Abstract
INTRODUCTION:
Ticks have evolved into a monophyletic group of highly adapted blood-feeding ectoparasites that originated from a clade of free-living scavenger mites nearly 225 million years ago. Unlike most geographically confined tick species that prefer a single vertebrate host, Ixodes spp. can parasitize many vertebrates and transmit diverse pathogens. Ixodid ticks undergo only three feeding events during their multiyear lifespan, ingesting blood meals that are nearly 100 times their weight. Their characteristic physiological adaptations were likely shaped by their sophisticated hematophagy and associations with coevolving vertebrate hosts. The molecular basis of how ticks maintain their complex postembryonic developmental program as well as their vectorial competence remains unclear. Ticks contain a functional JAK–STAT signaling cascade that induces robust antibacterial responses capable of limiting the proliferation of tick-borne pathogens. The pathway is activated in many arthropods by cytokine-like molecules such as Unpaired (UPD). However, the Ixodes scapularis genome is unusually devoid of recognizable UPD orthologs.
RATIONALE:
We recently discovered a cross-species cell signaling pathway in which a mammalian interferon (IFN-γ) ingested with the tick blood meal activates the Ixodes JAK–STAT pathway, generating potent microbicidal activities within the vector. To further elucidate a parallel interferon-like defense system in ticks, we have explored the identity and characteristics of the tick receptor for IFN-γ to uncover the mechanisms and biological significance of a cross-species cell signaling cascade that extends from mammals to their ectoparasites.
RESULTS:
We identified an I. scapularis receptor, Dome1, which binds with high selectivity and affinity to vertebrate (mouse, human, and avian) IFN-γ. Unlike Dome orthologs in other arthropods, including non-Ixodes ticks, Dome1 features unique extracellular regions found in vertebrate cytokine receptors. Dome1 colocalizes with IFN-γ on the luminal surface of the tick gut epithelium. The receptor is induced by ingested IFN-γ and regulates microbicidal responses orchestrated by the JAK–STAT pathway. Unexpectedly, Dome1 not only augmented tick immune responses, but was also required for I. scapularis development. Systemic Dome1 knockdown persisted from subadult to female I. scapularis and their offspring, and severely impaired tick fecundity and postembryonic development, reflected by failure to molt, loss of bilateral symmetry, and malformed internal organs and appendages, including missing legs, mouthparts, and anal pore structures. The Dome1–JAK–STAT pathway, present in most Ixodid tick genomes, directed the maintenance of the gut proteome and microbiome, in addition to supporting the regeneration and proliferation of gut cells, including stem cells. Dome1–JAK–STAT signaling dictated tick metamorphosis and organ development through the Hedgehog and Notch–Delta biochemical networks, ultimately impacting the ability of Ixodes to transmit Borrelia burgdorferi, the Lyme disease pathogen, to naïve murine hosts.
CONCLUSION:
We identify how certain conserved metazoan cell signaling pathways flexibly adapt vertebrate IFN-γ for use in arthropod immunity and development through the Dome1 receptor. Our studies highlight the evolutionary dependence of the major blood-feeding arthropod I. scapularis on mammalian hosts through cross-species signaling mechanisms that extend from mammals to their ectoparasites and influence arthropod immunity, vectorial competence, and development.
Ancestral signaling pathways serve critical roles in metazoan development, physiology, and immunity. We report an evolutionary interspecies communication pathway involving a central Ixodes scapularis tick receptor termed Dome1, which acquired a mammalian cytokine receptor motif exhibiting high affinity for interferon-gamma (IFN-γ). Host-derived IFN-γ facilitates Dome1-mediated activation of the Ixodes JAK–STAT pathway. This accelerates tick blood meal acquisition and development while upregulating antimicrobial components. The Dome1–JAK–STAT pathway, which exists in most Ixodid tick genomes, regulates the regeneration and proliferation of gut cells—including stem cells—and dictates metamorphosis through the Hedgehog and Notch–Delta networks, ultimately affecting Ixodes vectorial competence. We highlight the evolutionary dependence of I. scapularis on mammalian hosts through cross-species signaling mechanisms that dually influence arthropod immunity and development.
Graphical Abstract
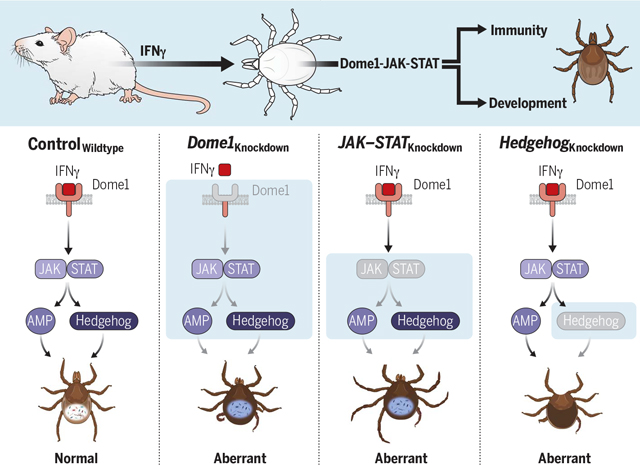
Mammalian-arthropodal crosstalk integrates vector immunity and development. The Ixodes scapularis protein Dome1 recruits mammalian IFN-γ from the tick’s blood meal. The interaction activates the tick JAK–STAT pathway, triggering signaling intermediaries, including an antimicrobial protein and Hedgehog protein, which affects tick metamorphosis, gut homeostasis, and pathogen transmission. Knockdowns of pathway components highlight the biological significance of cross-species signaling between mammals and their ectoparasites, which converge to regulate both arthropod immunity and development.
Ticks thrive in nature through prolonged periods of environmental extremes and metabolic quiescence (1). During their multiyear lifespan, Ixodes ticks undergo three feeding cycles, marked by a nearly 100-fold increase in body volume (2). Blood meal ingestion is aided by bursts of cellular proliferation, differentiation, and hypertrophy, which are central to organ development. Host specialization in ticks (3), including highly evolved hematophagy, supports the tick’s unique physiological adaptations, particularly blood meal ingestion and storage, survivability through extended intermolt periods, and the maintenance of complex reproductive and developmental programs (4), the molecular aspects of which remain unknown.
Arthropod vectors transmit diverse human pathogens, accounting for 17% of all infectious diseases around the globe (5). Among all arthropods, ticks constitute the most ancient and predominant group of vectors as they can parasitize almost any vertebrate class, including primeval reptiles (1). Although most tick species are confined within a given geographical region and prefer a single host, Ixodes spp. parasitize various hosts across multiple continents (6). In the US, ~95% of vector-borne diseases, including common infections similar to Lyme disease, are associated with ticks—particularly Ixodes scapularis (7). Despite their importance, the molecular aspects of Ixodes postembryonic development and biology remain enigmatic, partly because of their long life cycle, genetic intractability, and phylogenetic distance from model arthropods. Comparative genomics studies have also suggested unique molecular properties within ticks (8). The Ixodes genome encodes known components with puzzling omissions (9–11). For instance, the immunodeficiency (IMD) signaling pathway is operative in I. scapularis, but its central molecule, IMD, and a few other components do not appear to be present (10). Ticks also have a functional JAK–STAT pathway, which evolved in the earliest branching events of metazoan animals (12), comprising a few principal components (13). However, the biological effects of its activation are complicated by the use of several ligands, including cytokines (14), and its interaction with other pathways (15). Most arthropods produce their own cytokine-like molecules. Unpaired (UPD), for example, can bind a transmembrane receptor called Dome, which activates the JAK–STAT pathway that can support arthropod immunity (16) and development (17). Dome proteins, despite showing high sequence diversification, feature an extracellular region with a cytokine-binding homology module and multiple fibronectin type-III domains (18). Although flies encode multiple UPD ligands for Dome (17), there is a surprising absence of UPD orthologs in the I. scapularis genome (9, 19). Here we report that an I. scapularis Dome ortholog, which binds mammalian interferon-gamma (IFN-γ) acquired from the blood meal, triggers downstream tick JAK–STAT signaling cascades that integrate critical episodes of vector immunity and development. This study therefore highlights the adaptive flexibility of ancestral signal transduction pathways (12, 20) through the co-option of cross-species signaling between mammals and their ectoparasites.
Results
Identification of a mammalian IFN-γ-binding tick receptor
We previously found that mammalian IFN-γ acquired during a tick’s bloodmeal can trigger a potent antimicrobial response within the vector (21). As hematophagy in ticks originated with primeval vertebrates and evolved to include modern mammals (4), we explored whether the I. scapularis genome acquired a cytokine receptor that binds IFN-γ, such as through lateral gene transfer (22), with the ability to trigger immune signaling in ticks. We adopted an unbiased approach using coimmunoprecipitation (Co-IP) assay with whole tick lysates and mammalian IFN-γ as bait, coupled with mass spectrometry. We found that mouse IFN-γ bound a tick protein, annotated as B7P6I6 and termed hereafter as Dome1 (ISCW001458), which has similarity to Drosophila Dome (fig. S1). Basic Local Alignment Search Tool analysis revealed that the Ixodes genome encodes four additional Dome orthologs (23) bearing 23 to 67% identity to Dome1, termed hereafter as Dome2 (ISCW013495), Dome3 (ISCW013496), Dome4 (ISCW008121), and Dome5 (ISCW016699) (fig. S2) (9), although these were not identified in our assay. In addition to transmembrane and cytoplasmic regions, Dome1 features a large extracellular amino-terminal region containing a signal peptide, immunoglobulin (IG) domain, and three putative fibronectin type-III domains (Fig. 1A). The extracellular region also contains a mammalian cytokine-binding motif, the interfer-bind PF09294 (24, 25), present in rodents (natural hosts of I. scapularis) and humans (incidental hosts) but absent in non-Ixodes arthropods, including flies, mites, spiders, and other ticks (Fig. 1B). A ClustalW2 sequence alignment revealed homology between the Dome1 interfer-bind motif and bona fide IFN-γ receptors in mice and humans (Fig. 1C), as further supported by a phylogenetic tree analysis of basal eukaryotic lineages (Fig. 1D). The Dome1 interfer-bind motif was widespread across phyla, including a limited selection of bacterial species (fig. S3).
Fig. 1. Identification and characterization of Ixodes scapularis Dome1.
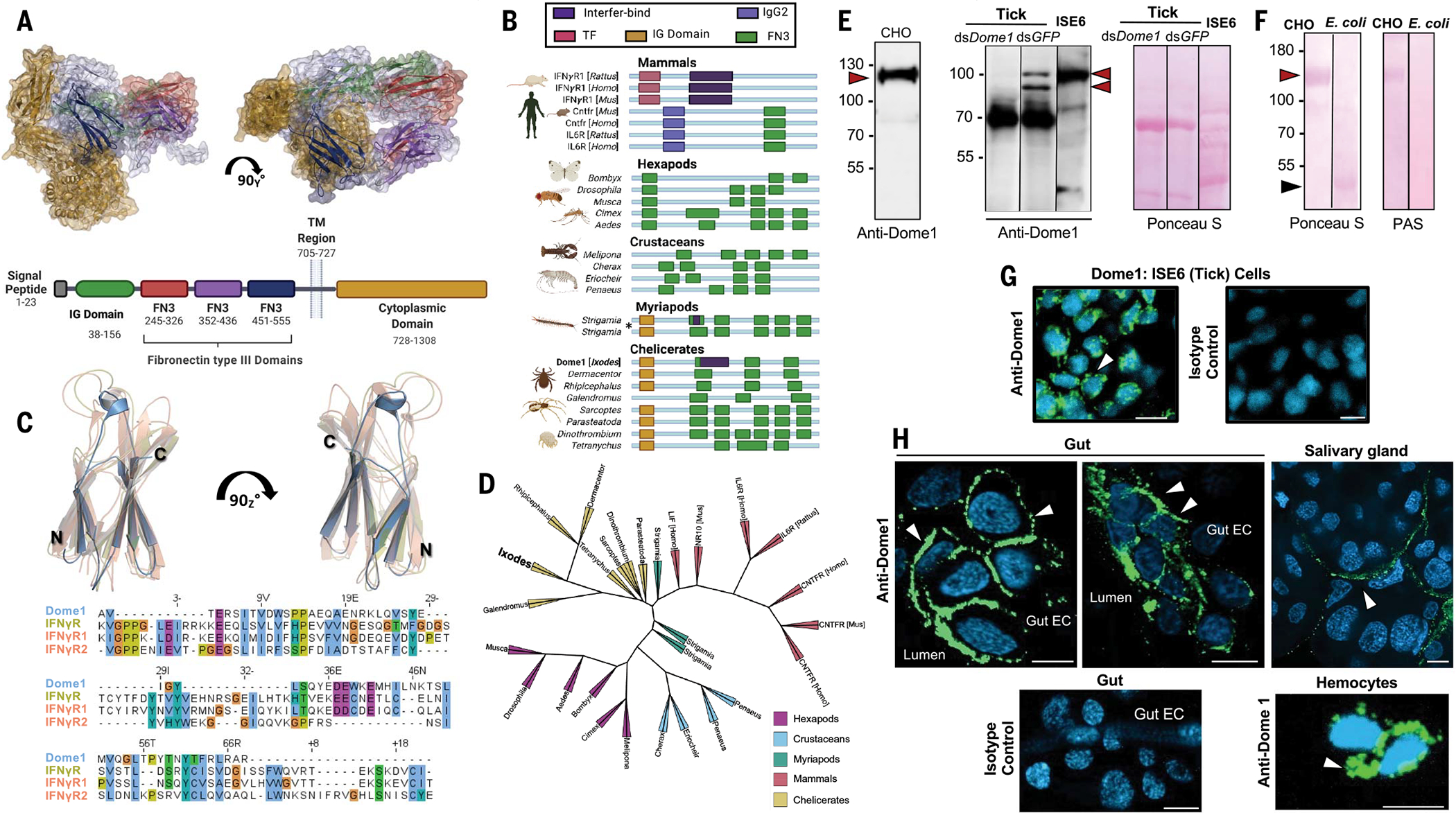
(A) A homology-guided model of Dome1 and a diagram showing its features and domains. The domains [IG, three fibronectin type-III (FN3), transmembrane (TM), and cytoplasmic] are color-coded in the 3D structure (top) and schematic diagram (bottom). (B) Acquisition of mammalian interfer-bind motif in Dome1. Sequence comparison of identifiable domains in mammalian cytokine receptors and Dome proteins from selected species of arthropod subphyla are shown (asterisk: two orthologs in the same species). (C) Predicted structure and sequence alignment of the cytokine (interfer-bind)–binding motifs from Dome1 (dark-colored helices) and IFN-γ receptors (IFNGR) from mice and humans (light-colored helices). IFNGR (Mus musculus) and IFNGR1 and IFNGR2 (Homo sapiens) are superimposed to show alignment. (D) Phylogenetic tree of basal eukaryotic lineage highlighting conservation of Dome1 through arthropod subphyla and selected mammals. (E) Dome1 in a mammalian cell line (CHO), I. scapularis cell line (ISE6), and nymphal tick lysates from partially (24 hour) fed ticks. Lysates from control (dsGFP-injected) and Dome1-knockdown (dsDome1-injected) ticks were used. Unlike Dome1 in CHO or ISE6 cells, native Dome1 in whole tick lysates appeared as multiple proteins (arrowheads). Protein loading is shown by Ponceau S staining. (F) Dome1 is a glycoprotein. Electrophoresed full or truncated ectodomains (red and black arrowheads, respectively) were visualized by periodic acid–Schiff (PAS) staining. Protein loading is indicated by Ponceau S. (G) Dome1 localization (arrowhead) in tick cells. Nuclei were labeled with DAPI. (H) Detection of Dome1 in nymphal guts, salivary glands, and hemocytes by confocal immunofluorescence microscopy. More Dome1 immunoreactivity (arrowheads) was detectable on the surface of gut epithelial cells facing the lumen (upper left and middle panels) and the surface of hemocytes (lower right panel). Results are representative of three independent biological replicates. For [(G) and (H)], IgG from normal mouse sera served as isotype controls; Dome1: green; DAPI: blue; scale bar: 10 μm.
As Dome1 was predicted to be a glycoprotein of 1308 amino acids with a large extracellular domain (~704 residues), we generated recombinant versions of the entire ectodomain and additional truncations, using mammalian and bacterial expression systems, respectively, and generated antibodies in mice (fig. S4, A to C). The recombinant Dome1 ectodomain produced in transfected mammalian cells migrated as a single 125-kDa protein. By contrast, native Dome1 in the ISE6 tick cell line or in fed nymphal tick guts was detected as multiple proteins of 70 to 100 kDa, suggesting additional post translational processing (Fig. 1E). The Dome1 ectodomain housed several N- and O-linked glycosylation sites (fig. S4A) and was glycosylated only when produced in mammalian cells, but not in Escherichia coli (Fig. 1F). Confocal immunofluorescence analysis indicated that native Dome1 was localized on the surfaces of ISE6 cells (Fig. 1G) and tick gut epithelial cells facing the lumen (Fig. 1H). The protein did not appear to be exclusive to the gut, as immunoreactivity was detected in other tick samples, including hemocytes and the salivary gland (Fig. 1H).
We next confirmed the interaction of recombinant Dome1 with mammalian IFN-γ. A yeast two-hybrid assay showed a selective interaction between mouse IFN-γ and Dome1 (Fig. 2A). In a pull-down assay, Dome1 interacted with IFN-γ (Fig. 2B). A microtiter well-based assay indicated that immobilized Dome1 bound IFN-γ, but not tumor necrosis factor (TNF) (Fig. 2C), reaching saturation with the addition of IFN-γ (Fig. 2D). The binding kinetics assessed by biolayer interferometry (BLI) showed that Dome1 bound IFN-γ with high affinity (KD of ~40 nM) (Fig. 2E). Notably, Dome1 also bound chicken and human IFN-γ with high affinities (Fig. 2, F to H). IFN-γ and Dome1 were found to colocalize on the surface of ISE6 cells as well as on the tick gut surface in vivo (Fig. 2I). Thus, Dome1 specifically binds mammalian IFN-γ on the luminal surface of the tick gut epithelium.
Fig. 2. Dome1 specifically interacts with IFN-γ.
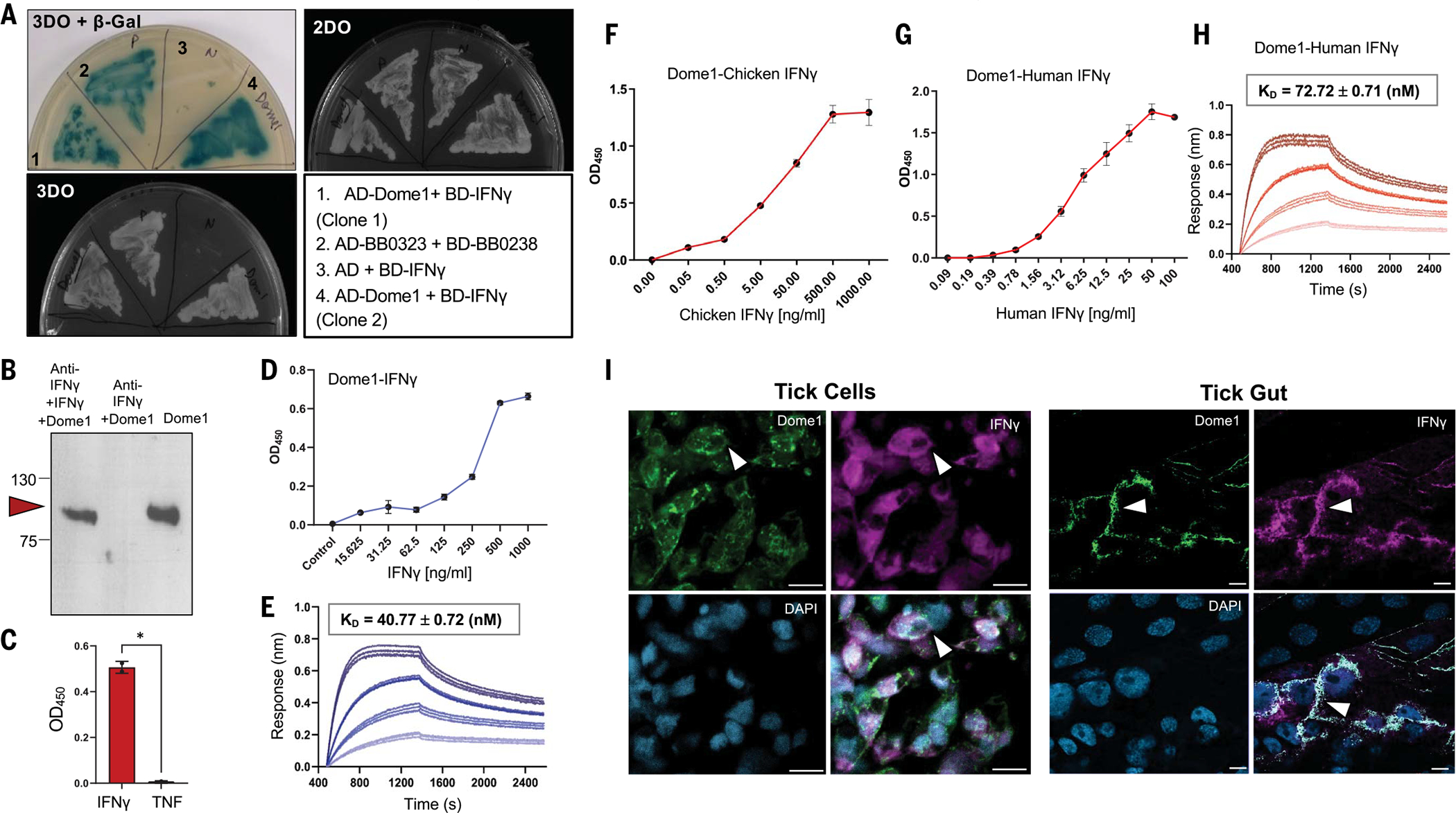
(A) Yeast two-hybrid assay demonstrates specific interaction between Dome1 and IFN-γ. Although all yeast cells grew on the double-dropout (2DO) media, only those with reporter gene activation grew on triple-dropout (3DO) media and expressed γ-Gal (blue color), confirming interactions between Dome1-IFN-γ or positive controls. (B) Anti-IFN-γ antibody pulls down soluble flag-tagged Dome1 in the presence of IFN-γ (left lane), but not in its absence (middle lane), confirming a Dome1–IFN-γ interaction. The pulled-down Dome1 was detected by immunoblot using anti-flag antibodies (right lane). (C) A microtiter well-based assay demonstrates a specific Dome1–mouse IFN-γ interaction. By contrast, no interaction was observed between mouse TNF and Dome1. Each data point shows the average of duplicate wells from one of three independent biological replicates, with similar results. (D) Dose-dependent binding of mouse IFN-γ to recombinant Dome1. The assay was performed as in (C), suggesting a dose-dependent increase in Dome1–IFN-γ interaction. (E) Bio-layer interferometry (BLI) sensorgrams of Dome1–IFN-γ interaction. The affinity constant of this interaction is presented above the sensorgram. (F and G) Dose-dependent binding of recombinant chicken IFN-γ (F) or human IFN-γ (G) to recombinant Dome1. These assays were performed as detailed in (C). (H) BLI sensorgrams of Dome1–human IFN-γ interaction, confirming a high-affinity interaction with human IFN-γ, analogous to those highlighted in the mouse BLI data. (I) Dome1 and IFN-γ colocalization on the surface of tick cells (left panel) and in tick gut cells (right panel). Recombinant mouse IFN-γ was incubated with ISE6 cells or dissected tick guts and probed with specific antibodies against Dome1 and IFN-γ, followed by FITC- or Alexa Fluor 568-labeled secondary antibodies. In both tick cells and gut tissues, IFN-γ colocalized with endogenous Dome1 protein (arrows). Error bars denote mean ± standard deviation (SD). Results are representative of 2 to 4 independent experiments. White bar: 10 μm. *P < 0.05, Student’s t test.
IFN-γ induces the microbicidal JAK–STAT signaling pathway in ticks through Dome1
The transmembrane Dome receptor activates the JAK–STAT signaling pathway (17). We therefore explored whether Dome1 induces the Ixodes JAK–STAT pathway, which can trigger microbicidal responses. Dome1 mRNA (Fig. 3A) and protein (fig. S4D) were both induced in feeding nymphs. Expression was enhanced during nymphal infestation on Borrelia burgdorferi-infected hosts (B. burgdorferi is a common Lyme disease agent) (fig. S4E), which produce systemic IFN-γ (Fig. 3A, rightmost panel and fig. S6A) acquired in the tick’s blood meal. The induction of Dome1 (Fig. 3A) and other known immune components (21) appeared specific to spirochetal infection as their expression remained unaltered in ticks exposed to another pathogen, Anaplasma phagocytophilum (fig. S5). Although Dome1 mRNA was detectable in multiple tick organs (Fig. 3B), protein expression was most obvious in the gut (fig. S6B). Using mutant mice that either could produce IFN-γ (Ifngr1-knockout) or were deficient in IFN-γ (Ifng-knockout), we found that Dome1 was induced by interferon in the tick gut but not in other organs (Fig. 3, C and D). Host IFN-γ influenced blood meal engorgement in ticks, as most nymphs that parasitized animals lacking the cytokine fed to repletion at least 24 hours later than those that engorged on IFN-γ–producing mice (Fig. 3E).
Fig. 3. Dome1, induced by host-derived IFN-γ, regulates tick borreliacidal responses through the JAK–STAT pathway.
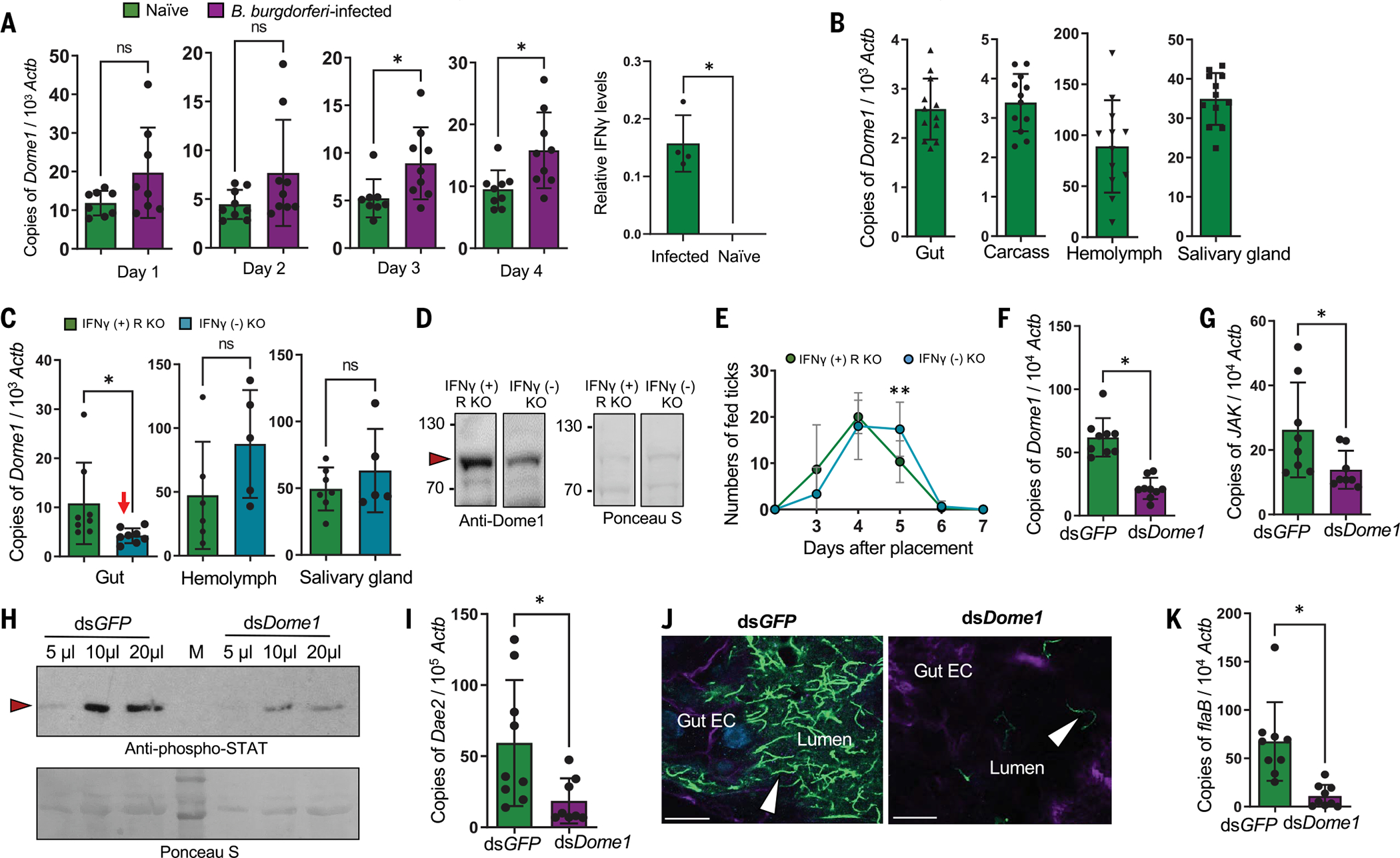
(A) Dome1 is induced during tick engorgement and B. burgdorferi infection. The reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis of Dome1 transcripts in nymphs fed on naïve or infected mice (left panel). The right panel shows IFN-γ levels in murine blood. (B) Expression of Dome1 in various tick tissues. Nymphs parasitized mice for 48 hours, and tissues were processed for Dome1 transcripts and normalized against tick Actb levels. (C) Ifng deficiency down-regulates Dome1 in tick guts (arrow). Groups of three Ifngr1-knockout mice, which produce the cytokine (IFN-γ + R KO) or Ifng-knockout mice (IFN-γ −KO) were parasitized by nymphs (30 ticks per group) for 48 hours. Dome1 transcripts were analyzed in tick tissues. (D) IFN-γ deficiency downregulates Dome1 in fed tick gut. Protein loading is indicated by Ponceau S staining. (E) Influence of host IFN-γ on blood meal acquisition by ticks. In the absence of IFN-γ, engorgement time is delayed at day 5, when nymphs ingest the major portion of the blood meal. **P < 0.05, determined using two-tailed Student’s t test. (F) RNAi-mediated Dome1 knockdown by microinjection of unfed nymphs (20 ticks per group), which then fed on Borrelia-infected mice for 48 hours. Dome1 mRNA levels were measured by RT-qPCR. (G to I) Dome1 knockdown reduces levels of JAK transcripts (G); STAT phosphorylation (“M” and arrowhead denote molecular weight markers and phosphorylated STAT protein, respectively) (H); and Dae2 transcripts (I) in 48-hour-fed ticks. (J) Dome1 knockdown affects colonization of B. burgdorferi (green, arrowhead) in the tick gut; nuclei and actin are labeled with DAPI (blue) and rhodamine phalloidin (violet). The image represents one of three biological replicates with similar results. (K) RT-qPCR analysis of B. burgdorferi, assessed by measuring flaB transcripts normalized to Actb levels. Results represent two to five independent experiments, where quantitative data are shown as individual data points; error bars show the means ± SDs (n = 9 to 30). White bar: 10 μm. *P < 0.05, determined using two-tailed Mann-Whitney U test; n.s., not significant.
To further understand the biological significance of interferon signaling in ticks—particularly the roles of its arthropod receptor, Dome1—we generated Dome1-knockdown ticks through RNA interference (RNAi) (fig. S6B). The microinjection of double-stranded RNA (dsRNA) targeting Dome1 in the tick gut substantially reduced its expression 48 hours after feeding in comparison to control (dsGFP) nymphs (Fig. 3F). Dome1 knockdown, which was concurrently observed in the salivary glands and hemolymph (fig. S6B), did not affect the expression of other Dome homologs, except for a small but detectable decrease in Dome5 transcripts (fig. S7). The impact on Dome5 protein levels was minimal, however (fig. S7). To assess whether Dome1 deficiency impaired the activation of the JAK–STAT pathway, we allowed nymphal ticks to parasitize B. burgdorferi-infected mice. The knockdown of Dome1 resulted in lower levels of JAK mRNA (Fig. 3G), STAT phosphorylation (Fig. 3H), and transcripts of antimicrobial gene Dae2 (Fig. 3I). In addition, Dome1 deficiency reduced B. burgdorferi levels in the guts of knockdown ticks (Fig. 3, J and K). Higher reactive oxygen species (ROS) activity was detected in Dome1-knockdown guts, as well as the altered expression of representative enzymes associated with oxidative stress, which may have contributed to the decreased spirochete levels (fig. S8). The regulation of tick microbicidal immune responses was specifically triggered by Dome1 and did not involve other homologs, such as Dome5. Indeed, a separate knockdown of Dome5 had no effect on tick immune responses, biology, or vectorial competence (fig. S9). Thus, Dome1 can recognize mammalian IFN-γ and trigger a cross-species immune signaling cascade through the JAK–STAT pathway, ultimately influencing B. burgdorferi levels.
Dome1 contributes to tick intermolt development through the JAK–STAT pathway
Dome1-knockdown ticks, despite complete blood meal acquisition (Fig. 4A), began to display reduced body weights and discolored cuticles 10 to 20 days after feeding (Fig. 4B). Dome1 mRNA and protein deficiency was maintained throughout the intermolt stage and in newly molted adults (fig. S6C). Most of the Dome1-knockdown nymphs were unable to molt after feeding, in contrast to control ticks (Fig. 4C). This suggested that Dome1—in addition to orchestrating mammalian interferon–induced microbicidal immune responses within the vector—also plays a critical role in tick intermolt development. To better understand the role of Dome1 in subadult ticks, we performed systemic RNAi in newly engorged larvae. Dome1-knockdown intermolt larvae showed severe growth defects 20 days after dsRNA treatment and blood meal engorgement, with most ticks unable to molt (Fig. 4D). By contrast, nearly all control larvae molted to nymphs. The few Dome1-knockdown ticks that were able to molt displayed a loss of bilateral symmetry and morphological defects, ranging from malformed mouthparts (palps and/or hypostome) to leg abnormalities, including unequal numbers of limbs on either side of the body (three versus four limbs) and rudimentary or underdeveloped legs without a base (coxa), in addition to darker abdomens and incomplete ecdysis (Fig. 4, E and F, and fig. S10).
Fig. 4. Dome1 is essential for optimal tick metamorphosis.
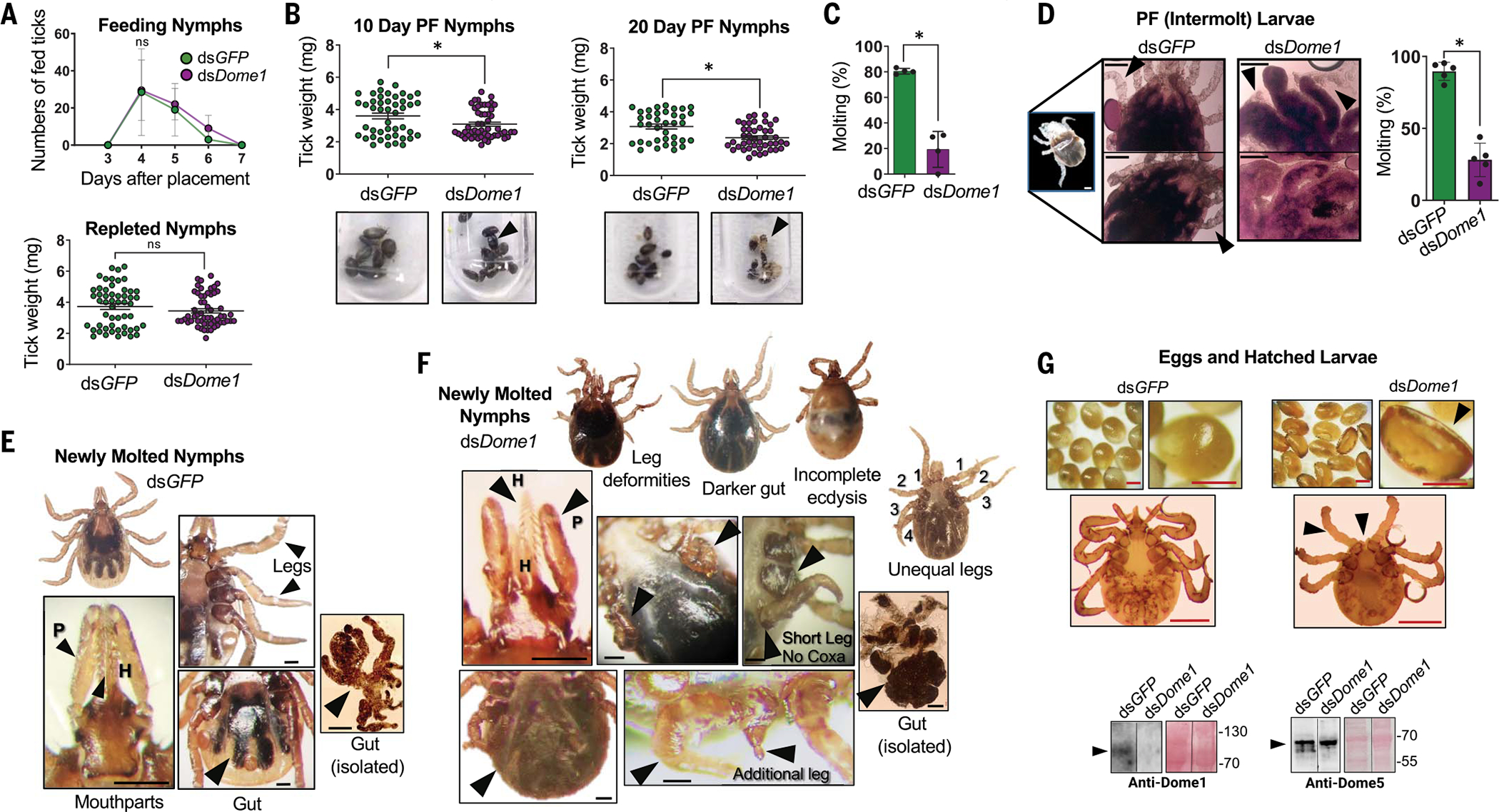
(A) Host blood meal acquisition in Dome1-knockdown ticks. Number of fed ticks (left panel) and tick engorgement weights (right panel) are shown. (B) Intermolt ticks. Compared with controls, Dome1-knockdown post-fed (PF) ticks displayed lower weights (upper panels) and different body colors with exposed exuviae or even death (lower panels, arrowheads). (C) Impairment of molting success in Dome1-knockdown nymphal ticks, as assessed by the percentage of molted ticks. (D) Dome1 silencing in fed larvae impairs development and molting. The control intermolt ticks (left panels) at 20 days post-fed (PF) reveal newly formed legs (arrowheads) which were absent or malformed in Dome1-knockdown groups (right panels, arrows). The percentage of molted ticks is shown in the rightmost panel. (E and F) Compared with controls (E), transstadial Dome1-knockdown ticks (F) showed deformities (arrows), including in hypostome (H) and palps (P), uneven legs with unequal lengths, stunted legs without coxa, and darker abdomens with incomprehensible gut diverticula. (G) Dome1 is essential for fecundity and larval development. The data represent an experiment where 80 adult ticks were microinjected with dsDome1 or dsGFP (control) RNA and allowed to engorge on groups of rabbits. Dome1 deficiency resulted in abnormal egg and larval development (top right panels), compared with the controls (top left panels). Dome1 knockdown was sustained in the mature eggs, which were analyzed for Dome1 (bottom left panels) or Dome5 (bottom right panels) protein levels by immunoblot. See additional results for hatched larvae in fig. S11. Results are representative of two to five independent experiments where quantitative data are shown as individual data points; error bars show the means ± SDs (n = 6 to 50). Black or white bar: 100 μm; red bar: 50 μm. *P < 0.05, determined using two-tailed Mann-Whitney U test; n.s., not significant.
To evaluate whether Dome1 was critical for fecundity and early larval development, Dome1-deficient adult females were allowed to parasitize rabbits and subsequently lay eggs. Similar to nymphs (Fig. 4A), a knockdown in Dome1 did not influence the repletion time or engorgement weight of fed adults, but did affect the quality, number, and size of the egg masses as well as the emergence of larvae, many of which were born with growth defects (Fig. 4G and fig. S11), similar to those previously observed in nymphs.
Dome1, but not Dome5, was the main driver of these phenotypic defects because eggs and emerged larvae from Dome1-knockdown females produced Dome5 protein at levels comparable to control ticks, whereas Dome1 protein remained largely undetectable (Fig. 4G and fig. S11). These Dome1-regulated developmental abnormalities were likely driven by the Ixodes JAK–STAT pathway, as the separate and systemic RNAi silencing of JAK or STAT transcripts yielded comparable effects on tick development (Fig. 5 and fig. S12, A to D). Although Dome1-knockdown ticks exhibited the most prominent body deformities (Fig. 4, E and F), some similarities were observed in the overall appearances of JAK- or STAT-knockdown ticks, including the presence of darkened abdomens (fig. S12B). Moreover, newly molted nymphs in which the expression of Dome1, JAK, or STAT was separately knocked down also displayed comparable developmental defects, including shorter hypostomes and palps, as well as deformities in the palp bases, legs, and anal pores (Fig. 5 and fig. S12, B to D). Cross-sections of whole nymphal ticks revealed a completely digested blood meal with normal development in control ticks, whereas Dome1-, JAK-, or STAT-knockdown ticks presented enlarged and abnormally developed bodies surrounding a large bolus of blood meal, suggesting impaired digestive activities in the gut (Fig. 5H). Thus, Dome1 contributes to tick intermolt development through the JAK–STAT pathway.
Fig. 5. The Ixodes JAK–STAT pathway contributes to Dome1-mediated tick metamorphosis.
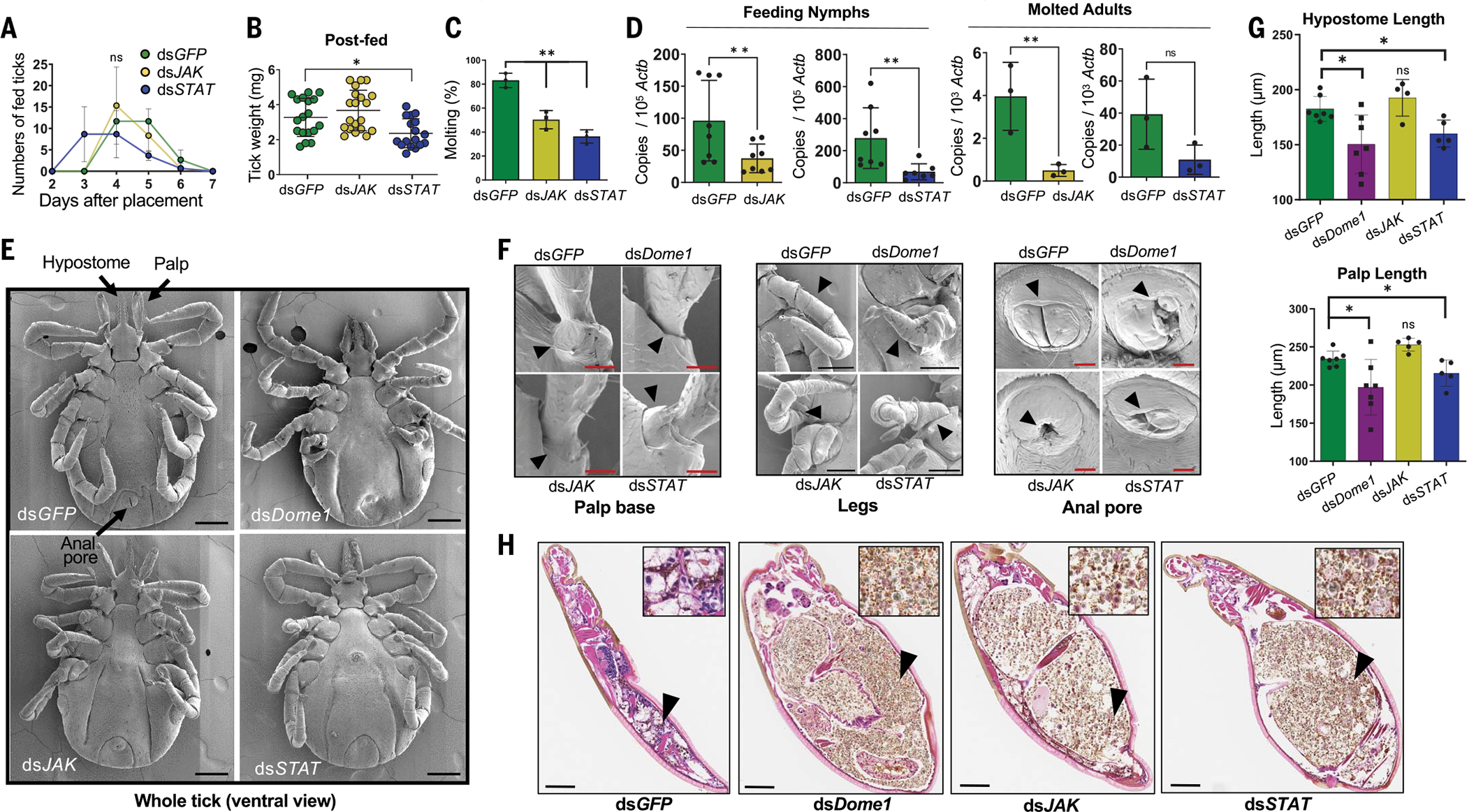
(A) Knockdown of STAT, but not JAK, impairs tick attachment. Ticks (25 nymphs per group) were microinjected with target dsRNA and placed on naïve mice. Detached fed ticks were enumerated. (B) STAT knockdown impairs tick weight. Ticks were weighed after feeding to repletion. (C) Molting success of knockdown ticks. Compared with controls, a reduction in molting was evident in JAK- and STAT-knockdown ticks. **P < 0.05, determined using two-tailed Student’s t test. (D) RNAi-mediated JAK and STAT knockdown effects are maintained transstadially. The ticks were analyzed as 48-hour-fed nymphs and as molted adults. **P < 0.05, determined using two-tailed Student’s t test. (E) Scanning electron micrographs of knockdown ticks. Unlike normal structures in control nymphs (arrows), Dome1-, JAK-, and STAT-knockdown ticks presented abnormal appearances, most noticeably in their malformed mouthparts, legs, and anal pores. (F) Close-up view of the morphological defects highlighted in (E), indicating defective palp bases, legs, and anal pores. (G) Quantitative assessment of morphological defects highlighted in (E), denoting shorter hypostome and palps in Dome1- and STAT-knockdown ticks. Additional results are presented in figs. S10 and S12. (H) Histological analysis of ticks. Engorged larvae were subjected to standard H&E staining. Enlarged and abnormally developed bodies surrounding a large bolus of improperly digested blood meal were seen in Dome1-, JAK-, or STAT-deficient ticks. The inset shows a magnified view of the gut contents (arrow), revealing the presence of the remnants of blood cells and microbes, which are predominant in all groups except for the control (dsGFP) ticks. Results are representative of two to three independent experiments where quantitative data are shown as individual data points; error bars show the means ± SDs (n = 3 to 30). Black bar: 100 μm; red bar: 20 mm. *P < 0.05, determined using two-tailed Mann-Whitney U test; n.s., not significant.
Gut biology and microbial homeostasis are maintained by Dome1
As Dome1 deficiency affected multiple aspects of tick immunity and development, we assessed possible alterations in the gut proteome and microbial homeostasis, in addition to the influence of B. burgdorferi on Dome1 signaling. Both Dome1 knockdown and B. burgdorferi infection affected the tick gut proteome (Fig. 6A and tables S1 to S4). Compared with naïve hosts, the impact of Dome1 knockdown in ticks feeding on B. burgdorferi-infected hosts was reflected by the more robust down-regulation of several proteins known to support organ development, homeostasis, replication, and immune responses (table S4). Thus, Dome1 signaling is more critical for the correct functioning of the gut when B. burgdorferi is present. Dome1 regulated pathogen levels and influenced the STAT protein (Fig. 3), which is known to affect the acellular gut barrier known as the peritrophic matrix (PM), important for gut homeostasis (26). Therefore, we examined the influence of Dome1 deficiency on PM function and the gut microbiome. The expression of gut peritrophin gene PM5 was reduced in Dome1-knockdown ticks (Fig. 6B). The PM barrier was also compromised, as indicated by the altered permeability of the fluorescein-conjugated high molecular weight dextran (Fig. 6C). Furthermore, Dome1 knockdown affected the diversity of the gut microbiome, including predominant bacterial genera such as Rickettsia, irrespective of B. burgdorferi infection (Fig. 6D). JAK–STAT-suppressed ticks showed similar effects on tick biology and gut microbial homeostasis (fig. S13), indicating that they constitute key drivers of Dome1-induced responses. Thus, Dome1 regulates gut homeostasis and microbial diversity.
Fig. 6. Dome1 supports gut homeostasis in feeding ticks.
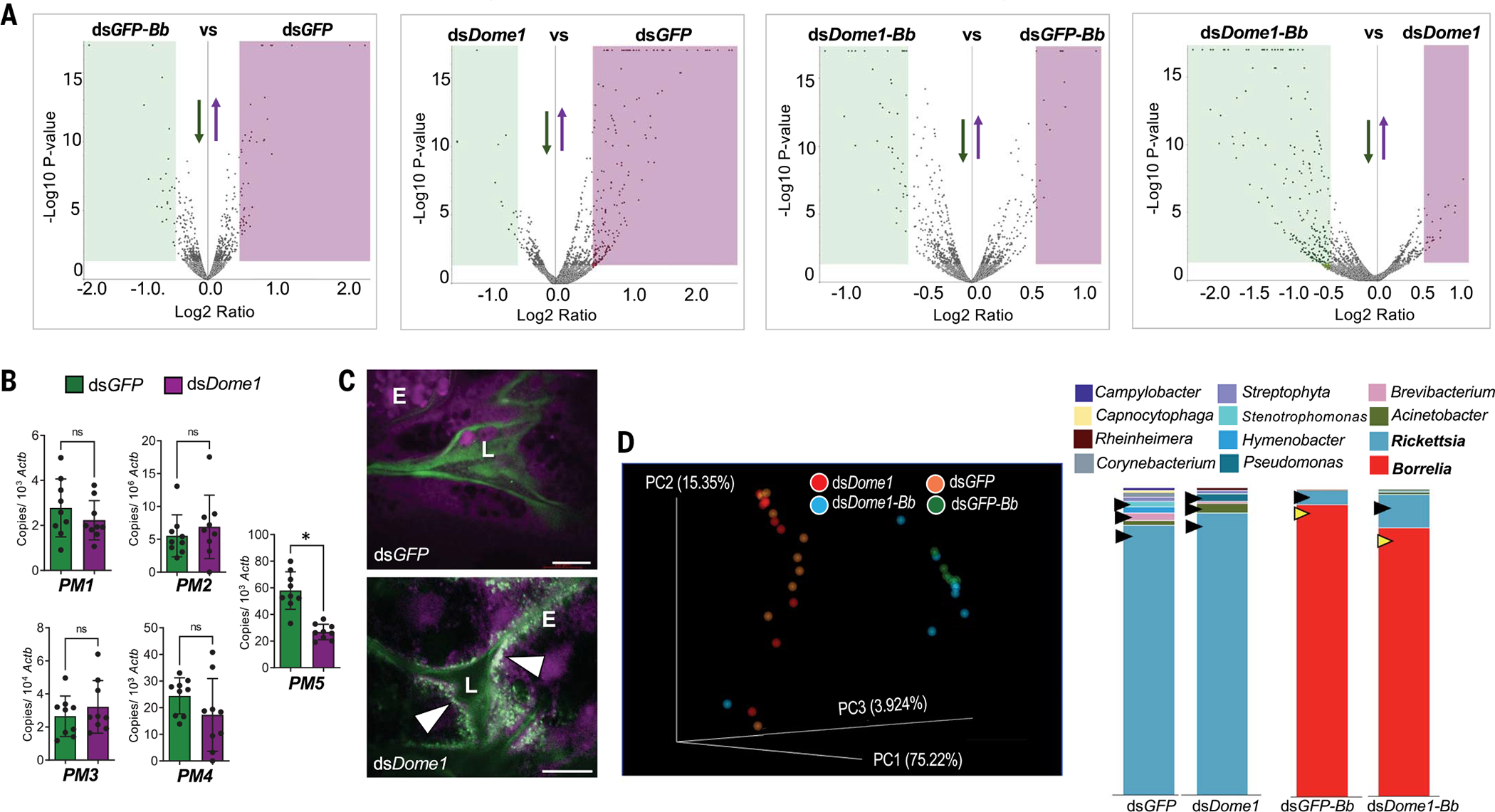
(A) Differential production of gut proteins in Dome1-knockdown ticks, in the presence or absence of B. burgdorferi, analyzed as follows: (i) B. burgdorferi-infected versus naïve control (dsGFP-Bb versus dsGFP), (ii) naïve Dome1-knockdown versus naïve control (dsDome1 versus dsGFP), (iii) B. burgdorferi-infected Dome1-knockdown versus B. burgdorferi-infected naïve control (dsDome1-Bb versus dsGFP-Bb), and (iv) B. burgdorferi-infected Dome1-knockdown versus naïve Dome1-knockdown (dsDome1-Bb versus dsDome1). The down- and up-regulated proteins are indicated by green or purple areas, respectively (see tables S1 to S4). (B) RT-qPCR assays show the down-regulation of a peritrophin gene, PM5, in Dome1-knockdown ticks. (C) Alteration of PM permeability in Dome1-knockdown ticks. Confocal microscopy showed the guts of 48-hour-fed nymphs, which had been microinjected with either dsDome1 or dsGFP RNA and were then capillary fed with fluorescein-conjugated 500,000 (green) or rhodamine red-conjugated 10,000 (violet) MW dextran molecules. The fluorescent beads are marked by arrowheads. L, lumen; E, epithelial cells. (D) 16S rRNA amplicon analysis of gut microbiota in Dome1-knockdown (dsDome1) and control (dsGFP) ticks, in the presence or absence of B. burgdorferi (32 nymphs per group), indicates alterations in microbial composition. The left panel denotes the principal coordinate analysis of weighted UniFrac distances of microbial communities; the right panel shows the genus-specific total bacterial abundance of Dome1-knockdown and control ticks in naïve (dsGFP versus dsDome1) or infected (dsGFP-Bb versus dsDome1-Bb) conditions. In naïve ticks, Dome1 deficiency altered the abundance of selected microbes (black arrowheads). In infected ticks, Dome1 knockdown decreased the abundance of Borrelia and enhanced the level of Rickettsia (yellow and black arrowheads, respectively). Results are representative of two to three independent experiments where quantitative data are shown as individual data points; error bars show the means ± SDs (n = 9 to 20). White bar: 20 μm. *P < 0.05, determined using two-tailed Mann-Whitney U test; n.s., not significant.
Dome1 supports gut regeneration and stem cell proliferation through the Notch–Delta and Hedgehog signaling pathways
We next investigated the cellular and molecular mechanisms by which the silencing of Dome1 affects gut homeostasis. During feeding, the tick gut undergoes bursts of developmental and remodeling events to acquire, ingest, and store a large volume of blood. To perturb this process, we induced experimental colitis in ticks by microinjection of 2% dextran sodium sulfate (DSS). DSS–treated ticks showed impaired feeding, as indicated by the reduced number of engorged ticks (Fig. 7A) and overall lower body weights (Fig. 7B). There were no differences in engorgement weights of ticks treated with only DSS and those coinjected with DSS and GFP dsRNA, but reduced weights were observed in ticks coinjected with DSS and Dome1 dsRNA (Fig. 7B), suggesting that Dome1 is required for the regeneration or remodeling of gut tissues.
Fig. 7. Dome1 maintains gut homeostasis through tissue regeneration and stem cell proliferation.
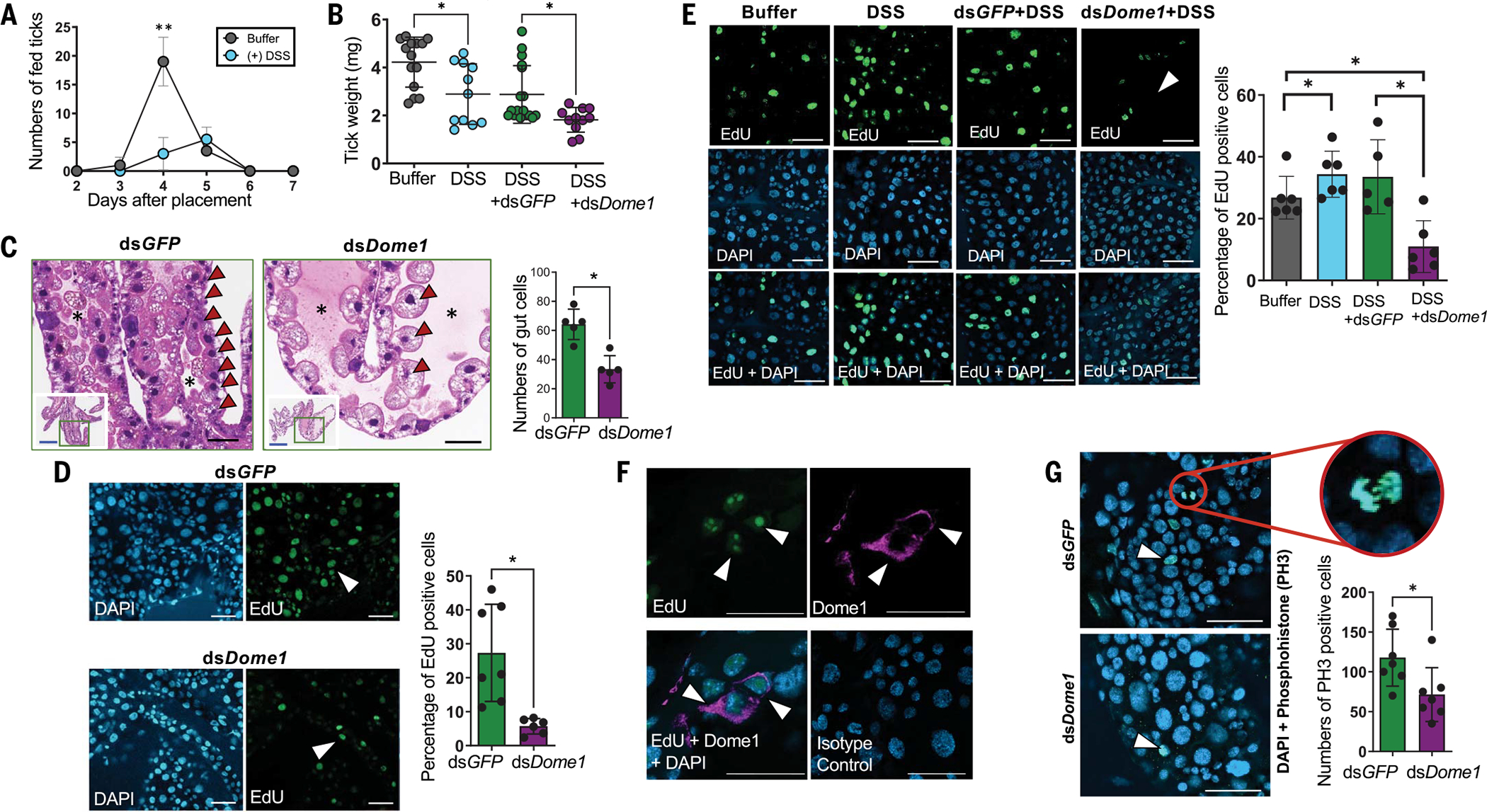
(A) Incomplete blood meal engorgement after induction of experimental colitis in DSS-treated ticks. **P < 0.05, determined using two-tailed Student’s t test. (B) Dome1 assists in tissue regeneration after injury. Ticks injected with DSS and coinoculated with dsDome1 RNA showed reductions in weight compared with the DSS-injected controls (dsGFP). (C) Dome1 maintains optimal gut cell population, as assessed by histology. The middle panel denotes the altered distribution of gut cells in Dome1-knockdown nymphs, showing cellular reduction (arrowheads) and an enlarged lumen (asterisk), compared with controls (left panel). The insets show the whole gut diverticula, with green boxes denoting the areas of magnified images. The right panel shows the microscopic enumeration of gut cells. (D) Cell proliferation in nymphal guts. Dome1-knockdown and control (dsGFP) nymphs fed on naïve mice for 72 hours to initiate gut cell proliferation. Proliferative EdU+ cells are marked by arrows (middle panels). The right panel shows the quantification of EdU+ cells. (E) Dome1 is essential for cell proliferation after gut injury. Nymphs were microinjected with buffer or DSS, in the presence or absence of dsRNA (dsDome1 or dsGFP). DSS-triggered gut cell proliferation activity was impaired by Dome1 deficiency (left panel, arrow), also shown by a reduction in cell counts (right panel). (F) Dome1 is expressed in proliferative cells. EdU+ cells were positive for Dome1 expression (violet). (G) Dome1 regulation of proliferative cells includes gut stem cells. Dome1-knockdown or control ticks fed on mice for 48 hours and were stained with anti-phospho-histone H3 (PH3) antibody (left panels, arrow), and enumerated (right panel). The circle represents a dividing stem cell nucleus in the control tick. Results are representative of two to three independent experiments where quantitative data are shown as individual data points; error bars show the means ± SDs (n = 6 to 25). For [(D) to (G)], nuclei are labeled with DAPI (blue) or EdU (green); white or black bars: 50 μm; blue bar: 300 μm. *P < 0.05, determined using two-tailed Mann-Whitney U test.
Dome1 contributed to the optimal maintenance of gut cell populations (Fig. 7C), likely by supporting their proliferation (Fig. 7D). Experimental tissue injury with DSS triggered extensive cell proliferation in nymphal guts, evidenced by a significant increase in 5-ethynyl-2′-deoxyuridine (EdU)-positive cells (Fig. 7E). The proliferative population, which highly expressed Dome1 (Fig. 7F), was decreased in DSS-treated Dome1-knockdown ticks (Fig. 7E, right panels), suggesting that Dome1 plays an essential role in cell proliferation after tissue injury. I. scapularis also displayed heightened gut cell proliferation when parasitizing IFN-γ–sufficient mice compared with IFN-γ–deficient mice (fig. S14), underscoring the influential roles of host IFN-γ in the up-regulation of Dome1 and the acceleration of blood meal engorgement in ticks (Fig. 3E). Dome1-mediated regulation of proliferative cells also involves gut stem cells, as their numbers were reduced in knockdown ticks (Fig. 7G).
The function of Dome1 likely involves developmental transcription factors, such as components of the Notch–Delta, epidermal growth factor receptor (EGFR), and Hedgehog signaling pathways. Enhanced expression of Dome1 was observed during early molting from larva to nymph (fig. S15A), whereas the knockdown of Dome1 impaired the expression of representative transcription factors from the Notch–Delta, EGFR, and Hedgehog pathways in nymphs (fig. S15B). Hox genes, a group of downstream transcription factors, were also induced after larval molting (fig. S15C). Notably, these genes were down-regulated in Dome1-knockdown ticks, compared with control ticks, whereas other transcription factors, such as SET and POU, remained unaffected (fig. S15D). Because Hox genes are regulated by the Hedgehog pathway (27), we created Hedgehog-knockdown ticks using RNAi. Similar to Dome1-, JAK-, or STAT-knockdown larvae, Hedgehog-silenced ticks were also unable to complete molting, displayed defects in multiple organs including the anal pore and legs, presented darkened abdomens, and often exhibited a complete loss of mouthparts (fig. S16). Furthermore, Hedgehog knockdown affected the expression of Notch, suggesting that their expression is connected during tick intermolt development. Thus, Dome1 regulates cellular proliferation and the homeostasis of gut tissues through JAK–STAT signaling through the Hedgehog and Notch–Delta pathways.
Ixodes Dome1–JAK–STAT pathway is required for optimal feeding of B. burgdorferi-infected ticks and pathogen transmission
We next assessed whether Dome1 or the JAK–STAT pathway affects pathogen survival throughout the tick intermolt stages as well as pathogen transmission to naïve hosts. In B. burgdorferi-infected larvae, the suppression of Dome1 mRNA and protein, along with JAK and STAT mRNA, was maintained in molted nymphs (Fig. 8A and fig. S12E). Notably, despite the substantial impact of Dome1–JAK–STAT on gut homeostasis and B. burgdorferi acquisition in ticks from infected hosts (Fig. 3 and Fig. 6), lower levels of spirochetes were detected throughout the intermolt period and in newly molted nymphs (Fig. 8B). When Dome1, JAK, or STAT were silenced in B. burgdorferi-infected ticks, only control (dsGFP) or JAK-knockdown ticks completed feeding on naïve C3H mice, whereas only a minority of the Dome1- or STAT-knockdown ticks (0 to 6%) were able to attach or engorge(fig.S12F).Control or JAK-knockdown ticks displayed regular hypostomes and normal engorgement, but some of the Dome1- or STAT-knockdown ticks that attempted host attachment exhibited malformed mouthparts, such as broken or distorted palps or hypostomes (Fig. 8C). Impaired feeding in Dome1- or STAT-knockdown ticks was further demonstrated by the reduced average time of tick attachment (Fig. 8D), numbers of fed ticks (Fig. 8E), and engorgement weights (Fig. 8F). After tick infestation, B. burgdorferi was undetectable in mice parasitized by Dome1-knockdown ticks, whereas low levels were recorded in STAT- and JAK-knockdown groups (Fig. 8, G and H, and fig. S12G). Despite the detection of spirochete-specific signals in the JAK-knockdown group (Fig.8H), culture analysis further confirmed that most animals remained uninfected (table S5).
Fig. 8. Ixodes Dome1 and JAK–STAT signaling pathway is required for optimal blood meal engorgement of B. burgdorferi-infected ticks and spirochete transmission to mice.
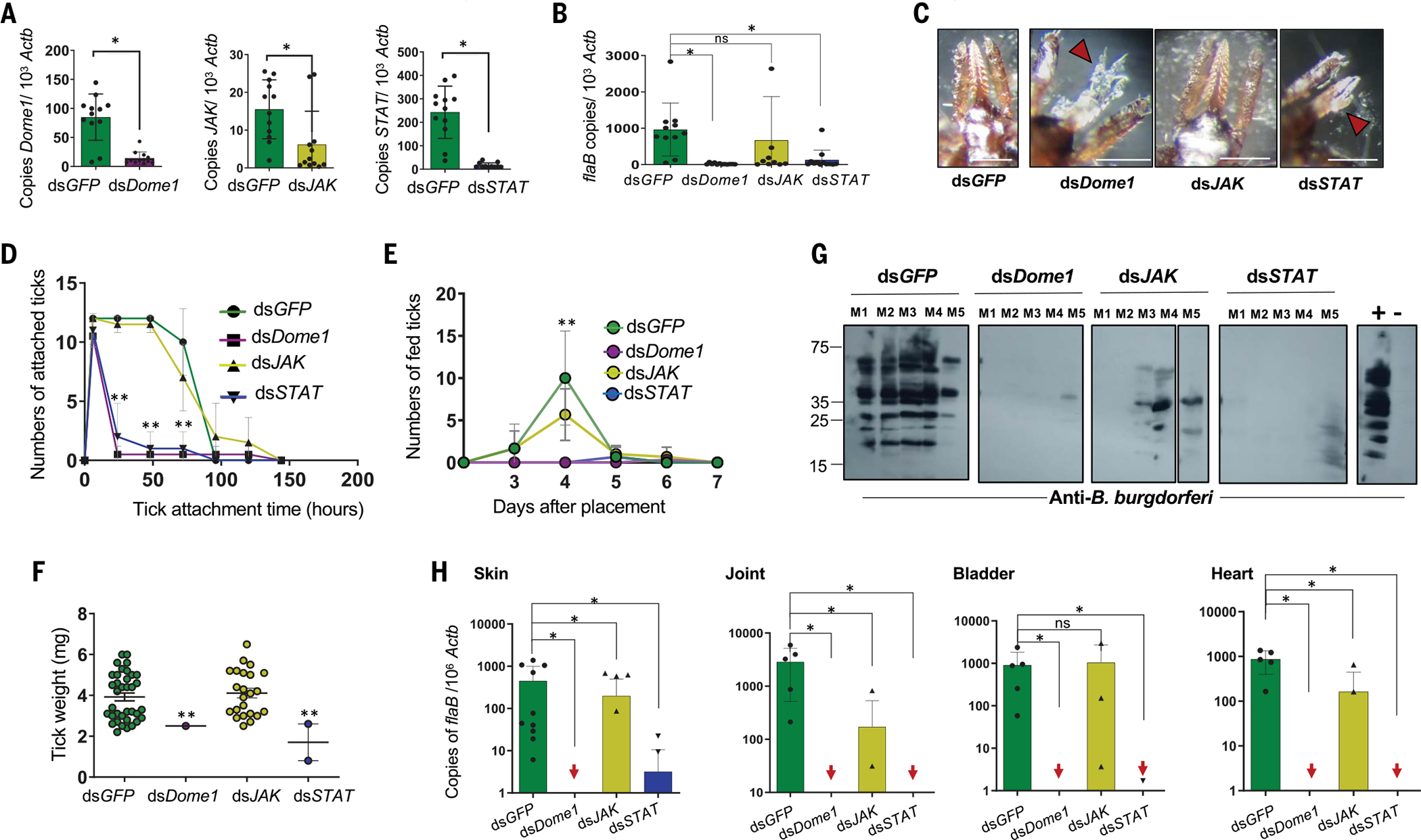
(A) Transstadial knockdown of Dome1, JAK, and STAT in infected unfed nymphs. Larvae that had engorged on B. burgdorferi-infected mice were microinjected with target dsRNA and allowed to molt. Transcripts in the infected nymphs were analyzed by RT-qPCR. (B) B. burgdorferi levels in unfed nymphs, as measured by the RT-qPCR assessment of flaB transcripts normalized against tick Actb levels. (C) Damaged tick mouthparts. Unlike the control or JAK-knockdown ticks, the detached Dome1- or STAT-knockdown ticks displayed distorted mouthparts (arrowheads). (D to F) The feeding parameters for the various groups are presented as the tick attachment time (D), number of fed ticks (E), and engorgement weight (F). Asterisks denote significant differences between dsDome1 or dsSTAT to other groups. **P < 0.05, determined using two-tailed Student’s t test. (G and H) Assessment of pathogen transmission to mice. 12 days after tick feeding, infection in individual animals was assessed by sera immunoblotting (G), or RT-qPCR assays (H) using one tissue sample per organ, except for proximal and distant skin samples relative to tick bite sites, by measuring copies of B. burgdorferi flaB transcripts normalized against mouse Actb levels. Arrows indicate murine tissue samples in dsDome1 or dsSTAT groups where flaB transcripts remain undetectable. For immunoblotting, sera from naïve mice and mice that were previously infected with B. burgdorferi were used as negative (−) and positive (+) controls, respectively. Loading controls are presented in fig. S12. Results are representative of three independent experiments where quantitative data are shown as individual data points; error bars show the means ± SDs (n = 9 to 36). White bar: 100μμm. *P < 0.05, determined using two-tailed Mann-Whitney U test; n.s., not significant.
Given the critical role of the I. scapularis Dome1–JAK–STAT pathway in the utilization of cross-species IFN-γ signaling mechanisms, we searched for these signaling components in the major tick genomes. Representative components of the Dome1–JAK–STAT pathway were present in all examined major tick vectors (fig. S17), providing support for their more widespread and evolutionary roles across diverse tick species. Despite the conservation of Dome1 orthologs, there was a high degree of diversification across major tick groups (fig. S18). However, the core Dome1 functions were likely maintained in ticks. For example, the interfer-bind motif was absent in Dome1 orthologs from most non-Ixodes tick genera (fig. S17) whereas the transcripts for Dome1 orthologs were detectable in the American dog tick (Dermacentor variabilis) and lone star tick (Amblyomma americanum), which were expressed during tick engorgement on mammalian hosts (fig. S19). Silencing of the Dome1 ortholog in D. variabilis showed that it supported cellular proliferation and microbial homeostasis in the gut (fig. S20), similar to its I. scapularis counterpart. Thus, the role of Dome1 is conserved across diverse tick genera.
Discussion
This study highlights the complex roles of Dome1 as a central receptor for a mammal–arthropod signaling cascade with converging effects on vector immunity and development. The protein is essential for the interspecies regulation of gut homeostasis, immunity, and blood meal digestion through the tick JAK–STAT pathway. These events, triggered by the incoming blood meal and expedited by the interaction between host-acquired IFN-γ and Dome1, are crucial for the tick’s postembryonic development and organogenesis, and are further supported by the Hedgehog and Notch–Delta biochemical networks. The integration of immunity and developmental functions in a single gut receptor enables ticks to sense danger signals emanating from infected hosts, thereby accelerating blood meal acquisition, which is essential for tick metamorphosis. Overall, this study underscores the notable flexibility of conserved cell signaling pathways, in addition to the commandeering of elements that can drive the complex cross-species regulation of immune responses, gut homeostasis, and organ development. It also emphasizes the intimate evolutionary relationship between ticks and their hosts at paramount points in the arthropod life cycle, marked by critical episodes of blood meal acquisition.
Mammalian cytokines affect the immunity and physiology of blood-feeding vectors (21, 28, 29), although the origin of cytokine receptors in arthropods remains unknown. Ticks, such as I. scapularis, are cosmopolitan in their selection of hosts, including lizards. As ancient ticks fed on primeval reptiles, it is possible that Dome1 originated early in the evolution of tick hematophagy. Indeed, Dome1 orthologs are present in representative reptilian, avian, and mammalian species (fig. S3), in addition to all examined tick groups (fig. S18). The occurrence of an IFN-γ–binding motif in Dome1 highlights how the evolution of vector parasitism offers opportunities for the acquisition of novel gene traits. The integration of Dome1’s interfer-bind motif in multiple tick species (figs. S17 and S18), together with its high-affinity interactions with mammalian or avian interferons, likely provides ticks with an ability to intercept immunity signals from a variety of infected mammals. Notably, although it remains unknown how I. scapularis evolved the ability to bind IFN-γ, the interfer-bind motif is detectable in several bacterial species, including Roseburia spp., which exist in the guts of both mammals and I. scapularis (30, 31), pointing to lateral gene transfer events as a possibility (22, 32). Regardless, IFN-γ-induced Dome1 controls the proliferation of I. scapularis gut cells, which would favor the intake of a large blood meal, augmenting tick survivability. Moreover, microbicidal responses triggered by Dome1–JAK–STAT signaling may support tick development by preserving essential nutrients, such as N-acetylglucosamine, the building block of tick chitin and bacterial peptidoglycan (33, 34). The silencing of the JAK–STAT pathway or the knockdown of Dome1 with multiple RNAi strategies (fig. S21) impairs organ development and tick metamorphosis, which suggests that these vectors reset their postembryonic development with each feeding cycle, revealing a critical role for a limited ancestral cell signaling pathway in the subadult development of ticks.
Because the role of the Dome1–JAK–STAT pathway on tick development is evident in the absence of IFN-γ or B. burgdorferi infection, it is likely that Dome1 binds endogenous tick ligands or other blood meal–derived factors. However, we were unable to identify additional Dome1 ligands in ticks, including UPD orthologs. Some other arthropods also lack genes encoding UPD, including non-Ixodes ticks (35), although some can alternatively produce endogenous ligands, such as Vago in Aedes mosquitoes (16) and novel C-type lectins in shrimp (14), that bind transmembrane Dome receptors and activate the JAK–STAT pathway, triggering antimicrobial immune responses. The biological significance of the expression of five Dome homologs in a single species such as I. scapularis remains puzzling, although the functions associated with Dome1 seem to be nonredundant to that of its homologs. The interference with tick development and metamorphosis through the knockdown of Dome1, JAK, or STAT impedes pathogen transmission from infected ticks to mammalian hosts, suggesting that these proteins may represent potential targets of anti-tick measures.
These data enhance our understanding of how eumetazoan cell signaling pathways (20, 36) have evolved. They highlight the flexibility and capacity of these pathways to adapt by integrating new components into existing networks that are likely important for the emergence of distinct immunological and physiological functions, which are in turn essential for animal survival in nature, particularly for ticks and other ectoparasites whose intimate host associations and complex hematophagy influence their persistence.
Materials and Methods
Bacteria, mice, and ticks
A clonal low-passage B. burgdorferi B31 isolate A3, grown in Barbour-Stoenner-Kelly H (BSK-H) media, was used throughout (21). A. phagocytophilum strain HZ was also used, as detailed (10). Four-to-six-week-old female C3H/HeN mice, Ifng-knockout (B6.129S7-Ifngtm1Ts/J) mice, and Ifngr1-knockout (B6.129S7-Ifngr1tm1Agt/J) mice, as well as 6-to-9-week-old female Dunkin Hartley guinea pigs and 6-week-old inbred female New Zealand white rabbits, were purchased from Charles River Laboratories or Jackson Laboratories. Ixodes scapularis, Dermacentor variabilis, and Amblyomma americanum ticks originating from the tick rearing facility at Oklahoma State University were maintained in the laboratory (21, 37, 38). All experiments were performed in accordance with the guidelines approved by the Institutional Animal Care and Use Committee and the Institutional Biosafety Committee.
Nucleic acid isolation and PCR
Total RNA isolation was performed as previously detailed (39, 40). Assessment of spirochete acquisition or transmission between ticks and C3H mice were performed as described (40, 41). The relative number of targeted gene transcripts in cDNA was measured using reverse transcription quantitative PCR (RT-qPCR) following published methods (42). The oligonucleotide primers used for the RT-qPCR analysis are indicated in table S6. Primers for the detection of various microbial species in the tick gut (43) and expression analyses of various peritrophin genes have been previously described (26). Primers for amplifying tick Actb from D. variabilis and A. americanum were designed as reported earlier (44, 45). The RNAi studies on D. variabilis were conducted on the Dome ortholog after cloning and sequencing the gene, which is submitted in GenBank with the nucleotide accession number OP503541.
Recombinant protein expression, purification, and antibody production
Protein induction and purification, as well as the production of polyclonal antibodies, were carried out using previously described methods (46, 47), with the following modifications. Secretory signal peptides and a Flag tag were added to the N-terminal, whereas a polyhistidine tag was linked to the C-terminal. The Dome1 gene was ligated to the pcDNA 3.0 vector using the Gibson Assembly Master Mix (New England Biolabs) following the manufacturer’s instructions and transfected into CHO cells using PEI reagent (Polysciences, Inc). G418-resistant clones were selected and analyzed for the secretion of the recombinant Dome1 protein.
Immunoprecipitation and pull-down assays
Recombinant mouse IFN-γ (BioLegend) was immobilized to anti-mouse IFN-γ monoclonal antibody (mAb) (clone R4-6A2, BioLegend) that was conjugated with Protein G-Sepharose 4B resin (Thermo Fisher Scientific). Precleaned whole tick lysates were incubated with the immobilized IFN-γ for 2 hours and bound proteins were then analyzed by mass spectrometry. The anti-mouse IFN-γ mAb conjugated with Protein G-Sepharose 4B resin binding proteins was used as a negative control. For the pulldown assay, purified Dome1-extracellular domain protein was incubated with immobilized mouse IFN-γ, followed by immunoblot analysis (48).
Protein–protein interaction assays
Analysis of IFN-γ interaction with Dome1 and other proteins was performed using a microtiter assay as reported (49). The ability of Dome1 to bind IFN-γ was measured by bio-layer interferometry (BLI) using the Octet Red384 system (ForteBio, Pall Corp.). Data were acquired in kinetics mode with standard settings and analyzed using the Data Analysis software v9.0 (ForteBio, Pall Corp.). Briefly, anti-Penta-HIS (HIS1K) biosensors (ForteBio, Pall Corp.) were prehydrated for 30 min in 200 μl of Kinetics Buffer (KB) (PBS+ 0.02% Tween 20, 0.1% BSA, and 0.05% NaN3). Dome1 was then loaded onto sensors at a final concentration of 25 nM. IFN-γ was diluted in KB to a final concentration of 5.5 nM to 0.7 μM and a kinetics assay was performed at 30°C. The results were analyzed with a global fit and 2:1 heterogeneous ligand-binding model using reference subtracted sensors with ligand or analyte-only controls, according to the manufacturer’s software platform. For protein–protein interactions in vivo, a yeast two-hybrid assay was used as detailed previously (50). AD-Dome1 and BD-IFN-γ constructs were cotransformed into yeast gold strains (Takara Bio Inc.) as per the manufacturer’s instructions and were plated on selective double dropout (2DO) media lacking leucine and trypsin and then incubated at 30°C for 3 to 4 days. The colonies were further restreaked on high stringency synthetic 3DO medium missing leucine, trypsin, and histidine. Colony growth was observed for 7 to 14 days.
Quantitative proteomics analysis
Samples for proteomics analysis were prepared as described with minor modifications (51). Processed tick proteins were labeled with the TMTPro 16-plex set (Thermo Fisher Scientific), fractionated with offline high pH, and analyzed using nanoscale liquid chromatography–tandem mass spectrometry (nano LC–MS/MS) with an orbitrap Fusion Lumos mass spectrometer interfaced to an Ultimate3000nano HPLC system.
Measurement of reactive oxygen species
ROS levels in the tick gut tissues were measured as described (52). Dissected tick samples were incubated with 60 μM dihydroethidium (DHE, Sigma-Aldrich), washed with PBS, and analyzed by confocal immunofluorescence microscopy (ZEISS LSM 800) for red fluorescence, using 10X lens and Zen imaging software (version 2.6), as previously reported (21).
Tick manipulation and infection studies
Gene silencing in ticks was performed according to previously published methods (21, 26) using primers detailed in table S6. For murine infection studies, wild-type C3H, Ifng-knockout, or Ifngr1-knockout mice were injected with 105B. burgdorferi and maintained for 14 days. Microinjected, control, or naïve ticks were allowed to feed on infected mice (25 ticks per mouse). For tick engorgement and transmission studies, each experimental group included two to five animals. The ticks were collected at various time points (24, 48, or 72 hours) or when fully fed, and were then analyzed using RT-qPCR (21). Similarly, groups of 25 adult I. scapularis were microinjected using microcapillaries carrying 5 to 10 μl of various dsRNA preparations (4 mg/ml), and infested on rabbit ears. For studies using D. variabilis or A. americanum, separate groups of nymphs (up to 50 per group) were placed on each individual guinea pig as detailed (53), and allowed to feed to repletion for 7 to 8 days. Parallel groups of ticks were also microinjected using microcapillaries carrying 5 ml of various dsRNA preparations (3 to 5 μg/ml). Some of the partially fed ticks were collected at various time points (24, 48, 72, and 96 hours) and were stored at −80°C before processing for cell proliferation assays, immunoblotting, or RT-qPCR assays. For cell proliferation experiments, partially fed (48 or 72 hours) nymphal ticks were microinjected using microcapillaries carrying 5 μl of 10 mM 5-ethynyl-2′-deoxyuridine (EdU) solution (Thermo Fisher Scientific), incubated for 4 to 5 hours and dissected to isolate gut samples, and fixed overnight with 4% paraformaldehyde in PBS. Following three washes with PBS, samples were permeabilized with acetone for 10 min at room temperature. The samples were subsequently blocked with 5% normal goat serum in PBS for two hours and incubated with Click-iT reaction cocktails (Thermo Fisher Scientific) for 45 min, and subsequently stained with DAPI and processed for confocal immunofluorescence microscopy. Artificial feeding of nymphs was performed as described in previously published methods (54). To induce colitis, groups of 25 nymphs, 2 to 4 weeks after molting, were microinjected using microcapillaries carrying 5 ml of 2% DSS (MP Biomedicals), then allowed to engorge on mice the following day, and processed accordingly.
Microscopy and histological analysis
To localize B. burgdorferi, Dome1, phosphor-histone H3 (PH3), or other structures including nuclei, within tick organs or cells, the samples were processed for confocal microscopy using published procedures (39). To avoid red—green juxtapositions in immunofluorescence images, the red Alexa Fluor 568 or rhodamine phalloidin signals in some of the samples were converted to an alternate pseudocolor (violet spectrum) in the ZEISS LSM 800 confocal microscope according to the manufacturer’s instructions. Dome1 or other primary antibodies were visualized using Alexa 488-labeled anti-rabbit IgG or Alexa 568-labeled anti-mouse or secondary antibodies. The specificity of these secondary antibodies was tested in samples without primary antibodies or in the presence of normal mouse IgG (isotype controls), which did not generate non-specific immunofluorescence signals, as shown in Fig. 1H (see table S7 for a list of all antibodies used in the study). Briefly, tick organs were fixed in 4% paraformaldehyde at 4°C overnight, and permeabilized with acetone for 10 min following three washes with PBS. Samples were blocked with 5% normal goat serum in PBS-T (PBS with 0.05% Tween 20) for 2 hours at room temperature and then incubated with primary antibodies at 4°C overnight. Following three washes with PBS-T, the tissues were incubated overnight with Alexa Fluor secondary antibodies. The cell nuclei or F-actin were stained with DAPI or rhodamine phalloidin, respectively (Thermo Fisher Scientific) and mounted with antifade mounting medium (Thermo Fisher Scientific). Confocal images were acquired using either a 10X dry, 40X water, or 63X oil objective lens and ZEN imaging software (version 2.6, ZEISS). Electron microscopic analyses were performed as detailed earlier (55). The images of whole ticks were captured using a Canon SX720HS digital camera under the Olympus SZ61 microscope. For histology, tick samples were cross-sectioned and subjected to standard hematoxylin and eosin (H&E) staining as reported (40). The stained slides were processed using a Leica Biosystems anatomic pathology scanner and captured using ImageScope software. For quantitative analysis, at least 10 random microscopic areas from three sections were enumerated for cell nuclei. All microscopic analyses were performed in a double-blinded manner.
Microbiome analysis
DNA from dsGFP- or dsDome1-microinjected nymphal ticks, either naïve or infected with B. burgdorferi (32 ticks per group), was prepared as described (56). Bacterial 16S rDNA amplicons were then generated using barcoded 16S universal primers (515F/806R), sequenced, and analyzed using the QIIME2, version 2020.11 (57). Operational Taxonomic Units were generated using Deblur (58) and assigned to taxonomic units using a pretrained Naïve Bayes classifier on the V3-V4 region of the 16S rRNA sequences from the Greengenes database, release 13.8 (59) at 99% similarity.
Bioinformatics and statistical analysis
Multiple sequence alignments were performed in Jalview 2.11.1.4 and aligned using TCoffee (60) with default running conditions. Homologs to the Dome1 protein were detected using HHPred (MPI Bioinformatics Toolkit) with three iterations of MSA sequence generation with at least 40% minimum coverage (61). UniProt accession IDs were obtained using R (4.0.1) and the packages MAPDB (v0.1) and UniProt.ws (v.2.30.0). Phylogenetic analysis (also see table S8) was performed using either the neighbor-joining method or the maximum likelihood method with 1000 replicate bootstrapping (62, 63) and Interactive Tree of Life (64). The I-TASSER automated modeling suite (65) was used on NIH HPC resources to construct all models. Structures were annotated and visualized using PyMol (Schrödinger, Inc.) and figures were created using BioRender (66). Dome orthologs were predicted by flyBase (67) and the domains and motifs were identified using the SMART (68) as well as Pfam and Motif Search (24, 25) databases.
Quantitative results are expressed as means ± standard deviation (SD). The statistical significance of the difference between the mean values of groups was analyzed using the two-tailed Mann–Whitney U or Student’s t test and one-way ANOVA methods. Results were analyzed using GraphPad Prism v9 (GraphPad Software Inc.); P < 0.05 was considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Nassar for her sincere assistance with the preparation of the manuscript. We are grateful to R. Barrio for her insightful discussions.
Funding:
This study was supported by the National Institute of Allergy and Infectious Diseases (award number AI138949 to U.P., J.H.F.P., and E.F.; AI080615, AI154542, and Steven & Alexandra Cohen Foundation to U.P.; AI116523 and AI134696 to J.H.F.P.; AI26033, AI165499, Steven & Alexandra Cohen Foundation, and in part by the Howard Hughes Medical Institute Emerging Pathogens Initiative to E.F.), the Intramural Research Program of the National Center for Advancing Translational Sciences (1ZIATR000413-01) to A.S., National Institute of Dental and Craniofacial Research Mass Spectrometry Facility (ZIA DE000751) to Y.W., and Grant PDI2021-124328OB-100 funded by MCIN/AEI/10.13039/501100011033 and by “ERDF A way of making Europe” to J.A. CIC bioGUNE thanks the Spanish Ministry of Science and Innovation for the Severo Ochoa Center of Excellence award (SEV-2016-0644).
Footnotes
Competing interests: The authors declare no competing interests.
SUPPLEMENTARY MATERIALS
Data and materials availability:
All data are available in the manuscript or in the supplementary materials.
REFERENCES AND NOTES
- 1.Parola P, Raoult D, Ticks and tickborne bacterial diseases in humans: An emerging infectious threat. Clin. Infect. Dis. 32, 897–928 (2001). doi: 10.1086/319347 [DOI] [PubMed] [Google Scholar]
- 2.Perner J et al. , RNA-seq analyses of the midgut from blood- and serum-fed Ixodes ricinus ticks. Sci. Rep. 6, 36695 (2016). doi: 10.1038/srep36695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCoy KD, Léger E, Dietrich M, Host specialization in ticks and transmission of tick-borne diseases: A review. Front. Cell. Infect. Microbiol. 3, 57 (2013). doi: 10.3389/fcimb.2013.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitsou C, Fikrig E, Pal U, Tick host immunity: Vector immunomodulation and acquired tick resistance. Trends Immunol. 42, 554–574 (2021). doi: 10.1016/j.it.2021.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO, Vector-borne diseases. https://www.who.int/mediacentre/factsheets/fs387/en/, (WHO, 2016). [Google Scholar]
- 6.Sonenshine DE, Range Expansion of Tick Disease Vectors in North America: Implications for Spread of Tick-Borne Disease. Int. J. Environ. Res. Public Health 15, 478 (2018). doi: 10.3390/ijerph15030478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paddock CD, Lane RS, Staples JE, Labruna MB, “Changing paradigms for tick-borne diseases in the Americas” in Global Health Impacts of Vector-Borne Diseases: Workshop Summary (National Academies Press, 2016), pp. 221–258 [Google Scholar]
- 8.Palmer WJ, Jiggins FM, Comparative Genomics Reveals the Origins and Diversity of Arthropod Immune Systems. Mol. Biol. Evol. 32, 2111–2129 (2015). doi: 10.1093/molbev/msv093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gulia-Nuss M et al. , Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat. Commun. 7, 10507 (2016). doi: 10.1038/ncomms10507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw DK et al. , Infection-derived lipids elicit an immune deficiency circuit in arthropods. Nat. Commun. 8, 14401 (2017). doi: 10.1038/ncomms14401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith AA, Pal U, Immunity-related genes in Ixodes scapularis—Perspectives from genome information. Front. Cell. Infect. Microbiol. 4, 116 (2014). doi: 10.3389/fcimb.2014.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nichols SA, Dirks W, Pearse JS, King N, Early evolution of animal cell signaling and adhesion genes. Proc. Natl. Acad. Sci. U.S.A. 103, 12451–12456 (2006). doi: 10.1073/pnas.0604065103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison DA, The Jak/STAT pathway. Cold Spring Harb. Perspect. Biol. 4, a011205 (2012). doi: 10.1101/cshperspect.a011205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun JJ, Lan JF, Zhao XF, Vasta GR, Wang JX, Binding of a C-type lectin’s coiled-coil domain to the Domeless receptor directly activates the JAK/STAT pathway in the shrimp immune response to bacterial infection. PLOS Pathog. 13, e1006626 (2017). doi: 10.1371/journal.ppat.1006626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aaronson DS, Horvath CM, A road map for those who don’t know JAK-STAT. Science 296, 1653–1655 (2002). doi: 10.1126/science.1071545 [DOI] [PubMed] [Google Scholar]
- 16.Paradkar PN, Trinidad L, Voysey R, Duchemin JB, Walker PJ, Secreted Vago restricts West Nile virus infection in Culex mosquito cells by activating the Jak-STAT pathway. Proc. Natl. Acad. Sci. U.S.A. 109, 18915–18920 (2012). doi: 10.1073/pnas.1205231109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrera SC, Bach EA, JAK/STAT signaling in stem cells and regeneration: From Drosophila to vertebrates. Development 146, dev167643 (2019). doi: 10.1242/dev.167643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown S, Hu N, Hombría JC, Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr. Biol. 11, 1700–1705 (2001). doi: 10.1016/S0960-9822(01)00524-3 [DOI] [PubMed] [Google Scholar]
- 19.Miller JR et al. , A draft genome sequence for the Ixodes scapularis cell line, ISE6. F1000 Res. 7, 297 (2018). doi: 10.12688/f1000research.13635.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pires-daSilva A, Sommer RJ, The evolution of signalling pathways in animal development. Nat. Rev. Genet. 4, 39–49 (2003). doi: 10.1038/nrg977 [DOI] [PubMed] [Google Scholar]
- 21.Smith AA et al. , Cross-Species Interferon Signaling Boosts Microbicidal Activity within the Tick Vector. Cell Host Microbe 20, 91–98 (2016). doi: 10.1016/j.chom.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boto L, Horizontal gene transfer in the acquisition of novel traits by metazoans. Proc. Biol. Sci. 281, 20132450 (2014). doi: 10.1098/rspb.2013.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flybase Consortium, Flybase: Gene: Dmel\dome (2022); http://flybase.org/reports/FBgn0043903.
- 24.Paysan-Lafosse T et al. , InterPro in 2022. Nucleic Acids Research, gkac993 (2022). doi: 10.1098/rspb.2013.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.GenomeNet, MOTIF Search (2022); https://www.genome.jp/tools/motif/.
- 26.Narasimhan S et al. , Gut microbiota of the tick vector Ixodes scapularis modulate colonization of the Lyme disease spirochete. Cell Host Microbe 15, 58–71 (2014). doi: 10.1016/j.chom.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodrigues AR et al. , Integration of Shh and Fgf signaling in controlling Hox gene expression in cultured limb cells. Proc. Natl. Acad. Sci. U.S.A. 114, 3139–3144 (2017). doi: 10.1073/pnas.1620767114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pakpour N, Akman-Anderson L, Vodovotz Y, Luckhart S,The effects of ingested mammalian blood factors on vector arthropod immunity and physiology. Microbes Infect. 15, 243–254 (2013). doi: 10.1016/j.micinf.2013.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luckhart S et al. , Mammalian transforming growth factor beta1 activated after ingestion by Anopheles stephensi modulates mosquito immunity. Infect. Immun. 71, 3000–3009 (2003). doi: 10.1128/IAI.71.6.3000-3009.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kasahara K et al. , Interactions between Roseburia intestinalis and diet modulate atherogenesis in a murine model. Nat. Microbiol. 3, 1461–1471 (2018). doi: 10.1038/s41564-018-0272-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan DT, A Metagenomic study of the tick midgut. The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences Dissertations and Theses, (2010). [Google Scholar]
- 32.Chou S et al. , Transferred interbacterial antagonism genes augment eukaryotic innate immune function. Nature 518, 98–101 (2015). doi: 10.1038/nature13965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vollmer W, Blanot D, de Pedro MA, Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167 (2008). doi: 10.1111/j.1574-6976.2007.00094.x [DOI] [PubMed] [Google Scholar]
- 34.Sze CW et al. , Study of the response regulator Rrp1 reveals its regulatory role in chitobiose utilization and virulence of Borrelia burgdorferi. Infect. Immun. 81, 1775–1787 (2013). doi: 10.1128/IAI.00050-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia N et al. , Large-Scale Comparative Analyses of Tick Genomes Elucidate Their Genetic Diversity and Vector Capacities. Cell 182, 1328–1340.e13 (2020). doi: 10.1016/j.cell.2020.07.023 [DOI] [PubMed] [Google Scholar]
- 36.Barolo S, Posakony JW, Three habits of highly effective signaling pathways: Principles of transcriptional control by developmental cell signaling. Genes Dev. 16, 1167–1181 (2002). doi: 10.1101/gad.976502 [DOI] [PubMed] [Google Scholar]
- 37.Schuijt TJ et al. , Identification and characterization of Ixodes scapularis antigens that elicit tick immunity using yeast surface display. PLOS ONE 6, e15926 (2011). doi: 10.1371/journal.pone.0015926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sajid A et al. , mRNA vaccination induces tick resistance and prevents transmission of the Lyme disease agent. Sci. Transl. Med. 13, eabj9827 (2021). doi: 10.1126/scitranslmed.abj9827 [DOI] [PubMed] [Google Scholar]
- 39.Yang X, Smith AA, Williams MS, Pal U, A dityrosine network mediated by dual oxidase and peroxidase influences the persistence of Lyme disease pathogens within the vector. J. Biol. Chem. 289, 12813–12822 (2014). doi: 10.1074/jbc.M113.538272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernard Q et al. , Plasticity in early immune evasion strategies of a bacterial pathogen. Proc. Natl. Acad. Sci. U.S.A. 115, E3788–E3797 (2018). doi: 10.1073/pnas.1718595115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye M et al. , HtrA, a Temperature- and Stationary Phase-Activated Protease Involved in Maturation of a Key Microbial Virulence Determinant, Facilitates Borrelia burgdorferi Infection in Mammalian Hosts. Infect. Immun. 84, 2372–2381 (2016). doi: 10.1128/IAI.00360-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Promnares K et al. , Borrelia burgdorferi small lipoprotein Lp6.6 is a member of multiple protein complexes in the outer membrane and facilitates pathogen transmission from ticks to mice. Mol. Microbiol. 74, 112–125 (2009). doi: 10.1111/j.1365-2958.2009.06853.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narasimhan S et al. , Modulation of the tick gut milieu by a secreted tick protein favors Borrelia burgdorferi colonization. Nat. Commun. 8, 184 (2017). doi: 10.1038/s41467-017-00208-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sunyakumthorn P, Petchampai N, Kearney MT, Sonenshine DE,Macaluso KR, Molecular characterization and tissue-specific gene expression of Dermacentor variabilis α-catenin in response to rickettsial infection. Insect Mol. Biol. 21, 197–204 (2012). doi: 10.1111/j.1365-2583.2011.01126.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bullard RL, Characterization of Glycine Rich Proteins From the Salivary Glands of the Lone Star Tick Amblyomma americanum. (The University of Southern Mississippi Dissertations 339, 2016); https://aquila.usm.edu/dissertations/339 [Google Scholar]
- 46.Kariu T et al. , BB0323 and novel virulence determinant BB0238: Borrelia burgdorferi proteins that interact with and stabilize each other and are critical for infectivity. J. Infect. Dis. 211, 462–471(2015). doi: 10.1093/infdis/jiu460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kariu T, Smith A, Yang X, Pal U, A chitin deacetylase-like protein is a predominant constituent of tick peritrophic membrane that influences the persistence of Lyme disease pathogens within the vector. PLOS ONE 8, e78376 (2013). doi: 10.1371/journal.pone.0078376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang X et al. , Characterization of multiprotein complexes of the Borrelia burgdorferi outer membrane vesicles. J. Proteome Res. 10, 4556–4566 (2011). doi: 10.1021/pr200395b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pal U et al. , Attachment of Borrelia burgdorferi within Ixodes scapularis mediated by outer surface protein A. J. Clin. Invest. 106, 561–569 (2000). doi: 10.1172/JCI9427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pal U et al. , TROSPA, an Ixodes scapularis receptor for Borrelia burgdorferi. Cell 119, 457–468 (2004). doi: 10.1016/j.cell.2004.10.027 [DOI] [PubMed] [Google Scholar]
- 51.Yang X et al. , Analysis of Borrelia burgdorferi Proteome and Protein-Protein Interactions. Methods Mol. Biol. 1690, 259–277 (2018). doi: 10.1007/978-1-4939-7383-5_19 [DOI] [PubMed] [Google Scholar]
- 52.Vaccaro A et al. , Sleep Loss Can Cause Death through Accumulation of Reactive Oxygen Species in the Gut. Cell 181, 1307–1328.e15 (2020). doi: 10.1016/j.cell.2020.04.049 [DOI] [PubMed] [Google Scholar]
- 53.Levin ML, Zemtsova GE, Killmaster LF, Snellgrove A,Schumacher LBM, Vector competence of Amblyomma americanum (Acari: Ixodidae) for Rickettsia rickettsii. Ticks Tick Borne Dis. 8, 615–622 (2017). doi: 10.1016/j.ttbdis.2017.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang X et al. , A novel tick protein supports integrity of gut peritrophic matrix impacting existence of gut microbiome and Lyme disease pathogens. Cell. Microbiol. 23, e13275 (2021). doi: 10.1111/cmi.13275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fischer ER, Hansen BT, Nair V, Hoyt FH, Dorward DW, Scanning electron microscopy. Curr Protoc Microbiol mc02b02s25 (2012). doi: 10.1002/9780471729259.mc02b02s25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh ST et al. , Diversity and phylogenetic analysis of endosymbiotic bacteria from field caught Bemisia tabaci from different locations of North India based on 16S rDNA library screening. Infect. Genet. Evol. 12, 411–419 (2012). doi: 10.1016/j.meegid.2012.01.015 [DOI] [PubMed] [Google Scholar]
- 57.Bolyen E et al. , Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019). doi: 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amir A et al. , Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems 2, e00191–e16 (2017). doi: 10.1128/mSystems.00191-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeSantis TZ et al. , Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006). doi: 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Notredame C, Higgins DG, Heringa J, T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 (2000). doi: 10.1006/jmbi.2000.4042 [DOI] [PubMed] [Google Scholar]
- 61.Gabler F et al. , Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr. Protoc. Bioinformatics 72, e108 (2020). doi: 10.1002/cpbi.108 [DOI] [PubMed] [Google Scholar]
- 62.Kumar S, Stecher G, Li M, Knyaz C, Tamura K, MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549 (2018). doi: 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stecher G, Tamura K, Kumar S, Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 37, 1237–1239 (2020). doi: 10.1093/molbev/msz312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Letunic I, Bork P, Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). doi: 10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Lab, Univ. of Michigan, I-TASSER: Protein Structure and Function Predictions (2022); https://zhanglab.ccmb.med.umich.edu/I-TASSER.
- 66.Biorender (2022); www.biorender.com.
- 67.Flybase Consortium, Flybase: A Database of Drosophila Genes & Genomes (2022); http://flybase.org. [DOI] [PMC free article] [PubMed]
- 68.Schultz J, Milpetz F, Bork P, Ponting CP, SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. U.S.A. 95, 5857–5864 (1998). doi: 10.1073/pnas.95.11.5857; [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the manuscript or in the supplementary materials.


