Abstract
Background
Aberrant expression of circular RNAs (circRNAs) contributes to the initiation and progression of pulmonary hypertension (PH). Hypoxia‐inducible factor (HIF) is a well‐known modulator of hypoxia‐induced PH. The role and underlying mechanism of circRNAs in the regulation of HIF expression remains elusive.
Methods and Results
We profiled pulmonary artery transcriptomes using RNA sequencing and screened circRNAs associated with hypoxia treatment. The expression of a novel circRNA, circ_chr11_67292179–67294612 (circ‐myh8), was increased by hypoxia in a time‐dependent manner. We evaluated the effects of circ‐myh8 overexpression by adeno‐associated virus or inhibition by short hairpin RNA on proliferation and cell cycling in mice and pulmonary artery smooth muscle cells. Overexpression of circ‐myh8 promotes PH under normoxia, and disruption of circ‐myh8 by short hairpin RNA mitigates PH in chronic hypoxic mice. Biologically, circ‐myh8 induces the proliferation and cell‐cycle progression of pulmonary artery smooth muscle cells in vivo and in vitro. Mechanistically, RNA pull‐down and RNA immunoprecipitation assays were used to examine the interaction of circRNAs with the binding protein KAT7 (lysine acetyltransferase 7). The acetylation level of lysine 5 of histone H4 in the transcriptional initiation region of HIF1α was determined by chromatin immunoprecipitation assay followed by reverse transcription‐quantitative polymerase chain reaction. Circ‐myh8 acts as a modular scaffold to recruit histone acetyltransferase KAT7 to the promoters of HIF1α, which elicits acetylation of lysine 5 of histone H4 in their promoters.
Conclusions
Our findings not only reveal the pivotal roles of circ‐myh8 in governing histone modification in anti‐PH treatment but also advocate triggering the circ‐myh8/KAT7/HIF1α pathway to combat PH.
Keywords: circular RNA, histone modification, KAT7, pulmonary hypertension
Subject Categories: Animal Models of Human Disease, Basic Science Research, Smooth Muscle Proliferation and Differentiation
Nonstandard Abbreviations and Acronyms
- CDK
cyclin‐dependent kinase
- circRNAs
circular RNAs
- FISH
fluorescent in situ hybridization
- H4K5
lysine 5 of histone H4
- HIF
hypoxia‐inducible factor
- PAH
pulmonary arterial hypertension
- PASMCs
pulmonary artery smooth muscle cells
- PCNA
proliferating cell nuclear antigen
- PDGFR
platelet‐derived growth factor receptor
- PH
pulmonary hypertension
- RIP
RNA immunoprecipitation
- RT‐qPCR
reverse transcription‐quantitative polymerase chain reaction
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
Clinical Perspective.
What Is New?
A novel circular RNA named circ‐myh8 was increased by hypoxia.
Circ‐myh8 induces the proliferation, cell‐cycle progression, and phenotype switch of pulmonary artery smooth muscle cells.
Circ‐myh8 acts as a modular scaffold to recruit histone acetyltransferase KAT7 to the promoters of hypoxia‐inducible factor‐1α genes.
What Are the Clinical Implications?
Hypoxia‐inducible factor‐1α is an important therapeutic target in pulmonary hypertension.
The circ‐myh8/KAT7 pathway could be used to combat pulmonary hypertension.
Pulmonary hypertension (PH) is a group of pathophysiological disorders that may involve multiple clinical conditions and progressive diseases, ultimately leading to premature death. A common pathological characteristic of PH is vascular remodeling, which manifests as medial thickening, mainly caused by uncontrolled proliferation and cell‐cycle progression of pulmonary artery smooth muscle cells (PASMCs). PH is invariably associated with reduced functional ability, impaired quality of life, greater oxygen requirements, and an increased risk of mortality. 1 Chronic hypoxia has emerged as a well‐known independent cause of vascular remodeling that also exacerbates PH caused by lung parenchyma or hypoxia. 2 , 3 At present, the application and benefits of existing drugs targeting pulmonary arterial hypertension (PAH) involving prostanoids, phosphodiesterase‐5 inhibitors, and endothelin receptor blockers in hypoxia‐induced PH remain unknown. These drugs provide symptomatic relief in some reports but suggest that they only modestly improve survival, as they could not prevent excessive PASMC proliferation and occlusive remodeling. Therefore, there is an urgent need to identify suitable targets that drive vascular remodeling to develop effective anti‐PH drugs.
Hypoxia‐inducible factor (HIF) is a well‐known modulator in the hypoxia signaling pathway and plays an important regulatory role in the pathogenesis of hypoxia‐induced PH. 4 The HIF family is mainly composed of 4 members—HIF1α, HIF1β, HIF2α, and HIF2β 5 —of which HIF1α is the key regulatory factor in the proliferation of PASMCs. 6 During the development of PH pathogenesis, aberrant HIF1α activation determines the PASMCs' proliferation and cell‐cycle progression and pulmonary vascular remodeling. 7 For instance, mice with a smooth muscle cell–specific HIF1α heterozygous inactivation or inducible deletion exhibit reduced vascular remodeling and blunted cell hypertrophy in mice with chronic hypoxic‐induced PH. 8 , 9 In addition, HIF‐1α also functions as an intrinsic pathogenic modulator in genetic forms and chemically induces PH. 10 These studies indicate that clarifying the HIF‐1α evokes cellular events, especially the uncontrolled proliferation and cell‐cycle progression of PASMCs, which may provide a promising avenue for PH pharmacotherapies.
Circular RNAs (circRNAs) are a novel class of noncoding RNAs that are characterized by a covalent closed‐loop structure. 11 , 12 Although circRNAs were first detected >20 years ago, their functions have not been explored until recently. 13 , 14 , 15 With the development of high‐throughput sequencing and bioinformatic analysis, large numbers of circRNAs have been successfully identified, and they are abundant, conserved, tissue specific, and development specific in mammalian cells. 16 , 17 Unlike their linear counterparts, circRNAs contain no 5′ to 3′ polarity or polyadenylation tails. CircRNAs are resistant to RNase R digestion and are highly stable in vivo compared with their parent genes. It is well established that inverted repeated Alu elements, exon skipping, and RNA‐binding proteins facilitate and regulate the formation of circRNAs. 18 , 19 , 20 Furthermore, circRNAs are widely involved in physiological and pathological processes, such as diabetes, 21 neurological disorders, 11 and especially cancer. 22
Most circRNAs function as microRNA sponges to regulate the expression of target genes, eventually promoting proliferation or inhibiting apoptosis of PASMCs. For example, has‐circ‐0016070 contains the binding site of miR‐942 and relieves the suppression of miR‐942 on cyclin D1. 23 Mmu‐circ‐0000790 competitively binds to miR‐374c and consequently upregulates the target gene FOXC1. 24 Besides the competitive endogenous RNA mechanism, circRNAs can serve as protein decoys, scaffolds, and recruiters to modulate protein–protein interactions. 25 For example, circ‐Foxo3 could prevent mouse double minute 2 from inducing Foxo3 ubiquitination and degradation, resulting in increased levels of the Foxo3 protein. 26 Circular catenin‐β1 promotes cancer progression by enhancing the transactivation of Yin Yang 1 by DDX3 and thus upregulating target genes involved with β‐catenin activation. 22 Moreover, it has been shown that circRNAs can be translated into polypeptides. 27 Previously, we have showed that circ‐Calm4 plays an important role in hypoxia‐induced pyroptosis and is dependent on the circ‐Calm4/miR‐124‐3p/Pdcd6 axis. 28 However, the functions of circRNAs in the regulation of HIF1α expression is yet to be defined.
In this study, genome‐wide circRNA expression profiles were initially examined in pulmonary artery tissue from mice exposed to normoxia and hypoxia. We characterized a novel circRNA, chr11_67292179–67294612, derived from exons 18, 19, 20, 21, and 22 of the myh8 gene circ_chr11_67292179–67294612 (circ‐myh8) and validated it by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). The role of circ‐myh8 in mouse PH models was examined by genetic overexpression and ablation using a lentivirus. Cell proliferation and cell‐cycle progression experiments were conducted, and it was determined that circ‐myh8 acts as a proliferation promoter gene targeted to HIF1 by recruiting hypoxia‐induced PH.
METHODS
The data supporting this study's findings are available from the corresponding author upon reasonable request.
Animal Models
Hypoxia PH mouse model: 40 male C57BL/6J mice (8‐week‐old) were randomly grouped into normoxia (21% FiO2), and hypoxia (10% FiO2) for 4, 8, 12, and 21 days. For the circ‐myh8 overexpression: thirty‐two 8‐week‐old male C57BL/6J mice were randomly divided into 2 groups. The overexpression circ‐myh8 group received lentiviral package circ‐myh8 in the nasal mucosa. The empty viral vector was used as the control group and received normal oxygen (21% FiO2) for 5 weeks. For the circ‐myh8 knockdown, lentiviral packaging short hairpin RNA targeted circ‐myh8 was inhaled through the nasal mucosa, with lentiviral packaging negative control as the control group, standard oxygen culture for 2 weeks, and hypoxia for 3 weeks. This animal study was approved by the ethics review board of Harbin Medical University ([2012]‐006). All experimental procedures were performed following the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health.
Plasma From Patients With PH
Plasma from 10 patients with PH was collected between May 2019 and July 2022 in the Second Affiliated Hospital of Harbin Medical University. Patients were subdivided into groups as follows: 6 patients with idiopathic PAH, 1 with connective tissue disease–associated PH, and 3 with PH caused by left heart disease (group 2 PH; Table S1). The ethics review board of Harbin Medical University approved the study protocol. All patients provided a written informed consent.
Cell Culture
The original mouse PASMCs were purchased from Procell Life Science&Technology Co. Ltd. (Wuhan, China) and maintained in a full‐growth medium (SMCM #1101; Sciencell Research Laboratories, Carlsbad, CA). The purity of the cells reached >90% identified by α‐smooth muscle actin immunofluorescence, and the cells are free of viruses, mycoplasma, bacteria, and the like. Cells were cultured in a humidified 37 °C, 5% CO2 incubator, split at 90% to 95% confluence by trypsin‐collagenase, and used between passages 5 and 7.
Plasmid and Lentivirus Construction
Plasmid
We synthesized the base sequence of 5 exons of the circ‐myh8 gene (exons 21–25) and inserted this fragment into pCDH‐CMV‐MCSEF1‐GFP+ Puro with restriction sites of BamHI and NotI Carrier (Abbots, Shanghai, China). The result of vector construction was verified by direct sequencing. Using Lipofectamine 2000 (Invitrogen, Waltham, MA) transfection manual, the overexpression pCDH‐CMV‐circ‐myh8 and the empty plasmid were transfected into PASMCs. The empty plasmid was pcDNA, and the overexpression plasmid was pcDNA‐circ.
Lentivirus
We cotransfected the constructed lentiviral vector of the circ‐myh8 base sequence and short hairpin RNA (shRNA) sequence and its auxiliary, packaging original vector plasmid into 293T cells, added enhancing buffer after 10 to 12 hours of transfection, and then changed to a new culture after 8 hours. After culturing for 48 hours, we collected the cell supernatant rich in lentiviral particles, and concentrated it to obtain a high‐titer lentiviral concentrate. The virus titer is measured and calibrated in 293T cells. The lentivirus control and overexpression were named vehicle and Lenti‐circ, respectively.
Small Interfering RNA and Gapmers
The small interfering RNA (siRNA) and the gapmers targeted to circ‐myh8 (containing the circ‐myh8 end‐to‐end junctions) were synthesized by Abbots. The siRNA and gapmers were transfected into PASMCs using Lipofectamine 2000 according to the manufacturer's protocols. The shRNA was named sh‐circ, and NC was used as a negative control. Sequences are listed in Table S2.
RNA Extraction and Reverse Transcription‐Quantitative Reverse Transcription Polymerase Chain Reaction
Invitrogen's Trizol was used to isolate total RNA, and 0.5 μg of total RNA was reverse transcribed into cDNA using ReverTra Ace qPCR RT Kit (Toyobo, Osaka, Japan). SYBR Green Realtime PCR Master Mix (Toyobo) and LightCycler 480 system (Roche, Basel, Switzerland) were used to measure circRNA and mRNA expression levels by qPCR. 18S was used as an internal control, and each experiment was repeated 6 to 8 times. The primers are listed in Table S3.
Western Blot
Tissue and cell samples were lysed with cell lysate (containing 1% phenylmethylsulfonyl fluoride), and SDS polyacrylamide gel electrophoresis was performed to separate proteins of different molecular weights and then transferred to negative control membranes. After being blocked with skimmed milk powder for 1 hour, the negative control membranes were incubated with KAT1 (1: 400), KAT5 (1:400), KAT7 (lysine acetyltransferase 7) (1:400), KAT8 (1:400), lysine 5 of histone H4 (H4K5)ac (1:400), H4K12ac (1:400), HIF1α (1:200), proliferating cell nuclear antigen (PCNA) (1:400), osteopontin (1:1000), calponin 1 (1:1000), platelet‐derived growth factor receptor (PDGFR)‐α (1:1000), PDGFR‐β (1:500), interleukin‐6 (1:1000), tumor necrosis factor‐α (1:1000), caspase 3 (1:1000), caspase 9 (1:1000), B‐cell lymphoma 2 (1:500), B‐cell lymphoma 2–like protein 4 (1:500), cyclin A (1:400), cyclin B (1:400), cyclin D (1:400), cyclin‐dependent kinase (CDK) 1 (1:400), CDK2 (1:400), and CDK4 (1:400) primary antibodies at 4 °C for overnight, washed with TBS‐T 5 times for 10 minutes each time, then incubated with secondary antibodies (1:8000) according to the source of the primary antibody. Membranes were developed using the enhanced chemiluminescence solution and x‐ray film in a dark room. Histone‐related antibodies were purchased from Cell Signaling Technology (Boston, MA), HIF1α was purchased from R&D Systems (Minneapolis, MN), and other antibodies were purchased from Boster Biological Technology Co. Ltd (Wuhan, China).
Immunofluorescence
Frozen sections of lung tissue or PASMCs were fixed with paraformaldehyde, 95 °C 1× saline–sodium citrate solution for antigen retrieval, 0.4% Triton treatment, goat serum blocking solution for 1 hour, incubation with ki67 (1:100), KAT7 (1:100), and H4K5ac (1:100) primary antibody at 37 °C overnight. The next day, after washing with PBS, the secondary antibody (1:100) was incubated at 37 °C for 2 hours, then mounted and imaged with a confocal microscope.
Scratch Assay
The mouse PASMCs were cultured on plates. Scratch was created by a pipette. Images were at 0 hour. The cells were treated with the indicated agents. Cells were cultured for another 24 hours before capturing the images, and the migration rate was analyzed.
Chromatin Immunoprecipitation
Using the Pierce Magnetic ChIP Kit from Thermo Scientific (Waltham, MA), PASMCs were cross‐linked with formaldehyde, and the cross‐linking reaction was terminated by glycine. After cross‐linking, the cells are digested with the RNase enzyme. Ultrasonic breaks the chromatin into 500‐ to 1000‐bp fragments. The chromatin fragments are subjected to immunoprecipitation with specific antibodies of KAT7, H4K5ac, H4K12ac, and H3K14ac. DNA is separated from the eluate of the immunoprecipitation reaction. The fragments were amplified by qPCR using primers from the HIF1α promoter.
RNA Pull‐Down
Synthetic RNA probe (Mmu‐circ‐myh8 antisense strand segment), using Pierce Magnetic RNA probe in the Pull‐Down Kit to probe, and cell lysates were incubated to form RNA‐protein complexes. The complex uses the RNase R enzyme to digest linear RNA. After eluting the RNA‐binding protein complex, RNA‐binding proteins were detected by western blot assay.
RNA Immunoprecipitation
The Magna RNA immunoprecipitation (RIP) RNA Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA) was used for RIP analysis, PASMCs and complete RIP Lysis Buffer (Millipore) was used for cracking; 10 μg of primary antibody (contrast rabbit IgG, Kat1, Kat5, Kat7, and Kat8) was incubated with magnetic beads at room temperature for 30 minutes, and the cell lysates were incubated with the magnetic bead antibody complex overnight at 4 °C. The beads were washed 6 times with a cold Ice‐Cold RIP Wash Buffer and resuspended in the protease K buffer. The immunoprecipitation RNA was purified by phenol‐chloroform extraction and ethanol precipitation. RT‐qPCR analysis was performed for downstream RNA detection.
Chromatin Isolation by RNA Purification
A biotinylated circ‐myh8 probe (circ‐myh8 antisense chain) was mixed with PASMC lysates. The chromatin complexes were purified using magnetic streptomycetes antibiotin beads and then rigorously washed by the ChIRP Kit (Guangzhou Saicheng Biotechnology Co. Ltd, Guangzhou, China) according to the manufacturer's protocol. The circ‐myh8–bound DNA was eluted with a mixture of RNase A and H for RT‐qPCR analysis.
Fluorescent in Situ Hybridization
Frozen sections of mouse lung tissues or PASMCs were digested with 0.25% pepsin at 37 °C for 30 minutes, fixed with 4% paraformaldehyde for 20 minutes, treated with 0.5% Triton X‐100 for 15 minutes, and incubated with a circ‐myh8 probe hybridization solution at 4 °C overnight. Then circ‐myhH8 probe hybridization fluid was aspirated, the nuclear was labeled with DAPI, and the changes of circ‐myh8 expression were imaged with confocal microscopy.
Masson's Trichrome Staining
Masson's trichrome staining was performed following the manufacturer's protocol (SOLARBIO, Beijing, China). The mice's lung tissues were fixed in 10% buffered formalin and embedded in paraffin. Transverse sections of the lung tissue were cut (6 μm), deparaffinized, and fixed in Mordant in Bouin's solution for 1 hour at 60 °C. Sections were stained sequentially at room temperature with hematoxylin, ferric chloride solution, Lichunred acid fuchsin, phosphotungstic/phosphomolybdic acid, and aniline blue. Sections were washed, dehydrated, and mounted with a xylene‐based mounting medium.
EdU Solution
PASMCs were cultured in 12‐well plates. After treatment with different factors, EdU solution of 50 μM was added to each well, the cells were incubated in a cell incubator for 2 hours and fixed with 4% paraformaldehyde, the reaction was stopped with glycine solution, 0.5% TritonX‐100 was permeated, Apollo staining reaction solution was added, and Hoechst33342 reaction solution was added to stain the nucleus. The number of positively staining cells was counted under a confocal microscope.
Luciferase Assay
The HIF1α promoter fragment was inserted into the front of the luciferase expression sequence to construct pGL3‐HIF1α reporter plasmid, and circ‐myh8 plasmid and pGL3‐HIF1α were cotransfected into 293T cells, the specific luciferase substrate was added, and the luciferase reacted with the substrate to produce fluorescence. The luciferase activity was measured by detecting fluorescence intensity, and the binding ability of circ‐myh8 to HIF1α promoter was evaluated.
RESULTS
Expression Profile of circRNAs in Pulmonary Artery From Hypoxic PH Mice
The expression profiles of circRNA in the pulmonary arteries (PAs) of hypoxic PH mice were analyzed by circRNA sequencing. Compared with the control mice, our previous work confirmed that a total of 67 circRNAs were differentially expressed in hypoxic PAs, of which 34 circRNAs were upregulated and 33 circRNAs were downregulated. 28 RT‐qPCR analysis further confirmed that the expression of a candidate circRNA, circ_chr11_67292179–67294612 (circ‐myh8), which was derived from exons 18, 19, 20, 21, and 22 of the myh8 gene, was significantly upregulated. Circ‐myh8 is a 723bp nucleotide located on mouse chromosome 11, GRCm38.p4 site, 67292179–67294612.
Changes of Circ‐myh8 Expression in Hypoxic PAs
To further clarify the characteristics of the novel circRNA circ‐myh8, the head‐to‐tail splices of exons 18 and 22 of the myh8 gene were verified by Sanger sequencing (Figure S1A and S1B). Since circRNA does not have the characteristic poly (A) tail, random and oligo dT primers were used in the reverse transcription process. Figure S1C shows that the reverse transcription of circ‐myh8 with oligo dT primers was significantly less than that of random primers. Figure S1D shows that circ‐myh8 is easily digested by RNase R compared with the linear mRNA of myh8. These data confirmed that circ‐myh8 is a stable circular RNA.
To detect circ‐myh8 expression in PAs during the development of group 3 PH mice, PA sections from different treatments were hybridized with the circ‐myh8 fluorescent in situ hybridization (FISH) probe. Compared with the control, circ‐myh8 expression was significantly increased in PAs from hypoxic PH mice at 12 and 21 days (Figure 1A). Circ‐myh8 expression was mainly localized in smooth muscle cells, as confirmed by double staining with the circ‐myh8 FISH probe and α‐smooth muscle actin (Figure 1A). RT‐qPCR results confirmed that the increasing expression of circ‐myh8 was initiated from 4 days, with a significant increase at 12 and 21 days (Figure 1B). We also detected circ‐myh8 expression in other tissues. RT‐qPCR results showed that the expression of circ‐myh8 increased in the hypoxic lung, heart, right ventricle, and kidney (Figure S2A), while FISH results showed that the expression of circ‐myh8 in the carotid artery and aorta was not affected by hypoxia (Figure S2B). As in situ hybridization results showed that circ‐myh8 is mainly expressed in PAs, we next detected circ‐myh8 expression in cultured PASMCs under hypoxic conditions. The expression of circ‐myh8 in PASMCs was significantly increased at 24 and 48 hours compared with that in the normoxic group (Figure 1C). We detected the expression of circ‐myh8 in the plasma of patients with PH using RT‐qPCR. The results showed that circ‐myh8 was increased in patients with PH compared with that in healthy controls (Figure 1D). Finally, the subcellular localization of circ‐myh8 was detected by FISH and cytoplasmic nuclear separation using RT‐qPCR. FISH results showed that the expression of circ‐myh8 increased in the nucleus after 24 hours of hypoxia (Figure 1E). RT‐qPCR results showed that circ‐myh8 increased 3.84±0.54‐fold (Figure 1F) in the whole cell lysate, 1.33±0.35‐fold in the cytoplasm fraction (Figure 1G), and 5.48±0.64‐fold in the nucleus fraction (Figure 1H) after 24 hours of hypoxia.
Figure 1. Circ‐myh8 elevation in pulmonary artery smooth muscle cells (PASMCs) correlates with hypoxia‐induced pulmonary hypertension (PH).
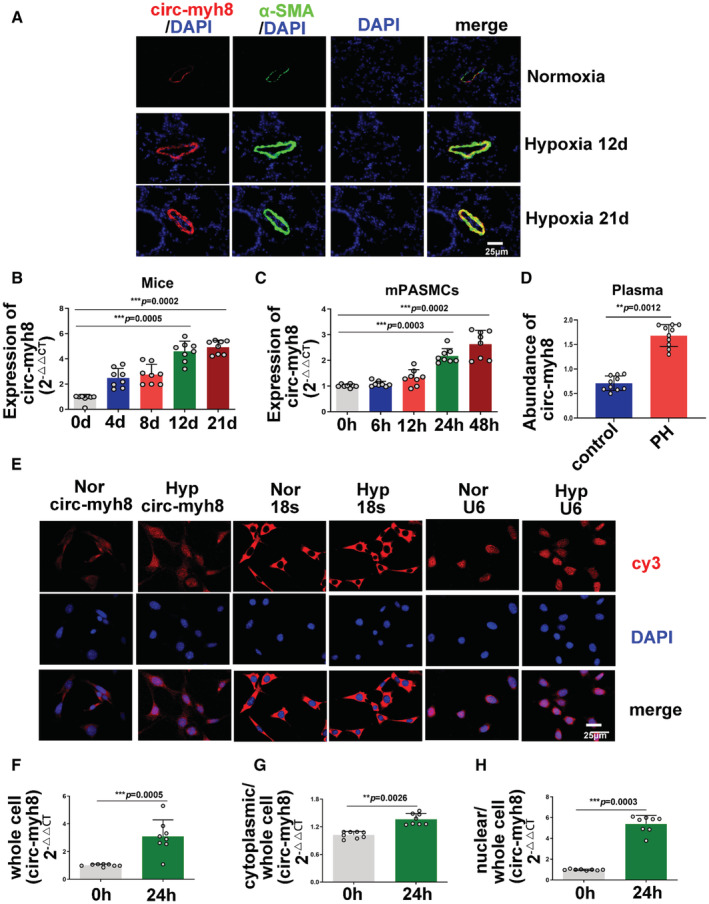
A, Fluorescent in situ hybridization (FISH) for circ‐myh8 (red) and immunofluorescence for α‐smooth muscle actin (αSMA, green) in the pulmonary artery (PA) from mice during development of hypoxia‐induced pulmonary hypertension (PH). The profiles of colocalization were also provided. Nuclei are counterstained with DAPI (blue). Scale bar, 25 μm. B, Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) assay for circ‐myh8 expression in PAs from mice during development of hypoxia‐induced PH. C, RT‐qPCR assay for circ‐myh8 expression in pulmonary artery smooth muscle cells (PASMCs) exposed to normoxia or hypoxia for indicated time points. D, RT‐qPCR assay for circ‐myh8 expression in plasma from patients with PH. E, FISH for circ‐myh8, 18s, and U6 expression and cellular localization in PASMCs exposed to normoxia or hypoxia for 24 hours, with the nuclei staining by DAPI (blue). 18s is a cytoplasm marker, and U6 serves as a marker for nucleus. Scale bar, 25 μm. F through H, RT‐qPCR assay for circ‐myh8 expression in whole cell lysate (F), cytoplasm (G), and nucleus (H) fraction in PASMCs exposed to normoxia or hypoxia for 24 hours. Data represent means±SEM from indicated independent experiments. Student's t test (for 2 means) or 1‐way ANOVA followed by Dunnett's test (for >2 means). 18s indicates 18S rRNA; Hyp, hypoxia; Nor, normoxia; and U6, U6 snRNA. *P<0.05, **P<0.01, ***P<0.001.
Circ‐myh8 Promotes Pulmonary Vascular Remodeling In Vivo
To further clarify the role of circ‐myh8 in vivo, circ‐myh8 was overexpressed with lentivirus and knocked out with lentivirus shRNA in mice. The 8‐week‐old mice were then treated as illustrated in Figure 2A. Both FISH (Figure 2B and 2C) and RT‐qPCR (Figure 2D) results showed that circ‐myh8 was successfully overexpressed by the lentivirus or knocked down by shRNA under hypoxic conditions. Meanwhile, right ventricular systolic pressure (Figure 2E), Felton index (Figure 2F), and hemodynamics index, including the pulmonary artery acceleration time and the pulmonary artery velocity time integral, assessed by echocardiography (Figure 2G), demonstrated that circ‐myh8 overexpression induced PH under normoxic conditions. However, knockdown of circ‐myh8 attenuated hypoxia‐induced PH. Next, the expression of the proliferation indicator Ki67 was detected by immunofluorescence staining (Figure 2H) and PCNA expression was measured by western blotting (Figure 2I). Both results showed that overexpression of circ‐myh8 promoted cell proliferation under normoxia, and knockdown of circ‐myh8 prevented hypoxia‐induced cell proliferation. We also examined vascular remodeling using Masson's trichrome and hematoxylin and eosin staining. The results showed that fibrosis and thickness of the vascular wall increased after circ‐myh8 overexpression under normoxia but was inhibited during circ‐myh8 knockdown under hypoxic conditions (Figure 2J). Taken together, these results verify that circ‐myh8 contributes to hypoxia‐induced PH in vivo.
Figure 2. Circ‐myh8 promotes the development of hypoxia‐induced pulmonary hypertension.
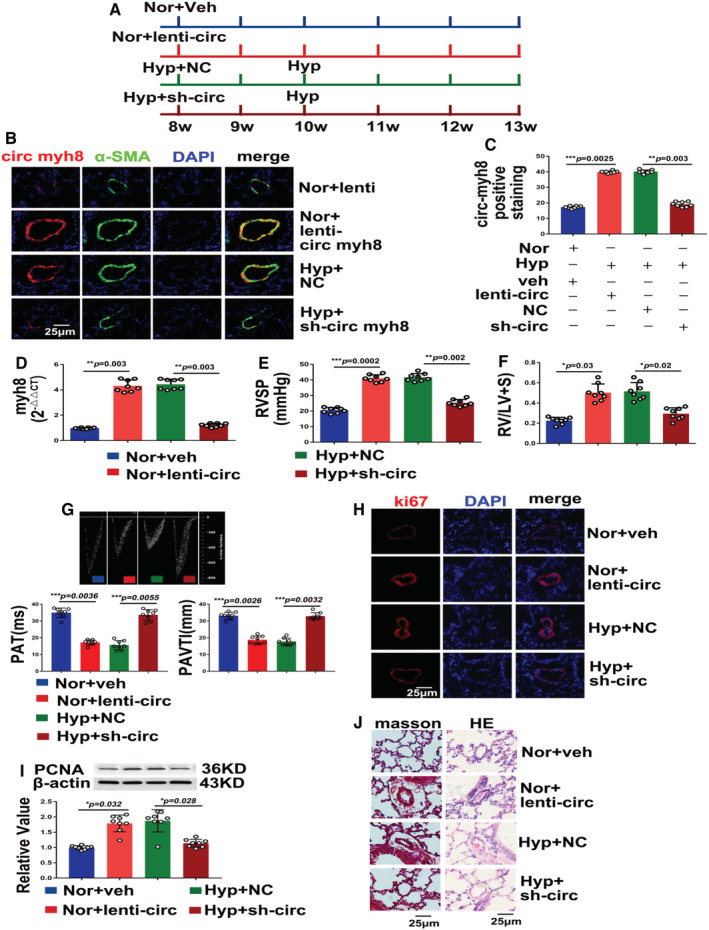
A, Schematic diagram of circ‐myh8 overexpression and knockdown mice models under normoxia or hypoxia. B, Fluorescent in situ hybridization (FISH) for circ‐myh8 (red) and immunofluorescence (IF) for α‐smooth muscle actin (αSMA, green) in the pulmonary artery (PA). Mice were overexpressed with circ‐myh8 lentivirus plasmid (lenti‐circ myh8), empty vector (lenti), or knockdown with sh‐circ myh8 or negative control (NC) under normoxia (21% FiO2) or hypoxia (10% FiO2) for indicated times. The profiles of colocalization were also provided. Nuclei are counterstained with DAPI (blue). Scale bar, 25 μm. C, Statistical analysis of circ‐myh8 expression in (B). D, Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) assay for circ‐myh8 expression in PAs from mice at the indicated treatment. E, Right ventricular systolic pressure (RVSP) was measured by right heart catheter. F, The Felton index was calculated as the ratio of right ventricular and left ventricular plus spatial weight. G, Representative image of hemodynamic (top panel), summarized data of PAT (left bottom) and PAVTI (right bottom) measured by ultrasound. H, immunofluorescence for ki67α (red) in pulmonary artery (PAs) section. The profiles of colocalization were also provided. Nuclei are counterstained with DAPI (blue). Scale bar, 25 μm. I, Representative images and summarized data of PCNA expression analysis by western blot. J, Representative images of Masson trichrome and hematoxylin and eosin staining in lung from the indicated treatment mice. Scale bar, 25 μm. Data represent means±SEM from indicated independent experiments. Student's t test (for 2 means) or 1‐way ANOVA followed by Dunnett's test (for >2 means). Hyp indicates hypoxia; lenti‐circ, lenti‐circ‐myh8; NC, negative control; Nor, normoxia; PAT, pulmonary artery acceleration time; PAVTI, pulmonary artery velocity time integral; PCNA, proliferating cell nuclear antigen; and veh, vehicle. *P<0.05, **P<0.01, ***P<0.001.
Circ‐myh8 Promotes PASMC Proliferation, Cell‐Cycle Progression, and Phenotype Switch In Vitro
As the circ‐myh8 is mainly expressed in the smooth muscle layer of Pas, we hypothesized that circ‐myh8 participates in PASMC proliferation and cell‐cycle progression. To evaluate this hypothesis, an siRNA targeting the end‐to‐end junction of circ‐myh8 (Figure S3A) was designed. The siRNA effectively silenced circ‐myh8 expression but had no effect on myh8 mRNA expression (Figure S3B and S3C). In contrast, a silencing siRNA targeting myh8 mRNA did not affect circ‐myh8 expression (Figure S3D). These data indicated that siRNA targeting the end‐to‐end junction of circ‐myh8 was able to knockdown circ‐myh8 expression. The EdU result showed (Figure 3A) that knockdown of circ‐myh8 knockdown prevented hypoxia‐induced PASMC proliferation. Moreover, both PCNA expression (Figure 3B) and the ki67 expression (Figure 3C) displayed similar effects. Next, we investigated the role of circ‐myh8 in the cell‐cycle progression. As expected, hypoxia promoted PASMCs to enter the S + G2/M phase, whereas circ‐myh8 knockdown attenuated these effects (Figure 3D). Furthermore, knockdown of circ‐myh8 prevented the increased expression of cyclins A, B, and D (Figure 3D), and coordinated the activation of important cell‐cycle proteins, including CDKs (CDK1, CDK2, and CDK4) (Figure 3E), which were induced by hypoxia. We revealed that circ‐myh8 promotes the proliferation of PASMCs.
Figure 3. Silencing of circ‐myh8 reverses hypoxia‐induced proliferation and cell‐cycle progression of pulmonary artery smooth muscle cells (PASMCs) in vitro.
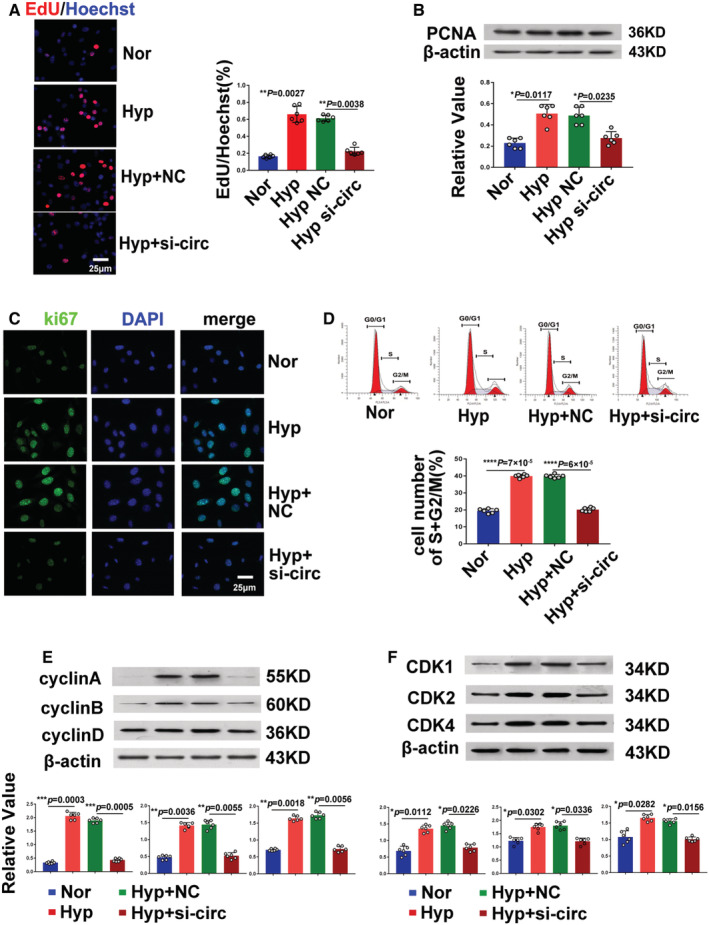
A, Representative images and summarized data EdU staining in PASMCs. PASMCs were transfected with circ‐myh8 siRNA or NC, and then exposed to hypoxia (0.3% FiO2) for 24 hours, with the nuclei stained by Hoechst (blue). Scale bar, 25 μm. B, Representative images and summarized data of PCNA expression analysis by western blot. C, Immunofluorescence for ki67α (red) in PASMCs. The profiles of colocalization with DAPI (blue) were also provided. Scale bar, 25 μm. D, Flow cytometry of the cell cycle of PASMCs. E, Representative images and summarized data of cyclin A, cyclin B, and cyclin D expression by western blot. F, Representative images and summarized data of cell‐cycle–dependent kinases (CDK1, CDK2, and CDK4) by western blot. Data represent means±SEM from indicated independent experiments. Student's t test (for 2 means) or 1‐way ANOVA followed by Dunnett's test (for >2 means). CDK indicates cyclin‐dependent kinase; Hyp, hypoxia; NC, negative control; Nor, normoxia; and PCNA, proliferating cell nuclear antigen. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Since the expression of circ‐myh8 in the nucleus was increased during hypoxia, and the efficiency of siRNA silencing of nuclear RNA was low, gapmers targeting circ‐myh8 expression (namely, gapmers‐circ) were designed, and the interference efficiency was tested. Furthermore, circ‐myh8 was overexpressed in a plasmid (Figure 4A). When circ‐myh8 was overexpressed under normoxia, the percentage of EdU‐positive cells increased, and when circ‐myh8 was knocked down by gapmers, the increase in the percentage of EdU‐positive cells induced by hypoxia was reversed (Figure 4B). Gapmers silencing circ‐myh8 in PASMCs were used to change the expression of EdU, PCNA, ki67, cell cycle, cyclin, and cell‐cycle–dependent protein kinase. PCNA protein expression (Figure 4C), Ki67 staining (Figure 4D), flow cytometry (Figure 4E), cyclin expression (cyclins A, B, and D; Figure 4F), and CDKs (CDK1, CDK2, and CDK4; Figure 4G) increased when circ‐myh8 was overexpressed and decreased when circ‐myh8 was knocked down by gapmers under hypoxic conditions. Moreover, we assessed the relationship between circ‐myh8 and phenotype switch. Expression of the contractile phenotype markers SM22α‐actin and calponin 1, and the synthetic phenotype marker osteopontin were examined. The expression of SM22α and calponin 1 was downregulated, but that of osteopontin was upregulated by circ‐myh8 overexpression and exposure to hypoxic conditions. The effect of hypoxia on the expression of SM22α, calponin 1, and osteopontin was reversed by circ‐myh8 knockdown in the gapmers (Figure 4H). To investigate the role of circ‐myh8 in PASMC migration, a wound‐healing assay was performed (Figure 4I). Hypoxia and circ‐myh8 overexpression increased PASMC migration, but knockdown of circ‐myh8 inhibited the increase in migration induced by hypoxia (Figure 4I).
Figure 4. Circ‐myh8 promotes pulmonary artery smooth muscle cell proliferation, cell‐cycle progression, and phenotype switch in vitro.
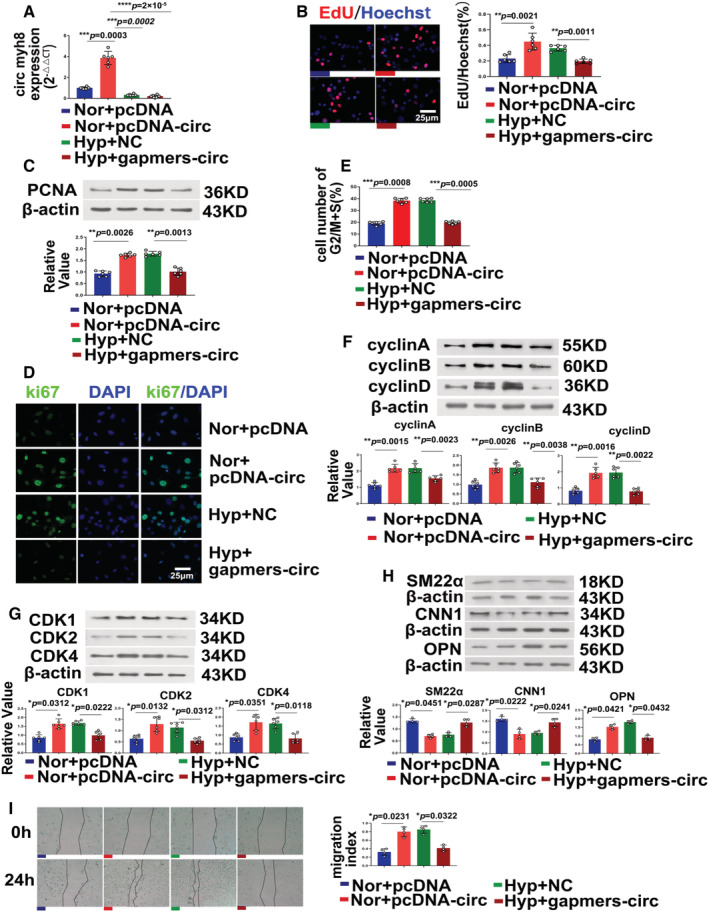
Pulmonary artery smooth muscle cells (PASMCs) were transfected with circ‐myh8 plasmid or pcDNA following cultured in normoxia condition for 24 hours. For the knockdown experiments, PASMCs were transfected with circ‐myh8 gapmers or NC and then exposed to hypoxia (0.3% FiO2) for 24 hours. A, Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) assay for circ‐myh8 expression in PASMCs. B, Representative images and summarized data of EdU staining. Scale bar, 25 μm. C through H, Representative images and summarized data of PCNA expression (C), Ki67 expression (D), cell cytometry (E), cyclins expression (F), cell‐cycle–dependent kinases expression (G), and phenotype markers SM22α, CNN1 and OPN (H). I, Representative images and summarized data of scratch assay. Data represent means ± SEM from indicated independent experiments. Student's t test (for 2 means) or 1‐way ANOVA followed by Dunnett's test (for >2 means). Scale bar, 25 μm. CDK indicates cyclin‐dependent kinase; CNN1, calponin 1; Hyp, hypoxia; NC, negative control; Nor, normoxia; OPN, osteopontin; PCNA, proliferating cell nuclear antigen; SM22α, smooth muscle 22 alpha; and veh, vehicle. *P<0.05, **P<0.01, ***P<0.001.
Furthermore, we examined the role of other PH‐related factors, such as apoptosis, inflammatory factors, and PDGFRs signaling. The results indicated that Bax, B‐cell lymphoma 2, caspase 3, and caspase 9 were not affected by circ‐myh8 (Figure S4A). However, the expression of tumor necrosis factor‐α, interleukin‐6, PDGFRα, and PDGFRβ increased when circ‐myh8 was overexpressed and decreased when circ‐myh8 was knocked down by gapmers or siRNA under hypoxic conditions (Figure S4B and S4C).
Gapmers of circ‐myh8 displayed similar effects as shRNA of circ‐myh8 in preventing cell proliferation and cell‐cycle progression induced by hypoxia. These data suggest that circ‐myh8 overexpression evokes, whereas knockdown prevents, hyperproliferation, increased cell‐cycle progression, and a phenotype switch from contractile to synthetic PASMCs induced by hypoxia.
Circ‐myh8 Functions Through Activating HIF1α Transcription
To explore the involvement of circ‐myh8 in promoting the development of hypoxia‐induced PH we checked the gene ontology enrichment and the Kyoto Encyclopedia of Genes and Genomes pathway analysis according to the circRNA sequencing data, which indicated that the hypoxia signaling pathway is the most important pathway related to pulmonary hypertension regulated by circRNAs (Figure 5A). Therefore, circ‐myh8 was either overexpressed or silenced in PASMCs, and changes in the expression of the 4 HIF subtypes were detected. The results showed that only HIF1α mRNA and protein expression increased when circ‐myh8 was overexpressed in normoxia, but decreased when circ‐myh8 was knocked down by shRNA or gapmers (Figure 5B and 5C). However, HIF1β, HIF2α, and HIF2β mRNA expression levels did not change (Figure 5D through 5F). In addition, inhibition of HIF1α expression by siRNA prevented circ‐myh8 overexpression‐evoked EdU positive staining of PASMCs under hypoxic conditions (Figure 5G). Knockdown of HIF1α also reduced circ‐myh8 overexpression‐induced PCNA (Figure 5H), ki67 positive staining (Figure 5I), and cyclin expression (cyclins A, B, and D, Figure 5J). In addition, knockdown of HIF1α inhibited the increased expression of cyclin A, B, and D proteins of circ‐myh8. Double luciferase reporter gene analysis showed that circ‐myh8 overexpression significantly enhanced the promoter activity of HIF1α, whereas HIF1α promoter activity decreased when circ‐myh8 was knocked down using shRNA or gapmers (Figure 5K). These results suggest that circ‐myh8 promotes the proliferation and cell‐cycle progression of PASMCs through the transcriptional activation of HIF1α.
Figure 5. Circ‐myh8 promotes the pulmonary artery smooth muscle cells (PASMCs) proliferation and cell‐cycle progression by upregulating the transcription activity of HIF1α.
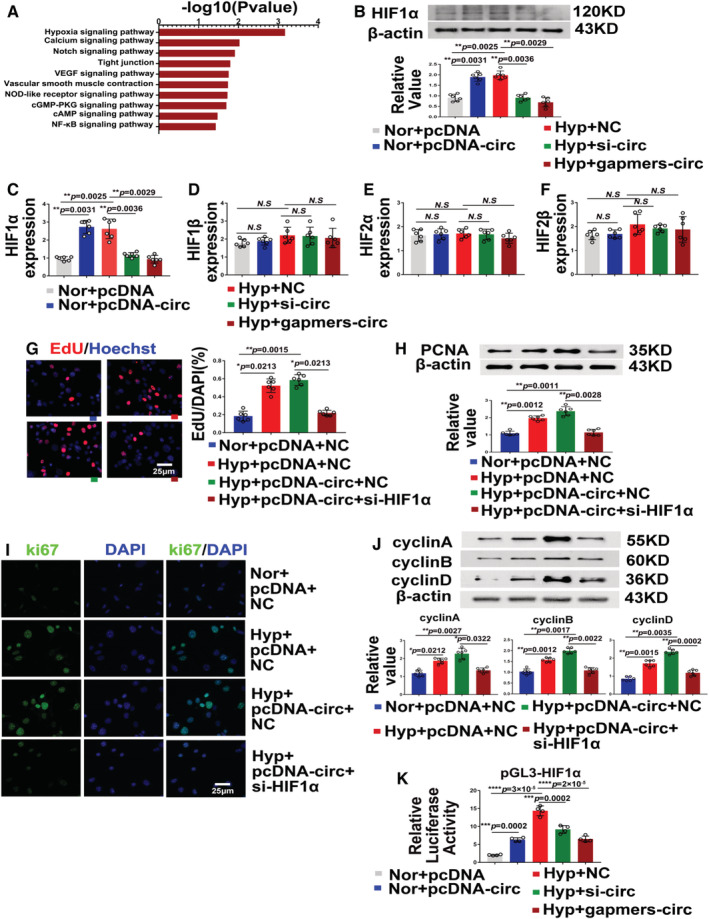
A, Circular RNA (circRNA) sequencing and pathway analysis of circ‐myh8 mediated mRNA expression profiles. B, Representative images and summarized data of HIF1α protein expression after overexpression or knockdown of circ‐myh8. For the circ‐myh8 overexpression, PASMCs were transfected with circ‐myh8 plasmid or pcDNA following culture in normoxia condition for 24 hours. For the circ‐myh8 knockdown, PASMCs were transfected with circ‐myh8 siRNA, gapmers, or NC, then exposed to hypoxia (0.3% FiO2) for 24 hours. C through F, Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) assay for HIF1α (C) HIF1β (D), HIF2α (E), and HIF2β (F) mRNA expression in the above cells in (B). The normal overexpression of circ‐myh8 can upregulate mRNA and protein levels, while the hypoxic knockdown of circ‐myh8 inhibits HIF1α mRNA and protein expression. G, Representative images and summarized data of EdU staining in cells. PASMCs were transfected with circ‐myh8 overexpression plasmid and control plasmid, with siRNA targeting HIF1α and siNC, then exposed to hypoxia (0.3% FiO2) for 24 hours. H through J, Representative images and summarized data of PCNA expression (H), Ki67 expression (I), and cyclins expression (J) of the above cells in (G). Scale bar, 25 μm. K, The luciferase activity of HIF1α promoters after circ‐myh8 overexpression or knockdown. PASMCs in 24‐well plates were transfected with circ‐myh8 plasmids or control, together with pGL3‐basic‐HIF1α (pGL3‐HIF1α) plasmid. The luciferase activity was measured 24 hours later. For the circ‐myh8 knockdown, PASMCs were transfected with circ‐myh8 small interfering RNA, gapmers, or NC, together with pGL3‐basic‐HIF1α (pGL3‐HIF1α) plasmid, then exposed to hypoxia (0.3% FiO2) for 24 hours. Data represent means±SEM from indicated independent experiments. Student's t test (for 2 means) or 1‐way ANOVA followed by Dunnett's test (for >2 means). HIF1α indicates hypoxia‐inducible factor alpha; Hyp, hypoxia; NC, negative control; Nor, normoxia; PCNA, proliferating cell nuclear antigen; and veh, vehicle. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Circ‐myh8 Acts as a Molecular Scaffold to Recruit KAT7 Into the Promoter of HIF1α
To further study the mechanism by which circ‐myh8 activates HIF1α transcription, we first searched the HIF1α promoter using the University of California–Santa Cruz website (genome.ucsc.edu) and found histone acetylation modification at the promoter site. Biotinylated circ‐myh8 probes were used to pull down the RNA‐protein complex following western blot analysis to evaluate candidate proteins, including lysine acetyltransferases KAT1, KAT5, KAT7, and KAT8, which may participate in transcriptional regulation (Figure 6A). The results showed that only KAT7 binds to circ‐myh8, whereas KAT1, KAT5, and KAT8 do not bind circ‐myh8 (Figure 6B and 6C). In addition, RIP was performed with the antibodies of KAT1, KAT5, KAT7, and KAT8. The results indicated that only KAT7 was able to bind with circ‐myh8 (Figure 6D and Figure S5). Moreover, FISH and immunocytofluorescence results showed that circ‐myh8 and KAT7 were colocated in the nuclei of PASMCs (Figure 6E). Molecular docking showed that circ‐myh8 fragments (131 bases at the end‐to‐end junction, upstream and downstream) formed multiple hydrogen bond interactions with KAT7 residues, including Tyr‐351, Lys‐401, Asp‐430, His‐431, Tyr‐470, Ser‐473, Cys‐474, Arg‐483, and Lys‐488 (Figure 6F). Subsequently, different fragments of KAT7 with a Flag‐tag and wild‐type KAT7 plasmids were synthesized and transfected into PASMCs. The RNA‐protein complex was pulled down using the circ‐myh8 probe antibody following examination with the Flag‐tag antibody. The results confirmed that the C2HC domain (336–390 aa) is crucial for the interaction of KAT7 with circ‐myh8 (Figure 6G). Previous studies have shown that KAT7 tends to acetylate at lysine5 and lysine12 of histone H4, and lysine 14 of histone H3. We used chromatin immunoprecipitation to detect the histone acetylation levels in the HIF1α promoter region. Chromatin immunoprecipitation assays were performed using 3 candidate histone antibodies: H4K5ac, H4K12ac, and H3K14ac. The results showed that only H4K5 acetylation was detected in the HIF1α promoter region, whereas H4K12 and H3K14 did not undergo acetylation (Figure 6H, Figure S6). Figure 6I shows that circ‐myh8 and H4K5ac were colocalized in the nucleus of PASMCs. The interaction between H4K5ac and circ‐myh8 was further demonstrated using an RNA pull‐down assay (Figure 6J). The binding modes of circ‐myh8, KAT7, and H4K5ac were further confirmed by molecular docking. The binding mode of the complex and H4 is shown in Figure 6K, and the docking score of HDOCK is −240.25. H4 forms hydrogen bonds with bases C20, A121, and U110 of RNA, Lys338, Glu346, and Arg344 of KAT7. H4 forms ionic interactions, such as salt bridges with bases C112 and U123 of RNA and Arg348 of KAT7. In addition, chromatin isolation by RNA purification experiments showed that circ‐myh8 could directly bind to the HIF1α promoter region (Figure 6L). The above results confirm that circ‐myh8 can act as a modular scaffold to recruit KAT7 into the promoter of the HIF1α gene and enhance the acetylation of histone H4K5. In addition, KAT7 silencing attenuated circ‐myh8‐induced H4K5ac levels in the promoter region of HIF1α (Figure 6M). The circ‐myh8–induced HIF1α mRNA and protein expression was reduced after KAT7 knockdown by siRNA (Figure 6N and 6O). Taken together, these data indicated circ‐myh8 increases the acetylation level of H4K5 in HIF1α promoter region by recruiting KAT7.
Figure 6. Circ‐myh8 acts as a modular scaffold to recruit KAT7 to the promoters of the HIF1α gene.
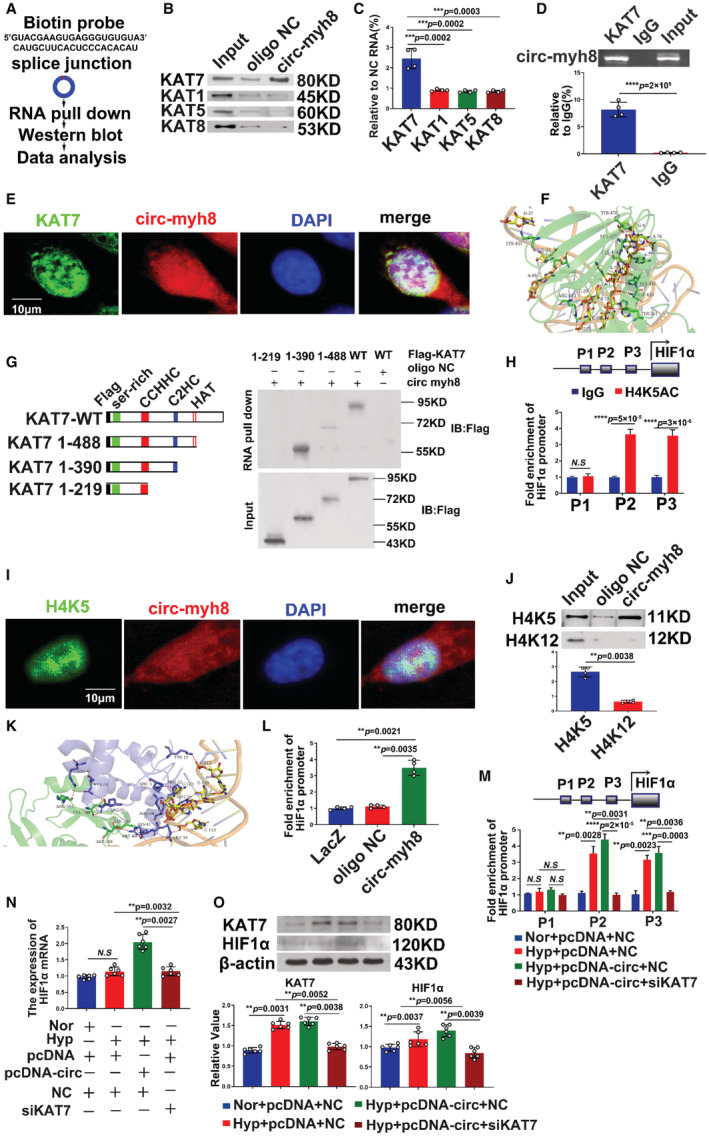
A, The experimental design for RNA pull‐down assay. RNA pull‐down was performed using a biotinylated circ‐myh8 probe, followed by western blot. B and C, Representative image (B) and summarized data (C) of RNA pull‐down assay followed by western blot for candidate proteins KAT1, KAT5, KAT7, and KAT8 in pulmonary artery smooth muscle cells (PASMCs). D, RIP assay for the binding of three candidate proteins with circ‐myh8. RIP was performed using KAT1, KAT5, KAT7 and KAT8 antibodies, followed by RT‐qPCR assay for circ‐myh8 expression in PASMCs. E, Fluorescent in situ hybridization (FISH) for circ‐myh8 (red) and immunofluorescence (IF) for KAT7 (green) in PASMCs, with the nuclei staining by DAPI (blue). The profiles of colocalization were also provided. Scale bar, 5 μm. F, The molecular docking of the interaction between circ‐myh8 fragment (131 base sequence upstream and downstream of that end‐to‐end junction, marked yellow cartoon) and KAT7 (shown as green). G, RNA pull‐down assay after transfection of wild type and truncated KAT7 expression plasmids using biotin‐labeled oligo or circ‐myh8 probes in PASMCs (right panel). The design of the truncated KAT7 expression plasmids (left panel). H, ChIP assay for H4K5ac level in HIF1α promoter regions. Final DNA extractions were PCR amplified using primers that cover P1 (−5837 to −5675 bp), P2 (−803 to −554 bp) and P3 (−403 to −172 bp) sites in HIF1α promoter. I, FISH for circ‐myh8 (red), and immunofluorescence for H4K5ac (green) in PASMCs, with the nuclei staining by DAPI (blue). The profiles of colocalization were also provided. Scale bar, 5 μm. J, RNA pull‐down assay followed by western blot for candidate proteins H4K5 and H4K12 in PASMCs. K, Z‐DOCK of prediction of the trimer complex structure of circ‐myh8 fragment (marked yellow cartoon), H4K5ac (marked blue cartoon), and KAT7 (marked green cartoon). A red dotted line marked the significant binding interaction. L, Chromatin isolation by RNA purification analysis for the binding of circ‐myh8 to HIF1α promoter in PASMCs. LacZ and oligo NC served as negative controls. M, ChIP assay for H4K5ac levels in HIF1α promoter promoters after KAT7 knockdown. PASMCs were transfected with circ‐myh8 or control plasmids, with KAT7 siRNA and the corresponding control. Cells were harvested for ChIP assay 24 hours later. N, RT‐qPCR for HIF1α mRNA expression in PASMCs treated as in (M). O, Western blot assay for KAT7 and HIF1α protein levels in PASMCs treated as in (M). Cells were exposured to hypoxia for 24 hours, and then harvested for western blot analysis. Data represent means±SEM from indicated independent experiments. Student's t test (for two means) or one‐way ANOVA followed by Dunnett's test (for >2 means). H4K12 indicates histone H4 lysine 12; H4K5, histone H4 lysine 5; HIF1α, hypoxia inducible factor alpha; Hyp, hypoxia; KAT, lysine acetyltransferase; LacZ, beta‐galactosidase; NC, negative control; Nor, normoxia; RT‐qPCR, reverse transcription‐quantitative polymerase chain reaction; and veh, vehicle. *P<0.05; **P<0.01, ***P<0.001.
DISCUSSION
In this study, we identified a novel circRNA, circ‐myh8, with an important biological impact in a hypoxia‐induced PH mouse model. The characteristics of circ‐myh8 were confirmed using RNase R treatment and Sanger sequencing. The expression of circ‐myh8 was significantly increased in hypoxic PASMCs compared with that in control PASMCs and was correlated with the time of exposure to hypoxic conditions. Overexpression of circ‐myh8 significantly promoted, while knockdown of circ‐myh8 significantly inhibited the proliferation and cell‐cycle progression of PASMCs and vascular remodeling in vivo and in vitro. Circ‐myh8 increases H4K5 acetylation associated with the promoter regions of HIF1α via the recruitment of KAT7, which eventually leads to the upregulation of HIF1α (Figure 7). Altogether, our study not only reveals the pivotal roles of circ‐myh8 in governing histone modification to activate HIF1α pathways, but also reveals that circ‐myh8 may be a promising diagnostic marker and therapeutic target to combat PH.
Figure 7. Schematic model of Circ‐myh8 in the regulation of HIF1α expression in PH.
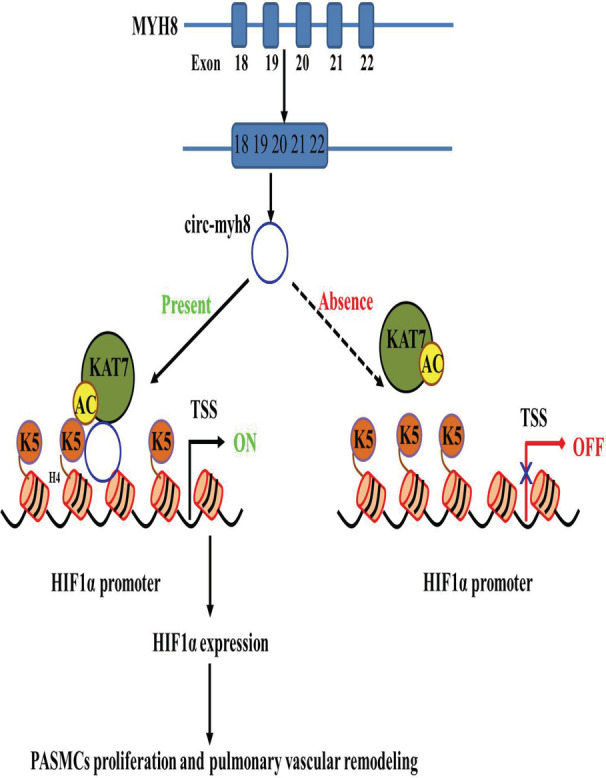
In this model, circ‐myh8 alters the histone modification pattern via recruiting KAT7 to HIF1α gene promoter regions as a modular scaffold, thereby increasing H4K5 acetylation in their promoter regions, which ultimately alters the HIF1α expression and promotes the proliferation of PASMCs and pulmonary vascular remodeling. H4K5 indicates histone H4 lysine 5; HIF1α, hypoxia‐inducible factor alpha; KAT, lysine acetyltransferase; PH, pulmonary hypertension; and TSS, transcription start site.
The expression patterns of circRNAs in lung tissues from hypoxia‐induced PH animal models have been profiled. 29 , 30 Our previous study found that circ‐calm4 serves as an miR‐337‐3p sponge to regulate myosin 10. 31 In this study, we demonstrated that circ‐myh8 biologically promotes the proliferation and cell‐cycle progression of PASMCs in vivo and in vitro. With increasing numbers of circRNAs discovered, the role of circRNAs in PH should be paid more attention in further studies.
We identified the proteins binding to circ‐myh8 and found that circ‐myh8 was a scaffold of KAT7. Moreover, we elucidated that the C2HC domain of KAT7 (336–390 aa) is crucial for its interaction with circ‐myh8. Interestingly, our study is consistent with recent research, which has revealed that circMRPS35 directly participates in H4K5 acetylation through recruited KAT7. 32 The phenomenon of KAT7 binding with 2 different circRNAs can be explained by a study that demonstrated 1 RNA binding protein can also combine with a subgroup of circRNAs and form circRNA‐protein complex families. 33 With increasing functions of circRNAs discovered, the use of bioinformatic approaches coupled with novel biochemical enrichment strategies will further provide a comprehensive understanding of circRNAs in PH.
Environmental stimuli may influence the reactive oxygen species level or DNA damage to affect the epigenetic stage or change the level of metabolites, such as nicotinamide adenine dinucleotide, acetyl‐coenzyme A, or S‐adenosylmethionine, to regulate the activity of chromatin‐modifying enzymes. 34 The 3‐dimensional architecture of chromatin is regulated by DNA methylation, histone posttranscriptional modifications, and chromatin remodelers. These chromatin modifications act in coordination to control RNA transcription. RNA products, including long noncoding RNAs (lncRNAs), microRNAs, and circRNAs regulate chromatin remodeling, gene transcription, and mRNA processing and modifications. 35
Chromatin remodeling is the dynamic modification of chromatin structure to allow access of regulatory elements to condensed genomic DNA. 36 Studies have mainly focused on the role of protein complexes in chromatin remodeling, including histone‐modifying enzymes, DNA‐modifying enzymes, and ATP‐dependent chromatin remodeling complexes. 37 Histone modifications include methylation, acetylation, phosphorylation, ubiquitylation, and sumoylation. 38 Histone methylation can be associated with either gene activation or repression, depending on the amino acid residue modified and the extent of methylation. Histone acetylation is dynamically regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). Histone acetyltransferases are categorized into at least 4 families: Gcn5/PCAF, MYST, p300/CBP, and the Rtt109 family. KAT7, also known as HBO1 and MYST2, is a member of the MYST family. 39 KAT7 prefers to acetylate H4 at lysine 5 and lysine 12 and H3 at lysine 14. Here, we show that circ‐myh8, a new scaffold partner of KAT7, specifically increases H4K5 acetylation levels in HIF‐1α gene promoter regions. Interestingly, recent research has revealed that long noncoding RNAs, such as long noncoding RNA HOTAIR and GClnc1, directly participate in histone modification and modulate target gene transcription. 40 , 41 Our study found that circRNAs directly affect the acetylation of the regulatory region of target genes and “unlock” their expression; all of these studies suggests that noncoding RNAs play a significant role in the histone modification of gene promoters.
This study revealed that increased circ‐myh8 was detected in PH mice, hypoxia‐exposed PASMCs, and plasma from patients with PH. This indicates that circ‐myh8 potentially acts as a diagnostic biomarker and therapeutic target for PH. However, there are some limitations that should be addressed in future studies. First, the expression of circ‐myh8 was increased in the lung, heart, right ventricle, and kidney but not in the carotid artery and aorta of hypoxic PH mice. Consistent with this, circ‐myh8 expression was positively associated with FOXO1 and FOXO3a expression in patients with gastric cancer. It is necessary to elucidate the disease‐specific and tissue‐specific circ‐myh8 expression pattern before acting as a diagnostic biomarker of PH. Second, because only 10 blood samples were obtained from patients with PH, we could not clarify the implication of increasing circ‐myh8 in the subtype of PAH (group 1 PH) or in other groups of patients with PH. Third, we could not exclude other mechanisms of circ‐myh8, such as acting as competitive endogenous RNAs and other protein scaffolds.
In conclusion, our study demonstrated that circ‐myh8 recruits histone acetyltransferase KAT7 as a modular scaffold, which subsequently increases the level of H4K5ac in the promoter regions of HIF1α and ultimately induces the proliferation and cell‐cycle progression of PASMCs. Taken together, our study is the first to demonstrate that circ‐myh8 and its associated pathway might be crucial targets for the diagnosis and treatment of hypoxia‐induced PH.
Sources of Funding
This work was supported by the National Natural Science Foundation of China (31771276 and 31471095 to Dr Zhu, 31701010 and 32171122 to Dr Xing, 31500936 and 32071120 to Dr Zheng); the Natural Science Foundation of Heilongjiang Province (LH2021H019 to Dr Xing); Supporting Certificate of Heilongjiang Postdoctoral Scientific Research Developmental Fund (LBH‐Q19121); the Fundamental Research Funds for the Provincial Universities (JFYWH202002); and Construction Project of Scientific Research and Innovation Team of Harbin Medical University–Daqing (HD‐CXTD‐202001). Central Finance supports Local Colleges and Universities Talent Development Funding from Finance Department of Heilongjiang Provincial (Excellent Young Talent Support Project to Dr Zheng).
Disclosures
None.
Supporting information
Tables S1–S3
Figures S1–S6
Acknowledgments
The blood samples from patients with PAH were provided by Professor Bingxiang Wu from the Second Affiliated Hospital of Harbin Medical University.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/JAHA.122.028299
For Sources of Funding and Disclosures, see page 15.
Contributor Information
Daling Zhu, Email: zhudaling@hrbmu.edu.cn.
Xiaodong Zheng, Email: zhengxiaodong@hmudq.edu.cn.
References
- 1. Qi J, Xing Y, Zhao X, Zhu D, Zheng X. Advanced diagnosis and therapy for pulmonary arterial hypertension. Nano Life. 2020;10:10. doi: 10.1142/S1793984420400036 [DOI] [Google Scholar]
- 2. Nathan SD, Barbera JA, Gaine SP, Harari S, Martinez FJ, Olschewski H, Olsson KM, Peacock AJ, Pepke‐Zaba J, Provencher S, et al. Pulmonary hypertension in chronic lung disease and hypoxia. Eur Respir J. 2019;53:53. doi: 10.1183/13993003.01914-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wilkins MR, Ghofrani HA, Weissmann N, Aldashev A, Zhao L. Pathophysiology and treatment of high‐altitude pulmonary vascular disease. Circulation. 2015;131:582–590. doi: 10.1161/CIRCULATIONAHA.114.006977 [DOI] [PubMed] [Google Scholar]
- 4. Pullamsetti SS, Mamazhakypov A, Weissmann N, Seeger W, Savai R. Hypoxia‐inducible factor signaling in pulmonary hypertension. J Clin Investig. 2020;130:5638–5651. doi: 10.1172/JCI137558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smith KA, Waypa GB, Dudley VJ, Budinger GRS, Abdala‐Valencia H, Bartom E, Schumacker PT. Role of hypoxia‐inducible factors in regulating right ventricular function and remodeling during chronic hypoxia‐induced pulmonary hypertension. Am J Respir Cell Mol Biol. 2020;63:652–664. doi: 10.1165/rcmb.2020-0023OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guignabert C, Humbert M. Targeting transforming growth factor‐beta receptors in pulmonary hypertension. Eur Respir J. 2021;57:57. doi: 10.1183/13993003.02341-2020 [DOI] [PubMed] [Google Scholar]
- 7. Semenza GL. Hypoxia‐inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yu AY, Shimoda LA, Iyer NV, Huso DL, Sun X, McWilliams R, Beaty T, Sham JS, Wiener CM, Sylvester JT, et al. Impaired physiological responses to chronic hypoxia in mice partially deficient for hypoxia‐inducible factor 1alpha. J Clin Investig. 1999;103:691–696. doi: 10.1172/JCI5912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ball MK, Waypa GB, Mungai PT, Nielsen JM, Czech L, Dudley VJ, Beussink L, Dettman RW, Berkelhamer SK, Steinhorn RH, et al. Regulation of hypoxia‐induced pulmonary hypertension by vascular smooth muscle hypoxia‐inducible factor‐1alpha. Am J Respir Crit Care Med. 2014;189:314–324. doi: 10.1164/rccm.201302-0302OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Veith C, Schermuly RT, Brandes RP, Weissmann N. Molecular mechanisms of hypoxia‐inducible factor‐induced pulmonary arterial smooth muscle cell alterations in pulmonary hypertension. J Physiol. 2016;594:1167–1177. doi: 10.1113/JP270689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 12. Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome‐wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age‐dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single‐stranded covalently closed circular RNA molecules existing as highly base‐paired rod‐like structures. Proc Natl Acad Sci USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci USA. 1996;93:6536–6541. doi: 10.1073/pnas.93.13.6536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis‐splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559 [DOI] [PubMed] [Google Scholar]
- 16. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rybak‐Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal‐Fluss R, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027 [DOI] [PubMed] [Google Scholar]
- 18. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kelly S, Greenman C, Cook PR, Papantonis A. Exon skipping is correlated with exon circularization. J Mol Biol. 2015;427:2414–2417. doi: 10.1016/j.jmb.2015.02.018 [DOI] [PubMed] [Google Scholar]
- 20. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- 21. Zhao Z, Li X, Jian D, Hao P, Rao L, Li M. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre‐diabetes and type 2 diabetes mellitus. Acta Diabetol. 2017;54:237–245. doi: 10.1007/s00592-016-0943-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang F, Fang E, Mei H, Chen Y, Li H, Li D, Song H, Wang J, Hong M, Xiao W, et al. Cis‐acting circ‐CTNNB1 promotes beta‐catenin signaling and cancer progression via DDX3‐mediated transactivation of YY1. Cancer Res. 2019;79:557–571. doi: 10.1158/0008-5472.CAN-18-1559 [DOI] [PubMed] [Google Scholar]
- 23. Zhou S, Jiang H, Li M, Wu P, Sun L, Liu Y, Zhu K, Zhang B, Sun G, Cao C, et al. Circular RNA hsa_circ_0016070 is associated with pulmonary arterial hypertension by promoting PASMC proliferation. Mol Ther Nucleic Acids. 2019;18:275–284. doi: 10.1016/j.omtn.2019.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang L, Liang H, Meng X, Shen L, Guan Z, Hei B, Yu H, Qi S, Wen X. mmu_circ_0000790 is involved in pulmonary vascular remodeling in mice with HPH via microRNA‐374c‐mediated FOXC1. Mol Ther Nucleic Acids. 2020;20:292–307. doi: 10.1016/j.omtn.2019.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19:172. doi: 10.1186/s12943-020-01286-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, Yang BB. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Misir S, Wu N, Yang BB. Specific expression and functions of circular RNAs. Cell Death Differ. 2022;29:481–491. doi: 10.1038/s41418-022-00948-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang Y, Liu H, Yu H, Zhou Y, Zhang J, Xin W, Li Y, He S, Ma C, Zheng X, et al. Circular RNA Calm4 regulates hypoxia‐induced pulmonary arterial smooth muscle cells pyroptosis via the Circ‐Calm4/miR‐124‐3p/PDCD6 axis. Arterioscler Thromb Vasc Biol. 2021;41:1675–1693. doi: 10.1161/ATVBAHA.120.315525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang J, Zhu MC, Kalionis B, Wu JZ, Wang LL, Ge HY, Chen CC, Tang XD, Song YL, He H, et al. Characteristics of circular RNA expression in lung tissues from mice with hypoxiainduced pulmonary hypertension. Int J Mol Med. 2018;42:1353–1366. doi: 10.3892/ijmm.2018.3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu SL, Deng YS, Liu J, Xu SY, Zhao FY, Wei L, Tian YC, Yu CE, Cao B, Huang XX, et al. Regulation of circular RNAs act as ceRNA in a hypoxic pulmonary hypertension rat model. Genomics. 2021;113:11–19. doi: 10.1016/j.ygeno.2020.11.021 [DOI] [PubMed] [Google Scholar]
- 31. Zhang J, Li Y, Qi J, Yu X, Ren H, Zhao X, Xin W, He S, Zheng X, Ma C, et al. Circ‐calm4 serves as an miR‐337‐3p sponge to regulate Myo10 (myosin 10) and promote pulmonary artery smooth muscle proliferation. Hypertension. 2020;75:668–679. doi: 10.1161/HYPERTENSIONAHA.119.13715 [DOI] [PubMed] [Google Scholar]
- 32. Jie M, Wu Y, Gao M, Li X, Liu C, Ouyang Q, Tang Q, Shan C, Lv Y, Zhang K, et al. CircMRPS35 suppresses gastric cancer progression via recruiting KAT7 to govern histone modification. Mol Cancer. 2020;19:56. doi: 10.1186/s12943-020-01160-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schneider T, Hung LH, Schreiner S, Starke S, Eckhof H, Rossbach O, Reich S, Medenbach J, Bindereif A. CircRNA‐protein complexes: IMP3 protein component defines subfamily of circRNPs. Sci Rep. 2016;6:31313. doi: 10.1038/srep31313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Etchegaray JP, Mostoslavsky R. Interplay between metabolism and epigenetics: a nuclear adaptation to environmental changes. Mol Cell. 2016;62:695–711. doi: 10.1016/j.molcel.2016.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang W, Song M, Qu J, Liu GH. Epigenetic modifications in cardiovascular aging and diseases. Circ Res. 2018;123:773–786. doi: 10.1161/CIRCRESAHA.118.312497 [DOI] [PubMed] [Google Scholar]
- 36. Lee HS, Park YY, Cho MY, Chae S, Yoo YS, Kwon MH, Lee CW, Cho H. The chromatin remodeller RSF1 is essential for PLK1 deposition and function at mitotic kinetochores. Nat Commun. 2015;6:7904. doi: 10.1038/ncomms8904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mashtalir N, D'Avino AR, Michel BC, Luo J, Pan J, Otto JE, Zullow HJ, McKenzie ZM, Kubiak RL, St Pierre R, et al. Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell. 2018;175:1272–1288.e20. doi: 10.1016/j.cell.2018.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zarzour A, Kim HW, Weintraub NL. Epigenetic regulation of vascular diseases. Arterioscler Thromb Vasc Biol. 2019;39:984–990. doi: 10.1161/ATVBAHA.119.312193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miotto B, Struhl K. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by geminin. Mol Cell. 2010;37:57–66. doi: 10.1016/j.molcel.2009.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu TC, Tang JY, Bao YJ, Hu Y, Lin Y, et al. LncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov. 2016;6:784–801. doi: 10.1158/2159-8290.CD-15-0921 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1–S3
Figures S1–S6


