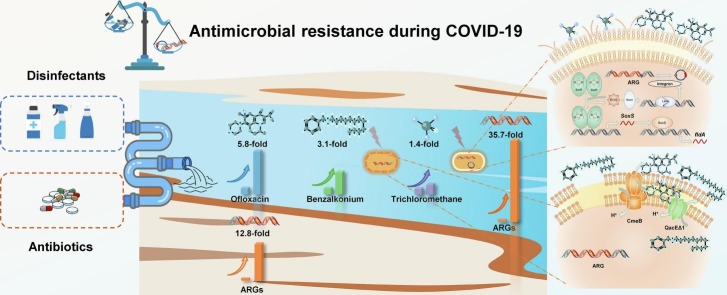
Abstract
During COVID-19 pandemic, chemicals from excessive consumption of pharmaceuticals and disinfectants i.e., antibiotics, quaternary ammonium compounds (QACs), and trihalomethanes (THMs), flowed into the urban environment, imposing unprecedented selective pressure to antimicrobial resistance (AMR). To decipher the obscure character pandemic-related chemicals portrayed in altering environmental AMR, 40 environmental samples covering water and soil matrix from surroundings of Wuhan designated hospitals were collected on March 2020 and June 2020. Chemical concentrations and antibiotic resistance gene (ARG) profiles were revealed by ultra-high-performance liquid chromatography-tandem mass spectrometry and metagenomics. Selective pressure from pandemic-related chemicals ascended by 1.4–5.8 times in March 2020 and then declined to normal level of pre-pandemic period in June 2020. Correspondingly, the relative abundance of ARGs under increasing selective pressure was 20.1 times that under normal selective pressure. Moreover, effect from QACs and THMs in aggravating the prevalence of AMR was elaborated by null model, variation partition and co-occurrence network analyses. Pandemic-related chemicals, of which QACs and THMs respectively displayed close interaction with efflux pump genes and mobile genetic elements, contributed >50 % in shaping ARG profile. QACs bolstered the cross resistance effectuated by qacEΔ1 and cmeB to 3.0 times higher while THMs boosted horizon ARG transfer by 7.9 times for initiating microbial response to oxidative stress. Under ascending selective pressure, qepA encoding quinolone efflux pump and oxa-20 encoding β-lactamases were identified as priority ARGs with potential human health risk. Collectively, this research validated the synergistic effect of QACs and THMs in exacerbating environmental AMR, appealing for the rational usage of disinfectants and the attention for environmental microbes in one-health perspective.
Keywords: Antimicrobial resistance, Quaternary ammonium compounds, Trihalomethanes, Cross resistance, Horizon gene transfer, COVID-19
Graphical abstract
1. Introduction
Disinfection related chemicals were associated with the emergence and dissemination of antimicrobial resistance (AMR). Quaternary ammonium compounds (QACs), the major constituent in biocides, promoted proliferation of antibiotic resistance genes (ARGs) and stimulated AMR through cross-resistance (Nordholt et al., 2021). Abundance of sul1 and blaTEM in microbial community significantly ascended by 5.4 and 19.2 times under QAC exposure (Harrison et al., 2020). Invoking transporters of QACs, microorganisms eject antibiotics from intracellular (Tezel and Pavlostathis, 2015). Trihalomethanes (THMs), typical by-products of chlorination (Srivastav et al., 2020), accelerated the dissemination of ARGs (Zhang et al., 2022b). Triggered by THM-induced reactive oxygen species (ROSs), microbial SOS response promoted the spread of ARGs for alleviating the repression to integrase gene expressing (Cai et al., 2021). From the one-health perspective, the effect of QACs and THMs to AMR is still urgent to be disclosed in the environment despite the laboratory evidence drawn under high exposure concentration.
Pandemic prevention supplies (PPSs) were consumed with an inordinate scale since COVID-19 pandemic (hereinafter the pandemic). To treat co-infection from bacteria and prevent the spread of pandemic, antibiotic and chlorine/QACs containing disinfectants were massively used. In stark contrast to the relatively low bacterial infection rate at 7 % (Lansbury et al., 2020), 74.6 % of worldwide COVID-19 patients took antibiotics while the prevalence was 76.2 % in China (Langford et al., 2021). Quinolones (20.0 %) represented by Ofloxacin (OFL), macrolides (18.9 %) represented by clarithromycin (CTM), and β-lactam (15.0 %) represented by cephalosporins (CEP) were the most commonly used antibiotics in COVID-19 treatment (Langford et al., 2021). According to the daily defined dose (DDD), consumption of antibiotics during pandemic rose by 4.6 times from 2715 g (Text S1) to 12,509 g (Table S1) in Wuhan (Han et al., 2022; Van Laethem et al., 2022). Based on the number of prescriptions, 1.6-fold increase was observed in the antibiotic-taking population size during pandemic (Text S2) (Li et al., 2019). In the meantime, disinfection has become a daily routine since pandemic. Benzalkonium Chloride (BAC), a type of QACs, was recommended by China (NHC, 2020), U.S. (EPA, 2020), and German (VAH, 2022) to inactivate COVID-19 virus in hospitals and households for its efficiency in destroying the lipid envelope (Ogilvie et al., 2021). In hotspots like hospitals, residential areas, and waste water treatment plants (WWTPs) of Wuhan, disinfection was respectively reinforced by 3.3 times (He et al., 2020), 4.3 times (Guo et al., 2021), and 2.1 times (Wuhan Water Affairs Bureau, 2020). Eventually, pandemic prevention chemicals i.e., leftovers and by-products of PPSs, arrived at the same sink — environment — via WWTPs and surface runoff, resulting in the “co-existence” of antibiotics, QACs, and THMs.
Antimicrobial resistance (AMR) in the context of pandemic was worthy of attention (Knight et al., 2021). Apart from the inducing of AMR from overused antibiotics (Sun et al., 2019; Zainab et al., 2020), excessive disinfection would promote the spread of AMR during pandemic (Lu and Guo, 2021). Under empirical antibiotic treatment, the proportion of multidrug-resistant bacteria among pathogens isolated from COVID-19 patients increased by 4.5 times (Temperoni et al., 2021). The flux of chemicals derived from antibiotics and disinfectants into environment jointly imposed selective pressure to AMR. To what extent the disinfection intensified AMR in the environment and the mechanism behind remained to be excavated urgently.
To decode the effect of disinfection related chemicals in altering environmental AMR, sampling was conducted in surroundings of Wuhan COVID-19 designated hospitals, the hotspot for disinfection, and downstream of WWTPs, the recipient of related chemicals. Two sampling campaigns were carried out on March 2020 when selective pressure from chemicals rose with the excessive disinfection and June 2020 when the pressure descended to normal level of pre-pandemic period. Alternation of ARGs when the ascending selective pressure declined to normal level was comprehensively deciphered through metagenomics. Contributions of disinfection related chemicals to environmental AMR were unraveled by means of null model, variation partition and co-occurrence network analyses. Moreover, synergistical effects of QACs and THMs in aggravating environmental AMR were calculated. Additionally, ARGs 1) with access to human and 2) promoted by related chemicals were defined as priority ARGs with potential human health risk under excessive disinfection. This work aimed to elaborate the interaction between disinfection and environmental AMR, providing instructions for the proper use of disinfectants.
2. Materials and methods
2.1. Sample collection
Wuhan city, located in the middle and lower reaches of Yangtze River basin, was one of the typical cities combating the initial wave of pandemic. Two sampling campaigns were conducted in Wuhan on March 24th and June 12th, 2020. On March 24th, there were still existing COVID-19 cases and PPSs were massively used to suppress the development of pandemic. On June 12th, COVID-19 cases have been completely cleared for 48 days and the usage of PPSs returned to normal with lockdown lifted.
As the main consumer of PPSs during pandemic, designated hospitals and household discharged related chemicals to environment through WWTPs and runoff. Based on previous research, chemicals from PPSs were enriched in waters around designated hospitals during pandemic (Hu et al., 2022). Therefore, samples from lakes near JinYinTan Hospital, HuoShenShan Hospital, and LeiShenShan Hospital were collected. Effluent recipients of SanJinTan WWTP, CaiDian WWTP, and HuangJiaHu WWTP that disposed of wastewater from the designated hospitals were also taken into account. Considering the high carbon-water partition constant of OFL (LogKoc = 4.64) (Cycoń et al., 2019) and BAC (LogKoc = 4.5–5.15) (Khan et al., 2017), soil would be the ultimate reservoir of these chemicals. Thus, soil in riparian area of the above waters was simultaneously collected. Overall, 20 surface water samples at depth of 0.5 m (1.0 L) and 20 topsoil samples at depth of 0–10 cm (500 g) were obtained at 10 sites in the two sampling campaigns (Table 1 ). For subsequent microbial analysis, water samples were filtered by 0.22 μm microporous membrane.
Table 1.
Detailed information of sampling sites.
| Nearest designated hospital | Sampling site | Geographic coordinate | Samples | Description |
|---|---|---|---|---|
| JinYinTan Hospital | HuangTan Lake | 30°39′50″N 114°16′36″E | HT | 500 m from JinYinTan Hospital |
| FuHe River | 30°39′50″N 114°21′3″E | FH | 1.5 km from outfall of SanJinTan WWTP | |
| HuoShenShan Hospital | ZhiYin Lake | 30°31′25″N 114°4′59″E | ZY | 500 m from HuoShenShan Hospital |
| HouGuan Lake | 30°33′3″N 114°3′36″E | HG | 2500 m from HuoShenShan Hospital | |
| HanJiang River | 30°33′59″N 114°17′5″E | HJ | 1.5 km from outfall of CaiDian WWTP | |
| LeiShenShan Hospital | HuangJia Lake | 30°26′25″N 114°17′7″E | HJH | 500 m from LeiShenShan Hospital |
| TangXun Lake | 30°26′21″N 114°19′39″E | TX | 2500 m from HuoShenShan Hospital | |
| XunSi River | 30°28′58″N 114°18′23″E | XS | 1.5 km from outfall of HuangJiaHu WWTP | |
| DongHu Lake | 30°33′5″N 114°20′56″E | DH | Inland lake | |
| NanHu Lake | 30°30′5″N 114°20′20″E | NH |
2.2. Detection of ARGs and ROS-response related genes
Information about ARGs and ROS-response related genes were accessed in virtue of metagenomics. DNA extracted by PowerSoil DNA Isolation Kit (Qiagen, German) was sequenced on Illumina HiSeq 2500 platform (Liu et al., 2020). For each sample, an aggregate comprising 10 G data was acquired. To start with downstream analyses, inferior raw data was eliminated by Trimmomatic (Bolger et al., 2014). Clean data was then assembled with de-novo method via Megahit tool (Li et al., 2016). After assembling, average 406,004 scaftigs with N50 at 990 bp were attained per sample. Predicted by Prodigal (Hyatt et al., 2010), open reading frames were clustered to form non-redundant unigenes by Linclust (Steinegger and Söding, 2018). Eventually, a catalogue of 22,046,813 unigenes with average length at 515 bp was constructed. The relative abundance of unigene was calculated by , in which rk represented number of reads aligned to gene k, Lk represented length of gene k.
ARGs were annotated by ARGs-OAP (V 2.0) (e ≤ 10−7, similarity >80 %, length > 75 %) (Yin et al., 2018) while mobile genetic elements (MGEs) were recognized by a published database (e ≤ 10−7) (Pärnänen et al., 2018). A catalogue consisted of ARGs targeted to frequently used antibiotics in pandemic i.e., quinolones, macrolides, and β-lactam was involved in this research. To verify the relationship between antibiotic consumption and ARGs, genes resistant to less used sulfonamides were also analyzed. In addition, genes regulated by SoxRS system were screened (Fig. S1) through eggNOG database (e ≤ 10−3) to reveal microbial response to oxidative stress (Pomposiello et al., 2001; Imlay, 2013; Huerta-Cepas et al., 2019). To discern whether ARGs were carried by pathogens, genes belonging to bacteria infecting primate were distinguished through PHI database (e ≤ e−10) (Urban et al., 2020).
2.3. Determination of pandemic prevention chemicals
Environmental concentrations of PPS-generated chemicals were determined via ultra-high-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) (Chen et al., 2021b, Chen et al., 2021a). Thirty-one types of antibiotics including the frequently used quinolones, macrolides as well as the less used sulfonamides were detected. Four types of disinfection by-products including trichloromethane (TCM), tribromomethane (TBM), dibromochloromethane (DBCM), and bromodichloromethane (BDCM) as well as two types of QAC leftovers including dodecyl dimethyl benzyl ammonium chloride (BAC-12) and tetradecyl dimethyl benzyl ammonium chloride (BAC-14) were quantified.
2.4. Data analyses
Difference of ARG abundance between two campaigns was tested using STAMP (V 2.1.3) software (Parks et al., 2014). To ascertain the assembly mechanism of ARG profile, null model analysis was carried out to calculate the stochasticity ratio based on difference between expected and observed similarity (Hou et al., 2021). To evaluate the role of chemicals, microbial community, and MGEs in shaping ARG profiles, variation partition analysis (VPA) was conducted with Vegan package of R software (V 4.0.5) (Hu et al., 2021). Co-occurrence network among chemicals, ARGs, and MGEs were constructed with ggClusterNet package of R software (V 4.0.5) (Wen et al., 2022). Pearson correlation coefficients between chemicals and ARGs were calculated via SPSS (V 20). Randomforest (RF) package was applied to identify effect of chemicals on specific ARG (Ke et al., 2022). In RF models with which the concentration of chemicals from PPSs was distinguished by ARGs, IncMSE (Increase in Mean Squared Error) measured the response degree of each ARG (Wright et al., 2020). The abundance of ARGs with IncMSE >1 % was more likely to fluctuate under effect of pandemic prevention chemicals.
3. Results
3.1. Dynamic of pandemic prevention chemicals and ARG profiles in the environment
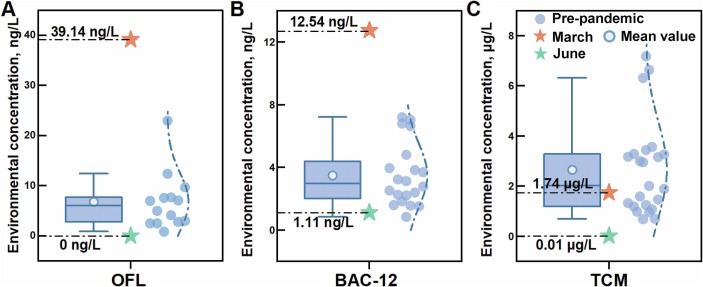
To evaluate the selective pressure from pandemic prevention chemicals, concentration of chemicals in March 2020/June 2020 were compared with that in pre-pandemic period. The concentration of OFL at Xunsi River in March 2020 was 5.8 times higher than that in Yangtze river basin before pandemic (Fig. 1A). The concentration of BAC-12 in Xunsi River ascended by 3.1 times compared to that before pandemic (Fig. 1B). TCM was detected at Xunsi River in March 2020 and its concentration increased by a factor of 1.4 times (Fig. 1C). In stark contrast, OFL was not detectable in June 2020 at the same site. Concentrations of BAC-12 and TCM reduced to the range of previous study. The excessive consumption of PPSs was related to the rise in environmental concentration of antibiotic and disinfection related chemicals, leading to the surging selective pressure in March 2020. With the usage of PPSs returning to normal, the selective pressure in June 2020 decreased to pre-pandemic level.
Fig. 1.
Comparison between concentration of pandemic prevention chemicals in March 2020/June 2020 and that in pre-pandemic period. Environmental concentrations of (A) ofloxacin (OFL) (Yan et al., 2013; Tong et al., 2014), (B) dodecyl dimethyl benzyl ammonium chloride (BAC-12) (Li et al., 2020a, Li et al., 2020b; Li et al., 2021), and (C) trichloromethane (TCM) (Jiang, 2006; Niu et al., 2017; Zhang et al., 2017a). The left was boxplot of concentration before pandemic. The right was fitted normal distribution curve based on concentration before pandemic.
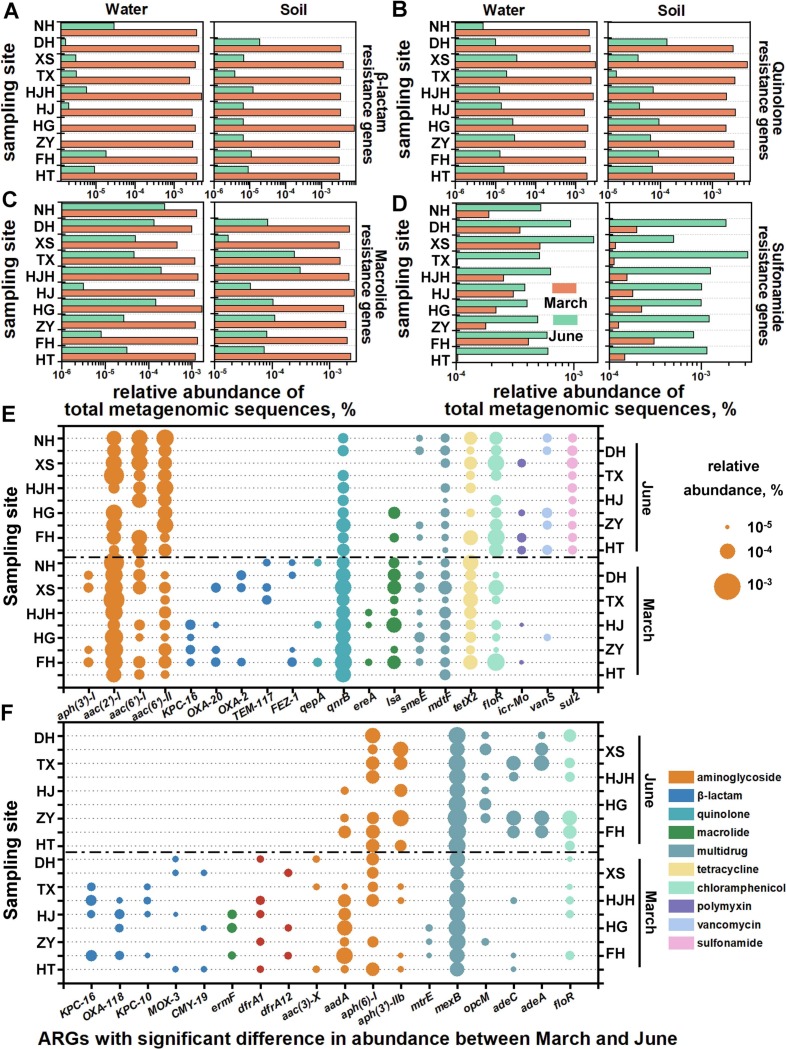
Temporal dynamics of ARG profiles were depicted when surging selective pressure of pandemic prevention chemicals in March 2020 descended to normal level in June 2020. In water samples, 300 ARG subtypes affiliated to 17 antibiotics were detected. With selective pressure alleviated, the number of ARG subtypes and the abundance of ARGs respectively decreased by 24.2 % (p < 0.01) and 97.2 % (p < 0.001) (Fig. S2A). FuHe River (173 ARG subtypes), HanJiang River (165 ARG subtypes) and XunSi River (141 ARG subtypes) that received effluent from WWTPs during pandemic were characterized by diverse ARG subtypes. Water samples from ZhiYin Lake and HouGuan Lake located nearby HuoShenShan hospital held the highest abundance of ARGs at 7.32 × 10−4 and 6.91 × 10−4 to total metagenomic sequences. A catalogue of 354 ARG subtypes resistant to 20 antibiotics were obtained from soil samples. Number of ARG subtypes in soil was similar between March (134 ARG subtypes) and June (123 ARG subtypes). In correspondence with ARG dynamic of water samples, the abundance of ARG descended by 92.2 % from March 2020 to June 2020 in soil samples (p < 0.001) (Fig. S2B). Holding the most ARG subtypes (150 ARG subtypes), soil sample from HouGuan Lake possessed the highest ARG abundance (6.09 × 10−4 to total metagenomic sequences). Echoing with the consumption rate, abundance of ARGs resistant to β-lactam, quinolone, and macrolide in March 2020 exceeded that in June 2020 respectively by 503 (water) / 439 (soil) (Fig. 2A), 13/16 (Fig. 2B), and 12/38 times (Fig. 2C). As for ARGs targeted to less frequently used sulfonamide in COVID-19 treatment, their abundance went through 2.5-fold rise in water and 7.7-fold rise in soil from March 2020 to June 2020 (Fig. 2D).
Fig. 2.
ARG profile in the environment under different selective pressure from pandemic prevention chemicals. Abundance of ARGs targeted to (A) β-lactam, (B) quinolone, (C) macrolide and (D) sulfonamide in March 2020 and June 2020. Abundance of ARGs with significant difference between March and June 2020 in (E) water, (F) soil samples.
To pinpoint ARGs responsible for temporal dynamics of ARG profile, difference in ARG abundance was tested under different selective pressure. Overall, 20 ARG subtypes including 5 β-lactam ARG subtypes, 2 quinolone ARG subtypes, and 2 macrolide ARG subtypes demonstrated significant discrepancy (p < 0.05) in water samples between March 2020 and June 2020 (Fig. 2E). Among 7 ARG subtypes with abundance ratio > 100 between March 2020 and June 2020, KPC-16 targeted to β-lactam in March was 225 times the abundance of that in June. In soil samples, abundance of 18 ARG subtypes represented by 5 β-lactam ARG subtypes and 5 multidrug ARG subtypes significantly diverged between two sampling campaigns (Fig. 2F). Out of 5 ARG subtypes with fold change of abundance >100 between March 2020 and June 2020, KPC-16 was 357 times more abundant under increasing selective pressure.
3.2. Contributions of pandemic prevention chemicals in shaping ARG profile
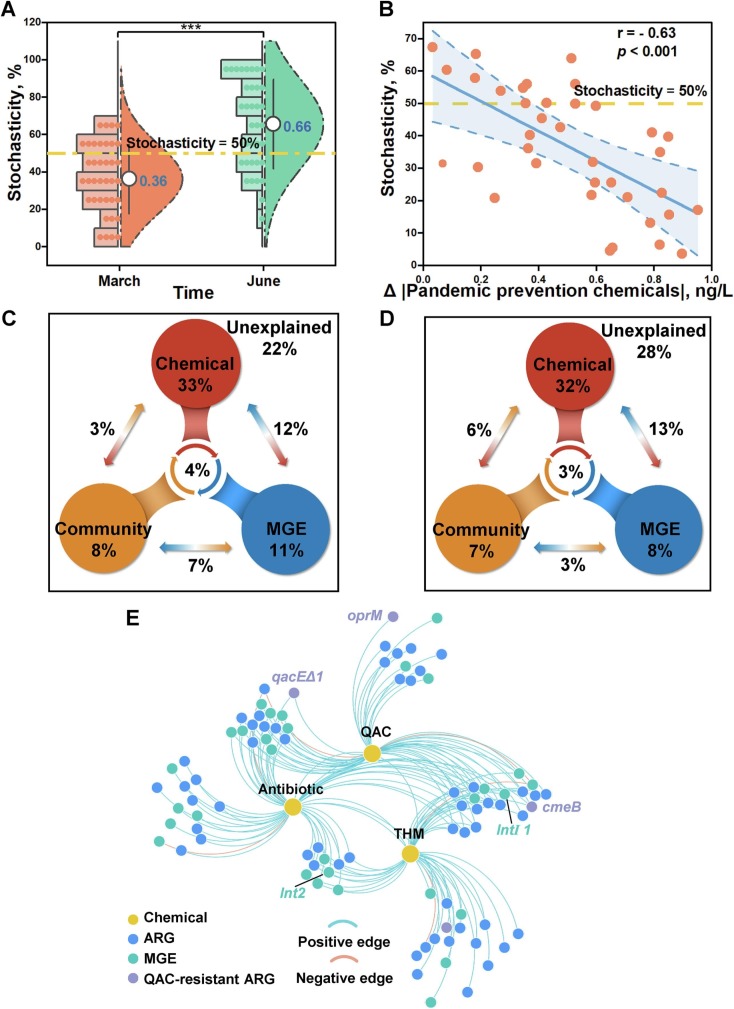
To determine the assembly mechanism of ARG profile, stochasticity ratio was calculated via null model. The stochasticity ratio of ARG profile in March 2020 was 36 % while it rose to 66 % in June 2020 (p < 0.001), indicating the transform from deterministic (< 50 %) assembly to stochastic (> 50 %) assembly (Fig. 3A). With the selective pressure from pandemic prevention chemicals intensifying, the stochasticity ratio sharply declined (r = −0.63; p < 0.001) (Fig. 3B). Accordingly, pandemic prevention chemicals considerably shaped ARG profile during pandemic.
Fig. 3.
Effect of pandemic prevention chemicals on ARGs. (A) Assembly of ARG profile in March 2020 and June 2020. (B) Relationship between stochasticity ratio in ARG assembly and pandemic prevention chemicals. Variation partition analysis among chemicals, microbial community, mobile genetic elements (MGEs) and (C) ARGs, (D) ARGs resistant to quinolones, macrolides, and β-lactam. Percentage was interpretation degree of each factor to ARG variation; arrow represented the combined interpretation of 2 factors; the middle circle represented the combined interpretation of 3 factors. (E) Co-occurrence network of pandemic prevention chemicals, ARGs, and MGEs.
To measure the effect of pandemic prevention chemicals in shaping ARG profile, VPA was conducted to compare the interpretability of chemicals with that of microbial community and MGE. Chemicals along with the biotic factors i.e., microbial community and MEG accounted for 78 % variation of ARGs (Fig. 3C). Notably, the interpretability of chemicals reached up to 52 %. Besides, the combined explanation rate of chemicals and MGEs was 12 %. As for ARGs resistant to quinolone, β-Lactam, and macrolide that were frequently used during pandemic, chemicals interpreted 54 % of variation in which the joint explanation rate with MGE was 13 % (Fig. 3D). It was speculated that pandemic prevention chemicals shaped the ARG profile through environmental filtration and interactions with MGEs.
To further decipher the relationship between pandemic prevention chemicals and ARGs, the co-occurrence network was built among chemicals, ARGs, and MGEs. Among 154 edges established on 3 chemicals, 56 ARG subtypes and 34 MGEs, 96 % of them were positive (Fig. 3E). Efflux pump genes involved in QAC-resistance e.g., qacEΔ1 encoding small multidrug resistance (SMR) protein and cmeB encoding resistance-nodulation-cell division (RND) protein, were closely related to antibiotics, implying the cross-resistance induced by QACs. Integrons i.e., intI1 and int2 simultaneously displayed positive interaction with THMs and antibiotics, suggesting that THMs promoted the horizonal gene transfer.
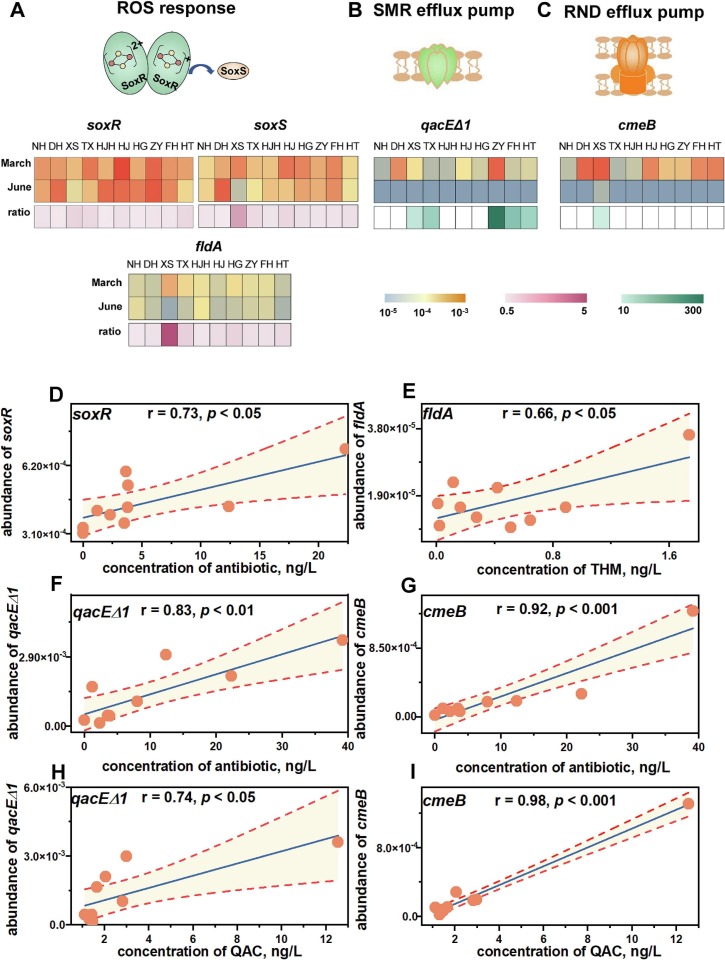
3.3. Mechanism of THMs and QACs in exacerbating environmental AMR
To delve into the role of THM and QAC in exacerbating environmental AMR, mechanism was probed through gene relevant with oxidative stress and cross resistance. THMs initiated microbial response to ROS and thus accelerating the dissemination of ARGs during pandemic. The abundance of gene soxR that encoded proteins to perceive oxidative stress was observed to significantly rise with antibiotic (r = 0.73, p < 0.05) (Fig. 4A) (Fig. 4D). At Xunsi River where the concentration of THMs was highest, the abundance of soxS that activated expression of downstream genes to combat oxidative stress in March 2020 was 5.5 times that in June 2020 (Fig. 4A). As for genes regulated by SoxRS system, fldA (r = 0.66, p < 0.05) associated with relief of oxidative stress by flavodoxin and recA (r = 0.68, p < 0.05) involved in the repair of DNA damage positively correlated with THMs (Fig. 4E). Compared to June, March saw the 10.8-fold abundance of fldA in Xunsi River (Fig. 4A). Besides, relationships between THMs and SoxRS-regulated cell membrane gene ompW (r = 0.69, p < 0.05), competence gene pilU (r = 0.68, p < 0.05) were significantly positive. The dynamic of intI 1 abundance echoed with alternations of THM concentration (r = 0.92, p < 0.001) (Fig. S4B). Also, ompW as well as genes in SoxRS system i.e., soxS and fldA displayed positive correlations in pace with intI 1 (Fig. S4C) (Fig. S4D). In accordance to the general least square regression model (Fig. S6), the possibility of HGT represented by abundance of MGEs averagely escalated by 7.9 times when the concentration of THMs increased by 1.4 times compared to pre-pandemic period.
Fig. 4.
Effect of THMs and QACs in boosting AMR. Comparisons of (A) genes related with ROS response, (B) qacEΔ1 gene, (C) cmeB gene between March 2020 and June 2020. (D) Relationship between antibiotic and soxR that encoded homodimeric regulatory protein to initiate ROS response. (E) Relationship between THM and soxRS system regulated fldA that encoded flavodoxin to relieve oxidative stress. Relationship between antibiotic and (F) qacEΔ1 encoding SMR efflux pump, (G) cmeB encoding RND efflux pump. Relationship between QAC and (F) qacEΔ1, (G) cmeB.
QAC induced microbial cross-resistance to antibiotic, intensifying environmental AMR during pandemic. As ARGs encoding efflux pump, qacEΔ1 (small multidrug resistance, SMR) and cmeB (resistance-nodulation-cell division, RND) were abundant under ascending selective pressure. Compared to June 2020, the abundance of qacEΔ1 and cmeB were respectively 150.7 (p < 0.05) and 57.2 (p < 0.001) times higher in March 2020 (Fig. 4B) (Fig. 4C). Apart from antibiotics, QACs could initiate AMR effectuated by qacEΔ1 and cmeB. Notably, explanation rates of QACs along with antibiotic in interpreting dynamics of qacEΔ1 (Fig. 4F) (Fig. 4H) and cmeB (Fig. 4G) (Fig. 4I) were > 80 %. Based on the general least square regression model (Fig. 4H) (Fig. 4I), 3.1-time rise in QAC concentration compared to pre-pandemic period corresponded to 2.4-time and 3.6-time ascendence in cross-resistance effect from qacEΔ1 and cmeB.
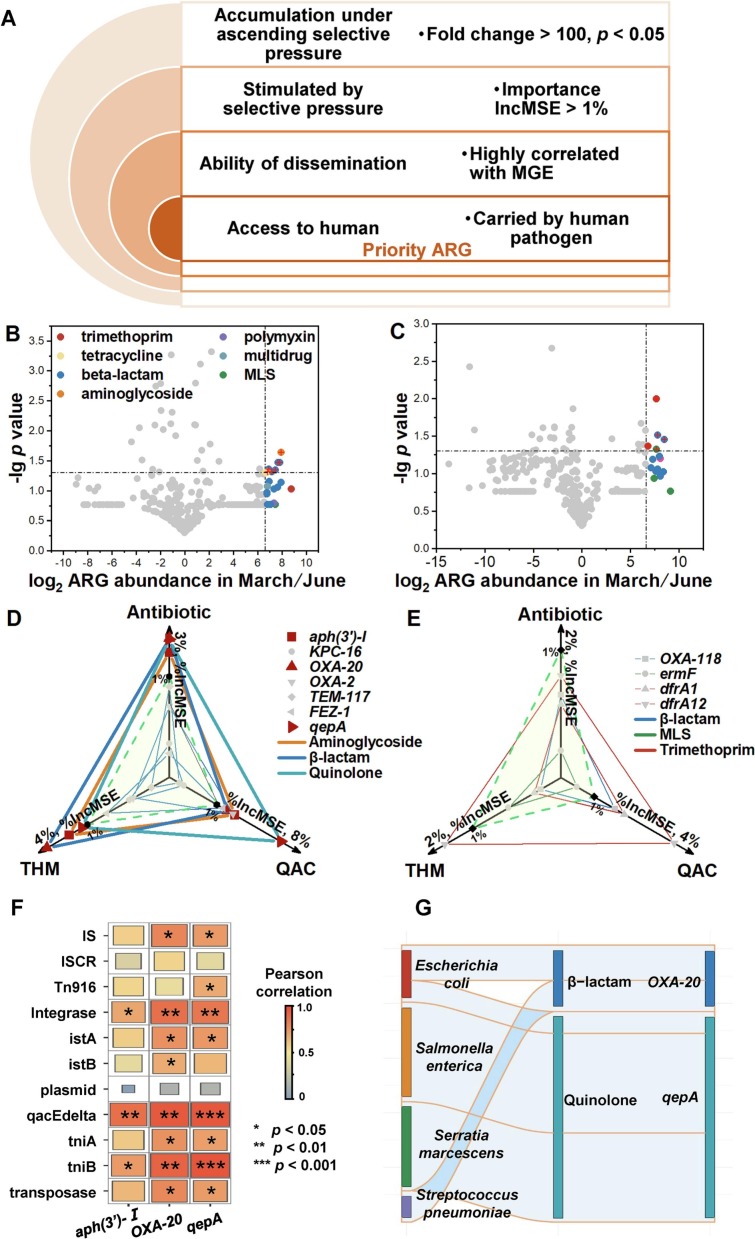
3.4. Identification of priority ARGs under excessive disinfection
To identify the effect of aggravated AMR on human health under excessive disinfection, priority ARGs with potential risk were screened. A framework covering the following criteria was established to discern priority ARGs in the context of excessive disinfection: i) fold change of ARG abundance under different selective pressure > 100 (p < 0.05); ii) response degree of ARG represented by IncMSE in RF model to antibiotic, THM, and QAC > 1 %; iii) horizontally transferable; iv) accessible to human beings. (Fig. 5A). ARGs that catered to all the standards were likely to affect human health under excessive disinfection and should be listed as priority. According to criterion i), 8 ARG subtypes, 5 of which were resistant to β-lactam, were chosen in water samples (Fig. 5B); 5 ARG subtypes consisted of 2 β-lactam, 2 trimethoprim, and 1 macrolide resistant genes were selected in soil samples (Fig. 5C). Aminoglycoside aph(3′)-I, β-lactam oxa-20, and quinolone qepA in water samples fulfilled criterion ii) (Fig. 5D). As for soil, none of ARGs conformed to the criterion ii) (Fig. 5E). oxa-20 located in class 1 integrons and plasmid-mediated qepA were positively related with MGEs such as insertion sequence, integron, and transposon, verifying their ability of dissemination (Fig. 5F). Carried by opportunistic pathogenic bacteria such as Escherichia coli, Streptococcus pneumoniae, and Streptococcus pneumoniae, oxa-20 and qepA were ultimately evaluated as priority ARGs (Fig. 5G).
Fig. 5.
Identification of priority ARGs with potential human health under excessive disinfection. (A) The framework for identifying priority ARGs. Fold changes of ARG abundance in (B) water samples, (C) soil samples under different selective pressure. Importance values of ARGs from (D) water samples and (E) soil samples in random forest model. (E) Correlation coefficient between ARGs and MGEs. (G) Abundance of ARGs that were carried by pathogenic bacteria.
4. Discussion
4.1. ARGs induced by massively used antibiotics
Massively used antibiotics from anthropogenic activities increased AMR burden in the environment. Compared to natural antibiotics in the environment, man-made antibiotics added more intense and extensive pressure to environmental microorganisms (Davies and Davies, 2010). The pollution level of antibiotics was observed to positively correspond with ARGs in soil (Sun et al., 2015), surface water (Zhang et al., 2020), and WWTPs (Gao et al., 2012). Nevertheless, these sentiments were refuted based on the fact that concentration of antibiotics in unpolluted environment was too low to select for AMR. Predicted antibiotic concentrations to induce resistance were mostly close to 1 ng/L, but the concentration of antibiotic in the environment was merely 10–100 μg/L (Bengtsson-Palme and Larsson, 2016; Qiao et al., 2018). Hence, the collinearity between ARGs and antibiotics was perceived to only indicate their same origin (Larsson and Flach, 2022). Excretions from human, the major source of antibiotic and ARGs in the environment, explained the high correlation between each other (Karkman and Parnanen, 2019). In this research, abundance of ARGs in the environment positively correlated with the concentration of antibiotic (r = 0.92, p < 0.001) (Fig. S7) as well as the usage of antibiotics (r = 0.75, p < 0.05) (Fig. S5). According to the above paradox, ARGs could be introduced to environment because of the usage of antibiotic and were also likely to be induced by residue of antibiotics in situ.
4.2. Synergistic effect from disinfection: complex impact of THMs and QACs
The synergistic effect from antibiotics, QACs, and THMs substantially boosted AMR in the environment. As typical by-products of chlorination, THMs were speculated to alter the AMR in a similar way as chlorine containing disinfectants. Chlorination destroyed antibiotic resistance bacteria but bolstered horizon transfer of ARGs in the meantime (Wang et al., 2020). Responding to ROSs provoked by chlorination, microorganisms launched SOS reaction which was in favor of conjugation and transformation of ARGs (Zhang et al., 2017b; Tang et al., 2022). In this research, the AMR burden from antibiotics became heavier due to additional effect of THMs. Along with antibiotics, THMs stimulated oxidative stress of microorganisms. fldA gene, encoding flavodoxin that tolerated ROSs pressure (de la Pena et al., 2013; Moyano et al., 2014) through reducing oxidized SoxR (Ziegelhoffer and Donohue, 2009), was positively correlated with THMs (Fig. 4E). THMs further modified abundance of gene associated with cell membrane and pilus, facilitating the horizontal transfer of ARGs (Zhang et al., 2021b). At Xunsi river where the concentration of THMs reached peak among all sampling sites, regulators i.e., ompW and ompR of outer membrane proteins related with molecule transport (Beketskaia et al., 2014) were 4.0 and 2.6 times more abundant under increasing selective pressure (Fig. S1C). At the same site, the abundance of pilL and pilU involved with pilus protein synthesis (Akahane et al., 2005) in March 2020 was respectively 4.4 and 6.7 times higher than that in June 2020 (Fig. S1B). Consequently, THMs benefited dissemination of antibiotic resistance through horizon transfer. Similarly, experiments on E. coli under exposure of THMs indicated the same mechanism that THMs advocated HGT as a result of ROS response (He et al., 2022). Besides, multidrug resistant gene i.e., qacEΔ1 and cmeB, thrived when confronting synergetic stress of antibiotics and QACs (Fig. 4B) (Fig. 4C). qacEΔ1 and cmeB respectively encode efflux transporters of small multidrug resistance (SMR) family (Kermani et al., 2020) and resistance-nodulation-cell division (RND) superfamily (Su et al., 2017), assisting microbes to survive adversity of QACs (Buffet-Bataillon et al., 2012). Also, antibiotics were discharged through the same efflux pump regulated by qacEΔ1 and cmeB through cross resistance (Poole, 2005). Under environment overloaded with antibiotics and QACs, cross resistance was conceived to considerably contribute to AMR.
4.3. Human health risk from ARG during the pandemic
As biological pollutant, ARG in the environment especially those acquired by clinical human pathogens were not controllable (Larsson and Flach, 2022). Consequently, the rise of environmental antimicrobial resistance was an emerging risk to human health. For the purpose of measuring human-originated pollution, ARGs cloud be selected as indicator. The abundance of qnrS gene in wastewaters accurately represented the usage of quinolones (Castrignano et al., 2020) while ermF in extreme habitats reflected the worldwide effect of anthropogenic activities (Yang et al., 2021). For predicting impact of AMR on human health, a subgroup was distinguished as ARGs with high risk (Zhang et al., 2021a). In this research, qepA gene resistant to quinolone and oxa-20 gene resistant to β-lactam were selected as ARGs that should be preferentially controlled for their high probability to affect human health under excessive disinfection. qepA was most likely to be carried by Escherichia coli and Streptococcus pneumoniae while Escherichia coli, Salmonella enterica, and Serratia marcescens were assumed as hosts of oxa-20 (Fig. 5F). qepA gene, usually located in plasmids, encoded major facilitator superfamily (MFS) protein that manipulates the outflow of quinolone antibiotics (Yamane et al., 2007). oxa-20 associated with class 1 integron regulated the expression of Class D β-lactamases (Poirel et al., 2010). Since the discovery of this quinolone resistant gene, qepA has been continuously found in human related environment. In poultry farms, the predominance of qepA gene reached up to 88.66 % (El-Aziz et al., 2021). During the pandemic, qepA and oxa-20 corresponded to quinolone and β-lactam antibiotics which were clinically consumed at rate of 20.0 % and 18.9 % (Langford et al., 2021). Indeed, initiating of efflux pump was deemed to be a universal microbial response when combating adverse conditions including but not limited to antibiotics and QACs (Henderson et al., 2021). Nowadays, the thriving of efflux pump related genes has become a peril to human health (Silver, 2011). The prevalence of quinolone resistant isolates from food soared from 82.4 % to 100.0 % with qepA gene as one of the determinants (Zhang et al., 2022a). Overall, the emergence of qepA and oxa-20 qualified to symbolize human health risk during the pandemic. Despite the expanding concerns from chemicals generated by pandemic prevention supplies, the potential human health risk derived in AMR promoted by these chemicals was overlooked. The framework put up in this research provided an approach to monitor this risk with indicator ARG that can be efficiently and precisely detected. To make the result more convincing, human health risk evaluation from qepA and oxa-20 should be conducted in the future. If possible, pathogens in hotspots like designated hospitals and WWTPs should be isolated to affirm the prevalence of qepA and oxa-20.
5. Conclusion
To summarize, this work emphasized the co-selection resulted from disinfection related chemicals in aggravating the AMR of urban environment. Besides the effect of antibiotics in inducing proliferation of ARGs, QACs and THMs synergistically promoted the spread of AMR for eliciting cross resistance and boosting ARG dissemination. qepA and oxa-20 were screened as priority ARGs with high probability to affect human health under excessive disinfection. Even though the prevalence of COVID-19 declined sharply nowadays, but disinfectants were still massively used in some places e.g., hospitals and elevator. These results underlined the proper use of disinfectants especially in hotspots and the attention for environmental microbiology in the framework of one-health perspective.
CRediT authorship contribution statement
Z.C.H. conducted the sampling, carried out experiments, analyzed study data, visualized research data, and wrote the initial draft. L.H.Y. conducted the sampling, carried out experiments, and analyzed study data. Z.S.L. analyzed study data, and reviewed and edited the original draft. J.H. conducted the sampling. Y.X.Z. carried out experiments. Y.H.J. carried out experiments. Y.Q.S. coordinated research activity. L.Z.Z. conceived of the study. B.L.H. conceived of the study, reviewed and edited the original draft and supervised the research.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 22193061) and the consulting research project of Chinese Academy of Engineering (2020-ZD-15).
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2023.163598.
Appendix A. Supplementary data
Supplementary material
Data availability
Data will be made available on request.
References
- Akahane K., Sakai D., Furuya N., et al. Analysis of the pilU gene for the prepilin peptidase involved in the biogenesis of type IV pili encoded by plasmid R64. Mol. Gen. Genomics. 2005;273(4):350–359. doi: 10.1007/s00438-005-1143-8. [DOI] [PubMed] [Google Scholar]
- Beketskaia M.S., Bay D.C., Turner R.J. Outer membrane protein OmpW participates with small multidrug resistance protein member EmrE in quaternary cationic compound efflux. J. Bacteriol. 2014;196(10):1908–1914. doi: 10.1128/JB.01483-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson-Palme J., Larsson D.G.J. Concentrations of antibiotics predicted to select for resistant bacteria: proposed limits for environmental regulation. Environ. Int. 2016;86:140–149. doi: 10.1016/j.envint.2015.10.015. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffet-Bataillon S., Tattevin P., Bonnaure-Mallet M., et al. Emergence of resistance to antibacterial agents: the role of quaternary ammonium compounds-a critical review. Int. J. Antimicrob. Agents. 2012;39(5):381–389. doi: 10.1016/j.ijantimicag.2012.01.011. [DOI] [PubMed] [Google Scholar]
- Cai Y.W., Sun T., Li G.Y., et al. Traditional and emerging water disinfection technologies challenging the control of antibiotic-resistant bacteria and antibiotic resistance genes. ACS ES&T Eng. 2021;1(7):1046–1064. [Google Scholar]
- Castrignano E., Yang Z.E., Feil E.J. Enantiomeric profiling of quinolones and quinolones resistance gene qnrS in European wastewaters. Water Res. 2020;175 doi: 10.1016/j.watres.2020.115653. [DOI] [PubMed] [Google Scholar]
- Chen B., Han J., Dai Han, et al. Biocide-tolerance and antibiotic-resistance in community environments and risk of direct transfers to humans: unintended consequences of community-wide surface disinfecting during COVID-19? Environ. Pollut. 2021;283 doi: 10.1016/j.envpol.2021.117074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.P., Lei L., Liu S.T., et al. Occurrence and risk assessment of pharmaceuticals and personal care products (PPCPs) against COVID-19 in lakes and WWTP-river-estuary system in Wuhan, China. Sci. Total Environ. 2021;792 doi: 10.1016/j.scitotenv.2021.148352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cycoń M., Mrozik A., Piotrowska-Seget Z. Antibiotics in the soil environment—degradation and their impact on microbial activity and diversity. Front. Microbiol. 2019;10, 338. doi: 10.3389/fmicb.2019.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J., Davies D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010;74(3):417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Pena T.C., Redondo F.J., Fillat M.F., et al. Flavodoxin overexpression confers tolerance to oxidativestress in beneficial soil bacteria and improves survival inthe presence of the herbicides paraquat and atrazine. J. Appl. Microbiol. 2013;115(1):236–246. doi: 10.1111/jam.12224. [DOI] [PubMed] [Google Scholar]
- El-Aziz N.K., Tartor Y.H., Gharieb R.M.A., et al. Extensive drug-resistant Salmonella enterica isolated from poultry and humans: prevalence and molecular determinants behind the co-resistance to ciprofloxacin and tigecycline. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.738784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPA List N: disinfectants for coronavirus (COVID-19) 2020. https://cfpub.epa.gov/wizards/disinfectants/
- Jiang F. Beijing University of Chemical Technology; 2006. Character Analysis of Organic Pollutants in Estuary Area. (in Chinese) [Google Scholar]
- Gao P., Munir M., Xagoraraki I. Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant. Sci. Total Environ. 2012;412–422:173–183. doi: 10.1016/j.scitotenv.2012.01.061. [DOI] [PubMed] [Google Scholar]
- Guo J., Liao M., He B., et al. Impact of the COVID-19 pandemic on household disinfectant consumption behaviors and related environmental concerns: a questionnaire-based survey in China. J. Environ. Chem. Eng. 2021;9(5) doi: 10.1016/j.jece.2021.106168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Wang Z., Huang H., et al. Estimating antibiotics use in major cities in China through wastewater-based epidemiology. Sci. Total Environ. 2022;826 doi: 10.1016/j.scitotenv.2022.154116. [DOI] [PubMed] [Google Scholar]
- Harrison K.R., Kappell A.D., McNamara P.J. Benzalkonium chloride alters phenotypic and genotypic antibiotic resistance profiles in a source water used for drinking water treatment. Environ. Pollut. 2020;257 doi: 10.1016/j.envpol.2019.113472. [DOI] [PubMed] [Google Scholar]
- He J., Zhang J., Gao J., et al. Investigation and analysis of the use of disinfectants in hospitals in Wuhan during the period of COVID-19. Chin. J. Disinfect. 2020;37, 611–613+617. [Google Scholar]
- He K., Xue B., Yang X.B., et al. Low-concentration of trichloromethane and dichloroacetonitrile promote the plasmid-mediated horizontal transfer of antibiotic resistance genes. J. Hazard. Mater. 2022;425 doi: 10.1016/j.jhazmat.2021.128030. [DOI] [PubMed] [Google Scholar]
- Henderson P.J.F., Maher C., Elbourne L.D.H., et al. Physiological functions of bacterial “Multidrug” efflux pumps. Chem. Rev. 2021;121(9):5417–5478. doi: 10.1021/acs.chemrev.0c01226. [DOI] [PubMed] [Google Scholar]
- Hou L., Zhang L., Li F., et al. Urban ponds as hotspots of antibiotic resistome in the urban environment. J. Hazard. Mater. 2021;403 doi: 10.1016/j.jhazmat.2020.124008. [DOI] [PubMed] [Google Scholar]
- Hu J.J., Zhao Y.X., Yao X.W., et al. Dominance of comammox Nitrospira in soil nitrification. Sci. Total Environ. 2021;780 doi: 10.1016/j.scitotenv.2021.146558. [DOI] [PubMed] [Google Scholar]
- Hu Z., Yang L., Han J., et al. Human viruses lurking in the environment activated by excessive use of COVID-19 prevention supplies. Environ. Int. 2022;163 doi: 10.1016/j.envint.2022.107192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Szklarczyk D., Heller D., et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47(D1):D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D., Chen G.L., LoCascio P.F., et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay J.A. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol. 2013;11(7):443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkman A., Parnanen K. Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat. Commun. 2019;10:80. doi: 10.1038/s41467-018-07992-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke M., Xu N., Zhang Z., et al. Development of a machine-learning model to identify the impacts of pesticides characteristics on soil microbial communities from high-throughput sequencing data. Environ. Microbiol. 2022:1–13. doi: 10.1111/1462-2920.16175. [DOI] [PubMed] [Google Scholar]
- Kermani A.A., Macdonald C.B., Burata O.E., et al. The structural basis of promiscuity in small multidrug resistance transporters. Nat. Commun. 2020;11(1):6064. doi: 10.1038/s41467-020-19820-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A.H., Macfie S.M., Ray M.B. Sorption and leaching of benzalkonium chlorides in agricultural soils. J. Environ. Manag. 2017;196:26–35. doi: 10.1016/j.jenvman.2017.02.065. [DOI] [PubMed] [Google Scholar]
- Knight G.M., Glover R.E., Mcquaid C.F., et al. Antimicrobial resistance and COVID-19: intersections and implications. eLife. 2021;10 doi: 10.7554/eLife.64139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford B.J., So M., Raybardhan S., et al. Antibiotic prescribing in patients with COVID-19: rapid review and meta-analysis. Clin. Microbiol. Infect. 2021;27(4):520–531. doi: 10.1016/j.cmi.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbury L., Lim B., Baskaran V., et al. Co-infections in people with COVID-19: a systematic review and meta-analysis. J. Infect. 2020;81(2):266–275. doi: 10.1016/j.jinf.2020.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson D.G.J., Flach C.F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022;20:257–269. doi: 10.1038/s41579-021-00649-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D.S., Sangion A., Li L. Evaluating consumer exposure to disinfecting chemicals against coronavirus disease 2019 (COVID-19) and associated health risks. Environ. Int. 2020;145 doi: 10.1016/j.envint.2020.106108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Yan S.J., Li D.D., et al. Trends and patterns of outpatient and inpatient antibiotic use in China's hospitals: data from the Center for Antibacterial Surveillance, 2012–16. J. Antimicrob. Chemother. 2019;74(6):1731–1740. doi: 10.1093/jac/dkz062. [DOI] [PubMed] [Google Scholar]
- Li D., Luo R., Liu C.M., et al. MEGAHIT v1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. doi: 10.1016/j.ymeth.2016.02.020. [DOI] [PubMed] [Google Scholar]
- Li W., Zhang Z., Li Y., et al. Assessing the distributions and fate of household and personal care chemicals (HPCCs) in the Songhua Catchment, Northeast China. Sci. Total Environ. 2021;786 doi: 10.1016/j.scitotenv.2021.147484. [DOI] [PubMed] [Google Scholar]
- Liu H., Hu Z.C., Zhou M., et al. Airborne microorganisms exacerbate the formation of atmospheric ammonium and sulfate. Environ.Pollut. 2020;263 doi: 10.1016/j.envpol.2020.114293. [DOI] [PubMed] [Google Scholar]
- Li W., Zhang Z., Sparham C., et al. Validation of sampling techniques and SPE-UPLC/MS/MS for home and personal care chemicals in the Songhua Catchment, Northeast China. Sci. Total Environ. 2020;707 doi: 10.1016/j.scitotenv.2019.136038. [DOI] [PubMed] [Google Scholar]
- Lu J., Guo J.H. Disinfection spreads antimicrobial resistance. Science. 2021;371(6528):474. doi: 10.1126/science.abg4380. [DOI] [PubMed] [Google Scholar]
- Moyano A.J., Tobares R.A., Rizzi Y.S., et al. A long-chain flavodoxin protects Pseudomonas aeruginosa from oxidative stress and host bacterial clearance. PLoS Genet. 2014;10(2) doi: 10.1371/journal.pgen.1004163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NHC Guidelines for the use of disinfectants. 2020. http://www.nhc.gov.cn/cms-search/xxgk/getManuscriptXxgk.htm?id=b9891e8c86d141a08ec45c6a18e21dc2
- Niu Z., Li X., Zhang Ying. Composition profiles, levels, distributions and ecological risk assessments of trihalomethanes in surface water from a typical estuary of Bohai Bay, China. Mar. Pollut. Bull. 2017;117(1):124–130. doi: 10.1016/j.marpolbul.2017.01.041. [DOI] [PubMed] [Google Scholar]
- Nordholt N., Kanaris O., Schmidt S.B.I., et al. Persistence against benzalkonium chloride promotes rapid evolution of tolerance during periodic disinfection. Nat. Commun. 2021;12(1):6792. doi: 10.1038/s41467-021-27019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie B.H., Solis-Leal A., Lopez J.B., et al. Alcohol-free hand sanitizer and other quaternary ammonium disinfectants quickly and effectively inactivate SARS-CoV-2. J. Hosp. Infect. 2021;108:142–145. doi: 10.1016/j.jhin.2020.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks D.H., Tyson G.W., Hugenholtz P., et al. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30(21):3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pärnänen K., Karkman A., Hultman J., et al. Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nat. Commun. 2018;9:3891. doi: 10.1038/s41467-018-06393-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Naas T., Nordmann P. Diversity, Epidemiology, and Genetics of Class D β-Lactamases. Antimicrob. Agents Chemother. 2010;54(1):24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomposiello J.P., Bennik M.H.J., Demple B. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 2001;183(13):3890–3902. doi: 10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 2005;56(1):20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- Qiao M., Ying G.G., Singer A.C., et al. Review of antibiotic resistance in China and its environment. Environ. Int. 2018;110:160–172. doi: 10.1016/j.envint.2017.10.016. [DOI] [PubMed] [Google Scholar]
- Silver L.L. Challenges of antibacterial discovery. Clin. Microbiol. Rev. 2011;24(1):71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastav L.A., Patel N., Chaudhary K.V. Disinfection by-products in drinking water: occurrence, toxicity and abatement. Environ. Pollut. 2020;267 doi: 10.1016/j.envpol.2020.115474. [DOI] [PubMed] [Google Scholar]
- Steinegger M., Söding J. Clustering huge protein sequence sets in linear time. Nat. Commun. 2018;9:2542. doi: 10.1038/s41467-018-04964-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C.C., Yin L.X., Kumar N., et al. Structures and transport dynamics of a Campylobacter jejuni multidrug efflux pump. Nat. Commun. 2017;8:171. doi: 10.1038/s41467-017-00217-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H.Y., Chen R.H., Jiang W., et al. QSAR-based investigation on antibiotics facilitating emergence and dissemination of antibiotic resistance genes: a case study of sulfonamides against mutation and conjugative transfer in Escherichia coli. Environ. Res. 2019;173:87–96. doi: 10.1016/j.envres.2019.03.020. [DOI] [PubMed] [Google Scholar]
- Sun M.M., Ye M., Wu J., et al. Positive relationship detected between soil bioaccessible organic pollutants and antibiotic resistance genes at dairy farms in Nanjing, Eastern China. Environ. Pollut. 2015;206:421–428. doi: 10.1016/j.envpol.2015.07.022. [DOI] [PubMed] [Google Scholar]
- Tang H.M., Liu Z.S., Hu B.L., et al. Effects of iron mineral adhesion on bacterial conjugation: interfering the transmission of antibiotic resistance genes through an interfacial process. J. Hazard. Mater. 2022;435 doi: 10.1016/j.jhazmat.2022.128889. [DOI] [PubMed] [Google Scholar]
- Temperoni C., Caiazzo L., Barchiesi F. High prevalence of antibiotic resistance among opportunistic pathogens isolated from patients with COVID-19 under mechanical ventilation: results of a single-center study. Antibiotics. 2021;10(9):1080. doi: 10.3390/antibiotics10091080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezel U., Pavlostathis S.G. Quaternary ammonium disinfectants: microbial adaptation, degradation and ecology. Curr. Opin. Biotechnol. 2015;33:296–304. doi: 10.1016/j.copbio.2015.03.018. [DOI] [PubMed] [Google Scholar]
- Tong L., Huang S., Wang Y., et al. Occurrence of antibiotics in the aquatic environment of Jianghan Plain, central China. Sci. Total Environ. 2014;497–498:180–187. doi: 10.1016/j.scitotenv.2014.07.068. [DOI] [PubMed] [Google Scholar]
- Urban M., Cuzick A., Seager J., et al. PHI-base: the pathogen-host interactions database. Nucleic Acids Res. 2020;48(D1):D613–D620. doi: 10.1093/nar/gkz904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAH VAH list of disinfectants. 2022. https://vah-liste.mhp-verlag.de/en/
- Van Laethem J., Wuyts S., Van Laere S., et al. Antibiotic prescriptions in the context of suspected bacterial respiratory tract superinfections in the COVID-19 era: a retrospective quantitative analysis of antibiotic consumption and identification of antibiotic prescription drivers. Intern. Emerg. Med. 2022;17:141–151. doi: 10.1007/s11739-021-02790-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.C., Wang J., Li S.M., et al. Synergistic effect of UV/chlorine in bacterial inactivation, resistance gene removal, and gene conjugative transfer blocking. Water Res. 2020;185 doi: 10.1016/j.watres.2020.116290. [DOI] [PubMed] [Google Scholar]
- Wen T., Xie P., Yang S., et al. ggClusterNet: an R package for microbiome network analysis and modularity-based multiple network layouts. iMeta. 2022;1(3) doi: 10.1002/imt2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R.J., Langille M.G.I., Walker T.R. Food or just a free ride? A meta-analysis reveals the global diversity of the Plastisphere. ISME J. 2020;15:789–806. doi: 10.1038/s41396-020-00814-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuhan Water Affairs Bureau Wuhan put into nearly 2000 tons of disinfectants to promote the strengthening of disinfection in urban drainage and sewage facilities. 2020. http://swj.wuhan.gov.cn/swyw/jcss/202004/t20200426_1126979.html
- Yamane K., Wachino J.I., Suzuki S., et al. New Plasmid-Mediated Fluoroquinolone Efflux Pump, QepA, Found in an Escherichia coli Clinical Isolate. 51(9) 2007. pp. 3354–3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Yang Y., Zhou J., et al. Antibiotics in the surface water of the Yangtze Estuary: occurrence, distribution and risk assessment. Environ. Pollut. 2013;175:22–29. doi: 10.1016/j.envpol.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Yang H.Y., Liu R.L., Liu H.F., et al. Evidence for long-term anthropogenic pollution: the hadal trench as a depository and indicator for dissemination of antibiotic resistance genes. Environ.Sci.Technol. 2021;55(22):15136–15148. doi: 10.1021/acs.est.1c03444. [DOI] [PubMed] [Google Scholar]
- Yin X., Jiang X.T., Chai B., et al. ARGs-OAP v2.0 with an expanded SARG database and Hidden Markov Models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes. Bioinformatics. 2018;34(13):2263–2270. doi: 10.1093/bioinformatics/bty053. [DOI] [PubMed] [Google Scholar]
- Zainab S.M., Junaid M., Xu N., et al. Antibiotics and antibiotic resistant genes (ARGs) in groundwater: a global review on dissemination, sources, interactions, environmental and human health risks. Water Res. 2020;187 doi: 10.1016/j.watres.2020.116455. [DOI] [PubMed] [Google Scholar]
- Zhang A.N., Gaston J.M., Dai C.Z.L., et al. An omics-based framework for assessing the health risk of antimicrobial resistance genes. Nat. Commun. 2021;12(1):4765. doi: 10.1038/s41467-021-25096-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.D., Lu S.Y., Wang Y.Q., et al. Occurrence of antibiotics and antibiotic resistance genes and their correlations in lower Yangtze River, China. Environ. Pollut. 2020;257 doi: 10.1016/j.envpol.2019.113365. [DOI] [PubMed] [Google Scholar]
- Zhang S., Wang Y., Lu J., et al. Chlorine disinfection facilitates natural transformation through ROS-mediated oxidative stress. ISME J. 2021;15(10):2969–2985. doi: 10.1038/s41396-021-00980-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Gu A.Z., He M., et al. Subinhibitory Cconcentrations of disinfectants promote the horizontal transfer of multidrug resistance genes within and across genera. Environ. Sci. Technol. 2017;51(1):570–580. doi: 10.1021/acs.est.6b03132. [DOI] [PubMed] [Google Scholar]
- Zhang Z.F., He S.K., Xu X.B., et al. Vol. 131. 2022. Antimicrobial susceptibility and molecular characterization of Salmonella enterica serovar Indiana from foods, patients, and environments in China during 2007–2016. [Google Scholar]
- Zhang Z.Y., Zhang Q., Lu T., et al. Residual chlorine disrupts the microbial communities and spreads antibiotic resistance in freshwater. J. Hazard. Mater. 2022;423 doi: 10.1016/j.jhazmat.2021.127152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.X., Zhao Q., Liu Y.L., et al. Determination of twelve kinds of volatile disinfection by-products in drinking water by gas chromatography with electron capture detect. Chin. J. Anal. Chem. 2017;45:1203–1208. [Google Scholar]
- Ziegelhoffer E.C., Donohue T.J. Bacterial responses to photo-oxidative stress. Nat. Rev. Microbiol. 2009;7(12):856–863. doi: 10.1038/nrmicro2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.