Abstract
Hepatocellular carcinoma (HCC) is one of the most common and deadly cancers worldwide and is characterized by complex molecular carcinogenesis. Neuropilins (NRPs) NRP1 and NRP2 are the receptors of multiple proteins involved in key signaling pathways associated with tumor progression. We aimed to systematically review all the available findings on their role in HCC. We searched the Scopus, Web of Science (WOS), PubMed, Cochrane and Embase databases for articles evaluating NRPs in preclinical or clinical HCC models. This study was registered in PROSPERO (CRD42022349774) and include 49 studies. Multiple cellular and molecular processes have been associated with one or both NRPs, indicating that they are potential diagnostic and prognostic biomarkers in HCC patients. Mainly NRP1 has been shown to promote tumor cell survival and progression by modulating several signaling pathways. NRPs mainly regulate angiogenesis, invasion and migration and have shown to induce invasion and metastasis. They also regulate the immune response and tumor microenvironment, showing a crucial interplay with the hypoxia response and microRNAs in HCC. Altogether, NRP1 and NRP2 are potential biomarkers and therapeutic targets, providing novel insight into the clinical landscape of HCC patients.
Keywords: Biomarker, Hepatocellular carcinoma, Neuropilins, Systematic review
INTRODUCTION
Liver tumors are common and deadly cancer, with increasing incidence and mortality rates [1]. Hepatocellular carcinoma (HCC) is the main primary liver tumor type, accounting for approximately 90% of all cases [2]. HCC is a heterogeneous cancer mainly diagnosed at advanced stages. Its survival rate remains very low despite the available systematic therapies that only increase the survival probability of patients 1–2 years [2,3]. It is characterized by unique molecular carcinogenesis involving multiple modulators, signaling pathways, and mechanisms [2,4,5]. All of these factors contribute to the difficulty in understanding HCC development and progression. Therefore, further studies are required to provide clearer insights into its molecular mediators to develop effective targeted therapies and improve the outcomes of HCC patients [2,4].
The molecular pathogenesis of HCC is generally complex, with increasing numbers of molecules reported to participate in the development, progression, and drug sensitivity of HCC cells [2,6]. Neuropilins (NRPs) have recently been the focus of several studies due to their involvement in numerous cellular processes in cancer, such as cell proliferation, migration, invasion, and angiogenesis [7,8]. NRPs are 130–140 kDa type-1 membrane glycoproteins [7] with extracellular, transmembrane, and cytoplasmic domains. They act as co-receptors for proteins such as vascular endothelial growth factor (VEGF), fibroblast growth factor (FGF), hepatocyte growth factor (HGF), and transforming growth factor β (TGF-β) [7,9]. There are two NRPs (NRP1 and NRP2) encoded by different genes on independent chromosomes but with a common domain structure [8,9]. While both NRPs share some characteristics and ligands, they differ in their tissue distribution and modulated signaling pathways [8,10]. Moreover, while NRP1 has been widely studied and characterized, fewer studies have focused on NRP2 [9]. Nevertheless, the role played by both NRPs in cancer, including HCC, has recently become of interest due to their strong correlation with key cellular and molecular mechanisms involved in tumor progression [8,9].
Interestingly, despite increasing studies evaluating the role of NRP1 and NRP2 in human HCC, no attempt has been made to compile the main findings of their potential roles in the HCC tumor landscape. Therefore, we aimed to summarize through a systematic literature review the findings of published studies using preclinical and/or clinical HCC models and evaluating one or both NRPs.
MATERIALS AND METHODS
Protocol and registration
This systematic review was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidelines [11] (Supplementary Tables 1 and 2) and was registered in the International Prospective Register of Systematic Reviews (PROSPERO) database (CRD4 2022349774).
Literature search strategy
A comprehensive literature search was performed in the Web of Science, Scopus, PubMed, Embase and Cochrane Library databases of articles published up to August 31, 2022, identifying 293 articles (Fig. 1). This search strategy combined the search terms “NRP”, “NRP1”, “NRP2”, and “HCC”, in the search queries used for each database (Supplementary Table 3).
Figure 1.
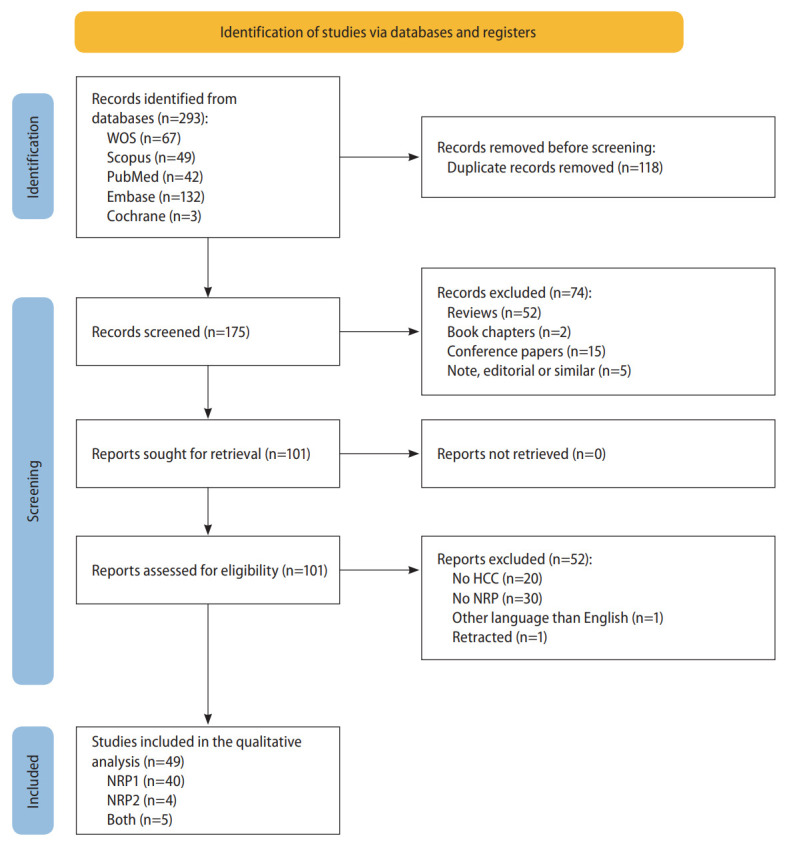
PRISMA flowchart of the study selection process. HCC, hepatocellular carcinoma; NRP, neuropilin; PRISMA, Preferred Reporting Items for Systematic Reviews and Meta-analyses; WOS, Web of Science.
Inclusion and exclusion criteria
Studies that met the following eligibility criteria were included in this systematic review: (1) they involved human patients diagnosed with HCC, animal HCC models, or in vitro primary or genetically-modified HCC cell line models; (2) they determined NRPs expression or derived effects from modifying NRPs; (3) they evaluated tumor-associated processes or characteristics. Studies that met the following exclusion criteria were removed during the study selection process: (1) they involved human patients without an HCC diagnosis, or unsuitable or poorly-described HCC models; (2) they were reviews or similar articles; (3) they did not evaluate or report NRPs expression or derived effects from modifying NRPs; (4) their full-text was not available in English.
Study selection
Two authors independently performed the study selection process, and discrepancies were resolved by a third author based on a consensus.
Duplicates among the original articles identified in the initial search of all databases were removed. Next, the articles were subjected to an initial screening by title and abstract based on exclusion criteria. Then, the remaining articles’ full texts were screened against the eligibility criteria, and those meeting the inclusion criteria were identified and included in the qualitative analysis.
Data collection and analysis
Three authors independently extracted data from the included studies, compiling the information in separate tables for preclinical and human studies.
The following information was included from preclinical studies: the first author’s name, publication year, model used, sample type, NRP subtype, measurement method, NRP alteration, associated cellular process, and alterations observed.
The following information was included from human studies: the first author’s name, publication year, case and control number, related etiology, mean age, sample type, NRP sub-type, determination sample type, tumor tissue expression, and clinical involvement.
RESULTS
Study selection and characteristics
The complete literature search and study selection process was conducted as described in Figure 1. The comprehensive search identified 293 unique articles after removing 118 duplicates between the five databases. Both inclusion and exclusion criteria were used to select studies that fit this study’s scope, identifying 49 eligible articles for inclusion in the systematic review.
The number of published articles assessing the NRPs’ role in different HCC models has increased in recent years, with most studies conducted since 2010 (Supplementary Fig. 1A). Curiously, in vitro studies have been replaced by in vivo studies using animal models or human samples. Moreover, when separately considering published studies on HCC patients (Supplementary Fig. 1B) and animals (Supplementary Fig. 1C), we observed an appreciable increase in published studies involving human patients in the last three years, along with increases in the number of patients used (Supplementary Fig. 1B). However, while in vivo models have been constantly used over time, their sample sizes have increased in recent years (Supplementary Fig. 1C). The main characteristics of all included articles are summarized separately for studies involving preclinical models (Table 1A) and human HCC samples (Table 1B).
Table 1.
Main characteristics of the included studies employing (A) preclinical and (B) clinical models
| A. Preclinical studies | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Study | Year | Model | Sample type | NRP | Method of measurement | NRP expression | NRP alteration | Cellular process associated | Specific alterations observed |
| Liao et al. [52] | 2008 | In vitro | Mahlavu, Huh-7, SK-Hep1 and HEK293T cell lines | NRP1 | RT-PCR | - | NRP1 silencing | Cell migration | ↓ Cell migration |
| Bergé et al. [36] | 2010 | In vitro | HepG2, SK-HEP-1 and PLC/PRF/5 cell lines | NRP1 | - | - | NRP1 silencing | Angiogenesis | No alterations |
| In vivo | Transgenic HCC C57BL/6 mice | NRP1 | ICC | - | NRP1 silencing | Tumor progression/development | ↓ Tumor growth | ||
| - | Angiogenesis | Inhibition of tumor vasculature remodeling | |||||||
| - | Immune-related response | ↑ IFN-γ | |||||||
| - | No changes in IFN-β and IL-12 | ||||||||
| Lee et al. [48] | 2010 | In vitro | HepG2 cell line | NRP1 | qRT-PCR | - | TCDD exposure | Xenobiotic toxicity associated to cancer | ↑ NRP1 |
| PRMT1 and PRMT4 co-inhibition by silencing | ↓ NRP1 | ||||||||
| Raskopf et al. [37] | 2010 | In vitro | Mouse Hepa129 cell line | NRP1 | Western blot | - | NRP1 silencing | Cell proliferation | No effects |
| - | Apoptosis | No effects | |||||||
| - | Cell migration | ↓ Cell migration | |||||||
| - | ↓ Tube formation ability | ||||||||
| In vivo | C3H mice with Hep129-derived tumor | NRP1 | qRT-PCR | - | NRP1 silencing | Tumor progression/development | ↓ Tumor growth by both siR NRP1 and siR Control | ||
| - | Cell proliferation | ↓ Cell proliferation | |||||||
| - | Apoptosis | ↓ Apoptosis | |||||||
| - | Angiogenesis | No changes in proangiogenic factors | |||||||
| - | Immune-related response | ↓ TNF-α | |||||||
| - | ↑ IFN-β | ||||||||
| Bergé et al. [12] | 2011 | In vitro | HepG2, SK-HEP-1 and PLC/PRF/5 cell lines | NRP1 | Western blot | - | Peptide N-derived inhibition | Cell proliferation | ↓ Cell viability |
| Apoptosis | ↑ Cleaved caspase-3 | ||||||||
| Angiogenesis | ↓ Capillary-like structure formation | ||||||||
| ↓ Total tube length | |||||||||
| ↓ Tubular network area | |||||||||
| In vivo | ASV-B transgenic C57BL/6 mice | NRP1 | qRT-PCR | Upregulated in HCC animals | Peptide N-derived inhibition | Tumor progression/development | ↓ Liver volume and weight | ||
| ↓ Nodule size and Ki67 staining | |||||||||
| Apoptosis | ↑ TUNEL staining | ||||||||
| Tumor vasculature/Invasion | Inhibition of tumor vasculature remodeling | ||||||||
| ↓ Microvessels number | |||||||||
| ↓ Total microvessels length | |||||||||
| ↓ Mean blood flow velocity of hepatic and mesenteric arteries | |||||||||
| Devbhandari et al. [54] | 2011 | In vitro | HCCLM3 cell line | NRP1 | Western blot and mass spectrometry | - | - | Migration | CD151-NRP1 complex-dependent migration |
| Lee et al. [38] | 2011 | In vitro | HepG2 and Huh-7 cell lines | NRP1 | Western blot | - | NRP1 silencing | Cell death | No alterations |
| Raskopf et al. [53] | 2012 | In vitro | Mouse Hepa129 cell line | NRP1 | qRT-PCR | - | Downregulation derived from VEGF silencing | Angiogenesis | VEGF silencing decreased NRP1 expression |
| NRP2 | qRT-PCR | - | Downregulation derived from VEGF silencing | Angiogenesis | VEGF silencing decreased NRP2 expression | ||||
| Yaqoob et al. [29] | 2012 | In vitro | HepG2 cell line | NRP1 | Western blot | - | HSC overexpressing NRP1 | Cell proliferation | Conditioned matrix from HSC increased Ki67 staining |
| Chishti et al. [14] | 2013 | In vivo | Sprague Dawley rats with drug-induced HCC | NRP1 | qRT-PCR | Upregulated in HCC animals | - | NRP expression | ↑ NRP1 in HCC |
| Xu and Xia [39] | 2013 | In vitro | HCCLM6 cell line | NRP1 | Western blot | - | NRP1 silencing | Cell proliferation | ↓ Cell growth rate |
| In vivo | HCCLM6 xenograft nude mice | NRP1 | qRT-PCR, Western blot and ICC | - | NRP1 silencing | Tumor progression/development | ↓ Tumor size | ||
| - | ↓ Tumor weight | ||||||||
| - | Tumor vasculature/Invasion | ↓ Neovascularization | |||||||
| Horwitz et al. [57] | 2014 | In vivo | Hep3B xenograft Mdr2-/- mice | NRP1 and NRP2 | qPCR | - | NR | Immune-related response | ↑ NRP1 and NRP2 in macrophages |
| Zhuang et al. [15] | 2014 | In vitro | HCCLM3 HCC cell line and L02 healthy liver cell line | NRP1 | ICC and qRT-PCR | - | CoCl2 treatment | Hypoxia response | ↓ NRP1 in L02 cells |
| In vivo | HCCLM3 orthotopic implantation in BALB/c nu/nu nude mice | NRP1 | qRT-PCR | - | - | Hypoxia response | ↓ NRP1 time-dependent in peritumoral tissue while ↑Hypoxia | ||
| Liu et al. [40] | 2015 | In vitro | PLC/PRF/5 and Huh-7 cell lines | NRP1 | qRT-PCR and Western blot | - | Overexpression derived from Inh-148b | Angiogenesis | ↑ Tube formation |
| Downregulation derived from miR-148b | Angiogenesis | ↓ Tube formation | |||||||
| - | MicroRNA modulation | NRP1 is a target of miR-148b | |||||||
| - | CSC properties | ↑ NRP1 expression in side population cells of HCC cell lines | |||||||
| In vivo | PLC/PRF/5 xenograft BALB/c nude mice | NRP1 | IHC | - | Overexpression derived from Inh-148b | Tumor progression/development | ↑ Cell division, tumor weight, tumor volume | ||
| - | Downregulation derived from miR-148b | Tumor progression/development | ↓ Cell division, tumor weight, tumor volume | ||||||
| In silico | - | NRP1 | Three computational algorithms to identify target genes | - | - | MicroRNA modulation | NRP1 is a target of miR-148b | ||
| Wittmann et al. [35] | 2015 | In vitro | 3sp, SNU-398, SNU-423, SNU-449, SNU-475, FLC-4 cell lines | NRP2 | qRT-PCR and Western blot | - | - | Mesenchymal phenotype | NRP2 was correlated with mesenchymal phenotype |
| 3sp, SNU-449 cell lines | - | NRP2 silencing | Migration and invasion | ↓ Migration and invasion abilities | |||||
| 3p, 3sp, Hep3B, PLC, SNU-423, SNU-449 cell lines | - | - | TGF-β signaling | NRP2 correlated with TGF-β | |||||
| 3p, 3sp, Hep3B, PLC, SNU-423, SNU-449 cell lines | - | NRP2 silencing | TGF-β signaling | No alterations | |||||
| 3p, 3sp, Hep3B, PLC, SNU-423, SNU-449 | - | TGF-β treatment | TGF-β signaling | ↑ NRP2 | |||||
| 3p, 3sp, Hep3B, PLC, SNU-423, SNU-449 | - | LY2109761 - TGF-β inhibitor | TGF-β signaling | ↓ NRP2 | |||||
| Kisseleva et al. [58] | 2016 | In vivo | Hepatoma 22a C3HA mice | NRP1 | qRT-PCR and flow cytometry | - | - | - | ↑ NRP1 in thymocytes |
| Sharma et al. [27] | 2016 | In vitro | Hep3B and HepG2 cell lines | NRP1 | qRT-PCR | - | - | - | ↑ NRP1 |
| Zhang et al. [16] | 2016 | In vitro | Bel-7402, SMMC-7721 and HepG2 cell lines, and L02 healthy liver cell line | NRP1 | qRT-PCR, Western blot and ICC | - | - | - | ↑ NRR1 in HCC cell lines |
| Bel-7402, SMMC-7721 and HepG2 - L02 cell lines | NRP1 | qRT-PCR, Western blot and ICC | - | - | Metastasis | ↑ NRR1 in high-metastatic cell lines | |||
| Wang et al. [64] | 2017 | In vitro | HepG2 cell line | NRP1 | GO functional enrichment analysis | - | miRNA-124 transfection | Axon guidance pathway | Enrichment of NRP1 |
| Xu et al. [42] | 2017 | In vitro | HepG2 and LX2 co-culture | NRP1 | Western blot and ICC | - | NRP1 silencing | Cell proliferation | ↓ Cell proliferation |
| Migration and invasion | ↓ Cell migration and invasion | ||||||||
| In vivo | HepG2 and LX2 xenograft nude mice | NRP1 | IHC | - | NRP1 knockdown | Tumor progression/development | ↓ Tumor volume | ||
| ↓ α-SMA staining | |||||||||
| Lin et al. [17] | 2018 | In vitro | Bel-7402 and SMMC- 7721 cell lines | NRP1 | qRT-PCR and Western blot | - | - | NRP1 targeting | NRP1 was regulated by TEAD |
| NRP1 silencing | Cell viability | ↓ Cell viability and colony formation | |||||||
| ↑ Caspase-3/7 activity | |||||||||
| Cheng et al. [51] | 2019 | In vitro | Huh-7 cell line | NRP1 | qRT-PCR | - | miR-148b overexpression | Migration and invasion | ↓ Cell migration |
| Lv et al. [43] | 2019 | In vitro | HepG2 cell line | NRP1 | Western blot | - | NRP1 silencing + SSd | Cell viability | ↓ Cell viability |
| - | Migration and invasion | ↓ Cell migration | |||||||
| In silico | - | NRP1 | HIT and TCMID databases | - | - | - | NRP1 is a target of SSd | ||
| Xu et al. [44] | 2019 | In vitro | HepG2 cell line | NRP1 | Confocal microscopy and flow cytometry | - | - | Targeted therapy | Succesfull detection of NRP1 antibody in the HCC cell surface |
| In vivo | HepG2 xenograft BALB/c nude mice | NRP1 | - | NR | - | Targeted therapy | Localization of the tTF- anti-NRP1 in the tumor after 2 h of intravenous administration | ||
| - | NR | - | Tumor progression/development | ↓ Tumor growth and progression | |||||
| Arab et al. [45] | 2020 | In vitro | HepG2Cyp2E1* cell line | NRP1 | - | - | Supernatant from HSC with knockdown of NRP1 | Cell proliferation | ↓ Lipid droplet formation |
| ↓ IGFBP3 | |||||||||
| ↑ SerpinA12 | |||||||||
| Yang [65] | 2020 | In vitro | Hep3B cell line | NRP1 | qRT-PCR and Western blot | - | Circ-ABCB10 overexpression | NRP1 | ↑ NRP1 expression |
| - | miR-340-5p/miR-452- 5p overexpression | NRP1 | ↓ NRP1 expression | ||||||
| In vivo | BALB/c athymic nude mice injected with Hep3B | NRP1 | Western blot | NR | Circ-ABCB10 overexpression | NRP1 | ↑ NRP1 expression | ||
| Ye et al. [28] | 2020 | In vitro | HepG2, SK-Hep and Bel-7404 cell lines, and L02 healthy liver cell line | NRP2 | qRT-PCR | NRP2 overexpression | - | - | ↑ NRP2 expression in HCC lines |
| Li et al. [26] | 2021 | In vitro | HCCLM3 and Huh-7 cell lines | NRP1 | qRT-PCR and Western blot | - | NRP1 silencing | Stem cell properties | ↓ Liver CSC population |
| Cell proliferation | ↓ Colony formation ability and sphere diameter | ||||||||
| EMT pathway | ↓ N-cadherin and vimentin | ||||||||
| HepG2, HCCLM3 and Huh-7 cell lines, and L02 healthy liver cell line | NRP1 | qRT-PCR and Western blot | NRP1 overexpression | NRP1 silencing | Cell migration | ↑ E-cadherin | |||
| In vivo | HCCLM3 xenograft nude mice | NRP1 | - | - | NRP1 silencing | Metastasis | ↓ Cell migration | ||
| Zhang et al. [21] | 2021 | In vivo | Wistar rats injected with rat hepatoma cell line CBRH-7919 or hepatoma cell line RH-35 | NRP2 | LC-MS/MS | NR | - | NRP2 | Pulmonary metastasis in 1 out of 5 grafts vs 5/5 |
| Li and Bao [49] | 2022 | In vivo | H22 tumor-bearing mouse model | NRP1 | Western blot | NR | IPE high dose treatment (TG group) | NRP1 | ↓ NRP2 in urinary samples |
| ↓ NRP1 expression in TG group | |||||||||
|
B. Clinical studies
| |||||||||
| Study | Year | Number (Case/Controls) | Etiology related | Mean age | Sample type | NRP | Type of determination sample | NRP expression in tumor sample | Clinical involvement |
| Beckebaum et al. [60] | 2004 | 65/70 | 54 Cirrhosis, from which: 21 HCV, 10 HBV, 9 Alcohol, 13 Cryptogenic, 1 Autoimmune and 3 no cirrhosis (2 HCV and 1 HBV) | 60±12.27 (Healthy: 57±21.56) | Freshly isolated perypheral blood mononuclear cells | NRP1 | Serum biomarker | Downregulated | Inversely correlated with IL-10 |
| Bergé et al. [12] | 2011 | 308/31 | NR | NR | Liver tissue | NRP1 | Tissue | Upregulated | Overexpression in HCC |
| Yaqoo et al. [29] | 2012 | 139/139 | NR | NR | Liver tissue | NRP1 | Tissue | NR | NRP1 overexpression correlated with shorter OS |
| Kitagawa et al. [13] | 2013 | 12 | 6 cirrhosis, 5 chronic hepatitis, 1 normal - 3 HBV, 7 HCV, 1 both, 1 negative | 51–81 | Liver tissue | NRP1 and NRP2 | Tissue | NRP1: No changes | Expression in HCC tissue |
| NRP2: downregulated | |||||||||
| Chishti et al. [14] | 2013 | 126/7 | NR | NR | Liver tissue | NRP1 | Tissue | Upregulated | Overexpression in HCC |
| Zhuang et al. [15] | 2014 | 214 | 168 cirrhosis | 50.45±12.4 | Liver tissue | NRP1 | Tissue | Upregulated in peritumoral | High peritumoral NRP1: lower TTR and OS |
| 176 HBsAg+ | |||||||||
| Villa et al. [41] | 2015 | 132 | 132 Cirrhosis | 68.25 (32–88) | Liver tissue | NRP | Tissue | NR | NRP is part of a hepatic signature that constitutes an independent factor for rapid tumor growth and mortality |
| 74 HCV | |||||||||
| 16 HBV | |||||||||
| 18 Alcohol | |||||||||
| 20 Dysmetabolic | |||||||||
| Wittman et al. [35] | 2015 | 133 | NR | NR | Liver tissue | NRP2 | Tissue | NR | NRP2 overexpression correlated wth higher tumor grading |
| Zhang et al. [16] | 2016 | 16/16† and 105/105† | 84 HBV | NR | Liver tissue | NRP1 | Tissue | Upregulated | NRP1 overexpression correlated with intrahepatic metastasis, Edmondson grade, TNM, portal vein invasion, shorter OS and RFS |
| Dong et al. [30] | 2017 | 190/190 | 154 Cirrhosis | 23-89 | Liver tissue | NRP2 | Tissue | NR | NRP2 overexpression correlated with higher histological grade, absence of cirrhosis, shorter OS and DFS |
| 152 HBV | |||||||||
| Lin et al. [17] | 2018 | 40/30 | NR | NR | Liver tissue | NRP1 | Tissue | Upregulated | Overexpression in HCC |
| 104/80 | NR | 55.37±8.63 | Serum sample | NRP1 | Serum biomarker | Upregulated | Correlated with serum AFP, γ-GT, Alb, bile acid, ALT, AST, ALP and pre-Alb | ||
| Morin et al. [18] | 2018 | 11 | NR | NR | Liver tissue | NRP1 | Tissue | NR | Marked NRP1 staining |
| Lyu et al. [19] | 2019 | 371/50 | NR | NR | Liver tissue | NRP1 | Tissue | Upregulated | Overexpression in HCC |
| Ono et al. [31] | 2020 | 41 | 7 HBV | 72 (46-84) | Serum sample | NRP1 | Serum biomarker (9 circulating cytokines and angiogenic factors signature) | NR | 9 circulating cytokines and angiogenic factors signature associated to lower PFS, OS and early PD |
| 9 HCV | |||||||||
| 9 HCV post SVR | |||||||||
| 6 Alcohol | 9 circulating cytokines and angiogenic factors signature positively correlated with AST and ALT, and negatively with Alb | ||||||||
| Liu et al. [67] | 2020 | NR | NR | NR | Liver tissue | NRP1 | Tissue | NR | TFAP4 was correlated with NRP1 as an immune marker in dendritic cells |
| Abdel Ghafar et al. [25] | 2021 | 50/50 | NR | 59.2±6.7 / 57.5±7.1 | Serum sample | NRP1 | Serum biomarker | Upregulated | Correlated with OS, BCLC stages B and C, tumor number (>3), tumor size (≥5 cm), vascular invasion and distant metastasis |
| Li et al. [26] | 2021 | 81 (cohort 1) | NR | NR | Liver tissue | NRP1 | Tissue | Upregulated | Overexpression in HCC |
| 16 (cohort 2) | NR | NR | Liver tissue | NRP1 | Tissue | Upregulated | Overexpression in HCC | ||
| 239 (cohort 3) | 89 Cirrhosis 75 HBV | NR | Liver tissue | NRP1 | Tissue | NR | NRP1 overexpression correlated with shorter OS and vascular invasion | ||
| 16 (cohort 4) | NR | NR | Liver tissue | NRP1 | Tissue | NR | NRP1 overexpression in patients with recurrence | ||
| 20 (cohort 5) | NR | NR | Liver tissue | NRP1 | Tissue | NR | NRP1 overexpression in patients with recurrence | ||
| Savier et al. [20] | 2021 | 14 | NR | 62.11 (47.1–78.8) | Primary human hepatocytes from HCC patients | NRP1 | Primary cells | Upregulated | Overexpression in HCC - NRP1 overexpression in the most aggressive tumors |
| Significant correlation with peptide internalization and tumor aggressiveness | |||||||||
| Xu et al. [34] | 2021 | 371 | NR | NR | Dendritic cells | NRP1 | Dendritic cells | NR | NRP1 expression on dendritic cells was correlated with Rad51, a valuable prognosis marker in HCC |
| Chen et al. [46] | 2021 | NR‡ | NR | NR | Liver tissue | NRP1 | Tissue | NR | NRP1 was correlated with CD36 |
| Li et al. [22] | 2022 | 374/50 | NR | NR | Liver tissue | NRP1 | Tissue | Upregulated | High NRP1 expression in the high-risk group of HCC patients |
| Liu et al. [47] | 2022 | 5/5 | NR | NR | Liver tissue | NRP1 and NRP2 | Tissue | NR | NRP1/NRP2-VEGFA interaction is involved in HCC tumorigenesis |
| Li et al. [23] | 2022 | NR‡ | NR | NR | Liver tissue | NRP1 | Tissue | Upregulated | Overexpression in HCC |
| Cheng et al. [61] | 2022 | NR | NR | NR | Liver tissue | NRP1 | Tissue | NR | NRP1 (as immune-related gene) was not correlated with KLRB1 |
| Fernández-Palanca et al. [24] | 2022 | 1,156 | NR | NR | Liver tissue | NRP1 | Tissue | Upregulated | Negatively correlated with OS and RFS |
| 149 | NR | NR | Serum sample | NRP1 | Serum biomarker | Upregulated | Directly associated to higher venous invasion and metastasis | ||
| Huang et al. [32] | 2022 | 247/241 | NR | NR | Liver tissue | NRP1 | Tissue | Upregulated | Correlated with higher recurrence |
| Li et al. [33] | 2022 | 156 | NR | NR | Liver tissue | NRP1 | Tissue | No altered | Correlated with higher recurrence |
| NRP1 was specifically expressed in CAF and/or TEC | |||||||||
α-SMA, α-smooth muscle actin; AFP, alpha fetoprotein; Alb, albumin; ALP, alkaline phosphatase; ALT, alanine transaminase; AST, aspartate aminotransferase; CAF, cancer-associated fibroblast; CSC, cancer stem cell; DFS, disease-free survival; γ-GT, gamma-glutamyl transpeptidase; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; HSC, hepatic stellate cell; ICC, immunocytochemistry; IFN, interferon; IGFBP3, insulin-like growth factor binding protein-3; IHC, immunohistochemistry; IL, interleukin; IPE, Inonotus hispidus petroleum ether extract; KLRB1, killer cell lectin-like receptor B1; LC-MS/MS, liquid chromatography-tandem mass spectrometry; miRNA, microRNA; NR, not reported; NRP, neuropilin; OS, overall survival; PD, progressive disease; PFS, progression-free survival; PRMT1, protein arginine methyltransferase 1; qRT-PCR, real-time reverse transcription polymerase chain reaction; RFS, recurrence-free survival; SSd, Saikosaponin d; SVR, sustained virologic response; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin; TEAD, TEA domain transcription factor; TEC, tumor-associated endothelial cell; TFAP4, transcription factor activating enhancer binding protein 4; TGF-β, transforming growth factor β; TNF-α, tumor necrosis factor-α; TTR, time to recurrence; VEGF, vascular endothelial growth factor.
HepG2Cyp2E1, cell line overexpressing ethanol-metabolizing enzyme cytochrome P450 2E1.
For differential expression in normal and HCC tissue by qRT-PCR in the 16 samples and for differential expression in normal and HCC tissue, and the remaining analysis with the 105 samples.
Sample data from different public databases without reporting total number of samples included in the study.
All 49 of the included studies evaluated the role of NRP1 and/or NRP2 in HCC, of which 11 used only in vitro models (22.45%), four only animal models (8.16%), seven both in vitro and in vivo models (14.29%), 19 only human HCC patient samples (38.78%) and eight preclinical and clinical samples (16.33%). There was a notable difference in the number of studies examining each NRP subtype. Only four of the 49 included studies examined NRP2 (8.16%), compared to 40 that examined NRP1 (81.63%), five examined both NRPs (10.20%). Most (22/27 [81.48%]) clinical articles on human patients evaluated NRP protein expression directly in tumor tissue. While detecting potential tumor markers in serum samples is an important tool for improving HCC diagnosis, only five studies analyzed serum NRP1 levels in human patients.
NRP measurements in preclinical models used different approaches. The most common was the real-time reverse transcription polymerase chain reaction (qRT-PCR; 33.33%), followed by western blot (16.67%), qRT-PCR and Western blot (20.00%), immunofluorescence microscopy (6.67%), and all three techniques (10.00%). In addition, some individual studies used different methodologies including qRT-PCR and immunofluorescence microscopy (3.33%), western blot and immunofluorescence microscopy (3.33%), and mass spectrometry (3.33%). One of the 30 preclinical studies did not assess NRP expression (3.33%).
In addition, most included studies targeted one NRP to assess the derived effects. However, their targeting strategies varied widely, from genetic silencing to using inhibitors or antibodies (Table 2). Given the many articles evaluating the NRPs’ role in various cellular and molecular processes in HCC, their main findings have been organized and described in the following sections.
Table 2.
Strategies employed for NRP targeting in the different studies included
| Targeting strategy | Specifications | NRP | Method of measurement | Model | Sample type | Outcome | Study | Year |
|---|---|---|---|---|---|---|---|---|
| shRNA silencing | NRP1 shRNA VSV-lentivirus | NRP1 | RT-PCR | In vitro | Mahlavu, Huh-7, SK-Hep1 and HEK293T cell lines | Migration | Liao et al. [52] | 2008 |
| NRP1 shRNA in lentivirus-based RNAi vector pLVTHM | NRP1 | qRT-PCR, Western blot and ICC | In vitro | Human hepatoma-derived HCCLM6 cell line HCCLM6 xenograft nude mice | Proliferation | Xu and Xia [39] | 2013 | |
| In vivo | ||||||||
| Lentiviral-based NRP1 shRNA from Origene | NRP1 | qRT-PCR and Western blot | In vitro | Bel-7402 and SMMC-7721 cell lines | Cell viability and apoptosis | Lin et al. [17] | 2018 | |
| Lentivirus pGCSIL-RFPshNRP1 self-constructed | NRP1 | Western blot and ICC | In vitro | HepG2 | Tumor progression and migration | Xu et al. [42] | 2017 | |
| NRP1 shRNA produced by GeneChem | NRP1 | qRT-PCR and Western blot | In vitro | HepG2, HCCLM3 and Huh-7 | CSC properties, proliferation, migration, EMT and metastasis | Li et al. [26] | 2021 | |
| In vivo | HCCLM3 xenograft nude mice | |||||||
| siRNA silencing | siRNA targeting mouse NRP1 (ID #155679, Ambion) | NRP1 | ICC | In vivo | Transgenic HCC C57BL/6 mice | Tumor progression and vascular remodeling | Bergé et al. [36] | 2010 |
| ON-TARGETplus NRP1 siRNA | NRP1 | Western blot | In vitro | Mouse Hepa129 cell line C3H mice with Hep129-derived tumor | Proliferation, apoptosis, inflammation and migration | Raskopf et al. [37] | 2010 | |
| In vivo | ||||||||
| NRP1 siRNA from Bioneer with Effectene reagent | NRP1 | Western blot | In vitro | HepG2 and Huh-7 | Cell death | Lee et al. [38] | 2011 | |
| ON-TARGETplus NRP2 siRNA | NRP2 | qRT-PCR and Western blot | In vitro | 3sp, SNU-398, SNU-423, SNU-449, SNU-475, FLC-4 cell lines | Migration, mesenchymal properties, TGF-β signaling | Wittmann et al. [35] | 2015 | |
| NRP1 siRNA and lipofectamine 2000 | NRP1 | Western blot | In vitro | HepG2 | Cell viability and migration | Lv et al. [43] | 2019 | |
| NRP1 siRNA from Qiagen | NRP1 | NR | In vitro | HepG2 | Cell proliferation | Arab et al. [45] | 2020 | |
| Inhibitors | TCDD | NRP1 | qRT-PCR | In vitro | HepG2 | Xenobiotic toxicity | Lee et al. [48] | 2010 |
| Peptide N | NRP1 | qRT-PCR and Western blot | In vitro | HepG2, SK-HEP-1 and PLC/PRF/5 cell lines | Cell proliferation, apoptosis and invasion | Bergé et al. [12] | 2011 | |
| In vivo | ASV-B transgenic C57BL/6 mice | |||||||
| miRNAs overexpression | miR-148b | NRP1 | IHC | In vivo | PLC/PRF/5 xenograft | Tumor progression, angiogenesis, microRNA modulation and CSC properties | Liu et al. [40] | 2015 |
| BALB/c nude mice | ||||||||
| miR-340-5p/miR-452-5p | NRP1 | qRT-PCR and Western blot | In vitro | Hep3B | microRNAs and circRNAs modulation | Yang [65] | 2020 | |
| In vivo | BALB/c athymic nude mice injected with Hep3B | |||||||
| Antibody | Truncated tissue factor anti-NRP1 monoclonal antibody | NRP1 | NR | In vivo | HepG2 xenograft BALB/c nude mice | Tumor progression | Xu et al. [44] | 2019 |
circRNA, circular RNA; CSC, cancer stem cell; EMT, epithelial-to-mesenchymal transition; HCC, hepatocelular carcinoma; ICC, immunocytochemistry; IHC, immunohistochemistry; miRNA, microRNA; NR, not reported; NRP, neuropilin; qRT-PCR, real-time reverse transcription polymerase chain reaction; shRNA, short hairpin RNA; siRNA, small interference RNA; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin; TGF-β, transforming growth factor β.
NRPs as diagnostic tissue or serum biomarkers
While NRPs were initially identified in the nervous system as axon guidance regulators [10], recent studies have reported multiple molecular functions [7,10]. Late diagnosis is one of the leading causes of HCC patients’ high mortality rate due to the absence of highly sensitive and specific biomarkers [2]. Therefore, an increasing number of studies have focused on the potential use of NRPs as tissue or serum biomarkers evaluating their expression levels in tumor and healthy liver samples from HCC patients to improve the current diagnostic tools for HCC diagnosis [12–26].
Studies have determined NRP protein levels in tumor tissue from HCC patients, finding NRP1 to be overexpressed compared to healthy liver tissue [12,14,16-20,23,24,26] and in high-risk HCC patients [22]. In contrast, Kitagawa et al. [13] did not find a difference, while lower NRP1 levels in tumor compared to peritumoral tissue have also been reported [15]. Similarly, NRP1 levels were increased in three HCC cell lines compared to the normal liver L02 [26], and Hep3B and HepG2 [27]. Contrariwise, NRP2 expression in HCC tissue has been analyzed to a lesser extent. Lower NRP2 expression was found in the HCC nodules versus the healthy liver tissue [13]. However, contradictory results have been reported for NRP2. While its urinary levels were decreased in a rat HCC model [21], an in vitro study found it was overexpressed in three human HCC cell lines [28]. These find ings highlight the necessity of further studies to clarify NRP2’s exact role in HCC.
Early HCC diagnosis is one of the main objectives of current clinical studies, where identifying useful biomarkers is an exciting area [2]. While all tissue and serum biomarkers are valuable tools for diagnosis at any tumor stage, non-invasive methods are usually preferred [3]. Only two studies have analyzed the potential use of either NRP1 or NRP2 as serum biomarkers, reporting that NRP1 could be a useful protein for HCC diagnosis based on its elevated levels in serum samples from patients with advanced HCC [17,25].
NRPs as prognostic biomarkers
Consistent with previous evidence, numerous studies have evaluated the prognostic role of NRP1 and NRP2 in HCC patients [15,16,24-26,29-33]. NRP1 overexpression was significantly correlated with shorter overall (OS) [16,24-26,29] and recurrence-free survival (RFS) [16,24] survival in patients, while NRP2 overexpression correlated with their lower OS and disease-free survival [30]. Moreover, NRP1 expression was correlated with RAD51 in hepatic stellate cells (HSCs), a valuable prognostic biomarker [34], and with an increased recurrence rate [32,33]. Peritumoral NRP1 levels have also been evaluated in HCC human samples. Patients with higher peritumoral NRP1 expression had higher OS, higher time to recurrence, and lower early recurrence incidence [15]. Intriguingly, a serum signature of nine circulating cytokines and angiogenic factors (NRP1, VEGF receptor 3 [VEGFR3], FGF23, Fas ligand [FasL], HGF, VEGFD, interleukin 1 receptor 2 [IL1R2], platelet-derived growth factor-BB [PDGF-BB], and Met tyrosine-protein kinase [c-MET]), correlated significantly with OS, progression-free survival, and early progression disease in advanced HCC [31].
In addition, associations between NRP overexpression and other clinical characteristics related to tumor aggressiveness have been reported. Both NRPs correlated strongly with advanced HCC stages. However, the staging method used differed [16,35], with NRP1 correlating with Edmondson grade and TNM classification [16], and NRP2 correlating with higher tumor grading [35]. Curiously, several studies found no association between both NRPs and age, sex, tumor size, and hepatitis B virus (HBV) infection [16,24,30]. Nevertheless, NRP1 overexpression was positively correlated with alpha-fetoprotein and other liver function markers and negatively correlated with albumin (Alb) and pre-Alb [17]. Moreover, increased serum NRP1 levels correlated with advanced Barcelona Clinic Liver Cancer stages (B and C), a tumor number ≥3, and a tumor size ≥ 5cm [25], highlighting the potential roles of NRPs in prognosis and different tumor-associated characteristics in human patients with HCC.
NRP effects on tumor progression and associated signaling pathways
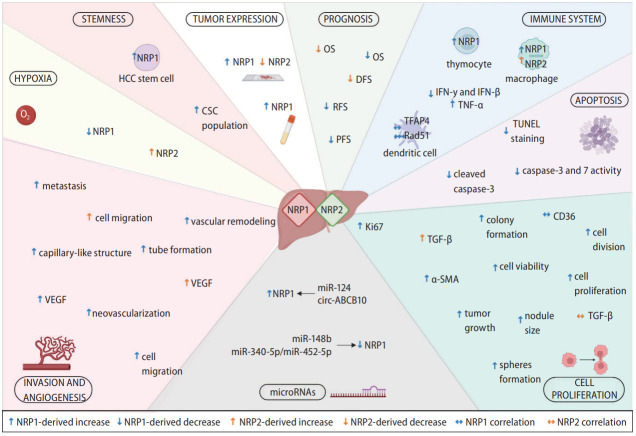
NRPs have been described as relevant oncology proteins due to their modulation of axon guidance and cell survival, migration, angiogenesis, and invasion [10]. This designation is directly associated with numerous articles evaluating their role on different cellular processes and molecular signaling pathways involved in HCC progression (Fig. 2) [12,15,17,26,35-48].
Figure 2.
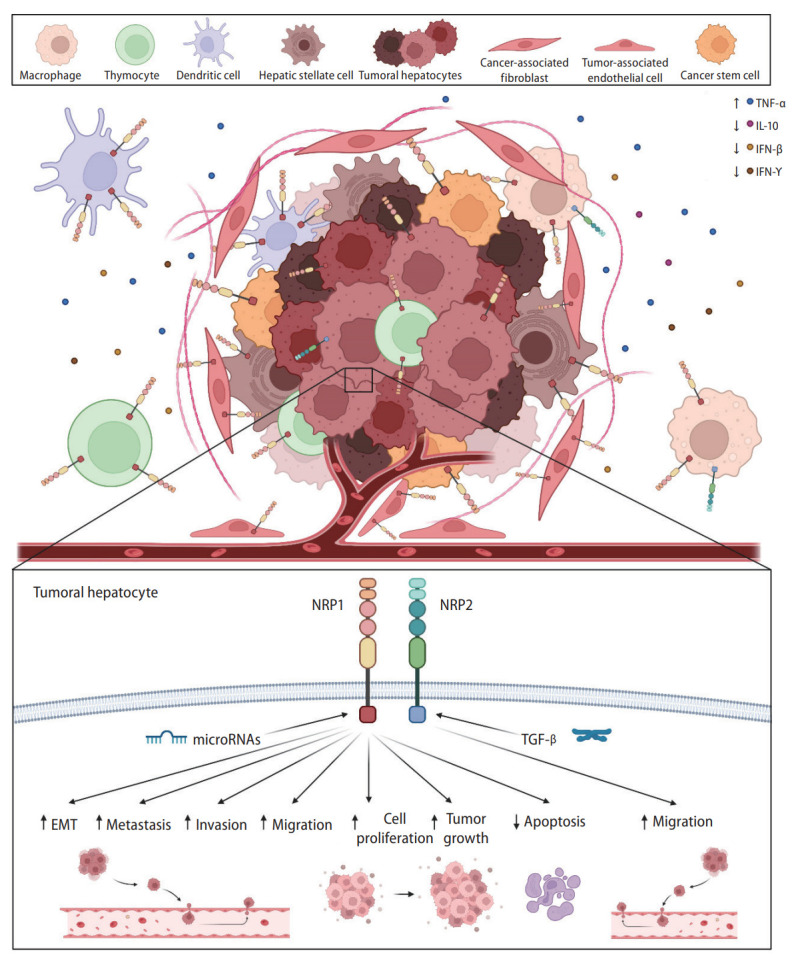
Main cellular and molecular mechanisms modulated by NRP1 and NRP2. NRPs are expressed in tumor cells and other tumor-associated populations that constitute the tumor microenvironment and participate in the immune response. Both NRP1 and NRP2 are expressed in a broad number of cell types and are involved in different cellular and molecular mechanisms responsible for HCC development and progression, modulating several cellular processes. EMT, epithelial-to-mesenchymal transition; IFN-β, interferon beta; IFN-γ, interferon gamma; IL-10, interleukin-10; NRP, neuropilin; TGF-β, transforming growth factor β; TNF-α, tumoral necrosis factor-α.
Both NRPs promote tumor progression by modulating cell proliferation, viability and apoptosis, with NRP1 being the most characterized. Numerous preclinical investigations showed that NRP1 downregulation decreased the growth and viability of HepG2 [12,42,43], SK-HEP-1 [12], PLC/PRF/5 [12,40], HCCLM6 [39], Huh-7 [26,40], Bel-7402, SMMC-7721 [17] and HCCLM3 [26] cells. However, no effects were observed in mouse Hepa129 cells [37]. Similar effects have been described in animal models, where a marked tumor growth inhibition was observed after NRP1 knockdown [12,36,37,39,42,44,49]. A five-gene signature that included NRP1 was identified as an independent risk factor for a faster tumor growth rate in HCC patients [41]. Furthermore, an interaction between NRP1 and VEGFA promoted HCC tumorigenesis by dysregulating cell-to-cell interactions in patients with HCC [47].
Similarly, apoptosis was altered when NRP1 expression was abolished in both in vitro and in vivo models, leading to an increase in cleaved caspase-3 levels and TUNEL-positive cells [12]. However, a previous study did not report cell death changes after NRP1 silencing in two HCC cell lines [38]. Peritumoral NRP1 expression was decreased and correlated with increased tumor size and other associated characteristics [15], highlighting NRP1’s crucial role in HCC tumor progression. In addition, NRP1 correlated positively with the immune marker cluster of differentiation 36 (CD36), a potential prognostic and immunologic marker in different cancers, including HCC [46]. Intriguingly, a recent study treated HepG2 cells with the supernatant from NRP1-downregulated HSCs, causing a decrease in lipid droplet content and insulin-like growth factor binding protein-3 (IGFBP3) expression, and an increase in serpin family A member 12 (SERPINA12) levels [45].
Only one study has analyzed the processes modulated by NRP2 [35]. Specifically, it was directly modulated by TGF-β signaling and correlated with a mesenchymal-like phenotype in vitro. Curiously, while TGF-β overexpression increased NRP2 levels, NRP2 silencing did not exhibit significantly altered TGF-β expression [35].
Role of NRP in migration and invasion-related processes
Invasion and migration processes are well-described events that characterizes the aggressiveness of solid tumors and are highly associated with tumor progression and recurrence [50].
The NRPs’ main effects in human pathologies are generally related to the modulation of signaling pathways that drive angiogenesis and cancer cell migration [10]. Numerous studies evaluated the NRPs’ role in these cellular processes in preclinical models [12,26,35-37,39,40,42,43,51-54] and human patients (Fig. 2) [24,25]. NRP2 downregulation significantly decreased the migration of two different HCC cell lines [35]. However, most studies have described similar results after NRP1 knockdown, finding lower cell migration ability in different cellular models, including the HCCLM3 [26,54], Huh-7 [26,51,52], HepG2 [42,43], Mahlavu, SK-Hep1, and HEK293T human lines [52] and mouse Hepa129 cell line [37].
NRP1 was also overexpressed in the highly metastatic Bel-7402 cell line, but was underexpressed in the less metastatic HepG2 and SMMC-7721 cell lines [16]. A recent study evaluated NRP1 function in metastasis, generating an HCC mouse model using NRP1-silenced and non-silenced HCCLM3 cells [26]. Curiously, they found that NRP1 downregulation greatly diminished the number of grafted mice with lung metastasis (1 of 5), compared to control mice (5 of 5) [26]. Similar findings have been reported in studies on HCC patients, where those with elevated NRP1 levels had a higher probability of venous invasion and metastasis [24,25].
At the molecular level, the NRPs’ role in angiogenesis-related signaling has been analyzed by two studies [26,53]. After VEGF silencing in the mouse Hepa129 HCC cell line, both NRP1 and NRP2 levels were markedly downregulated [53], supporting their interplay with the VEGF family ligands described in other tumor types [10]. Moreover, a recent study found that NRP1 modulated the epithelial-to-mesenchymal transition (EMT) pathway in two HCC cell lines, with decreased N-cadherin and vimentin expression and increased E-cadherin levels observed after NRP1 silencing [26].
Other relevant clinical aspects associated with tumor invasion and metastasis have been evaluated and associated with NRPs (Fig. 2) [12,36,37,39,40]. Two independent studies performed by the same research group successfully inhibited NRP1 activity through genetic silencing or inhibitor treatment, significantly reducing tumor vascular remodeling [12,36]. Moreover, an in vitro matrigel assay analysis of capillary-like structures showed that NRP1 inhibition with peptide N decreased the formation of capillary-like structures and other tumor-associated characteristics [12]. In this line, while the tubeforming ability of mouse Hepa129 cells decreased after NRP1 silencing [37]. NRP1 overexpression via microRNA (miRNA)-148b (miR-148b) silencing raised the tube-forming ability of human PLC/PRF/5 HCC cells [40]. Moreover, lower neovascularization was observed in an in vivo HCC mouse model after NRP1 silencing [39].
Studies on human HCC patients have described similar results. NRP1 overexpression was significantly correlated with intrahepatic metastasis [16] and vascular invasion [16,26] in two independent HCC cohorts, supporting the potential roles for NRP1 and NRP2 in the mechanisms underlying HCC progression.
NRPs and the immune response
HCC is a complex and heterogeneous tumor in which malignant hepatocytes and other tumor-associated cells affect the development, progression, and drug responsiveness of tumor cells [55,56]. Both innate and adaptive immune cells have been strongly associated with the modulation of cellular responses to chronic inflammation, fibrosis, or cirrhosis contributing to hepatocarcinogenesis and HCC progression [56]. Several investigations have focused on the tumor immunological microenvironment as a key mechanism in HCC (Fig. 2) [34,36,37,57-61].
A preclinical study exploring NRP1 and NRP2 in Mdr2 deficient HCC mice, found their expression higher in macrophages than in hepatocytes [57]. Additionally, thymocytes isolated from mice bearing a transplantable hepatoma 22a had higher NRP1 levels than control animals [58]. Among liver immune cells, dendritic cells expressed NRP1, correlating with RAD51 [34] and transcription factor activating enhancer binding protein 4 (TFAP4) [59], valuable prognosis and immune response markers, respectively, in patients with HCC [34,59].
Otherwise, several studies have described significant correlations between NRP1 and different immune response mediators in the HCC tumor microenvironment. One study found NRP1 was inversely correlated with interleukin-10 (IL-10) in patients with HCC [60]. However, another study found no significant correlation between NRP1 and killer cell lectin-like receptor B1 (KLRB1) [61]. Similarly, NRP1 downregulation via siRNA silencing increased interferon gamma (IFN-γ) [36] and IFN-β levels [37], and decreased tumor necrosis factor-α (TNF-α) levels [37] in two independent studies employing in vivo HCC models [36,37]. Therefore, while the exact mechanisms remain unclear, NRPs might exert an interesting function in the tumor-associated immune response in HCC.
NRPs and miRNAs
MiRNAs are highly conserved small noncoding RNAs that modulate gene transcription by binding to target messenger RNAs (mRNAs) [62,63]. They are differentially expressed among tissues and cancer stages, and are frequently dysregulated during oncogenesis [62]. The role of miRNAs in cancer development and progression gained attention given their broad underlying modulating processes [62,63]. In HCC, several miRNAs have been associated with tumor progression through their control of cell proliferation, invasion, migration and HCC development acting either as tumor suppressors or promoters [62,63]. Moreover, various miRNAs have shown to directly modulate target genes involved in critical tumor-associated processes [62].
Several studies have evaluated the interplay between miRNAs and NRP1, and the underlying mechanisms (Fig. 2) [40,51,64,65]. NRP1 has shown to be a target of miR-148b [40,51] and miR-340-5p/miR-452-5p [65], in both in vitro and in vivo HCC models. In addition, miR-124 or circular RNA (circRNA) circ-ABCB10 overexpression increased NRP1 levels in the HepG2 [64] and Hep3B [65] HCC cell lines, respectively. Interestingly, miR-148b-induced NRP1 downregulation strongly decrease cell migration in vitro [51], and tube formation and cell division in an HCC mouse model [40]. However, despite these novel findings, further studies should be performed to clarify the interplay between miRNAs and their associated effects in HCC.
Association between NRPs and the tumor microenvironment
The tumor microenvironment is a key factor in cancer development and progression that undergoes dynamic changes involving various components [66]. Cancer stem cells (CSCs), HSCs, cancer-associated fibroblasts (CAFs), cytokines and growth factors are tumor microenvironment components strongly associated with HCC progression [66-68]. Numerous studies have assessed the modulatory effects of the tumor microenvironment on NRP expression and activity, recently reporting some exciting findings (Fig. 2) [15,26,29,33,40,53].
CSCs are a small population of tumor cells that control differentiation, tumorigenicity, metastasis, and therapeutic resistance in HCC [67], with NRP1 appearing to play a key role [26,40]. Specifically, NRP1 downregulation in an in vitro HCC model significantly decreased the liver CSC population [26]. In contrast, NRP1 was overexpressed in the CSC population of two HCC cell lines [40] and expressed explicitly in CAFs and tumor-associated endothelial cells in HCC patients [33]. Among the cell types influencing the tumor microenvironment, HSCs have been broadly associated with HCC progression [66]. However, only one study evaluated NRP1 expression in HSCs, finding increased HepG2 cell proliferation in matrix conditioned with HSCs overexpressing NRP1 [29].
Low oxygen conditions, hypoxia, are crucial in HCC development, progression and chemoresistance [68-70]. A hypoxic microenvironment modulated both NRPs in vitro, decreasing NRP1 levels but increasing NRP2 levels (Fig. 2) [53]. Intriguingly, this study found that hypoxia decreased NRP expression when VEGF was silenced [53]. Similarly, cobalt chloride-induced hypoxia decreased NRP1 expression in the liver L02 cells [15]. Moreover, hypoxia directly modulated NRP1 in an in vivo HCC model, with NRP1 levels decreased in peritumoral tissue and negatively correlated with hypoxia-inducible factor 1-alpha (HIF-1α) levels [15]. Overall, both NRPs appear to contribute to tumor microenvironment modulation, showing key interactions with different tumor-associated cell types and the oxygen conditions in HCC.
DISCUSSION
While NRPs were described as key proteins in the nervous system through axon guidance modulation [10], recent findings also indicate an intriguing role in HCC and other cancer types [7,8].
Numerous studies have indicated a potential role for NRP1 as a tumor biomarker in HCC [12,14,16-20,23,24,26], while fewer and contradictory results have been reported for NRP2 [13,21,28]. Studies have found both NRPs overexpressed in tumor samples from different cancer types, including cholangiocarcinoma [18,71-75], supporting mainly NRP1, but also NRP2, as potential biomarkers in HCC. However, further studies are needed to clarify their exact role and improve the diagnostic tools available for human HCC. Moreover, both NRPs are strongly associated with worse prognoses and different tumor-associated parameters in HCC patients [15,16,24-26,29-33,35]. Similar findings were found with different solid tumors, where NRPs negatively correlated with prognosis and higher invasion and metastasis risk [18,71,73-75], highlighting their potential role as diagnostic tools for HCC patient prognosis.
Most studies evaluating NRP function in HCC have used preclinical models, where mainly NRP1, but also NRP2, exhibited an interesting modulatory roles by promoting cell survival, tumor progression, invasion and migration, and inhibiting apoptosis [12,15,17,26,35-48,51-54]. Similar findings were reported for other tumor types, describing TGF-β and other signaling pathways as potential targets of NRP regulation [76-79]. Additionally, invasion, migration and metastasis were also found to be strongly modulated by both NRPs in different solid tumor models [24,80,81], supporting roles for NRP1 and NRP2 in mechanisms underlying tumor progression and spread. Nevertheless, while these findings highlight interesting function for NRPs in HCC tumor progression and invasion abilities, further studies are needed to identify the exact mechanisms and signaling pathways modulated by NRP1 and NRP2.
Based on solid tumor heterogeneity and multiple processes involved in tumor cell adaptation and progression, numerous studies have described key functions for NRP1, and to a lesser extent NRP2, in modulating the immune response against tumor hepatocytes [34,36,37,57-61], that are associated with the potential interplay between NRPs, miRNAs [40,51,64,65], and the tumor microenvironment [15,26,29,33,40,53]. Other studies have also reported a correlation between NRP1 [82-85] and NRP2 [86] expression in different immune cells and immune response suppression in other cancers [82-86]. Specifically, NRPs had a crucial role in modulating the immune response [10,82], where NRP1 appeared to be associated with immune response suppression in cancer [82]. Regulatory T cells with depleted NRP1 exhibited decreased function, restoring the antitumor immunity and TNF-α production [83]. Moreover, NRP1 expression in macrophages, dendritic cells and other associated cell populations was associated with a restrained inflammatory response [84,85]. NRP2 expression in tumor-associated macrophages promoted tumor growth by regulating macrophage phagocytosis [86]. Therefore, based on these findings in different HCC models, both NRPs potentially play a key role in modulating the tumor-associated immune response, making them potential biomarkers in the HCC tumor landscape.
Consistent with growing evidence reinforcing the key role of miRNAs in cancer, these non-codifying RNAs modulate NRP1 in HCC, increasing tumor hepatocyte proliferation and migration [40,51,64,65]. Different miRNAs (e.g., miR-376a) or circRNAs (e.g., circ-LDLRAD3) regulated tumor progression in other cancer models [76,78]. While few studies have provided clearer results on the mechanisms underlying the potential interplay between miRNAs and NRPs, these findings suggest that miRNAs could be potential modulators of NRPs expression and activity, but mainly of NRP1, controlling key processes involved in HCC progression and invasion.
Several investigations have described that NRPs, primarily NRP1, are highly influenced by the tumor microenvironment, with tumor-associated cell populations playing a crucial role [87-89]. Among them, CAFs and CSCs could be modulated by NRP1 or NRP2 in different cancer types [87-89]. Indeed, NRP1 appears to have an interesting role in the response of different cell types in the tumor microenvironment, acting as a potential modulator of tumor adaptation and progression. Moreover, the interplay between hypoxia and NRPs has recently been explored in other cancers [90-92]. However, these results showed an opposite hypoxia effect to HCC, with increased NRP1 [91] but decreased NRP2 expression under hypoxic conditions [92]. Together with the studies on HCC, these results indicate that further investigations are needed to obtain a clearer understanding of the exact mechanism through which hypoxia and NRPs might contribute to tumor development.
Limitations
This review aimed to provide a clear and complete understanding of the main mechanisms modulated by NRPs in HCC development and progression. Nevertheless, some limitations exist that are mostly associated to the high heterogeneity among studies. The main limitation was that most articles examined only one NRP, with 40 articles focused on NRP1 but only four on NRP2; five examined both NRPs. This limitation led to greater discordance in the results obtained, mainly for NRP2, increasing the uncertainty of the conclusions drawn. Moreover, as shown in Table 1A, the methods employed for determining NRP1 or NRP2 levels were inconsistent, with most articles focusing on one NRP, using different targeting strategies. Discrepancies between studies could be explained by the different methods used to measure NRPs and the chosen targeting strategy.
Additionally, while multiple cellular and molecular processes had been evaluated, the number of studies analyzing each mechanism was highly heterogeneous. The diagnostic and prognostic values of NRP1 and NPR2, and their crucial role in invasion and migration, were the main processes studied, with few articles focusing on their interplay with miRNAs or the tumor microenvironment. While these interactions are key mechanisms in cancer development and progression, only five and six studies have explored them in HCC, respectively.
Finally, although increasing numbers of human studies have been published, they have not always described the main characteristics of HCC patients, with etiology, age, or country missing in some articles. Moreover, public databases hindered data extraction by not stating the number of patients included in the analysis. In summary, some important limitations should be considered when understanding and interpreting the main conclusions of this systematic review.
CONCLUSIONS AND FUTURE PERSPECTIVES
To the best of our knowledge, this article is the first systematic review focusing on the role of NRPs in HCC, summarizing all the results obtained from preclinical and clinical studies (Fig. 3). Increasing evidence suggests vital roles for these receptors (NRP1 and NRP2) in tumor-associated processes. The results summarized here suggest that NRP1 could act as a potential diagnostic biomarker and, with NRP2, an interesting prognostic biomarker in HCC patients. The NRPs have modulatory effects on different signaling pathways that promote tumor progression and are crucial mediators of the HCC cell invasion and migration abilities. The tumor-associated immune response is also strongly associated with NRPs, mainly NRP1, and the tumor microenvironment, in which different tumor cell populations have higher NRP1 levels. The interplay between miRNAs and NRPs has gained interest since several miRNAs directly modulate NPR1, restraining tumor cell proliferation. In summary, NRPs appear to have critical roles in various processes involved in tumor development and progression, suggesting the potential of both, but mainly NRP1, as tumor biomarkers and potential targets for improving the HCC patient outcomes.
Figure 3.
Main findings from the studies included in this systematic review describing modulatory effects associated to NRP1 and NRP2 in HCC. Specific modulatory effects exerted by both NRPs are graphically shown, together with correlations observed in different cellular processes and molecular mechanisms. α-SMA, α smooth muscle actin; CSC, cancer stem cell; DFS, disease-free survival; IFN-β, interferon beta; IFN-γ, interferon gamma; OS, overall survival; PFS, progression-free survival; RFS, recurrence-free survival; TFAP4, transcription factor activating enhancer binding protein 4; TGF-β, transforming growth factor beta; TNF-α, tumoral necrosis factor-α; VEGF, vascular endothelial growth factor.
Acknowledgments
This work was supported by the Ministry of Science and Innovation (MCIN/AEI/10.13039/501100011033) [project PID2020-119164RB-I00]. CIBERehd is funded by Instituto de Salud Carlos III (ISCIII), Spain. P.F.-P. is supported by the Ministry of Education (MCIN/AEI/10.13039/501100011033) [grant FPU17/01995] and T.P.-S. by the Asociación Española Contra el Cáncer (AECC)-Junta Provincial de León, Spain.
Abbreviations
- α-SMA
α smooth muscle actin
- Alb
albumin
- CAFs
cancer-associated fibroblasts
- CD36
cluster of differentiation 36
- CRC
colorectal cancer
- CSCs
cancer stem cells
- EMT
epithelial-to-mesenchymal transition
- FasL
Fas ligand
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HIF-1α
hypoxia-inducible factor 1-alpha
- HSCs
hepatic stellate cells
- IFN-γ
interferon gamma
- IGFBP3
insulin-like growth factor binding protein-3
- IL1R2
interleukin 1 receptor 2
- IL-10
interleukin-10
- KLRB1
killer cell lectin-like receptor B1
- miR-148b
microRNA-148b
- mRNA
messenger RNAs
- NRP
neuropilin
- OS
overall survival
- PDGF-BB
platelet-derived growth factor-BB
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-analyses: PROSPERO
- RFS
recurrence-free survival
- SERPINA12
serpin family A member 12
- TFAP4
transcription factor activating enhancer binding protein 4
- TGF-β
transforming growth factor beta
- TNF-α
tumor necrosis factor-α
- VEGFA
vascular endothelial growth factor A
- VEGFR3
VEGF receptor 3
Footnotes
Authors’ contribution
All authors were responsible for study conception and design, interpretation of the data, and drafting of the manuscript. Systematic literature review, data extraction, and data analysis were performed by P.F.-P., T.P.-S., C.M.-B. and B.S.-M. In addition, M.J.T., J.G.-G. and J.L.M. supervised the study, and carried out the review and editing of the paper. The final version of the manuscript was approved by all authors.
Conflicts of Interest
The authors have no conflicts to disclose.
Supplementary materials
Supplementary material is available at Clinical and Molecular Hepatology website (http://www.e-cmh.org).
PRISMA 2020 checklist
PRISMA 2020 for abstracts checklist
Full search strategy employed for each online database (up to and including August 31st, 2022)
Evaluation of the published articles. (A) Temporal distribution of the number of articles published employing in vitro, in vivo or human models, or a combination of them. Comparison of the number of studies conducted with (B) human or (C) animal models with the mean of the human or animals examined per year, respectively.
REFERENCES
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- 3.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–1314. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 4.Alqahtani A, Khan Z, Alloghbi A, Said Ahmed TS, Ashraf M, Hammouda DM. Hepatocellular carcinoma: Molecular mechanisms and targeted therapies. Medicina (Kaunas) 2019;55:526. doi: 10.3390/medicina55090526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fondevila F, Méndez-Blanco C, Fernández-Palanca P, González-Gallego J, Mauriz JL. Anti-tumoral activity of single and combined regorafenib treatments in preclinical models of liver and gastrointestinal cancers. Exp Mol Med. 2019;51:1–15. doi: 10.1038/s12276-019-0308-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cucarull B, Tutusaus A, Rider P, Hernáez-Alsina T, Cuño C, García de Frutos P, et al. Hepatocellular carcinoma: Molecular pathogenesis and therapeutic advances. Cancers (Basel) 2022;14:621. doi: 10.3390/cancers14030621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dumond A, Pagès G. Neuropilins, as relevant oncology target: Their role in the tumoral microenvironment. Front Cell Dev Biol. 2020;8:662. doi: 10.3389/fcell.2020.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Napolitano V, Tamagnone L. Neuropilins controlling cancer therapy responsiveness. Int J Mol Sci. 2019;20:2049. doi: 10.3390/ijms20082049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broz M, Kolarič A, Jukič M, Bren U. Neuropilin (NRPs) related pathological conditions and their modulators. Int J Mol Sci. 2022;23:8402. doi: 10.3390/ijms23158402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niland S, Eble JA. Neuropilins in the context of tumor vasculature. Int J Mol Sci. 2019;20:639. doi: 10.3390/ijms20030639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Syst Rev. 2021;10:89. doi: 10.1186/s13643-021-01626-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergé M, Allanic D, Bonnin P, de Montrion C, Richard J, Suc M, et al. Neuropilin-1 is upregulated in hepatocellular carcinoma and contributes to tumour growth and vascular remodelling. J Hepatol. 2011;55:866–875. doi: 10.1016/j.jhep.2011.01.033. [DOI] [PubMed] [Google Scholar]
- 13.Kitagawa K, Nakajima G, Kuramochi H, Ariizumi SI, Yamamoto M. Lymphatic vessel endothelial hyaluronan receptor-1 is a novel prognostic indicator for human hepatocellular carcinoma. Mol Clin Oncol. 2013;1:1039–1048. doi: 10.3892/mco.2013.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chishti MA, Kaya N, Binbakheet AB, Al-Mohanna F, Goyns MH, Colak D. Induction of cell proliferation in old rat liver can reset certain gene expression levels characteristic of old liver to those associated with young liver. Age (Dordr) 2013;35:719–732. doi: 10.1007/s11357-012-9404-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhuang PY, Wang JD, Tang ZH, Zhou XP, Yang Y, Quan ZW, et al. Peritumoral neuropilin-1 and VEGF receptor-2 expression increases time to recurrence in hepatocellular carcinoma patients undergoing curative hepatectomy. Oncotarget. 2014;5:11121–11132. doi: 10.18632/oncotarget.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Liu P, Jiang Y, Dou X, Yan J, Ma C, et al. High expression of neuropilin-1 associates with unfavorable clinicopathological features in hepatocellular carcinoma. Pathol Oncol Res. 2016;22:367–375. doi: 10.1007/s12253-015-0003-z. [DOI] [PubMed] [Google Scholar]
- 17.Lin J, Zhang Y, Wu J, Li L, Chen N, Ni P, et al. Neuropilin 1 (NRP1) is a novel tumor marker in hepatocellular carcinoma. Clin Chim Acta. 2018;485:158–165. doi: 10.1016/j.cca.2018.06.046. [DOI] [PubMed] [Google Scholar]
- 18.Morin E, Sjöberg E, Tjomsland V, Testini C, Lindskog C, Franklin O, et al. VEGF receptor-2/neuropilin 1 trans-complex formation between endothelial and tumor cells is an independent predictor of pancreatic cancer survival. J Pathol. 2018;246:311–322. doi: 10.1002/path.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyu Z, Jin H, Yan Z, Hu K, Jiang H, Peng H, et al. Effects of NRP1 on angiogenesis and vascular maturity in endothelial cells are dependent on the expression of SEMA4D. Int J Mol Med. 2020;46:1321–1334. doi: 10.3892/ijmm.2020.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savier E, Simon-Gracia L, Charlotte F, Tuffery P, Teesalu T, Scatton O, et al. Bi-functional peptides as a new therapeutic tool for hepatocellular carcinoma. Pharmaceutics. 2021;13:1631. doi: 10.3390/pharmaceutics13101631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Gao Y, Wei J, Gao Y. Dynamic changes of urine proteome in rat models inoculated with two different hepatoma cell lines. J Oncol. 2021;2021:8895330. doi: 10.1155/2021/8895330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Zhang S, Zhang S, Kuang W, Tang C. Inflammatory responserelated long non-coding RNA signature predicts the prognosis of hepatocellular carcinoma. J Oncol. 2022;2022:9917244. doi: 10.1155/2022/9917244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li XY, Ma WN, Su LX, Shen Y, Zhang L, Shao Y, et al. Association of angiogenesis gene expression with cancer prognosis and immunotherapy efficacy. Front Cell Dev Biol. 2022;10:805507. doi: 10.3389/fcell.2022.805507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernández-Palanca P, Payo-Serafín T, Fondevila F, Méndez-Blanco C, San-Miguel B, Romero MR, et al. Neuropilin-1 as a potential biomarker of prognosis and invasive-related parameters in liver and colorectal cancer: A systematic review and meta-analysis of human studies. Cancers (Basel) 2022;14:3455. doi: 10.3390/cancers14143455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdel Ghafar MT, Elkhouly RA, Elnaggar MH, Mabrouk MM, Darwish SA, Younis RL, et al. Utility of serum neuropilin-1 and angiopoietin-2 as markers of hepatocellular carcinoma. J Investig Med. 2021;69:1222–1229. doi: 10.1136/jim-2020-001744. [DOI] [PubMed] [Google Scholar]
- 26.Li X, Zhou Y, Hu J, Bai Z, Meng W, Zhang L, et al. Loss of neuropilin1 inhibits liver cancer stem cells population and blocks metastasis in hepatocellular carcinoma via epithelial-mesenchymal transition. Neoplasma. 2021;68:325–333. doi: 10.4149/neo_2020_200914N982. [DOI] [PubMed] [Google Scholar]
- 27.Sharma BK, Srinivasan R, Chawla YK, Chakraborti A. Vascular endothelial growth factor: Evidence for autocrine signaling in hepatocellular carcinoma cell lines affecting invasion. Indian J Cancer. 2016;53:542–547. doi: 10.4103/0019-509X.204765. [DOI] [PubMed] [Google Scholar]
- 28.Ye K, Ouyang X, Wang Z, Yao L, Zhang G. SEMA3F promotes liver hepatocellular carcinoma metastasis by activating focal adhesion pathway. DNA Cell Biol. 2020;39:474–483. doi: 10.1089/dna.2019.4904. [DOI] [PubMed] [Google Scholar]
- 29.Yaqoob U, Cao S, Shergill U, Jagavelu K, Geng Z, Yin M, et al. Neuropilin-1 stimulates tumor growth by increasing fibronectin fibril assembly in the tumor microenvironment. Cancer Res. 2012;72:4047–4059. doi: 10.1158/0008-5472.CAN-11-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong X, Guo W, Zhang S, Wu T, Sun Z, Yan S, et al. Elevated expression of neuropilin-2 associated with unfavorable prognosis in hepatocellular carcinoma. Onco Targets Ther. 2017;10:3827–3833. doi: 10.2147/OTT.S139044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ono A, Aikata H, Yamauchi M, Kodama K, Ohishi W, Kishi T, et al. Circulating cytokines and angiogenic factors based signature associated with the relative dose intensity during treatment in patients with advanced hepatocellular carcinoma receiving lenvatinib. Ther Adv Med Oncol. 2020;12:1758835920922051. doi: 10.1177/1758835920922051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang ZL, Xu B, Li TT, Xu YH, Huang XY, Huang XY. Integrative analysis identifies cell-type-specific genes within tumor microenvironment as prognostic indicators in hepatocellular carcinoma. Front Oncol. 2022;12:878923. doi: 10.3389/fonc.2022.878923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li JH, Tao YF, Shen CH, Li RD, Wang Z, Xing H, et al. Integrated multi-omics analysis identifies ENY2 as a predictor of recurrence and a regulator of telomere maintenance in hepatocellular carcinoma. Front Oncol. 2022;12:939948. doi: 10.3389/fonc.2022.939948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu H, Xiong C, Chen Y, Zhang C, Bai D. Identification of Rad51 as a prognostic biomarker correlated with immune infiltration in hepatocellular carcinoma. Bioengineered. 2021;12:2664–2675. doi: 10.1080/21655979.2021.1938470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wittmann P, Grubinger M, Gröger C, Huber H, Sieghart W, Peck-Radosavljevic M, et al. Neuropilin-2 induced by transforming growth factor-β augments migration of hepatocellular carcinoma cells. BMC Cancer. 2015;15:909. doi: 10.1186/s12885-015-1919-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergé M, Bonnin P, Sulpice E, Vilar J, Allanic D, Silvestre JS, et al. Small interfering RNAs induce target-independent inhibition of tumor growth and vasculature remodeling in a mouse model of hepatocellular carcinoma. Am J Pathol. 2010;177:3192–3201. doi: 10.2353/ajpath.2010.100157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raskopf E, Vogt A, Standop J, Sauerbruch T, Schmitz V. Inhibition of neuropilin-1 by RNA-interference and its angiostatic potential in the treatment of hepatocellular carcinoma. Z Gastroenterol. 2010;48:21–27. doi: 10.1055/s-0028-1109907. [DOI] [PubMed] [Google Scholar]
- 38.Lee J, Lee J, Yu H, Choi K, Choi C. Differential dependency of human cancer cells on vascular endothelial growth factor-mediated autocrine growth and survival. Cancer Lett. 2011;309:145–150. doi: 10.1016/j.canlet.2011.05.026. [DOI] [PubMed] [Google Scholar]
- 39.Xu J, Xia J. NRP-1 silencing suppresses hepatocellular carcinoma cell growth in vitro and in vivo. Exp Ther Med. 2013;5:150–154. doi: 10.3892/etm.2012.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Q, Xu Y, Wei S, Gao W, Chen L, Zhou T, et al. miRNA-148b suppresses hepatic cancer stem cell by targeting neuropilin-1. Biosci Rep. 2015;35:e00229. doi: 10.1042/BSR20150084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villa E, Critelli R, Lei B, Marzocchi G, Cammà C, Giannelli G, et al. Neoangiogenesis-related genes are hallmarks of fast-growing hepatocellular carcinomas and worst survival. Results from a prospective study. Gut. 2016;65:861–869. doi: 10.1136/gutjnl-2014-308483. [DOI] [PubMed] [Google Scholar]
- 42.Xu ZC, Shen HX, Chen C, Ma L, Li WZ, Wang L, et al. Neuropilin-1 promotes primary liver cancer progression by potentiating the activity of hepatic stellate cells. Oncol Lett. 2018;15:2245–2251. doi: 10.3892/ol.2017.7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lv Y, Hou X, Zhang Q, Li R, Xu L, Chen Y, et al. Untargeted metabolomics study of the in vitro anti-hepatoma effect of saikosaponin d in combination with NRP-1 knockdown. Molecules. 2019;24:1423. doi: 10.3390/molecules24071423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu P, Zou M, Wang S, Wang L, Wang L, Luo F, et al. Preparation of truncated tissue factor antineuropilin-1 monoclonal antibody conjugate and identification of its selective thrombosis in tumor blood vessels. Anticancer Drugs. 2019;30:441–450. doi: 10.1097/CAD.0000000000000767. [DOI] [PubMed] [Google Scholar]
- 45.Arab JP, Cabrera D, Sehrawat TS, Jalan-Sakrikar N, Verma VK, Simonetto D, et al. Hepatic stellate cell activation promotes alcohol-induced steatohepatitis through Igfbp3 and SerpinA12. J Hepatol. 2020;73:149–160. doi: 10.1016/j.jhep.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen YJ, Liao WX, Huang SZ, Yu YF, Wen JY, Chen J, et al. Prognostic and immunological role of CD36: A pan-cancer analysis. J Cancer. 2021;12:4762–4773. doi: 10.7150/jca.50502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Z, Zhang S, Ouyang J, Wu D, Chen L, Zhou W, et al. Single-Cell RNA-seq analysis reveals dysregulated cell-cell interactions in a tumor microenvironment related to HCC development. Dis Markers. 2022;2022:4971621. doi: 10.1155/2022/4971621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee J, Lee E, Kwon D, Lim Y, Oh S, Oh M, et al. Up-regulation of cancer-related genes in HepG2 cells by TCDD requires PRMT I and IV. Mol Cell Toxicol. 2010;6:111–118. [Google Scholar]
- 49.Li Z, Bao H. Deciphering key regulators of Inonotus hispidus petroleum ether extract involved in anti-tumor through whole transcriptome and proteome analysis in H22 tumor-bearing mice model. J Ethnopharmacol. 2022;296:115468. doi: 10.1016/j.jep.2022.115468. [DOI] [PubMed] [Google Scholar]
- 50.Papaconstantinou D, Tsilimigras DI, Pawlik TM. Recurrent hepatocellular carcinoma: Patterns, detection, staging and treatment. J Hepatocell Carcinoma. 2022;9:947–957. doi: 10.2147/JHC.S342266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng C, Zhang Z, Wang S, Chen L, Liu Q. Reduction sensitive CC9-PEG-SSBPEI/miR-148b nanoparticles: Synthesis, characterization, targeting delivery and application for anti-metastasis. Colloids Surf B Biointerfaces. 2019;183:110412. doi: 10.1016/j.colsurfb.2019.110412. [DOI] [PubMed] [Google Scholar]
- 52.Liao YL, Sun YM, Chau GY, Chau YP, Lai TC, Wang JL, et al. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008;27:5578–5589. doi: 10.1038/onc.2008.168. [DOI] [PubMed] [Google Scholar]
- 53.Raskopf E, Vogt A, Decker G, Hirt S, Daskalow K, Cramer T, et al. Combination of hypoxia and RNA-interference targeting VEGF induces apoptosis in hepatoma cells via autocrine mechanisms. Curr Pharm Biotechnol. 2012;13:2290–2298. doi: 10.2174/138920112802502088. [DOI] [PubMed] [Google Scholar]
- 54.Devbhandari RP, Shi GM, Ke AW, Wu FZ, Huang XY, Wang XY, et al. Profiling of the tetraspanin CD151 web and conspiracy of CD151/integrin β1 complex in the progression of hepatocellular carcinoma. PLoS One. 2011;6:e24901. doi: 10.1371/journal.pone.0024901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giraud J, Chalopin D, Blanc JF, Saleh M. Hepatocellular carcinoma immune landscape and the potential of immunotherapies. Front Immunol. 2021;12:655697. doi: 10.3389/fimmu.2021.655697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruf B, Heinrich B, Greten TF. Immunobiology and immunotherapy of HCC: Spotlight on innate and innate-like immune cells. Cell Mol Immunol. 2021;18:112–127. doi: 10.1038/s41423-020-00572-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Horwitz E, Stein I, Andreozzi M, Nemeth J, Shoham A, Pappo O, et al. Human and mouse VEGFA-amplified hepatocellular carcinomas are highly sensitive to sorafenib treatment. Cancer Discov. 2014;4:730–743. doi: 10.1158/2159-8290.CD-13-0782. [DOI] [PubMed] [Google Scholar]
- 58.Kisseleva EP, Krylov AV, Lyamina IV, Kudryavtsev IV, Lioudyno VI. Role of vascular endothelial growth factor (VEGF) in thymus of mice under normal conditions and with tumor growth. Biochemistry (Mosc) 2016;81:491–501. doi: 10.1134/S0006297916050060. [DOI] [PubMed] [Google Scholar]
- 59.Liu JN, Kong XS, Sun P, Wang R, Li W, Chen QF. An integrated pan-cancer analysis of TFAP4 aberrations and the potential clinical implications for cancer immunity. J Cell Mol Med. 2021;25:2082–2097. doi: 10.1111/jcmm.16147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beckebaum S, Zhang X, Chen X, Yu Z, Frilling A, Dworacki G, et al. Increased levels of interleukin-10 in serum from patients with hepatocellular carcinoma correlate with profound numerical deficiencies and immature phenotype of circulating dendritic cell subsets. Clin Cancer Res. 2004;10:7260–7269. doi: 10.1158/1078-0432.CCR-04-0872. [DOI] [PubMed] [Google Scholar]
- 61.Cheng X, Cao Y, Wang X, Cheng L, Liu Y, Lei J, et al. Systematic pan-cancer analysis of KLRB1 with prognostic value and immunological activity across human tumors. J Immunol Res. 2022;2022:5254911. doi: 10.1155/2022/5254911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oura K, Morishita A, Masaki T. Molecular and functional roles of microRNAs in the progression of hepatocellular carcinoma-A review. Int J Mol Sci. 2020;21:8362. doi: 10.3390/ijms21218362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khare S, Khare T, Ramanathan R, Ibdah JA. Hepatocellular carcinoma: The role of microRNAs. Biomolecules. 2022;12:645. doi: 10.3390/biom12050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang G, Liu H, Wei Z, Jia H, Liu Y, Liu J. Systematic analysis of the molecular mechanism of microRNA-124 in hepatoblastoma cells. Oncol Lett. 2017;14:7161–7170. doi: 10.3892/ol.2017.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang W, Ju HY, Tian XF. Circular RNA-ABCB10 suppresses hepatocellular carcinoma progression through upregulating NRP1/ABL2 via sponging miR-340-5p/miR-452-5p. Eur Rev Med Pharmacol Sci. 2020;24:2347–2357. doi: 10.26355/eurrev_202003_20501. [DOI] [PubMed] [Google Scholar]
- 66.Novikova MV, Khromova NV, Kopnin PB. Components of the hepatocellular carcinoma microenvironment and their role in tumor progression. Biochemistry (Mosc) 2017;82:861–873. doi: 10.1134/S0006297917080016. [DOI] [PubMed] [Google Scholar]
- 67.Liu YC, Yeh CT, Lin KH. Cancer stem cell functions in hepatocellular carcinoma and comprehensive therapeutic strategies. Cells. 2020;9:1331. doi: 10.3390/cells9061331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bao MH, Wong CC. Hypoxia, metabolic reprogramming, and drug resistance in liver cancer. Cells. 2021;10:1715. doi: 10.3390/cells10071715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Méndez-Blanco C, Fondevila F, García-Palomo A, González-Gallego J, Mauriz JL. Sorafenib resistance in hepatocarcinoma: Role of hypoxia-inducible factors. Exp Mol Med. 2018;50:1–9. doi: 10.1038/s12276-018-0159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Méndez-Blanco C, Fondevila F, Fernández-Palanca P, García-Palomo A, Pelt JV, et al. Stabilization of hypoxia-inducible factors and BNIP3 promoter methylation contribute to acquired sorafenib resistance in human hepatocarcinoma cells. Cancers (Basel) 2019;11:1984. doi: 10.3390/cancers11121984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tse BWC, Volpert M, Ratther E, Stylianou N, Nouri M, McGowan K, et al. Neuropilin-1 is upregulated in the adaptive response of prostate tumors to androgen-targeted therapies and is prognostic of metastatic progression and patient mortality. Oncogene. 2017;36:3417–3427. doi: 10.1038/onc.2016.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Onsurathum S, Haonon O, Pinlaor P, Pairojkul C, Khuntikeo N, Thanan R, et al. Proteomics detection of S100A6 in tumor tissue interstitial fluid and evaluation of its potential as a biomarker of cholangiocarcinoma. Tumour Biol. 2018;40:1010428318767195. doi: 10.1177/1010428318767195. [DOI] [PubMed] [Google Scholar]
- 73.Yasuoka H, Kodama R, Tsujimoto M, Yoshidome K, Akamatsu H, Nakahara M, et al. Neuropilin-2 expression in breast cancer: Correlation with lymph node metastasis, poor prognosis, and regulation of CXCR4 expression. BMC Cancer. 2009;9:220. doi: 10.1186/1471-2407-9-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Keck B, Wach S, Taubert H, Zeiler S, Ott OJ, Kunath F, et al. Neuropilin-2 and its ligand VEGF-C predict treatment response after transurethral resection and radiochemotherapy in bladder cancer patients. Int J Cancer. 2015;136:443–451. doi: 10.1002/ijc.28987. [DOI] [PubMed] [Google Scholar]
- 75.Rushing EC, Stine MJ, Hahn SJ, Shea S, Eller MS, Naif A, et al. Neuropilin-2: A novel biomarker for malignant melanoma? Hum Pathol. 2012;43:381–389. doi: 10.1016/j.humpath.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang L, Chen Y, Wang H, Zheng X, Li C, Han Z. miR-376a inhibits breast cancer cell progression by targeting neuropilin-1 NR. Onco Targets Ther. 2018;11:5293–5302. doi: 10.2147/OTT.S173416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dong Y, Hao L, Shi ZD, Fang K, Yu H, Zang GH, et al. Solasonine induces apoptosis and inhibits proliferation of bladder cancer cells by suppressing NRP1 expression. J Oncol. 2022;2022:7261486. doi: 10.1155/2022/7261486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Y, Yin H, Chen X. Circ-LDLRAD3 enhances cell growth, migration, and invasion and inhibits apoptosis by regulating miR-224-5p/NRP2 axis in gastric cancer. Dig Dis Sci. 2021;66:3862–3871. doi: 10.1007/s10620-020-06733-1. [DOI] [PubMed] [Google Scholar]
- 79.Grandclement C, Pallandre JR, Valmary Degano S, Viel E, Bouard A, Balland J, et al. Neuropilin-2 expression promotes TGF-β1-mediated epithelial to mesenchymal transition in colorectal cancer cells. PLoS One. 2011;6:e20444. doi: 10.1371/journal.pone.0020444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhan QQ, Liu QY, Yang X, Ge YH, Xu L, Ding GY, et al. Effects of silencing neuropilin-2 on proliferation, migration, and invasion of colorectal cancer HT-29. Bioengineered. 2022;13:11042–11049. doi: 10.1080/21655979.2022.2068363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hang C, Yan HS, Gong C, Gao H, Mao QH, Zhu JX. MicroRNA-9 inhibits gastric cancer cell proliferation and migration by targeting neuropilin-1. Exp Ther Med. 2019;18:2524–2530. doi: 10.3892/etm.2019.7841. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 82.Chuckran CA, Liu C, Bruno TC, Workman CJ, Vignali DA. Neuropilin-1: A checkpoint target with unique implications for cancer immunology and immunotherapy. J Immunother Cancer. 2020;8:e000967. doi: 10.1136/jitc-2020-000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Overacre-Delgoffe AE, Chikina M, Dadey RE, Yano H, Brunazzi EA, Shayan G, et al. Interferon-γ drives Treg fragility to promote anti-tumor immunity. Cell. 2017;169:1130–1141.e11. doi: 10.1016/j.cell.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Casazza A, Laoui D, Wenes M, Rizzolio S, Bassani N, Mambretti M, et al. Impeding macrophage entry into hypoxic tumor areas by Sema3A/Nrp1 signaling blockade inhibits angiogenesis and restores antitumor immunity. Cancer Cell. 2013;24:695–709. doi: 10.1016/j.ccr.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 85.Chaudhary B, Elkord E. Novel expression of Neuropilin 1 on human tumor-infiltrating lymphocytes in colorectal cancer liver metastases. Expert Opin Ther Targets. 2015;19:147–161. doi: 10.1517/14728222.2014.977784. [DOI] [PubMed] [Google Scholar]
- 86.Roy S, Bag AK, Dutta S, Polavaram NS, Islam R, Schellenburg S, et al. Macrophage-derived neuropilin-2 exhibits novel tumor-promoting functions. Cancer Res. 2018;78:5600–5617. doi: 10.1158/0008-5472.CAN-18-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Glinka Y, Mohammed N, Subramaniam V, Jothy S, Prud’homme GJ. Neuropilin-1 is expressed by breast cancer stem-like cells and is linked to NF-κB activation and tumor sphere formation. Biochem Biophys Res Commun. 2012;425:775–780. doi: 10.1016/j.bbrc.2012.07.151. [DOI] [PubMed] [Google Scholar]
- 88.Elaimy AL, Guru S, Chang C, Ou J, Amante JJ, Zhu LJ, et al. VEGF-neuropilin-2 signaling promotes stem-like traits in breast cancer cells by TAZ-mediated repression of the Rac GAP β2-chimaerin. Sci Signal. 2018;11:eaao6897. doi: 10.1126/scisignal.aao6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen C, Zhang R, Ma L, Li Q, Zhao YL, Zhang GJ, et al. Neuropilin-1 is up-regulated by cancer-associated fibroblast-secreted IL-8 and associated with cell proliferation of gallbladder cancer. J Cell Mol Med. 2020;24:12608–12618. doi: 10.1111/jcmm.15825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen XJ, Wu S, Yan RM, Fan LS, Yu L, Zhang YM, et al. The role of the hypoxia-Nrp-1 axis in the activation of M2-like tumor-associated macrophages in the tumor microenvironment of cervical cancer. Mol Carcinog. 2019;58:388–397. doi: 10.1002/mc.22936. [DOI] [PubMed] [Google Scholar]
- 91.Jin Y, Che X, Qu X, Li X, Lu W, Wu J, et al. CircHIPK3 promotes metastasis of gastric cancer via miR-653-5p/miR-338-3p-NRP1 axis under a long-term hypoxic microenvironment. Front Oncol 2020;10:1612. Erratum in: Front Oncol. 2021;11:783320. doi: 10.3389/fonc.2021.783320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Coma S, Shimizu A, Klagsbrun M. Hypoxia induces tumor and endothelial cell migration in a semaphorin 3F- and VEGF-dependent manner via transcriptional repression of their common receptor neuropilin 2. Cell Adh Migr. 2011;5:266–275. doi: 10.4161/cam.5.3.16294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA 2020 checklist
PRISMA 2020 for abstracts checklist
Full search strategy employed for each online database (up to and including August 31st, 2022)
Evaluation of the published articles. (A) Temporal distribution of the number of articles published employing in vitro, in vivo or human models, or a combination of them. Comparison of the number of studies conducted with (B) human or (C) animal models with the mean of the human or animals examined per year, respectively.