ABSTRACT
STUDY QUESTION
What are the trends and developments in preimplantation genetic testing (PGT) in 2018 as compared to previous years?
SUMMARY ANSWER
The main trends observed in this 21st dataset on PGT are that the implementation of trophectoderm biopsy with comprehensive whole-genome testing is most often applied for PGT-A and concurrent PGT-M/SR/A, while for PGT-M and PGT-SR, single-cell testing with PCR and FISH still prevail.
WHAT IS KNOWN ALREADY
Since it was established in 1997, the ESHRE PGT Consortium has been collecting and analysing data from mainly European PGT centres. To date, 20 datasets and an overview of the first 10 years of data collections have been published.
STUDY DESIGN, SIZE, DURATION
The data for PGT analyses performed between 1 January 2018 and 31 December 2018 with a 2-year follow-up after analysis were provided by participating centres on a voluntary basis. Data were collected using an online platform, which is based on genetic analysis and has been in use since 2016.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Data on biopsy method, diagnostic technology, and clinical outcome were submitted by 44 centres. Records with analyses for more than one PGT for monogenic disorders (PGT-M) and/or PGT for chromosomal structural rearrangements (PGT-SR), or with inconsistent data regarding the PGT modality, were excluded. All transfers performed within 2 years after the analysis were included, enabling the calculation of cumulative pregnancy rates. Data analysis, calculations, and preparation of figures and tables were carried out by expert co-authors.
MAIN RESULTS AND THE ROLE OF CHANCE
The current data collection from 2018 covers a total of 1388 analyses for PGT-M, 462 analyses for PGT-SR, 3003 analyses for PGT for aneuploidies (PGT-A), and 338 analyses for concurrent PGT-M/SR with PGT-A.
The application of blastocyst biopsy is gradually rising for PGT-M (from 19% in 2016–2017 to 33% in 2018), is status quo for PGT-SR (from 30% in 2016–2017 to 33% in 2018) and has become the most used biopsy stage for PGT-A (from 87% in 2016–2017 to 98% in 2018) and for concurrent PGT-M/SR with PGT-A (96%). The use of comprehensive, whole-genome amplification (WGA)-based diagnostic technology showed a small decrease for PGT-M (from 15% in 2016–2017 to 12% in 2018) and for PGT-SR (from 50% in 2016–2017 to 44% in 2018). Comprehensive testing was, however, the main technology for PGT-A (from 93% in 2016–2017 to 98% in 2018). WGA-based testing was also widely used for concurrent PGT-M/SR with PGT-A, as a standalone technique (74%) or in combination with PCR or FISH (24%). Trophectoderm biopsy and comprehensive testing strategies are linked with higher diagnostic efficiencies and improved clinical outcomes per embryo transfer.
LIMITATIONS, REASONS FOR CAUTION
The findings apply to the data submitted by 44 participating centres and do not represent worldwide trends in PGT. Details on the health of babies born were not provided in this manuscript.
WIDER IMPLICATIONS OF THE FINDINGS
The Consortium datasets provide a valuable resource for following trends in PGT practice.
STUDY FUNDING/COMPETING INTEREST(S)
The study has no external funding, and all costs are covered by ESHRE. There are no competing interests declared.
TRIAL REGISTRATION NUMBER
N/A.
Keywords: PGT, structural rearrangements, monogenic disorders, aneuploidy, human embryo, registry, data collection, comprehensive genetic testing
Introduction
Since ESHRE established the preimplantation genetic testing (PGT) Consortium in 1997, its main objectives have been to collect prospective and retrospective data on PGT treatments and their outcome, to provide guidance and network opportunities to PGT centres and to promote best practice. Twenty sets of data on PGT cycles, analyses, pregnancies, deliveries, and children have been published to date (Geraedts et al., 1999, 2000; ESHRE PGD Consortium Steering Committee, 2002; Sermon et al., 2005; Harper et al., 2006; Sermon et al., 2007; Goossens et al., 2008; Harper et al., 2008; Goossens et al., 2009; Harper et al., 2010; Goossens et al., 2012; Moutou et al., 2014; De Rycke et al., 2015, 2017; Coonen et al., 2020; Van Montfoort et al., 2021). An overview of the first 10 years of data collection was published in 2012 (Harper et al., 2012). Overall, the data collections provide a valuable resource for data mining and for following trends in PGT practice.
Up to data collection XVI (2013), data submission from the participating centres has been retrospective, relying on pre-designed Excel and later FileMakerPro (Claris International, Cupertino, CA, USA) files. The years 2013–2015 were a transition period and were covered by summary data, collected in Excel (Coonen et al., 2020). A new online data registration platform was then used from data collection XIX (2016) onwards. The online database has been adapted to accommodate the increased complexity and changes in the overall timeline of PGT treatments and offers the opportunity for real-time data registration. Data from embryo biopsy, genetic analysis, and embryo transfer, possibly occurring in completely separate time frames, are registered in connected modules. The genetic analysis is the central module to which multiple oocyte collection modules and multiple embryo transfer modules can be linked. The new database structure therefore differs from the previous data collection systems, which were based on cycles with one oocyte collection followed by one analysis and one embryo transfer.
The classification of PGT cycles with sexing for X-linked diseases and for chromosomal numerical aberrations of high genetic risk within the PGT-SR indication group has been maintained in the new data collection systems. The implementation of comprehensive genetic testing has enabled concurrent PGT-M/SR and PGT-A, and data from such double indications were considered as a distinct category. It also allowed us to better monitor the incidence of aneuploidies, including mosaic aneuploidies.
This article presents data from PGT treatments registered in the online database during the year 2018.
Materials and methods
The report includes PGT analyses conducted between 1 January 2018 and 31 December 2018 and covers data on PGT indication, biopsy method, diagnostic technology, the efficiency of the different procedures, and (cumulative) clinical PGT outcome in terms of positive hCG and live births of all fresh and frozen embryo transfers reported up until 2 years after the analysis date. Data on PGT treatments were provided by 44 Consortium members, mainly based in European countries (39), covering the following treatment modalities: PGT for monogenic/single gene defects (PGT-M), PGT for chromosomal structural rearrangements (PGT-SR), PGT for aneuploidies (PGT-A), and concurrent PGT-M or PGT-SR with PGT-A. Centres used a unique login account to upload the data in an anonymized format onto an online platform designed for the specific requirements of this data collection (Dynamic Solutions, Barcelona, Spain). The database was exported to SQL Server Management Studio Version 15 (Microsoft Corporation, Redmond, WA, USA) and Structured Query Language (SQL) was used to retrieve the data per analysis from the database. These data were exported to Excel 2016 (Microsoft Corporation, Redmond, WA, USA) where the tables with the numbers and percentages as well as the figures were generated. A total of 5670 genetic analyses conducted within 2018 were entered into the database. In total, 408 (7.2%) were excluded because no indication for PGT was filled in and 71 (1.2%) analyses had to be excluded because of inconsistent data regarding the PGT modality or the number of indications. Within the included PGT analyses (n = 5191), missing data were left blank and are reported as ‘not reported’. Following curation, in-depth data analysis was carried out by expert members of the ESHRE PGT Consortium Steering Committee.
The data from 2018 are compared to the data from 2016 to 2017 (Van Montfoort et al., 2021) or to means from earlier datasets. The terminology used in this report was based on the revised glossary for infertility care (Zegers-Hochschild et al., 2017). A clinical pregnancy was defined as the presence of at least one positive heartbeat. An ongoing pregnancy was defined as the presence of at least one positive heartbeat at 12 weeks of gestation and a live birth was defined as a liveborn child after 20 weeks of gestation.
Results
The current data collection covers a total of 5191 analyses initiated in 2018 with PGT-M accounting for 27%, PGT-SR for 9%, PGT-A for 58%, and concurrent PGT-M/SR with PGT-A for 6% of analyses. Overall data per PGT modality are presented in Figs 1, 2, and 3, Tables I and II and the text, while detailed results, per modality and per sub-indication, can be found in Supplementary Tables SI, SII, SIII, SIV, SV, SVI, SVII, SVIII, SIX, SX, SXI, and SXII.
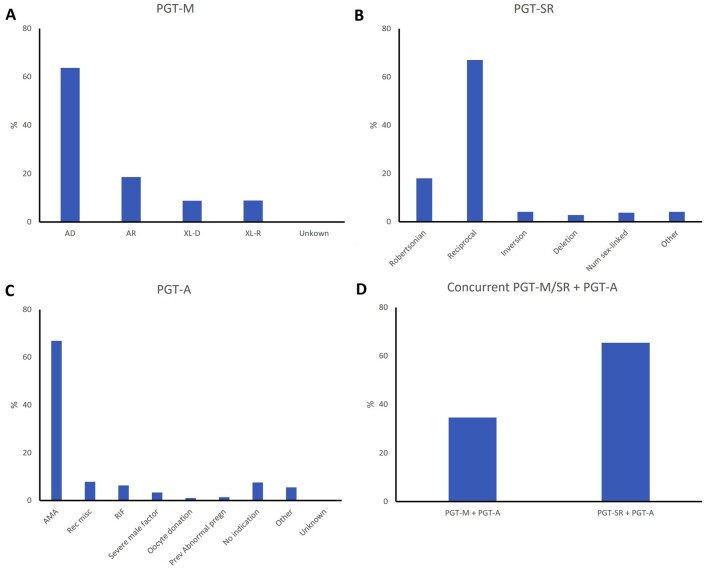
Figure 1.
Distribution of PGT indications in 2018. (A) Preimplantation genetic testing for monogenic/single gene defects (PGT-M), (B) PGT for chromosomal structural rearrangements (PGT-SR), (C) PGT for aneuploidies (PGT-A), and (D) concurrent PGT-M/SR with PGT-A. AD, autosomal dominant; AR, autosomal recessive; XL-D, X-linked dominant; XL-R, X-linked recessive; AMA, advanced maternal age; Rec misc, recurrent miscarriage; RIF, repeated implantation failure; Prev abnormal pregn, previous abnormal pregnancy.
Figure 2.
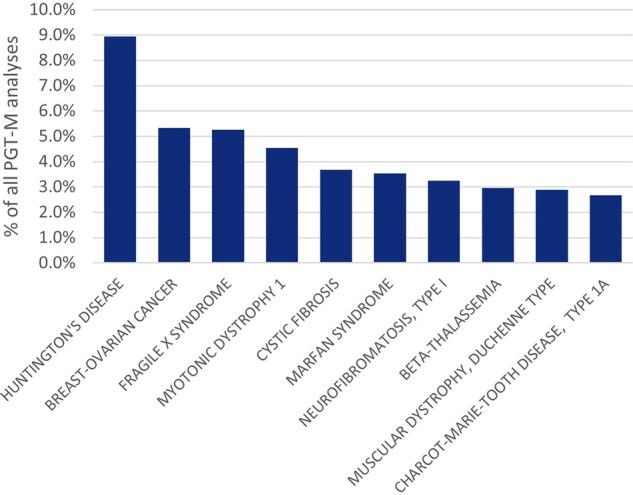
Top 10 of the indications for which PGT-M was applied in 2018. PGT-M: preimplantation genetic testing for monogenic/single gene defects.
Figure 3.
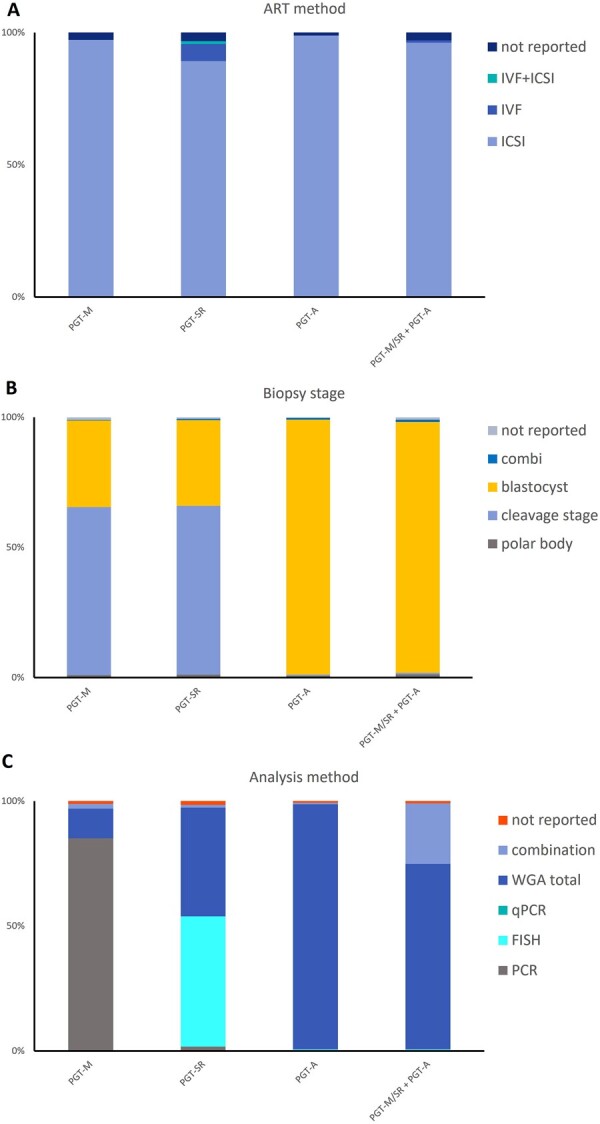
Distribution of methods used among the different PGT modalities in 2018. (A) ART method, (B) biopsy stage, and (C) analysis method. PGT-M: preimplantation genetic testing for monogenic/single gene defects; PGT-SR: PGT for chromosomal structural rearrangements; PGT-A: PGT for aneuploidies; PGT-M/SR + PGT-A: concurrent PGT-M/SR with PGT-A; Combi: combination; WGA: whole-genome amplification; qPCR: quantitative PCR.
Table I.
Data on biopsy and analysis at embryo level.
| PGT-M |
PGT-SR |
PGT-A |
PGT-M/SR + PGT-A |
Total |
Total/# biopsied |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| # biopsied (initial) | 7519 | 100% | 2253 | 100% | 9416 | 100% | 1810 | 100% | 20 998 | 100% | 100% |
| # fresh | 7121 | 95% | 2034 | 90% | 9236 | 98% | 1739 | 96% | 20 130 | 96% | 96% |
| # thawed/warmed | 398 | 5% | 219 | 10% | 180 | 2% | 71 | 4% | 868 | 4% | 4% |
| # analysed | 7396 | 100% | 2212 | 100% | 9355 | 100% | 1729 | 100% | 20 692 | 100% | 99% |
| # not diagnosed | 942 | 13% | 180 | 8% | 192 | 2% | 63 | 4% | 1377 | 7% | 7% |
| # failed | 507 | 7% | 74 | 3% | 88 | 1% | 35 | 2% | 704 | 3% | 3% |
| # inconclusive | 435 | 6% | 106 | 5% | 104 | 1% | 28 | 2% | 673 | 3% | 3% |
| # diagnosed | 6454 | 87% | 2032 | 92% | 9163 | 98% | 1666 | 96% | 19 315 | 93% | 92% |
| # immediate transfer | 722 | 223 | 76 | 11 | 1032 | 5% | |||||
| # frozen | 1918 | 319 | 5133 | 876 | 8246 | 39% | |||||
PGT-M: preimplantation genetic testing for monogenic/single gene defects; PGT-SR: PGT for chromosomal structural rearrangements; PGT-A: PGT for aneuploidies; PGT-M/SR + PGT-A: concurrent PGT-M/SR with PGT-A.
Table II.
Data on data transfer and pregnancy outcome (% are related to number of transfers).
| PGT-M |
PGT-SR |
PGT-A |
PGT-M/SR + PGT-A |
Total |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| # analyses | 1388 | 27% | 462 | 9% | 3003 | 58% | 338 | 7% | 5191 | |
| # analyses without transfer* | 409 | 29% | 194 | 42% | 1515 | 50% | 131 | 39% | 2249 | 43% |
| # analyses with transfer* | 979 | 71% | 268 | 58% | 1488 | 50% | 207 | 61% | 2942 | 57% |
| # transfers | 1375 | 100% | 326 | 100% | 1862 | 0 | 260 | 100% | 3823 | 100% |
| # with fresh embryos | 488 | 36% | 130 | 40% | 17 | 1% | 3 | 1% | 638 | 17% |
| # with thawed/warmed embryos | 884 | 64% | 196 | 60% | 1845 | 99% | 256 | 98% | 3181 | 83% |
| # with fresh and thawed/warmed embryos | 3 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 3 | 0% |
| # not reported | 0 | 0% | 0 | 0% | 0 | 0% | 1 | 0% | 1 | 0% |
| # SET | 1260 | 92% | 295 | 90% | 1741 | 94% | 250 | 96% | 3546 | 93% |
| # DET | 109 | 8% | 31 | 10% | 121 | 6% | 10 | 4% | 271 | 7% |
| # >DET | 6 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 6 | 0% |
| # SET_unknown | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% | 0 | 0% |
| # positive hCG test | 561 | 41% | 114 | 35% | 1145 | 61% | 151 | 58% | 1971 | 52% |
| # lost to FU after positive hCG test | 5 | 0% | 6 | 2% | 15 | 1% | 4 | 2% | 30 | 1% |
| # PUL | 86 | 6% | 15 | 5% | 73 | 4% | 9 | 3% | 183 | 5% |
| # blighted ovum | 29 | 2% | 5 | 2% | 37 | 2% | 4 | 2% | 75 | 2% |
| # ectopic pregnancies | 5 | 0% | 1 | 0% | 4 | 0% | 0 | 0% | 10 | 0% |
| # clinical pregnancies | 436 | 32% | 87 | 27% | 1016 | 55% | 134 | 52% | 1673 | 44% |
| # lost to FU after clinical pregnancy | 76 | 6% | 15 | 5% | 155 | 8% | 20 | 8% | 266 | 7% |
| # pregnancy loss <12 weeks | 19 | 1% | 0 | 0% | 41 | 2% | 5 | 2% | 65 | 2% |
| # ongoing pregnancy (>12 weeks) | 341 | 25% | 72 | 22% | 820 | 44% | 109 | 42% | 1342 | 35% |
| # singletons | 327 | 24% | 69 | 21% | 786 | 42% | 103 | 40% | 1285 | 34% |
| # multiples | 14 | 1% | 3 | 1% | 34 | 2% | 6 | 2% | 57 | 1% |
| # lost to FU after ongoing pregnancy | 0 | 0% | 0 | 0% | 1 | 0% | 0 | 0% | 1 | 0% |
| # pregnancy loss 12–20 weeks | 6 | 0% | 0 | 0% | 20 | 1% | 2 | 1% | 28 | 1% |
| # pregnancy with no live birth | 1 | 0% | 0 | 0% | 5 | 0% | 0 | 0% | 6 | 0% |
| # pregnancy with at least one live birth | 334 | 24% | 72 | 22% | 794 | 43% | 107 | 41% | 1307 | 34% |
| # live born children | 352 | 77 | 838 | 113 | 1380 | |||||
*% expressed per # analyses
SET: single embryo transfer; DET: double embryo transfer; PUL: pregnancy of unknown location; FU: follow-up; PGT-M: preimplantation genetic testing for monogenic/single gene defects; PGT-SR: PGT for chromosomal structural rearrangements; PGT-A: PGT for aneuploidies; PGT-M/SR + PGT-A: concurrent PGT-M/SR with PGT-A.
PGT-M
During 2018, 1388 PGT-M analyses were reported, the majority of which were linked with a single oocyte collection (93%) or biopsy event (97%), with a mean female age of 32.9 years (Supplementary Table SI). Nearly two-thirds of PGT-M analyses were performed for an autosomal dominant disease (64%), followed by autosomal recessive and X-linked indications, which accounted for 19% and 18%, respectively (Fig. 1A), in line with the previous datasets. The top 10 of the indications for which PGT-M is performed are depicted in Fig. 2. Nine percent of the analyses are performed for Huntington’s disease, followed by hereditary breast cancer type 1 (5.4%), Fragile-X syndrome (5.3%), myotonic dystrophy type 1 (4.5%), and cystic fibrosis (3.7%).
In line with previous data, ICSI was the main fertilization method (97%), and cleavage-stage biopsy was, in 2018, still the most widely used biopsy stage (65%) (Fig. 3). PCR remained the most widely used first-line method of DNA amplification (85%). Compared to other modalities, the application of blastocyst biopsy is only gradually increasing in PGT-M (8% in 2013, 12% in 2015, 19% in 2016–2017, and 33% in 2018). The implementation of comprehensive, whole-genome amplification (WGA) diagnostic technology (9% in 2013, 12% in 2015, 15% in 2016–2017, and 12% in 2018) remains status quo. Further detailed results on ART method, biopsy stage and analysis method for PGT-M analyses can be found in Supplementary Table SI.
Data on biopsy and analysis at embryo level for PGT-M are presented in Table I and Supplementary Table SII. Of the 7519 embryos for biopsy, 5% were thawed/warmed embryos. Most of the embryos analysed gave a diagnostic result (87%) of which 50% were genetically transferable. This is concordant with previous datasets: 88% of analysed embryos gave a diagnostic result and 47% were genetically transferable in dataset XIX–XX. Of the 13% of embryos without diagnosis, 6% showed an inconclusive result, while in 7%, the analysis failed. Of all transferable embryos, 22% were freshly transferred, 60% were cryopreserved, and 16% could not be used for the patient, most likely because of poor embryo development between biopsy and transfer stage. When expressed per biopsied embryo, 43% were genetically transferable, and 10% and 26% were transferred and cryopreserved, respectively.
Within the 2 years following the 1388 analyses reported, 1375 transfers were performed, 92% of which were single embryo transfers, 36% were fresh, and 64% were frozen transfers (Table II, Supplementary Table SIII).
Overall, a positive hCG was obtained in 41% of transfers, yielding a clinical pregnancy rate of 32% per embryo transfer procedure. This is in line with the clinical outcome reported in the 2016–2017 dataset (35% per embryo transfer), however, with a small reduction in the outcome. When expressed per analysis (n = 1388) and per analysis with at least one transfer (n = 979), the cumulative positive hCG and clinical pregnancy rate 2 years after the analysis were 40%/57% (positive hCG) and 31%/44% (clinical pregnancy), respectively. In 6% of the transfers, the pregnancy was lost to follow-up after the clinical pregnancy. Of the remaining 360 clinical pregnancies, 341 (95%) went on to an ongoing pregnancy and 334 (98%) ended in at least one live birth. The number of reported live born children in the remaining transfers was 352.
PGT-SR
For 2018, 462 PGT-SR analyses were reported, the majority of which were linked with a single oocyte collection (92%) or biopsy event (97%). The mean age of women undergoing PGT-SR was 34.2 years (Supplementary Table SIV). PGT for reciprocal translocations was performed more often than for any other type of structural rearrangement (67%), while Robertsonian translocations accounted for 18% of PGT-SR analyses (Fig. 1B). For reciprocal translocations, the number of cycles performed for female carriers (48%) equalled that for male carriers (51%), whereas for Robertsonian translocations, the number of cycles performed for male carriers was about 2-fold that of female carriers. Figures are similar to those of previous datasets. ICSI was the preferred fertilization method (89%) (Fig. 3). Cleavage-stage biopsy was most applied (65%) and remained at the same level as was reported in previous datasets (73% in 2014, 63% in 2015, and 67% in 2016–2017). Biopsy at the blastocyst stage was performed in 33% of cases. In more than half of the analyses (52%), FISH remained the preferred methodology. Overall, the use of comprehensive, WGA-based technology has slightly increased when compared with the previous dataset (44 to 50%), comprising a greater proportion of cycles applying next-generation sequencing (NGS) (14 to 26%) at the expense of the proportion of cycles applying array comparative genomic hybridization (array CGH) (35 to 16%).
Further detailed results on ART method, biopsy stage and analysis method for PGT-SR analyses can be found in Supplementary Table SIV.
Data on biopsy and analysis at embryo level for PGT-SR are presented in Table I and Supplementary Table SV. Ten percent of the 2253 embryos for biopsy were thawed/warmed embryos. A diagnosis was assigned in 92% of the analysed embryos. Of all transferable embryos, 36% were freshly transferred, 52% were cryopreserved, and 12% could not be used for the patient because of insufficient embryo morphology.
Of all analyses, 58% were linked with at least one embryo transfer procedure within the 2 years following the analysis, either in a fresh (40%) or a frozen (60%) cycle (Table II and Supplementary Table SVI). A single embryo was transferred in 90% of embryo transfers.
A positive hCG was obtained in 35% of transfers, with a clinical pregnancy rate of 27% per embryo transfer (Table II), which is slightly lower than the outcome reported in the 2016–2017 dataset (30% per embryo transfer). Per analysis, the cumulative positive hCG and clinical pregnancy rates were 25% and 19%, respectively, while per analysis with at least one transfer, the rates were 42% and 32%. Five percent of the transfers with a clinical pregnancy were lost to follow-up. The remaining 72 clinical pregnancies resulted in 72 (100%) ongoing pregnancies and 72 (100%) pregnancies with at least one live birth (Table II).
PGT-A
During 2018, 3003 PGT-A analyses were reported, the majority of which were linked with a single oocyte collection (95%) or biopsy event (90%). The average female age of women applying for PGT-A was 38.6 years. As expected, the age of women referred for advanced maternal age (AMA) (40.6 years) and oocyte donation (41.2 years) were the highest (Supplementary Table SVII). The most common indication for PGT-A was AMA (67%), followed by recurrent miscarriage (8%) and repeated implantation failure (6%) and severe male factor (3%) previous abnormal pregnancy (1.4%) and oocyte donation (1%) (Fig. 1C). In 8% of analyses, PGT-A was performed without a reported medical indication.
The trend of performing PGT-A as a common IVF add-on was noted for the first time in the datasets 2013–2015 and 2016–2017 (5–9% without indication).
In 99% of all analyses, ICSI was used for fertilization (Fig. 3A). Almost all the biopsies were performed at the blastocyst stage (98%). Cleavage-stage biopsy, which was 8% in 2016–2017, accounted for only 0.6% in 2018 (Fig. 3B). For the genetic analysis, WGA-based methodologies, coupled with array CGH, accounted for only 3%, while analysis with NGS was performed in 95% of the cases (Fig. 3C). This was a further increase from the 93% figure from 2016–2017. FISH, which is not recommended for PGT-A (ESHRE PGT-SR/PGT-A Working Group et al., 2020), was used only in 0.7% of analyses, showing a reduction of 2.3% since the data of 2016–2017. Further detailed results on ART method, biopsy stage and analysis method for PGT-A analyses can be found in Supplementary Table SVII.
Data on biopsy and analysis at embryo level for PGT-A are presented in Table I and Supplementary Table SVIII. Only a minority of the 9416 embryos had been cryopreserved prior to biopsy (2%). Of the embryos analysed, 98% gave a diagnosis: 54% of these diagnosed embryos were aneuploid, 39% were euploid, and 7% of embryos were diagnosed as mosaic embryos. As expected, the majority of aneuploid embryos (64%) were found in AMA cycles and the lowest number (20%) was found in the oocyte donation group. Within these two groups, mosaic embryos accounted for 5% and 17%, respectively. Most of the euploid and mosaic embryos were cryopreserved (98% and 96%, respectively) (Supplementary Table SVIII). This was expected, given the frequent use of blastocyst biopsy with comprehensive analysis, which is usually coupled with embryo cryopreservation and transfer in a later cycle. The majority of the aneuploid embryos were discarded (80%) and 20% were cryopreserved.
At least one embryo transfer procedure was carried out for 50% of analyses, almost all (99%) being a frozen embryo transfer and involving a single embryo (94%), which was either euploid (94%) or mosaic (5%) (Table II and Supplementary Table SIX). A positive hCG was obtained in 61% of transfers, leading to a clinical pregnancy rate of 55% per embryo transfer. The 2016–2017 dataset showed a clinical pregnancy rate of 52% per embryo transfer, indicating a slightly increased outcome for the current dataset. Per analysis, a cumulative positive hCG rate of 38% and a clinical pregnancy rate of 34% was reached, while after including only the analyses with a transfer, these rates were 77% and 68%, respectively. With a lost to follow-up after a clinical pregnancy of 8% of the transfers, a total of 838 live born children were reported (Supplementary Table SIX).
Concurrent PGT-M/PGT-SR and PGT-A
For 2018, 338 analyses for the concurrent indication group were reported (Table II). One-third concerned PGT-M with PGT-A (35%) and two-thirds involved PGT-SR with PGT-A (65%) (Fig. 1D). A minority of analyses were linked with more than one oocyte collection (15%) or biopsy event (17%), pointing to the practice of pooling of oocytes/embryos before analysis. The finding that only 4% of the biopsied embryos were thawed/warmed before biopsy indicates that embryos are biopsied before cryopreservation and the biopsied samples are stored and pooled into one analysis. Pooling of more than a single cohort of embryos likely yields a larger series of samples for testing and ensures a better chance that a genetically transferable embryo ensues after selection for both PGT-M/SR and PGT-A. Pooling at the level of analysis circumvents multiple vitrification-warming of embryos compared to the practice of accumulation at the level of biopsy. The practice of pooling seems more applied for the concurrent indication group than for other PGT modalities.
The mean female age of women applying for concurrent PGT-M/PGT-SR and PGT-A was 34.5 years (Supplementary Table SX). Most cycles were performed with ICSI (96%) (Fig. 3) and biopsy was primarily performed at the blastocyst stage (96%). For the PGT-SR/PGT-A subgroup, comprehensive WGA methods were applied in 96% of analyses with the majority WGA followed by NGS (86%). For the PGT-M/PGT-A subgroup, the most frequent methods were WGA followed by NGS (27%), WGA and SNP array (5%), and combined methods (67%). Further detailed results on ART method, biopsy stage and analysis method for concurrent PGT-M/SR and PGT-A analyses can be found in Supplementary Table SX.
Data on biopsy and analysis at embryo level for PGT-M/SR with PGT-A are presented in Table II and Supplementary Table SXI. Here again, most embryos for biopsy were fresh embryos (96%). Genetic diagnosis was successful for 96% of analysed embryos.
Within the group of embryos which are genetically transferable and euploid for PGT-M/PGT-SR (n = 599), many embryos were cryopreserved: 97% of euploid embryos, 69% of mosaic embryos, and even 34% of aneuploid embryos. As can be expected, most of the latter embryo group is discarded (66%), and so are 28% of mosaic embryos.
Within the group of non-genetically transferable embryos (n = 584), a large part is discarded: 57% of euploid embryos, 30% of mosaic embryos, 67% of aneuploid embryos, and 70% of embryos without diagnosis for PGT-A. Remarkably, a part of the non-genetically transferable embryos is still cryopreserved (43% euploid embryos, 70% mosaic embryos, 33% aneuploid embryos, and 30% of embryos without diagnosis for PGT-A). In total, 35.4% (207/584) of non-genetically transferable embryos remain cryopreserved. It is unclear whether this relates to data from centres which are obliged to cryopreserve all tested embryos because of legal regulations or whether the centres’ policy considers the possibility of transferring these embryos. In 483 embryos (29%), only a PGT-A diagnosis was entered into the database instead of a combined PGT-M/SR and PGT-A diagnosis. These were therefore labelled as missing in Supplementary Table SXI.
At least one embryo transfer procedure was feasible for 61% of analyses (Table II and Supplementary Table SXII). Nearly all transfers involved a frozen embryo transfer (98%) and a single embryo (96%). Embryos effectively transferred were usually genetically transferable plus euploid (62%), mosaic (2%), or diagnosis for PGT-A was not reported (3%). Four percent of the transferred embryos were euploid/mosaic, genetically non-transferable and 25% were euploid with unreported genetic transferability.
A positive hCG was obtained in 58% of transfers leading to a clinical pregnancy rate of 52% per embryo transfer. Per analysis, the cumulative positive hCG rate was 45% and the clinical pregnancy rate 40%. Per analysis with transfer these rates are 73% and 65%. One hundred and thirteen live born children have been reported. These data are similar to those of the previous data collection.
Discussion
The ESHRE PGT Consortium was established almost simultaneously with the start of PGT treatment and has, since 1997, published 20 extensive data collection reports. This allows the Consortium to have continuous monitoring of the evolution of PGT in European and non-European countries. The data reported in the present article involve data from the year 2018. As in the 2016–2017 report, this report also analysed data from PGT-M/SR and PGT-A double indications allowing monitoring of the incidence of aneuploidies, including mosaic aneuploidies, in embryos analysed for PGT-M and PGT-SR since 2016. This indicates that the analysis of the entire chromosome set-up of the embryo is becoming increasingly widespread.
The relative contributions of PGT-M (37%), PGT-SR (9%), PGT-A (58%), and concurrent PGT-M/SR/A (6%) have shifted slightly in favour of PGT-A, now reaching more than half of the cycles. This may, however, be a coincidence as the cohort of contributing centres differs with each dataset, or this may point to an increased application of PGT-A. The major indication of each PGT modality (AMA for PGT-A, reciprocal translocation for PGT-SR, and autosomal dominant disease for PGT-M) has not changed compared to previous datasets. Compared to the overview of the first 10 years of data collection (Harper et al., 2012), the top five PGT-M indications from this 2018 dataset and the ranking of the 10 years’ overview paper have four indications in common: cystic fibrosis, Huntington’s disease, myotonic dystrophy type 1, and fragile X. Testing for breast cancer gene 1 (BRCA) (likely) pathogenic variants, not mentioned in the list of 15 most frequent indications in the 10 years’ review, is the second most frequent PGT-M indication in the current dataset.
Although trophectoderm biopsy and comprehensive genetic testing have emerged as preferential methods for PGT-A and concurrent PGT-M/SR/A in previous years, the implementation of these methods for PGT-M or PGT-SR remains limited.
The efficiency of diagnostic testing is acceptable for PGT-M (87%) and PGT-SR (92%), Genetic testing for these modalities is mostly based on single cell-testing (65%) which may explain the failed and inconclusive results. The diagnostic efficiency of the modalities where mainly a small number of trophectoderm cells are biopsied for genetic testing is higher (98% for PGT-A and 96% for concurrent PGT-M/SR/A).
The percentages of mosaic embryos for PGT-A and PGT-M/SR/A reported in this data collection seem lower than the percentages currently reported in the literature. As only 27 out of 44 centres reported mosaic embryos, these data are limited and should be confirmed with additional data. Before the first paper on the transfer of mosaic embryos leading to healthy babies in 2015 (Greco et al., 2015), mosaic embryos were not transferred (Viotti, 2020). It is likely that PGT centres were still adapting their transfer policy in 2018. Based on the recent survey on mosaic embryos of the ESHRE Working Group on Chromosomal Mosaicism et al. (2022), in which it is reported that 80% of centres that biopsy three or more cells report mosaicism, we might expect that in future Consortium data collections, there will be an increase in centres reporting mosaics.
The historical mean clinical pregnancy rate was stable at around 30% per embryo transfer for all PGT modalities. Clinical outcomes per embryo transfer for PGT-M are slightly above this mean in the recent data collections: 34%, 35%, and 32% for the 2013–2015, 2016–2017, and the 2018 datasets, respectively. For PGT-SR, the clinical outcomes are still in line with the historical mean: 34%, 30%, and 27% for the 2013–2015, 2016–2017, and the 2018 datasets, respectively. The enhanced outcomes for PGT-A (47% and 50% in datasets 2013–2015 and 2016–2017, respectively) are confirmed in the current dataset with a clinical pregnancy outcome of 55% per transfer. As the implementation of trophectoderm biopsy (98%), comprehensive testing (98%), and frozen embryo transfer (99%) is the most advanced for this indication group, the question arises of whether these practices are somehow associated with improved clinical outcomes. There were no big changes reported for PGT-M or PGT-SR without PGT-A concerning biopsy and testing policies (trophectoderm biopsy in 30% of all cases, in both modalities; the use of comprehensive testing in 12% and 44%, respectively). There is a small shift towards transfer after cryopreservation instead of fresh embryo transfer (from 58% to 64% for PGT-M and from 54% to 60% for PGT-SR) but none of these has an impact on clinical outcomes. Similar good clinical pregnancy rates were found for concurrent PGT-M/SR and PGT-A, a modality with similar practices of mainly trophectoderm biopsy (96%), comprehensive testing (74%), or comprehensive testing in combination with PCR or FISH (24%) and frozen embryo transfer (98%). The clinical pregnancy rate was 50% for PGT-M/A cycles and 32% in PGT-M cycles without PGT-A. The difference in clinical outcome for the PGT-SR group (27%) versus the PGT-SR/A group (53%) is 2-fold. It remains to be seen whether further advances in ART practices for PGT-M and PGT-SR lead to better clinical outcomes.
For concurrent PGT-M/SR and PGT-A groups, it was also possible to evaluate the incidence of mosaic embryos and the frequency of aneuploidy in genetically transferable embryos. Similar to observations in the 2016–2017 collection, for PGT-M + PGT-A, 7% of the analysed embryos were reported to be mosaic, while only 2% were reported in the PGT-SR + PGT-A group. After PGT-A, 19% of genetically transferable embryos for PGT-M and 8% for PGT-SR were aneuploid. These are similar to values reported in the 2016–2017 collection in which for PGT-M and PGT-SR, 23% and 13% of genetically transferable embryos were aneuploid, respectively, confirming a high percentage of aneuploid embryos in these groups.
This report presents the second dataset from the online platform. A particular benefit of this new database is that its structure allows calculation of cumulative outcome rates from the multiple fresh/frozen transfers following an analysis. This is an asset, given that since the implementation of vitrification, and the introduction of a freeze-all strategy related to the introduction of trophectoderm biopsies and comprehensive testing, ART/PGT outcome measurements have shifted to cumulative success rates.
However, with the implementation of the new online platform, the number of registered treatments has decreased (5191 analyses in 2018 and 8803 analyses in 2016–2017 versus 11 120 cycles to oocyte retrieval in 2015), while the number of participating centres has also decreased compared to previous years (44 as compared to the average number of 62 from 2010–2017). This decreasing trend conflicts with data from the ESHRE European IVF-Monitoring Consortium, showing an increase in PGT practice (Wyns et al., 2021); the latter data more likely reflect the true PGT activity given that IVF/ICSI data submission is compulsory in some countries. Detailed data registration to the PGT database, done on a voluntary basis, is time-consuming and this may particularly form a burden for large PGT centres. Therefore, it was accepted that some centres registered partial data. In this way, by focusing on data quality and not on quantity, the data presented are reliable and still reflect the major trends in these PGT centres. The ESHRE PGT Consortium Steering Committee greatly acknowledges the effort of all centres for their contribution.
The new online platform has a specific module for follow-up of children born; however, these data were not included in the current data report. It was decided to collect these data over a longer period and report them later in a separate article.
Supplementary Material
Acknowledgements
The Steering Committee greatly acknowledges the effort of all contributing centres.
List of the centres participating in one or more data collections discussed in this report:
Argentina: Fecunditas, Dept. of Genetics and IVF, Buenos Aires; Austria: Institut für Medizinische Genetik, Kepler Universitätsklinikum, MC IV, Linz; Belgium: Brussels Free University, Centre of Medical Genetics, Brussels; Ghent University Hospital, Infertility Centre, Ghent; LIFE-URG, Leuven; Reproductive Genetics Unit—CME, UZ Leuven; Czech Republic: Institute Pronatal, Genetics, Praha; Denmark: Center for Preimplantation Genetic Testing, Aalborg University Hospital; Finland: HUS/Reproductive Medicine Unit, Helsinki; France: C.M.C.O.-SIHCUS, CECOS Alsace, Unité de Diagnostic pré-implantatoire, Service de Biologie de la Reproduction, Strasbourg; Germany: Zentrum Für Humangenetik, Humangenetisches Labor, Regensburg; Kinderwunsch centrum München, IVF Labor, Munich; Kinderwunschzentrum an der Gedächteniskirche, Berlin; Heidelberg University Hospital, Heidelberg; Centrum für Gynäkologische Endokrinologie und Reproduktionsmedizin Freiburg, CERF—Frauenärztinnen, Freiburg; Medizinisch Genetisches Zentru, MGZ München; Greece: Laboratory of Medical Genetics, National and Kapodistrian University of Athens, Choremio Research Laboratory, St Sophia Children’s Hospital; HYGEIA IVF EMBRYOGENESIS, Thessaloniki; GENESIS ATHENS CLINIC, Athens; Hungary: Versys Clinics—Human Reproduction Institute Ltd., Budapest; Ireland: Waterstone Clinic, Cork; Israel: Lis Maternity Hospital, Dept. of IVF, Tel Aviv; Italy: S.I.S.M.E.R. Bologna; Villa Mafalda, Rome; Eurofins GENOMA Group srl, Rome; Juno Genetics, Rome; Fondazione IRCCS, Ca’ Granda Ospedale, Maggiore Policlinico, Milano; Japan: St. Mother Hospital, Kitakyushu; Takeuchi Ladies Clinic/Infertility Center, Aira-shi, Kagoshima; Poland: INVICTA Fertility Clinic, Gdansk; Portugal: Genetics, Department of Pathology, Faculty of Medicine, University of Porto; Scotland: Glasgow Royal Infirmary, Assisted Conception Services, Glasgow; Spain: Women’s Health Dexeus, Barcelona; Instituto Valenciano de Infertilidad (IVI); Hospital Universitario Quironsalud Madrid; Fundacion Puigvert, Seminologia i Reproduccio, Barcelona; Sweden: Karolinska University Hospital, Department of Clinical Genetics; Sahlgrenska University Hospital, Department of Ob/Gyn, Göteborg; Taiwan: Chang Gung Memorial Hospital, Obstetrics and Gynecology; The Netherlands: PGT Working Group MUMC+, Dept. of Clinical Genetics, Maastricht; University Medical Center Groningen, IVF/Fertility Laboratory, Groningen; University Medical Center Utrecht, Utrecht; Turkey: Istanbul Memorial Hospital, ART & Reproductive Genetics Center, Istanbul; Istenhegyi Gene Diagnostic Center; Istanbul.
Contributor Information
F Spinella, Eurofins GENOMA Group srl, Molecular Genetics Laboratories, Rome, Italy.
F Bronet, IVIRMA—IVI Madrid, Madrid, Spain.
F Carvalho, Genetics—Department of Pathology, Faculty of Medicine, University of Porto, Porto, Portugal; i3s—Instituto de Investigação e Inovação em Saúde, University of Porto, Porto, Portugal.
E Coonen, Department of Clinical Genetics, GROW School for Oncology and Developmental Biology, Maastricht University Medical Centre, Maastricht, The Netherlands.
M De Rycke, Centre for Medical Genetics, UZ Brussel, Brussels, Belgium.
C Rubio, PGT-A Research, Igenomix, Valencia, Spain.
V Goossens, ESHRE Central Office, Strombeek-Bever, Belgium.
A Van Montfoort, Department of Clinical Genetics, GROW School for Oncology and Developmental Biology, Maastricht University Medical Centre, Maastricht, The Netherlands.
Data availability
All data are incorporated into the article and its Supplementary Material.
Authors’ roles
F.S., M.D.R., F.C., F.B., and C.R. drafted and edited the article. A.V.M. was responsible for raw data curation, contributed to the tables, and designed the figures. E.C. edited the article. V.G. was responsible for the data collection, contributed to the tables and figures and was responsible for final editing of the article. All authors revised and approved the final manuscript.
Funding
The study has no external funding, and all costs are covered by the European Society of Human Reproduction and Embryology.
Conflict of interest
There are no competing interests to declare.
References
- Coonen E, van Montfoort A, Carvalho F, Kokkali G, Moutou C, Rubio C, De Rycke M, Goossens V.. ESHRE PGT Consortium data collection XVI-XVIII: cycles from 2013 to 2015. Hum Reprod Open 2020;2020:hoaa043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rycke M, Belva F, Goossens V, Moutou C, SenGupta SB, Traeger-Synodinos J, Coonen E.. ESHRE PGD Consortium data collection XIII: cycles from January to December 2010 with pregnancy follow-up to October 2011. Hum Reprod 2015;30:1763–1789. [DOI] [PubMed] [Google Scholar]
- De Rycke M, Goossens V, Kokkali G, Meijer-Hoogeveen M, Coonen E, Moutou C.. ESHRE PGD Consortium data collection XIV-XV: cycles from January 2011 to December 2012 with pregnancy follow-up to October 2013. Hum Reprod 2017;32:1974–1994. [DOI] [PubMed] [Google Scholar]
- ESHRE PGD Consortium Steering Committee. ESHRE Preimplantation Genetic Diagnosis Consortium: data collection III (May 2001). Hum Reprod 2002;17:233–246. [DOI] [PubMed] [Google Scholar]
- ESHRE PGT-SR/PGT-A Working Group, Coonen E, Rubio C, Christopikou D, Dimitriadou E, Gontar J, Goossens V, Maurer M, Spinella F, Vermeulen N, De Rycke M.. ESHRE PGT Consortium good practice recommendations for the detection of structural and numerical chromosomal aberrations. Hum Reprod Open 2020;2020:hoaa017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ESHRE Working Group on Chromosomal Mosaicism, De Rycke M, Capalbo A, Coonen E, Coticchio G, Fiorentino F, Goossens V, Mcheik S, Rubio C, Sermon K, Sfontouris I. et al. ESHRE survey results and good practice recommendations on mamaging chromosomal mosaicism. Hum Reprod Open 2022;4:hoac044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraedts J, Handyside A, Harper J, Liebaers I, Sermon K, Staessen C, Thornhill A, Vanderfaeillie A, Viville S; ESHRE PGD Consortium Steering Committee. ESHRE Preimplantation Genetic Diagnosis (PGD) Consortium: preliminary assessment of data from January 1997 to September 1998. Hum Reprod 1999;14:3138–3148. [DOI] [PubMed] [Google Scholar]
- Geraedts J, Handyside A, Harper J, Liebaers I, Sermon K, Staessen C, Thornhill A, Viville S, Wilton L; European Society of Human Reproduction and Embryology Preimplantation Genetic Diagnosis Consortium Steering Committee. ESHRE preimplantation genetic diagnosis (PGD) consortium: data collection II (May 2000). Hum Reprod 2000;15:2673–2683. [DOI] [PubMed] [Google Scholar]
- Goossens V, Harton G, Moutou C, Scriven PN, Traeger-Synodinos J, Sermon K, Harper JC; European Society of Human Reproduction and Embryology PGD Consortium. ESHRE PGD Consortium data collection VIII: cycles from January to December 2005 with pregnancy follow-up to October 2006. Hum Reprod 2008;23:2629–2645. [DOI] [PubMed] [Google Scholar]
- Goossens V, Harton G, Moutou C, Traeger-Synodinos J, Van Rij M, Harper JC.. ESHRE PGD Consortium data collection IX: cycles from January to December 2006 with pregnancy follow-up to October 2007. Hum Reprod 2009;24:1786–1810. [DOI] [PubMed] [Google Scholar]
- Goossens V, Traeger-Synodinos J, Coonen E, De Rycke M, Moutou C, Pehlivan T, Derks-Smeets IA, Harton G.. ESHRE PGD Consortium data collection XI: cycles from January to December 2008 with pregnancy follow-up to October 2009. Hum Reprod 2012;27:1887–1911. [DOI] [PubMed] [Google Scholar]
- Greco E, Minasi MG, Fiorentino F.. Healthy babies after intrauterine transfer of mosaic aneuploid blastocysts. N Engl J Med 2015;373:2089–2090. [DOI] [PubMed] [Google Scholar]
- Harper JC, Boelaert K, Geraedts J, Harton G, Kearns WG, Moutou C, Muntjewerff N, Repping S, SenGupta S, Scriven PN. et al. ESHRE PGD Consortium data collection V: cycles from January to December 2002 with pregnancy follow-up to October 2003. Hum Reprod 2006;21:3–21. [DOI] [PubMed] [Google Scholar]
- Harper JC, Coonen E, De Rycke M, Harton G, Moutou C, Pehlivan T, Traeger-Synodinos J, Van Rij MC, Goossens V.. ESHRE PGD Consortium data collection X: cycles from January to December 2007 with pregnancy follow-up to October 2008. Hum Reprod 2010;25:2685–2707. [DOI] [PubMed] [Google Scholar]
- Harper JC, de Die-Smulders C, Goossens V, Harton G, Moutou C, Repping S, Scriven PN, SenGupta S, Traeger-Synodinos J, Van Rij MC. et al. ESHRE PGD consortium data collection VII: cycles from January to December 2004 with pregnancy follow-up to October 2005. Hum Reprod 2008;23:741–755. [DOI] [PubMed] [Google Scholar]
- Harper JC, Wilton L, Traeger-Synodinos J, Goossens V, Moutou C, SenGupta SB, Pehlivan Budak T, Renwick P, De Rycke M, Geraedts JP. et al. The ESHRE PGD Consortium: 10 years of data collection. Hum Reprod Update 2012;18:234–247. [DOI] [PubMed] [Google Scholar]
- Moutou C, Goossens V, Coonen E, De Rycke M, Kokkali G, Renwick P, SenGupta SB, Vesela K, Traeger-Synodinos J.. ESHRE PGD Consortium data collection XII: cycles from January to December 2009 with pregnancy follow-up to October 2010. Hum Reprod 2014;29:880–903. [DOI] [PubMed] [Google Scholar]
- Sermon K, Moutou C, Harper J, Geraedts J, Scriven P, Wilton L, Magli MC, Michiels A, Viville S, De Die C.. ESHRE PGD Consortium data collection IV: May-December 2001. Hum Reprod 2005;20:19–34. [DOI] [PubMed] [Google Scholar]
- Sermon KD, Michiels A, Harton G, Moutou C, Repping S, Scriven PN, SenGupta S, Traeger-Synodinos J, Vesela K, Viville S. et al. ESHRE PGD Consortium data collection VI: cycles from January to December 2003 with pregnancy follow-up to October 2004. Hum Reprod 2007;22:323–336. [DOI] [PubMed] [Google Scholar]
- Van Montfoort A, Carvalho F, Coonen E, Kokkali G, Moutou C, Rubio C, Goossens V, De Rycke M.. ESHRE PGT Consortium data collection XIX-XX: cycles from 2016 to 2017. Hum Reprod Open 2021;2021:hoab024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti M. Preimplantation genetic testing for chromosomal abnormalities: aneuploidy, mosaicism, and structural rearrangements. Genes (Basel) 2020;11:602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyns C, De Geyter C, Calhaz-Jorge C, Kupka MS, Motrenko T, Smeenk J, Bergh C, Tandler-Schneider A, Rugescu IA, Vidakovic S. et al. ART in Europe, 2017: results generated from European registries by ESHRE. Hum Reprod Open 2021;202:hoab026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegers-Hochschild F, Adamson GD, Dyer S, Racowsky C, de Mouzon J, Sokol R, Rienzi L, Sunde A, Schmidt L, Cooke ID. et al. The international glossary on infertility and fertility care, 2017. Hum Reprod 2017;32:1786–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are incorporated into the article and its Supplementary Material.