Abstract
Background
Artemisinin resistance mutations in Plasmodium falciparum kelch13 (Pfk13) have begun to emerge in Africa, with Pfk13-R561H being the first reported in Rwanda in 2014, but limited sampling left questions about its early distribution and origin.
Methods
We genotyped P. falciparum positive dried blood spot (DBS) samples from a nationally representative 2014–2015 Rwanda Demographic Health Surveys (DHS) HIV study. DBS were subsampled from DHS sampling clusters with >15% P. falciparum prevalence, as determined by rapid testing or microscopy done during the DHS study (n clusters = 67, n samples = 1873).
Results
We detected 476 parasitemias among 1873 residual blood spots from a 2014–2015 Rwanda Demographic Health Survey. We sequenced 351 samples: 341/351 were wild-type (97.03% weighted), and 4 samples (1.34% weighted) harbored R561H that were significantly spatially clustered. Other nonsynonymous mutations found were V555A (3), C532W (1), and G533A (1).
Conclusions
Our study better defines the early distribution of R561H in Rwanda. Previous studies only observed the mutation in Masaka as of 2014, but our study indicates its presence in higher-transmission regions in the southeast of the country at that time.
Keywords: artemisinin, antimalarial resistance, drug resistance, k13, kelch13, r561H
Artemisinin-based combination therapies (ACTs) are currently the key antimalarials used in Africa, and emerging resistance threatens decades of public health gains. Concerningly, mutations in the Plasmodium falciparum kelch13 (Pfk13) gene causing in vitro artemisinin resistance are emerging in Africa [1, 2]. Additionally, recent reports have shown delayed parasite clearance in Rwandan patients infected with R561H mutants after treatment with ACT [3]. While clinical resistance to ACT remains absent in Africa, decreased sensitivity to artemisinin has led to ACT treatment failure in Southeast Asia (SEA), making the emergence of validated Pfk13 resistance mutations a grave concern [4–6].
Early in the emergence of artemisinin resistance in SEA, multiple mutations in Pfk13 were found, with the eventual emergence of C580Y as the dominant genotype. A similar pattern may be occuring in Africa with multiple validated or candidate resistance mutations emerging including R561H, C469F/Y, and A675V [1, 2, 7, 8]. Understanding trends in resistance allele frequencies can only be done through intensive genomic surveillance.
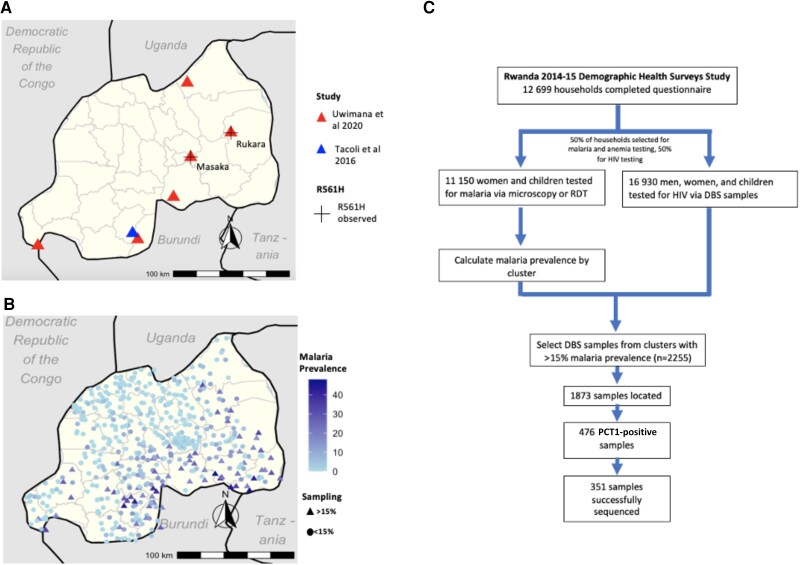
The first report of artemisinin-resistant Pfk13 mutants in Africa occurred in Rwanda in 2014, with 7.4% of samples collected in Masaka between 2014 and 2015 harboring R561H [1]. By 2018, the reported R561H prevalence increased to 19.6% in Masaka and 22% in Rukara [9]. While ring-stage survival assays confirmed in vitro artemisinin resistance, these initial studies found no association between clinical outcomes and Pfk13 mutations. However, a more recent study has demonstrated an association with R561H and delayed parasite clearance after treatment, the clinical metric that drives World Health Organization policy to change therapy [9]. The expansion of a validated resistance mutation is highly concerning, and the lack of broader sampling in previous studies makes it difficult to trace R561H's spread through the country. To our knowledge, only 2 Pfk13 genotyping studies were done in Rwanda between 2010 and 2015, the years preceding and including the first report of R561H (Figure 1A) [1, 10]. These studies were limited to a few sampling sites, leaving questions about the true distribution of R561H early in its emergence.
Figure 1.
Previous studies and our study design. A, Sampling sites of P. falciparum genotyping studies done in Rwanda, 2010–2015. Sites included Masaka, Ruhuha, Bugarama, Kibirizi, Nyarurema and Rukara [1], and the Huye district, 2010–2015 [10]. B, DHS 2014 sampling clusters. We sequenced samples from clusters with >15% malaria prevalence. C, Study design. We used DBS samples from the HIV arm of a 2014–2015 DHS study, selecting 2255 samples taken from 67 clusters with >15% malaria, as calculated in the malaria/anemia arm. Abbreviations: DBS, dried blood spot; DHS, Rwanda Demographic Health Surveys; PCR, polymerase chain reaction; RDT, rapid diagnostic test.
Here we used an existing Demographic Health Surveys (DHS) sample set, originally designed as a random representative sample of the Rwandan population for HIV prevalence, conducted between 2014 and 2015. Repurposing this nationally representative data set provides an opportunity for a nationwide baseline study of R561H prevalence in Rwanda early in the emergence timeline (Figure 1B). This study also employs a cost-conscious approach for genotyping discarded samples that can be employed in resource-limited settings.
METHODS
Sample Collection and Preparation
Sample collection for 2014–2015 DHS in Rwanda included a subset of participants from whom dried blood spots (DBS) were collected on filter paper for HIV testing. Residual DBS were provided by the Rwanda Biomedical Center. The DHS included 12 699 GPS-tagged households in 492 clusters in 30 regions who completed the initial questionnaire. Of these households, 50% were selected for malaria and anemia testing in women and children, and the other 50% were selected for HIV testing via DBS sample collection (Figure 1C). Complete survey methods are described by the DHS [11]. The final anonymized DHS data set was downloaded from https://dhsprogram.com. All data cleaning and analysis was done in R, version 4.1.2, using packages detailed here: https://github.com/bailey-lab/Rwanda-DHS-2014-15. Analyses utilizing parasite genomes from de-identified samples were deemed nonhuman subjects research at the University of North Carolina at Chapel Hill (NC, USA) and Brown University (RI, USA).
From the DHS survey database, malaria prevalence for each cluster was calculated based on either positive rapid diagnostic test (RDT) or microscopy. Given that we could not simply identify positive DBS samples based on RDT testing as they were different groups, we focused on clusters where we would expect to detect a reasonable number of positives to genotype, selecting clusters with >15% malaria prevalence (n clusters = 67, n samples = 2255). Regional malaria prevalence was calculated in R using survey-weighted means, with DHS household survey weights. We located 1873 of these samples and placed three 6-mm punches into wells of a 2-mL plate. DNA was extracted using Chelex [12].
Amplification of Short Polymerase Chain Reaction Amplicons Including R561H Site
A 2-step polymerase chain reaction (PCR) was optimized to amplify short amplicons with sample barcodes covering amino acid positions 516–572 on the Pfk13 gene (Supplementary Methods). PCR step 1 product was resolved on an agarose gel, and the presence of a faintly visible or stronger band was used to select samples for sequencing. First-step primers were based on previous designs [13] and contained molecular inversion probe (MIP)–compatible 5' linking sequences, enabling barcoding [12]. Positive samples (n = 476) were replated using an OT2 robot (Opentrons, Brooklyn, NY, USA). PCR step 2 was used to add unique barcodes to each sample, and these products were resolved on a gel.
Library Preparation
To reduce oversequencing of high-parasitemia samples, 5 μL of each PCR step 2 reaction was sorted into high- and low-parasitemia pools based on the PCR step 2 band intensity using an Opentrons OT2. To prepare libraries for sequencing, a 1.2× SPRI bead cleanup was performed on the 2 pools eluted in 30 μL of TE low EDTA pH = 8 (Thermo Fisher Scientific, Waltham MA, USA). The eluted pools were combined for a total volume of 20 μL, and amplicons were resolved on a 1.0% agarose gel. Fragments of the correct size were excised, and DNA was purified using the NEB DNA Gel Extraction kit T1020S (Ipswich, MA, USA). Library quality was assessed using a fragment analyzer (Agilent, Santa Clara, CA, USA); the library was pooled with other libraries and sequenced on an Illumina Nextseq 550 using a 300-cycle mid output kit at 5.3 pM.
Analysis
Samples were demultiplexed using MIPtools software available at https://github.com/bailey-lab/MIPTools and analyzed using the SeekDeep pipeline available at https://github.com/bailey-lab/SeekDeep [14]. Samples that had <50 reads on the first pass of sequencing (n = 243) were repooled, gel-extracted, and resequenced. Samples with ≥20 total combined reads were included in subsequent analysis (n = 351). Mutation prevalence was calculated using HIV survey weights.
Spatial correlation of R561H was calculated in GeoDa (https://geodacenter.github.io) using Moran's I with empirical Bayes to take varying cluster sample sizes into account. Significance was calculated in Geoda with 999 permutations. A 97.5% confidence interval of inferred R561H prevalence was created using a spatial binomial logistic model.
Map figures were created in RStudio using spatial data downloaded from the DHS website.
RESULTS
By genotyping parasites from discarded DHS samples, we were able to investigate the distribution of a small range of Pfk13 mutations (positions 516–572) in Rwanda early in the emergence of R561H.
Short Amplicon PCR Was an Effective Way to Determine Infections
Amplification identified 25.4% of samples as positive (476/1873), similar to the expected 25.7% positivity found by DHS by RDT. On a per-cluster level, we found that amplicon success had moderate correlation with the expected P. falciparum prevalence (Pearson coefficient = 0.357).
Targeted Deep Sequencing of Amplicons Revealed R561H and Other Pfkelch13 Mutations
Of the 476 PCR-positive samples, 351 (73.7%) were successfully sequenced for the 516–572 amino acid region, revealing a majority of wild-type parasites (341/351, 97.03%) (Supplementary Table 1). One sample (0.27%) had a synonymous mutation (G533G), and 9 samples (2.70%) had nonsynonymous Pfk13 mutations, including R561H (4), V555A (3), C532W (1), and G533A (1) (Supplementary Table 1).
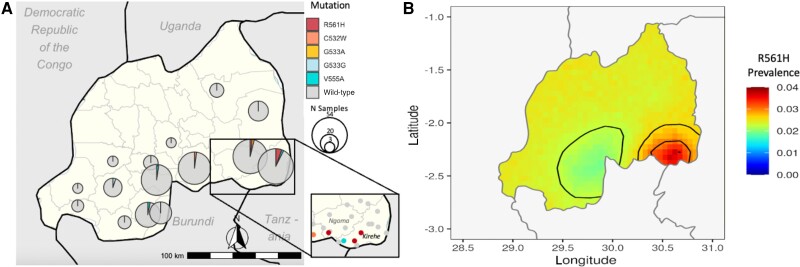
The 4 R561H isolates were spatially clustered across 3 DHS clusters in the southeast part of the country, representing 5.47% of samples in Kirehe and 2.74% in Ngoma. (Figure 2A; Supplementary Figure 1, Supplementary Table 2). The Moran's I value of spatial correlation of clusters with R561H was 0.133 (P = .046), indicating significant spatial clustering. Additionally, a 97.5% confidence interval of inferred R561H prevalence based on the number of mutant isolates, sampling scheme, and geographic proximity to other isolates highlighted localization in the southeast and higher confidence of low or no prevalence in the southwest, but lower of confidence in unsampled northern regions (Figure 2B).
Figure 2.
Geographic distribution of Pfk13 mutations in high–malaria prevalence demographic health survey clusters in Rwanda. A, Mutations confirmed by sequencing amplicons corresponding to Pfk13 amino acids 516–572 are presented as pie charts, the center of each corresponding to the center of the DHS survey district. Mutation presence at the DHS cluster level shown for the Kirehe and Ngoma districts. B, The 97.5% confidence interval of inferred R561H prevalence using a spatial binomial logistic model. Abbreviation: DHS, Rwanda Demographic Health Surveys.
DISCUSSION
Previous Pfk13 genotyping in Rwanda in the years leading up to the first observation of R561H failed to capture the larger picture across the country. This study provides expanded geographic sampling and an improved baseline of a small range of Pfk13 genotypes in relatively high-prevalence regions early in the R561H emergence timeline. Previous studies with limited sampling schemes only observed the mutation in Masaka as of 2014, but our study demonstrates that it was also present and significantly clustered in high-transmission regions in the southeast of the country. In keeping with prior reports, we found a small number of clinically unvalidated polymorphisms, 3 previously not observed in Rwanda as of 2014–2015 (Supplementary Table 3). Though unvalidated, these mutations should continue to be monitored for potential resistance. Of note, G533A, which we observed at 1.63% in Ngoma, has been associated with treatment failure in India, but cannot be directly linked to ACT resistance [15].
The presence of R561H in 3 spatially clustered high-transmission clusters outside of previously reported areas is significant for control efforts. While additional data are needed to better understand the origin and spread of the mutation, it is concerning that the resistant parasites were already present in high-transmission areas as of 2014. This was unexpected early in the emergence timeline, as it is hypothesized that low-transmission areas drive selection of resistant parasites due to low levels of host immunity and reduced within-host competition [16, 17]. A possible explanation for the current findings could be periods of low and high transmission due to intermittent indoor residual spraying in these regions. Additionally, it is possible that these mutants were imported from elsewhere in Rwanda or the neighboring Tanzania or Burundi, though R561H was not observed in these countries as of 2014–2015 [18]. Our sampling of only high-transmission regions could have biased these results as well, and further analysis of low-transmission settings from the DHS should be pursued. Regardless of cause, the presence of mutant parasites in high-transmission zones is of significant concern for the control of malaria.
Though these samples allowed for retrospective genotyping, working with old, discarded samples poses challenges. Some bags were not fully sealed, some had water damage, and the desiccant was exhausted in most sample bags. The DNA was thus variably degraded, forcing us to focus on just a short amplicon (516–572) containing the R561H mutation site. While this did not capture all mutations of interest in Rwanda (ie, C469F, P574L, and A675V), these could be pursued with additional amplicons [1, 7, 10].
Our amplicon approach was not perfect, with some clusters falling above or below the expected value of Pf positivity based on DHS RDT data due to inherent variance from testing different subsets of individuals such as differences in average age (RDT was 18 years compared with 22 years for PCR). While PCR is normally more sensitive than RDT, sample degradation likely decreased our sensitivity, and our assays did not leverage multicopy genes. Additionally, RDTs can be false positives due to the persistence of HRP2 protein despite clearance of the parasite.
Relatively low sequencing success can likely be attributed to selection of PCR step 1 positive amplicons, as any sign of a band was classified as positive. The presence of nonspecific bands could have led to misidentification of positives at this step.
The sampling scheme should also be taken into consideration. This method led to samples from older patients, with sequenced samples from patients averaging 22 years. As factors such as preexisting immunity are thought to have an impact on the evolution of parasites, the relative proportion of mutant parasites may be different from studies with a lower median age such as Uwimana et al. (2020), who only took samples from children age 1–14 years [1]. Evaluation of antimalarial resistance from population surveys that target children as a high-burden group for malaria, such as Malaria Indicator Surveys (MIS), should help fill in gaps in this important demographic group.
This approach requires significant manual labor for large studies. Sorting through thousands of samples, punching dried blood spots, and running gels to identify Plasmodium-positive samples were not a small task. This in practice required us to sample from high–malaria prevalence areas to not waste resources and time on unknown samples in low-prevalence areas for little output. This is a key limitation, as low-prevalence areas are historically the emergence sites of resistant parasites, so it is possible that R561H existed in 2014–2015 in areas we did not sample. Due to the sampling scheme, this study demonstrates the mutation's presence in higher transmission regions, but it cannot suggest that it was present solely or predominantly in these regions. Finally, we did not sample directly from Masaka, where R561H was first observed in 2014, and thus this does not contradict those previous findings. However, our sampling scheme included multiple clusters near Masaka and did not find R561H. When possible, future national sampling should coordinate HIV and RDT sampling for ease of genotyping residual samples as molecular genomic techniques can cost-effectively layer on additional questions relevant to public health.
Our study significantly expands the examination of early distribution of artemisinin-resistant Pfk13 R561H parasites in Rwanda using samples derived from a representative sample of circulating parasites. These data will allow for better modeling of the initial rate of spread when combined with other retrospective analyses and current surveys.
Supplementary Material
Acknowledgments
Financial support. This project was supported by the U.S. National Institutes of Health (R01AI156267 to J.A.B., J.B.M., and J.J.J. and K24AI134990 to J.J.J.). The funder had no role in the implementation or interpretation of the project.
Contributor Information
Rebecca Kirby, Brown University, Providence, Rhode Island, USA.
David Giesbrecht, Brown University, Providence, Rhode Island, USA.
Corine Karema, Quality Equity Health Care, Kigali, Rwanda; Swiss Tropical and Public Health Institute, University of Basel, Basel, Switzerland.
Oliver Watson, MRC Centre for Global Infectious Disease Analysis, Imperial College London, London, United Kingdom; London School of Hygiene and Tropical Medicine, London, United Kingdom.
Savannah Lewis, Brown University, Providence, Rhode Island, USA.
Tharcisse Munyaneza, National Reference Laboratory, Rwanda Biomedical Center, Kigali, Rwanda.
Jean De Dieu Butera, National Reference Laboratory, Rwanda Biomedical Center, Kigali, Rwanda.
Jonathan J Juliano, UNC, Chapel Hill, North Carolina, USA.
Jeffrey A Bailey, Brown University, Providence, Rhode Island, USA.
Jean-Baptiste Mazarati, INES-Ruhengeri, Ruhengeri, Rwanda.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
References
- 1. Uwimana A, Legrand E, Stokes BH, et al. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat Med 2020; 26:1602–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Asua V, Conrad MD, Aydemir O, et al. Changing prevalence of potential mediators of aminoquinoline, antifolate, and artemisinin resistance across Uganda. J Infect Dis 2021; 223:985–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Straimer J, Gandhi P, Renner KC, Schmitt EK. High prevalence of Plasmodium falciparum K13 mutations in Rwanda is associated with slow parasite clearance after treatment with artemether-lumefantrine. J Infect Dis 2022; 225:1411–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ashley EA, Dhorda M, Fairhurst RM, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 2014; 371:411–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ariey F, Witkowski B, Amaratunga C, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 2014; 505:50–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takala-Harrison S, Jacob CG, Arze C, et al. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in Southeast Asia. J Infect Dis 2015; 211:670–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bergmann C, van Loon W, Habarugira F, et al. Increase in Kelch 13 polymorphisms in Plasmodium falciparum, Southern Rwanda. Emerg Infect Dis 2021; 27:294–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van Loon W, Oliveira R, Bergmann C, et al. In vitro confirmation of artemisinin resistance in Plasmodium falciparum from patient isolates, Southern Rwanda, 2019. Emerg Infect Dis 2022; 28:852–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Uwimana A, Umulisa N, Venkatesan M, et al. Association of Plasmodium falciparum kelch13 R561H genotypes with delayed parasite clearance in Rwanda: an open-label, single-arm, multicentre, therapeutic efficacy study. Lancet Infect Dis 2021; 21:1120–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tacoli C, Gai PP, Bayingana C, et al. Artemisinin resistance–associated K13 polymorphisms of Plasmodium falciparum in Southern Rwanda, 2010–2015. Am J Trop Med Hyg 2016; 95:1090–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. National Institute of Statistics of Rwanda, Rwanda, DHS Program , eds. Rwanda Demographic and Health Survey, 2014–15: Final Report. National Institute of Statistics of Rwanda, Ministry of Finance and Economic Planning: Ministry of Health; The DHS Program, ICF International; 2016.
- 12. Aydemir O, Janko M, Hathaway NJ, et al. Drug-resistance and population structure of Plasmodium falciparum across the democratic Republic of Congo using high-throughput molecular inversion probes. J Infect Dis 2018; 218:946–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Makunin A, Korlević P, Park N, et al. A targeted amplicon sequencing panel to simultaneously identify mosquito species and Plasmodium presence across the entire Anopheles genus. Mol Ecol Resour 2022; 22:28–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hathaway NJ, Parobek CM, Juliano JJ, Bailey JA. Seekdeep: single-base resolution de novo clustering for amplicon deep sequencing. Nucleic Acids Res 2018; 46:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mishra N, Prajapati SK, Kaitholia K, et al. Surveillance of artemisinin resistance in Plasmodium falciparum in India using the kelch13 molecular marker. Antimicrob Agents Chemother 2015; 59:2548–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. White NJ. Antimalarial drug resistance. J Clin Invest 2004; 113:1084–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Whitlock AOB, Juliano JJ, Mideo N. Immune selection suppresses the emergence of drug resistance in malaria parasites but facilitates its spread. PLoS Comput Biol 2021; 17:e1008577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stokes BH, Dhingra SK, Rubiano K, et al. Plasmodium falciparum K13 mutations in Africa and Asia impact artemisinin resistance and parasite fitness. eLife 2021; 10:e66277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.