Abstract
Background
Statins are guideline‐recommended medications that reduce cardiovascular events in patients with diabetes. Yet, statin use is concerningly low in this high‐risk population. Identifying reasons for statin nonuse, which are typically described in unstructured electronic health record data, can inform targeted system interventions to improve statin use. We aimed to leverage a deep learning approach to identify reasons for statin nonuse in patients with diabetes.
Methods and Results
Adults with diabetes and no statin prescriptions were identified from a multiethnic, multisite Northern California electronic health record cohort from 2014 to 2020. We used a benchmark deep learning natural language processing approach (Clinical Bidirectional Encoder Representations from Transformers) to identify statin nonuse and reasons for statin nonuse from unstructured electronic health record data. Performance was evaluated against expert clinician review from manual annotation of clinical notes and compared with other natural language processing approaches. Of 33 461 patients with diabetes (mean age 59±15 years, 49% women, 36% White patients, 24% Asian patients, and 15% Hispanic patients), 47% (15 580) had no statin prescriptions. From unstructured data, Clinical Bidirectional Encoder Representations from Transformers accurately identified statin nonuse (area under receiver operating characteristic curve [AUC] 0.99 [0.98–1.0]) and key patient (eg, side effects/contraindications), clinician (eg, guideline‐discordant practice), and system reasons (eg, clinical inertia) for statin nonuse (AUC 0.90 [0.86–0.93]) and outperformed other natural language processing approaches. Reasons for nonuse varied by clinical and demographic characteristics, including race and ethnicity.
Conclusions
A deep learning algorithm identified statin nonuse and actionable reasons for statin nonuse in patients with diabetes. Findings may enable targeted interventions to improve guideline‐directed statin use and be scaled to other evidence‐based therapies.
Keywords: artificial intelligence, cardiovascular disease, diabetes, electronic health records, medication adherence, natural language processing, statins
Subject Categories: Cardiovascular Disease; Diabetes, Type 1; Diabetes, Type 2; Primary Prevention; Compliance/Adherence
Nonstandard Abbreviations and Acronyms
- Clinical BERT
Clinical Bidirectional Encoder Representations from Transformers
Clinical Perspective.
What Is New?
Nearly 1 in 2 patients with diabetes in our cohort lacked guideline‐directed statin prescriptions, with gaps observed in younger, female, and Black individuals.
A novel deep learning approach accurately identified statin nonuse and classified reasons for statin nonuse from unstructured electronic health record data, including patient reasons (side effects/contraindications and statin hesitancy), clinician reasons (guideline‐discordant practice), and system reasons (clinical inertia) for statin nonuse, which further varied by patient race and ethnicity.
What Are the Clinical Implications?
By better understanding the real‐world patient, clinician, and system barriers to guideline‐directed statin use across a health system, we can design tailored interventions to address modifiable barriers to improve statin use and atherosclerotic cardiovascular disease treatment.
Patients with diabetes are at high risk for major adverse cardiovascular events including premature cardiovascular mortality. 1 , 2 , 3 , 4 Statins significantly reduce these cardiovascular events and are therefore recommended for patients with diabetes across major practice guidelines. 5 , 6 , 7 , 8 However, despite compelling evidence of benefit and safety, guideline‐directed statin use remains concerningly low in practice. In 2015 to 2018, approximately half of a nationally representative cohort of individuals with diabetes in the United States were not on statins. 9 Statin nonuse is more common in underrepresented populations including women and minority groups, can contribute to excess cardiovascular events, and represents an important public health gap for cardiovascular disease prevention. 10 , 11 , 12 , 13 , 14 , 15
Identifying reasons for statin nonuse can guide targeted interventions to improve guideline‐directed statin use. Such reasons may include patient, clinician, and system factors, may drive practice variation around statin use in diabetes, and are usually documented in unstructured, narrative portions of the electronic health record (EHR). 13 , 16 , 17 To understand statin nonuse in diabetes across a health system, it is therefore necessary to analyze unstructured EHR data at scale. In patients with diabetes, prior work has shown suboptimal real‐world documentation regarding statin use, so innovative approaches are needed to overcome this limitation and accurately understand statin nonuse from unstructured EHR data. 17 Artificial intelligence approaches to natural language processing (NLP) may help analyze such large‐scale unstructured data. We previously demonstrated that a deep learning NLP approach accurately identified statin nonuse and reasons for statin nonuse from complex, unstructured EHR data of patients with atherosclerotic cardiovascular disease in a Northern California health system. 16
In this study, we aimed to develop and validate a deep learning NLP approach using Clinical Bidirectional Encoder Representations from Transformers (Clinical BERT) to identify statin nonuse and actionable reasons for statin nonuse from unstructured EHRs of individuals with diabetes. 16 , 18 The goal of this approach was to develop an artificial intelligence–based pipeline to characterize statin nonuse across a health system using comprehensive EHR data and ultimately guide targeted interventions to improve guideline‐directed statin use.
RESEARCH DESIGN AND METHODS
The data sets analyzed during the current study are not publicly available because of reasonable privacy and security concerns. The underlying EHR data are not easily redistributable to researchers other than those engaged in the Institutional Review Board‐approved research collaborations in the current project. The corresponding author may be contacted for access to EHR data for an Institutional Review Board‐approved collaboration.
Design and Study Cohort
This study was approved by the Stanford University Institutional Review Board. Informed consent was waived under Exemption 4 (research on existing data). For this retrospective cohort study, we identified patients at Stanford Health Care Alliance, a large, multisite health system in Northern California. Stanford Health Care Alliance consists of an academic hospital, a community hospital, and a multisite community practice network. The cohort included patients who were diagnosed with type 1 or type 2 diabetes between January 1, 2014 and December 31, 2020 in ambulatory settings in Primary Care, Cardiology, and Endocrinology clinics at Stanford Health Care Alliance. The first diabetes diagnosis was considered the index diagnosis. Patients were between ages 20 and 89 years at time of diagnosis and did not have atherosclerotic cardiovascular disease (ASCVD). International Classification of Diseases Ninth and Tenth Editions were used to identify diabetes and ASCVD.
Statin prescriptions were identified using RxNorm codes (Table S1). Patients were classified according to the presence or absence of statin prescriptions documented in their structured EHR medication data. Prescriptions at diagnosis or a new prescription within 1 month after index diagnosis were included. In the case of a prescription that had no recorded end date, the prescription was considered active if it had started within 6 months before diagnosis. Patients who lacked statin prescriptions were classified according to whether they had documented statin allergies in structured EHR fields. Patients who lacked allergies were then assessed for any mention of statin terms (Table S2) in unstructured notes dated up to 30 days after diagnosis. A 30‐day period was chosen because encounter documentation is expected to be completed within 14 days in this health system, so we allowed additional time, up to 1 month, after a diabetes diagnosis for providers to document statin use or nonuse. Patients who had any statin terms in unstructured notes were included in the deep learning NLP data set used for the development of NLP models to identify statin nonuse and reasons for statin nonuse.
Demographic and clinical characteristics at the time of index diagnosis were abstracted from the EHR including age, sex, race and ethnicity, medical history, laboratory studies, diagnosing encounter specialty, number of hospitalizations in the year before diagnosis, and patient insurance payor type (private, Medicare, or Medicaid). Laboratory studies including total cholesterol, low‐density lipoprotein cholesterol, and creatinine kinase or creatinine phosphokinase, were obtained within 6 months of initial diagnosis, and the value closest to the diagnosis date was used. Statin prescriptions were categorized based on intensity as high, moderate, or low, as previously described. 19
Outcomes
The primary outcome was statin nonuse. The secondary outcome was the reason for statin nonuse according to categories derived from manual annotation.
NLP Model Development
From the deep learning NLP data set as described above, all sentences that contained statin terms were concatenated in 1 document per patient for model training and evaluation. We used Clinical BERT, a benchmark deep learning approach consisting of models pretrained on a large set of clinical notes that can be fine‐tuned to a text classification task in a process called transfer learning. 18 The Clinical BERT model was pretrained on notes from MIMIC III, a database containing EHRs from intensive care unit patients in Boston, MA. 20
Manual Annotation
We created a ground‐truth data set for model training and evaluation through manual annotation of a randomly selected sample of patients (N=1000). Four authors (AS, CW, SN, FR) manually annotated this data set to determine documentation of active statin use versus nonuse and annotate reasons for statin nonuse. Overlapping review was performed in a set of 203 patients. For overlapping review, reviewers were paired into 2 groups of 2 reviewers, and 103 patients' documents were annotated by the first group, and 100 documents were annotated by the second group. Discrepancies were resolved by further clinician review (AS or FR).
Five categories for reasons for statin nonuse were finalized after manual annotation: statin‐associated side effects/contraindication, guideline‐discordant clinician practice, clinical inertia, statin hesitancy, and nonspecific reasons. In addition to side effects attributed to prior statin use, if statins were avoided based on pre‐existing comorbidities (such as liver disease) or perceived contraindications (including pregnancy) without a trial or challenge, these reasons were categorized within the side effects/contraindication category. This was done because of low individual frequencies for some of these reasons and to comply with privacy regulations that prevent the reporting of groups with fewer than 10 patients. Clinicians avoiding statin use based on lipid levels was considered guideline‐discordant practice, given that statin indications for diabetes are primarily independent of lipid levels. Deferral of statin decisions to future visits despite their indications was considered clinical inertia. Statin hesitancy was noted when patients expressed a preference to avoid statins despite discussion of their indication. Nonspecific documentation was recorded when statin nonuse was documented without additional explanation.
NLP to Identify Statin Nonuse
We first developed a Clinical BERT model for binary classification between patients with and without documented statin use from unstructured EHR documentation. 16
NLP to Identify Reasons for Statin Nonuse
We then developed a multiclass Clinical BERT model to determine reasons for statin nonuse using our previously developed framework. 16 For this, we fine‐tuned the Clinical BERT pretrained model to perform multiclass classification with our data set to identify the reason for statin nonuse according to 1 of the 5 categories as previously described.
NLP Training and Evaluation
For training and evaluation, we split the manually annotated cohort into an 80% training set and 20% test set. We used the “transformers” library (Python software, version 3.7) to the train the BERT models, and its built‐in “train” methods. We opted for the Adam gradient descent algorithm (with epsilon=1e‐6) on a cross‐entropy loss to quantify the misclassification error. We used 10‐fold cross‐validation to validate the model and tune the hyperparameters (learning rate, number of epochs, batch size, strength of weight decay, and Adam's epsilon value). To evaluate model performance, we assessed precision, recall, area under curve (AUC), score, and F1 score. Precision (or positive predictive value) measures the fraction of correct positive predictions divided by all documents predicted as positive. Recall (or true positive rate) measures the fraction of correct positive predictions divided by the number of results that should have been predicted as positive. AUC score corresponds to the probability that a classifier will rank a positive document higher than a negative one. Finally, F1 score is defined as the harmonic mean of precision and recall. For the multiclass classifiers for the reasons for statin nonuse, we used a one‐versus‐rest strategy and reported the weighted macro‐averages of these metrics based on their proportions in the evaluation set. For example, if we have k possible prediction classes (that is, reasons for statin nonuse) for the one‐versus‐rest strategy, we compute the metric of interest a total of k times as if in a binary case each time for each class (that is, each time, labeling the class of interest as 1 and all other classes as 0, computing metrics, and repeating this process for each class). After obtaining all k metrics, these are aggregated in a weighted manner based on the proportion of each class in the evaluation set. After completing training and evaluation, the binary NLP model and highest performing multiclass NLP model were applied across the deep learning NLP data set.
Alternative NLP Approaches
We developed several contemporary alternative NLP methods for comparison with Clinical BERT following the same pipeline as above, namely: Convolutional Neural Network on word2vec‐like embeddings (word2vec+CNN, a context‐free model), base BERT (a pretrained model), and bioBERT (a pretrained model). 21 , 22 The word2vec+CNN model is a nonpretrained NLP model. Base BERT is an NLP model that is pretrained on Wikipedia and BookCorpus, and thus has broad exposure to human language. BioBERT is a model that is initialized with base BERT weights and then re‐trained on PubMed abstracts, and thus has exposure to broad scientific terminology.
Statistical Analysis
For comparisons of baseline characteristics, we performed an unpaired t test for numeric data and χ2/Fisher exact tests for categorical variables. We used Cohen's kappa coefficients to assess concordance between reviewers. All statistical tests were 2‐sided with the threshold of P≤0.05 for statistical significance. We calculated 95% CIs for each metric of NLP performance (Precision, Recall, F1 score, and AUC) by performing a bootstrap resampling of 10 000 iterations to compute each metric and chose the interval between the 2.5% and 97.5% percentiles of these analyses. We compared unadjusted and adjusted odds ratios (ORs) of receiving high‐intensity statin prescriptions (versus other statin intensities) and of receiving any statin prescriptions (versus no prescriptions) through multivariable logistic regression. Analyses were performed using Python software, version 3.7, including with transformers and scikit‐learn packages. 16
RESULTS
Study Cohort
There were 33 461 patients with diabetes in the study cohort (Table 1, Figure 1). Patients were on average 59±15 years old, and 49% were women, 36% were Non‐Hispanic White, 8% were non‐Hispanic Black (Black), 15% were Hispanic, and 24% were non‐Hispanic Asian (Asian).
Table 1.
Baseline Characteristics by the Presence or Absence of a Statin Prescription
| Characteristic at index date | Total (N=33 461) | Statin prescription absent (N=15 880) | Statin prescription present (N=17 581) | P value |
|---|---|---|---|---|
| Age, y, mean (SD) | 59.0 (14.9) | 54.6 (16.0) | 62.9 (12.5) | <0.001 |
| Female | 16 540 (49.4) | 8279 (52.1) | 8261 (47.0) | <0.001 |
| Race or ethnicity | ||||
| Non‐Hispanic White | 12 127 (36.2) | 5536 (34.9) | 6591 (37.5) | <0.001 |
| Non‐Hispanic Black | 2751 (8.2) | 1365 (8.6) | 1386 (7.9) | |
| Hispanic | 5027 (15.0) | 2711 (17.1) | 2316 (13.2) | |
| Non‐Hispanic Asian | 8067 (24.1) | 3528 (22.2) | 4539 (25.8) | |
| Other | 4026 (12.0) | 2017 (12.7) | 2009 (11.4) | |
| Diabetes type | ||||
| Type I | 2434 (7.3) | 1740 (11.0) | 694 (3.9) | <0.001 |
| Type II | 31 027 (92.7) | 14 140 (89.0) | 16 887 (96.1) | |
| Body mass index, kg/m2, mean (SD) | 31.0 (7.4) | 31.0 (7.8) | 31.0 (7.0) | 0.341 |
| Current smoking | 1991 (6.0) | 907 (5.7) | 1084 (6.2) | 0.084 |
| Hemoglobin A1c, %, mean (SD) | 7.6 (2.0) | 7.7 (2.0) | 7.6 (2.0) | <0.001 |
| Total cholesterol, mg/dL, mean (SD) | 179.9 (44.7) | 188.8 (41.2) | 173.6 (46.0) | <0.001 |
| LDL‐cholesterol, mg/dL, mean (SD) | 101.7 (36.3) | 110.9 (33.3) | 95.2 (37.0) | <0.001 |
| HDL‐cholesterol, mg/dL, mean (SD) | 49.3 (15.4) | 49.6 (16.2) | 49.0 (14.8) | 0.027 |
| Creatine kinase level, U/L, mean (SD) | 133.1 (143.2) | 132.5 (152.5) | 133.4 (138.5) | 0.937 |
| Ezetimibe use | 447 (1.3) | 126 (0.8) | 321 (1.8) | <0.001 |
| 2‐y Charleson Comorbidity Index score, mean, (SD) | 2.0 (1.8) | 1.9 (1.8) | 2.1 (1.8) | <0.001 |
| Liver disease | 2630 (7.9) | 1447 (9.1) | 1183 (6.7) | <0.001 |
| Hospitalizations in prior 1 y, N (%) | 1302 (3.9) | 718 (4.5) | 584 (3.3) | <0.001 |
| Provider location | ||||
| SHC, academic center | 10 084 (30.1) | 5182 (32.6) | 4902 (27.9) | <0.001 |
| UHA, community network | 22 461 (67.1) | 10 032 (63.2) | 12 429 (70.7) | |
| ValleyCare, community hospital | 779 (2.3) | 604 (3.8) | 175 (1.0) | |
| Insurance status | ||||
| Private | 12 345 (36.9) | 6677 (42.0) | 5668 (32.2) | <0.001 |
| Medicare | 10 561 (31.6) | 3965 (25.0) | 6596 (37.5) | |
| Medicaid | 1683 (5.0) | 869 (5.5) | 814 (4.6) | |
| Other | 5636 (16.8) | 2817 (17.7) | 2819 (16.0) | |
| Encounter specialty issuing the diabetes diagnosis | ||||
| Cardiology | 3769 (11.3) | 1387 (8.7) | 2382 (13.5) | <0.001 |
| Endocrinology | 5526 (16.5) | 2847 (17.9) | 2679 (15.2) | |
| Primary care | 24 166 (72.2) | 11 646 (73.3) | 12 520 (71.2) | |
Data are presented as n (%) unless otherwise noted. HDL indicates high‐density lipoprotein; LDL, low‐density lipoprotein; SHC, Stanford Health Care (academic hospital); UHA, University Health Alliance (community practice network); and ValleyCare, ValleyCare Hospital (community hospital).
Figure 1. Study cohort.
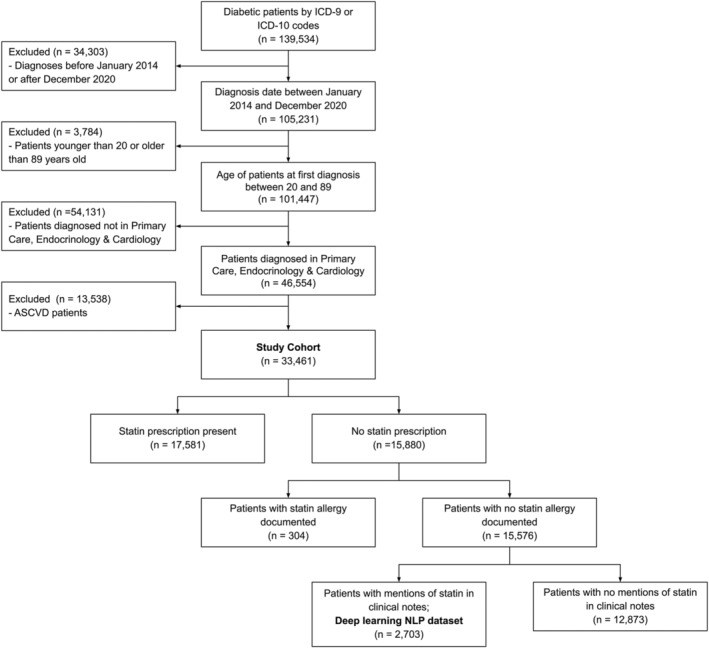
ASCVD indicates atherosclerotic cardiovascular disease; ICD‐9, International Classification of Diseases, Ninth Revision; ICD‐10, International Classification of Diseases, Tenth Revision; NLP, natural language processing; and SHA, Stanford Health Care Alliance (consisting of an academic hospital, a community hospital, and a community practice clinic network).
Within the cohort, a total of 15 880 (47%) individuals did not have statin prescriptions of any intensity in their structured EHRs. Only 304 (2%) of these individuals had statin allergies listed in structured EHRs (Figure 1). Individuals lacking statin prescriptions were more likely to be younger, female, Black, and have higher baseline low‐density lipoprotein cholesterol and were less likely to have been seen in a cardiology practice compared with those with statin prescriptions (Table 1, Table S3). Among those who received statin prescriptions, patients who were under 40 years of age, female, of Asian race, or had type 1 diabetes were less likely to receive high‐intensity statins versus low‐ or moderate‐intensity statins (Table S4).
Of 15 576 individuals without statin prescriptions or structured statin allergies, 12 873 (83%) had no mention of any statin terms in unstructured clinical notes. The remaining 2703 patients formed the deep learning NLP data set for model training and validation from unstructured data (Figure 2).
Figure 2. Development, validation, and application of a deep learning NLP approach (Clinical BERT) to identify statin nonuse and reasons for statin nonuse from unstructured data of individuals with diabetes.
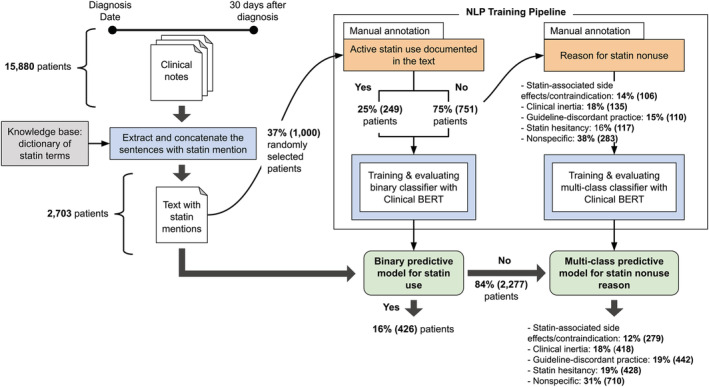
BERT indicates Bidirectional Encoder Representations from Transformers; and NLP, natural language processing.
Manual Annotation
Within the manually annotated subset of the deep learning NLP data set (Figure 2), 249 (25%) patients were statin users per unstructured data despite no structured prescription data. Kappa coefficients for overlapping manual review of patients was 0.86 for the 103 documents annotated by the first group of 2 reviewers and 0.76 for the 100 documents annotated by the second group of reviewers, indicating good concordance.
NLP Evaluation
In the held‐out test set, the binary Clinical BERT model classified statin nonuse with an overall AUC of 0.99 (95% CI, 0.98–1.00; Table 2). Among statin nonusers, the multiclassification Clinical BERT model classified reasons for statin nonuse with an overall weighted average AUC of 0.90 (95% CI, 0.85–0.93).
Table 2.
Performance of Deep Learning NLP Models to Characterize Statin Nonuse From Unstructured Clinical Notes
| Task | Dataset | Model | Precision | Recall | F1‐score | AUC |
|---|---|---|---|---|---|---|
| Binary classification of statin nonuse |
Training: N=800 documents Test: N=200 documents |
word2vec+CNN | Unable to do better than a constant classifier (labels everything as the majority class of the training set) | |||
| BaseBERT | 0.92 (0.85–0.98) | 0.90 (0.82–0.97) | 0.91 (0.85–0.96) | 0.96 (0.93–1.00) | ||
| BioBERT | 0.87 (0.77–0.95) | 0.90 (0.85–0.93) | 0.88 (0.82–0.94) | 0.98 (0.96–1.00) | ||
| ClinicalBERT | 0.92 (0.85–0.99) | 0.92 (0.86–0.98) | 0.92 (0.87–0.96) | 0.99 (0.98–1.00) | ||
| Multilabel classification of reasons for statin nonuse |
Training: N=600 documents Test: N=151 documents |
word2vec+CNN | 0.14 (0.11–0.19) | 0.38 (0.15–0.44) | 0.21 (0.17–0.27) | 0.45 (0.40–0.52) |
| BaseBERT | 0.59 (0.51–0.66) | 0.60 (0.50–0.69) | 0.59 (0.52–0.66) | 0.83 (0.79–0.87) | ||
| BioBERT | 0.66 (0.60–0.73) | 0.66 (0.59–0.73) | 0.66 (0.59–0.72) | 0.87 (0.83–0.91) | ||
| ClinicalBERT | 0.68 (0.62–0.77) | 0.68 (0.61–0.76) | 0.68 (0.61–0.76) | 0.90 (0.85–0.93) | ||
AUC indicates area under the curve; BERT, Bidirectional Encoder Representations from Transformers; and NLP, natural language processing.
NLP Model Application
After NLP model development and evaluation, the binary statin nonuse model and the multiclass model for reasons for statin nonuse were applied to the full deep learning NLP data set (Figure 2). Clinical BERT found that 426 (16%) were statin users based on unstructured data despite no documented statin prescriptions. Among statin nonusers, Clinical BERT identified the reasons for statin nonuse (Figure 2). Statin hesitancy (19%), guideline‐discordant practice (19%), and clinical inertia (18%) were more common than statin‐associated side effects/contraindications (12%).
Reasons for statin nonuse also varied by patient characteristics including age, race and ethnicity, insurance, and diabetes type (Table 3). In comparison with younger individuals, older individuals (>75 years of age) were more likely to experience statin‐associated side effects/contraindications (23.4%) as compared with statin hesitancy, clinical inertia, or guideline‐discordant practice (P<0.05 for comparisons). Hispanic individuals were most likely to experience guideline‐discordant practice (24.7%, P<0.05 for comparisons) versus the other reasons (except for nonspecific documentation), while Black patients were most likely to experience clinical inertia (24.0%, P<0.05 for comparisons). Individuals on Medicaid were most likely to experience guideline‐discordant practice as compared with the other reasons (P<0.05). Those with type 1 diabetes were most likely to experience guideline‐discordant practice (22.0%, P<0.05) as compared with the other reasons (except nonspecific documentation). The prevalence of statin‐associated side effects/contraindications was higher in those with type 2 diabetes (12.6%) as compared with those with type 1 diabetes (8.6%). Excerpts from notes demonstrating the various reasons for statin nonuse are outlined in Table 4.
Table 3.
Reasons for Statin Nonuse, Stratified by Patient Characteristics, in the Deep Learning NLP Cohort
| Category | All | Statin nonusers per NLP | Statin nonusers stratified by reason for nonuse (N, % of statin nonusers) | P value | ||||
|---|---|---|---|---|---|---|---|---|
| Statin‐associated side effects, contraindications | Statin hesitancy | Clinical inertia | Guideline‐discordant practice | Nonspecific | ||||
| Total | 2703 | 2277 | 279 (12.2) | 428 (18.8) | 418 (18.4) | 442 (19.4) | 710 (31.1) | |
| Age, y | ||||||||
| 20–39 | 376 | 357 | 28 (7.8) | 38 (10.6) | 63 (17.6) | 93 (26.1) | 135 (37.8) | <0.001 |
| 40–75 | 2077 | 1753 | 212 (12.1) | 362 (20.7) | 338 (19.3) | 325 (18.5) | 516 (29.4) | |
| >75 | 250 | 167 | 39 (23.4) | 28 (16.8) | 17 (10.2) | 24 (14.4) | 59 (35.3) | |
| Sex | ||||||||
| Female | 1347 | 1140 | 163 (14.3) | 219 (19.2) | 192 (16.8) | 200 (17.5) | 366 (32.1) | <0.007 |
| Male | 1356 | 1137 | 116 (10.2) | 209 (18.4) | 226 (19.9) | 242 (21.3) | 344 (30.3) | |
| Race and ethnicity | ||||||||
| Non‐Hispanic White | 899 | 769 | 121 (15.7) | 173 (22.5) | 134 (17.4) | 127 (16.5) | 214 (27.8) | <0.001 |
| Non‐Hispanic Black | 218 | 192 | 17 (8.9) | 33 (17.2) | 46 (24.0) | 31 (16.1) | 65 (33.9) | |
| Hispanic | 430 | 352 | 48 (13.6) | 42 (11.9) | 69 (19.6) | 87 (24.7) | 106 (30.1) | |
| Non‐Hispanic Asian | 670 | 567 | 56 (9.9) | 110 (19.4) | 109 (19.2) | 103 (18.2) | 189 (33.3) | |
| Insurance provider | ||||||||
| Private | 1083 | 963 | 104 (10.8) | 193 (20.0) | 204 (21.2) | 161 (16.7) | 301 (31.3) | <0.001 |
| Medicare | 746 | 568 | 99 (17.4) | 119 (20.9) | 83 (14.6) | 91 (16.0) | 176 (31.0) | |
| Medicaid | 193 | 139 | 10 (7.2) | 16 (11.5) | 15 (10.8) | 64 (46.0) | 34 (24.5) | |
| Other | 402 | 360 | 35 (9.7) | 55 (15.3) | 69 (19.2) | 81 (22.5) | 120 (33.3) | |
| Diabetes type | ||||||||
| Type 1 | 202 | 186 | 16 (8.6) | 28 (15.0) | 27 (14.5) | 41 (22.0) | 74 (39.8) | 0.001 |
| Type 2 | 2501 | 2091 | 263 (12.6) | 400 (19.1) | 391 (18.7) | 401 (19.2) | 636 (30.4) | |
NLP indicates natural language processing.
Table 4.
Excerpts from Unstructured Data Demonstrating Documentation of Reasons for Statin Nonuse
| Category | Excerpt(s) |
|---|---|
| Statin‐associated side effects/contraindications |
“…diffuse myalgias developed…” “…statins are contraindicated in pregnancy” |
| Statin hesitancy |
“…patient is deferring” “…continues to express hesitancy…” |
| Clinical inertia | “…plan on discussing…statin at a future visit” |
| Guideline‐discordant practice | “LDL is borderline… hold off on a statin” |
| Nonspecific | “Statin: no” |
DISCUSSION
In a multiethnic, multisite EHR cohort of patients with diabetes, nearly half lacked guideline‐directed statin prescriptions. A deep learning NLP approach leveraged unstructured EHR data to accurately identify statin nonuse and potentially actionable reasons for statin nonuse, which spanned patient‐level (side effects/contraindications, statin hesitancy), clinician‐level (guideline‐discordant practice), and system‐level (clinical inertia) factors, including differences by race and ethnicity.
Although statins conclusively reduce cardiovascular events in diabetes, real‐world statin utilization remains poor despite guideline recommendations, representing an important and well‐recognized target for population interventions. 9 Prior work from the American College of Cardiology's PINNACLE (Practice Innovation and Clinical Excellence) registry with >200 000 patients with diabetes showed significant practice variation regarding statin use in patients with diabetes along with poor documentation of statin prescriptions. 17 Our deep learning NLP approach to understand statin nonuse may help address these crucial public health gaps and inform strategies to bridge them through several implications.
First, our findings clearly demonstrated gaps in structured statin prescriptions in patients with diabetes. Nearly half of our cohort with diabetes lacked guideline‐directed statin prescriptions, with disparities observed in younger, female, and Black individuals. These results add to prior literature demonstrating statin prescription gaps and disparities in diabetes 9 and emphasize the need to understand reasons for nonuse to ultimately improve guideline adherence in a disease that represents a major public health burden.
Second, research and quality improvement efforts to understand medication utilization in a health system, including for diabetes, often rely on structured EHR data. The present findings add to our prior work underscoring the importance of analyzing unstructured EHR data to mitigate inaccurate structured medication information in studies of diabetes. 16 Of patients without structured prescriptions, our deep learning NLP approach found that 16% were statin users who would have otherwise been misclassified as nonusers based on structured data alone. Multimodal analytic approaches that use comprehensive EHR data may thus improve ways to track, monitor, and improve diabetes medication adherence within health systems.
Third, our deep learning NLP approach analyzed large volumes of unstructured notes to identify reasons for statin nonuse in diabetes that are potentially actionable through evidence‐based clinical decision support tools, clinician and patient education, and systems‐level interventions. Statin‐associated side effects, for instance, may include nocebo effects that can respond favorably to re‐challenge protocols that could be nudged. 23 Guideline‐discordant practice patterns, such as deferring statin use based on perceived low‐density lipoprotein control, may be responsive to clinician education or point‐of‐care decision support, and clinical inertia may benefit from systemwide interventions to encourage timely statin prescriptions. 24 Addressing such clinician and system‐level reasons may help address previously observed real‐world variation in statin use in patients with diabetes. 17
Our prior work assessed reasons for statin nonuse in patients with ASCVD. 16 In the present study, we observed that side effects accounted for a lower percentage of the reasons for nonuse (12% with diabetes versus 33% with ASCVD previously). 16 This may be related to a younger population with diabetes (especially with type 1 diabetes) and fewer comorbidities as suggested by a lower proportion of participants with hospitalization in the preceding year. In addition, clinical inertia accounted for ≈18% of the documented reasons for nonuse in the present study but was not prevalent enough to be a separate category in prior work with patients with ASCVD. This suggests that systems interventions to encourage timely statin prescription and overcome clinical inertia may be particularly important in those with diabetes. Additionally, pregnancy was cited as a reason for statin nonuse in the present study (included within the side effects/contraindications group) and may contribute to the observed sex‐based disparities in guideline‐directed statin use. This was not seen in prior work with patients with ASCVD, potentially because cohorts with diabetes are more likely to include younger patients. 16 Identifying such cases may be of high contemporary interest given recent changes in statin labeling by the US Food and Drug Administration and the resulting discussions around whether statins remain absolutely or relatively contraindicated in pregnant individuals. 25 NLP‐guided approaches thus offer the ability to capture high‐volume clinical data to identify real‐world gaps in the use of lifesaving therapies across different high‐risk cohorts including those with diabetes. NLP‐guided interventions should be studied prospectively to assess effectiveness to improve statin utilization.
Fourth, there are well‐established disparities in statin utilization, including in diabetes, by key factors such as race and ethnicity, which may link with disparities in adverse cardiovascular events. 10 , 11 , 12 , 13 , 14 , 15 Our approach may help delineate the underlying differences in reasons for statin nonuse that could contribute to such disparities. For instance, based on their relative representation across the various reasons, we observed that Hispanic patients experienced guideline‐discordance practice more frequently versus the other reasons, pointing to a potentially actionable clinician‐level issue in this data set. Such findings may inform targeted interventions to address disparities in statin utilization in diabetes by tackling structural biases that may explain these findings.
Clinical BERT generally demonstrated better performance in comparison with other NLP models. This may be related to Clinical BERT being pretrained on a data set of clinical notes (MIMIC III) before additional re‐training in our study. The word2vec‐CNN model is a nonpretrained model and demonstrated poorer performance. Base BERT is pretrained broadly on human language from Wikipedia and BookCorpus rather than clinical notes specifically. BioBERT is a model that was initialized with base BERT weights and re‐trained on PubMed abstracts, and thus has broad exposure to scientific terminology and performed nearly as well as Clinical BERT. Clinical BERT was initialized with base BERT weights and pretrained on clinical notes, and it may therefore be expected to perform better than the above NLP approaches. For the multiclass task of Clinical BERT to predict reasons for statin nonuse, we chose a one‐versus‐rest strategy because the various classes (reasons for statin nonuse) were relatively well‐balanced in our data set, and accounting for class proportions to obtain a weighted average for each metric may help account for any slight imbalances.
Our study has certain limitations. Our Northern California–based cohort was multisite and diverse but may not be generalizable across the United States, given that it included primarily insured patients. However, our Clinical BERT models were pretrained on external notes from a different health system in Boston, Massachusetts, which we fine‐tuned at our health system in California with favorable performance, thus indicating a potentially generalizable pipeline across systems and populations. Care fragmentation may result in missing outside medication data in single health‐system analyses, but our EHR system reconciles medications prescribed outside of our health system and medications that are self‐reported by patients, thus potentially mitigating this limitation. We were unable to disaggregate race and ethnicity groups further or include certain socioeconomic information such as household income because of data limitations. Because of low overall documentation, we observed small group sizes when separating reasons for statin nonuse by characteristics such race and ethnicity as in Table 3, so these findings should be considered hypothesis‐generating. Muscle and nonmuscle side effects were collapsed into a single category based on their prevalence. These sizes may increase as documentation practices are standardized.
CONCLUSION
In conclusion, in a multiethnic cohort of individuals with diabetes, nearly half lacked guideline‐directed statin prescriptions. A deep learning NLP approach identified statin nonuse and potentially actionable reasons for statin nonuse including key patient, clinician, and system factors from unstructured EHRs. Findings may help inform targeted interventions to improve real‐world statin utilization in high‐risk patients.
Sources of Funding
Dr Ashish Sarraju received support from the American Heart Association (Grant 20SFRN35360178). Dr Fatima Rodriguez received support from the National Heart, Lung, and Blood Institute, National Institutes of Health (1K01HL144607), the American Heart Association/Robert Wood Johnson Harold Amos Medical Faculty Development Program, and Grant #2022051 from the Doris Duke Charitable Foundation. Dr Hernandez‐Boussard was supported by the National Library Of Medicine of the National Institutes of Health under Award Number R01LM013362. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The study sponsors/funders were not involved in the design of the study; the collection, analysis, and interpretation of data; or writing the report; and did not impose any restrictions regarding the publication of the report.
Disclosures
F. Rodriguez has received consulting fees from Novartis, Novo Nordisk, and HealthPals. T. Hernandez‐Boussard has received consulting fees from Verantos and Institute for Systems Biology. The remaining authors have no competing interests to declare.
Supporting information
Tables S1–S4
Acknowledgments
None.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/JAHA.122.028120
For Sources of Funding and Disclosures, see page 10.
References
- 1. Baena‐Díez JM, Peñafiel J, Subirana I, Ramos R, Elosua R, Marín‐Ibañez A, Guembe MJ, Rigo F, Tormo‐Díaz MJ, Moreno‐Iribas C, et al. Risk of cause‐specific death in individuals with diabetes: a competing risks analysis. Diabetes Care. 2016;39:1987–1995. doi: 10.2337/dc16-0614 [DOI] [PubMed] [Google Scholar]
- 2. Raghavan S, Vassy JL, Ho YL, Song RJ, Gagnon DR, Cho K, Wilson PWF, Phillips LS. Diabetes mellitus‐related all‐cause and cardiovascular mortality in a national cohort of adults. J Am Heart Assoc. 2019;8:e011295. doi: 10.1161/JAHA.118.011295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mulnier HE, Seaman HE, Raleigh VS, Soedamah‐Muthu SS, Colhoun HM, Lawrenson RA, de Vries CS. Risk of myocardial infarction in men and women with type 2 diabetes in the UK: a cohort study using the General Practice Research Database. Diabetologia. 2008;51:1639–1645. doi: 10.1007/s00125-008-1076-y [DOI] [PubMed] [Google Scholar]
- 4. Soedamah‐Muthu SS, Fuller JH, Mulnier HE, Raleigh VS, Lawrenson RA, Colhoun HM. High risk of cardiovascular disease in patients with type 1 diabetes in the U.K.: a cohort study using the General practice research database. Diabetes Care. 2006;29:798–804. doi: 10.2337/diacare.29.04.06.dc05-1433 [DOI] [PubMed] [Google Scholar]
- 5. Colhoun HM, Betteridge DJ, Durrington PN, Hitman GA, Neil HA, Livingstone SJ, Thomason MJ, Mackness MI, Charlton‐Menys V, Fuller JH, et al. Primary prevention of cardiovascular disease with atorvastatin in type 2 diabetes in the Collaborative Atorvastatin Diabetes Study (CARDS): multicentre randomised placebo‐controlled trial. Lancet. 2004;364:685–696. doi: 10.1016/S0140-6736(04)16895-5 [DOI] [PubMed] [Google Scholar]
- 6. Collins R, Armitage J, Parish S, Sleigh P, Peto R; Heart Protection Study Collaborative Group . MRC/BHF Heart Protection Study of cholesterol‐lowering with simvastatin in 5963 people with diabetes: a randomised placebo‐controlled trial. Lancet. 2003;361:2005–2016. doi: 10.1016/S0140-6736(03)13636-7 [DOI] [PubMed] [Google Scholar]
- 7. de Vries FM, Denig P, Pouwels KB, Postma MJ, Hak E. Primary prevention of major cardiovascular and cerebrovascular events with statins in diabetic patients: a meta‐analysis. Drugs. 2012;72:2365–2373. doi: 10.2165/11638240-000000000-00000 [DOI] [PubMed] [Google Scholar]
- 8. Sever PS, Poulter NR, Dahlöf B, Wedel H, Collins R, Beevers G, Caulfield M, Kjeldsen SE, Kristinsson A, McInnes GT, et al. Reduction in cardiovascular events with atorvastatin in 2,532 patients with type 2 diabetes: Anglo‐Scandinavian Cardiac Outcomes Trial‐lipid‐lowering arm (ASCOT‐LLA). Diabetes Care. 2005;28:1151–1157. doi: 10.2337/diacare.28.5.1151 [DOI] [PubMed] [Google Scholar]
- 9. Leino AD, Dorsch MP, Lester CA. Changes in statin use among U.S. adults with diabetes: a population‐based analysis of NHANES 2011–2018. Diabetes Care. 2020;43:3110–3112. doi: 10.2337/dc20-1481 [DOI] [PubMed] [Google Scholar]
- 10. Vinogradova Y, Coupland C, Brindle P, Hippisley‐Cox J. Discontinuation and restarting in patients on statin treatment: prospective open cohort study using a primary care database. BMJ. 2016;353:i3305. doi: 10.1136/bmj.i3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maddox TM, Borden WB, Tang F, Virani SS, Oetgen WJ, Mullen JB, Chan PS, Casale PN, Douglas PS, Masoudi FA, et al. Implications of the 2013 ACC/AHA cholesterol guidelines for adults in contemporary cardiovascular practice: insights from the NCDR PINNACLE registry. J Am Coll Cardiol. 2014;64:2183–2192. doi: 10.1016/j.jacc.2014.08.041 [DOI] [PubMed] [Google Scholar]
- 12. Rodriguez F, Lin S, Maron DJ, Knowles JW, Virani SS, Heidenreich PA. Use of high‐intensity statins for patients with atherosclerotic cardiovascular disease in the Veterans Affairs Health System: practice impact of the new cholesterol guidelines. Am Heart J. 2016;182:97–102. doi: 10.1016/j.ahj.2016.09.007 [DOI] [PubMed] [Google Scholar]
- 13. Zhang H, Plutzky J, Skentzos S, Morrison F, Mar P, Shubina M, Turchin A. Discontinuation of statins in routine care settings: a cohort study. Ann Intern Med. 2013;158:526–534. doi: 10.7326/0003-4819-158-7-201304020-00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Booth JN III, Colantonio LD, Chen L, Rosenson RS, Monda KL, Safford MM, Kilgore ML, Brown TM, Taylor B, Dent R, et al. Statin discontinuation, reinitiation, and persistence patterns among Medicare beneficiaries after myocardial infarction: a cohort study. Circ Cardiovasc Qual Outcomes. 2017;10:e003626. doi: 10.1161/CIRCOUTCOMES.117.003626 [DOI] [PubMed] [Google Scholar]
- 15. Rodriguez F, Maron DJ, Knowles JW, Virani SS, Lin S, Heidenreich PA. Association of statin adherence with mortality in patients with atherosclerotic cardiovascular disease. JAMA Cardiol. 2019;4:206–213. doi: 10.1001/jamacardio.2018.4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sarraju A, Coquet J, Zammit A, Chan A, Ngo S, Hernandez‐Boussard T, Rodriguez F. Using deep learning‐based natural language processing to identify reasons for statin nonuse in patients with atherosclerotic cardiovascular disease. Commun Med (Lond). 2022;2:88. doi: 10.1038/s43856-022-00157-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pokharel Y, Gosch K, Nambi V, Chan PS, Kosiborod M, Oetgen WJ, Spertus JA, Ballantyne CM, Petersen LA, Virani SS. Practice‐level variation in statin use among patients with diabetes: insights from the PINNACLE registry. J Am Coll Cardiol. 2016;68:1368–1369. doi: 10.1016/j.jacc.2016.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alsentzer E, Murphy JR, Boag W, Weng WH, Jin D, Naumann T, McDermott MBA. Publicly available clinical BERT embeddings. arXiv. Preprint posted online April 6, 2019. 2019;1904:03323. doi: 10.48550/arXiv.1904.03323 [DOI] [Google Scholar]
- 19. Rodriguez F, Maron DJ, Knowles JW, Virani SS, Lin S, Heidenreich PA. Association between intensity of statin therapy and mortality in patients with atherosclerotic cardiovascular disease. JAMA Cardiol. 2017;2:47–54. doi: 10.1001/jamacardio.2016.4052 [DOI] [PubMed] [Google Scholar]
- 20. Johnson AE, Pollard TJ, Shen L, Lehman LW, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, Mark RG. MIMIC‐III, a freely accessible critical care database. Sci Data. 2016;3:160035. doi: 10.1038/sdata.2016.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mikolov T, Sutskever I, Chen K, Corrado G, Dean J. Distributed representations of words and phrases and their compositionality. Preprint posted online October 16, 2013. arXiv. 2013;1310.4546. [Google Scholar]; doi: 10.48550/arXiv.1310.4546 [DOI]
- 22. Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, Kang J. BioBERT: a pre‐trained biomedical language representation model for biomedical text mining. Bioinformatics. 2022;36:1234–1240. doi: 10.1093/bioinformatics/btz682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wood FA, Howard JP, Finegold JA, Nowbar AN, Thompson DM, Arnold AD, Rajkumar CA, Connolly S, Cegla J, Stride C, et al. N‐of‐1 trial of a statin, placebo, or no treatment to assess side effects. N Engl J Med. 2020;383:2182–2184. doi: 10.1056/NEJMc2031173 [DOI] [PubMed] [Google Scholar]
- 24. Adusumalli S, Westover JE, Jacoby DS, Small DS, VanZandbergen C, Chen J, Cavella AM, Pepe R, Rareshide CAL, Snider CK, et al. Effect of passive choice and active choice interventions in the electronic health record to cardiologists on statin prescribing: a cluster randomized clinical trial. JAMA Cardiol. 2021;6:40–48. doi: 10.1001/jamacardio.2020.4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maricio R, Khera A. Statin use in pregnancy: is it time for a paradigm shift? Circulation. 2022;145:496–498. doi: 10.1161/CIRCULATIONAHA.121.058983 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1–S4


