Highlights
-
•
Ovalbumin (OVA)-EGCG conjugates were fabricated by an ultrasound-assisted method.
-
•
OVA-EGCG conjugates were studied by experimental assays and computer simulations.
-
•
The interaction of OVA and EGCG altered the protein conformation.
-
•
The emulsifying property of the conjugates was better than that of native OVA.
-
•
Conjugates induced by ultrasound-assisted method showed superiority in antioxidant properties.
Keywords: Protein, Polyphenol, Ultrasound, Molecular docking simulation, Covalent binding
Abstract
The influence of ultrasound-assisted free radical modification on the structure and functional properties of ovalbumin-epigallocatechin gallate (OVA-EGCG) conjugates was investigated by experimental measurements and computer simulations. Compared with the traditional free radical condition, the ultrasonic-assisted processing significantly increased the conjugating efficiency of OVA and EGCG and shortened the conjugating from 24 h to 1 h without affecting the equivalent amount of EGCG conjugating. The sodium dodecyl sulfate–polyacrylamide gel electrophoresis and multi-spectroscopy analysis (Fourier transform infrared spectroscopy, intrinsic fluorescence spectroscopy, and UV spectroscopy) indicated that the covalent conjugates could be formed between OVA and EGCG. And modification in the conformation of OVA was induced by EGCG. Furthermore, molecular docking results demonstrated the possession of high-affinity EGCG binding location on OVA, supporting and clarifying the experimental results. In addition, the functional properties of OVA including emulsification (emulsifying activity and emulsion stability) and antioxidant properties (DPPH scavenging capacity and ABTS scavenging capacity) were significantly improved after conjugation with EGCG, especially in ultrasound-assisted conditions. Overall, OVA-EGCG conjugates produced by ultrasound-assisted free radical treatment could be applied as a potential emulsifier and antioxidant, thereby expanding the application of OVA as a dual-functional ingredient.
1. Introduction
The development of dual-functional protein-based conjugates with outstanding emulsifying and antioxidant capacity is of particular attention since the increasing demand for functional composite food [1], [2]. Ovalbumin (OVA), the main component of egg white protein, was considered as an excellent source of animal protein and a promising functional food ingredient because of its high nutritional value and easy isolation and purification from egg white [3], [4], [5]. Nevertheless, more than half of the amino acids in native OVA were hydrophobic amino acids, and the intramolecular hydrophobic interaction, results in the formation of a highly aggregated conformation of OVA, thus exhibiting inferior functional properties and limiting the application as a functional ingredient [6], [7], [8].
Covalent conjugation between protein and polyphenol has been verified to be extremely efficient in improving the functional properties of proteins, especially in antioxidant stability [9], [10], [11], [12]. Generally, enzymatic catalysis [13], free radical [14], and alkaline reaction [15], [16] were widely applied for covalent fabricating protein–polyphenol conjugates. Among them, protein–polyphenol conjugates prepared by the enzymatic catalysis method were relatively expensive. Moreover, the products were accompanied by obvious browning, which is a hurdle to the industrial application [17]. Unlike enzymatic catalysis, the free radical and alkaline treatments involve reaction conditions that was relatively simple, mild, and economic. Furthermore, previous studies have proved that protein–polyphenol conjugates prepared by the free radical method have higher functional properties and bio-safety than the conjugates prepared by the alkali method [18], [19]. Eepigallocatechin-3-gallate (EGCG), a predominant active ingredient in green tea, has been considered as an ideal polyphenol material for the development of protein-based conjugates due to its widely biological activity, especially excellent antioxidant activities [20], [21]. Moreover, several studies have been evidenced that EGCG possesses high-affinity for protein [22], [23]. Based on the above comprehensive analysis, the OVA-EGCG conjugates were prepared by the free radical method in this study.
However, one of the primary disadvantages of the conventional free radical method for preparing protein–polyphenol conjugates was the relatively longer reaction time needed (more than 24 h), which was an obstacle to the massive commercialized production. In theory, this restriction could be solved by combining free radical treatment with ultrasonic technology. As is well-known, ultrasound, a non-thermal processing technology, has been widely applied in the food industry [24], [25], [26], [27], due to its operability, time-saving, and inexpensive. As previously described, the combination of mechanical, cavitation, turbulence and microstreaming effects generated by ultrasound could promote the unfolding of the protein structure and the exposure of reacting groups, thus increasing the possibility of conjugation between protein and other molecules [28], [29]. For instance, Zhang et al. found that ultrasound-assisted alkaline treatment could remarkably decrease the conjugated time from 24 h to 40 min without affecting the binding degree [30]. Similarly, Geng et al. reported that the ultrasonic method could form a more stable soy protein isolate-epigallocatechin gallate system with preferable functional properties [20]. Currently, the application of different ultrasonic time-assisted free radical preparation of protein–polyphenol conjugates in the development of dual-functional protein-based conjugates is still limited. Particularly, there has been limited information on the comprehensive analysis of the protein–polyphenol binding underlying mechanism by experimental determination and computer simulation.
Therefore, OVA-EGCG conjugates were fabricated by traditional or different ultrasonic time-assisted free radical treatments in this study. And the effects of different ultrasonic treatments on the structural and functional properties of the EGCG-OVA conjugates were comprehensively analyzed by experimental detection and computer simulation. The amount of total EGCG content was used to evaluate the degree of combination of OVA and EGCG. Variations in the physicochemical and structural properties of OVA were assessed by the sulfhydryl group, SDS-PAGE, FTIR spectroscopy, intrinsic fluorescence spectroscopy, and UV spectroscopy. Further, molecular docking was used to explore potential binding sites between OVA and EGCG. In addition, the influence of ultrasound and EGCG on the functional properties of OVA were assessed by emulsifying capacity (emulsifying activity and emulsion stability) and antioxidant capacity (DPPH scavenging capacity and ABTS scavenging capacity). Therefore, the results of this study will provide a novel insight and theoretical basis for developing dual-functional OVA ingredients with superior emulsification and antioxidant capacity.
2. Material and methods
2.1. Material
Ovalbumin from chicken egg white (OVA, A-5253) was purchased from Sigma Chemical Co. (St. Louis, Mo, USA). Epigallocatechin-3-gallate (EGCG, purity ≥98%) and dialysis membranes (MW: 8000–14,000 KDa) were purchased from Yuanye Reagent Co. (Shanghai, China). Folin-Ciocalteu phenol reagent, Coomassie Brilliant Blue Fast Staining solution (P1300), and ColorMixed Protein Marker (11–245 KD) were purchased from Solarbio Science and Technology Co., Ltd. (Beijing, China). 2,2-Diphenyl-1-picrylhydrazyl (DPPH, purity ≥ 96%) and 2,2′-Azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS, purity ≥ 98%) were purchased from Macklin Biochemical Co., Ltd (Shanghai, China). Corn oil was purchased from Golden dragon fish grain oil Food Co. LTD (Shanghai, China). All other chemicals were of analytical grade and purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China).
2.2. Fabrication of the OVA-EGCG conjugates
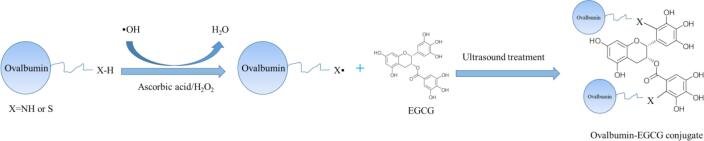
The OVA-EGCG conjugates by ultrasound-assisted free radical treatment were fabricated according to the method by Geng et al. with slight modifications [20]. Briefly, 1 g OVA powder was dissolved in 100 mL deionized water and was fully hydrated at 4 ℃ overnight, and then the hydrated solution was centrifuged at 4 ℃ and 10,000g for 20 min to obtain a fully soluble OVA solution. Afterward, 1 mL of the hydrogen peroxide (5.0 mol/L) and 0.25 g of ascorbic acid was introduced to the OVA solution as the radical initiator systems. Subsequently, the mixtures were incubated with magnetic stirring at 25 ℃ for 2 h, and then the EGCG was added and sonicated (50%, 5 on, and 3 off) for different reacted times (30 min, 60 min, 90 min, and 120 min) using ultrasound equipment (Scientz Sonifier II D, Xin Zhi Ultrasonics Co., Ltd., China). Also, the whole ultrasonic process was carried out in an ice bath to avoid heating during the ultrasonic reaction. After the different times of ultrasound treatment, the conjugates were dialyzed at 4 ℃ for 48 h to remove unreacted EGCG by dialysis with 8000–14,000 KDa molecular weight cut-off membrane and then vacuum lyophilized for later analysis. The ultrasound-assisted free radical of OVA-EGCG conjugates prepared under different ultrasonic response times were named as U-OVA-EGCG30, U-OVA-EGCG60, U-OVA-EGCG90, and U-OVA-EGCG120. For the preparation of OVA-EGCG conjugate by traditional free radical methods, the mixed solution containing EGCG was incubated with continuous stirring for 24 h and other step was performed as described above for the preparation of the U-OVA-EGCG conjugates but without ultrasound treatment. Native soluble OVA without EGCG and OVA-EGCG conjugate prepared by traditional free radical methods (T-OVA-EGCG) were used as a comparison. The covalent conjugation of ovalbumin and EGCG by the ultrasound-assisted free radical method was exhibited in Fig. 1.
Fig. 1.
Covalent conjugation of ovalbumin and EGCG by the ultrasound-assisted free radical method.
2.3. Characterization of OVA-EGCG conjugates
2.3.1. Total EGCG contents of the OVA-EGCG conjugates
The Folin–Ciocalteu assay was used to measure the total EGCG contents of T-OVA-EGCG, and U-OVA-EGCG conjugates based on the Zhang et al with a slight modifications [30]. Briefly, 0.5 mL of each OVA-EGCG conjugates sample diluted 10 times was added with 2.5 mL of 0.2 M Folin–Ciocalteu reagent (Solarbio, F8060). The mixtures were mixed thoroughly and incubated in the dark for 3 min. Next, 2 mL Na2CO3 solution was introduced and incubated in the dark for 120 min. Finally, a Varioskan Flash Spectra (Thermo Fisher Scientific Co., Ltd., USA) was used to record the absorbance of the mixture solution at 760 nm. The total EGCG content of each T-OVA-EGCG and U-OVA-EGCG conjugate was expressed as the same amount of EGCG based on a standard curve (Abs = 3.7363*Concentration(EGCG) + 0.0641, R2 = 0.9991) prepared by determined the absorbance value of different concentrations of EGCG (0, 0.05, 0.1, 0.15, 0.2, 0.25, and 0.3 mg/mL).
2.3.2. Sulfhydryl group (SH) contents of the OVA-EGCG conjugates
The free sulfhydryl groups were measured by Ellman’s reagent method as reported by Chen et al. with slight modifications [31]. Briefly, 0.5 mL of each native OVA, T-OVA-EGCG, and U-OVA-EGCG conjugates sample solution was mixed with 4.5 mL of standard buffer solutions (0.5% (w/v) SDS, 0.086 mol/L Tris, 0.092 mol/L Glycine and 0.004 mol/L EDTA, pH 8.0). Subsequently, the above reaction solutions were added to 50 uL of Ellman’s reagent (4 mg/mL DTNB in Tris-Glycine buffer, pH 8.0). The absorbance of the mixture solutions was read at 412 nm on a Varioskan Flash Spectra (Thermo Fisher Scientific Co., Ltd., USA) after reacting for 15 min in a dark environment.
Where: A412: the absorbance value of the determined sample at the wavelength of 412 nm, D is the dilution index, and C is the concentration of the determined samples.
2.3.3. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) of the OVA-EGCG conjugates
The molecular weight distribution changes of native OVA, T-OVA-EGCG, and U-OVA-EGCG conjugates were determined by SDS–PAGE after different ways of conjugation with EGCG. The SDS-PAGE was performed referring to Luo et al. with slight modifications [32]. The SDS-PAGE was performed using 5% stacking gel and 12% separating gel. The different sample solutions were fully mixed with 5× SDS-PAGE loading buffer containing β-mercaptoethanol, followed by heating the mixtures solutions at 100 ℃ for 5 min, and centrifuged at 10,000g and 4 ℃ for 5 min. The supernatant of each sample (10 μL) was loaded into a gel (5% stacking gel and 12% separating gel), and migration was performed at 100 V for 95 min. Finally, the Gels were stained and decolorized according to the instructions with Coomassie Brilliant Blue Fast Staining solution (Solarbio, P1300).
2.3.4. Fourier-transform infrared (FTIR) spectroscopy of the OVA-EGCG conjugates
The FTIR of native OVA, T-OVA-EGCG, and U-OVA-EGCG conjugates were acquired with an Bruker Tensor-27 FTIR spectrometer (Bruker, Germany) according to the method reported by Zheng et al [8]. Briefly, 4 mg of freeze-dried samples were fully mixed with 600 mg of potassium bromide (KBr) in an agate mortar, and pressed into KBr pellets for measurement. The FTIR spectroscopy was recorded in the scanning ranges of 400–4000 cm−1.
2.3.5. Intrinsic fluorescence spectroscopy of the OVA-EGCG conjugates
The intrinsic fluorescence emission spectroscopy analysis for native OVA, T-OVA-EGCG, and U-OVA-EGCG conjugates was determined using a RF-5301 fluorescence spectrophotometer (Shimadzu Co., Ltd., Tokyo, Japan). The diluted sample solutions were performed at the excitation wavelength of 280 nm and the emission spectra of 300–500 nm.
2.3.6. Ultraviolet absorption (UV) spectroscopy of the OVA-EGCG conjugates
The UV absorption spectra of native OVA, T-OVA-EGCG, and U-OVA-EGCG conjugates were recorded by putting diluted corresponding sample solution into a 1.0 cm quartz cuvette and scan wavelength range from 200 to 800 nm at 25 ℃ using a Cary 60 UV–Vis spectrophotometer (Agilent technologies Inc. Co., Ltd., USA).
2.4. Molecular docking of OVA and EGCG
The OVA and EGCG were docked and visualized using the Autodock Vina and PyMol software to further predict potential binding sites and understand the molecular interaction mechanism. The structure of OVA (PDB: 1OVA) and EGCG (PubChem CID:65064) were collected from the RCSB Protein Data Bank database (https://www.rcsb.org/) and PubChem database (https://pubchem.ncbi.nlm.nih.gov/), respectively. The mode with the lowest binding energy of Autodock Vina software was selected and further analyzed by PyMol software.
2.5. Antioxidant activity of OVA-EGCG conjugates
2.5.1. DPPH radical scavenging assay
The DPPH radical scavenging assay was carried out according to the method described by Geng et al. with a slightly modified [20]. The different samples were combined with a DPPH-ethanol solution (0.0039 g DPPH was fully dissolved into anhydrous ethanol, diluted into 100 mL, and stirred away from light for 2 h) at the ratio of 1–4. Subsequently, the absorbance of the mixture solution was read at 517 nm after 30 min in the dark using a Varioskan Flash Spectra (Thermo Fisher Scientific Co., Ltd., USA). The anhydrous ethanol was a control.
Where, A0: the absorbance value of water. A1: the absorbance value of samples. A2: The absorption value of DPPH substituted by ethanol.
2.5.2. ABTS free radical scavenging assay
The ABTS free radical scavenging assay was carried out according to the method described by Geng et al. with a slightly modified [20]. The original ABTS solution was prepared by the mixture of ABTS and potassium persulfate solution at the ratio of 1 to 1 and was placed in the dark for 12 h–16 h. The original ABTS solution was diluted with phosphate buffer solution (pH 7.2–7.4, 0.01 M) until the absorption value was 0.7 ± 0.05. Subsequently, the samples were mixed with the diluted ABTS solution at a ratio of 1–19, and the absorbance value was recorded at 734 nm after incubation for 6 min using a Varioskan Flash Spectra (Thermo Fisher Scientific Co., Ltd., USA).
2.6. Emulsifying activity index (EAI) and emulsion stability index (ESI) of OVA-EGCG conjugates emulsion
The emulsifying properties (EAI and ESI) for the emulsions of native OVA, T-OVA-EGCG, and U-OVA-EGCG conjugates were measured according to the method described by Li et al [33] with slight adjustments. 5 mL of soybean oil was added to 15 mL of each sample solution and then was emulsified to O/W emulsions using an Ultra-Turrax homogenizer (T18, IKA, Germany) at 12,000 rpm for 3 min. Subsequently, 50 uL of each homogenized emulsion was taken from the bottom at 0 min and 10 min, and dilution with sodium dodecyl sulfate to 5 mL. Finally, the absorbance of the dilution emulsion was determined at 500 nm using a Varioskan Flash Spectra (Thermo Fisher Scientific Co., Ltd., USA).
Here, DF: the dilution factor; C: the concentration of protein (g/mL); Ψ, the optical path (1 cm); θ: the volume fraction of oil (0.25); A0 and A10: the absorbance of emulsions at 0 and 10 min, respectively.
2.7. Statistical analysis
All the experiments were carried out in triplicate. Figures were generated by Origin software (Version 8.5). The significant difference was checked by Duncan’s multiple range test (p < 0.05) using DPS Statistics software (Version 7.05).
3. Results and discussion
3.1. Determination of total EGCG contents of OVA-EGCG conjugates
The total EGCG contents of OVA-EGCG conjugates were determined to evaluate the degree of EGCG binding to the OVA molecules. Table 1 presents the total EGCG of corresponding OVA-EGCG conjugates prepared by traditional or different ultrasonic time-assisted free radical treatments. The total EGCG content of OVA-EGCG conjugates with traditional free radical treatments alone for 24 h was 21.96 ± 0.53 mg/g, while the total EGCG content acquired by U-OVA-EGCG conjugates was 22.48 ± 1.15 mg/g after a shorter reaction time (60 min) under ultrasonic-assisted treatment. It exhibited no significant difference from the traditional free radical treatment, suggesting that the ultrasonic treatment could markedly accelerate the interaction between OVA and EGCG, and shorten the time required for a free radical time from 24 h to 1 h without affecting the binding degree of OVA and EGCG. For ultrasound-assisted treatment, the total EGCG content of OVA-EGCG conjugates first increased and then decreased over the ultrasonic time, reaching the highest total EGCG content at 90 min (31.99 ± 0.70 mg/g). These increased results of total EGCG content may be due to the mechanical and cavitation effects generated by ultrasonic treatment. Both effects could accelerate the production of free hydroxyl radicals [20] and the unfolding of protein molecules [30], thus increasing the reaction possibility of OVA and EGCG. Moreover, the microstreaming induced by ultrasound treatment results in the high-speed collision between protein and polyphenol, and the decrease in particle size and increase in the specific surface area [20], which attributes to improving the efficiency of combining the OVA with EGCG. The decrease in total EGCG content was observed with the extension of ultrasound time to 120 min, which might be because the excess energy generated by ultrasound promoted the aggregation of proteins, thereby reducing the reaction sites with polyphenols. Similarly, previous reports have suggested that excessive ultrasound treatment leads to the denaturation and aggregation of proteins and the decomposition of polyphenols, thereby reducing the ability of polyphenols to bind to proteins [34], [35].
Table 1.
Total EGCG content in T-OVA-EGCG and U-OVA-EGCG conjugates.
| Samples | Total EGCG content |
|---|---|
| mg/g | |
| T-OVA-EGCG | 21.96 ± 0.53c |
| U-OVA-EGCG30 | 17.35 ± 0.77d |
| U-OVA-EGCG60 | 22.48 ± 1.15c |
| U-OVA-EGCG90 | 31.99 ± 0.70a |
| U-OVA-EGCG120 | 26.47 ± 0.84b |
Note: Dates are given as means ± standard deviations. Values with superscript letters in the same column indicate significantly different at P < 0.05. T-OVA-EGCG: ovalbumin-epigallocatechin-3-gallate conjugates prepared by traditional free radical treatment; U-OVA-EGCG30, U-OVA-EGCG60, U-OVA-EGCG90, and U-OVA-EGCG120: the ultrasound-assisted free radical of OVA-EGCG conjugates prepared under different ultrasonic response times of 30 min, 60 min, 90 min, and 120 min, respectively.
3.2. Sulfhydryl group (SH) contents of the OVA-EGCG conjugates
The sulfhydryl group of protein was a representative group to express the side chain groups [23], and was also susceptible to phenolic compounds [31]. As shown in Table 2, the contents of free sulfhydryl group in both T-OVA-EGCG conjugates and U-OVA-EGCG conjugates were markedly decreased compared with that of native OVA. Similarly, Meng et al. found sulfhydryl groups were lost when whey protein isolate was bound with three polyphenols [19]. These reasons for a decrease in sulfhydryl groups could be explained by the fact that the free sulfhydryl group in the protein could interact with the hydroxyl group provided by the introduced phenolic compound, thus reducing the amount of sulfhydryl group in the protein [20]. In addition, the loss of sulfhydryl in the U-OVA-EGCG conjugates was more pronounced under ultrasonic assistance (P < 0.05), suggesting that ultrasound treatment facilitated the binding of OVA molecules to EGCG, due to ultrasound-induced cavitation and mechanical effects [36]. These effects were conducive to the expansion of the internal structure of protein and the lengthening of the peptide chain, providing more possibilities for binding to EGCG, as demonstrated by the total EGCG content (Table 1).
Table 2.
Free sulfhydryl group in T-OVA-EGCG and U-OVA-EGCG conjugates.
| Samples | Free sulfhydryl group |
|---|---|
| μmol/g | |
| OVA | 8.60 ± 0.47a |
| T-OVA-EGCG | 4.36 ± 0.12c |
| U-OVA-EGCG30 | 6.29 ± 0.19b |
| U-OVA-EGCG60 | 4.31 ± 0.05c |
| U-OVA-EGCG90 | 3.88 ± 0.04d |
| U-OVA-EGCG120 | 4.07 ± 0.11 cd |
Note: Dates are given as means ± standard deviations. Values with superscript letters in the same column indicate significantly different at P < 0.05. T-OVA-EGCG: ovalbumin-epigallocatechin-3-gallate conjugates prepared by traditional free radical treatment; U-OVA-EGCG30, U-OVA-EGCG60, U-OVA-EGCG90, and U-OVA-EGCG120: the ultrasound-assisted free radical of OVA-EGCG conjugates prepared under different ultrasonic response times of 30 min, 60 min, 90 min, and 120 min, respectively.
3.3. Evidence for the formation of OVA-EGCG conjugates
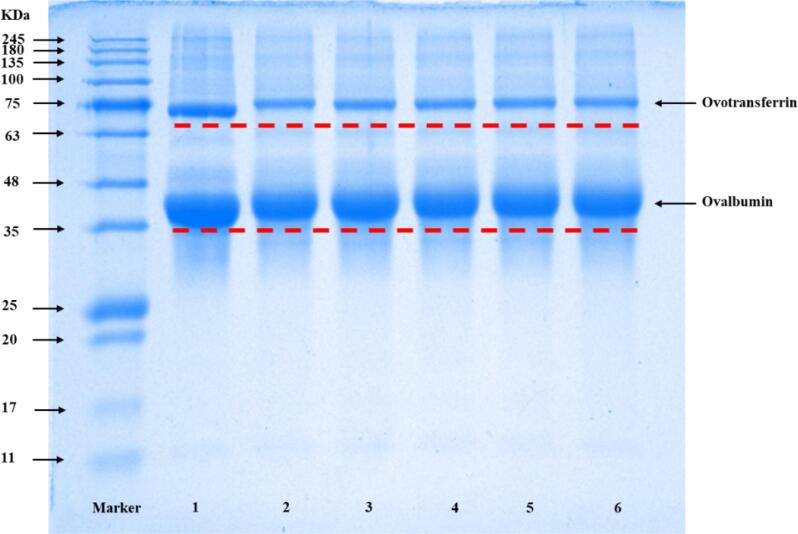
The molecular weight distribution of the native OVA and OVA-EGCG conjugates by conventional and ultrasound-assisted free radical methods was monitored by SDS-PAGE. As shown in Fig. 2, the native OVA (land 1) mainly had bands at around 75 and 45 kDa, which represented ovotransferrin and ovalbumin, respectively [18]. The OVA-EGCG conjugates obtained by traditional free radical and ultrasound-assisted free radical methods also exhibited the same bands at about 75 and 45 kDa, but the corresponding bands slightly shifted migration, especially in U-OVA-EGCG conjugates, indicating that ovotransferrin and ovalbumin might be involved in binding to EGCG. The SDS-PAGE loading buffer mainly contains SDS, β-mercaptoethanol, bromophenol blue, etc. SDS contributed to dissociating hydrogen-bonded structures with unfolding of ordered molecular conformation, while disulfide bonds could be broken by β-mercaptoethanol, which eliminated the differences between protein samples. Thus, these results indicated that the covalent bonds were formed in OVA-EGCG conjugates between OVA and EGCG. This upward migration trend of bands was consistent with the above results of the total phenol contents (Table 1), since ultrasonic treatment helped more proteins to be covalently modified by EGCG to form higher relative molecule weight conjugates.
Fig. 2.
The sodium dodecyl sulfate–polyacrylamide gel electrophoresis patterns of ovalbumin during glycosylation. 1: Native ovalbumin; 2: T-OVA-EGCG conjugates (ovalbumin-epigallocatechin-3-gallate conjugates prepared by traditional free radical treatment); 3–6: U-OVA-EGCG conjugates (the ultrasound-assisted free radical of OVA-EGCG conjugates prepared under different ultrasonic response times of 30 min, 60 min, 90 min, and 120 min, respectively).
3.4. FTIR spectroscopy analysis of the secondary structure changes in OVA-EGCG conjugates
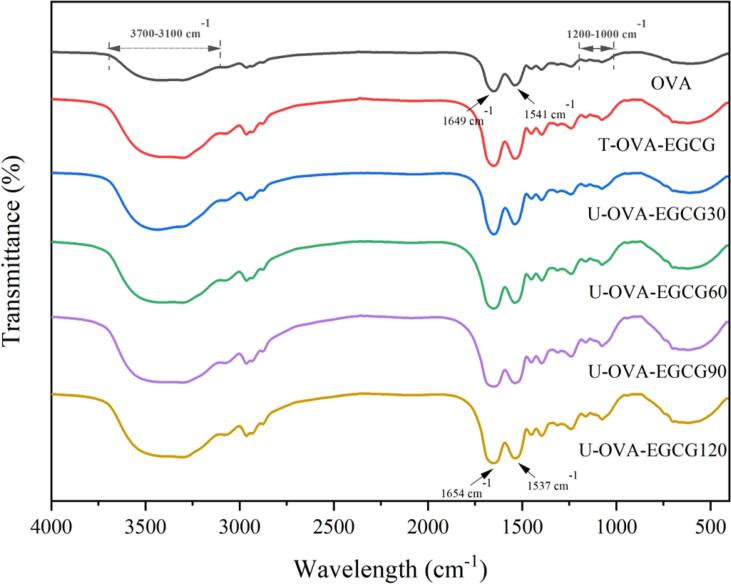
To further prove whether the covalent-linking was formed and to obtain information about the changes in protein conformation after traditional free radical or different ultrasonic time-assisted free radical methods, FTIR spectroscopy was investigated. Fig. 3 illustrated the FTIR spectra of native OVA, T-OVA-EGCG conjugate prepared by traditional free radical treatment, and U-OVA-EGCG conjugates prepared by different ultrasonic time-assisted free radical treatments. The characteristic absorption band locations and intensities of all OVA-EGCG conjugates were remarkably different from those of the untreated OVA, suggesting that the conjugated reaction of EGCG with OVA resulted in the rearrangement and conformation alterations of the protein. For instance, the absorption intensities of 3700–3100 cm−1, which corresponds to amide A and related to the stretching vibration of N–H and hydrogen band [3], [30], enhanced remarkably in all the OVA-EGCG conjugates, which might ascribe to the addition of abundant O-H bands from the EGCG molecular and the formation of hydrogen bonding of the OVA-EGCG conjugates when using free radicals or ultrasonic time-assisted free radical methods. Similarly, Yang et al. suggested the formation of covalent conjugation between pumpkin (Cucurbita sp.) seed protein isolate and pyrogallic acid (1,2,3-benzene-triol) polyphenol [37]. Moreover, the changes in the absorption peak at 1700–1600 cm−1 (amide I) and 1600–1500 cm−1(amide II) were attributed to C O stretching vibrations and N-H bending vibrations, respectively [38]. The introduction of EGCG caused a pronounced redshift from 1649 cm−1 to 1654 cm−1 in the amide I band, possibly because EGCG could be connected to the C O group of protein by hydrophobic interaction [20], [36]. Additionally, all absorption peaks of OVA-EGCG conjugates exhibited a blueshift in amide II from 1541 cm−1 to 1537 cm−1, which is possibly due to EGCG binding to the amino acid in OVA involvement in the C-N and N-H vibrations. These changes in the amide I and amide II bands indicated that electrostatic interaction occurred between OVA and EGCG [38]. Furthermore, the fingerprint region range from 1200 to 1000 cm−1 of OVA-EGCG conjugates changes significantly following the binding EGCG, indicating possible involvement in hydrogen bonding or other poor electron interactions, such as π–π and dipole interactions [3], [39].
Fig. 3.
The Fourier transform infrared spectroscopy of native OVA, T-OVA-EGCG conjugates, and U-OVA-EGCG conjugates. OVA: native ovalbumin; T-OVA-EGCG: ovalbumin-epigallocatechin-3-gallate conjugates prepared by traditional free radical treatment; U-OVA-EGCG30, U-OVA-EGCG60, U-OVA-EGCG90, and U-OVA-EGCG120: the ultrasound-assisted free radical of OVA-EGCG conjugates prepared under different ultrasonic response times of 30 min, 60 min, 90 min, and 120 min, respectively.
3.5. Spectroscopy analysis of the conformation and micro-environment changes in OVA-EGCG conjugates
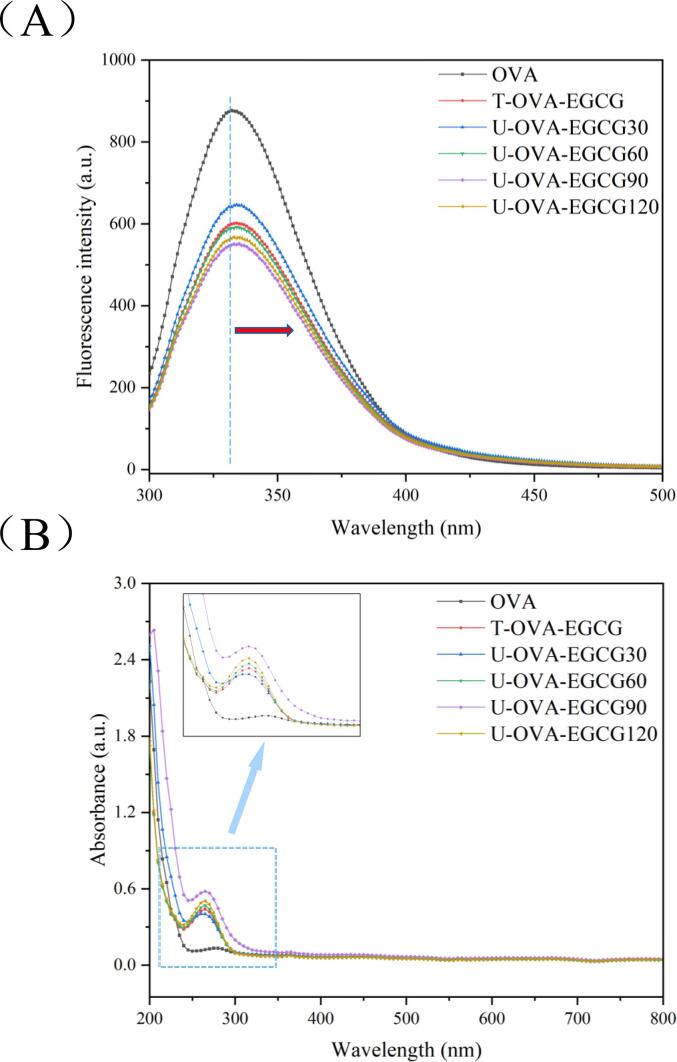
Since the chromophores of proteins were particularly sensitive to changes in the polarity of the microenvironment, the introduction of small molecules could change the microenvironment, leading to changes in the inherent fluorescence intensity of protein [31]. In addition, changes in fluorescence intensity could suggest the accessibility of small molecules to chromophores of proteins, which contributed to understanding the binding mechanism of proteins and ligands [22]. Thus, the intrinsic fluorescence emission spectrum for native OVA and its OVA-EGCG conjugates obtained by different free radical methods was employed. As exhibited in Fig. 4 A, the fluorescence intensity was higher for native OVA than all OVA-EGCG conjugates, with redshifts from 332 nm to 335 nm after grafting with EGCG using the traditional and ultrasound-assisted free radical methods. The results described above we consistent with previous studies as reported by Meng et al. [20] and He et al. [40]. For OVA-EGCG conjugates, the quenching in fluorescence intensity and the redshift in the maximum absorption wavelength suggested that the hydrophobic group (Trp, Tyr, and Phe resides) within the OVA possibly participated in the covalent binding. Moreover, the conjugation result in distinct changes of protein, exposing more chromosomal groups to a hydrophilic environment [40], [41]. Interestingly, the fluorescence intensity was negatively correlated with the total EGCG content of conjugates (Table 1). The fluorescence intensity of conjugates with ultrasound-assisted treatment decreased the most, suggesting that ultrasound could promote the interaction between OVA and EGCG.
Fig. 4.
The intrinsic fluorescence spectra (A) and UV spectra (B) of native OVA, T-OVA-EGCG conjugates, and U-OVA-EGCG conjugates. OVA: native ovalbumin; T-OVA-EGCG: ovalbumin-epigallocatechin-3-gallate conjugates prepared by traditional free radical treatment; U-OVA-EGCG30, U-OVA-EGCG60, U-OVA-EGCG90, and U-OVA-EGCG120: the ultrasound-assisted free radical of OVA-EGCG conjugates prepared under different ultrasonic response times of 30 min, 60 min, 90 min, and 120 min, respectively.
Changes in the intensity and location of the UV absorption spectra could also evaluate the changes in the conformation of proteins and the microenvironment of the hydrophobic amino acid residues caused by phenolic introduction [22], [31]. Fig. 4 B shows the UV absorption spectra for native OVA, T-OVA-EGCG conjugates, and U-OVA-EGCG conjugated induced at different ultrasonic response time-assisted free radical treatments. The maximum absorption peak wavelength (λmax) of native OVA was around 280 nm. However, all OVA-EGCG conjugates (T-OVA-EGCG conjugate and different U-OVA-EGCG conjugates) not only exhibited a significant enhancement in absorption intensity but also a remarkable blue-shift wavelength. In general, an increase in absorption strength represents the exposure of the hydrophobic groups (Trp and Tyr), while the blue shift in λmax represents an enhancement in the polarity of the microenvironment around hydrophobic groups [31]. In addition, compared with the OVA-EGCG conjugates by the conventional free radical method, the blue shift and intensity increasement caused by the ultrasonic-assisted free radical method was more obvious, suggesting increasing the conjugation between the OVA and EGCG. Moreover, extending the ultrasonic response time was conducive to exposing aromatic amino acids and thus made the microenvironment more hydrophilic, which was consistent with the results of the intrinsic fluorescence emission spectrum (Fig. 4A).
3.6. Molecular docking of OVA and EGCG
The combination of computer simulations and experimental measurements was conducive to understanding comprehensive detailed insight into the potential molecular forces between receptors and ligands involved in the binding interaction [42]. Thus, Autodock Vina software was applied to perform molecular docking of OVA and EGCG to locate the preferred binding region. Subsequently, two kinds of programs, the Pymol software and the Protein-Ligand Interaction Profiler (PLIP) webpage [43], analyzed the protein–ligand interaction to verify the experimental results. Previous studies have found that OVA possesses potential binding sites for flavonoids, such as glabridin [7], naringenin, genistein, naringin, puerarin, and daidzein [44]. In this study, Autodock Vina software was used to select the best-docked conformation of OVA and EGCG based on the lowest binding energy, and obtain the amino residues, the numbers of and distances of interaction force (hydron bonds, hydrophobic interactions, and π-Stacking, etc.) that interact with OVA and EGCG.
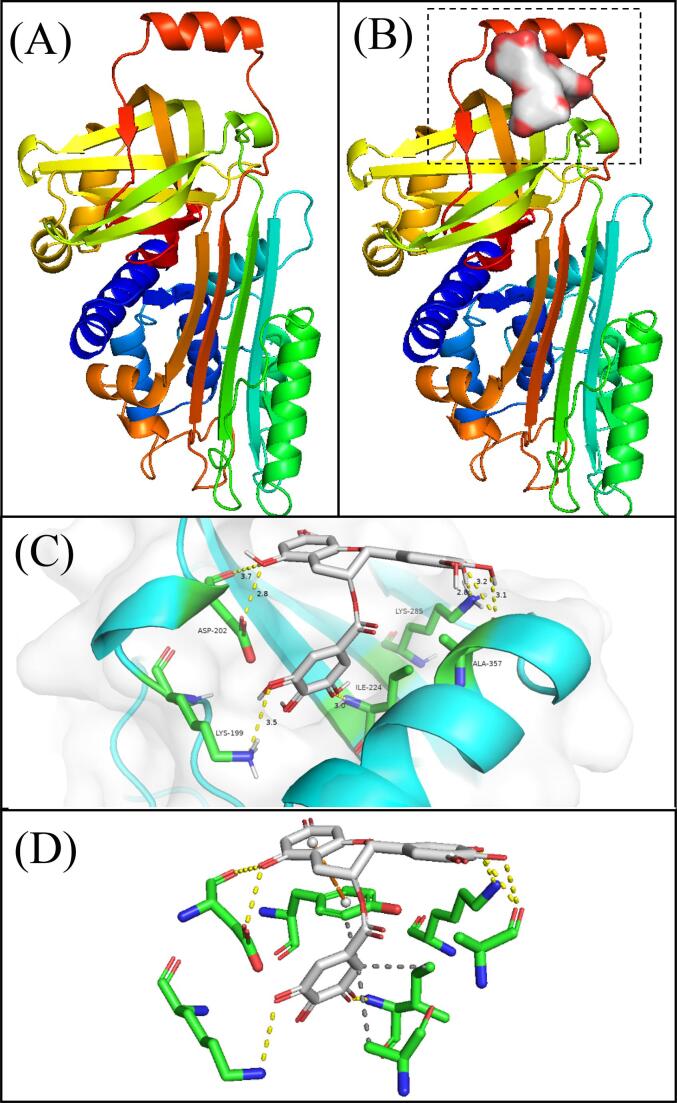
Fig. 5A shows the Three-dimensional structure of OVA collected from the RCSB Protein Data Bank database. Fig. 5B exhibited that the docked conjugate of EGCG and OVA with the lowest energy conformer (ΔG = −7.6 kcal/mol) obtained from Autodock Vina software, revealing that the EGCG may conjugate with the special region on the protein surface. The best-docked conformation of the EGCG binging site to OVA was identified between α-helix, β-sheets domain, and random coil (Fig. 5B). Thereinto, EGCG established hydrogen bond contacts with Lys-199, Asp-202, Ile-224, Lys-285, and Ala-357 amino acid residues with a distance range from 2.80 Å to 3.55 Å (Fig. 5C). This finding validates the experiment result of FTIR (Fig. 3), which indicated that hydroxyl groups on EGCG could form abundant hydrogen bonds with the surface of OVA. Moreover, the detailed molecular docking results including hydrogen bonding (yellow dotted lines), hydrophobic interaction (gray dotted lines), and π-stacking (orange dotted lines) were exhibited in Fig. 5D and summarized in Table 4. Furthermore, EGCG has been found to form hydrophobic interactions with Tyr-222, Ile-224, and Ala-353 amino acid residues of OVA (Fig. 5D and Table 3). This finding testifies to the experiment result of the intrinsic fluorescence emission spectrum, and UV absorption spectra (Fig. 4), which suggested that hydrophobic interactions may be involved in the formation of the OVA-EGCG conjugates.
Fig. 5.
Molecular docking analysis results of ovalbumin (OVA) and epigallocatechin-3-gallate (EGCG). (A) Three-dimensional structure of OVA collected from the RCSB Protein Data Bank database; (B) the best-docked position between OVA and EGCG; (C) the best-docked conformation for OVA and EGCG. The blue represents OVA, the green stick structure represents amino acid resides, the red-gray represents EGCG, and the yellow dotted line represents hydrogen-bonding. (D) Protein-ligand docking simulation for the determination of OVA-EGCG interaction using protein–ligand interaction profiler. Yellow dotted lines represent hydrogen bonding, gray dotted lines represent hydrophobic interaction, and orange dotted lines represent π stacking.
Table 4.
Antioxidant properties of T-OVA-EGCG and U-OVA-EGCG conjugates.
| Samples | DPPH scavenging capacity/% | ABTS scavenging capacity/% |
|---|---|---|
| OVA | 23.16 ± 2.90e | 17.16 ± 2.18e |
| T-OVA-EGCG | 68.15 ± 1.35c | 61.02 ± 1.83c |
| U-OVA-EGCG30 | 46.34 ± 3.0d | 30.08 ± 2.60d |
| U-OVA-EGCG60 | 68.61 ± 2.75c | 60.51 ± 1.61c |
| U-OVA-EGCG90 | 89.50 ± 3.31a | 78.01 ± 0.90a |
| U-OVA-EGCG120 | 75.38 ± 1.46b | 70.92 ± 1.83b |
Note: Dates are given as means ± standard deviations. Values with superscript letters in the same column indicate significantly different at P < 0.05. T-OVA-EGCG: ovalbumin-epigallocatechin-3-gallate conjugates prepared by traditional free radical treatment; U-OVA-EGCG30, U-OVA-EGCG60, U-OVA-EGCG90, and U-OVA-EGCG120: the ultrasound-assisted free radical of OVA-EGCG conjugates prepared under different ultrasonic response times of 30 min, 60 min, 90 min, and 120 min, respectively.
Table 3.
The interaction force between OVA and EGCG at the best-docked position.
| Index | Residue | AA | Distance D-H |
Distance D-A |
|
|---|---|---|---|---|---|
| Hydrogen Bonds | 1 | 199A | Lys | 2.60 | 3.55 |
| 2 | 202A | Asp | 2.20 | 2.84 | |
| 3 | 202A | Asp | 3.29 | 3.71 | |
| 4 | 224A | Ile | 2.11 | 2.96 | |
| 5 | 285A | Lys | 2.13 | 2.80 | |
| 6 | 357A | Ala | 2.36 | 3.17 | |
| 7 | 357A | Ala | 2.18 | 3.14 | |
| Hydrophobic interactions | 1 | 222A | Tyr | 3.28 | |
| 2 | 224A | Ile | 3.77 | ||
| 3 | 353A | Ala | 3.74 | ||
| π-Stacking | 1 | 222A | Tyr | 3.97 | |
3.7. Functional properties of OVA-EGCG conjugates
3.7.1. Emulsifying properties analysis of OVA-EGCG conjugates
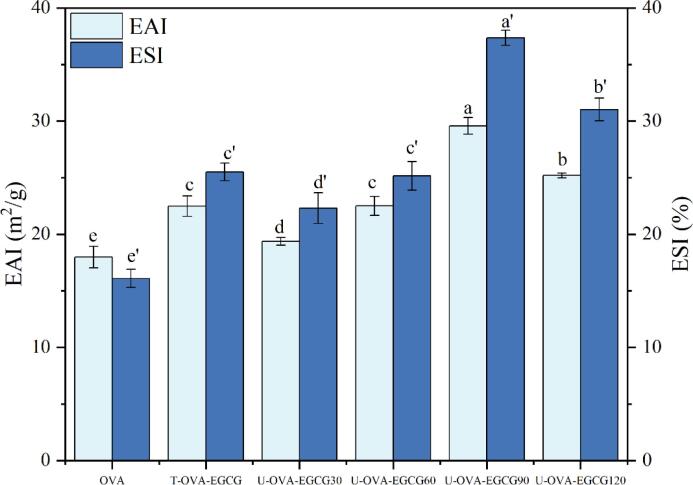
The emulsifying activity index (EAI) and emulsion stability index (ESI) are representative indexes for the assessment of emulsifying properties [8], [33]. The EAI and ESI results of native OVA, and OVA-EGCG conjugates by conventional and ultrasound-assisted free radical methods were exhibited in Fig. 6. It was worth noting that all OVA-EGCG conjugates by the introduction of EGCG were remarkably greater than those of native OVA. The EAI and ESI values of OVA-EGCG90 conjugates were 1.64 times and 2.32 times that of untreated OVA, respectively. These findings suggested that the conjugation with polyphenols could enhance emulsifying properties of OVA. Similarly, Han et al. found that whey protein isolate-EGCG conjugates induced by the covalent and non-covalent combination had higher emulsifying properties including the EAI and ESI than untreated whey protein isolate [45]. The improvement of emulsifying properties may be due to the exposure aromatic resides and the reduction of the surface hydrophobicity of the protein by binding to polyphenols [19], [46], as described in the intrinsic fluorescence emission spectrum measurements of Fig. 4 A, thus improving the affinity of the protein to the O/W interface. Moreover, the adsorption capacity of protein on the O/W interface was susceptible to the change in protein conformation [45]. After conjugation with polyphenols, the molecular flexibility of OVA was enhanced, resulting in improved emulsification properties. Interestingly, compared with T-OVA-EGCG conjugate treated by traditional free radical treatment, the emulsifying properties of U-OVA-EGCG conjugates prepared by ultrasound-assisted free radical treatment were better, which might account for cavitation and mechanical effects caused by ultrasound to accelerate the conjugation and reduce the particle size.
Fig. 6.
Changes in the emulsifying activity index (EAI) and emulsion stability index (ESI) of native OVA, T-OVA-EGCG conjugates, and U-OVA-EGCG conjugates. OVA: native ovalbumin; T-OVA-EGCG: ovalbumin-epigallocatechin-3-gallate conjugates prepared by traditional free radical treatment; U-OVA-EGCG30, U-OVA-EGCG60, U-OVA-EGCG90, and U-OVA-EGCG120: the ultrasound-assisted free radical of OVA-EGCG conjugates prepared under different ultrasonic response times of 30 min, 60 min, 90 min, and 120 min, respectively. Values are given as the means ± standard deviation. Bars denoted with different lowercase letters are significantly different (P < 0.05).
3.7.2. Antioxidant properties of OVA-EGCG conjugates
The antioxidant properties of nature OVA, T-OVA-EGCG conjugate induced by traditional free radical treatment, and U-OVA-EGCG conjugates induced by different ultrasonic time-assisted free radical treatments were assessed using the DPPH scavenging capacity and ABTS scavenging capacity. As summarized in Table 4, native OVA also had a little capacity for radical scavenging, owing to amino acid residues like Try, Tyr, and Met [3], [47]. For all OVA-EGCG conjugates, the antioxidant capacity was remarkably higher (P < 0.05) than that of OVA alone, especially those prepared by ultrasound-assisted free radical treatments. The result indicated that the antioxidant capacity of OVA could be enhanced significantly by conjugating with EGCG. Because of the six ortho-phenolic hydroxyl groups in the structure of EGCG, it has excellent antioxidant activity, which could scavenge free radicals and metal ion chelating and delay the production of reactive oxygen species [20], [48].Thus, the introduction of EGCG enhanced the antioxidant ability of the system. Similarly, Chen et al. [23] and Tong et al. [41] have also reported that the binding of protein to EGCG enhanced the antioxidant capacity of protein. In addition, the disparity in the antioxidant capacity of different samples could be ascribed to the different amounts of EGCG attached to OVA molecules. This assumption was evidenced by the measurement of levels of EGCG content (Table 1). Noteworthy, the topmost antioxidant capacity (DPPH:89.50 ± 3.31 % and ABTS:78.01 ± 0.90 %) was collected in the U-OVA-EGCG90 conjugate, which may have happened because the ultrasound-assisted induced modified protein exhibited superior affinity with EGCG [20]. Moreover, cavitation and mechanical effects generated during ultrasound facilitated the partial unfolding of protein and exposure of more aromatic amino acids, which contribute to the enhancement of antioxidant capacity [28], [30].
4. Conclusions
In conclusion, the OVA-EGCG conjugates were successfully prepared by traditional free radical treatment and different ultrasonic time-assisted free radical treatments. Compared with traditional free radical treatment, the ultrasound-assisted greatly reduced the response time from 24 h to 60 min without affecting the equivalent amount of EGCG conjugating. Moreover, the content of EGCG increased and free sulfhydryl groups decreased with the prolonged ultrasonic time. The UV and intrinsic fluorescence spectroscopy analysis suggested that ultrasound was conducive to exposing aromatic amino acids and accelerating the interaction between OVA and EGCG. The molecular docking testified that hydrogen bonding, hydrophobic interaction, and π-stacking may be involved in the formation of the OVA-EGCG conjugates. Additionally, the enhancement in the emulsifying properties (EAI and ESI) and antioxidant properties (DPPH and ABTS) of OVA-EGCG conjugates certified the ultrasound and the addition of EGCG contributed to functional properties. According to the above results, it was hypothesized that OVA-EGCG conjugates could be applicable as emulsifiers and antioxidants to broaden the value and application of OVA in functional foods.
CRediT authorship contribution statement
Yimei Zheng: Conceptualization, Methodology, Investigation, Writing – original draft. Boyu Chen: Investigation, Validation. Xuanxiang Huang: Investigation, Validation. Hui Teng: Supervision, Writing – review & editing, Project administration, Funding acquisition. Chao Ai: Supervision, Writing – review & editing. Lei Chen: Conceptualization, Project administration, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work is supported by the National Natural Science Foundation of China (NSFC, Grant No. 32272315, 32072209, 32061160477), the Natural Science Foundation of Guangdong Province (2022A1515010694), the Innovative Team Program of High Education of Guangdong Province (2021KCXTD021).
Contributor Information
Hui Teng, Email: tenghui850610@126.com.
Chao Ai, Email: acworke_mail@163.com.
Lei Chen, Email: chenlei841114@hotmail.com.
References
- 1.Liu C., Lv N., Xu Y.Q., Tong H., Sun Y., Huang M., Ren G., Shen Q., Wu R., Wang B., Cao Z., Xie H. pH-dependent interaction mechanisms between beta-lactoglobulin and EGCG: Insights from multi-spectroscopy and molecular dynamics simulation methods. Food Hydrocoll. 2022;133 doi: 10.1016/j.foodhyd.2022.108022. [DOI] [Google Scholar]
- 2.Buitimea-Cantua N.E., Gutierrez-Uribe J.A., Serna-Saldivar S.O. Phenolic-protein interactions: effects on food properties and health benefits. J. Med. Food. 2018;21:188–198. doi: 10.1089/jmf.2017.0057. [DOI] [PubMed] [Google Scholar]
- 3.Razzak M.A., Cho S.J. Molecular characterization of capsaicin binding interactions with ovalbumin and casein. Food Hydrocoll. 2022;133 doi: 10.1016/j.foodhyd.2022.107991. [DOI] [Google Scholar]
- 4.Kovacs-Nolan J., Phillips M., Mine Y. Advances in the value of eggs and egg components for human health. J. Agric. Food Chem. 2005;53:8421–8431. doi: 10.1021/jf050964f. [DOI] [PubMed] [Google Scholar]
- 5.Jalili-Firoozinezhad S., Filippi M., Mohabatpour F., Letourneur D., Scherberich A. Chicken egg white: hatching of a new old biomaterial. Mater. Today. 2020;40:193–214. doi: 10.1016/j.mattod.2020.05.022. [DOI] [Google Scholar]
- 6.Ai M., Xiao N., Jiang A. Molecular structural modification of duck egg white protein conjugates with monosaccharides for improving emulsifying capacity. Food Hydrocoll. 2021;111 doi: 10.1016/j.foodhyd.2020.106271. [DOI] [Google Scholar]
- 7.Razzak M.A., Choi S.S. Delineating the interaction mechanism of glabridin and ovalbumin by spectroscopic and molecular docking techniques. Food Chem. 2021;347:128981. doi: 10.1016/j.foodchem.2020.128981. [DOI] [PubMed] [Google Scholar]
- 8.Zheng Y., Chang Y., Luo B., Teng H., Chen L. Molecular structure modification of ovalbumin through controlled glycosylation with dextran for its emulsibility improvement. Int. J. Biol. Macromol. 2022;194:1–8. doi: 10.1016/j.ijbiomac.2021.11.130. [DOI] [PubMed] [Google Scholar]
- 9.Teng H., Zheng Y., Cao H., Huang Q., Xiao J., Chen L. Enhancement of bioavailability and bioactivity of diet-derived flavonoids by application of nanotechnology: A review. Crit. Rev. Food Sci. Nutr. 2023;63(3):378–393. doi: 10.1080/10408398.2021.1947772. [DOI] [PubMed] [Google Scholar]
- 10.Sun X., Sarteshnizi R.A., Udenigwe C.C. Recent advances in protein–polyphenol interactions focusing on structural properties related to antioxidant activities. Curr. Opin. Food Sci. 2022;45 doi: 10.1016/j.cofs.2022.100840. [DOI] [Google Scholar]
- 11.Zhang C., He Y., Zheng Y., Ai C., Cao H., Xiao J., El-Seedi H., Chen L., Teng H. Effect of carboxymethyl cellulose (CMC) on some physico-chemical and mechanical properties of unrinsed surimi gels. LWT. 2023;180 doi: 10.1016/j.lwt.2023.114653. [DOI] [Google Scholar]
- 12.Cao H., Saroglu O., Karadag A., Diaconeasa Z., Zoccatelli G., Conte‐Junior C.A., Gonzalez‐Aguilar G.A., Ou J., Bai W., Zamarioli C.M., Freitas L.A.P., Shpigelman A., Campelo P.H., Capanoglu E., Hii C.L., Jafari S.M., Qi Y., Liao P., Wang M., Zou L., Bourke P., Simal‐Gandara J., Xiao J. Available technologies on improving the stability of polyphenols in food processing. Food Front. 2021;2(2):109–139. [Google Scholar]
- 13.Wang H., You S., Wang W., Zeng Y., Su R., Qi W., Wang K., He Z. Laccase-catalyzed soy protein and gallic acid complexation: Effects on conformational structures and antioxidant activity. Food Chem. 2022;375 doi: 10.1016/j.foodchem.2021.131865. [DOI] [PubMed] [Google Scholar]
- 14.Feng J., Cai H., Wang H., Li C., Liu S. Improved oxidative stability of fish oil emulsion by grafted ovalbumin-catechin conjugates. Food Chem. 2018;241:60–69. doi: 10.1016/j.foodchem.2017.08.055. [DOI] [PubMed] [Google Scholar]
- 15.Guo Y., Bao Y.H., Sun K.F., Chang C., Liu W.F. Effects of covalent interactions and gel characteristics on soy protein-tannic acid conjugates prepared under alkaline conditions. Food Hydrocoll. 2021;112 doi: 10.1016/j.foodhyd.2020.106293. [DOI] [Google Scholar]
- 16.Liu F., Ma C., Gao Y., McClements D.J. Food-grade covalent complexes and their application as nutraceutical delivery systems: a review. Compr. Rev. Food Sci. Food Saf. 2017;16:76–95. doi: 10.1111/1541-4337.12229. [DOI] [PubMed] [Google Scholar]
- 17.Jing H., Sun J., Mu Y., Obadi M., McClements D.J., Xu B. Sonochemical effects on the structure and antioxidant activity of egg white protein–tea polyphenol conjugates. Food Funct. 2020;11:7084–7094. doi: 10.1039/D0FO01636E. [DOI] [PubMed] [Google Scholar]
- 18.Sun J., Zhang F., Liu T., Jing H., Huang Y., Obadi M., Xu B. Ultrasound-enhanced egg white proteins conjugated with polyphenols: The structure of the polyphenols on their functional properties. LWT-Food Sci. Technol. 2022;164 doi: 10.1016/j.lwt.2022.113600. [DOI] [Google Scholar]
- 19.Meng Y., Li C. Conformational changes and functional properties of whey protein isolate-polyphenol complexes formed by non-covalent interaction. Food Chem. 2021;364 doi: 10.1016/j.foodchem.2021.129622. [DOI] [PubMed] [Google Scholar]
- 20.Geng M., Feng X., Yang H., Wu X., Li L., Li Y., Teng F. Comparison of soy protein isolate-(-)-epigallocatechin gallate complexes prepared by mixing, chemical polymerization, and ultrasound treatment. Ultrason. Sonochem. 2022;90 doi: 10.1016/j.ultsonch.2022.106172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu T., Lin L., Zhang X., Wang X., Ding J. Covalent modification of soy protein hydrolysates by EGCG: Improves the emulsifying and antioxidant properties. Food Res. Int. 2023;164 doi: 10.1016/j.foodres.2022.112317. [DOI] [PubMed] [Google Scholar]
- 22.Han S., Cui F., McClements D.J., Xu X., Ma C., Wang Y., Liu X., Liu F. Structural characterization and evaluation of interfacial properties of pea protein isolate-EGCG molecular complexes. Foods. 2022;11:2895. doi: 10.3390/foods11182895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J., Zhang X., Chen X., Bassey A.P., Zhou G., Xu X. Phenolic modification of myofibrillar protein enhanced by ultrasound: The structure of phenol matters. Food Chem. 2022;386 doi: 10.1016/j.foodchem.2022.132662. [DOI] [PubMed] [Google Scholar]
- 24.Cao G., Chen X., Hu B., Yang Z., Wang M., Song S., Wang L., Wen C. Effect of ultrasound-assisted resting on the quality of surimi-wheat dough and noodles. Ultrason. Sonochem. 2023;94 doi: 10.1016/j.ultsonch.2023.106322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J., Zhang X., Fu M., Chen X., Pius B.A., Xu X. Ultrasound-assisted covalent reaction of myofibrillar protein: The improvement of functional properties and its potential mechanism. Ultrason. Sonochem. 2021;76 doi: 10.1016/j.ultsonch.2021.105652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song Y., Lu Y., Bi X., Chen L., Liu L., Che Z. Inactivation of staphylococcus aureus by the combined treatments of ultrasound and nisin in nutrient broth and milk. eFood. 2021;2(3):140–146. [Google Scholar]
- 27.Srivastava S., Pandey V.K., Singh P., Bhagya Raj G.V.S., Dash K.K., Singh R. Effects of microwave, ultrasound, and various treatments on the reduction of antinutritional factors in elephant foot yam: A review. eFood. 2022;3:e40. [Google Scholar]
- 28.Nwachukwu I.D., Aluko R.E. Structural and functional properties of food protein-derived antioxidant peptides. J. Food Biochem. 2019;43(1) doi: 10.1111/jfbc.12761. [DOI] [PubMed] [Google Scholar]
- 29.Tang S.Q., Du Q.H., Fu Z. Ultrasonic treatment on physicochemical properties of water-soluble protein from moringa oleifera seed. Ultrason. Sonochem. 2021;71 doi: 10.1016/j.ultsonch.2020.105357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang S., Li X., Yan X., McClements D.J., Ma C., Liu X., Liu F. Ultrasound-assisted preparation of lactoferrin-EGCG conjugates and their application in forming and stabilizing algae oil emulsions. Ultrason. Sonochem. 2022;89 doi: 10.1016/j.ultsonch.2022.106110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y., Ma M. Foam and conformational changes of egg white as affected by ultrasonic pretreatment and phenolic binding at neutral pH. Food Hydrocoll. 2020;102 doi: 10.1016/j.foodhyd.2019.105568. [DOI] [Google Scholar]
- 32.Luo X., Lu J., Wu Y., Duan W., An F., Huang Q., Chen L., Wei S. Reducing the potential allergenicity of amandin through binding to (−)-epigallocatechin gallate. Food Chemistry: X. 2022;16 doi: 10.1016/j.fochx.2022.100482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li R., Wang X., Liu J., Cui Q., Wang X., Chen S., Jiang L. Relationship between molecular flexibility and emulsifying properties of soy protein isolate-glucose conjugates. J. Agric. Food Chem. 2019;67:4089–4097. doi: 10.1021/acs.jafc.8b06713. [DOI] [PubMed] [Google Scholar]
- 34.Gogate P.R. Cavitational reactors for process intensification of chemical processing applications: a critical review. Chem. Eng. Process.- Process Intensification. 2008;47:515–527. doi: 10.1016/j.cep.2007.09.014. [DOI] [Google Scholar]
- 35.Hu H., Wu J., Li-Chan E.C.Y., Zhu L., Zhang F., Xu X., Fan G., Wang L., Huang X., Pan S. Effects of ultrasound on structural and physical properties of soy protein isolate (SPI) dispersions. Food Hydrocoll. 2013;30:647–655. doi: 10.1016/j.foodhyd.2012.08.001. [DOI] [Google Scholar]
- 36.Sun J., Huang Y., Liu T., Jing H., Zhang F., Obadi M., Xu B. Evaluation of crossing-linking sites of egg white protein-polyphenol conjugates: Fabricated using a conventional and ultrasound-assisted free radical technique. Food Chem. 2022;386 doi: 10.1016/j.foodchem.2022.132606. [DOI] [PubMed] [Google Scholar]
- 37.Yang C., Wang B., Wang J., Xia S., Wu Y. Effect of pyrogallic acid (1,2,3-benzenetriol) polyphenol-protein covalent conjugation reaction degree on structure and antioxidant properties of pumpkin (Cucurbita sp.) seed protein isolate, LWT-Food. Sci. Technol. 2019;109:443–449. doi: 10.1016/j.lwt.2019.04.034. [DOI] [Google Scholar]
- 38.Sun F., Li B., Guo Y., Wang Y., Cheng T., Yang Q., Liu J., Fan Z., Guo Z., Wang Z. Effects of ultrasonic pretreatment of soybean protein isolate on the binding efficiency, structural changes, and bioavailability of a protein-luteolin nanodelivery system. Ultrason. Sonochem. 2022;88 doi: 10.1016/j.ultsonch.2022.106075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Freitas G.B.L., De Almeida D.J., Carraro E., Kerppers I.I., Martins G.A.G., Mainardes R.M., Khalil N.M., Messias-reason I.J.T. formulation, characterization, and in vitro/in vivo studies of capsaicin-loaded albumin nanoparticles. Mater. Sci. Eng.: C-Mater. Biol. Appl. 2018;93:70–79. doi: 10.1016/j.msec.2018.07.064. [DOI] [PubMed] [Google Scholar]
- 40.He W., Xu H., Lu Y., Zhang T., Li S., Lin X., Xu B., Wu X. Function, digestibility and allergenicity assessment of ovalbumin-EGCG conjugates. J. Funct. Foods. 2019;61 doi: 10.1016/j.jff.2019.103490. [DOI] [Google Scholar]
- 41.Tong X., Cao J., Tian T., Lyu B., Miao L., Lian Z., Cui W., Liu S., Wang H., Jiang L. Changes in structure, rheological property and antioxidant activity of soy protein isolate fibrils by ultrasound pretreatment and EGCG. Food Hydrocoll. 2022;122 doi: 10.1016/j.foodhyd.2021.107084. [DOI] [Google Scholar]
- 42.Barroso da Silva F.L., Carloni P., Cheung D., Cottone G., Donnini S., Foegeding E.A., Gulzar M., Jacquier J.C., Lobaskin V., MacKernan D., Mohammad Hosseini Naveh Z., Radhakrishnan R., Santiso E.E. Understanding and controlling food protein structure and function in foods: perspectives from experiments and computer simulations. Annu. Rev. Food Sci. Technol. 2020;11(1):365–387. doi: 10.1146/annurev-food-032519-051640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adasme M.F., Linnemann K.L., Bolz S.N., Kaiser F., Salentin S., Haupt V.J., Schroeder M. PLIP 2021: Expanding the scope of the protein–ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021;49(2021):W530–W534. doi: 10.1093/nar/gkab294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong Y., Yang L., Dai T., Zhu Z., Chen H., Wu J., Gong E.S. Flavonoids enhance gel strength of ovalbumin: Properties, structures, and interactions. Food Chem. 2022;387 doi: 10.1016/j.foodchem.2022.132892. [DOI] [PubMed] [Google Scholar]
- 45.Han X., Liang Z., Tian S., Liu L., Wang S. Epigallocatechin gallate (EGCG) modification of structural and functional properties of whey protein isolate. Food Res. Int. 2022;158 doi: 10.1016/j.foodres.2022.111534. [DOI] [PubMed] [Google Scholar]
- 46.Afizah M.N., Rizvi S.S.H. Functional properties of whey protein concentrate texturized at acidic pH: Effect of extrusion temperature. LWT-Food Sci. Technol. 2014;57:290–298. doi: 10.1016/j.lwt.2014.01.019. [DOI] [Google Scholar]
- 47.Liu Y., Ying D., Cai Y., Le X. Improved antioxidant activity and physicochemical properties of curcumin by adding ovalbumin and its structural characterization. Food Hydrocoll. 2017;72:304–311. doi: 10.1016/j.foodhyd.2017.06.007. [DOI] [Google Scholar]
- 48.Gu L., Peng N., Chang C., McClements D.J., Su Y., Yang Y. Fabrication of surface-active antioxidant food biopolymers: Conjugation of catechin polymers to egg white proteins. Food Biophys. 2017;12:198–210. doi: 10.1007/s11483-017-9476-5. [DOI] [Google Scholar]