SUMMARY
It is widely believed that hematopoiesis after birth is established by hematopoietic stem cells (HSCs) in the bone marrow and that HSC-independent hematopoiesis is limited only to primitive erythro-myeloid cells and tissue-resident innate immune cells arising in the embryo. Here, surprisingly, we find that significant percentages of lymphocytes are not derived from HSCs, even in 1-year-old mice. Instead, multiple waves of hematopoiesis occur from embryonic day 7.5 (E7.5) to E11.5 endothelial cells, which simultaneously produce HSCs and lymphoid progenitors that constitute many layers of adaptive T and B lymphocytes in adult mice. Additionally, HSC lineage tracing reveals that the contribution of fetal liver HSCs to peritoneal B-1a cells is minimal and that the majority of B-1a cells are HSC independent. Our discovery of extensive HSC-independent lymphocytes in adult mice attests to the complex blood developmental dynamics spanning the embryo-to-adult transition and challenges the paradigm of HSCs exclusively underpinning the postnatal immune system.
Graphical abstract
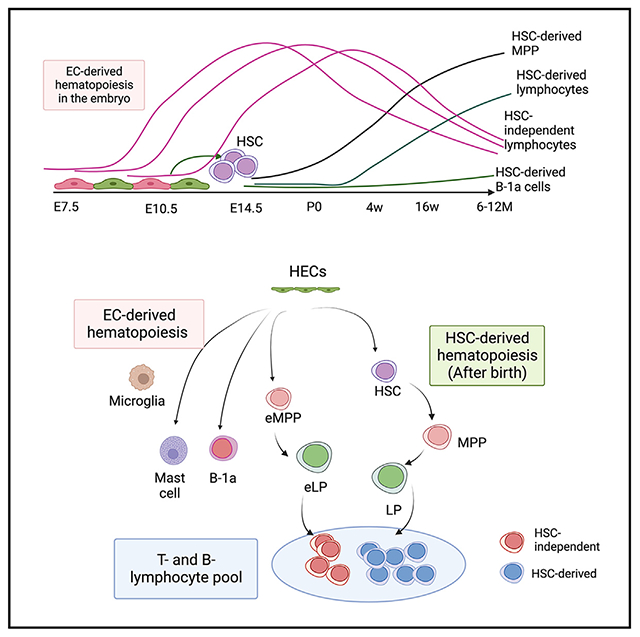
In brief
Kobayashi et al. show that multiple waves of HSC-independent lymphopoiesis occur in the embryo, which persist into adults and are not fully replaced by HSC-derived lymphocytes even in 12-month-old mice. The presence of HSC-independent adaptive lymphocytes in adult mice challenges the stem cell theory in the hematology field.
INTRODUCTION
All blood cells are derived from specialized endothelial cells (ECs), referred to as hemogenic ECs (HECs) in the extraembryonic yolk sac (YS) and aorta-gonad-mesonephros (AGM) region of the mouse embryo during a narrow time window.1–5 Before the emergence of hematopoietic stem cells (HSCs) from HECs in the AGM regions at embryonic day 11 (E11), multiple waves of blood cell production occur directly from HECs and contribute to transient erythro-myelopoiesis.4 During this time, in vitro T and B lymphoid and multipotent progenitor (MPP) potentials of HECs in the early YS and embryo have been reported.6–12 However, the physiological presence of HSC-independent MPPs and lymphoid progenitors (LPs) has yet to be unequivocally determined. Although a few B progenitors have been found in the HSC-deficient fetal liver (FL), whether they persist into adult life remains unknown because of the neonatal lethality of the knockout animals.13,14 Therefore, even if such HSC-independent MPPs and LPs exist, how long and to what extent they persist into postnatal life remains elusive. On the other hand, it is not well known when HSCs initiate and dominate hematopoiesis (e.g., differentiate into various blood lineages). Because very few HSCs are produced in the AGM region at E10–E11, HSCs need to expand in number in the FL and placenta to maintain hematopoiesis throughout life.15–17 Recently, contrasting results have been reported in mice and fish regarding HSCs’ contribution to hematopoiesis in the embryo.18,19 HSC lineage tracing studies in adult mice have shown their slow contribution to lymphoid cells.20–22 In vivo barcoding studies have demonstrated that MPPs, not HSCs, are the major providers of blood cells in adult mice.23,24 These studies raised the question of whether HSCs are the sole drivers of hematopoiesis, particularly lymphopoiesis. For instance, whether HSCs in the FL are the main providers of peritoneal innate-like B-1a cells is still controversial.
B lymphocytes are mainly categorized into three subsets: bone marrow (BM)-derived B-2 cells (e.g., splenic follicular [FO] B cells), marginal zone (MZ) B cells, and innate-like B-1 cells, which reside mainly in the body cavities. CD5+ B-1a cells are not replenished by BM HSCs and have generally been considered to be derived from FL HSCs,25–27 whereas other reports have shown that FL MPPs, not HSCs, repopulated B-1a cells more efficiently.28,29
In this study, we asked when HSCs initiate lymphopoiesis to generate T, B-2, and B-1a cells using HSC and EC lineage tracing mouse models. We found that fetal and neonatal HSCs did not contribute to lymphopoiesis until 4 weeks after birth. BM HSCs gradually produced lymphocytes but did not completely replace HSC-independent T and B lymphocytes even at 1 year of age. Interestingly, fetus-derived MPPs were promptly replaced by HSC-derived MPPs but minimally persisted until 1 year of age. For B-1a cells, the HSC lineage tracing assays showed that fetal and neonatal HSCs did not contribute to the peritoneal B-1a cell pool and that HSC-independent HEC-derived B-1a cells were maintained throughout life. Transplantation assays of E11.5 HSC precursors without co-culture demonstrated the presence of transplantable MPPs and B-1a progenitors, in addition to HSCs, in the AGM region. Our study provides definitive evidence of a prolonged presence of HSC-independent lymphocytes in adult mice and identifies the major source of B-1a cells in the mouse embryo.
RESULTS
Fetal and postnatal HSCs do not fully contribute to lymphopoiesis
Fgd5 is expressed exclusively in long-term HSCs (LT-HSCs; lin−Sca-1+c-kit+ [LSK] CD150+CD48− cells) in the FL and BM. Fgd5CreERT2:Rosa-TdTomato (iFgd5) mice enable us to label HSCs at the time of tamoxifen (TAM) injection.20,30 To examine the contribution of perinatal HSCs to hematopoiesis in post-natal life, we labeled HSCs in post-natal day 2 (P2) BM by a single TAM injection and traced Tomato+ cells in various hematopoietic subsets at different time points after birth (Figures 1A and S1). There were variations of Tomato labeling efficiency among animals; therefore, to be accurate despite variations in labeling efficiency in a consistent manner, we calculated the Tomato ratio of each blood cell type to LT-HSCs (Tomato ratio = Tomato% of a defined cell type/Tomato% of LT-HSCs) and normalized the variation as described previously.20 If a cell population is HSC derived, then the Tomato ratio will converge to 1.0 over time.20 When we looked at the Tomato ratio of spleen FO B cells, CD4+, and CD8+ T cells 4 weeks after birth, the Tomato ratios of these lymphoid cell populations were extremely low (close to zero) (Figure 1B), while the Tomato ratios of MPP was around 0.5. The Tomato ratio of splenic macrophages was as low as 0.3, indicating that HSC-derived hematopoiesis had just started and had not yet provided mature blood cells. At 8–12 weeks of age, the Tomato ratios of BM MPPs and spleen macrophages became around 0.75, while spleen FO, CD4+, and CD8+ T cells showed around 0.3. When mice were examined at 6–12 months of age, the Tomato ratios of MPPs and spleen myeloid cells were around 0.8, while spleen B and T cells were still around 0.5, indicating that HSC-derived lymphocytes did not replace all spleen lymphocytes even in 12-month-old mice. To eliminate the possibility that the variations in labeling efficiency across experiments may cause Tomato ratios to deviate from 1, we generated “fate-mapping scatterplots” that depicted the relationship between Tomato % measured in HSCs and each blood subset (Figures 1C–1E). These plots showed that Tomato% in HSCs exhibited some variations due to differences in labeling efficiency across experiments. Nevertheless, the Tomato% of many blood subsets was highly correlated with the Tomato% in HSCs, consistent with the derivations of these cell types from HSCs. Also, even though HSC labeling showed some variations, the labeling efficiency was high; the absolute Tomato% of HSCs was 70.4% ± 4.3% within 4 weeks (n = 3) and 77.6% ± 16.5% after 8 weeks (n = 15). In the case of MPPs and spleen macrophages, strong correlations were observed after 8 weeks, as shown by closed red circles in Figure 1C (R-squared [R2] = 0.98 and 0.98, slope = 0.81 and 0.76, respectively). Spleen FO B cells and CD4 and CD8 T cells also exhibited a strong correlation with HSCs after 6 months of age (R2 = 0.95, 0.95, and 0.94, respectively) (shown by closed circles in Figure 1D). Before 4 months of age, these B and T cells showed some correlation with HSCs (R2 = 0.74, 0.68, and 0.70, respectively; slope = 0.23, 0.23, and 0.21, respectively) (shown by open circles in Figure 1D). When the Tomato% of HSCs was approximately 100%, these lymphoid subsets showed around 60% Tomato+, also indicating that P2 HSCs were not the sole source of mature lymphoid cells even at 1 year of age (Figure 1D). Importantly, the Tomato% of B-1a cells and mast cells remained low irrespective of the high Tomato% of HSCs and the time of analysis (Figure 1E), which suggested that B1-a and mast cells are HSC independent after birth.
Figure 1. HSCs start lymphopoiesis in mice after 4 weeks of age and do not completely replace fetus-derived lymphocytes.
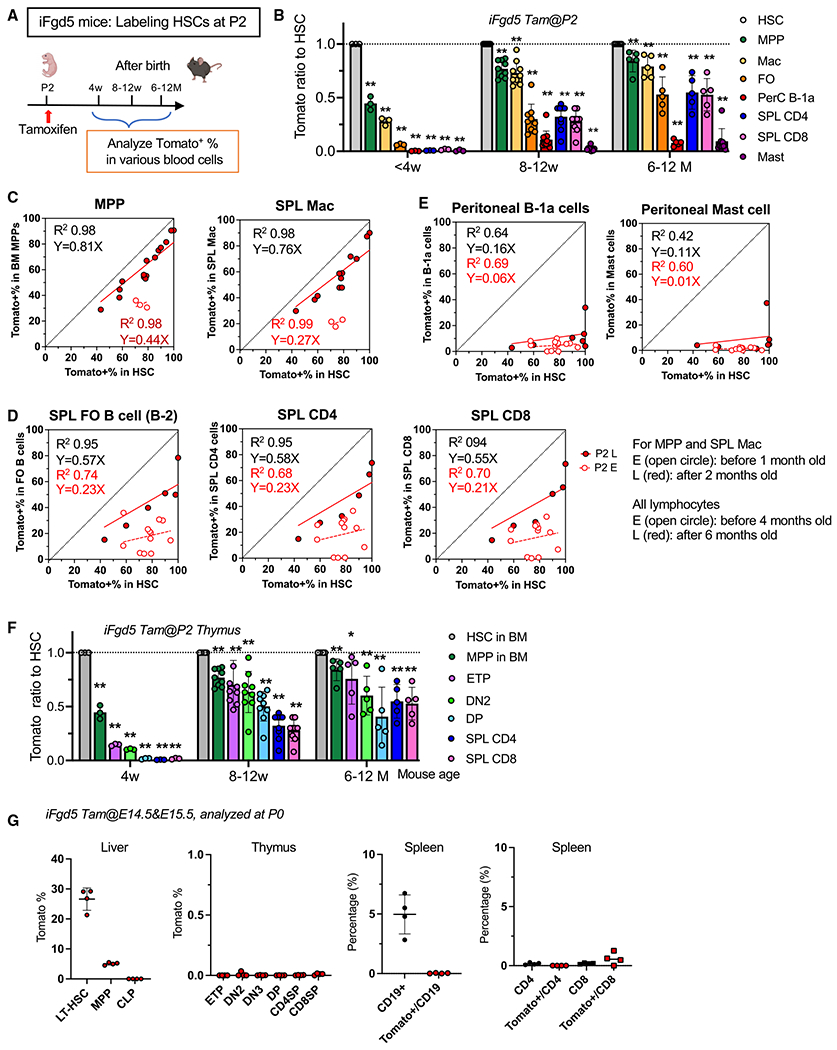
(A) Experimental design. TAM was injected once on P2 into iFgd5 mice to label HSCs. Tomato+ blood cells in the BM, spleen, and peritoneal cavity were examined at different time points.
(B) The Tomato ratio of each blood cell subset to HSCs (<4 weeks, n = 3; 8–12 weeks, n = 9; >6 months, n = 5–6).
(C–E) “Fate mapping scatterplots” showing the correlation between Tomato% in HSCs (x axis) and Tomato% of a target cell population (y axis). The equation and R2 values were calculated using Prism.
(C) Scatterplots of MPP (n = 18) and spleen macrophages (n = 15). A strong correlation was seen as early as 8 weeks after birth (late, red dots). Blue dots represent correlation when analyzed before 4 weeks of age.
(D) Scatterplots of spleen FO (n = 18), CD4 (n = 18), and CD8 T cells (n = 18). Blue dots, less than 4 months; red dots, more than 6 months.
(E) Scatterplots of peritoneal B-1a cells (n = 18) and mast cells (n = 17). Blue dots, less than 4 months; red dots, more than 6 months.
(F) The Tomato ratio of T progenitor populations in the thymus at 4 weeks (n = 4), 8–12 weeks (n = 9), and 5–12 months (n = 5) after birth.
(G) Actual Tomato% in various hematopoietic populations in the P0 liver, thymus, and spleen when TAM was injected at E14.5 and E15.5 (n = 4). **p < 0.01, *p < 0.05 (unpaired Student’s t test).
We also examined the Tomato% ratios of T progenitor cells in the thymus, which reflects recent replenishment by BM HSPCs. When we labeled HSCs in P2 neonatal BM, extremely low Tomato ratios were observed in the early thymic progenitors (ETPs), CD4CD8 double-negative (DN) 2 to 3, and double-positive (DP) T cells in the thymus 4 weeks after birth (Figure 1F). The Tomato ratios of ETPs, DN2, and DP T cells after 6 months were still significantly lower than 1.0. These data further indicate that HSCs do not produce B and T lymphocytes for the first 4 weeks after birth and gradually contribute to lymphopoiesis thereafter and that a majority of lymphocytes in early life are derived from progenitors in the fetus. These results raised the question of which cells are the origin of lymphocytes in early life.
To address this question, we tested whether FL HSCs produce lymphocytes in neonates. We labeled FL HSCs by injecting TAM at E14.5 and E15.5, and up to 30% of FL HSCs were Tomato+ at birth (P0) (Figure 1G). However, no Tomato+ lymphocytes were detected in the neonatal liver, thymus, and spleen, indicating that neonatal lymphocytes were not derived from FL HSCs. We further traced FL HSC-derived hematopoiesis after birth (Figure S2). Similar to P2 labeling results, FL-HSC-derived lymphopoiesis was not observed until 8 weeks after birth and did not fully contribute to spleen B and T cells even 6–12 months after birth (Figure S2B). Strong correlations between FL HSCs and progenitors/lymphocytes, including MPPs, spleen FO B cells, and CD4+ and CD8+ T cells, were observed after 6–12 months of age (Figure S2C, closed red circles). Thus, HSCs in E14.5 FL and P2 BM exhibited similar kinetics of lymphopoiesis, and FL HSCs did not initiate lymphopoiesis in the embryo.
Fetal and postnatal HSCs show limited contributions to the peritoneal B-1a cell pool for life
As mentioned above, while FO B-2 cells were gradually produced by HSCs after 8 weeks of age, the Tomato ratio of peritoneal B-1a cells to HSCs stayed very low (<0.2) up to 1 year after birth (Figures 1B, 1E, S2B, and S2C) when HSCs were labeled at E14.5 FL or P2 BM. Therefore, unlike B-2 cells, a majority of B-1a cells were HSC independent for life, and the contribution of HSCs to the peritoneal B-1a cell pool was minimal for life. This result was similar to mast cell development, which was completely independent of HSCs (Figures 1B and 1E) and originated from ECs in the embryos.31–33
Fetus-derived lymphopoiesis persists for life
Given the results that HSCs do not fully contribute to adult lymphocytes, we aimed to determine the origin of these cells. Traditionally, lymphoid potentials have been detected in ECs of the embryo and YS as early as E8.5 using in vitro culture.6,7,9 To determine the embryonic origins of postnatal lymphoid cells in a physiological setting, we utilized the EC lineage tracing mouse model, Cdh5CreERT2:Rosa-TdTomato (iCdh5), in which ECs are specifically labeled at the time of TAM injection. We administered a single dose of TAM into pregnant mice at E7.5, E8.5, E9.5, E10.5, or E11.5 and examined at which embryonic stage ECs produced the lymphoid subsets in mice at less than 4 months and more than 6 months of age. (Figures 2A, and S3). As shown in Figure 2B, EC labeling during E7.5-E11.5 marks HSC-independent progenitors and a portion of HSC-derived blood cells because some ECs marked at E7.5 may not immediately produce blood cells and may contribute to producing HSCs at E10–E11. Therefore, to identify HSC-independent lymphopoiesis, we need to subtract the Tomato% of HSC-derived cells from that of EC-derived cells. As we obtained equations for the Tomato% of HSC-derived lymphocytes (Figures 1C–1E), we calculated the “adjusted EC-derived Tomato%,” in which the estimated Tomato% of the HSC-derived population was subtracted from the Tomato% of the EC-derived population (Figures 1C, 1D, 2, and S3). This calculation was based on the results that HSCs at E14.5 and P2 start lymphopoiesis 4 weeks after birth and will represent HSC-independent hematopoiesis (Figures 1 and S2). We noted that E7.5 ECs contributed to all lymphoid subsets in young mice (<4 months old) and that their contribution diminished in mice older than 6 months (Figures 1C–1F), while ECs at E8.5–E10.5 contributed to all lymphoid subsets and MPPs before 4 months and after 6 months of age. This HSC-independent progenitor production was completed by E11.5 (Figures 2C–2G), while HSC production in E11.5 ECs was evident (Figure S3B), indicating that E11.5 ECs exclusively marked HSCs.
Figure 2. HECs produce MPP and B and T lymphocytes that persist in old mice.
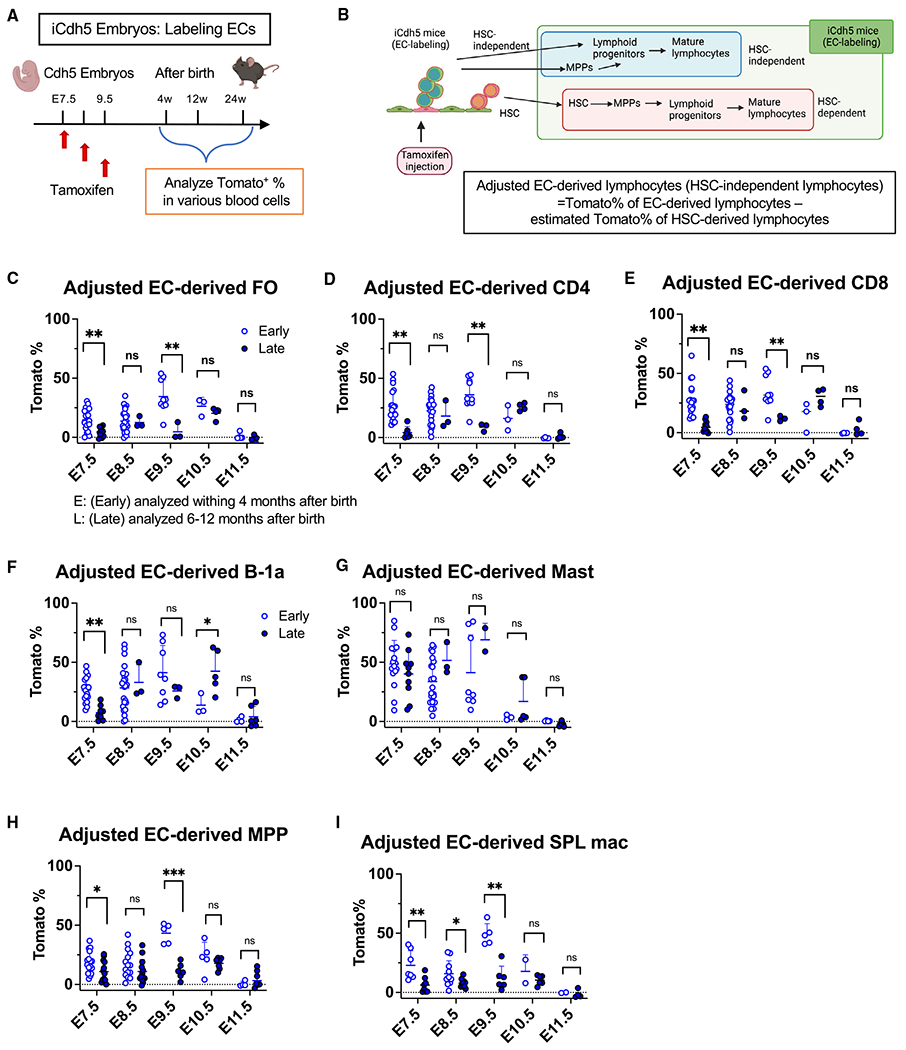
(A) Experimental design. A single dose of TAM was injected into iCdh5 pregnant mice at E7.5–E11.5, respectively, and Tomato+ HSPCs were examined at different time points after birth.
(B) Cdh5Cre marks HSC-independent and HSC-dependent lineages and defined “adjusted EC-derived lymphocytes.” The estimated Tomato% of HSC-derived cells was determined using the equations in each fate mapping plot in Figure 1.
(C–I) Adjusted EC-derived spleen FO B cells (C), CD4 (D), CD8 (E), peritoneal B-1a cells (F), mast cells (G), BM MPPs (H), and spleen macrophages (mac) (I) when TAM was injected at each embryonic time point. Shown are (E) early time points (<4 months old) and (L) late time points (>6 months old). The numbers of analyzed mice and actual Tomato% of these cells are shown in Figure S3. ***p < 0.001, **p < 0.01, *p < 0.05, unpaired Student’s t test. The numbers of analyzed mice are listed in Figure S3A.
MPPs and LPs in the FL originate independent of HSCs from ECs as early as E7.5
Because E14.5 FL HSCs did not contribute to progenitors in P0 neonates (Figure 1G), we next determined the origin of MPPs and LPs in the FL and precisely examined which stage of ECs produced HSCs and HSPCs in the FL. We injected a single dose of TAM into pregnant iCdh5 mice at E7.5, E9.5, or E11.5, respectively, and examined Tomato+ HSPCs in E15.5 FL (Figure 3A). When TAM was injected at E7.5 and embryos were examined at E10.5, at the time of HSC emergence from ECs, CD45+c-kit+ HPCs and vascular endothelial (VE)-cadherin (VC)+ ECs in the YS exhibited approximately 80% Tomato+ labeling (Figure 3B), while ECs in the AGM region were only 10% ± 6% Tomato+ (n = 5). When E15.5 FL was examined following TAM injection at E7.5, 5% ± 2% of HSCs were labeled with Tomato, while MPP, common LPs (CLPs), and CD19+ B progenitors were around 20% Tomato+ (p < 0.01, n = 4), indicating that these HPCs in the E15.5 FL were neither derived from E10.5 AGM ECs nor HSCs but likely from E7.5 ECs via HPCs at E10.5 or even YS ECs at E10.5 (Figures 3B and 3C). When TAM was injected at E9.5 and embryos were analyzed at E10.5, 74.8% ± 2.9% of ECs in the AGM and 93.3% ± 2.1% of ECs in the YS were Tomato+, while the Tomato% of HPCs was very low (Figure 3E, n = 3). When E15.5 FL was examined following TAM injection at E9.5, the Tomato% of HSCs and all other HPCs was around 80% (Figure 3F), suggesting that these cells were all derived from ECs at E10.5. When TAM was injected at E11.5 and E15.5 FL was examined, the Tomato% of MPPs was significantly lower than that of HSCs (MPPs, 3.0% ± 3.6%; HSCs, 21.6% ± 11.6%), indicating that E11.5 ECs exclusively marked the first HSCs (Figure 3G).
Figure 3. HECs produce the majority of HPCs in the FL.
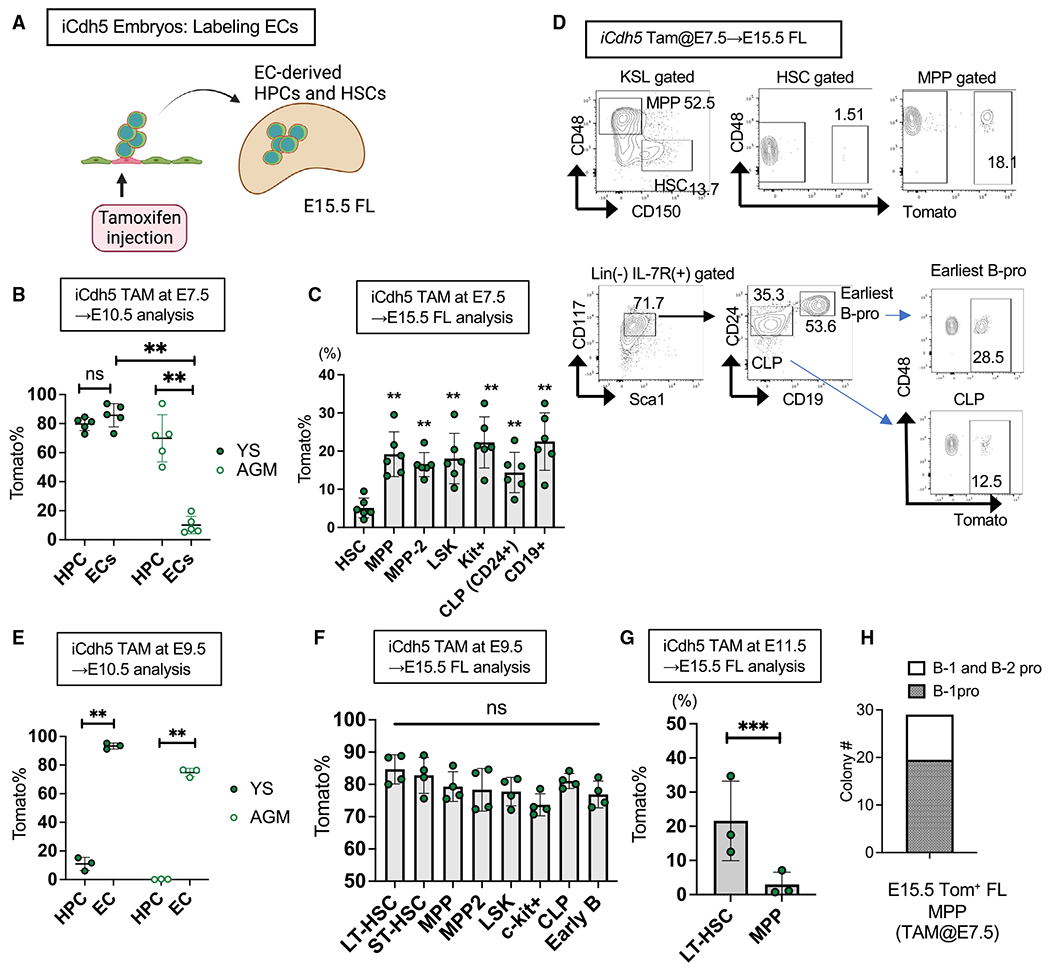
(A) Experimental design. TAM was injected at E7.5, E9.5, or E11.5, and FL progenitors were examined at E15.5.
(B) Tomato% in CD45+c-kit+ HPCs and CD45−VC+ ECs in the YS and AGM region at E10.5 following TAM injection at E7.5 (n = 5).
(C) Tomato% in HSCs and HPCs in the E15.5 FL following TAM injection at E7.5 (n = 6).
(D) Representative fluorescence-activated cell sorting (FACS) plots of (C).
(E) Tomato% in HPCs and ECs in the YS and AGM region at E10.5 following TAM injection at E9.5 (n = 3).
(F) Tomato% in HSCs and HPCs in the E15.5 FL following TAM injection at E9.5 (n = 4).
(G) Tomato% in HSCs and MPPs in the E15.5 FL following TAM injection at E11.5 (n = 3).
(H) B-1 and/or B-2 progenitor colony-forming cell numbers. Tomato+ MPPs in E15.5 FL following TAM injection at E7.5 were sorted and used for the B-progenitor colony formation assay. Eight days after plating, all colonies were individually picked up and subjected to FACS analysis for B-1 (CD93+CD19+B220−) and/or B-2 progenitors (CD93+CD19−B220+). **p < 0.01, *p < 0.05, unpaired Student’s t test.
Taken together, these data revealed that multiple waves of EC-derived progenitors contributed to HPCs in the FL, while HSCs were derived specifically from ECs at E10.5–E11.5.
FL MPPs contain HSC-independent B-1 and B-2 common progenitors
Previous studies have reported that FL MPPs, but not LT-HSCs, most efficiently produced B-1a cells upon transplantation.28,29 Because fetal HSCs are not the major provider of peritoneal B-1a cells under a steady-state condition (Figures S2B and S2C), FL MPPs that have B-1a potential must be derived from precursors at earlier stages than FL HSCs, such as HECs that can produce various hematopoietic cells.10,34 Hence, these data strongly indicated that ECs as early as E7.5 produced MPPs and LPs in E15.5 FL (Figures 3C and 3D). Because Tomato+ E15.5 FL MPPs contain B-1a precursors upon transplantation,29 we examined their B-1 and B-2 progenitor potential using modified B cell colony assays.29 From 500 Tomato+ MPPs marked at E7.5 in iCdh5 mice, 29 B progenitor colonies were detected (Figure 3H). Among them, we found 20 B-1 progenitor colonies and 9 B-1 and B-2 mixed progenitor colonies. These data suggest that E15.5 FL HSC-independent MPPs contain common progenitors for B-1 and B-2 lymphocytes.
HSC-independent MPPs and B-1-biased repopulating cells are present in E11.5 embryos
Although EC labeling at E9.5–E11.5 efficiently marked HSCs in the FL, it is challenging to detect HSCs at E10.5 by transplantation assays without ex vivo culture. It is well accepted that HSC precursors (pre-HSCs), which are intermediate precursors linking HECs and adult repopulating HSCs, are present during E10.5–E11.5 and express c-kit and VC, encoded by Cdh5.35 The first adult repopulating cells are detectable at E11.5 without co-culture, and the E11.5 AGM region also contains pre-HSCs that become HSCs at later stages. We have reported previously that the CD45−VC+c-kit+ phenotypic pre-HSC population in the E10.5 AGM region contains more B-1a-biased precursors than multilineage repopulating cells.29 This result led us to assume that the phenotypic pre-HSC population was heterogeneous, including progenitor cells. In this study, we tested whether we could detect HSC-independent LPs and HSCs separately by transplanting phenotypic pre-HSCs from the E11.5 AGM region into immunodeficient non-obese diabetic (NOD)/severe combined immunodeficiency (SCID)/Il2γcnull (NSG) neonates. We injected 5–50 cells of CD45+Ter119−VC+c-kit+ endothelial protein C receptor (EPCR)+type II phenotypic pre-HSC population36 isolated from the E11.5 AGM region (Figure S4A) into sublethally irradiated NSG neonates. Of 25 recipient mice, 15 showed donor-derived CD45.2+ cells (>0.1%) in the peripheral blood (PB) (Figures 4A–4C). When we examined the transplant recipients 4–6 months post transplantation, we found three types of engraftment patterns: multilineage engraftment with BM LSK cells (HSC engraftment, mice 1–5; Figures 4A and 4D–4F), multilineage engraftment without BM LSK cells (MPP engraftment, mice 6–11; Figures 4B, 4D, and 4G), and only B-1 with minimum B-2 cell engraftment (mice 12–15; Figures 4C and 4D).
Figure 4. LT-HSCs, MPPs, and B-1 repopulating cells arise independent from VC+c-kit+EPCR+ cells in the E11.5 AGM region.
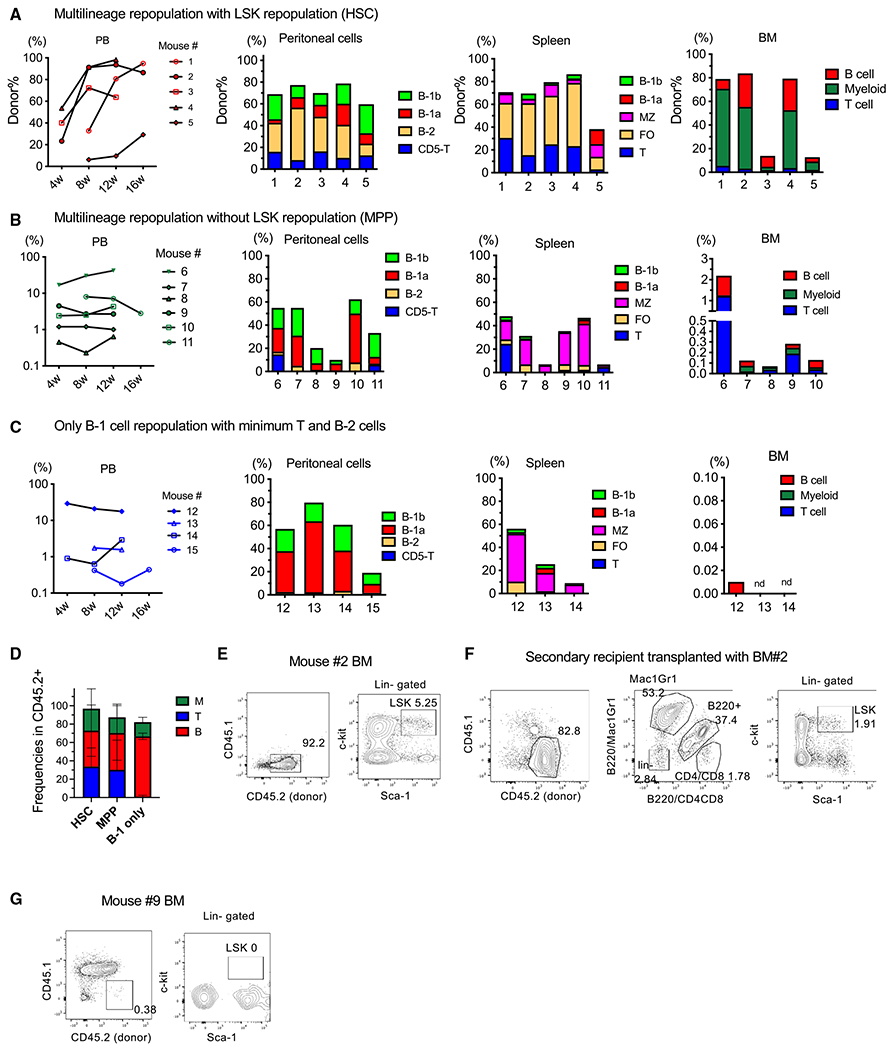
(A–C) Five to 50 pre-HSCs from the E11.5 AGM region were injected into sublethally irradiated NSG neonates. Shown is the CD45.2+ donor cell percentage in the peripheral blood (PB) of the recipient mice 4–6 weeks after transplantation (left panels). Donor cell percentage and their composition within the lymphoid subsets in the peritoneal cells, spleen, and BM are depicted. The engraftment patterns were categorized into three groups: (A) multi-lineage engraftment with BM LSK repopulation (HSC engraftment), (B) multi-lineage engraftment without BM LSK repopulation (MPP engraftment), and (C) only B-1 with minimal B-2 cell engraftment without BM repopulation (B-1 cell engraftment).
(D) Donor-derived cell lineage distribution in the PB of the recipient mice in each group. HSC, HSC-engraftment group (A), n = 5; MPP, multi-lineage engraftment group (B), n = 6; B-1 only, only B-1 with minimal B-2 cell engraftment group (C), n = 4.
(E) Representative FACS plots of the BM of an HSC-engrafted mouse (mouse 2).
(F) Representative FACS plots of the BM of the secondary transplanted mice with BM cells from mouse 2.
(G) Representative FACS plots of the BM of an MPP engrafted mouse (mouse 9).
Mice 1–5 exhibited LT multi-lineage repopulation with a significant donor cell percentage in the PB, peritoneal cavity, spleen, and BM (Figures 4A and 4D) and predominant B-2 cell engraftment with B-1a and B-1b cells. In the recipient BM, successful donor-derived LSK cell repopulation was also confirmed (Figure 4E). Thus, these mice were categorized as HSC-repopulated mice. Importantly, the secondary recipient BM also showed donor-derived LSK repopulation (Figure 4F), indicating that LSK-repopulating cells were the functional HSCs that harbored self-renewal ability. In contrast, mice 6–11 showed multilineage repopulation in the PB (Figure 4D) and LT reconstitution without donor LSK cells in the BM (Figures 4B and 4G), indicating that they were engrafted with HSC-independent progenitors; thus, we named them MPP-engrafted mice. These mice showed predominant B-1 and MZ B cell engraftment with minimal B-2 cells in the peritoneal cavity and spleen (Figure 4B) and B-2, T, and myeloid cell engraftment in the PB and BM (Figures 4B and 4D), although the donor percentage in the BM was very low (<0.5%) (Figure 4B, right panel). Mice 12–15 showed B-1a, B-1b, and MZ B cell engraftment in the peritoneal cavity and spleen but did not show donor-derived cells in the BM (Figure 4C), indicating HSC-independent B-1 cell engraftment. One mouse (12) showed only B-1 and B-2 cell engraftment (Figure 4C spleen), suggesting the presence of common B-1 and B-2 progenitors.
These results indicate that the phenotypic pre-HSC population in the E11.5 AGM region is heterogeneous, containing HSC-independent LT engraftable MPPs and B-1 precursors. Importantly, all engrafted mice exhibited B-1a cell repopulation, whereas B-2 cells were dominant in HSC-engrafted mice. Thus, it seems that B-1a potential is the default property within the E11.5 phenotypic pre-HSC population and that B-2 cell dominant generation is a hallmark of functional HSCs.
scRNA-seq showed heterogeneity in the VC+c-kit+EPCR+ phenotypic pre-HSC population with HSC and B-lymphoid signatures
Based on the transplantation assays, we found that the VC+c-kit+EPCR+ phenotypic pre-HSC population in the E11.5 AGM region contained multilineage repopulating HSCs, HSC-independent MPPs, and HSC-independent B-1-biased progenitors (Figure 4); therefore, we refer to phenotypic pre-HSCs as pre-HSPCs. We hypothesized that different hematopoietic potentials as above could be identified using gene expression profiles among pre-HSPCs. To verify our hypothesis, we performed single-cell RNA sequencing (scRNA-seq) of sorted VC+c-kit+EPCR+ pre-HSPCs from E11.5 AGM and YS, E12.5 FL HSCs (CD45+LSK+EPCR+), and E14.5 FL HSCs (CD45+LSK+CD150+CD48−) (Figure S4). In parallel, the selected sorted subsets were transplanted into sublethally irradiated NSG neonates to validate their hematopoietic capabilities (Figure S4D). Because VC+c-kit+EPCR+ cells constitute an extremely small population in the AGM region (0.02% ± 0.01% in our data, more than 3 experiments), we sorted individual cells from the abovementioned populations and generated single-cell full-length transcriptomes using SMART-Seq. We distributed the genes (read counts > 10) in each cell and excluded cells that expressed fewer than 2,000 genes and genes that were detected in less than 10 cells (Figure S4E). At last, 95 cells and 11,814 genes were used for further analysis. Principal-component analysis (PCA) of scRNA-seq showed clear separation of FL HSCs from pre-HSPCs (Figure 5A). Unbiased sc consensus clustering (SC3) of the whole transcriptomes separated these cells into four distinct clusters (Figure S4F). The top 10 differentially expressed genes are depicted in Figure S4G. Notch-related genes were highly expressed in many AGM cells but downregulated in FL HSCs (Figure S5A), similar to the previously reported transitional requirement of Notch signaling.37
Figure 5. scRNA-seq analysis showed HSC and B lymphoid signatures in E11.5 pre-HSCs and E12 and E14 FL HSCs.
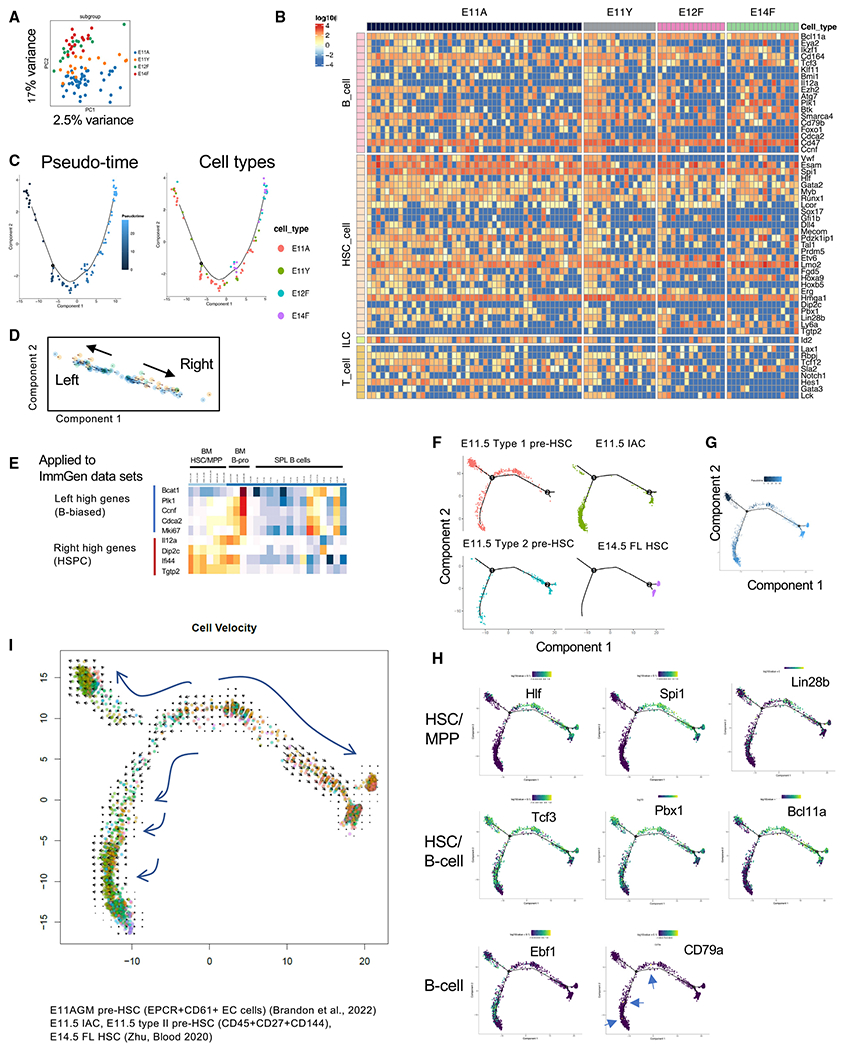
(A) Dimensionality reduction of scRNA-seq data using PCA colored by cell type. E11A, E11.5 AGM pre-HSCs; E11Y, E11.5 YS pre-HSCs; E12F, E12.5 FL HSCs; and E14F, E14.5 FL HSCs.
(B) A heatmap depicting the expression of HSC-, B cell-, T cell-, and ILC-related genes in the E11 AGM and YS pre-HSC populations and E12.5 and E14.5 FL HSCs. Red, blue, and yellow intensities indicate high, low, and intermediate expression levels, respectively.
(C) Pseudo-time analysis of our scRNA-seq data.
(D) Velocity analysis of the 96-cell data, indicating two directions (right arrow and left arrow).
(E) Genes that were highly expressed at both ends of the arrows in Figure 1D were applied to the gene expression subsets of HSC and B progenitors in the ImmGen database (https://www.immgen.org/Databrowser19/DatabrowserPage.html).
(F and G) Pseudo-time analysis of the public data on E11.5 pre-HSCs (types I and II), intra-aortic clusters (IACs), and E14.5 FL HSCs.38,39 Each cell type was also mapped in the pseudo-time analysis.
(H) Selected HSC- and lymphoid cell-related genes were plotted in the pseudo-time analysis of the public data. Genes expressed and/or essential for HSC/MPP (top panel), HSC/B progenitors (center panel), and B progenitors (bottom panel) are depicted.
(I) Velocity analysis of the public datasets.
We then focused on expression of HSC- and lymphoid cell-related genes in each cell (Figure 5B) and examined the trajectory of cell states with the order of cells from the E11.5 AGM to E14.5 FL HSCs using pseudo-time analysis. The trajectory map indicated progression of E11.5 AGM pre-HSPCs to E12.5 and E14.5 FL HSCs (Figure 5C), although we did not see clear branches leading to HSC and B lymphoid lineages. Almost all phenotypic pre-HSPCs and HSCs expressed essential genes for B cell development, such as Ikzf1 and Tcf3, in addition to various HSC-related genes (Figures 5B, and S5B). Next, we performed velocity analysis to identify an unbiased trajectory based on the gene splicing status.40,41 Interestingly, the velocity analysis of our data showed two directions of the gene expression status: right and left directions (Figure 5D arrows). We compared gene expression between the right and left end of Figure 5D (Table S1) and applied them to ImmGen datasets for expressed genes in BM HSCs and B progenitors. The listed genes showed similar expression as that seen in BM B progenitors (highly expressed on the left) and BM HSCs (highly expressed on the right) (Figure 5E) in the ImmGen database, suggesting that the pre-HSPC population may mature into two ways. To validate these findings, we analyzed public scRNA-seq large data of pre-HSPC populations or intra-aortic clusters that were populations equivalent to our E11.5 AGM pre-HSPCs (Figures 5F–5I and S5C–S5E).38,39 Uniform manifold approximation and projection (UMAP) showed 13 clusters, and FL HSCs were distributed to several clusters (Figure S5D). Similar to our data, HSC or lymphoid genes were expressed in all clusters (Figure S5E). The pseudo-time analysis revealed a differentiation trajectory from type I pre-HSPCs to type II pre-HSPC, intra-aortic clusters (IACs), and E14.5 FL HSCs (Figures 5F and 5G). Importantly, there were two branches, one of which was connected to FL HSCs and the other to the status with more IACs, suggesting the presence of two different directions of differentiation from type1 pre-HSPCs (Figure 5F). The genes primarily expressed in fetal HSC/MPPs, including Hlf, Spi1, and Lin28b,42–44 were mainly mapped in the branch connected to FL HSCs (Figure 5H, top panel). Tcf3, Pbx1, and Bcl11a, genes critical for HSC maintenance and B cell development,45–50–were mapped to branches connected to FL HSCs and AGM IACs (Figure 5H, center panel). Ebf1, essential for B cell development,51 was exclusively expressed in the left branch originating from pre-HSPCs to IACs (Figure 5H, bottom panel). These expression patterns reflected our transplantation results in which the pre-HSC population matures into HSCs or B-1-biased progenitors.
We also performed a velocity analysis of these large public datasets, which indicated the following three directions of the velocities (Figure 5I): one connected to HSCs, the second connected to IACs, and the third connected to pre-HSPCs. These results were compatible with our velocity analysis in Figure 5D because our scRNA-seq data represented E11.5 type II pre-HSPCs and E14.5 FL HSCs, which were matched with the branches connected to HSCs and IACs of the public data (Figures 5D and 5F, bottom two panels, and 5I). Thus, scRNA-seq analysis of our data and public large datasets supported the presence of HSC-independent LP development from the VC+c-kit+ phenotypic “pre-HSC population,” which was also shown by our transplantation assays (Figure 4).
DISCUSSION
In this study, we demonstrated that (1) HSCs do not fully contribute to all of the lymphocytes, (2) B-2 and T cells derived from fetal ECs constitute major populations in postnatal mice and significantly persist in adult mice up to 12 months old, (3) HSC-independent MPPs exist in a physiological setting, and a small portion of MPPs can persist up to 12 months after birth, and (4) innate-like peritoneal B-1a cells are rarely produced by HSCs, including in the FL stage, and fetal-EC-derived B-1a cells are maintained for life.
Although the presence of HSC-independent immune cells has recently been recognized, these reported HSC-independent cells found in adult mice are tissue-resident immune cells, including microglia, subsets of γδT cells, and mast cells.31–33,52–54 Therefore, contrary, adaptive immune cells (e.g., CD4+ or CD8+ T cells and B-2 cells) have been believed to be derived from HSCs. Our data showed that the majority of MPPs, T, and B-2 cells within 4 weeks after birth were fetal EC-derived and that a portion of these cells persisted LT in adult life. These results challenge the current paradigm of HSC-derived hematopoiesis in the adult BM.
Our results indicating the persistence of fetus-derived lymphopoiesis in mice 12 months of age agree with the recent inducible Flt3 lineage tracing study by Patel et al.55 that showed that FL Flt3+ MPPs provide a significant percentage of lymphocytes in adult mice. In their study, the origin of FL MPPs remained unknown; however, our data clearly indicated that FL MPPs are derived from HECs at earlier stages. Thus, their findings and our study complementarily explain the fetal origin of T and B lymphoid cells in adult mice. Our results of fetal-EC-derived lymphopoiesis in adult mice are also in line with previous reports showing that postnatal HSC labeling showed a gradual increase of HSC-derived-lymphocytes over 32 weeks18,20–22,56 and a recent report showing a minimal contribution of definitive HSCs to fetal hematopoiesis in a fish model.19 In addition, the presence of HSC-independent MPPs in the pre-HSC population has also been reported using the AGM-EC co-culture system.10 Therefore, together with our results, the phenotypic pre-HSC population contains cells that mature to HSCs and HSC-independent LPs, both of which support hematopoiesis in postnatal life.
A few caveats have to be considered, though. In our study, MPPs showed a strong correlation with HSCs in iFgd5 mice older than 8 weeks, and around 80% of MPPs were derived from HSCs (Figures 1B and 1C). Therefore, the majority of MPPs that support hematopoiesis in adult life must be derived from HSCs with a minimal contribution from fetus-derived MPPs. Indeed, spleen macrophages showed a strong correlation with Fgd5Cre+ MPPs. These results are contrasted with the barcoding data showing MPP-derived native hematopoiesis.24 Because it is known that Fgd5 also marks a small portion of MPPs, we also examined whether short-term (ST)-HSCs and MPP populations were labeled by TAM injection into P2 iFdg5 neonates (Figures S2D and S2E). At P3 (24 h after TAM injection), around 16% of LT-HSCs were labeled, while 6% and 1% of ST-HSCs and MPPs were labeled, respectively (Figure S2D). In the same littermates at P5 (72 h after TAM injection), 70% of LT-HSCs were labeled, while 30% and 8% of ST-HSCs and MPPs were labeled, respectively (Figure S2E). These results indicated that TAM injection at P2 labeled a portion of ST-HSCs and MPPs in addition to a majority of LT-HSCs by P5, suggesting that Tomato+ blood cells in iFgd5 mice contain some ST-HSC-derived (up to 35%) and MPP-derived (up to 10%) cells. Therefore, it is possible that the Tomato% of HSC-derived MPPs might have been overestimated by including directly labeled HSC-independent ST-HSCs and MPPs at P2. Also, the barcoding study in adult mice cannot identify the origin of MPPs that have different barcodes from HSCs when those MPPs were already differentiated from HSCs before barcoding.
Another important finding, that most B-1a cells are HSC independent for life, has finally resolved the long-standing controversy in the field about the primary source of the peritoneal B-1a cells. It is widely accepted that peritoneal B-1a cells are derived from FL precursors but not adult BM HSCs.25,57,58 Transplantation assays using cellular barcoding or Flk2-Cre mice showed the B-1a cell repopulation ability of HSCs.26,27 However, our lineage tracing mouse models revealed that even FL HSCs did not produce B-1a cells efficiently; rather, HECs are the major source of B-1a cells, and transplantation assays showed the transient B-1a cell repopulating capacity of HSCs in the AGM region only at E10.5–E11.5. These results displayed the unique ontogeny of peritoneal B-1a cells. Of note, BM progenitors can reportedly generate B-1a cells with extensive N additions to the immunoglobulin (Ig) V-D junction.59,60 These previous reports are in line with our results that up to 10%–20% of B-1a cells are Tomato+ upon HSC labeling (Figures 1B and 1E).
In our EC lineage tracing study, one question remained about which blood cells E9.5 ECs produced. EC labeling at E7.5 resulted in more Tomato+ HPCs than HSCs in the E15.5 FL, while EC labeling at E11.5 resulted in exclusively labeled HSCs in the E15.5 FL. Evidently, E7.5 ECs produced HSC-independent progenitors in the FL, while E11.5 ECs produced only HSCs. However, EC labeling at E9.5 marked 80% Tomato+ ECs in the AGM region and YS at E10.5 and a similar Tomato% in HSCs and other HPCs in the E15.5 FL (Figure 3F). This result raised the question of whether Tomato+ MPPs and LPs in the E15.5 FL were derived directly from ECs, produced simultaneously with HSCs, or through the first HSCs at E10.5. Is it possible for the very few HSCs arising from ECs at E10.5 to produce all MPPs and LPs in E15.5 FL while expanding for only 5 days? The numbers of Tomato+ phenotypic HSCs, MPPs, and CLPs in E15.5 FL were 4,291 ± 790, 99,222 ± 25,018, and 44,683 ± 30,116, respectively, in our data (Figures 3C and 3F). Using these numbers, we utilized the approximate Bayesian computation (ABC) model for HSC differentiation to learn parameters that reproduced the observed cell counts (Figure S6). To explain the observed HSC and progenitor cell counts at E15.5, the ABC simulation indicated that at least 57 (confidence interval [CI]: 7–98) HSCs were required at E9.5. This was a conservative estimate because we allowed the HSCs to have much higher proliferation rates in our simulation than previously estimated.17,56 If we had constrained HSC proliferation rates to be lower (to be more in line with prior publications), we would need an even higher number of HSCs at E9.5. In this regard, Rybtsov et al.61 precisely calculated the number of pre-HSCs and HSCs during the transition from E9 to E12 and estimated that, by late E11.5, the embryo contains ~70 pre-HSCs in total. However, the estimated numbers of pre-HSC and HSCs at E9.5 were almost zero. A previous report also showed that the frequencies of HSCs in the embryo estimated by direct transplantation into neonates were 1/44.8 embryo equivalent (e.e.) at E9.5 and 1/2.84 e.e. at E10.5.62 Our recent study showed that the frequency of multilineage repopulating HSCs among highly purified VC+c-kit+EPCR+ cells at E10.5 was 1/10 e.e. All of these numbers are much lower than the estimated HSC number that was calculated by the ABC simulation for HSCs to produce MPPs and LPs in E15.5 FL. Thus, it will be more reasonable to consider that MPPs and LPs in E15.5 FL are HSC-independent progenitors. A recent paper by Yokomizo et al.63 reported that HSCs minimally contribute to hematopoietic progenitors in the FL, which also supports our data that HSCs do not start hematopoiesis during the FL stage.
Taken together, we propose a new paradigm where multiple hematopoietic progenitors are produced from ECs during E7.5–E11.5 and arise simultaneously with HSCs at E10–E11. These HSC-independent fetus-derived lymphocytes constitute a major population, even in mice 6–12 months old (Figure 6A). HSCs are produced from E10–E11 HECs and seed the FL (Figure 6B) but do not start lymphopoiesis until 4 weeks of age and do not fully produce all of the mature lymphocytes even at 1 year of age (up to 60%). B-1a cells are produced from HECs during E7.5–E11.5 and persist for life, while HSCs’ contribution to this population is minimal (Figure 6A). Thus, embryonic EC-derived lymphopoiesis continues much longer than traditionally considered, and these results challenge the paradigm of HSC dogma in hematology.
Figure 6. Our working model proposing EC-derived multiple waves of hematopoiesis.
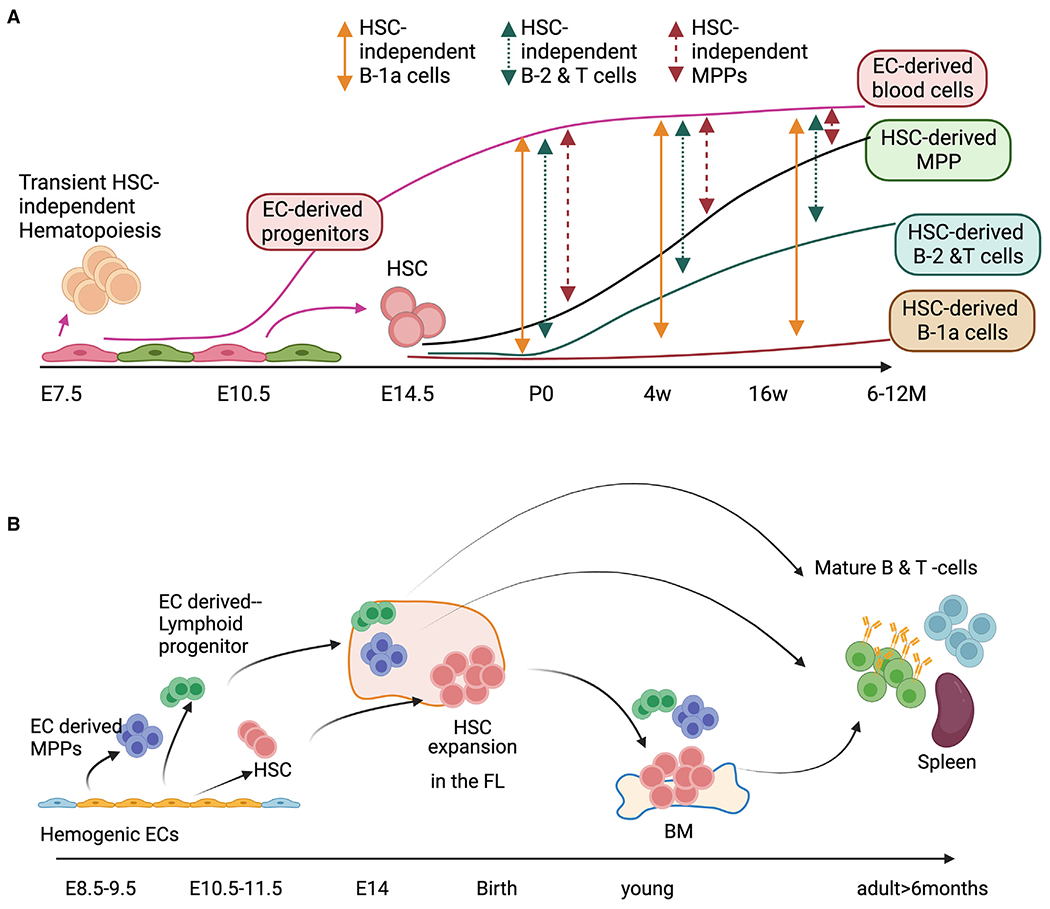
(A) EC-derived (HSC-independent) and HSC-derived lymphopoiesis show how we can estimate the percentages of each wave.
(B) Multiple waves of hematopoiesis produce MPPs, LPs, B-1a cells, and HSCs during E7.5–E11.5. EC-derived HSPCs are the major cell populations in young animals. While postnatal HSC-derived lymphoid cells replace these embryonic EC-derived blood cells with age, a significant percentage of embryonic EC-derived lymphocytes persist in old mice.
Limitations of the study
There are some limitations of the study using the iCdh5 and iFgd5 lineage tracing mouse models. As mentioned above, although Fgd5 specifically marks LT-HSCs in the BM, it still marks a portion of phenotypic ST-HSCs and MPPs. Therefore, the percentage of HSC-derived cells could have been slightly overestimated. Additionally, in iFgd5 mice, the labeling efficiency of FL HSCs was relatively low (up to 30%) compared with neonatal HSCs in the BM (>70% labeled); therefore, we cannot completely rule out the possibility that Tomato− HSCs in the FL might produce lymphocytes during the perinatal period. In iCdh5 mice, it is challenging to avoid labeling HSCs emerging at E10–E11 even when TAM is injected at an earlier stage (E7.5 or E8.5). Therefore, we calculated the adjusted EC labeling percentage as shown in Figure 2 as our best estimate. To better understand the relationship between HSCs, MPPs, and HSC-dependent/independent lymphocytes, further investigations will be required, using a combination of lineage tracing and barcoding in animals.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Momoko Yoshimoto (Momoko.Yoshimoto@uth.tmc.edu).
Materials availability
Mouse lines generated in this study were originally obtained from the following.
Cdh5(PAC)-CreERT2 mice from Dr. Ralf Adams.
Fgd5CreERT2 mice from Jackson Laboratory (Stock No: 027789).
Rosa-TdTomato mice from Jackson Laboratory (Stock No: 007909).
Data and code availability
Sequencing data have been deposited in the GEO database under the accession number GSE182206.
All original code has been deposited at github and is publicly available from the date of publication of the article. URLs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD3e 145-2C11 | eBioscience | 14-0031-82; RRID:AB_2621636 |
| CD4 GK1.5 | Tonbo, BD Bioscience | 20-0041; RRID:AB_11205236 |
| CD5 53-7.3 | eBioscience | 17-0051-82; RRID: AB_469331 |
| CD8a 53-6.7 | Tonbo | 50-0081; RRID: AB_10596646 |
| CD11b M1/70 | Biolegend, Tonbo, BD Bioscience | 101206; RRID: AB_312789, 35-0112; RRID: AB_2621676, 563553; RRID: AB_2738276 |
| CD19 1D3 | eBioscience, Tonbo | 50-0193; RRID: AB_2621752 |
| CD21 7E9, 4E3 | Biolegend, eBioscicence | 123407; RRID: AB_940403, 11-0212; RRID: AB_464977 |
| CD24 M1/69 | eBioscience | 45-242-82; RRID:AB_1210701 |
| CD25 PC61.5 | Biolegend, eBioscience | 102005; RRID: AB_312854, 12-0251-82; RRID: AB_465607 |
| CD41 MWReg30 | eBioscience | 110411-82; RRID: AB_10639937 |
| CD43 S11 | Biolegend | 143219; RRID: AB_2800666 |
| CD44 IM7 | eBioscience, BD Bioscience | 25-0441-82; RRID: AB_469623, 560569; RRID: AB_1727484 |
| CD93 AA4.1 | eBioscience, Biolegend | 17-5892-82; RRID: AB_469466, 136511; RRID: AB_2275879 |
| CD117 2B8 | eBioscience, Biolegend | 25-1171-82, 105826; RRID: AB_1626278 |
| CD150 TC15-12F12.2 | Biolegend | 115910; RRID: AB_493461 |
| CD45 30-F11 | BD Bioscience, Biolegend, Tonbo | 552848; RRID: AB_394489, 103112; RRID: AB_312977, 35-0451; RRID: AB_469717 |
| CD45.1 A20 | BD Bioscience, eBioscience | 565212; RRID:AB_2722493, 47-0453-82; RRID: AB_1582228 |
| CD45.2 104 | BD Bioscience, Biolegend | 564616; RRID: AB_2738867, 109806; RRID: AB_313443 |
| IgM II/41, RMM-1 | BD Bioscience, Biolegend | 743329; RRID: AB_2741430, 406506; RRID: AB_315056 |
| IgD 11-26c.2a | Biolegend | 405720; RRID: AB_2561876 |
| B220 RA3-6B2 | Tonbo, Biolegend | 60-0452; RRID: AB_2621849, 103207; RRID: AB_312992 |
| FcERI MAR-1 | Biolegend, Tonbo | 134319; RRID: AB_10641135, 35-5898; RRID: AB_2621719 |
| F4/80 BM8 | eBioscience | 47-4801-82; RRID: AB_1548745 |
| CCR6 140706 | BD Bioscience | 561751; RRID: AB_10896986 |
| Gr-1 RB6-8C5 | Biolegend | 108428; RRID: 893558 |
| EPCR 1560 | eBioscience | 12-2012-82; RRID: AB_914317 |
| CD144 11D4.1 | BD Bioscience | 562242; RRID: AB_2737608 |
| CD48 HM48-1 | Biolegend, BD Bioscience | 103404; RRID: AB_313019, 740236; RRID: AB_2739984 |
| CD23 B3B4 | eBioscience | 25-0232-82; RRID: AB_469604 |
| CD127 A7R34 | Biolegend, eBioscience | 135012; RRID: AB_1937216, 12-1271-82; RRID: AB_465844 |
| Ter119 TER-119 | Tonbo, Biolegend, eBioscience | 65-5921; RRID: AB_2935630, 116223; RRID:AB_2137788, 11-5921-82; RRID:AB_465311 |
| Sca-1 D7 | eBioscience, Biolegend | 25-5981-82; RRID: AB_469669, 108124; RRID:AB_893615 |
| CD34 RAM34 | eBioscience | 11-0341-82; RRID: AB_465021 |
| Thy1.2 30-H12 | Biolegend, eBioscience | 105328; RRID: AB_10613293, 11-0902-82; RRID:AB_465154 |
| CD184 2B11 | eBioscience | 12-9991-82; RRID: 891391 |
| Chemicals, peptides, and recombinant proteins | ||
| Tamoxifen | Sigma | T5648 |
| Progesterone | Sigma | P0130 |
| Recombinant mouse IL-7 | PeproTech | 217-17 |
| Recombinant mouse SCF | PeproTech | 250-03 |
| Recombinant mouse Flt3-ligand | PeproTech | 250-31L |
| Collagenase Type I (0.25%) | STEMCELL Technologies | 07902 |
| Critical commercial assays | ||
| SMART-Seq V4 Ultra Low RNA Kit | Takara | # 634898 |
| MethoCult M3630 | STEMCELL Technologies | 03630 |
| Deposited data | ||
| scRNA-sequencing | GEO database | GSE182206 |
| Experimental models: Cell lines | ||
| OP-9 stromal cells | Dr. Toru Nakano at Osaka University, Japan | ATCC CRL-2749 |
| Experimental models: Organisms/strains | ||
| Fgd5ZsGreen-CreERT2 | C57BU6N-Fgd5tm3(cre/ERT2)Djr/J | Jackson no. 027789 |
| Cdh5(PAC)-CreERT2 | C57BL/6-Tg(Cdh5-cre/ERT2) | Taconic no. 13073 |
| Rosa-TdTomato | B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | Jackson no. 007909 |
| NSG | NOD.Cg-PrkdcscidIl2rgtm1wjl/Szj | Jackson no. 005557 |
| Boy/J | B6.SJL-PtprcaPepcb/BoyJ | Jackson no. 002014 |
| Software and algorithms | ||
| Fastqc | Babraham Institute | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| STAR | PMID: 23104886 | https://github.com/alexdobin/STAR |
| Htseq-count | PMID: 25260700 | https://github.com/htseq/htseq |
| SC3 | PMID: 28346451 | http://bioconductor.org/packages/release/bioc/vignettes/SC3/inst/doc/SC3.html |
| Monocle2 | PMID: 28825705 | http://cole-trapnell-lab.github.io/monocle-release/docs/ |
| CellRanger | PMID: 28091601 | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/using/tutorials |
| Scikit-learn | https://scikit-learn.org/stable/ | |
| Velocyto.py | PMID: 30089906 | http://velocyto.org/velocyto.py/tutorial/analysis.html |
| velocyto.R | https://github.com/velocyto-team/velocyto.R | |
| scVelo | Volker Bergen | https://scvelo.readthedocs.io |
| Script used for Figures 5A and 5D | This paper | https://github.com/seitalab/hsc_independent |
| Approximate Bayesian Computation (Figure S6) | This paper | https://github.com/djhshih/analysis-lineage-tracing-b1a |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Experimental animals
Cdh5(PAC)-CreERT2 mice were crossed with Rosa-TdTomato mice (Jackson Laboratory Stock No: 007,909) and Cdh5CreE2: Rosa-Tomato mice were generated. Similarly, Fgd5CreERT2 mice were crossed with Rosa-TdTomato mice and Fgd5CreERT2: Rosa-Tomato mice were generated. These mice were timed mated with Rosa-Tomato mice and the vaginal plugs were confirmed in the following morning. The noon on the day that the plug was found was counted as embryonic day 0.5. Tamoxifen (15ng/mother body weight) was administrated into timed mated pregnant iCdh5 dams at E7.5, 8.5, 9.5, 10.5, 11.5 respectively, or into timed mated pregnant Fgd5 dams at E14.5, or P2 neonates. 10-15 mice for each Tam injection date were examined.
Cdh5(PAC)-CreERT2: Td-Tomato mice were timed mated with Rosa-DNMAML-GFP mice. Tamoxifen was injected into the pregnant mice at E7.5 and mice were examined after birth.
For transplantation assays, C57BL/6 mice were timed mated and embryos at E11.5 were harvested from the pregnant dams for donor cells. The embryonic age was confirmed by the somite numbers and developmental features of the embryos.
NOD/SCID/Il2γcnull mice (NSG mice. Jackson Laboratory Stock No: 005,557, OD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice) were mated and day2–5 neonates were used for recipients of transplantation assays. Recipient NSG neonates were sublethally irradiated (150rad) before donor cells were injected into facial vein.
For secondary transplantation, lethally irradiated adult B6.SJL-Ptprca Pepcb/BoyJ mice (BoyJ mice, Stock No: 002,014, B6 Cd45.1) were used for recipients.
Mice were kept in specific pathogen free condition and all the experimental procedures using the mice were approved by Animal Welfare Committee at UTHealth.
METHOD DETAILS
Lineage tracing experiments
Cdh5CreERT2: Rosa-Tomato (iCdh5) mice were timed mated. A single dose of Tamoxifen (TAM: Sigma) 15ng/mother body weight together with Progesterone (7.5 ng) solved in corn oil was administrated to the pregnant dam by oral gavage at E7.5, 8.5, 9.5, 10.5, and 11.5 respectively. TAM usually makes the delivery difficult, therefore, Cesarean section was performed on day 19 pregnant dams to rescue the embryos and these pups were taken care of by a surrogate mother prepared in advance. Fgd5CreERT2: Rosa-Tomato (iFgd5) mice were used for marking HSCs. TAM was injected into E14.5 pregnant dam or P2 neonatal mice. We harvested various hematopoietic tissues including peritoneal cells, spleen, thymus, and bone marrow from TAM administrated embryos/mice and examined Tomato+ percentages in each hematopoietic subset. 10-15 mice for each TAM injection date were examined. We performed 3-4 experiments (biological replicates) per each TAM injection.
Cell harvesting, sorting and transplantation
E11.5 embryos were harvested from C57BL/6 pregnant mice. The embryonic age was confirmed by the somite numbers and developmental features of the embryos. AGM region was dissected and digested with 0.25% collagenase (Stem cell technology) for 30 min at 37°C to make a single cell suspension. Cells were then stained with anti-CD11b, Ter119, CD144, c-kit, and CD206 antibodies and pre-HSC population was sorted by single cell sorting mode on BD FACS Melody. Five to 60 cells were injected to sublethally irradiated NSG neonates (day 2-5). The peripheral blood was collected from the recipient mice every month starting at 6 weeks after transplantation. Four to 6 months after transplantation, all the hematopoietic tissues were collected and examined for CD45.2+ donor cells by flow cytometry.
For secondary transplantation, 1-2 million BM cells from the primary recipient mice were injected into lethally irradiated adult BoyJ mice. Blood cells of NSG and BoyJ mice express CD45.1.
B-progenitor colony forming assay
Five hundred FL multipotent progenitors (MPPs) were plated to Methocult M3630 (Stemcell Technologies) containing 10 ng/mL IL-7 with 105 OP-9 cells. Eight days after plating, colony numbers were counted and each colony was picked up and stained with antimouse CD45, AA4.1, CD19, B220, and CD11b to identify B-1 and B-2 progenitors using flow cytometry.
scRNA-sequencing
E11.5 pre-HSCs, E12.5 and 14.5 FL HSCs were single cell sorted into 96 well plate (1 cell/well). RNA were extracted from each well and converted into cDNA using SAMRT-Seq Single cell kit (Takara). DNA library was made and sequenced at Single Cell Genomic Core at the Baylor College of Medicine. Briefly, following cDNA synthesis, Nextera XT DNA library preparation kit (Illumina) was used to prepare library.120pg of cDNA was simultaneously fragmented and tagged with adapter sequences by transposome. The product was then amplified using 12 cycles of PCR and purified. Final library was sequenced using Illumina Novaseq 600.
Bioinformatics analysis of scRNA-sequencing
The quality of all sequenced samples was analyzed using FastQC. Raw reads were aligned to the GRCm38 reference genome using STAR with default parameters. The expression count matrix was generated using htseq-count. We filtered genes whose read counts less than 10 and cells that were less than 2000 genes. Read counts were normalized using DEseq2 with default parameters. The normalized matrix was clustered by SC3. We chose K = 2 for SC3 as the best represented the heterogeneity in our dataset. Marker genes in each cluster were filtered by the area under the ROC curve (auroc) > 0.85 and the adjusted p values<0.01. Trajectory and pseudotime analysis were performed by monocle2 package with default parameters.
PCA analysis was performed using scikit-learn software package.64
RNA Velocity analysis was performed according to the original article.40,41 In brief, unspliced pre-mRNA counts and mature spliced mRNA counts for each cell were computed from BAM file, then RNA velocity was computed using stochastic model. The result was visualized onto the PCA plot based on the expression profile of mature spliced mRNA for each cell. Each arrow represents a direction of a cell transition based on the RNA Velocity.
scRNA-seq analysis of published data
E11 AGM data from Hadland, B et al.38 (GSE145886) and E11.5/E14.5 data from Zhu Q. et al.39 (GSE137117) are downloaded in fastq format from NCBI GEO dataset (Figure 5C). Raw scRNA-Seq reads of samples were processed individually according to 10x Genomics CellRanger software65 and mapped to the mm10 genome with default parameters. We analyzed the scRNA-seq and filtered the cells with at least 400 genes and less 10% of raw reads belong to mitochondrial genes. Samples were merged by Seurat v4 packages. The integration of data was performed using Seurat CCA method. Clustering was then performed by Seurat’s FindClusters function, with resolution res = 0.5 and then visualization was performed by 2D UMAP plots. The Monocle platform was used for pseudotime analysis with default parameters. Velocyto.py41 was used for detecting the expression of spliced and unspliced transcripts of genes. “velocyto.R”(https://github.com/velocyto-team/velocyto.R) was performed RNA velocity analysis. All the cells and genes used in velocity analysis were consistent with the Seurat results. Velocity fields were then projected onto UMAP generated by Seurat.
Statistical analysis
Non-parametric student-t test was used for statistical analysis in Figures 1, 2, and 3.
The simulation model to produce output stem and progenitor cell numbers
Approximate Bayesian Computation was used to learn parameters for the HSC differentiation model proposed by Busch et al. 2015,56 using observed cell counts at E15.5, in order to infer the initial HSC number at E9.5. Net proliferation rates and differentiation rates were each independently sampled from Uniform (0, 4), based on the bounds previously determined by Busch et al. Under this model, a net proliferation rate of 3 per day corresponds to 2 cell divisions per day. Initial HSC number was sampled from 1 to 100 uniformly. Model simulations were performed for with time steps of 1 h. A total of 10,000,000 samples of model parameters were generated, and sample parameters were accepted only if the predicted cell numbers were all within the 99% confidence interval of the observed data, resulting in an acceptance rate of about 0.01%.
Supplementary Material
Highlights.
Multiple waves of lymphopoiesis occur in mouse embryos and persist into adult life
HSCs do not produce all of the lymphocytes in the adult
HSC-independent MPPs are produced simultaneously with HSCs from ECs
HSCs minimally contribute to the peritoneal B-1a cell pool in adult mice
ACKNOWLEDGMENTS
We are grateful to Dr. Mervin C. Yoder at Indiana University and Dr. Brian R. Davis at the University of Texas Health Science Center at Houston for their comments and critical review of the manuscript. The scRNA-seq work was performed at the Single Cell Genomics Core at BCM, partially supported by NIH shared instrument grants (S10OD023469 and S10OD025240) and P30EY002520. This work was supported by NIH R01AI121197 (to M.Y.) and R01AI147685 (to J.K.). H.W. and J.Q.W. were supported by grants from the NIH (R01 NS088353 and R21 NS113068), and the Amy and Edward Knight Fund – the UTHSC Senator Lloyd Bentsen Center for Stroke Research. This work was also partly supported by the NIH through grants 1UL1TR003167 and 1R01AG066749 and the Cancer Prevention and Research Institute of Texas through grant RP170668 (to W.J.Z). D.J.S. was supported by the HKU-100 start-up research grant. Figure 6 and the graphical abstract were created with BioRender.com.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112239.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, and Speck NA (2009). Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457, 887–891. 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zovein AC, Hofmann JJ, Lynch M, French WJ, Turlo KA, Yang Y, Becker MS, Zanetta L, Dejana E, Gasson JC, et al. (2008). Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell 3, 625–636. 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tober J, Yzaguirre AD, Piwarzyk E, and Speck NA (2013). Distinct temporal requirements for Runx1 in hematopoietic progenitors and stem cells. Development 140, 3765–3776. 10.1242/dev.094961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dzierzak E, and Bigas A (2018). Blood development: hematopoietic stem cell dependence and independence. Cell Stem Cell 22, 639–651. 10.1016/j.stem.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 5.Ganuza M, Hall T, Finkelstein D, Chabot A, Kang G, and McKinney-Freeman S (2017). Lifelong haematopoiesis is established by hundreds of precursors throughout mammalian ontogeny. Nat. Cell Biol 19, 1153–1163. 10.1038/ncb3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cumano A, Dieterlen-Lievre F, and Godin I (1996). Lymphoid potential, probed before circulation in mouse, is restricted to caudal intra-embryonic splanchnopleura. Cell 86, 907–916. 10.1016/s0092-8674(00)80166-x. [DOI] [PubMed] [Google Scholar]
- 7.Nishikawa SI, Nishikawa S, Kawamoto H, Yoshida H, Kizumoto M, Kataoka H, and Katsura Y (1998). In vitro generation of lymphohematopoietic cells from endothelial cells purified from murine embryos. Immunity 8, 761–769. 10.1016/s1074-7613(00)80581-6. [DOI] [PubMed] [Google Scholar]
- 8.Yokota T, Huang J, Tavian M, Nagai Y, Hirose J, Zúñiga-Pflücker JC, Péault B, and Kincade PW (2006). Tracing the first waves of lymphopoiesis in mice. Development 133, 2041–2051. 10.1242/dev.02349. [DOI] [PubMed] [Google Scholar]
- 9.Yoshimoto M, Montecino-Rodriguez E, Ferkowicz MJ, Porayette P, Shelley WC, Conway SJ, Dorshkind K, and Yoder MC (2011). Embryonic day 9 yolk sac and intra-embryonic hemogenic endothelium independently generate a B-1 and marginal zone progenitor lacking B-2 potential. Proc. Natl. Acad. Sci. USA 108, 1468–1473. 10.1073/pnas.1015841108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dignum T, Varnum-Finney B, Srivatsan SR, Dozono S, Waltner O, Heck AM, Ishida T, Nourigat-McKay C, Jackson DL, Rafii S, et al. (2021). Multipotent progenitors and hematopoietic stem cells arise independently from hemogenic endothelium in the mouse embryo. Cell Rep. 36, 109675. 10.1016/j.celrep.2021.109675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godin I, Dieterlen-Lièvre F, and Cumano A (1995). Emergence of multipotent hemopoietic cells in the yolk sac and paraaortic splanchnopleura in mouse embryos, beginning at 8.5 days postcoitus. Proc. Natl. Acad. Sci. USA 92, 773–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshimoto M, Porayette P, Glosson NL, Conway SJ, Carlesso N, Cardoso AA, Kaplan MH, and Yoder MC (2012). Autonomous murine T-cell progenitor production in the extra-embryonic yolk sac before HSC emergence. Blood 119, 5706–5714. 10.1182/blood-2011-12-397489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi M, Shelley WC, Seo W, Vemula S, Lin Y, Liu Y, Kapur R,Taniuchi I, and Yoshimoto M (2014). Functional B-1 progenitor cells are present in the hematopoietic stem cell-deficient embryo and depend on Cbfbeta for their development. Proc. Natl. Acad. Sci. USA 111, 12151–12156. 10.1073/pnas.1407370111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller J, Horner A, Stacy T, Lowrey C, Lian JB, Stein G, Nuckolls GH, and Speck NA (2002). The core-binding factor beta subunit is required for bone formation and hematopoietic maturation. Nat. Genet 32, 645–649. 10.1038/ng1049. [DOI] [PubMed] [Google Scholar]
- 15.Kumaravelu P, Hook L, Morrison AM, Ure J, Zhao S, Zuyev S, Ansell J, and Medvinsky A (2002). Quantitative developmental anatomy of definitive haematopoietic stem cells/long-term repopulating units (HSC/RUs): role of the aorta-gonad-mesonephros (AGM) region and the yolk sac in colonisation of the mouse embryonic liver. Development 129, 4891–4899. [DOI] [PubMed] [Google Scholar]
- 16.Gekas C, Dieterlen-Lièvre F, Orkin SH, and Mikkola HKA (2005). The placenta is a niche for hematopoietic stem cells. Dev. Cell 8, 365–375. 10.1016/j.devcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 17.Ema H, and Nakauchi H (2000). Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood 95, 2284–2288. [PubMed] [Google Scholar]
- 18.Zhang Y, McGrath KE, Ayoub E, Kingsley PD, Yu H, Fegan K, McGlynn KA, Rudzinskas S, Palis J, and Perkins AS (2021). Mds1(CreERT2), an inducible Cre allele specific to adult-repopulating hematopoietic stem cells. Cell Rep. 36, 109562. 10.1016/j.celrep.2021.109562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ulloa BA, Habbsa SS, Potts KS, Lewis A, McKinstry M, Payne SG, Flores JC, Nizhnik A, Feliz Norberto M, Mosimann C, and Bowman TV (2021). Definitive hematopoietic stem cells minimally contribute to embryonic hematopoiesis. Cell Rep. 36, 109703. 10.1016/j.celrep.2021.109703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Säwen P, Eldeeb M, Erlandsson E, Kristiansen TA, Laterza C, Kokaia Z, Karlsson G, Yuan J, Soneji S, Mandal PK, et al. (2018). Murine HSCs contribute actively to native hematopoiesis but with reduced differentiation capacity upon aging. Elife 7, e41258. 10.7554/eLife.41258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Upadhaya S, Sawai CM, Papalexi E, Rashidfarrokhi A, Jang G, Chattopadhyay P, Satija R, and Reizis B (2018). Kinetics of adult hematopoietic stem cell differentiation in vivo. J. Exp. Med 215, 2815–2832. 10.1084/jem.20180136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawai CM, Babovic S, Upadhaya S, Knapp DJHF, Lavin Y, Lau CM, Goloborodko A, Feng J, Fujisaki J, Ding L, et al. (2016). Hematopoietic stem cells are the major source of multilineage hematopoiesis in adult animals. Immunity 45, 597–609. 10.1016/j.immuni.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez-Fraticelli AE, Wolock SL, Weinreb CS, Panero R, Patel SH, Jankovic M, Sun J, Calogero RA, Klein AM, and Camargo FD (2018). Clonal analysis of lineage fate in native haematopoiesis. Nature 553, 212–216. 10.1038/nature25168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun J, Ramos A, Chapman B, Johnnidis JB, Le L, Ho YJ, Klein A, Hofmann O, and Camargo FD (2014). Clonal dynamics of native haematopoiesis. Nature 514, 322–327. 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardy RR, and Hayakawa K (1991). A developmental switch in B lymphopoiesis. Proc. Natl. Acad. Sci. USA 88, 11550–11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kristiansen TA, Jaensson Gyllenbäck E, Zriwil A, Björklund T, Daniel JA, Sitnicka E, Soneji S, Bryder D, and Yuan J (2016). Cellular barcoding links B-1a B cell potential to a fetal hematopoietic stem cell state at the single-cell level. Immunity 45, 346–357. 10.1016/j.immuni.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 27.Beaudin AE, Boyer SW, Perez-Cunningham J, Hernandez GE, Derderian SC, Jujjavarapu C, Aaserude E, MacKenzie T, and Forsberg EC (2016). A transient developmental hematopoietic stem cell gives rise to innate-like B and T cells. Cell Stem Cell 19, 768–783. 10.1016/j.stem.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosn EEB, Waters J, Phillips M, Yamamoto R, Long BR, Yang Y, Gerstein R, Stoddart CA, Nakauchi H, and Herzenberg LA (2016). Fetal hematopoietic stem cell transplantation fails to fully regenerate the B-lymphocyte compartment. Stem Cell Rep. 6, 137–149. 10.1016/j.stemcr.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi M, Tarnawsky SP, Wei H, Mishra A, Azevedo Portilho N, Wenzel P, Davis B, Wu J, Hadland B, and Yoshimoto M (2019). Hemogenic endothelial cells can transition to hematopoietic stem cells through a B-1 lymphocyte-biased state during maturation in the mouse embryo. Stem Cell Rep. 13, 21–30. 10.1016/j.stemcr.2019.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gazit R, Mandal PK, Ebina W, Ben-Zvi A, Nombela-Arrieta C, Silberstein LE, and Rossi DJ (2014). Fgd5 identifies hematopoietic stem cells in the murine bone marrow. J. Exp. Med 211, 1315–1331. 10.1084/jem.20130428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gentek R, Ghigo C, Hoeffel G, Bulle MJ, Msallam R, Gautier G, Launay P, Chen J, Ginhoux F, and Bajénoff M (2018). Hemogenic endothelial fate mapping reveals dual developmental origin of mast cells. Immunity 48, 1160–1171.e5. 10.1016/j.immuni.2018.04.025. [DOI] [PubMed] [Google Scholar]
- 32.Li Z, Liu S, Xu J, Zhang X, Han D, Liu J, Xia M, Yi L, Shen Q, Xu S, et al. (2018). Adult connective tissue-resident mast cells originate from late erythro-myeloid progenitors. Immunity 49, 640–653.e5. 10.1016/j.immuni.2018.09.023. [DOI] [PubMed] [Google Scholar]
- 33.Yoshimoto M, Kosters A, Cornelius S, Valiente N, Cheng H, Latorre A, Nishida C, Ghosn EEB, and Kobayashi M (2022). Mast cell repopulating ability is lost during the transition from pre-HSC to FL HSC. Front. Immunol 13, 896396. 10.3389/fimmu.2022.896396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hadland B, and Yoshimoto M (2018). Many layers of embryonic hematopoiesis: new insights into B cell ontogeny and the origin of hematopoietic stem cells. Exp. Hematol 60,1–9. 10.1016/j.exphem.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rybtsov S, Sobiesiak M, Taoudi S, Souilhol C, Senserrich J, Liakhovitskaia A, Ivanovs A, Frampton J, Zhao S, and Medvinsky A (2011). Hierarchical organization and early hematopoietic specification of the developing HSC lineage in the AGM region. J. Exp. Med 208, 1305–1315. 10.1084/jem.20102419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou F, Li X, Wang W, Zhu P, Zhou J, He W, Ding M, Xiong F, Zheng X, Li Z, et al. (2016). Tracing haematopoietic stem cell formation at single-cell resolution. Nature 533, 487–492. 10.1038/nature17997. [DOI] [PubMed] [Google Scholar]
- 37.Souilhol C, Lendinez JG, Rybtsov S, Murphy F, Wilson H, Hills D, Batsivari A, Binagui-Casas A, McGarvey AC, MacDonald HR, et al. (2016). Developing HSCs become Notch independent by the end of maturation in the AGM region. Blood 128, 1567–1577. 10.1182/blood-2016-03-708164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hadland B, Varnum-Finney B, Dozono S, Dignum T, Nourigat-McKay C, Heck AM, Ishida T, Jackson DL, Itkin T, Butler JM, et al. (2022). Engineering a niche supporting hematopoietic stem cell development using integrated single-cell transcriptomics. Nat. Commun 13, 1584. 10.1038/s41467-022-28781-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Q, Gao P, Tober J, Bennett L, Chen C, Uzun Y, Li Y, Howell ED, Mumau M, Yu W, et al. (2020). Developmental trajectory of prehematopoietic stem cell formation from endothelium. Blood 136, 845–856. 10.1182/blood.2020004801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bergen V, Lange M, Peidli S, Wolf FA, and Theis FJ (2020). Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol 38, 1408–1414. 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- 41.La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yokomizo T, Watanabe N, Umemoto T, Matsuo J, Harai R, Kihara Y, Nakamura E, Tada N, Sato T, Takaku T, et al. (2019). Hlf marks the developmental pathway for hematopoietic stem cells but not for erythro-myeloid progenitors. J. Exp. Med 216, 1599–1614. 10.1084/jem.20181399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Copley MR, Babovic S, Benz C, Knapp DJHF, Beer PA, Kent DG, Wohrer S, Treloar DQ, Day C, Rowe K, et al. (2013). The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat. Cell Biol 15, 916–925. 10.1038/ncb2783. [DOI] [PubMed] [Google Scholar]
- 44.Iwasaki H, Somoza C, Shigematsu H, Duprez EA, Iwasaki-Arai J, Mizuno SI, Arinobu Y, Geary K, Zhang P, Dayaram T, et al. (2005). Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood 106, 1590–1600. 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, et al. (2010). A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol 11, 635–643. 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Semerad CL, Mercer EM, Inlay MA, Weissman IL, and Murre C (2009). E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc. Natl. Acad. Sci. USA 106, 1930–1935. 10.1073/pnas.0808866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ficara F, Murphy MJ, Lin M, and Cleary ML (2008). Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell 2, 484–496. 10.1016/j.stem.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanyal M, Tung JW, Karsunky H, Zeng H, Selleri L, Weissman IL, Herzenberg LA, and Cleary ML (2007). B-cell development fails in the absence of the Pbx1 proto-oncogene. Blood 109, 4191–4199. 10.1182/blood-2006-10-054213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luc S, Huang J, McEldoon JL, Somuncular E, Li D, Rhodes C, Mamoor S, Hou S, Xu J, and Orkin SH (2016). Bcl11a deficiency leads to hematopoietic stem cell defects with an aging-like phenotype. Cell Rep. 16, 3181–3194. 10.1016/j.celrep.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu Y, Wang J, Khaled W, Burke S, Li P, Chen X, Yang W, Jenkins NA, Copeland NG, Zhang S, and Liu P (2012). Bcl11a is essential for lymphoid development and negatively regulates p53. J. Exp. Med 209, 2467–2483. 10.1084/jem.20121846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin H, and Grosschedl R (1995). Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature 376, 263–267. 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 52.Gentek R, Ghigo C, Hoeffel G, Jorquera A, Msallam R, Wienert S, Klauschen F, Ginhoux F, and Bajénoff M (2018). Epidermal gamma-delta T cells originate from yolk sac hematopoiesis and clonally self-renew in the adult. J. Exp. Med 215, 2994–3005. 10.1084/jem.20181206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F, and Rodewald HR (2015). Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547–551. 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spidale NA, Sylvia K, Narayan K, Miu B, Frascoli M, Melichar HJ, Zhihao W, Kisielow J, Palin A, Serwold T, et al. (2018). Interleukin-17-Producing gammadelta T Cells originate from SOX13(+) progenitors that are independent of gammadeltaTCR signaling. Immunity 49, 857–872.e5. 10.1016/j.immuni.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patel SH, Christodoulou C, Weinreb C, Yu Q, da Rocha EL, Pepe-Mooney BJ, Bowling S, Li L, Osorio FG, Daley GQ, and Camargo FD (2022). Lifelong multilineage contribution by embryonic-born blood progenitors. Nature 606, 747–753. 10.1038/s41586-022-04804-z. [DOI] [PubMed] [Google Scholar]
- 56.Busch K, Klapproth K, Barile M, Flossdorf M, Holland-Letz T, Schlenner SM, Reth M, Höfer T, and Rodewald HR (2015). Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature 518, 542–546. 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- 57.Ghosn EEB, Sadate-Ngatchou P, Yang Y, Herzenberg LA, and Herzenberg LA (2011). Distinct progenitors for B-1 and B-2 cells are present in adult mouse spleen. Proc. Natl. Acad. Sci. USA 108,2879–2884. 10.1073/pnas.1019764108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Montecino-Rodriguez E, Leathers H, and Dorshkind K (2006). Identification of a B-1 B cell-specified progenitor. Nat. Immunol 7, 293–301. 10.3389/fimmu.2022.946202. [DOI] [PubMed] [Google Scholar]
- 59.Holodick NE, Vizconde T, and Rothstein TL (2014). B-1a cell diversity: nontemplated addition in B-1a cell Ig is determined by progenitor population and developmental location. J. Immunol 192,2432–2441. 10.4049/jimmunol.1300247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holodick NE, Repetny K, Zhong X, and Rothstein TL (2009). Adult BM generates CD5+ B1 cells containing abundant N-region additions. Eur. J. Immunol 39, 2383–2394. 10.1002/eji.200838920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rybtsov S, Ivanovs A, Zhao S, and Medvinsky A (2016). Concealed expansion of immature precursors underpins acute burst of adult HSC activity in foetal liver. Development 143, 1284–1289. 10.1242/dev.131193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arora N, Wenzel PL, McKinney-Freeman SL, Ross SJ, Kim PG, Chou SS, Yoshimoto M, Yoder MC, and Daley GQ (2014). Effect of developmental stage of HSC and recipient on transplant outcomes. Dev. Cell 29, 621–628. 10.1016/j.devcel.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yokomizo T, Ideue T, Morino-Koga S, Tham CY, Sato T, Takeda N, Kubota Y, Kurokawa M, Komatsu N, Ogawa M, et al. (2022). Independent origins of fetal liver haematopoietic stem and progenitor cells. Nature 609, 779–784. 10.1038/s41586-022-05203-0. [DOI] [PubMed] [Google Scholar]
- 64.Pedregosa E, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. (2011). Scikit-learn: machine learning in Python. J. Mach. Learn. Res 12, 2825–2830. [Google Scholar]
- 65.Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049. 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited in the GEO database under the accession number GSE182206.
All original code has been deposited at github and is publicly available from the date of publication of the article. URLs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD3e 145-2C11 | eBioscience | 14-0031-82; RRID:AB_2621636 |
| CD4 GK1.5 | Tonbo, BD Bioscience | 20-0041; RRID:AB_11205236 |
| CD5 53-7.3 | eBioscience | 17-0051-82; RRID: AB_469331 |
| CD8a 53-6.7 | Tonbo | 50-0081; RRID: AB_10596646 |
| CD11b M1/70 | Biolegend, Tonbo, BD Bioscience | 101206; RRID: AB_312789, 35-0112; RRID: AB_2621676, 563553; RRID: AB_2738276 |
| CD19 1D3 | eBioscience, Tonbo | 50-0193; RRID: AB_2621752 |
| CD21 7E9, 4E3 | Biolegend, eBioscicence | 123407; RRID: AB_940403, 11-0212; RRID: AB_464977 |
| CD24 M1/69 | eBioscience | 45-242-82; RRID:AB_1210701 |
| CD25 PC61.5 | Biolegend, eBioscience | 102005; RRID: AB_312854, 12-0251-82; RRID: AB_465607 |
| CD41 MWReg30 | eBioscience | 110411-82; RRID: AB_10639937 |
| CD43 S11 | Biolegend | 143219; RRID: AB_2800666 |
| CD44 IM7 | eBioscience, BD Bioscience | 25-0441-82; RRID: AB_469623, 560569; RRID: AB_1727484 |
| CD93 AA4.1 | eBioscience, Biolegend | 17-5892-82; RRID: AB_469466, 136511; RRID: AB_2275879 |
| CD117 2B8 | eBioscience, Biolegend | 25-1171-82, 105826; RRID: AB_1626278 |
| CD150 TC15-12F12.2 | Biolegend | 115910; RRID: AB_493461 |
| CD45 30-F11 | BD Bioscience, Biolegend, Tonbo | 552848; RRID: AB_394489, 103112; RRID: AB_312977, 35-0451; RRID: AB_469717 |
| CD45.1 A20 | BD Bioscience, eBioscience | 565212; RRID:AB_2722493, 47-0453-82; RRID: AB_1582228 |
| CD45.2 104 | BD Bioscience, Biolegend | 564616; RRID: AB_2738867, 109806; RRID: AB_313443 |
| IgM II/41, RMM-1 | BD Bioscience, Biolegend | 743329; RRID: AB_2741430, 406506; RRID: AB_315056 |
| IgD 11-26c.2a | Biolegend | 405720; RRID: AB_2561876 |
| B220 RA3-6B2 | Tonbo, Biolegend | 60-0452; RRID: AB_2621849, 103207; RRID: AB_312992 |
| FcERI MAR-1 | Biolegend, Tonbo | 134319; RRID: AB_10641135, 35-5898; RRID: AB_2621719 |
| F4/80 BM8 | eBioscience | 47-4801-82; RRID: AB_1548745 |
| CCR6 140706 | BD Bioscience | 561751; RRID: AB_10896986 |
| Gr-1 RB6-8C5 | Biolegend | 108428; RRID: 893558 |
| EPCR 1560 | eBioscience | 12-2012-82; RRID: AB_914317 |
| CD144 11D4.1 | BD Bioscience | 562242; RRID: AB_2737608 |
| CD48 HM48-1 | Biolegend, BD Bioscience | 103404; RRID: AB_313019, 740236; RRID: AB_2739984 |
| CD23 B3B4 | eBioscience | 25-0232-82; RRID: AB_469604 |
| CD127 A7R34 | Biolegend, eBioscience | 135012; RRID: AB_1937216, 12-1271-82; RRID: AB_465844 |
| Ter119 TER-119 | Tonbo, Biolegend, eBioscience | 65-5921; RRID: AB_2935630, 116223; RRID:AB_2137788, 11-5921-82; RRID:AB_465311 |
| Sca-1 D7 | eBioscience, Biolegend | 25-5981-82; RRID: AB_469669, 108124; RRID:AB_893615 |
| CD34 RAM34 | eBioscience | 11-0341-82; RRID: AB_465021 |
| Thy1.2 30-H12 | Biolegend, eBioscience | 105328; RRID: AB_10613293, 11-0902-82; RRID:AB_465154 |
| CD184 2B11 | eBioscience | 12-9991-82; RRID: 891391 |
| Chemicals, peptides, and recombinant proteins | ||
| Tamoxifen | Sigma | T5648 |
| Progesterone | Sigma | P0130 |
| Recombinant mouse IL-7 | PeproTech | 217-17 |
| Recombinant mouse SCF | PeproTech | 250-03 |
| Recombinant mouse Flt3-ligand | PeproTech | 250-31L |
| Collagenase Type I (0.25%) | STEMCELL Technologies | 07902 |
| Critical commercial assays | ||
| SMART-Seq V4 Ultra Low RNA Kit | Takara | # 634898 |
| MethoCult M3630 | STEMCELL Technologies | 03630 |
| Deposited data | ||
| scRNA-sequencing | GEO database | GSE182206 |
| Experimental models: Cell lines | ||
| OP-9 stromal cells | Dr. Toru Nakano at Osaka University, Japan | ATCC CRL-2749 |
| Experimental models: Organisms/strains | ||
| Fgd5ZsGreen-CreERT2 | C57BU6N-Fgd5tm3(cre/ERT2)Djr/J | Jackson no. 027789 |
| Cdh5(PAC)-CreERT2 | C57BL/6-Tg(Cdh5-cre/ERT2) | Taconic no. 13073 |
| Rosa-TdTomato | B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | Jackson no. 007909 |
| NSG | NOD.Cg-PrkdcscidIl2rgtm1wjl/Szj | Jackson no. 005557 |
| Boy/J | B6.SJL-PtprcaPepcb/BoyJ | Jackson no. 002014 |
| Software and algorithms | ||
| Fastqc | Babraham Institute | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| STAR | PMID: 23104886 | https://github.com/alexdobin/STAR |
| Htseq-count | PMID: 25260700 | https://github.com/htseq/htseq |
| SC3 | PMID: 28346451 | http://bioconductor.org/packages/release/bioc/vignettes/SC3/inst/doc/SC3.html |
| Monocle2 | PMID: 28825705 | http://cole-trapnell-lab.github.io/monocle-release/docs/ |
| CellRanger | PMID: 28091601 | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/using/tutorials |
| Scikit-learn | https://scikit-learn.org/stable/ | |
| Velocyto.py | PMID: 30089906 | http://velocyto.org/velocyto.py/tutorial/analysis.html |
| velocyto.R | https://github.com/velocyto-team/velocyto.R | |
| scVelo | Volker Bergen | https://scvelo.readthedocs.io |
| Script used for Figures 5A and 5D | This paper | https://github.com/seitalab/hsc_independent |
| Approximate Bayesian Computation (Figure S6) | This paper | https://github.com/djhshih/analysis-lineage-tracing-b1a |


