Abstract
Toxin-Antitoxin (TA) systems are abundant in prokaryotes and play an important role in various biological processes such as plasmid maintenance, phage inhibition, stress response, biofilm formation, and dormant persister cell generation. TA loci are abundant in pathogenic intracellular micro-organisms and help in their adaptation to the harsh host environment such as nutrient deprivation, oxidation, immune response, and antimicrobials. Several studies have reported the involvement of TA loci in establishing successful infection, intracellular survival, better colonization, adaptation to host stresses, and chronic infection. Overall, the TA loci play a crucial role in bacterial virulence and pathogenesis. Nonetheless, there are some controversies about the role of TA system in stress response, biofilm and persister formation. In this review, we describe the role of the TA systems in bacterial virulence. We discuss the important features of each type of TA system and the recent discoveries identifying key contributions of TA loci in bacterial pathogenesis.
Keywords: Toxin-antitoxin, Infection, Virulence, Intracellular survival, Host stress, Biofilm formation
1. Introduction
Initially, toxin-antitoxin (TA) systems were identified on plasmids and termed plasmid maintenance systems. Later on, numerous TA systems were also identified on chromosomes [[1], [2], [3]]. Generally, TA systems exist in an operon and encode a stable toxin and an unstable antitoxin. Toxin and antitoxin can be a protein or an RNA. Toxin targets the cellular processes leading to cell growth arrest or cell death, whereas antitoxin reverses the toxic effect of the toxin. Toxins are involved in the regulation of various biological processes. For example, plasmid-encoded TA loci play a crucial role in plasmid maintenance through a mechanism called post-segregational killing (PSK) [4]. In PSK, toxin-mediated lysis of cells eliminates plasmid-free cells as they are unable to express antitoxins to neutralize the remaining toxin activity. Toxin activity causes plasmid addiction to the cells therefore plasmid-encoded TA systems were termed plasmid addiction modules. Chromosomal TA systems of bacteria are involved in various biological processes such as stress response [5], biofilm formation [6], phage inhibition [7], virulence [8], and persistence [9]. It shows the functional diversity of the chromosomal TA system and its importance in adaptation to different stress conditions. However, the role of TA systems in stress response, biofilm formation, and antibiotic persister formation has been debated [[10], [11], [12], [13], [14], [15]]. Further, TA loci are abundant in pathogenic organisms [16,17] and their involvement in bacterial pathogenesis has been confirmed by several studies. TA systems have been shown to be important for the establishment of successful infection, intracellular survival, colonization, adaptation to host stresses, and chronic infection [6,[18], [19], [20], [21], [22], [23], [24]]. Overall, in this review, we focus on the important features of each type of TA system and recent studies on their contribution to bacterial pathogenesis.
2. Types of TA system
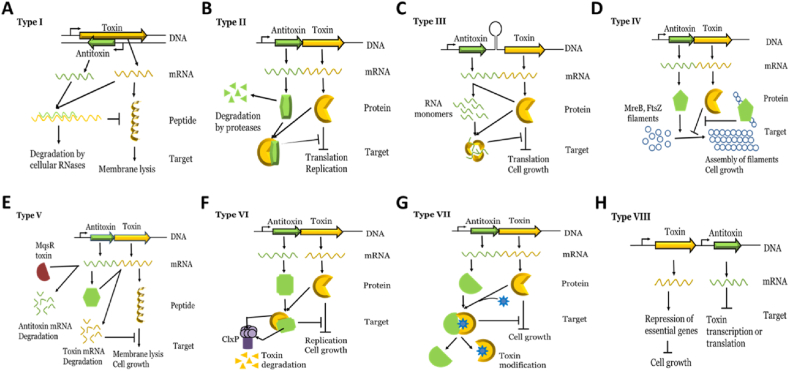
TA systems are categorized into eight (I-VIII) types according to the mode of inhibition of toxin activity by antitoxin (Fig. 1). In type I, antitoxin RNA binds to toxin mRNA and inhibits toxin mRNA translation. In type II, antitoxin protein interacts with toxin protein, forms complex, and inhibits toxin activity. In type III, antitoxin RNA binds to toxin protein and inhibits its activity. In type IV, antitoxin protein binds to a substrate of toxin protein and inhibits toxin activity indirectly. In type V, antitoxin protein cleaves toxin mRNA and inhibits its translation. In type VI, antitoxin protein interacts with toxin protein and acts as a proteolytic adapter for the degradation of toxin by proteases. In type VII, antitoxin protein interacts with toxin protein and inhibits toxin activity via post-translational modification of toxin. In type VIII, antitoxin RNA inhibits the transcription of toxins or interacts with toxin RNA leading to toxin RNA degradation. Details of each type of TA system have been discussed in brief in the following sections.
Fig. 1.
Types of TA system- TA system has been categorized in eight types based on the antitoxin mode of inhibition of toxin. A. In type I, antitoxin RNA binds to toxin mRNA and inhibits toxin mRNA translation. B. In type II, antitoxin interacts with toxin and inhibits toxin activity. C. In type III, antitoxin RNA binds to toxin and inhibits its activity. D. In type IV antitoxin binds to the substrate of toxin and inhibits toxin activity indirectly. E. In type V, antitoxin cleaves toxin mRNA and inhibits its translation. F. In type VI, antitoxin interacts with toxin and act as proteolytic adapter for degradation for toxin. G. In type VII, antitoxin interacts with toxin and inhibits toxin activity via post-translational modification. H. In type VIII, antitoxin RNA inhibits the transcription of toxin or promotes toxin RNA degradation. Toxins of all types of TA system are either bacteriostatic or bactericidal in nature.
2.1. Type I - (Fig. 1A)
Type I TA genes have been shown to be arranged as either overlapping convergently transcribed gene pairs or as divergently transcribed gene pairs located apart [25,26]. The TA pair encodes a toxin peptide and an antitoxin RNA (Fig. 1A). Mostly, toxins are small hydrophobic proteins and act as lytic peptide except SymE and RalR. SymE is a RNA endonuclease [27] and RalR is a DNA endonuclease [28]. Overexpression of toxin (lytic peptide) causes growth arrest or cell death by forming pores in the membrane that results in membrane disintegration or loss of membrane potential [[29], [30], [31], [32], [33], [34]]. The membrane disintegrition leads to ATP loss with consequences for replication, transcription, and translation [35]. For example, overexpression of hok, relF, srnB, pndA, fst, ibsC, shoB, tisB and dinQ toxins cause the destruction of membrane potential or membrane itself leading to cell death [[29], [30], [31], [32], [33], [34]]. Membrane disintegration leads to the formation of ‘ghost’ cells (lysed cells with damaged membrane) and eventually cell death [29]. Overexpression of antitoxin prevents the toxic effect of the toxin. Antitoxin inhibits toxin activity either by inhibiting the toxin mRNA translation or by promoting toxin mRNA degradation. To inhibit the toxin mRNA translation, antitoxin functions in multiple ways. For example, in Escherichia coli symE-symR TA system, antitoxin binds to the overlapping region of the ribosome binding site (RBS) of toxin mRNA and prevents the ribosomes from binding directly [27]. In E. coli hok-sok and ldrD-rdlD TA systems, antitoxin RNA binds to the Shine-Dalgarno sequence of the leader peptide which is translationally coupled to the toxin, and thereby inhibits the translation of toxin indirectly [36,37]. In E. coli tisB1-istR1 and zorO-orzO TA systems, antitoxin RNA binds to 5′ UTR and inhibits toxin translation [38,39]. In Enterococcus faecalis RNAI-RNAII TA system, both toxin and antitoxin RNA interact with each other and form a partial duplex structure that results in toxin translation inhibition [40]. Taken together, antitoxin RNA inhibits the translation of toxin mRNA by binding to the overlapping UTR region or toxin mRNA region. Whereas in the case of antitoxin mediated degradation of toxin mRNA, both toxin and antitoxin RNAs form a duplex, that is cleaved by RNase III. For example, in txpA-ratA of Bacillus subtilis, antitoxin RNA interacts with toxin RNA via base pairing and forms a duplex. Cleavage of the duplex by the RNase III results in the degradation of both RNAs [41]. In bsrG-SR4 of B. subtilis, antitoxin RNA overlaps with the 3′ end of the toxin and forms a duplex. Cleavage of the duplex by RNase III at the downstream position is followed by complete degradation of both RNAs by endonuclease Y and the 3′–5′ exoribonuclease R [41]. Interestingly, SR4 antitoxin RNA is bifunctional as it inhibits the translation of toxin mRNA by forming RNA duplex with toxin mRNA as well as by inducing conformational modulation around the toxin ribosome binding site that obstructs ribosome binding [42].
2.2. Type II- (Fig. 1B)
Among all TA systems, type II TA systems are well-characterized TA systems. They are abundant and exist in multiple copies on the chromosomes. They exist in operons and are transcriptionally coupled. Both toxin and antitoxin are proteins. Overexpression of toxin causes growth arrest by targeting replication, translation, cell wall synthesis and small metabolites [43] (Fig. 1B). Currently, type II toxins have been categorized into nine superfamilies based on their structural characteristics: ParE/RelE, MazF, HicA, VapC, HipA, FicT/Doc, AtaT/TacT, Zeta and MbcT [44](Table 1). Toxins belonging to one family can sometimes have a different mode of action. For example, ParE is a gyrase poison that causes the accumulation of DNA breaks [45] whereas RelE exhibits ribosome-dependent mRNA endonuclease activity [46]. MazF is an endonuclease that cleaves RNAs including mRNA [47], rRNA [48] and tRNA [49] whereas CcdB act as a gyrase inhibitor [50]. HicA acts as an endonuclease that cleaves mRNA [51]. The VapC contains a typical PIN domain structure and cleaves different tRNAs including tRNAfMet, tRNALeu, tRNASer, tRNATrp and also 23S rRNA [52,53]. The HipA and HipT act as a kinase that phosphorylates the glutamyl-tRNA synthetase and tryptophanyl-tRNA synthetase, respectively [54,55]. Doc is a kinase that phosphorylates the translation elongation factor EF-Tu leading to the inability of EF-Tu to bind aminoacylated tRNAs [56]. The FicT adenylylates DNA gyrase and topoisomerase IV at their ATP-binding sites [57,58]. AtaT and TacT, both act as an acetyltransferase enzyme where AtaT efficiently acetylates Gly-tRNAGly, Trp-tRNATrp, Tyr-tRNATyr and Phe-tRNAPhe isoacceptors in addition to Met-tRNAfMet [59,60], and TacT acetylates Gly-tRNAGly, Trp-tRNATrp, Leu-tRNALeu and Ser-tRNASer [61]. Zeta is a UDP-N-acetylglucosamine kinase that causes inhibition of cell wall biosynthesis [62]. MbcT with its ADP-ribosyl transferase activity depletes the intracellular NAD+ pool [63]. Interestingly, PhoH2 and EzeT are bifunctional. The N-terminal of PhoH2 consists of a PIN domain with ribonuclease activity while the C-terminal has RNA helicase domain [64]. The helicase activity is coupled to unwind RNA and facilitate RNA cleavage. Whereas in the case of EzeT, the C-terminal domain catalyzes the phosphorylation UDP-N-acetylglucosamine while the N-terminal domain strongly attenuates kinase activity and keeps EzeT in an autoinhibited state [65]. Overall, toxins are RNases, kinases, and acetyltransferases which arrest cell growth when overexpressed. To counter the toxin activity, antitoxins bind to active sites of their cognate toxin and inhibit activity [66,67]. For example, MazE binds to the active site of cognate toxin and neutralizes its RNase activity [68]. The binding of RelB to its cognate toxin, leads to displacement of the c-terminal region essential for toxin activity [69]. Epsilon and PezA creates steric hindrance to the ATP/GTP binding sites and inhibits the activity of their respective cognate toxins [62,70]. However, few antitoxins inhibit their cognate toxin activity without blocking the active site of the toxin. For example, HipB inhibits cognate toxin by facilitating confirmation of toxin to an inactive state [71]. In the case of mqsR-mqsA and higB-higA TA systems, antitoxin binds to a site other than the active site to inhibit the toxin activity [67,72]. Taken together, antitoxin inhibits toxin activity directly by binding to active sites or indirectly by binding to other sites as well.
Table 1.
Nine superfamilies and other identified novel superfamilies of type II toxins (Adapted from Ref. [44]).
| S.N. | Family | Toxin | Activity | Target | References |
|---|---|---|---|---|---|
| 1 | ParE/RelE | ParE | DNA gyrase inhibition | Replication | [45] |
| RelE, HigB, YoeB, YafQ, MqsR, YafO | Ribosome-dependent mRNA cleavage | Translation | [46,72,[73], [74], [75], [76]] | ||
| 2 | MazF | MazF, YdcE, PemK, ChpBK | mRNA, rRNA and tRNA cleavage | Translation | [[47], [48], [49],[77], [78], [79]] |
| CcdB | DNA gyrase inhibition | Replication | [50] | ||
| 3 | HicA | HicA | mRNA cleavage | Translation | [51] |
| 4 | VapC | VapC, PhoH2 | Cleavage in anticodon region of tRNAs Cleavage of 23S rRNA at the sarcin-ricin loop | Translation | [52,53,64] |
| 5 | HipA | HipA, HipT | Phosphorylation of glutamyl-tRNA synthetase and tryptophanyl-tRNA synthetase | Translation | [54,55] |
| 6 | FicT/Doc | FicT | Adenylylation of Topoisomerase IV and DNA gyrase | Replication | [57,58] |
| Doc | Phosphorylation of EF-Tu elongation factor | Translation | [56] | ||
| 7 | AtaT/TacT | AtaT | Acetylation of Gly-tRNAGly, Trp-tRNATrp, Tyr-tRNATyr, Phe-tRNAPhe and Met-tRNAfMet | Translation | [59,60] |
| TacT | Acetylation of Gly-tRNAGly, Trp-tRNATrp, Leu-tRNALeu and Ser-tRNASer | Translation | [61] | ||
| 8 | Zeta | Zeta, PezT, EzeT | Phosphorylation of peptidoglycan precursor uridine diphosphate-N-acetylglucosamine (UNAG) | Cell wall synthesis | [62,65,70] |
| 9 | MbcT | MbcT | Phosphorylation of NAD+ | Metabolic stress | [63] |
| Other identified novel toxin superfamilies | |||||
| 12 | VapD | VapD | RNA cleavage | Not defined | [80] |
| 13 | RnlA | RnlA | mRNA cleavage | Translation | [81] |
2.3. Type III- (Fig. 1C)
Type III TA pair of genes exist in an operon and both genes are separated by a weak transcriptional terminator. The operon encodes toxin protein and antitoxin RNA (Fig. 1C). Till now, very few type III TA systems have been identified. For example, the first identified type III TA system is toxN-toxI of Erwinia carotovora [82] and the other one is abiQ-antiQ of Lactococcus lactis [83]. TA operon encodes full-length antitoxin RNA (precursor RNA) consisting of repeats of small nucleotide sequences and toxin protein. When a toxin is overexpressed, it causes bacteriostasis [82,84]. The toxin is a sequence specific endoribonuclease that cleaves repetitive precursors of antitoxin RNA into individual RNA pseudoknot units [85]. RNA pseudoknots are a dominant and compact form of structured RNAs with available surfaces for interaction with other molecules [86,87]. Pseudoknot units of RNA antitoxin bind to toxin and form inactive complex, leading to toxin inactivation [85].
2.4. Type IV- (Fig. 1D)
Type IV TA systems exist in operons and encode both toxin and antitoxin in protein form. To date, only a few type IV TA systems have been reported. The first identified type IV TA system is cbtA-cbeA of E. coli and two more homologs, ykfI-yafW and ypjF-yfjZ have also been identified in E. coli. Toxin CbtA exerts its toxic effect by inhibiting the polymerization of cytoskeletal proteins FtsZ and MreB leading to alteration in cell shape, consequently inhibiting the replication and causing cell-growth arrest. Antitoxin CbeA does not interact with toxin for inactivation but inhibits toxin action indirectly by enhancing the bundling of cytoskeletal polymers of MreB and FtsZ (Fig. 1D) [88].
2.5. Type V- (Fig. 1E)
To date, only one type V TA system, ghoT-ghoS has been identified in E. coli. ghoT-ghoS exists in an operon and encodes toxin GhoT and antitoxin GhoS, small proteins. GhoT is a lytic peptide that causes ghost cell formation. GhoS prevents GhoT toxin-mediated toxicity by cleaving the ghoT mRNA and inhibiting its translation. In the absence of GhoS, the GhoT is lethal (Fig. 1E) [89,90].
2.6. Type VI- (Fig. 1F)
To date, only one type VI TA system, socB-socA of Caulobactercres centus has been identified [91]. socB-socA exists in an operon and unlike the canonical TA system, SocB toxin is unstable and susceptible to protease ClpXP whereas SocA antitoxin acts as a proteolytic adapter for the degradation of SocB (Fig. 1F). SocA binds to its substrate SocB and delivers SocB to proteases for degradation. SocB exerts its toxic effect by inhibiting DNA replication elongation via direct interaction with DnaN. DnaN is a beta sliding clamp protein thatr forms a ring around the dsDNA. During replication, DnaN binds to DNA polymerase III and increases its processivity [92,93]. SocB competes with RNA polymerase III for binding to DnaN and disrupts the synthesis of both strands leading to replication fork collapse [91].
2.7. Type VII- (Fig. 1G)
Type VII TA systems exist as operons and encode both toxin and antitoxin in protein form. Overexpression of the toxin causes growth arrest, and the toxin is inactivated by antitoxin mediated modification of the toxin. Antitoxin interacts with toxin and causes post-translational modification such as phosphorylation, oxidation and AMPlyation (Fig. 1G). To date, three subtypes of VII TA systems, namely, tgIT-takA/menT3-menA3, hha-tomB, and HEPN-MNT have been identified. tgIT-takA/menT3-menA3 (Rv1044-Rv1045) of Mycobacterium tuberculosis encodes toxin TgIT/MenT3, a nucleotidyltransferase [94,95] and antitoxin TakA/MenA3, a serine protein kinase [94]. TgIT/MenT3 inhibits protein synthesis by preventing the charging of tRNAs, preferentially tRNASer by adding pyrimidines (C or U) not purines, to the 3′-CCA acceptor stems of uncharged tRNAs [95]. Contrarily, Yu et al. have shown that TgIT/MenT3 binds to GTP specifically [94]. TgiT/MenT3 inhibits the growth of bacteria and its cognate antitoxin TakA/MenA3 antagonizes its toxic activity [94,95]. TakA/MenA3 interacts with TgiT/MenT3 [94,95] and inactivates by phosphorylation of TgIT [94]. Further, in hha-tomB of E. coli, overexpression of Hha is bacteriolytic due to repression of rare tRNAs, leading to alteration in translation, induction of proteases and activation of lytic cryptic prophage genes [96]. TomB mediated oxidation of Hha causes loss of Hha structure that results in Hha inactivation [97]. The third TA system, HEPN-MNT exists as an operon and encodes HEPN toxin protein and MNT antitoxin protein [98]. HEPN toxin act as ribonuclease which cleaves mRNA and tRNA thereby affecting translation. Interestingly, in Shewanella oneidensis, HEPN toxin cleaves mRNAs not tRNA or rRNA while in Aphanizomenon flos-aquae, HEPN cleaves tRNA at a specific position, not mRNA or rRNA [99]. MNT inactivates HEPN by di-AMPlylation in A. flos-aquae while by tri-AMPlylation in S. oneidensis [100,101].
2.8. Type VIII- (Fig. 1H)
Type VIII TA gene pair encodes both toxin and antitoxin in RNA form (Fig. 1H). To date, only two type VIII TA systems, creT-creA in Haloarcula hispanica and sdsR-ryeA in E. coli have been reported. Interestingly, creT-creA system is embedded within diverse CRISPR-Cas loci where the CRISPR cascade regulates the transcription of creT-creA pair. creT-creA system consists of CreT (Cascade-repressed toxin) toxin and CreA (CRISPR RNA–resembling antitoxin RNA) antitoxin. Expression of CreT causes bacteriostasis by sequestering the rare tRNAArg . Sequestration of the tRNAArg impairs the translation of some essential genes leading to growth arrest. CreA mimics a CRISPR RNA and inhibits CreT transcription in association with cascade. CreA requires Cas6 for maturation, thus CreA becomes antitoxic only in the presence of cascade. creA RNA and creT promoter have partial complementarity which directs the cascade for repression of creT transcription. creT-creA system forms a symbiosis with CRISPR to make CRISPR addictive for the host [102]. The other TA system, sdsR-ryeA of E. coli encodes small sdsR toxin RNA and ryeA antitoxin RNA which are transcribed through the opposite strand. Expression of sdsR causes Hfq-dependant cell death. SdsR regulates the expression of many genes including yhcB, an inner membrane protein whose repression by SdsR leads to cell death. To counter the SdsR toxic effect, RyeA interacts with SdsR via base pairing and covers the entire SdsR leading to degradation of both RNAs by RNase III [103].
2.9. New type of TA systems
Recently few TA systems have been identified with unique characteristics or similarities with more than one type of TA system. Some examples are briefly described below.
2.9.1. darT-darG TA system
Jankevicius et al. identified a new TA system, darT-darG in M. tuberculosis [104]. darT-darG TA pair encodes DarT toxin and DarG antitioxin, and both are in protein form. Expression of DarT is bacteriostatic which is reversed by DarG expression. DarT acts as DNA ADP-ribosyltransferase that specifically modifies thymidines on single-stranded DNA in a sequence-specific manner thereby affecting DNA replication. The modification of thymidines can be removed by DarG indicating that darT-darG pair acts via reversible DNA ADP-ribosylation. Further, darT-darG TA system also shows stable protein-protein interaction which might be additional toxin inhibition activity of DarG [104].
2.9.2. ToxSAS–antiToxSAS TA system
Jimmy et al. discovered five small alarmone synthetase (SAS subfamilies), FaRel, FaRel2, PhRel, PhRel2, and CapRel with TA-like arrangements and showed that four of them exist in two gene operon while FaRel is three gene operon. ToxSAS toxin exert their toxicity through the production of toxic nucleotide alarmones, ppGpp and ppApp which at high concentrations inhibits bacterial growth via targeting transcription, translation, and ribosome assembly [105,106]. Further, Jimmy et al. showed that out of five subfamilies, four of them encode an antitoxin that neutralizes only their cognate antitoxin. However, in the case of faRel system (three gene operon system), the toxSAS gene is flanked by two antitoxin genes and each gene is sufficient to counter the toxSAS toxicity. One antitoxin works as a type II TA system antitoxin which interacts with toxSAS via protein-protein interaction, whereas, another antitoxin acts as a type IV antitoxin which encodes a (p)ppGpp degrading enzyme, small alarmone hydrolase (SAH) and degrades the molecular product of toxSAS [106].
Characteristics of both TA systems indicate that there is a crosstalk among different type of TA systems . They are sharing the characteristics of two TA systems, type II and type IV, suggesting that the TA system should be reclassified based on other parameters such as sequence similarity or organism specific (bacterial or archaeal) or localisation (chromosomal or plasmid) rather than the mode of toxin inhibition by antitoxin.
2.10. Biological functions of TA systems
Most of the TA systems have been proposed to play roles in phage inhibition and stress response. For example, several type I TA systems of B. subtilis are located on prophages and they have been shown to play an important role in phage maintenance [41]. Type II mazF-mazE and rnlA-rnlB are involved in phage inhibition as they significantly block infection of phage P1 and phage T4, respectively [,107,108]. Type II PemK toxin induces dormant state and inhibits phage infection [109]. Type III toxins have been shown to be involved in abortive phage infection [[83], [84]] [,]. The newly identified ToxSAS-antiToxSAS and darT-darG TA systems also have been shown to be involved in the inhibition of phage infection [110,111]. Moreover, the role of TA systems have also been proposed in stress response, biofilm formation and persister formation. For example, in type I TA systems, SOS induces the transcription of toxins, tisB, dinQ and symE [27]. In E. coli, TisB toxin induces persister cell formation after the administration of ciprofloxacin [112]. Persister cells are dormant cells that show tolerance to antibiotics without undergoing genetic change [113]. Later, TisB-induced persistence was shown as phase dependent as TisB induces antibiotic persistance in exponantial phase not in stationary phase cultures [112,114] or condition dependent [115]. Type II TA systems are the most abundant and characterized TA system, and they have been shown to play important role in stress response, biofilm and persister cell formation [116,[117], [118], [119]].. However, their roles have been challenged by many recent studies [[10], [11], [12], [13], [14], [15]]. Moreover, under adverse conditions like nutrient starvation, oxidative stress, and antibiotic challenge, they are often transcriptionally upregulated [[120], [121], [122]] but does not lead to toxin activation [123]. Type IV TA systems have been shown to be involved in resistant to oxidative stress and biofilm formation [124]. Type V TA system, ghoT-ghoS has been shown to be involved in persister cell formation [89] and adaptation of growth under the unfavourable condition [90]. Role of type VI TA systems has been proposed in DNA damage response as the socB-socA operon is induced by the DNA damaging agent, mitomycin [91]. Type VII TA system, hha-tomB is highly induced in biofilms [125] and presumably controls the biofilm formation and virulence [126]. Type VIII TA systems, creT-creA makes cells addicted to CRISPR whereas sdsR might be involved in RpoS-mediated stress-survival response [127,128]. Overall, these studies suggest that TA systems play significant role in phage inhibition. Although TA systems have been implicated in stress response, their role has remained controversial.
3. TA loci in pathogenic bacteria
The abundance of TA systems in pathogens suggests that better understanding of the mechanisms of action of TA systems in pathogens may enable the development of new lines of treatment for infections caused by them. Hospital-acquired infections are an important concern as infections caused by multidrug-resistant (MDR) bacteria (superbugs) lead to high mortality. ESKAPE is a group of antibiotic-resistant bacteria that can escape the biocidal action of antibiotics and represents new paradigms in pathogenesis, transmission, and resistance [129]. Several type II TA systems are present in ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter spp.) as well as in other pathogen including E. coli, Burkholderia spp., Streptococcus pneumoniae and M. tuberculosis [130]. Moreover, other studies have also reported the presence or abundance of TA loci in pathogenic organisms such as in M. leprae, M. tuberculosis, Rickettsia prowazekii, Corynebacterium diphtheriae, Treponema pallidum, Yersinia pestis, Bordetella pertussis, S. pneumoniae, S. pyogenes, Salmonella typhimurium, Shigella dysenteriae, Vibrio cholerae, Klebsiella pneumoniae, Bacillus anthracis and Pseudomonas putida [16,17,[131], [132], [133], [134], [135]]. Besides that, TA systems are present on virulence plasmids of pathogenic bacteria for their stabilization. For example, three TA systems, mvpT-mvpA/vapC-vapB, ccdB-ccdA and gmvT-gmvA present on pINV plasmid of Shigella species [136,137] and two vapC-capB and ccdB-ccdA TA systems on virulence plasmid pSLT of S. typhimurium [138]. Moreover, multiple TA systems have been detected on a plasmid encoding extended-spectrum β-lactamases (ESBLs) that are most important acquired resistance determinants in members of the Enterobacteriaceae [[139], [140], [141]]. Pathogenic E. coli producing CTX-M ESBLs have become a major cause of infections in both the community and hospitals [139]. TA systems, pemK-pemI, ccdB-ccdA, vagD-vagC, Hok–Sok and srnB-srnC are the most frequently represented TA systems in E. coli producing CTX-M ESBLs [141] that probably contribute to plasmid maintenance in their host [142,143]. Taken together, these studies shows the abundance of TA loci in pathogenic organisms including antibiotic resistant organisms and suggest the possible link between the TA systems and pathogenicity.
4. Contribution of TA system in bacterial pathogenesis
Many studies have provided evidence of the involvement of the TA system in bacterial pathogenesis and virulence using infection models. To date, type I, II, and VII TA systems have been found to be involved in bacterial virulence. Examples of TA systems's contribution to bacterial virulence are discussed in the following sections and summarized in Table 2.
Table 2.
TA systems in bacterial pathogenesis.
| TA system | Pathogen | Role of TA system | Infection model | Contribution to pathogenesis | Reference |
|---|---|---|---|---|---|
| Type I TA system | |||||
| sprG1-sprF1 | Staphylococcus aureus | Hemolysis | Erythrocytes | Host cell lysis | [144] |
| ef0409-ef0408 | Enterococcus faecalis | Adaptation to host stresses, regulation of expression of metabolic enzymes | larvae Galleria mellonella, macrophages and mice | Hypervirulence, Intracellular survival, colonization | [18] |
| tisB-istR, ldrA-rdlD and hok-sok | Salmonella typhimurium | Regulation of growth of bacteria inside the host | Fibroblast | Fitness,Intracellular survival | [19] |
| Type II TA system | |||||
| yoeB-yefM | Extraintestinal pathogenic Escherichia coli (ExPEC) | Regulation of growth of bacteria inside the host | Mice | Colonization | [20] |
| pasT-pasI | ExPEC | Adaptation to host stresses, Tolerance to antibiotics, Biofilm and persister formation | Mice | Fitness, Intracellular survival | [20] |
| vapC-vapB and vapD-vapX | Haemophilus influenzae | Persister formation | Epithelial tissue | Survival inside the host, Colonization, Persistent infection | [21] |
| vapB2-vapC2 and T4-A4 | S. typhimurium | Regulation of growth of bacteria inside the host, Metabolic readjustment | Macrophage, Fibroblast, and Epithelial cell | Fitness, Intracellular survival | [19] |
| mazF-mazE | S. aureus | MazF cleaves SraP mRNA, a virulence determinant | – | Regulation of virulence factors | [145] |
| Mycobacterium tuberculosis | Regulation of growth of bacteria inside the host | Guinea pig | Pathology in organs | [22] | |
| higB1-higA1 | M. tuberculosis | Regulation of expression of the genes involved in virulence, detoxification, and adaptation | Guinea pig | Infection, Fitness | [23] |
| fitB-fitA | Neisseria gonorrhoeae | Regulates rate of intracellular replication and growth of bacteria | Epithelial cell | Intracellular survival | [146] |
| relE-relB | Vibrio cholerae | Biofilm and persister cell formation | Mice | Intracellular survival colonization | [147] |
| Type VII TA system | |||||
| hha-ybaJ | ExPEC | Regulation of growth of bacteria inside the host | Mice | Colonization | [20] |
| menT2-menA2 | M. tuberculosis | Regulation of growth of bacteria inside the host | Guinea pig | Colonization | [148] |
4.1. Type I TA systems in bacterial virulence
4.1.1. sprG1-sprF1
sprG1-sprF1 TA system has been shown to play role in the virulence of S. aureus [144]. S. aureus is a human pathogen that causes life-threatening community-associated infections [149]. The sprG1-sprF1 of S. aureus encodes antitoxin RNA and a small hydrophobic peptide toxin. The sprG1 toxin gene possesses two internal initiation codons which result in the generation of two different peptides, a short and a long peptide. The shorter peptide is more effective in bacterial growth inhibition whereas the longer peptide is more active against human erythrocytes. The longer peptide is primarily expressed and secreted into the extracellular medium. After secretion, the longer peptide is supposed to cause hemolysis as synthetic SprG1 peptides have been shown to be able to lyse erythrocytes efficiently. Longer peptides are rich in positively chargeed aa which is the probable reason for their higher secretion and thereby high cytolytic effect against erythrocytes [144]. Therefore, it was suggested that longer peptides can be effective for host cell lysis. However, the shorter peptide has been shown to be effective in causing interspecies death, such as in E. coli and P. aeruginosa, which are frequently associated with staphylococcal infections [144]. Thus, if both peptides are produced in sufficient quantity, they can cause host cell lysis leading to the release of virulence factor, facilitating the spread of infection and allowing growth advantage over other bacterial species as well.
4.1.2. ef0409-ef0408
In E. faecalis, the ef0408 sRNA of ef0409-ef0408 is intricately involved in the infection [18]. E. faecalis an inhabitant of the gastrointestinal tracts of humans and animals is a common hospital-acquired infection associated with catheter-associated urinary tract infections, endocarditis, and surgical and burn wound infections [150]. Chromosome encoded ef0409-ef0408 [151,152] is homologous to the RNAI-RNAII TA system, and ef0408 is supposed to act as an antitoxin through interaction with ef0409 [[152], [153], [154]]. Michaux et al. selected 6 sRNA candidates including ef0408 sRNA of E. faecalis and assessed their role in pathogenesis as well as in stress response. To determine their involvement in the infection, they created sRNA deletion mutants and tested their effect on the infection process using three infection models: the larvae Galleria mellonella, macrophages, and mice. Among all 6 mutants, three mutants including Δef0605, Δef1368, and Δef3314 showed less virulent phenotype than the wild type as it decreased the killing of G. mellonella larvae. However, Δef0408 mutant showed the hypervirulence phenotype as it increased the G. mellonella larvae killing rate significantly. Further, in Δef0408 infected mice, bacterial load was higher in the liver and kidneys. Additionally, in the macrophage infection model, the mutant survived better than the wild type. Moreover, to confirm the involvement of ef0409-ef0408 in virulence, they investigated the phenotype of mutant, wild-type and complemented strains under conditions that might be relevant in the gastrointestinal tract or during the infection process such as oxidative-, osmotic-, detergent stress conditions and serum. The mutant Δef0408 showed better survival under such conditions [18]. Taken together, authors suggested that these sRNAs stabilize the homeostasis of the cells specifically under environmental changes. Commensal bacteria such as E. faecalis may need an equilibrium between favorable colonization (by repressing virulence) and pathogenicity according to the host environment. Therefore, sRNA may act as a key regulator in the transition from a commensal relationship to virulence. Besides that, transcriptional analysis of Δef0408 mutant and wild type showed low transcription level of ef0409 in both cases [18]. Even at low level, free toxin ef0409 might be contributed to its hypervirulent phenotype. ef0408 sRNA could be acting as sensor and suppressor of ef0409 toxin activity to control growth and virulence. Overall, it suggests that more research is needed to gain insight into the mechanism of sRNA mediated regulation of bacterial virulence.
4.1.3. tisB-istR, ldrA-rdlD and hok-sok
Lobato-Marquez et al. investigated the activity of different TA modules in the regulation of the growth of S. typhimurium inside the host [19]. They showed that a selected group of type I toxins, TisB, LdrA and Hok are required for the survival of intracellular S. typhimurium. They impact the fitness of bacteria inside the fibroblast by negatively regulating bacterial growth. The controlled action of these toxins may assist S. typhimurium in acquiring the metabolic dormant state in fibroblasts. To confirm the role of toxins, they analyzed whether S. typhimurium produces these toxins in response to the host environment. S. typhimurium isolated from human fibroblast showed upregulation of these three toxins indicating S. typhimurium upregulates these toxins in response to the intracellular environment of fibroblast which impacts the growth negatively [19]. Importantly, they also showed that ldrA-rdl and hok-sok are absent in non-pathogenic species S. bongori. Furthermore, ldrA is conserved in all S. enterica serovars indicating the importance of the ldrA-rdlD TA system for S. enterica pathogenesis.
4.2. Type II TA systems in bacterial virulence
4.2.1. yoeB-yefM and pasT-pasI
Norton et al. identified the role of yoeB-yefM and pasT-pasI in the virulence of extraintestinal pathogenic E. coli (ExPEC) strain and showed that each system acts independently to promote colonization and persistence [20]. ExPEC colonizes in diverse niches and causes sepsis, meningitis, and urinary tract infections. Only a subset of the known TA system of E. coli is associated with the ExPEC strain [155,156]. To address their role in pathogenesis, they constructed deletion mutants of TA systems and tested them in a murine infection model. They found that yoeB-yefM and pasT-pasI are required for the colonization and survival of ExPEC inside the host. yoeB-yefM is independently involved in the colonization of ExPEC in the bladder, whereas pasT-pasI is required for the survival of ExPEC inside the kidneys. Moreover, pasT-pasI also increases the persister cell formation thereby tolerance to antibiotics. Low-level expression of pasT protects ExPEC from stresses such as nutrient limitation, and oxidative and nitrosative stress [20]. Thus, these observations suggest both TA systems provide a fitness advantage to the pathogen depending on the level of toxin expression and environmental stress. However, a recent study has shown that pasT-pasI is not a TA system but a bacterial homolog of the mitochondrial Coq10 that acts as an accessory factor in the ubiquinone-dependent electron transport chain [157].
4.2.2. vapC1-vapB1 and vapD-vapX
Ren et al. have reported that during extended infection of non-typeable Haemophilus influenzae (NTHi), vapC1-vapB1 and vapD-vapX TA loci maintain its survival and persistent infection [21]. NTHi is a common commensal of the upper respiratory tract and causes respiratory tract infections in humans. NTHi is the most common cause of infection in the middle ear (otitis media) [158]. During infection, NHTi cells are exposed to adverse conditions such as host immune responses, nutrient deprivation, and antibiotic treatment. NTHi forms biofilms that possibly play a role in resistance to host immune response and antibiotics, thus favoring recurrent otitis media [159,160]. After countering those stresses, a subpopulation of NTHi causes persistent infection. As TA systems are known to be involved in biofilm formation and adaptation to environmental stresses, Ren et al. investigated the role of vapC1-vapB1 and vapD-vapX in NTHi pathogenesis [21]. They created deletion mutants of these TA loci and tested the survival in the respiratory epithelial tissue model during long-term infections. They showed both loci, vapC1-vapB1 and vapD-vapX contribute significantly to NTHi persistence and allow its survival during extended infections. Furthermore, to confirm the role of both loci in persistence and survival, they tested the mutants in a chinchilla model of otitis media and showed that both loci were independently involved in the survival of NTHi during extended infection [21]. Overall, both loci seem to be important for the intracellular survival of NTHi for a longer period that allows persistent infections.
4.2.3. vapC2-vapB2 and T4-A4
Lobato et al. have shown that VapC2 and T4 toxins impact the fitness of S. typhimurium inside fibroblasts [19]. S. enterica is an intracellular pathogen causing persistent infections in humans [161]. To study the pathogenesis of S. typhimurium, murine models have been extensively used as they mimic acute and chronic human infections [162]. During infection, S. typhimurium shows limited proliferation inside macrophages [163] and it attenuates growth in cultured fibroblasts [164] as well as in non-phagocytic cells of the intestinal lamina propria [165]. They analyzed the proteomes of isolated S. typhimurium from macrophages and found the upregulation of different toxins of type II TA systems including the VapC2 with the highest increase. To determine the contribution of toxins in S. typhimurium dormancy during chronic and persistent infection, they constructed deletion mutants of upregulated type II toxins and tested them in macrophage, fibroblast, and epithelial cell infection models. Among toxin mutants, ΔvapC2 mutant showed a strong phenotype with an 80% decrease in survival inside fibroblasts. Furthermore, in all three infection models (macrophage, fibroblast, and epithelial cells), apart from vapC2, the T4 mutant also decreased the survival and limited the intracellular proliferation of S. typhimurium. Taken together both VapC2 and T4 toxins impact the fitness of S. typhimurium inside the host cell. T4 has a Gcn5-related acetyl transferase (GNAT) domain [19] and in S. typhimurium, GNAT domain-containing protein has been related to the control of carbon utilization and metabolic flux via acetylation of several metabolic enzymes [166]. Thus, the T4 mutant phenotype indicates an important role of the metabolic readjustment in the fitness of S. typhimurium inside the host cell. Overall, both vapC2-vapB2 and T4-A4 TA systems contribute to the survival of S. typhimurium inside the host during chronic and persistent infection.
4.2.4. mazF-mazE
mazF-mazE TA systems have been shown to be involved in the pathogenesis of different micro-organisms. Some notable examples are S. aureus and M. tuberculosis which are briefly discussed below.
4.2.4.1. mazF-mazE of S. aureus
S. aureus is the most common cause of hard-to-treat infections at hospitals and healthcare facilities. S. aureus causes an endovascular infection where interaction with human platelets is required for its pathogenesis. In cardiovascular infections, SraP, a glycoprotein, has been reported as a key virulence determinant that contains a cell wall-anchoring motif (LPXTG) [167,168]. Zhu et al. have shown that SraP mRNA is highly susceptible to the ribonuclease activity of MazF toxin as it contains a high number of MazF cleavage-specific sequences. Therefore, when mazF was induced under stress conditions, the synthesis of SraP was significantly reduced. Further, they have shown that the MazF cleavage sequence is significantly abundant in the mRNAs of other pathogenic factors of S. aureus as well [145] which suggests an important regulatory role for mazF-mazE TA system in the pathogenicity of S. aureus.
Recently, another group, Ma et al. have shown the role of the mazF-mazE system in S. aureus chronic infection [6]. They examined the role of mazF-mazE in virulence using a murine model and showed that mazF increases antibiotic tolerance and allow the transition of bacteria from acute to chronic infection [6]. Therefore, mazF-mazE not only makes the S. aureus more tolerant to antibiotics but more tolerant to the host.
4.2.4.2. mazF-mazE of M. tuberculosis
M. tuberculosis is one of the most dreadful human pathogens [169] which causes tuberculosis by infecting primarily the lungs and then spreading to other organs such as kidneys, spine and brain. M. tuberculosis may persist in the small fraction of infected individual for a longer period and can be reactivated at any time during life [170]. Thus, latency in the host is an important factor for M. tuberculosis pathogenesis. TA loci have been proposed as one of the latency-inducing factors as they are involved in adaptation to host stresses, biofilm formation and persistence. M. tuberculosis possesses nine different mazF-mazE like TA loci where three TA loci, namely, Rv1102c (mazF3), Rv1991c (mazF6) and Rv2801c (mazF9) encode functional toxin [22]. Tiwari et al. had determined the contribution of three MazF toxins in adaptation to host stresses where all three MazFs contribute cumulatively to M. tuberculosis growth and adaptation to oxidative stress and nutrient deprivation in macrophages. Furthermore, mazF-mazE TA systems were differentially induced in M. tuberculosis persisters which are important for the adaptation as well as the virulence of M. tuberculosis [22]. Moreover, Kaushal et al. used the guinea pig experimental model of tuberculosis to study the contribution of MazFs in M. tuberculosis virulence. They showed that mazF triple deletion mutant (mazF3, mazF6 and mazF9) causes less pathology in lung and liver sections of guinea pigs. Additionally, the triple mutant showed more growth defects in the spleen and liver when compared to the lungs suggesting MazF might be involved in the dissemination of the disease from the lungs to the spleen and liver [22]. Overall, observations from both studies suggest that MazFs contribute to the ability of M. tuberculosis to adapt to oxidative and nutrient-limiting conditions as well as regulate the growth of M. tuberculosis inside the host.
4.2.5. higB1-higA1
HigB1-HigA1 of M. tuberculosis is not a classical two component TA system but a tripartite system (Rv1955-Rv1956-Rv1957), named TAC (Toxin-Antitoxin-Chaperone), composed of Rv1955-Rv1956 encoding a higB1-higA1 pair coupled to Rv1957 encoding a SecB like molecular chaperone [17,171]. Rv1957 chaperone controls higB1-higA1 by directly acting on the antitoxin by preventing its aggregation and protecting it from degradation. Moreover, HigA1 not alone but along with Rv1957 counteracts the toxin activity of HigB1 [171]. Like the MazF toxin, the HigB1 toxin of M. tuberculosis, has also been shown to contribute to its fitness, survival under host conditions and pathogenesis in a guinea pig model [23]. Deletion mutants of higB1 significantly reduced bacterial loads and pathological damage in the infected guinea pigs. Furthermore, transcriptome analysis of mutants showed the repression of the genes involved in virulence, detoxification and adaptation [23]. Therefore, it suggests that HigB1 might be required for the establishment of successful infection and fitness of M. tuberculosis inside the host.
4.2.6. fitB-fitA
Hopper et al. have reported the involvement of fitB-fitA in Neisseria gonorrhoeae pathogenesis [146]. N. gonorrhoeae is a pathogen that causes the sexually transmitted disease gonorrhoeae by infecting the mucosal epithelium of the urogenital tract. N. gonorrhoeae survive and grow within epithelial cells. Hopper et al. conducted a study where they screened different TA mutants in an epithelial cell model and showed that mutants of fitB-fitA operon exhibit a fast intracellular trafficking (fit) phenotype across polarized epithelial monolayers. Furthermore, the fitB-fitA mutant showed an accelerated rate of intracellular replication [146]. It indicates that components of fit-B-fitA operon act as intracellular growth regulators in N. gonorrhoeae and contribute to its persistency inside the host.
4.2.7. relE-relB
Wang et al. have reported the contribution of relE-relB TA system in the survival of Vibrio cholerae inside the host [24]. V. cholerae colonizes inside the intestine of a human host [147]. Wang et al. investigated the role of relE-relB loci in V. cholerae pathogenesis [24]. V. cholerae possesses 7 relE-relB loci and 6 out of 7 encode functional toxins. All seven relE-relB loci have been shown to be induced under virulence-inducing conditions in vitro. However, inside the host, these loci might help in biofilm formation to adapt to the host's stresses. Therefore, to assess their role in biofilm formation, Wang et al. constructed deletion mutants of each relE-relB locus and showed that relE7-relB7 is important for biofilm formation. Further, Wang et al. tested mutants in an infant mouse model also and showed that relE4-relB4 and relE7-relB7 contribute to V. cholerae colonization [24]. Therefore, these observations suggest that relE-relB TA systems play redundant roles in regulating biofilm formation and colonization, and contribute to V. cholerae survival and its virulence.
4.3. Type VII TA systems in bacterial virulence-
4.3.1. hha-ybaJ
hha-ybaJ TA system promotes colonization and persistence in E. coli strain, ExPEC [20]. Deletion mutant of hha-ybaJ in a murine infection model showed that Hha-ybaJ TA system is required for the colonization of ExPEC in the bladder [20]. Therefore, it suggests that hha-ybaJ might be involved in ExPEC colonization and survival inside the host.
4.3.2. menT2-menA2
MenT2 toxin of menT-menA2 plays important role in mycobacterial pathogenesis as deletion of menT2 significantly reduced the bacterial count in the lung and spleen of mutant infected guinea pigs. Lungs of guinea pigs infected with wild type strain showed severe tissue damage however, the tissue damage was considerably reduced in ΔmenT2 strain infected guinea pigs [148]. These observations suggest that MenT2 plays important role in successful colonization or bacterial dissemination from the lungs to the spleens.
5. Discussion
Earlier TA systems were regarded as selfish systems ensuring their maintenance in the cell. Later, several studies demonstrated that TA loci modulate important functions of the cells in response to different stressors and may act as stress response managers. Under stress conditions, TA loci regulate bacterial growth and lead to biofilm formation and persister cell formation, both are equally important for the adaptation of bacteria. However, many recent studies have shown discrepancies in the role of TA systems in stress, biofilm, and pesister formation. For example, a previous study on type I toxin, TisB reported the it induces persister formation [112]. However, later it was shown that TisB is not essential for persistence [115]. To clear the discrepancy, Goormaghtigh et al. repeated the previous study results of Dorr T et al., and found that the results are reproducible. Therefore, they concluded that TisB-induced persistence to fluoroquinolones is condition dependent or specific to experimental conditions [115]. This suggest that studying role of TA systems in persistence is technically challenging as different group report different results using the same methodologies. Therefore, special attention needs to be paid to the limitation and drawbacks of these studies. Similarly, there is debate over the role of type II TA systems in stress response, biofilm formation, and persister formation. For example, the type II mqsR-mqsA system has previously been shown to play a role in stress response and biofilm formation [[172], [173], [174], [175], [176]]. A study by Fraikin et al. demonstrated that mqsR-mqsA system is not involved in the core biological functions of E. coli, such as stress response and biofilm formation [177]. However, a recent study described the role of the mqsR-mqsA-mqsC, a TAC system in bacterial defence against phage [178]. Interestingly, a previous study from K. Gerdes lab showed that deletions of 10 type II TA systems in E. coli decrease the level of persistence to antibiotics [179,180] and type II hipB-hipA TA system induces persistence through activation of 10 TA systems [181]. Later, Gerdes lab discovered that the 10 TA systems deleted strain was severely compromised by infection of φ80 prophages and the observed decrease in persistence was due to these phage infections, not the TA systems [15]and this observation led to retraction of previous findings [[182], [183], [184]]. Recently, a similar observation was also reported by other groups that deletions of 10 TA systems in E. coli do not affect the persistence to antibiotics [185,186]. Similarly, in S. enterica, deletion of 12 TA systems showed no significant effect on persistence [187]. In P. putida also, TA systems are not important elements in stress tolerance as deletion of 13 TA systems did not affect the phenotypes associated to tolerance to different stress factors, the abundance of persister cells, and biofilm formation [188]. Another discrepancy was reported about pasT-pasI of ExPEC which was shown as TA pair and PasT induces persister formation leading to antibiotic tolerance in ExPEC [20]. Moreover, another study also showed that RatA/PasT/YfjG of E. coli is a TA toxin that inhibits protein synthesis by inhibiting ribosome assembly [189]. However, Fino et al. showed that there is no link of PasT to either TA systems or ribosomes, but PasT is a bacterial homolog of mitochondrial Coq10 that acts as an accessory factor in the ubiquinone-dependent electron transport chain (ETC) [157]. Further, the downstream gene, YfjF/RatB/PasI also did not show antitoxic activity [157,189]. Regarding the role of PasT in antibiotic tolerance, Fino et al. deny the direct role of PasT as it seems to be deeply wired into bacterial redox balance and energy metabolism through its role as a facilitator of ubiquinone-dependent respiration. Therefore, the link of PasT in antibiotic tolerance is indirect and mediates through broad distortions of bacterial physiology caused by defective aerobic respiration. Similarly, another study also showed that the expression of the PasT/RatA ortholog of S. typhimurium does not affect bacterial growth [19]. Interestingly, the PasT of S. typhimurium has a difference of five amino acids from the first ten amino acids of E. coli PasT that are required for toxicity. Replacement of this region by the E. coli PasT sequence partially restored anti-proliferative activity. Moreover, they identified some type II TA systems including PasTI in S. typhimurium which did not behave like bonafide TA system as they did not show toxicity or neutralization effect. For example, CcdB toxin of S. typhimurium did not show toxicity and when the sequence of CcdB of S. typhimurium and E. coli was compared, R99W amino acid substitution was found in the CcdB of S. typhimurium and that residue is crucial for the toxicity in E. coli. The reversion of that R99W amino acid substitution restored the toxin activity of CcdB in S. typhimurium [19]. Therefore, these observations suggest that a few TA modules are diverging and losing their properties with respect to functional homologs. Taken together, these studies strongly emphasize the need of a review of the TA systems and the reassessment of the biological functions of TA systems. Further, TA studies need to be performed with standard and common protocols using well-characterized strains as many groups are producing contradictory results using same methodologies.
6. Concluding remarks
The involvement of several TA loci in bacterial pathogenesis has been established using infection models. During bacterial pathogenesis, they play important role in the establishment of infection, survival, fitness, and colonization inside the host (Fig. 2). Based on their contribution to virulence, the TA system can be also considered a new class of virulence factors. TA modules contribute to the fitness of bacteria, and they help bacteria to survive longer inside the host during chronic infection. Moreover, TA loci are significantly induced under virulence-inducing conditions or in intracellular bacteria isolated from the infected host. Therefore, it suggests that TA modules play a crucial role in bacterial pathogenesis. However, the underlying mechanism of their contribution in pathogenesis is not well understood as there are many unanswered questions surrounding the mechanism of action of the TA system during bacterial infection. Some questions are, what is the level of expression of toxins during the infection? What are the factors that activate TA loci inside the host? What are the pathways involved in signaling TA loci in response to host alarms? How antitoxin degradation by proteases is regulated? etc. Answers to these questions will help in better understanding of the mechanism of action of each component of the TA system in bacterial virulence. In summary, TA systems contribute to bacterial pathogenesis and more research is needed to gain mechanistic insights into the working of TA systems. It could allow better disease control and management.
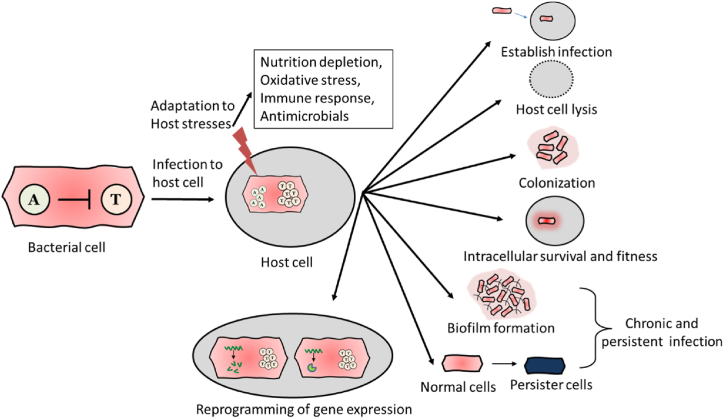
Fig. 2.
Contribution of TA systems in bacterial pathogenesis. During infection, bacteria get exposed to host stresses such as nutrient deprivation, oxidation, immune response and antimicrobials. Induction of toxins helps bacteria to establish successful infection, adapt host environment, intracellular survival and fitness, host cell lysis, and better colonization in organs. Toxins increase biofilm formation and persister cell formation which assist choronic and persistant infection. TA loci reprogramme the metabolic genes to adapt the host stesses and enhance bacterial virulence.
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
No data was used for the research described in the article.
Declaration of interest's statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Gerdes K., Christensen S.K., Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 2.Pandey D.P., Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Melderen L., Saavedra De Bast M. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerdes K., Rasmussen P.B., Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. U. S. A. 1986;83:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hazan R., Sat B., Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 2004;186:3663–3669. doi: 10.1128/JB.186.11.3663-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma D., Mandell J.B., Donegan N.P., Cheung A.L., Ma W., Rothenberger S., et al. The toxin-antitoxin MazEF drives Staphylococcus aureus biofilm formation, antibiotic tolerance, and chronic infection. mBio. 2019;10 doi: 10.1128/mBio.01658-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pecota D.C., Wood T.K. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J. Bacteriol. 1996;178:2044–2050. doi: 10.1128/jb.178.7.2044-2050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown J.S., Gilliland S.M., Spratt B.G., Holden D.W. A locus contained within a variable region of pneumococcal pathogenicity island 1 contributes to virulence in mice. Infect. Immun. 2004;72:1587–1593. doi: 10.1128/iai.72.3.1587-1593.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rycroft J.A., Gollan B., Grabe G.J., Hall A., Cheverton A.M., Larrouy-Maumus G., et al. Activity of acetyltransferase toxins involved in Salmonella persister formation during macrophage infection. Nat. Commun. 2018;9:1993. doi: 10.1038/s41467-018-04472-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Melderen L., Wood T.K. Commentary: what is the link between stringent response, endoribonuclease encoding type II toxin-antitoxin systems and persistence? Front. Microbiol. 2017;8:191. doi: 10.3389/fmicb.2017.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramisetty B.C., Ghosh D., Roy Chowdhury M., Santhosh R.S. What is the link between stringent response, endoribonuclease encoding type II toxin-antitoxin systems and persistence? Front. Microbiol. 2016;7:1882. doi: 10.3389/fmicb.2016.01882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shan Y., Brown Gandt A., Rowe S.E., Deisinger J.P., Conlon B.P., Lewis K. ATP-dependent persister formation in Escherichia coli. mBio. 2017;8 doi: 10.1128/mBio.02267-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harms A., Maisonneuve E., Gerdes K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science. 2016;354 doi: 10.1126/science.aaf4268. [DOI] [PubMed] [Google Scholar]
- 14.Harms A., Brodersen D.E., Mitarai N., Gerdes K. Toxins, targets, and triggers: an overview of toxin-antitoxin biology. Mol. Cell. 2018;70:768–784. doi: 10.1016/j.molcel.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Harms A., Fino C., Sorensen M.A., Semsey S., Gerdes K. Prophages and growth dynamics confound experimental results with antibiotic-tolerant persister cells. mBio. 2017;8 doi: 10.1128/mBio.01964-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgiades K., Raoult D. Genomes of the most dangerous epidemic bacteria have a virulence repertoire characterized by fewer genes but more toxin-antitoxin modules. PLoS One. 2011;6 doi: 10.1371/journal.pone.0017962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sala A., Bordes P., Genevaux P. Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins. 2014;6:1002–1020. doi: 10.3390/toxins6031002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michaux C., Hartke A., Martini C., Reiss S., Albrecht D., Budin-Verneuil A., et al. Involvement of Enterococcus faecalis small RNAs in stress response and virulence. Infect. Immun. 2014;82:3599–3611. doi: 10.1128/IAI.01900-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lobato-Marquez D., Moreno-Cordoba I., Figueroa V., Diaz-Orejas R., Garcia-del Portillo F. Distinct type I and type II toxin-antitoxin modules control Salmonella lifestyle inside eukaryotic cells. Sci. Rep. 2015;5:9374. doi: 10.1038/srep09374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norton J.P., Mulvey M.A. Toxin-antitoxin systems are important for niche-specific colonization and stress resistance of uropathogenic Escherichia coli. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren D., Walker A.N., Daines D.A. Toxin-antitoxin loci vapBC-1 and vapXD contribute to survival and virulence in nontypeable Haemophilus influenzae. BMC Microbiol. 2012;12:263. doi: 10.1186/1471-2180-12-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tiwari P., Arora G., Singh M., Kidwai S., Narayan O.P., Singh R. MazF ribonucleases promote Mycobacterium tuberculosis drug tolerance and virulence in Guinea pigs. Nat. Commun. 2015;6:6059. doi: 10.1038/ncomms7059. [DOI] [PubMed] [Google Scholar]
- 23.Sharma A., Sagar K., Chauhan N.K., Venkataraman B., Gupta N., Gosain T.P., et al. HigB1 toxin in Mycobacterium tuberculosis is upregulated during stress and required to establish infection in Guinea pigs. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.748890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., Wang H., Hay A.J., Zhong Z., Zhu J., Kan B. Functional RelBE-family toxin-antitoxin pairs affect biofilm maturation and intestine colonization in Vibrio cholerae. PLoS One. 2015;10 doi: 10.1371/journal.pone.0135696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brantl S. Bacterial chromosome-encoded small regulatory RNAs. Future Microbiol. 2009;4:85–103. doi: 10.2217/17460913.4.1.85. [DOI] [PubMed] [Google Scholar]
- 26.Brantl S. Acting antisense: plasmid- and chromosome-encoded sRNAs from Gram-positive bacteria. Future Microbiol. 2012;7:853–871. doi: 10.2217/fmb.12.59. [DOI] [PubMed] [Google Scholar]
- 27.Kawano M., Aravind L., Storz G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 2007;64:738–754. doi: 10.1111/j.1365-2958.2007.05688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo Y., Quiroga C., Chen Q., McAnulty M.J., Benedik M.J., Wood T.K., et al. RalR (a DNase) and RalA (a small RNA) form a type I toxin-antitoxin system in Escherichia coli. Nucleic Acids Res. 2014;42:6448–6462. doi: 10.1093/nar/gku279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerdes K., Bech F.W., Jorgensen S.T., Lobner-Olesen A., Rasmussen P.B., Atlung T., et al. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986;5:2023–2029. doi: 10.1002/j.1460-2075.1986.tb04459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ono T., Akimoto S., Ono K., Ohnishi Y. Plasmid genes increase membrane permeability in Escherichia coli. Biochim. Biophys. Acta. 1986;867:81–88. doi: 10.1016/0167-4781(86)90067-9. [DOI] [PubMed] [Google Scholar]
- 31.Weaver K.E., Weaver D.M., Wells C.L., Waters C.M., Gardner M.E., Ehli E.A. Enterococcus faecalis plasmid pAD1-encoded Fst toxin affects membrane permeability and alters cellular responses to lantibiotics. J. Bacteriol. 2003;185:2169–2177. doi: 10.1128/JB.185.7.2169-2177.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fozo E.M., Hemm M.R., Storz G. Small toxic proteins and the antisense RNAs that repress them. Microbiol. Mol. Biol. Rev. : MMBR (Microbiol. Mol. Biol. Rev.) 2008;72:579–589. doi: 10.1128/MMBR.00025-08. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Unoson C., Wagner E.G. A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol. Microbiol. 2008;70:258–270. doi: 10.1111/j.1365-2958.2008.06416.x. [DOI] [PubMed] [Google Scholar]
- 34.Weel-Sneve R., Kristiansen K.I., Odsbu I., Dalhus B., Booth J., Rognes T., et al. Single transmembrane peptide DinQ modulates membrane-dependent activities. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brantl S., Jahn N. sRNAs in bacterial type I and type III toxin-antitoxin systems. FEMS Microbiol. Rev. 2015;39:413–427. doi: 10.1093/femsre/fuv003. [DOI] [PubMed] [Google Scholar]
- 36.Thisted T., Gerdes K. Mechanism of post-segregational killing by the hok/sok system of plasmid R1. Sok antisense RNA regulates hok gene expression indirectly through the overlapping mok gene. J. Mol. Biol. 1992;223:41–54. doi: 10.1016/0022-2836(92)90714-u. [DOI] [PubMed] [Google Scholar]
- 37.Gerdes K., Wagner E.G. RNA antitoxins. Curr. Opin. Microbiol. 2007;10:117–124. doi: 10.1016/j.mib.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Darfeuille F., Unoson C., Vogel J., Wagner E.G. An antisense RNA inhibits translation by competing with standby ribosomes. Mol. Cell. 2007;26:381–392. doi: 10.1016/j.molcel.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 39.Wen J., Won D., Fozo E.M. The ZorO-OrzO type I toxin-antitoxin locus: repression by the OrzO antitoxin. Nucleic Acids Res. 2014;42:1930–1946. doi: 10.1093/nar/gkt1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greenfield T.J., Franch T., Gerdes K., Weaver K.E. Antisense RNA regulation of the par post-segregational killing system: structural analysis and mechanism of binding of the antisense RNA, RNAII and its target. RNAI. Mol. Microb. 2001;42:527–537. doi: 10.1046/j.1365-2958.2001.02663.x. [DOI] [PubMed] [Google Scholar]
- 41.Durand S., Jahn N., Condon C., Brantl S. Type I toxin-antitoxin systems in Bacillus subtilis. RNA Biol. 2012;9:1491–1497. doi: 10.4161/rna.22358. [DOI] [PubMed] [Google Scholar]
- 42.Jahn N., Brantl S. One antitoxin--two functions: SR4 controls toxin mRNA decay and translation. Nucleic Acids Res. 2013;41:9870–9880. doi: 10.1093/nar/gkt735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jurenas D., Fraikin N., Goormaghtigh F., Van Melderen L. Biology and evolution of bacterial toxin-antitoxin systems. Nat. Rev. Microbiol. 2022 doi: 10.1038/s41579-021-00661-1. [DOI] [PubMed] [Google Scholar]
- 44.Zhang S.-P., Wang Q., Quan S.-W., Yu X.-Q., Wang Y., Guo D.-D., et al. vol. 6. 2020. pp. 68–79. (Type II Toxin–Antitoxin System in Bacteria: Activation, Function, and Mode of Action). [Google Scholar]
- 45.Jiang Y., Pogliano J., Helinski D.R., Konieczny I. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 2002;44:971–979. doi: 10.1046/j.1365-2958.2002.02921.x. [DOI] [PubMed] [Google Scholar]
- 46.Pedersen K., Zavialov A.V., Pavlov M.Y., Elf J., Gerdes K., Ehrenberg M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell. 2003;112:131–140. doi: 10.1016/s0092-8674(02)01248-5. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y., Zhang J., Hoeflich K.P., Ikura M., Qing G., Inouye M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell. 2003;12:913–923. doi: 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 48.Mets T., Lippus M., Schryer D., Liiv A., Kasari V., Paier A., et al. Toxins MazF and MqsR cleave Escherichia coli rRNA precursors at multiple sites. RNA Biol. 2017;14:124–135. doi: 10.1080/15476286.2016.1259784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schifano J.M., Cruz J.W., Vvedenskaya I.O., Edifor R., Ouyang M., Husson R.N., et al. tRNA is a new target for cleavage by a MazF toxin. Nucleic Acids Res. 2016;44:1256–1270. doi: 10.1093/nar/gkv1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dao-Thi M.H., Van Melderen L., De Genst E., Afif H., Buts L., Wyns L., et al. Molecular basis of gyrase poisoning by the addiction toxin CcdB. J. Mol. Biol. 2005;348:1091–1102. doi: 10.1016/j.jmb.2005.03.049. [DOI] [PubMed] [Google Scholar]
- 51.Turnbull K.J., Gerdes K. HicA toxin of Escherichia coli derepresses hicAB transcription to selectively produce HicB antitoxin. Mol. Microbiol. 2017;104:781–792. doi: 10.1111/mmi.13662. [DOI] [PubMed] [Google Scholar]
- 52.Dienemann C., Boggild A., Winther K.S., Gerdes K., Brodersen D.E. Crystal structure of the VapBC toxin-antitoxin complex from Shigella flexneri reveals a hetero-octameric DNA-binding assembly. J. Mol. Biol. 2011;414:713–722. doi: 10.1016/j.jmb.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winther K., Tree J.J., Tollervey D., Gerdes K. VapCs of Mycobacterium tuberculosis cleave RNAs essential for translation. Nucleic Acids Res. 2016;44:9860–9871. doi: 10.1093/nar/gkw781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaspy I., Rotem E., Weiss N., Ronin I., Balaban N.Q., Glaser G. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat. Commun. 2013;4:3001. doi: 10.1038/ncomms4001. [DOI] [PubMed] [Google Scholar]
- 55.Vang Nielsen S., Turnbull K.J., Roghanian M., Baerentsen R., Semanjski M., Brodersen D.E., et al. Serine-threonine kinases encoded by split hipA homologs inhibit tryptophanyl-tRNA synthetase. mBio. 2019;10 doi: 10.1128/mBio.01138-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Castro-Roa D., Garcia-Pino A., De Gieter S., van Nuland N.A.J., Loris R., Zenkin N. The Fic protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat. Chem. Biol. 2013;9:811–817. doi: 10.1038/nchembio.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harms A., Stanger F.V., Scheu P.D., de Jong I.G., Goepfert A., Glatter T., et al. Adenylylation of gyrase and topo IV by FicT toxins disrupts bacterial DNA topology. Cell Rep. 2015;12:1497–1507. doi: 10.1016/j.celrep.2015.07.056. [DOI] [PubMed] [Google Scholar]
- 58.Lu C., Nakayasu E.S., Zhang L.Q., Luo Z.Q. Identification of Fic-1 as an enzyme that inhibits bacterial DNA replication by AMPylating GyrB, promoting filament formation. Sci. Signal. 2016;9 doi: 10.1126/scisignal.aad0446. ra11. [DOI] [PubMed] [Google Scholar]
- 59.Jurenas D., Chatterjee S., Konijnenberg A., Sobott F., Droogmans L., Garcia-Pino A., et al. AtaT blocks translation initiation by N-acetylation of the initiator tRNA(fMet) Nat. Chem. Biol. 2017;13:640–646. doi: 10.1038/nchembio.2346. [DOI] [PubMed] [Google Scholar]
- 60.Yashiro Y., Sakaguchi Y., Suzuki T., Tomita K. Mechanism of aminoacyl-tRNA acetylation by an aminoacyl-tRNA acetyltransferase AtaT from enterohemorrhagic E. coli. Nat. Commun. 2020;11:5438. doi: 10.1038/s41467-020-19281-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheverton A.M., Gollan B., Przydacz M., Wong C.T., Mylona A., Hare S.A., et al. A Salmonella toxin promotes persister formation through acetylation of tRNA. Mol. Cell. 2016;63:86–96. doi: 10.1016/j.molcel.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meinhart A., Alonso J.C., Strater N., Saenger W. Crystal structure of the plasmid maintenance system epsilon/zeta: functional mechanism of toxin zeta and inactivation by epsilon 2 zeta 2 complex formation. Proc. Natl. Acad. Sci. U. S. A. 2003;100:1661–1666. doi: 10.1073/pnas.0434325100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Freire D.M., Gutierrez C., Garza-Garcia A., Grabowska A.D., Sala A.J., Ariyachaokun K., et al. An NAD(+) phosphorylase toxin triggers Mycobacterium tuberculosis cell death. Mol. Cell. 2019;73:1282–12891 e8. doi: 10.1016/j.molcel.2019.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andrews E.S., Arcus V.L. The mycobacterial PhoH2 proteins are type II toxin antitoxins coupled to RNA helicase domains. Tuberculosis. 2015;95:385–394. doi: 10.1016/j.tube.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 65.Rocker A., Meinhart A. A cis-acting antitoxin domain within the chromosomal toxin-antitoxin module EzeT of Escherichia coli quenches toxin activity. Mol. Microbiol. 2015;97:589–604. doi: 10.1111/mmi.13051. [DOI] [PubMed] [Google Scholar]
- 66.Boggild A., Sofos N., Andersen K.R., Feddersen A., Easter A.D., Passmore L.A., et al. The crystal structure of the intact E. coli RelBE toxin-antitoxin complex provides the structural basis for conditional cooperativity. Structure. 2012;20:1641–1648. doi: 10.1016/j.str.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schureck M.A., Maehigashi T., Miles S.J., Marquez J., Cho S.E., Erdman R., et al. Structure of the Proteus vulgaris HigB-(HigA)2-HigB toxin-antitoxin complex. J. Biol. Chem. 2014;289:1060–1070. doi: 10.1074/jbc.M113.512095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kamada K., Hanaoka F., Burley S.K. Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol. Cell. 2003;11:875–884. doi: 10.1016/s1097-2765(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 69.Li G.Y., Zhang Y., Inouye M., Ikura M. Inhibitory mechanism of Escherichia coli RelE-RelB toxin-antitoxin module involves a helix displacement near an mRNA interferase active site. J. Biol. Chem. 2009;284:14628–14636. doi: 10.1074/jbc.M809656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khoo S.K., Loll B., Chan W.T., Shoeman R.L., Ngoo L., Yeo C.C., et al. Molecular and structural characterization of the PezAT chromosomal toxin-antitoxin system of the human pathogen Streptococcus pneumoniae. J. Biol. Chem. 2007;282:19606–19618. doi: 10.1074/jbc.M701703200. [DOI] [PubMed] [Google Scholar]
- 71.Schumacher M.A., Piro K.M., Xu W., Hansen S., Lewis K., Brennan R.G. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science. 2009;323:396–401. doi: 10.1126/science.1163806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brown B.L., Grigoriu S., Kim Y., Arruda J.M., Davenport A., Wood T.K., et al. Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hurley J.M., Woychik N.A. Bacterial toxin HigB associates with ribosomes and mediates translation-dependent mRNA cleavage at A-rich sites. J. Biol. Chem. 2009;284:18605–18613. doi: 10.1074/jbc.M109.008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y., Inouye M. The inhibitory mechanism of protein synthesis by YoeB, an Escherichia coli toxin. J. Biol. Chem. 2009;284:6627–6638. doi: 10.1074/jbc.M808779200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Prysak M.H., Mozdzierz C.J., Cook A.M., Zhu L., Zhang Y., Inouye M., et al. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol. Microbiol. 2009;71:1071–1087. doi: 10.1111/j.1365-2958.2008.06572.x. [DOI] [PubMed] [Google Scholar]
- 76.Zhang Y., Yamaguchi Y., Inouye M. Characterization of YafO, an Escherichia coli toxin. J. Biol. Chem. 2009;284:25522–25531. doi: 10.1074/jbc.M109.036624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pellegrini O., Mathy N., Gogos A., Shapiro L., Condon C. The Bacillus subtilis ydcDE operon encodes an endoribonuclease of the MazF/PemK family and its inhibitor. Mol. Microbiol. 2005;56:1139–1148. doi: 10.1111/j.1365-2958.2005.04606.x. [DOI] [PubMed] [Google Scholar]
- 78.Pimentel B., Madine M.A., de la Cueva-Mendez G. Kid cleaves specific mRNAs at UUACU sites to rescue the copy number of plasmid R1. EMBO J. 2005;24:3459–3469. doi: 10.1038/sj.emboj.7600815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y., Zhu L., Zhang J., Inouye M. Characterization of ChpBK, an mRNA interferase from Escherichia coli. J. Biol. Chem. 2005;280:26080–26088. doi: 10.1074/jbc.M502050200. [DOI] [PubMed] [Google Scholar]
- 80.Bertelsen M.B., Senissar M., Nielsen M.H., Bisiak F., Cunha M.V., Molinaro A.L., et al. Structural basis for toxin inhibition in the VapXD toxin-antitoxin system. Structure. 2021;29:139–150 e3. doi: 10.1016/j.str.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Naka K., Qi D., Yonesaki T., Otsuka Y. RnlB antitoxin of the Escherichia coli RnlA-RnlB toxin-antitoxin module requires RNase HI for inhibition of RnlA toxin activity. Toxins. 2017;9 doi: 10.3390/toxins9010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fineran P.C., Blower T.R., Foulds I.J., Humphreys D.P., Lilley K.S., Salmond G.P. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. U. S. A. 2009;106:894–899. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Samson J.E., Spinelli S., Cambillau C., Moineau S. Structure and activity of AbiQ, a lactococcal endoribonuclease belonging to the type III toxin-antitoxin system. Mol. Microbiol. 2013;87:756–768. doi: 10.1111/mmi.12129. [DOI] [PubMed] [Google Scholar]
- 84.Blower T.R., Pei X.Y., Short F.L., Fineran P.C., Humphreys D.P., Luisi B.F., et al. A processed noncoding RNA regulates an altruistic bacterial antiviral system. Nat. Struct. Mol. Biol. 2011;18:185–190. doi: 10.1038/nsmb.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Short F.L., Pei X.Y., Blower T.R., Ong S.L., Fineran P.C., Luisi B.F., et al. Selectivity and self-assembly in the control of a bacterial toxin by an antitoxic noncoding RNA pseudoknot. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E241–E249. doi: 10.1073/pnas.1216039110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brierley I., Pennell S., Gilbert R.J. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat. Rev. Microbiol. 2007;5:598–610. doi: 10.1038/nrmicro1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Peselis A., Serganov A. vol. 5. Wiley Interdiscip Rev RNA; 2014. pp. 803–822. (Structure and Function of Pseudoknots Involved in Gene Expression Control). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Masuda H., Tan Q., Awano N., Wu K.P., Inouye M. YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol. Microbiol. 2012;84:979–989. doi: 10.1111/j.1365-2958.2012.08068.x. [DOI] [PubMed] [Google Scholar]
- 89.Wang X., Lord D.M., Cheng H.Y., Osbourne D.O., Hong S.H., Sanchez-Torres V., et al. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat. Chem. Biol. 2012;8:855–861. doi: 10.1038/nchembio.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cheng H.Y., Soo V.W., Islam S., McAnulty M.J., Benedik M.J., Wood T.K. Toxin GhoT of the GhoT/GhoS toxin/antitoxin system damages the cell membrane to reduce adenosine triphosphate and to reduce growth under stress. Environ. Microbiol. 2014;16:1741–1754. doi: 10.1111/1462-2920.12373. [DOI] [PubMed] [Google Scholar]
- 91.Aakre C.D., Phung T.N., Huang D., Laub M.T. A bacterial toxin inhibits DNA replication elongation through a direct interaction with the beta sliding clamp. Mol. Cell. 2013;52:617–628. doi: 10.1016/j.molcel.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Johnson A., O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 93.Maki S., Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. II. A novel complex including the gamma subunit essential for processive synthesis. J. Biol. Chem. 1988;263:6555–6560. [PubMed] [Google Scholar]
- 94.Yu X., Gao X., Zhu K., Yin H., Mao X., Wojdyla J.A., et al. Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism. Commun. Biol. 2020;3:216. doi: 10.1038/s42003-020-0941-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cai Y., Usher B., Gutierrez C., Tolcan A., Mansour M., Fineran P.C., et al. A nucleotidyltransferase toxin inhibits growth of Mycobacterium tuberculosis through inactivation of tRNA acceptor stems. Sci. Adv. 2020;6:eabb6651. doi: 10.1126/sciadv.abb6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Garcia-Contreras R., Zhang X.S., Kim Y., Wood T.K. Protein translation and cell death: the role of rare tRNAs in biofilm formation and in activating dormant phage killer genes. PLoS One. 2008;3:e2394. doi: 10.1371/journal.pone.0002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marimon O., Teixeira J.M., Cordeiro T.N., Soo V.W., Wood T.L., Mayzel M., et al. An oxygen-sensitive toxin-antitoxin system. Nat. Commun. 2016;7 doi: 10.1038/ncomms13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yao J., Guo Y., Zeng Z., Liu X., Shi F., Wang X. Identification and characterization of a HEPN-MNT family type II toxin-antitoxin in Shewanella oneidensis. Microb. Biotechnol. 2015;8:961–973. doi: 10.1111/1751-7915.12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jia X., Yao J., Gao Z., Liu G., Dong Y.H., Wang X., et al. Structure-function analyses reveal the molecular architecture and neutralization mechanism of a bacterial HEPN-MNT toxin-antitoxin system. J. Biol. Chem. 2018;293:6812–6823. doi: 10.1074/jbc.RA118.002421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yao J., Zhen X., Tang K., Liu T., Xu X., Chen Z., et al. Novel polyadenylylation-dependent neutralization mechanism of the HEPN/MNT toxin/antitoxin system. Nucleic Acids Res. 2020;48:11054–11067. doi: 10.1093/nar/gkaa855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Songailiene I., Juozapaitis J., Tamulaitiene G., Ruksenaite A., Sulcius S., Sasnauskas G., et al. HEPN-MNT toxin-antitoxin system: the HEPN ribonuclease is neutralized by OligoAMPylation. Mol. Cell. 2020;80:955–970 e7. doi: 10.1016/j.molcel.2020.11.034. [DOI] [PubMed] [Google Scholar]
- 102.Li M., Gong L., Cheng F., Yu H., Zhao D., Wang R., et al. Toxin-antitoxin RNA pairs safeguard CRISPR-Cas systems. Science. 2021;372 doi: 10.1126/science.abe5601. [DOI] [PubMed] [Google Scholar]
- 103.Choi J.S., Kim W., Suk S., Park H., Bak G., Yoon J., et al. The small RNA, SdsR, acts as a novel type of toxin in Escherichia coli. RNA Biol. 2018;15:1319–1335. doi: 10.1080/15476286.2018.1532252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jankevicius G., Ariza A., Ahel M., Ahel I. The toxin-antitoxin system DarTG catalyzes reversible ADP-ribosylation of DNA. Mol. Cell. 2016;64:1109–1116. doi: 10.1016/j.molcel.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hauryliuk V., Atkinson G.C., Murakami K.S., Tenson T., Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 2015;13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jimmy S., Saha C.K., Kurata T., Stavropoulos C., Oliveira S.R.A., Koh A., et al. A widespread toxin-antitoxin system exploiting growth control via alarmone signaling. Proc. Natl. Acad. Sci. U. S. A. 2020;117:10500–10510. doi: 10.1073/pnas.1916617117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Alawneh A.M., Qi D., Yonesaki T., Otsuka Y. An ADP-ribosyltransferase Alt of bacteriophage T4 negatively regulates the Escherichia coli MazF toxin of a toxin-antitoxin module. Mol. Microbiol. 2016;99:188–198. doi: 10.1111/mmi.13225. [DOI] [PubMed] [Google Scholar]
- 108.Koga M., Otsuka Y., Lemire S., Yonesaki T. Escherichia coli rnlA and rnlB compose a novel toxin-antitoxin system. Genetics. 2011;187:123–130. doi: 10.1534/genetics.110.121798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bleriot I., Blasco L., Pacios O., Fernandez-Garcia L., Ambroa A., Lopez M., et al. The role of PemIK (PemK/PemI) type II TA system from Klebsiella pneumoniae clinical strains in lytic phage infection. Sci. Rep. 2022;12:4488. doi: 10.1038/s41598-022-08111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dedrick R.M., Jacobs-Sera D., Bustamante C.A., Garlena R.A., Mavrich T.N., Pope W.H., et al. Prophage-mediated defence against viral attack and viral counter-defence. Nat. Microb. 2017;2 doi: 10.1038/nmicrobiol.2016.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.LeRoux M., Srikant S., Teodoro G.I.C., Zhang T., Littlehale M.L., Doron S., et al. The DarTG toxin-antitoxin system provides phage defence by ADP-ribosylating viral DNA. Nat. Microb. 2022;7:1028–1040. doi: 10.1038/s41564-022-01153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dorr T., Vulic M., Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lewis K. Persister cells. Annu. Rev. Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 114.Volzing K.G., Brynildsen M.P. Stationary-phase persisters to ofloxacin sustain DNA damage and require repair systems only during recovery. mBio. 2015;6:e00731–e00815. doi: 10.1128/mBio.00731-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Goormaghtigh F., Van Melderen L. Single-cell imaging and characterization of Escherichia coli persister cells to ofloxacin in exponential cultures. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aav9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim Y., Wood T.K. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem. Biophys. Res. Commun. 2010;391:209–213. doi: 10.1016/j.bbrc.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Semanjski M., Germain E., Bratl K., Kiessling A., Gerdes K., Macek B. The kinases HipA and HipA7 phosphorylate different substrate pools in Escherichia coli to promote multidrug tolerance. Sci. Signal. 2018;11 doi: 10.1126/scisignal.aat5750. [DOI] [PubMed] [Google Scholar]
- 118.Harrison J.J., Wade W.D., Akierman S., Vacchi-Suzzi C., Stremick C.A., Turner R.J., et al. The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm. Antimicrob. Agen. Chemother. 2009;53:2253–2258. doi: 10.1128/AAC.00043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tripathi A., Dewan P.C., Siddique S.A., Varadarajan R. MazF-induced growth inhibition and persister generation in Escherichia coli. J. Biol. Chem. 2014;289:4191–4205. doi: 10.1074/jbc.M113.510511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Muthuramalingam M., White J.C., Bourne C.R. Toxin-antitoxin modules are pliable switches activated by multiple protease pathways. Toxins. 2016;8 doi: 10.3390/toxins8070214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ronneau S., Helaine S. Clarifying the link between toxin-antitoxin modules and bacterial persistence. J. Mol. Biol. 2019;431:3462–3471. doi: 10.1016/j.jmb.2019.03.019. [DOI] [PubMed] [Google Scholar]
- 122.Christensen-Dalsgaard M., Jorgensen M.G., Gerdes K. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 2010;75:333–348. doi: 10.1111/j.1365-2958.2009.06969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.LeRoux M., Culviner P.H., Liu Y.J., Littlehale M.L., Laub M.T. Stress can induce transcription of toxin-antitoxin systems without activating toxin. Mol. Cell. 2020;79:280–292 e8. doi: 10.1016/j.molcel.2020.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Brown J.M., Shaw K.J. A novel family of Escherichia coli toxin-antitoxin gene pairs. J. Bacteriol. 2003;185:6600–6608. doi: 10.1128/JB.185.22.6600-6608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Barrios A.F., Zuo R., Ren D., Wood T.K. Hha, YbaJ, and OmpA regulate Escherichia coli K12 biofilm formation and conjugation plasmids abolish motility. Biotechnol. Bioeng. 2006;93:188–200. doi: 10.1002/bit.20681. [DOI] [PubMed] [Google Scholar]
- 126.Hong S.H., Lee J., Wood T.K. Engineering global regulator Hha of Escherichia coli to control biofilm dispersal. Microb. Biotechnol. 2010;3:717–728. doi: 10.1111/j.1751-7915.2010.00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gutierrez A., Laureti L., Crussard S., Abida H., Rodriguez-Rojas A., Blazquez J., et al. beta-Lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat. Commun. 2013;4:1610. doi: 10.1038/ncomms2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Frohlich K.S., Papenfort K., Berger A.A., Vogel J. A conserved RpoS-dependent small RNA controls the synthesis of major porin OmpD. Nucleic Acids Res. 2012;40:3623–3640. doi: 10.1093/nar/gkr1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rice L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 2008;197:1079–1081. doi: 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 130.Garcia C.R., Oliva J., Romero M.T., Diaz-Torres L.A. Enhancing the photocatalytic activity of Sr4 Al14 O25 : Eu(2+) , Dy(3+) persistent phosphors by codoping with Bi(3+) ions. Photochem. Photobiol. 2016;92:231–237. doi: 10.1111/php.12570. [DOI] [PubMed] [Google Scholar]
- 131.Ramage H.R., Connolly L.E., Cox J.S. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zaychikova M.V., Zakharevich N.V., Sagaidak M.O., Bogolubova N.A., Smirnova T.G., Andreevskaya S.N., et al. Mycobacterium tuberculosis type II toxin-antitoxin systems: genetic polymorphisms and functional properties and the possibility of their use for genotyping. PLoS One. 2015;10 doi: 10.1371/journal.pone.0143682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Molina L., Udaondo Z., Duque E., Fernandez M., Bernal P., Roca A., et al. Specific gene loci of clinical Pseudomonas putida isolates. PLoS One. 2016;11 doi: 10.1371/journal.pone.0147478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wei Y.Q., Bi D.X., Wei D.Q., Ou H.Y. Prediction of type II toxin-antitoxin loci in Klebsiella pneumoniae genome sequences. Interdiscip. Sci. 2016;8:143–149. doi: 10.1007/s12539-015-0135-6. [DOI] [PubMed] [Google Scholar]
- 135.Verma S., Bhatnagar R. MoxT toxin of Bacillus anthracis exhibits sequence specific ribonuclease activity. Biochem. Biophys. Res. Commun. 2014;450:998–1004. doi: 10.1016/j.bbrc.2014.06.092. [DOI] [PubMed] [Google Scholar]
- 136.McVicker G., Tang C.M. Deletion of toxin-antitoxin systems in the evolution of Shigella sonnei as a host-adapted pathogen. Nat. Microb. 2016;2 doi: 10.1038/nmicrobiol.2016.204. [DOI] [PubMed] [Google Scholar]
- 137.Sayeed S., Brendler T., Davis M., Reaves L., Austin S. Surprising dependence on postsegregational killing of host cells for maintenance of the large virulence plasmid of Shigella flexneri. J. Bacteriol. 2005;187:2768–2773. doi: 10.1128/JB.187.8.2768-2773.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lobato-Marquez D., Molina-Garcia L., Moreno-Cordoba I., Garcia-Del Portillo F., Diaz-Orejas R. Stabilization of the virulence plasmid pSLT of Salmonella typhimurium by three maintenance systems and its evaluation by using a new stability test. Front. Mol. Biosci. 2016;3:66. doi: 10.3389/fmolb.2016.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Doumith M., Dhanji H., Ellington M.J., Hawkey P., Woodford N. Characterization of plasmids encoding extended-spectrum beta-lactamases and their addiction systems circulating among Escherichia coli clinical isolates in the UK. J. Antimicrob. Chemother. 2012;67:878–885. doi: 10.1093/jac/dkr553. [DOI] [PubMed] [Google Scholar]
- 140.Mnif B., Harhour H., Jdidi J., Mahjoubi F., Genel N., Arlet G., et al. Molecular epidemiology of extended-spectrum beta-lactamase-producing Escherichia coli in Tunisia and characterization of their virulence factors and plasmid addiction systems. BMC Microbiol. 2013;13:147. doi: 10.1186/1471-2180-13-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mnif B., Vimont S., Boyd A., Bourit E., Picard B., Branger C., et al. Molecular characterization of addiction systems of plasmids encoding extended-spectrum beta-lactamases in Escherichia coli. J. Antimicrob. Chemother. 2010;65:1599–1603. doi: 10.1093/jac/dkq181. [DOI] [PubMed] [Google Scholar]
- 142.Boyd D.A., Tyler S., Christianson S., McGeer A., Muller M.P., Willey B.M., et al. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto. Antimicrob. Agent. Chemother. 2004;48:3758–3764. doi: 10.1128/AAC.48.10.3758-3764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Woodford N., Carattoli A., Karisik E., Underwood A., Ellington M.J., Livermore D.M. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob. Agen. Chemother. 2009;53:4472–4482. doi: 10.1128/AAC.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pinel-Marie M.L., Brielle R., Felden B. Dual toxic-peptide-coding Staphylococcus aureus RNA under antisense regulation targets host cells and bacterial rivals unequally. Cell Rep. 2014;7:424–435. doi: 10.1016/j.celrep.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 145.Zhu L., Inoue K., Yoshizumi S., Kobayashi H., Zhang Y., Ouyang M., et al. Staphylococcus aureus MazF specifically cleaves a pentad sequence, UACAU, which is unusually abundant in the mRNA for pathogenic adhesive factor SraP. J. Bacteriol. 2009;191:3248–3255. doi: 10.1128/JB.01815-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hopper S., Wilbur J.S., Vasquez B.L., Larson J., Clary S., Mehr I.J., et al. Isolation of Neisseria gonorrhoeae mutants that show enhanced trafficking across polarized T84 epithelial monolayers. Infect. Immun. 2000;68:896–905. doi: 10.1128/IAI.68.2.896-905.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Faruque S.M., Albert M.J., Mekalanos J.J. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. : MMBR (Microbiol. Mol. Biol. Rev.) 1998;62:1301–1314. doi: 10.1128/MMBR.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gosain T.P., Singh M., Singh C., Thakur K.G., Singh R. Disruption of MenT2 toxin impairs the growth of Mycobacterium tuberculosis in Guinea pigs. Microbiology. 2022;168 doi: 10.1099/mic.0.001246. [DOI] [PubMed] [Google Scholar]
- 149.Anstead G.M., Cadena J., Javeri H. Treatment of infections due to resistant Staphylococcus aureus. Methods Mol. Biol. 2014;1085:259–309. doi: 10.1007/978-1-62703-664-1_16. [DOI] [PubMed] [Google Scholar]
- 150.Murray B.E. The life and times of the Enterococcus. Clin. Microbiol. Rev. 1990;3:46–65. doi: 10.1128/CMR.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Weaver K.E. The par toxin-antitoxin system from Enterococcus faecalis plasmid pAD1 and its chromosomal homologs. RNA Biol. 2012;9:1498–1503. doi: 10.4161/rna.22311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Weaver K.E., Reddy S.G., Brinkman C.L., Patel S., Bayles K.W., Endres J.L. Identification and characterization of a family of toxin-antitoxin systems related to the Enterococcus faecalis plasmid pAD1 par addiction module. Microbiology. 2009;155:2930–2940. doi: 10.1099/mic.0.030932-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Weaver K.E., Jensen K.D., Colwell A., Sriram S.I. Functional analysis of the Enterococcus faecalis plasmid pAD1-encoded stability determinant par. Mol. Microbiol. 1996;20:53–63. doi: 10.1111/j.1365-2958.1996.tb02488.x. [DOI] [PubMed] [Google Scholar]
- 154.Weaver K.E. Emerging plasmid-encoded antisense RNA regulated systems. Curr. Opin. Microbiol. 2007;10:110–116. doi: 10.1016/j.mib.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 155.Alteri C.J., Smith S.N., Mobley H.L. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Blango M.G., Mulvey M.A. Persistence of uropathogenic Escherichia coli in the face of multiple antibiotics. Antimicrob. Agen. Chemother. 2010;54:1855–1863. doi: 10.1128/AAC.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fino C., Vestergaard M., Ingmer H., Pierrel F., Gerdes K., Harms A. PasT of Escherichia coli sustains antibiotic tolerance and aerobic respiration as a bacterial homolog of mitochondrial Coq10. Microbiol. 2020;9 doi: 10.1002/mbo3.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Murphy T.F. Respiratory infections caused by non-typeable Haemophilus influenzae. Curr. Opin. Infect. Dis. 2003;16:129–134. doi: 10.1097/00001432-200304000-00009. [DOI] [PubMed] [Google Scholar]
- 159.Hall-Stoodley L., Hu F.Z., Gieseke A., Nistico L., Nguyen D., Hayes J., et al. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA. 2006;296:202–211. doi: 10.1001/jama.296.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Slinger R., Chan F., Ferris W., Yeung S.W., St Denis M., Gaboury I., et al. Multiple combination antibiotic susceptibility testing of nontypeable Haemophilus influenzae biofilms. Diagn. Microbiol. Infect. Dis. 2006;56:247–253. doi: 10.1016/j.diagmicrobio.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 161.Ruby T., McLaughlin L., Gopinath S., Monack D. Salmonella's long-term relationship with its host. FEMS Microbiol. Rev. 2012;36:600–615. doi: 10.1111/j.1574-6976.2012.00332.x. [DOI] [PubMed] [Google Scholar]
- 162.Crawford R.W., Rosales-Reyes R., Ramirez-Aguilar Mde L., Chapa-Azuela O., Alpuche-Aranda C., Gunn J.S. Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4353–4358. doi: 10.1073/pnas.1000862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mastroeni P., Grant A., Restif O., Maskell D. A dynamic view of the spread and intracellular distribution of Salmonella enterica. Nat. Rev. Microbiol. 2009;7:73–80. doi: 10.1038/nrmicro2034. [DOI] [PubMed] [Google Scholar]
- 164.Garcia-del Portillo F. Salmonella intracellular proliferation: where, when and how? Microb. Infect. 2001;3:1305–1311. doi: 10.1016/s1286-4579(01)01491-5. [DOI] [PubMed] [Google Scholar]
- 165.Nunez-Hernandez C., Tierrez A., Ortega A.D., Pucciarelli M.G., Godoy M., Eisman B., et al. Genome expression analysis of nonproliferating intracellular Salmonella enterica serovar Typhimurium unravels an acid pH-dependent PhoP-PhoQ response essential for dormancy. Infect. Immun. 2013;81:154–165. doi: 10.1128/IAI.01080-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Thao S., Escalante-Semerena J.C. Biochemical and thermodynamic analyses of Salmonella enterica Pat, a multidomain, multimeric N(epsilon)-lysine acetyltransferase involved in carbon and energy metabolism. mBio. 2011;2 doi: 10.1128/mBio.00216-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fitzgerald J.R., Foster T.J., Cox D. The interaction of bacterial pathogens with platelets. Nat. Rev. Microbiol. 2006;4:445–457. doi: 10.1038/nrmicro1425. [DOI] [PubMed] [Google Scholar]
- 168.Siboo I.R., Chambers H.F., Sullam P.M. Role of SraP, a serine-rich surface protein of Staphylococcus aureus, in binding to human platelets. Infect. Immun. 2005;73:2273–2280. doi: 10.1128/IAI.73.4.2273-2280.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.North R.J., Jung Y.J. Immunity to tuberculosis. Annu. Rev. Immunol. 2004;22:599–623. doi: 10.1146/annurev.immunol.22.012703.104635. [DOI] [PubMed] [Google Scholar]
- 170.Stewart G.R., Robertson B.D., Young D.B. Tuberculosis: a problem with persistence. Nat. Rev. Microbiol. 2003;1:97–105. doi: 10.1038/nrmicro749. [DOI] [PubMed] [Google Scholar]
- 171.Bordes P., Cirinesi A.M., Ummels R., Sala A., Sakr S., Bitter W., et al. SecB-like chaperone controls a toxin-antitoxin stress-responsive system in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 2011;108:8438–8443. doi: 10.1073/pnas.1101189108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang X., Kim Y., Hong S.H., Ma Q., Brown B.L., Pu M., et al. Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 2011;7:359–366. doi: 10.1038/nchembio.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kwan B.W., Lord D.M., Peti W., Page R., Benedik M.J., Wood T.K. The MqsR/MqsA toxin/antitoxin system protects Escherichia coli during bile acid stress. Environ. Microbiol. 2015;17:3168–3181. doi: 10.1111/1462-2920.12749. [DOI] [PubMed] [Google Scholar]
- 174.Gonzalez Barrios A.F., Zuo R., Hashimoto Y., Yang L., Bentley W.E., Wood T.K. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022) J. Bacteriol. 2006;188:305–316. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Soo V.W., Wood T.K. Antitoxin MqsA represses curli formation through the master biofilm regulator CsgD. Sci. Rep. 2013;3:3186. doi: 10.1038/srep03186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kim Y., Wang X., Zhang X.S., Grigoriu S., Page R., Peti W., et al. Escherichia coli toxin/antitoxin pair MqsR/MqsA regulate toxin CspD. Environ. Microbiol. 2010;12:1105–1121. doi: 10.1111/j.1462-2920.2009.02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fraikin N., Rousseau C.J., Goeders N., Van Melderen L. Reassessing the role of the type II MqsRA toxin-antitoxin system in stress response and biofilm formation: mqsA is transcriptionally uncoupled from mqsR. mBio. 2019;10 doi: 10.1128/mBio.02678-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Vassallo C.N., Doering C.R., Littlehale M.L., Teodoro G.I., Laub M.T. A functional selection reveals previously undetected anti-phage defence systems in the E. coli pangenome. Nat. Microb. 2022;7:1568–1579. doi: 10.1038/s41564-022-01219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Maisonneuve E., Shakespeare L.J., Jorgensen M.G., Gerdes K. Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. U. S. A. 2011;108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 180.Maisonneuve E., Castro-Camargo M., Gerdes K. ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell. 2013;154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 181.Germain E., Roghanian M., Gerdes K., Maisonneuve E. Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc. Natl. Acad. Sci. U. S. A. 2015;112:5171–5176. doi: 10.1073/pnas.1423536112. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 182.Retraction for Maisonneuve, et al. Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E2901. doi: 10.1073/pnas.1803278115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Retraction for Germain, et al. Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc. Natl. Acad. Sci. U. S. A. 2019;116 doi: 10.1073/pnas.1906160116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Maisonneuve E., Castro-Camargo M., Gerdes K. ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell. 2018;172:1135. doi: 10.1016/j.cell.2018.02.023. [DOI] [PubMed] [Google Scholar]
- 185.Goormaghtigh F., Fraikin N., Putrins M., Hallaert T., Hauryliuk V., Garcia-Pino A., et al. Reassessing the role of type II toxin-antitoxin systems in formation of Escherichia coli type II persister cells. mBio. 2018;9 doi: 10.1128/mBio.00640-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Svenningsen M.S., Veress A., Harms A., Mitarai N., Semsey S. Birth and resuscitation of (p)ppGpp induced antibiotic tolerant persister cells. Sci. Rep. 2019;9:6056. doi: 10.1038/s41598-019-42403-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Pontes M.H., Groisman E.A. Slow growth determines nonheritable antibiotic resistance in Salmonella enterica. Sci. Signal. 2019;12 doi: 10.1126/scisignal.aax3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Rosendahl S., Tamman H., Brauer A., Remm M., Horak R. Chromosomal toxin-antitoxin systems in Pseudomonas putida are rather selfish than beneficial. Sci. Rep. 2020;10:9230. doi: 10.1038/s41598-020-65504-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhang Y., Inouye M. RatA (YfjG), an Escherichia coli toxin, inhibits 70S ribosome association to block translation initiation. Mol. Microbiol. 2011;79:1418–1429. doi: 10.1111/j.1365-2958.2010.07506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.