Abstract
Oral cancer is a prevalent malignancy that affects the oral cavity in the region of head and neck. The study of oral malignant lesions is an essential step for the clinicians to provide a better treatment plan at an early stage for oral cancer. Deep learning based computer-aided diagnostic system has achieved success in many applications and can provide an accurate and timely diagnosis of oral malignant lesions. In biomedical image classification, getting large training dataset is a challenge, which can be efficiently handled by transfer learning as it retrieves the general features from a dataset of natural images and adapted directly to new image dataset. In this work, to achieve an effective deep learning based computer-aided system, the classifications of Oral Squamous Cell Carcinoma (OSCC) histopathology images are performed using two proposed approaches. In the first approach, to identify the best appropriate model to differentiate between benign and malignant cancers, transfer learning assisted deep convolutional neural networks (DCNNs), are considered. To handle the challenge of small dataset and further increase the training efficiency of the proposed model, the pretrained VGG16, VGG19, ResNet50, InceptionV3, and MobileNet, are fine-tuned by training half of the layers and leaving others frozen. In the second approach, a baseline DCNN architecture, trained from scratch with 10 convolution layers is proposed. In addition, a comparative analysis of these models is carried out in terms of classification accuracy and other performance measures. The experimental results demonstrate that ResNet50 obtains substantially superior performance than selected fine-tuned DCNN models as well as the proposed baseline model with an accuracy of 96.6%, precision and recall values are 97% and 96%, respectively.
Keywords: Transfer learning, Deep learning, Oral cancer, Oral squamous cell carcinoma, Convolutional neural network, Histopathology
1. Introduction
Oral cancer belongs to the class of head and neck cancers that account for around 3% of all types of cancers diagnosed worldwide [1]. It is listed as the sixth most predominant cancer happening world wide [2]. The primary causes of this disease are the habits of chewing tobacco, betel nuts, and smoking, which affect the oral cavity, nasopharynx, and pharynx regions [3]. Oral cancer is common among people, mostly from developing countries, due to unawareness, the limited facility for clinical diagnosis, and lack of oral specialists especially in south Asian countries [4]. A patient with oral cancer has various symptoms such as difficulties while eating, speaking, inflammations within the mouth, and visible marks on the face. Rapid progression of the disease leads to lining formation in squamous cells resulting in OSCC that leads to a threat to life [4]. Thus, an early diagnosis of these oral malignant lesions is required to reduce the chance of cancerous transformation. In literature, it has been reported that oral cancer is having a high risk of recurrence [5]. Therefore, an in-depth analysis of its occurrence and progression is necessary for its prognosis. A five-year oral cancer survival report indicates a prognostic rate of around 35% to 50% which includes preferable analysis of different pathological aspects relevant to OSCC for increasing the rate of disease survival [6]. Consequently, from the pathologist's point of view, it is essential to provide a precise histological classification of oral lesions.
In traditional approaches, pathologist's findings are subjective due to various factors such as microscope model, staining quality, time taken by a pathologist for analysis of each slide, and his experience over the time [5]. In consequence, the diagnostic findings may lead to error prone and delay in follow up procedure. In contrast to this, the deep learning-based approaches do not depend on the domain experts for hand crafted feature engineering and yield high classification accuracy [7], [8]. In the domain, for a precise diagnosis, biopsy images considered as the pathological gold standard are limited; in such cases, training deep learning models for prominent features using a limited number of image samples is not preferable. Dealing with this problem, researchers have successfully applied transfer learning with deep learning models with a limited number of biopsy images, which signifies the successful implementation of the pretrained models for the classification task [9]. Besides, transfer learning reduces the training time as training a deep neural network from scratch on a complex problem can often take days or even weeks [10]. The previous study proposed by Navarun et al. [11], has verified that by applying transfer learning with replacing the classification layers only, leads to lower the performance of the pretrained models compared to the CNN model trained from scratch. In this study, we have incorporated the notion of half training the layers of pretrained models to increase the efficiency of transfer learning with the small dataset.
In this work, the histopathological images of oral lesions are considered to analyze the potential malignancy by applying deep learning models utilizing transfer learning. This paper aims to utilize the information from a source dataset to increase the learning effectiveness or to decrease the training sample size in the target dataset. This study focuses on the efficiency of transfer learning by fine-tuning the existing deep learning models with the OSCC images for binary classification in comparison to a model built from scratch. Fine-tuning has been done by considering 50% of the frozen layers of the DCNN models. Thus half of the model's layers is allowed to train with the OSCC images. Besides, a baseline DCNN with a less complex archetecture is proposed which can efficiently differentiate between malignant and benign tumors.
The following are the key contributions of this article:
-
1.
Transfer learning assisted deep convolutional neural networks (DCNNs), such as pretrained VGG16, VGG19, ResNet50, InceptionV3, and MobileNet, are fine-tuned by freezing initial half of the layers and training the remaining half of it with OSCC data.
-
2.
A baseline DCNN architecture, trained from scratch with 10 convolution layers is proposed.
-
3.
A comparative analysis of these models is carried out in terms of classification accuracy and other performance measures.
-
4.
Time taken for each deep learning model for training and prediction is compared to find out optimal model.
The remainder of the article is systematically arranged as follows. In section 2, the background of the transfer learning and types of DCNN are presented. Related works for OSCC classification are discussed in section 3. The material and methods for experimentation as well as model structures and parameters are represented in section 4. Comparison among different deep learning models and experimental results are represented in section 5. Section 6 represents the discussion of the results and the time taken for training and prediction of the models. Finally, the section 7 represents conclusion with future work.
2. Background
Recent advancements in Artificial Intelligence (AI) have proven that deep learning models provide promising outcomes in comparison with conventional (shallow) machine learning algorithms in different domains [12], [13]. In the ImageNet Large Scale Visual Recognition Competition (ILSVRC), an annual competition for object classification tasks [14], the winners have mostly used deep neural network architectures to classify ImageNet data, especially convolutional neural networks (CNNs). The exceptional performance of CNNs at the competition, to classify ImageNet data, has been proven its magnificent ability [14], [15]. Through ILSVRC, many state-of-the-art DCNN models have been evolved for different computer vision applications, such as AlexNet [16], GoogLeNet [17], Xception [18], VGGNet [19], ResNet50 [20], MobileNet [21]. These DCNNs are useful for learning the patterns from the images and are able to classify using these patterns, thereby eliminating the need for manual feature extraction [22], [23], [24]. We have explored most studied CNN architectures and selected VGG16, VGG19, ResNet50, InceptionV3 and MobileNet which are the widely used models for the image recognition tasks. The selection of these models is based on the work proposed by the authors previously and found that these models are performing better. These models are similar to those of the successful CNN architectures on the ImageNet challenge, with extensive research in several computer vision benchmarks. The considered architectures have gained popularity for applying transfer learning to save training time with small image dataset. The selected relevant architectures related to our work are described briefly in following subsections.
VGG16 and VGG19
VGG16 and VGG19 are variants of VGGNet proposed by the Visual Geometry Group [19]. VGG16 and VGG19 having 16 and 19 layers, are the most commonly used VGG models. The convolutional layers of these networks use size kernels and max-pooling layers are of size. VGG19 has 16 convolutional layers, 3 fully connected layers, 5 MaxPool layers, and 1 SoftMax layer; however, VGG16 has 13 convolutional layers, 3 fully connected layers, 5 MaxPool layers, and 1 SoftMax layer. Thus, the architectural difference between these two is that in each of the three convolutional blocks, VGG19 has one more convolutional layer.
InceptionV3
Szegedy et al. [25] have developed a different type of CNN architecture called inception model. It differs from the regular CNN as it contains Inception blocks which means adding the same input tensor with several filter sizes and combining their outputs. InceptionV3 is an enhanced variant of InceptionV1 and InceptionV2 proposed by Szegedy et al. with additional parameters. It consists of parallel convolutional layers with 3 different filter sizes (, , ) and MaxPool layer. The outputs of each parallel convolutional layer are combined and delivered to the succeeding inception module. This enables the model to take the advantage of multilevel feature extraction (extracts general () and local () features at the same time) with multiple filters thereby enhancing the performance of the model.
ResNet50
ResNet50 [20] is an acronym for the residual network proposed by He et al. in 2015 and he won the ILSVRC (ImageNet) competition. It is a type of convolutional neural network used for image classification. The main purpose of this network is to introduce a new residual layer architecture. ResNet50 consists of five stages and each of which includes a convolution block and an identity block. Each of the convolution block and the identity block consists of three convolution layers. In literature, deeper CNN architectures are developed for solving more complex tasks and improved classification accuracy. However, developing a deeper network increases the complexity of the training process resulting in accuracy degradation or saturation. These issues are resolved by the residual network which learns from residuals. ResNet50 does this using a direct connection from the nth layer's input to some ()th layer, which makes training easier.
MobileNet
MobileNet [21] is a CNN model used for image classification and mobile vision. It needs less computational power to run and apply transfer learning. This makes it ideal for mobile devices, embedded systems, and GPU-free computers or low computationally efficient systems with better performance. The key difference between MobileNet and typical CNN architecture is that instead of a single 3x3 convolution layer, the batch norm, and ReLU are used. It splits the convolution into a 3x3 depth-wise convolution and a 1x1 pointwise convolution to build lightweight deep neural networks. Mainly, It conducts one convolution for each color channel instead of merging all three and flattening them.
3. Related works
In the last decade, the use of DCNNs in medical image processing and applications has been an emerging area of research for disease diagnosis and prognosis [26], [27], [28].
Based on the capability of handling natural image classification, some of the pretrained DCNN models are selected for this study to handle medical images (see Table 1). These models have learned to extract potent and valuable features from the ImageNet database of 1000 image categories. As the models are trained on huge datasets, a better depiction of low-level features such as edges, rotation, lights, shapes have been learned and can be shared in order to facilitate knowledge transfer. As a result, these models work effectively as feature extractors for new images in a variety of computer vision applications [14], [16]. Table 1 depicts the number of layers (depth) and parameters of the selected DCNN models. Here, the depth signifies the number of convolutional layers present in the network. The models support RGB images for varied width (W), height (H), and a fixed depth (D) of 3. The effectiveness of transfer learning for detecting oral lesions by using these pretrained DCNN models is shown in this analysis.
Table 1.
Abridgment of pretrained DCNNs models employed for this study.
In literature, researchers have performed oral cancer classification by applying machine learning and deep learning techniques. Kim et al. [29] and Mohd et al. [30] have done the retroactive study for early detection of oral cancer and predicted the survival rate. Most of the research have been carried out using machine learning techniques to detect oral submucous fibrosis (OSF) [31], [32], [33], [34]. Studies done by T. Y. Rahman et al. [35], [36] are based on the binary classification of OSCC. The previous studies have reported that by using Support Vector Machine (SVM) and Linear Discriminant Classifier (LDA), 100% classification accuracy was achieved. This accuracy is due to the handcrafted feature extraction technique and the dataset being used is very small (in the range of 10 to 100).
Navarun et al. [11] have used the transfer learning notion for the categorization of four types of OSCC with four pretrained DCNN models and compared them with a proposed 10-layered CNN model. They have replaced the fully connected layers for random weight initialization to relearn from the oral cancer histopathological images. Thus, performance of the model trained from scratch surpassed the pretrained models. Since the authors have replaced only the classification layers of the pretrained models, it is observed that the performance of the transfer learning based models is lower in comparison to the base CNN model. In this study, we have focused on binary categorizations of oral histopathological images using transfer learning technique by freezing the initial half of the layers of each network and training the remaining half. Freezing of 50% is done to achieve the benefit of both i.e., training half of the layers, exploring the benefits of learning from scratch, as well as, availing of the benefits of transfer learning. As per our knowledge, no such work on histopathological images of oral cancer exists.
4. Materials and methods
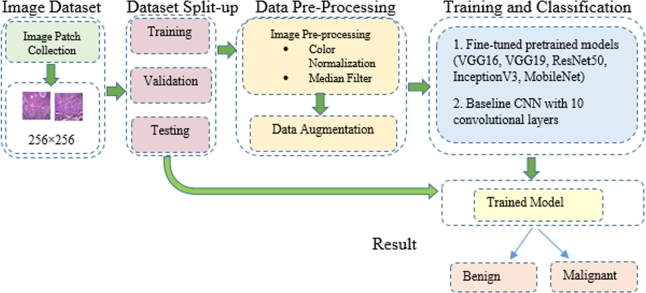
The present work focuses on binary classification of oral histopathological images through the transfer learning technique and a suggested CNN model. The detection process is carried out in four stages as shown in Fig. 1. Initially, histopathological images of OSCC were collected and then split into training, validation and testing. The training dataset was preprocessed followed by data augmentation. In the final stage, ImageNet pretrained networks were fine-tuned for the OSCC image dataset. To handle binary classification, the fully connected layers of DNN architectures are modified accordingly to achieve binary predicted labels of oral histopathological images as benign or malignant. The proposed CNN model was also trained for the same dataset.
Figure 1.
Proposed approach for binary classification of oral lesions.
4.1. Image dataset
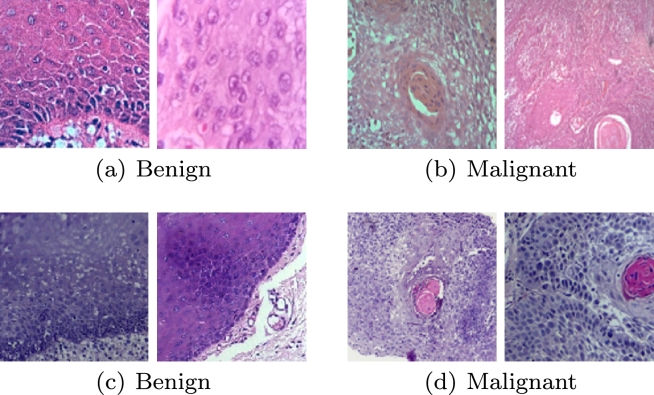
Histological hematoxylin and eosin-stained sections of normal mucosa and cancerous oral lesion were collected from the Institute of Dental Sciences (IDS), SUM Hospital, Bhubaneswar, India. The data have been collected taking into account of the patient concern and ethical committee clearance (Ref No./DMR/IMS.SH/SOA/1800040). The images were acquired under 100× magnification. A total of 1035 sample patches of normal mucosa (Benign) and 1154 sample patches of OSCC (Malignant) cases of size were collected for this study. Moreover, to include variations in the dataset, 965 patches of normal mucosa and 846 patches of OSCC of size are taken which were of 100× magnification from Mendeley datasets [37], resulting in 2000 patches for each class (Table 2). The Mendeley dataset consists of 89 normal histopathological images and 439 OSCC images in 100× magnification. An expert pathologist performed ground-truth labeling by identifying the region of interest (ROI), out of which image patches were taken to create balanced dataset. Sample image patches are shown in Fig. 2.
Table 2.
Dataset used for this study.
| Type | No. of patches in the dataset |
Total no. of patches collected | |
|---|---|---|---|
| IDS | Mendeley | ||
| Benign | 1035 | 965 | 2,000 |
| Malignant | 1154 | 846 | 2,000 |
Figure 2.
Sample benign and malignant image patches from IDS (first row, (a) and (b)) and Mendeley dataset (second row, (c) and (d)).
4.2. Dataset split-up
The total volume of image patches from Table 2 is divided into two groups: training and testing in the proportion of 80% and 20% respectively, as per the train-test split technique widely adopted. The training dataset is again divided into train and validation set. The number of training, validation and testing images are 2800, 400 and 800, respectively.
4.3. Image preprocessing
4.3.1. Color normalization and median filter
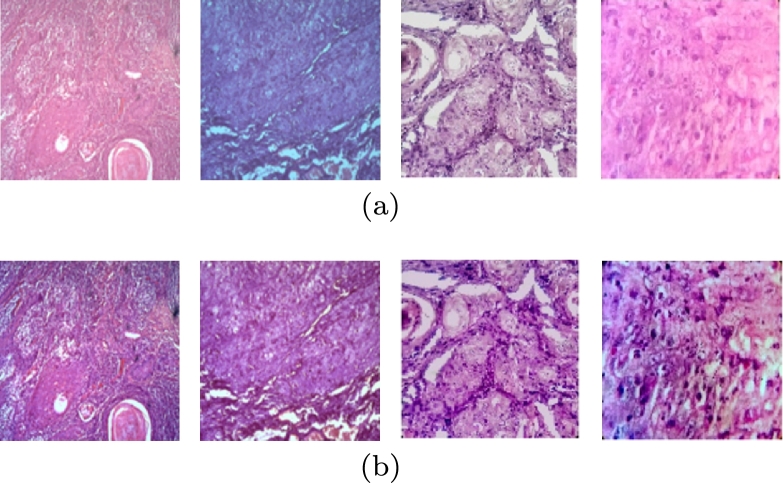
As we have collected image patches from two different databases, they are different in color and shade, possibly due to some variability conditions and staining protocols. To avoid the effects of varying color staining and make the model independent of color features, stain normalization has been applied using the Reinhards technique [38] for the training set. Thereafter, the median filter has been applied for further noise reduction as certain bright and dark pixels are associated with microscope while capturing the image [39], [40]. The histopathological image patches of OSCC before and after preprocessing are shown in Figs. 3(a) and 3(b), respectively.
Figure 3.
Histopathological image patches of OSCC (a) before preprocessing and (b) after preprocessing (stain normalization and median filtering).
4.3.2. Data augmentation
The collected image patches of benign and malignant cases are limited and hence not sufficient to generalize the DCNN models for classification. Consequently, to increase the number of image patches, data augmentation is adopted which will overcome the overfitting issue by providing good generalization of the DCNN models during testing. The data augmentation facilitates producing significantly higher number of training data for each class [41], which involves image manipulation by various techniques. In this work, data augmentation involves flipping, inverting, scaling, and rotation (, ) of the original image leading to 14,000 image patches.
4.4. Training and classification
The above-mentioned two classification approaches such as pretrained models that uses transfer learning concept and proposed baseline CNN model, trained from scratch are elucidated in more detail below.
4.4.1. Fine-tuning of pretrained models
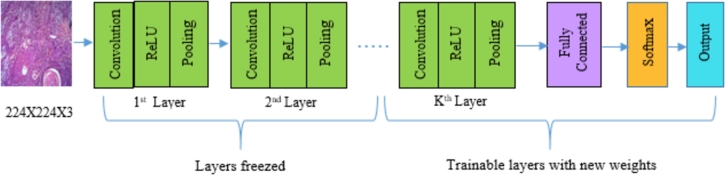
Despite the fact that the pretrained models VGG16, VGG19, ResNet50, InceptionV3, and MobileNet are already trained on the ImageNet dataset, the models are further fine-tuned using oral cancer dataset with little modification to the architecture (freezing half of the layers and training the remaining). Fine-tuning of transfer learning involves the transfer of training knowledge acquired by addressing a particular classification problem using a large set of labeled images to other domain classification task. The oral cancer dataset contains entirely different categories than the ImageNet dataset; however, the pretrained models can extract significant features from oral cancer histopathological images depending on the transfer learning concept. Usually, training a DCNN from scratch needs a vast number of training samples, and training time is more compared to transfer learning. Pretrained weights of ImageNet are considered for this study as a relatively limited number of training images are available for oral cancer dataset. Before fine-tuning, the final fully connected layer with a thousand number of output nodes of selected models is replaced with two output nodes to handle binary classification problem. Thereafter, the pretrained weights of ImageNet dataset are loaded to fine-tune the selected DCNN models. Essentially, 50% layers of the pretrained DCNN models are freezed, and the rest of the layers are allowed to fine-tune on the oral cancer dataset (see Fig. 4). To obtain the optimal number of frozen layers, an experiment was conducted by freezing 75%, 50%, 25% of the total number of layers. We found that 50% of the frozen layers gave promising results for the classification problem addressed in this paper. The number of layers freezed for each model, are provided in Table 3.
Figure 4.
Block diagram of transfer learning of DCNNs for binary class prediction.
Table 3.
Number of DCNN layers freezed for the study.
| Pretrained DCNN models | Layers freezed upto depth |
|---|---|
| VGG16 | 8 |
| VGG19 | 9 |
| InceptionV3 | 24 |
| ResNet50 | 26 |
| MobileNet | 15 |
4.4.2. Baseline CNN model
To achieve a simple and sufficiently efficient model, a baseline CNN architecture is proposed, as shown in Fig. 5, which is trained from scratch with the collected image patches. Architecture of the proposed model is based on the structure of VGGNet with kernel size , which accepts an image patch of size as an input (based on the base paper [11]). It consists of 10 convolution layers, 4 max-pooling layers, and two fully connected layers. The max-pooling layer performs subsampling by reducing the size of the feature map to half. After each convolution layer, the activation function ReLU is applied to prevent the gradient vanishing problem in CNN by proficiently spreading the gradient [42]. Batch normalization is followed by the ReLU activation, which normalizes the distribution of the features. In addition, some negative features are truncated by the non-linearity layer ReLU. Dropout [43] is used as a regularization parameter to boost network generalization capability and prevent overfitting issues. The flatten dense layer connects all the input neurons in a layer to the output neurons in the next layer. In the last layer, softmax is used which provides the probabilities of oral lesion being malignant or benign.
Figure 5.
Architecture of the Proposed CNN model.
4.5. Hyperparameter setup
To make a fair comparison between the outcomes of all the models, standardization of the hyperparameters has been considered across all the experiments. After performing different experiments, the following values were chosen for standard hyperparameters for all the models. These are:
-
•
Optimizer: Adam
-
•
Base learning rate: 0.001
-
•
Learning rate strategy: Early Stopping, for every 5 epochs, step reduces by a factor of 0.5
-
•
Momentum: 0.9
-
•
Weight decay: 0.0005
-
•
Batch size: 32
4.6. Experimental setup
All the experiments are performed in a system (Quadro P5200) with a six-core i7 processor, 32 GB of GDDR5 RAM, and NVIDIA-2560 CUDA processing cores, 16 GB GPU (32 GB GDDR5 graphics memory and 2560 CUDA cores). Keras (high-level neural network library run on TensorFlow or Theano) based on the python interface is used to implement the experimental framework of oral lesion classification of DCNN models. Model training on the Quadro P5200 with 16 GB GPU is substantially faster due to the parallel architecture that uses 2560 CUDA cores.
4.7. Performance metrics
To assess the suggested model's effectiveness, from various classification performance measures, the benchmark metrics namely accuracy, precision, recall, and F-measure are used in this study. The recall or true positive rate (TPR) of a classifier indicates the proportion of correctly positive classified images to the overall number of positive images. Precision or positive predictive value (PPV) indicates the number of positive images accurately classified to the overall number of positive predicted images. The harmonic mean of precision and recall is represented by F-measure or F-score. The following equations (1) to (4) represent the performance metrics used in this study.
| (1) |
| (2) |
| (3) |
| (4) |
5. Results
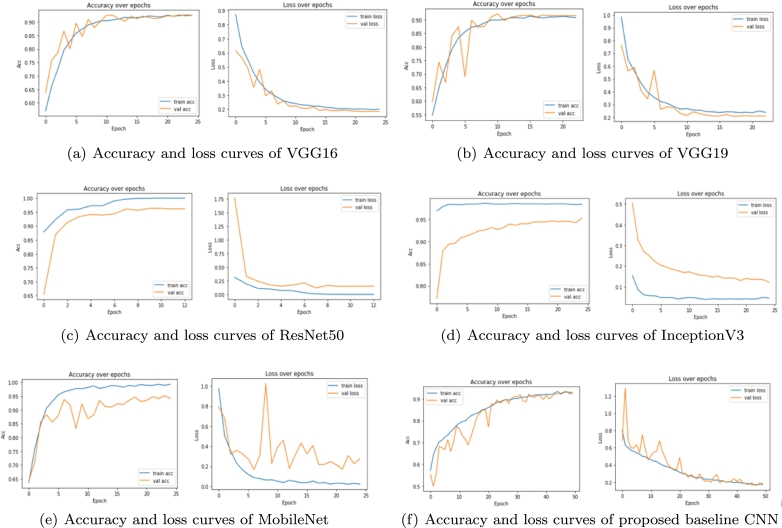
The image patches of Oral Squamous Cell Carcinoma (OSCC) from training set are used to fine-tune the VGG16, VGG19, Inceptionv3, ResNet50, and MobileNet models; however, the proposed baseline CNN model is trained from scratch. Within the training process, the validation set is used to retain the best fine-tuned model in each epoch in terms of validation accuracy. The test set was further used to evaluate the trained network's prediction abilities in terms of different performance measures and time taken. Accuracy and loss curves (also called dual learning curves) for VGG16 are shown in Fig. 6 (a) for both training and validation set. The figure shows that the pretrained model VGG16 learns the characteristics of OSCC image patches effectively and performs well on the validation set. A similar observation can also be noticed in case of different pretrained models considered in this study, as shown in Figs. 6(b) to 6(f). The dual learning curves are further discussed in more detail for individual models.
Figure 6.
The learning curves of the deep learning models used for this study: (a) VGG16, (b) VGG19, (c) ResNet50, (d) InceptionV3, (e) MobileNet and (f) Proposed baseline CNN.
Moreover, to determine the best suitable model for classification of oral lesion patches, a comparative assessment has been made among fine-tuned DCNN models and the suggested baseline CNN model in terms of learning curves and performance measures.
The results of each fine-tuned DCNN models and the suggested baseline CNN model are evaluated and compared based on the following evaluation metrics.
-
•
Model accuracy and loss curves
-
•
Confusion matrix
-
•
Quantitative evaluation
-
•
Statistical analysis with McNemar's test
5.1. Model accuracy and loss curves
In this section, accuracy and loss curves of the DCNN models are discussed, where each curve comprises both training and validation curves. The training curve is determined from the training set, which tells how well the model is able to learn. Conversely, the validation curve is obtained from a validation set that demonstrates how well the model generalizes itself. Alternatively, the error on the training dataset is termed as the training loss, while the error after running the validation dataset through the trained network is given as the validation loss.
Based on the study [11], our experiments have been carried out for 10 epochs then raised up to 25 epochs. The selection of 25 epochs was done based on the empirical observation that learning mostly converges well within 25 epochs in all of these experiments except the proposed baseline CNN model.
Figs. 6(a) and 6(b) show the training accuracy and loss curve of VGG16 and VGG19, respectively. From the figures, it can be observed that the accuracy of the models increases exponentially prior to 12th epoch then starts saturating. Comparably, the loss curves are decreasing as observed for training data as well as validation data, then a relative stability can be observed. The decreasing loss indicates that VGG16 and VGG19 can learn the features but their accuracy did not considerably improve from 91.5% and 92.65%, respectively. The training and validation accuracy of ResNet50 are increasing up to 8th epoch as seen from Fig. 6 (c). After 8 epochs, it becomes stable by achieving the plateau. Thus, the training stops early at 12 epochs by attaining the highest validation accuracy of 96.6%. There is a sudden fall in the first epoch for the validation loss curve, and after 8 epochs, it becomes stable with a loss value of 0.1520. The ResNet50 model outperforms the rest of the models by achieving an accuracy of 96.6% and a loss rate of 0.1520 at epoch 8, which represents the best result. From Figs. 6 (d) and 6 (e), it can be observed that InceptionV3 and MobileNet have achieved the accuracy of 95.25% and 95.02%, but there is a substantial gap between training and validation curves. Moreover, the validation curve is fluctuating or not stable for MobileNet. Fig. 6 (f) represents our proposed baseline CNN model's accuracy and loss curves. As this CNN is training the data from scratch, within 25 epochs, the model was not stable. So, the model was trained for 50 epochs. Both the training and validation accuracy values increase up to 40 epochs then become stable with values 93.18% and 93.15%, respectively. The model fits well as it is adapting the training set. The validation loss curve shows a good fit with the training loss. The loss values are 0.1784 and 0.1714 for validation and training, respectively. But the pretrained ResNet50 exceeds this performance by giving the best result for our experiment. It can be observed from the figures that none of the studied models have shown overfitting.
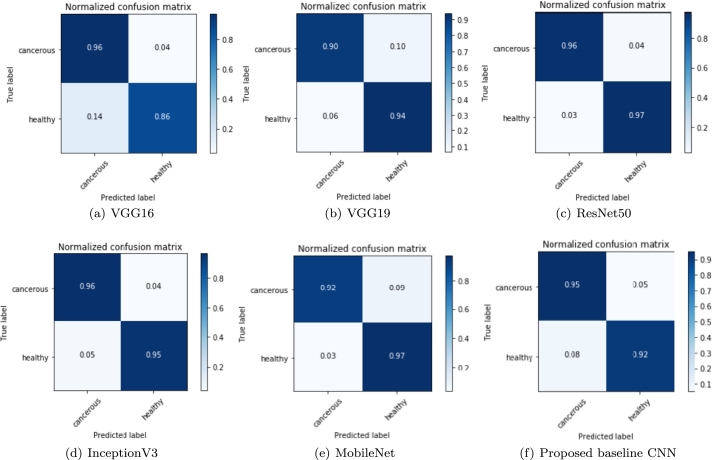
5.2. Confusion matrix
The confusion matrix illustrates the comprehensive representation of the prediction results after classification. The confusion matrices of Figs. 7(a) to 7(f) represent outcome of the test data.
Figure 7.
Confusion matrix reflecting the performance of each individual classifier for the evaluation of TP, FP, FN and TN labels: (a) VGG16, (b) VGG19, (c) ResNet50, (d) InceptionV3, (e) MobileNet and (f) Proposed baseline CNN.
Here, the normalized confusion matrix of all the DCNN models and the proposed baseline CNN model is presented. The True-Negative (TN), True-Positive (TP), False-Positive (FP), and False-Negative (FN) values are demonstrated here. Although the highest TN and TP values are always needed, the FP and FN values are as well essential in the biomedical area. When a person is well but is mistakenly diagnosed as sick, we have an FP case, which involves undesired mental anguish, financial outlay, and potentially hazardous health side effects from unneeded therapy. On the other side, we have an FN situation when a person is unwell but is labeled as healthy, resulting in wrong diagnosis. This may increase the severity of the disease condition and patient will die. In the long run, this involves more mental anguish and financial outlay for more expensive therapies. For a classifier model, the values in the matrices reflect the predictable proportion for the corresponding classes in row and column. The TPs for the class being evaluated are represented by the diagonal values and the other values signifying the error rates. It is seen from Fig. 7(c), the confusion matrix of the pretrained ResNet50 model indicates the highest TPs, and the minimal misclassification values for FP and FN.
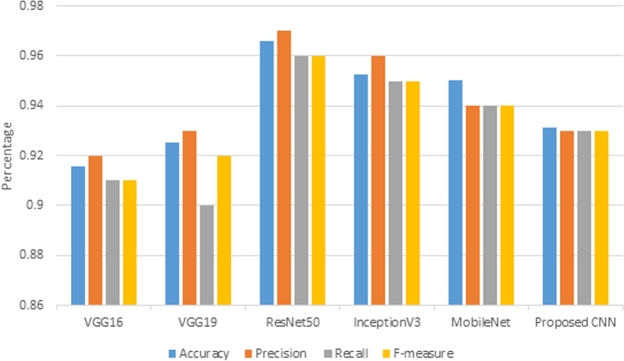
5.3. Quantitative evaluation
The classification report can be produced from the performance evaluation metrics namely accuracy, precision, recall, and F-measure. These measures are useful for successful prediction. High precision signifies a low false positive rate, and high recall signifies a low false negative rate. Higher values of precision and recall lead to higher F-measure values, which indicates how well the models are predicting. The performance measures of different DCNN models as well as proposed CNN are presented in Table 4. The accuracy values are denoted with ± standard deviation whereas other fields display the mean values over several execution steps of the DCNN models.
Table 4.
Predictive results of DCNN models for binary classification of oral lesion.
| Models | Accuracy | Precision | Recall | F-measure | p-value |
|---|---|---|---|---|---|
| VGG16 | 0.91 ± .02 | 0.92 | 0.91 | 0.91 | 0.018 |
| VGG19 | 0.92 ± .03 | 0.93 | 0.90 | 0.92 | 0.31 |
| InceptionV3 | 0.95 ± .03 | 0.96 | 0.95 | 0.95 | 0.70 |
| ResNet50 | 0.96 ± .02 | 0.97 | 0.96 | 0.96 | 0.76 |
| MobileNet | 0.95 ± .03 | 0.94 | 0.94 | 0.94 | 0.083 |
| Proposed baseline CNN | 0.93 ± .06 | 0.93 | 0.93 | 0.93 | 0.40 |
Fig. 8 shows that ResNet50 outperforms other DCNN models with the highest accuracy, precision, and recall. It is observed that an average accuracy of more than 95% was offered by ResNet50, InceptionV3, and MobileNet. The accuracies of VGG16 and VGG19 are less in comparison to the proposed baseline CNN. It is observed that, in terms of accuracy, precision, and recall, VGG16 shows the least result. Thus, the fine-tuned version of VGG16 is the worst performing model.
Figure 8.
Performance comparison of different DCNN models for detecting oral malignancy lesion with accuracy, precision, recall and F-measure.
5.4. Statistical analysis with McNemar's test
McNemar's test confirms the statistical significance of the classification models. The standardized normal test is the basis for this test [44]. Considering the true cases only while evaluating the classification model is the major mistake. False predictions also have to be considered before rendering any decision. We want to keep these predictions as minimum as possible. Although precision and recall slightly explain the performance of the positive classes (or, negative) by considering the false cases too, there is no comparison between the false positives and the false negatives. In this scenario, McNemar's test should be applied to determine the probability of difference between false negative and false positive cases. The test is widely used in medical domain to compare the effect of a treatment against a control. This test checks whether there is a statistical difference in false predictions or the model equally performs for the wrong (false) predictions [45].
In this article, McNemar's test is used for contingency tables to find whether row and column marginal frequencies are equal for paired samples. In our case, the row and column marginal frequencies meant for confusion matrices, are the number of false predictions for both positive and negative classes. The Chi-Square distribution is used to calculate the probability of difference. Discordant pairs from the confusion matrix of each model are the current interest of our analysis. We need to obtain a p-value from the Chi-Square test by using the McNemar's statistic. The null and alternate hypothesis are:
H0: The marginal proportions of the discordant are not significantly different from each other. (There is no difference between the marginal frequencies.)
H1: The marginal proportions of the discordant are significantly different from each other. (There is a significant difference between the marginal frequencies.)
The McNemar's test statistic is as follows:
where b and c are the discordant pair from the confusion matrix.
This test measures the skewness between the FPs and FNs. The degree of freedom is determined by the product of row minus one and column minus one i.e., . The commonly used 5% significance level is considered for this study. If the p-value>0.05, it is considered that there is no significant difference between false negatives and false positives; and, p-value ≤ 0.05, means there is a significant difference.
The calculated p-values for each model are provided in Table 4. The p-value obtained for VGG16 was 0.018 which is less than the significant level. Thus the null hypothesis is rejected, indicating that there is substantial difference between the marginal frequencies of the false positive and false negatives. On the other hand, the p-values for all other networks are greater than the significant level. Thus, the null hypothesis can not be rejected.
5.5. Hyperparameter tuning for the proposed baseline CNN model
This section explains the tuning of hyperparameters conducted for the suggested baseline CNN model to achieve the simple and optimal model for training the oral cancer dataset. Hyperparameter tuning is essential as it controls the complete behavior of a deep learning model. The main purpose was to find an optimal hyperparameter combination that minimizes the loss function to give better performances. This experiment was performed with the same dataset split-up formerly utilized in the experiments. The proposed configuration with ten convolution layers provided the maximum classification accuracy of 93.15% for 50 epochs. Adam optimizer with max pooling was better by retaining the kernel size as .
6. Discussion
Transfer learning of pretrained DCNN models on natural pictures, is a feasible approach to assist in medical image classification. As the models are applied on a new problem, we need to adapt it to suit the problem. Hence, fine-tuning is required for the pretrained models where training process starts with some learned weights that come from pretrained models. For this study, the weights of 50% layers of each pretrained models were freezed (not allowed to update during training), and the remaining layers were fine-tuned with OSCC histopathological images.
We have tried with more generalized models so that more variation in the data that may come from different geographical regions, could be accommodated. Freezing of 50% is done to achieve the benefit of both i.e., training half of the layers and availing of the benefit of transfer learning. If half of the layer is frozen, and we try to train the model, it will take about half of the time as compared to a fully trainable model.
From the Table 5 it is observed that even though there is little enhanced performance for 25% freezing of layers, it is taking on an average 30% more time, compared to freezing 50% layers. For 75% freezing of layers, the time taken is less, on an average 200 secs compared to 50% freezing; but there is decrease in performance. On the other hand, if we freeze it too early (i.e., 25%), it will give inaccurate predictions, (not widely used also). Thus, for further analysis authors have considered 50% freezing of layers.
Table 5.
Performance based on 25%, 50% and 75% freezing of layers.
| Models | 25% freezing |
50% freezing |
75% freezing |
|||
|---|---|---|---|---|---|---|
| Time (sec) | Accuracy | Time (sec) | Accuracy | Time (sec) | Accuracy | |
| VGG16 | 7175.4 | 0.92 ± .02 | 4795.6 | 0.91 ± .02 | 4101.2 | 0.91 ± .01 |
| VGG19 | 7395.1 | 0.93 ± .06 | 4930.3 | 0.92 ± .03 | 4320.6 | 0.91 ± .09 |
| InceptionV3 | 4765.5 | 0.97 ± .03 | 3163.6 | 0.96 ± .02 | 2822.4 | 0.95 ± .06 |
| ResNet50 | 3246.5 | 0.95 ± .05 | 2163.1 | 0.95 ±.03 | 2001.2 | 0.94 ± .08 |
| MobileNet | 2813.7 | 0.95 ± .05 | 1855.8 | 0.95 ± .03 | 1508.7 | 0.95 ± .01 |
Overfitting [43] is a significant problem with the deep learning models. This typically occurs when the DCNN model does not adapt well to new data but fits the training data well. This is more obvious with the limited data for training. In this study, all the pretrained DCNN and proposed baseline CNN models are trained with 14000 images (moderate range of dataset) and dropout, regularization, and batch normalization layers are used to avoid this issue [16], [17], [43]. From the accuracy and loss curves, we have observed that the training and validation accuracy, as well as training loss and validation loss, are alike, which specifies that models have fitted well to the training dataset. In the case of overfitting, the training accuracy would have been significantly greater than the validation accuracy; and, the training loss would have been substantially lower than validation loss. Additionally, the used augmented technique aided the deep learning models to generalize well and delivered reasonably precise outcomes.
Table 6 shows that MobileNet has taken the least time for training whereas, VGG16 and VGG19 required more time compared to all other pretrained and baseline CNN models. The variation of the training time depends on the learnable parameters and the depth of the model. VGG16 and VGG19 are taking more time due to the large number of learnable parameters as shown in Table 1. For a single image prediction, VGG16, as the most straightforward architecture, takes the lowest time than other models.
Table 6.
Training, Computation time and Parameters of the DCNN models.
| Pretrained DCNN models | Training time in sec | Time for a single image prediction in sec | Number of model parameters |
|---|---|---|---|
| VGG16 | 4795.6 | 0.657 | 134,268,738 |
| VGG19 | 4930.3 | 0.821 | 138,357,544 |
| InceptionV3 | 2163.1 | 2.1 | 23,903,010 |
| ResNet50 | 3163.6 | 3.12 | 23,788,418 |
| MobileNet | 1855.8 | 1.44 | 5,853,890 |
| Proposed CNN | 3460.4 | 1.46 | 3,420,610 |
In [11], Navarun et al. have suggested replacing the fully connected layers for DCNN models to relearn from the histopathological images of oral cancer. Their proposed CNN with 8 convolution layers exceeds all the pretrained models presented in their work, which exhibits that the model trained from scratch is performing better compared to the transfer learning approaches.
Based on the above fact, and seeing the difference in the nature of the ImageNet dataset and oral histopathological image dataset, it is observed that only replacing the final fully connected layer of the pretrained models may not extract all the relevant underlying features for the classification task to provide promising results. Thus, we have considered half freezed layers for random weights, and remaining layers are trained with oral histopathological image dataset. The results show that the pretrained models based on half frozen transfer learning perform better in comparison to the existing baseline model.
Even if we are increasing two more layers making CNN a 10 layered architecture, due to the smaller dataset it is not performing well compared to half trained ResNet model. The comparison with other state-of-the-art works is presented below.
6.1. Comparison with other cutting-edge models
First, to determine the best performing architecture, individual models’ accuracy and loss results have been compared in Figs. 6(a) to 6(f). After that, the confusion matrix of different deep learning architectures and proposed CNN are compared. In terms of different performance measures, Table 6 and Fig. 8 illustrate the comparison between different deep learning models used in this experiment. For each model, the accuracy curve for training and validation set increases to reach a stable phase (as seen from Figs. 6(a) to 6(f)). The fine-tuned versions of ResNet50, InceptionV3, and MobileNet have shown satisfactory performance, and the accuracy increases for both training and validation sets (as seen from Figs. 6(c), 6(d) and 6(e)). Performance of these models exceeds the low performance of the baseline CNN, VGG16, and VGG19. The training and validation accuracies of proposed CNN, VGG16, and VGG19 are above 90% after 15 epochs and these values become steady till the end of the training. However, ResNet50, InceptionV3, and MobileNet have produced the validation accuracy above 95%, as seen from Table 4.
For this study, the predictive models produced by InceptionV3 and MobileNet algorithms do not fit well to the training set. The simple 10 convolutional layer deep model is able to achieve 93.15% accuracy. It is shallower compared to other pretrained models used in this study. With such a simple architecture it achieved comparable accuracy. As the model is trained from scratch, it may require more training dataset, more epoch and fine tuning of huge parameters to obtain comprehensive prediction. It achieved the accuracy of 3% less compared to the ResNet50 pretrained model. So it increases the false positive and false negative cases, which in turn increases risk of the diagnostic consequences from the patient's perspective. Overall classification performance shows that ResNet50 is the most efficient model for the binary classification of oral lesions.
On the other hand, CNN, and other deep learning models are trained from scratch with more than 100 epochs, requiring more time to achieve the highest performance. With very little training (after 8 epochs, it gets saturated), our ResNet50 performs better compared to the deep learning models trained from scratch. This study has focused on the efficiency of transfer learning by fine-tuning the existing deep learning models with the OSCC images for binary classification. From this perspective, we should deduce from Table 7 that our method is highly comparable with current work done in the field of OSCC.
Table 7.
Comparison of various methods with proposed system for the classification of oral histopathological images.
| Methodology | Image size | Features | Classification methods | Accuracy (%) |
|---|---|---|---|---|
| Krishnan et al. [31] | Normal-90 OSFWD-42 OSFD-26 | 71 features of wavelet family (LBP, Gabor, BMC) | SVM | 88.38 |
| Krishnan et al. [32] | Normal-90 OSFWD-42 OSFD-26 | HOS, LBP, LTE | Fuzzy | 95.7 |
| Krishnan et al. [40] | Normal-341 OSF-429 | Morphological and textural features | SVM | 99.66 |
| Belvin et al. [46] | 16 malignant images with 192 patches | Texture features and Run Length features | Back propagation based ANN | 97.92 |
| Anuradha. K. et al. [47] | Not mentioned | Energy, Entropy, Contrast, Correlation, Homogeneity | SVM | 92.5 |
| DevKumar et al. [48] | High grade-15 Low grade-25 Healthy-2 | Identification of various layers –epithelial, subepithelial, keratin region and keratin pearls | Random Forest | 96.88 |
| Navarun et al. [11] | Benign-1656 WDSCC-2634 MDSCC-2110 PDSCC - 1921 | Image's raw pixel data | CNN | 97.5 |
| Nanditha B R et al. [49] | Benign-63 Malignant-269 | Image's raw pixel data | Ensemble model (ResNet50 and VGG16) | 96.2 |
| Proposed method | WDSCC-400 MDSCC-400 PDSCC-400 | Image's raw pixel data | Residual Network | 96.6 |
Powerful image classification models have been built using transfer learning of pretrained models, which have boosted the accuracy with less time and epochs to converge than the model trained from scratch. The fine-tuned models get a kind of head start as the pretrained models have already learned high-level features. We have considered small dataset and applied the rigid transformations of data augmentation.
Very deep neural network (ResNet150, EfficientNet (200 to 800 layers)) are not considered for this study and may be considered in future).
7. Conclusion
In this study, the concept of transfer learning over pretrained deep convolution neural networks (DCNN) has been successfully employed for binary classification of oral histopathological images into benign and malignant lesions. Moreover, a baseline DCNN model has been proposed for the classification of oral lesions. The performances of fine-tuned pretrained DCNN models (VGG16, VGG19, ResNet50, InceptionV3, MobileNet) have been evaluated and compared with the efficacy of the proposed baseline CNN model. From the experimental results, we have observed that the 50% frozen version of ResNet50 is the most efficient model among selected DCNN architectures with the highest accuracy of 96.6%; however, MobileNet has taken the least time to get trained to achieve saturated accuracy of 95.1%. Furthermore, the performance evaluation of the proposed baseline CNN showed that the model is simple and able to classify the oral lesions with accuracy of 93.15%. This work represents an effective and inexpensive way of screening Oral Squamous Cell Carcinoma to assist the doctors/pathologists in their clinical practice to diagnose patients for effective clinical treatment. The baseline CNN model can be further improved by replacing each convolution layer with a residual block, and it is our future target. By considering large dataset, more deep dense layers and high performance GPU systems, this work can be further scaled up.
CRediT authorship contribution statement
Santisudha Panigrahi: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper. Bhabani Sankar Nanda: Performed the experiments; Analyzed and interpreted the data. Ruchi Bhuyan: Analyzed and interpreted the data. Kundan Kumar; Susmita Ghosh; Tripti Swarnkar: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare the following conflict of interests: Dr. Susmita Ghosh; [One of the authors is an Associate editor].
Contributor Information
Santisudha Panigrahi, Email: santisudha.nanda@gmail.com.
Bhabani Sankar Nanda, Email: bhabanisankar.nanda@carrier.com.
Ruchi Bhuyan, Email: ruchibhuyan30@soa.ac.in.
Kundan Kumar, Email: kundankumar@soa.ac.in.
Susmita Ghosh, Email: susmitaghoshju@gmail.com.
Tripti Swarnkar, Email: triptiswarnakar@soa.ac.in.
Data availability
Data associated with this study has been deposited at 10.17632/ftmp4cvtmb.1.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Dhanuthai K., Rojanawatsirivej S., Thosaporn W., Kintarak S., Subarnbhesaj A., Darling M., Kryshtalskyj E., Chiang C.P., Shin H.I., Choi S.Y., et al. Oral cancer: a multicenter study. Med. Oral Patol. Oral Cir. Bucal. 2018;23(1):e23. doi: 10.4317/medoral.21999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar M., Nanavati R., Modi T.G., Dobariya C., et al. Oral cancer: etiology and risk factors: a review. J. Cancer Res. Ther. 2016;12(2):458. doi: 10.4103/0973-1482.186696. [DOI] [PubMed] [Google Scholar]
- 4.Nayak V.N., Donoghue M., Selvamani M. Oral squamous cell carcinoma: a 5 years institutional study. J. Med., Radiol., Pathol. Surg. 2015;1(5):3–6. [Google Scholar]
- 5.Weckx A., Riekert M., Grandoch A., Schick V., Zöller J.E., Kreppel M. Time to recurrence and patient survival in recurrent oral squamous cell carcinoma. Oral Oncol. 2019;94:8–13. doi: 10.1016/j.oraloncology.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Ganpathi Iyer S., Pradhan S., Pai P., Patil S. Surgical treatment outcomes of localized squamous carcinoma of buccal mucosa. Head & Neck: Journal for the Sciences and Specialties of the Head and Neck. 2004;26(10):897–902. doi: 10.1002/hed.20096. [DOI] [PubMed] [Google Scholar]
- 7.O'Mahony N., Campbell S., Carvalho A., Harapanahalli S., Hernandez G.V., Krpalkova L., Riordan D., Walsh J. Science and Information Conference. Springer; 2019. Deep learning vs. traditional computer vision; pp. 128–144. [Google Scholar]
- 8.Hussein I.J., Burhanuddin M.A., Mohammed M.A., Benameur N., Maashi M.S., Maashi M.S. Fully-automatic identification of gynaecological abnormality using a new adaptive frequency filter and histogram of oriented gradients (hog) Expert Systems. 2021 [Google Scholar]
- 9.Kassani S.H., Kassani P.H., Wesolowski M.J., Schneider K.A., Deters R. Classification of histopathological biopsy images using ensemble of deep learning networks. 2019. arXiv:1909.11870 arXiv preprint.
- 10.Xie J., Liu R., Luttrell J., IV, Zhang C. Deep learning based analysis of histopathological images of breast cancer. Front. Genet. 2019;10:80. doi: 10.3389/fgene.2019.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das N., Hussain E., Mahanta L.B. Automated classification of cells into multiple classes in epithelial tissue of oral squamous cell carcinoma using transfer learning and convolutional neural network. Neural Netw. 2020;128:47–60. doi: 10.1016/j.neunet.2020.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Ar Mohamed, Dahl G.E., Hinton G. Acoustic modeling using deep belief networks. IEEE Trans. Audio Speech Lang. Process. 2011;20(1):14–22. [Google Scholar]
- 13.Vincent P., Larochelle H., Bengio Y., Manzagol P.A. Proceedings of the 25th International Conference on MachineLlearning. 2008. Extracting and composing robust features with denoising autoencoders; pp. 1096–1103. [Google Scholar]
- 14.Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M., et al. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015;115(3):211–252. [Google Scholar]
- 15.Smirnov E.A., Timoshenko D.M., Andrianov S.N. Comparison of regularization methods for imagenet classification with deep convolutional neural networks. AASRI Proc. 2014;6:89–94. [Google Scholar]
- 16.Krizhevsky A., Sutskever I., Hinton G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM. 2017;60(6):84–90. [Google Scholar]
- 17.Szegedy C., Liu W., Jia Y., Sermanet P., Reed S., Anguelov D., Erhan D., Vanhoucke V., Rabinovich A. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015. Going deeper with convolutions; pp. 1–9. [Google Scholar]
- 18.Chollet F. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Xception: deep learning with depthwise separable convolutions; pp. 1251–1258. [Google Scholar]
- 19.Simonyan K., Zisserman A. Very deep convolutional networks for large-scale image recognition. 2014. arXiv:1409.1556 arXiv preprint.
- 20.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 21.Howard A.G., Zhu M., Chen B., Kalenichenko D., Wang W., Weyand T., Andreetto M., Adam H. MobileNets: Efficient convolutional neural networks for mobile vision applications. 2017. arXiv:1704.04861 arXiv preprint.
- 22.Litjens G., Kooi T., Bejnordi B.E., Setio A.A.A., Ciompi F., Ghafoorian M., Van Der Laak JA Van Ginneken B., Sánchez C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Roth H.R., Lu L., Liu J., Yao J., Seff A., Cherry K., Kim L., Summers R.M. Improving computer-aided detection using convolutional neural networks and random view aggregation. IEEE Trans. Med. Imaging. 2016;35(5):1170–1181. doi: 10.1109/TMI.2015.2482920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin H.C., Roth H.R., Gao M., Lu L., Xu Z., Nogues I., Yao J., Mollura D., Summers R.M. Deep convolutional neural networks for computer-aided detection: Cnn architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging. 2016;35(5):1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Rethinking the inception architecture for computer vision; pp. 2818–2826. [Google Scholar]
- 26.Ker J., Wang L., Rao J., Lim T. Deep learning applications in medical image analysis. IEEE Access. 2017;6:9375–9389. [Google Scholar]
- 27.Mehmood A., Iqbal M., Mehmood Z., Irtaza A., Nawaz M., Nazir T., Masood M. Prediction of heart disease using deep convolutional neural networks. Arab. J. Sci. Eng. 2020:1–14. [Google Scholar]
- 28.Shamim M.Z.M., Syed S., Shiblee M., Usman M., Ali S. Automated detection of oral pre-cancerous tongue lesions using deep learning for early diagnosis of oral cavity cancer. 2019. arXiv:1909.08987 arXiv preprint.
- 29.Kim D.W., Lee S., Kwon S., Nam W., Cha I.H., Kim H.J. Deep learning-based survival prediction of oral cancer patients. Sci. Rep. 2019;9(1):1–10. doi: 10.1038/s41598-019-43372-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohd F., Noor N., Bakar Z.A., Rajion Z.A. The 7th International Conference on Information Technology (ICIT 2015) 2015. Analysis of oral cancer prediction using features selection with machine learning; pp. 283–288. [Google Scholar]
- 31.Krishnan M.M.R., Chakraborty C., Ray A.K. Wavelet based texture classification of oral histopathological sections. Int. J. Microsc., Sci. Technol. Appl. Educ. 2010;2(4):897–906. [Google Scholar]
- 32.Krishnan M., Acharya U., Chakraborty C., Ray A. Automated diagnosis of oral cancer using higher order spectra features and local binary pattern: a comparative study. Technol. Cancer Res. Treat. 2011;10(5):443–455. doi: 10.7785/tcrt.2012.500221. [DOI] [PubMed] [Google Scholar]
- 33.Patra R., Chakraborty C., Chatterjee J. Textural analysis of spinous layer for grading oral submucous fibrosis. Int. J. Comput. Appl. 2012;47:975–8887. [Google Scholar]
- 34.Krishnan M.M.R., Shah P., Choudhary A., Chakraborty C., Paul R.R., Ray A.K. Textural characterization of histopathological images for oral sub-mucous fibrosis detection. Tissue Cell. 2011;43(5):318–330. doi: 10.1016/j.tice.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Rahman T., Mahanta L., Chakraborty C., Das A., Sarma J. Textural pattern classification for oral squamous cell carcinoma. J. Microsc. 2018;269(1):85–93. doi: 10.1111/jmi.12611. [DOI] [PubMed] [Google Scholar]
- 36.Rahman T.Y., Mahanta L.B., Das A.K., Sarma J.D. Automated oral squamous cell carcinoma identification using shape, texture and color features of whole image strips. Tissue Cell. 2020;63 doi: 10.1016/j.tice.2019.101322. [DOI] [PubMed] [Google Scholar]
- 37.Rahman T.Y. A histopathological image repository of normal epithelium of oral cavity and oral squamous cell carcinoma, mendeley data, v1. 2019. https://doi.org/10.17632/ftmp4cvtmb.1https://data.mendeley.com/datasets/ftmp4cvtmb/1
- 38.Reinhard E., Adhikhmin M., Gooch B., Shirley P. Color transfer between images. IEEE Comput. Graph. Appl. 2001;21(5):34–41. [Google Scholar]
- 39.Shih F.Y. John Wiley & Sons; 2010. Image Processing and Pattern Recognition: Fundamentals and Techniques. [Google Scholar]
- 40.Krishnan M.M.R., Chakraborty C., Paul R.R., Ray A.K. Hybrid segmentation, characterization and classification of basal cell nuclei from histopathological images of normal oral mucosa and oral submucous fibrosis. Expert Syst. Appl. 2012;39(1):1062–1077. [Google Scholar]
- 41.Mikołajczyk A., Grochowski M. 2018 International Interdisciplinary PhD Workshop (IIPhDW) IEEE; 2018. Data augmentation for improving deep learning in image classification problem; pp. 117–122. [Google Scholar]
- 42.Nair V., Hinton G.E. ICML. 2010. Rectified linear units improve restricted Boltzmann machines. [Google Scholar]
- 43.Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014;15(1):1929–1958. [Google Scholar]
- 44.Agresti A. vol. 482. John Wiley & Sons; 2003. Categorical Data Analysis. [Google Scholar]
- 45.Smith M.Q.P., Ruxton G.D. Effective use of the mcnemar test. Behav. Ecol. Sociobiol. 2020;74(11):1–9. [Google Scholar]
- 46.Thomas B., Kumar V., Saini S. 2013 IEEE International Conference on Signal Processing, Computing and Control (ISPCC) IEEE; 2013. Texture analysis based segmentation and classification of oral cancer lesions in color images using ann; pp. 1–5. [Google Scholar]
- 47.Anuradha K., Sankaranarayanan K. Detection of oral tumors using marker controlled segmentation. Int. J. Comput. Appl. 2012;52(2):15–18. [Google Scholar]
- 48.Das D.K., Bose S., Maiti A.K., Mitra B., Mukherjee G., Dutta P.K. Automatic identification of clinically relevant regions from oral tissue histological images for oral squamous cell carcinoma diagnosis. Tissue Cell. 2018;53:111–119. doi: 10.1016/j.tice.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 49.Nanditha B., Geetha A., Chandrashekar H., Dinesh M., Murali S. An ensemble deep neural network approach for oral cancer screening. Int. J. Online Biomed. Eng. 2021 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data associated with this study has been deposited at 10.17632/ftmp4cvtmb.1.