Abstract
Rationale and objectives
Radiomics is a promising, non-invasive method for determining the prognosis of high-grade glioma (HGG). The connection between radiomics and the HGG prognostic biomarker is still insufficient.
Materials and methods
In this study, we collected the pathological, clinical, RNA-sequencing, and enhanced MRI data of HGG from TCIA and TCGA databases. We characterized the prognostic value of CSF3. Kaplan–Meier (KM) analysis, univariate and multivariate Cox regression, subgroup analysis, Spearman analysis, and gene set variation analysis enrichment were used to elucidate the prognostic value of the CSF3 gene and the correlation between CSF3 and tumor features. We used CIBERSORT to analyze the correlation between CSF3 and cancer immune infiltrates. Logistic regression (LR) and support vector machine methods (SVM) were used to build the radiomics models for the prognosis prediction of HGG based on the expression of CSF3.
Results
Based on the radiomics score calculated from LR model, 182 patients with HGG from TCGA database were divided into radiomics score (RS) high and low groups. CSF3 expression varied between tumor and normal group tissues. CSF3 expression was found to be a significant risk factor for survival outcomes. A positive association was found between CSF3 expression and immune infiltration. The radiomics model based on both LR and SVM methods showed high clinical practicability.
Conclusion
The results showed that CSF3 has a prognostic value in HGG. The developed radiomics models can predict the expression of CSF3, and further validate the predictions of the radiomics models for HGG.
Keywords: Radiomics, CSF3, Prognosis, MRI, High-grade glioma
1. Introduction
Glioma is a common primary tumor of the central nervous system [1]. In general, high-grade gliomas (HGGs) are considered as World Health Organization (WHO) grade 3–4 diffuse gliomas, which are classified based on histology and are characterized by malignancy and a dismal prognosis [2,3]. According to the WHO 2021 Classification of CNS, HGGs are also considered as grade 3–4 diffuse gliomas. The latest edition of WHO classification criteria is not only based on histological types, but also adopts a new integrated diagnostic criterion that combines histological phenotype and molecular typing (such as IDH, 1p/19q, TERT, EGFR, CDKN2A/B and so on) to more accurately assess the prognosis of gliomas. At present, the therapeutic approaches for gliomas include surgical treatment, radiotherapy, chemotherapy, targeted therapy, and immunotherapies [4]. Despite these treatments, overall survival (OS) has not considerably improved. Some prognostic biomarkers in gliomas have previously been explored to improve management and personalized treatment [5]. However, effective prognostic factors are still sought to improve the prognosis of HGG and inform individualized glioma management strategies.
Colony-stimulating factor 3 (CSF3) encodes the glycoprotein granulocyte colony-stimulating factor (G-CSF), which is closely associated with the hematopoietic function of the body, and plays an important regulatory role in the proliferation, differentiation, and progression of neutrophil cell lineages. G-CSF is synthesized and released by various cells, including normal neurons and immune cells, along various types of advanced, solid malignancies, including glioblastomas. However, the impact of G-CSF on tumor biology remains controversial. G-CSF plays a crucial role in neuroprotection against neurodegenerative disorders [6] and shows major value in chemotherapy [7]. The G-CSF secreted by mutant isocitrate dehydrogenase 1 (mIDH1) glioma stem/progenitor-like cells can induce myeloid cell reprogramming and produce non-immunosuppressive myeloid cells, which were shown to improve immunotherapeutic efficacy in patients with mIDH1 low-grade glioma [8]. G-CSF can consequently promote tumor metastasis and progression by intensifying the immunosuppressive environment of the tumor microenvironment [9,10]. Accordingly, it will be beneficial to determine the correlation between CSF3 and the prognosis of HGG and the effect of CSF3 on the tumor microenvironment in HGG.
Radiomics is a high-throughput method of mining quantitative image features in conjunction with machine learning [11], which has great importance in cancer research and can improve the accuracy of cancer prognosis determination. Radiomics studies conducted on glioma have shown promising results. Ita et al. [12] found that glioma radiographic tumor burden is positively correlated with CSF3 expression in the plasma. Analyzing HGG prognosis using radiomics models based on machine-learning methods may provide promising results.
Toward this end, the aim of this study was to construct radiomics models using data collected from The Cancer Genome Atlas (TCGA) and The Cancer Imaging Archive (TCIA) databases to non-invasively predict the mRNA expression of CSF3 in HGG via magnetic resonance imaging (MRI), which could in turn indirectly reflect the prognosis of HGG patients. Bioinformatic analysis was integrated to explore the underlying molecular mechanisms of CSF3 and its association with the immune microenvironment.
2. Materials and methods
2.1. Public data sources and data analysis
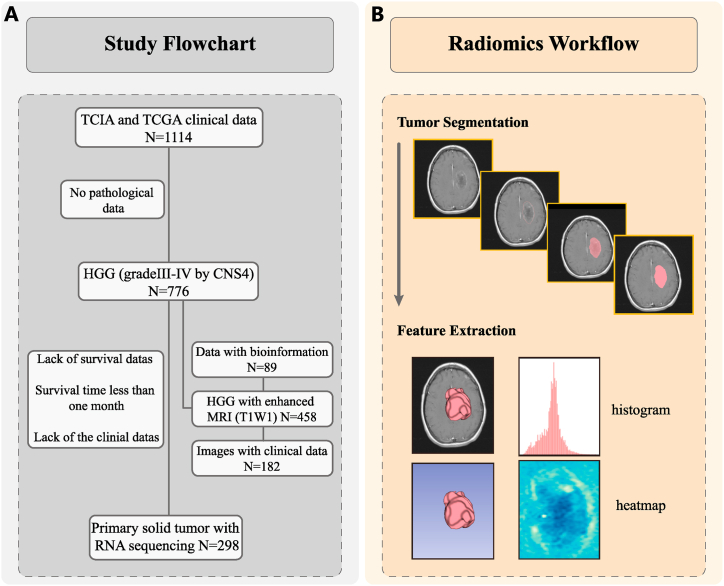
To explore the prognostic value of the CSF3 gene and the radiomics prediction model, pathological data were collected from a total of 298 HGG patients, and medical imaging data (n = 182) and transcriptomic sequencing data (n = 89), including clinical and follow-up data, were downloaded from TCIA and TCGA databases, respectively. The study flowchart was shown in Fig. 1A. To explore the differences in CSF3 expression between tumor and normal tissues, the RNA-sequencing (RNA-seq) data of 689 tumor and 1189 normal samples were collected from TCGA low-grade glioma datasets, TCGA-glioblastoma multiform datasets, and the GTEx database via the University of California Santa Cruz (UCSC). The data of each sample were transformed to transcripts per million (TPM) values for normalization.
Fig. 1.
(A) Data summary. (B) Radiomics workflow.
2.1.1. Prognosis assessment
The patients were divided into low and high groups based on CSF3 gene expression values using the “survminer” R package. The Kaplan–Meier (KM) method was used to calculate the OS, and the differences in survival rates between two groups were evaluated using a log-rank test. Cox regression was performed to investigate the relationship between CSF3 and OS. The role of CSF3 as an independent indicator of OS was assessed using the multivariate Cox model. Subsequently, a subgroup analysis was used to evaluate the correlation between CSF3 expression and OS in each subgroup using a forest plot. Subgroups were evaluated by age, gender, grade, isocitrate dehydrogenase (IDH) mutation status, 1p19q co-deletion, methylguanine methyltransferase (MGMT) promoter status, chemotherapy, and radiotherapy. Interactions between CSF3 and each of the aforementioned subgroups were evaluated. The relationships between CSF3 expression and clinical parameters were visualized via a heat map of Spearman’s rank correlation coefficients.
2.2. Immune microenvironment analysis
CIBERSORT was used to identify an immune cell according to gene expression levels and determine the correlation between immune cell infiltrations in HGG samples based on CSF3 expression. Normalized gene expression data of the glioma samples were uploaded to the CIBERSORTx website to characterize the abundance of 22 immune cell types. A further correlation analysis of CSF3 expression and immune cell infiltration was performed using Spearman’s correlation coefficient. Statistical significance was set at P < 0.05.
2.3. Enrichment analysis of differentially expressed genes between the CSF3-High and CSF3-low-expression groups
Gene set variation analysis was used in the R software to calculate the pathway enrichment of glioma patients for the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Hallmark gene sets. The top 20 pathways were visualized using the “limma” R package. Heat maps were drawn using the R package “pheatmap” for unsupervised classification to explore whether samples with similar pathway activities had specific biological implications.
2.4. Radiomics feature extraction and selection
The volume of interest (VOI) segmentation of each image was performed using the 3D slicer software to extract radiomics features. Another radiologist, not involved in this study, randomly selected the images of 30 patients for analysis. The reproducibility of VOI was evaluated using the intraclass correlation coefficient (ICC). If the index is high, it can be assumed that the corresponding stability is strong; ICC ≥0.8 indicates high, 0.5–0.79 medium, and <0.5 low consistency. The features meeting the requirement of ICC ≥0.8 were retained. The minimum redundancy maximum relevance (mRMR) [13] method was applied to select the features with maximum correlation with CSF3 expression and eliminate the redundancy between features. To further select features with higher distinguishing power for CSF3 gene expression, recursive feature elimination was used to select features by gradually reducing the examined feature set size. After the prediction model is established, it is trained by the original feature data, and the corresponding weights are set for each feature. Then, the feature with the minimum absolute weight is gradually deleted so that the best expected feature is finally obtained. The radiomics workflow was shown in Fig. 1B.
2.5. Radiomics model construction and validation
The selected features were applied to build radiomics prediction models of CSF3 expression based on logistic regression (LR) and support vector machine (SVM) methods. The assessment indices of area under the receiver operating characteristic (ROC) curve (AUC), sensitivity, specificity, and accuracy were determined for the dataset. The diagnostic performance of the radiomics models was assessed via the ROC curve, AUC, precision–recall curve, calibration curve, and decision curve analysis on the training and validation data. The radiomics score (RS) was then computed. The potential association between the RS and CSF3 gene expression was evaluated using the Wilcoxon rank-sum test. Correlation between the RS based on the LR model and M1/M2 macrophage-related genes were analyzed by Spearman correlation analysis using the R package “stats.” The genes with a p-value <0.05 were selected.
2.6. Connection between radiomics models and clinical prognosis
The 182 patients with HGG in TCGA database included in the survival analysis were classified into high and low groups according to the RS. The optimal cut-off of the RS was determined from the radiomics model based on LR. The KM curves and the log-rank test were used for survival analysis. The prognostic significance of clinical scores and RS was determined using the Cox regression analysis method. A subgroup analysis was used to compare RSs in each subgroup and visualized with forest plots. Subgroups were evaluated by age, gender, grade, IDH mutation status, 1p19q co-deletion, MGMT promoter status, chemotherapy, and radiotherapy. Spearman’s correlation coefficients between the RS and M1/M2 macrophage-related genes were calculated.
2.7. Statistical analysis
All analyses were conducted using the R software (v. 4.1.0). Visualization was performed using the ggplot2, pheatmap, or survminer packages. KM curve analysis was performed using the survival and survminer packages. Cox regression was performed using the survival and forestplot software packages. The radiomics models were constructed using the pROC, measures, ResourceSelection, modEvA, and ggpubr tools. To generate figures, multiple R packages, including survminer, CBCgrps, limma, caret, irr, mRMRe, glm, and stats, were used.
3. Results
3.1. Data summary
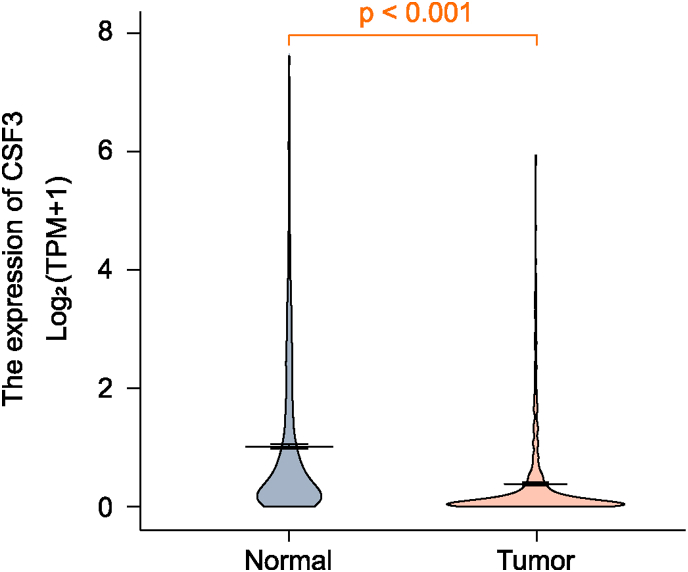
Based on CSF3 expression levels, the patients were divided into CSF3-high (n = 161) and CSF3-low-expression groups (n = 137) by the optimal cut-off value of 0.0516. The relationship between CSF3 levels and the clinical characteristics of glioma patients in TCGA dataset is shown in Table 1. CSF3 levels were significantly associated with age, WHO grade, IDH mutation status, 1p19q co-deletion, and MGMT promoter status (all P < 0.001). CSF3 expression varied between tumor and normal group tissues, with a median difference of −0.24 (−0.287 to −0.201), which was statistically significant (P < 0.001). The results were displayed in Fig. 2.
Table 1.
Correlations of CSF3 expression with clinical features of high-grade glioma in The Cancer Genome Atlas cohort.
| Variables | Total (n = 298) | Low (n = 137) | High (n = 161) | P-value |
|---|---|---|---|---|
| Age (years), n (%) | <0.001 | |||
| <60 | 197 (66) | 107 (78) | 90 (56) | |
| ≥60 | 101 (34) | 30 (22) | 71 (44) | |
| Gender, n (%) | 0.048 | |||
| Female | 120 (40) | 64 (47) | 56 (35) | |
| Male | 178 (60) | 73 (53) | 105 (65) | |
| Grade, n (%) | <0.001 | |||
| III | 170 (57) | 114 (83) | 56 (35) | |
| IV | 128 (43) | 23 (17) | 105 (65) | |
| IDH status, n (%) | <0.001 | |||
| Wild type | 169 (57) | 54 (39) | 115 (71) | |
| Mutant | 129 (43) | 83 (61) | 46 (29) | |
| Chr 1p_19q co-deletion, n (%) | 0.043 | |||
| No | 248 (83) | 107 (78) | 141 (88) | |
| Yes | 50 (17) | 30 (22) | 20 (12) | |
| MGMT promoter status, n (%) | <0.001 | |||
| Unmethylated/Unknown | 118 (40) | 34 (25) | 84 (52) | |
| Methylated | 180 (60) | 103 (75) | 77 (48) | |
| Chemotherapy, n (%) | 0.125 | |||
| No | 79 (27) | 30 (22) | 49 (30) | |
| Yes | 219 (73) | 107 (78) | 112 (70) | |
| Radiotherapy, n (%) | 0.023 | |||
| No | 70 (23) | 41 (30) | 29 (18) | |
| Yes | 228 (77) | 96 (70) | 132 (82) |
Fig. 2.
Comparison of CSF3 expression between normal and tumor tissues.
3.2. Correlation of CSF3 expression with the prognosis of glioma patients
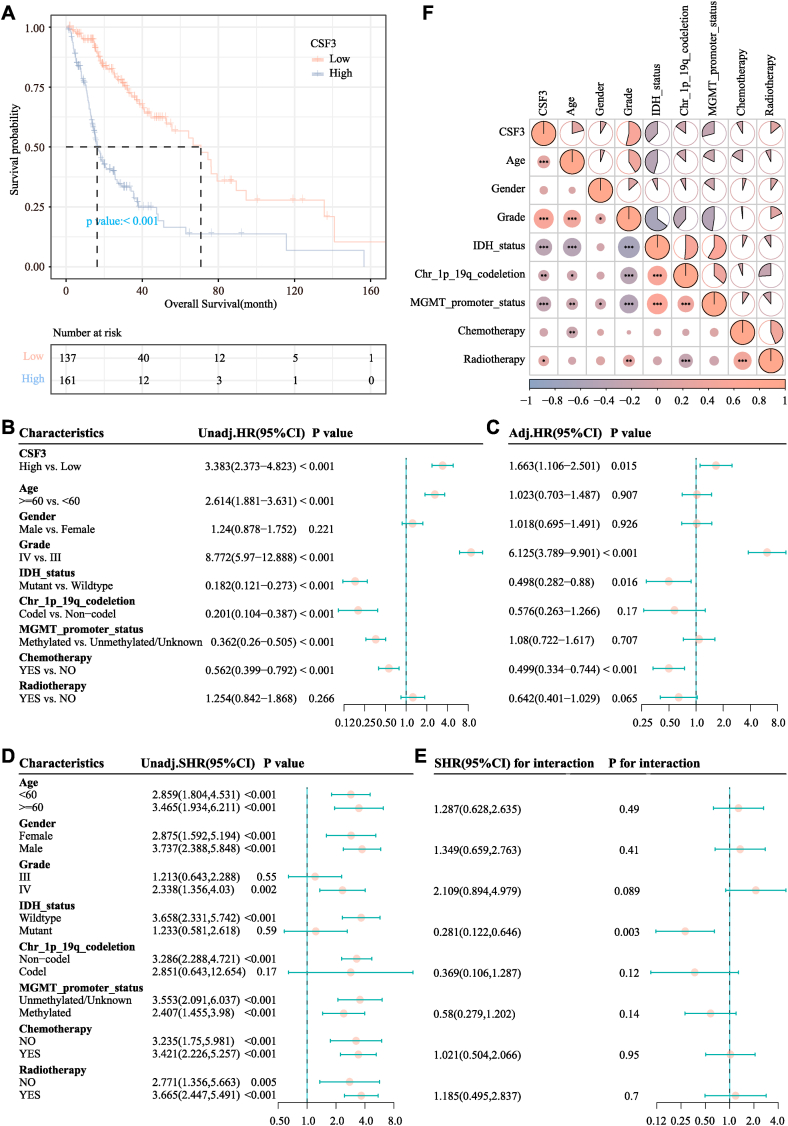
The median OS in the low-CSF3 expression and high-CSF3 expression groups was 70.87 months [95% confidence interval (CI) = 52.6–94.5] and 16.3 months (95% CI = 14.4–20.07) as shown in Fig. 3A, respectively. The KM curve showed that higher CSF3 levels were related with a worse survival outcome (P < 0.001). In the univariate analysis, CSF3 was a risk factor for survival outcome [hazard ratio (HR) = 3.383; 95% CI = 2.373–4.823; P < 0.001)], which was shown in Fig. 3B. According to the regression results, CSF3 expression (HR = 1.663; 95% CI = 1.106–2.501; P = 0.015) was a significant risk factor for survival outcomes, which was shown in Fig. 3C. In the subgroup analysis, in the age groups of both <60 years (HR = 2.859; 95% CI = 1.804–4.531; P < 0.001) and ≥60 years (HR = 3.465; 95% CI = 1.934–6.211; P = 0.49), no significant interaction was observed between CSF3 and age. The corresponding results were displayed in Fig. 3D and E. Fig. 3F correlation heat map showed an obvious positive correlation between CSF3 expression and tumor grade (P < 0.05) and a negative correlation between CSF3 expression and MGMT promoter methylation status (P < 0.05), whereas no significant correlation was observed between CSF3 expression and gender (P > 0.05).
Fig. 3.
Association of CSF3 expression and the prognosis of high-grade glioma (HGG) patients. (A) Kaplan–Meier survival curves for the effects of CSF3 expression. (B) Forest plot of the univariate Cox regression model. (C) Forest plot of the multivariate Cox regression model. (D) Subgroup analysis of the association of CSF3 with overall survival (OS). (E) Interaction analysis of CSF3 expression and other variables. (F) Relationship of CSF3 expression with the clinical characteristics of HGG patients.
3.3. Association between CSF3 expression and the immune infiltration profile
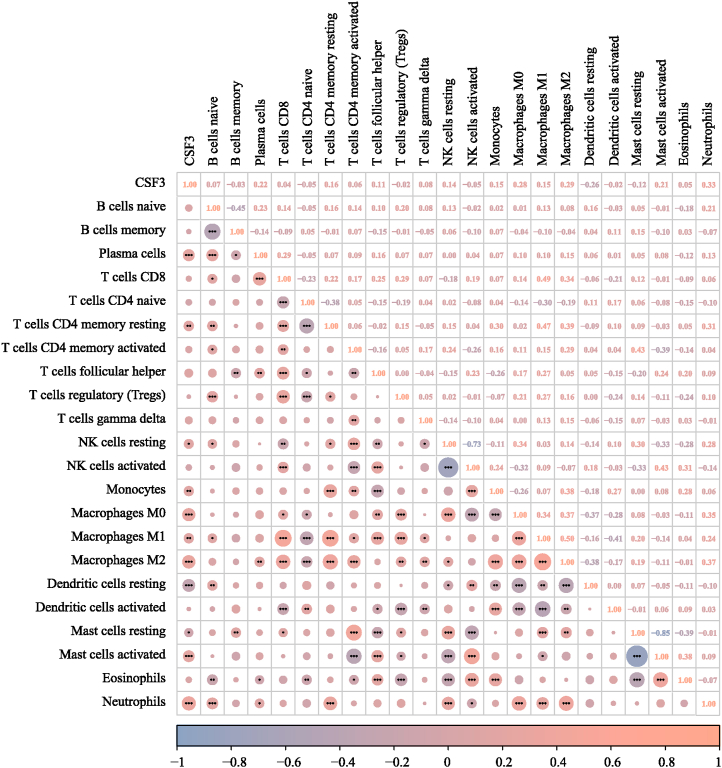
Immune cell infiltration analysis in HGG which was displayed in Fig. 4 showed a significant positive correlation between CSF3 expression and the infiltration of M0, M1, and M2 macrophages; plasma cells; and neutrophils (P < 0.05).
Fig. 4.
Heat map showing the correlation of immune cell infiltration with CSF3 expression.
3.4. Association between CSF3 expression and tumor progression pathways
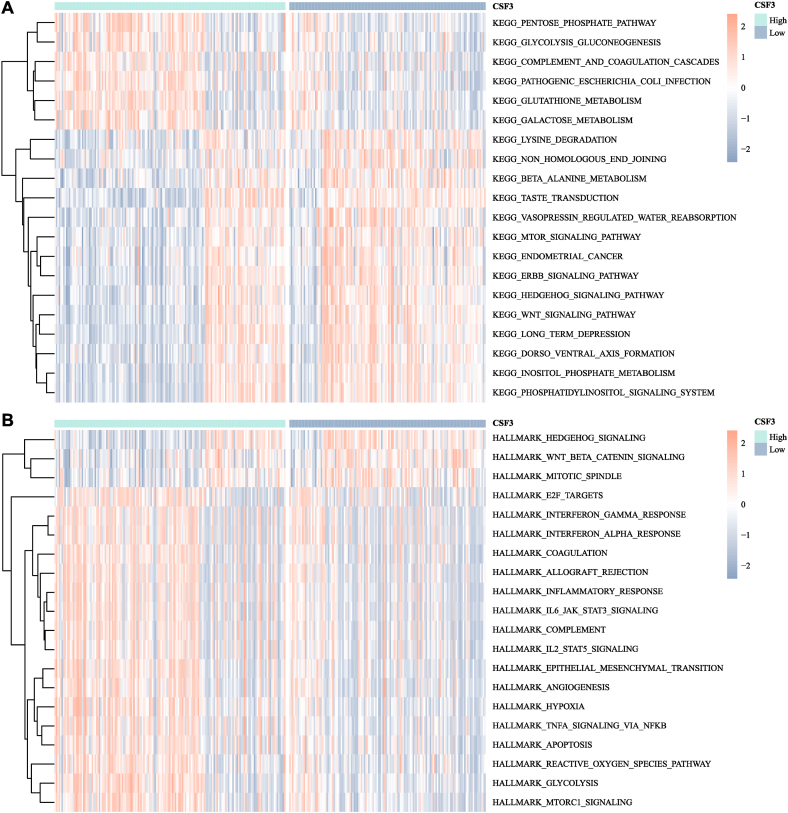
In the KEGG gene set, the CSF3-high expression group was significantly enriched in glutathione metabolism and the glycolysis and gluconeogenesis pathways. The CSF3-low-expression group was significantly enriched in the WNT signaling pathway and phosphatidylinositol signaling system pathway. In the Hallmark gene set, the CSF3-high expression group was significantly enriched in various pathways, including tumor necrosis factor (TNF)α signaling via nuclear factor kappa-B (NF-κB) and epithelial–mesenchymal transition, while the CSF3-low-expression group was significantly enriched in diverse pathways, including WNT signaling and mitotic spindle. The results were displayed in Fig. 5A and B.
Fig. 5.
Heat maps of CSF3 expression in different gene sets. (A) KEGG pathway analysis; (B) Hallmark pathway analysis.
3.5. Construction of the radiomics model based on LR and SVM methods
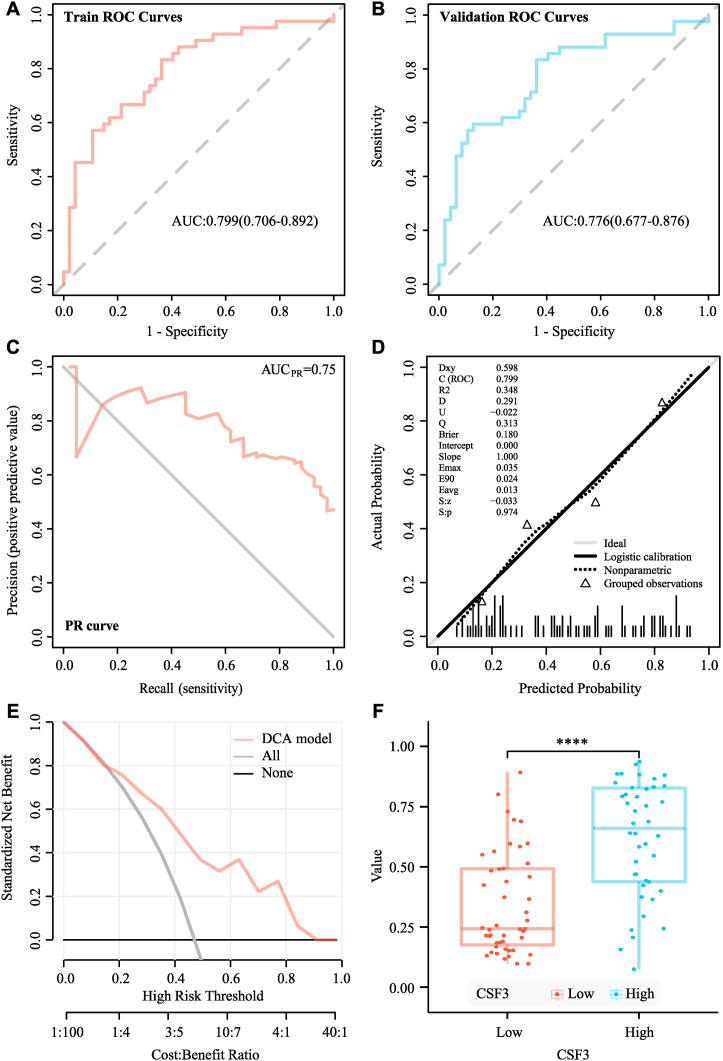
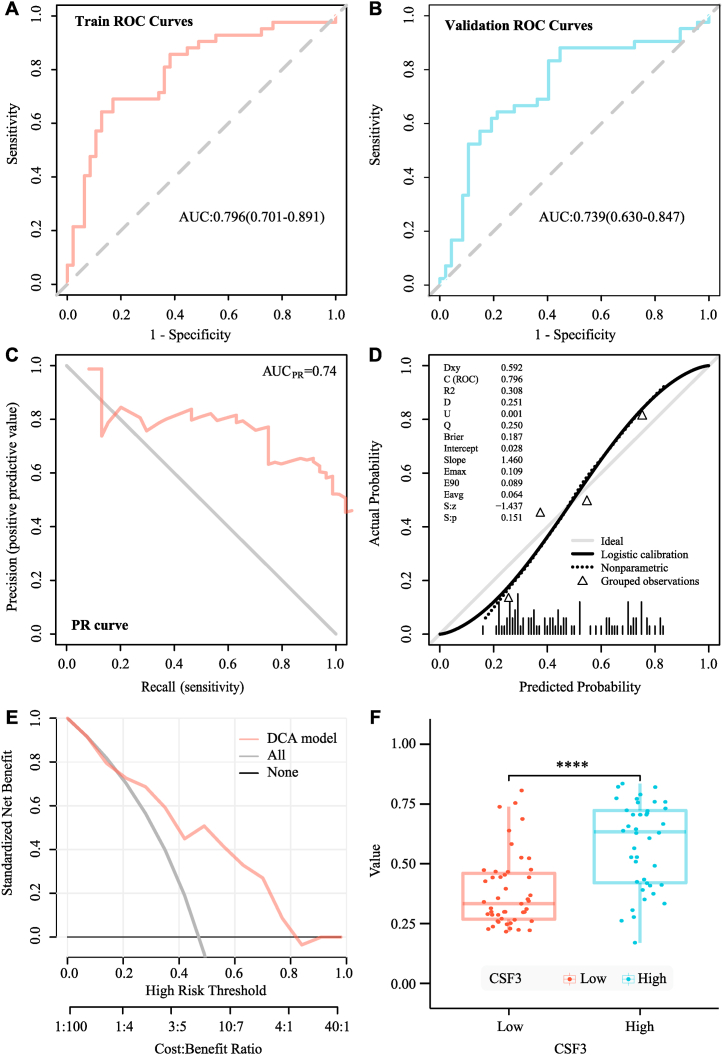
The median ICC value of the 89 imaging features was 0.971 (83.2% of all features); these features were entered into the subsequent feature screening. Finally, seven features were selected. They were glrlm_GrayLevelNonUniformityNormalized, glrlm_RunVariance, firstorder_InterquartileRange, glcm_Idn, firstorder_Skewness, glszm_SizeZoneNonUniformity and glcm_Imc1. The radiomics model was then analyzed using the LR model. Based on the ROC curve shown in Fig. 6A and B, the AUC value of the model in the training set was 0.799 and the cross-verified AUC value was 0.776. The AUC value of the precision-recall curve shown in Fig. 6C was 0.75. The calibration curve and Hosmer-Lemeshow goodness-of-fit test displayed in Fig. 6D were used to assess the prediction probability and true value of the imaging omics prediction model (P = 0.971). The decision curve analysis display model shown in Fig. 6E indicated high clinical practicability. SVM model analysis also showed that the radiomics model had a good prediction effect. Based on the ROC curve displayed in Fig. 7A and B, the AUC value of the model in the training set was 0.796, while the AUC value of cross-validation was 0.739. The AUC value of the precision-recall curve shown in Fig. 7C was 0.74. The calibration curve and Hosmer-Lemeshow goodness-of-fit test displayed in Fig. 7D showed consistency between the prediction probability and true value of the radiomics model (P = 0.27). The decision curve analysis display model shown in Fig. 7E also indicated high clinical practicability. The AUC value of the LR model was slightly higher than that of the SVM model. However, DeLong’s test showed no significant difference between the AUC values of the SVM and LR models in the training set (P = 0.767), and both models had good predictive efficiencies. The evaluations of the logistic regression radiomics model and the SVM radiomics model in the training and validation cohorts are shown in Table 2 and Table 3. The probability of predicting CSF3 expression levels using the RS was significantly different between the CSF3-high and CSF3-low-expression groups (P < 0.001), the corresponding results were displayed in Fig. 6, Fig. 7F. CD80, IL-6 expression (P < 0.001) was positively associated with the RS, whereas TLR4 (P = 0.022) expression was negatively associated with the radiomics score.
Fig. 6.
Evaluation of the logistic regression (LR) model. (A) Receiver operating characteristic (ROC) curve of the training cohort. (B) ROC curve of the validation cohort. (C) Precision–recall (PR) curve. (D) Calibration curve. (E) Decision curve analysis (DCA). (F) Differences in the radiomics scores between the CSF3 high- and low-expression groups.
Fig. 7.
Evaluation of the support vector machine (SVM) model. (A) Receiver operating characteristic (ROC) curve of the training cohort. (B) ROC curve of the validation cohort. (C) Precision–recall (PR) curve. (D) Calibration curve. (E) Decision curve analysis (DCA). (F) The radiomics scores between the CSF3 high- and low-expression groups.
Table 2.
Evaluation of the logistic regression radiomics model in the training and validation cohorts.
| Cohort | AUC (95% CI) | THRE | Accuracy | Sensitivity | Specificity | PPV | NPV | Brier score |
|---|---|---|---|---|---|---|---|---|
| Training | 0.799 (0.706–0.892) | 0.374 | 0.73 | 0.833 | 0.638 | 0.673 | 0.811 | 0.18 |
| Validation | 0.766 (0.677–0.876) | 0.387 | 0.73 | 0.833 | 0.638 | 0.673 | 0.811 | 0.191 |
AUC, area under the receiver operating characteristic curve; CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value; THRE, threshold.
Table 3.
Evaluation of the SVM radiomics model in the training and validation cohorts.
| Cohort | AUC (95% CI) | THRE | Accuracy | Sensitivity | Specificity | PPV | NPV | Brier score |
|---|---|---|---|---|---|---|---|---|
| Training | 0.796 (0.701–0.891) | 0.483 | 0.764 | 0.69 | 0.83 | 0.784 | 0.75 | 0.187 |
| Validation | 0.739 (0.63–0.847) | 0.374 | 0.708 | 0.881 | 0.553 | 0.638 | 0.839 | 0.216 |
AUC, area under the receiver operating characteristic curve; CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value; THRE, threshold.
3.6. Predictive performance of the radiomics models
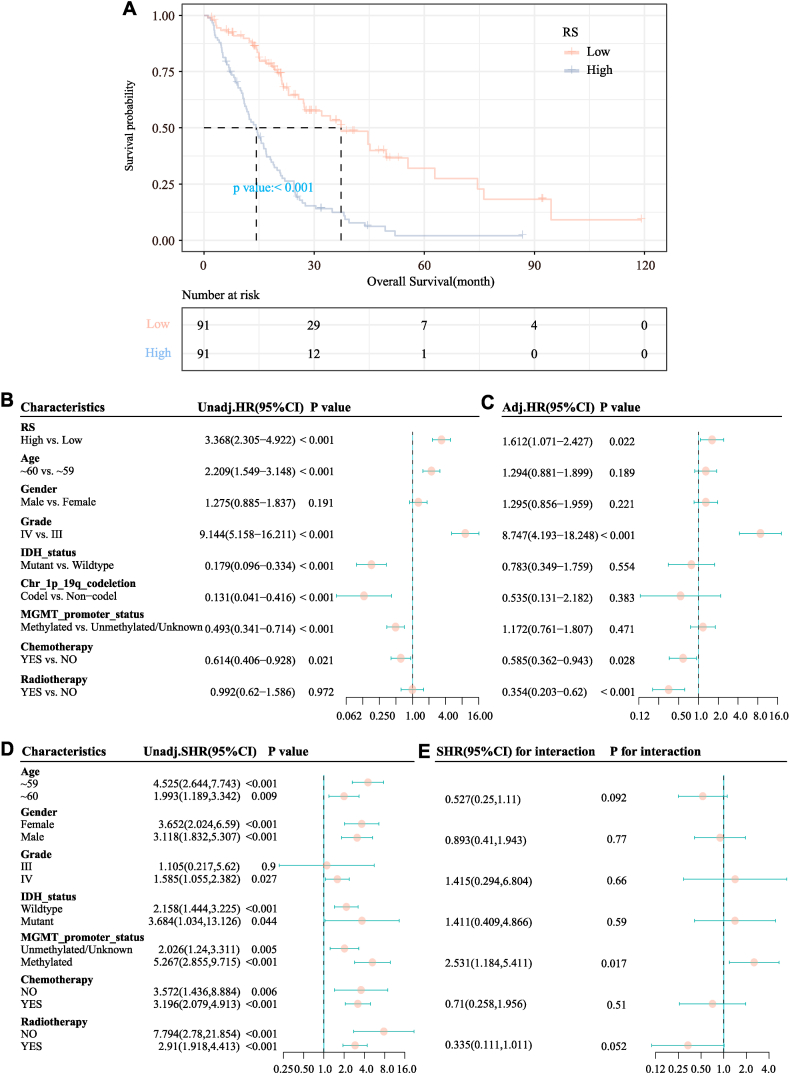
The 182 patients with HGG in TCGA database included in the survival analysis were divided into radiomics score (RS)-high (n = 91) and -low groups (n = 91) based on a cut-off score of 0.519; the clinical information of the patients in the two groups is shown in Table 4. Clear differences were observed in age distribution between the two groups (P < 0.001). The median OS in the RS-low and RS-high groups was 37.36 months (95% CI = 27.3–62.86) and 14.26 months (95% CI = 11.53–17.03), respectively. The KM curve in Fig. 8A showed that a higher RS level was significantly related to the shortening of OS (P < 0.001). The RS was a significant risk factor for OS in the univariate analysis (P < 0.001) as shown in Fig. 8B. Other variables of interest can be described with reference to the same pattern. In the multivariate analysis as displayed in Fig. 8C, after multi-factor adjustment, RS (HR = 1.612; 95% CI = 1.071–2.427; P = 0.022) was a statistically significant risk factor for OS. In the subgroup analysis as shown in Fig. 8D and E, a high RS was a risk factor for OS in both the age groups of <60 years (HR = 4.525; 95% CI = 2.644–7.743) and ≥60 years (HR = 1.993; 95% CI = 1.189–3.342), with no significant interaction between RS and age (interaction test P = 0.092).
Table 4.
Clinical characteristics of patients divided according to the radiometrics score.
| Variables | Total (n = 182) | Low (n = 91) | High (n = 91) | P-value |
|---|---|---|---|---|
| Age (years), n (%) | <0.001 | |||
| <60 | 112 (62) | 69 (76) | 43 (47) | |
| ≥60 | 70 (38) | 22 (24) | 48 (53) | |
| Gender, n (%) | 0.011 | |||
| Female | 80 (44) | 49 (54) | 31 (34) | |
| Male | 102 (56) | 42 (46) | 60 (66) | |
| Grade, n (%) | <0.001 | |||
| III | 62 (34) | 56 (62) | 6 (7) | |
| IV | 120 (66) | 35 (38) | 85 (93) | |
| IDH status, n (%) | <0.001 | |||
| Wild type | 134 (74) | 47 (52) | 87 (96) | |
| Mutant | 48 (26) | 44 (48) | 4 (4) | |
| Chr 1p19q co-deletion, n (%) | <0.001 | |||
| No | 163 (90) | 72 (79) | 91 (100) | |
| Yes | 19 (10) | 19 (21) | 0 (0) | |
| MGMT promoter status, n (%) | <0.001 | |||
| Unmethylated/Unknown | 94 (52) | 32 (35) | 62 (68) | |
| Methylated | 88 (48) | 59 (65) | 29 (32) | |
| Chemotherapy, n (%) | 0.148 | |||
| No | 39 (21) | 15 (16) | 24 (26) | |
| Yes | 143 (79) | 76 (84) | 67 (74) | |
| Radiotherapy, n (%) | 0.568 | |||
| No | 34 (19) | 19 (21) | 15 (16) | |
| Yes | 148 (81) | 72 (79) | 76 (84) |
Fig. 8.
Association of the radiomics score based on the logistic regression (LR) model and the prognosis of high-grade glioma (HGG) patients. (A) Kaplan–Meier curves for the effects of the radiomics score on overall survival (OS). (B) Forest plot of the univariate Cox regression model. (C) Forest plot of the multivariate Cox regression model. (D) Subgroup analysis of the associations of radiomics scores with OS. (E) Interaction analysis between the radiomics score and other variables.
4. Discussion
Substantial prior research has shown that G-CSF is important in the progression of tumor growth, angiogenesis, and migration [10]. According to previous findings, G-CSF can promote the proliferation of cancer cells while accelerating the migration of glioma cells to improve their invasion performance [14,15]. Analysis of G-CSF expression in triple-negative breast cancer [16], human non-small cell lung cancer [17,18], and cervical cancer [19] revealed that high G-CSF expression correlates with lower OS. High serum G-CSF levels in malignant tumors were related to an inferior OS [20]. However, data regarding the association between CSF3 expression and glioma survival outcomes are sparse. Many biomarkers in gliomas have been studied to date. IDH mutations, ATRX, MYB, MN1, and MGMT are considered positive prognostic markers, whereas H3F3A, TERT, CDKN2A, and epidermal growth factor receptor vIII (EGFR) are considered negative prognostic markers [5]. CSF3 has emerged as a promising new prognostic biomarker in HGG.
MRI combined with radiomics analysis has attracted research attention in potential clinical value for evaluating the status and prognosis of HGG patients in a non-invasive manner. Furthermore, radiomics has the potential to predict CSF3 expression levels, thereby increasing prognostic information and contributing to making better clinical decisions. Ita et al. [12] reported a positive correlation between the fold change in CSF3 levels and the tumor burden based on glioma imaging. Alghamri et al. [8] further showed that CSF3 expression is related to a favorable prognosis in glioma patients with mIDH1 [8]. In the present study, we mainly used the SVM and LR methods to construct a model based on enhanced MRI radiomics, which could non-invasively predict the levels of CSF3 and determine the prognosis of HGG patients. Based on this analysis, it can be inferred that the levels of CSF3 affect the prognosis of HGG (HR = 3.383; 95% CI = 2.373–4.823). Patients with increased CSF3 expression had poor prognosis with shorter survival periods. Multivariate analyses indicated that CSF3 was an independent risk factor for OS outcome in HGG. Moreover, the effect of CSF3 on OS was similar among different age groups. CSF3 expression had a significantly positive correlation with the tumor grade and a significantly negative correlation with the MGMT promoter methylation status. Overall, these findings indicate that non-invasive prediction of CSF3 expression may contribute to improved individualized clinical decision-making.
Kim et al. [21] showed that the heterogeneity of the tumor immune microenvironment of HGG can be identified by absolute quantification of tumor-expanding immune cells, which can be non-invasively predicted by radiomic features. Alghamri et al. [12] further showed that glioma-derived G-CSF induces the expansion of neutrophils, which is beneficial for immunotherapy. Furthermore, in the present study, CSF3 expression was positively correlated to tumor grade (P < 0.05). Moreover, in HGG, CSF3 expression was positively correlated to the immune cell infiltration of M0, M1, and M2 macrophages; plasma cells; and neutrophils (P < 0.05). The CSF3 high-expression group was mainly enriched in pathways of glutathione metabolism, glycolysis, gluconeogenesis, TNFα signaling via NF–B, and epithelial–mesenchymal transition. However, the CSF3 low-expression group was mainly enriched in the WNT signaling, phosphatidylinositol signaling system, and mitotic spindle pathways. Gao et al. [22] showed that CSF3 is involved in the inflammatory response, is highly expressed and secreted in inflammation-related pathways and is a potential plasma marker for glioma. Inflammation has connections with angiogenesis with regard to tumor growth and migration [23].
Radiomic data derived from the entire tumor can provide important information regarding gene expression and mutation patterns, thus aiding in diagnosis, prognosis, and OS predictions [24]. MRI relies on morphological features for a diagnosis, but an objective and quantitative method is lacking. Radiomics can extract high-throughput features by imaging the images, identify deep information that cannot be obtained directly by eye, and enable quantitative analysis. Wang et al. [25] extracted 3D MRI and clinical predictors of HGG patients, from which radiomics and depth features were extracted and integrated into prognostic models. Tian et al. [26] used the radiomics analysis method to extract MRI image features and built a prediction model that could effectively predict telomerase reverse transcriptase (TERT) mutations in HGG (AUC = 0.955). We delineated the radiomics features extracted by VOI at the intersection of TCIA–TCGA data. The best feature subsets were selected successively using the mRMR and recursive feature elimination algorithms, and seven radiomics features were finally extracted. These features were significantly related to the OS of HGG patients (HR = 3.368; 95% CI = 2.305–4.922) and successfully predicted CSF3 expression levels. The radiomic score showed a significant positive correlation with CSF3 expression, along with the expression of CD80, IL-6, and CD163. A higher radiomic score was related to a worse OS. Moreover, the radiomic score was a risk factor for OS in multivariate analysis.
Both the LR- and SVM-based models had good prediction effects, with AUC values of 0.799 and 0.796, respectively. Moreover, the radiomics models outperformed clinical data in determining the prognosis of HGG patients, and even higher model performance was achieved upon combining the radiomics model with clinical features. Pasquini et al. [27] showed that machine learning radiomics models that employ ensemble classifiers can predict relevant tasks such as OS, IDH mutation, EGFR amplification, and Ki-67 levels for HGG in the best-performing models. The high-scoring radiomics profile revealed a possible correlation between MRI and tumor histology [27].
Although these newly developed radiomics models performed well, several limitations of the current study need to be addressed. First, advanced MRI methods such as perfusion and diffusion weighted imaging were not considered, and the accuracy of radiomics needs to be further improved. Second, manual delineation of VOI may cause human bias. Although manual delineation of a region of interest is the most accurate approach among currently available lesion segmentation methods, it is affected by subjective factors. Semi-automatic or automatically delineated target area extraction of imaging features may be more objective, although it is time-consuming.
In conclusion, a predictive model of HGG was constructed based on enhanced MRI radiomics, which showed good stability and diagnostic efficacy. The MRI radiomics characteristics in HGG patients were significantly associated with CSF3 expression, which could indirectly reflect the prognosis. With the rapid development of big data technologies, advances in radiomics, and the demand for precision medicine, radiomics combined with genomics and proteomics will become a new research direction with great clinical value.
Declarations
Author contribution statement
Leina li: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Meidan Hou: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data.
Shaobo Fang: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Data availability statement
Data included in article/supp. material/referenced in article.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e15325.
Abbreviations
- HGGs
high-grade gliomas
- WHO
World Health Organization
- CSF3
Colony-stimulating factor 3
- G-CSF
granulocyte colony-stimulating factor
- IDH
isocitrate dehydrogenase
- mIDH1
mutant isocitrate dehydrogenase 1
- TCGA
The Cancer Genome Atlas
- TCIA
Cancer Imaging Archive
- MRI
magnetic resonance imaging
- RNA-seq
RNA-sequencing
- GBM
glioblastoma multiform
- TPM
transcripts per million
- KM
Kaplan–Meier
- IDH
isocitrate dehydrogenase
- MGMT
methylguanine methyltransferase
- GSVA
Gene set variation analysis
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- VOI
volume of interest
- ICC
intraclass correlation coefficient
- mRMR
minimum redundancy maximum relevance
- LR
logistic regression
- SVM
support vector machine
- ROC
receiver operating characteristic
- AUC
area under curve
- HR
hazard ratio
- RS
radiomics score
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Ostrom Q.T., Bauchet L., Davis F.G., et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2014;16:896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nayak L., Reardon D.A. High-grade gliomas. Continuum (Minneap Minn) 2017;23:1548–1563. doi: 10.1212/CON.0000000000000554. [DOI] [PubMed] [Google Scholar]
- 3.Tan Y., Mu W., Wang X.C., et al. Improving survival prediction of high-grade glioma via machine learning techniques based on MRI radiomic, genetic and clinical risk factors. Eur. J. Radiol. 2019;120 doi: 10.1016/j.ejrad.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Bush N.A., Chang S.M., Berger M.S. Current and future strategies for treatment of glioma. Neurosurg. Rev. 2017;40:1–14. doi: 10.1007/s10143-016-0709-8. [DOI] [PubMed] [Google Scholar]
- 5.Śledzińska P., Bebyn M.G., Furtak J., et al. Prognostic and predictive biomarkers in gliomas. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms221910373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahi V., Jamwal S., Kumar P. Neuroprotection through G-CSF: recent advances and future viewpoints. Pharmacol. Rep. 2021;73:372–385. doi: 10.1007/s43440-020-00201-3. [DOI] [PubMed] [Google Scholar]
- 7.Yeo B., Redfern A.D., Mouchemore K.A., et al. The dark side of granulocyte-colony stimulating factor: a supportive therapy with potential to promote tumour progression. Clin. Exp. Metastasis. 2018;35:255–267. doi: 10.1007/s10585-018-9917-7. [DOI] [PubMed] [Google Scholar]
- 8.Alghamri M.S., McClellan B.L., Avvari R.P., et al. G-CSF secreted by mutant IDH1 glioma stem cells abolishes myeloid cell immunosuppression and enhances the efficacy of immunotherapy. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abh3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kast R.E., Hill Q.A., Wion D., et al. Glioblastoma-synthesized G-CSF and GM-CSF contribute to growth and immunosuppression: potential therapeutic benefit from dapsone, fenofibrate, and ribavirin. Tumour. Biol. 2017;39 doi: 10.1177/1010428317699797. [DOI] [PubMed] [Google Scholar]
- 10.Theron A.J., Steel H.C., Rapoport B.L., et al. Contrasting immunopathogenic and therapeutic roles of granulocyte colony-stimulating factor in cancer. Pharmaceuticals (Basel) 2020;13:406. doi: 10.3390/ph13110406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambin P., Leijenaar R.T.H., Deist T.M., et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 12.Ita M.I., Wang J.H., Toulouse A., et al. The utility of plasma circulating cell-free messenger RNA as a biomarker of glioma: a pilot study. Acta Neurochir. 2022;164:723–735. doi: 10.1007/s00701-021-05014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albaradei S., Thafar M., Alsaedi A., et al. Machine learning and deep learning methods that use omics data for metastasis prediction. Comput. Struct. Biotechnol. J. 2021;19:5008–5018. doi: 10.1016/j.csbj.2021.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gay A.N., Chang S., Rutland L., et al. Granulocyte colony stimulating factor alters the phenotype of neuroblastoma cells: implications for disease-free survival of high-risk patients. J. Pediatr. Surg. 2008;43:837–842. doi: 10.1016/j.jpedsurg.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J., Yao L., Zhao S., et al. Granulocyte-colony stimulating factor promotes proliferation, migration and invasion in glioma cells. Cancer Biol. Ther. 2012;13:389–400. doi: 10.4161/cbt.19237. [DOI] [PubMed] [Google Scholar]
- 16.Hollmén M., Karaman S., Schwager S., et al. G-CSF regulates macrophage phenotype and associates with poor overall survival in human triple-negative breast cancer. OncoImmunology. 2016;5 doi: 10.1080/2162402X.2015.1115177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei L.X., Chang W.L., Guo A.T., et al. Expression of granulocyte colony stimulating factor in patients with non-small cell lung cancer and its clinicopathological significance. Zhonghua Bing Li Xue Za Zhi. 2011;40:721–725. [PubMed] [Google Scholar]
- 18.Nakamura M., Oshika Y., Abe Y., et al. Gene expression of Granulocyte colony stimulating factor (G-CSF) in non-small cell lung cancer. Anticancer Res. 1997;17:573–576. [PubMed] [Google Scholar]
- 19.Du H., Zhang H., Zhang Y., et al. Expression of G-CSF and clinical pathological significance in cervical cancer. Zhonghua Yixue Zazhi. 2016;96:358–360. doi: 10.3760/cma.j.issn.0376-2491.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Stathopoulos G.P., Armakolas A., Tranga T., et al. Granulocyte colony-stimulating factor expression as a prognostic biomarker in non-small cell lung cancer. Oncol. Rep. 2011;25:1541–1544. doi: 10.3892/or.2011.1226. [DOI] [PubMed] [Google Scholar]
- 21.Kim A.R., Choi K.S., Kim M.S., et al. Absolute quantification of tumor-infiltrating immune cells in high-grade glioma identifies prognostic and radiomics values. Cancer Immunol. Immunother. 2021;70:1995–2008. doi: 10.1007/s00262-020-02836-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao Y., Zhang E., Liu B., et al. Integrated analysis identified core signal pathways and hypoxic characteristics of human glioblastoma. J. Cell Mol. Med. 2019;23:6228–6237. doi: 10.1111/jcmm.14507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broekman M.L., Maas S.L.N., Abels E.R., et al. Multidimensional communication in the microenvirons of glioblastoma. Nat. Rev. Neurol. 2018;14:482–495. doi: 10.1038/s41582-018-0025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gillies R.J., Kinahan P.E., Hricak H.H.H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y., Shao Q., Luo S., et al. Development of a nomograph integrating radiomics and deep features based on MRI to predict the prognosis of high grade gliomas. Math. Biosci. Eng. 2021;18:8084–8095. doi: 10.3934/mbe.2021401. [DOI] [PubMed] [Google Scholar]
- 26.Tian H., Wu H., Wu G., et al. Noninvasive prediction of tert promoter mutations in high-grade glioma by radiomics analysis based on multiparameter MRI. BioMed Res. Int. 2020;2020 doi: 10.1155/2020/3872314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasquini L., Napolitano A., Lucignani M., et al. AI and high-grade glioma for diagnosis and outcome prediction: do all machine learning models perform equally well? Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.601425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. material/referenced in article.