Abstract
The adeno-associated virus (AAV) is one of the most potent vectors in gene therapy. The experimental profile of this vector shows its efficiency and accepted safety, which explains its increased usage by scientists for the research and treatment of a wide range of diseases. These studies require using functional, pure, and high titers of vector particles. In fact, the current knowledge of AAV structure and genome helps improve the scalable production of AAV vectors. In this review, we summarize the latest studies on the optimization of scalable AAV production through modifying the AAV genome or biological processes inside the cell.
Keywords: AAV, AAV production, ITR, ITRs, AAP, MAAP
1. Introduction
Adeno-associated viruses (AAV) are small viruses able to infect humans and other primate species. They are non-enveloped viruses belonging to the genus Dependoparvovirus, which belongs to the family Parvoviridae. This small (20 nm) virus was first identified as an Adenovirus (Ad) preparation contaminant in 1965 by Atchison et al. Atchison, Casto [1]. Several natural AAV serotypes are already known, with AAV2 being the best characterized and most commonly used. These serotypes differ in their capsid structure, leading to differences in their tropism and immunogenicity in host organisms.
AAV has a relatively simple genome and is a non-pathogenic virus, which is why AAV vectors are promising viral vectors for gene therapy. Moreover, AAV has shown a low frequency of integration into the host genome [2,3], which indicates its safety to be used in clinical studies. Another advantage of AAV vectors is their ability to provide stable transgene expression, which lasts for over a year in some in vivo experiments. AAV has already been used to treat CNS [4] and retinal degenerative diseases, various types of muscular dystrophy, as well as heart, lung, and liver diseases. In addition to natural AAV serotypes, recombinant AAV (rAAV) serotypes are also included in pre- and clinical trials to treat different diseases, e.g., hemophilia B [[5], [6], [7]].
The first AAV-based gene therapy drug, Glybera, was for patients with lipoprotein lipase (LPL) deficiency [8]. Glybera was approved by the European Medicines Agency (EMA) in 2012, but later, in 2017, it was withdrawn from the market due to commercial failure. The current AAV-based gene therapy market has two FDA-approved AAV-based gene therapies: 1) Luxturna (voretigene neparvovec), approved in 2017 for Leber congenital amaurosis treatment, with subsequent European Commission approval in November 2018, and 2) Zolgensma, which was approved in 2019 for spinal muscular atrophy. The approval of these medications has dramatically impacted the field of AAV-based gene therapies. In addition, AAV has also been studied as a gene therapy tool for cancer treatment. The modification of AAV capsids to achieve a higher transduction efficiency toward cancer cells is one of the strategies used for cancer cells targeting using AAV vectors [9]. Another strategy is the use of suicide gene therapy methods using AAV vectors. This approach triggers cell death processes caused by the delivery of transgenes to cancer cells [10,11].
AAV vectors are also used in the non-invasive chemogenetic technology called DREADDs (Designer Receptors Exclusively Activated by Designer Drugs) technology [12,13]. This technology is widely applied in behavioral neurosciences, where AAV- delivered DREADDs modulate G Protein Coupled Receptor (GPCR) activity in vivo. This GPCR modulation helps researchers understand the links between brain activity and behavior and the pathogenesis of the neural disease. AAV-DREADDs are successfully used to perform alcohol consumption studies in mice models [14] and in vitro studies of cellular neural networks [15]. Similarly, AAV vectors are used in different optogenetic approaches to cure different retinal degenerative diseases and glaucoma; with already clinical trials of retinal gene therapies going on, all of them are AAV delivery-based [16]. Another example of AAV vector application in optogenetics is using AAV vectors for hearing restoration studies in deaf gerbils and mice models [17,18]. These applications of AAV vectors in different scientific fields emphasize the importance of AAV production optimization to cover research needs.
The increasing number of clinical and preclinical studies using AAV-based gene therapy necessitates using cost-effective methods for AAV production. Many studies have been conducted to optimize the AAV production in both upstream and downstream stages [19,20]. Upstream optimization includes plasmid modifications, using different cell lines and transfection methods. Currently, the most widely used and available method for assembling AAV particles is the transfection of adherent HEK293 cell culture. However, the difficulty in scaling and the lack of efficiency in producing viral particles, even at a high percentage of transfection [21], has become an impetus for studying other assembly systems. The mainly used today are 1) HEK293 suspension culture [22,23], 2) SF9 insect cells with recombinant Baculovirus (rBV) expression vectors [24,25], 3) stable producer HeLa cells [26,27], and 4) platforms using replication-deficient herpes simplex virus (HSV) [28,29]. However, each platform has drawbacks and limitations regarding safety, flexibility, and versatility. For example, although suspension cultures are well scalable, not all cells receive the optimal number and ratio of plasmids during transfection, which can lead to a decrease in the proportion of full capsids. Utilization of insect cells Sf9 and rBV results in a different ratio of viral proteins than the wild type [25]. In addition, post-translational capsid modifications and viral DNA methylation in AAVs assembled in non-mammalian cells are different from AAVs assembled in HEK293 [30] and may account for treatment side effects. In the case of producer cell lines, creating new subclones for each serotype and transgene is time-consuming and challenging to manufacture. Thus, although these systems seem very promising, many optimizations will be required to overcome the existing difficulties and create a robust and universal platform. Another solution could be to modify the genome and structure of AAVs to improve production, such as increased yield, purity, and effectiveness. However, limitations still exist since the detailed biological processes of AAV are not fully understood. In our review, we focus on the most recent peer-reviewed articles from 2020 to 2023 in the field of AAV genome modifications to provide new and up-to-date information and results that could be used to achieve a better AAV production yield with increased purity, safety, and efficacy. We also mention articles from earlier years to give a complete overview of what has already been done.
2. Brief background about AAV
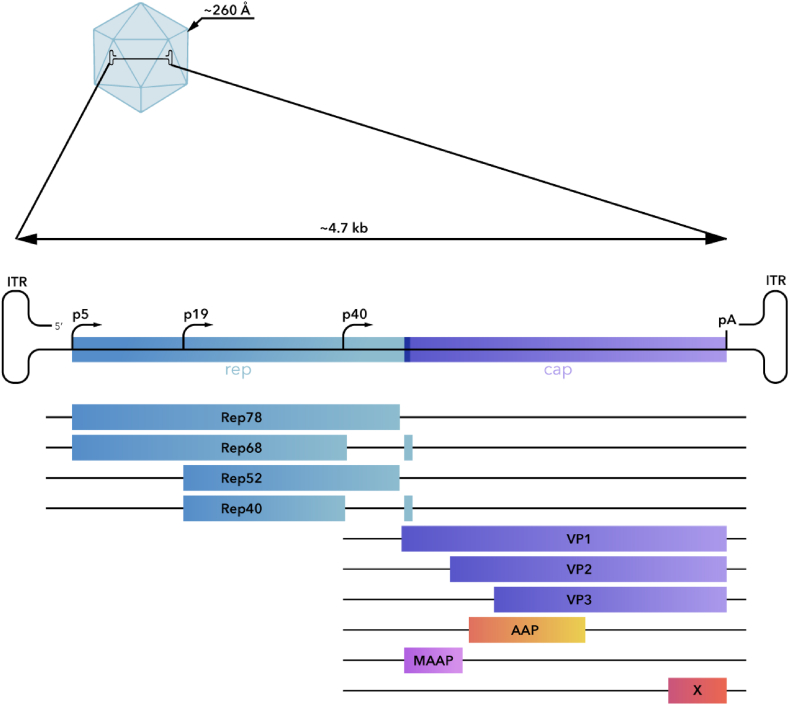
AAV particles consist of a 4.7-kb ssDNA genome packaged in the viral capsid which is ∼260A in diameter [31]. This icosahedral capsid of ∼3.8 MDa consists of 60 subunits of 3 distinct viral proteins (VPs), which vary only in their N-terminus [32] (Fig. 1). The small packaging capacity of AAV is considered one of the main limitations of using AAV vectors in gene therapy.
Fig. 1.
AAV genome flanked by ITRs, coding for Cap and Rep AAV genes.
The AAV genome is flanked by two inverted terminal repeats (ITRs), each of which is of approximately 145bp in length [33] (Fig. 2). ITRs play a primary role in AAV replication, transcription regulation, and genome packaging [34]. The AAV genome contains two main open reading frames (ORFs), under the control of 3 promoters (Fig. 1). The first ORF, located in the Cap gene, encodes for the three viral capsid proteins VP1, VP2, and VP3 (87 kDa, 72 kDa, 62 kDa, respectively) by alternative start codons, one for each protein [35]. Cap ORF also codes for the assembly activating protein (AAP), membrane-associated accessory protein (MAAP), and the X protein, which is involved in the AAV life cycle, particularly in increasing AAV2 autonomous DNA replication (no helper) in differentiating keratinocytes, its natural host tissue, and in AAV2 DNA replication in Ad5-infected 293 cells [36].
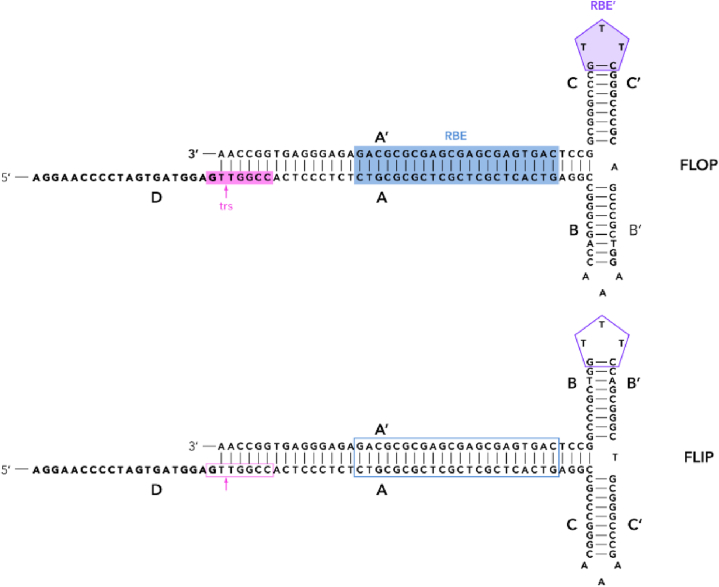
Fig. 2.
wt ITR structure.
The transcription of the second ORF, Rep ORF, is initiated at the P5 and P19 starting sites, and it results in the expression of AAV's four non-structural proteins (Rep78, Rep68, Rep52, and Rep 40) using the alternative splicing of the transcripts. The large Reps (Rep78 and Rep68) have been shown to possess site-specific [5] and strand specific endonuclease activity, DNA-RNA and DNA-DNA helicase activities [[37], [38], [39], [40]], and DNA binding [39,41] functions, all of which appear to be required for viral replication, and site-specific integration into the host genome [[42], [43], [44], [45]]. Small Reps (Rep52 and Rep40) are necessary for genome packaging. Although these Rep proteins are important for replication, they are not sufficient to generate a productive infection [5]. AAV requires coinfection with a helper virus, either adenovirus (Ad) or herpes simplex virus (HSV), for productive infection [46].
The first step in the AAV infection process is binding to cell surface receptors. AAV2, AAV3, and AAV6 bind to Heparan sulfate proteoglycan (HSPG) [47]. Other cell surface receptors for AAV binding are sialic acid for AAV1, AAV4, AAV5, and AAV6, and N-terminal galactose for AAV9 [48,49], while AAVrh10 binds to cell surface glycans with terminal galactose [50]. The binding of AAV to cell receptors is initiated by binding to attachment proteins on the cell surface, of which KIAA0319L (named AAVR) is well known [51]. Different types of attachment proteins and receptors are associated with different serotypes, and this diversity in receptor binding between serotypes defines the different tropisms of different serotypes. Although the available data about the interaction between all AAV serotypes, cell receptors, and proteins are not abundant, these data can be used for optimizing the production cell line of specific AAV serotypes. It may be helpful to conduct the AAV production process of a specific serotype with cell lines unfavorable for the serotype; with this, we can reduce the re-transduction of newly produced vectors into the manufacturing cells, which might lead to higher virus titer. The next step in AAV infection is endocytosis toward the nucleus. Different AAV endocytosis pathways were defined. For example, AAV2 infection has all the hallmarks of CLIC/GEEC endocytosis, which is a clathrin-independent endocytic pathway [52], while AAV5 infection pathway predominantly by clathrin-coated vesicles toward the Golgi apparatus [53]. AAV endosomal escape could be mediated by the ph drop inside this compartment, which leads to the exposure of the hidden phospholipase A2 (PLA2) domain located in the unique N terminus of capsid protein VP1 (VP1u) [3]. After that, AAV particles enter the nuclear pore complex [54], and the genome release happens during nuclear entry or within the nucleus [3,55] and localized around the nuclei co-localizing with euchromatin, where the active transcription and DNA repair occur [54]. For wild-type AAVs, the genome replication starts if a helper virus is present. In fact, this replication is mediated by the AAV-encoded Rep proteins, helper virus-encoded proteins, the host cell replication proteins RPA, RFC, PCNA, and DNA polymerase delta. [54]; and AAV ITRs serve as the origin of transcription to start the conversion of ssAAV into dsAAV. This is a rate-limiting step during AAV transduction, and that is why the use of self-complementary AAV (scAAV) genomes can bypass the synthesis of the second strand [3,56] and simultaneously escape ssDNA degradation, which enables the rapid onset of transgene expression by scAAV vectors [56]. However, due to robust gene expression, transgene products delivered via scAAV elicit a more robust immune response than transgenes delivered via a single-stranded AAV vector [57]. After uncoating, the single-stranded genomes convert to double-stranded multimeric circular concatemeric episomal forms that persist long-term in post-mitotic cells [54]. Notably, a short period of vector genome instability after dsDNA conversion leads to a significant loss of gene expression. Losses at each of these steps contribute to the overall efficiency of the vector in terms of the dose of VGP required to achieve each transduction event. The detailed mechanisms of AAV uncoating and trafficking toward the nucleolus stay of interest since these steps can affect the AAV new particle synthesis and the efficiency of viral genome packaging. That is why understanding these underlying mechanisms is essential, especially in AAV vector production protocols and AAV tropism studies in vivo.
3. Modification of the AAV genome and structure for scalable, optimized AAV production
3.1. Viral titer, transgene expression and encapsidation
Testing AAV-based gene therapies during the preclinical and clinical phases requires large-scale production with significantly higher titers of vector particles than for in vitro experiments. Currently, AAV yield is increasing by using various cell lines, assembly systems, transfection types, and downstream stage optimization. However, several other approaches based on the modification of the rAAV genome or structure can be used to obtain a higher titer of pure and functional AAV viral particles. Examples of these approaches are discussed in the following sections.
3.1.1. ITRs structure modifications
Sequence changes in ITRs can significantly affect the replication and encapsidation of AAV, which leads to more efficient production of viral particles or improved vector variants. Invention of a self-complementary AAV (scAAV) [58], which is a crucial component of a treatment for spinal muscular atrophy named Zolgensma [59], was made by ITR sequence modification. This structure is obtained by removing the trs from one of the ITRs (Table 1), thereby preventing Rep68/78 cleavage, resulting in duplication of vector genomes linked by the altered internal ITR [58]. Delivery of a duplicated genome to the nucleus promotes self-annealing with the formation of a double-stranded DNA structure without the need to complete the second strand. Thus, transcription of the gene of interest and, as a result, the expression of the encoded protein, starts sooner and sustains at higher levels [60].
Table 1.
ITR modifications and their effects on AAV production. Gray letters, nucleotides deleted in the ITR structure. Red letters, nucleotides modified in the ITR sequence. Changes are relative to wild-type ITR.
| Modifications |
Effects |
|||||
|---|---|---|---|---|---|---|
| Name | Scheme | Titer | Transduction efficiency | Transgene expression | Genome encapsidation | Contaminants |
| ITRΔtrs (scAAV) | 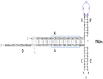 |
– | An increase in vitro from 5- to 140-fold | A higher level | – | – |
| ITRΔBC | 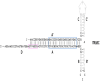 |
A reduction by 75% | – | An increase in vitro and in vivo | Similar levels as wtITR | – |
| CpG-free ITR | 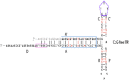 |
A 3-fold reduction | – | Similar levels as wtITR | Similar levels as wtITR | – |
| L-AD/L-A’D’ | 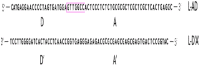 |
A 3-fold reduction | 70% of wt ITR in vitro | – | – | A lower level of contaminating helper and packaging plasmid backbones |
Another approach to modifying the ITR sequence is to remove or mutate non-essential ITR structural elements. Most often, attempts to alter the ITR sequence led to disruption of replication and/or encapsidation of viral genome [58,[61], [62], [63], [64]]. However, some deletions in ITR sequences, or even the AAV genomes containing only one ITR structure, do not affect the ability to package the AAV genome [[65], [66], [67]]. In 2017, Zhou et al. continued to study the effect of ITR region deletion on rAAV packaging and expression. Suggesting that the BB’ and CC’ regions may not be essential for rAAV replication and encapsidation [68,69], a modified ITR (ITRΔBC rAAV) [64] (Table 1) was created. It was shown that deletions of these regions in two ITRs did not affect encapsidation, but reduced rAAV titer by 75%. The presence of the Rep binding element (RBE) and trs in the AA’ region recognizable by Rep68/78 [[70], [71], [72]] explains the preserved packaging of rAAV. The reason for the reduced titer of ITRΔBC rAAV may be related to absence of RBE’, which is also necessary for interaction of large Rep proteins, leading to unwinding of ITRs and formation of a “stem-loop'' structure on the trs [63,73]. The absence of RBE’ in rAAV plasmids lead to a lower replication rate compared to wild type [74], which is consistent with an 8-fold decrease in viral genomic DNA replication of ITRΔBC rAAV. Therefore, it could be a cause of reduced viral titer. Surprisingly, transgene expression of the ITRΔBC rAAV system was higher than wild type, both in vitro and in vivo [64]. It was shown that DNA molecules with palindromic terminal repeats constrained in a T-shaped hairpin conformation at the ends are liable to a loss of gene expression mediated by ataxia telangiectasia mutated (ATM) [75]. The deletion of the BB’ and CC’ regions lead to replacing a transcriptionally silent T-shaped hairpin [75] with a plain U-shaped hairpin unaffected by ATM-dependent silencing [75,76]. Therefore, the U-shaped HP ends of ITRΔBC that remained intact by ATM may cause increased expression of rAAV, which is consistent with reported rAAV transduction enhancement due to loss of ATM function [77]. In addition, the Mre11 (MRN) complex, which also recognizes and interacts with AAV ITRs during AAV infection [78,79], limits AAV transduction. In the presence of MRN inhibitor a change in transduction efficiency was observed, always lower with ITRΔBC rAAV, in comparison to rAAV with a wt ITR, suggesting that deletion of the BB’ and CC’ regions decreases the inhibition effect of the MRN complex on AAV expression.
The most recent modification of the ITR sequence was based on the similarity between ITR and promoter structures. In 2020, Earley and colleagues discovered multiple transcription start sites (TSS) primarily concentrated in a 40 bp region that contained the RBE [80]. TSS presence in the ITR sequence, in addition to high content of cytosine and guanine (64–70%), and high CpG dinucleotide frequency (83–95%) resemble transcriptionally active CpG islands (CGI), which are the most common type of promoters in vertebrates [[81], [82], [83], [84]]. It can be assumed that ITRs act as CGI type promoters and possibly play a role in the life cycle of wild-type AAV. In addition, unmethylated CGIs are immunostimulants [85], the elimination of which can lead to a decrease in the immune response to gene therapy [[86], [87], [88], [89]]. In a recent study, Pan lab found that ITRs completely free of CpG motifs (Table 1) reduce vector yield by about 3-fold, but contribute to the same genome encapsidation level as wild type ITRs [90]. The lower titer of vectors without CGIs can be explained by reduction of replication of the vector genome, which is predictable, as CGIs are usually the origin of replication [[91], [92], [93]]. Surprisingly, wild-type ITR vectors and CpG-free ITR vectors showed similar levels of transgene expression and copy number of the vector genome in vivo [90].
3.1.2. MAAP modification
In 2021, Galibert et al. examined the biology of MAAP2 in AAV production in more detail. It turned out that truncation and inactivation of MAAP led to higher levels of expression of Rep and AAP proteins. The presence of full-length MAAP demonstrated a specific degradation product of VP proteins. The absence of at least the last 10 amino acids at the C-terminus of the MAAP protein reduced the degradation of the capsid and increased the expression of capsid proteins, while other truncated forms of MAAP completely prevented the degradation of the AAV2 capsid, which led to a 3.5-fold increase in assembled capsids for some variants. It can be assumed that MAAP somehow affects the processes of capsid degradation and the stability of VPs [94]. It also can be assumed that, since truncated forms or inactivated MAAP leads to increased expression of the AAP, this protein directly leads to the prevention of degradation of VPs and an increase in their stability.
3.1.3. The role of AAP
In 2010, Assembly Activating Protein (AAP) was identified in the AAV2 genome [95]. It is one of the Cap gene proteins, resulting from an additional nested, alternative ORF (ORF2) and translation initiation at a nonconventional translation starting site. This study showed that this 23 KDa protein plays a positive role in co-transporting newly synthesized VPs to the nucleolus, where it is located, and plays a positive role in capsid assembly.
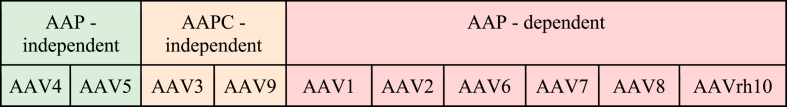
Although the study of AAP2 demonstrated that this protein is essential for capsid assembly, such a trend is only occasionally observed. Analysis of different AAV serotypes showed that the AAP requirement varies significantly depending on the virus serotype. In 2017, Grosse et al. and Early et al. demonstrated that the absence of AAP decreased the titers of viral particles from about 2.5 to 3000 times compared to the wild type [96,97], These studies revealed the independence of AAV4, AAV5, and AAV11 from AAP in capsid assembly. Further research in 2018 suggested that serotype dependence on AAP fell into three categories: (i) AAP-independent, (ii) AAPC - independent, or (iii) AAP-dependent [98] (Table 2).
Table 2.
Dependence of AAV serotype on AAP. Green boxes indicate AAP-independent serotypes (complete absence of AAP resulted in » 1% of WT titer). Yellow boxes indicate AAPC-independent serotypes (absence of the C-terminal two-thirds of AAP resulted in >10% of WT titer). Red boxes indicate AAP-dependent serotypes (complete absence of AAP resulted in ∼1% of WT titer) [98].
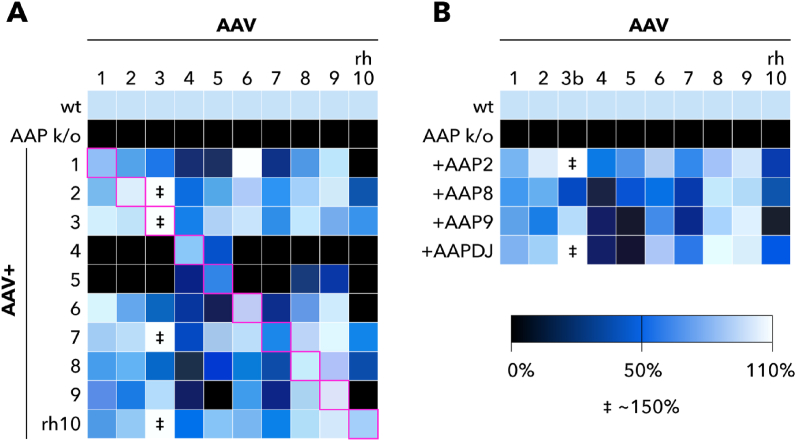
In 2017, Grosse et al. demonstrated that production of functional vector particles, which was determined as the numbers of GFP-expressing cells after transduction, in the absence of AAP could be restored by co-expression of trans AAP from different serotypes [96] (Fig. 3A). Interestingly, AAV3 production increased by about 1.5 times with the co-expression of several AAP serotypes, which indicates a direct AAV3 assembly dependency on AAP preservation. Additionally, AAP4 and AAP5 more efficiently restore the production of AAV4 and AVV5, likely due to the significant difference in capsid and AAP sequences of these serotypes from all others [96,99]. Interestingly, AAP overexpression in AAP-expressing AAV variants does not increase the yields of functional particles, whether wild type or chimeric variants [96,100]. Therefore, if artificial AAV variants do not produce functional particles or have a low yield titer, this is not necessarily associated with alerted AAP. Instead, this may reflect the presence of inherent deficiencies in the capsid for assembly into virions that cannot be overcome with AAP [100].This is consistent with another experiment, where co-expression of recombinant AAP from AAVDJ, a chimeric capsid created by DNA shuffling of eight different serotypes (primarily AAV2, 8, and 9) was performed [100] (Fig. 3B). It was revealed that AAPDJ rescued the production of these three AAVs without AAP to an almost similar degree as each AAP itself. In addition, it successfully worked with other serotypes, except for AAV4 and AAV5. Moreover, a subsequent study of 60 different chimeric AAP demonstrated the retention of at least some of their activity to rescue AAP knock-out mutants. Therefore, generation of chimeric AAP during chimeric AAV production is most likely not a limiting factor.
Fig. 3.
Trans-complementation of 10 AAP-depleted helper plasmids with 10 AAP variants [96,100].
However, in 2018, Viney and colleagues studied AAV chimeric capsids, in which the VP3 sequence was taken from AAV6, and VP1 and VP2 from different serotypes. They found that, although these chimeras have, on average, 3 logs lower viral titer, they show an enhanced transducing ability, compared to wild-type AAV6 (from 1.5 to 2 logs). Assuming that this could be caused by AAP knock-out due to a change in its sequence, the key amino acid sequence of the wild-type AAP6 N-terminus (amino acids from 13 to 27) was restored. As a result, it led to complete recovery of the virus titer and reduction of transduction efficiency to the levels of wild type AAV6. In addition, chimeras with an altered AAP genome demonstrate reduced expression of all three capsid proteins [101]. Although it is impossible to deny the essential role of AAP for the stable production of the capsid, whether additional expression of AAP will affect the assembly and functionality of chimeric capsids needs to be further studied. Additional studies of AAPs of various serotypes are required to understand the biology of AAV assembly better and further apply this knowledge to produce recombinant viral particles.
3.2. Modification of AAV production dynamics; full vs empty capsids
3.2.1. Dynamics of AAV replication and packaging
Understanding biological processes during AAV production helps optimize the conditions and materials used in production for better yield. In 2021, the first mechanistic-based kinetic model of rAAV production was presented by Nguyen et al. They made the model on HEK293 cells. According to this model and literature data, AAV DNA replication in cells begins about 12 h post-transfection (hpt), much later than capsid assembly. The replication peaks at 24 hpt and reaches a plateau up to 48 hpt. In contrast, capsid formation begins in the first hours after transfection and reduces by 24 hpt. As a result, capsids, which are made first, leave the nucleus without DNA and the packaging of AAV genome is limited due to the lack of available capsids. That is why changes in DNA replication kinetics at initial points could impact the production of complete virions. To facilitate DNA replication along with capsid formation, the Rep and Cap proteins may be split into two different plasmids to ensure earlier gene expression of the Rep genes. In 2016, Emmerling and colleagues described a system with split and modified Rep and Cap genes. They have shown that this system allowed an increase of the titer by over two times compared to standard plasmids. Moreover, a shortened helper plasmid (10 236 bp instead of 15 263 bp) significantly increased vector yield (2,7 × 10^5 VG/cell), which was explained by increase in transfection efficiency and copy number of required genes per given amount of transfected DNA in the case of the short plasmid [102].
3.2.2. Rep genes modifications
The Rep gene of AAV2 is widely used for manufacturing different AAV serotypes with varied efficiency. Using the Rep gene corresponding to the produced serotype instead of Rep2 may increase the number of complete viral particles. The utilization of AAV1, 6, or 8 Rep genes demonstrated low or absent VP expression and reduced protein expression of spliced Rep proteins, Rep68 and Rep40, compared to Rep2 construction, which could be explained by the different activity of the Cap gene promoter p40, located at the 3′ end of the Rep gene, or by some mechanisms at the stage of mRNA splicing. To restore the expression of capsid and Rep proteins, the 3′-end of the AAV2 Rep (part of the 3′-end responsible for the zinc finger domain) was introduced into AAV1 or AAV6. Surprisingly, this strategy did not work with AAV8, where expression of the VPs was restored using the 3′-end region of AAV1 Rep encoding the DNA-binding domain.
Based on these results, Mckenna and colleagues created chimeric Rep genes consisting of AAV1, AAV2, AAV6, and AAV8 domains with a common 3′-end of the AAV2 Rep gene. These chimeric combinations increased the proportion of total capsids by 2–4 times for several tested serotypes [103]. Thus, using chimeric Rep genes instead of the standard AAV2 Rep for producing viral particles may be a promising approach to reducing the proportion of empty virions, thereby increasing transduction efficiency.
3.2.3. The role of MAAPMAAP
As a viral egress factor, can also play a role in AAV production dynamics. In a study published in 2021, Elmore and colleagues conducted a series of experiments that demonstrated that reduced expression of MAAP8 resulted in a 2-day delay in virus secretion with minimal change in total virus titer. This influence of MAAP expression on AAV dynamics affects the harvest time point of assembled AAV vectors from cell lysate and medium (supernatant) [104].
3.3. Reducing AAV production contaminants
3.3.1. P5-promoter modification
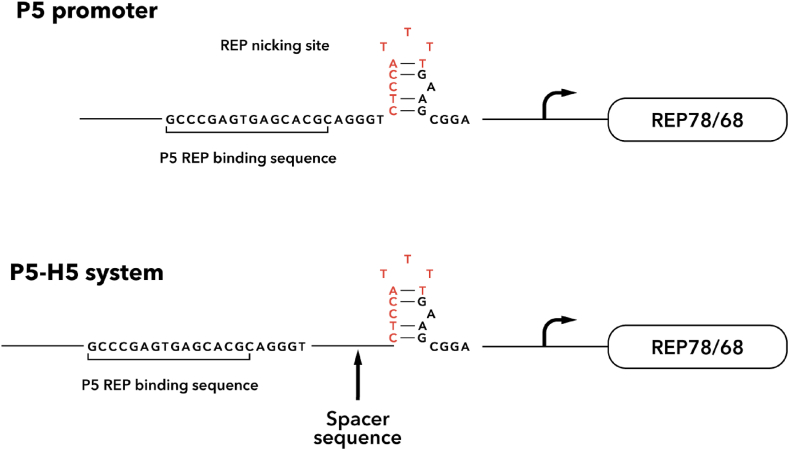
In addition to the role of the Rep gene modification in reducing the proportion of empty capsids, some modifications can also help remove non-viral DNA impurities in the assembled virions. In 2022, Brimble and colleagues [105] identified packaged contaminant sequences from upstream of the AAV P5 promoter on the Rep-Cap production plasmid into rAAV vectors. In this study, researchers demonstrated that DNA contaminants associated with the P5 promoter can be transcribed and efficiently translated in vitro and in vivo. The contaminants could also establish antigen-specific immunity of CD8+ T cells in mice. Thus, these contaminants negatively affect the efficiency of AAV vector usage in gene therapy. A possible reason for the packaging of these contaminants is the proximity of a Rep binding site (RBS) and a Rep nicking site of the P5 promoter, which may be the cause for packaging sequences of impurities adjacent to P5. (Fig. 4). Although the in vivo activity of the upstream P5 promoter was shown in liver cells, there is still a need to study such activity in other tissues. This is especially important with the ongoing engineering of new AAV capsids, where the transduction efficiency of a serotype toward an organ could be so high that these contaminants could cause a severe negative effect.
Fig. 4.
P5 promoter structure and modified promoter structure.
To overcome this problem, a modified promoter was obtained by physically separating the Rep nicking site from the RBS in P5 by spacer sequences of about 5bp or 100bp. In the same study, the system of separating the Rep nicking site from RBS was tested for its efficiency in large-scale AAV production. The results showed that this system led to lower contaminants with the same viral titer and that these vectors were potent when injected in mice at the same level as vectors produced by the conventional P5 plasmid.
Thus, overall, this system provided a decrease in Rep-Cap plasmid contamination, regardless of the serotype and transgene used, while maintaining the virus titer [105]. Therefore, the Rep gene modifications could significantly improve the quality of the produced viral vectors [106].
3.3.2. ITR modification
Using a PCR-amplified transgene with one copy of the AD sequence (Table 1) instead of the vector plasmid is also efficient for packaging rAAV, which allows for longer transgenes (∼230 nucleotides), and also reduces the amount of DNA contaminants in viral particles [[107], [108], [109]]. The packaging efficiency of such rAAVs (rAAV-L-AD/DA) became higher than rAAV-pAD, and also contaminating helper and packaging plasmid backbones were found at deficient levels, which means that L-AD does not recombine with plasmids to form chimeric products. The rAAV-L-AD provided in vitro transduction efficiency by ∼70% (10^5 VG/cell), while the transduction efficiency of rAAV-pAD was negligible because viral particles with non-vector genomes and empty particles compete for receptor binding with full particles, inhibiting its transduction efficiency [110].
3.3.3. MAAP modification for less contaminants
The effect of MAAP on the packaging of production plasmids was also found. At 72 h post-transfection, wild-type AAV2 was observed to be contaminated with the antibiotic resistance gene at a level of 3.5%. Lack of MAAP expression resulted in contamination reaching 47%, compared to the AAV genome. Moreover, in the absence of MAAP, most of the contaminating DNA comes from the AAV2 genomic plasmid. MAAP may be responsible for recognizing the AAV genome by the D-sequence of AAV ITR, which is not involved in forming the Holliday structure. Only the stable truncated form of MAAP at the C-terminus showed levels of contamination similar to that of the wild type. In addition, this variant contributed to an increased level of viral particles containing the AAV genome [94]. Thus, MAAP plays a significant role in the biology of AAV and the production of viral particles, thereby representing a promising object for research and optimization of the assembly process.
4. Conclusion
The fact that AAVs have different and wild range tropism to organs and tissues makes them applicable in studies relating to different diseases. AAV vectors are an excellent tool for the infection of many animal models, including rodents, rabbits, dogs, minipigs, and non-human primates. These models enable the researchers to find the optimal animal model for their studies and help to test different gene therapy methods, vector efficiency and safety, the lifespan of a gene therapy trial, the immunogenicity of the delivered vector and transgene, and possible side effects after a gene therapy procedure [111]. It is vital to mention that for some AAV serotypes, the tropism varies between species, making it challenging to transfer the therapy from pre-clinical to clinical trials. The increased interest in using AAV as a platform for disease treatment makes it essential to improve vectors to achieve higher transduction efficiency, more specific tropism, less immune response, and more stable transgene expression. As previously mentioned in this review, introducing specific modifications to the AAV genome can help improve the production yield with higher titer and fewer contaminants. These modifications can also improve the safety of the gene therapy, as increasing the full/empty capsid ratio will reduce the immunogenicity of the vector dose and make it more concentrated and, thus, more functional. All mentioned modifications can be effectively used in in vivo studies when a qualified scalable production of AAV is established. In the last decade, a significant number of discoveries have been made regarding the AAV genome. However, the mechanisms and fundamental biology of AAV, with the consequences on the productivity and integrity of the vector after genome modifications, still need to be fully understood. The modifications of the AAV genome is a part of an extensive range of changes to be applied during the AAV production cascade, from the upstream to downstream stages. A further understanding of the recently discovered MAAP and X proteins, the chimeric structures of the REP and AAP, and AAV biological processes during particle assembly will help in more effective and faster optimization of AAV vectors production to reach the ultimate goal of finding the most efficient gene therapy vectors for different diseases.
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Funding statement
Professor Pavel Volchkov was supported by Ministry of Science and Higher Education of the Russian Federation (agreement # 075-03-2022-107/10).
Data availability statement
There is no additional data available for this study
Declaration of interest’s statement
The authors declare no conflict of interest.
References
- 1.Atchison R.W., Casto B.C., Hammon W.M. Adenovirus-associated defective virus particles. Science. 1965;149:754–756. doi: 10.1126/science.149.3685.754. [DOI] [PubMed] [Google Scholar]
- 2.Berns K.I., Muzyczka N. AAV: an overview of unanswered questions. Hum. Gene Ther. 2017;28:308–313. doi: 10.1089/hum.2017.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dhungel B.P., Bailey C.G., Rasko J.E.J. Journey to the center of the cell: tracing the path of AAV transduction. Trends Mol. Med. 2021;27:172–184. doi: 10.1016/j.molmed.2020.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Lykken E.A., et al. Recent progress and considerations for AAV gene therapies targeting the central nervous system. J. Neurodev. Disord. 2018;10:16. doi: 10.1186/s11689-018-9234-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drouin L.M., Agbandje-McKenna M. Adeno-associated virus structural biology as a tool in vector development. Future Virol. 2013;8:1183–1199. doi: 10.2217/fvl.13.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manno C.S., et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 7.Nathwani A.C., et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaudet D., Méthot J., Kastelein J. Gene therapy for lipoprotein lipase deficiency. Curr. Opin. Lipidol. 2012;23:310–320. doi: 10.1097/MOL.0b013e3283555a7e. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X., Xu W. Aminopeptidase N (APN/CD13) as a target for anti-cancer agent design. Curr. Med. Chem. 2008;15:2850–2865. doi: 10.2174/092986708786242840. [DOI] [PubMed] [Google Scholar]
- 10.Santiago-Ortiz J.L., Schaffer D.V. Adeno-associated virus (AAV) vectors in cancer gene therapy. J. Contr. Release. 2016;240:287–301. doi: 10.1016/j.jconrel.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zi-Bo L.I., et al. Recombinant AAV-mediated HSVtk gene transfer with direct intratumoral injections and Tet-On regulation for implanted human breast cancer. BMC Cancer. 2006;6:66. doi: 10.1186/1471-2407-6-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonaventura J., et al. High-potency ligands for DREADD imaging and activation in rodents and monkeys. Nat. Commun. 2019;10:4627. doi: 10.1038/s41467-019-12236-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menard C., Russo S.J. Non-invasive chemogenetics. Nat. Biomed. Eng. 2018;2:467–468. doi: 10.1038/s41551-018-0269-z. [DOI] [PubMed] [Google Scholar]
- 14.Belmer A., et al. Neural serotonergic circuits for controlling long-term voluntary alcohol consumption in mice. Mol. Psychiatr. 2022;27:4599–4610. doi: 10.1038/s41380-022-01789-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weir J.S., et al. bioRxiv; 2023. Selective Inhibition of Excitatory Synaptic Transmission Alters the Emergent Bursting Dynamics of in Vitro Neural Networks; p. 2022. 09.08.507095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prosseda P.P., et al. Advances in ophthalmic optogenetics: approaches and applications. Biomolecules. 2022;12:269. doi: 10.3390/biom12020269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bali B., et al. Analyzing efficacy, stability, and safety of AAV-mediated optogenetic hearing restoration in mice. Life Sci. Alliance. 2022;5 doi: 10.26508/lsa.202101338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wrobel C., et al. Optogenetic stimulation of cochlear neurons activates the auditory pathway and restores auditory-driven behavior in deaf adult gerbils. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aao0540. eaao0540. [DOI] [PubMed] [Google Scholar]
- 19.Kotin R.M. Large-scale recombinant adeno-associated virus production. Hum. Mol. Genet. 2011;20:R2–R6. doi: 10.1093/hmg/ddr141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams B., Bak H., Tustian A.D. Moving from the bench towards a large scale, industrial platform process for adeno-associated viral vector purification. Biotechnol. Bioeng. 2020;117:3199–3211. doi: 10.1002/bit.27472. [DOI] [PubMed] [Google Scholar]
- 21.Dash S., et al. 2022. Only A Small Fraction Of Cells Produce Assembled Capsids During Transfection-Based Manufacturing Of Adeno-Associated Virus Vectors; pp. 1685–1690. 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park J.Y., et al. Scalable production of adeno-associated virus type 2 vectors via suspension transfection. Biotechnol. Bioeng. 2006;94:416–430. doi: 10.1002/bit.20776. [DOI] [PubMed] [Google Scholar]
- 23.Grieger J.C., Soltys S.M., Samulski R.J. Production of recombinant adeno-associated virus vectors using suspension HEK293 cells and continuous harvest of vector from the culture media for GMP FIX and FLT1 clinical vector. Mol. Ther. 2016;24:287–297. doi: 10.1038/mt.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith R.H., Levy J.R., Kotin R.M. A simplified baculovirus-AAV expression vector system coupled with one-step affinity purification yields high-titer rAAV stocks from insect cells. Mol. Ther. 2009;17:1888–1896. doi: 10.1038/mt.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mietzsch M., et al. OneBac 2.0: Sf9 cell lines for production of AAV1, AAV2, and AAV8 vectors with minimal encapsidation of foreign DNA. Hum. Gene Ther. Methods. 2017;28:15–22. doi: 10.1089/hgtb.2016.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiao C., et al. Feasibility of generating adeno-associated virus packaging cell lines containing inducible adenovirus helper genes. J. Virol. 2002;76:1904–1913. doi: 10.1128/JVI.76.4.1904-1913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark K.R., et al. Cell lines for the production of recombinant adeno-associated virus. Hum. Gene Ther. 1995;6:1329–1341. doi: 10.1089/hum.1995.6.10-1329. [DOI] [PubMed] [Google Scholar]
- 28.Adamson-Small L., et al. A scalable method for the production of high-titer and high-quality adeno-associated type 9 vectors using the HSV platform. Mol. Ther. Meth. Clin. Dev. 2016;3 doi: 10.1038/mtm.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clément N., Knop D.R., Byrne B.J. Large-scale adeno-associated viral vector production using a herpesvirus-based system enables manufacturing for clinical studies. Hum. Gene Ther. 2009;20:796–806. doi: 10.1089/hum.2009.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rumachik N.G., et al. Methods matter: standard production platforms for recombinant AAV produce chemically and functionally distinct vectors. Mol. Ther. Meth. Clin. Develop. 2020;18:98–118. doi: 10.1016/j.omtm.2020.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chapman M., Agbandje-Mckenna M. 2005. Atomic Structure of Viral Particles; pp. 107–123. [Google Scholar]
- 32.Srivastava A., Lusby E.W., Berns K.I. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spear I.S., et al. Evidence for two nucleotide sequence orientations within the terminal repetition of adeno-associated virus DNA. J. Virol. 1977;24:627–634. doi: 10.1128/jvi.24.2.627-634.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiorini J.A., et al. In: Adeno-Associated Virus (AAV) Vectors in Gene Therapy. Berns K.I., Giraud C., editors. Springer Berlin Heidelberg; Berlin, Heidelberg: 1996. The roles of AAV rep proteins in gene expression and targeted integration; pp. 25–33. [Google Scholar]
- 35.Rose J.A., et al. Structural proteins of adenovirus-associated viruses. J. Virol. 1971;8:766–770. doi: 10.1128/jvi.8.5.766-770.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao M., You H., Hermonat P.L. The X gene of adeno-associated virus 2 (AAV2) is involved in viral DNA replication. PLoS One. 2014;9 doi: 10.1371/journal.pone.0104596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young S.M., Jr., et al. Roles of adeno-associated virus Rep protein and human chromosome 19 in site-specific recombination. J. Virol. 2000;74:3953–3966. doi: 10.1128/jvi.74.9.3953-3966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Im D.S., Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 39.Im D.S., Muzyczka N. Partial purification of adeno-associated virus Rep78, Rep52, and Rep40 and their biochemical characterization. J. Virol. 1992;66:1119–1128. doi: 10.1128/jvi.66.2.1119-1128.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wonderling R.S., Kyöstiö S.R., Owens R.A. A maltose-binding protein/adeno-associated virus Rep68 fusion protein has DNA-RNA helicase and ATPase activities. J. Virol. 1995;69:3542–3548. doi: 10.1128/jvi.69.6.3542-3548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Im D.S., Muzyczka N. Factors that bind to adeno-associated virus terminal repeats. J. Virol. 1989;63:3095–3104. doi: 10.1128/jvi.63.7.3095-3104.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balagúe C., Kalla M., Zhang W.W. Adeno-associated virus Rep78 protein and terminal repeats enhance integration of DNA sequences into the cellular genome. J. Virol. 1997;71:3299–3306. doi: 10.1128/jvi.71.4.3299-3306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLaughlin S.K., et al. Adeno-associated virus general transduction vectors: analysis of proviral structures. J. Virol. 1988;62:1963–1973. doi: 10.1128/jvi.62.6.1963-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shelling A.N., Smith M.G. Targeted integration of transfected and infected adeno-associated virus vectors containing the neomycin resistance gene. Gene Ther. 1994;1:165–169. [PubMed] [Google Scholar]
- 45.Surosky R.T., et al. Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J. Virol. 1997;71:7951–7959. doi: 10.1128/jvi.71.10.7951-7959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pereira D.J., McCarty D.M., Muzyczka N. The adeno-associated virus (AAV) Rep protein acts as both a repressor and an activator to regulate AAV transcription during a productive infection. J. Virol. 1997;71:1079–1088. doi: 10.1128/jvi.71.2.1079-1088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Summerford C., Samulski R.J. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bell C.L., et al. The AAV9 receptor and its modification to improve in vivo lung gene transfer in mice. J. Clin. Invest. 2011;121:2427–2435. doi: 10.1172/JCI57367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen S., et al. Terminal N-linked galactose is the primary receptor for adeno-associated virus 9. J. Biol. Chem. 2011;286:13532–13540. doi: 10.1074/jbc.M110.210922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mietzsch M., et al. Structural study of Aavrh.10 receptor and antibody interactions. J. Virol. 2021;95:e01249. doi: 10.1128/JVI.01249-21. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pillay S., et al. An essential receptor for adeno-associated virus infection. Nature. 2016;530:108–112. doi: 10.1038/nature16465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nonnenmacher M., Weber T. Adeno-associated virus 2 infection requires endocytosis through the CLIC/GEEC pathway. Cell Host Microbe. 2011;10:563–576. doi: 10.1016/j.chom.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bantel-Schaal U., Hub B., Kartenbeck J. Endocytosis of adeno-associated virus type 5 leads to accumulation of virus particles in the Golgi compartment. J. Virol. 2002;76:2340–2349. doi: 10.1128/jvi.76.5.2340-2349.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Büning H., et al. Engineering the AAV capsid to optimize vector-host-interactions. Curr. Opin. Pharmacol. 2015;24:94–104. doi: 10.1016/j.coph.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 55.Bijlani S., et al. The role of recombinant AAV in precise genome editing. Front. Genome Editing. 2022;3 doi: 10.3389/fgeed.2021.799722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horowitz E.D., et al. Biophysical and ultrastructural characterization of adeno-associated virus capsid uncoating and genome release. J. Virol. 2013;87:2994–3002. doi: 10.1128/JVI.03017-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu T., et al. Self-complementary AAVs induce more potent transgene product-specific immune responses compared to a single-stranded genome. Mol. Ther. 2012;20:572–579. doi: 10.1038/mt.2011.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCarty D.M., et al. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. 2003;10:2112–2118. doi: 10.1038/sj.gt.3302134. [DOI] [PubMed] [Google Scholar]
- 59.Mendell J.R., et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med. 2017;377:1713–1722. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 60.McCarty D.M., Monahan P.E., Samulski R.J. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 2001;8:1248–1254. doi: 10.1038/sj.gt.3301514. [DOI] [PubMed] [Google Scholar]
- 61.Ryan J.H., Zolotukhin S., Muzyczka N. Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J. Virol. 1996;70:1542–1553. doi: 10.1128/jvi.70.3.1542-1553.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ling C., et al. Enhanced transgene expression from recombinant single-stranded D-sequence-substituted adeno-associated virus vectors in human cell lines in vitro and in murine hepatocytes in vivo. J. Virol. 2015;89:952–961. doi: 10.1128/JVI.02581-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brister J.R., Muzyczka N. Mechanism of Rep-mediated adeno-associated virus origin nicking. J. Virol. 2000;74:7762–7771. doi: 10.1128/jvi.74.17.7762-7771.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou Q., et al. Deletion of the B-B' and C-C' regions of inverted terminal repeats reduces rAAV productivity but increases transgene expression. Sci. Rep. 2017;7:5432. doi: 10.1038/s41598-017-04054-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Savy A., et al. Impact of inverted terminal repeat integrity on rAAV8 production using the baculovirus/sf9 cells system. Hum. Gene Ther. Methods. 2017;28:277–289. doi: 10.1089/hgtb.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Samulski R.J., et al. Rescue of adeno-associated virus from recombinant plasmids: gene correction within the terminal repeats of AAV. Cell. 1983;33:135–143. doi: 10.1016/0092-8674(83)90342-2. [DOI] [PubMed] [Google Scholar]
- 67.Wang X.S., Ponnazhagan S., Srivastava A. Rescue and replication of adeno-associated virus type 2 as well as vector DNA sequences from recombinant plasmids containing deletions in the viral inverted terminal repeats: selective encapsidation of viral genomes in progeny virions. J. Virol. 1996;70:1668–1677. doi: 10.1128/jvi.70.3.1668-1677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chiorini J.A., et al. Sequence requirements for stable binding and function of Rep68 on the adeno-associated virus type 2 inverted terminal repeats. J. Virol. 1994;68:7448–7457. doi: 10.1128/jvi.68.11.7448-7457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Musatov S., et al. A cis-acting element that directs circular adeno-associated virus replication and packaging. J. Virol. 2002;76:12792–12802. doi: 10.1128/JVI.76.24.12792-12802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bishop B.M., et al. Role of the terminal repeat GAGC trimer, the major Rep78 binding site, in adeno-associated virus DNA replication. FEBS Lett. 1996;397:97–100. doi: 10.1016/s0014-5793(96)01149-0. [DOI] [PubMed] [Google Scholar]
- 71.Hüser D., et al. Integration preferences of wildtype AAV-2 for consensus rep-binding sites at numerous loci in the human genome. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCarty D.M., et al. Identification of linear DNA sequences that specifically bind the adeno-associated virus Rep protein. J. Virol. 1994;68:4988–4997. doi: 10.1128/jvi.68.8.4988-4997.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ling C., et al. The adeno-associated virus genome packaging puzzle. J. Mol. Genet. Med. 2015;9 doi: 10.4172/1747-0862.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bohenzky R.A., LeFebvre R.B., Berns K.I. Sequence and symmetry requirements within the internal palindromic sequences of the adeno-associated virus terminal repeat. Virology. 1988;166:316–327. doi: 10.1016/0042-6822(88)90502-8. [DOI] [PubMed] [Google Scholar]
- 75.Cataldi M.P., McCarty D.M. Hairpin-end conformation of adeno-associated virus genome determines interactions with DNA-repair pathways. Gene Ther. 2013;20:686–693. doi: 10.1038/gt.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cataldi M.P., McCarty D.M. Differential effects of DNA double-strand break repair pathways on single-strand and self-complementary adeno-associated virus vector genomes. J. Virol. 2010;84:8673–8682. doi: 10.1128/JVI.00641-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sanlioglu S., Benson P., Engelhardt J.F. Loss of ATM function enhances recombinant adeno-associated virus transduction and integration through pathways similar to UV irradiation. Virology. 2000;268:68–78. doi: 10.1006/viro.1999.0137. [DOI] [PubMed] [Google Scholar]
- 78.Schwartz R.A., et al. The Mre11/Rad50/Nbs1 complex limits adeno-associated virus transduction and replication. J. Virol. 2007;81:12936–12945. doi: 10.1128/JVI.01523-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lentz T.B., Samulski R.J. Insight into the mechanism of inhibition of adeno-associated virus by the Mre11/Rad50/Nbs1 complex. J. Virol. 2015;89:181–194. doi: 10.1128/JVI.01990-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Earley L.F., et al. Adeno-associated virus serotype-specific inverted terminal repeat sequence role in vector transgene expression. Hum. Gene Ther. 2020;31:151–162. doi: 10.1089/hum.2019.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu J., et al. On the nature of human housekeeping genes. Trends Genet. 2008;24:481–484. doi: 10.1016/j.tig.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 82.Saxonov S., Berg P., Brutlag D.L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Juven-Gershon T., et al. The RNA polymerase II core promoter - the gateway to transcription. Curr. Opin. Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carninci P., et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 85.Weiner G.J., et al. Immunostimulatory oligodeoxynucleotides containing the CpG motif are effective as immune adjuvants in tumor antigen immunization. Proc. Natl. Acad. Sci. U. S. A. 1997;94:10833–10837. doi: 10.1073/pnas.94.20.10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Faust S.M., et al. CpG-depleted adeno-associated virus vectors evade immune detection. J. Clin. Invest. 2013;123:2994–3001. doi: 10.1172/JCI68205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wright J.F. Codon modification and PAMPs in clinical AAV vectors: the tortoise or the hare? Mol. Ther. 2020;28:701–703. doi: 10.1016/j.ymthe.2020.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bertolini T.B., et al. Effect of CpG depletion of vector genome on CD8(+) T cell responses in AAV gene therapy. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.672449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Konkle B.A., et al. BAX 335 hemophilia B gene therapy clinical trial results: potential impact of CpG sequences on gene expression. Blood. 2021;137:763–774. doi: 10.1182/blood.2019004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pan X., et al. 2022. Rational Engineering of a Dunctional CpG-free ITR for AAV Gene Therapy; pp. 333–345. 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Delgado S., et al. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J. 1998;17:2426–2435. doi: 10.1093/emboj/17.8.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sequeira-Mendes J., et al. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Antequera F., Bird A. CpG islands as genomic footprints of promoters that are associated with replication origins. Curr. Biol. 1999;9:R661–R667. doi: 10.1016/s0960-9822(99)80418-7. [DOI] [PubMed] [Google Scholar]
- 94.Galibert L., et al. Functional roles of the membrane-associated AAV protein MAAP. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-01220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sonntag F., Schmidt K., Kleinschmidt J.A. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10220–10225. doi: 10.1073/pnas.1001673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Grosse S., et al. Relevance of assembly-activating protein for adeno-associated virus vector production and capsid protein stability in mammalian and insect cells. J. Virol. 2017;91 doi: 10.1128/JVI.01198-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Earley L.F., et al. Adeno-associated virus (AAV) assembly-activating protein is not an essential requirement for capsid assembly of AAV serotypes 4, 5, and 11. J. Virol. 2017;91 doi: 10.1128/JVI.01980-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maurer A.C., et al. The assembly-activating protein promotes stability and interactions between AAV's viral proteins to nucleate capsid assembly. Cell Rep. 2018;23:1817–1830. doi: 10.1016/j.celrep.2018.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sonntag F., et al. The assembly-activating protein promotes capsid assembly of different adeno-associated virus serotypes. J. Virol. 2011;85:12686–12697. doi: 10.1128/JVI.05359-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Herrmann A.K., et al. Impact of the assembly-activating protein on molecular evolution of synthetic adeno-associated virus capsids. Hum. Gene Ther. 2019;30:21–35. doi: 10.1089/hum.2018.085. [DOI] [PubMed] [Google Scholar]
- 101.Viney L., et al. Adeno-associated virus (AAV) capsid chimeras with enhanced infectivity reveal a core element in the AAV genome critical for both cell transduction and capsid assembly. J. Virol. 2021;95 doi: 10.1128/JVI.02023-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Emmerling V.V., et al. Rational plasmid design and bioprocess optimization to enhance recombinant adeno-associated virus (AAV) productivity in mammalian cells. Biotechnol. J. 2016;11:290–297. doi: 10.1002/biot.201500176. [DOI] [PubMed] [Google Scholar]
- 103.Mietzsch M., et al. 2021. Improved Genome Packaging Efficiency of Adeno-associated Virus Vectors Using Rep Hybrids. 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Elmore Z.C., et al. The membrane associated accessory protein is an adeno-associated viral egress factor. Nat. Commun. 2021;12:6239. doi: 10.1038/s41467-021-26485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brimble M.A., et al. Preventing packaging of translatable P5-associated DNA contaminants in recombinant AAV vector preps. Mol. Ther. Meth. Clin. Dev. 2022;24:280–291. doi: 10.1016/j.omtm.2022.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brimble M.A., et al. Preventing packaging of translatable P5-associated DNA contaminants in recombinant AAV vector preps. Mol. Ther. Meth. Clin. Develop.. 2022;24:280–291. doi: 10.1016/j.omtm.2022.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Adachi K., Tomono T. 2022. A PCR-Amplified Transgene Fragment Flanked By A Single Copy Of A Truncated Inverted Terminal Repeat For Recombinant Adeno-Associated Virus Production Prevents Unnecessary Plasmid DNA Packaging; pp. 449–457. 29. [DOI] [PubMed] [Google Scholar]
- 108.Okada, H., et al.
- 109.Okada, T.
- 110.Gao K., et al. Empty virions in AAV8 vector preparations reduce transduction efficiency and may cause total viral particle dose-limiting side-effects. Mol. Ther. Meth. Clin. Dev. 2014;1 doi: 10.1038/mtm.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Son Y.W., et al. Advances in selecting appropriate non-rodent species for regulatory toxicology research: policy, ethical, and experimental considerations. Regul. Toxicol. Pharmacol. 2020;116 doi: 10.1016/j.yrtph.2020.104757. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There is no additional data available for this study