Abstract
Background:
Fexofenadine is a recommended in vivo probe drug for phenotypic P-glycoprotein (P-gp) and organic anion transporting polypeptide (OATP) 1B1/3 transporter activities. This study evaluated a limited sampling strategy using a population pharmacokinetic approach to estimate plasma fexofenadine exposure as an index of P-gp and OATP activities.
Methods:
In previous studies, a single oral dose of fexofenadine (120 mg) was administered alone or in combination with grapefruit juice, Panax ginseng, or Echinacea purpurea to healthy adult participants. Serial plasma samples were collected up to 72 h after administration and fexofenadine concentrations were measured. A population pharmacokinetic model was developed using nonlinear mixed-effects modeling. Limited sampling models (LSMs) using single- and 2-timepoint fexofenadine concentrations were compared with full profiles from intense sampling using empirical Bayesian post hoc estimations of systemic exposure derived from the population pharmacokinetic model. Predefined criteria for LSM selection and validation included a coefficient of determination (R2) ≥ 0.90, relative percentage mean prediction error (%MPE) ≥ −5 to ≤ 5%, relative percentage mean absolute error (%MAE) ≤ 10%, and relative percentage root mean square error (%RMSE) ≤ 15%.
Results:
Fexofenadine concentrations (n = 1520) were well described using a two-compartment model. Grapefruit juice decreased the relative oral bioavailability of fexofenadine by 25%, whereas P. ginseng and E. purpurea had no effect. All the evaluated single timepoint fexofenadine LSMs showed unacceptable %MPE, %MAE, and/or %RMSE. Although adding a second time point improved precision, the predefined criteria were not met.
Conclusions:
Identifying novel fexofenadine LSMs to estimate P-gp and OATP1B1/3 activities in healthy adults for future transporter-mediated drug-drug interaction studies remains elusive.
Keywords: Drug-drug interaction, fexofenadine, limited sampling strategy, natural product, transporter
Background
Pharmacokinetic drug-drug interactions (DDIs) mediated by cytochrome P450 (CYP) enzymes are a long-standing pharmacotherapeutic concern. Although transporter-mediated DDIs are increasingly being recognized, the clinical ramifications of these interactions are not well understood. The US Food and Drug Administration (FDA) and European Medicines Agency (EMA) currently list fexofenadine as a recommended in vivo probe drug to assess transporter-mediated DDI studies involving the efflux transporter P-glycoprotein (P-gp) and uptake transporter organic anion transporting polypeptides (OATP) 1B1/3.1,2 Perturbations in the fexofenadine area under the plasma concentration versus time curve (AUC) and/or systemic or apparent oral clearance are assumed to reflect P-gp and OATP1B1/3 activities.1,2 Furthermore, the design and conduct of in vitro and clinical DDI studies are detailed in the updated 2020 FDA guidance documents.3,4 For all drugs, accurate determination of fexofenadine AUC and/or oral clearance requires intense sample collection. However, such a determination can be inconvenient for fexofenadine based on a half-life of approximately 14 h, necessitating sample collection to 48–72 h, or three to five half-lives.5-7 In addition, intense sampling may not be ideal in certain study populations, including cancer, geriatric, and pediatric patients.
A limited sampling strategy is a validated alternative method8,9 for intense sampling to estimate systemic drug exposure. This strategy has been applied in the context of therapeutic drug monitoring for mycophenolic acid, tacrolimus, and cyclosporine, as well as for prednisolone in kidney, liver, and hematopoietic stem cell transplant recipients.10-15 A limited sampling strategy involving a population pharmacokinetic approach has been evaluated to estimate CYP2C9, CYP2C19, and CYP3A activities with various probe drugs.16-18 An advantage of such an approach is flexibility in plasma collection times. A traditional limited sampling strategy using non-compartmental analysis does not allow for deviations in sample collection time points.
The rationale of the current study was to assess whether a limited sampling strategy using a population pharmacokinetic approach can be applied to fexofenadine. Using data from previously published studies, we developed a population pharmacokinetic model for healthy adult participants who were administered a single oral dose of fexofenadine alone or in combination with the natural product grapefruit juice, Panax ginseng, or Echinacea purpurea. The objective of this study was to develop single- and 2-timepoint limited sampling models (LSMs) using a population pharmacokinetic approach for potential future applications in transporter-mediated DDI studies to estimate P-gp and OATP1B1/3 activities.
Methods
STUDY PARTICIPANTS
An institutional review board exemption was obtained from the University of California, San Diego Human Research Protections Program (201959). Fexofenadine concentration-time data from four previous pharmacokinetic studies were obtained (Table 1).19-22 Healthy adult participants were administered a single oral 120 mg dose of fexofenadine alone in each study. A single oral 120 mg fexofenadine dose was administered a second time concurrently with a single glass of 240 mL grapefruit juice22, after Panax ginseng 500 mg twice daily for 28 days19, or after Echinacea purpurea 500 mg three times daily for 28 days.21 Specifically, for the P. ginseng and E. purpurea studies, fexofenadine was administered on the morning of day 28. Only healthy volunteer data from the study by Nolin et al.20 were used in the current analysis.
Table 1.
Summary of published studies on oral fexofenadine used in the analysis
| Study | Number of participants |
Fexofenadine collection timepoints (h) |
Study intervention a |
|---|---|---|---|
| 120 | 10 (4 females) | 0, 0.5, 1, 2, 3, 4, 5, 6, 8, 12 | Baseline |
| 222 | 18 (9 females) | 0, 1, 1.5, 2, 3, 4, 5, 6, 8, 10, 12, 24, 36, 48, 72 | Baseline, baseline plus grapefruit juice |
| 319 | 12 (4 females) | 0, 0.5, 1, 1.5, 2, 2.5, 3, 3.5, 4, 5, 6, 8, 24 | Baseline, baseline plus Panax ginseng |
| 421 | 13 (5 females) | 0, 0.5, 1, 1.5, 2, 2.5, 3, 3.5, 4, 5, 6, 8, 24 | Baseline, baseline plus Echinacea Purpurea |
Healthy, adult participants received 120 mg of fexofenadine alone (baseline) or in combination with grapefruit juice (240 mL taken with fexofenadine once), Panax ginseng (500 mg twice daily for 28 days), or Echinacea purpurea (500 mg three times a day for 4 weeks).
Individual age, weight, and self-identified race demographics were not available for all studies. Inclusion criteria included individuals who were at least 18 years of age, able to provide written informed consent, had no clinically significant medical history and/or physical examination findings, and abstained from any medications and/or dietary supplements at least 7 days prior to the study intervention. Individuals were excluded if they had clinically significant (greater than 2× the upper and/or lower normal limits) blood and urine laboratory tests, were current nicotine smokers, had a known history of a fexofenadine adverse reaction, and were pregnant or breastfeeding.
PLASMA COLLECTION AND FEXOFENADINE QUANTIFICATION
Serial plasma samples were collected from each participant at various study-specific time points up to 72 h after fexofenadine administration (Table 1). Plasma fexofenadine concentrations were measured using ultra-performance or high-performance liquid chromatography-tandem mass spectrometry. The details of each method are provided elsewhere.19-22
POPULATION PHARMACOKINETIC MODEL DEVELOPMENT
Population pharmacokinetic modeling was conducted using nonlinear mixed-effects modeling in NONMEM (v. 7.3; ICON Development Solutions, Hanover, MD, USA). RStudio R (version 4.0.3; Boston, MA, USA) was used to visualize the data. The fexofenadine plasma concentration-time data obtained when fexofenadine was administered alone or in combination with grapefruit juice, P. ginseng, or E. purpurea were used for model development. A first-order conditional estimation method with interaction was used throughout model development.
STRUCTURAL AND STATISTICAL MODEL
A 2-compartment (ADVAN 4, TRANS 4 subroutine) model was used to describe the fexofenadine concentration-time profiles. Interindividual variability in pharmacokinetic parameters was assumed to be log-normally distributed. The interaction between intersubject variability for clearance, volume of distribution (individually assessed for central [V2] and peripheral [V3] compartments), and first-order absorption rate constant (ka) was characterized. The residual variability was assessed using a combined proportional and additive error model.
MODEL SELECTION AND INTERNAL MODEL EVALUATION
Model development and selection were guided by comparing the objective function values (i.e., −2 log likelihood [−2LL]). A decrease in the objective function value by 3.84 for 1 degree of freedom was considered statistically significant (P < 0.05). Exposure to grapefruit juice, P. ginseng, or E. purpurea was evaluated as a covariate for clearance and relative oral bioavailability. Potential covariates were added individually on a univariate screen. All covariates that were significant in this phase were evaluated using a stepwise addition approach, and covariates that improved the objective function values by at least 8 (P < 0.005) were retained in the final model. A 1000-sample bootstrap validation was performed using Wings for NONMEM. The total AUC (0→ꝏ) for the final population pharmacokinetic model was calculated using NONMEM as the dose divided by CL/F.
LIMITED SAMPLING STRATEGY
LSMs using single- or 2-timepoint concentrations were assessed to estimate the fexofenadine systemic exposure (total AUC). The time points (1, 2, 3, 4, 5, 6, or 8 h) were selected because they were available for all participants in all studies. Individual Bayesian post hoc estimates for total fexofenadine AUC were determined using the final population pharmacokinetic model with data from single- or 2-timepoint LSMs for each participant by fixing model parameters to those of the estimates derived from the final composite model. Fexofenadine AUCs estimated from the LSMs were then compared with fexofenadine AUCs from the full concentration-time profiles. Predefined criteria for LSM selection and validation included a coefficient of determination (R2) ≥ 0.9, relative percent mean prediction error (%MPE) ≥ −5% to ≤ 5%, relative percent mean absolute error (%MAE) ≤ 10%, and relative percent root mean squared prediction error (%RMSE) ≤ 15%.24,25
Results
FEXOFENADINE POPULATION PHARMACOKINETICS
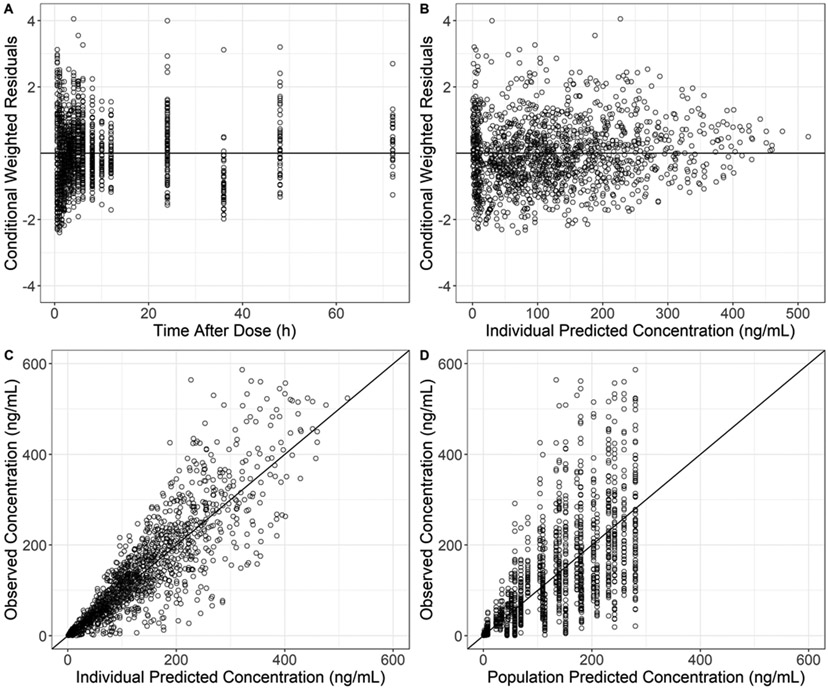
Fexofenadine plasma concentrations (n = 1,520) were obtained from 53 participants (22 females). The lower limit of quantification ranged from 0.1–1.0 ng/mL, with inter- and intra-assay precision and accuracy of <15%. Each participant in Studies 2-4 was administered 120 mg of fexofenadine (Table 1). Fexofenadine plasma concentration-time profiles were adequately described using a 2-compartment model with sequential zero/first-order absorption (Figure 1 Supplemental Digital Content). The effects of grapefruit juice and E. purpurea on the relative oral bioavailability of fexofenadine, grapefruit juice, and P. ginseng on fexofenadine clearance were significant in univariate analysis. The effect of the grapefruit juice on the relative oral bioavailability of fexofenadine was retained in the final model. The population pharmacokinetic parameters for fexofenadine from the final model and the corresponding standard error estimates are summarized in Table 2. Relative oral bioavailability decreased by 25% in the presence of grapefruit juice. P. ginseng and E. purpurea had no effect. The goodness-of-fit plots indicate no structural bias (Figure 1). Post hoc CL/F values for the control arms were not statistically different among the studies. The bootstrap evaluation of the final model successfully converged 84% of the time (Table 2). The population pharmacokinetic parameter estimates of the final model were within the 95% confidence intervals and close to the median bootstrap estimates, demonstrating that the final population pharmacokinetic model adequately represented the study population.
Table 2.
Fexofenadine population pharmacokinetic parameter estimates from the final model and parameter estimates from bootstrap validation
| Parameter | Final Estimate | Standard error of estimate |
Bootstrap Estimate (median and 95% CI)* |
|---|---|---|---|
| V2/F (L) | 27.3 | 10.8 | 28.8 (17.7–47.8) |
| CL/F (L/h) | 69.7 | 3.65 | 69.8 (62.3–78.6) |
| Ka (h−1) | 0.282 | 0.0192 | 0.282 (0.241–0.332) |
| V3/F (L) | 269 | 28.6 | 264 (214–324) |
| Q/F (L/h) | 19.7 | 2.00 | 19.5 (15.6–24.9) |
| D1 (h) | 1.59 | 0.106 | 1.59 (1.17–2.12) |
| GFJ on F | 0.748 | 0.0448 | 0.746 (0.668–0.834) |
| Between Subject Variability | |||
| V2/F | 151% | 0.234 | 147% (111–191) |
| CL/F | 40% | 0.0382 | 40% (32–47) |
| Ka | 33% | 0.0342 | 32% (0.244–0.389) |
| V3/F | 47% | 0.0919 | 44% (22–61) |
| Residual Variability | |||
| Proportional Error | 38.4% | 0.0153 | 38.2% (35–42) |
| Additive Error | 0.48 ng/mL | 0.146 | 0.46 ng/mL (0.002–1.72) |
Bootstrap converged 84% of the time.
/(0.748 with coadministration of grapefruit juice)
Figure 1.
Diagnostic plots for the fexofenadine population pharmacokinetic model. Conditional weighted residuals versus time after dosing (A) and individual predicted concentrations (B). Fexofenadine concentrations versus individually predicted concentrations (C) and population-predicted concentrations (D).
LIMITED SAMPLING STRATEGY
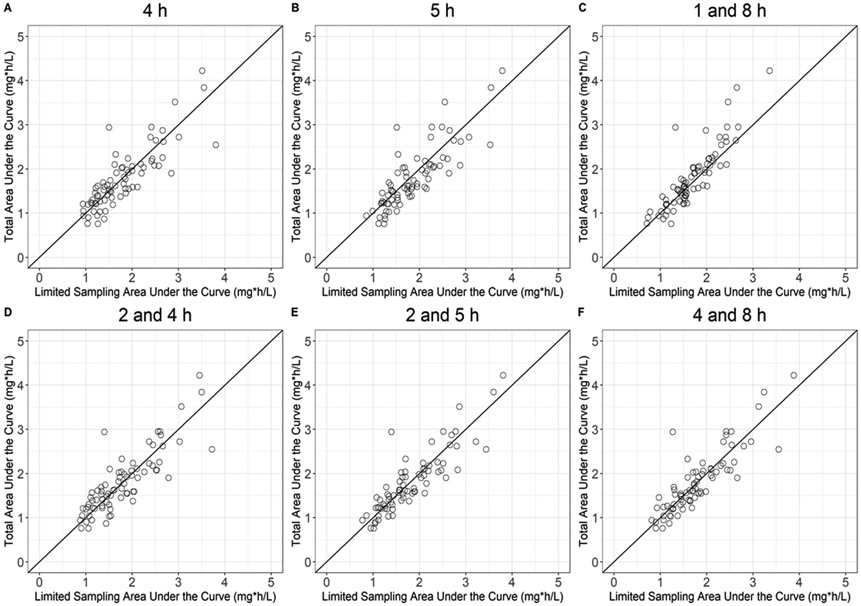
Fexofenadine single- and 2-timepoint LSMs are summarized in Table 3. None of the evaluated single timepoint fexofenadine LSMs met the predefined criteria for R2, %MPE, %MAE, and %RMSE. The addition of a second time point, such as the 2- plus 4-h, 2- plus 5-h, and 4- plus 8-h LSMs met the criteria for %MPE, with improvement in %RMSE and R2 values. However, neither %RMSE nor R2 reached acceptable limits. The plots comparing the observed and estimated AUCs for the LSMs are presented in Figure 2.
Table 3.
Single- and two-time point-limited sampling models to estimate fexofenadine exposure
| Timepoint(s) to estimate AUC |
R2 | %MPE | %MAE | %RMSE |
|---|---|---|---|---|
| 1-h | 0.08 | 9% | 27% | 37% |
| 2-h | 0.23 | 18% | 26% | 35% |
| 4-h | 0.73 | −2% | 15% | 20% |
| 5-h | 0.72 | −8% | 16% | 21% |
| 8-h | 0.60 | 9% | 18% | 26% |
| 1 plus 8 h | 0.80 | 10% | 13% | 19% |
| 2 plus 4 h | 0.72 | 0.05% | 15% | 20% |
| 2 plus 5 h | 0.78 | −2% | 13% | 18% |
| 2 plus 8 h | 0.81 | 21% | 14% | 21% |
| 4 plus 8 h | 0.77 | −1% | 13% | 18% |
| Acceptable criteria | ≥ 0.9 | −5% to +5% | ≤ 10% | ≤ 15% |
Abbreviations: AUC, area under the plasma concentration versus time curve; R2, Coefficient of Determination; %MAE, relative percent mean absolute error; %MPE, relative percent mean prediction error; %RMSE, relative percent root mean squared error.
Bold values denote that the model is within acceptable limits.
Figure 2.
Fexofenadine total observed area under the plasma concentration versus time curve (AUC) versus selected single- and 2-timepoint-limited sampling estimated AUC. The solid line indicates unity.
Discussion
The study rationale was to develop a fexofenadine LSM to accurately estimate P-gp and OATP1B1/3 activities for future general use in transporter-mediated DDI studies. This study used a limited sampling strategy based on a population pharmacokinetic model with a Bayesian estimation approach. Fexofenadine plasma concentration-time profiles were adequately described using a 2-compartment model with sequential zero/first-order absorption. As previously reported, grapefruit juice significantly decreased the relative oral bioavailability of fexofenadine, whereas P. ginseng and E. purpurea had no effect. The pharmacokinetic parameter estimates for fexofenadine V2 and CL were consistent with previously published information.26 Most fexofenadine (60–70%) is bound to plasma proteins, and 80% of the dose is eliminated unchanged in the feces.27 In this study, participants were administered fexofenadine in the fasted state because the Cmax and AUC of multiple fexofenadine formulations (e.g., immediate-release, oral suspension, and oral disintegrating tablet) decrease when combined with food.27
We did not evaluate subject demographics as possible covariates in the population pharmacokinetic model for two reasons. First, kidney impairment and individuals aged 65 years or older impact fexofenadine pharmacokinetics.27 These covariates do not apply to the current study, as all subjects had normal kidney function and were younger than 65 years. Second, demographic data were evaluated in a population pharmacokinetic model of patients with seasonal allergic rhinitis (n = 548). Age, weight, height, body surface area, sex, and race had no impact and were excluded from the final population pharmacokinetic model.26
A limited sampling strategy based on a population pharmacokinetic approach provides several advantages over the traditional limited sampling strategy approaches. Flexibility is provided because it accommodates the variability in the timing of blood sample collection. In addition, the incorporation of information from an entire population combined with individual data increases the ability to estimate pharmacokinetic parameters. This approach contrasts with non-compartmental analysis LSMs, which are restricted to specific time points and do not allow for sample collection deviation.28,29 Non-compartmental analysis LSMs randomly divide participant data into a training set for model development and a validation set for model validation.25,28,29 Consequently, the portion of participant data used during model development may not be reflective of a broader population.
Collectively, results from the current and previous studies suggest that LSM development remains CYP- and transporter-probe-specific. In a study of healthy adult participants (n = 152), a 4- plus 6-h midazolam LSM was developed with a population pharmacokinetic approach that adequately estimated CYP3A inhibitory, but not induced, conditions.18 Another study recommended several single- and 2-timepoint LSMs to estimate S-warfarin exposure during CYP2C9 constitutive conditions (n = 100).17 In contrast, LSMs using a population pharmacokinetic approach were unable to accurately estimate CYP2C19 activity using omeprazole as the probe drug.16
The 1-, 2-, 8-, and 2 plus 8-h LSMs showed unacceptable R2 values and/or exceeded the acceptable limits of bias and precision. One confounding factor was the influence of distribution at early time points, impacting LSMs at 1 and/or 2 h. Another possible confounding factor was the observed inter-individual pharmacokinetic variability in a given study. Variability was expected, as the data were pooled from four studies. However, the extent of variability was unexpected with a pooled dataset, as observed with the final population pharmacokinetic model estimates (Table 2). The V2/F showed a high final estimate for between-participant variability (150%). Between-participant variability in CL/F was also high (40%). The high variability associated with the pharmacokinetic data may have limited the predictive ability of the final model, leading to a less accurate estimate of the fexofenadine AUC.
The current results contrast with those of previous studies recommending fexofenadine LSMs.30,31 In one study, a single time point using Cmax was strongly correlated (r = 0.97, n= 145) with fexofenadine AUC.30 Interpretation of correlations for model development and/or validation is problematic, as correlations are unable to determine whether independent variable(s) (e.g., Cmax) are true causes of dependent variable (e.g., AUC) changes. Correlations are also unable to ascertain bias and precision or whether LSMs can be improved using transformation methods.24,32 In another study, a 1.5- plus 4-h and a 2- plus 4-h fexofenadine LSM was developed and validated for healthy adults.31 Appropriate model development and validation steps were conducted, specifically jackknife analysis for model validation owing to the small sample size (n = 16). One potential reason for the conflicting results for the 2- plus 4-h LSM is the impact of clinical study site specificity. Site specificity is a limitation of a limited sampling strategy whereby the observed LSMs are only applicable to the participants and conditions from which the models were developed.33 The 2- plus 4-h LSM was susceptible to site specificity, as the study was conducted using participant data collected from a single site and exclusively during constitutive P-gp and OATP1B1/3 conditions. In the present study, fexofenadine LSMs were derived from multiple sites and altered P-gp and OATP1B1/3 conditions. Furthermore, comparisons regarding the 1.5- plus 4-h LSM31 were not possible because the 1.5-h timepoint was not collected in all studies used for our analysis (Table 1).
The current study used conservative %MPE, %MAE, and %RMSE acceptance limits similar to those of previous studies.25,34,35 If the same acceptance limits were applied to the aforementioned 2- plus 4-h LSM,31 then the reported R2 = 0.88 and %MAE = 10.3% would not be acceptable. We acknowledge that the conservative acceptance limits are arbitrary, but were chosen because of the anticipated decreased variability in fexofenadine systemic exposure in healthy participants. Currently, there are no universally accepted %MAE, %MPE, or %RMSE values. Previous studies used less conservative acceptance limits to evaluate bias and precision (e.g., %MPE < 20%, %RMSE < 25%) in patients and children.36,37 If such acceptance limits were applied in the current study, then several of the aforementioned LSMs would be acceptable (e.g., 2- plus 4-h, 4- plus 8-h). For future transporter-mediated DDI studies evaluating P-gp/OATP1/3 inhibitory and/or induction conditions, less conservative acceptable limits for bias and precision, as well as for R2, are needed, given that the pharmacokinetic variability of the probe drug is higher than that of constitutive P-gp conditions.28,37
Although fexofenadine poses challenges as a P-gp probe substrate owing to transporter non-selectivity, its use has several advantages. P-gp induction and inhibition mainly affect the oral bioavailability of P-gp substrates, thus using a P-gp substrate that is sensitive to changes in intestinal P-gp activity is desirable.1,2 P-gp-mediated efflux from the small intestine and blood-brain barrier are primary determinants of fexofenadine systemic exposure, and given its low oral bioavailability, fexofenadine represents an ideal choice of substrate.27,38 Dabigatran etexilate is another drug commonly used as a P-gp probe substrate. However, dabigatran etexilate is a prodrug that is hydrolyzed by carboxylesterase, which may confound DDI results if the concomitantly administered study drug modulates this enzyme.39 Lastly, fexofenadine is widely available, is not a substrate for CYP enzymes, and has a favorable safety profile.
This study has several limitations. The contribution of P-gp and OATP1B1/3 to the pharmacokinetic variability of fexofenadine is unknown. LSMs were derived from healthy participants and do not apply to special populations, such as children, older adults, or patients with hepatic impairment or kidney disease with known altered P-gp and/or OATP1B1/3 function. Another limitation is the lack of ABCB1 (or MDR1) genotyping, as previous studies have reported that fexofenadine plasma AUC or plasma concentrations are lower in participants harboring the ABCB1 3435CC genotype.40,41
Fexofenadine concentration data used for this analysis were obtained using liquid chromatography-tandem mass spectrometry analytical methods across all study sites. Differences in analytical specificity and techniques between study sites may have affected the variability in fexofenadine concentration. The extent of this contribution to the current study is unknown. However, this was evaluated using the CYP3A probe drug, midazolam. Concentration data (n = 73) were obtained from different institutions and used different analytical methods.35 A single-, 2-, and 3-timepoint midazolam LSM was generated combining concentrations across all institutions and regardless of assay method. Using the same %MPE, %MAE, and %RMSE limits as in the current study, validation of midazolam LSMs revealed acceptable bias and precision.35
Conclusion
This study evaluated a fexofenadine-limited sampling strategy based on a population pharmacokinetic model using a Bayesian estimation approach. Fexofenadine pharmacokinetics in healthy adult participants have been well described by a 2-compartment population pharmacokinetic model. Grapefruit juice, but not P. ginseng or E. purpurea, significantly decreased the relative oral bioavailability of fexofenadine. The evaluated single- and 2-timepoint LSMs were unable to accurately estimate the fexofenadine AUC and thus were unable to estimate P-gp and OATP1B1/3 activities. A semiphysiological model may be a more appropriate methodological approach for describing the data and should be considered in future studies.
Supplementary Material
Fexofenadine concentration versus time profiles by treatment group for each study. Black dots represent the observed data and the single thick line denotes the median concentration-time profile.
Acknowledgments
Part of this work was presented virtually at the 123rd American Society for Clinical Pharmacology & Therapeutics Annual Meeting, March 2022.
Conflicts of Interest and Source of Funding
All authors declare no conflicts of interest. Dr. Piscitelli is a post-doctoral fellow with funding supported by the Skaggs School of Pharmacy and Pharmaceutical Sciences and Pfizer, Inc. Funding support was provided by a Research in Pediatric and Developmental Pharmacology NIH grant (1U54HD090259-01 to E.V.C.) and a National Center for Complementary and Integrative Health grant (U54 AT008909 to M.F.P.).
References
- 1.Food and Drug Adminstration. Drug Development and Drug Interactions: Table of Substrates, Inhibitors and Inducers. Available from: https://www.fda.gov/drugs/developmentapprovalprocess/developmentresources/druginteractionslabeling/ucm093664.htm. Accessed August 18, 2022.
- 2.European Medicines Agency. Guideline on the investigation of drug interactions. [Available from: https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-investigation-drug-interactions-revision-1_en.pdf Accessed August 18, 2022.
- 3.Food and Drug Administration. Clinical Drug Interaction Studies – Cytochrome P450 Enzyme- and Transporter-Mediated Drug Interactions. Guidance for Industry. Available from: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/clinical-drug-interaction-studies-cytochrome-p450-enzyme-and-transporter-mediated-drug-interactions Accessed August 18, 2022.
- 4.Food and Drug Administration. In Vitro Drug Interaction Studies — Cytochrome P450 Enzyme- and Transporter-Mediated Drug Interactions Guidance for Industry. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/in-vitro-drug-interaction-studies-cytochrome-p450-enzyme-and-transporter-mediated-drug-interactions Accessed August 18, 2022.
- 5.Bedada SK, Boga PK, Kotakonda HK. The effect of diosmin on the pharmacokinetics of fexofenadine in healthy human volunteers. Xenobiotica. 2017;47(3):230–235. [DOI] [PubMed] [Google Scholar]
- 6.Calvo E, Lee JS, Kim SW, et al. Modulation of fexofenadine pharmacokinetics by osimertinib in patients with advanced EGFR-mutated non-small cell lung cancer. J Clin Pharmacol. 2019;59(8):1099–1109. [DOI] [PubMed] [Google Scholar]
- 7.Hamman MA, Bruce MA, Haehner-Daniels BD, Hall SD. The effect of rifampin administration on the disposition of fexofenadine. Clin Pharmacol Ther. 2001;69(3):114–121. [DOI] [PubMed] [Google Scholar]
- 8.Chaobal HN, Kharasch ED. Single-point sampling for assessment of constitutive, induced, and inhibited cytochrome P450 3A activity with alfentanil or midazolam. Clin Pharmacol Ther. 2005;78(5):529–539. [DOI] [PubMed] [Google Scholar]
- 9.Mueller SC, Drewelow B. Evaluation of limited sampling models for prediction of oral midazolam AUC for CYP3A phenotyping and drug interaction studies. Eur J Clin Pharmacol. 2013;69(5):1127–1134. [DOI] [PubMed] [Google Scholar]
- 10.Li H, Mager DE, Bemer MJ, et al. A limited sampling schedule to estimate mycophenolic Acid area under the concentration-time curve in hematopoietic cell transplantation recipients. J Clin Pharmacol. 2012; 52(11):1654–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barraclough KA, Isbel NM, Kirkpatrick CM, et al. Evaluation of limited sampling methods for estimation of tacrolimus exposure in adult kidney transplant recipients. Br J Clin Pharmacol 2011; 71(2):207–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barraclough KA, Isbel NM, McWhinney BC, et al. Evaluation of limited sampling strategies for total and free prednisolone in adult kidney transplant recipients. Eur J Clin Pharmacol 2011; 67(12):1243–1252. [DOI] [PubMed] [Google Scholar]
- 13.Chaabane A, Aouam K, Ben Fredj N, et al. Limited sampling strategy of mycophenolic acid in adult kidney transplant recipients: influence of the post-transplant period and the pharmacokinetic profile. J Clin Pharmacol 2013; 53(9):925–933. [DOI] [PubMed] [Google Scholar]
- 14.Zhu L, Wang H, Rao W, Qu W, Sun L. A limited sampling strategy for tacrolimus in liver transplant patients. Int J Clin Pharmacol Ther 2013; 51(6):509–512. [DOI] [PubMed] [Google Scholar]
- 15.Meier-Kriesche HU, Alloway R, Gaber AO, Canafax DM, Kaplan B. A limited sampling strategy for the estimation of 12-hour SangCya and neoral AUCs in renal transplant recipients. J Clin Pharmacol 1999; 39(2):166–171. [DOI] [PubMed] [Google Scholar]
- 16.Lin S, Nikanjam M, Capparelli EV, et al. Evaluation of omeprazole limited sampling strategies to estimate constitutive cytochrome P450 2C19 activity in healthy adults. Ther Drug Monit. 2018;40(6):754–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tran L, Nikanjam M, Capparelli EV, et al. S-warfarin limited sampling strategy with a population pharmacokinetic approach to estimate exposure and cytochrome P450 (CYP) 2C9 activity in healthy adults. Eur J Clin Pharmacol. 2021;77(9):1349–1356. [DOI] [PubMed] [Google Scholar]
- 18.Yang J, Nikanjam M, Capparelli EV, et al. Midazolam limited sampling strategy with a population pharmacokinetic approach to simultaneously estimate cytochrome P450 (CYP) 3A constitutive, inhibition, and induction/activation conditions in healthy adults. J Clin Pharmacol. 2019;59(11):1495–1504. [DOI] [PubMed] [Google Scholar]
- 19.Malati CY, Robertson SM, Hunt JD, et al. Influence of Panax ginseng on cytochrome P450 (CYP)3A and P-glycoprotein (P-gp) activity in healthy participants. J Clin Pharmacol. 2012;52(6):932–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nolin TD, Frye RF, Le P, et al. ESRD impairs nonrenal clearance of fexofenadine but not midazolam. J Am Soc Nephrol. 2009;20(10):2269–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penzak SR, Robertson SM, Hunt JD, et al. Echinacea purpurea significantly induces cytochrome P450 3A activity but does not alter lopinavir-ritonavir exposure in healthy subjects. Pharmacotherapy. 2010;30(8):797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Won CS, Lan T, Vandermolen KM, et al. A modified grapefruit juice eliminates two compound classes as major mediators of the grapefruit juice-fexofenadine interaction: an in vitro-in vivo "connect". J Clin Pharmacol. 2013;53(9):982–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beal SL. Ways to fit a PK model with some data below the quantification limit. J Pharmacokinet Pharmacodyn. 2001;28(5):481–504. [DOI] [PubMed] [Google Scholar]
- 24.Sheiner LB, Beal SL. Some suggestions for measuring predictive performance. J Pharmacokinet Biopharm. 1981;9:503–512. [DOI] [PubMed] [Google Scholar]
- 25.Al-Khatib M, Shapiro RJ, Partovi N, Ting LS, Levine M, Ensom MH. Limited sampling strategies for predicting area under the concentration-time curve of mycophenolic acid in islet transplant recipients. Ann Pharmacother. 2010;44(1):19–27. [DOI] [PubMed] [Google Scholar]
- 26.Center For Drug Evaluation and Research. Application Number: 20872. Clinical Pharmacology and Biopharmacetics Review(s). [Available from: https://www.accessdata.fda.gov/drugsatfda_docs/nda/2000/20-872_Allegra_biopharmr_P1.pdf Accessed June 28, 2022.
- 27.Allegra (Fexofenadine). Prescribing Information. [Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2008/020872s018,021963s002lbl.pdf Accessed June 28, 2022.
- 28.David OJ, Johnston A. Limited sampling strategies for estimating cyclosporin area under the concentration-time curve: review of current algorithms. Ther Drug Monit. 2001;23(2):100–114. [DOI] [PubMed] [Google Scholar]
- 29.Ting LS, Villeneuve E, Ensom MH. Beyond cyclosporine: a systematic review of limited sampling strategies for other immunosuppressants. Ther Drug Monit. 2006;28(3):419–430. [DOI] [PubMed] [Google Scholar]
- 30.Srinivas NR. Prediction of area under the curve for a p-glycoprotein, a CYP3A4 and a CYP2C9 substrate using a single time point strategy: assessment using fexofenadine, itraconazole and losartan and metabolites. Drug Dev Ind Pharm. 2016;42(6):945–957. [DOI] [PubMed] [Google Scholar]
- 31.Coelho EB, Cusinato DAC, Ximenez JP, Lanchote VL, Struchiner CJ, Suarez-Kurtz G. Limited sampling modeling for estimation of phenotypic metrics for CYP enzymes and the ABCB1 transporter using a cocktail approach. Front Pharmacol. 2020;11:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagelkerke N A note on a general definition of the coefficient of determination. Biometrika. 1991;78(3):691–692. [Google Scholar]
- 33.Mahmood I Center specificity in the limited sampling model (LSM): can the LSM developed from healthy subjects be extended to disease states? Int J Clin Pharmacol Ther. 2003;41(11):517–523. [DOI] [PubMed] [Google Scholar]
- 34.Ignjatovic AR, Miljkovic B, Todorovic D, Timotijevic I, Pokrajac M. Evaluation of single-point sampling strategies for the estimation of moclobemide exposure in depressive patients. J Clin Pharmacol. 2011;51(5):661–671. [DOI] [PubMed] [Google Scholar]
- 35.Kim JS, Nafziger AN, Tsunoda SM, et al. Limited sampling strategy to predict AUC of the CYP3A phenotyping probe midazolam in adults: application to various assay techniques. J Clin Pharmacol. 2002;42(4):376–382. [PubMed] [Google Scholar]
- 36.Mahmood I Evaluation of a morphine maturation model for the prediction of morphine clearance in children: how accurate is the predictive performance of the model? Br J Clin Pharmacol. 2011;71(1):88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Musuamba FT, Rousseau A, Bosmans JL, et al. Limited sampling models and Bayesian estimation for mycophenolic acid area under the curve prediction in stable renal transplant patients co-medicated with ciclosporin or sirolimus. Clin Pharmacokinet. 2009;48(11):745–758. [DOI] [PubMed] [Google Scholar]
- 38.Tahara H, Kusuhara H, Fuse E, Sugiyama Y. P-glycoprotein plays a major role in the efflux of fexofenadine in the small intestine and blood-brain barrier, but only a limited role in its biliary excretion. Drug Metab Dispos. 2005;33(7):963–968. [DOI] [PubMed] [Google Scholar]
- 39.Chu X, Galetin A, Zamek-Gliszczynski MJ, Zhang L, Tweedie DJ, International transporter C. dabigatran etexilate and digoxin: Comparison as clinical probe substrates for evaluation of P-gp inhibition. Clin Pharmacol Ther. 2018;104(5):788–792. [DOI] [PubMed] [Google Scholar]
- 40.Akamine Y, Miura M, Sunagawa S, Kagaya H, Yasui-Furukori N, Uno T. Influence of drug-transporter polymorphisms on the pharmacokinetics of fexofenadine enantiomers. Xenobiotica. 2010;40(11):782–789. [DOI] [PubMed] [Google Scholar]
- 41.Yi SY, Hong KS, Lim HS, et al. A variant 2677A allele of the MDR1 gene affects fexofenadine disposition. Clin Pharmacol Ther. 2004;76(5):418–427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fexofenadine concentration versus time profiles by treatment group for each study. Black dots represent the observed data and the single thick line denotes the median concentration-time profile.