Abstract
Purpose:
We developed and validated two parsimonious algorithms to predict the time of diagnosis of any stage of acute kidney injury (any-AKI) or moderate-to-severe AKI in clinically actionable prediction windows.
Materials and Methods:
In this retrospective single-center cohort of adult ICU admissions, we trained two gradient-boosting models: 1) any-AKI model, predicting the risk of any-AKI at least 6 hours before diagnosis (50,342 admissions), and 2) moderate-to-severe AKI model, predicting the risk of moderate-to-severe AKI at least 12 hours before diagnosis (39,087 admissions). Performance was assessed before disease diagnosis and validated prospectively.
Results:
The models achieved an area under the receiver operating characteristic curve (AUROC) of 0.756 at six hours (any-AKI) and 0.721 at 12 hours (moderate-to-severe AKI) prior. Prospectively, both models had high positive predictive values (0.796 and 0.546 for any-AKI and moderate-to-severe AKI models, respectively) and triggered more in patients who developed AKI vs. those who did not (median of 1.82 [IQR 0-4.71] vs. 0 [IQR 0-0.73] and 2.35 [IQR 0.14-4.96] vs. 0 [IQR 0-0.8] triggers per 8 hours for any-AKI and moderate-to-severe AKI models, respectively).
Conclusions:
The two AKI prediction models have good discriminative performance using common features, which can aid in accurately and informatively monitoring AKI risk in ICU patients.
Keywords: Acute Kidney Injury, Artificial Intelligence, Machine Learning, Prediction
Introduction
Acute kidney injury (AKI) affects up to 20% of hospitalized patients and is associated with a myriad of short- and long-term adverse outcomes [1, 2]. Hospitalized patients with AKI experience longer lengths of stay, higher mortality, and increased costs [3-6]. After discharge, the comorbidity burden for AKI survivors increases, including chronic kidney disease [7] and cardiovascular disease [8]. Alongside these complications is a significant decline in the patients' quality of life [3, 9, 10]. Thus, there is an urgent need to develop strategies to limit the incidence and severity of AKI to minimize these deleterious effects.
Enhanced modeling of individualized risk has been proposed to address the burden of AKI in hospitalized patients. Early identification of high-risk patients, either based on clinical or laboratory features, could lead to the deployment of bundled interventions designed to limit the incidence, severity, and complications of the syndrome [11-14]. Risk prediction models developed for AKI have ranged from relatively simple (a select group of static comorbidities) [15, 16] to very complex (artificial intelligence-based with hundreds of features) [17-21]. While the simple models may be operationally more attractive, they fail to leverage the robust data obtained from contemporary electronic health records [22]. For instance, in the scenario where an otherwise stable patient underwent major thoracic surgery and experienced acute blood loss anemia with hemodynamic instability intraoperatively, the preoperative risk based on age, sex, or comorbid conditions would indicate minimal postoperative AKI risk. However, intraoperative data, including blood product, fluid, vasopressor use, laboratory parameters, or vital signs, would yield a higher postoperative risk than would otherwise have been expected [20].
Several artificial intelligence/machine learning models have been developed for AKI prediction [22]. These models vary in their enrolled population (ICU vs. ward vs. all hospitalized patients, key disease states like surgery or burns), choice of endpoint (any stage AKI, stages 2/3 AKI, need for kidney replacement therapy), the time horizon for prediction (e.g., 6-, 12-, 24-, 48-, 72-h), and level of complexity. The objective of this study was to develop an easily deployable machine learning model to predict AKI development or progression with clinically relevant lead times, ensuring enough time to institute meaningful clinical actions. To achieve this objective, we included two distinct aims for model development, including 1) predicting the risk of developing any stage of AKI 6 hours before its occurrence, which could be used for screening purposes, and 2) predicting the risk of developing moderate-to-severe AKI, 12 hours before its occurrence, to allow appropriate time for implementation of more preventive measures. In addition to these more mathematically complex models, we developed simpler linear regression approximations to ensure their generalizability, interpretation, and implementation.
Materials and Methods
Study population
We screened all intensive care unit (ICU) admissions to the Mayo Clinic Hospital (Rochester, MN) from January 1, 2005, to December 31, 2017. All adult, non-pregnant subjects who provided research authorization were included. Multiple admissions for each patient <10 hours apart were combined into a single encounter. We excluded encounters with 1) baseline creatinine > 5 mg/dL, 2) no recorded urine output or creatinine, 3) ICU admissions shorter than 24 hours, or 4) AKI stage 3 defined based solely on the need for renal replacement therapy (Appendix A, Supplementary Figure 1). AKI electronic alerts identified patients who met the AKI definition criteria[23]. For those who presented with AKI or developed AKI within the prediction windows, we used electronic alerts to exclude them as a prediction of AKI for those with AKI or individuals on the verge of AKI development was not the target of our study. For those who developed AKI during their ICU admission, AKI electronic alerts were used to adjudicate AKI as the outcome of interest for the models. Data from electronic health records (EHR), patient monitors, and patient outcomes were extracted for the cohort of interest.
This study was reviewed and approved by the Mayo Clinic institutional review board (IRB# 07-001380). The need for informed consent was waived due to the retrospectative nature and minimal-risk of this study.
Data Extraction and Definitions
We extracted expert-identified predictor features (vital signs, laboratory measurements, medications, and clinical interventions) and outcome variables (hourly urine outputs, serum creatinine, and continuous weight measurements) from the above described dataset (Appendix B). Variables were filtered for plausible values (Supplementary Table 1) and carried forward if missing (Supplementary Figure 2). Only 90 variables with prevalences of >5% in both modeling cohorts were used in the final analysis (Supplementary Table 2).
AKI stages throughout ICU stay were calculated using an existing electronic implementation of the KDIGO criteria [23]. The diagnosis time of AKI was defined as the first time the urine-output- or creatinine-based AKI criteria for any (stages 1, 2, or 3) or moderate-to-severe (stages 2 or 3) AKI were met. Encounters not meeting the AKI criteria were considered the control group (i.e., no AKI for any-AKI cohort and no AKI or AKI stage 1 for moderate-to-severe AKI cohort).
Model Training
Two models were trained to predict two distinct AKI outcomes: an any-AKI model to predict any-AKI (stage ≥1); and a separate moderate-to-severe AKI model to predict moderate-to-severe AKI (stage > 1). The models were trained at a specific time window in advance (the training time): six hours for any-AKI model and twelve hours for the moderate-to-severe AKI model. AKI diagnosis times for the control groups were randomly generated (Appendix C).
To ensure adequate input data for model training, we included encounters with an AKI diagnosis time greater than one hour after the prediction window, i.e., 6 and 12 hours for our two models. Thus, all encounters whose AKI diagnosis time was less than 7 hours from ICU admission were excluded from the any-AKI model. In addition, all encounters with moderate-to-severe AKI diagnosis times less than 13 hours from ICU admission were excluded from the moderate-to-severe AKI model. Finally, encounters with possible AKI at admission were also excluded from both models (Supplementary Figure 1).
The data for each model were split randomly into training (63%), testing (27%), and validation (10%), stratified by the maximum AKI stage. Both models were trained using gradient boosting (python package xgboost version 1.2.0) and used the same initial feature set of 90 features. A maximum tree depth of 3, 50 estimators, and a learning rate of 0.1 were used for both models, chosen using 5-fold cross-validation (Appendix D, Supplementary Figure 3). Boruta feature selection [24] was used to determine parsimonious feature sets for the final training of the models.
Model evaluation
Thresholds for each model were chosen to achieve a specificity of 90% at the training time.
Model performance was assessed with two time-frames: relative to AKI diagnosis and relative to ICU admission (Supplementary Figure 4). Performance relative to AKI diagnosis was quantified using the areas under the receiver operating characteristic (AUROC) and precision-recall (AUPRC) curves, and false/true positive/negative rates (Appendix E.1). Prospective performance (relative to ICU admission) was assessed using false/true positive/negative rates calculated based on timely model triggers (scores above the threshold) and model triggering rates over time from ICU admission until AKI diagnosis (Appendix E.2).
Feature importance was evaluated using SHAP (SHapley Additive exPlanations) values [25], calculated using the python package shap, version 0.30.0.
Simplified Models
For each outcome, a linear regression was fit to scaled gradient-boosting scores using only the top 10 features as input (Appendix F).
Results
Cohort Characteristics
A total of 50,342 encounters were included in the any-AKI cohort (58.3% prevalence of AKI), and 39,087 in the moderate-to-severe cohort (31.5% prevalence of moderate-to-severe AKI) (Appendix A). The two cohorts had broadly similar demographics and outcomes (Table 1). The intensity and stage of AKI were progressively associated with poorer outcomes. In any-AKI model cohort, ICU mortality rates in patients without AKI and patients with AKI stage 3 were 1.6% and 13.9%, respectively. In the same cohort, the ICU length of stay increased from a median of 43 hours (IQR 40 hours) among patients with no AKI to 127 hours (IQR 237 hours) among patients with AKI stage 3. The results for ICU mortality and length of stay for the moderate-to-severe AKI model cohort were similar. Patients with AKI stage 3 tended to have AKI diagnosis later in their stay: in any-AKI model cohort, patients with stage 3 first experience AKI an average of 47.78 hours after admission [SD 71.41 hours] versus an average 33.79 hours after admission [SD 42.75 hours] for patients with AKI stage 1.
Table 1:
Demographic and outcome information for encounters used in model training, testing, or validation
| Any AKI model | Mod/severe AKI model | |||||||
|---|---|---|---|---|---|---|---|---|
| Max AKI stage | No AKI | 1.0 | 2.0 | 3.0 | No AKI | 1.0 | 2.0 | 3.0 |
| # Encounters | 21,026 | 17,785 | 5,548 | 5,983 | 15,566 | 11,211 | 5,983 | 6,327 |
| Female * | 8,998 (42.8%) | 7,269 (40.9%) | 2,472 (44.6%) | 2,452 (41.0%) | 6,661 (42.8%) | 4,524 (40.4%) | 2,633 (44.0%) | 2,586 (40.9%) |
| Black * | 248 (1.2%) | 191 (1.1%) | 66 (1.2%) | 116 (1.9%) | 177 (1.1%) | 124 (1.1%) | 73 (1.2%) | 122 (1.9%) |
| BMI at admission | 28.11 (6.84) | 29.99 (7.44) | 33.44 (9.34) | 29.3 (8.44) | 28.04 (6.84) | 29.75 (7.59) | 33.46 (9.34) | 29.34 (8.39) |
| Death in ICU * | 328 (1.6%) | 697 (3.9%) | 448 (8.1%) | 831 (13.9%) | 263 (1.7%) | 483 (4.3%) | 524 (8.8%) | 928 (14.7%) |
| Death in Hospital * | 765 (3.6%) | 1,254 (7.1%) | 701 (12.6%) | 1,101 (18.4%) | 595 (3.8%) | 887 (7.9%) | 815 (13.6%) | 1,223 (19.3%) |
| ICU LOS (hours) # | 43.0 (40.0) | 63.0 (70.0) | 80.0 (104.0) | 127.0 (237.0) | 46.0 (43.0) | 72.0 (82.0) | 81.0 (106.0) | 132.0 (239.0) |
| Hospital LOS (hours) # | 149.0 (151.0) | 197.0 (206.0) | 240.0 (273.25) | 263.0 (428.0) | 153.0 (164.0) | 225.0 (251.0) | 248.0 (289.0) | 277.0 (455.0) |
| Hours to first stage > 0 | NaN | 33.79 (42.75) | 23.46 (29.72) | 47.78 (71.41) | NaN | 30.33 (93.62) | 20.97 (30.26) | 44.31 (70.99) |
| Hours to first stage > 1 | NaN | NaN | 46.86 (64.9) | 88.07 (130.14) | NaN | NaN | 47.28 (65.98) | 87.95 (129.26) |
Some encounters are used in both models. Variables marked with * are summarized as # (%); variables marked with # are summarized with median (IQR); all other variables are summarized with means (sd).
Performance Relative to AKI Diagnosis
At the training time of six hours before any-AKI, the any-AKI model had AUCs of 0.743 and 0.756 in the test and validation cohorts, respectively (Table 2). The AUPRC values were also good (0.796 in the test and 0.807 in the validation cohorts). At 90% specificity, the any-AKI model detected 38% of cases (sensitivity) in the testing and validation cohorts. The high specificity allowed for high positive predictive values: over 84% of the patients flagged by the model (having scores above the threshold 6 hours before AKI) developed AKI.
Table 2:
Model performance at the training windows
| Model | Cohort | AUC | AUPRC | Se (Sp=90%) | Sp (Sp=90%) | PPV (Sp=90%) |
|---|---|---|---|---|---|---|
| Any AKI | Training | 0.754 | 0.807 | 0.388 | 0.904 | 0.849 |
| Testing | 0.743 | 0.796 | 0.381 | 0.900 | 0.843 | |
| Validation | 0.756 | 0.807 | 0.385 | 0.905 | 0.851 | |
| Moderate/severe AKI | Training | 0.732 | 0.584 | 0.357 | 0.904 | 0.631 |
| Testing | 0.715 | 0.564 | 0.342 | 0.900 | 0.614 | |
| Validation | 0.721 | 0.559 | 0.341 | 0.903 | 0.613 |
Overall model performance at training window (6 hours for any AKI, 12 hours for moderate/severe AKI). Results for each model are shown for the training, testing, and validation sets. The thresholds for both models were chosen to yield a specificity of 90% in the test set. AUC=area under the receiver operating characteristic curve; AUPRC=area under the precision-recall curve; Se=sensitivity; Sp=specificity; PPV=positive predictive value.
At the training time of twelve hours before moderate-to-severe AKI, the moderate-to-severe AKI model had AUC values of 0.715 and 0.721 in the test and validation sets, respectively. The lower prevalence of moderate-to-severe AKI (31% vs. 58% in any-AKI) resulted in lower AUPRC values, 0.564 and 0.557, in the testing and validation sets, respectively. As with the any-AKI model, the required high specificity resulted in low sensitivity of 34% in both the testing and validation sets. However, positive predictive values (PPV) remained higher than the prevalence (31% prevalence vs. 61% PPV) in the testing and validation sets.
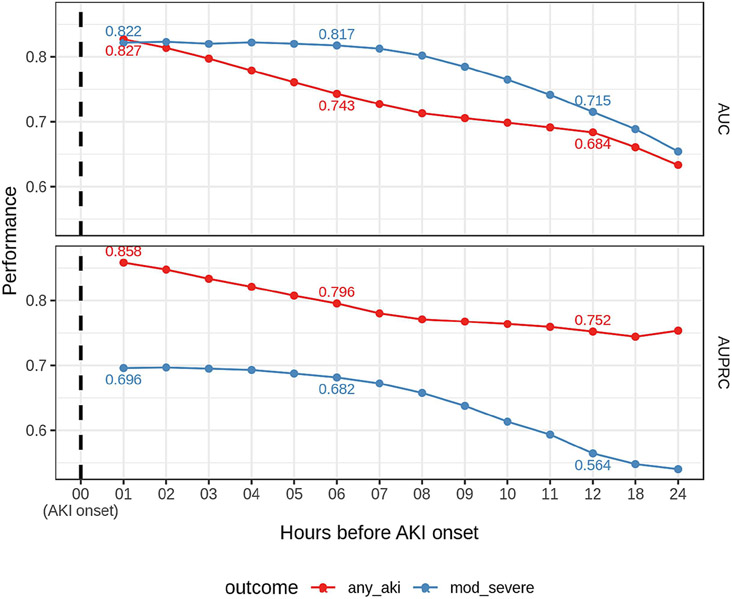
Figure 1 demonstrates the models' performances over time before AKI diagnosis. As expected, the closer to the event, the better the model performance. The moderate-to-severe AKI model demonstrated more consistent performance up to six hours before disease diagnosis than the any-AKI model, which displays a linear increase over time. Additionally, at each time point except one hour before AKI, the moderate-to-severe AKI model has a higher AUC. However, the moderate-to-severe model has lower AUPRC values than the any-AKI model due to the lower prevalence of the outcome, as also observed at the training window (Table 2).
Figure 1: Performance over time relative to AKI onset.
The area under the receiver operating characteristic curve (top panel) and the area under the precision-recall curve (bottom panel) over time relative to AKI onset. 'AKI onset' is the first time the stage is > 0 for the any-AKI model and the first time the stage is > 1 for the moderate-to-severe AKI model. Time zero represents AKI onset, hour 01, 1 hour before AKI onset, etc. Red indicates the performance of any-AKI model, and blue performance of the moderate-to-severe AKI model. Performance is shown in the test dataset.
Prospective model performance
In practice, the time of AKI diagnosis is unknown in advance, so we also examined how the models might perform at identifying AKI patients when run prospectively from ICU admission. We calculated scores for each model every hour after ICU admission. Patients were classified as true/false positive/negative based on how many times the model was triggered (had a score above the threshold) before AKI diagnosis (Supplementary Table 3). Thus, the performance of the prospective model was evaluated: how likely the models are to identify AKI patients in practice (Table 3).
Table 3:
Prospective model performance
| Model | Cohort | Se | Sp | PPV |
|---|---|---|---|---|
| Any AKI | Training | 0.610 | 0.775 | 0.790 |
| Testing | 0.605 | 0.776 | 0.791 | |
| Validation | 0.619 | 0.777 | 0.796 | |
| Moderate/severe AKI | Training | 0.689 | 0.734 | 0.543 |
| Testing | 0.687 | 0.733 | 0.544 | |
| Validation | 0.694 | 0.739 | 0.546 |
The model performance when evaluated prospectively. Encounters are counted as 'positive' if the model triggers in a 'timely' manner before onset (see Methods) and negative if not. Results for each model are shown for the training, testing, and validation sets. Se=sensitivity; Sp=specificity; PPV=positive predictive value.
Compared to performance at the training times (Table 2), both models demonstrated higher sensitivity, detecting a higher percentage of patients who developed AKI (38% vs. 60% for the any-AKI model; 34% vs. 68% for the moderate-to-severe AKI model). There was a corresponding decrease in the positive predictive value, from 85.1% to 79.6% and from 61.3% to 54.6% for the any-AKI model and moderate-to-severe AKI model, respectively.
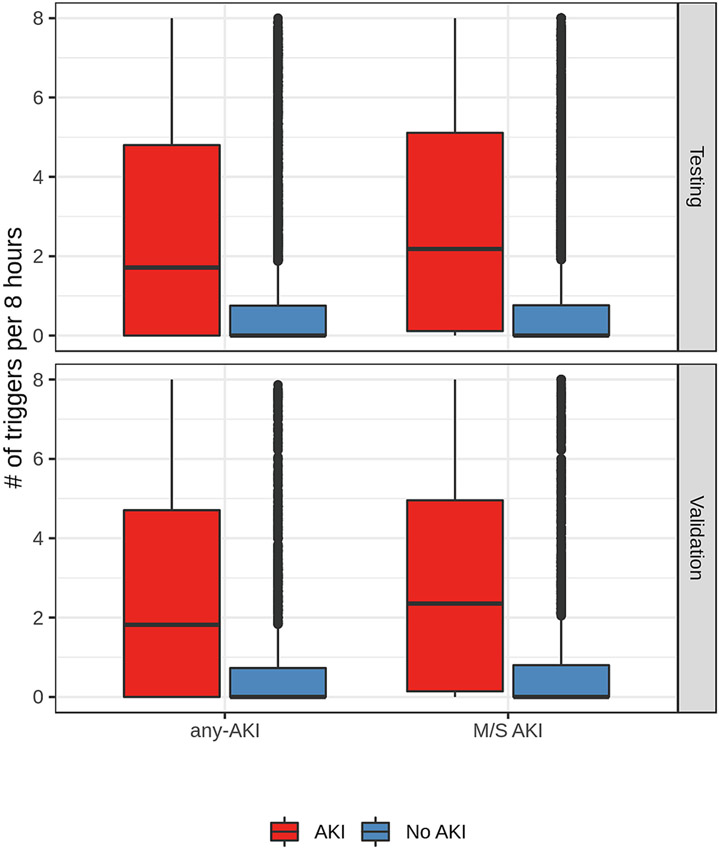
The models demonstrated good discrimination regarding the number of triggers among cases and controls (Figure 2). In both the test and validation datasets, AKI cases had a median of about two triggers every eight hours (any-AKI model 1.71 [IQR 0-4.8] in the test cohort and 1.82 [IQR 0-4.71] in the validation cohort; moderate-to-severe AKI model 2.18 [IQR 0.11-5.11] in the test cohort and 2.35 [IQR 0.14-4.96] in the validation cohort, while the control group had a median of zero triggers (any-AKI 0 [IQR 0-0.75] in the test cohort and 0 [IQR 0-0.73] in the validation cohort; moderate-to-severe AKI 0 [IQR 0-0.76] in the test cohort and 0 [IQR 0-0.8] in the validation cohort).
Figure 2: Number of triggers before AKI onset.
The distribution of the number of times the any-AKI model (left) and moderate-to-severe AKI model (right) trigger before AKI onset among cases (red) and controls (blue). Results are shown in the test (bottom) and validation (top) samples. AKI onset times for control patients are randomly generated.
Feature Importance
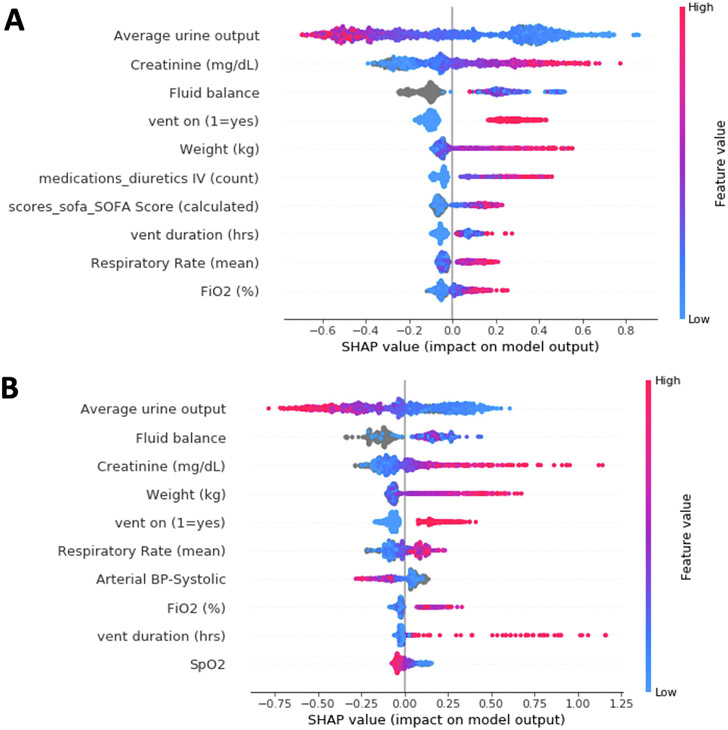
The feature importance ranking using SHaP values for both models is shown in Figure 3. While the two models share several critical elements, they also use some unique features. Urine output and measured serum creatinine values from six or 12 hours before AKI diagnosis were highly predictive for both models. Fluid balance, the need for invasive mechanical ventilation, and the most recent weight measure were also in the top five features for both models. The duration of invasive mechanical ventilation and average respiratory rate in the previous 24 hours were predictive for any-AKI and moderate-to-severe AKI. While the duration of invasive mechanical ventilation was more important for predicting any-AKI, the average respiratory rate was a more important feature in predicting moderate-to-severe AKI.
Figure 3: Features important in model prediction.
The top 10 features contributed to predictions for the any-AKI model (A) or moderate-to-severe AKI model (B).
There were also some distinctions in the predictive features. Physiological scores such as the Glasgow Coma scale and SOFA score were critical features for any-AKI but not for moderate-to-severe AKI. The use of IV diuretics in the previous 72 hours was an essential feature for any-AKI but not moderate-to-severe AKI. Conversely, vital signs such as heart rate, blood pressure, and respiratory rate were more important for predicting moderate-to-severe AKI.
Simplified model performance
The simplified any-AKI model performance was lower than the gradient boosting model measured by AUC and AUPRC (Supplementary Table 4). Unlike the gradient boosting model, with its more-sophisticated modeling framework, the simplified model assigned a positive weight to urine output rather than a negative one (Supplementary Table 5). Further, although the range of values for urine output is higher than for creatinine, the weight for creatinine assigned by the model was much higher than that for urine output (785 vs. 1).
The simplified moderate-to-severe AKI model (see Methods) had only a slight decrease in performance relative to the gradient boosting model as measured by AUC and AUPRC (Supplementary Table 4). The simplified moderate-to-severe model had a negative association with urine output, but a higher weight was assigned to creatinine despite the lower range of values (Supplementary Table 5).
Discussion and Conclusions
As the value of early recognition and prevention of AKI is progressively recognized, the interest in developing and validating AKI predictive models has grown over the past two decades [26-28]. While some models are designed to be calculated at or within the first 48 hours of ICU admission to evaluate AKI risk once (static models), other models can change their prediction based on incoming data (dynamic models). Most static predictions employ regression models using mostly baseline characteristics and some exposure data, including the reason for admission, laboratory information, and initial vital signs. However, dynamic models primarily rely on machine learning techniques and use features that change over time, like continuous vital signs and laboratory information [29]. In a systematic review of the static and dynamic hospital-acquired AKI prediction models, the AUROC range was reported as 0.66-0.80 in the internal validation cohorts and 0.65-0.71 in the external validation cohorts [26].
In this large-scale retrospective study, we developed and validated two dynamic models that use ICU data to predict any AKI stage within 6 hours before its diagnosis and moderate-to-severe AKI within 12 hours before it begins using data from the original cohort of 131,873 admissions. In addition, we demonstrated that the models' performance continually improves, starting from an AUC of 0.74 at 6 hours before 0.83 at 1 hour before diagnosis for any-AKI model and an AUC of 0.72 at 12 hours before 0.82 at 1 hour before diagnosis for moderate-to-severe AKI.
Our models have several significant strengths. First, we use a large heterogenous dataset, including more than 50,000 and 30,000 patients from multiple ICUs, to develop the any-AKI or moderate-to-severe AKI models. The cohort contains a good mix of medical and surgical ICU patients. Most models use small retrospective datasets with fewer than ten events per predictor, limiting the optimal performance of machine learning tools and potentially leading to over- or under-fitting [26]. Secondly, we used serum creatinine and urine output criteria to identify patients with AKI, capturing both presentations of AKI and thereby increasing the sensitivity of our alerts. Thirdly, we developed two models with different thresholds of AKI intensity: any-AKI stage vs. moderate-to-severe AKI, allowing clinicians to choose the appropriate AKI threshold for risk score calculation based on their clinical application and resource availability. Furthermore, our models start calculating risk scores immediately after ICU admission, enhancing their utility and lowering the incidence of missing opportunities to prevent AKI. Finally, the features we chose for our models are clinically relevant and physiologically associated with AKI.
The top five features were the same in both models, even though their orders differed. Both models ranked urine output, fluid balance, and weight as important, with urine output being the most important feature in both models. This finding reflects the high prevalence of AKI due to low urine output in the population. High creatinine values were associated with a higher risk of AKI. For the any-AKI model, the SOFA score indicative of the severity of illness and the number of diuretics administered in the previous 72 hours were important predictors. Patients on the mechanical ventilator, duration of ventilation, the fraction of inspired oxygen, and respiratory rate were also included in the top 10 features for both models. These features are typically measured frequently in ICU patients and are readily available in most EHRs.
Currently, available models are often very complex. They include many features to calculate the risk score, requiring advanced computations. This often limits their utility at the bedside unless their outputs are automatically calculated and incorporated within the medical records [26]. Therefore, in feature selection for inclusion in the final models, we limited the number of features to generate parsimonious models with minimal impact on their performance. While this strategy does not eliminate the need to compute the risk scores, it alleviates the risk of missing values. Further, we provided simple linear approximations that can be used in place of the gradient-boosting models when all ten inputs are available. Due to the more straightforward utility of the simplified models at the bedside, despite their lower performance, we suggest priority be given to the simplified moderate-to-severe AKI model for initial screening purposes. After identifying higher-risk patients using simplified models, full gradient-boosting models could be used for further details. This approach is particularly very practical for the model predicting moderate-to-severe AKI.
Many available dynamic models recalculate AKI risks in time blocks of 4-6 hours [17]. As the golden hours for AKI prevention and management are limited, the time lag between calculations could impact their potential benefits. Our models can handle missing data and show a risk score at ICU admission when minimal data becomes available. Moreover, our models are designed to recalculate the risk scores as soon as any additional information becomes available to patients' medical records, minimizing the risk of missing golden hours. In addition, the models are trained to predict patient risk 6 hours before any-AKI stage and 12 hours before moderate-to-severe AKI, which are clinically relevant time windows. The models' first triggers happened at a median of 15 (Any-AKI) and 25 (moderate-to-severe AKI) hours before disease diagnosis (Supplementary Table 8, Supplementary Figure 5).
Most currently available models are designed to alert clinicians of any change in AKI risk, leading to provider fatigue. We chose an alert cutoff with higher specificity to mitigate the risk of alarm fatigue, increase providers' trust in the risk prediction model, and assist with resource allocation to higher-risk patients in the case of limited resource availability. This choice was at the expense of achieving lower sensitivity in both models. Nevertheless, this is not unprecedented in the literature, as the model by Tomasev and colleagues had lower sensitivity of about 56% [17]. The models have a sensitivity of 38% (at 6 hours before any AKI) and 34% (at 12 hours before moderate-to-severe AKI) for a specificity of 90%. This indicates that the models will have very few false positives. Since this is a continuously running algorithm, it generates multiple alerts over time. Therefore, we defined a more realistic definition of timely alerts as the number of AKI alerts in 2 to 10 hours preceding the AKI stage change and moderate-to-severe AKI alerts in 2 to 18 hours preceding. This definition gave a higher sensitivity at the expense of lower specificity and positive predictive values (Table 3). Therefore, in a real-time deployment, the models can identify more patients while they have more false positives. We also evaluated the models by decreasing the targeted specificity, which resulted in higher sensitivity but lowered positive predictive values for both models (Supplementary Tables 6 and 7).
The two models can be used together to identify and manage AKI effectively. We propose selecting a high-sensitivity threshold for the any-AKI model. This will result in the model identifying most of the potential AKI cases. These patients can be put on an advanced monitoring protocol. The moderate-to-severe AKI model can be run on patients identified by any-AKI model and have a threshold optimized for high specificity. This will identify patients most likely to develop moderate-to-severe AKI with a low chance of false positives. These patients can be more intensively managed.
Few models are validated externally [26]. Furthermore, as curated retrospective datasets are used for model development and validation, performance often declines when applied to other datasets. Our models also validated performance internally. However, considering the inclusion of readily available features in most ICU data sets, we believe the impact of this limitation is alleviated to the greatest possible extent.
Current models are primarily based on retrospective datasets and are rarely validated prospectively. In a recent study, the authors used a prospective simulated strategy to validate the results of models based on deep learning techniques and showed poor performance in predicting acute events among hospitalized patients [30]. This may indicate a dire need for prospective validation of the models that have been developed. In our study, we simulated the models' prospective behavior, which did not result in a decline in performance.
A potential limitation to applying this model to all ICU patients comes from the cohort selection criteria. During the model training phase, we excluded patients who started RRT for reasons other than AKI; therefore, the models do not apply to that cohort. In addition, we also excluded patients who presented with community-acquired AKI or developed AKI soon after ICU admission from the model training. While our models can still calculate the risk of AKI development in these patients, their clinical applications remain uncertain.
AKI as a syndrome is an umbrella term for many etiologies, limiting the performance of these models in identifying the primary reason for each AKI episode. Like many other predictive models, our models cannot differentiate particular etiology resulting in AKI.
While the progress in the field has been palpable, the models have not been implemented in clinical practice for several reasons, including lack of external validation [26], lack of prospective or clinical validation, small training datasets, lack of cause determination, model complexity, burdensome alerting frameworks, and infrequent assessment of risk. We have taken steps to alleviate these limitations as much as possible. However, these drawbacks remain due to the lack of external data and prospective testing.
In conclusion, we developed and internally validated machine learning predictors of any moderate-to-severe AKI in a large cohort of adult ICU patients that provides high specificity with clinically relevant lead time to implement individualized preventive measures for patients at higher risk of AKI. The subsequent steps in evolving these models include the external and real-time prospective validations of their performance and assessing their impacts on the patients' clinical outcomes in large-scale multicenter clinical trials.
Supplementary Material
Highlights.
We developed and evaluated separate models for any stage of acute kidney injury (any-AKI or stage ≥ 1) and moderate-to-severe AKI (stage > 1)
The models are parsimonious, use commonly-available features, employ clinically-actionable lead times, and handle missing inputs
The models identified 80% of any-AKI patients and 55% of moderate-to-severe AKI patients between admission and KDIGO- based diagnosis.
Funding:
This project was supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number K23AI143882 (PI; EFB)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: Emma Schwager, Erina Ghosh, and Larry Eshelman are Philips Research North America employees. All other authors have disclosed that they do not have any conflicts of interest.
References:
- [1].Susantitaphong P, Cruz DN, Cerda J, Abulfaraj M, Alqahtani F, Koulouridis I, et al. World incidence of AKI: a meta-analysis. Clin J Am Soc Nephrol 2013;8(9):1482–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kashani K, Shao M, Li G, Williams AW, Rule AD, Kremers WK, et al. No increase in the incidence of acute kidney injury in a population-based annual temporal trends epidemiology study. Kidney international 2017;92(3):721–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nisula S, Vaara ST, Kaukonen KM, Reinikainen M, Koivisto SP, Inkinen O, et al. Six-month survival and quality of life of intensive care patients with acute kidney injury. Crit Care 2013;17(5):R250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jones J, Holmen J, De Graauw J, Jovanovich A, Thornton S, Chonchol M. Association of complete recovery from acute kidney injury with incident CKD stage 3 and all-cause mortality. Am J Kidney Dis 2012;60(3):402–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Silver SA, Chertow GM. The Economic Consequences of Acute Kidney Injury. Nephron 2017;137(4):297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dasta JF, Kane-Gill S. Review of the Literature on the Costs Associated With Acute Kidney Injury. J Pharm Pract 2019;32(3):292–302. [DOI] [PubMed] [Google Scholar]
- [7].Heung M, Steffick DE, Zivin K, Gillespie BW, Banerjee T, Hsu CY, et al. Acute Kidney Injury Recovery Pattern and Subsequent Risk of CKD: An Analysis of Veterans Health Administration Data. Am J Kidney Dis 2016;67(5):742–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Odutayo A, Wong CX, Farkouh M, Altman DG, Hopewell S, Emdin CA, et al. AKI and Long-Term Risk for Cardiovascular Events and Mortality. J Am Soc Nephrol 2017;28(1):377–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Johansen KL, Smith MW, Unruh ML, Siroka AM, O'Connor TZ, Palevsky PM. Predictors of health utility among 60-day survivors of acute kidney injury in the Veterans Affairs/National Institutes of Health Acute Renal Failure Trial Network Study. Clin J Am Soc Nephrol 2010;5(8):1366–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Villeneuve PM, Clark EG, Sikora L, Sood MM, Bagshaw SM. Health-related quality-of-life among survivors of acute kidney injury in the intensive care unit: a systematic review. Intensive care medicine 2016;42(2):137–46. [DOI] [PubMed] [Google Scholar]
- [11].Kashani K, Rosner MH, Haase M, Lewington AJP, O'Donoghue DJ, Wilson FP, et al. Quality Improvement Goals for Acute Kidney Injury. Clin J Am Soc Nephrol 2019;14(6):941–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Meersch M, Schmidt C, Hoffmeier A, Van Aken H, Wempe C, Gerss J, et al. Prevention of cardiac surgery-associated AKI by implementing the KDIGO guidelines in high-risk patients identified by biomarkers: the PrevAKI randomized controlled trial. Intensive care medicine 2017;43(11):1551–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gocze I, Jauch D, Gotz M, Kennedy P, Jung B, Zeman F, et al. Biomarker-guided Intervention to Prevent Acute Kidney Injury After Major Surgery: The Prospective Randomized BigpAK Study. Annals of surgery 2018;267(6):1013–20. [DOI] [PubMed] [Google Scholar]
- [14].Selby NM, Casula A, Lamming L, Stoves J, Samarasinghe Y, Lewington AJ, et al. An Organizational-Level Program of Intervention for AKI: A Pragmatic Stepped Wedge Cluster Randomized Trial. J Am Soc Nephrol 2019;30(3):505–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Malhotra R, Kashani KB, Macedo E, Kim J, Bouchard J, Wynn S, et al. A risk prediction score for acute kidney injury in the intensive care unit. Nephrol Dial Transplant 2017;32(5):814–22. [DOI] [PubMed] [Google Scholar]
- [16].Shawwa K, Ghosh E, Lanius S, Schwager E, Eshelman L, Kashani KB. Predicting acute kidney injury in critically ill patients using comorbid conditions utilizing machine learning. Clin Kidney J 2021;14(5):1428–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tomasev N, Glorot X, Rae JW, Zielinski M, Askham H, Saraiva A, et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature 2019;572(7767):116–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Churpek MM, Carey KA, Edelson DP, Singh T, Astor BC, Gilbert ER, et al. Internal and External Validation of a Machine Learning Risk Score for Acute Kidney Injury. JAMA Netw Open 2020;3(8):e2012892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Koyner JL, Carey KA, Edelson DP, Churpek MM. The Development of a Machine Learning Inpatient Acute Kidney Injury Prediction Model. Critical care medicine 2018;46(7):1070–7. [DOI] [PubMed] [Google Scholar]
- [20].Adhikari L, Ozrazgat-Baslanti T, Ruppert M, Madushani R, Paliwal S, Hashemighouchani H, et al. Improved predictive models for acute kidney injury with IDEA: Intraoperative Data Embedded Analytics. PLoS One 2019;14(4):e0214904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chiofolo C, Chbat N, Ghosh E, Eshelman L, Kashani K. Automated Continuous Acute Kidney Injury Prediction and Surveillance: A Random Forest Model. Mayo Clin Proc 2019;94(5):783–92. [DOI] [PubMed] [Google Scholar]
- [22].De Vlieger G, Kashani K, Meyfroidt G. Artificial intelligence to guide management of acute kidney injury in the ICU: a narrative review. Curr Opin Crit Care 2020;26(6):563–73. [DOI] [PubMed] [Google Scholar]
- [23].Ahmed A, Vairavan S, Akhoundi A, Wilson G, Chiofolo C, Chbat N, et al. Development and validation of electronic surveillance tool for acute kidney injury: A retrospective analysis. J Crit Care 2015;30(5):988–93. [DOI] [PubMed] [Google Scholar]
- [24].Kursa MB, Rudnicki WR. Feature Selection with the Boruta Package. J Stat Softw 2010;36(11):13. [Google Scholar]
- [25].Lundberg SM, Erion G, Lee S-I. Consistent Individualized Feature Attribution for Tree Ensembles. ArXiv 2018;abs/1802.03888. [Google Scholar]
- [26].Hodgson LE, Sarnowski A, Roderick PJ, Dimitrov BD, Venn RM, Forni LG. Systematic review of prognostic prediction models for acute kidney injury (AKI) in general hospital populations. BMJ Open 2017;7(9):e016591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wilson T, Quan S, Cheema K, Zarnke K, Quinn R, de Koning L, et al. Risk prediction models for acute kidney injury following major noncardiac surgery: systematic review. Nephrol Dial Transplant 2016;31(2):231–40. [DOI] [PubMed] [Google Scholar]
- [28].Van Acker P, Van Biesen W, Nagler EV, Koobasi M, Veys N, Vanmassenhove J. Risk prediction models for acute kidney injury in adults: An overview of systematic reviews. PLoS One 2021;16(4):e0248899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Barracca A, Contini M, Ledda S, Mancosu G, Pintore G, Kashani K, et al. Evolution of Clinical Medicine: From Expert Opinion to Artificial Intelligence. Journal of Translational Critical Care Medicine 2020;2(4):78–82. [Google Scholar]
- [30].Shah PK, Ginestra JC, Ungar LH, Junker P, Rohrbach JI, Fishman NO, et al. A Simulated Prospective Evaluation of a Deep Learning Model for Real-Time Prediction of Clinical Deterioration Among Ward Patients*. Crit Care Med 2021;49(8):1312–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.