Abstract
Background/Purpose:
Adequate image registration of anatomic and functional MRI scans is necessary for MR-guided head and neck cancer (HNC) adaptive radiotherapy planning. Despite the quantitative capabilities of diffusion-weighted imaging (DWI) MRI for treatment plan adaptation, geometric distortion remains a considerable limitation. Therefore, we systematically investigated various deformable image registration (DIR) methods to co-register DWI and T2-weighted (T2W) images.
Materials/Methods:
We compared three commercial (ADMIRE, Velocity, Raystation) and three open-source (Elastix with default settings [Elastix Default], Elastix with parameter set 23 [Elastix 23], Demons) post-acquisition DIR methods applied to T2W and DWI MRI images acquired during the same imaging session in twenty immobilized HNC patients. In addition, we used the non-registered images (None) as a control comparator. Ground-truth segmentations of radiotherapy structures (tumor and organs at risk) were generated by a physician expert on both image sequences. For each registration approach, structures were propagated from T2W to DWI images. These propagated structures were then compared with ground-truth DWI structures using the Dice similarity coefficient and mean surface distance.
Results:
19 left submandibular glands, 18 right submandibular glands, 20 left parotid glands, 20 right parotid glands, 20 spinal cords, and 12 tumors were delineated. Most DIR methods took < 30 seconds to execute per case, with the exception of Elastix 23 which took ~458 seconds to execute per case. ADMIRE and Elastix 23 demonstrated improved performance over None for all metrics and structures (Bonferroni-corrected p < 0.05), while the other methods did not. Moreover, ADMIRE and Elastix 23 significantly improved performance in individual and pooled analysis compared to all other methods.
Conclusions:
The ADMIRE DIR method offers improved geometric performance with reasonable execution time so should be favored for registering T2W and DWI images acquired during the same scan session in HNC patients. These results are important to ensure the appropriate selection of registration strategies for MR-guided radiotherapy.
Keywords: deformable image registration, magnetic resonance, adaptive radiotherapy, quality assurance
1. Introduction:
Radiation therapy (RT) is an essential treatment modality for head and neck cancer (HNC) 1. Conventionally, RT has relied on radiographic images to enable pre-treatment segmentation of target volumes and nearby organs at risk (OAR) to plan intensity-modulated doses 2,3. However, throughout RT, the dynamic changes in target volumes and OARs coupled to patient-specific changes (e.g., weight loss) can lead to unintended doses of radiation to OARs and subsequent debilitating side effects 4. These potential unintended doses are particularly relevant for HNC because the head and neck region is home to various complex, highly radiosensitive structures and tissue interfaces that can drastically change during RT 4,5.
Image-guided RT, during which radiation dose can be administered in tandem with onboard imaging, has become a promising alternative to conventional intensity-modulated RT, in part due to increasingly ubiquitous image-guided technology, such as MR-Linac devices 6,7. MR-guided treatment also affords the ability to capture distinct patient anatomy with varying contrasts via weighted sequence acquisitions, such as T2-weighted (T2W) images, and functional information, such as through diffusion-weighted imaging (DWI). DWI has shown particular benefit in aiding treatment adaptation through improved detection of target volumes and assessment of treatment response 8. Therefore, combined T2W and DWI acquisition enable the gathering of anatomic and functional information that can be used for adaptive MR-guided personalized RT.
Anatomical and functional sequences acquired in the same imaging session for MR-guided treatment typically have minimal variation in patient position and geometry between sequence acquisitions, often due to careful patient immobilization 9. However, these multisequence acquisitions can be misaligned by motion artifacts from respiration or swallowing 4, susceptibility artifacts, chemical shift artifacts, ghosting artifacts 8, and geometric distortions 10. Post-acquisition image registration, the process by which homologous image voxels from multi-temporal or multi-modal image sets are mapped to each other 11,12, is an important approach to align anatomical and functional sequences. Rigid image registration involves global matching between image sets, while deformable image registration (DIR) uses optimization algorithms to adjust image transformation models. Most implementations of DIR involve a transformation that establishes a geometric correspondence between fixed and moving images, an objective function, and an optimization approach to maximize the similarity between images 13–15. Importantly, even minor differences in patient anatomy can result in devastating improper dose administration in HNC 4,16, highlighting the need for consistent image co-registration when propagating segmentations of target volumes and OARs for radiotherapy treatment planning. Therefore, determining the impact of post-acquisition registration techniques (i.e., DIR) on multisequence MRI acquisitions is crucial for MR-guided treatment of HNC.
While we have previously investigated intra-modality CT to CT registration 17 and inter-modality CT to MRI registration 18, to our knowledge, there are no studies that investigate registration techniques for intra-acquisition MRI in HNC. Therefore, to facilitate further development and optimization of MR-guided RT adaptive planning technologies, we systematically analyzed DIR methods in T2W and DWI MRI sequences acquired during the same imaging session.
2. Methods:
We developed a quality assurance workflow for evaluating and benchmarking the performance of different image registration methods for T2W and DWI images (described in subsection 2.2) of 20 HNC patients (2.1). We evaluated different DIR methods provided by commercial RT treatment planning software, as well as open-source DIR implementations (2.3). The deformation vector field (DVF) generated by each method was used to propagate the manually segmented structures (2.2) from T2W to DWI images. The structures propagated by different methods to DWI images were compared to the ground-truth segmentations for performance evaluation (2.4).
2.1. Patient Characteristics:
Twenty patients with HNC who had undergone RT in a clinical trial (NCT03145077) were included in this analysis. All clinical and imaging data were generated between May 30, 2017 and April 1, 2019 and were retrospectively collected under a HIPAA-compliant protocol (PA16–0302) that was approved by The University of Texas MD Anderson Cancer Center’s institutional review board. All patients provided study-specific informed consent. The median patient age was 54 years, with a male predominance (80%). Primary tumor sites included the oropharynx, nasopharynx, and oral cavity. Full patient clinical and demographic characteristics are summarized in Table 1.
Table 1.
Patient clinical and demographic characteristics.
| Characteristic | Value |
|---|---|
|
| |
| Age (median, range) | 54 (32–77) |
| Sex | |
| Male | 16 |
| Female | 4 |
| Race | |
| Asian | 1 |
| White/Caucasian | 19 |
| Tumor subsite | |
| Base of tongue | 10 |
| Tonsil | 5 |
| Buccal mucosa | 1 |
| Floor of mouth | 1 |
| Nasal cavity | 1 |
| Nasopharynx | 1 |
| Unknown | 1 |
| T-category | |
| T0 | 1 |
| T1 | 5 |
| T2 | 7 |
| T3 | 2 |
| T4 | 5 |
| N-category | |
| Nx | 1 |
| N1 | 11 |
| N2a | 1 |
| N2b | 6 |
| N2c | 1 |
| AJCC stage | |
| I | 2 |
| II | 6 |
| III | 2 |
| IVA | 9 |
| IVB | 1 |
| Therapeutic combination | |
| RT alone | 3 |
| RT + CC | 8 |
| RT + surgery | 1 |
| RT + CC + IC | 5 |
| RT + CC + surgery | 1 |
| RT + CC + IC + surgery | 2 |
Unless otherwise indicated, data shown correspond to patient number counts. AJCC, American Joint Committee on Cancer; RT, radiotherapy; CC, concurrent chemotherapy; IC, induction chemotherapy.
2.2. Imaging Data:
Pre-RT T2W and DWI MRI sequences in Digital Imaging and Communications in Medicine (DICOM) format for each of the 20 patients were curated from our imaging databases. T2W images and DWI images with a b value of 0 (b0) were collected in the same imaging session while the patient was immobilized in a thermoplastic mask using a 1.5 Tesla Siemens MRI simulator. b0 images were selected for the analysis, as opposed to higher b value images, principally for two reasons: (1) The contrast of the T2W image is more similar to the b0 image, so intensity-based registration algorithms would perform better on these images 19, (2) The longer echo time at higher b values causes a loss in signal-to-noise ratio 20, which can degrade registration performance 21. Characteristics of the imaging sequences are shown in Table 2. For each image set (T2W image and DWI image), ground-truth segmentations for the left and right submandibular glands, left and right parotid glands, cervical spinal cord, and primary gross tumor volume were manually generated by a trained physician expert (radiologist with > 5 years of experience in HNC). Notably, primary gross tumor volumes that were not visible on imaging secondary to induction chemotherapy or surgical excision were not segmented; some submandibular glands were also excluded due to surgical excision. Moreover, some patients had oral metallic implants which led to minor signal-loss artifacts, but these artifacts did not obscure regions of interest used for segmentation. All segmentations were generated in Velocity AI (v.3.0.1; Varian Medical Systems; Palo Alto, CA, USA) in DICOM RT structure format. The anonymized image sets and structure files are publicly available online through Figshare (doi: 10.6084/m9.figshare.17162435).
Table 2.
MRI sequence acquisition parameters. Median value displayed with range of values shown in parenthesis. No parenthesis indicates all values were the same for all patients.
| Acquisition Parameter | T2W | DWI |
|---|---|---|
|
| ||
| Repetition time (ms) | 4800 | 5000 (1500–7000) |
| Echo time (ms) | 80 | 65 (50–102) |
| Echo train length | 15 | 15 (0–63) |
| Flip angle (°) | 180 (166–180) | 120 (90–180) |
| Slice thickness (mm) | 2 | 4 |
| In-plane resolution (mm) | 0.5 | 2.0 (1.0–2.0) |
| Slice gap (mm) | 2 | 4 |
| Acquisition matrix | 256×230 | 128×128 |
| Pixel bandwidth (Hz/px) | 300 | 870 (750–1220) |
| Number of averages | 1 | 2 |
| Number of axial slices | 120 | 28 (24–48) |
T2W, T2-weighted magnetic resonance imaging; DWI, diffusion-weighted magnetic resonance imaging.
2.3. Image Registration:
For this analysis, we investigated several DIR registration methods from different commercial radiotherapy software packages and open-source implementations. Specifically, the following DIR methods were utilized:
ADMIRE: A proprietary approach from the commercial software package ADMIRE (v.3.29; Elekta AB; Stockholm, Sweden) that implements an atlas-based approach with head pose correction, dense mutual-information, and refinement using a deformable surface model.
Velocity: A proprietary approach from the commercial software package Velocity AI (v.3.0.1; Varian Medical Systems; Palo Alto, CA, USA) that implements a 3-pass (coarse-medium-fine resolution) modified B-spline.
Elastix Default: An open-source approach from the popular medical image registration library Elastix (SimpleElastix Python interface 22) that utilizes a multi-resolution B-spline (default parameter map). Specifically, the algorithm uses an adaptive stochastic gradient descent optimizer, linear interpolator, FixedImagePyramidSchedule = [8, 8, 8, 4, 4, 4, 2, 2, 2, 1, 1, 1], MovingImagePyramidSchedule = [8, 8, 8, 4, 4, 4, 2, 2, 2, 1, 1, 1], number of resolutions = 4, and 4096 spatial samples. More information on the algorithm can be found in the Elastix documentation 23.
Elastix 23: An open-source approach from the popular medical image registration library Elastix (SimpleElastix Python interface 22) that utilizes localized mutual information combined with a bending energy penalty in a B-spline transformation (referred to as parameter map 23 in the Elastix Zoo) 24. Specifically, the algorithm uses an adaptive stochastic gradient descent optimizer, B-spline interpolator, Image Pyramid Schedule = [8, 8, 2, 4, 4, 1, 1, 1, 0.5], number of resolutions = 3, and 10000 spatial samples. This algorithm was selected because it was explicitly developed for multi-modality head and neck registration. More information on the algorithm can be found in the Elastix documentation 23.
Demons: An open-source approach based on the Demons 25 family of algorithms available in SimpleITK 26. Specifically, the algorithm uses a multi-resolution framework with shrink factors of [4,2,1] and smoothing sigmas of [8,4,0], a linear interpolator, and a gradient descent optimizer (learning rate = 1.0, number of iterations = 20, convergence minimum value = 1e−6, convergence window size = 10).
Raystation: A proprietary approach from the commercial software package RayStation Research (RaySearch Laboratories, Stockholm, Sweden) that implements a modified ANAtomically CONstrained Deformation Algorithm 27 using only intensity-based registration.
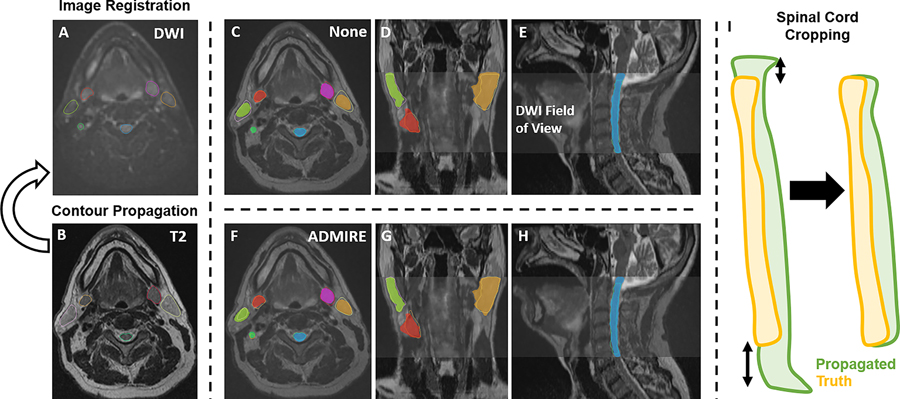
For all cases, the DWI image was used as the fixed image, and the T2W image was used as the moving image; T2W images were resampled to the DWI image. To maintain adequate comparisons between structures generated on T2W images and DWI images, before registration all structures were cropped to the image with the smaller field of view, i.e., DWI image. For additional deformable vector field (DVF) visualization and analysis, Jacobian determinant matrices were derived from DVF files of each method using SimpleITK or as direct output from ADMIRE. As a control comparator for all cases, we also analyzed the raw images with no post-acquisition registration applied, i.e., an implicit rigid registration as a byproduct of patient immobilization (labeled as “None”). After the registration process, for each method, we propagated the ground-truth segmentations from the T2W images to the DWI images using the corresponding transformations to generate propagated structures (Figure 1A–H). These propagated structures were then compared to the ground-truth structures on the DWI image in the subsequent analysis. Before the analysis, all images and structure files were transformed into Neuroimaging Informatics Technology Initiative format. Finally, because there were small variations in the inferior and superior slices of the cervical spinal cord, we cropped these structures so that the heights of the propagated segmentation and ground-truth segmentation were equal (Figure 1I).
Figure 1.
Study workflow. Contours are propagated from the moving image (B, T2-weighted image [T2]) to the fixed image (A, diffusion-weighted image [DWI]) for each registration method. C-E and F-H show propagated and ground-truth structures for no registration (labeled as “None”) and ADMIRE registration approaches, respectively. The spinal cord was also cropped so that the height of the propagated segmentation and ground-truth segmentation were equal (I).
2.4. Statistical Analysis:
Several evaluation metrics were used to compare the propagated structure sets after registration to the ground-truth structures delineated on the DWI images. Specifically, for each individual structure, the Dice similarity coefficient (DSC) and mean surface distance (MSD) were calculated as they are well-established and ubiquitous metrics for measuring volumetric and surface distance information, respectively 28. Additional volumetric and surface distance metrics were calculated for supplementary analyses (Appendix A). Metrics were calculated using the surface-distance Python package 29 and in-house Python code. After performing a Shapiro-Wilk test 30, we found that our data were not normally distributed (p<0.05). Therefore, we used nonparametric statistical tests for our analysis. For each metric and each structure, we compared registration methods against ‘None’ using one-sided Wilcoxon signed-rank tests (alternative hypothesis of ‘greater’ for DSC and alternative hypothesis of ‘less’ for MSD) with Bonferroni adjustments for multiple comparisons 31. Similarly, we pooled metrics for OARs for sub-analysis and performed pair-wise analysis using previously described Wilcoxon signed-rank tests with Bonferroni corrections. Interobserver variability, dosimetric, target registration error (TRE), and DVF Jacobian matrix analysis were performed in Appendix B, Appendix C, Appendix D, and Appendix E, respectively. For all statistical analyses, p-values less than 0.05 were considered significant. All statistical analyses were performed in Python v.3.7 32.
3. Results:
3.1. Quantitative Comparison:
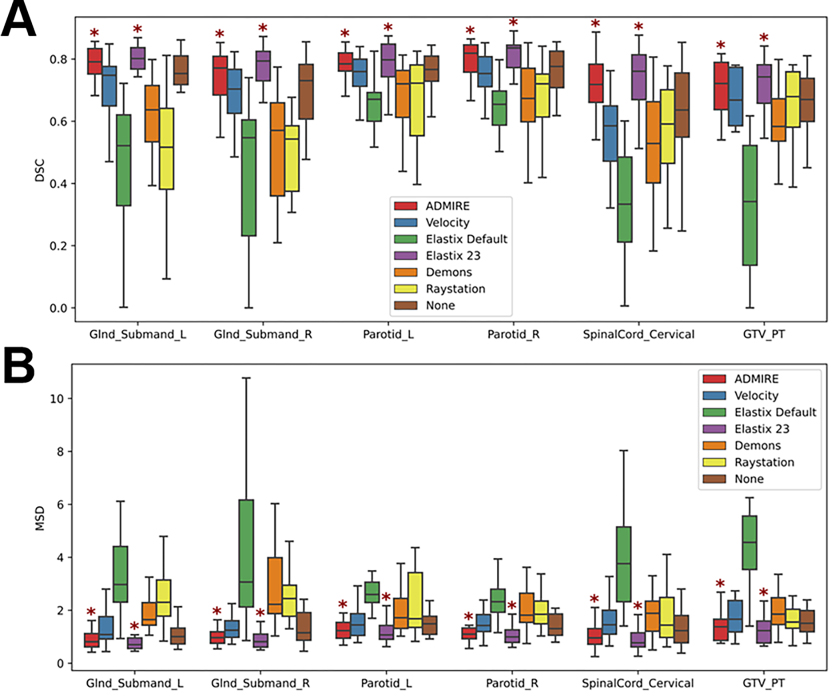
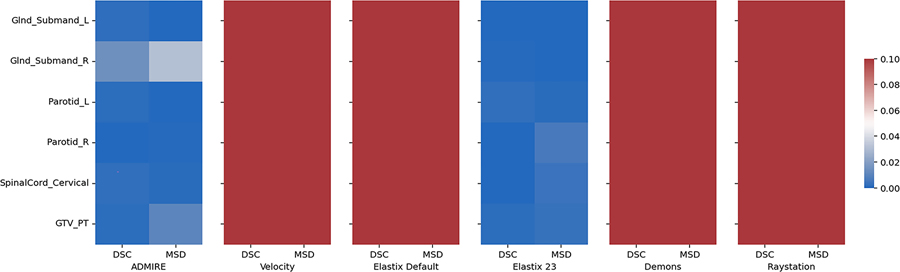
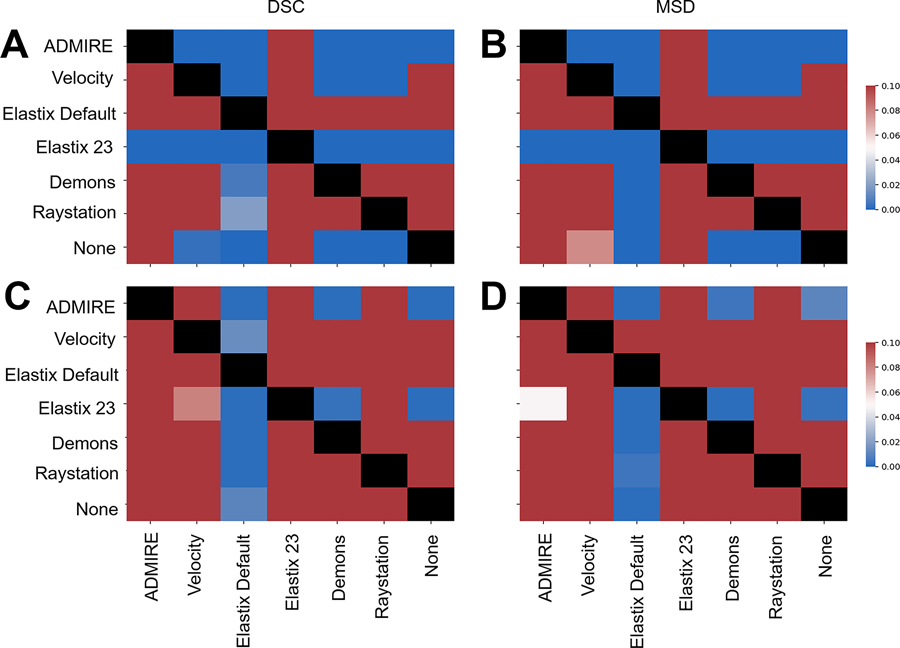
19 left submandibular glands, 18 right submandibular glands, 20 left parotid glands, 20 right parotid glands, 20 cervical spinal cords, and 12 primary tumors in both T2W and DWI images were used in the analysis. ADMIRE, Velocity, Elastix Default, Elastix 23, Demons, and Raystation each took approximately 23, 7, 46, 458, 4, and 13 seconds to complete one case, respectively. For each method, most OAR structures had similar performance across the multiple metrics except for the cervical spinal cord which was notably worse (Figure 2); we therefore performed additional analysis to investigate spinal cord structures in individual cases in Appendix F. Compared to the structures generated by no registration (“None”), all structures demonstrated an improvement with the ADMIRE and Elastix 23 methods, and worsened with all other methods (Figure 2). Specifically, the ADMIRE and Elastix 23 methods showed significant improvements (p<0.05 on one sided Wilcoxon signed rank test) for both DSC and MSD metrics for all structures (Figure 3). When metrics were pooled across structures, similar trends emerged where the ADMIRE and Elastix 23 methods demonstrated the best performance compared to the other methods, with DSC gains over None of up to .05 and .07, respectively in the OARs, and up to .08 and .09, respectively, in the tumor (Table 3). Moreover, pair-wise comparisons of pooled OAR structures and the tumor demonstrated that Elastix 23 offered significantly improved performance over ADMIRE (p<0.05), and that Elastix Default, Demons, and Raystation were consistently outperformed by the other methods (Figure 4). Dosimetric (Appendix C) and TRE (Appendix D) analysis revealed no significant improvements of any DIR methods compared to None.
Figure 2.
Box plots of evaluation metrics for each structure according to the registration method for Dice similarity coefficient (DSC) [A] and mean surface distance (MSD) [B]. Asterisks indicate a significant improvement between the registration method and no registration (None). Glnd_Submand, submandibular gland; L, left; R, right; GTV_PT, primary gross tumor volume.
Figure 3.
Heatmap of Bonferroni corrected p-values for one-way Wilcoxon-signed rank tests between various registration methods and no registration (None) indicating significant improvement across evaluation metrics and structures. Blue colors correspond to significant p-values (p<0.05) while red colors correspond to non-significant values (p>0.05). Glnd_Submand, submandibular gland; L, left; R, right; GTV_PT, primary gross tumor volume; DSC, Dice similarity coefficient; MSD, mean surface distance.
Table 3.
Evaluation metrics (mean ± standard deviation) across pooled structures according to each registration method. DSC, Dice similarity coefficient; MSD, mean surface distance.
| Metric | ADMIRE | Velocity | Elastix Default | Elastix 23 | Demons | Raystation | None | |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| OARs | DSC | 0.76±0.09 | 0.67±0.16 | 0.51±0.21 | 0.78±0.08 | 0.60±0.16 | 0.58±0.18 | 0.71±0.13 |
| MSD (mm) | 1.12±0.51 | 1.68±1.25 | 3.68±2.92 | 1.01±0.49 | 2.19±1.16 | 2.39±1.92 | 1.46±0.92 | |
|
| ||||||||
| Tumor | DSC | 0.70±0.09 | 0.59±0.25 | 0.33±0.20 | 0.71±0.09 | 0.60±0.12 | 0.63±0.16 | 0.62±0.21 |
| MSD (mm) | 1.38±0.55 | 3.63±6.62 | 5.49±3.59 | 1.25±0.52 | 2.00±0.82 | 1.88±1.03 | 2.64±3.56 | |
OARs, organs at risk; DSC, Dice similarity coefficient; MSD, mean surface distance.
Figure 4.
Heatmap of Bonferroni corrected p-values for one-way Wilcoxon-signed rank tests for pair-wise comparisons between various registration methods indicating significant improvement of method in row vs. method in column. Top plots correspond to all pooled organ at risk structures using Dice similarity coefficient (DSC) (A) and mean surface distance (MSD) (B). Bottom plots correspond to tumor using DSC (C) and MSD (D). Blue colors correspond to significant p-values (p<0.05) while red colors correspond to non-significant values (p>0.05). Comparisons between the same method (diagonal entries) are blacked out.
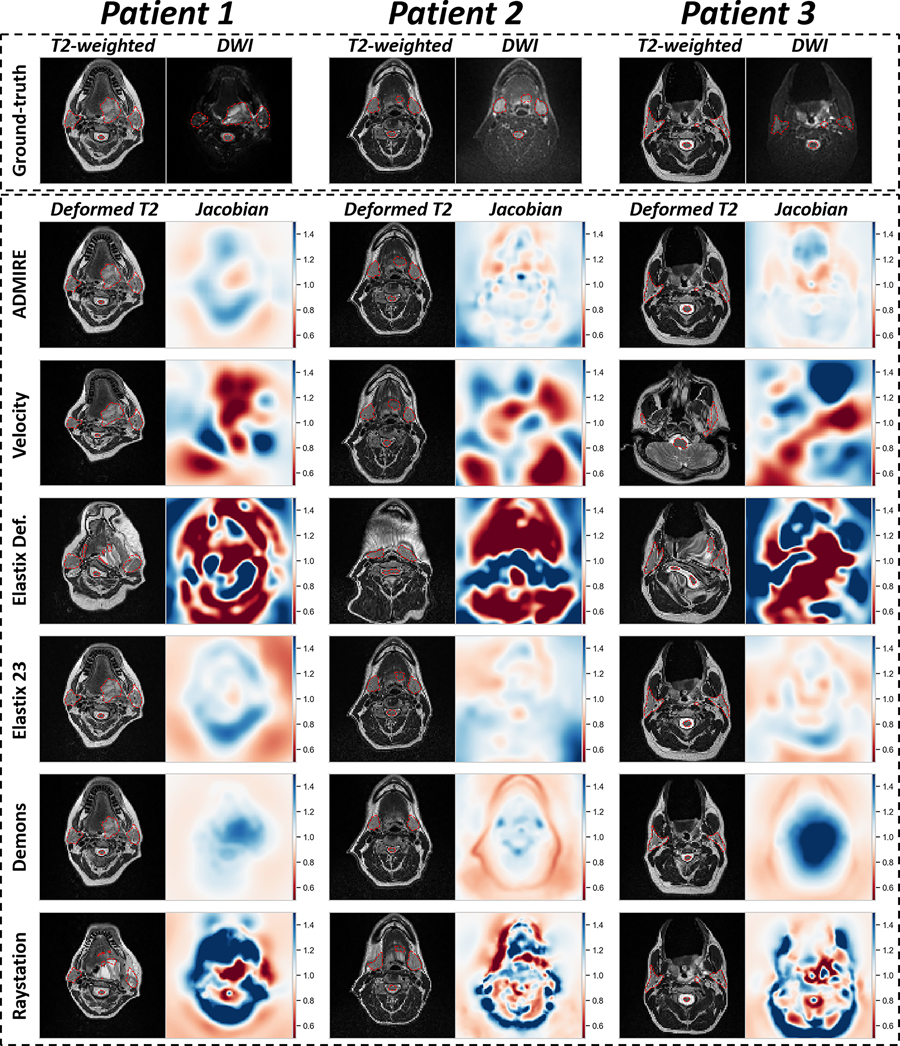
3.2. Qualitative Comparison:
We visually compared the deformed T2W images of the various DIR methods and their corresponding Jacobian determinant matrices in Figure 5. Generally, most methods often yielded visually similar deformed image outputs as the ground-truth T2W image. However, large deviations in DVF warping could sometimes be observed for Velocity and Elastix Default. Additional quantitative analysis of Jacobian determinants for the various methods can be found in Appendix E.
Figure 5.
Visualization of deformable image registration method outputs for 3 example patients. The top row shows original T2-weighted image and diffusion weighted image (DWI) with ground-truth segmentations overlaid (red dotted outline). The deformed T2-weighted image with overlaid propagated segmentation (red dotted outline) and corresponding Jacobian determinant is shown for each image registration method. Jacobian determinant plots are all shown on the same color bar scale from 0 to 1.5. Jacobian determinants greater than 1 (blue colors) indicate volume expansion, between 0 and 1 (red colors) indicate volume reduction, and equal to 1 (white color) indicates no change.
4. Discussion
In this study, we systematically analyzed a variety of DIR methods and compared them to non-registered images from multisequence MRI acquisitions for HNC image-guided treatment applications. Our results highlight that specific DIR methods can improve upon pre-established head and neck immobilization, as shown by measuring the similarity of propagated ground-truth segmentations from T2W images to DWI images compared to ground-truth segmentations on DWI images (Figures 2 and 3).
The best overall results were obtained using ADMIRE and Elastix 23 (Table 3), with all metric and structure combinations having significantly better performance than None (Figure 3). While we tested other deformable methods (i.e., Velocity, Demons, Elastix Default, and Raystation), they were significantly worse for most metric and structure combinations when compared to None. Moreover, Velocity and Elastix Default sometimes yielded implausible DVFs, as indicated by qualitative and quantitative analysis of Jacobian determinants (Figure 4, Appendix E), which may be due to these DIR algorithmic implementations being unable to accommodate large variations in intensity domains of the T2W and DWI images. Importantly, almost all structures individually and on pooled analysis showed improved DSC and MSD for the ADMIRE and Elastix 23 methods (Table 3). These results indicate that these methods provide significantly improved volumetric and surface distance overlap, which may warrant their use during intra-acquisition MRI sequences for MR-guided treatment. Moreover, while dosimetrically there were no significant improvements for any structures for ADMIRE and Elastix 23 compared to None (Appendix C), these differences may still be clinically significant. Similarly, for TRE analysis on a subset of methods, ADMIRE often offered decreased registration error compared to no registration (Appendix D), but was nonsignificant, likely secondary to the already minimal registration errors induced through the use of an immobilization mask. Notably, Elastix 23 provided significantly improved volumetric and surface distance performance compared to ADMIRE. However, these improvements may not be clinically significant (DSC gains of ~1%) and come at the cost of a much longer execution time (~7 minutes longer), therefore, ADMIRE should likely be preferred for workflows where time is a limiting factor, i.e., adaptive radiotherapy. It is also worth noting the spinal cord is especially sensitive to distortion-causing artifacts 33, making it a particularly challenging structure to co-register adequately. While the general performance for the spinal cord was lower than that of other OAR structures, the ADMIRE based methods were still able to offer significantly improved performance compared to None (Figures 2 and 3); cases with lower performance tended to have a larger degree of spinal curvature than cases with higher performance (Appendix F). Therefore, while the ADMIRE method should still be preferred over no registration, special caution should be used in quality assurance when used for spinal cord segmentations. Notably, there were no significant differences using segmentations generated by different observers for any DIR method (Appendix B), indicating that our data are not confounded by interobserver variability.
While several previous studies have investigated the relative performance of registration methods in various anatomical sites 34–36, there is a general lack of investigations of head and neck imaging. However, a few recent important studies have investigated registration quality assessment in head and neck imaging using radiotherapy structure analysis similar to our current study 17,18. For example, Mohamed et al. 17 investigated the registration quality of diagnostic CT to simulation CT in HNC where images were acquired at different time points and with different scan settings and found that certain DIR methods demonstrated improved performance over a control group (rigid registration) for OAR and target conformance for most comparison metrics, similar to our study. Oppositely, Kiser et al. 18 showed that for CT and T2W MRI scans acquired with standard treatment immobilization techniques, MRI to CT DIR was not superior to rigid registration, with neither technique producing clinically satisfactory results (DSCs of 0.62 – 0.65). Importantly, the ADMIRE method investigated in our study produce potentially clinically meaningful results as we observe significant performance gains across various structures that may impact MR-guided treatments with a reasonable execution time.
To date, no systematic anatomical to functional MRI registration studies have been performed for HNC. However, intra-acquisition MRI registration techniques have been investigated in other anatomical sites. Specifically, several studies have compared registration techniques for various MRI sequences in the prostate 37–39. For example, Buerger et al. compared the performance of five state-of-the-art DIR image registration techniques for accurate image fusion of DWI with T2W images and found fast elastic image registration provided improved performance compared to other deformable techniques such as B-spline and Demons 38. This result was further echoed in Eriksson et al., which confirmed that fast elastic image registration was the best technique for T1-weighted to T2W anatomic sequence registration 39. Our results are consistent with these observations that selecting appropriate deformable techniques offers significantly improved performance for intra-acquisition registration.
There are several limitations to our study. We limited our analysis of intra-acquisition registration techniques in MRI to T2W and DWI sequences since these are the most germane to current MR-guided RT applications. However, several additional sequences can be studied to investigate these phenomena. In this study, we only tested b0 images, which were readily available and common for DWI workflows in HNC. Furthermore, as no images suffered from major geometric distortion, we did not address geometric distortion in this study. Future iterations of this study should investigate other DWI-derived images and the influence of geometric distortion on DIR. Notably, we have found dosimetric improvements for certain DIR methods (though statistically non-significant), but the clinical significance of these improvements is unknown and should be confirmed through additional experiments such as normal tissue complication probability calculations. Moreover, we anticipate structure-level examination of average apparent diffusion coefficient (ADC) values would yield similar results to our dosimetric evaluation and have therefore opted to forgo further studies based on ADC, however, determination of voxel-wise differences in ADC map generation induced by registration errors could be useful for dose-painting applications and should be the subject of future investigations. Additionally, we have limited our investigation to a few critical RT HNC structures of interest using one expert observer; future studies should investigate a greater number of structures in a greater number of patients with a larger number of observers. It should also be noted that while most contoured structures were not expected to vary considerably between T2 and DWI, pathologic structures may be interpreted differently on these images, thus caution should be used when interpreting results for the tumor. Finally, we have limited our analysis to intra-acquisition images collected during the same image acquisition session. However, for MR-guided RT applications, registration techniques are also relevant for images taken at different time points. Therefore, future studies should investigate these registration techniques applied to different imaging time points in an MR-guided RT workflow.
5. Conclusions
In summary, this is the first study to investigate intra-acquisition MRI registration quality in HNC patients. We identify a deformable registration technique from the ADMIRE software package that offers the most significant gains in registration quality with reasonable execution time for T2W to DWI image registration compared to other methods. Our results are a crucial first step towards registration quality assurance for MR-guided treatment approaches that implement multi-sequence acquisitions combining anatomical and functional imaging.
Supplementary Material
Acknowledgments
The authors thank Ann Sutton, Scientific Editor, and Ashli Nguyen-Villarreal, Associate Scientific Editor, in the Research Medical Library at The University of Texas MD Anderson Cancer Center, for editing this article. The authors also acknowledge the following people for their contributions to the NIH-funded academic-industrial partnership grant (R01DE028290) that funded this work and for their general support and feedback regarding this project: Spencer Marshall, Hafid Akhiat, Michel Moreau, Nathan Cho, Edyta Bubula-Rehm, Chunhua Men, and Etienne Lessard of Elekta and Alex Dresner of Philips.
Funding Statement
This work was supported by the National Institutes of Health (NIH) through a Cancer Center Support Grant (P30-CA016672–44) and an Academic-Industrial Partnership Award (R01 DE028290). K.A. Wahid is supported by the American Legion Auxiliary Fellowship in Cancer Research, the Dr. John J. Kopchick Fellowship through The University of Texas MD Anderson UTHealth Graduate School of Biomedical Sciences, and a NIDCR F31 fellowship (1 F31 DE031502–01). B.A. McDonald receives research support from an NIH NIDCR Award (F31DE029093) and the Dr. John J. Kopchick Fellowship through The University of Texas MD Anderson UTHealth Graduate School of Biomedical Sciences. T.C. Salzillo is supported by a training fellowship from The University of Texas Health Science Center at Houston Center for Clinical and Translational Sciences TL1 Program (TL1TR003169). C.D. Fuller received funding from an NIH NIDCR Award (1R01 DE025248–01/R56 DE025248) and Academic-Industrial Partnership Award (R01 DE028290); the National Science Foundation (NSF), Division of Mathematical Sciences, Joint NIH/NSF Initiative on Quantitative Approaches to Biomedical Big Data (QuBBD) Grant (NSF 1557679); the NIH Big Data to Knowledge (BD2K) Program of the National Cancer Institute (NCI) Early Stage Development of Technologies in Biomedical Computing, Informatics, and Big Data Science Award (1R01 CA214825); the NCI Early Phase Clinical Trials in Imaging and Image-Guided Interventions Program (1R01 CA218148); the NIH/NCI Cancer Center Support Grant (CCSG) Pilot Research Program Award from the UT MD Anderson CCSG Radiation Oncology and Cancer Imaging Program (P30 CA016672); the NIH/NCI Head and Neck Specialized Programs of Research Excellence (SPORE) Developmental Research Program Award (P50 CA097007); and the National Institute of Biomedical Imaging and Bioengineering (NIBIB) Research Education Program (R25 EB025787).
Abbreviations:
- T2W
T2-weighted
- DWI
diffusion-weighted imaging
- RT
Radiation therapy
- HNC
head and neck cancer
- OAR
organs at risk
- DIR
deformable image registration
Footnotes
Conflict of Interest: C.D.F. has received direct industry grant support, speaking honoraria, and travel funding from Elekta AB. D.T., N.O., V.W., and J.P.C. are employees of Elekta AB. The other authors have no conflicts of interest to disclose.
Contributor Information
Joint Head and Neck Radiotherapy-MRI Development Cooperative:
Mohamed A. Naser, Kareem A. Wahid, Sara Ahmed, Vivian Salama, Cem Dede, Benjamin W. Edwards, Ruitao Lin, Brigid McDonald, Travis C. Salzillo, Renjie He, Yao Ding, Moamen Abobakr Abdelaal, Daniel Thill, Nicolette O’Connell, Virgil Willcut, John P. Christodouleas, Stephen Y Lai, Clifton D. Fuller, and Abdallah S. R. Mohamed
Data Availability Statement:
The anonymized image sets and structure files are publicly available online through Figshare (doi: 10.6084/m9.figshare.17162435).
References
- 1.Koyfman SA, Brizel DM, Posner MR. General principles of radiation therapy for head and neck cancer. UpToDate; Post, TW, Ed.; UpToDate: Waltham, MA, USA. Published 2018. Accessed August 20, 2021. https://www.uptodate.com/contents/general-principles-of-radiation-therapy-for-head-and-neck-cancer
- 2.Bortfeld T IMRT: a review and preview. Phys Med Biol. 2006;51(13):R363. [DOI] [PubMed] [Google Scholar]
- 3.Marta GN, Silva V, de Andrade Carvalho H, et al. Intensity-modulated radiation therapy for head and neck cancer: systematic review and meta-analysis. Radiother Oncol. 2014;110(1):9–15. [DOI] [PubMed] [Google Scholar]
- 4.Mali SB. Adaptive radiotherapy for head neck cancer. J Maxillofac Oral Surg. 2016;15(4):549–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan HE, Sher DJ. Adaptive radiotherapy for head and neck cancer. Cancers Head Neck. 2020;5(1):1. doi: 10.1186/s41199-019-0046-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollard JM, Wen Z, Sadagopan R, Wang J, Ibbott GS. The future of image-guided radiotherapy will be MR guided. Br J Radiol. 2017;90(1073):20160667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choudhury A, Budgell G, MacKay R, et al. The future of image-guided radiotherapy. Clin Oncol. 2017;29(10):662–666. [DOI] [PubMed] [Google Scholar]
- 8.Chawla S, Kim S, Wang S, Poptani H. Diffusion-weighted imaging in head and neck cancers. Futur Oncol. 2009;5(7):959–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohamed ASR, Hansen C, Weygand J, et al. Prospective analysis of in vivo landmark point-based MRI geometric distortion in head and neck cancer patients scanned in immobilized radiation treatment position: Results of a prospective quality assurance protocol. Clin Transl Radiat Oncol. 2017;7:13–19. doi: 10.1016/j.ctro.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weygand J, Fuller CD, Ibbott GS, et al. Spatial precision in magnetic resonance imaging–guided radiation therapy: the role of geometric distortion. Int J Radiat Oncol Biol Phys. 2016;95(4):1304–1316. [DOI] [PubMed] [Google Scholar]
- 11.Rong Y, Rosu-Bubulac M, Benedict SH, et al. Rigid and Deformable Image Registration for Radiation Therapy: A Self-Study Evaluation Guide in YYYY Clinical Trial Participation. Pract Radiat Oncol Published online 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill DLG, Batchelor PG, Holden M, Hawkes DJ. Medical image registration. Phys Med Biol. 2001;46(3):R1. [DOI] [PubMed] [Google Scholar]
- 13.Kessler ML. Image registration and data fusion in radiation therapy. Br J Radiol. 2006;79(SPEC. ISS.). doi: 10.1259/bjr/70617164 [DOI] [PubMed] [Google Scholar]
- 14.Sotiras A, Davatzikos C, Paragios N. Deformable medical image registration: A survey. IEEE Trans Med Imaging. 2013;32(7):1153–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliveira FPM, Tavares JMRS. Medical image registration: a review. Comput Methods Biomech Biomed Engin. 2014;17(2):73–93. [DOI] [PubMed] [Google Scholar]
- 16.Schwartz DL, Garden AS, Shah SJ, et al. Adaptive radiotherapy for head and neck cancer—dosimetric results from a prospective clinical trial. Radiother Oncol. 2013;106(1):80–84. [DOI] [PubMed] [Google Scholar]
- 17.Mohamed ASR, Ruangskul M-N, Awan MJ, et al. Quality assurance assessment of diagnostic and radiation therapy–simulation CT image registration for head and neck radiation therapy: anatomic region of interest–based comparison of rigid and deformable algorithms. Radiology. 2015;274(3):752–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiser K, Meheissen MAM, Mohamed ASR, et al. Prospective quantitative quality assurance and deformation estimation of MRI-CT image registration in simulation of head and neck radiotherapy patients. Clin Transl Radiat Oncol. 2019;18:120–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhushan C, Haldar JP, Choi S, Joshi AA, Shattuck DW, Leahy RM. Co-registration and distortion correction of diffusion and anatomical images based on inverse contrast normalization. Neuroimage. 2015;115:269–280. doi: 10.1016/j.neuroimage.2015.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higaki T, Nakamura Y, Tatsugami F, et al. Introduction to the Technical Aspects of Computed Diffusion-weighted Imaging for Radiologists. RadioGraphics. 2018;38(4):1131–1144. doi: 10.1148/rg.2018170115 [DOI] [PubMed] [Google Scholar]
- 21.Kale SC, Lerch JP, Henkelman RM, Chen XJ. Optimization of the SNR-resolution tradeoff for registration of magnetic resonance images. Hum Brain Mapp. 2008;29(10):1147–1158. doi: 10.1002/hbm.20453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marstal K, Berendsen F, Staring M, Klein S. SimpleElastix: A user-friendly, multi-lingual library for medical image registration. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops.; 2016:134–142. [Google Scholar]
- 23.Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW. Elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging. 2009;29(1):196–205. [DOI] [PubMed] [Google Scholar]
- 24.Leibfarth S, Mönnich D, Welz S, et al. A strategy for multimodal deformable image registration to integrate PET/MR into radiotherapy treatment planning. Acta Oncol (Madr). 2013;52(7):1353–1359. doi: 10.3109/0284186X.2013.813964 [DOI] [PubMed] [Google Scholar]
- 25.Pennec X, Cachier P, Ayache N. Understanding the “demon’s algorithm”: 3D non-rigid registration by gradient descent. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 1999:597–605. [Google Scholar]
- 26.Lowekamp B, Chen D, Ibanez L, Blezek D. The Design of SimpleITK. Front Neuroinformatics. 2013;7. https://www.frontiersin.org/article/10.3389/fninf.2013.00045 [DOI] [PMC free article] [PubMed]
- 27.Weistrand O, Svensson S. The ANACONDA algorithm for deformable image registration in radiotherapy. Med Phys. 2015;42(1):40–53. doi: 10.1118/1.4894702 [DOI] [PubMed] [Google Scholar]
- 28.Sherer MV, Lin D, Elguindi S, et al. Metrics to evaluate the performance of auto-segmentation for radiation treatment planning: A critical review. Radiother Oncol. Published online 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikolov S, Blackwell S, Zverovitch A, et al. Clinically Applicable Segmentation of Head and Neck Anatomy for Radiotherapy: Deep Learning Algorithm Development and Validation Study. J Med Internet Res. 2021;23(7):e26151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shapiro SS, Wilk MB. An analysis of variance test for normality (complete samples). Biometrika. 1965;52(3/4):591–611. [Google Scholar]
- 31.Wilcoxon F Individual comparisons by ranking methods. In: Breakthroughs in Statistics. Springer; 1992:196–202. [Google Scholar]
- 32.Van Rossum G, Drake FL. Python 3 Reference Manual. Scotts Valley, CA: CreateSpace; 2009. [Google Scholar]
- 33.Andre JB, Bammer R. Advanced diffusion-weighted magnetic resonance imaging techniques of the human spinal cord. Top Magn Reson imaging TMRI. 2010;21(6):367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ou Y, Akbari H, Bilello M, Da X, Davatzikos C. Comparative evaluation of registration algorithms in different brain databases with varying difficulty: results and insights. IEEE Trans Med Imaging. 2014;33(10):2039–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein A, Andersson J, Ardekani BA, et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46(3):786–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fabri D, Zambrano V, Bhatia A, et al. A quantitative comparison of the performance of three deformable registration algorithms in radiotherapy. Z Med Phys. 2013;23(4):279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Luca M, Giannini V, Vignati A, et al. A fully automatic method to register the prostate gland on T2-weighted and EPI-DWI images. In: 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE; 2011:8029–8032. [DOI] [PubMed] [Google Scholar]
- 38.Buerger C, Sénégas J, Kabus S, et al. Comparing nonrigid registration techniques for motion corrected MR prostate diffusion imaging. Med Phys. 2015;42(1):69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eriksson M Comparison of five methods for deformable, multi-modal image registration in prostate and pelvic area. Published online 2015. https://aaltodoc.aalto.fi/handle/123456789/15209
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The anonymized image sets and structure files are publicly available online through Figshare (doi: 10.6084/m9.figshare.17162435).