Abstract
Pancreatic ductal adenocarcinoma (PDAC) is a lethal malignancy and is largely refractory to available treatments. Identifying key pathways associated with disease aggressiveness and therapeutic resistance may characterize candidate targets to improve patient outcomes. We used a strategy of examining the tumors from a subset of PDAC patient cohorts with the worst survival to understand the underlying mechanisms of aggressive disease progression and to identify candidate molecular targets with potential therapeutic significance. Non-negative matrix factorization (NMF) clustering, using gene expression profile, revealed three patient subsets. A 142-gene signature specific to the subset with the worst patient survival, predicted prognosis and stratified patients with significantly different survival in the test and validation cohorts. Gene-network and pathway analysis of the 142-gene signature revealed dysregulation of Clusterin (CLU) in the most aggressive patient subset in our patient cohort. Hepatocyte nuclear factor 1 b (HNF1B) positively regulated CLU, and a lower expression of HNF1B and CLU was associated with poor patient survival. Mechanistic and functional analyses revealed that CLU inhibits proliferation, 3D spheroid growth, invasiveness and epithelial-to-mesenchymal transition (EMT) in pancreatic cancer cell lines. Mechanistically, CLU enhanced proteasomal degradation of EMT-regulator, ZEB1. In addition, orthotopic transplant of CLU-expressing pancreatic cancer cells reduced tumor growth in mice. Furthermore, CLU enhanced sensitivity of pancreatic cancer cells representing aggressive patient subset, to the chemotherapeutic drug gemcitabine. Taken together, HNF1B/CLU axis negatively regulates pancreatic cancer progression and may potentially be useful in designing novel strategies to attenuate disease progression in PDAC patients.
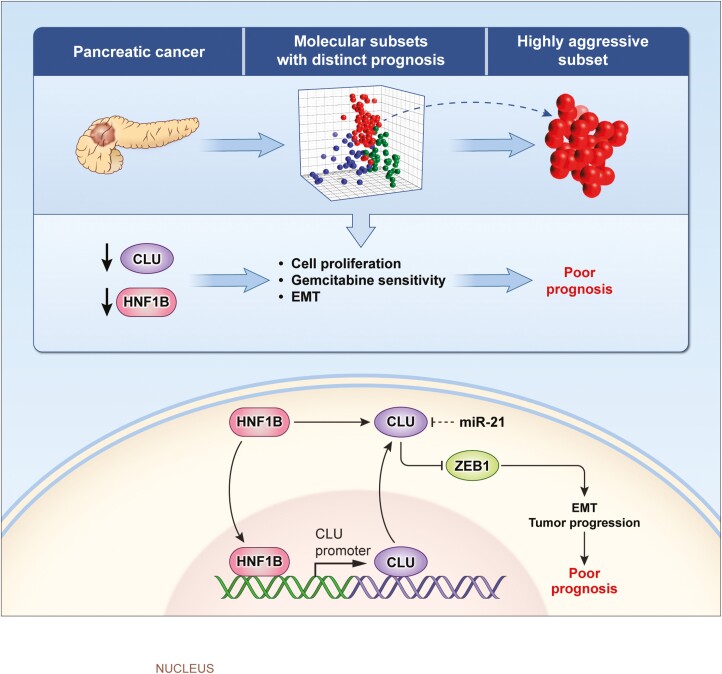
This study illustrates a novel HNF1B/Clusterin axis negatively regulating disease aggressiveness by enhancing proteasomal degradation of ZEB1 and regulating EMT in a highly aggressive subset of PDAC. Therapies targeting HNF1B/Clusterin may provide a novel approach to potentially improve disease outcome in PDAC patients.
Graphical abstract
Dysregulation of HNF1B/CLU Axis enhances disease progression in PDAC. A decrease in CLU level enhances cell proliferation and EMT, and reduces sensitivity to gemcitabine resulting in enhanced progression and poor patients outcome.
Introduction
Pancreatic cancer is one of the most lethal malignancies and is ranked as the third leading cause of death due to cancer with an estimated 62 210 new cases and 49 830 deaths in 2022 in the USA (1). The 5-year survival rate in pancreatic cancer is still less than 10%. Disturbingly, a consistent rise in incidence and death in pancreatic cancer is estimated to make it the second leading cause of cancer-related death by 2030 (2). Pancreatic ductal adenocarcinoma (PDAC) constitutes a major lethal type of pancreatic cancer and is largely refractory to available treatments with heterogenous chemotherapeutic response in subsets of patients (3–8). Therefore, identifying key biological pathways associated with disease aggressiveness and therapeutic resistance is crucial to characterize candidate targets for improving outcome in PDAC patients.
Genomic, transcriptomics and metabolomic analyses have identified distinct subtypes that are associated with differences in overall survival and relative response to treatments (3–10). However, the growing consensus favors the existence of two major subtypes of PDAC. The classical or pancreatic progenitor subtype identifies PDAC patients with a relatively better prognosis and is characterized by differentiated ductal marker such as PDX1, whereas, the basal-like, squamous or quasi-mesenchymal (QM) subtype is characterized by the expression of basal-like markers such as cytokeratin 81 (KRT81), which predicts poorer prognosis in PDAC patients (4). A number of studies have highlighted the differences in the transcriptional network (7,11,12), therapy resistance (13) and stromal alterations (14) underlying these subtypes for further characterization. However, the etiology and molecular mechanisms of disease progression in these subtypes are still largely unclear and has not yet contributed to the development of an effective strategy for clinical intervention. In a recent study, the cell of origin is defined as a critical determinant of molecular subtypes in PDAC (15). Therefore, further delineation of underlying mechanisms of disease progression in aggressive subtypes may provide key insights for improving overall disease management.
In this study, we hypothesized that transcriptomics, and the mechanistic and functional analyses of potentially relevant genes and pathways in the most aggressive subset of human PDAC may reveal key mechanisms of disease progression and identify candidate targets for intervention in pancreatic cancer patients. Therefore, we examined the most aggressive molecular subset of PDAC patients in our test and validation cohorts to understand the underlying mechanisms of disease progression and to identify candidate molecular targets with potential therapeutic significance (schema shown in Supplementary Figure S1, available at Carcinogenesis Online). Integrative gene expression analysis of tumors in our test cohort (N = 139) of PDAC patients identified a 148 specific gene signature that identified patients with the worst prognosis. Furthermore, the identified 148 candidate genes and related pathways associated with the highly aggressive subsets of pancreatic cancer were mechanistically, functionally, and clinically characterized by using cell lines, preclinical mouse model and PDAC patients’ cohorts. Our findings resulted in the identification of a novel hepatocyte nuclear factor 1 b (HNF1B)/Clusterin (CLU) axis, which regulates the disease progression in pancreatic cancer.
Materials and methods
Human samples
Human primary pancreatic tumor tissues from PDAC patients were collected at the University of Maryland Medical System at Baltimore, MD through National Cancer Institute (NCI)-UMD resource contract and the Department of Surgery at University of Medicine, Gottingen, Germany. The characteristics of the patients in test cohort (N = 139) are described in Supplementary Table S1, available at Carcinogenesis Online. Use of clinical specimens was reviewed and permitted by the NCI-Office of Human Subject Research Protection (OHSRP, Exempt# 4678) at the National Institutes of Health, Bethesda, MD.
Cell lines
All the cell lines used in this study were purchased from American Type Culture Collection (ATCC), Rockville, MD in July 2015 and were authenticated using short tandem repeat analysis prior to their use in different experiments. All cells were reauthenticated by short tandem repeat analysis in the last 1 year at ATCC.
Microarray gene expression profiling and analysis
Messenger RNA (mRNA) expression profiling was performed at the microarray core facility of the NCI, Frederick, MD. Affymetrix GeneChip Human 1.0 ST arrays were used for mRNA expression profiling according to the manufacturer’s protocol. The mRNA microarray expression data have been deposited in the National Center for Biotechnology Information’s (NCBIs) Gene Expression Omnibus (GSE 183795). To analyze mRNA expression profile all arrays were first Robust Multiarray Averaging (RMA) normalized. We plotted samples using principal component analysis and remove the batch effect using Empirical Bayes methods implemented in ComBat suite. We selected probes with standard deviation (SD) > 0.6 as the intrinsically variable genes and performed non-negative matrix factorization (NMF) analysis. The analysis revealed a stable consensus cluster to represent subtypes of the pancreatic adenocarcinoma with a cophenetic coefficient > 0.94 when proposing the number of groups from 2 to 5. Gene signature for each subset was analyzed by comparing a subset with the other two subsets with a strict cut-off of 2-fold change and Benjamini-Hochberg adjusted P-value of 0.01. We visualized data sets with the Hierarchical Clustering Viewer using Partek genomic suite.
Additional details of all the material and methods are provided as supplementary information.
Statistical analysis
Significant differences in the mRNA expression, migration and invasion, colony formation ability, 3D spheroid formation were determined using Student t-test. One-way analysis of variance (ANOVA) was used for more than two groups of comparison. Data are presented as mean values ± SD of triplicates. Kaplan-Meier analysis and Log Rank test were performed to evaluate the difference in survival between two groups of patients using Graphpad Prism 8.0. Pearson’s correlation was used for examining the correlation between the expressions of two genes. P < 0.05 was considered statistically significant.
Results
Gene expression profile identified subsets of PDAC patients with a distinct outcome
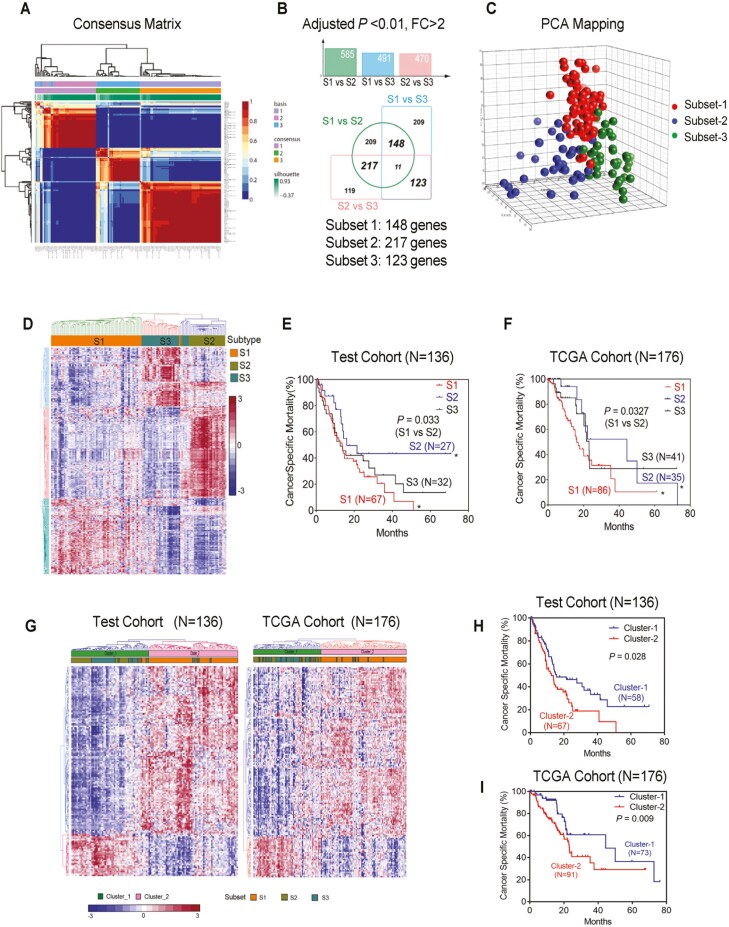
Global gene expression profiling has identified distinct subtypes in different human cancers, which are useful for guidance of cancer prevention, diagnosis, and therapy. We first performed gene expression profiling of 139 tumor samples from PDAC patients to separate the subgroups of patients with different prognosis. We selected probes with SD >0.6 as the intrinsically variable genes and performed non-NMF analysis with consensus clustering to identify subsets of PDAC with a cophenetic coefficient > 0.94. This analysis discovered three molecular subsets (Figure 1A). Class comparison analysis between each subset was performed to generate subset-specific gene signatures. After strict cut-off of 2-fold change and Benjamini-Hochberg adjusted P-value < 0.01, we obtained three sets of gene signatures which could represent three subsets of PDAC patients denoted as subset-1 (S1) with 148 genes, subset-2 (S2) with 217 genes and subset-3 (S3) with 123 genes as shown in the Venn diagram (Figure 1B; Supplementary Table S2, available at Carcinogenesis Online). Principal component analysis of mRNA expression profile with these gene signatures revealed three distinct subsets for the three consensus clusters (Figure 1C). Unsupervised hierarchical clustering of the patients using these three gene signatures stratified three molecular subsets and the patient assignment to each subset is very similar with NMF analysis (Figure 1D). Kaplan-Meier survival analysis was then performed on these three subsets of patients, which showed that patients in S2 subset has significantly better survival (Log Rank test, P = 0.033) as compared with patients in S1 subset (Figure 1E), which was further validated in patients from The Cancer Genome Atlas (TCGA) cohort (N = 176) (Log Rank test, P = 0.0327) (Figure 1F). We next mined publicly available PDAC gene expression datasets (GSE16515, N = 37; GSE15471, N = 39) and concordantly found the above-defined three subsets (Supplementary Figure S2, available at Carcinogenesis Online). However, the survival information in the two of these cohorts were not available. QM (3), or basal-like (4), are the most commonly identified subtypes of pancreatic cancer with the worst survival. Consistently, we found that subset-1 patients in our test cohort overlap with 86.2% QM-PDA, and 71.8% Basal-like subtype (Supplementary Figure S3, available at Carcinogenesis Online), indicating the robust nature of our gene signature to classify PDAC. Next, we investigated if the 148 gene signature in S1 subset, the worst prognosis group in our cohort, can stratify PDAC patients with distinct prognosis. The 148 S1 subset-specific gene signature stratified patients in two clusters and predicted prognosis in a test (N = 136, P = 0.028) as well as validation cohorts (N = 176, P = 0.009) with significantly different survival (Figure 1G–I). Furthermore, Clusters predicted by 148 genes signature could predict S1 subset with 91.1% and 89.2% accuracy in the test and validations cohort, respectively (percentage of subset-1 patients in Cluster-2 with poor survival) (Figure 1G). Taken together, these findings identified a specific gene signature that could stratify patients with the worst outcome for further investigation into the molecular insights of disease aggressiveness.
Figure 1.
Gene expression profile identified subsets of PDAC patients with distinct prognosis. (A) Non-negative matrix factorization (NMF) analysis of 139 PDAC cases from microarray dataset after selecting for genes with SD greater than 0.6. Consensus matrix for cophenetic coefficient occurred for k = 3 are shown. (B) Subset specific signature (subset-1, S1, 148 genes; subset-2, S2, 217 genes; subset-3, S3, 123 genes) showed by Venn diagram illustrating the overlapped genes among S1, S2, S3 subsets. Differentially expressed genes between each of the subsets were identified by using ANOVA (adjusted P < 0.01, fold change >2). (C) Principal components analysis of mRNA expression profile using subset signature reveals three molecular subsets of tumors as shown by consensus clusters. (D) Hierarchical clustering analysis showing distinct gene expression profiles, using three specific subset gene-signature. (E and F) Kaplan-Meier survival curve comparing survival of patients in S1, S2 and S3 subsets in the test and TCGA cohort. P value is obtained by Log-rank test. (G) Hierarchical clustering analysis using 148 S1 gene-signature showing two distinct clusters (cluster-1 and cluster-2) of PDAC cases in both our test cohort (N = 136) and TCGA validation cohort (N = 176). (H and I) Kaplan-Meier survival curve comparing survival of PDAC cases in cluster-1 and cluster-2, in the test and validation cohorts. P values were obtained using Log Rank test.
CLU is a candidate gene associated with disease aggressiveness in patients with the worst survival
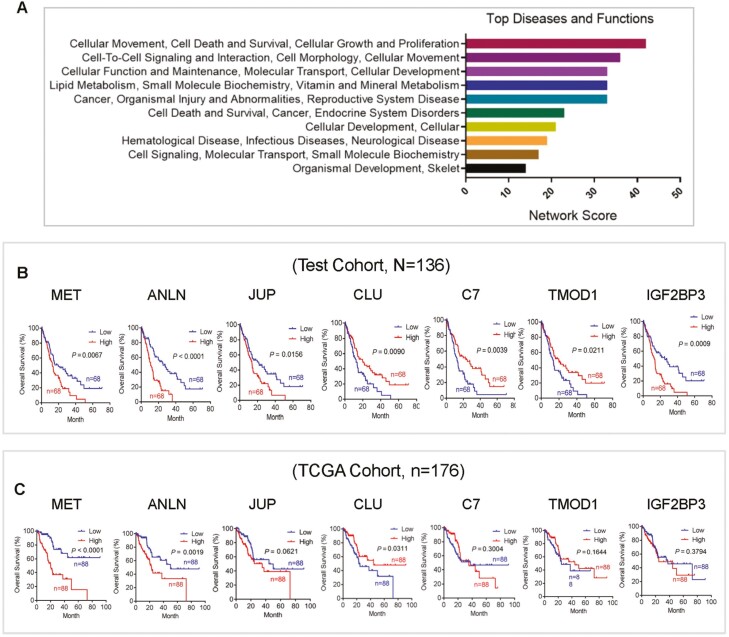
Ingenuity Pathway Analysis of 148, S1-Specific genes, identified top network genes that are associated with cellular movement, growth and proliferation, cell death and survival ranked by network score (Figure 2A). Cox regression analysis of subset-1 specific top network genes revealed that MET proto-oncogene (MET), anillin actin binding protein (ANLN), junction plakoglobin (JUP), CLU, complement C7 (C7), tropomodulin 1, insulin-like growth factor 2 mRNA binding protein 3 (IGF2BP3) could predict patients’ outcome in PDAC (Supplementary Table S3, available at Carcinogenesis Online). Additionally, Kaplan-Meier survival analysis of these genes indicated that MET, ANLN and CLU were associated with survival in the test (N = 136) as well as validation cohorts (N = 176) (Figure 2B and C). The functional role of MET and ANLN has been extensively described in pancreatic cancer (16,17). CLU is a molecular chaperone responsible for the stability of secreted proteins. It encodes secretory CLU (sCLU) and nuclear CLU (nCLU) protein which are differentially implicated in pro- and antiapoptotic processes in different cancer types (18,19). However, the mechanistic and functional role of CLU is not clearly understood in pancreatic cancer. We therefore pursued our further investigation into the mechanistic and functional role of CLU for its potential biological relevance in disease progression and its regulation in pancreatic cancer.
Figure 2.
CLU is one of the candidate genes associated with patient outcome. (A) Ingenuity Pathway Analysis of 148 S1-Specific genes identified top network genes associated with cellular movement, growth and proliferation, cell death and survival ranked by network score. (B and C) Kaplan-Meier survival analysis of the top network genes associated with survival (Cox regression analysis), including MET, ANLN, JUP, CLU, C7, TMOD1, IGF2BP3, in test (N = 136) and validated cohort (N = 176). A lower expression of CLU and higher expression of MET and ANLN associated with poor survival in both test and validation cohorts.
CLU inhibits proliferation and 3D spheroid growth in pancreatic cancer cell lines with gene expression profile representing subset-1 patient cohort
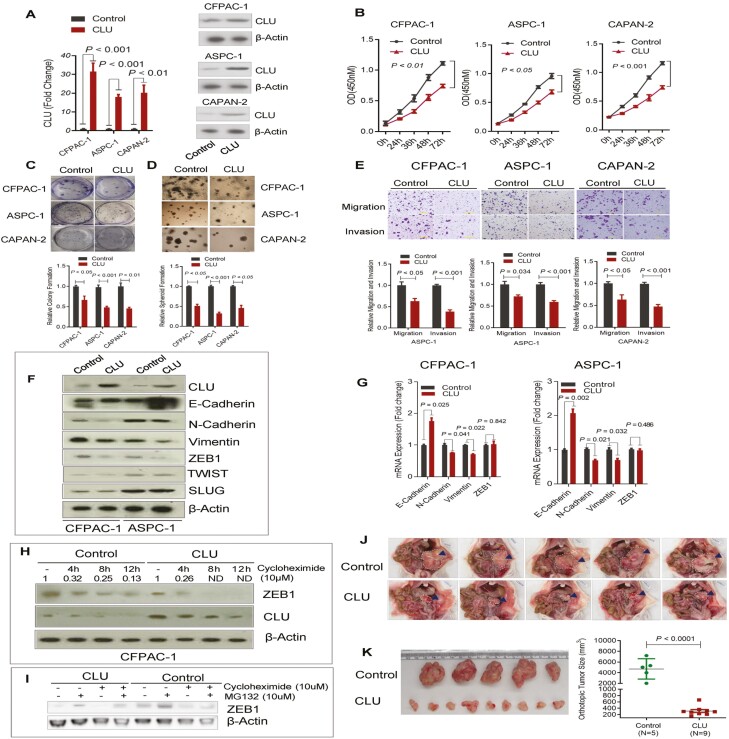
To investigate the function of CLU, we first screened the human pancreatic cancer cell-lines representing subset 1 with characteristic gene expression profile. Gene expression profile of pancreatic cancer cell lines from Cancer Cell Line Encyclopedia dataset (20) (https://portals.broadinstitute.org/ccle) was merged into our test cohort gene expression profile. Unsupervised hierarchical clustering and subclass mapping analysis identified CFPAC-1, ASPC-1 and CAPAN-2 cell lines as representatives of S1 subset patient tumors (Supplementary Figure S4, available at Carcinogenesis Online). We then generated lentiviral-mediated CLU-overexpressing stable pancreatic cancer cell lines (Figure 3A). CLU-overexpression resulted in a significant decrease in proliferation index of these pancreatic cancer cells (Figure 3B). The effect of CLU on the growth characteristics was further measured by clonogenicity and spheroid formation assay, which showed 50–75% reduction in the colony formation and spheroid growth (Figure 3C and D). On the contrary, knockdown of CLU in Su86.86 cells significantly enhanced proliferation and colony formation (Supplementary Figure S5A–C, available at Carcinogenesis Online).
Figure 3.
CLU inhibits proliferation, 3D spheroid growth, migration and invasion and suppresses epithelial-to-mesenchymal transition. (A) Lenti-viral mediated generation of stable pancreatic cancer cell lines overexpressing CLU as determined by quantitative real-time PCR (qRT-PCR) (left panel) and immunobloting (right panel). (B) CLU overexpression suppresses the proliferation of CFPAC-1, ASPC-1 and CAPAN-2 cells. (C) CLU-overexpressing cells showed reduced colony formation as compared with control cells. (D) CLU significantly suppresses spheroid formation in CFPAC-1, ASPC-1 and CAPAN-2 cell lines. (E) CLU overexpression suppresses migration and invasion as assessed by in vitro transwell assays. (F–H) CLU regulates EMT through promoting ZEB1 degradation. F, CLU-overexpression resulted in an increase in E-cadherin and decrease in N-cadherin, Vimentin and ZEB1 expression at the protein level as determined by immunoblotting. G, CLU enhances E-cadherin and suppresses N-Cadherin and Vimentin mRNA level as assessed by qRT-PCR. CLU overexpression showed no marked effect on the mRNA level of ZEB1. H, CLU overexpressing or control cells were treated with the 10 µm protein synthesis inhibitor cycloheximide for indicated time period, the protein expression of ZEB1 was measured by Western blot analysis and normalized to actin loading control (ND, not detectable). (I) CFPAC-1 control or CLU-overexpressing cells were treated with 10 μM proteasome inhibitor MG132 and Cycloheximide either alone or in combination for 8 h. ZEB1 expression was determined by western blot. (J–K) CLU inhibits pancreatic tumor growth in orthotopic mouse model. CLU significantly inhibited orthotopic tumor growth of CLU-overexpressing cells as compared with controls. (J and K) Orthotopic xenograft of 1 million cells implanted in the pancreas of nude mice showed a significant decrease in the growth of tumors arising from CLU expressing pancreatic cancer cells as compared with control cells. Data represent Mean ± SD.
CLU inhibits migration, invasion, and suppresses epithelial-to-mesenchymal transition through promoting ZEB1 degradation
We further investigated the potential role of CLU in the mobility and invasiveness of tumor cell lines representing subset-1 patients’ cohort, which is largely a prerequisite for invasion and metastasis. CLU overexpression significantly reduced the migration and invasion ability of these pancreatic cancer cells (Figure 3E). Furthermore, the Knockdown of CLU significantly enhanced migration and invasion in Su86.86 cell line (Supplementary Figure S5D, available at Carcinogenesis Online). Mechanistically, CLU expression led to the acquisition of epithelial features in pancreatic cancer cells. CLU expression resulted in an increase in E-cadherin and a decrease in N-cadherin and vimentin mRNA as well as protein expression in pancreatic cancer cell lines (Figure 3F and G). ZEB1 is a key epithelial-to-mesenchymal transition (EMT) regulator and functions as a transcriptional factor to enhance cell plasticity and promote metastasis in pancreatic cancer (21). Although ZEB1 protein expression was reduced in CLU over-expressing cells, there was no significant difference at the mRNA level in CLU overexpressing cells as compared to control cells (Figure 3F and G). It is conceivable that CLU may regulate ZEB1 at the post-translational level by affecting ZEB1 protein stability. It is hypothesized that CLU enhances proteasomal degradation (22). Therefore, we evaluated whether CLU reduces ZEB1 by enhancing proteasomal degradation. Treatment of CLU-overexpressing pancreatic cancer cell line CFPAC-1 with protein synthesis inhibitor cycloheximide resulted in the substantial inhibition of ZEB1 protein expression after 8 h. However, similar treatment in vector-control CFPAC cells showed the retention of about 25% of ZEB1 protein expression as compared to the untreated control cells (Figure 3H). Furthermore, MG-132, the inhibitor of proteasomal degradation, could restore the expression of ZEB1 in CLU overexpressing cells at a higher level than in control (Figure 3I). Collectively, these findings indicated that CLU may inhibit EMT in part by attenuating ZEB1 through proteasomal degradation in pancreatic cancer cells which may potentially contribute to its tumor-inhibitory role in pancreatic cancer.
CLU inhibits pancreatic tumor growth in a preclinical mouse model
To further investigate the role of CLU in pancreatic cancer growth and progression in vivo, we extended our investigation to orthotopic implantation of CLU-overexpressing stable pancreatic cancer cell line in mice. CLU-overexpressing Capan-2 cells, orthotopically transplanted in nude mice, showed a significant decrease in tumor growth as compared with control (P < 0.0001) xenografts (Figure 3J and K). This in vivo finding further supports the tumor inhibitory function of CLU in pancreatic cancer.
CLU enhances sensitivity to gemcitabine in pancreatic cancer
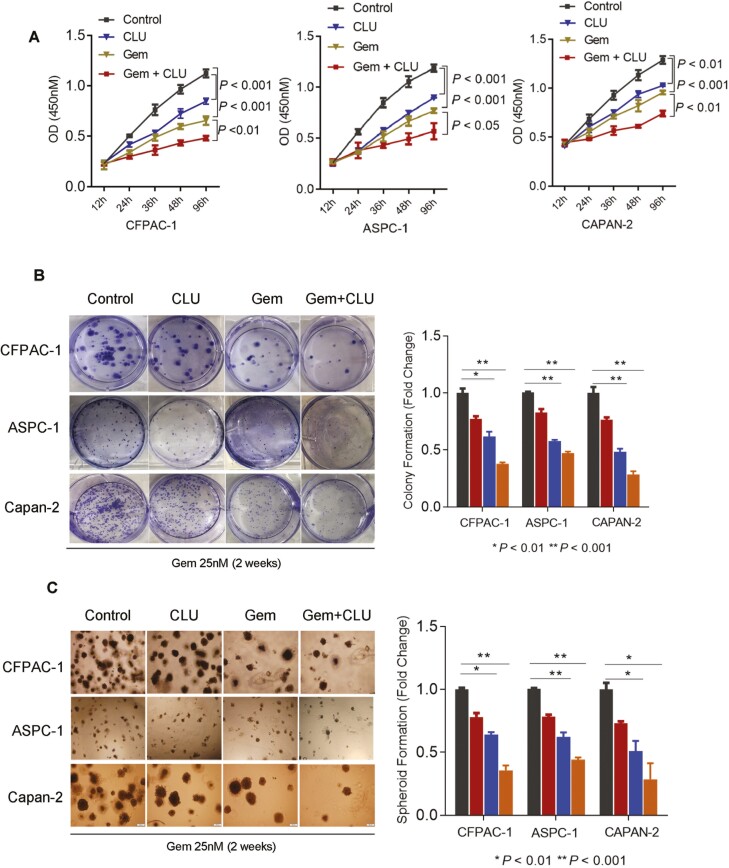
Gemcitabine has been used as a first-line standard chemotherapeutic drug against PDAC for more than a decade and efforts are being made to enhance its effectiveness as combination therapy. Therefore, we investigated if an enhanced level of CLU improves the sensitivity of pancreatic cancer cells to gemcitabine in pancreatic cancer. CLU overexpressing human pancreatic cancer cell lines showed a significant reduction in survival as compared to control cells following gemcitabine treatment (Figure 4A). These findings were further supported by clonogenicity and spheroid formation assays, which showed a significantly lower colony-forming ability (Figure 4B) and spheroid formation (Figure 4C) in CLU-overexpressing cells as compared with control cells following gemcitabine treatment. Earlier studies have described that nCLU is proapoptotic in contrast to sCLU. Our CLU overexpressing cell lines showed nuclear accumulation of CLU and increased apoptosis as indicated by enhanced Caspase-3 activity (Supplementary Figure S6, available at Carcinogenesis Online). Taken together these findings are consistent with the hypothesis that CLU enhances sensitivity to Gemcitabine in pancreatic cancer cell lines representing subset-1 cohort of patients with worse survival.
Figure 4.
CLU enhances sensitivity to Gemcitabine in pancreatic cancer cell lines. (A–C) CLU overexpressing cells showed a higher sensitivity to chemotherapeutic drug gemcitabine (25 nM) as compared with control cells, determined by cell survival index (A), colony formation assay (B) and spheroid formation (C). Data are presented as means ± SD from three independent experiments.
HNF1B regulates CLU in pancreatic cancer
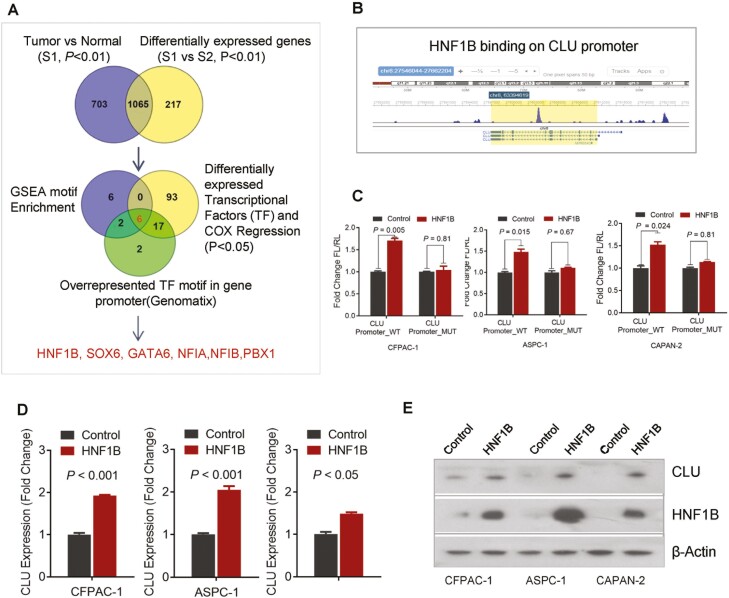
To further investigate the regulatory mechanisms of CLU in pancreatic cancer, we first investigated the potential key transcriptional regulators for subset-1 gene signature. Pancreatic cancer-specific genes from subset-1 patients (tumor versus normal, Pv< 0.01) were compared with genes differentially expressed between subset S1 and S2, which resulted in the identification of 1065 specific genes for S1 subset (Figure 5A; Supplementary Table S4, available at Carcinogenesis Online). We then used an integrative strategy, which combines Gene Set Enrichment Analysis (GSEA) motif enrichment (23), overrepresented transcriptional factor motif analysis by Genomatix software suite, and differentially expressed transcriptional factors associated with survival (cox regression) in PDAC cohort. This approach identified HNF1B, SOX6, GATA6, NF1A, NF1B and PBX1 as potential candidate regulators for subset-1 gene signature (Figure 5A). Further assessment of the datasets in the literature led us to identify ChIP-seq peak for the binding of HNF1B transcription factor to CLU promoter (Figure 5B>) (24). Luciferase reporter-based activity assay showed the interaction of HNF1B to CLU promoter resulting in an increase in reporter activity following the expression of HNF1B. However, these effects were abolished by the mutation of a core binding region on CLU promoter (Figure 5C). Furthermore, the overexpression of HNF1B enhanced both mRNA and protein expression of CLU (Figure 5D and E), which is consistent with the hypothesis that HNF1B upregulates CLU in pancreatic cancer.
Figure 5.
HNF1B regulates CLU in pancreatic cancer. (A) Schema depicting integrative approach to identify putative key transcriptional regulator for subset-1 gene signature. Venn diagram showing the integrative bioinformatics strategy, which resulted in the identification of putative transcriptional factors. Pancreatic cancer specific genes from subset-1 patients (Tumor versus normal, P < 0.01) were compared with genes differentially expressed between subsets S1 and S2, which resulted in the identification of 1037 genes, these genes were further subjected to Gene Set Enrichment Analysis (GSEA) motif enrichment assessment, overrepresented transcriptional factor motif analysis by Genomatix, and overlaped with differentially expressed transcriptional factors associated with survival (Cox regression analysis). HNF1B, SOX6, GATA6, NF1A, NF1B and PBX1 were identified as candidate regulators for subset-1 gene signature. (B) Identification of the HNF1B-binding site in the promoter regions of CLU using publicly available data set (18). (C) HNF1B enhanced the luciferase reporter activity of pGL-4-Basic construct containing wild-type CLU promoter sequence but had no effect on mutant construct in CFPAC-1, ASPC-1 and CAPAN-2 cell lines. (D, E) An increase in endogenous CLU mRNA and protein expression by HNF1B overexpression as shown by qRT-PCR and immunoblotting in pancreatic cancer cell lines.
Clinical validation of HNF1B/CLU axis in human pancreatic cancer
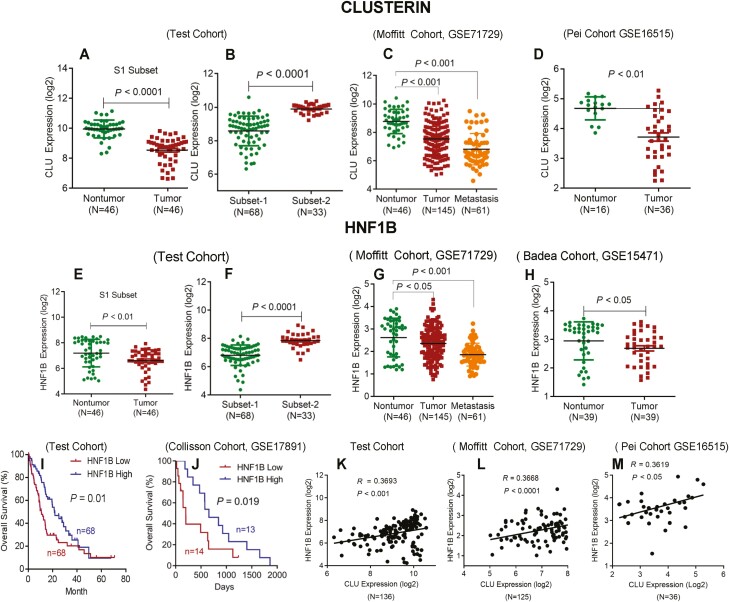
Next, we examined the relevance of HNF1B/CLU axis in the disease aggressiveness in human pancreatic cancer using multiple independent cohorts of PDAC patients. A lower expression of CLU was found in tumors as compared to the adjacent nontumor pancreas (Figure 6A). Tumors from subset-1 showed a reduced expression of CLU in test cohort (Figure 6B). Furthermore, we validated these findings in publicly available dataset (GSE71729) (5). A significantly lower expression of CLU was found in metastatic and primary tumors as compared with nontumors pancreas (Figure 6C). A lower expression of CLU in tumors as compared with nontumors pancreas was also identified in a second validation dataset (GSE16515) (Figure 6D). Additionally, we also evaluated the expression of CLU in paraffin embedded tumor and nontumor tissues from distinct subsets. Immunohistochemical staining showed a lower expression of CLU in tumors as compared to nontumors (P = 0.005), and tumors representing subset-1 showed a lower expression of CLU as compared with tumors from subset-2 patients (P = 0.022) (Supplementary Figure S7, available at Carcinogenesis Online). We next sought to identify the clinical relevance of HNF1B in PDAC cases. A lower expression of HNF1B was found in tumor as compared with nontumor pancreas, and also in tumors from Subset-1 cases as compared with subset 2, which was also validated in an additional gene expression dataset (GSE 71729, GSE15471) (Figure 6E–H, available at Carcinogenesis Online). Furthermore, a lower expression of HNF1B was associated with poor survival in our test cohort, which was further validated in an independent cohort (GSE17891) (Figure 6I and J). Finally, CLU expression in the tumors positively correlated with HNF1B in the test cohort and two additional validation cohorts (Figure 6K–M). Taken together our findings with multiple validations in independent patient cohorts demonstrate the clinical relevance of HNF1B/CLU axis in disease aggressiveness and patient outcome in the highly aggressive subset of patients with PDAC.
Figure 6.
Clinical relevance of HNF1B/CLU axis in human pancreatic cancer. (A) A reduced expression of CLU was found in tumor as compared with nontumor tissue. (B) A lower expression of CLU was detected in subset-1 as compared with subset-2 patients in the test cohort (left two panels). (C and D) Validation of CLU expression in publicly available datasets. Metastatic tumors expressed a lower level of CLU as compared with primary tumors and adjacent nontumors pancreas in GSE71729 dataset (C). A lower expression of CLU was found in tumor versus nontumor in GSE16515 Dataset. Dot plots represent the normalized log 2 transformed CLU expression values obtained by gene expression microarray analysis (D). (E) A lower level of HNF1B was found in tumors as compared with adjacent nontumor pancreas. (F) A lower expression of HNF1B in subset-1 as compared with subset-2 patients in the test cohort (left two panels). (G and H) Validation of HNF1B expression in publicly available datasets. A lower level of CLU was found in primary tumors and metastatic tumors as compared with nontumor pancreas in GSE71729 dataset (G); a lower expression of CLU was found in tumor as compared with nontumor pancreas in GSE15471 Dataset (H). (I and J) Kaplan-Meier survival analysis showing a significantly poorer survival in patients with a lower (<median) as compared to patients with a higher HNF1B expression (>median) in test and validation cohorts (Log Rank test). (K–M) Pearson correlation analysis showing a positive correlation between CLU and HNF1B expression level in the test and validation cohorts (publicly available datasets GSE71729 and GSET16515). Each data point represents an individual patient with PDAC.
Discussion
Recent studies have significantly contributed to our understanding of pancreatic tumor biology and revealed that the underlying mechanisms of pancreatic cancer progression and disease aggressiveness are highly complex and need further investigation using novel approaches (25). Furthermore, the continuously evolving insights into pancreatic tumor biology have not yet contributed to the effective treatment in patients with this lethal malignancy. Among several recent advancements, one is the classification of PDAC into subgroups that are associated with distinct patient prognoses (3–5,7). The existence of distinct subgroups with difference in survival among patients with PDAC, a uniformly lethal disease, provide an opportunity to understand the underlying mechanisms of disease aggressiveness. Although an increasing number of studies have identified molecular subtypes which associate with the time of survival following diagnosis in pancreatic cancer patients, our understanding of the mechanisms of disease aggressiveness and their clinical implication remains inadequate. Recently, the cells of origin have been implicated in the development of distinct subtypes (15). In the present study, molecular analysis of a highly aggressive subset of PDAC in multiple independent patient cohorts, followed by the mechanistic and functional investigation using preclinical models, have identified CLU as a potential inhibitor of tumor growth, which is regulated by HNF1B, attenuating disease progression by negatively regulating ZEB1 and EMT, and enhancing sensitivity to chemotherapeutic drug gemcitabine. Furthermore, dysregulation of HNF1B/CLU axis was found to be relevant in the clinical outcome of PDAC patients and may potentially be exploited for the management of this lethal cancer.
CLU is a glycoprotein with multiple physiological and pathophysiological roles. It is implicated in several physiological functions including immune regulation, inflammation, lipid transport, cellular adhesion, differentiation and remodeling, stabilization of stressed protein, apoptosis, proliferation and survival (26,27). CLU is described to play a role in human pathologies including cancer with evidence of its context-dependent and often paradoxical roles in many human cancer (28,29). Although both tumor-suppressing and tumor-promoting functions of CLU have been described, the exact role of CLU in cancer and in particular pancreatic cancer needs further investigation. A lower expression of CLU is described in head and neck cancer and is targeted by oncogenic miRNA-21 (30). Consistent with these findings, a negative correlation was found between miR-21 and CLU gene expression in tumors from our PDAC patients’ cohort (Supplementary Figure S8, available at Carcinogenesis Online). Moreover, an increased expression of CLU is reported in breast cancer (31). In line with these evidences, genetic deletion of CLU enhanced neuroblastoma growth and metastasis suggesting a tumor-inhibitory role of CLU (32). Two major isoforms of CLU, nCLU and sCLU, have been described and are attributed to its diverse functions depending on the context and pattern shift of these isoforms as described in colorectal cancer (33). In our gene expression profiling of a large cohort of PDAC patients (N = 136), a lower expression of CLU (<median) was associated with a subset of patients with worst survival. This observation was also validated in an independent cohort of PDAC patients in TCGA dataset. Consistently, in a small cohort of PDAC patients, CLU positive tumors as determined by immunohistochemistry associated with better prognosis as compared to CLU-negative tumors (34). Our findings indicated that CLU may have potential biological relevance in impeding pancreatic cancer progression. Consistent with this hypothesis, CLU inhibited proliferation, migration, invasion, and spheroid growth in pancreatic cancer cells. Knockdown of CLU expression significantly enhanced proliferation, colony formation, migration and invasion. Furthermore, CLU significantly reduced orthotopic tumor growth in mice. Taken together these findings showed that CLU plays a tumor-inhibitory role in pancreatic cancer. Further pharmacological approaches may highlight its potential therapeutic significance. Furthermore, earlier studies have described the role of CLU in modulating the sensitivity of chemotherapeutic drugs (35). As anticipated, CLU was found to be associated with both enhancement and reduction of sensitivity of tumor cells to chemotherapeutic agents (36–38). This discrepancy was explained as the result of the existence of nuclear and secretory isoforms of CLU with pro- and anti-apoptotic functions, respectively. An increased nCLU/sCLU ratio in cancer cells enhanced the sensitivity of chemotherapeutic agents (39). Consistently, overexpression of CLU in pancreatic cancer cell lines, representing a highly aggressive patient subset in our cohort, enhanced the sensitivity of the standard of care drug gemcitabine with an increased accumulation of nCLU and caspase-3 activity (Figure 4; Supplementary Figure S6, available at Carcinogenesis Online). Therefore, it should be investigated if an enhanced level of CLU improves the sensitivity of pancreatic cancer to gemcitabine and enhances outcome in resected patients of pancreatic cancer.
CLU gene expression is regulated by multiple mechanisms including epigenetic modulation, oncogenic signaling, transcription factors, inflammatory signaling molecules and their interactive pathways, further suggesting a highly complex and context-dependent function of CLU (28). One example is oncomiR-21, which specifically suppresses proapoptotic tumor suppressive nCLU in head and neck squamous cell carcinoma (30). Likewise, miR-378 targets anti-apoptotic sCLU and reduces the growth of lung adenocarcinoma (40). Similarly, NF-kB (41), Myc (41), STAT and cAMP (42) are some of the other factors that are reported to modulate CLU expression. However, the regulation of CLU in pancreatic cancer and its clinical relevance, to the best of our knowledge, is previously not described.
By using an integrative strategy as described above, we identified HNF1B transcription factor as a regulator of CLU in pancreatic cancer. HNF1B bound to the CLU promoter and enhanced its expression at mRNA and protein level in multiple pancreatic cancer cell lines. HNF1B plays a role in the development and progression of human cancer with evidence suggesting both tumor-suppressing and tumor-promoting functions (43). A number of single nucleotide polymorphisms in HNF1B genes are recognized, which are linked to an increased risk of cancer (44,45). A recent genome-wide meta-analysis identifies a new susceptibility HNF1B locus rs4795218 at 17q12 for pancreatic cancer (46). A higher HNF1B predicts a better prognosis and inhibits disease progression by binding to SLUG in prostate cancer (47). A lower expression of HNFB1 was found in tumor as compared to nontumor pancreas in our PDAC patient cohort. Furthermore, a significantly lower HNF1B was observed in subset 1 patients with worse survival. These findings were also validated in other independent datasets (Figure 6G and H) showing a significantly reduced HNF1B in metastatic disease as compared to patients without metastasis. Additionally, a lower HNF1B predicted poorer survival in test and validation cohorts. Our findings identified HNF1B as a novel regulator of CLU in pancreatic cancer patients.
Taken together these findings provided mechanistic and clinical evidence of a novel HNF1B/CLU axis, which negatively regulates pancreatic cancer progression and disease aggressiveness and may potentially be relevant in designing strategies for improving patient outcome.
Supplementary Material
Acknowledgments
The authors would like to thank Ms. Elise Bowman LHC, CCR, NCI and Dr Dean Mann, MD/PhD, University of Maryland School of Medicine and NCI-University of Maryland study coordinators for the procurement of clinical biospecimens from pancreatic cancer patients.
Abbreviations
- ANLN
anillin actin binding protein
- CLU
Clusterin
- C7
complement C7
- EMT
epithelial-to-mesenchymal transition
- HNF1B
HNF1 homeobox B
- IGF2BP3
insulin-like growth factor 2 mRNA binding protein 3
- JUP
junction plakoglobin
- MET
MET proto-oncogene, receptor tyrosine kinase
- NFM
non-negative matrix factorization
- PDAC
pancreatic ductal adenocarcinoma
- qRT-PCR
quantitative real-time PCR
- mRNA
messenger RNA
- TMOD1
tropomodulin 1
Contributor Information
Shouhui Yang, Pancreatic Cancer Section, Laboratory of Human Carcinogenesis, CCR, NCI, NIH, Bethesda, MD, USA.
Wei Tang, Laboratory of Human Carcinogenesis, CCR, NCI, NIH, Bethesda, MD, USA.
Azadeh Azizian, Department of General, Visceral and Pediatric Surgery, University Medical Center Göttingen, Göttingen, Germany.
Jochen Gaedcke, Department of General, Visceral and Pediatric Surgery, University Medical Center Göttingen, Göttingen, Germany.
Philipp Ströbel, Department of General, Visceral and Pediatric Surgery, University Medical Center Göttingen, Göttingen, Germany.
Limin Wang, Laboratory of Human Carcinogenesis, CCR, NCI, NIH, Bethesda, MD, USA.
Helen Cawley, Pancreatic Cancer Section, Laboratory of Human Carcinogenesis, CCR, NCI, NIH, Bethesda, MD, USA.
Yuuki Ohara, Pancreatic Cancer Section, Laboratory of Human Carcinogenesis, CCR, NCI, NIH, Bethesda, MD, USA.
Paloma Valenzuela, Pancreatic Cancer Section, Laboratory of Human Carcinogenesis, CCR, NCI, NIH, Bethesda, MD, USA.
Lin Zhang, Pancreatic Cancer Section, Laboratory of Human Carcinogenesis, CCR, NCI, NIH, Bethesda, MD, USA.
Trisha Lal, Howard University College of Medicine, Washington, DC, USA.
Sanju Sinha, Cancer Data Science Laboratory, CCR, NCI, Bethesda, MD, USA.
Eythan Rupin, Cancer Data Science Laboratory, CCR, NCI, Bethesda, MD, USA.
Nader Hanna, Division of Surgical Oncology, University of Maryland School of Medicine, Baltimore, MD, USA.
B Michael Ghadimi, Department of General, Visceral and Pediatric Surgery, University Medical Center Göttingen, Göttingen, Germany.
S Perwez Hussain, Pancreatic Cancer Section, Laboratory of Human Carcinogenesis, CCR, NCI, NIH, Bethesda, MD, USA.
Funding
Intramural Research Program of the Center for Cancer Research, NCI, NIH.
Conflict of interest
The authors declare no competing financial interest.
Authors contributions
S.Y., S.P.H. (study concept, data acquisition, writing manuscript); H.C., W.T., A.A., J.G., P.S., L.W., J.G., Y.O., P.V., L.Z., T.L., E.R., N.H., B.M.G. (data acquisition, statistical analysis, technical and material support).
Data availability
The microarray expression data underling this article have been deposited in the National Center for Biotechnology Information’s (NCBIs) Gene Expression Omnibus (GSE 183795).
References
- 1. Siegel, R.L., et al. (2022) Cancer statistics, 2022. CA Cancer J. Clin., 72, 7–33. [DOI] [PubMed] [Google Scholar]
- 2. Rahib, L., et al. (2014) Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res., 74, 29134006–29132921. [DOI] [PubMed] [Google Scholar]
- 3. Bailey, P., et al. (2016) Genomic analyses identify molecular subtypes of pancreatic cancer. Nature, 531, 47–52. [DOI] [PubMed] [Google Scholar]
- 4. Cancer Genome Atlas Research Network. et al. (2017) Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell, 32, 185–203.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Collisson, E.A., et al. (2011) Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med., 17, 500–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Daemen, A., et al. (2015) Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors. Proc. Natl. Acad. Sci. U.S.A., 112, E4410–E4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moffitt, R.A., et al. (2015) Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet., 47, 1168–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Puleo, F., et al. (2018) Stratification of pancreatic ductal adenocarcinomas based on tumor and microenvironment features. Gastroenterology, 155, 19991999–1992013.e3. [DOI] [PubMed] [Google Scholar]
- 9. Kalimuthu, S.N., et al. (2020) Morphological classification of pancreatic ductal adenocarcinoma that predicts molecular subtypes and correlates with clinical outcome. Gut, 69, 317–328. [DOI] [PubMed] [Google Scholar]
- 10. Mehla, K., et al. (2020) Metabolic subtyping for novel personalized therapies against pancreatic cancer. Clin. Cancer Res., 26, 6–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Adams, C.R., et al. (2019) Transcriptional control of subtype switching ensures adaptation and growth of pancreatic cancer. Elife, 8, e45313, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hamdan, F.H., et al. (2018) DeltaNp63-dependent super enhancers define molecular identity in pancreatic cancer by an interconnected transcription factor network. Proc. Natl. Acad. Sci. U.S.A., 115, E12343–E12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Noll, E.M., et al. (2016) CYP3A5 mediates basal and acquired therapy resistance in different subtypes of pancreatic ductal adenocarcinoma. Nat. Med., 22, 278–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Somerville, T.D.D., et al. (2020) Squamous trans-differentiation of pancreatic cancer cells promotes stromal inflammation. Elife, 9, e53381, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Flowers, B.M., et al. (2021) Cell of origin influences pancreatic cancer subtype. Cancer Discov., 11, 660–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang, A., et al. (2019) ANLN-induced EZH2 upregulation promotes pancreatic cancer progression by mediating miR-218-5p/LASP1 signaling axis. J. Exp. Clin. Cancer Res., 38, 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ebert, M., et al. (1994) Coexpression of the c-met proto-oncogene and hepatocyte growth factor in human pancreatic cancer. Cancer Res., 54, 5775–5778. [PubMed] [Google Scholar]
- 18. Koltai, T. (2014) Clusterin: a key player in cancer chemoresistance and its inhibition. Onco Targets Ther., 7, 447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shannan, B., et al. (2006) Challenge and promise: roles for clusterin in pathogenesis, progression and therapy of cancer. Cell Death Differ., 13, 12–19. [DOI] [PubMed] [Google Scholar]
- 20. Barretina, J., et al. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature, 483, 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krebs, A.M., et al. (2017) The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol., 19, 518–529. [DOI] [PubMed] [Google Scholar]
- 22. Satapathy, S., et al. (2021) The dual roles of clusterin in extracellular and intracellular proteostasis. Trends Biochem. Sci., 46, 652–660. [DOI] [PubMed] [Google Scholar]
- 23. Subramanian, A., et al. (2005) Gene Set Enrichment Analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A., 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Diaferia, G.R., et al. (2016) Dissection of transcriptional and cis-regulatory control of differentiation in human pancreatic cancer. EMBO J., 35, 595–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beatty, G.L., et al. (2021) The biological underpinnings of therapeutic resistance in pancreatic cancer. Genes Dev., 35(13-14), 940–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trougakos, I.P., et al. (2009) Advances and challenges in basic and translational research on clusterin. Cancer Res., 69, 403–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garcia-Aranda, M., et al. (2018) Regulation of clusterin gene expression. Curr. Protein Pept. Sci., 19, 612–622. [DOI] [PubMed] [Google Scholar]
- 28. Garcia-Aranda, M., et al. (2018) Regulation of clusterin gene expression. Curr. Protein Pept. Sci., 19, 612–622. [DOI] [PubMed] [Google Scholar]
- 29. Tellez, T., et al. (2016) The role of clusterin in carcinogenesis and its potential utility as therapeutic target. Curr. Med. Chem., 23, 4297–4308. [DOI] [PubMed] [Google Scholar]
- 30. Mydlarz, W., et al. (2014) Clusterin is a gene-specific target of microRNA-21 in head and neck squamous cell carcinoma. Clin. Cancer Res., 20, 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Redondo, M., et al. (2000) Overexpression of clusterin in human breast carcinoma. Am. J. Pathol., 157, 393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chayka, O., et al. (2009) Clusterin, a haploinsufficient tumor suppressor gene in neuroblastomas. J. Natl. Cancer Inst., 101, 663–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pucci, S., et al. (2004) Modulation of different clusterin isoforms in human colon tumorigenesis. Oncogene, 23, 2298–2304. [DOI] [PubMed] [Google Scholar]
- 34. Xie, M.J., et al. (2002) Expression of clusterin in human pancreatic cancer. Pancreas, 25, 234–238. [DOI] [PubMed] [Google Scholar]
- 35. Garcia-Aranda, M., et al. (2017) Clusterin inhibition mediates sensitivity to chemotherapy and radiotherapy in human cancer. Anticancer Drugs, 28, 702–716. [DOI] [PubMed] [Google Scholar]
- 36. Kevans, D., et al. (2012) Clusterin and chemotherapy sensitivity under normoxic and graded hypoxic conditions in colorectal cancer. J. Gastrointest. Cancer, 43, 305–313. [DOI] [PubMed] [Google Scholar]
- 37. Xu, M., et al. (2015) Clusterin silencing sensitizes pancreatic cancer MIA-PaCa-2 cells to gmcitabine via regulation of NF-kB/Bcl-2 signaling. Int. J. Clin. Exp. Med., 8, 12476–12486. [PMC free article] [PubMed] [Google Scholar]
- 38. Zhong, B., et al. (2010) Induction of clusterin by AKT—role in cytoprotection against docetaxel in prostate tumor cells. Mol. Cancer Ther., 9, 1831–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Essabbani, A., et al. (2013) Exon-skipping strategy by ratio modulation between cytoprotective versus pro-apoptotic clusterin forms increased sensitivity of LNCaP to cell death. PLoS One, 8, e54920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen, X.S., et al. (2016) miRNA-378 reverses chemoresistance to cisplatin in lung adenocarcinoma cells by targeting secreted clusterin. Sci. Rep., 6, 19455,1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sala, A., et al. (2009) Regulation of CLU gene expression by oncogenes and epigenetic factors: implications for tumorigenesis. Adv. Cancer Res., 105, 115–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pignataro, O.P., et al. (1992) Cyclic adenosine-3ʹ,5ʹ-monophosphate negatively regulates clusterin gene-expression in Leydig tumor-cell lines. Endocrinology, 130, 2745–2750. [DOI] [PubMed] [Google Scholar]
- 43. Chandra, S., et al. (2021) Hepatocyte nuclear factor 1 beta: a perspective in cancer. Cancer Med., 10, 1791–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sun, J., et al. (2008) Evidence for two independent prostate cancer risk-associated loci in the HNF1B gene at 17q12. Nat. Genet., 40, 1153–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thomas, G., et al. (2008) Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet., 40, 310–315. [DOI] [PubMed] [Google Scholar]
- 46. Klein, A.P., et al. (2018) Genome-wide meta-analysis identifies five new susceptibility loci for pancreatic cancer. Nat. Commun., 9, 556, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang, J.Q., et al. (2020) HNF1B-mediated repression of SLUG is suppressed by EZH2 in aggressive prostate cancer. Oncogene, 39, 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray expression data underling this article have been deposited in the National Center for Biotechnology Information’s (NCBIs) Gene Expression Omnibus (GSE 183795).