Abstract
When the sequence of the Euglena gracilis chloroplast genome was reported in 1993 the α-subunit gene (rpoA) of RNA polymerase appeared to be missing, based on a comparison of all putative reading frames to the then known rpoA loci. Since there has been a large increase in known rpoA sequences, the question of a Euglena chloroplast rpoA gene was re-examined. A previously described unknown reading frame of 161 codons was found to be part of an rpoA gene split by a single group III intron. This rpoA gene, which is highly variable from species to species, was then isolated and characterized in five other euglenoid species, Euglena anabaena, Euglena granulata, Euglena myxocylindracea, Euglena stellata and Euglena viridis, and in the Astasia longa plastid genome. All seven Euglena rpoA genes have either one or three group III introns. The rpoA gene products in Euglena spp. appear to be the most variable in this gene family when compared to the rpoA gene in other species of bacteria, algae and plants. Additionally, Euglena rpoA proteins lack a C-terminal domain required for interaction with some regulatory proteins, a feature shared only with some chlorophyte green algae. The E.gracilis rpoA gene is the distal cistron of a multigene cluster that includes genes for carbohydrate biosynthesis, photosynthetic electron transport, an antenna complex and ribosomal proteins. This study provides new insights into the transcription system of euglenoid plastids, the organization of the plastid genome, group III intron evolution and euglenoid phylogeny.
INTRODUCTION
Two types of RNA polymerases are known to exist in chloroplasts (1). One is a single subunit, nuclear encoded enzyme related to various phage RNA polymerases. This polymerase is involved primarily in expression of the transcriptional and translational systems of the organelle, but not the photosynthetic machinery. The other polymerase, largely responsible for the transcription of photosynthetic genes, is closely related to multisubunit eubacterial RNA polymerases. Genes for four subunits, rpoA, rpoB, rpoC1 and rpoC2, are normally encoded by the plastid genome (2). These chloroplast genes correspond to only three RNA polymerase core genes of eubacteria because the rpoC1 and rpoC2 polypeptides are enclosed in a single bacterial rpoC gene. The fifth subunit, σ, is encoded by the nuclear genome. Some chloroplast operons containing both photosynthetic genes and ribosomal genes are probably transcribed by both enzymes (3). The two types of chloroplast RNA polymerases use three different types of promoters. Two of these promoters appear to operate with only one of the two enzymes, while the third class can operate with both.
When the sequence of the Euglena gracilis chloroplast genome was reported in 1993 (4), 82 genes and 149 introns were identified. Although the rpoB, rpoC1 and rpoC2 genes were identified, the α-subunit gene (rpoA) appeared to be missing, based on a comparison of all putative reading frames to the known rpoA loci.
In the ensuing years there has been a large increase in known rpoA sequences. The description of an rpoA consensus sequence has also appeared in the PFAM database (5) of conserved protein domains. We decided to re-examine the question of a Euglena chloroplast rpoA gene. A previously described unknown reading frame (orf161) was found to have significant similarity to the rpoA gene of Helicobacter pylori (accession no. P56001). orf161 was found to be the C-terminal exon of an rpoA gene split by a single group III intron, co-transcribed with at least six upstream genes. This rpoA gene, which is highly variable from species to species, was then isolated and characterized in five other euglenoid species, Euglena anabaena, Euglena granulata, Euglena myxocylindracea, Euglena stellata and Euglena viridis, and in the Astasia longa plastid genome (6). This study provides new insights into the transcription system of euglenoid plastids, the organization of the plastid genome, group III intron evolution and euglenoid phylogeny.
MATERIALS AND METHODS
Euglena cultures and nucleic acid extraction
The following euglenoid strains were obtained from the University of Texas Culture Collection: E.gracilis var. Z strain (UTEX 753), E.stellata (UTEX 327), E.viridis (UTEX 85), E.myxocylindracea (UTEX 1989), E.anabaena (UTEX 373) and E.granulata (UTEX 453).
Euglena spp. liquid cultures were maintained in either heterotrophic (Euglena broth; Sigma) or photoautotrophic medium (Cramer-Myers medium) (7) under continuous illumination. The growth rate of E.gracilis exceeded the growth rates of the other cultures under these conditions.
Total nucleic acid (TNA) extracts were prepared as previously described (8), either directly from cultures obtained from the UTEX culture collection grown on solid slants or following additional growth in liquid culture.
RNA was isolated using TRIAZOL reagent (Gibco BRL) or double purified using an Ambion Mini-Plant purification kit (Ambion).
PCR amplification, cDNA synthesis and sequencing
The rpoA gene was isolated from the euglenoid species E.gracilis, E.anabaena, E.granulata, E.myxocylindracea, E.stellata and E.viridis by PCR amplification from primers targeted to the flanking ycf9 and trnS genes. Aliquots of 0.05–0.2 µg/ml TNA extracts were amplified with the synthetic oligonucleotides P3 and P4. Primer P3 corresponds to a redundant version of coordinates 72239–72258 (5′-CCWGTTGTWTTTGCWTCTCC, accession no. X70810) of E.gracilis ycf9. Primer P4 is specific for the trnP gene (coordinates 70977–70993, 5′-GAATCCTGTCATYCCGA). To establish exon–intron boundaries within the six putative rpoA loci, the following species-specific primers within rpoA were used for cDNA synthesis, PCR amplification and as primers for DNA sequencing. For E.gracilis: P1, cDNA primer, coordinates 71611–71637, 5′-CTCTTCTAAACCTAAAAAGTT-GTGAAC; P2, PCR primer, coordinates 71945–71971, 5′-GAAATATTTAAAAATATATGTTTTAAG. For E.viridis (accession no. AY047487): cDNA primer, coordinates 1085–1103, 5′-CAACCAACGTCTTTTATCAG; PCR primer, coordinates 333–350, 5′-CTAATCAACTTAGGCGTA. For E.granulata (accession no. AY047485): cDNA primer, coordinates 1081–1097, 5′-CTTGTGCTTATATCCAG; PCR primer, coordinates 353–370, 5′-CTTTCGTCAGTTTTTGCT. For E.anabaena (accession no. AY047483): cDNA primer, coordinates 1125–1143, 5′-CAGTGAAAGTTTTTGCT; PCR primer, coordinates 349–366, 5′-CCCTCTTGTAATAGGTAC. For E.myxocylindracea (accession no. AY047485): cDNA primer, coordinates 1095–1115, 5′-CCAATTTAATTATCTTTAGAG; PCR primer, coordinates 348–366, 5′-ACTTAATTCGTCGTATTCT. For E.stellata (accession no. AY047487): cDNA primer, coordinates 1097–1114, 5′-AACTAGCGTAAGGAATGC; PCR primer, coordinates 360–377, 5′-TGTGATAGAGTAGCTTGG.
Reverse transcription and PCR reactions were performed as previously described (8).
Polycistronic transcripts
Oligonucleotide primers for analysis of genes that may be co-transcribed with the putative rpoA gene were selected based on the known sequence of the chloroplast DNA of E.gracilis (4). All primer pairs were designed to have introns within the amplified areas because cDNA without an intron is a control for DNA contamination. The cDNA primer (P1) for cDNA synthesis was designed in the rpoA gene of E.gracilis. The cDNA was amplified from PCR primers in upstream genes. ycf9: P5, 72237–72256, 5′-CCAGTTATTTTTGCTTCTCC. rpl12: P6, 72619–72639, 5′-GTTCCAAAGGTTGTATGTGAG. rps9: P7, 73126–73145, 5′-GCGCCACAATTCTCAAAAAG. All coordinates refer to GenBank accession no. X70810. Two other cDNA primers in the rps9 gene (P8, 74781–74802, 5′-GAAATCCATAACTTACCTATTAC) and psaC gene (P9, 74959–74989, 5′-GACTTGTTTCAGAACCTAAATAAATAAACAC) were employed. Their corresponding PCR primers were in the rpl32 gene (P10, 76049–76070, 5′-CTCGTAGAAACTCAAGGAAAAG) and rbcL gene (P11, 76299–76318, 5′-TAAATGGAGTCCAGAACTTG).
Computer analysis
orf161 from E.gracilis was first shown to have a region similar to the rpoA gene by a PSI-BLAST search (NTBES) (9). Open reading frame analysis and determination of putative amino acid sequences were done with the computer program DNA Strider. Nucleotide and protein sequence alignments were carried out using the PILEUP and Clustal X programs (GCG Sequence Analysis Package v.8.0; GCG, Madison, WI).
A consensus sequence was calculated both with the GCG program PRETTY and by hand. The amino acid with the majority occurrence or similarity was used in the consensus. In other cases which were not clearly defined for Euglena spp. the amino acid is outlined, as shown.
Phylogenetic studies
All rpoA sequences are from the GenBank database (accession nos are provided in parentheses). For evolutionary comparison representatives of different taxa were taken. These included the bacteria Thermus thermophilus (BAA75549), Thermotoga maritima (AAD36540), Leptospira interrogans (AAD40610), Borrelia burgdorferi (O51455), Mycobacterium leprae (CAB39833), Campylobacter jejuni (CAB73583), Helicobacter pylori (P56001) and Escherichia coli (X53843), the blue-green algae Synechococcus (O24710) and Synechocystis pcc6803 (P73297), the glaucocystophyte Cyanophora paradoxa (AAA81287), the red algae Porphyra purpurea (AAC08179) and Cyanidium caldarium (AAF12928), the diatom Odontella sinensis (P49465), the cryptophyte algae Guillardia theta (AF0411468.1) and Pyrenomonas salina (X65872 S53396), the green algae Nephroselmis olivacea (AAD54788), Chlorella vulgaris (NP 045920), Mesostigma viride (AAF43801) and Spirogyra maxima (AAC95318.1) and the land plants Arabidopsis thaliana (NP051090), Pinus thunbergii (NP042438) and Nicotiana tabacum (CAA77376.1).
The nucleotide and putative amino acid sequences of the rpoA genes were aligned in the Clustal X (1.8) multiple alignment program. Phylogenetic trees on 30 amino acid sequences were constructed using the distance [neighbor joining (NJ)] method of Saitou and Nei (10). Boostrap analysis was done with 1000 replicates of the starting tree.
Parsimony analysis was done using PAUP 4.0b8, with 1000 bootstrap replicates. A heuristic method was used on 30 amino acid sequences and a branch-and-bound method was used on a smaller set of nucleotide sequences (Euglena spp. plus H.pylori as an outgroup). Maximum likelihood (ML) analysis based on the nucleotide sequences of eight proteins found in Euglena spp. and H.pylori was done using settings which correspond to the HKY85 model. The number of bootstrap replicates was 100 and the total number of sites used in ML was 759.
RESULTS
Identification of the putative E.gracilis chloroplast rpoA gene
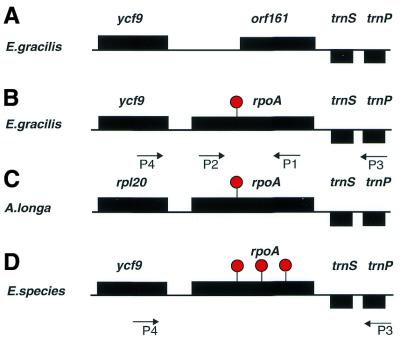
An open reading frame of 161 codons (orf161, GenBank accession no. X70810) and unknown function was previously identified in the E.gracilis chloroplast genome. This putative gene is distal to a rbcL–rpl32–psaC–rps9–rpl12–ycf9 gene cluster (4). Two tRNA genes, trnP and trnS, located at the 3′-end of orf161, are transcribed from the opposite DNA strand (Fig. 1A). When this putative orf161 coding region was first described there was no significant similarity with any known protein in the public databases.
Figure 1.
Schematic diagram of the ycf9–rpoA–trnS region. Primers P1 and P2 were used for amplification of this region. Black boxes represent the genes for ycf9, rpoA and trnS. Group III introns are indicated by red lollipops. (A) The previously reported E.gracilis gene organization (4) and the primers used for PCR and RT–PCR. (B) The E.gracilis rpoA locus as characterized in this study. (C) The postulated A.longa rpoA coding locus. (D) The rpoA coding loci for five Euglena species. One intron is present in each of E.gracilis and A.longa. Two additional introns are present in E.anabaena, E.granulata, E.myxocylindracea, E.stellata and E.viridis.
Using PSI-BLAST searches and current databases, significant similarity was identified between orf161 and the C-terminal domain of an rpoA gene for the DNA-directed RNA polymerase α-chain in H.pylori. The similarity to the bacterial rpoA gene could be extended to more than 200 codons if an upstream region separated by a single group III intron was postulated.
To determine whether orf161 is transcribed and spliced, RT–PCR was performed. Primer P4 (Fig. 1) was used to prime reverse transcription from within the putative orf161 RNA. Primer P3 (Fig. 1) was used to amplify the resulting cDNA(s) from the postulated upstream exon using PCR. The resulting RT–PCR product was cloned and sequenced. The deduced amino acid sequence of the cDNA corresponds to the two exons of a putatively split rpoA gene of 217 codons, which is interrupted by a single group III intron of 123 nt between codons 45 and 46 (Fig. 1B).
The rpoA-like gene belongs to the bacterial RNA polymerase α-chain family
The deduced 217 amino acid sequence of the split rpoA gene was used for a RPS-BLAST (9) search of the PFAM database of conserved protein domains (5). The only match was a highly significant alignment with the bacterial RNA polymerase α-chain family (accession no. PF01000). Members of this family include the α-subunit of the RNA polymerase from eubacteria and chloroplasts. The α-subunit consists of two independently folded domains (11–13). The larger N-terminal domain (αNTD) is involved in interactions with the other subunits of the enzyme. Only the αNTD is required for subunit assembly and basal transcription. The smaller C-terminal domain (αCTD) interacts with the template DNA and transcriptional activators. The E.gracilis rpoA aligned only with the αNTD. The αCTD is absent in this chloroplast RNA polymerase subunit.
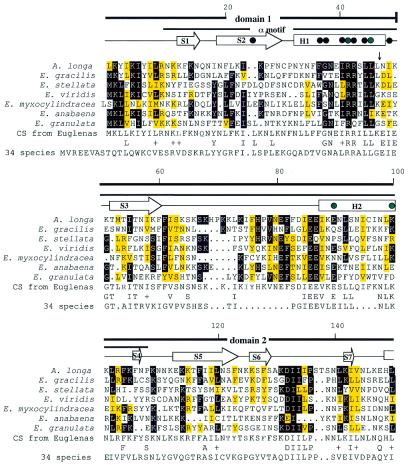
The structure of the αNTD of the E.coli RNA polymerase α-subunit has been determined to 2.5 Å resolution by X-ray crystallography (14). In this structure a pair of helices (H1 and H3) from one monomer interact with cognate helices of the other to form a hydrophobic dimer interface. Sequence alignment of the bacterial and E.gracilis chloroplast rpoA αNTD, combined with secondary structure predictions, are consistent with the prediction that the chloroplast rpoA subunit forms a homodimer containing the key sequence elements required for RNA polymerase assembly and basal transcription (Fig. 2).
Figure 2.
An rpoA amino acid sequence alignment using PILEUP (GCG Sequence Analysis Package v.8.0). Conserved amino acids in Euglena spp. are indicated by a black background, while similar amino acids are indicated by a yellow background. Similar amino acids are defined as E and D, F and Y, R and K, L, V, I and M, and S and T. Locations of introns are indicated by vertical arrows. The amino acids that were chosen randomly for the Euglena consensus sequence are shown outlined. The consensus of the N-terminal sequence from 34 species of bacteria, algae and land plants is shown. Conserved amino acids between the two consensus sequences are shown and similar amino acids are indicated by a + sign. The secondary structure of the αNTD is indicated schematically above the α sequences; helices H1–H3 are indicated by rectangles; strands S1–S11 are indicated by arrows. Black dots denote residues that participate in the hydrophobic core of the α dimer interface and green dots indicate mutations that cause defects in binding to other subunits that were previously reported for E.coli (14).
Identification of the rpoA genes and introns from the plastid genomes of five additional Euglena protists
In order to confirm the identification of a new Euglena plastid gene and to identify conserved protein sequence motifs, rpoA loci from other Euglena species were isolated. PCR primers P1 and P2, chosen to complement predicted conserved regions in the flanking ycf9 and trnP genes (Fig. 1), were used for amplification of DNAs from E.anabaena, E.granulata, E.myxocylindracea, E.stellata and E.viridis. A single major product 1.2–1.4 kb in size was obtained from each PCR reaction. This product was cloned and sequenced (GenBank accession nos AY047483–AY047487). Each DNA sequence encoded the 3′-end of the ycf9 gene, a complete rpoA locus and the trnS gene, distal to rpoA and coded on the opposite strand.
To identify rpoA introns, RT–PCR reactions were carried out with primers specific for each rpoA gene. The resulting cDNAs were cloned and sequenced on both strands. Locations for all splice boundaries were determined (GenBank accession nos AY047483–AY047487). The rpoA loci in the five new sequences are each interrupted by three group III introns at conserved locations within the coding region (Fig. 1D). The single E.gracilis group III intron corresponds in location to the first intron in the other five species.
Identification of an rpoA gene in A.longa
Astasia longa is a colorless protist with a plastid genome closely related to that of E.gracilis. The 73 kb genome of A.longa encodes nearly the same set of non-photosynthetic genes as E.gracilis (6). To search for an A.longa plastid rpoA locus, a BLAST search of A.longa plastid DNA (accession no. ALO294725) with the E.gracilis rpoA cDNA sequence as the query was performed. The A.longa rpoA locus was identified as part of a small operon distal to the rpl20 ribosomal protein gene. As in all other Euglena species described above (Fig. 1C), the trnP and trnS cistrons flank rpoA at the 3′-end. The A.longa gene is split by a single group III intron of 111 nt, located at the same position within the coding region as the single E.gracilis group III intron. Astasia longa lacks the genes for the photosynthetic proteins ycf9 and psaC that are upstream of E.gracilis rpoA.
Identification of the rpoA genes from seven Euglena protists
The derived amino acid sequences from the putative rpoA genes of E.anabaena, E.granulata, E.myxocylindracea, E.stellata, E.viridis and A.longa were each used to perform a BLAST query of both the non-redundant protein database and the PFAM database of conserved protein domains. Significant alignments (E value between 7e-03 and 5e-05) were found between E.granulata, E.myxocylindracea, E.stellata and E.viridis and both eubacterial and plastid rpoA genes (not shown). In the PFAM search, all sequences except E.anabaena rpoA showed a highly significant alignment only with the bacterial RNA polymerase α-chain family (accession no. PF01000). The E values for these alignments (9) in increasing order are E.myxocylindracea (4e-09), E.stellata (5e-09), E.granulata (3e-07), E.viridis (5e-06), E.gracilis (6e-04) and A.longa (2.2e-02). For the four species with the highest E values, there were 23–26% identical residues compared to the conserved PFAM domain over a span of 180–197 residues. When similar residues were considered, the match was 41–46% positive for these same four sequences.
Comparative analysis of the rpoA-like gene from seven Euglena protists
An alignment of the amino acid sequences of all seven rpoA genes was created using Clustal W. All sequences aligned with each other, in agreement with their individual alignments with the PFAM consensus sequence. Each protist sequence is predicted to begin with a methionine codon, except A.longa and E.myxocylindracea rpoA, which appear to begin with a leucine codon. The conceptual translation products from the Euglena species contained between 204 and 221 amino acids.
The multiple alignment for the seven Euglena plastid rpoA subunit sequences is shown in Figure 2. For comparison, the conserved αNTD of the bacterial RNA polymerase α-chain family and the predicted secondary structure for this family (14) are also shown. All Euglena plastid sequences, including E.anabaena, align with each other and with the conserved domain sequence. Among the seven protist sequences, there are only five invariant residues. As with E.gracilis rpoA, the most notable difference between the euglenoid rpoA sequences and the PFAM consensus is the absence of a C-terminal domain and a linker between the two domains.
Group III introns
Analysis of the rpoA loci from A.longa and the six Euglena species has led to the identification of 17 new group III introns. All seven species have an intron at the same location within the coding region for helix 1 of domain 1 (Fig. 2). The five new Euglena sequences all have two additional introns at the same locations within the coding regions for β-sheets 9 and 10. All 17 introns are typical group III introns of the chloroplast genomes of Euglena. The lengths of the 17 introns vary from 93 to 121 nt, with an average of 99 nt. The base compositions range from 79 to 93% A + U, with an average of 85%. All but two of the introns have the conserved 5′-splice boundary motif 5′-NUNNG, with the G at position +5 being invariant. All 17 introns have a group II intron-like domain 6 at their 3′-end, with the bulging A involved in branch point formation during splicing found at position –8 or –7 from the 3′-splice site (not shown). The remaining internal sequences between the 5′-splice boundary and 3′ domain 6 are highly variable, both among introns in the same species and among the same intron in different species.
Identification of evolutionarily conserved genes flanking rpoA
The rpoA locus of five different Euglena species was amplified from primers within the flanking trnP and ycf9 genes. As a result, all of the new DNA sequences also contain the coding region for the 32 C-terminal codons of five ycf9 loci. The ycf9 locus encodes a novel structural component of the photosystem II (PSII) light-harvesting complex (LHC), which appears to be required for stable integration of the pigment-binding LHC protein CP26 into the antenna of PSII (15–17). As is typical for photosynthetic membrane proteins, the partial ycf9 gene is much more highly conserved than rpoA (data not shown). The PCR primer site in the trnP gene is within a dicistronic trnP–trnS cluster found in both E.gracilis (4) and A.longa (6). The same dicistronic trnP–trnS locus was also present in the five additional Euglena species. Each of the six Euglena and the single Astasia trnS gene has a UGA anticodon and large variable loop of 16–18 nt.
Euglena gracilis rpoA RNA transcript analysis
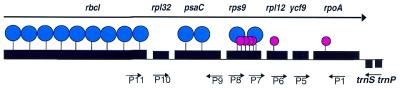
To determine if the E.gracilis rpoA gene is transcribed from a promoter in the ycf9–rpoA intercistronic region, experiments designed to detect the 5′-end of a primary transcript were performed. These experiments all gave negative results (data not shown). In order to determine if rpoA could be transcribed with an upstream cistron(s), a series of RT–PCR experiments were performed. Each of the reactions was designed with at least one intron internal to the target sequences to ensure that spliced cDNA and not genomic DNA was being amplified. A map of the cDNA and PCR primer locations for this experiment is shown in Figure 3. cDNA synthesized from primer P1 in rpoA exon 2 was amplified with primers P5 in ycf9, P6 in exon 2 of rpl12 and P7 in exon 7 of rps9. Each reaction gave a product corresponding in size to the corresponding fully spliced di-, tri- and tetracistronic cDNAs. The identities of the fully spliced ycf9–rpoA, rpl12–ycf9–rpoA and rps9–rpl12–ycf9–rpoA cDNAs were confirmed by cloning and sequencing on both strands (data not shown). To determine if any additional upstream genes could also be co-transcribed with rpoA, cDNA was synthesized from primer P8 in rps9 exon 1 and P9 in psaC exon 3. These cDNAs were amplified, respectively, with primers P10 in rpl32 and P11 in rbcL. The products of these reactions, confirmed by cloning and sequencing (data not shown), corresponded to the expected, spliced segments of the rpl32–psaC–rps9 cistrons and the rbcL–rpl32–psaC polycistronic mRNAs. From these RT–PCR experiments it appears that rpoA can be transcribed and spliced as the distal cistron of a polycistronic mRNA containing the rbcL–rpl32–psaC–rps9–rpl12–ycf9–rpoA genes. Although large operons and operons encoding genes for different functional complexes have previously been described for chloroplast genomes from Euglena and other species, this RNA is unusual for the diversity of functional components. We cannot rule out the possibility that the large, fully spliced, polycistronic mRNA detected by RT–PCR represents a minor fraction of rpoA transcripts and exists due to inefficient termination of upstream transcripts.
Figure 3.
The E.gracilis chloroplast rbcL–rpoA operon. The Euglena chloroplast gene is encoded in the rbcL operon, which is ∼10.6 kb in size. The arrow indicates the direction of transcription. Boxes denote genes of the operon. Large blue lollipops indicate Group II introns, while small red lollipops indicate Group III introns.
Phylogenetic analysis
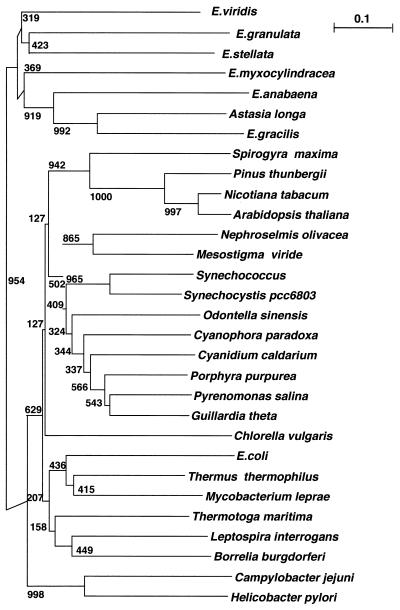
The amino acid sequences encoding the α-subunits of RNA polymerase in seven euglenoid species and 23 species of bacteria, algae and land plants were aligned using Clustal W and then manually aligned with Euglena spp. Within RNA polymerase, 360 characters (278 parsimony informative) were found. Phylogenetic relationships were determined using the NJ distance. The NJ tree is displayed, with the bootstrap values shown near the nodes (Fig. 4). The full tree showed the presence of four clades, Euglena spp., bacteria, algae and land plants. The Euglena rpoA clade is well resolved from all other rpoA genes. Based on phylogeny, E.gracilis and A.longa are very closely related and represent the most derived branch in the euglenoid phylogeny. Euglena anabaena and E.myxocylindracea are basal to E.gracilis and A.longa. The highest bootstrap numbers appear for the E.gracilis, A.longa and E.anabaena branches. The branching order in Euglena spp. was not well resolved due to low bootstrap numbers. The separation of bacteria, algae, plants and Euglena spp. into different clades using rpoA phylogeny is supported by strong bootstrap numbers. The closest bacterium to Euglena spp. was H.pylori, which also had a high BLAST score when compared to E.gracilis. Green algae were the only phylogenetic group which belonged to two clades. One cryptophyte green alga, S.maxima, was basal in the plant clade. Chlorella vulgaris was the closest branched alga to Euglena spp. and has separate branching in the algae clade. Two other green algae, N.olivacea and M.viride, were branched together in the clade with other algae.
Figure 4.
Phylogenetic NJ tree inferred from a rpoA sequence comparison. Branch lengths are proportional to the expected mean number of substitutions per site along the branch, as quantified by the scale bar. The tree was generated using CLUSTAL X (GCG Sequence Analysis Package v.8.0). Boostrap numbers for the 1000 boostrap replications conducted are indicated.
Due to the large data set, parsimony and maximum likelihood analysis using the branch-and-bound method were performed only for Euglena spp., with H.pylori as the outgroup. Euglena gracilis, A.longa and E.anabaena had the same branching as in the NJ tree, but for other Euglena spp. branching was not resolved (not shown).
DISCUSSION
Protein structure
The best studied rpoA gene product is the RNA polymerase α-subunit from E.coli, which consists of two structural domains. The αNTD contains determinants for dimerization and is an initiator for assembly into RNA polymerase (14,18–21). The C-terminal domain plays a role in transcription initiation (22–26).
When the RNA polymerase α-subunit PFAM consensus sequence and the domain and secondary structure elements from E.coli are compared (Fig. 3), the key features of the rpoA gene products of Euglena chloroplasts can be described. Despite the highly variable sequences, the αNTD is conserved in all seven Euglena species, which have all the main structural motifs. Domain 1, which functions in the α–α interaction (14), is better conserved in Euglena spp. than domain 2. The least conserved region occurs at the end of domain 2, which had high variability among the species. Only the green algae, specifically C.vulgaris and N.olivacea, had an additional 28 and 45 amino acids, respectively, in this region, whereas Euglena spp. were missing this variable region completely (∼20 amino acids). Because 10 of the 14 amino acids participating in the dimer interaction in E.coli are present in Euglena spp., we predict dimer formation in Euglena spp. as well. The presence of amino acids known to interact with other subunits is in accord with a multisubunit structure of RNA polymerase in Euglena spp.
The absence of a C-terminal domain in the rpoA gene of Euglena spp. is a novel feature, previously reported only for C.vulgaris. Based on absence of the C-terminal domain in C.vulgaris and possible absence of the rpoA gene in Euglena and Epifagus, it had been suggested that rpoA may not be functional (27) in chloroplasts. Of the two types of RNA polymerase, we now know that the one with an α-subunit is responsible for transcription of photosynthetic genes. Therefore, the absence of rpoA in Epifagus, a parasitic plant whose plastid genome is missing photosynthetic genes, can be explained by the absence of a target promoter for this type of RNA polymerase. The rpoA in Euglena spp. has been the missing component of the transcription system for photosynthetic genes. rpoA is clearly present in chloroplast genomes, including that of A.longa (6).
As was shown for E.coli, the αCTD participates in transcription regulation by interaction with a group of protein transcription factors and DNA enhancer (UP) elements. Absence of the αCTD in C.vulgaris but not in other studied algae and Euglena spp. and presence of the αCTD in other chloroplast genomes could suggest that some recent changes have occurred in the chlorophyte branch, as Euglena spp. are the sister branch to green algae (28). The linker between the two domains was also lost as its function was associated with flexibility and binding of the C-terminal domain (13).
Some functional changes may have preceded loss of the C-domain. Escherichia coli contains different types of promoters. Those associated with higher activity have two properties linked to the αCTD: (i) binding of the αCTD to the promoter UP element distal to the –35 sequence (23,29); (ii) binding of the αCTD to CRP proteins, which bind as a dimer to 22 bp sequences that are located at a variety of positions upstream of the promoter –35 element, centered near position –61.5, –71.5, –81.5 or –91.5 (30,31).
Although rpoA genes are highly variable, the amino acid sequence of the rpoA gene was a good candidate for phylogenetic studies in Poaceae spp. (32). On the basis of a phylogenetic tree, separate clades for bacteria, algae and higher plants and Euglena spp. were obtained. In this study we find that the Euglena rpoA genes form a fourth, distinct clade in the rpoA phylogenetic tree (Fig. 4). The most distinguishing feature was that the Euglena rpoA clade had the most variability when compared to other clades. Previously, more robust chloroplast phylogenies had been obtained when many genes were analyzed together. Mesostigma was positioned as the earliest divergence in the phylogeny of green plants (33). In our study green algae have the most variable standing in the rpoA-based phylogenetic tree. Spirogyra maxima is the only green alga which is in the same clade as plants. In a study of 45 chloroplast proteins, NJ analysis resulted in a grouping of Euglena and chlorophytes, diatoms and rhodophytes (28), which is similar to our rpoA tree. The cryptophyte alga G.theta was shown to have a common ancestry with the red algae, which are also grouped together in our analysis (34).
The bootstrap numbers for euglenoid branching are low in the rpoA tree (Fig. 4). Euglena gracilis and A.longa have the highest bootstrap number branching and are sister taxa. The close evolutionary relationship between E.gracilis and A.longa is supported by all previous studies (8,35,36). Astasia longa, a non-photosynthetic organism, is missing all photosynthetic genes except rbcL. The organization of common genes is completely different in the two species. A comparison of this gene order is commonly considered more reliable to determine evolutionary relationships among plastids than sequence comparisons because genes can have different mutation rates (37). However, the preservation of gene order in closely related species does not apply to E.gracilis and A.longa, possibly due to the extensive rearrangements that accompanied loss of photosynthetic genes in A.longa. In most land plants the chloroplast genomes display great similarities in gene order, although occasional variations appear due to inversions of large sequences (38). Again, green algae show high variability of gene order (39).
Euglena anabaena has relatively high bootstrap branching in the Euglena clade, while the other branches displayed low bootstrap numbers due to the high variability of this gene. Previous phylogenetic studies with euglenoids based on rbcL also gave weak bootstrap support (8). The phylogenetic tree based on the rpoA gene has some differences from our previous results based on other genes (35,36). The low bootstrap numbers do not support any preferred phylogeny. However, some common features were present in all studies: branching of E.myxocylindracea is close to E.gracilis and branching of E.stellata is one of the most basal.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Jane Dugas for technical assistance with the manuscript. Thanks are also due to Nesrin Kuscuoglu and Richard De Armond for their technical assistance. This work has been supported by NIH grant GM35665.
REFERENCES
- 1.Greenberg B.M., Narita,J.O., DeLuca-Flaherty,C., Gruissem,W., Rushlow,K.A. and Hallick,R.B. (1984) Evidence for two RNA polymerase activities in Euglena gracilis chloroplasts. J. Biol. Chem., 259, 14880–14887. [PubMed] [Google Scholar]
- 2.Hess W.R. and Borner,T. (1999) Organellar RNA polymerases of higher plants. Int. Rev. Cytol., 190, 1–59. [DOI] [PubMed] [Google Scholar]
- 3.Santis-MacIossek G., Kofer,W., Bock,A., Schoch,S., Maier,R.M., Wanner,G., Rudiger,W., Koop,H.U. and Herrmann,R.G. (1999) Targeted disruption of the plastid RNA polymerase genes rpoA, B and C1: molecular biology, biochemistry and ultrastructure. Plant J., 18, 477–489. [DOI] [PubMed] [Google Scholar]
- 4.Hallick R.B., Hong,L., Drager,R.G., Favreau,M.R., Montfort,A., Orsat,B., Spielmann,A. and Stutz,E. (1993) Complete sequence of Euglena gracilis chloroplast DNA. Nucleic Acids Res., 15, 3537–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Finn,R.D. and Sonnhammer,E.L.L. (1999) Pfam 3.1: 1313 multiple alignments match the majority of proteins. Nucleic Acids Res., 27, 260–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gockel G. and Hachtel,W. (2000) Complete gene map of the plastid genome of the nonphotosynthetic euglenoid flagellate Astasia longa. Protist, 151, 347–351. [DOI] [PubMed] [Google Scholar]
- 7.Edelman M., Hallick,R. and Chua,N. (1982) Methods in Chloroplast Biology. Elsevier Biomedical Press, Amsterdam, The Netherlands, p. 42.
- 8.Thompson M.D., Copertino,D.W., Thompson,E., Favreau,M.R. and Hallick,R.B. (1995) Evidence for the late origin of introns in chloroplast genes from an evolutionary analysis of the genus Euglena. Nucleic Acids Res., 23, 4745–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saitou N. and Nei,M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol., 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 11.Blatter E.E., Ross,W., Tang,H., Gourse,R.L. and Ebright,R.H. (1994) Domain organization of RNA polymerase alpha subunit: C-terminal 85 amino acids constitute a domain capable of dimerization and DNA binding. Cell, 78, 889–896. [DOI] [PubMed] [Google Scholar]
- 12.Negishi T., Fujita,N. and Ishihama,A. (1995) Structural map of the alpha subunit of Escherichia coli RNA polymerase: structural domains identified by proteolytic cleavage. J. Mol. Biol., 248, 723–728. [DOI] [PubMed] [Google Scholar]
- 13.Meng W., Savery,N.J., Busby,S.J. and Thomas,M.S. (2000) The Escherichia coli RNA polymerase alpha subunit linker: length requirements for transcription activation at CRP-dependent promoters. EMBO J., 19, 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang G. and Darst,S.A. (1998) Structure of the Escherichia coli RNA polymerase alpha subunit amino-terminal domain. Science, 281, 262–266. [DOI] [PubMed] [Google Scholar]
- 15.Ruf S., Biehler,K. and Bock,R. (2000) A small chloroplast-encoded protein as a novel architectural component of the light-harvesting antenna. J. Cell Biol., 149, 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maenpaa P., Gonzalez,E.B., Chen,L., Khan,M.S., Gray,J.C. and Aro,E.M. (2000) The ycf 9 (orf 62) gene in the plant chloroplast genome encodes a hydrophobic protein of stromal thylakoid membranes. J. Exp. Bot., 51, 375–382. [DOI] [PubMed] [Google Scholar]
- 17.Swiatek M., Kuras,R., Sokolenko,A., Higgs,D., Olive,J., Cinque,G., Muller,B., Eichacker,L.A., Stern,D.B., Bassi,R., Herrmann,R.G. and Wollman,F.A. (2001) The chloroplast gene ycf9 encodes a photosystem II (PSII) core subunit, PsbZ, that participates in PSII supramolecular architecture. Plant Cell, 13, 1347–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Igarashi K., Hanamura,A., Makino,K., Aiba,H., Aiba,H., Mizuno,T., Nakata,A. and Ishihama,A. (1991) Functional map of the alpha subunit of Escherichia coli RNA polymerase: two modes of transcription activation by positive factors. Proc. Natl Acad. Sci. USA, 88, 8958–8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura M., Fujita,N. and Ishihama,A. (1994) Functional map of the alpha subunit of Escherichia coli RNA polymerase. Deletion analysis of the amino-terminal assembly domain. J. Mol. Biol., 242, 107–115. [DOI] [PubMed] [Google Scholar]
- 20.Kimura M. and Ishihama,A. (1995) Functional map of the alpha subunit of Escherichia coli RNA polymerase: insertion analysis of the amino-terminal assembly domain. J. Mol. Biol., 248, 756–767. [DOI] [PubMed] [Google Scholar]
- 21.Kimura M. and Ishihama,A. (1995) Functional map of the alpha subunit of Escherichia coli RNA polymerase: amino acid substitution within the amino-terminal assembly domain. J. Mol. Biol., 254, 342–349. [DOI] [PubMed] [Google Scholar]
- 22.Igarashi K. and Ishihama,A. (1991) Bipartite functional map of the E. coli RNA polymerase alpha subunit: involvement of the C-terminal region in transcription activation by cAMP-CRP. Cell, 65, 1015–1022. [DOI] [PubMed] [Google Scholar]
- 23.Ross W., Gosink,K.K., Salomon,J., Igarashi,K., Zou,C., Ishihama,A., Severinov,K. and Gourse,R. (1993) A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science, 262, 1407–1413. [DOI] [PubMed] [Google Scholar]
- 24.Jeon Y.H., Negishi,T., Shirakawa,M., Yamazaki,T., Fujita,N., Ishihama,A. and Kyogoku,Y. (1995) Solution structure of the activator contact domain of the RNA polymerase alpha subunit. Science, 270, 1495–1497. [DOI] [PubMed] [Google Scholar]
- 25.Gaal T., Ross,W., Blatter,E.E., Tang,H., Jia,X., Krishnan,V.V., Assa-Munt,N., Ebright,R.H. and Gourse,R.L. (1996) DNA-binding determinants of the alpha subunit of RNA polymerase: novel DNA-binding domain architecture. Genes Dev., 10, 16–26. [DOI] [PubMed] [Google Scholar]
- 26.Kainz M. and Gourse,R.L. (1998) The C-terminal domain of the alpha subunit of Escherichia coli RNA polymerase is required for efficient rho-dependent transcription termination. J. Mol. Biol., 284, 1379–1390. [DOI] [PubMed] [Google Scholar]
- 27.Wakasugi T., Nagai,T., Kapoor,M., Sugita,M., Ito,M., Ito,S., Tsudzuki,J., Nakashima,K., Tsudzuki,T., Suzuki,Y., Hamada,A., Ohta,T., Inamura,A., Yoshinaga,K. and Sugiura,M. (1997) Complete nucleotide sequence of the chloroplast genome from the green alga Chlorella vulgaris: the existence of genes possibly involved in chloroplast division. Proc. Natl Acad. Sci. USA, 94, 5967–5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adachi J., Waddell,P.J., Martin,W. and Hasegawa,M. (2000) Plastid genome phylogeny and a model of amino acid substitution for proteins encoded by chloroplast DNA. J. Mol. Evol., 50, 348–358. [DOI] [PubMed] [Google Scholar]
- 29.Tagami H. and Aiba,H. (1999) An inactive open complex mediated by an UP element at Escherichia coli promoters. Proc. Natl Acad. Sci. USA, 96, 7202–7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishihama A. (1993) Protein-protein communication within the transcription apparatus. J. Bacteriol., 175, 2483–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Busby S. and Ebright,R.H. (1999) Transcription activation by catabolite activator protein (CAP). J. Mol. Biol., 293, 199–213. [DOI] [PubMed] [Google Scholar]
- 32.Petersen G. and Seberg,O. (1997) Phylogenetic analysis of the Triticeae (Poaceae) based on rpoA sequence data. Mol. Phylogenet. Evol., 7, 217–230. [DOI] [PubMed] [Google Scholar]
- 33.Lemieux C., Otis,C. and Turmel,M. (2000) Ancestral chloroplast genome in Mesostigma viride reveals an early branch of green plant evolution. Nature, 403, 649–652. [DOI] [PubMed] [Google Scholar]
- 34.Douglas S.E. and Penny,S.L. (1999) The plastid genome of the cryptophyte alga, Guillardia theta: complete sequence and conserved synteny groups confirm its common ancestry with red algae. J. Mol. Evol., 48, 236–244. [DOI] [PubMed] [Google Scholar]
- 35.Doetsch N.A., Thompson,M.D. and Hallick,R.B. (1998) A maturase-encoding group III twintron is conserved in deeply rooted euglenoid species: are group III introns the chicken or the egg? Mol. Biol. Evol., 15, 76–86. [DOI] [PubMed] [Google Scholar]
- 36.Doetsch N.A., Thompson,M.D., Favreau,M.R. and Hallick,R.B. (2001) Comparison of psbK operon organization and group III intron content in chloroplast genomes of 12 Euglenoid species. Mol. Gen. Genet., 64, 682–690. [DOI] [PubMed] [Google Scholar]
- 37.Kowallik K.V. (1997) Origin and evolution of chloroplasts: current status and future perspectives. In Schenk,H.E.A., Herrmann,R.G., Jeon,K.W., Müller,N.E. and Schwemmler,W. (eds), Eukaryotism and Symbiosis: Intertaxonic Combination versus Symbiotic Adaptation. Springer Verlag, Berlin, Germany, pp. 3–23.
- 38.Palmer J.D. (1991) Plastid chromosomes: structure and evolution. In Bogorad,L. and Vasil,I.K. (eds), Cell Culture and Somatic Cell Genetics of Plants: The Molecular Biology of Plastids. Academic Press, San Diego, CA, Vol. 7a, pp. 5–53.
- 39.Boudreau E., Otis,C. and Turmel,M. (1994) Conserved gene clusters in the highly rearranged chloroplast genomes of Chlamydomonas moewsii and Chlamydomonas reinhardtii. Plant Mol. Biol., 24, 585–602. [DOI] [PubMed] [Google Scholar]