SUMMARY
The C9orf72 hexanucleotide repeat expansion (HRE) is the most frequent genetic cause of the neurodegenerative diseases amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Here, we describe the pathogenic cascades that are initiated by the C9orf72 HRE DNA. The HRE DNA binds to its protein partner DAXX and promotes its liquid-liquid phase separation, which is capable of reorganizing genomic structures. An HRE-dependent nuclear accumulation of DAXX drives chromatin remodeling and epigenetic changes such as histone hypermethylation and hypoacetylation in patient cells. While regulating global gene expression, DAXX plays a key role in the suppression of basal and stress-inducible expression of C9orf72 via chromatin remodeling and epigenetic modifications of the promoter of the major C9orf72 transcript. Downregulation of DAXX or rebalancing the epigenetic modifications mitigates the stress-induced sensitivity of C9orf72-patient-derived motor neurons. These studies reveal a C9orf72 HRE DNA-dependent regulatory mechanism for both local and genomic architectural changes in the relevant diseases.
INTRODUCTION
Nucleotide repeat elements, including microsatellites or short tandem repeats, are common in eukaryotic genomes.1 Over 50 different types of genetic disorders, primarily neurological and neuromuscular, have been linked to expanded short nucleotide repeats.2 However, understanding the native functions of these repeat elements and their roles in human diseases is still at an early stage. In normal human populations, GGGGCC repeats are typically 2-10 units long in a non-coding region of the C9orf72 gene. This hexanucleotide repeat expansion (HRE) ranges from several hundreds to thousands of units of (GGGGCC)n and is the most common genetic cause for amyotrophic lateral sclerosis (ALS), which is characterized by motor neuron neurodegeneration, as well as for frontotemporal dementia (FTD), which affects the frontal and temporal lobes of the brain.3,4 The pathogenic mechanisms by which the HRE mutation leads to neurodegeneration remain a focus of investigation for the relevant neurodegenerative diseases.
The etiology of C9orf72 HRE-linked neurodegeneration has thus far involved several non-mutually exclusive hypotheses, including loss of C9orf72 functions, aberrant-repeat-containing RNAs, and repeat-associated non-ATG-dependent (RAN) translation.5 In loss-of-function disease mechanisms, both C9orf72 RNA and protein levels are reduced in patient cells and tissues; however, the regulation of C9orf72 expression under physiological or pathological conditions is not well understood. C9orf72 is a DENN-domain-containing protein that functions in several or- ganelles and processes, including lysosomes, mitochondria, and autophagy.6-11 The deficiency in C9orf72 decreases the fitness of human-patient-derived motor neurons or perturbs immune systems in animal models.12-14 In gain-of-function disease mechanisms, the repeat-containing RNAs sequester RNA-binding proteins,15,16 and the RAN translation generates poly-dipeptide repeats,17-19 thus leading to RNA and protein toxicity. Loss of C9orf72 functions synergizes with gain-of-function mechanisms in models of C9orf72 HRE-linked diseases.20 Unlike the extensive studies on the C9orf72 HRE RNA and its translational products, the pathogenic mechanisms arising from the repeat-containing DNA remain poorly understood, and it is unclear how the expanded repeat DNA and its protein partners contribute to either loss- or gain-of-function disease mechanisms.
Chromatin conformations and epigenetic modifications are critical for the regulation of genomic landscapes and gene expression and thus underlie transcriptional dysregulation in many neurodegenerative diseases.21 The eukaryotic genome is organized hierarchically and spatially, and chromosomes can fold into units of tens to hundreds of kilobases known as topologically associating domains (TAD),22 a conserved feature of genome organization that enables preferential local or longrange interactions within the domains. The dynamics of chromatin structure are closely related to epigenetic regulations such as modifications of DNA and histones. Liquid-liquid phase separation (LLPS) of chromatin-interacting proteins such as chromatin remodeling complexes and transcription mediators can drive the formation of chromatin compartmentation and long-range interactions,23-25 and aberrant chromatin remodeling and epigenetic modifications have been implicated in the etiology of ALS.26,27 In C9orf72 HRE-linked ALS/FTD, hypermethylated DNA and altered histone modifications have been observed in the repeat DNA and nearby sequences.28-30 However, the genome-wide landscapes of chromatin status in C9orf72 HRE patients are largely unexplored, and the mechanisms underlying the changes in the HRE site or the whole genome remain unknown.
Here, we report the identification of a key DNA-binding protein (DBP), DAXX, that recognizes the C9orf72 HRE DNA and regulates global chromatin structure and epigenetic modification as well as the transcription of the C9orf72 gene. The HRE mutations induce a nuclear accumulation of DAXX, which undergoes G4C2 repeat DNA-dependent LLPS and drives local and global changes in chromatin structure and epigenetic modification. We have uncovered a stress-inducible feature of C9orf72 gene expression, a biologically significant stress-responsive mechanism that is lost in the C9orf72 HRE patient cells in a DAXX-dependent manner. Our findings reveal mechanisms by which DAXX and its condensates shape genomic landscapes through chromatin remodeling and epigenetic modifications and illustrate pathogenic cascades initiated from the HRE DNA that affect both loss- and gain-of-function disease processes.
RESULTS
DBP DAXX marks C9orf72 HRE-dependent pathology in patient cells
To identify DBPs specifically associated with the C9orf72 HRE (C9HRE) DNA, we employed a quantitative proteomic analysis using stable isotope labeling with amino acids (SILAC). HEK293 cells were metabolically labeled with SILAC isotopes to saturation and then lysed and subjected to a pull-down assay using biotinylated double-stranded DNAs (dsDNA) of (G4C2)6 or a length-matched random sequence control (Figure S1A). Subsequent mass spectrometry analysis identified 309 proteins in the precipitates isolated by the (G4C2)6 probes (Table S1). Among them, 22 proteins were found to be highly enriched by the C9HRE DNA, of which 9 are transcriptional factors (Figure S1B), indicating a successful pull-down of DBPs. Notably, the most enriched protein that selectively recognized C9HRE DNA, as ranked by SILAC ratio (Table S1), was found to be DAXX, an epigenetic factor involved in transcriptional regulation.31,32
Several assays were used to validate the interaction between DAXX and the G4C2 repeat dsDNA. First, we confirmed that DAXX was significantly enriched in the nuclear precipitates pulled down by the (G4C2)6 dsDNA probe in an immunoblotting assay (Figure 1A). Next, to test whether DAXX protein and G4C2 repeat dsDNA interact directly, we purified human DAXX protein (Figure S1D) and performed an electrophoresis mobility shift assay (EMSA) using 50 Alex Fluor 488-labeled (G4C2)10 dsDNA or a length-matched random sequence control. The EMSA results demonstrated a significantly preferential binding of DAXX to (G4C2)10 dsDNA when compared with the random control probe (Figure S1C), confirming the specific recognition of the C9orf72 DNA repeat by DAXX. In addition, to establish the association of endogenous DAXX with the expanded C9orf72 G4C2 DNA repeats in situ, we carried out DNA fluorescent in situ hybridization (FISH) with DAXX immunostaining on multiple lines of ALS/FTD patient lymphocytes carrying C9orf72 HRE mutations. Consistent with the heterozygosity of the C9orf72 HRE, the DNA FISH probe revealed one single HRE locus in each labeled patient cell but not in the control cells. Moreover, each HRE-positive FISH dot was co-localized with one of the DAXX-positive granules, suggesting that the HRE DNA was associated with condensed DAXX in the nuclei (Figures 1B and S1F).
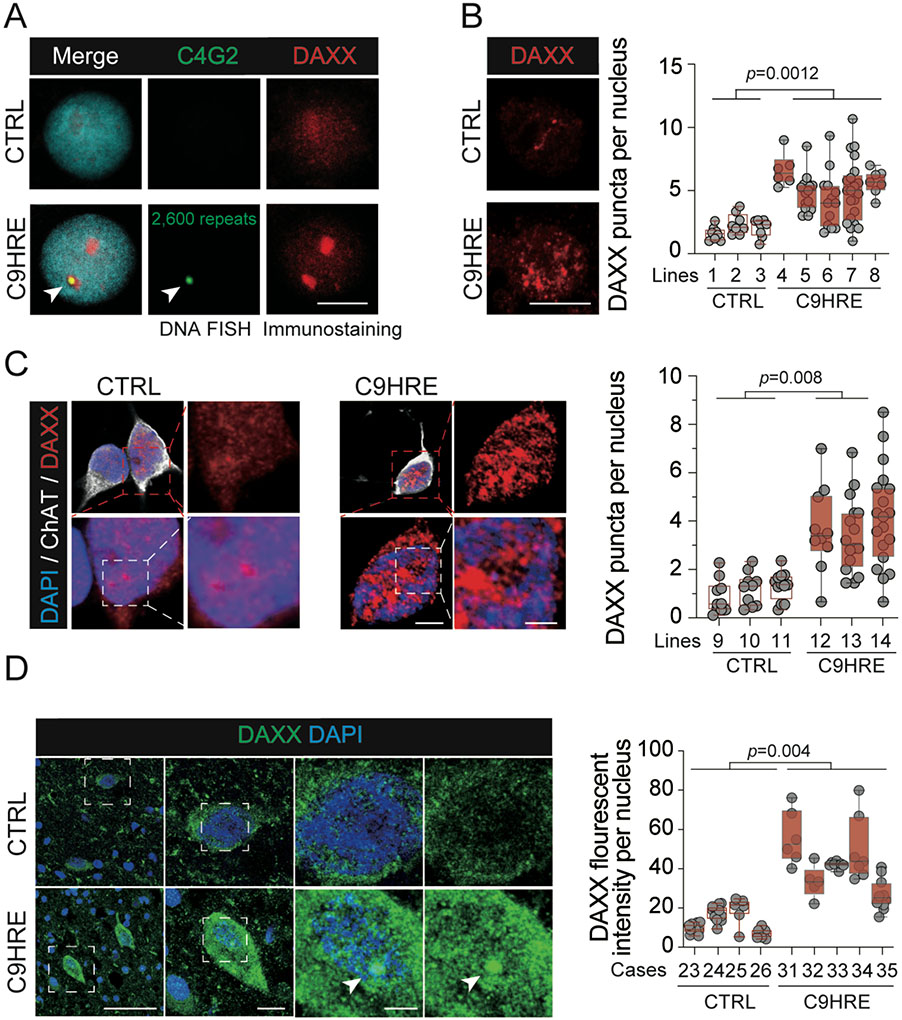
Figure 1. C9orf72 HRE-associated DAXX condensates in ALS patient cells.
(A) Immunoblotting of DAXX in the precipitates pulled down by a random or (G4C2)6 dsDNA biotin-labeled probe from nuclear fractions (n = 3 biological replicates).
(B) Co-localization of the C9HRE DNA locus with one of the DAXX condensates. Representative images show DNA FISH analysis of the HRE locus using an Alexa Fluor 488-labeled (C4G2)4 ssDNA probe, with co-immunostaining for DAXX, for C9-ALS patient B lymphocytes harboring a $2,600 G4C2 repeat and control cells without the expanded repeat. A focal plane in which the repeat locus and one of the DAXX puncta co-localize (arrowhead) is shown. Scale bar, 10 mm.
(C) Increased DAXX condensation in C9HRE ALS patient-derived motor neurons. Representative immunostaining images of DAXX in control and C9HRE motor neurons are shown. Ten fields containing 72–117 neurons from two independent slides for each cell line were examined (n = 3 independent pairs of iMN lines). Scale bars, 3 (left) and 1 (right) mm.
(D) Increased immunostaining of DAXX in C9HRE patient spinal cord neurons. Representative DAXX immunostaining images are shown, and neurons are identified through their characteristic shapes. Each dot represents the average intensity of DAXX immunofluorescence per nucleus in a field of view (n = 5 C9HRE cases and 4 control cases). Scale bars, 100, 10, and 5 mm (from left to right).
See also Figure S1.
In addition to the DAXX accumulation at the expanded G4C2 repeat locus, we observed an aberrant distribution pattern of DAXX throughout the nuclei in the C9HRE patient cells and tissues. Immunostaining of endogenous DAXX in multiple lines of B lymphocytes, induced pluripotent stem cell (iPSC)-differentiated motor neurons (iMNs), and spinal cord tissues showed significantly increased DAXX signals and enlarged DAXX-positive granules in the C9HRE cells and tissues when compared with those in controls (Figures 1C, 1D, and S1E). In the control B lymphocytes, iMNs, and spinal cord tissues, the signals for DAXX showed a largely diffuse pattern or occasional appearance of small dots in both the nucleus and cytoplasm (Figures 1C, 1D, and S1E). Consistent with the presence of nuclear localization signals in DAXX, it had a predominantly nuclear localization, but the motor neurons showed relatively more cytoplasmic distribution than the B lymphocytes. However, in the C9HRE cells and tissues, the immunostaining signal levels of DAXX were significantly increased in the nuclei of the B lymphocytes and in both the nuclei and the cytoplasm of the motor neurons, where the DAXX granules were either clearly demarcated or connected into a network (Figures 1C, 1D, and S1E). The consistent nuclear accumulation of DAXX suggests widespread nuclear changes in the patient cells and tissues.
Phase separation of DAXX remodels chromatin structures
In accordance with the granular structures formed by DAXX in the cells, an extended intrinsically disordered region identified at its C-terminal half (Figure S2A), and a previous report of DAXX undergoing phase separation,33 we observed that overexpression of DAXX enhanced its condensation in the nucleus (Figure S2C) and the fast fluorescence recovery after photo-bleaching of DAXX-GFP granules confirmed the LLPS feature of DAXX condensates in live cells (Figure S2B). Moreover, we demonstrated the intrinsic ability of purified DAXX protein to form liquid droplets in vitro (Figure 2A). Next, to test the role of the HRE dsDNA in DAXX condensation, we added (G4C2)10 or a size-matched control dsDNA probe to an LLPS-inducing solution containing DAXX protein. Compared with the control probe, the (G4C2)10 dsDNA significantly promoted formation of DAXX droplets (Figure 2A). Furthermore, to study the effects of longer G4C2 repeats on DAXX LLPS, we generated dsDNA fragments with 21 or 70 units of G4C2 repeats by restriction digestion of respective DNA plasmids. The longer (G4C2)n dsDNAs led to formation of significantly more DAXX droplets, while size-matched dsDNA controls lacked any effect (Figure 2B), indicating a repeat-length-dependent effect of C9HRE dsDNA on the LLPS of DAXX.
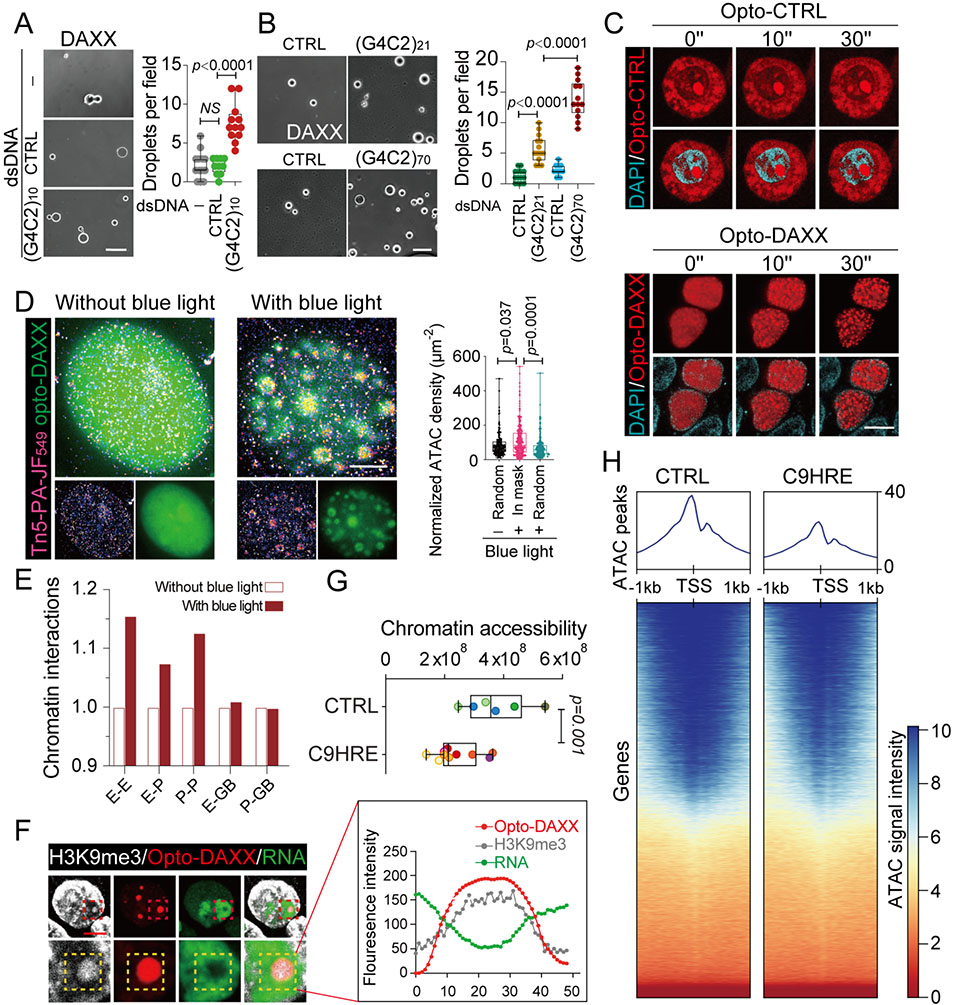
Figure 2. Liquid-liquid phase separation of DAXX reorganizes chromatin topology and spatial transcription.
(A) The HRE dsDNA promotes the liquid-liquid phase separation of DAXX. Purified DAXX was incubated in LLPS buffer (1 M NaCl) without dsDNA or with (G4C2)10 dsDNA or a size-matched control dsDNA at room temperature for 0.5 h. 10–12 fields of view in each group were quantified and statistically analyzed. Scale bars, 10 mm.
(B) The HRE repeat-length-dependent effects on the liquid-liquid phase separation of DAXX. Purified DAXX was incubated in LLPS buffer (0.5 M NaCl) with G4C2 dsDNAs of different repeat lengths or length-matched control dsDNAs at room temperature for 0.5 h. 9–13 fields of view in each group were quantified and statistically analyzed. Scale bar, 10 mm.
(C) Time-lapse images of the liquid-liquid phase separation of nuclear Opto-DAXX. Upon exposure to blue-light illumination, DAXX-mCherry-CRY2 is trans- located to the nucleus and forms discrete yet fusible condensates, while the mCherry-CRY2 control remains mostly diffuse in the cytoplasm in HEK293 cells. Scale bars, 10 mm.
(D) The 2D ATAC-PALM images show the accessible chromatins spatially restructured by the liquid-liquid phase separation of Opto-DAXX, activated by blue-light illumination. Approximately 200 condensates were statistically analyzed in each group. Scale bar, 3 mm.
(E) Genome interactions profiled by HiChIP among regulatory regions including promoters (P), enhancers (E), and gene bodies (GBs), with or without liquid-liquid phase separation of Opto-DAXX as a result of blue-light illumination for 10 min.
(F) Representative images and quantification of signals for H3K9me3 and nascent RNA at Opto-DAXX droplets after a 6-h illumination with blue light. Scale bar, 5 mm.
(G) Quantification of global chromatin accessibility, measured by ATAC-seq in six C9HRE and four control iMN lines (n = 6–10 biological replicates; different colors represent samples from independent iMN lines).
(H) Heatmaps of peak coverage 1 kb upstream and downstream of all TSSs for the ATAC-seq data from the C9HRE and control iMN lines in (G).
See also Figure S2.
To visualize the phase separation of DAXX in live cells, we developed an “Opto-DAXX” protein model by fusing DAXX with mCherry and a CRY2 domain, the latter being the photolyase homology region (PHR) of Arabidopsis thaliana that self-clusters upon blue-light illumination.34,35 In contrast to the control mCherry-CRY2 protein, which remained diffuse in the cytoplasm without detectable changes in its distribution, DAXX proteins with either the C-terminal or N-terminal fusion showed an exclusively nuclear distribution after exposure to blue light (Figures 2C and S2E) and formed abundant liquid droplets in a time-dependent manner when activated by the blue light (Figures 2C and S2D). These DAXX droplets were formed through either separation from a dispersed phase or fusion of existing droplets (Figures 2C and S2D). While a subset of the DAXX droplets were static, others showed dynamic behaviors of fusion and fission (Figures 2C and S2D). Next, we utilized the optogenetic system to study the functional consequences of DAXX condensation in live cells. Using time-lapse photography, we recorded the dynamic changes in Opto-DAXX and chromatin signals during the light-induced phase separation of Opto-DAXX. We observed that the condensation of Opto-DAXX profoundly changed the shape and intensity of chromatin, as visualized by DAPI staining (Figure S2F and S2G), suggesting that DAXX phase separation restructures the chromatin conformation.
To understand the effects of DAXX phase separation on genome topology, we employed HiChIP, a technology that combines in situ high-throughput chromosome conformation capture (Hi-C) with chromatin immunoprecipitation (ChIP),36 to profile three-dimensional chromatin architectures. We first exposed HEK293 cells expressing Opto-DAXX to blue-light illumination to induce the DAXX phase separation, and then subjected the cells to in situ Hi-C contact generation and ChIP analysis using an antibody against DAXX, with the same cells without blue-light illumination serving as a control. When the HiChIP sequencing results were analyzed, we found that the DAXX condensation had increased the long-range chromatin interactions among the regulatory regions, such as enhancers and promoters, throughout the genome, whereas the interactions associated with the gene bodies were unaffected (Figure 2E). The enhanced long-range interactions among the regulatory regions suggest that DAXX condensation may alter gene expression by modulating the chromatin spatial architecture.
To better visualize the changes in chromatin accessibility occurring as a result of DAXX phase separation, we employed a super-resolution chromatin-imaging technology, the assay for transposase-accessible chromatin-photoactivated localization microscopy (ATAC-PALM), which enables the visualization of the entire accessible genome on a nanometer scale through the use of a DNA probe comprising Tn5 transposase conjugated to bright photoactivatable Janelia Fluor 549 (Tn5-JF549) (Fig- ure S2H).37,38 In the absence of blue light, Opto-DAXX distribution was diffuse and the Tn5-JF549 probe was scattered evenly across the nuclei (Figures 2D and S2I). With a brief illumination by blue light for 5 min, however, DAXX became condensed into granules, and the Tn5-JF549 probe assembled onto these granules with a pattern of significant co-localization (Figures 2D and S2I). These data indicate that the acutely induced DAXX condensates were associated with accessible chromatins, consistent with the observation that DAXX condensation promoted interactions among the gene regulatory regions (Figure 2E), since these regulatory regions are typically located at accessible chromatin domains.39 These data also indicate that the DAXX condensation is a strong regulator of chromatin structures.
Having established the function of DAXX as an anchor for the interactions of regulatory sequences, we examined the transcription activity associated with the DAXX condensates. We first probed the co-localization of RNA Pol II with the DAXX condensates in Opto-DAXX-expressing HEK293 cells after the blue-light illumination. With a short exposure to blue light for 5 min, immunostaining analysis revealed that endogenous RNA Pol II was partially co-localized with the Opto-DAXX drop- lets; however, with a relatively long exposure to blue light for 4 h, RNA Pol II was excluded from the Opto-DAXX droplets (Figure S2J), suggesting low levels of transcription activity at these sites. Furthermore, by using 5-ethynyl uridine and click chemistry to visualize newly synthesized RNAs, we found that the signals for nascent RNAs were significantly decreased in the Opto-DAXX droplets after the long blue-light exposure (Figure 2F). Together, these data suggest that the phase separation of DAXX drives the assembly of distant open chromatins enriched in regulatory sequences and inhibits this transcription of these genomic regions (Figure S2K).
Increased DAXX dysregulates genome-wide gene expression in C9HRE cells
The phase separation of DAXX is expected to be influenced by its protein concentrations. In accordance with the increase in DAXX condensates in the C9HRE cells and spinal cord tissues (Figures 1C, 1D, and S1E), we found that the protein levels of DAXX on immunoblots were consistently higher in the C9HRE patient iMNs (Figures 3A and 3B), spinal cords (Figures 3F and 3G), and B lymphocytes (Figure S3A) than in the control samples. Subcellular nucleocytoplasmic fractionation analysis of the B lymphocytes confirmed the nuclear accumulation of DAXX in the patient cells (Figure S3B), consistent with the increase observed in the nuclear DAXX condensates (Figure S1E). Moreover, deletion of the expanded hexanucleotide repeats in C9HRE iMNs led to significantly reduced DAXX protein levels in the resulting isogenic control iMNs, confirming that the aberrant DAXX accumulation was a consequence of the C9HRE mutation (Figure S3C).
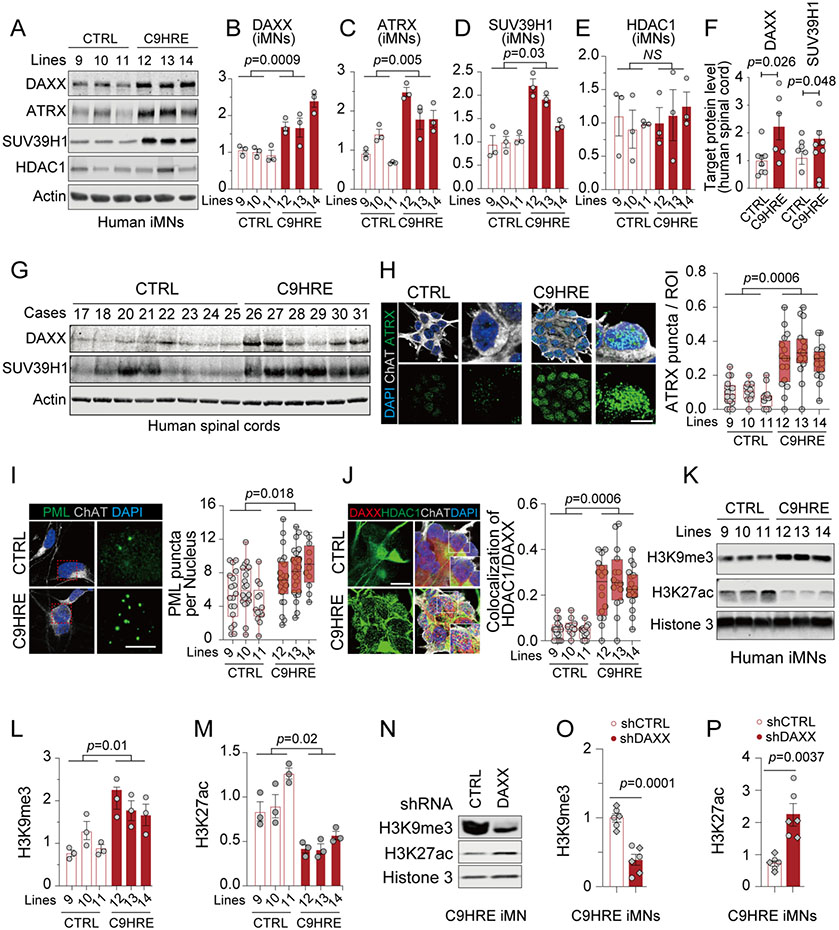
Figure 3. Increases in nuclear DAXX and its condensation are associated with epigenetic dysregulation in C9HRE patient cells.
(A–E) Immunoblotting of DAXX and related epigenetic regulators in control and C9HRE iMNs (n = 3 independent pairs of iMN lines; each dot represents a biological replicate).
(F and G) Immunoblotting of DAXX and SUV39H1 in the cervical spinal cords from C9HRE patients and controls (n = 6–8).
(H) ATRX condensates in control and C9HRE iMNs, visualized by immunostaining (n = 3 independent pairs of iMN lines; each dot represents the percentage of punctum area in regions of interest [ROIs]). Scale bars, 10 (left) and 5 (right) mm.
(I) Immunostaining of nuclear PML puncta in control and C9HRE iMNs (n = 3 independent pairs of iMN lines; each dot represents the average number of puncta per nucleus in a field of view). Scale bars, 10 (left) and 5 (right) mm.
(J) Association of HDAC1 with DAXX in the nuclei of control and C9HRE iMNs, as shown by co-immunostaining (n = 3 independent pairs of iMN lines; each dot in the graph represents the percentage of DAXX condensates co-localized with HDAC1 in a nucleus). Scale bar, 10 mm.
(K–M) Immunoblotting of H3K9me3 and H3K27ac normalized against total Histone 3 in control and C9HRE iMNs (n = 3 independent pairs of iMN lines; each dot represents a biological replicate).
(N–P) Knockdown of DAXX reduces the level of H3K9me3 but increases that of H3K27ac in the C9HRE iMNs (n = 6 biological replicates; different shapes of dots represent independent iMN lines). The level of total Histone 3 was used for normalization.
See also Figure S3.
Given the increase in nuclear levels of DAXX protein and its condensates in the C9HRE patient cells, we asked how DAXX might affect genome-wide gene expression in these cells. Consistent with the nuclear enrichment of DAXX in C9HRE cells, DAXX ChIP-seq analysis showed more DAXX-binding sites in the genomes of the HRE-harboring B lymphocyte cells (1,162 peaks) than in the control cells (836 peaks) (Figure S3D). To obtain a comprehensive understanding of the chromatin status in the C9HRE cells, we performed the assay for transposase-acces- sible chromatin with high-throughput sequencing (ATAC-seq) using iMNs from C9HRE patients and healthy individuals. By combining our ATAC-seq data with those from the NEUROLINCS database, we compiled an ATAC-seq dataset derived from iMNs of ten C9HRE patients and six controls. As compared with the control iMNs, the C9HRE iMNs generally showed lower ATAC-seq peak signals, suggesting a more compact genomic state under these conditions (Figure 2G). Since the chromatin accessibility of transcription start sites (TSSs) is essential for gene expression, we performed a more detailed analysis of the ATAC-seq data around TSSs and observed that the chromatin accessibility at TSSs in the ge- nomes of C9HRE patient iMNs was lower than that in control iMNs (Figure 2H).
To explore whether DAXX globally represses gene transcrip- tion in C9HRE cells, we performed RNA-seq analysis of C9HRE iMNs. Compared with isogenic control iMNs, we identified ~2,000 genes that were downregulated in C9HRE iMNs. When DAXX was partially knocked down in the C9HRE iMNs, the expression of a fraction of the downregulated genes (358 out of ~2,000 genes) was restored (Figure S3E, Supplementary Table 2), indicating that DAXX globally suppresses gene transcription in C9HRE iMNs. By analyzing the ATAC-seq data of the 358 DAXX-regulated genes, we identified 141 genes that exhibited reduced chromatin accessibility and no genes with increased chromatin accessibility in the C9HRE iMNs (Figure S3F). Thus, beyond the C9orf72 locus, there were at least hundreds of genes that were dysregulated by the aberrant DAXX accumulation in the C9HRE iMNs.
DAXX alters epigenetic regulations in C9HRE patient cells
Histone post-translational modifications are important epigenetic markers that induce changes in chromatin structures and transcriptional regulation. DAXX has been reported to form a complex with the transcriptional regulator ATRX and then recruit the histone lysine methyltransferase SUV39H1 to methylate H3K9 into H3K9me3.31 Along with the increase in nuclear DAXX proteins shown by immunoblotting analysis, we observed that the levels of ATRX in the patient-derived iMNs were higher than those in control iMNs (Figures 3A and 3C). Moreover, the levels of SUV39H1 were consistently higher in the C9HRE ALS patient iMNs (Figures 3A and 3D) and spinal cords (Figures 3F and 3G) than those of the controls. Immunostaining analysis indicated that the quantity and size of the ATRX granules were significantly increased in the nuclei of the patient iMNs when compared with those in control iMNs (Figure 3H). Similarly, C9HRE B lymphocytes showed more ATRX granules than did the controls; interestingly, a subset of the ATRX granules co-localized with the DAXX granules, suggesting that ATRX co-condensed with DAXX in the granules (Figures S3G-S3I). The PML nuclear bodies (PML-NB) serve as a scaffold where DAXX and ATRX shuttle in and out to modify chromatins,40 and we found that the number of PML-NBs was significantly higher in the C9HRE patient iMNs than in control iMNs (Figure 3I). In addition, DAXX was reported to inhibit the acetylation of H3K27 via the histone deacetylase HDAC1.41,42 Although the total levels of HDAC1 were unchanged in the patient cells, we observed a significant increase in the nuclear localization of HDAC1, which showed a significant co-localization with DAXX condensates in the C9HRE iMNs (Figure 3J) and B lymphocytes (Figures S3J-S3L) when compared with that in the control cells, suggesting that DAXX phase separation could alter HDAC1 activity at specific genomic regions in the patient cell lines.
One of the main functions of DAXX in the nucleus is related to histone modifications, specifically promoting H3K9me3 and suppressing H3K27ac to regulate gene expression.31,41 Given the observation that DAXX was increased in C9HRE patient cells (Figures 3A and 3B), especially in the nuclei (Figures 1B, 1C, S1E, and S3B), we examined the global levels of H3K9me3 and H3K27ac in the C9HRE patient iMNs by immunoblotting. We found that the C9HRE iMNs displayed significantly higher levels of H3K9me3 and lower levels of H3K27ac than did the control iMNs (Figure 3K-M). Importantly, the knockdown of DAXX rebalanced the global dysregulation of H3K9me3 and H3K27ac in the patient iMNs (Figures 3N-3P), demonstrating that DAXX plays a critical role in the dysregulation of the epigenetic histone markers in the C9HRE patient cells.
DAXX suppresses chromatin accessibility at the C9orf72 locus in patient cells
To gain further insights into the DAXX-mediated regulation of gene expression, we focused our analysis on the C9orf72 gene, which harbors the HRE mutation that is a prominent binding site of DAXX. The C9orf72 gene uses alternative start sites and splicing to produce at least three transcriptional variants V1-V3, and the HRE mutation is located either in the intron 1 region of transcripts V1 (NM_145005.5) and V3 (NM_001256054.1) or in the putative promoter region of the transcript V2 upstream of its TSS (NM_018325.3) (Figure S4A). Consistent with previous reports,5,43 V2 was the predominant transcript among the three variants, accounting for approximately 80%-90% of the C9orf72 transcripts (Figures S4B and S4C). We searched the Eu- karyotic Promoter Database44 and identified two GC box sites within 5 kb upstream of the V2 TSS (Figure S4D). The first GC box, a 10-bp segment 257 bp upstream of the V2 TSS, is located within exon 1a of the V1/V3 transcript; the second GC box, a 49-bp segment immediately upstream of the V2 TSS, comprises the entire intron 1b of the V1/V3 transcript (Figure S4D). Given the second GC box’s significant length and proximity to the TSS, we postulated that this region has the promoter function for the V2 transcript. Indeed, when we fused intron 1b with an EGFP-coding sequence, it drove a robust expression of EGFP protein and mRNA (Figures S4E-S4G), confirming the promoter activity of the region, hereafter termed the C9V2 promoter. The G4C2 repeat is located immediately upstream of the C9V2 promoter (Figure S4D), and expansion of the repeats could disrupt V2 expression. By analyzing the ATAC-seq dataset derived from iMNs of C9HRE patients and controls, we found that the patient iMNs exhibited a significant reduction in the chromatin accessibility at the C9V2 promoter region when compared with that of the control iMNs, (Figures 4A and 4B). Consistently, B lympho- cytes harboring the C9HRE mutation exhibited a similar loss of chromatin accessibility at the C9orf72 promoter region (Figure S5B). Since V2 is the predominant variant among C9orf72 transcripts, these data are consistent with the notion that a reduction in the V2 expression underlies the loss of C9orf72 expression in patient cells with HRE mutations.3,45
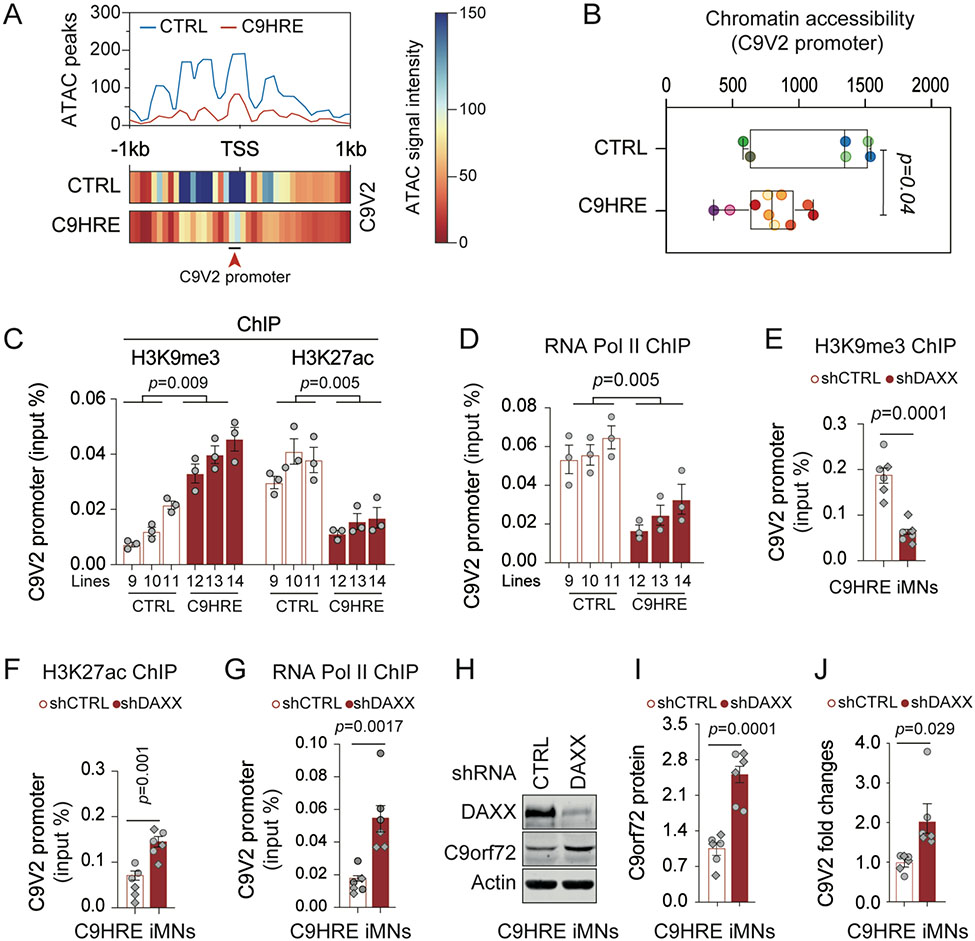
Figure 4. DAXX mediates HRE-associated chromatin abnormalities and transcriptional repression at the C9V2 promoter in C9HRE iMNs.
(A and B) Chromatin accessibility at the C9V2 promoter site, as indicated by peak coverage and heatmaps (A) and the quantification of the ATAC signals (B) in control and C9HRE iMNs (n = 6–10 biological replicates; different colors represent independent iMN lines).
(C and D) ChIP-qPCR analysis of the occupancies of endogenous H3K9me3, H3K27ac, and RNA Pol II at the C9V2 promoter region in control and C9HRE iMNs (n = 3 independent pairs of iMN lines).
(E–G) ChIP-qPCR analysis of the occupancies of H3K9me3, H3K27ac, and RNA Pol II at the C9V2 promoter region in C9HRE iMNs upon the knockdown of DAXX (n = 6 biological replicates; different shapes of dots represent independent iMN lines).
(H–J) Immunoblotting of C9orf72 protein (H and I) and qRT-PCR analysis of the C9V2 mRNA (J) in C9HRE iMNs upon the knockdown of DAXX (n = 6 biological replicates; different shapes of dots represent independent iMN lines).
See also Figures S4 and S5.
To understand the epigenetic mechanism through which the expansion of the G4C2 repeats influences V2 expression, we examined the occupancy of H3K9me3 or H3K27ac, the markers for transcription suppression or activation, respectively, at the V2 promoter in C9HRE patient iMNs. Using ChIP-qPCR with antibodies specifically against H3K9me3 or H3K27ac, we observed a profound increase in H3K9me3 occupancy as well as a substantial decrease in H3K27ac occupancy at the V2 promoter in the C9HRE patient iMNs as compared with those in control iMNs (Figure 4C), suggesting reduced transcription activity at the promoter region containing the HRE mutation. Furthermore, ChIP-qPCR analysis using antibodies against RNA polymerase II showed a significant reduction in the polymerase occupancy at the V2 promoter in the C9HRE patient iMNs when compared with that in the control iMNs (Figure 4D), confirming the repres- sive transcription status of this region containing the HRE mutation. In addition, B lymphocytes from C9HRE patients also showed a much lower occupancy of RNA polymerase II at the V2 promoter than did control cells (Figure S5C).
To understand the role of DAXX in HRE-dependent genomic regulation, we evaluated the effects of DAXX knockdown on epigenetic and transcriptional changes in C9HRE patient iMNs. A partial knockdown of DAXX significantly reduced the occupancy of H3K9me3 at the C9V2 promoter but increased the occupancy of H3K27ac at this site, as indicated by ChIP-qPCR analysis (Figures 4E and 4F). Furthermore, the DAXX knockdown led to a significant increase in the recruitment of RNA polymerase II to the C9orf72 V2 promoter in the C9HRE iMNs (Figure 4G) and B lymphocytes (Figure S5D). Accordingly, we found that the DAXX knockdown significantly increased the expression of both C9orf72 protein and V2 mRNA in the C9HRE iMNs (Figures 4H-4J) as well as in iPSCs (Figures S5E-S5H) and B lymphocytes (Figures S5I-S5L) carrying the HRE mutations. These results collectively indicate that the HRE mutations alter the epigenetic modifications and reduce the chromatin accessibility of the locus, thereby repressing the expression of the C9orf72 gene and demonstrate that DAXX plays a critical role in mediating the HRE-dependent transcriptional suppression.
Stress-dependent induction of C9orf72 expression is lost in HRE-harboring patient cells
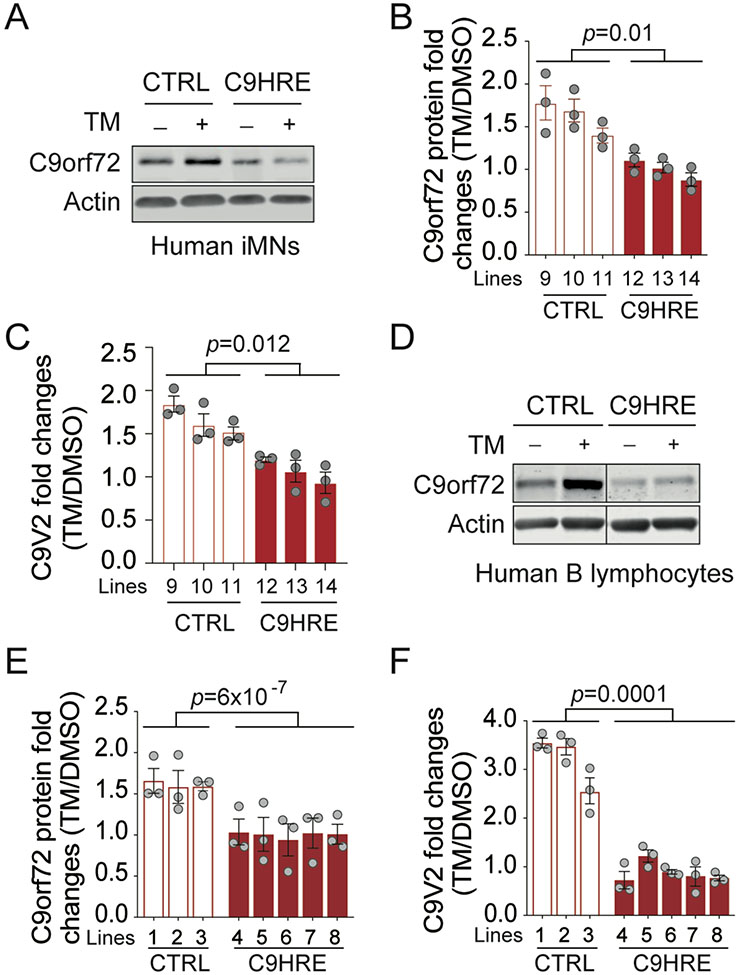
In addition to the reduction in the basal levels of C9orf72 protein and mRNA (Figures S6A-S6D), we found that the expression of C9orf72 was inducible by stress in normal cells, but this dynamic regulation was lost in patient cells harboring the HRE mutations. We therefore examined the transcriptional regulation of C9orf72 under neurodegeneration-associated stress conditions, such as the stress by tunicamycin, which inhibits protein N-glycosylation to disrupt ER-associated degradation,46 or thapsigargin, which induces ER stress by selectively inhibiting ER Ca+-ATPases.47 When the iMNs from healthy individuals were stressed by exposure to tunicamycin, both C9orf72 protein and V2 mRNA levels were found to be significantly increased (Figures 5A-5C). In contrast, such tunicamycin-induced upregulation of C9orf72 expression was absent from the iMNs derived from C9HRE patients (Figures 5A-5C). Similarly, the stress-responsive induction of C9orf72 expression at both the protein and mRNA levels was observed in normal B lymphocytes treated with either tunicamycin or thapsigargin, but the dynamic regulation of C9orf72 expression was lost in multiple lines of B lymphocytes from C9HRE patients (Figures 5D-5F and S6E-S6I).
Figure 5. Stress-dependent induction of C9orf72 is impaired in C9HRE ALS patient cells.
(A–C) Immunoblotting of C9orf72 protein (A and B) and qRT-PCR analysis of the C9V2 mRNA (C) in control and C9HRE iMNs treated with 5 mg/mL tu- nicamycin (TM) or DMSO for 24 h (n = 3 independent iMN lines; each dot represents a biological repli- cate).
(D–F) Immunoblotting of C9orf72 protein (D and E) and qRT-PCR analysis of the C9V2 mRNA (F) in control and C9HRE B lymphocytes treated with 1 mg/mL TM or DMSO for 24 h (n = 3–5 independent cell lines; each dot represents a biological replicate). See also Figure S6.
The stress-responsive induction of the C9orf72 V2 transcript and the corresponding protein was also observed in other cell types, such as HEK293 cells (Figure S6J) and human RPE1 cells (Figures S6K-S6M). We further observed that the expression of C9orf72 could be induced by stress from toxic proteins associated with neurodegenerative diseases. Proline-arginine (PR) poly-dipeptides are one type of proteotoxic products significantly increased the levels of the C9orf72 pre-mRNA translated from expanded G4C2 repeat RNAs. By expressing and protein (Figures S6N and S6O). Notably, only the construct with randomized codons encoding 82 repeats of C9orf72 V2 transcript, and not the V1 or V3 transcript, was poly-PR dipeptides (PR82) in RPE1 cells, we found that PR82 significantly changed, indicating that the stress-dependent regulation of C9orf72 expression is specific to the V2 transcript (Figure S6P).
DAXX mediates HRE-dependent inhibition of stress-induced transcription of C9orf72
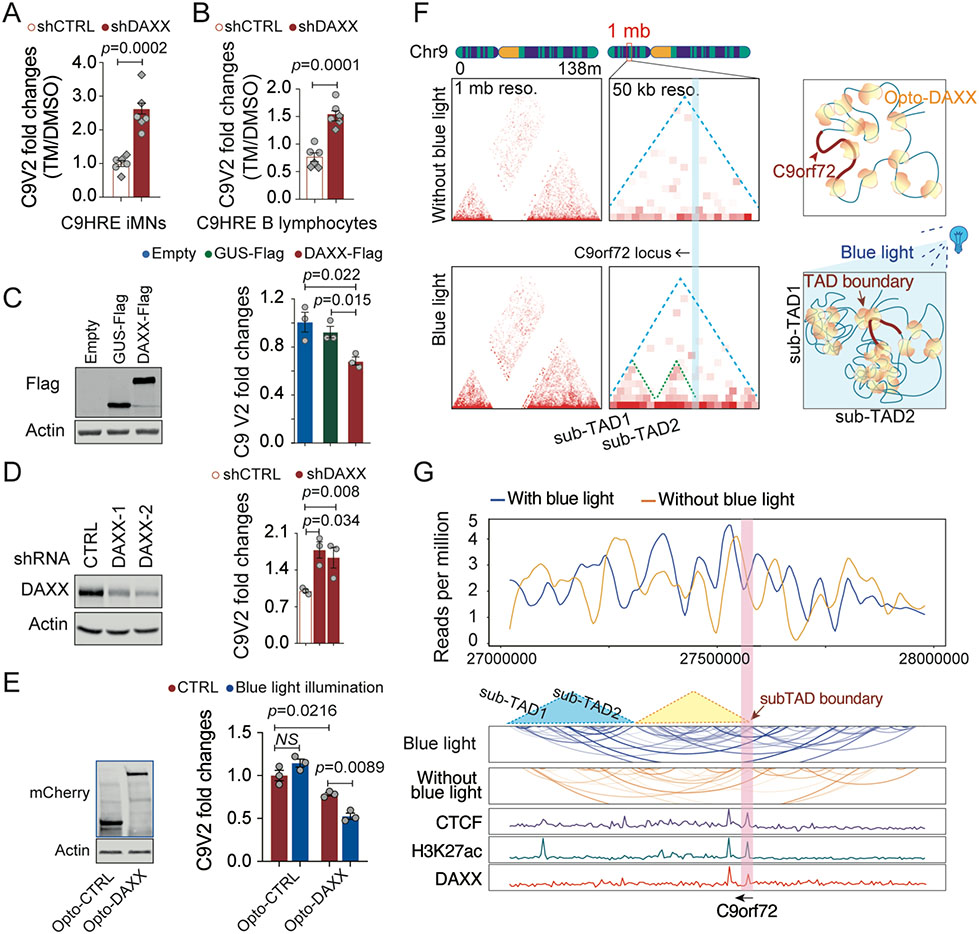
Since DAXX was found to accumulate in the nuclei of cells harboring the C9orf72 HRE mutation, we asked whether DAXX mediates the inhibition of stress-induced C9orf72 transcription in patient cells. We performed qRT-PCR analysis to quantify the stress-induced expression of the C9orf72 V2 transcript in pa- tient cells after knockdown of DAXX. We found that knockdown of DAXX in either C9HRE iMNs or C9HRE B lymphocytes restored the tunicamycin-induced upregulation of the V2 mRNA levels, which did not occur in the control shRNA-treated C9HRE iMNs (Figures 6A and 6B). Together, these data demonstrate that DAXX mediates HRE-dependent inhibition of the stress-induced transcription of C9orf72.
Figure 6. DAXX phase separation mediates HRE-associated C9orf72 suppression.
(A and B) qRT-PCR analysis of stress-induced expression of the C9V2 mRNA in C9HRE iMNs and human B lymphocytes upon knockdown of DAXX by shRNAs. The cells were treated with 5 mg/mL tunicamycin (TM) or DMSO for 24 h (n = 6 biological replicates; different shapes of dots represent independent iMN lines). (C) Fold changes in the C9V2 mRNA levels upon DAXX-FLAG overexpression in human RPE1 cells. An empty vector and a GUS-FLAG overexpression served as controls (n = 3 biological replicates).
(D) Fold changes in the C9V2 mRNA levels in human RPE1 cells expressing a control shRNA or shRNAs targeting DAXX (n = 3 biological replicates).
(E) Fold changes in the C9V2 mRNA levels in HEK293 cells expressing Opto-control or Opto-DAXX with or without exposure to blue light for 8 h (n = 3 biological replicates).
(F) TAD analysis of DAXX HiChIP for chromosome 9 and the C9orf72 locus in HEK293 cells expressing Opto-DAXX with or without blue-light illumination. Resolution is set to 1 mb or 50 kb for the whole chromosome 9 or the C9orf72 locus, respectively.
(G) Virtual chromatin contact profiles derived from the DAXX HiChIP analysis of the C9orf72 locus, with references to the ChIP-seq data for DAXX, CTCF, and H3K27ac in the region.
See also Figure S5.
The diseases linked to the C9orf72 HRE mutations are dominantly inherited, and most patient cells carry one expanded repeat allele and one wild-type allele. Since the patient cells exhibited a complete loss of stress-induced C9orf72 expression, we investigated the mechanism through which the stress-induced transcription at the wild-type allele is suppressed in the patient cells. Since the HRE mutations induce the accumulation of DAXX in the nuclei of patient cells, we asked whether elevated levels of DAXX suppress C9orf72 expression at the wild-type allele. Since the C9orf72 V2 transcript specifically underlies the stress-induced expression of the gene, we tested the potential association of DAXX with the C9orf72 promoters in wild-type cells. ChIP-qPCR analysis in human RPE1 cells showed enrichment of DAXX at the C9orf72 V2 promoter but not at the promoter regions for V1/3 (Figure S5M), demonstrating the intrinsic occupancy of DAXX specifically on the V2 promoter at the wild-type allele. To test the effects of elevated levels of DAXX on C9orf72 expression, we over-expressed DAXX in wild-type cells and observed a significant reduction in C9orf72 V2 transcript levels as compared with the cells expressing the empty vector or b-glucuronidase (GUS) as a control gene (Figure 6C). The role of DAXX in negatively regulating C9orf72 transcription was further confirmed by the in- crease in V2 transcript levels upon DAXX knockdown in wild- type cells (Figure 6D). Together, these data support the notion that the nuclear accumulation of DAXX as a result of the HRE mutations in patient cells can lead to a suppression of V2 transcribed from the wild-type allele.
To gain a mechanistic understanding of the role of DAXX phase separation in the regulation of C9orf72 transcription, we utilized the optogenetic system to study the regulation of the C9orf72 V2 expression. Upon an 8h exposure to blue light, a significant reduction in the C9orf72 V2 mRNA was observed in HEK293 cells expressing Opto-DAXX but not in the cells expressing the control protein when normalized against the levels before the blue-light illumination (Figure 6E). Furthermore, we used the HiChIP-seq analysis to examine the 3D chromatin structural shifts at the C9orf72 locus as a result of the inducible phase separation of DAXX. Consistent with our genome-wide observations, we found that the condensation of Opto-DAXX promoted 3D chromatin interactions across chromosome 9, as indicated by the enhanced signals in the interaction matrix plots (Figure 6F). TADs are the architectural units of 3D chromatins, in which there is a high frequency of chromatin interactions.22 With a 50-kb resolution for the TAD analysis, we found that the phase separation of Opto-DAXX led to the formation of two new sub-TADs (Figures 6F and 6G). TAD boundaries are frequently located in and around the TSSs that are enriched with CTCF, a chromatin insulator involved in boundary establishment.22 Disruption of topological boundaries can cause dysregulation of gene expression at the boundaries.27 The TAD anal- ysis revealed a new boundary formed at the C9orf72 promoter region as a result of Opto-DAXX condensation (Figures 6F and 6G). An analysis of previously reported ChIP-seq data indicated that the peaks of CTCF and DAXX overlap at the site of the newly formed sub-TAD boundary at the C9orf72 promoter (Figures 6F and 6G), suggesting that both CTCF and DAXX participated in the establishment of the boundary. Furthermore, an H3K27ac peak also co-localized with the CTCF and DAXX peaks and the newly formed TAD boundary at the C9orf72 promoter locus (Figure 6G), suggesting that both chromatin conformation and histone modification may contribute to the DAXX-dependent regulation of transcription activity at the C9orf72 locus. Thus, these data together suggest that DAXX acts as a key regulator of transcription at the C9orf72 locus through its modulation of 3D chromatin interactions.
DAXX regulates the susceptibility of C9orf72 HRE motor neurons to stress
We have observed that HRE-dependent accumulation of DAXX suppresses the expression of C9orf72 by acting on the promoter of the predominant V2 transcript. Unlike V2, which has the hexanucleotide repeats in its promoter (Figure S4A), the C9orf72 V1 and V3 transcripts contain the repeats in their first intron and thus are responsible for the disease-associated RNA toxicity and RAN translation, both of which originate from the repeat-containing V1/3 transcripts. Consistent with the lack of DAXX enrichment at the promoter of V1/3 (Figure S5M), we found that knockdown of DAXX did not significantly affect the levels of V1/V3 transcripts, as shown by qRT-PCR analysis of these transcripts in C9HRE iMNs with or without DAXX knockdown (Figure S7A). We also performed RNA FISH to directly detect the G4C2 repeat RNA foci, which serve as a marker for the repeat RNAs in C9HRE iMNs.48 With the loss of DAXX, there was no significant change in the quantity or size of the RNA foci, as indicated by the FISH analysis (Figures S7B and S7C). These results indicate that DAXX regulates the expression of C9orf72 primarily through its action on the V2 promoter and has little effect on the transcription of the repeat-containing V1/3 transcripts.
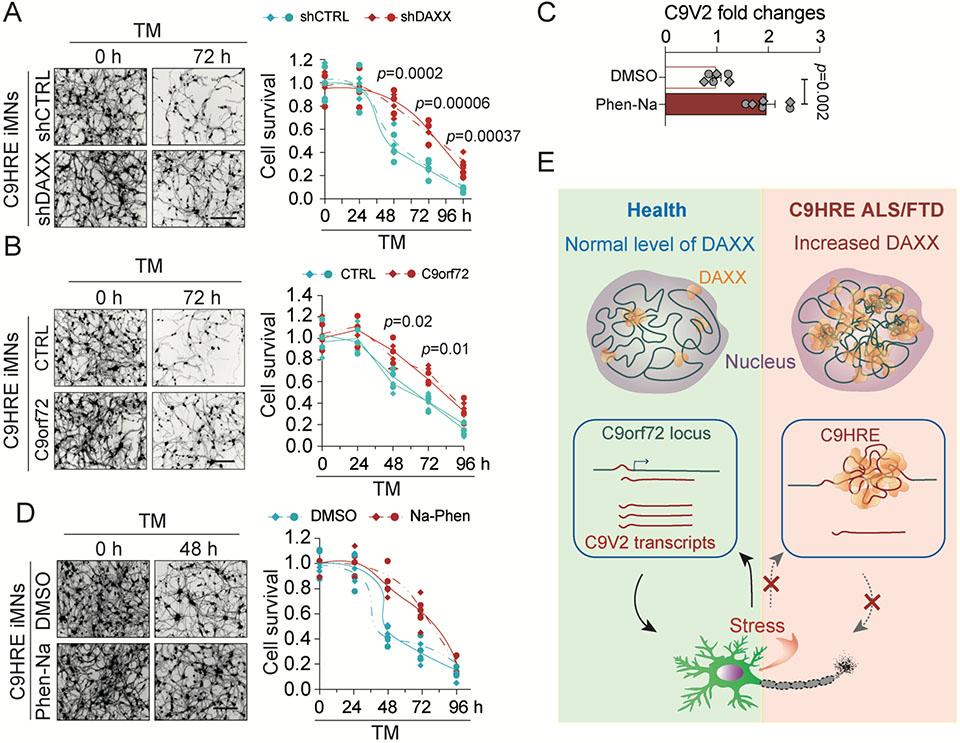
Motor neurons differentiated from iPSCs carrying the C9orf72 HRE mutation are susceptible to the stress induced by tunicamycin.16 Given the function of DAXX in regulating the expression of C9orf72 under stress, we investigated the role of DAXX or C9orf72 in mediating the sensitivity of C9HRE iMNs. Whereas tunicamycin induced a time-dependent loss of C9HRE iMNs, a partial knockdown of DAXX led to significantly higher survival rates for the C9HRE iMNs than for the controls (Figure 7A); furthermore, restoration of C9orf72 expression significantly enhanced the survival of the C9HRE iMNs (Figures 7B and S7F), indicating that the reduction in DAXX or the increase in C9orf72 had a neuroprotective effect on C9HRE patient neurons. At the same time, neither DAXX knockdown nor C9orf72 overexpression protected wild-type iMNs from the tuni- camycin-induced toxicity (Figures S7D-S7G), indicating that the regulatory effects of DAXX or C9orf72 is specific to C9HRE patient neurons where DAXX accumulation and C9orf72 deficiency are part of the pathological changes.
Figure 7. DAXX regulates the susceptibility of C9orf72 HRE iMNs to proteotoxic stress.
(A) Knockdown of DAXX increases the survival of C9HRE iMNs under the stress of tunicamycin (TM) treatment (5 mg/mL). Neuronal survival was measured by calcein-AM staining at the indicated time points (n = 6 biological replicates; different colors or point chapes represent independent iMN lines). Scale bar, 100 mm. (B) Increased expression of C9orf72 promoted the survival of C9HRE iMNs under stress. C9HRE iMNs expressing an empty vector or human C9orf72 were treated with TM (5 mg/mL), and neuronal survival was measured by calcein-AM staining at the indicated time points (n = 6 biological replicates; different colors or point shapes represent independent iMN lines). Scale bar, 100 mm.
(C and D) C9HRE iMNs were stressed with TM (5 mg/mL) and simultaneously treated with Na-Phen (10 mM) or DMSO. The Na-Phen treatment increased the C9orf72 V2 mRNA expression as measured by qPCR (C) and promoted neuronal survival as measured by calcein-AM staining at the indicated time points (D) (n = 6 biological replicates; different colors or point chapes represent independent iMN lines). Scale bar, 100 mm.
(E) Pathological cascades of chromatin architectural and epigenetic abnormalities initiated by C9orf72 HRE-dependent DAXX condensation in patient cells. Abnormal accumulation of nuclear DAXX condensates, as a result of the expanded hexanucleotide repeats, drives genome-wide chromatin structural changes and epigenetic dysregulation in C9orf72 HRE ALS/FTD patient cells. At the C9orf72 locus, the major C9orf72 transcript is stress-inducible at the transcriptional level, but the HRE mutation blocks the stress-dependent induction of C9orf72 in patient cells through DAXX-mediated chromatin remodeling. The loss of transcriptional plasticity of the C9orf72 gene compromises the survival fitness of neurons under stress and may therefore contribute to the neurodegeneration in ALS/FTD and relevant diseases.
See also Figure S7.
Given that the HRE-dependent loss of C9orf72 V2 expression was associated with DAXX-induced histone hypoacetylation at the C9orf72 V2 promoter, we evaluated the effects of increasing histone acetylation levels on C9orf72 V2 expression and neuronal health in C9HRE iMNs. To reverse the decrease in the levels of H3K27ac, we treated the C9HRE iMNs with sodium phenylbutyrate (Na-Phen), an HDAC inhibitor that increases the acetylation of H3K27.49 This pro-acetylation treatment yielded an increase in C9orf72 V2 expression as well as a robust protec- tion of the C9HRE iMNs against the tunicamycin-induced toxicity (Figures 7C and 7D). Taken together, these data suggest that DAXX, C9orf72, and the associated epigenetic event can be modulated to alleviate the sensitivity of C9orf72 HRE motor neurons to stress.
DISCUSSION
The expansion of repetitive DNA sequences has been linked to an increasing number of human neurological disorders. The molecular functions of the C9orf72 hexanucleotide repeat, especially at the DNA level, remain largely undefined. In this study, we have identified DAXX as a key DBP that recognizes the C9orf72 HRE DNA and undergoes HRE-dependent phase separation and protein condensation, leading to global chromatin remodeling and epigenetic dysregulation in patient cells (Figure 7E). The nuclear accumulation of DAXX in HRE-harboring cells suppresses a stress-dependent induction of the expression of C9orf72 through the regulation of chromatin structures and epigenetic modifications. These results have revealed the mechanisms through which C9orf72 HRE DNA and its key protein partner reshape genomic architectures and initiate pathogenic cas- cades that lead to compromised cell fitness.
We identified DAXX as a key C9HRE DBP that recognizes the G4C2 hexanucleotide repeat DNA and undergoes a series of changes in an HRE-dependent manner. Through its interaction with the HRE DNA and the resulting increase in its nuclear concentration, DAXX undergoes enhanced LLPS and molecular condensation. Likely existing in an equilibrium between its different phases, DAXX accumulates and condenses at the C9orf72 HRE site and throughout the nuclei in HRE-containing patient cells. By modeling the phase separation of DAXX using an optogenetic system, we observed that the phase separation of DAXX drives chromatin remodeling, epigenetic changes, and transcriptional regulation in the whole genome. The condensation of DAXX promotes the three-dimensional interactions among gene regulatory sequences, including promoters and enhancers. With the recruitment of histone modification pro- teins such as ATRX, SUV39H1, and HDAC1, DAXX promotes an increase in the transcription suppression marker H3K9me3 and a decrease in the transcription activation marker H3K27ac in the gene regulatory regions. Consequently, the oc- cupancy of RNA polymerases is decreased, and transcription is suspended. These results provide a mechanistic basis for the alterations in genomic topology in patient cells harboring the C9orf72 HRE mutations, consistent with the architectural changes in chromatin observed in C9orf72-ALS/FTD patient tissues.27
The HRE-dependent nuclear accumulation of DAXX has a profound effect on the expression of the C9orf72 gene, which exem- plifies the roles of DAXX in genomic and epigenetic regulation and provides insights into the disease mechanisms related to the expression of C9orf72. One of the surprising findings in this study was the stress-inducible expression of C9orf72, which could be important for the maintenance of cell fitness, especially under disease-associated stress conditions. DAXX plays a critical role in the stress-dependent induction of C9orf72 by recognizing the GC-rich promoter of the predominant V2 transcript. Our HiChIP analysis revealed that the phase separation of DAXX drives the changes in the 3D genomic interactions of the C9orf72 V2 promoter and renders the promoter inactive in reorganized chromatin structures. In the patient cells, the stress-dependent induction of C9orf72 was blocked, with elevated levels of DAXX suppressing the activity of the C9orf72 V2 promoter on both the HRE mutant and wild-type alleles. Since the heterozygous HRE mutation induced an increase in nuclear DAXX, thereby leading to development of the phenotype from both alleles, these results provide a dominant-negative mechanism that may underlie the contributions of the haploinsufficiency of C9orf72 to the diseases. Notably, DAXX did not influence the levels of either the V1 or V3 transcript, both of which start upstream of the C9orf72 V2 promoter region, but it specifically regulated the expression of the predominant V2 transcript. Therefore, knockdown of DAXX would not influence V1/V3-dependent RNA toxicity or RNA-dependent RAN translation, consistent with the observation of the absence of change in the levels of HRE RNA foci upon DAXX knockdown. However, the DAXX knockdown significantly enhanced the resistance of patient-derived iMNs to stress-induced toxicity, demonstrating the protective effects of derepressing the DAXX-mediated sup- pression of C9orf72 V2 expression under stress.
In addition to a role for DAXX in suppressing the C9orf72 transcription as part of the loss-of-function disease mecha- nisms, the HRE-dependent elevation of DAXX-induced genome-wide pathologic changes that contributed to the gain-of-function disease mechanisms. These HRE-dependent DAXX-mediated pathological changes include genome-wide perturbations in epigenetic regulation and global gene expres- sion in C9orf72 ALS/FTD patients. Other functions of DAXX could also be perturbed as a consequence of its accumulation in C9orf72 patient cells. Transcriptional dysregulation associated with changes in chromatin structure or epigenetic modifications are a widespread feature in ALS patients, including those linked to SOD1, FUS, TDP43, or the polydipeptide repeats.50-53 We have found that reducing DAXX or rebalancing the histone hypoacetylation mitigates the sensitivity of C9orf72 ALS patient-derived iMNs to stress. Taken together, our studies suggest that DAXX-mediated pathogenic cascades, including those causing epigenetic dysregulation, provide new strategies and potential targets for interventions to prevent or treat C9orf72 HRE-associated diseases.
METHODS
Human cell lines
Eight human B lymphocyte cell lines were obtained from NINDS Human Cell and Data Repository, including three control and five C9HRE patient lines. Eight human iPS cell lines were employed, including three controls, three C9HRE patient lines, and two isogenic controls. The two pairs of C9HRE iPSCs and isogenic controls were obtained from Justin Ichida’s Lab at the University of Southern California, and the rest of iPSCs were obtained from NINDS Human Cell and Data Repository. The patients’ information for these human cell lines refers to Table S3. All the human motor neurons used in the study were differentiated from human iPSCs. Human HEK293 cells and human RPE1 cells were obtained from ATCC. For shRNA or drug administration, cells were randomly divided into groups for different treatments. All cellular assays were repeated at least three times, and cellular data were collected and analyzed in a blind manner. Sequencing data were processed and analyzed by a person who were completely blind to the sample information.
Human tissues
Four controls and five C9HRE patient spinal cord tissues were used for immunostaining analysis. Eight controls and six C9HRE patient spinal cord tissues were employed for immunoblotting assays. All the human tissues used in this study were obtained from The Target ALS Multicenter Postmortem Tissue Core, The VA Biorepository Brain Bank, and The Johns Hopkins ALS Postmortem Tissue Core. Data collection for tissue staining and immunoblotting was performed in a blind manner. The detailed information for the patients were provided in Table S3.
Plasmids
To construct an optogenetic DAXX system, DAXX cDNA from Flag-Daxx/pRK5 (a gift from Xiaolu Yang; Addgene plasmid # 27974)54 was subcloned into pHR-mCh-Cry2WT (a gift from Clifford Brangwynne; Addgene plasmid # 101221)35 or pHR-sfGFP-Cry2WT using a Gibson Assembly Cloning (NEB, E5510S) method to create pHR-DAXX-mCherry-Cry2WT and pHR-DAXX-sfGFP-Cry2WT. For the promoter activity test, the human C9orf72 intron 1b or intron 1a sequence (Figure S4D) was cloned into a pGL4-uPAter-EGFP vector without a promoter element. The PR82 lentiviral expression construct (pLenti-PR82) expressing 82 proline-arginine dipeptide repeats was cloned using the Gateway cloning system into the pLenti-puro-CMV (w118) vector (a gift from Eric Campeau and Paul Kaufman; Addgene plasmid # 17452).55 The PR82 coding sequence was derived from a previous sequence with randomized codons designed to produce only the proline-arginine dipeptide repeat (a gift from Adrian Isaacs).61
Cell culture, drug treatment, lentiviral shRNA knockdown, and live-cell imaging
Human B lymphocytes were cultured in RPMI 1640 medium containing 15% FBS. Human iPSCs were maintained in StemFlex medium (Gibico, A3349401), with medium exchange every other day. Human HEK293 cells and human RPE1 cells were obtained from ATCC. HEK293 cells were cultured in DMEM containing 10% FBS. Human retinal pigment epithelial 1 (RPE1) cells were cultured in DMEM/F12 containing 10% FBS and 0.01 mg/ml hygromycin B. All cell lines were checked regularly for mycoplasma contamina- tion. To induce cell stress, RPE1 cells were treated with thapsigargin (30 nM) or tunicamycin (5 mg/ml) for 24 h, and B lymphocytes were treated with thapsigargin (40 nM) or tunicamycin (1 mg/ml) for 24 h. To express PR82, RPE1 cells were transfected with the pLenti-PR82 plasmid and cultured for 48 h.
All knockdown experiments were carried out via transduction with lentiviruses expressing various shRNAs. The shRNAs targeting human DAXX were TRCN0000003800, TRCN0000279733, and TRCN0000003801 (Sigma). Treatments with the various shRNAs yielded similar results, and therefore representative data are presented. To produce lentiviral particles, 53106 HEK293 cells were seeded onto a 10-cm dish coated with PEI. The next day, the DAXX shRNA-expressing pLKO.1 or pLKO.5 lentiviral plasmid and viral packaging vectors (psPAX2 and pMD2G) were co-transfected using Lipofectamine 2000 (Thermo Fisher, 11668500) for 8 h before the Opti-MEM transfection medium was changed to fresh DMEM containing 10% FBS. After 72 h of lentiviral production, the culture medium was collected and filtered through a 0.45-mm cellulose acetate membrane to remove debris. Lentiviral particles were concentrated by precipitation with 40% PEG 8000 and centrifugation (1,600 g) and then resuspended in 1 × PBS. The concentrated lentiviral particles were used in the transduction of B lymphocytes or iPSCs for 48 h, followed by puromycin selection (3 mg/ml for B lymphocytes and 0.5-1.5 mg/ml for iPSCs).
For live-cell imaging of Opto-DAXX clustering, 0.8×105 HEK293 cells were seeded onto a FluoroDish (World Precision Instruments, FD35-100) 1 day before transfection with pHR-DAXX-mCherry-Cry2WT or pHR-mCh-Cry2WT. At 24 h after the transfection, live cells were exposed to blue light, and time-lapse images of each channel were captured at the indicated intervals by using an SP8 confocal microscope (Leica). DNA was visualized by DAPI staining in live cells.
Motor neuron differentiation of human iPS cells and experimental analyses
All the eight human iPS cell lines were differentiated into motor neurons as previously described.62 In brief, iPSCs were seeded onto Matrigel-coated plates and differentiated into neuroepithelial progenitor cells (NEPCs) by culturing 6 days in neural medium (1:1 DMEM/F12:neurobasal medium, GlutaMax, N2 supplement, B27 supplement, and ascorbic acid) containing 3 mM CHIR99021, 2 mM SB431542, and 2 mM DMH-1. NEPCs were dissociated with dispase (1 U/ml) and split into new plates coated with Matrigel with a ratio of about 1:6. NEPCs were differentiated into motor neuron progenitor cells (MNPCs) using neural medium supplemented with 1 mM CHIR99021, 2 mM SB431542, 2 mM DMH-1, 0.1 mM retinoic acid (RA), and 0.5 mM purmorphamine. After culturing for 6 days, MNPCS were dissociated and cultured in suspension using neural medium containing 0.5 mM RA and 0.1 mM purmorph- amine. Six days later, neural spheres were dissociated and plated onto a Matrigel-coated plate. Adherent neural spheres were cultured in neural medium supplemented with 0.5 mM RA, 0.1 mM purmorphamine, and 0.1 mM compound E. After 12 days, mature motor neurons were acquired for experiments. The medium was changed every other day during the entire differentiation period.
For knockdown experiments on motor neurons, iMNs were randomly divided into groups and each group was infected twice with the lentiviruses expressing different shRNAs, on day 10 and day 12 at the final differentiation stage, and the cells were used on day 15 to allow for efficient DAXX knockdown. Mature iMNs were stressed with tunicamycin (5 mg/ml) in the presence of Na-Phen (10 mM) for the indicated times, and neuronal survival at each time point was measured by calcein-AM staining. In brief, iMNs were stained with 3 mM calcein-AM (Invitrogen, C1430), and the fluorescence of surviving iMNs was read using a plate reader (Synergy H1 Hybrid) with excitation/emission at 485 nm/535 nm. Optical fields with calcein-positive iMNs were randomly selected and images captured using a fluorescence microscope (Nikon Eclipse TS100).
C9orf72 V2 promoter analysis
The C9orf72 promoter region (Eukaryotic Promoter Database ID C9orf72_1) between -5000 bp to +100 bp was examined for the presence of the GC box motif using the Eukaryotic Promoter Database. Two putative GC boxes, located in introns 1b and 1a, were identified with a p-value less than 10-5. To test the promoter activity of the intron 1b and 1a, HEK293 cells were transfected with pGL4-uPAter-EGFP plasmids containing intron 1b or intron 1a. One day after the transfection, the cells were analyzed for EGFP expression by western blotting, fluorescence detection, and qPCR analysis. qPCR EGFP expression was normalized using the mRNA of the ampicillin resistance gene (AmpR) from the pGL4-uPAter-EGFP plasmid, together with resident GAPDH mRNA.
SILAC quantitative proteomic analysis
To identify DNA G4C2 repeat-interacting proteins, DNA probes were used to pull down their target proteins in SILAC-labeled cells. The DNA oligonucleotide 5’-(G4C2)6AACAAC-biotin-3’ and a randomized control, 5’-GACTGACTGATAGATCCTAAGTACTGAT- TACTGACTAACAAC-biotin-3’, together with their complementary oligonucleotides without a biotin label were synthesized (Integrated DNA Technologies). Each pair of complementary strands were incubated at 95 °C for 5 min and ramped down to 20 °C in 5oC/min increments to anneal in 23 annealing buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 2 mM EDTA). Annealed dsDNAs were phosphorylated at 37 °C for 2 h using T4 polynucleotide kinase (NEB, M0201S) and then ligated with T4 DNA ligase at room temperature for 4 h. The dsDNAs were purified via phenol–chloroform extraction and stored at −20 °C.
HEK293 cells were cultured in SILAC medium with either light lysine and arginine, or heavy lysine (13C615N2) and arginine (13C6 15N4), along with 10% dialyzed fetal bovine serum (Thermo Fisher Scientific, 88440) and penicillin/streptomycin. Following 10 days of metabolic labelling, nuclear proteins were extracted using Nuclear and Cytoplasmic Extraction Reagent (ThermoFisher Scientific). Biotin-labeled G4C2 repeats or random dsDNAs (20 mg each) were incubated with streptavidin beads (Dynabeads MyOne Streptavidin C1, Invitrogen, 65001) in DNA binding buffer (2 M NaCl, 10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.01% Tween-20) at room temperature for 1 h with rotation. The dsDNA-bead complexes were washed twice using DNA binding buffer and then twice with pro- tein binding buffer (PBS [pH 7.4] containing 0.01% Tween-20). The dsDNA-bead probes were then incubated with 400 mg of nuclear protein in protein binding buffer at 4 °C for 2 h with rotation, washed three times with protein binding buffer, and heated at 95 °C for 5 min in NuPAGE loading buffer (Invitrogen, NP0007) containing 20 mM DTT. The resulting immunoprecipitates were separated on a 4 - 12% gradient gel and digested with trypsin. The digested peptide samples were analyzed and quantified with an LTQ-Orbitrap-Velos mass spectrometer.
Protein purification and EMSA
DAXX protein was expressed and purified from human cells. HEK293 cells were transfected with Flag-DAXX-pRK5 (Addgene 27974) and then lysed by sonication in ice-cold lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% TritonX-100, 2.5 mM sodium pyrophosphate, 1 mM beta-glycerophosphate, 1 mM Na3VO4, 1 mg/ml leupeptin, 1 mM PMSF [APExBIO, A2587], and Protease Inhibitor Cocktail [1:200, Millipore Sigma, P8340]) with a Diagenode Bioruptor at high power with an on/off cycle of 30 sec for 20 min. Cell lysates were centrifuged at 12,000 g at 4 °C for 20 min, and the supernatants were har- vested for immunoprecipitation. After incubation with anti-Flag M2 magnetic beads (Sigma, M8823) at room temperature for 1 h, the beads were washed three times with a buffer (50 mM Tris-HCl [pH 7.4] and 150 mM NaCl) and then eluted with a buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 150 ng/ml 33Flag peptides (Sigma, F4799) at 4 °C for 30 min with rotation. Purified protein was concentrated with a centrifugal filter (Millipore, 50 kDa, UFC805096) and stored at −80oC in buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM DTT, 50% glycerol, 1 mM PMSF (APExBIO, A2587), and Protease Inhibitor Cocktail (1:200, Millipore Sigma, P8340).
For EMSA analysis, a ssDNA probe, (CCCCGG)10, and a size-matched control probe (GACTGACTGATAGATCCTAAGTACTG ATTACTGACTATAGATCTAAGTCATGATCAGTTA) were synthesized and labeled with an Alexa Fluor 488 fluorescent tag at the 5’ terminus (Integrated DNA Technologies). To generate dsDNA probes, the fluorescence-labeled ssDNA probe was annealed with its complementary strand. Increasing concentrations of purified protein were incubated with the probes (50 nM) in binding buffer containing 10 mM Tris (pH 7.5), 1 mM EDTA, 0.1 mM DTT, 10 mg/ml BSA, and 5% glycerol. After incubation at room temperature for 30 min, the mixture was run on a non-denaturing polyacrylamide gel in TBE buffer. Gel shift images were captured on an Amersham Typhoon Imager 9200, and the ratio of bound DNA to free DNA was analyzed with ImageJ (version 2.0.0-rc-59/1.51k).
In vitro liquid-liquid phase separation (LLPS)
To purify DAXX protein for in vitro LLPS analysis, the cDNA of human DAXX (Addgene 27947) was cloned into the pET28a plasmid and expressed in Rosetta E. coli. Bacteria were cultured in 50 ml LB medium at 37 °C with kanamycin (50 mg/ml) overnight. Next day, 30 mL of the bacterial suspension was added into 1 L of LB medium and grown at 37 °C until OD600 reached 0.6. Bacteria were cultured with IPTG (0.4 mM) induction at 16 °C overnight to express DAXX and then pelleted and lysed in buffer (50 mM NaH2PO4, 300 mM NaCl, 0.05% tween-20, 10 mM imidazole, pH=8.0). Lysates were centrifuged, and supernatants were collected to pull down His-tagged DAXX with Ni NTA agarose beads incubated at 4 °C for 1 h. After washing twice with buffer (50 mM NaH2PO4, 300 mM NaCl, 0.05% tween-20, 20 mM imidazole, pH=8.0), beads were eluted with buffer (50 mM NaH2PO4, 300 mM NaCl, 0.05% tween-20, 250 mM imidazole, pH=8.0). Elution buffer was exchanged by a centrifugal filter (Millipore, 30 kD, UFC803024) and the purified proteins were aliquoted and stored at −80 °C in buffer containing 20 mM Tris-HCl (pH=7.5), 150 mM NaCl, 1 mM DTT, 50% glycerol, PMSF, and protease inhibitor.
Purified DAXX protein at a final concentration of 500 ng/ml was incubated in LLPS buffer (1 M NaCl) for 30 min at room temperature to allow the formation of protein droplets. To examine the effects of DNA on the LLPS of DAXX, (G4C2)10 or a size-matched control dsDNA probe (at a final concentration of 200 nM) was added into LLPS buffer (1 M NaCl) with 500 ng/ml of DAXX. After incubation at room temperature for 30 min, images of protein droplets were captured and analyzed. To test an HRE size-dependent effect on DAXX’s phase separation in vitro, G4C2 dsDNA fragments were cut from DNA plasmids16 and then gel-purified. DAXX protein was added into NaCl (0.5 M) buffer at a final concentration of 500 ng/ml, and dsDNA probes were added into the buffer at a final concentration of 200 nM. After incubation at room temperature for 1 h, DAXX droplets were imaged for analysis.
ATAC-PALM
HEK293 cells were plated onto 8-well Lab-TEK chambers (Thermo Fisher #155409) pre-coated with fibronectin (Millipore FC010, 7.5 ml/ml). At 24 h after plating, cells were transfected with pHR-DAXX-sfGFP-Cry2WT using Lipofectamine 3000 (Thermo Fisher, L3000001), incubated for 48 h, and light-activated by a blue-light LED transilluminator (BLooK, GeneDireX, model: BK001) for 5 min before fixation in 4% paraformaldehyde solution (Electron Microscopy Sciences #15710) for 10 min at room temperature. Cells were washed, permeabilized, and incubated with Tn5 PA-JF549 transposase as described.37 For a single well of the 8-well Lab-TEK chamber, a 20 ml reaction mix (10 mM Tris-HCl [pH 7.6], 5 mM MgCl2, 10% dimethylformamide, and 25 nM Tn5 PA-JF549 transposase) was spread over the entire well using a sheet of Parafilm, and the cells were incubated at 37 °C for 1 h. Following the incubation with the transposase, the cells were washed 3 times with 1× PBS containing 0.01% SDS and 50 mM EDTA for 8 min each at 55 °C, then washed 2 times with 1× PBS, and kept in 1× PBS during imaging.
2D ATAC-PALM single-molecule imaging was performed on a Nikon Eclipse Ti microscope equipped with a 100X oil-immersion objective lens (Nikon, N.A. = 1.49), a Lumencor light source, two filter wheels (Lambda 10-3, Sutter Instrument, Novato, CA), perfect focusing systems and EMCCD (iXon3, Andor, Belfast, United Kingdom). Emission filters (Semrock, Rochester, New York) were switched in front of the cameras for GFP or JF549 emission, and a band mirror (405/488/561/633 BrightLine quad-band bandpass filter, Semrock) was used to reflect the laser into the objective. Cells were first excited with a 10% 488 nm laser to acquire an epifluorescence GFP channel image. Then Tn5 PA-JF549 transposase single molecules were detected using a 405 nm laser (10% power) for photo-activation and a 561 nm laser (100% power) for excitation. The acquisition time was 30 ms. A given plane of the cell was imaged for 10,000-20,000 iterations to exhaust single-molecule detections.
Single molecules were localized using the ThunderSTORM plugin of ImageJ and visualized using the Normalized Gaussian method. To calculate the Tn5 PA-JF549 transposase localization density per Opto-GFP-DAXX droplet in the light-activated sample, a GFP channel image of the same cell was first used to select droplets as regions of interest (ROIs). For localizations, 20 × 1-μm2 areas of the nucleus were randomly chosen and stored as ROIs. Localization density per ROI was calculated by dividing the number of localizations per ROI by the area of the ROI. Localization density was normalized by the total number of localizations per nucleus, accounting for cellular variation in Tn5 PA-JF549 labeling.
ATAC-seq
One pair of human B lymphocytes (~1×105) and two pairs of progenitor-differentiated motor neurons (~1.6×106) were used for ATAC-seq as described.63 Cells were pelleted in 1× PBS (500 g for 5 min, 4 °C) and resuspended in 50 ml of cold lysis buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, and 0.1% IGEPAL CA-630) with gentle pipetting. Cell lysates were centrifuged (5003g for 10 min, 4 °C) to pellet the nuclei, which were then incubated with 50 ml of transposition reaction buffer (25 ml of 23 Tagment DNA reaction buffer [Illumina FC-121-1030], 2.5 ml of Nextera Tn5 Transposase [Illumina FC-121-1030], and 22.5 ml of nuclease-free H2O) at 37 °C for 30 min with gentle rotation. After the reaction, genomic DNA was purified with a Qiagen MinElute PCR Purification Kit (Qiagen, 28204) and eluted in 10 ml of elution buffer (10 mM Tris buffer [pH 8.0]).
To amplify the transposed DNA fragments, the DNA sample was mixed with PCR amplification buffer containing nuclease-free H2O, Nextera PCR Primer 1, Nextera PCR Primer 2 (barcode), and NEBNext High-Fidelity 23 PCR Master Mix (New England Labs, M0541), and then amplified by 1 cycle of 72 °C incubation for 5 min and 98 °C for 30 sec, followed by 5 cycles of 98 °C for 10 sec, 63 °C for 30 sec, and 72 °C for 1 min. To avoid PCR saturation of a library and reduce the GC and size bias, 5 ml of the first PCR reaction were amplified in a side qPCR reaction to select an appropriate cycle number for the second-round PCR reaction, which was set to 1 cycle of 98 °C incubation for 30 sec, followed by 20 cycles of 98 °C for 10 sec, 63 °C for 30 sec, and 72 °C for 1 min. The cycle number corresponding to a quarter of the maximum fluorescence intensity was used in the final PCR amplification. After the final amplification, PCR products were purified using a Qiagen MinElute PCR Purification Kit (Cat. NO. 28204) and analyzed by deep sequencing.
ATAC-seq data were processed on Galaxy.56 In brief, adapters were removed using Cutadapt and only reads with a size of more than 20 bp were retained for analysis. The quality of the reads was measured by FastQC. The trimmed reads were sequentially mapped to the human reference genome (hg19 version) using Bowtie in an end-to-end model. The maximum fragment length (distance between read pairs) was set to 1,000 bp, and the parameters were set as highly sensitive. Reads with low mapping quality, inappropriate pairing, or mapping to mitochondrial DNA were filtered out. Duplicate fragments were removed with Picard MarkDuplicates. An insert size of Tn5 was plotted with a paired-end histogram for fragment length distribution, with peaks at 200 bp, 400 bp, and/or 600 bp. The ATAC-seq datasets (LINCS ID: LDG-1394: LDS-1501, LDS-1502) from NEUROLINCS were analyzed with the data generated in this study. MACS2 was used to find peak callings (extension size, 200 and shift size, 100), and the MACS2 file was used to generate a bigwig file and a heatmap of coverage at TSSs of interest. The ATAC-seq peaks were defined as the accessible chromatin domains, and the signals of all genomic peaks presented as RPM (reads per million) and normalized with the background were combined together to indicate chromatin accessibility.
HiChIP
HiChIP was performed as previously described.36 HEK293 cells were plated in 10-cm dishes 1 day before pHR-DAXX-mCherry- Cry2WT transfection, followed by a 2-day incubation. The plates with >80% of cells expressing Opto-DAXX were randomly divided into two groups, with each containing 1×107 cells. One group was activated by blue-light illumination for 10 min, and the other served as a negative control. Cells were detached, pelleted, resuspended at 1×106 cells/ml in freshly prepared 1% formaldehyde, and incubated at room temperature for 10 min with gentle rotation, with constant blue-light illumination for the activated group during this step. The formaldehyde was quenched with glycine at a final concentration of 125 mM for 5 min. Following three washes with ice-cold PBS, 1×107 cells were lysed in 500 ml ice-cold lysis buffer (10 mM Tris-HCl [pH 8.0], 10 mM NaCl, 0.2% NP-40, and Protease Inhibitor Cocktail [1:200, Millipore Sigma, P8340]), incubated at 4 °C for 30 min, and centrifuged at 2,500 g for 5 min to collect nuclei. The nuclear pellets were washed once with 500 ml of ice-cold lysis buffer, suspended in 100 ml 0.5% SDS, and incubated at 62 °C for 10 min. Triton X-100 (10%, 50 ml) was added into the pellets to quench the SDS, followed by the addition of 285 ml H2O and incubation at 37 °C for 15 min. In situ chromatins were digested with 375 U of MboI restriction enzyme in 500 ml of 13 NEB buffer 2 at 37 °C for 2 h before heat inactivation (65 °C, 20 min). The overhangs of the cut DNA fragments were filled in with biotin-labeled dATP (Thermo, 19524016) using 5 U DNA polymerase I large (Klenow) fragment (NEB, M0210) at 37 °C for 1 h. The blunt-ended DNA fragments were ligated with NEB T4 DNA ligase buffer, 10% Triton X-100, BSA, and T4 DNA ligase (NEB, M0202) at room temperature for 4 h with rotation. After the incubation, nuclei were pelleted at 2,500 g for 5 min and subjected to sonication (Diagenode Bioruptor: 30-sec cycles, high power for 15 min) in nuclear lysis buffer (50 mM Tris-HCl [pH 7.5], 10 mM EDTA, 1% SDS, and Protease Inhibitor Cocktail [1:200, Millipore Sigma, P8340]). The sonicated samples were centrifuged at 16,100 g for 15 min at 4 °C and used for chromatin immunoprecipitation with an antibody against DAXX (Sigma, D7810), following the same protocol described in the section on chromatin immunoprecipitation qPCR below. A sample (150 ng) of the DNA pulled down from the ChIP was used to capture biotin- labeled DNAs using 5 ml of Streptavidin C-1 beads in 20 ml of 1× biotin binding buffer (5 mM Tris-HCl [pH 7.5], 0.5 mM EDTA, and 1 M NaCl) at room temperature for 15 min with rotation. Beads were washed twice with 500 ml Tween wash buffer (5 mM Tris-HCl [pH 7.5], 0.05% Tween-20, 0.5 mM EDTA, and 1 M NaCl) at 55 °C for 2 min with shaking and once with 100 ml of 23 Tagment DNA buffer (20 mM Tris-HCl [pH 7.5], 10 mM MgCl2, and 20% dimethylformamide). The washed beads were added into 25 ml of 23 Tagment DNA buffer with 4 ml Tn5 Transposase (Illumina, FC-121-1030) and incubated at 55 °C for 10 min. After the incubation, the beads were removed and washed twice in 50 mM EDTA at 50 °C (first for 30 min and then 3 min), twice in Tween wash buffer at 55 °C for 2 min, and once in 10 mM Tris. The washed beads were used for PCR amplification and sequencing as described in the section on ATAC-seq.
For HiChIP data analysis, Illumina and Nextera adapter sequences were removed using Trim Galore (version 0.6.6).57 The raw reads were aligned to the human hg19 genome using HiC-Pro (version 2.11.4)58 with bin sizes of 5 kb, 50 kb, and 100 kb. The MboI restriction site, followed by fill-in, and the ligation was specified as GATCGATC. The processed data were analyzed, and contact maps were generated using the HiTC R package (version 1.23.0).59
The enhancers were defined as the genomic binding sites of H3K27ac. The processed BAM files and peak files from the H3K27ac experiments in HEK293 cells were downloaded directly from ENCODE website.64 The raw fastq file of Myc-DAXX ChIP-seq experiments in HEK293 was downloaded from Gene Expression Omnibus (GEO) with accession code GSE107348. The raw reads were aligned to the human hg19 genome using Bowtie 2 (version 2.3.5.1).60
Chromatin immunoprecipitation sequencing and qPCR
Chromatin immunoprecipitation (ChIP) was performed by using a ChIP assay kit (Millipore Sigma, 17-295). In brief, protein complexes were crosslinked with 1% formaldehyde at 37 °C for 10 min and quenched with glycine at a final concentration of 125 mM at 37 °C for 5 min. After three washes with cold 1× PBS containing 1 mM PMSF (APExBIO, A2587) and Protease Inhibitor Cocktail (1:200, Milli- pore Sigma, P8340), the cells were pelleted and lysed in SDS lysis buffer (50 mM Tris [pH 8.1], 1% SDS, 10 mM EDTA, Protease In- hibitor Cocktail [1:200, Millipore Sigma, P8340], and 1 mM PMSF [APExBIO, A2587]). The cell lysates were sonicated on ice to achieve an average DNA length of 200–1,000 bp. After centrifugation, the supernatants were harvested and diluted 10 times with buffer (0.01% SDS, 1.1% Triton x-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], 167 mM NaCl, Protease Inhibitor Cocktail [1:200, Millipore Sigma, P8340], and 1 mM PMSF [APExBIO, A2587]) and pre-cleared with salmon sperm DNA/Protein A agarose-50% slurry (Millipore Sigma, 16-157C) at 4 °C for 30 min. The pre-cleared samples were randomly divided into new tubes and incubated with respective antibodies against RNA polymerase II (Millipore Sigma, 17-620), DAXX (Sigma, D7810), or IgG (Millipore Sigma, 17-620; Cell Signaling Technology, 2729) at 4 °C overnight with rotation. The beads were washed sequentially with low-salt immune complex wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], and 150 mM NaCl), high-salt immune com- plex wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], and 500 mM NaCl), LiCl salt immune complex wash buffer (0.25 M LiCl, 1% IGEPAL-CA630, 1% sodium deoxycholate, 1 mM EDTA, and 10 mM Tris [pH 8.1]), and TE buffer (10 mM Tris [pH 8.0] and 1 mM EDTA). Beads were eluted with elution buffer (1% SDS and 0.1M NaHCO3) at room temperature for 15 min with rotation. NaCl (20 ml, 5 M) and RNase A (2 ml of 10 mg/ml, ThermoFisher Scientific, EN0531) were added to 500 ml of each eluate and incubated overnight at 65 °C with rotation. Proteinase K (2 ml of 20 mg/ml, ThermoFisher Scientific, EO0491) was added and incubated at 60 °C for 1 h with rotation. DNA was purified using the phenol-chloroform method.
The ChIP-seq libraries were prepared using an NEBNext Ultra II FS DNA Library Prep kit (NEB, E6177S). Briefly, DNAs were fragmented into sizes of ~350 bp, ligated with adaptors, and amplified by PCR. The amplification products were purified and analyzed by deep sequencing. Raw reads were processed with TrimGalore v0.6.757 and controlled for quality with FastQC v.0.11.9. The trimmed reads were aligned to the human genome (hg19) using Bowtie2 v2.3.5.1.65 Uniquely mapped reads were retained to identify DAXX- associated peaks using MACS266 v2.2.6 with standard parameters (q-value cutoff 0.05). Peaks were annotated and visualized with an R package, ChIPseeker.67 For ChIP-qPCR, two primer sets were designed for C9orf72 intron 1b: 5’-TCTGGAACTCAG GAGTCGCG-3’ (forward 1), 5’-GCAGCCGAACCCCAAACAGC-3’ (reverse 1), 5’-TGGCGAGTGGGTGAGTGAGG-3’ (forward 2), and 5’-GAGGGAAAGTAAAAATGCGTCGA-3’ (reverse 2). Subsequently, qPCR analysis was performed, and fold changes were normalized with IgG controls and inputs in statistical analysis.
RNA-seq
Two pairs of C9HRE iMNs with isogenic controls were employed in RNA-seq analysis. Total RNAs were extracted by an RNeasy Plus mini kit (Qiagen, 74136), and RNA libraries for sequencing were prepared using a commercial kit (Invitrogen, A39005096). Extracted RNAs were hybridized with the probe mixture at 75 °C for 5 min and 60 °C for 30 min, and then incubated with rRNA depletion beads to remove rRNA. After purification, the rRNA-depleted samples were fragmented by RNase III to acquire an average size of ~300 bp, and then purified by Dynabeads Cleanup Beads for adaptor ligation. The adaptor-ligated RNA mixtures were reversely transcribed to synthesize cDNA. Purified cDNA samples were amplified by PCR and analyzed by sequencing. Raw reads were processed with TrimGalore v0.6.757 and controlled for quality with FastQC v.0.11.9. The processed reads were mapped to the human genome (hg19) and GENCODE v19 annotated transcripts using Hisat265 v2.2.1 and quantified with featureCounts68 v2.0.1. Raw gene counts were loaded into R to perform differential expression analysis with edgeR69 and generate plots and heatmaps.
Fluorescence in situ hybridization and co-immunostaining
DNA FISH was performed with modifications of a previously described method.70 (CCCCGG)4 with an Alexa Fluor 488 fluorescence tag at the 3’ terminus was synthesized (Integrated DNA Technologies) and used as a probe for G4C2 repeats as previously reported.4 Cells were fixed with 2% paraformaldehyde in 1× PBS (pH 7.4) at room temperature for 15 min, washed, and permeabilized with ice-cold 0.4% Triton X-100/1× PBS for 10 min. Cells were blocked in buffer containing 2.5% BSA, 10% goat serum, and 0.1% Tween-20 at room temperature for 1 h, incubated with anti-DAXX antibody (Cell Signaling Technology, 4533) at 4 °C overnight, and the with a fluorescent secondary antibody at room temperature for 2 h. Cells were treated with RNase A (0.1 mg/ul) at 37 °C for 1 h, washed, and permeabilized with ice-cold 0.7% Triton X-100 and 0.1 M HCl for 10 min. Genomic DNA and the probe were denatured in 50% formamide, 23 SSC, and 10% dextran sulfate at 95 °C for 30 min, incubated at 37 °C for 1 h, and washed with 0.43 SSC and 0.3% Tween-20 at room temperature. Slides were stained with DAPI and sealed for imaging.
RNA FISH for foci containing G4C2 repeat RNAs was performed as previously described.19 In brief, C9HRE iMNs were fixed in 3.75% formaldehyde in PBS at room temperature for 10 min and permeabilized in prechilled 70% ethanol on ice for 30 min. Cells were rehydrated in wash buffer (40% deionized formamide in 23 SSC) for 10 min, followed by rehydration with 40% formamide in 23 SSC for 10 min. After blocking in hybridization buffer (40% formamide, 23 SSC, 20 mg/ml BSA, 100 mg/ml dextran sulfate, 10 mg/ml yeast tRNA, and 2 mM vanadyl sulfate ribonucleosides) at 55 °C for 10 min, cells were incubated at 55 °C for 2 h in the hybridization buffer containing 125 nM (C4G2)4-Cy3 probes, which had been denatured at 95 °C for 5 min and chilled on ice. Cells were washed three times with the wash buffer at 55 °C for 10 min and sealed with Prolong Gold Antifade reagent containing DAPI (Invitrogen, P36931). All DNA and RNA FISH images were captured using an SP8 confocal microscope (Leica).
Immunostaining and nascent RNA visualization
Cells were seeded onto a glass slide coated with PEI, cultured for 24 h, washed, and fixed with 4% formaldehyde for 20 min at room temperature. After washes, the cells were permeabilized and blocked in buffer containing 5% normal serum and 0.3% Triton X-100 in 1× PBS for 2 h at room temperature, followed by incubation with a primary antibody: anti-DAXX (Cell Signaling Technology, 4533), anti-PML (Santa Cruz, sc-966), anti-ATRX (Santa Cruz, sc55584), anti-HDAC1 (Santa Cruz, sc81598), anti-H3K9me3 (Active Motif, 61013), anti-RNA polymerase II (Cell Signaling Technology, 2629), or anti-ChAT (Millipore Sigma, AB144P) at 4 °C overnight. The slides were washed and incubated with a fluorescent secondary antibody at room temperature for 2 h, followed by PBS washes. The slides were then sealed with a Prolong Gold Antifade reagent with DAPI (Invitrogen, P36931). For human tissue staining, slides were sequentially rinsed in xylene, 100% ethanol, 95% ethanol, 70% ethanol, 50% ethanol, and H2O, and then antigen retrieval was performed in buffer (0.1 M citrate acid and 0.1 M sodium citrate) at 95 °C for 30 min. After washes in 13 PBS, slides were incubated with a block buffer (2% normal donkey serum, 1% BSA, 0.05% Tween 20) at room temperature for 1 h, followed by incubation with the anti-DAXX antibody (1:100, Cell Signaling Technology, 4533) at 4 °C overnight. Slides were washed in 1× PBS and incubated with a secondary antibody for 1 h at room temperature. After washing in 1× PBS three times, the slides were sealed with a Prolong Gold Antifade reagent with DAPI (Invitrogen, P36931). All fluorescent images were captured with an SP8 confocal microscope (Leica). Puncta with sizes over 500 nM without any limit on circularity were quantified using ImageJ and statistically analyzed.
Nascent RNAs in HEK293 cells were labeled using an Alexa Fluor 488-tagged nucleoside and click chemistry as described (Invitrogen, C10329). In brief, HEK293 cells were plated onto glass slides and the transfected with Opto-DAXX the next day. One day after transfection, the cells were exposed to blue light for 6 h. During the last hour, the cells were incubated with 5-ethynyl uridine to label nascent RNAs, then fixed and permeabilized, and subjected to click chemistry to visualize fluorescently labeled nascent RNAs using an SP8 confocal microscope (Leica). Finally, the fluorescence intensity of 5-ethynyl uridine inside or outside the DAXX droplets was measured and analyzed.
Quantitative PCR
Total RNAs were extracted with an RNeasy Plus mini kit (Qiagen, 74136), and cDNAs were synthesized using QuantiTect reverse transcription reagents (Qiagen, 205313). qPCR reactions were carried out on a Bio-Rad thermal cycler using PowerUp SYBR Green Master Mix (ThermoFisher Scientific). The primer sets for C9orf72 transcripts V1–3 were described previously.71 The expression of C9orf72 pre-mRNA was measured with a pair of primers targeted to the junction region between exon 2 and intron 3. The mRNA expression levels were analyzed by the ΔΔCt method and normalized against housekeeping genes. In the promoter analysis, the EGFP transcription level was normalized to the GAPDH transcript and the plasmid-encoded ampicillin resistance (AmpR) transcript to exclude any bias induced by differential transfection efficiency.
Immunoblotting
Cultured cells were washed twice with PBS and lysed in cold RIPA buffer containing 50 mM Tris (pH 7.5), 0.5% SDS, 150 mM NaCl, 0.5% NP40, 20 mM EDTA, 1 mM PMSF, and Protease Inhibitor Cocktail (1:200, Millipore Sigma, P8340). Cytoplasmic and nuclear fractions of B lymphocytes were isolated with a subcellular fractionation kit (Thermo Fisher, 78840). Human spinal cord tissues were homogenized and lysed in a modified RIPA buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% NP40, 0.1% SDS, 100 mM NaF, 17.5 mM b-glycerophosphate, 2.5% sodium deoxycholate, and 10% glycerol) containing phosphatase inhibitors 2 and 3 (1:100; Millipore Sigma), 1 mM PMSF, 2 mM NaVO4, and Protease Inhibitor Cocktail (1:200, Millipore Sigma, P8340). After sonication on ice, the sam- ples were centrifuged at 12,000 g for 10 min at 4 °C, and supernatants were collected for analysis.
Protein concentrations were measured by the bicinchoninic acid assay (ThermoFisher, 23225). Following SDS-PAGE and western blotting, membranes were incubated with primary antibodies at 4 °C overnight, including anti-DAXX (Cell Signaling Technology, 4533), anti-C9orf72 (BioRad, VMA00065), anti-SUV39H1 (Cell Signaling Technology, D11B6), anti-ATRX (Santa Cruz, sc55584), anti-HDAC1 (Santa Cruz, sc81598), anti-H3K9me3 (Active Motif, 61013), anti-H3K27ac (Cell Signaling Technology, 8173), anti-H3 (Cell Signaling Technology, 4499), anti-PARP (Cell Signaling Technology, 9542), anti-GFP (Invitrogen, A-11122), anti-Flag (Sigma, F3165), anti-GAPDH (Invitrogen, TAB1001), and anti-actin (Santa Cruz, sc-47778). After washes with TBST, the membranes were incubated with a fluorescent secondary antibody at room temperature for 2 h. Western blot images were captured by an Odyssey scanner (LI-COR) and analyzed with Image Studio (version 5.2.5). Protein expression levels were normalized with actin for the total protein, GAPDH for the cytoplasmic fraction, and PARP for the nuclear fraction.
Data and code availability
All the processed and raw data for HiChIP, RNA-seq, ChIP-seq, and ATAC-seq have been deposited in NCBI Gene Expression Omnibus(GEO) and can be accessed from the GEO number: GSE223401.
QUANTIFICATION AND STATISTICAL ANALYSIS
Experiments on human RPE1 cells, B lymphocytes, HEK293 cells, iPSCs, and iPSC-differentiated motor neurons were performed with triplicate samples in at least three independent experiments. Experiments on iPSC-differentiated motor neurons were repeated on three independent cell lines for each group of control and patient cells. Average values for each group were used for statistical analysis. Statistical analysis was performed on Prism 9.0 software. The unpaired Student’s t-test was applied for single comparisons, and one-way analysis of variance (ANOVA) for multiple comparisons with Bonferroni’s test. All data are presented as means ± SEM unless otherwise indicated.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from NIH (NS089616, NS110098, NS074324, and NS128494), Packard Center for ALS Research at Johns Hop- kins, Target ALS Foundation, Maryland Stem Cell Research Fund, and the U.S. Department of Defense. Bioinformatic analysis in this work was performed at the Advanced Research Computing at Hopkins (ARCH) core facility (rockfish. jhu.edu), which is supported by the National Science Foundation (NSF) grant number OAC1920103. We would like to thank Justin Ichida at the University of Southern California for providing iPSC lines, NINDS and NIGMS Cell Repos- itory, Ian Robey at the VA Biorepository Brain Bank (VA merit review BX002466), Lyle Ostrow, Kathleen Wilsbach, and Kathyrn Gallo at the Johns Hopkins ALS Postmortem Tissue Core, and the Target ALS Multicenter Post- mortem Tissue Core for providing patient cells and tissues, Howard Hughes Medical Institute for the ATAC-PALM facility, and the members of Wang lab fordiscussion.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Tóth G, Gáspári Z, and Jurka J (2000). Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res 10, 967–981. 10.1101/gr.10.7.967 Genome Res. 2000. 10: 967–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malik I, Kelley CP, Wang ET, and Todd PK (2021). Molecular mechanisms underlying nucleotide repeat expansion disorders. Nat Rev Mol Cell Biol 22, 589–607. 10.1038/s41580-021-00382-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeJesus-Hernandez M, Mackenzie Ian R., Boeve Bradley F., Boxer Adam L., Baker M, Rutherford Nicola J., Nicholson Alexandra M., Finch NiCole A., Flynn H, Adamson J, et al. (2011). Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron 72, 245–256. 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renton Alan E., Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick Jennifer C., Laaksovirta H, van Swieten John C., Myllykangas L, et al. (2011). A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron 72, 257–268. 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balendra R, and Isaacs AM (2018). C9orf72-mediated ALS and FTD: multiple pathways to disease. Nature Reviews Neurology 14, 544–558. 10.1038/s41582-018-0047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ugolino J, Ji YJ, Conchina K, Chu J, Nirujogi RS, Pandey A, Brady NR, Hamacher-Brady A, and Wang J (2016). Loss of C9orf72 Enhances Autophagic Activity via Deregulated mTOR and TFEB Signaling. PLoS Genet 12, e1006443. 10.1371/journal.pgen.1006443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amick J, Roczniak-Ferguson A, and Ferguson SM (2016). C9orf72 binds SMCR8, localizes to lysosomes and regulates mTORC1 signaling. Mol Biol Cell 27. 3040–3051. 10.1091/mbc.e16-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sellier C, Campanari ML, Julie Corbier C, Gaucherot A, Kolb-Cheynel I, Oulad-Abdelghani M, Ruffenach F, Page A, Ciura S, Kabashi E, and Charlet-Berguerand N (2016). Loss of C9ORF72 impairs autophagy and synergizes with polyQ Ataxin-2 to induce motor neuron dysfunction and cell death. EMBO J 35, 1276–1297. 10.15252/embj.201593350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan PM, Zhou X, Robins AM, Paushter DH, Kim D, Smolka MB, and Hu F (2016). The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy-lysosome pathway. Acta Neuropathol Commun 4, 51. 10.1186/s40478-016-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang T, Liu H, Itoh K, Oh S, Zhao L, Murata D, Sesaki H, Hartung T, Na CH, and Wang J (2021). C9orf72 regulates energy homeostasis by stabilizing mitochondrial complex I assembly. Cell metabolism 33, 531–546. 10.1016/j.cmet.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang M, Liang C, Swaminathan K, Herrlinger S, Lai F, Shiekhattar R, and Chen JF (2016). A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Sci Adv 2, e1601167. https://DOI: 10.1126/sciadv.1601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Y, Lin S, Staats KA, Li Y, Chang WH, Hung ST, Hendricks E, Linares GR, Wang Y, Son EY, et al. (2018). Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat Med 24, 313–325. 10.1038/nm.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Rourke JG, Bogdanik L, Yanez A, Lall D, Wolf AJ, Muhammad AK, Ho R, Carmona S, Vit JP, Zarrow J, et al. (2016). C9orf72 is required for proper macrophage and microglial function in mice. Science 351, 1324–1329. https://DOI: 10.1126/science.aaf1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burberry A, Suzuki N, Wang JY, Moccia R, Mordes DA, Stewart MH, Suzuki-Uematsu S, Ghosh S, Singh A, Merkle FT, et al. (2016). Loss-of-function mutations in the C9ORF72 mouse ortholog cause fatal autoimmune disease. Science translational medicine 8, 347ra393. https://DOI: 10.1126/scitranslmed.aaf6038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donnelly CJ, Zhang PW, Pham JT, Heusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, et al. (2013). RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 80, 415–428. 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, et al. (2014). C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507, 195–200. 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW 3rd, Rademakers R, et al. (2013). Unconventional Translation of C9ORF72 GGGGCC Expansion Generates Insoluble Polypeptides Specific to c9FTD/ALS. Neuron 77, 639–646. 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. (2013). The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338. https://DOI: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 19.Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, et al. (2013). RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA 110, E4968–4977. 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Q, Jiang J, Gendron TF, McAlonis-Downes M, Jiang L, Taylor A, Diaz Garcia S, Ghosh Dastidar S, Rodriguez MJ, King P, et al. (2020). Reduced C9ORF72 function exacerbates gain of toxicity from ALS/FTD-causing repeat expansion in C9orf72. Nat Neurosci 23, 615–624. 10.1038/s41593-020-0619-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berson A, Nativio R, Berger SL, and Bonini NM (2018). Epigenetic Regulation in Neurodegenerative Diseases. Trends Neurosci 41, 587–598. 10.1016/j.tins.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, and Ren B (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin Y, Chang YC, Lee DSW, Berry J, Sanders DW, Ronceray P, Wingreen NS, Haataja M, and Brangwynne CP (2018). Liquid Nuclear Condensates Mechanically Sense and Restructure the Genome. Cell 175, 1481–1491. 10.1016/j.cell.2018.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huo X, Ji L, Zhang Y, Lv P, Cao X, Wang Q, Yan Z, Dong S, Du D, Zhang F, et al. (2020). The Nuclear Matrix Protein SAFB Cooperates with Major Satellite RNAs to Stabilize Heterochromatin Architecture Partially through Phase Separation. Molecular Cell 77, 368–383. 10.1016/j.molcel.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Fasciani A, D’Annunzio S, Poli V, Fagnocchi L, Beyes S, Michelatti D, Corazza F, Antonelli L, Gregoretti F, Oliva G, et al. (2020). MLL4-associated condensates counterbalance Polycomb-mediated nuclear mechanical stress in Kabuki syndrome. Nat Genet 52, 1397–1411. 10.1038/s41588-020-00724-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paez-Colasante X, Figueroa-Romero C, Sakowski SA, Goutman SA, and Feldman EL (2015). Amyotrophic lateral sclerosis: mechanisms and therapeutics in the epigenomic era. Nat Rev Neurol 11, 266–279. 10.1038/nrneurol.2015.57. [DOI] [PubMed] [Google Scholar]
- 27.Sun JH, Zhou L, Emerson DJ, Phyo SA, Titus KR, Gong W, Gilgenast TG, Beagan JA, Davidson BL, Tassone F, and Phillips-Cremins JE (2018). Disease-Associated Short Tandem Repeats Co-localize with Chromatin Domain Boundaries. Cell 175, 224–238. 10.1016/j.cell.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belzil VV, Bauer PO, Prudencio M, Gendron TF, Stetler CT, Yan IK, Pregent L, Daughrity L, Baker MC, Rademakers R, et al. (2013). Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol 126, 895–905. 10.1007/s00401-013-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xi Z, Zinman L, Moreno D, Schymick J, Liang Y, Sato C, Zheng Y, Ghani M, Dib S, Keith J, et al. (2013). Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. Am J Hum Genet 92, 981–989. 10.1016/j.ajhg.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu EY, Russ J, Wu K, Neal D, Suh E, McNally AG, Irwin DJ, Van Deerlin VM, and Lee EB (2014). C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD. Acta Neuropathol 128, 525–541. 10.1007/s00401-014-1286-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He Q, Kim H, Huang R, Lu W, Tang M, Shi F, Yang D, Zhang X, Huang J, Liu D, and Zhou S (2015). The Daxx/Atrx Complex Protects Tandem Repetitive Elements during DNA Hypomethylation by Promoting H3K9 Trimethylation. Cell Stem Cell 17, 273–286. 10.1016/j.stem.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin D-Y, Huang Y-S, Jeng J-C, Kuo H-Y, Chang C-C, Chao T-T, Ho C-C, Chen Y-C, Lin T-P, Fang H-I, et al. (2006). Role of SUMO-Interacting Motif in Daxx SUMO Modification, Subnuclear Localization, and Repression of Sumoylated Transcription Factors. Molecular Cell 24, 341–354. 10.1016/j.molcel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 33.Bouchard JJ, Otero JH, Scott DC, Szulc E, Martin EW, Sabri N, Granata D, Marzahn MR, Lindorff-Larsen K, Salvatella X, et al. (2018). Cancer Mutations of the Tumor Suppressor SPOP Disrupt the Formation of Active, Phase-Separated Compartments. Mol Cell 72, 19–36 e18. 10.1016/j.molcel.2018.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taslimi A, Vrana JD, Chen D, Borinskaya S, Mayer BJ, Kennedy MJ, and Tucker CL (2014). An optimized optogenetic clustering tool for probing protein interaction and function. Nature communications 5, 4925. 10.1038/ncomms5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, and Brangwynne CP (2017). Spatiotemporal Control of Intracellular Phase Transitions Using Light-Activated optoDroplets. Cell 168, 159–171. 10.1016/j.cell.2016.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, and Chang HY (2016). HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nature Methods 13, 919–922. 10.1038/nmeth.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie L, Dong P, Chen X, Hsieh TS, Banala S, De Marzio M, English BP, Qi Y, Jung SK, Kieffer-Kwon KR, et al. (2020). 3D ATAC-PALM: super-resolution imaging of the accessible genome. Nat Methods 17, 430–436. 10.1038/s41592-020-0775-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai D, Feliciano D, Dong P, Flores E, Gruebele M, Porat-Shliom N, Sukenik S, Liu Z, and Lippincott-Schwartz J (2019). Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat Cell Biol 21, 1578–1589. 10.1038/s41556-019-0433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. (2012). The accessible chromatin landscape of the human genome. Nature 489, 75–82. 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lallemand-Breitenbach V, and de Thé H (2018). PML nuclear bodies: from architecture to function. Current opinion in cell biology 52, 154–161. 10.1016/j.ceb.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 41.Martire S, Gogate AA, Whitmill A, Tafessu A, Nguyen J, Teng Y-C, Tastemel M, and Banaszynski LA (2019). Phosphorylation of histone H3.3 at serine 31 promotes p300 activity and enhancer acetylation. Nature Genetics 51, 941–946. 10.1038/s41588-019-0428-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Leo C, Zhu J, Wu X, O’Neil J, Park E-J, and Chen JD (2000). Sequestration and Inhibition of Daxx-Mediated Transcriptional Repression by PML. Molecular and Cellular Biology 20, 1784–1796. 10.1128/MCB.20.5.1784-1796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tran H, Almeida S, Moore J, Gendron Tania F., Chalasani U, Lu Y, Du X, Nickerson Jeffrey A., Petrucelli L, Weng Z, and Gao F-B (2015). Differential Toxicity of Nuclear RNA Foci versus Dipeptide Repeat Proteins in a Drosophila Model of C9ORF72 FTD/ALS. Neuron 87, 1207–1214. 10.1016/j.neuron.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dreos R, Ambrosini G, Périer RC, and Bucher P (2014). The Eukaryotic Promoter Database: expansion of EPDnew and new promoter analysis tools. Nucleic Acids Research 43, D92–D96. 10.1093/nar/gku1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Blitterswijk M, Gendron TF, Baker MC, DeJesus-Hernandez M, Finch NA, Brown PH, Daughrity LM, Murray ME, Heckman MG, Jiang J, et al. (2015). Novel clinical associations with specific C9ORF72 transcripts in patients with repeat expansions in C9ORF72. Acta neuropathologica 130, 863–876. 10.1007/s00401-015-1480-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cherepanova N, Shrimal S, and Gilmore R (2016). N-linked glycosylation and homeostasis of the endoplasmic reticulum. Current Opinion in Cell Biology 41, 57–65. 10.1016/j.ceb.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lytton J, Westlin M, and Hanley MR (1991). Thapsigargin inhibits the sarcoplasmic or endoplasmic reticulum Ca-ATPase family of calcium pumps. Journal of Biological Chemistry 266, 17067–17071. 10.1016/S0021-9258(19)47340-7. [DOI] [PubMed] [Google Scholar]
- 48.Sareen D, O’Rourke JG, Meera P, Muhammad AKMG, Grant S, Simpkinson M, Bell S, Carmona S, Ornelas L, Sahabian A, et al. (2013). Targeting RNA Foci in iPSC-Derived Motor Neurons from ALS Patients with a C9ORF72 Repeat Expansion. Science Translational Medicine 5, 208ra149. DOI: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warrell RP Jr., He LZ, Richon V, Calleja E, and Pandolfi PP (1998). Therapeutic targeting of transcription in acute promyelocytic leukemia by use of an inhibitor of histone deacetylase. J Natl Cancer Inst 90, 1621–1625. 10.1093/jnci/90.21.1621. [DOI] [PubMed] [Google Scholar]
- 50.Yang L, Gal J, Chen J, and Zhu H (2014). Self-assembled FUS binds active chromatin and regulates gene transcription. Proc Natl Acad Sci USA 111, 17809–17814. 10.1073/pnas.1414004111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berson A, Sartoris A, Nativio R, Van Deerlin V, Toledo JB, Porta S, Liu S, Chung CY, Garcia BA, Lee VM, et al. (2017). TDP-43 Promotes Neurodegeneration by Impairing Chromatin Remodeling. Curr Biol 27, 3579–3590. 10.1016/j.cub.2017.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barbosa LF, Cerqueira FM, Macedo AF, Garcia CC, Angeli JP, Schumacher RI, Sogayar MC, Augusto O, Carri MT, Di Mascio P, and Medeiros MH (2010). Increased SOD1 association with chromatin, DNA damage, p53 activation, and apoptosis in a cellular model of SOD1-linked ALS. Biochim Biophys Acta 1802, 462–471. 10.1016/j.bbadis.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y-J, Guo L, Gonzales PK, Gendron TF, Wu Y, Jansen-West K, O’Raw AD, Pickles SR, Prudencio M, Carlomagno Y, et al. (2019). Heterochromatin anomalies and double-stranded RNA accumulation underlie C9orf72 poly(PR) toxicity. Science 363, eaav2606. https://DOI: 10.1126/science.aav2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang J, Qu LK, Zhang J, Wang W, Michaelson JS, Degenhardt YY, El-Deiry WS, and Yang X (2006). Critical role for Daxx in regulating Mdm2. Nat Cell Biol 8, 855–862. 10.1038/ncb1442. [DOI] [PubMed] [Google Scholar]
- 55.Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, Campisi J, Yaswen P, Cooper PK, and Kaufman PD (2009). A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One 4, e6529. 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IO, Pietrzyk J, et al. (2014). C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345, 1192–1194. https://DOI: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y, Wang T, Ji YJ, Johnson K, Liu H, Johnson K, Bailey S, Suk Y, Lu Y-N, Liu M, and Wang J (2018). A C9orf72–CARM1 axis regulates lipid metabolism under glucose starvation-induced nutrient stress. Genes & Development 32, 1380–1397. 10.1101/gad.315564.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buenrostro JD, Wu B, Chang HY, and Greenleaf WJ (2015). ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21.29.1–21.29.9. 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Batut B, Hiltemann S, Bagnacani A, Baker D, Bhardwaj V, Blank C, Bretaudeau A, Brillet-Guéguen L, Čech M, Chilton J, et al. (2018). Community-Driven Data Analysis Training for Biology. Cell Syst 6, 752–758. 10.1016/j.cels.2018.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Felix Krueger FJ, Ewels Phil, Afyounian Ebrahim, & Schuster-Boeckler Benjamin (2021). FelixKrueger/TrimGalore: v0.6.7. Zenodo. 10.5281/zenodo.5127899 [DOI] [Google Scholar]
- 61.Servant N, Varoquaux N, Lajoie BR, Viara E, Chen C-J, Vert J-P, Heard E, Dekker J, and Barillot E (2015). HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biology 16, 259. 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Servant N, Lajoie BR, Nora EP, Giorgetti L, Chen CJ, Heard E, Dekker J, and Barillot E (2012). HiTC: exploration of high-throughput ‘C’ experiments. Bioinformatics 28, 2843–2844. 10.1093/bioinformatics/bts521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Consortium EP (2004). The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306, 636–640. Thhps:// doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 64.Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360. 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu G, Wang LG, and He QY (2015). ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383. 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 68.Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 69.Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chaumeil J, Micsinai M, and Skok JA (2013). Combined immunofluorescence and DNA FISH on 3D-preserved interphase nuclei to study changes in 3D nuclear organization. J Vis Exp 3, e50087. https:// DOI: 10.3791/50087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gendron TF, Chew J, Stankowski JN, Hayes LR, Zhang YJ, Prudencio M, Carlomagno Y, Daughrity LM, Jansen-West K, Perkerson EA, et al. (2017). Poly(GP) proteins are a useful pharmacodynamic marker for C9ORF72-associated amyotrophic lateral sclerosis. Science translational medicine 9, eaai7866. https:// DOI: 10.1126/scitranslmed.aai786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the processed and raw data for HiChIP, RNA-seq, ChIP-seq, and ATAC-seq have been deposited in NCBI Gene Expression Omnibus(GEO) and can be accessed from the GEO number: GSE223401.