Heart failure (HF) is a major public health problem and beta-blockers (BB) are cornerstone therapy for HF with reduced ejection fraction (HFrEF). However, individual patients’ responses to BB vary substantially and clinical characteristics are not powerful enough predictors to target therapy, thus consensus guidelines recommend BB for all HFrEF patients without contraindication.1 Previous pharmacogenetic associations of BB showed proof of concept, but lacked validation and clinical utility.2 We recently created and validated, in ~1,200 European ancestry HFrEF patients, a novel polygenic response predictor (PRP) for BB in HFrEF.3 The PRP identified ~25% of patients with substantial survival benefit of BB (HR=0.19, 95%CI 0.06–0.65), distinct from the remaining patients with little survival benefit (HR=0.84, 95%CI 0.53–1.32). This provocative finding requires additional validation prior to prospective testing, so we sought replication using another large, independent dataset. The United Kingdom Biobank (UKB) contains health and genetic data on hundreds of thousands of participants from the UK, including medication prescription data. We tested the BB-PRP in the UKB using similar approaches as the original publication.3
We identified white patients with HF diagnosis, genetic data, vital status, and prescription data in the UKB. BB exposure was tabulated using dose, number of tablets, and prescription frequency, to generate an exposure metric that is a proportion of target exposure (based on HF clinical trials) similar to previously work.3, 4 The PRP is tabulated from genotype at 44 loci across the genome and was calculated exactly as previously described including using the original beta-coefficients.3 Overall, there was 2% missingness of genotypes needed for score calculation, which were imputed by random draw from the distribution of each SNP. UKB enrollment date was the index date for patients with prevalent HF, otherwise the index date was the date of first HF diagnosis. Time from index date to all-cause mortality was tabulated. The statistical analysis used proportional hazards models adjusted for Meta-analysis Global Group in Chronic Heart Failure (MAGGIC) score,5 BB propensity score, PRP, BB exposure, and BB exposure*PRPcategory interaction, with BB as a continuous time-updating variable, PRP main effect as a continuous variable, and PRPcategory dichotomized at the original cutoff value (≤68 predicted responder vs. > 68 non-responder).3 Since LVEF was only available in 255 patients, it was omitted from the MAGGIC calculation, and we included all participants regardless of LVEF. We tested similar models stratified by PRPcategory. The assumption of proportional hazards was assessed by visual inspection of plots of survival, survival time, and log(-log(survival time), which were consistent with proportionality. We depicted survival curves separated by PRPcategory and high vs. low BB exposure (first vs. third quartile, respectively, which were no BB and 54% target exposure). All analyses were performed in SAS 9.4 (SAS Institute, Cary, NC)
Among 7141 HF patients, 34% were women, 54% had coronary disease, 33% had atrial fibrillation, and 51% had baseline BB usage. The cohort mean PRP was 81 (± 18), with n=1642 (23%) responders and n=5499 non-responders. No baseline characteristics differed significantly across PRPcategory. Over a median follow-up of 5.2 years there were 1014 deaths (14.2%), 212 (12.9%) in responders and 802 (14.6%) in non-responders. The results are summarized in the Figure. In the total cohort, the PRP main effect was not significant (HR=0.99, p=0.92) while the interaction of PRPcategory*BB-exposure was significant (p=0.044, ß=0.576). In stratified analyses, PRP-responders had strong survival benefit associated with BB (HR=0.57, 95%CI 0.33–1.00, p=0.052), while the PRP-non-responders showed little BB effect (HR=0.90, 95%CI 0.70–1.15, p=0.388). Exploratory analyses in a subgroup of patients with diagnostic codes indicative of low LVEF subtype or a known low LVEF (n=1107, 135 deaths) showed no significant PRPcategory*BB interaction (p =0.34), despite a larger interaction effect estimate (ß=0.85, HR=0.32 in PRP-responders vs. 0.75 in PRP-non-responders).
Figure 1.
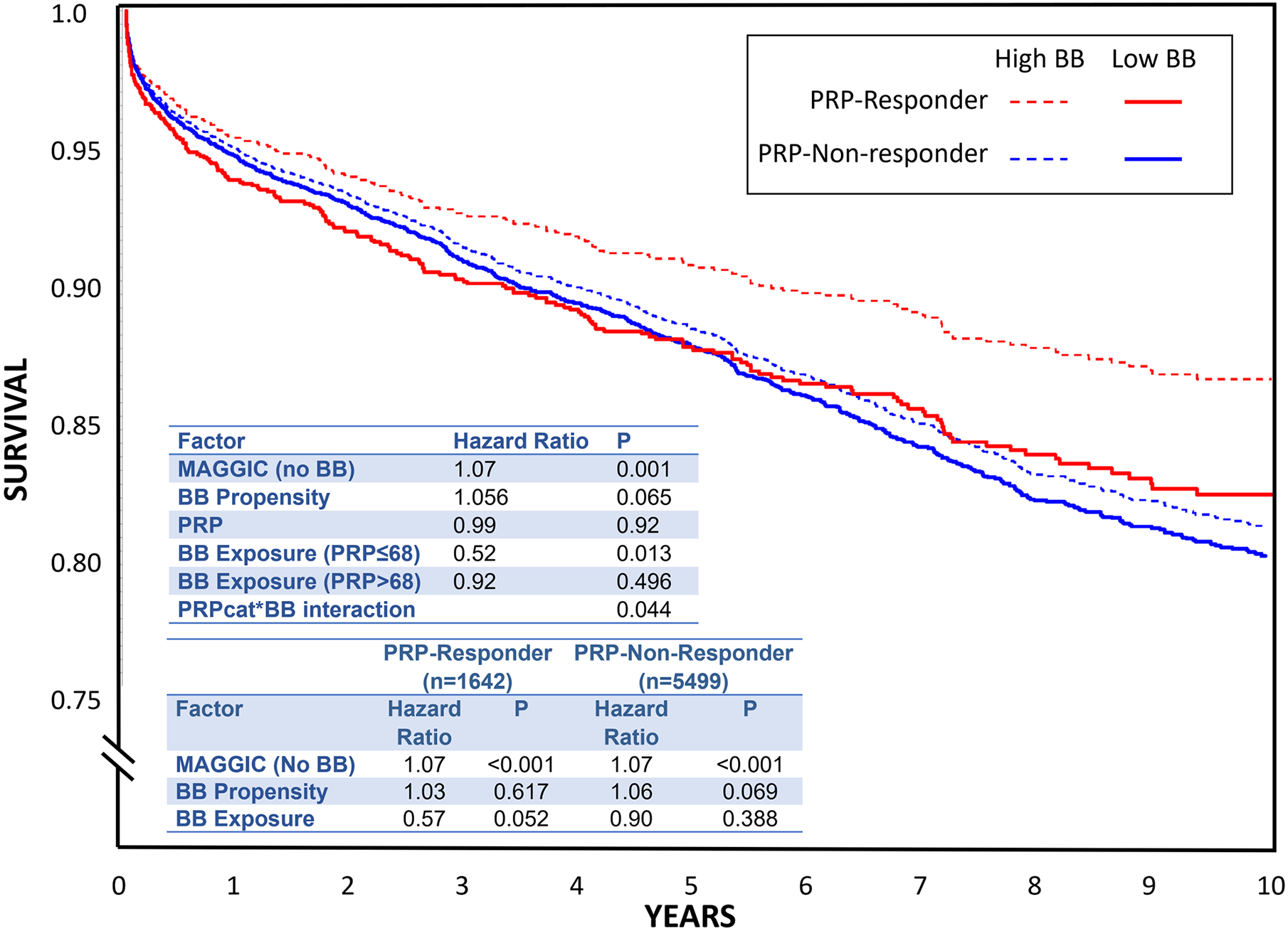
Survival curves from Cox modeling depicting first quartile (solid) versus third quartile (dotted) of beta blocker exposure, separated by PRP categories (red are predicted responders and blue are predicted non-responders). The space between the dotted and solid lines (of the same color) represents the survival impact of beta blocker exposure. Top embedded table is the output of the primary analysis. The bottom table shows results stratified by PRP category (predicted responder vs. predicted non-responder).
Overall, these findings are consistent with our earlier work, indicating that the PRP-predicted responders had substantial survival benefit with BB while the PRP-predicted non-responders did not. There are some limitations, such as lack of LVEF in most UKB participants and lack of medication dispensing data. However, these should bias toward the null, and the large sample size should compensate for some potential misclassification. Further, unrestricting LVEF potentially broadens the relevance of these findings. Since only survival was examined, other benefits of BB (e.g. heartrate or reverse remodeling) remain to be investigated. Finally, the most important limitation is that this score pertains solely to European ancestry, and there remains urgent need for improvements in polygenic scores for diverse populations. In conclusion, the BB-PRP replicated in HF patients from the UKB regardless of LVEF. This innovative tool could be the first effective genomic guide for BB therapy in HF, and now requires testing in a clinical trial. While not yet widely applied in many medications or disease states, the polygenic score approach to drug response is theoretically generalizable, warranting additional investigation.
Sources of Funding:
This research was supported by the National Heart, Lung, and Blood Institute (Lanfear R01HL103871, R01HL132154; Williams R01HL118267, R01HL141845; Luzum K08HL146990, L30HL110279). Dr. Lanfear is also supported in part by the National Institute for Minority Health and Disparities (P50MD017351). Dr. Williams is also supported in part by the National Institute of Allergy and Infectious Diseases (R01AI079139) and the National Institute of Diabetes and Digestive and Kidney Diseases (R01DK064695, R01DK113003).
Disclosures:
David E. Lanfear is a consultant for Abbott Laboratories, Amgen, Astra Zeneca, Cytokinetics, Illumina, Janssen, Martin Pharmaceuticals, Vicardia, and DCRI (Novartis) and has participated in clinical research from Amgen, AstraZeneca, Bayer, Critical Diagnostics, Lilly, Somalogic, and Janssen, and has a patent (held by Henry Ford Health System) for the beta blocker polygenic score. Hani N. Sabbah is consultant to Arena Pharmaceuticals, ViCardia, Stealth Biotherapeutics and Impulse Dynamics, Inc., and has received research funding from Novartis, Arena Pharmaceuticals, and Impulse Dynamics, Inc. Jasmine A. Luzum, Ruicong She, Jia Li, Bin Liu, Edward Peterson and. L. Keoki Williams have nothing to disclose.
References:
- 1.Writing C, Maddox TM, Januzzi JL Jr., Allen LA, Breathett K, Butler J, Davis LL, Fonarow GC, Ibrahim NE, Lindenfeld J, Masoudi FA, Motiwala SR, Oliveros E, Patterson JH, Walsh MN, Wasserman A, Yancy CW, Youmans QR. 2021 update to the 2017 acc expert consensus decision pathway for optimization of heart failure treatment: Answers to 10 pivotal issues about heart failure with reduced ejection fraction: A report of the american college of cardiology solution set oversight committee. J Am Coll Cardiol. 2021;77:772–810 [DOI] [PubMed] [Google Scholar]
- 2.Talameh JA, Lanfear DE. Pharmacogenetics in chronic heart failure: New developments and current challenges. Curr Heart Fail Rep. 2012;9:23–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lanfear DE, Luzum JA, She R, Gui H, Donahue MP, O’Connor CM, Adams KF, Sanders-van Wijk S, Zeld N, Maeder MT, Sabbah HN, Kraus WE, Brunner-La Rocca HP, Li J, Williams LK. Polygenic score for beta-blocker survival benefit in european ancestry patients with reduced ejection fraction heart failure. Circ Heart Fail. 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lanfear DE, Hrobowski T, Peterson EL, Wells K, Swadia T, Spertus JA, Williams LK. Association of beta blocker exposure with outcomes in heart failure differs between african american and white patients. Circ Heart Fail. 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pocock SJ, Ariti CA, McMurray JJ, Maggioni A, Kober L, Squire IB, Swedberg K, Dobson J, Poppe KK, Whalley GA, Doughty RN, Meta-Analysis Global Group in Chronic Heart F. Predicting survival in heart failure: A risk score based on 39 372 patients from 30 studies. Eur Heart J. 2013;34:1404–1413 [DOI] [PubMed] [Google Scholar]


