESULTS
The abnormal up-regulation of A1R in AD depends on tau pathology
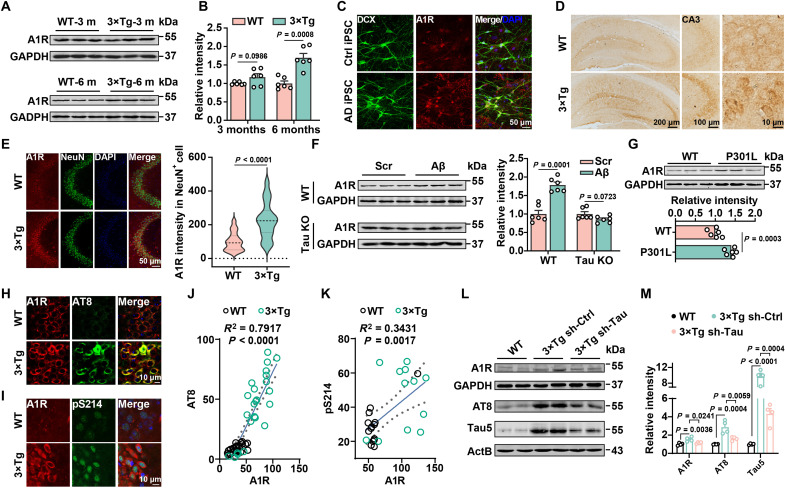
To investigate the potential role of A1R in AD, we first examined the expression of A1R in hippocampal homogenates of 3×Tg mice. Western blot results showed that the level of the A1R protein was notably increased in 3×Tg mice at 6 months but not at 3 months of age (Fig. 1, A and B). We reprogrammed fibroblasts from patients with AD into iPSCs and further differentiated them into glutamatergic neurons to examine whether the A1R protein is up-regulated in patients with AD (fig. S1, A to D). Neurons derived from patients with AD displayed significant morphological and functional synaptic deficits (fig. S1, E to J). Meanwhile, the A1R protein level was up-regulated in iPSC-derived neurons from patients with AD (Fig. 1C and fig. S2A). Consistently, the up-regulation of A1R was also apparent in the frontal cortex of patients with AD (fig. S2B), which accompanied and positively correlated with tau pathology (fig. S2, C and D). Using immunohistochemical approaches, we found that the increased A1R expression in AD mice was mainly localized in the soma of neurons but not astrocytes or microglia of the hippocampal CA3 region in both 3×Tg (Fig. 1, D and E, and fig. S2, E and F) and P301L mice (fig. S2, G to I). A slight increase in expression was also detected in the hippocampal dentate gyrus (DG) of 3×Tg mice (Fig. 1D and fig. S2J). The up-regulation of A1R in AD seems to depend on tau pathology because 1 μM Aβ1–42 oligomer insult only led to an increase in A1R expression in the primary hippocampal cultures from wild-type (WT) mice but not those from tau knockout (KO) mice (Fig. 1F). Furthermore, A1R was expressed at much higher levels in the hippocampus of P301L mice (Fig. 1G), in which tau pathology is prominent (29). The A1R protein level was also substantially increased in the hippocampus of APP/PS1 mice at 12 months of age, when abnormal tau hyperphosphorylation was apparent (fig. S3, A and B) (30). The A1R expression was positively correlated with phosphorylated tau levels in 3×Tg mice and P301L mice (Fig. 1, H to K, and fig. S3, C to F). Consistent with this finding, overexpression of WT or P301L mutant human Tau (hTau) in Neuro-2a (N2a) cells significantly increased A1R expression (fig. S3, G to J). To confirm that the increase of A1R in 3×Tg mice is tau pathology dependent, shRNA of hTau (sh-Tau) by lentivirus was injected into the hippocampus of 5-month-old 3×Tg mice, and after 25 days, we found that the A1R protein level and the tau hyperphosphorylation had decreased (Fig. 1, L and M). Moreover, the A1R protein level was not changed in tau KO mice compared to WT controls mice (fig. S3, K and L). Together, these data suggest that the abnormal increase in A1R expression in AD depends on tau pathology.
Fig. 1. The elevation of A1R in AD is tau pathology dependent.
(A) Immunoblot of A1R and glyceraldehyde phosphate dehydrogenase (GAPDH) in the hippocampi of 3- and 6-month-old 3×Tg mice and WT mice. (B) Quantification of A1R level relative to GAPDH. n = 6 for each group. (C) Immunofluorescence for A1R in the iPSC-derived neurons of patients with AD and the health control (Ctrl). n = 6 for each group. DAPI, 4′,6-diamidino-2-phenylindole. DCX, Doublecortin antibody. (D) Immunostaining for A1R in hippocampal slices from 6-month-old 3×Tg mice and WT mice. CA3 neurons are amplified. (E) Left: Immunofluorescence for A1R (red) and NeuN (green) in the hippocampal CA3 region of 3×Tg mice and WT mice. Right: The fluorescent intensities of A1R in NeuN+ cells. n = 34 to 38 neurons from six to eight mice per group. (F) Primary hippocampal neurons [12 days in vitro (DIV)] from WT or Tau knockout (Tau KO) mice were treated with Aβ1–42 (Aβ) oligomers (1 μM) or the scrambled control (Scr) for 48 hours, and the A1R protein was detected by Western blotting. n = 6 for each group. (G) Immunoblot of A1R in the hippocampi of 6-month-old P301L mice and WT mice. n = 6 for each group. (H and I) Immunofluorescence for A1R (red) and AT8 or pS214 (green) in the hippocampal CA3 region of 3×Tg mice and WT mice. (J and K) The correlation analysis between A1R and phosphorylated tau (p-tau) was analyzed. (L and M) Immunoblot of A1R and tau phosphorylation in 6-month-old 3×Tg by sh-Tau or vector infection. n = 4 mice for each group.
Increased A1R expression is caused by the inhibition of miR-133a-3p via Mef2c suppression
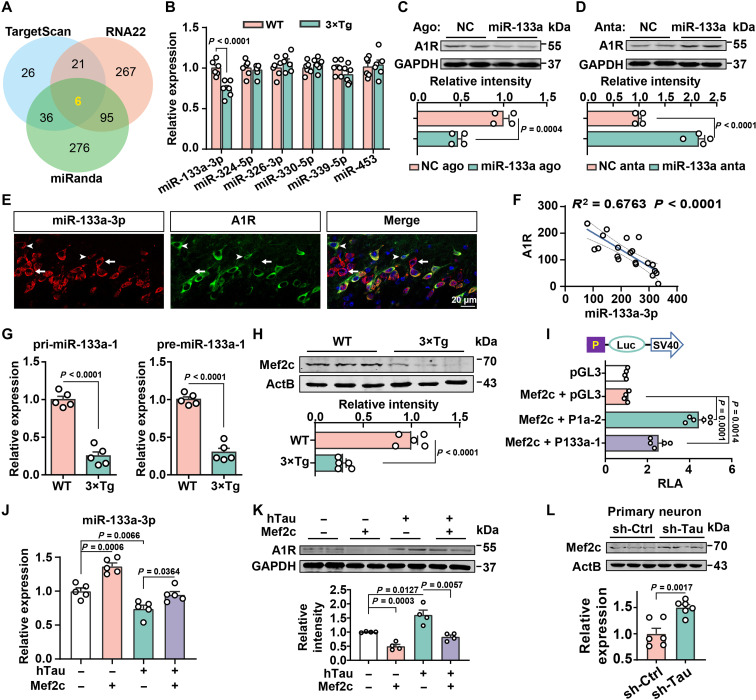
We then sought to decipher the underlying molecular mechanism of aberrant A1R up-regulation in AD. We first examined the degradation of A1R because tau has been reported to interrupt protein degradation (31, 32). A cycloheximide (CHX) chase analysis revealed that tau overexpression did not extend the A1R half-life (2 hours) compared with that of the vector control (fig. S3M), which excluded the possibility of the disruption of A1R degradation by tau overexpression. We then examined the Adora1 mRNA level but did not observe any differences in patients with AD (fig. S3, N and O), 3×Tg mice (fig. S3P), or tau-overexpressing N2a cells (fig. S3Q). These data implied that the elevated A1R level in individuals with AD may be due to posttranscriptional regulation, which is mainly suppressed by miRNAs (33). By combining the predicted outcomes from a series of bioinformatics tools (TargetScan, miRanda, and RNA22), six miRNAs were identified as candidates that regulate A1R: miR-133a-3p, miR-324-5p, miR-326-3p, miR-330-5p, miR-339-5p, and miR-453 (Fig. 2A and table S1). The expression of these miRNAs was examined in 3×Tg mice at the ages of 3 and 6 months using quantitative reverse transcription polymerase chain reaction (qRT-PCR), and only the expression of miR-133a-3p was reduced in 3×Tg mice and tau-overexpressing cells (Fig. 2B and fig. S3R). In addition, a decreasing trend was observed in patients with AD (fig. S3, S and T). Notably, miRNAs regularly bind with the 3′ untranslated region (3′UTR) of their specific targets via their seed region. By analyzing the 3′UTR of Adora1, we identified two conserved miR-133a-3p binding sites among various species (fig. S3U). To verify the posttranscriptional regulation of A1R by miR-133a-3p, we constructed WT and target site–mutated 3′UTRs of Adora1, which were inserted into dual-luciferase reporter vectors (fig. S3U) and then transfected them into 293T cells. We found that miR-133a-3p agomirs significantly inhibited luciferase activity in WT 3′UTR-transfected cells but not in mutant cells (fig. S3V). Moreover, transfection of specific agomirs or antagomirs for miR-133a-3p (fig. S4A) into N2a cells caused the down-regulation or up-regulation, respectively, of the A1R protein (Fig. 2, C and D) but not its mRNA (fig. S4B). In addition, both the miR-133a-3p and A1R signals were detected in the hippocampus, while their expression levels were negatively correlated in AD mice (Fig. 2, E and F, and fig. S4C). These data strongly suggest that A1R is a direct target of miR-133a-3p and that the aberrant neuronal expression of A1R is caused by miR-133a-3p loss in individuals with AD.
Fig. 2. The elevation of A1R in AD is mediated by the inhibition of miR-133a caused by Mef2c suppression.
(A) Venn diagram displaying the predicted miRNAs that target A1R translation. (B) qPCR for six candidate miRNAs in the hippocampi of mice as indicated. n = 6 mice per group. (C and D) Immunoblot for A1R level in N2a cells that transfected with miR-133a-3p agomir (miR-133a ago) or scrambled control (NC ago) and miR-133a-3p antagomir (miR-133a anta) or scrambled control (NC anta). n = 4 independent sets of cultures. (E and F) Fluorescence of miR-133a-3p (red) and A1R (green) in the hippocampal CA3 of C57 mice (E) and the correlation analysis (F). Arrows and arrowheads indicate neurons with higher and lower miR-133a-3p expression. n = 21 cells from four to six slices (G) qPCR for miR-133a-1 precursors in the hippocampi of mice as indicated. n = 5 mice. (H) Immunoblot of Mef2c in the hippocampi of mice as indicated; n = 5 mice. (I) Top: Diagram for the luciferase reporter vectors for pri-miR-133a-1 (P133a-1) or pri-miR-1a-2 (P1a-2) promoter. Bottom: The luciferase activity was analyzed in human embryonic kidney (HEK) 293T cells transfected with different luciferase reporter plasmids with pcDNA-Mef2c. n = 4 to 6 independent sets of cultures. (J and K) qPCR for the miR-133a-3p levels (J) and immunoblot of A1R (K) in N2a cells transfected with hTau or control plasmid with or without the pcDNA-Mef2c vector. n = 4 to 5 independent sets of cultures. (L) Immunoblot of Mef2c in primary neurons from 3×Tg mice by sh-Tau or sh-Ctrl. n = 6 independent sets of cultures.
To understand the cause of miR-133a-3p loss in AD, we measured the primary and precursor transcript levels of miR-133a-3p and found that the expression of pri-miR-133a-1 and pre-miR-133a-1 was significantly reduced in 3×Tg mice (Fig. 2G). A slight reduction in pre-miR-133a-2 levels and no change in pri-miR-133a-2 levels were also detected (fig. S4D), suggesting that the loss of miR-133-3p in individuals with AD may be because of the transcriptional suppression of miR-133a-1. As miR-133a-3p and miR-1a belong to the same cluster and display the same expression pattern and because pri-miR-1 was also decreased in the 3×Tg mice (fig. S4E), we identified seven transcription factors localized upstream of the miR-133a-1 gene and eight transcription factors for the miR-1a-2 gene (tables S2 and S3) using online bioinformatics tools such as MotifMap, TFBIND, and Jaspar. We then examined the expression of three common transcription factors (Mef2c, Arnt, and SRY) in the hippocampus of 3×Tg mice at 5 months of age. We detected a substantial decrease in Mef2c expression but no change in Arnt mRNA levels in 3×Tg mice (fig. S4F). SRY was not detected (fig. S4F), consistent with the Allen Brain Atlas (https://mouse.brain-map.org/gene/show/21435). Moreover, the MEF2c mRNA level was significantly down-regulated in patients with AD (fig. S4G). The Mef2c protein level was also reduced in the 3×Tg hippocampus (Fig. 2H) and N2a cells overexpressing hTau (fig. S4H). Further analysis revealed 5 binding sites in the promoter of the miR-133a-1 gene and 12 binding sites in the miR-1a-2 gene for the Mef2c transcription factor (fig. S4I). Overexpression of Mef2c in N2a cells substantially increased the expression of miR-133a-3p (fig. S4J), as well as pri-miR-133a-1, premiR-133a-1, pri-miR-1a-2, and pre-miR 1a-2 (fig. S4K) but not pre-miR-133a-2 and pre-miR-1a-1 (fig. S4L). Overexpression of Mef2c markedly increased the luciferase intensity in the cells transfected with the promoter regions of miR-133a-1 (p-miR-133-1) and miR-1a-2 (p-miR-1a-2). Consistently, upon Mef2c overexpression, the luciferase activity of p-miR-133-1 was lower than that of p-miR-1a-2 because there were fewer binding sites, as described above (Fig. 2I). To rule out the possible role of miR-1a-2 in mediating the miR-133a-3p increase by Mef2c, we overexpressed the miR-1a-2 precursor in N2a cells and found that neither the levels of the miR-133a-3p transcripts nor A1R protein were changed (fig. S4, M and N). In N2a cells, transfection of Mef2c notably blocked the reduction in miR-133a-3p expression and increase in A1R levels induced by tau (Fig. 2, J and K). Last, in sh-Tau–infected hippocampal neurons, Mef2c levels were restored (Fig. 2L). Collectively, these data strongly suggest that the abnormal increase in A1R expression caused by tau in individuals with AD is due to posttranscriptional dysregulation of a Mef2c-mediated miR-133a-3p pathway.
Activation of neuronal A1R signaling promotes glial activation and the inflammatory response
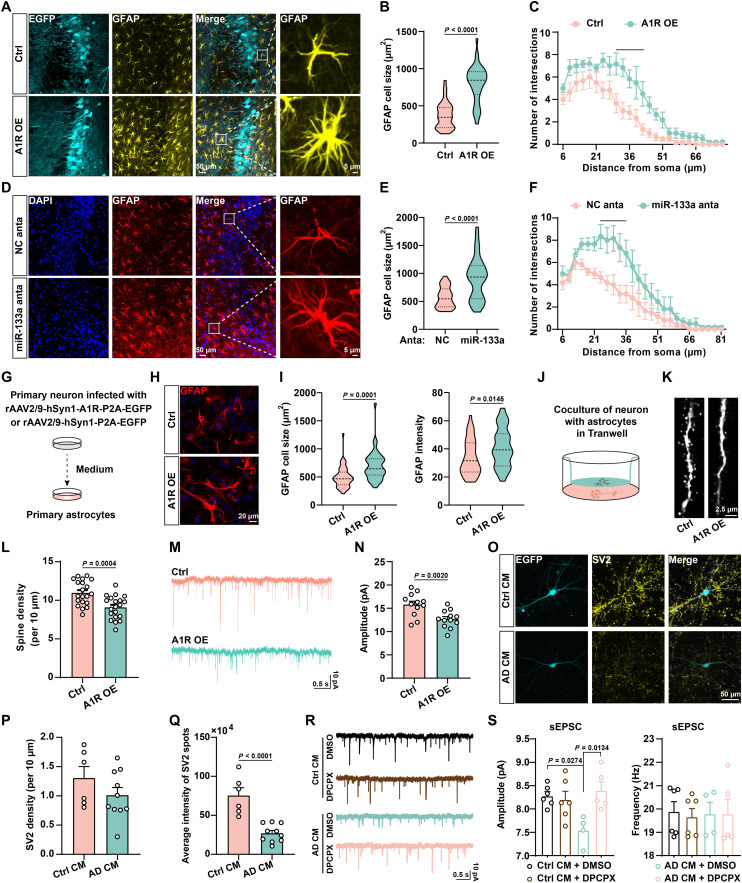
To decipher how the aberrant miR-133a-3p/A1R signal contributes to the pathogenesis of AD, we first injected miR-133a-3p antagomirs or adeno-associated virus (AAV) packaged with full-length mouse Adora1 cDNA driven by the human synapsin 1 (hSyn1) promoter into the hippocampal CA3 region of WT mice at 5 months of age (fig. S5, A and B) to replicate the aberrant miR-133a-3p/A1R signal identified in the AD brain (fig. S5, C and D). One month later, the mice were subjected to the Morris water maze and Y maze to test learning and memory. Both miR-133a-3p inhibition and A1R overexpression impaired learning and memory, as indicated by the longer latency to reach the platform (fig. S5E), the reduced number of platform crossings and time spent in the target quadrant in the Morris water maze tasks (fig. S5, F and G), and the shorter exploration time of the new arm in the Y maze tasks than in the control groups (fig. S5H). Thus, artificially introducing aberrant miR-133a-3p/A1R signaling in WT mice recapitulates the learning/memory deficits observed in individuals with AD. We observed significantly greater numbers of reactive astrocytes and microglia in the mice with A1R overexpression or miR-133a-3p inhibition (Fig. 3, A to F, and fig. S5, I and J). A similar result was also observed in the 3×Tg mice (fig. S5, K and L). We then raised the question of whether neuronal A1R up-regulation might promote glial activation. Therefore, we cultured primary hippocampal neurons and then infected them with an AAV for the overexpression of A1R (rAAV2/9-hSyn1-A1R-P2A-EGFP) or a control AAV (rAAV2/9-hSyn1-P2A-EGFP). After 14 days of infection, we transferred the neuron-conditioned medium to primary astrocytes or primary microglia (Fig. 3G). We found that conditioned medium from rAAV2/9-hSyn1-A1R-P2A-EGFP–infected neurons in which the A1R-related signaling pathway is activated (fig. S5M) significantly promoted the activation of astrocytes and inflammatory responses (Fig. 3, H and I, and fig. S5N). Furthermore, naïve neurons cocultured with astrocytes pretreated with conditioned medium from A1R-overexpressing neurons displayed detrimental effects on both dendritic morphology and synaptic transmission (Fig. 3, J to N). To understand how altered neuron-glial cross-talk in patients with AD leads to synaptic abnormalities, we collected medium from AD iPSC-derived neurons (AD), in which A1R is up-regulated, as shown in Fig. 1C, and conditioned medium from unaffected control neurons (Ctrl). We then treated astrocytes with neuron-conditioned medium for 1 week and cocultured pretreated astrocytes with Ctrl neurons for another 4 weeks. We found that astrocytes pretreated with AD neuron–conditioned medium significantly impaired the synaptic protein distribution and spontaneous excitatory postsynaptic currents (sEPSC) amplitude of naïve neurons compared to those pretreated with Ctrl neuron–conditioned medium (Fig. 3, O to S). The reduced sEPSC amplitude of AD iPSC neurons was rescued by DPCPX, an A1R antagonist, suggesting a critical role for A1R activation in synaptic dysfunction in individuals with AD (Fig. 3, R and S). These data strongly suggested that up-regulation of neuronal A1R signaling may trigger glial activation and inflammation, which, in turn, leads to synaptic disorders in both AD mice and iPSC models.
Fig. 3. Activation of neuronal A1R signaling promotes glial activation and inflammatory responses.
(A to F) Immunofluorescence of astrocyte [glial fibrillary acidic protein (GFAP)] in AAV for A1R overexpression (A1R OE) or control (Ctrl) infected (A) and antagomir of miR-133a-3p (miR-133a anta) or scrambled control (NC anta) injected (D) the hippocampal CA3 region of 3-month-old C57 mice. The cell size (B) and (E) and number of process intersections (C) and (F) of astrocytes were analyzed. n = 32 to 38 neurons from six mice per group. (G) Diagram illustrating primary astrocytes cultured in medium from primary neurons infected with virus as indicated. (H and I) Representative images of primary astrocytes stained with anti-GFAP (H). The cell sizes and intensity of GFAP-positive astrocytes were analyzed in (I). n = 39 to 44 neurons from six mice per group. (J) A diagram of cocultures of neurons and astrocytes. (K and L) Representative images (K) and density (L) of neuronal dendritic spines. n = 20 neurons from five mice per group. (M and N) Representative traces (M) and quantitative analysis for the amplitude (N) of mEPSCs from neurons that cocultured with astrocytes treated as (J). n = 12 cells from three mice per group. (O to Q) Immunofluorescence (O), density (P), and average intensity of synaptic vesicle proteins 2 (anti-SV2) (Q) in human iPSC–derived neurons were treated with control conditional medium (Ctrl CM)– or AD conditional medium (AD CM)–treated astrocytes. n = 6 to 10 independent sets of cultures. EGFP, enhanced green fluorescent protein. (R and S) Representative traces (R) and quantification for the amplitude and frequency (S) of sEPSCs from Ctrl or AD CM–treated neurons with dimethyl sulfoxide (DMSO) or 8-Cyclopentyl-1,3-dipropylxanthine (DPCPX). n = 4 to 6 neurons per group.
Restoration of the miR-133a-3p/A1R signaling pathway rescues memory deficits and dendritic abnormalities in 3×Tg mice
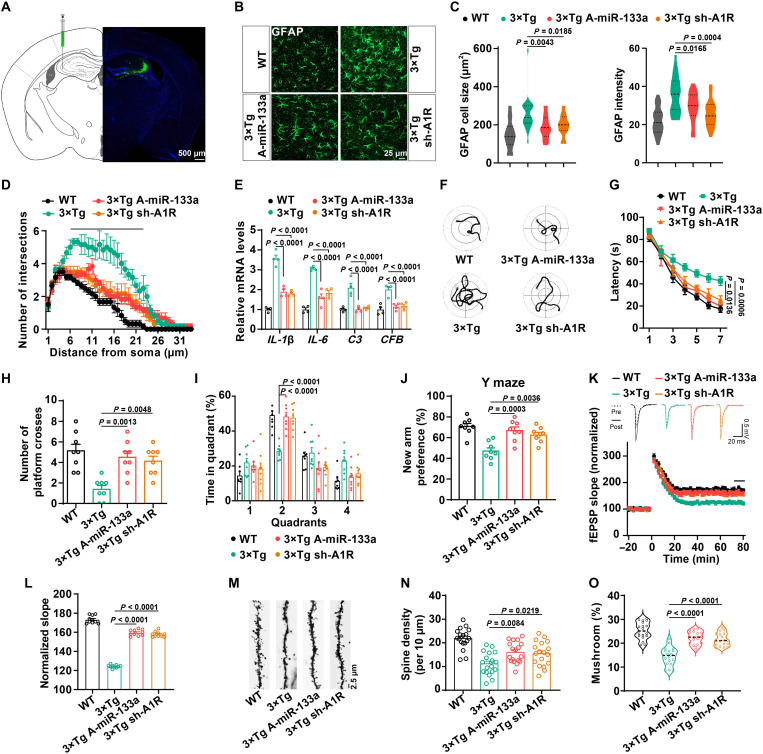
Next, we sought to delineate whether correcting the abnormalities in the miR-133a-3p/A1R signal would rebuild normal neuron-glial cross-talk and rescue learning/memory impairments in an AD model. We injected miR-133a-3p agomirs or lentivirus-packed A1R shRNA into the hippocampal CA3 area of 5-month-old 3×Tg mice (Fig. 4A and fig. S6, A and B). We showed that miR-133a-3p was expressed mainly in neurons but not astrocytes or microglia using fluorescence in situ hybridization (FISH) (fig. S6, C to F). Treatment with miR-133a-3p agomirs decreased the A1R protein level in 3×Tg mice compared with the scrambled control (NC) agomir group 3 weeks later (fig. S6, G and H). Both miR-133a-3p agomirs and A1R silencing effectively suppressed astrocyte activation and the inflammatory response in the hippocampus of 3×Tg mice (Fig. 4, B to E). Meanwhile, correcting miR-133a-3p/A1R signaling significantly reduced the latency to reach the platform in the training stage and increased the number of platform crossings and the duration that the mice stayed in the target quadrant during the probe trials of the Morris water maze (Fig. 4, F to I). In the Y maze experiment, the 3×Tg-miR-133a-3p-Ago or 3×Tg-sh-A1R mice exhibited better performance in exploring the new arm than the 3×Tg group (Fig. 4J). We also evaluated the synaptic plasticity of mossy fiber (MF)–CA3 synapses in 3×Tg mice after miR-133a-3p agomir or A1R shRNA treatment. Both treatments effectively reversed the impairments in long-term potentiation (LTP) in 3×Tg mice (Fig. 4, K and L). Moreover, the density of dendritic spines and the percentage of mature spines were also restored upon miR-133a-3p administration or A1R silencing (Fig. 4, M to O). Thus, correcting the abnormalities of neuronal miR-133a-3p/A1R signaling might ameliorate glial reactivity and inflammatory responses, as well as impaired synaptic plasticity and learning/memory deficits.
Fig. 4. Rebuild miR-133a-3p/A1R signaling pathway rescues memory/synaptic deficits in AD mice.
Lentivirus-packed shRNA-A1R or the agomir of miR-133a-3p was injected into the hippocampal CA3 region of 5-month-old 3×Tg mice. The lentivirus-packed EGFP vector was injected into WT mice. (A) Diagram (left) and representative confocal image (right) of sh-A1R lentivirus infection. (B to D) Immunofluorescence of GFAP with the pseudo color (B), the cell size and GFAP intensity (C), and the number of process intersections of astrocytes (D) in the hippocampal CA3 of different mice treated as indicated. n = 21 to 27 from six mice per group. The line in (D) indicates statistical significance. (E) qPCR for RNA levels of interleukin-1β (IL-1β), interleukin-6 (IL-6), complement C3 (C3), and complement factor B (CFB). n = 4 per group. (F to I) The representative swimming path on day 7 (F), latency to reach a hidden platform from days 1 to 7 (G), number of crossings to the platform region in day 9 (H), and the percent of time spent in each quadrant of day 9 (I) in the Morris water maze tasks of different groups. n = 8 per group. (J) The percentage of time spent in the novel arm in the Y maze task. n = 8 per group. (K and L) Electrophysiological recordings of field excitatory postsynaptic potential (fEPSP) slope (K) and quantitative analysis (L) to evaluate the CA3-CA1 LTP. n = 10 slices from four mice per group. (M to O) Representative images (M), spine density (N), and percent of mushroom type spines (O) in CA3 neurons of different groups. n = 18 neurons from three mice per group.
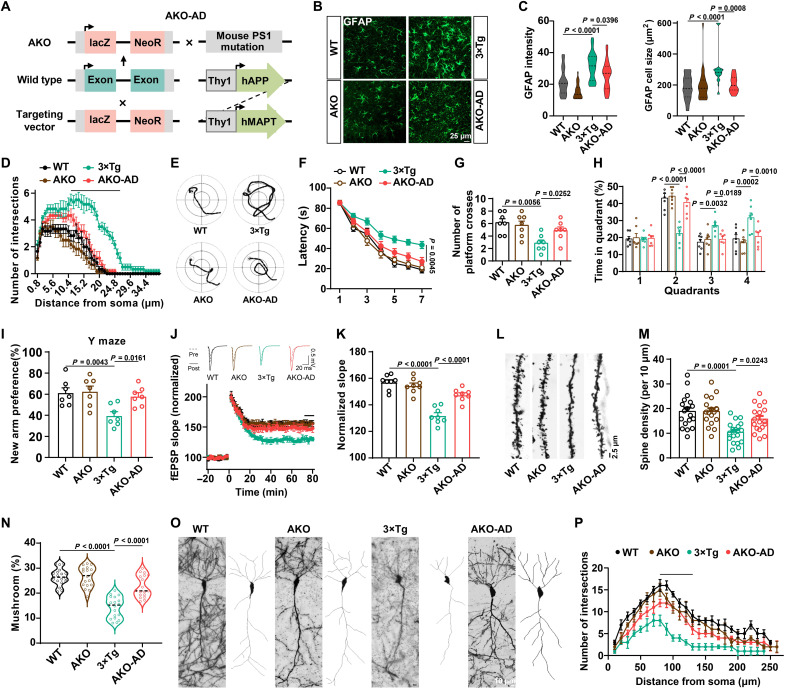
Genetic deletion of A1R in 3×Tg mice rescues learning/memory impairments and synaptic dysfunction in an AD model
To verify the protective effects of A1R suppression on neuron-glial cross-talk, synaptic plasticity, and high-order brain function in AD mice, we generated a mouse strain (AKO-AD) by crossing AKO mice with 3×Tg mice (Fig. 5A and fig. S7, A to D). The body sizes and brain weights of AKO-AD mice were comparable to those of control mice (fig. S7, E and F). Astrocyte reactivation and inflammatory responses were reduced in AKO-AD mice (Fig. 5, B to D). The results of the Morris water maze showed that AKO-AD mice displayed shorter latencies on the last 3 days of training (Fig. 5, E and F) than 3×Tg mice. In the probe trial, the number of platform crossings and the duration spent in the target quadrant were significantly restored in AKO-AD mice (Fig. 5, G and H), with no difference in their movement speed observed (fig. S7G). Compared to the 3×Tg mice, the AKO-AD mice showed a greater preference for new arms in the Y maze test, similar to the WT mice (Fig. 5I). Consistent with our previous study, AKO mice did not display any memory impairments (34). We then examined synaptic plasticity in hippocampal slices from these mice. LTP from the MF-CA3 projection had recovered in AKO-AD mice (Fig. 5, J and K). In addition, the abnormalities of dendritic trees and dendritic spines were also rescued in AKO-AD mice (Fig. 5, L to P). Furthermore, we found that hyperphosphorylation of tau at the Thr231, Ser262, Ser396, and Ser202/Thr205 sites was significantly attenuated in AKO-AD mice compared with 3×Tg mice (fig. S7, H and I). Meanwhile, A1R deletion prevented the overproduction of Aβ (fig. S7, J to L). Collectively, these data strongly suggested that genetic deletion of A1R reduces memory/synaptic deficits, tau pathology, and Aβ pathology in 3×Tg mice.
Fig. 5. A1R deletion rescues learning/memory impairments and synaptic dysfunction in AD mice.
(A) The diagram for the generation of AKO-AD mice. (B to D) Immunofluorescence of GFAP (B), the cell size and GFAP intensity (C), and the number of process intersections of astrocytes (D) in the hippocampal CA3 of different strains. n = 23 to 29 cells from six mice per group. (E to H) The representative swimming path on day 7 (E), latency to reach a hidden platform from days 1 to 7 (F), number of crossings to the platform region in day 9 (G), and the percent of time spent in each quadrant of day 9 (H) in the Morris water maze tasks of different groups. n = 7 per group. (I) The percentage of time spent in the novel arm in the Y maze task. n = 7 per group. (J and K) Electrophysiological recordings of fEPSP slope (J) and quantitative analysis (K) to evaluate the CA3-CA1 LTP. n = 8 slices from four mice per group. (L to N) Representative images (L), spine density (M), and percent of mushroom type spines (N) in CA3 neurons of different groups. n = 18 neurons from three mice per group. (O and P) Representative images (left) and two-dimensional (2D) reconstructions (right) for one neuron (O) and Sholl analysis (P) to evaluate dendritic complexity in CA3 neurons. n = 5 to 6 neurons from four mice per group. The line means statistical significance.
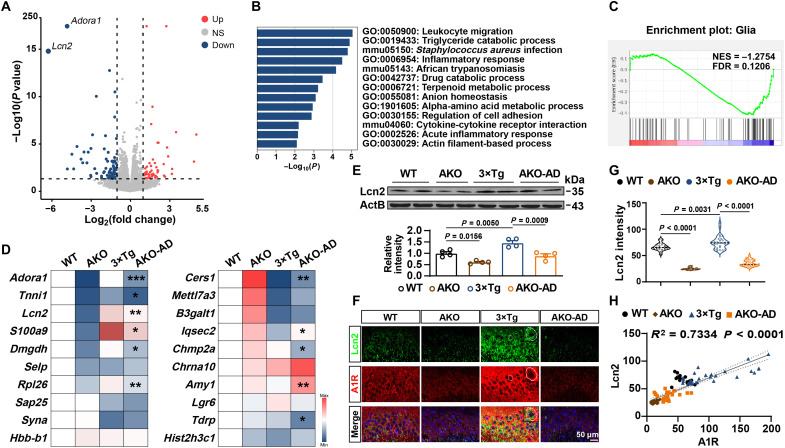
AKO suppresses the neuronal expression and release of Lcn2
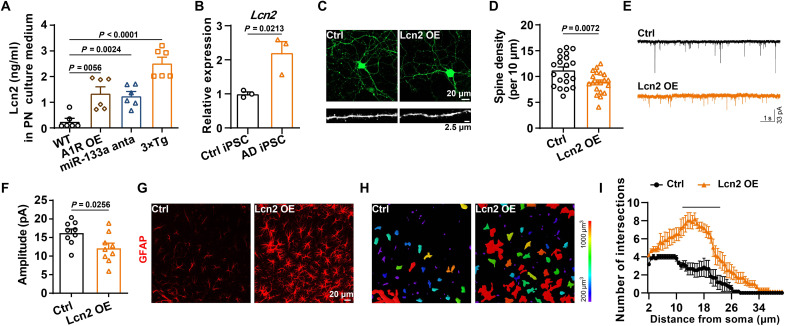
To investigate how neuronal A1R remodeled neuron-glial cross-talk and the underlying molecular mechanism, we then performed RNA-seq on RNAs isolated from the bulk hippocampal CA3 region of AKO and WT mice (fig. S8A). We found that 51 mRNAs were up-regulated, whereas 77 mRNAs were down-regulated in AKO mice (Fig. 6A and table S4). We found that many genes related to the inflammatory response were deregulated in AKO mice by performing Gene Ontology (GO) analysis, including the inflammatory response (GO: 0006954), acute inflammatory response (GO: 0002526) (Fig. 6B), and negative regulation of glial cell differentiation (GO: 0045686) pathways (fig. S8B). Given the important role of neuroinflammation in synaptic/memory disorders in individuals with AD (35), we performed gene set enrichment analyses (GSEAs) and observed a substantial reduction in a set of reactive glial genes (Fig. 6C and fig. S8C). We further performed qPCR assays for these genes, and the results indicated that the expression of some was significantly reduced in AKO-AD mice, such as Adora1, Tnnil1, Lcn2, S100a9, Dmgdh, and Rpl26 (Fig. 6D). Among them, Lcn2 was most interesting because (i) the down-regulation of Lcn2 in AKO mice is most prominent, (ii) it plays an important role in neuroinflammation (36, 37), and (iii) it is involved in dendritic morphology and spine maturation (38). Consistently, the Lcn2 levels were increased in the 3×Tg mice and suppressed to the normal level in AKO-AD mice (Fig. 6E). Furthermore, Lcn2 colocalized with A1R in the CA3 regions and positively correlated with A1R (Fig. 6, F to H). Lcn2 expression was up-regulated in neurons in 3×Tg mice more compared with astrocytes or microglia (fig. S9, A to C). In the medium of primary cultured neurons from 3×Tg mice or neurons transfected with the A1R overexpression vector or miR-133a-3p inhibitor, the Lcn2 level was increased (Fig. 7A), suggesting excessive neuronal release. The levels of the Lcn2 transcripts were substantially increased in AD iPSC-derived neurons (Fig. 7B). Overexpression of Lcn2 driven by the hSyn1 promoter in pure neuron cultures (fig. S9, D and E) not only reduced the dendritic tree, the spine maturation, and the amplitude of miniature excitatory postsynaptic currents (mEPSCs) (Fig. 7, C to F) but also promoted the activation of astrocytes (Fig. 7, G to I). These data suggest that neuronal Lcn2 produced by A1R pathways may lead to glia-dependent inflammatory responses and eventually result in dysfunction of the dendritic spines and transmission, both of which are apparent in AD mouse models.
Fig. 6. AKO suppresses Lcn2 expression.
(A) Volcano plot showing the fold change (FC) and significance of differences in the expression of mRNAs in the hippocampal CA3 region of AKO mice compared to WT mice at 4 months of age. FC > 1 and P < 0.05 are indicated by the dashed lines. NS, not significant. (B and C) GO enrichment (B) and GSEA enrichment (C) analysis of deregulated genes in AKO mice compared with WT mice. NES, normal enrichment score; FDR, false discovery rate. (D) qPCR of top 20 deregulated genes in different strains. n = 3 to 5 mice per group. *P < 0.05, **P < 0.01, and ***P < 0.001 for AKO-AD versus 3×Tg. (E) Immunoblot of Lcn2 in the hippocampus of different strains at 6 months of age. n = 4 mice per group. (F and G) Representative images of immunofluorescence staining with an anti-Lcn2 antibody (green) and anti-A1R antibody (red) in the hippocampal CA3 region of the four groups are shown in (F). A magnified image was shown in the top right corner. The intensity of Lcn2 staining was statistically analyzed in (G). n = 19 to 34 cells from six mice per group. (H) The results from the correlation analysis between the intensities of Lcn2 and A1R staining are shown. n = 19 to 34 cells from six mice per group.
Fig. 7. Up-regulation of neuronal Lcn2 impairs synaptic plasticity and promotes astrocytes activation.
(A) The Lcn2 levels in the culture medium of primary neurons from WT mice and 3×Tg mice or treated with A1R OE and miR-133a anta were detected. n = 6 independent sets of cultures. (B) The Lcn2 mRNA levels were examined in iPSC-derived neurons treated with CM, as shown in Fig. 3J. n = 3 independent sets of cultures. (C and D) Representative images of morphology (top) and dendritic spines (bottom) (C), as well as quantification of the spine density (D) in the neurons that overexpressed Lcn2 or the control virus. n = 20 neurons from four independent cultures per group. (E and F) Representative traces (E) and quantitative analysis for the amplitude (F) of mEPSCs from neurons that overexpressed Lcn2 or the control virus. n = 12 cells from three mice per group. (G to I) Immunofluorescence of GFAP (G), the 3D reconstruction images (H), and the number of process intersections (I) of astrocytes in the hippocampal CA3 with or without Lcn2 overexpression (rAAV2/9-hSyn1-Lcn2-P2A-EGFP or rAAV2/9-hSyn1-P2A-EGFP). n = 6 cells from three mice per group. The underline indicates a significant difference between the Lcn2 OE and Ctrl groups.
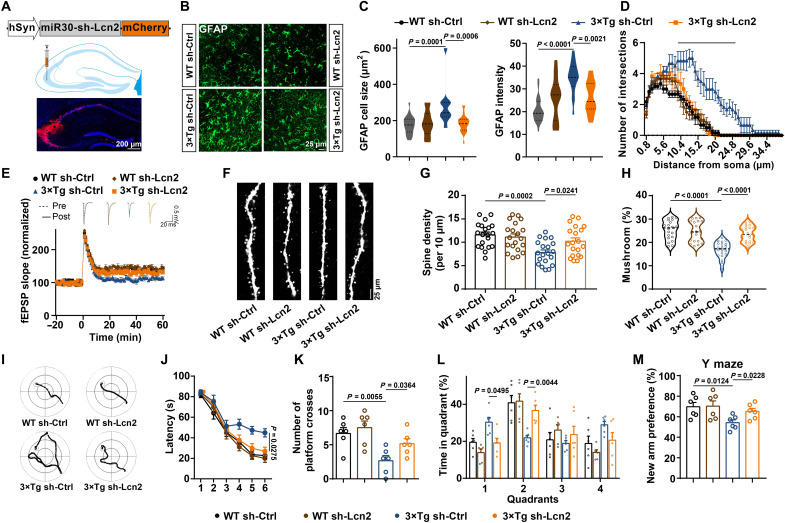
Knockdown of Lcn2 in neurons alleviates glial activation, memory impairments, and synaptic abnormalities in 3×Tg mice
We then explored whether the suppression of neuronal Lcn2 would reduce abnormal neuron-glial cross-talk and attenuate synaptic/memory impairments in AD mice. We generated an AAV packed with an effective Lcn2 shRNA under a neuronal-specific promoter and injected it into the hippocampus of 3×Tg mice at 6 months of age (Fig. 8A and fig. S10, A and E). One month later, we found that sh-Lcn2 effectively reduced the activation of astrocytes in 3×Tg mice (Fig. 8, B to D). The expression of the proinflammatory cytokines IL-1β, IL-6, and TNF-α was down-regulated upon silencing Lcn2 (fig. S10F). Furthermore, Lcn2 silencing efficiently restored LTP in the hippocampus of AD mice (Fig. 8E). We further examined dendritic spines on green fluorescent protein (GFP)–positive neurons and found that a reduction in neuronal Lcn2 expression rescued the impairments of dendritic spines in AD mice (Fig. 8, F to H). Moreover, sh-Lcn2 alleviated the spatial memory impairment, as indicated by the shorter latencies, greater number of platform crossings, and longer durations spent in the target quadrant (Fig. 8, I to L). Similar effects were also detected in the Y maze task (Fig. 8M). Collectively, these data suggest that neuronal Lcn2 knockdown may dampen the immune cross-talk between neurons and glial cells and then rescue memory and synaptic deficits in an AD model.
Fig. 8. Silencing of Lcn2 alleviates memory impairments and synaptic abnormalities in 3×Tg mice.
AAV packed shRNA-Lcn2 or the scrambled control (Ctrl) was injected into the hippocampal CA3 region of 6-month-old 3×Tg mice and WT mice. (A) Schematic illustrating the neuron-specific Lcn2 shRNA silencing with the miR30 vector (top) and the diagram (middle) and representative confocal image (bottom) for virus infection. (B to D) Immunofluorescence of GFAP (B), the cell size and GFAP intensity (C), and the number of process intersections of astrocytes (D) in the hippocampal CA3 area. n = 18 to 22 cells from six mice per group. (E) LTP was recorded in the four groups of mice. n = 6 slices from three mice in each group. (F to H) Representative images (F), the spine density (G), and percent of mushroom type spines (H) in CA3 neurons. n = 20 neurons from five mice. (I to L) The representative swimming path on day 6 (I), latency to reach a hidden platform from days 1 to 6 (J), number of crossings to the platform region in day 8 (K), and the percent of time spent in each quadrant of day 8 (L) in the Morris water maze tasks of different groups. n = 7 mice per group. (M) The percentage of time spent in the novel arm in the Y maze task. n = 6 mice per group.
DISCUSSION
Our study reports that the expression of the A1R protein but not mRNA was increased in the hippocampus of 3×Tg AD mouse models, AD brains, and AD iPSC-derived neurons, consistent with a recent study (21). This result raised the question of whether this aberrant alteration of A1R in AD is mediated by posttranscriptional mechanisms. In subsequent studies, we revealed that the up-regulation of A1R is caused by the loss of miR-133a-3p in the AD brain. We identified two conserved binding sites located in the 3′UTR of Adora1 that might be directly modulated by miR-133a-3p. MiR-133 was first experimentally characterized in mice because of its critical role in modulating skeletal muscle proliferation and differentiation. In addition, miR-133 has been implicated in multiple human diseases, particularly in cancer and cardiovascular diseases (39–41). Both miR-133a and miR-133b are expressed in the brain, and the latter regulates the maturation and function of dopaminergic neurons in the midbrain (42). The level of miR-133b is reduced in both the plasma and midbrain tissues of patients with Parkinson’s disease (PD) and may serve as a PD biomarker (43). In addition, miR-133 enhances functional recovery after spinal cord injury and might regulate neurite outgrowth (44, 45). Decreased expression of miR-133b but not miR-133a was also detected in the serum of patients with AD (46). Here, we reported a decreased level of miR-133a-3p in the hippocampus of patients with AD and 3×Tg mice. Loss of miR-133a-3p up-regulated A1R in individuals with and models of AD. Artificially inhibiting miR-133a-3p in the hippocampus impaired synaptic plasticity and memory retention in WT mice. Moreover, an infusion of the agomir of miR-133a-3p into the hippocampus effectively ameliorated synaptic and memory disorders in AD mice. We further identified that the reduction in the expression of the transcription factor Mef2c might be responsible for the disruption of the miR-133a-3p/A1R signal in individuals with AD. Mef2c is expressed at high levels in the cortex, hippocampus, and cerebellum (47) and plays an important role in brain development and synapse formation (48). Whole-genome sequencing indicated that the single-nucleotide polymorphism mutation of the Mef2c gene is closely related to AD (49, 50). Consistent with our data, the level of the Mef2c mRNA was significantly reduced in the blood of patients with AD, and its expression level was negatively correlated with the Alzheimer’s Disease Assessment Scale score (51). Mef2c knockout mice displayed decreased neurogenesis, increased apoptosis, and an imbalance of excitatory to inhibitory neurotransmission (52), which were observed in the AD brain (53, 54). In contrast, overexpression of Mef2c in the prefrontal cortex promoted learning and memory in mice (55). We revealed that Mef2c was significantly down-regulated in 3×Tg mice and was involved in disrupting miR-133a-3p/A1R signaling, subsequently leading to synaptic and memory impairments. These data suggest a critical role for Mef2c/miR-133a-3p in brain function and provide further evidences for the miRNA-based regulatory axis involved in the pathogenic process of AD, consistent with our previous reports (56–59).
In addition, the up-regulation of A1R was mainly observed in the CA3 region of the hippocampus in the brains of 3×Tg mice and patients with AD. Moreover, ectopic expression of A1R in WT mice led to memory deficits, while genetic deletion of A1R in 3×Tg mice alleviated these memory and synaptic deficits. Restoration of the miR-133a-3p/A1R signaling pathway mitigated memory/synaptic malfunction in AD mice. Under normal conditions, A1R is predominantly expressed in the CA1 region of the hippocampus, and activation of A1R slows the release of acetylcholine and glutamate (60, 61) and suppresses LTP and long-term depression (19, 62). Moreover, A1R agonists have been shown to be able to impair learning and memory (20, 63), indicating the negative regulatory effect of A1R on synaptic plasticity. Although the results for the alteration in A1R levels in the AD brain are controversial, increased A1R immunoreactivity has been observed in neurons with neurofibrillary tangles (21), suggesting a possible link between the A1R-mediated signal transduction pathway and tau pathology–related disorders. Our study confirmed that the up-regulation of A1R depends on tau pathology because reduced tau pathology repressed the increase in A1R expression in 3×Tg mice. These data are consistent with previous studies showing that A1R knockdown inhibits tau phosphorylation levels in SH-SY5Y cells treated with Aβ25–35 (64) and that isoflurane exposure impairs spatial memory via the activation of A1R (65). Caffeine, a nonselective A1R antagonist, might exert a protective effect on AD (66, 67). Higher coffee intake may lower the risk of AD or cognitive decline by reducing pathological cerebral amyloid deposition (27). However, caffeine may trigger seizures and influence brain excitability, consistent with the imbalance of excitation-inhibition in individuals with AD (68, 69). These results remind us to use caffeine cautiously, and other A1R antagonists must be explored.
We next deciphered the underlying neuroprotective mechanisms of A1R knockdown in AD mouse models. By using the RNA-seq technique, we compared the alterations in gene expression in the hippocampus between AKO and WT mice. Among the differentially expressed genes, Lcn2 was down-regulated to the greatest extent in AKO mice. Moreover, the expression of Lcn2 was significantly increased in 3×Tg mice and suppressed in AKO-AD mice. Consistently, the Lcn2 level was elevated in patients with mild cognitive impairment (70) and those with AD (71). Lcn2 is important for dendritic spine formation (38), and overexpression of Lcn2 in primary neurons led to abnormal synaptic transmission and decreased spine density, as described here. Knockdown of Lcn2 in the hippocampus using a neuron-specific shRNA strategy attenuated learning/memory impairments and synaptic deficits in 3×Tg mice. We found that excess Lcn2 released from neurons with a tau pathology burden induced the abnormal activation of astrocytes and microglia, subsequently resulting in neuroinflammation and synaptic destruction in neurons. To our knowledge, this report shows that tau pathology initiates the neuronal release of Lcn2 and then promotes astrocyte activation. Some other specific molecules secreted by neurons into the extracellular fluid regulate the function and metabolism of astrocytes. Transferrin derived from neurons has been reported to stimulate glycolysis in astrocytes (72). In Drosophila, the release of fibroblast growth factor (FGF) by neurons has been shown to control the extension and elaboration of astrocyte processes (73). Despite the direct role of Lcn2 in regulating dendritic morphology, Lcn2 released from neurons might be an important factor that modulates neuron-glial interactions under pathological conditions. In ischemic stroke, Lcn2 is released by injured neurons to enhance distress signals that activate microglia and astrocytes (74). Consistent with this finding, genetic deletion of Lcn2 was reported to reduce gliosis and neuroinflammation and ameliorate cognitive dysfunction in diabetic mice (75). Moreover, the increase in Lcn2 levels may be induced by the activation of p38 mitogen-activated protein kinase (76, 77), which acts downstream of A1R and has been detected in the AD brain (78). Thus, targeting the increase in Lcn2 levels might be able to rebuild healthy cross-talk between neurons and glial cells in AD brains.
Collectively, our study not only describes a neuron-glial cross-talk mechanism underlying tau pathology–related synaptic disorders but also reveals that miR-133a-3p/A1R/Lcn2 signaling might be a potential therapeutic target for AD (Fig. 9).
Fig. 9. Schematic diagram of the current study.
In the AD brain, abnormally hyperphosphorylated tau suppresses the transcription factor Mef2c, subsequently reducing miR-133a-3p transcription. The loss of miR-133a-3p in the AD brain leads to an increase in A1R levels and induces the aberrant up-regulation of Lcn2, which ultimately causes synaptic and memory impairments in individuals with AD.
MATERIALS AND METHODS
Mice
The 3×Tg mice (catalog no. 34830-JAX; RRID: MMRRC_034830-JAX), P301L mice (JAX: catalog no. 024854; RRID: IMSR_JAX: 024854), and the C57BL/6J mice (JAX: catalog no. 000664; RRID: IMSR_JAX: 000664) were purchased from the Jackson Laboratory (Bar Harbor, ME). The 3×Tg mice had been crossed with C57BL/6 mice for more than 20 generations such that the offspring was on the C57BL/6 background in our laboratory. The AKO mice (MGI ID: MGI: 4940034, RRID: IMSR_JAX:014161) were housed in our laboratory and were a gift from J. Schnermann at the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (NIDDK/NIH) (56, 79). Genetic knockout mice of A1R in AD mice were generated by crossing 3×Tg mice and AKO mice. Tau KO mice were preserved in our laboratory, and genotyping of tau KO mice was performed as described in a previous study (80). And nontransgenic littermates were used as the WT mice. All mice were crossed with C57BL/6 mice, and the genetic background of those mice was C57BL/6. All animals were housed on a 12-hour light/dark cycle in a temperature-controlled room with food and water available ad libitum, and this study was approved by the Animal Care and Use Committee of Tongji Medical College (Wuhan, China).
Cell culture
Mouse N2a cells (N2a cell line) and human embryonic kidney (HEK) 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (high glucose and GlutaMAX) supplemented with 5% fetal bovine serum (FBS) and 1% penicillin/streptomycin and maintained at 37°C in 5% carbon dioxide (CO2). Cell transfection was performed with Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Primary neurons were isolated from AKO mice, 3×Tg mice, AKO-AD mice, and control mice and cultured as described previously (81). The cortices or hippocampi of embryos [embryonic day 16 (E16) and E17] were dissected, and the meninges were removed. Tissues were digested in trypsin at 37°C for 10 min and filtered through a 40-μm cell strainer. The collected neurons were plated onto poly-d-lysine–coated coverslips in a six-well plate containing plating medium (DMEM/F12 with 10% FBS and 1% penicillin/streptomycin) and incubated for 2 to 4 hours. The media were then replaced with maintenance medium (Neurobasal medium supplemented with 2% B-27, 1× GlutaMAX, and 1% penicillin/streptomycin) and changed every 3 days with maintenance medium containing cytarabine (2 μM). Two weeks later, the supernatant was collected from the primary neurons for subsequent biochemical analyses. For the rAAV infection of primary neurons, rAAV [1012 transducing units (TU)/ml] was added to the media on 7 day in vitro (DIV7), and the media were fully replaced on DIV15. Neuron-derived conditioned media were collected on DIV21 and centrifuged at 3000g for 10 min to remove cell debris. The supernatant was then passed through a 0.22-μm filter and used in subsequent experiments.
Primary astrocytes were prepared from the brains of newborn [postnatal day 0 (P0) to P2] C57BL/6J mice. Cortical tissues were dissociated, and cells were collected as described above. The collected cells were then seeded in dishes containing culture medium (DMEM supplemented with 10% FBS and 1% penicillin/streptomycin). The culture medium was replaced with fresh culture medium every 3 days. Eight to 10 days later, the astrocytes were digested with 0.25% trypsin and plated on poly-d-lysine–coated coverslips in 12-well plates. A six-well Transwell system with a pore size of 0.4 μm was used for the cocultures of primary neurons and astrocytes. Neurons were seeded in the lower compartment on coverslips, whereas astrocytes were cultured in the upper compartment on coverslips (82).
The generation, culture, and characterization of iPSC lines from patients with AD and unaffected controls are described below. Fibroblasts from patients with AD and age-, sex-, and race-matched unaffected controls (table S5) were obtained at Coriell. All studies followed approved Stem Cell Research Oversight Committee and Institutional Review Board protocols at the Cincinnati Children’s Hospital Medical Center. Mouse embryonic fibroblasts (MEFs) that were dissected from E13.5 CF-1 mouse embryos were cultured in DMEM/Nutrient Mixture F12 (DMEM/F12; Invitrogen) supplemented with 10% FBS (HyClone), 2 mM l-glutamine (Invitrogen), and 0.1 mM nonessential amino acids (NEAA) (Invitrogen), as previously described (83).
All iPSCs were generated with a nonintegrating, self-replicating RNA reprogramming vector (ReproRNA-OKSGM, STEMCELL Technologies). All iPSC colonies were manually selected under a dissecting microscope after 3 to 6 weeks for further expansion and characterization. For routine culture, iPSCs (passage < 40) were plated on irradiated MEFs in human iPSC medium consisting of DMEM/F12 (Invitrogen), 20% Knockout serum replacement (Invitrogen), 2 mM l-glutamine (Invitrogen), 0.1 mM minimum essential medium NEAA (Invitrogen), 0.1 mM β-mercaptoethanol (Invitrogen), and human basic FGF (10 ng/ml; PeproTech). Media were changed daily, and iPSC clones were lifted with collagenase (1 mg/ml in DMEM/F12 for 30 min at 37°C; Invitrogen) as described (83). For feeder-free culture of iPSCs, colonies were cultured on Matrigel-coated six-well plates (BD Biosciences) with mTeSR1 media (STEMCELL Technologies). Karyotyping analysis using the standard G-banding technique was performed at Cincinnati Children’s Hospital Medical Center, and the results were interpreted by clinical laboratory specialists.
The protocol for the differentiation of iPSCs into forebrain-specific neural progenitors and cortical glutamatergic neurons was described previously (83). In general, iPSC colonies were lifted from the feeder layer with collagenase treatment (1 mg/ml) for 30 min and suspended in embryoid body (EB) medium, consisting of FGF-2–free iPSC medium supplemented with 2 μM dorsomorphin and 2 μM A-83, in nontreated polystyrene plates for 4 days with a daily medium change. After 4 days, EB medium was replaced by neural induction medium (NIM) consisting of half DMEM/F12 and half Neurobasal medium, N2 supplement, and NEAA. The floating EBs were then transferred to Matrigel-coated six-well plates on day 7 to form neural tube–like rosettes. The attached rosettes were maintained for 15 days with half of the NIM changed every other day. On day 22, the rosettes were picked mechanically and transferred to low attachment plates (Corning) to form neurospheres in NIM for 3 days. The suspended neurospheres were dissociated with Accutase at 37°C for 10 min and seeded in Matrigel-coated six-well plates. The monolayer progenitors were propagated for two to three passages and cultured in NIM before neuronal differentiation. The medial ganglionic eminence (MGE) progenitors that had been isolated with Accutase at 37°C for 5 min were placed onto poly-d-lysine/laminin-coated coverslips in neuronal culture medium consisting of Neurobasal medium supplemented with 2 mM l-glutamine, B27, brain-derived neurotrophic factor (10 ng/ml; PeproTech), glial cell line–derived neurotrophic factor (10 ng/ml; PeproTech), ascorbic acid (200 ng/ml; Sigma-Aldrich), and 1 μM cyclic adenosine monophosphate (Sigma-Aldrich). Compound E (0.2 μM; EMD Biosciences) was applied to synchronize postmitotic neurons for 3 days. Half of the medium was replaced once a week during continuous culture. For immunostaining and electrophysiological recordings, neural progenitors were plated on a confluent layer of rodent astrocytes, as described previously (83). These cultures exhibited similar neuronal densities, and parallel cultures were used for the examination of different iPSC lines.
Human brain samples
Postmortem brain tissues from four patients with AD and five age-matched controls diagnosed by neurology were provided by the Tissue Bank of the Institute of Geriatrics, Chinese PLA General Hospital, and Chinese PLA Medical Academy, and a detailed description of the patient information is provided in previous studies (58, 84). Total RNA-seq data from entorhinal cortex samples from 90 patients with AD and 84 healthy donors were provided by the Oxford Brain Bank (85), and the use of these human tissue samples for this study was approved by the Ethics Committees of the University of Lübeck. This study was approved by the Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology.
RNA isolation and qRT-PCR
Total RNA was extracted from cells or tissues using TRIzol reagent (Invitrogen, CA, USA), and 1 μg of RNA was reverse-transcribed for mRNA and miRNA analyses using a first strand cDNA synthesis kit (TOYOBO, Osaka, Japan) and miRcute Plus miRNA first-strand cDNA kit (Tiangen, Beijing, China), respectively. The qPCR program was performed with ABI StepOne Plus system using SYBR Green Premix Ex Taq (Takara, Tokyo, Japan). The primers used to detect mRNAs and miRNAs are listed in table S6. The relative expression levels of mRNAs or miRNAs were determined using the 2−ΔΔCT method after normalization to β-actin or U6, respectively, as the loading control. All experiments included biological and technical replicates.
Immunoblotting
Mice were euthanized, and the hippocampi were immediately separated from the brains. The tissues were homogenized on ice with radioimmunoprecipitation assay lysis buffer (Beyotime, Shanghai, China). After boiling for 10 min, these samples were ultrasonically disrupted on ice 20 times, and the protein concentration was measured using the BCA Protein Assay Reagent (Thermo Fisher Scientific, Illinois, USA). Proteins were separated on 10% SDS–polyacrylamide gel electrophoresis gels and transferred to nitrocellulose membranes (GE HealthCare Life Sciences, Loughborough, UK). After blocking with 5% nonfat milk for 30 min, the membranes were incubated with primary antibodies overnight at 4°C, followed by washes with phosphate-buffered saline (PBS)–Tween 20. Then, the membranes were incubated with anti-rabbit or anti-mouse immunoglobulin G conjugated secondary antibody IRdye 800 (1:10,000; Rockland Immunochemicals) for 1 hour at room temperature. The protein bands were detected using the Odyssey Imaging System (LI-COR, Lincoln, NE, USA).
Immunocytochemistry
Immunofluorescence staining was performed according to the manufacturer’s instructions. After anesthetization with a mixture of ketamine (100 mg/kg) and dexmedetomidine (0.5 mg/kg), mice were perfused with 1× PBS, fixed with 4% (v/v) paraformaldehyde (PFA), and dehydrated with 30% sucrose for 24 hours at 4°C. Brain slices (30 μm thick) were obtained with a cryostat and rinsed with 1× PBS three times. Then, these slices were treated with 3% hydrogen peroxide for 30 min to block endogenous peroxidase activity. After incubation with 0.1% Triton X-100 for 20 min to permeabilize the membrane, the slices were blocked with 5% bovine serum albumin (BSA) for 30 min. The brain slices were incubated with the primary antibody overnight at 4°C. After washes with PBS, the slices were probed with the biotinylated secondary antibody and streptomycin-labeled peroxidase solution for 1 hour at room temperature and then stained with 3,3'-diaminobenzidine (DAB) reagent for 1 to 10 min at 37°C. After washing, the brain slices were dehydrated with different concentrations of alcohol (75, 80, 95, and 100%), rendered transparent in xylene, and sealed on glass slides. The digital images of all slices were captured using a Coolpix 5000 Nikon camera.
Immunofluorescence staining
The mice were anesthetized as described above and perfused with 0.9% (w/v) NaCl and 4% (v/v) PFA. Brain sections were cut with a cryostat to a thickness of 30 μm and then washed with PBS three times for 5 min each. Next, the membrane was permeabilized with 0.1% Triton X-100 for 15 min. After blocking with 3% BSA for 30 min, the brain slices were incubated with the primary antibody at 4°C overnight. After three washes with PBS, the brain slices were subsequently incubated with a fluorescent dye–conjugated secondary antibody for 1 hour at room temperature in a dark environment. After washing, the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) at room temperature for 10 min. Slices were imaged with a confocal laser scanning microscope LSM800 (Carl Zeiss, Oberkochen, Germany), and images were processed using ImageJ or Fiji software.
Fluorescence in situ hybridization
FISH was performed using a previously described protocol (86). First, the mice from all groups were perfused with 0.9% NaCl and 4% PFA. Brain tissues were fixed in PFA at 4°C for 24 hours and then dehydrated in 30% (w/v) sucrose in PFA at 4°C until complete dehydration was achieved. The brain slices were cut to a thickness of 20 μm using a cryostat. The probes for miRNAs were synthesized by Boster Co. Ltd. (Wuhan, China), and FISH was performed according to the manufacturer’s instructions. Afterward, the brain slices were blocked for 1 hour with 3% BSA in 1× PBS and stained with primary antibodies against A1R overnight at 4°C. After washes with 1× PBS, the slices were incubated with a fluorescent dye–conjugated secondary antibody for 1 hour at room temperature. Last, the nuclei were stained with DAPI for 10 min and then slices were washed three times. All images were captured using a confocal microscope (Zeiss, LSM 800).
Plasmids, agomir/antagomirs, viruses, and stereotaxic injection
The overexpression vector for Adora1 was generated by cloning the coding sequence (CDS) of mouse Adora1 (NM_001039510). The AAVs (rAAV2/9) for Adora1 overexpression and the lentivirus carrying the short hairpin RNA targeting Adora1 were packaged by Umibio Co. Ltd. (Shanghai, China). For Adora1 knockdown, the plasmid hSyn1-miR30-shRNA-P2A-eGFP was used with some modifications based on a previously described method (87, 88). The shRNA-seq of Adora1 and hTau were listed in table S6. The pcDNA-hTau-GFP plasmid was preserved for our experiments. The mmu-miR-133a-3p agomir and antagomir and the scrambled control were purchased from RiboBio (Guangzhou, China). The concentrations of the miR-133a agomir and antagomir were determined as described in our previous articles (56, 80). Mice were anesthetized, and two holes were generated in the skull above the hippocampal CA3 region. A total of 1.5 μl of rAAVs (1013 TU/ml), 2 μl of lentiviruses (109 TU/ml), and 2 μl of agomir or antagomir (100 μM) were stereotaxically injected at an infusion rate of 0.2 μl/min into the hippocampal CA3 (anterior/posterior, −2.0 mm; medial/lateral, ±2.5 mm; dorsal/ventral, −2.3 mm) using a Hamilton microsyringe. The needle was left in the animal’s brain for 10 min and then slowly withdrawn. Subsequently, the wound was sutured, and the mice were allowed to recover.
CHX chase assay
CHX was applied to evaluate the effect of hTau on the stability of A1R by inhibiting protein synthesis using previously described methods (86). HEK293T cells were transfected with the hTau or control plasmid (pcDNA) and treated with 10 μM CHX (Sigma-Aldrich, CA, USA) at 36 hours posttransfection. Then, the cells were collected, and Western blotting was performed to analyze A1R expression at the indicated time points.
Luciferase activity assay
The 3′UTR of Adora1 (NM_001039510) was cloned and inserted into psiCHECK2 within the Xho I and Not I restriction sites located downstream of the Renilla luciferase gene using the following primers: forward 5′-TGGCTGCCCTGTACCTTGG-3′ and reverse 5′-AGGGCTTCACAATCTTTTATTAGAC-3′. Mutation of the Adora1 3′UTR was performed using a Mut Express II fast mutagenesis kit V2 (catalog no. C214, Vazyme Biotech Co. Ltd.). The plasmids containing the WT or mutant Adora1 3′UTR were transfected into HEK293T cells with miR-133a or NC agomirs at a final concentration of 200 nM. For the promoter luciferase reporters, pGL3 containing the promoter sequences of miR-133a and miR-1a were cotransfected into HEK293T cells with pRL-TK and Mef2c. Cells were harvested 48 hours later, and luciferase assays were performed using a dual-luciferase reporter assay kit (Promega) according to the manufacturer’s instructions. The values of Renilla activity relative to firefly activity were used for analysis.
Behavioral assays
Morris water maze
The Morris water maze was performed as described previously (56). A circular pool was filled with opaque water, and a hidden platform was placed below the surface of the water in one of the quadrants. The movement of mice was recorded using a digital tracking device. Briefly, the mice were trained to find the hidden platform for seven consecutive days with three trials per day. On the ninth day, the probe trial was performed to test spatial memory. The hidden platform was removed, and the mice were tested for their ability to find the platform location within 90 s after starting from the opposite quadrant. The escape latency to reach the former location of the platform and swimming speed were measured, and the percent time spent in each quadrant was also analyzed.
Y maze
The Y maze test was also used to evaluate spatial learning and memory (25), especially hippocampus-dependent memory. The Y maze box is composed of three arms with an included angle of 120°, and the size of each arm is 30 cm by 8 cm by 15 cm (length by width by height). The mice were acclimated to the environment for 10 min before 2 days of training. The three arms of the Y maze were randomly set as the start arm, old arm, and novel arm. The novel arm was closed by a partition plate in the training period and opened in the testing period. Throughout the experiment, the start arm and old arm were always open, and the animals were placed in the start arm and allowed to freely explore for 10 min. After 24 hours, the novel arm was opened in the testing stage, and the mice were placed in the start arm and allowed to explore for 5 min to probe spatial memory. A video camera was placed 1.5 m above the maze to record the movement of the mice. The new arm preference was calculated as follows: percent time spent in the novel arm/total time spent in both the old and novel arms × 100.
Electrophysiological recording
Mice were anesthetized, and coronal brain slices (300 μm thick) were prepared in artificial cerebrospinal fluid (ACSF) bubbled with 95% O2 and 5% CO2, which contained 124 mM NaCl, 3.0 mM KCl, 2.0 mM CaCl2, 1.2 mM MgSO4, 1.25 mM KH2PO4, 26 mM NaHCO3, and 11 mM glucose, using previously described methods (56, 89). Slices were then immersed with oxygenated ACSF at 30°C for at least 30 min and transferred to a recording chamber filled with ACSF. Field excitatory postsynaptic potentials (fEPSPs) were recorded from CA3 neurons by stimulating mossy fibers. The MED64 system (Alpha Med Sciences, Tokyo, Japan) was used to record the fEPSPs. After a 30-min stable baseline, LTP was induced by tetanic stimulation with three trains of the 100-Hz stimulus (at 30-s intervals).
For whole-cell patch-clamp recordings (90–93), pipettes (5 megohm) were filled with an internal recording solution containing 140 mM potassium gluconate, 2 mM NaCl, 10 mM Hepes, 0.2 mM EGTA, 2 mM Mg–adenosine triphosphate (MgATP), and 0.3 mM Na–guanosine triphosphate (NaGTP; 290 mOsm). The solution for voltage clamping contained 17.5 mM CsCl, 0.05 mM EGTA, 10 mM Hepes, 2 mM MgATP, 0.2 mM GTP, and 5 mM QX-314 (pH 7.4) (292 mOsm). For mEPSCs, pyramidal neurons from hippocampal CA3 were recorded at −70 mV in the presence of 1 mM tetrodotoxin and 10 mM bicuculline. The electrophysiological data were filtered at l2 kHz and acquired at 10 kHz using ClampFit 10.2 software (Molecular Devices). MiniAnalysis software (Synaptosoft, Decatur, GA) was used to analyze the mEPSC events with an amplitude threshold of 5 pA. The data were analyzed using GraphPad Prism 8.0 software, and the figures were modified using Adobe Illustrator CS6.
Golgi staining
Golgi staining was performed using a FD Rapid GolgiStain Kit (catalog no. PK401, FD Neuro Technologies Inc.) according to the manufacturer’s instructions. Briefly, the mice were anesthetized and perfused with 0.9% NaCl for 1 min, and then the brains were quickly removed from the skull. After rinses with double-distilled water, the brains were immersed in impregnation solution (equal amounts of solutions A/B were mixed) for 2 weeks at room temperature in the dark. The impregnation solution was changed every 3 days. Subsequently, the brains were transferred into solution C and incubated at room temperature in the dark for 1 week. Brain slices were cut at a thickness of 100 μm using vibrating blade microtomes (Leica VT1000s). Afterward, the brain slices were mounted on high-adhesion glass slides with solution C. The brain sections were stained with a mixture of solutions D and E for 10 min. Then, the slices were washed, dehydrated with sequential ethanol solutions for 4 min, and cleared in xylene three times for 4 min each. The images were captured using a Coolpix 5000 Nikon camera. Sholl analysis and dendritic spines were analyzed using ImageJ or Fiji software, as previously described (81, 94).
Sholl analysis
Sholl analysis was performed to analyze the complexity of the astrocyte arbor and dendritic trees using previously described methods (95, 96). The dendritic spines were calculated in two segments on the main branches at 100 and 200 mm from the soma, as previously reported (59).
Construction of astrocyte three-dimensional morphology and analysis of astrocytes
Confocal images and z stacks with a step size of 1 mm were captured using a confocal laser microscope (Carl Zeiss). Images were processed with Imaris 9 software. Then, the three-dimensional (3D) reconstructions were obtained, and volumes of astrocytes were analyzed. At least four randomly selected brain sections per mouse were used for immunostaining (96, 97).
Laser capture microdissection, processing for RNA-seq, and data analysis
The hippocampal CA3 region was isolated from brain slices using laser capture microdissection according to previously described methods (98, 99). The mice were anesthetized as described above and perfused with cold ribonuclease-free PBS. The brain of each mouse was dissected and cryosectioned at 20 μm, and mounted sections were onto 20 glass slides. The hippocampal CA3 regions were selectively captured by laser capture microdissection (Leica LMD7) with the 7.5-μm laser setting, and the laser was set to a pulse of 90 mW for 1 ms. Approximately four to six mice were pooled as one sample for the AKO group or the WT group. Then, three samples from each group were stored on dry ice until RNA was extracted using the RNAqueous-Micro Kit (Ambion, catalog no. AM1931). RNA-seq was conducted using the Illumina HiSeq2000 platform and completed by Beijing GeneCode Biotech Co. Ltd. Ribosomal RNA was removed from the total RNA, which was blasted against the Mus musculus genome by HISAT, and gene expression was calculated as fragments per kilobase of transcript per million mapped reads (FPKM). Differentially expressed RNAs were analyzed using the DESeq2 package. The Metascape website (https://metascape.org/gp/index.html) was used for the GO analysis. GSEA was conducted on all expressed genes.
Enzyme-linked immunosorbent assay
Enzyme-linked immunosorbent assay (ELISA) was performed to quantify Lcn2 protein levels according to the manufacturer’s instructions. Aβ40 and Aβ42 levels were detected by homogenizing the hippocampus in buffer containing 50 mM NaCl, 10 mM tris-HCl, 1 mM EDTA, 2% SDS, and 0.5 mM Na3VO4. The homogenates were boiled for 10 min and then centrifuged at 10,000g for 10 min. The final supernatants were used for ELISA, and the cell culture supernatant samples were treated according to the manufacturer’s instructions.
Statistical analysis
All data are presented as the means ± SEM and were analyzed using GraphPad Prism software (version 8). A two-tailed Student’s t test was used to assess the variance between two groups, and the difference among multiple groups was determined using one- or two-way analysis of variance (ANOVA) followed by post hoc tests, as listed in table S7. Correlations were assessed using a linear regression model (chi-square test). P < 0.05 indicates statistical significance. Both P < 0.01 and P < 0.001 represent extremely significant differences.
Acknowledgments
The AKO mice were a gift from J. Schnermann at NIDDK/NIH. We thank all the technicians and core facility in the Analytical and Testing Center, Huazhong University of Science and Technology.
Funding: This study was partially supported by the National Key Research and Development Program of China (grant no. 2019YFE0121200), the National Natural Science Foundation of China (grant nos. 82030032, 82261138555, 32070960, 81761138043, 81901103, 31721002, and 81961128005), the Top-Notch Young Talents Program of China of 2014, the Academic Frontier Youth Team of Huazhong University of Science and Technology to L.-Q.Z., the Hubei Provincial Natural Science Foundation (2022CFA004 to L.-Q.Z.), the NIH (grant no. NS122169), the Cincinnati Children’s Hospital Medical Center (CCHMC) Trustee Grant Award, and the CCHMC Center for Pediatric Genomics Award to Z.G.
Author contributions: L.-Q.Z. initiated and designed the study. L.-Q.Z. and D.L. supervised the study. L.-T.Z., H.-C.K., H.-Z.H., W.-Q.A., Y.Z., and W.-F.Z. performed the molecular biological experiments and animal experiments. L.-T.Z. and Z.-Q.L. analyzed the RNA-seq data. M.-F.D., H.L., and L.-T.Z. performed the electrophysiological recordings. L.L., A.K.S., S.S., M.S., and Z.G. generated AD iPSC lines, performed biochemical and electrophysiological experiments using iPSCs, and analyzed the data. Y.-Z.H., Z.-T.H., H.-H.Z., and J.-J.J. provided the human tissue samples. L.B. and M.S. analyzed the expression of MEF2c and ADORA1 in human samples. L.-T.Z., H.-C.K., Y.L., D.L., J.W., H.-Y.M., and L.-Q.Z. analyzed the data. L.-Q.Z., L.-T.Z., D.L., and Z.G. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The RNA-seq data are accessible through GEO Series accession number GSE193826 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE193826). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Supplementary Methods
Legends for tables S1 to S4
Tables S5 to S8
Supplementary data full gels for all the Western blots in this study.
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S4
REFERENCES AND NOTES
- 1.H. W. Querfurth, F. M. LaFerla, Alzheimer's disease. N. Engl. J. Med. 362, 329–344 (2010). [DOI] [PubMed] [Google Scholar]
- 2.D. J. Selkoe, J. Hardy, The amyloid hypothesis of Alzheimer's disease at 25 years. EMBO Mol. Med. 8, 595–608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.L. S. Honig, B. Vellas, M. Woodward, M. Boada, R. Bullock, M. Borrie, K. Hager, N. Andreasen, E. Scarpini, H. Liu-Seifert, M. Case, R. A. Dean, A. Hake, K. Sundell, V. Poole Hoffmann, C. Carlson, R. Khanna, M. Mintun, R. DeMattos, K. J. Selzler, E. Siemers, Trial of solanezumab for mild dementia due to Alzheimer’s disease. N. Engl. J. Med. 378, 321–330 (2018). [DOI] [PubMed] [Google Scholar]
- 4.H. Kettenmann, A. Verkhratsky, Neuroglia: The 150 years after. Trends Neurosci. 31, 653–659 (2008). [DOI] [PubMed] [Google Scholar]
- 5.K. Richetin, P. Steullet, M. Pachoud, R. Perbet, E. Parietti, M. Maheswaran, S. Eddarkaoui, S. Begard, C. Pythoud, M. Rey, R. Caillierez, K. Q. Do, S. Halliez, P. Bezzi, L. Buee, G. Leuba, M. Colin, N. Toni, N. Deglon, Tau accumulation in astrocytes of the dentate gyrus induces neuronal dysfunction and memory deficits in Alzheimer's disease. Nat. Neurosci. 23, 1567–1579 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Z. Jiwaji, S. S. Tiwari, R. X. Aviles-Reyes, M. Hooley, D. Hampton, M. Torvell, D. A. Johnson, J. McQueen, P. Baxter, K. Sabari-Sankar, J. Qiu, X. He, J. Fowler, J. Febery, J. Gregory, J. Rose, J. Tulloch, J. Loan, D. Story, K. McDade, A. M. Smith, P. Greer, M. Ball, P. C. Kind, P. M. Matthews, C. Smith, O. Dando, T. L. Spires-Jones, J. A. Johnson, S. Chandran, G. E. Hardingham, Reactive astrocytes acquire neuroprotective as well as deleterious signatures in response to Tau and Ass pathology. Nat. Commun. 13, 135 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.R. Abeti, A. Y. Abramov, M. R. Duchen, Beta-amyloid activates PARP causing astrocytic metabolic failure and neuronal death. Brain 134, 1658–1672 (2011). [DOI] [PubMed] [Google Scholar]
- 8.M. Talantova, S. Sanz-Blasco, X. Zhang, P. Xia, M. W. Akhtar, S. Okamoto, G. Dziewczapolski, T. Nakamura, G. Cao, A. E. Pratt, Y. J. Kang, S. Tu, E. Molokanova, S. R. McKercher, S. A. Hires, H. Sason, D. G. Stouffer, M. W. Buczynski, J. P. Solomon, S. Michael, E. T. Powers, J. W. Kelly, A. Roberts, G. Tong, T. Fang-Newmeyer, J. Parker, E. A. Holland, D. Zhang, N. Nakanishi, H. S. Chen, H. Wolosker, Y. Wang, L. H. Parsons, R. Ambasudhan, E. Masliah, S. F. Heinemann, J. C. Pina-Crespo, S. A. Lipton, Aβ induces astrocytic glutamate release, extrasynaptic NMDA receptor activation, and synaptic loss. Proc. Natl. Acad. Sci. U.S.A. 110, E2518–E2527 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.R. E. Gonzalez-Reyes, M. O. Nava-Mesa, K. Vargas-Sanchez, D. Ariza-Salamanca, L. Mora-Munoz, Involvement of astrocytes in Alzheimer's disease from a neuroinflammatory and oxidative stress perspective. Front. Mol. Neurosci. 10, 427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.X. Li, J. Zhang, D. Li, C. He, K. He, T. Xue, L. Wan, C. Zhang, Q. Liu, Astrocytic ApoE reprograms neuronal cholesterol metabolism and histone-acetylation-mediated memory. Neuron 109, 957–970.e8 (2021). [DOI] [PubMed] [Google Scholar]
- 11.A. Badimon, H. J. Strasburger, P. Ayata, X. Chen, A. Nair, A. Ikegami, P. Hwang, A. T. Chan, S. M. Graves, J. O. Uweru, C. Ledderose, M. G. Kutlu, M. A. Wheeler, A. Kahan, M. Ishikawa, Y. C. Wang, Y. E. Loh, J. X. Jiang, D. J. Surmeier, S. C. Robson, W. G. Junger, R. Sebra, E. S. Calipari, P. J. Kenny, U. B. Eyo, M. Colonna, F. J. Quintana, H. Wake, V. Gradinaru, A. Schaefer, Negative feedback control of neuronal activity by microglia. Nature 586, 417–423 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.K. Carvalho, E. Faivre, M. J. Pietrowski, X. Marques, V. Gomez-Murcia, A. Deleau, V. Huin, J. N. Hansen, S. Kozlov, C. Danis, M. Temido-Ferreira, J. E. Coelho, C. Meriaux, S. Eddarkaoui, S. L. Gras, M. Dumoulin, L. Cellai, C. E. B. B. B. Neuro, I. Landrieu, Y. Chern, M. Hamdane, L. Buee, A. L. Boutillier, S. Levi, A. Halle, L. V. Lopes, D. Blum, Exacerbation of C1q dysregulation, synaptic loss and memory deficits in tau pathology linked to neuronal adenosine A2A receptor. Brain 142, 3636–3654 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.A. G. Orr, E. C. Hsiao, M. M. Wang, K. Ho, D. H. Kim, X. Wang, W. Guo, J. Kang, G. Q. Yu, A. Adame, N. Devidze, D. B. Dubal, E. Masliah, B. R. Conklin, L. Mucke, Astrocytic adenosine receptor A2A and Gs-coupled signaling regulate memory. Nat. Neurosci. 18, 423–434 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.F. Rijo-Ferreira, T. E. Bjorness, K. H. Cox, A. Sonneborn, R. W. Greene, J. S. Takahashi, Sleeping sickness disrupts the sleep-regulating adenosine system. J. Neurosci. 40, 9306–9316 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.A. Rahman, The role of adenosine in Alzheimer's disease. Curr. Neuropharmacol. 7, 207–216 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.M. Schindler, C. A. Harris, B. Hayes, M. Papotti, P. P. Humphrey, Immunohistochemical localization of adenosine A1 receptors in human brain regions. Neurosci. Lett. 297, 211–215 (2001). [DOI] [PubMed] [Google Scholar]
- 17.J. F. Chen, H. K. Eltzschig, B. B. Fredholm, Adenosine receptors as drug targets—What are the challenges? Nat. Rev. Drug Discov. 12, 265–286 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O. Pascual, K. B. Casper, C. Kubera, J. Zhang, R. Revilla-Sanchez, J. Y. Sul, H. Takano, S. J. Moss, K. McCarthy, P. G. Haydon, Astrocytic purinergic signaling coordinates synaptic networks. Science 310, 113–116 (2005). [DOI] [PubMed] [Google Scholar]
- 19.A. de Mendonca, T. Almeida, Z. I. Bashir, J. A. Ribeiro, Endogenous adenosine attenuates long-term depression and depotentiation in the CA1 region of the rat hippocampus. Neuropharmacology 36, 161–167 (1997). [DOI] [PubMed] [Google Scholar]
- 20.K. P. Corodimas, H. Tomita, Adenosine A1 receptor activation selectively impairs the acquisition of contextual fear conditioning in rats. Behav. Neurosci. 115, 1283–1290 (2001). [DOI] [PubMed] [Google Scholar]
- 21.E. Angulo, V. Casado, J. Mallol, E. I. Canela, F. Vinals, I. Ferrer, C. Lluis, R. Franco, A1 adenosine receptors accumulate in neurodegenerative structures in Alzheimer disease and mediate both amyloid precursor protein processing and tau phosphorylation and translocation. Brain Pathol. 13, 440–451 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.J. L. Albasanz, S. Perez, M. Barrachina, I. Ferrer, M. Martin, Up-regulation of adenosine receptors in the frontal cortex in Alzheimer's disease. Brain Pathol. 18, 211–219 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.S. M. Neuner, L. A. Wilmott, B. R. Hoffmann, K. Mozhui, C. C. Kaczorowski, Hippocampal proteomics defines pathways associated with memory decline and resilience in normal aging and Alzheimer's disease mouse models. Behav. Brain Res. 322, 288–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.J. Stockwell, E. Jakova, F. S. Cayabyab, Adenosine A1 and A2A receptors in the brain: Current research and their role in neurodegeneration. Molecules 22, 676 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.F. J. A. Dennissen, M. Anglada-Huguet, A. Sydow, E. Mandelkow, E.-M. Mandelkow, Adenosine A1 receptor antagonist rolofylline alleviates axonopathy caused by human Tau ΔK280. Proc. Natl. Acad. Sci. U.S.A. 113, 11597–11602 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R. A. Cunha, How does adenosine control neuronal dysfunction and neurodegeneration? J. Neurochem. 139, 1019–1055 (2016). [DOI] [PubMed] [Google Scholar]
- 27.J. W. Kim, M. S. Byun, D. Yi, J. H. Lee, S. Y. Jeon, G. Jung, H. N. Lee, B. K. Sohn, J. Y. Lee, Y. K. Kim, S. A. Shin, C. H. Sohn, D. Y. Lee; KBASE Research Group , Coffee intake and decreased amyloid pathology in human brain. Transl. Psychiatry 9, 270 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.T. Yang, X. Gao, M. Sandberg, C. Zollbrecht, X.-M. Zhang, M. Hezel, M. Liu, M. Peleli, E.-Y. Lai, R. A. Harris, A. E. Persson, B. B. Fredholm, L. Jansson, M. Carlström, Abrogation of adenosine A1 receptor signalling improves metabolic regulation in mice by modulating oxidative stress and inflammatory responses. Diabetologia 58, 1610–1620 (2015). [DOI] [PubMed] [Google Scholar]
- 29.J. Lewis, E. McGowan, J. Rockwood, H. Melrose, P. Nacharaju, M. Van Slegtenhorst, K. Gwinn-Hardy, M. Paul Murphy, M. Baker, X. Yu, K. Duff, J. Hardy, A. Corral, W. L. Lin, S. H. Yen, D. W. Dickson, P. Davies, M. Hutton, Neurofibrillary tangles, amyotrophy and progressive motor disturbance in mice expressing mutant (P301L) tau protein. Nat. Genet. 25, 402–405 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Y. Hong, C. B. Chan, I. S. Kwon, X. Li, M. Song, H. P. Lee, X. Liu, P. Sompol, P. Jin, H. G. Lee, S. P. Yu, K. Ye, SRPK2 phosphorylates tau and mediates the cognitive defects in Alzheimer's disease. J. Neurosci. 32, 17262–17272 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.J. F. Abisambra, U. K. Jinwal, L. J. Blair, J. C. O'Leary III, Q. Li, S. Brady, L. Wang, C. E. Guidi, B. Zhang, B. A. Nordhues, M. Cockman, A. Suntharalingham, P. Li, Y. Jin, C. A. Atkins, C. A. Dickey, Tau accumulation activates the unfolded protein response by impairing endoplasmic reticulum-associated degradation. J. Neurosci. 33, 9498–9507 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.N. Myeku, C. L. Clelland, S. Emrani, N. V. Kukushkin, W. H. Yu, A. L. Goldberg, K. E. Duff, Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nat. Med. 22, 46–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.W. Filipowicz, S. N. Bhattacharyya, N. Sonenberg, Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 9, 102–114 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Q. Zhou, S. Zhu, Y. Guo, L. Lian, Q. Hu, X. Liu, F. Xu, N. Zhang, H. Kang, Adenosine A1 receptors play an important protective role against cognitive impairment and long-term potentiation inhibition in a pentylenetetrazol mouse model of epilepsy. Mol. Neurobiol. 55, 3316–3327 (2018). [DOI] [PubMed] [Google Scholar]
- 35.M. T. Heneka, M. J. Carson, J. El Khoury, G. E. Landreth, F. Brosseron, D. L. Feinstein, A. H. Jacobs, T. Wyss-Coray, J. Vitorica, R. M. Ransohoff, K. Herrup, S. A. Frautschy, B. Finsen, G. C. Brown, A. Verkhratsky, K. Yamanaka, J. Koistinaho, E. Latz, A. Halle, G. C. Petzold, T. Town, D. Morgan, M. L. Shinohara, V. H. Perry, C. Holmes, N. G. Bazan, D. J. Brooks, S. Hunot, B. Joseph, N. Deigendesch, O. Garaschuk, E. Boddeke, C. A. Dinarello, J. C. Breitner, G. M. Cole, D. T. Golenbock, M. P. Kummer, Neuroinflammation in Alzheimer's disease. Lancet Neurol. 14, 388–405 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.E. Jang, J. H. Kim, S. Lee, J. H. Kim, J. W. Seo, M. Jin, M. G. Lee, I. S. Jang, W. H. Lee, K. Suk, Phenotypic polarization of activated astrocytes: The critical role of lipocalin-2 in the classical inflammatory activation of astrocytes. J. Immunol. 191, 5204–5219 (2013). [DOI] [PubMed] [Google Scholar]
- 37.E. Jang, S. Lee, J. H. Kim, J. H. Kim, J. W. Seo, W. H. Lee, K. Mori, K. Nakao, K. Suk, Secreted protein lipocalin-2 promotes microglial M1 polarization. FASEB J. 27, 1176–1190 (2013). [DOI] [PubMed] [Google Scholar]
- 38.M. Mucha, A. E. Skrzypiec, E. Schiavon, B. K. Attwood, E. Kucerova, R. Pawlak, Lipocalin-2 controls neuronal excitability and anxiety by regulating dendritic spine formation and maturation. Proc. Natl. Acad. Sci. U.S.A. 108, 18436–18441 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.A. Care, D. Catalucci, F. Felicetti, D. Bonci, A. Addario, P. Gallo, M. L. Bang, P. Segnalini, Y. Gu, N. D. Dalton, L. Elia, M. V. Latronico, M. Hoydal, C. Autore, M. A. Russo, G. W. Dorn II, O. Ellingsen, P. Ruiz-Lozano, K. L. Peterson, C. M. Croce, C. Peschle, G. Condorelli, MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 13, 613–618 (2007). [DOI] [PubMed] [Google Scholar]
- 40.J. F. Chen, E. M. Mandel, J. M. Thomson, Q. Wu, T. E. Callis, S. M. Hammond, F. L. Conlon, D. Z. Wang, The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.P. Pidikova, R. Reis, I. Herichova, miRNA clusters with down-regulated expression in human colorectal cancer and their regulation. Int. J. Mol. Sci. 21, 4633 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.M. P. Heyer, A. K. Pani, R. J. Smeyne, P. J. Kenny, G. Feng, Normal midbrain dopaminergic neuron development and function in miR-133b mutant mice. J. Neurosci. 32, 10887–10894 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.X. Zhang, R. Yang, B. L. Hu, P. Lu, L. L. Zhou, Z. Y. He, H. M. Wu, J. H. Zhu, Reduced circulating levels of miR-433 and miR-133b are potential biomarkers for Parkinson's disease. Front. Cell. Neurosci. 11, 170 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.T. Theis, M. Yoo, C. S. Park, J. Chen, S. Kugler, K. M. Gibbs, M. Schachner, Lentiviral delivery of miR-133b improves functional recovery after spinal cord injury in mice. Mol. Neurobiol. 54, 4659–4671 (2017). [DOI] [PubMed] [Google Scholar]
- 45.H. Xin, Y. Li, B. Buller, M. Katakowski, Y. Zhang, X. Wang, X. Shang, Z. G. Zhang, M. Chopp, Exosome-mediated transfer of miR-133b from multipotent mesenchymal stromal cells to neural cells contributes to neurite outgrowth. Stem Cells 30, 1556–1564 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Q. Yang, Q. Zhao, Y. Yin, miR-133b is a potential diagnostic biomarker for Alzheimer's disease and has a neuroprotective role. Exp. Ther. Med. 18, 2711–2718 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.G. E. Lyons, B. K. Micales, J. Schwarz, J. F. Martin, E. N. Olson, Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J. Neurosci. 15, 5727–5738 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.A. Assali, A. J. Harrington, C. W. Cowan, Emerging roles for MEF2 in brain development and mental disorders. Curr. Opin. Neurobiol. 59, 49–58 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.J. C. Lambert, C. A. Ibrahim-Verbaas, D. Harold, A. C. Naj, R. Sims, C. Bellenguez, A. L. DeStafano, J. C. Bis, G. W. Beecham, B. Grenier-Boley, G. Russo, T. A. Thorton-Wells, N. Jones, A. V. Smith, V. Chouraki, C. Thomas, M. A. Ikram, D. Zelenika, B. N. Vardarajan, Y. Kamatani, C. F. Lin, A. Gerrish, H. Schmidt, B. Kunkle, M. L. Dunstan, A. Ruiz, M. T. Bihoreau, S. H. Choi, C. Reitz, F. Pasquier, C. Cruchaga, D. Craig, N. Amin, C. Berr, O. L. Lopez, P. L. De Jager, V. Deramecourt, J. A. Johnston, D. Evans, S. Lovestone, L. Letenneur, F. J. Moron, D. C. Rubinsztein, G. Eiriksdottir, K. Sleegers, A. M. Goate, N. Fievet, M. W. Huentelman, M. Gill, K. Brown, M. I. Kamboh, L. Keller, P. Barberger-Gateau, B. McGuiness, E. B. Larson, R. Green, A. J. Myers, C. Dufouil, S. Todd, D. Wallon, S. Love, E. Rogaeva, J. Gallacher, P. S. George-Hyslop, J. Clarimon, A. Lleo, A. Bayer, D. W. Tsuang, L. Yu, M. Tsolaki, P. Bossu, G. Spalletta, P. Proitsi, J. Collinge, S. Sorbi, F. Sanchez-Garcia, N. C. Fox, J. Hardy, M. C. Deniz Naranjo, P. Bosco, R. Clarke, C. Brayne, D. Galimberti, M. Mancuso, F. Matthews; European Alzheimer's Disease Initiative (EADI); Genetic and Environmental Risk in Alzheimer's Disease; Alzheimer's Disease Genetic Consortium; Cohorts for Heart and Aging Research in Genomic Epidemiology, S. Moebus, P. Mecocci, M. Del Zompo, W. Maier, H. Hampel, A. Pilotto, M. Bullido, F. Panza, P. Caffarra, B. Nacmias, J. R. Gilbert, M. Mayhaus, L. Lannefelt, H. Hakonarson, S. Pichler, M. M. Carrasquillo, M. Ingelsson, D. Beekly, V. Alvarez, F. Zou, O. Valladares, S. G. Younkin, E. Coto, K. L. Hamilton-Nelson, W. Gu, C. Razquin, P. Pastor, I. Mateo, M. J. Owen, K. M. Faber, P. V. Jonsson, O. Combarros, M. C. O'Donovan, L. B. Cantwell, H. Soininen, D. Blacker, S. Mead, T. H. Mosley Jr., D. A. Bennett, T. B. Harris, L. Fratiglioni, C. Holmes, R. F. de Bruijn, P. Passmore, T. J. Montine, K. Bettens, J. I. Rotter, A. Brice, K. Morgan, T. M. Foroud, W. A. Kukull, D. Hannequin, J. F. Powell, M. A. Nalls, K. Ritchie, K. L. Lunetta, J. S. Kauwe, E. Boerwinkle, M. Riemenschneider, M. Boada, M. Hiltuenen, E. R. Martin, R. Schmidt, D. Rujescu, L. S. Wang, J. F. Dartigues, R. Mayeux, C. Tzourio, A. Hofman, M. M. Nothen, C. Graff, B. M. Psaty, L. Jones, J. L. Haines, P. A. Holmans, M. Lathrop, M. A. Pericak-Vance, L. J. Launer, L. A. Farrer, C. M. van Duijn, C. Van Broeckhoven, V. Moskvina, S. Seshadri, J. Williams, G. D. Schellenberg, P. Amouyel, Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 45, 1452–1458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.G. Davies, N. Armstrong, J. C. Bis, J. Bressler, V. Chouraki, S. Giddaluru, E. Hofer, C. A. Ibrahim-Verbaas, M. Kirin, J. Lahti, S. J. van der Lee, S. Le Hellard, T. Liu, R. E. Marioni, C. Oldmeadow, I. Postmus, A. V. Smith, J. A. Smith, A. Thalamuthu, R. Thomson, V. Vitart, J. Wang, L. Yu, L. Zgaga, W. Zhao, R. Boxall, S. E. Harris, W. D. Hill, D. C. Liewald, M. Luciano, H. Adams, D. Ames, N. Amin, P. Amouyel, A. A. Assareh, R. Au, J. T. Becker, A. Beiser, C. Berr, L. Bertram, E. Boerwinkle, B. M. Buckley, H. Campbell, J. Corley, P. L. De Jager, C. Dufouil, J. G. Eriksson, T. Espeseth, J. D. Faul, I. Ford, S. Generation, R. F. Gottesman, M. E. Griswold, V. Gudnason, T. B. Harris, G. Heiss, A. Hofman, E. G. Holliday, J. Huffman, S. L. Kardia, N. Kochan, D. S. Knopman, J. B. Kwok, J. C. Lambert, T. Lee, G. Li, S. C. Li, M. Loitfelder, O. L. Lopez, A. J. Lundervold, A. Lundqvist, K. A. Mather, S. S. Mirza, L. Nyberg, B. A. Oostra, A. Palotie, G. Papenberg, A. Pattie, K. Petrovic, O. Polasek, B. M. Psaty, P. Redmond, S. Reppermund, J. I. Rotter, H. Schmidt, M. Schuur, P. W. Schofield, R. J. Scott, V. M. Steen, D. J. Stott, J. C. van Swieten, K. D. Taylor, J. Trollor, S. Trompet, A. G. Uitterlinden, G. Weinstein, E. Widen, B. G. Windham, J. W. Jukema, A. F. Wright, M. J. Wright, Q. Yang, H. Amieva, J. R. Attia, D. A. Bennett, H. Brodaty, A. J. de Craen, C. Hayward, M. A. Ikram, U. Lindenberger, L. G. Nilsson, D. J. Porteous, K. Raikkonen, I. Reinvang, I. Rudan, P. S. Sachdev, R. Schmidt, P. R. Schofield, V. Srikanth, J. M. Starr, S. T. Turner, D. R. Weir, J. F. Wilson, C. van Duijn, L. Launer, A. L. Fitzpatrick, S. Seshadri, T. H. Mosley Jr., I. J. Deary, Genetic contributions to variation in general cognitive function: A meta-analysis of genome-wide association studies in the CHARGE consortium (N=53949). Mol. Psychiatry 20, 183–192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.T. Sao, Y. Yoshino, K. Yamazaki, Y. Ozaki, Y. Mori, S. Ochi, T. Yoshida, T. Mori, J. I. Iga, S. I. Ueno, MEF2C mRNA expression and cognitive function in Japanese patients with Alzheimer's disease. Psychiatry Clin. Neurosci. 72, 160–167 (2018). [DOI] [PubMed] [Google Scholar]
- 52.S. Tu, M. W. Akhtar, R. M. Escorihuela, A. Amador-Arjona, V. Swarup, J. Parker, J. D. Zaremba, T. Holland, N. Bansal, D. R. Holohan, K. Lopez, S. D. Ryan, S. F. Chan, L. Yan, X. Zhang, X. Huang, A. Sultan, S. R. McKercher, R. Ambasudhan, H. Xu, Y. Wang, D. H. Geschwind, A. J. Roberts, A. V. Terskikh, R. A. Rissman, E. Masliah, S. A. Lipton, N. Nakanishi, NitroSynapsin therapy for a mouse MEF2C haploinsufficiency model of human autism. Nat. Commun. 8, 1488 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.D. Bi, L. Wen, Z. Wu, Y. Shen, GABAergic dysfunction in excitatory and inhibitory (E/I) imbalance drives the pathogenesis of Alzheimer's disease. Alzheimers Dement. 16, 1312–1329 (2020). [DOI] [PubMed] [Google Scholar]
- 54.J. J. Palop, L. Mucke, Amyloid-beta-induced neuronal dysfunction in Alzheimer's disease: From synapses toward neural networks. Nat. Neurosci. 13, 812–818 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.A. C. Mitchell, B. Javidfar, V. Pothula, D. Ibi, E. Y. Shen, C. J. Peter, L. K. Bicks, T. Fehr, Y. Jiang, K. J. Brennand, R. L. Neve, J. Gonzalez-Maeso, S. Akbarian, MEF2C transcription factor is associated with the genetic and epigenetic risk architecture of schizophrenia and improves cognition in mice. Mol. Psychiatry 23, 123–132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.X. Wang, D. Liu, H. Z. Huang, Z. H. Wang, T. Y. Hou, X. Yang, P. Pang, N. Wei, Y. F. Zhou, M. J. Dupras, F. Calon, Y. T. Wang, H. Y. Man, J. G. Chen, J. Z. Wang, S. S. Hebert, Y. Lu, L. Q. Zhu, A novel MicroRNA-124/PTPN1 signal pathway mediates synaptic and memory deficits in Alzheimer's disease. Biol. Psychiatry 83, 395–405 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Y. S. Xiong, F. F. Liu, D. Liu, H. Z. Huang, N. Wei, L. Tan, J. G. Chen, H. Y. Man, C. X. Gong, Y. Lu, J. Z. Wang, L. Q. Zhu, Opposite effects of two estrogen receptors on tau phosphorylation through disparate effects on the miR-218/PTPA pathway. Aging Cell 14, 867–877 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.A. J. Xie, T. Y. Hou, W. Xiong, H. Z. Huang, J. Zheng, K. Li, H. Y. Man, Y. Z. Hu, Z. T. Han, H. H. Zhang, N. Wei, J. Z. Wang, D. Liu, Y. Lu, L. Q. Zhu, Tau overexpression impairs neuronal endocytosis by decreasing the GTPase dynamin 1 through the miR-132/MeCP2 pathway. Aging Cell 18, e12929 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D. Liu, H. Tang, X. Y. Li, M. F. Deng, N. Wei, X. Wang, Y. F. Zhou, D. Q. Wang, P. Fu, J. Z. Wang, S. S. Hebert, J. G. Chen, Y. Lu, L. Q. Zhu, Targeting the HDAC2/HNF-4A/miR-101b/AMPK pathway rescues tauopathy and dendritic abnormalities in Alzheimer's disease. Mol. Ther. 25, 752–764 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.R. Corradetti, G. Lo Conte, F. Moroni, M. B. Passani, G. Pepeu, Adenosine decreases aspartate and glutamate release from rat hippocampal slices. Eur. J. Pharmacol. 104, 19–26 (1984). [DOI] [PubMed] [Google Scholar]
- 61.T. V. Dunwiddie, B. B. Fredholm, Adenosine A1 receptors inhibit adenylate cyclase activity and neurotransmitter release and hyperpolarize pyramidal neurons in rat hippocampus. J. Pharmacol. Exp. Ther. 249, 31–37 (1989). [PubMed] [Google Scholar]
- 62.A. de Mendonca, J. A. Ribeiro, Endogenous adenosine modulates long-term potentiation in the hippocampus. Neuroscience 62, 385–390 (1994). [DOI] [PubMed] [Google Scholar]
- 63.W. Hauber, A. Bareiss, Facilitative effects of an adenosine A1/A2 receptor blockade on spatial memory performance of rats: Selective enhancement of reference memory retention during the light period. Behav. Brain Res. 118, 43–52 (2001). [DOI] [PubMed] [Google Scholar]
- 64.Z. Liu, F. Wang, M. Tang, Y. Zhao, X. Wang, Amyloid β and tau are involved in sleep disorder in Alzheimer's disease by orexin A and adenosine A(1) receptor. Int. J. Mol. Med. 43, 435–442 (2019). [DOI] [PubMed] [Google Scholar]
- 65.C. L. Zuo, C. M. Wang, J. Liu, T. Shen, J. P. Zhou, X. R. Hao, Y. Z. Pan, H. C. Liu, Q. Q. Lian, H. Lin, Isoflurane anesthesia in aged mice and effects of A1 adenosine receptors on cognitive impairment. CNS Neurosci. Ther. 24, 212–221 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.B. B. Fredholm, K. Battig, J. Holmen, A. Nehlig, E. E. Zvartau, Actions of caffeine in the brain with special reference to factors that contribute to its widespread use. Pharmacol. Rev. 51, 83–133 (1999). [PubMed] [Google Scholar]
- 67.G. W. Arendash, C. Cao, Caffeine and coffee as therapeutics against Alzheimer's disease. J. Alzheimers Dis. 20, S117–S126 (2010). [DOI] [PubMed] [Google Scholar]
- 68.R. R. van Koert, P. R. Bauer, I. Schuitema, J. W. Sander, G. H. Visser, Caffeine and seizures: A systematic review and quantitative analysis. Epilepsy Behav. 80, 37–47 (2018). [DOI] [PubMed] [Google Scholar]
- 69.F. Maestu, W. de Haan, M. A. Busche, J. DeFelipe, Neuronal excitation/inhibition imbalance: Core element of a translational perspective on Alzheimer pathophysiology. Ageing Res. Rev. 69, 101372 (2021). [DOI] [PubMed] [Google Scholar]
- 70.J. Choi, H. W. Lee, K. Suk, Increased plasma levels of lipocalin 2 in mild cognitive impairment. J. Neurol. Sci. 305, 28–33 (2011). [DOI] [PubMed] [Google Scholar]
- 71.E. Eruysal, L. Ravdin, H. Kamel, C. Iadecola, M. Ishii, Plasma lipocalin-2 levels in the preclinical stage of Alzheimer's disease. Alzheimers Dement. 11, 646–653 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.A. Zawislak, P. Jakimowicz, J. A. McCubrey, D. Rakus, Neuron-derived transthyretin modulates astrocytic glycolysis in hormone-independent manner. Oncotarget 8, 106625–106638 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.T. Stork, A. Sheehan, O. E. Tasdemir-Yilmaz, M. R. Freeman, Neuron-glia interactions through the Heartless FGF receptor signaling pathway mediate morphogenesis of Drosophila astrocytes. Neuron 83, 388–403 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.C. Xing, X. Wang, C. Cheng, J. Montaner, E. Mandeville, W. Leung, K. van Leyen, J. Lok, X. Wang, E. H. Lo, Neuronal production of lipocalin-2 as a help-me signal for glial activation. Stroke 45, 2085–2092 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.A. Bhusal, M. H. Rahman, I. K. Lee, K. Suk, Role of hippocampal lipocalin-2 in experimental diabetic encephalopathy. Front. Endocrinol. 10, 25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.S. Basu, N. Chaudhary, S. Shah, C. Braggs, A. Sawant, S. Vaz, R. Thorat, S. Gupta, S. N. Dalal, Plakophilin3 loss leads to an increase in lipocalin2 expression, which is required for tumour formation. Exp. Cell Res. 369, 251–265 (2018). [DOI] [PubMed] [Google Scholar]
- 77.R. Kido, Y. Hiroshima, J. I. Kido, T. Ikuta, E. Sakamoto, Y. Inagaki, K. Naruishi, H. Yumoto, Advanced glycation end-products increase lipocalin 2 expression in human oral epithelial cells. J. Periodontal Res. 55, 539–550 (2020). [DOI] [PubMed] [Google Scholar]
- 78.J. K. Lee, N. J. Kim, Recent advances in the inhibition of p38 MAPK as a potential strategy for the treatment of Alzheimer's disease. Molecules 22, 1287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.B. Johansson, L. Halldner, T. V. Dunwiddie, S. A. Masino, W. Poelchen, L. Gimenez-Llort, R. M. Escorihuela, A. Fernandez-Teruel, Z. Wiesenfeld-Hallin, X. J. Xu, A. Hardemark, C. Betsholtz, E. Herlenius, B. B. Fredholm, Hyperalgesia, anxiety, and decreased hypoxic neuroprotection in mice lacking the adenosine A1 receptor. Proc. Natl. Acad. Sci. U.S.A. 98, 9407–9412 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.K. Zheng, F. Hu, Y. Zhou, J. Zhang, J. Zheng, C. Lai, W. Xiong, K. Cui, Y. Z. Hu, Z. T. Han, H. H. Zhang, J. G. Chen, H. Y. Man, D. Liu, Y. Lu, L. Q. Zhu, miR-135a-5p mediates memory and synaptic impairments via the Rock2/Adducin1 signaling pathway in a mouse model of Alzheimer's disease. Nat. Commun. 12, 1903 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.D. Liu, N. Wei, H. Y. Man, Y. Lu, L. Q. Zhu, J. Z. Wang, The MT2 receptor stimulates axonogenesis and enhances synaptic transmission by activating Akt signaling. Cell Death Differ. 22, 583–596 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.U. De Simone, F. Caloni, L. Gribaldo, T. Coccini, Human co-culture model of neurons and astrocytes to test acute cytotoxicity of neurotoxic compounds. Int. J. Toxicol. 36, 463–477 (2017). [DOI] [PubMed] [Google Scholar]
- 83.Z. Wen, H. N. Nguyen, Z. Guo, M. A. Lalli, X. Wang, Y. Su, N. S. Kim, K. J. Yoon, J. Shin, C. Zhang, G. Makri, D. Nauen, H. Yu, E. Guzman, C. H. Chiang, N. Yoritomo, K. Kaibuchi, J. Zou, K. M. Christian, L. Cheng, C. A. Ross, R. L. Margolis, G. Chen, K. S. Kosik, H. Song, G. L. Ming, Synaptic dysregulation in a human iPS cell model of mental disorders. Nature 515, 414–418 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.W. D. Bao, P. Pang, X. T. Zhou, F. Hu, W. Xiong, K. Chen, J. Wang, F. Wang, D. Xie, Y. Z. Hu, Z. T. Han, H. H. Zhang, W. X. Wang, P. T. Nelson, J. G. Chen, Y. Lu, H. Y. Man, D. Liu, L. Q. Zhu, Loss of ferroportin induces memory impairment by promoting ferroptosis in Alzheimer's disease. Cell Death Differ. 28, 1548–1562 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.V. Dobricic, M. Schilling, J. Schulz, L. S. Zhu, C. W. Zhou, J. Fuss, S. Franzenburg, L. Q. Zhu, L. Parkkinen, C. M. Lill, L. Bertram, Differential microRNA expression analyses across two brain regions in Alzheimer's disease. Transl. Psychiatry 12, 352 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Y. Su, M. F. Deng, W. Xiong, A. J. Xie, J. Guo, Z. H. Liang, B. Hu, J. G. Chen, X. Zhu, H. Y. Man, Y. Lu, D. Liu, B. Tang, L. Q. Zhu, MicroRNA-26a/death-associated protein kinase 1 signaling induces synucleinopathy and dopaminergic neuron degeneration in Parkinson's disease. Biol. Psychiatry 85, 769–781 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.F. Stegmeier, G. Hu, R. J. Rickles, G. J. Hannon, S. J. Elledge, A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 102, 13212–13217 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.C. J. Shen, D. Zheng, K. X. Li, J. M. Yang, H. Q. Pan, X. D. Yu, J. Y. Fu, Y. Zhu, Q. X. Sun, M. Y. Tang, Y. Zhang, P. Sun, Y. Xie, S. Duan, H. Hu, X. M. Li, Cannabinoid CB(1) receptors in the amygdalar cholecystokinin glutamatergic afferents to nucleus accumbens modulate depressive-like behavior. Nat. Med. 25, 337–349 (2019). [DOI] [PubMed] [Google Scholar]
- 89.Y. Yang, X. Shu, D. Liu, Y. Shang, Y. Wu, L. Pei, X. Xu, Q. Tian, J. Zhang, K. Qian, Y. X. Wang, R. S. Petralia, W. Tu, L. Q. Zhu, J. Z. Wang, Y. Lu, EPAC null mutation impairs learning and social interactions via aberrant regulation of miR-124 and Zif268 translation. Neuron 73, 774–788 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.M. Deng, Q. Zhang, Z. Wu, T. Ma, A. He, T. Zhang, X. Ke, Q. Yu, Y. Han, Y. Lu, Mossy cell synaptic dysfunction causes memory imprecision via miR-128 inhibition of STIM2 in Alzheimer's disease mouse model. Aging Cell 19, e13144 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.M. Ma, W. Xiong, F. Hu, M. F. Deng, X. Huang, J. G. Chen, H. Y. Man, Y. Lu, D. Liu, L. Q. Zhu, A novel pathway regulates social hierarchy via lncRNA AtLAS and postsynaptic synapsin IIb. Cell Res. 30, 105–118 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.A. He, C. Zhang, X. Ke, Y. Yi, Q. Yu, T. Zhang, H. Yu, H. Du, H. Li, Q. Tian, L. Q. Zhu, Y. Lu, VGLUT3 neurons in median raphe control the efficacy of spatial memory retrieval via ETV4 regulation of VGLUT3 transcription. Sci. China Life Sci. 65, 1590–1607 (2022). [DOI] [PubMed] [Google Scholar]
- 93.X. Li, H. Yu, B. Zhang, L. Li, W. Chen, Q. Yu, X. Huang, X. Ke, Y. Wang, W. Jing, H. Du, H. Li, T. Zhang, L. Liu, L.-Q. Zhu, Y. Lu, Molecularly defined and functionally distinct cholinergic subnetworks. Neuron 110, 3774–3788.e7 (2022). [DOI] [PubMed] [Google Scholar]
- 94.H. Tang, M. Ma, Y. Wu, M. F. Deng, F. Hu, H. A. M. M. Almansoub, H.-Z. Huang, D.-Q. Wang, L.-T. Zhou, N. Wei, H. Man, Y. Lu, D. Liu, L.-Q. Zhu, Activation of MT2 receptor ameliorates dendritic abnormalities in Alzheimer's disease via C/EBPα/miR-125b pathway. Aging Cell 18, e12902 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.E. Shin, Y. Kashiwagi, T. Kuriu, H. Iwasaki, T. Tanaka, H. Koizumi, J. G. Gleeson, S. Okabe, Doublecortin-like kinase enhances dendritic remodelling and negatively regulates synapse maturation. Nat. Commun. 4, 1440 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.S. SheikhBahaei, B. Morris, J. Collina, S. Anjum, S. Znati, J. Gamarra, R. Zhang, A. V. Gourine, J. C. Smith, Morphometric analysis of astrocytes in brainstem respiratory regions. J. Comp. Neurol. 526, 2032–2047 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.A. M. Reeves, E. Shigetomi, B. S. Khakh, Bulk loading of calcium indicator dyes to study astrocyte physiology: Key limitations and improvements using morphological maps. J. Neurosci. 31, 9353–9358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.A. E. Jaffe, D. J. Hoeppner, T. Saito, L. Blanpain, J. Ukaigwe, E. E. Burke, L. Collado-Torres, R. Tao, K. Tajinda, K. R. Maynard, M. N. Tran, K. Martinowich, A. Deep-Soboslay, J. H. Shin, J. E. Kleinman, D. R. Weinberger, M. Matsumoto, T. M. Hyde, Profiling gene expression in the human dentate gyrus granule cell layer reveals insights into schizophrenia and its genetic risk. Nat. Neurosci. 23, 510–519 (2020). [DOI] [PubMed] [Google Scholar]
- 99.A. Roy, M. Deng, K. A. Aldinger, I. A. Glass, K. J. Millen, Laser capture micro-dissection (LCM) of neonatal mouse forebrain for RNA isolation. Bio Protocol 10, e3475 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Supplementary Methods
Legends for tables S1 to S4
Tables S5 to S8
Supplementary data full gels for all the Western blots in this study.
Tables S1 to S4