Abstract
The pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has highlighted the need for simple, low-cost, and scalable diagnostics that can be widely deployed for rapid testing. Clustered regularly interspaced short palindromic repeats (CRISPR)–based diagnostics have emerged as a promising technology, but its implementation in clinical laboratories has been limited by the requirement of a separate amplification step prior to CRISPR-associated (Cas) enzyme–based detection. This article reports the discovery of two novel Cas12 enzymes (SLK9 and SLK5-2) that exhibit enzymatic activity at 60°C, which, when combined with loop-mediated isothermal amplification (LAMP), enable a real-time, single-step nucleic acid detection method [real-time SHERLOCK (real-time SLK)]. Real-time SLK was demonstrated to provide accurate results comparable to those from real-time quantitative RT-PCR in clinical samples, with 100% positive and 100% negative percent agreement. The method is further demonstrated to be compatible with direct testing (real-time SLK Direct) of samples from anterior nasal swabs, without the need for standard nucleic acid extraction. Lastly, SLK9 was combined with either Alicyclobacillus acidoterrestris AacCas12b or with SLK5-2 to generate a real-time, multiplexed CRISPR-based diagnostic assay for the simultaneous detection of SARS-CoV-2 and a human-based control in a single reaction, with sensitivity down to 5 copies/μL and a time to result of under 30 minutes.
Clustered regularly interspaced short palindromic repeats (CRISPR)–based diagnostics such as SHERLOCK (real-time SLK) and DETECTR have emerged as technologies promising for simple, point-of-need detection of target nucleic acids.1, 2, 3, 4, 5 These methods typically leverage the high specificity of programmable CRISPR-associated (Cas) enzymes, which, when combined with isothermal nucleic acid amplification, achieve approximately PCR-level (aM) sensitivity. To date, only two methods have demonstrated the robust performance required for Emergency Use Authorization by the US Food and Drug Administration, one being the SHERLOCK CRISPR severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) kit, the first CRISPR-based diagnostics system with Emergency Use Authorization [SHERLOCK CRISPR SARS-CoV-2 Kit instructions for use; Sherlock Biosciences, Watertown, MA; and SARS-CoV-2 DETECTR Reagent Kit instructions for use; Mammoth Biosciences, Brisbane, CA (revoked)]. Both methods, SHERLOCK and DETECTR, require nucleic acid extraction, independent reactions for all targets, and a reverse-transcription loop-mediated isothermal amplification (RT-LAMP) step that is separate from the CRISPR-Cas detection step. Although a simplified, high-throughput method has been established for use in clinical laboratories,6 this requirement of separate amplification and detection reactions is not suitable for at-home testing.
Several studies have demonstrated simplified processes in which amplification and detection are combined into a single step.3 , 7, 8, 9 Several methods use lower-temperature isothermal amplification methods (eg, recombinase polymerase amplification) to allow for one-pot detection by previously identified mesophilic Cas enzymes, but these methods are typically less sensitive, resulting in a dramatic decrease of up to sixfold in the lower limit of detection (LoD).10 Other studies have achieved one-pot CRISPR-based detection with LAMP by leveraging Cas enzymes with greater thermostability, but these enzymes lose activity at temperatures optimal for LAMP, resulting in reduced sensitivity and/or increased time to result (TTR).1 , 3 Historically, these challenges have been overcome with an increase in the target concentration via sample concentration and enzyme stabilization reagents, resulting in a greater overall sensitivity, with a LoD of 100 copies (cp) per reaction in testing for SARS-CoV-2, which is still 20-fold less sensitive compared to the CDC's real-time quantitative RT-PCR (RT-qPCR) assay.3
Given the diversity of CRISPR-Cas systems, this study sought to identify novel, thermostable Cas enzymes that would enable robust performance at higher temperatures that are optimal for LAMP (>60°C). Candidate Cas enzymes were identified computationally and screened experimentally for thermostability. A novel Cas12a enzyme was discovered, SLK-9, from an unidentified spirochete species that has been shown to have various degrees of functionality at 60°C to 75°C. A novel Cas12b enzyme (SLK5-2) was also identified from an uncharacterized metagenome isolated from a hot spring; this enzyme is also able to function at 60°C. Previously, four Cas12b enzymes have been shown to be active at >55°C, with one of them demonstrating functionality in a one-pot RT-LAMP Cas reaction at 60°C to 62°C.3 , 7 , 11 , 12 During the preparation of the present article, two novel Cas12a enzymes with similar thermostability were described; with one of these enzymes, Yellowstone metagenome YmeCas12a, nuclease activity was reported at 62°C.13
SLK9 was used to develop a real-time SLK assay in which Cas-based detection occurs simultaneously with RT-LAMP. Due to reverse transcription, amplification, and detection occurring simultaneously, real-time SLK produces results in 10 to 30 minutes, threefold faster than conventional RT-qPCR. In the present study, the analytical LoD of real-time SLK was 0.5 cp/μL with the use of extracted RNA. Real-time SLK was evaluated using 26 positive and 30 negative clinical samples and demonstrated 100% positive and 100% negative percent agreement with RT-qPCR results.
Also, a simple thermal treatment of nasal swab samples was compatible with real-time SLK, eliminating the need for RNA extraction of samples. The real-time SLK assay was combined with a direct method for sample nucleic acid processing to generate an assay, hereafter referred to as real-time SLK Direct (SLK Direct). Analytical sensitivity with this method was demonstrated to be 5 and 20 cp/μL in samples of anterior nasal swabs in saline (ANS/saline). The present study also demonstrated 100% positive and 100% negative percent agreement with results from RT-qPCR using 36 clinical samples. A further advancement of CRISPR-based diagnostics was shown by demonstrating a real-time, multiplexed SLK assay, combining the isothermal amplification and Cas-based detection of SARS-CoV-2 and human-based internal control in a single reaction using extracted clinical samples. Lastly, by combining the two novel Cas enzymes described in this study, a LoD of 5 cp/μL was determined using nonextracted clinical samples, and by increasing the reaction temperature to 60°C, a TTR of under 30 minutes was achieved. The advancements described in this publication resulted in a robust, high-sensitivity, high-specificity, and simple CRISPR-based diagnostic platform.
Materials and Methods
Clinical and Contrived Samples
Remnant nasopharyngeal (NP) swab samples in saline and in viral and universal transport media were purchased from Boca Biolistics (Boca Raton, FL) and BioIVT (Westbury, NY) and stored at −80°C. All samples were purchased with a clinical result from SARS-CoV-2 assays from either Panther (Hologic, Marlborough, MA), Cobas (Roche Diagnostics, Indianapolis, IN), or PerkinElmer (Waltham, MA). Contrived samples were prepared for assay-performance validation using SARS-CoV-2 viral particles purchased from ZeptoMetrix (Buffalo, NY) and diluted to concentrations in indicated matrices before use.
Oligos, Probes, Reporters, and Guides
All oligos, probes, reporters, and guides used in this study were synthesized by Integrated DNA Technologies (Coralville, IA). Sequences can be found in Table 1 .
Table 1.
Sequences
| Target | Primer type | Sequence |
|---|---|---|
| N | F3 | 5′-GCTTCTACGCAGAAGGGA-3′ |
| B3 | 5′-GTGACAGTTTGGCCTTGT-3′ | |
| FIP | 5′-TACTGCTGCCTGGAGTTGAATTCCTCTTCTCGTTCCTCATC-3′ | |
| BIP | 5′-GCTTTGCTGCTGCTTGACAGTGTTGTTGGCCTTTACCA-3′ | |
| LF | 5′-CTTGAACTGTTGCGACTACGT-3′ | |
| LB | 5′-ATTGAACCAGCTTGAGAGCAAA-3′ | |
| Orf1ab | F3 | 5′-TGAAAATAGGACCTGAGCG-3′ |
| B3 | 5′-ACACCTAGTCATGATTGCA-3′ | |
| FIP | 5′-CCAATAGAATGATGCCAACAGGCGATAGACGTGCCACATGC-3′ | |
| BIP | 5′-GATTGATGTTCAACAATGGGGTTTCATTACCATGGACTTGACAAT-3′ | |
| LF | 5′-AAGTGTCTGAAGCAGTGGAAAA-3′ | |
| LB | 5′-GGTAACCTACAAAGCAACCATGAT-3′ | |
| RNaseP | F3 | 5′-TTGATGAGCTGGAGCCA-3′ |
| B3 | 5′-CACCCTCAATGCAGAGTC-3′ | |
| FIP | 5′-GTGTGACCCTGAAGACTCGGTTTTAGCCACTGACTCGGATC-3′ | |
| BIP | 5′-CCTCCGTGATATGGCTCTTCGTTTTTTTCTTACATGGCTCTGGTC-3′ | |
| LF | 5′-ATGTGGATGGCTGAGTTGTT-3′ | |
| LB | 5′-GGCATGCTGAGTACTGGACCTC-3′ | |
| Target | Guide target sequence | |
|---|---|---|
| N | 5′-GCCATTGCCAGCCATTCTAGCAGGAG-3′ | |
| Orf1ab | 5′-ATCAATCATAAACGGATTATAGACG-3′ | |
| RNaseP | 5′-CCAAGTAATTGAAAAGACACTCCTC-3′ | |
B3, backward outer primer; BIP, backward inner primer; F3, forward outer primer; FIP, forward inner primer; LB, loop backward primer; LF, loop forward primer; Orf1ab, open reading frame (ORF)1a and ORF1b.
Plasmid Construction and Protein Purification
Open reading frames of candidate Cas enzymes were synthesized (Twist Bioscience, South San Francisco, CA), amplified, and cloned into pET28a vector using the restriction enzymes NdeI and XhoI. Expression was induced in BL21(DE3) Escherichia coli through the addition of 200 μmol/L isopropyl β-D-1-thiogalactopyranoside, and proteins were purified from culture lysates using a HisTrap HP column (Cytiva, Marlborough, MA) in an AKTA pure fast protein liquid chromatography system (Cytiva). For further cation exchange, proteins were loaded onto a 5 mL HiTrap Heparin HP column (Cytiva) using fast protein liquid chromatography and eluted over a salt gradient from 500 mmol/L to 2 mol/L NaCl in elution buffer (20 mmol/L Tris-HCl, 2 mol/L NaCl, 5% glycerol, 1 mmol/L dithiothreitol, pH 8.0). Protein-of-interest–containing fractions were pooled and exchanged into 20 mmol/L Tris-HCl, 500 mmol/L NaCl, 5% glycerol, and 1 mmol/L dithiothreitol (pH 8.0) using PD10 (Cytiva). Protein concentration was measured using a bicinchoninic acid assay, and activity was tested using an in vitro Cas enzyme reaction assay.
Protein Thermal Shift Assay
Protein thermal shift assay was performed following the vendor's recommended protocol (Thermo Fisher Scientific, Waltham, MA). Briefly, 12.5 μL of each Cas enzyme (500 ng/μL) was mixed with equimolar amounts of specific guide, 2.5 μL protein thermal shift dye (8×), and 5 μL of protein thermal shift buffer. The mixture was placed in the QuantStudio 5 qPCR instrument (Applied Biosciences, Thermo Fisher Scientific) to track fluorescence changes while slowly increasing the temperature. Data analysis to extract melting temperature was performed by taking the first derivative of raw fluorescence intensity collected in the X4-M4 channel.
Alignment of the Primers and Guide RNAs (crRNAs) Used to Known SARS-CoV-2 Genomes
A bioinformatics analysis was performed to determine the in silico inclusivity against all recent SARS-CoV-2 genomes, including variants denoted by the World Health Organization as variants of interest and variants of concern (https://www.who.int/publications/m/item/historical-working-definitions-and-primary-actions-for-sars-cov-2-variants, last accessed November 28, 2021) (Supplemental Table S1). The SARS-CoV-2 sample sizes geographically representing the United States and the rest of the world were 4284 and 4804, respectively. Upon curation of viral genomes, a multiple sequence alignment was constructed using the Wuhan SARS-CoV-2 (NC_045512.2; https://www.ncbi.nlm.nih.gov/nuccore, last accessed November 11, 2021) as the reference anchor. All molecular components were subsequently aligned to the multiple sequence alignment in order to tabulate its homology. The inclusivity of a target was defined on the basis of all components having perfect homology to their intended target (Supplemental Table S2).
Modified CDC RT-qPCR Assay
The CDC RT-qPCR protocol was followed except for use of the QuantStudio 5 system instead of the 7500 Dx qPCR machine (Thermo Fisher Scientific) [CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel Instructions for Use; CDC, Atlanta GA]. In brief, the QIAamp viral RNA kit (Qiagen, Germantown, MD) was used to extract RNA from clinical samples, and CDC primer/probe sequences (Integrated DNA Technologies) were used for all assays. Three RT-qPCR reactions per sample were performed to detect N1, N2, and RNaseP (RP) targets. The means of the N1 and N2 CT values are reported in data tables (Supplemental Tables S3 and S4). For RNA-extracted samples, TaqPath 1-Step RT-qPCR Master Mix, CG (Thermo Fisher Scientific) was used. For the reverse-transcription stage, samples were heated at 25°C for 2 minutes, then at 50°C for 15 minutes and at 95°C for 2 minutes. The PCR stage followed 45 cycles at 95°C for 3 seconds then at 55°C for 30 seconds. For samples tested using the CDC's heat-inactivation protocol, the samples were heated to 95°C for 1 minute followed by a 4°C hold. The UltraPlex 1-Step ToughMix (4×) (Quantabio, Beverly, MA) was used and heat-inactivated samples were run on the QuantStudio 5 qPCR machine using the following steps: reverse-transcription stage, 50°C for 10 minutes and at 95°C for 3 minutes, followed by the PCR stage of 45 cycles at 95°C for 3 seconds, then at 55°C for 30 seconds.
Extraction Method
Samples were extracted and eluted in water using the PureLink Viral RNA extraction kit (Thermo Fisher Scientific) following the manufacturer's recommended protocol, with two additional steps: a 1-minute dry-spin step after the wash step, followed by a 15-minute air-dry step at room temperature before elution of RNA. Input sample volume was 200 μL and elution volume was 30 to 50 μL. The extracted material was stored at −80°C and thawed before use.
Direct Method
For pretreatment of nasal swabs eluted in 0.9% saline, 10 μL of clinical matrix was mixed with 1 μL of Proteinase K (New England BioLabs, Ipswich, MA) and 0.4 μL RNAsecure (InvitroGen, Waltham, MA). The mixture was heated for 6 minutes at 65°C and for 3 minutes at 98°C and cooled to 4°C to 10°C before direct addition to the SLK reaction. Samples were heated using a hotplate or thermocycler. Nonextracted materials were freshly pretreated before use.
Real-Time SLK Reaction
Real-time SLK was performed with 40 μL of total reaction volume. Final concentrations of components in the real-time SLK reaction mix were 1× N–replicase polyprotein (Orf)-1–RP primer mix, 1× WarmStart LAMP reaction mix (New England BioLabs), 292 nmol/L SLK-9 enzyme, 112.5 nmol/L N/O/RP guide RNA, and 125 nmol/L poly-C reporter. A total of 14 μL of extracted RNA or 8 μL of direct sample was added to the SLK reaction mix. Fluorescence was measured every 1 minute for 90 minutes using the QuantStudio 5 qPCR instrument set to 60°C.
Real-Time, Multiplexed SLK Reaction
Real-time SLK was performed with 40 μL of total reaction volume as described in the previous paragraph, with the addition of either 368 nmol/L Alicyclobacillus acidoterrestris AacCas12b or 292 nmol/L SLK5-2 and corresponding guides at 520 nmol/L for Aac and 292 nmol/L for SLK5-2.
Multiplex detection was performed by adding 10 or 5 μL of pretreated clinical sample directly into the multiplexed SLK reaction mix and then measured on the QuantStudio 5 qPCR instrument for fluorescence readout at 56°C (AacCas12b) or 60°C (SLK5-2).
RT-SLK Data Analysis
First derivative analysis was performed on the raw fluorescence values recorded every minute of the assay for each sample. The first derivative values indicate the slope of the fluorescence signal. The maximal slope value for the negative samples in an experiment was determined, and the SD of the slopes for the negative samples was calculated. The maximal slope value plus three SD values calculated from the negative samples was set as the cutoff value. The TTR (in minutes) was defined as the time at which the fluorescence of a sample surpassed the cutoff value in three consecutive recordings.
Data and Materials Availability
All data, code, and materials used in the analyses are available upon request. Some materials require material transfer agreements (MTAs). All data needed to evaluate the conclusions in the study can be found in the study and in the Supplemental Materials. Inquiries and requests for materials should be addressed to the corresponding author.
Results
Cas Enzyme Discovery and Characterization
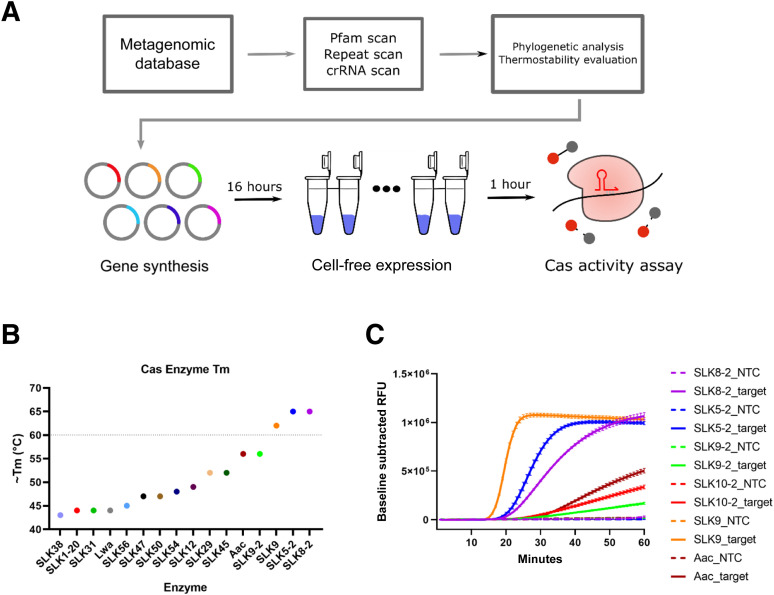
An initial computational metagenomic search for CRISPR arrays to identify novel Cas enzymes yielded approximately 7000 potential candidates. Of these, 96 candidates were selected based on phylogenetic analysis and deep-learning models designed to predict thermal stability (Figure 1 A). Of the 96 candidates predicted to be thermostable, one Cas12a enzyme, SLK9, had a melting temperature of >60°C (Figure 1B and Supplemental Figure S1A). Notably, the melting temperature of SLK9 was determined to be 61°C, which is greater than that of the commonly used CRISPR enzyme Leptotrichia wadei LwaCas13a14 and thermostable AacCas12b1 , 11 (Figure 1B). A second round of enzyme discovery was performed, resulting in the identification of seven additional thermostable Cas12a/b enzymes. Thermal shift assays revealed that two of the seven Cas enzymes, SLK5-2 and SLK8-2, had melting temperatures of 65°C (Figure 1B and Supplemental Figure S1B). The five Cas enzymes that had the highest melting temperatures were selected, and their functionality was evaluated further in an RT-LAMP reaction performed at 60°C. The combination of Cas detection of target in an RT-LAMP reaction was termed real-time SHERLOCK (RT-SLK). All five enzymes tested in the real-time SLK reaction demonstrated collateral cleavage activity in the presence of target RNA (Figure 1C). Two of the novel Cas enzymes, SLK9 and SLK5-2, showed specific detection of target and generated a high fluorescence signal within 25 minutes. SLK9 was selected to further optimize and validate the real-time SLK assay using contrived and clinical samples, and both SLK9 and SLK5-2 were evaluated in multiplexed Cas enzyme real-time SLK reactions.
Figure 1.
Thermostable Cas enzyme discovery and characterization. A: High-throughput novel Cas12 enzyme discovery pipeline. B: Melting temperature (Tm), as determined by thermal shift assay for 14 novel Cas12a/b enzymes, Leptotrichia wadei LwaCas13 and AacCas12. The horizontal line indicates the preferred Tm of 60°C. C: Real-time SLK assay run at 60°C, testing five novel Cas12a/b enzymes and AacCas12b. The baseline-subtracted RFU signal (relative fluorescence units) is a measure of collateral cleavage activity of each enzyme. SARS-CoV-2 genomic RNA at 200 cp/μL or 10 ng/μL human genomic RNA was used as target, and water, as the no-template control (NTC). Data are expressed as means ± SEM. n = 3 per sample. crRNA, guide RNA; Pfam, database of protein families.
Real-Time SLK and Real-Time SLK Direct SARS-CoV-2 Assays
Previously developed RT-LAMP primer sets for the N, Orf1ab, and RNaseP genes from the SLK CRISPR SARS-CoV-2 kit (SHERLOCK CRISPR SARS-CoV-2 Kit instructions for use; Sherlock Biosciences, Watertown, MA) were used to evaluate the performance of SLK9 in a real-time SLK reaction. The assay consisted of commercially available RT-LAMP mix, target-specific LAMP primers, SLK9, amplicon-specific crispr RNA, and a ssDNA fluorescence reporter. Primers and crispr RNAs for the detection of two SARS-CoV-2 targets, the N and Orf1ab genes, were combined in a single reaction to increase inclusivity and detection of known variants of SARS-CoV-2. In silico analysis showed that 97.1% and 100% of SARS-CoV-2 genomes had 100% homology across all primer and guide sequences for either the Orf1ab or N target in the assay for the Delta and Omicron variants, respectively (Supplemental Tables S1 and S2). Figure 2 A illustrates the process wherein the initial nucleic acid extraction was followed by a single reaction combining RT-LAMP and Cas-based detection reagents. Collateral cleavage of the ssDNA reporter occurs when the crispr RNA complexed with SLK9 binds the dsDNA target sequence.4
Figure 2.
Real-time SHERLOCK (RT-SLK). A: The real-time SLK Direct process. B–F: Time to result (TTR, in minutes) in each sample. Samples not detected are labeled ND (not determined). B: Titration of extracted SARS-CoV-2 genomic RNA, ranging from 100,000 to 0.1 cp/μL. Each concentration was tested in triplicate. C and D: TTR of RNaseP control signal (RP) versus SARS-CoV-2 N/Orf1ab target (NO) in ANS/saline samples spiked with indicated concentrations of SARS-CoV-2 viral particles. C: ANS/saline samples were purified using the PureLink viral DNA/RNA kit (Thermo Fisher Scientific, Waltham, MA) then added to real-time SLK reaction. D: ANS/saline samples were treated with Proteinase K (New England BioLabs, Ipswich, MA), RNAsecure (InvitroGen, Waltham, MA), and heat and directly added to real-time SLK Direct reaction. E and F: Positive and negative clinical samples were tested using real-time SLK and the CDC's RT-qPCR assay; results are reported as TTR. Dotted lines indicate cutoff values for either assay, and samples that were not detected are labeled ND and considered negative. E: Clinical samples—36 NP/saline, 10 NP/viral transport medium, and 10 NP/universal transport medium—were purified using the PureLink viral DNA/RNA kit and added to the real-time SLK reaction. F: A total of 36 NP/saline clinical samples were treated with Proteinase K, RNAsecure, and heat and directly added to a real-time SLK Direct reaction. gRNA, genomic RNA.
Initial evaluation of the real-time SLK method was performed using titrated amounts of SARS-CoV-2 genomic RNA ranging from 100,000 to 0.1 cp/μL (Figure 2B). After an initial validation using genomic RNA, RNA purified from ANS/saline that was spiked with ZeptoMetrix SARS-CoV-2 viral particles ranging from 100 to 0.1 cp/μL was evaluated (Supplemental Figure S2A). To confirm the LoD, 20 additional ANS/saline samples spiked with 0.5 cp/μL SARS-CoV-2 viral particles were extracted and tested with real-time SLK to assess analytical sensitivity. All 20 samples tested positive for SARS-CoV-2 and the RNaseP control within 25 minutes (Figure 2C). This LoD of 0.5 cp/μL is comparable to those of the highest-sensitivity SARS-CoV-2 assays currently available, with LoDs from 0.18 to 1 NAAT-detectable units per microliter (https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-reference-panel-comparative-data, last accessed January 17, 2020).
After it was demonstrated that real-time SLK could be used to reliably detect purified RNA from samples, the potential of the real-time SLK assay to be performed directly on a nasal swab eluted in saline after a simple thermal treatment of the specimen was evaluated. The approach to detecting SARS-CoV-2 directly from a sample was termed real-time SLK Direct. ANS/saline samples spiked with ZeptoMetrix SARS-CoV-2 viral particles at concentrations ranging from 100 to 2.5 cp/μL were treated with a mixture of Proteinase K (1.6 U/20-μL reaction) and the commercially available RNase-inhibition reagent RNAsecure (1:25 v/v), and heated for 6 minutes at 65°C, followed by a 3-minute incubation at 98°C. Proteinase K degrades nucleases and aids in the lysis of the viral particle along with the 95°C heating step, while RNAsecure inhibits RNases that can degrade the exposed viral RNA.15 These treated samples were then directly added to the real-time SLK Direct reaction mixture. All of three replicates with 10 cp/μL of SARS-CoV-2 were positive by real-time SLK Direct within 40 minutes (Supplemental Figure S2B). The LoD was confirmed with 20 additional samples at 5 cp/μL SARS-CoV-2, with all 20 testing positive by real-time SLK Direct (Figure 2D).
Clinical Sample Testing
The performance of real-time SLK and real-time SLK Direct was evaluated on clinical samples that were previously determined as positive or negative for SARS-CoV-2 by an external Clinical Laboratory Improvement Amendments (CLIA)–certified laboratory. The positive clinical samples used had a wide range of CT values (16 to 36), as determined by using the CDC's RT-qPCR assay protocol (Supplemental Tables S3 and S4). In total, 56 NP clinical samples (26 positive and 30 negative), consisting of swabs eluted in saline or viral or universal transport media, were purified using the PureLink viral RNA kit. Purified RNA was tested using real-time SLK, demonstrating 100% concordance with the results from the CLIA-certified laboratory and the RT-qPCR results derived in-house (Figure 2E and Supplemental Tables S3 and S4). TTR was less with all positive samples than with standard RT-qPCR. Next, the real-time SLK Direct method was clinically evaluated using 16 positive and 20 negative NP/saline clinical samples. All 16 positive samples were positive by real-time SLK Direct. Again, TTR was faster in all 16 positive clinical samples with real-time SLK Direct than with RT-qPCR (Figure 2F and Supplemental Table S3). Overall, it was demonstrated that the real-time SLK Direct method provided fast results, with no loss of clinical sensitivity or specificity.
Real-Time, Multiplexed SLK Direct
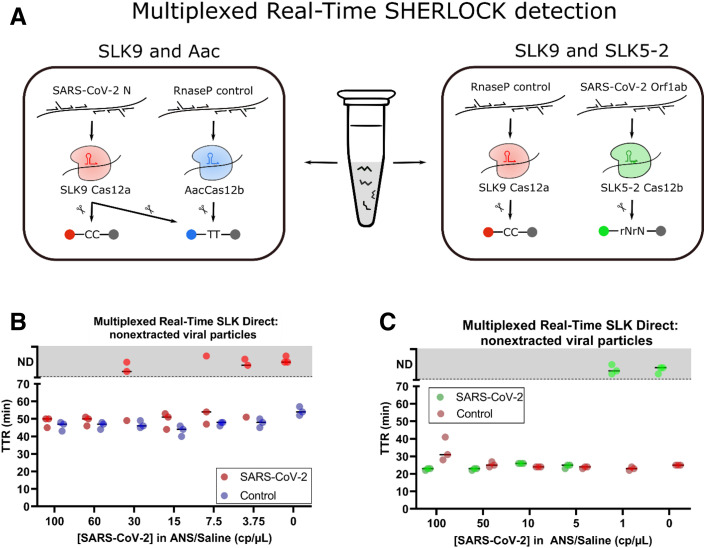
The discovery of thermostable Cas enzymes with orthogonal trans-cleavage preferences enabled the development of real-time, multiplexed, CRISPR-based diagnostics. After real-time SLK with SLK9 was established, a real-time, multiplexed method was developed by incorporating a second Cas enzyme that can be independently programed to detect a separate target sequence. AacCas12b has previously been shown to be compatible with LAMP reaction run at temperatures of up to 56°C and was combined with SLK9 to generate a real-time, multiplexed method.1 Although both SLK9 and AacCas12b have the capacity to cleave poly-T reporter molecules, only SLK9 was found to cleave poly-C reporters, allowing for the determination of which enzyme was activated (Figure 3 ). The real-time, multiplexed detection system contains, in a single reaction, RT-LAMP components for duplexed LAMP of the SARS-CoV-2 N gene and the human RP control, SLK9 enzyme with crispr RNA targeting the N gene, AacCas12b enzyme with crispr RNA targeting the human RP gene, a carboxyfluorescein–black hole quencher 1 (FAM-BHQ-1)–modified poly-T reporter, and Texas Red–BHQ-2–modified poly-C reporter.
Figure 3.
Multiplexed, real-time SLK. A: Multiplexed, real-time SLK reaction components using SLK9 with AacCas12b (left) and SLK9 with SLK5-2 (right). B and C: Samples of ANS/saline (B) or ANS/Tris-EDTA (C) spiked with indicated concentrations of SARS-CoV-2 viral particles were treated with Proteinase K (New England BioLabs, Ipswich, MA), RNAsecure (InvitroGen, Waltham, MA), and heat and directly added to multiplexed, real-time SLK Direct reaction. B: ANS/saline samples tested with multiplexed, real-time SLK SLK9/Aac assay. Fluorescence values greater than background values are reported in time to result (TTR) and were determined for red channel fluorescence (SARS-CoV-2) and green channel fluorescence (control). Samples that do not show increased fluorescence above the cutoff value are plotted in the gray zone in the ND (not determined) section. C: ANS/TE samples tested with multiplexed, real-time SLK SLK9/SLK5-2 assay. Increased fluorescence values greater than the background were determined for red channel fluorescence (SARS-CoV-2) and green channel fluorescence (RNaseP control). Samples that do not show increased fluorescence above the cutoff value are plotted in the gray zone in the ND section.
This real-time, multiplexed CRISPR diagnostic platform requires only a single transfer step in which a heat-lysed sample is added to the reaction; the only instrument requirements after sample addition are fluorescence-detection capability and temperature control to maintain 56°C. After sample addition, the N and RP targets are amplified by corresponding LAMP primer sets, followed by the activation of corresponding Cas enzymes. When SLK9 is activated, it will cleave both poly-C and poly-T reporters, enabling detection in both the FAM (green) and Texas Red (red) fluorescence channels. When AacCas12b is activated, it will cleave only poly-T reporters, enabling detection in the FAM fluorescence channel only. For results interpretation, positive samples show on signal in the Texas Red channel or on in both the FAM and Texas Red channels, and negative samples show on signal in the FAM channel and off signal in the Texas Red channel (Figure 3A). The assay is invalid if off signals are seen in both the FAM and Texas Red channels. Real-time, multiplexed SLK was validated using 41 extracted clinical samples, demonstrating 100% positive and 100% negative percent agreement with RT-qPCR results. Multiplexed real-time SLK Direct was validated with 10 NP/saline swab samples, demonstrating 100% positive and 100% negative percent agreement with RT-qPCR results (Supplemental Figure S3).
Also, SLK9 was paired with another novel Cas12 enzyme that was identified in this study, SLK5-2. Here, RNaseAlert (InvitroGen) was used in the FAM (green) channel to indicate activated SLK5-2 enzyme detecting SARS-CoV-2 and cleavage of poly-C–Texas Red in the red channel for the activation of SLK9 and the detection of the internal control gene human RP. Within the SLK5-2/SLK9 system, a positive sample is indicated by an on signal in the green channel, regardless of the red channel status, whereas a negative sample shows on only in the red channel. In the absence of an on signal in either a green or red channel, the sample is invalid. Due to the greater thermostability of both enzymes, multiplex detection using SLK5-2/SLK9 was performed at 60°C. Both multiplex approaches were compared side-by-side with the present novel real-time SLK Direct approach using heat/Proteinase K–treated ANS samples spiked with SARS-CoV-2 ZeptoMetrix particles ranging from 100 to 3.75 cp/μL (Figure 3, B and C). In the SLK9/Aac system, SARS-CoV-2 particles at 60 cp/μL were detected in three of three replicates within 50 minutes using real-time SLK Direct. The SLK5-2/SLK9 real-time SLK Direct approach had a sensitivity of 5 cp/μL and a TTR of under 30 minutes.
Discussion
PCR-based diagnostics are currently the gold standard method for nucleic acid detection of viruses and bacteria. The global pandemic caused by SARS-CoV-2 spurred increased efforts to develop alternative molecular diagnostics that do not require specialized thermocyclers.16 Isothermal amplification methods hold great potential for low-resource molecular diagnostics; however, a problem common to all isothermal amplification technologies is nonspecific amplification, which can result in false-positive reactions in systems that rely on non–sequence-specific detection methods.3 , 17, 18, 19 Many groups have developed strategies to overcome rates of false positivity by either optimizing the addition of components, reducing the read time to better discriminate real amplification from background amplification, using indirect detection strategies, or using probes to increase assay specificity and reduce nontemplated amplification.20, 21, 22, 23 Other groups have added a CRISPR-based detection step to increase the specificity of isothermal amplification. Recombinase polymerase amplification and LAMP have both been demonstrated to be compatible with Cas13 and Cas12 enzymes.1, 2, 3 RT-LAMP was chosen for nucleic acid amplification given that LAMP has been shown to be more tolerant than PCR to inhibitors present in clinical samples, which enables the detection of nucleic acids from lysates without nucleic acid isolation and concentration.24 , 25 Unfortunately, LAMP requires incubation at 56°C to 65°C, and only three commercially available Cas12 enzymes have been characterized to function at that temperature (Alicyclobacillus acidiphilus AapCas12b, AacCas12b, and Brevibacillus BrCas12b).3 , 11 , 12 Therefore, the present study aimed to identify novel Cas enzymes that could function within the temperatures used in a LAMP reaction.
This article presents the discovery of two thermostable Cas12 enzymes, SLK9 and SLK5-2. SLK9 was used to develop a rapid, sensitive, real-time, one-pot, CRISPR-based assay for the detection of SARS-CoV-2. The performance of SLK9 in a real-time SLK assay was validated using contrived, extracted ANS samples; a LoD of 0.5 viral particles per microliter was demonstrated, which is comparable to that of the gold standard method for SARS-CoV-2 detection, RT-qPCR [CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel Instructions for Use; CDC, Atlanta GA]. It was then demonstrated that real-time SLK can be used without extraction by treating samples with Proteinase K, RNAsecure, and heat. The LoD for the direct detection of viral particles in contrived ANS samples was 5 cp/μL. Both methods were then used to detect SARS-CoV-2 from archived clinical specimens. Positive and negative percent agreement with RT-qPCR results were 100% in both the extracted (real-time SLK) and nonextracted samples (real-time SLK Direct).
The discovery of thermostable Cas12 enzymes was essential for developing a multiplexed real-time SLK assay. Previously, only Cas12b enzymes had demonstrated thermostability at 60°C and were not compatible with multiplexing because they all utilized the same single-guide RNA sequences.3 The novel Cas12a enzyme, SLK9, was combined with either AacCas12b or SLK5-2 (a novel Cas12b) to develop a multiplexed real-time SLK assay, and performance was validated with the simultaneous detection of SARS-CoV-2 and the human RP internal control within a single reaction. Due to the presence of the less thermostable AacCas12b enzyme, these reactions were run at 56°C, slowing amplification and therefore increasing the overall TTR. Because of the greater thermostability of both enzymes and therefore more efficient LAMP, multiplexing of SLK9 and SLK5-2 resulted in a LoD of 5 cp/μL in contrived ANS/Tris-EDTA samples, with a TTR of under 30 minutes.
This method for SARS-CoV-2 detection is sensitive and rapid, omits expensive and time-consuming sample preparation, and, when combined with a second Cas enzyme, enables real-time, multiplexed detection, as demonstrated by the detection of the SARS-CoV-2 Orf1ab gene and internal control RP gene within a single reaction. Additional studies to identify more thermostable Cas enzymes can further increase multiplexing capabilities, increasing the value of this method for at-home and point-of-care diagnostics.
Footnotes
Supported by Good Venture Fund (GVF).
Disclosures: All authors are employees of Sherlock Biosciences, Inc.
Supplemental material for this article can be found at https://doi.org/10.1016/j.jmoldx.2023.03.009.
Author Contributions
J.M.P., X.L., M.K.W., P.R., B.J.M., H.B., and W.J.B. conceptualized the study; J.M.P., E.S.F., L.C., K.S., P.P.N., I.A., A.L., S.S., and M.C.D. performed experiments; P.R., M.K.W., B.J.M., H.B., and W.J.B. supervised the study; J.M.P., B.J.M., and H.B. wrote the manuscript; and J.M.P., M.K.W., P.R., E.S.F., B.J.M., and H.B. reviewed and edited the manuscript.
Supplemental Data
Protein thermal shift assay determining the melting temperature (Tm) values of 16 Cas enzymes. Tm was determined by peak intensity of the first derivative of fluorescence intensity. A: Derivative of fluorescence intensity from 25°C to 100°C for SLK9, SLK12, SLK29, SLK31, SLK38, SLK45, SLK47, SLK50, SLK54, and SLK56. B: Derivative of fluorescence intensity from 25°C to 100°C for LwaCas13a, AacCas12b, SLK10-2, SLK9-2, SLK8-2, and SLK5-2.
Real-time SLK LoD range finding, using extracted and nonextracted [Proteinase K (New England BioLabs, Ipswich, MA)/RNAsecure (InvitroGen, Waltham, MA), and heat treated] anterior nasal swabs in saline (ANS/saline) samples. A and B: Pooled ANS/saline were spiked with indicated concentrations of SARS-CoV-2 viral particles and purified using the PureLink viral DNA/RNA kit (Thermo Fisher Scientific, Waltham, MA) (A) or treated with Proteinase K/RNASecure and heated (B). All samples were tested for the detection of the SARS-CoV-2 N/Orf1ab (NO) gene and human RNaseP internal control (RP). Time to result (TTR, in minutes) is indicated for real-time SLK when the fluorescence signal surpasses the cutoff value, plotted on the y axis, and the concentration of SARS-CoV-2 in each sample, indicated on the x axis. Samples were considered negative if fluorescence values did not surpass background fluorescence and are labeled as ND (not determined).
Evaluation of multiplexed, real-time SLK SLK9/Aac clinical samples. A: A total of 41 NP swabs in universal or viral transport media, previously tested in an outside CLIA-certified laboratory and determined to be positive or negative, were purified using the PureLink viral DNA/RNA kit (Thermo Fisher Scientific, Waltham, MA) and then tested in a multiplexed, real-time SLK9/Aac assay and the CDC's RT-qPCR assay. Samples were considered negative if fluorescence values did not surpass background fluorescence and are labeled ND (not determined). B: A total of 10 clinical NP swabs in saline, previously tested in an outside CLIA-certified laboratory and determined to be positive or negative, were treated with RNASecure (InvitroGen, Waltham, MA), Proteinase K (New England BioLabs, Ipswich, MA), and heat and directly added to a multiplexed, real-time SLK RS9/Aac reaction; times to result (TTR, in minutes) with the real-time SLK assay are plotted on the y axis. The same 10 samples were also RNA extracted and tested using the CDC's RT-qPCR assay, with TTR values plotted on the x axis. Samples were considered negative if the fluorescence values did not surpass background fluorescence and are labeled as ND.
References
- 1.Li L., Li S., Wu N., Wu J., Wang G., Zhao G., Wang J. HOLMESv2: a CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth Biol. 2019;8:2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- 2.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A., Myhrvold C., Bhattacharyya R.P., Livny J., Regev A., Koonin E.V., Hung D.T., Sabeti P.C., Collins J.J., Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joung J., Ladha A., Saito M., Kim N.-G., Woolley A.E., Segel M., Barretto R.P.J., Ranu A., Macrae R.K., Faure G., Ioannidi E.I., Krajeski R.N., Bruneau R., Huang M.-L.W., Yu X.G., Li J.Z., Walker B.D., Hung D.T., Greninger A.L., Jerome K.R., Gootenberg J.S., Abudayyeh O.O., Zhang F. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N Engl J Med. 2020;383:1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li S.-Y., Cheng Q.-X., Wang J.-M., Li X.-Y., Zhang Z.-L., Gao S., Cao R.-B., Zhao G.-P., Wang J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018;4:20. doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J.S., Ma E., Harrington L.B., da Costa M., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manning B.J., Khan W.A., Peña J.M., Fiore E.S., Boisvert H., Tudino M.C., Barney R.E., Wilson M.K., Singh S., Mowatt J.A., Thompson H.J., Tsongalis G.J., Blake W.J. High-throughput CRISPR–Cas13 SARS-CoV-2 test. Clin Chem. 2021;68:172–180. doi: 10.1093/clinchem/hvab238. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen L.T., Macaluso N.C., Pizzano B.L.M., Cash M.N., Spacek J., Karasek J., Miller M.R., Lednicky J.A., Dinglasan R.R., Salemi M., Jain P.K. A thermostable Cas12b from Brevibacillus leverages one-pot discrimination of SARS-CoV-2 variants of concern. EBioMedicine. 2022;77 doi: 10.1016/j.ebiom.2022.103926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D. One-pot detection of COVID-19 with real-time reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay and visual RT-LAMP assay. bioRxiv. 2020 doi: 10.1101/2020.04.21.052530. [Preprint]. [DOI] [Google Scholar]
- 9.Wang R., Qian C., Pang Y., Li M., Yang Y., Ma H., Zhao M., Qian F., Yu H., Liu Z., Ni T., Zheng Y., Wang Y. opvCRISPR: one-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosens Bioelectron. 2021;172:112766. doi: 10.1016/j.bios.2020.112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arizti-Sanz J., Freije C.A., Stanton A.C., Petros B.A., Boehm C.K., Siddiqui S., Shaw B.M., Adams G., Kosoko-Thoroddsen T.S.F., Kemball M.E., Uwanibe J.N., Ajogbasile F.V., Eromon P.E., Gross R., Wronka L., Caviness K., Hensley L.E., Bergman N.H., MacInnis B.L., Happi C.T., Lemieux J.E., Sabeti P.C., Myhrvold C. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat Commun. 2020;11:5921. doi: 10.1038/s41467-020-19097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shmakov S., Abudayyeh O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E., Minakhin L., Joung J., Konermann S., Severinov K., Zhang F., Koonin E.V. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian Y., Liu R.R., Xian W.D., Xiong M., Xiao M., Li W.J. A novel thermal Cas12b from a hot spring bacterium with high target mismatch tolerance and robust DNA cleavage efficiency. Int J Biol Macromol. 2020;147:376–384. doi: 10.1016/j.ijbiomac.2020.01.079. [DOI] [PubMed] [Google Scholar]
- 13.Fuchs R.T., Curcuru J.L., Mabuchi M., Noireterre A., Weigele P.R., Sun Z., Robb G.B. Characterization of Cme and Yme thermostable Cas12a orthologs. Commun Biol. 2022;5:325. doi: 10.1038/s42003-022-03275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lalli M.A., Langmade J.S., Chen X., Fronick C.C., Sawyer C.S., Burcea L.C., Wilkinson M.N., Fulton R.S., Heinz M., Buchser W.J., Head R.D., Mitra R.D., Milbrandt J. Rapid and extraction-free detection of SARS-CoV-2 from saliva with colorimetric reverse-transcription loop-mediated isothermal amplification. Clin Chem. 2021;67:415–424. doi: 10.1093/clinchem/hvaa267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freije C.A., Sabeti P.C. Detect and destroy: CRISPR-based technologies for the response against viruses. Cell Host Microbe. 2021;29:689–703. doi: 10.1016/j.chom.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lobato I.M., O'sullivan C.K. Recombinase polymerase amplification: basics, applications and recent advances. Trends Analyt Chem. 2018;98:19–35. doi: 10.1016/j.trac.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Senarath K.D., Usgodaarachchi R.B., Navaratne V., Nagahawatte A., Wijayarathna C.D., Alvitigala J., Goonasekara C.L. Non specific amplification with the lamp technique in the diagnosis of tuberculosis in Sri Lankan settings. J Tuberc Res. 2014;2:168–172. [Google Scholar]
- 19.Wang D.-G., Brewster J.D., Paul M., Tomasula P.M. Two methods for increased specificity and sensitivity in loop-mediated isothermal amplification. Molecules. 2015;20:6048–6059. doi: 10.3390/molecules20046048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Tanner N.A. Efficient multiplexing and variant discrimination in reverse-transcription loop-mediated isothermal amplification with sequence-specific hybridization probes. Biotechniques. 2022;73:247–255. doi: 10.2144/btn-2022-0096. [DOI] [PubMed] [Google Scholar]
- 21.Mori Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 22.Hardinge P., Murray J.A.H. Reduced false positives and improved reporting of loop-mediated isothermal amplification using quenched fluorescent primers. Sci Rep. 2019;9:7400. doi: 10.1038/s41598-019-43817-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heithoff D.M., Barnes L., Mahan S.P., Fox G.N., Arn K.E., Ettinger S.J., Bishop A.M., Fitzgibbons L.N., Fried J.C., Low D.A., Samuel C.E., Mahan M.J. Assessment of a smartphone-based loop-mediated isothermal amplification assay for detection of SARS-CoV-2 and influenza viruses. JAMA Netw Open. 2022;4:5. doi: 10.1001/jamanetworkopen.2021.45669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edwards T., Burke P.A., Smalley H.B., Gillies L., Hobbs G. Loop-mediated isothermal amplification test for detection of Neisseria gonorrhoeae in urine samples and tolerance of the assay to the presence of urea. J Clin Microbiol. 2014;52:2163–2165. doi: 10.1128/JCM.00314-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaneko H., Kawana T., Fukushima E., Suzutani T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J Biochem Biophys Methods. 2007;70:499–501. doi: 10.1016/j.jbbm.2006.08.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein thermal shift assay determining the melting temperature (Tm) values of 16 Cas enzymes. Tm was determined by peak intensity of the first derivative of fluorescence intensity. A: Derivative of fluorescence intensity from 25°C to 100°C for SLK9, SLK12, SLK29, SLK31, SLK38, SLK45, SLK47, SLK50, SLK54, and SLK56. B: Derivative of fluorescence intensity from 25°C to 100°C for LwaCas13a, AacCas12b, SLK10-2, SLK9-2, SLK8-2, and SLK5-2.
Real-time SLK LoD range finding, using extracted and nonextracted [Proteinase K (New England BioLabs, Ipswich, MA)/RNAsecure (InvitroGen, Waltham, MA), and heat treated] anterior nasal swabs in saline (ANS/saline) samples. A and B: Pooled ANS/saline were spiked with indicated concentrations of SARS-CoV-2 viral particles and purified using the PureLink viral DNA/RNA kit (Thermo Fisher Scientific, Waltham, MA) (A) or treated with Proteinase K/RNASecure and heated (B). All samples were tested for the detection of the SARS-CoV-2 N/Orf1ab (NO) gene and human RNaseP internal control (RP). Time to result (TTR, in minutes) is indicated for real-time SLK when the fluorescence signal surpasses the cutoff value, plotted on the y axis, and the concentration of SARS-CoV-2 in each sample, indicated on the x axis. Samples were considered negative if fluorescence values did not surpass background fluorescence and are labeled as ND (not determined).
Evaluation of multiplexed, real-time SLK SLK9/Aac clinical samples. A: A total of 41 NP swabs in universal or viral transport media, previously tested in an outside CLIA-certified laboratory and determined to be positive or negative, were purified using the PureLink viral DNA/RNA kit (Thermo Fisher Scientific, Waltham, MA) and then tested in a multiplexed, real-time SLK9/Aac assay and the CDC's RT-qPCR assay. Samples were considered negative if fluorescence values did not surpass background fluorescence and are labeled ND (not determined). B: A total of 10 clinical NP swabs in saline, previously tested in an outside CLIA-certified laboratory and determined to be positive or negative, were treated with RNASecure (InvitroGen, Waltham, MA), Proteinase K (New England BioLabs, Ipswich, MA), and heat and directly added to a multiplexed, real-time SLK RS9/Aac reaction; times to result (TTR, in minutes) with the real-time SLK assay are plotted on the y axis. The same 10 samples were also RNA extracted and tested using the CDC's RT-qPCR assay, with TTR values plotted on the x axis. Samples were considered negative if the fluorescence values did not surpass background fluorescence and are labeled as ND.